Abstract
Background & Aims
Chronic hepatitis B is an incurable disease. Addressing the unmet medical need for therapies has been hampered by a lack of suitable cell culture models to investigate the HBV life cycle in a single experimental setup. We sought to develop a platform suitable to investigate all aspects of the entire HBV life cycle.
Methods
HepG2-NTCPsec+ cells were inoculated with HBV. Supernatants of infected cells were transferred to naïve cells. Inhibition of infection was determined in primary and secondary infected cells by high-content imaging of viral and cellular factors. Novel antivirals were triaged in cells infected with cell culture- or patient-derived HBV and in stably virus replicating cells. HBV internalisation and target-based receptor binding assays were conducted.
Results
We developed an HBV platform, screened 2,102 drugs and bioactives, and identified 3 early and 38 late novel HBV life cycle inhibitors using infectious HBV genotype D. Two early inhibitors, pranlukast (EC50 4.3 μM; 50% cytotoxic concentration [CC50] >50 μM) and cytochalasin D (EC50 0.07 μM; CC50 >50 μM), and 2 late inhibitors, fludarabine (EC50 0.1 μM; CC50 13.4 μM) and dexmedetomidine (EC50 6.2 μM; CC50 >50 μM), were further investigated. Pranlukast inhibited HBV preS1 binding, whereas cytochalasin D prevented the internalisation of HBV. Fludarabine inhibited the secretion of HBV progeny DNA, whereas dexmedetomidine interfered with the infectivity of HBV progeny. Patient-derived HBV genotype C was efficiently inhibited by fludarabine (EC50 0.08 μM) and dexmedetomidine (EC50 8.7 μM).
Conclusions
The newly developed high-content assay is suitable to screen large-scale drug libraries, enables monitoring of the entire HBV life cycle, and discriminates between inhibition of early and late viral life cycle events.
Lay summary
HBV infection is an incurable, chronic disease with few available treatments. Addressing this unmet medical need has been hampered by a lack of suitable cell culture models to study the entire viral life cycle in a single experimental setup. We developed an image-based approach suitable to screen large numbers of drugs, using a cell line that can be infected by HBV and produces large amounts of virus particles. By transferring viral supernatants from these infected cells to uninfected target cells, we could monitor the entire viral life cycle. We used this system to screen drug libraries and identified novel anti-HBV inhibitors that potently inhibit HBV in various phases of its life cycle. This assay will be an important new tool to study the HBV life cycle and accelerate the development of novel therapeutic strategies.
Keywords: HBV, HepG2-NTCP, Entry, Replication, Virion secretion, High-throughput screening, Small-molecule inhibitors, FDA-approved drugs, Supernatant transfer, Patient-derived HBV
Abbreviations: %CV, percent coefficient of variation; %Imax, percent maximum inhibition; CC50, 50% cytotoxic concentration; cccDNA, covalently closed circular DNA; CHB, chronic hepatitis B; HCC, hepatocellular carcinoma; CpAM, core protein allosteric modifiers; dpi, days post-infection; DRC, dose–response curve; FDA, Food and Drug Administration; GEq, genome equivalents; HBVpt, patient-derived HBV; HCS, high content screening; HID, N-hydroxyisoquinolinedione; HLCs, hepatocyte-like cells; HTS, high-throughput screening; IFA, immunofluorescence analysis; IFNα, interferon alpha; IFNλ, interferon lambda; iPSCs, induced pluripotent stem cells; LHB, HBV large surface protein; LMV, lamivudine; MoA, mechanism of action; MyrB, myrcludex B; NTCP, sodium taurocholate cotransporting polypeptide; p1, passage 1; p2, passage 2; PEG, polyethylene glycol; PF-rcDNA, protein-free relaxed circular DNA; pgRNA, pregenomic RNA; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SOP, standard operation procedure; TDF, tenofovir disoproxil fumarate; TI, therapeutic index
Graphical abstract
Highlights
-
•
We developed a high-content screening assay suitable to monitor the entire HBV life cycle and eligible to discriminate between early and late viral life cycle inhibition.
-
•
We screened FDA-approved drugs and bioactives.
-
•
We confirmed antiviral activity in primary and secondary assays, using stably virus replicating cells and cell culture- and patient-derived HBV.
-
•
Novel HBV inhibitors prevent receptor binding, virus internalisation, replication, or egress of viral progeny.
Introduction
Chronic hepatitis B (CHB) is one of the most widespread infectious diseases, with 257 million chronically infected patients worldwide, responsible for 887,000 deaths annually.1 CHB therapy with nucleos(t)ide analogues reduces the incidence of hepatocellular carcinoma (HCC) by blocking reverse transcription, overall viral replication, and subsequent HBV spread. Depending on the medical circumstances of the patient, the immune modulator interferon alpha (IFNα) is being used for HBV therapy. However, in the majority of cases, neither treatment regimen is curative, and patients depend on expensive, lifelong care. Because current HBV therapies do not achieve a functional cure, additional antiviral therapies are needed. A lack of suitable drug screening models has made developing new therapies difficult. Over the past 3 decades, cell-free, target-based screening approaches have been widely used in the pharmaceutical industry for first-in-class drug identification; however, the clinical relevance of research outcomes using these approaches has not met expectations. Hypothesis-driven approaches assume that a single molecular mechanism of action (MoA) can reflect the complex biology of a disease, which is a particularly problematic assumption.
In contrast, empirical analyses, including phenotypic assays, require fewer mechanistic assumptions and provide data that partially elucidate the molecular complexities of a drug's activity on a cellular level. The most important aspects to consider when developing a reliable cell-based screening assay are the model's ability to recapitulate host biology and the disease's physiopathologic relevance in vitro and in a large-scale screening context. The selected cell culture model for phenotypic cell-based screening should support all relevant molecular host–pathogen interactions required for the pathogen of interest. For diseases caused by intracellular pathogens, host–pathogen interactions—including cell invasion, replication, morphogenesis, egress, and spread of progeny—are important considerations. However, cell culture models that support all molecular mechanisms of the entire HBV life cycle are limited. HBV has a restricted host range, has high tissue tropism, and depends on the differentiation status of specific cellular factors. Therefore, only highly differentiated human hepatocyte-derived cells can be used for in vitro infection studies. Because none of the currently available, unmodified human hepatoma cell lines are susceptible to HBV, primary human hepatocytes remain the gold standard for in vitro infection experiments. However, primary cells have numerous limitations, including high cost, limited availability, requirements for delicate handling, and donor-to-donor variability.
Redifferentiated HepaRG immortalised hepatoma cells are an alternative in vitro HBV infection system.2 However, this system uses a laborious differentiation procedure requiring several weeks under delicate culture conditions and results in limited infection efficacy. Therefore, HepaRG cells are not suitable for high-throughput screening (HTS) campaigns. With the discovery of sodium taurocholate cotransporting polypeptide (NTCP) as a hepatocyte-specific cell membrane functional receptor for HBV, the development of improved HBV-susceptible hepatoma cell lines became possible.3 HepG2-NTCP cells are unlimited in supply, convenient to handle, and highly susceptible to HBV infection, which leads to enhanced assay reproducibility and suitability for drug discovery.4 However, because HBV-susceptible cell lines exhibit deficiencies in virus amplification and production of progeny viruses, hepatoma cells that can be transiently or stably transfected with HBV, such as HepAD38 and HepG2.2.15, have been used for decades to investigate the late steps of the viral life cycle (e.g. viral replication and secretion) and to conduct HTS studies.5,6
Recently, the use of human hepatocyte-like cells (HLCs) generated from induced pluripotent stem cells (iPSCs) or embryonic stem cells has been described to examine the entire HBV life cycle using a single in vitro system.7 However, only limited progeny virus secretion and spreading after initial infection have been reported in HLCs after a complex differentiation procedure spanning 21 days. Therefore, an appropriate and convenient in vitro phenotypic screening system that efficiently and robustly supports all steps of the HBV life cycle is still needed.
Our recently reported HepG2-NTCPsec+ cell clone supports the production of a high titre of infectious HBV progeny upon inoculation and thereby unifies the entire HBV life cycle in 1 in vitro system.8 Therefore, we utilised HepG2-NTCPsec+ cells to develop a phenotypic high-content screening (HCS) platform and screened United States Food and Drug Administration (FDA)-approved drugs and bioactives. Identified hits were confirmed by dose–response curve (DRC) analysis, and selected drugs were subjected to MoA studies.
Materials and methods
HBV infection and supernatant transfer assays
Infection of HepG2-NTCPsec+ cells and supernatant transfer were conducted as described previously.8 A detailed standard operation procedure (SOP) is provided in Table S1. Briefly, HepG2-NTCPsec+ passage 1 (p1) cells were plated in 384-well plates and inoculated with HBV. For inhibition of HBV infection, cells were pretreated with reference compounds 2 h before and during inoculation. For passage 2 (p2) cell infection, supernatants of p1 cells were supplemented with polyethylene glycol (PEG) 8000 and transferred to p2 cell cultures at 6 days post-infection (dpi). At 18 h post-infection, cells were washed repeatedly and incubated for 120 h. Further materials and methods are described in the Supplementary Data file.
Results
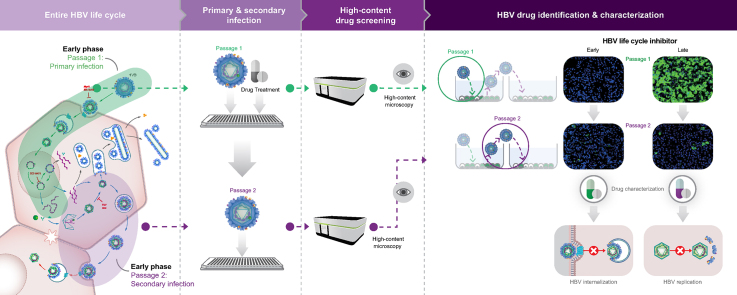
Development of a phenotypic HCS assay supporting interrogation of the entire HBV life cycle
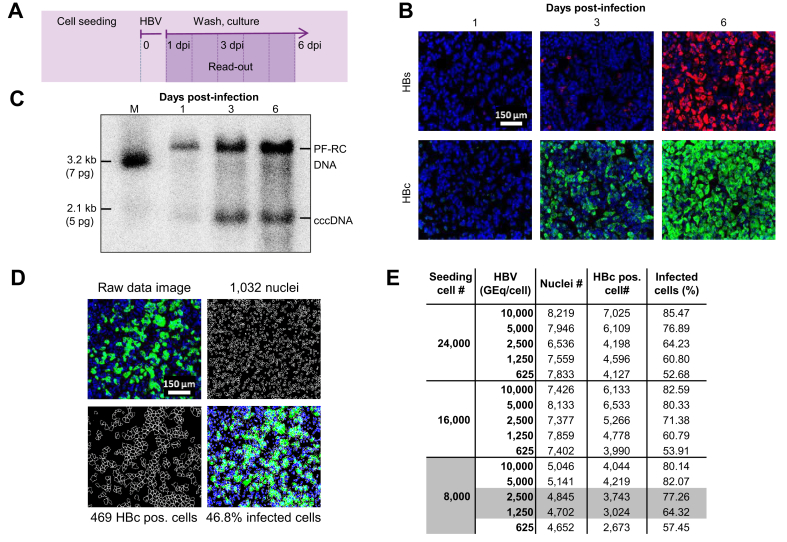
Utilizing the novel HBV-susceptible HepG2-NTCPsec+ cell line, which efficiently produces infectious HBV progeny, we devised a 384-well plate image-based HCS assay to identify inhibitors of early and late HBV life cycle steps. First, time-course infection experiments were performed to determine the appropriate time point and infection marker for assay read-out using immunofluorescence analysis (IFA) of the HBcAg and HBsAg (Fig. 1A,B). In parallel, the formation of covalently closed circular DNA (cccDNA) and protein-free relaxed circular DNA (PF-rcDNA) was monitored by Southern blotting (Fig. 1C). In both assays, no significant infection was detectable at 1 dpi, whereas all infection markers increased in a time-dependent manner until 6 dpi. In the IFA, the HBc showed higher signal intensity, robustness, and closely correlated kinetics with the generation of HBV cccDNA and PF-rcDNA compared with HBs. Because of the favourable HBc IFA signal-to-noise ratio, optimal HCS read-out was assigned at 6 dpi. Cell segmentation of raw data images accurately quantified the total number of cell nuclei as well as the number of HBc-positive cells. These data were used to calculate the infection ratio as a percentage (absolute number of infected cells/absolute number of cells × 100) (Fig. 1D). For further assay optimisation, the appropriate cell density and HBV DNA load (genome equivalents [GEq]/cell) for inoculation were evaluated in a matrix setting (Fig. 1E). HepG2-NTCPsec+ cells were seeded at 8,000, 16,000, and 24,000 cells/well, inoculated with 625, 1,250, 2,500, 5,000, or 10,000 HBV GEq/cell, and resulted in infection rates between 52% and 85%. Optimal HCS conditions were identified as seeding of 8,000 cells/well and inoculating with 1,250–2,500 GEq/cell, which resulted in robust HBV infection rates of 64–77%.
Fig. 1.
Development of a phenotypic HBV infection assay.
(A) Schematic illustration of the assay. Time-dependent analysis of HBV infection using (B) IFA of intracellular HBc and HBs expression and (C) cccDNA formation by Southern blotting at 1, 3, and 6 dpi of HepG2-NTCPsec+ cells. (D) Microscopic image processing and analysis for quantification of HBV-infected cells. Representative raw data images (top left), counting of individual nuclei (top right with 1,032 nuclei), and individual HBc-positive cells (lower left with 469 HBc-positive single cells). Absolute counts were used to determine relative values of ~47% of infected cells (lower right). (E) Assessment of cell density and HBV GEq/cell for phenotypic analysis of HBV infection. HepG2-NTCPsec+ cells were seeded at 8,000, 16,000, or 24,000 cells/well in 384-well plates and inoculated with 313–10,000 HBV GEq/cell. Cells were fixed at 6 dpi, and the number of nuclei, HBc-positive cells, and relative percent of infected cells were determined as described (D). A total of 6 individual pictures/well (in duplicate) were used for calculations. Nuclei (blue), HBs (red), and HBc (green). cccDNA, covalently closed circular DNA; dpi, days post-infection; GEq, genome equivalents; IFA, immunofluorescence analysis.
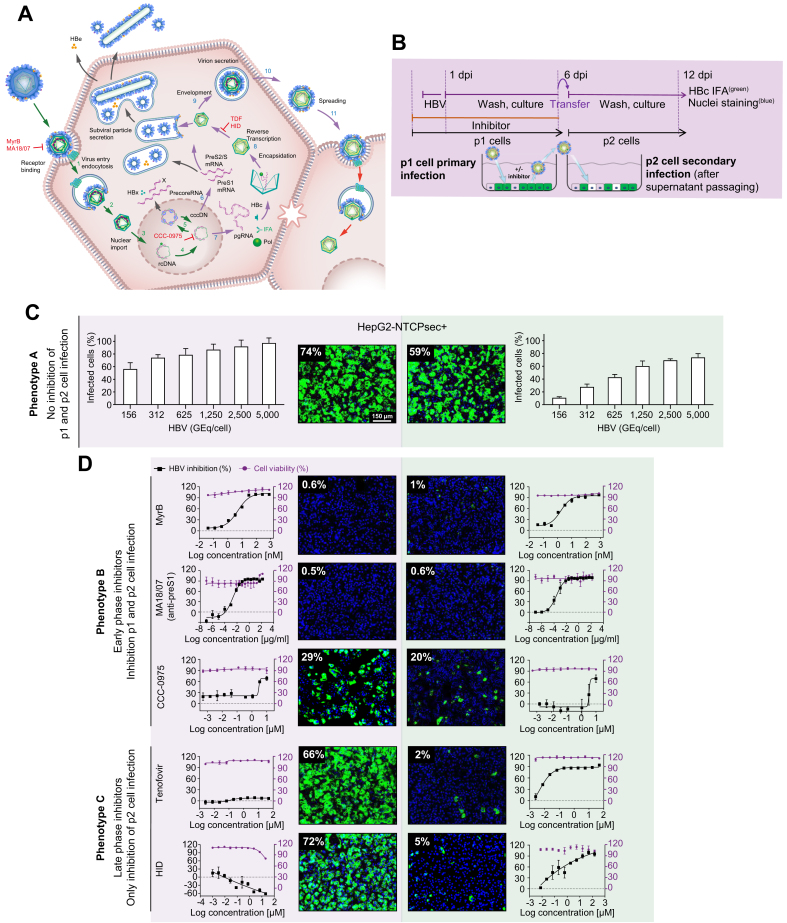
The image-based screening assay read-out of newly produced HBcAg in de novo infected p1 cells monitors all early steps of the HBV life cycle that lead to the establishment of infection (e.g. receptor binding, internalisation, nuclear import, and cccDNA formation) as well as the transcription of pregenomic RNA (pgRNA) and expression of HBcAg (Fig. 2A). Moreover, to monitor the late HBV life cycle steps of virion morphogenesis and egress (e.g. encapsidation, reverse transcription, HBsAg envelopment, and secretion of infectious viral particles), we harvested cell culture supernatants containing the newly produced progeny virus from infected p1 HepG2-NTCPsec+ cells and transferred them to naïve target p2 cells as a secondary infection step (Fig. 2A,B). However, the HCS assay does not monitor the production of HBeAg and the secretion of HBsAg in the form of subviral particles. To determine the optimal conditions for HepG2-NTCPsec+ cells to produce HBV progeny, we inoculated p1 cells with serially diluted HBV (150–5,000 GEq/cell), which resulted in a viral load-dependent p1 infection rate of approximately 55–95% (Fig. 2C, left). After a stringent washout of the inoculum, transferring supernatants enriched in newly produced HBV progeny resulted in an inoculum-dependent infection rate of 10–70% in p2 cells (Fig. 2C, right). Because p1 and p2 infection rates reached a plateau with a robust window of measurement at 2,500 HBV GEq/cell, we chose this inoculum concentration for the screening campaign.
Fig. 2.
Establishment of an HBV infection assay to monitor the entire HBV life cycle.
(A) HBV life cycle scheme. Early life cycle steps of infection establishment (1–5, green arrows) inhibited by reference inhibitors MyrB, MA18/07 (Step 1, receptor binding), and CCC-0975 (Step 5, cccDNA formation). Late HBV life cycle steps of morphogenesis and egress (6–12, blue arrows) inhibited by TDF and HID (Step 9, reverse transcription, RNase H). (B) Experimental setup of the 2-step HBV supernatant transfer infection assay. HepG2-NTCPsec+ p1 cells were seeded at 8,000 cells/well in 384-well plates and pre-incubated with the early or late life cycle phase reference inhibitor MyrB, MA18/07, CCC-0975, TDF, or HID for 2 h prior to inoculation with HBV for 18 h. After cells had been repeatedly washed at 1 dpi, they were treated with reference inhibitors and cultured until 6 dpi. Supernatants were harvested and transferred to naïve HepG2-NTCPsec+ p2 cells. Cells p1 and p2 were analysed using IFA (HBc) and counterstained with Hoechst (nuclei). Infected cells were identified microscopically and analysed as described in Fig. 1. (C) Infection with serially diluted HBV under untreated conditions (phenotype A). (D) HBV infection under treatment with early HBV life cycle inhibitor (phenotype B, top panel) or late HBV life cycle inhibitor (phenotype C, lower panel). HBc (green) and nuclei (blue); dose-response curve regression: red line = % cell viability (right y-axis); black line = % inhibition of infection (left y-axis). cccDNA, covalently closed circular DNA; dpi, days post-infection; HID, N-hydroxyisoquinolinedione; IFA, immunofluorescence analysis; MyrB, myrcludex B; pgRNA, pregenomic RNA; TDF, tenofovir disoproxil fumarate.
To validate the screening assay, we evaluated the activity of a multitude of reference inhibitors of the early and late HBV life cycle phases. The virus inoculum was pretreated with neutralising anti-preS1 antibody MA18/7, or p1 cells were pre-incubated with serially diluted myrcludex B (MyrB), CCC-0975, tenofovir disoproxil fumarate (TDF), or N-hydroxyisoquinolinedione (HID) before viral inoculation. The residual virus was washed out, and cells were pretreated and incubated with inhibitors for 6 days before the supernatants were harvested. Supernatants were promptly transferred to p2 cells for secondary infection. Three different phenotypes were observed from secondary infections and are described as follows: Phenotype A showed that inactive compounds produced no significant inhibition of p1 and p2 cell infection as compared with mock DMSO-treated control (Fig. 2C). Phenotype B demonstrated that early-phase inhibitors MA18/7, MyrB, and CCC-0975 inhibited HBV infection in p1 and p2 cells (Fig. 2D, upper panel). The p2 cell infection was inhibited because early-phase inhibitors prevent the establishment of p1 cell infection, causing the reduced formation of HBV cccDNA genomes, HBV replication, and secretion of progeny HBV. Phenotype C showed that late-phase inhibitors TDF and HID did not affect the infection status of p1 cells. However, by inhibiting viral replication in infected p1 cells, the production of HBV progeny was significantly impaired, as demonstrated by a reduced number of infected p2 cells (Fig. 2D, lower panel).
Based on DRC analyses with serially diluted inhibitors (Fig. 2D), the median effective and cytotoxic concentrations (EC50 and CC50), the therapeutic index (TI; CC50/EC50), and the percent maximum inhibition (%Imax) of each reference inhibitor were calculated for p1 and p2 infection (Table 1). All parameters for early and late HBV life cycle reference inhibitors were in the range of previously reported data using distinct in vitro assays. These data demonstrate that the entire HBV life cycle can be efficiently and conveniently analysed using a single screening platform with HepG2-NTCPsec+ cells.
Table 1.
DRC analysis of reference inhibitors.
| Early-phase inhibitors |
Early- and late-phase inhibitor |
Late-phase inhibitors |
|||||
|---|---|---|---|---|---|---|---|
| MyrB (nM) | MA18/07 (IU/ml) | CCC-0975 (μM) | Bay41-4109 (μM) | HID (μM) | LMV (μM) | TDF (μM) | |
| p1 infection | |||||||
| EC50 | 5.0 | 0.0008 | ~5.0 | 1.3 | >50 | >10 | >50 |
| CC50 | >50 | >50 | >10 | >20 | >50 | >10 | >50 |
| %Imax | 99 | 99 | 60 | 99 | ND | ND | ND |
| TI | >9.9 | >62,500 | >2.0 | >100 | ND | ND | ND |
| Published EC50 System PMID |
~1 HepG2-NTCP 24845614 |
ND | 10 HepDE19 22644022 |
ND | ND | ND | ND |
| p2 infection (after supernatant transfer) | |||||||
| EC50 | 1.6 | 0.00013 | ~8.0 | 0.16 | 3.1 | 0.017 | 4.0 |
| CC50 | >50 | >50 | >10 | >20 | >50 | >10 | >50 |
| %Imax | 99 | 99 | 70 | 99 | 99 | 93 | 81 |
| TI | >31.3 | >384,000 | >2.0 | >186 | >16.1 | >588 | >12.5 |
| Published EC50 System PMID |
ND | ND | ND | 202 HepG2.2.15 22162746 |
4.2 HepDES19 24858512 |
6.0 HepG2.2.15 15259898 |
1.1 HepG2.2.15 16801428 |
Calculated values of DRC regression analyses of EC50, CC50, %Imax, and TI for various classes of HBV inhibitors are represented for p1 and p2 cell infection. Information on EC50 values obtained in different published cell culture systems are provided. %Imax, percent maximum inhibition; CC50, 50% cytotoxic concentration; DRC, dose–response curve; HID, N-hydroxyisoquinolinedione; LMV, lamivudine; MyrB, myrcludex B; ND, no data; TDF, tenofovir disoproxil fumarate; Ti, therapeutic index.
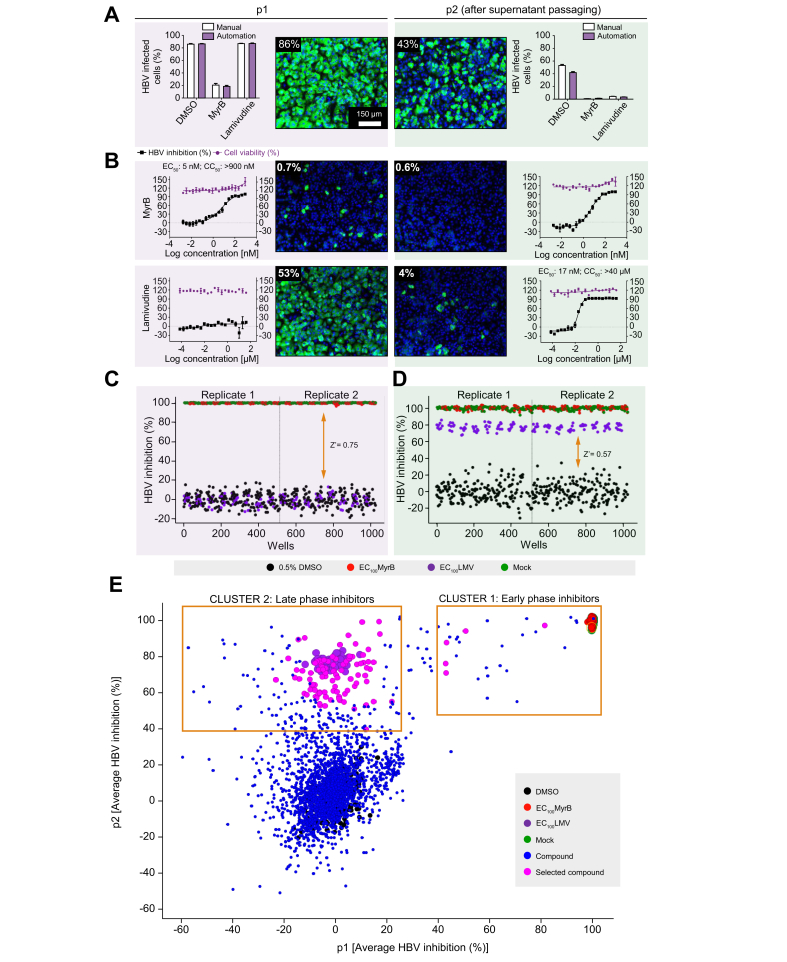
To increase the compound screening capacity, the assay was adapted for HTS automation, and an SOP was implemented (Table S1). Aside from manual preparation of the HepG2-NTCPsec+ single-cell suspension, automated instruments were used for cell plating, compound dilutions and treatment, HBV inoculation, PEG dispensing, cell washing, cell culture supernatant transfer, cell fixation, IFA, and assay read-out. For image analysis, in-house software was utilised to quantitatively measure thousands of images and automatically assign phenotypes. A side-by-side comparison of manual and automated workflows is shown in Fig. 3A. Using 2,000 HBV GEq/cell, we observed comparable infection rates in p1 and p2 cells (Fig. 3A). Under automated conditions, the reference inhibitors MyrB and lamivudine (LMV) exhibited the expected A and B phenotypes for early and late virus life cycle inhibition. Additionally, these inhibitors provided an acceptable signal-to-noise ratio of 85–20% and 40–5% in p1 and p2 cells, respectively. Reducing the inoculum resulted in a lower measurement window and was, therefore, not considered (Table S2A,B). Furthermore, DRC analysis revealed an EC50 of 6.3 nM (CC50 >900 nM) for MyrB in p1 cells and an EC50 of 17 nM (CC50 >40 μM) for LMV in p2 cells. These data were comparable with results obtained by manual infection assays and similar to values reported previously (Fig. 3B). To assess the robustness of the automated HCS assay, the entire 384-well plates were infected with 2,000 HBV GEq/cell, and intraplate variations were determined. Heat map analysis of p1 and p2 cells demonstrated plate uniformity at high infection rates of 66% and 31% on average, respectively. Only minor well-to-well variations were indicated by low SDs and percent coefficient of variation (%CV = [SD/mean infection] × 100) of 5% and 9%, respectively (Fig. S1).
Fig. 3.
Adaptation of HBV infection assay for HTS automation.
The HBV 2-step infection SOP was adapted to laboratory automated liquid handling devices for HTS studies. (A) Comparison of HBV infection efficiency under manual dispensing vs. automated liquid handling conditions. DMSO- (0.5%, v/v), MyrB- (900 nM), or LMV-treated (40 μM) HepG2-NTCPsec+ p1 cells were inoculated with 2,000 GEq/cell HBV before the supernatants were transferred to naïve p2 cells at 6 dpi. A representative image is shown for infected p1 and p2 cells treated with 0.5% DMSO (v/v). (B) DRC analysis with the early HBV life cycle reference inhibitor MyrB (starting from 900 nM), and the late life cycle reference inhibitor LMV (starting from 40 μM) with 20-points 2-fold serial dilutions. Representative images are shown. HBV inhibition (black squares) and cell viability (red circles) are shown as percentages indicated by the left and right y-axes, respectively. Control performance in (C) p1 and (D) p2 cells. Scatterplot distribution shows HBV inhibition (%) with 0.5% DMSO (v/v) (black), 900 nM MyrB (red), 40 μM LMV (purple), and mock infection (green). (E) Compound distribution of average HBV inhibition (%) in p1 and p2 cells. Clusters of compounds exhibiting different activity patterns are depicted within yellow squares. CC50, 50% cytotoxic concentration; cccDNA, covalently closed circular DNA; dpi, days post-infection; DRC, dose–response curve; HTS, high-throughput screening; GEq, genome equivalents; LMV, lamivudine; MyrB, myrcludex B; PF-rcDNA, protein-free relaxed circular DNA; SOP, standard operation procedure.
Primary HCS campaign, hit selection, and confirmation
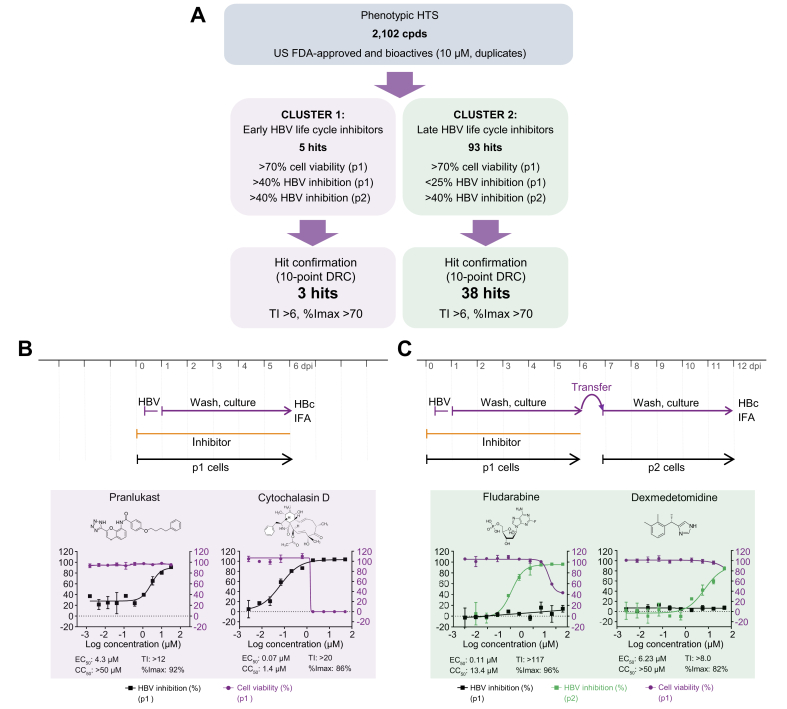
After successful HCS assay validation, we screened a collection of 2,102 small molecules, including bioactives with known targets (77%) and FDA-approved drugs (23%), in duplicate at a final concentration of 10 μM in DMSO. In total, 2 replicate sets were screened, and each set included a single well for all 2,102 test compounds. To monitor assay performance throughout the screen, each 384-well plate included 24 wells of DMSO-treated negative controls, 16 wells of reference inhibitor LMV- or MyrB-treated positive controls, and 16 wells of DMSO mock-infection controls. Inter-plate means and SDs for the DMSO-treated vs. MyrB- or LMV-treated wells from 16 individual 384-well plates (n = 224 wells) were used to calculate Z′ values (0.75 and 0.57 in p1 and p2 cells, respectively; Fig. 3C,D). These data indicate a sufficient measurement window and robust plate-to-plate uniformity for the HCS campaign. Furthermore, for each screening set, 2 DRC reference plates for MyrB and LMV were included as controls to monitor assay sensitivity. These results exhibited sigmoidal curves and expected results for EC50 values (Fig. S2A). Duplicate screening results of percent HBV inhibition from Sets 1 and 2 were analysed as scatterplots and used to determine R2 values (0.95 and 0.89 for p1 and p2 cells, respectively). These data indicate the high robustness and reproducibility of the HCS assay (Fig. S2B,C). To select for novel HBV hits, compounds causing p1 cell viability of <70% were excluded from further analysis. The remaining compounds were plotted as HBV inhibition in p1 cells vs. inhibition in p2 cells (Fig. 3E). Clusters of compounds exhibiting inhibitory activity patterns of early- and late-phase inhibitors (phenotypes B and C) could be distinguished. Early-phase inhibitors clustered with the reference inhibitor MyrB were gated at >40% inhibition in p1 and p2 cells, and 5 hits exhibiting preferable low cytotoxic effects were selected. In contrast, 93 late-phase inhibitors clustered with the reference inhibitor LMV were gated at <25% inhibition in p1 cells and >40% inhibition in p2 cells were selected.
We selected 4.9% of the screening library (98 inhibitors) for follow-up via several rounds of DRC confirmation using internal library compound stocks and fresh vendor-ordered stocks. Of the 98 selected compounds tested, 3 early and 38 late HBV life cycle inhibitors were confirmed using DRC analysis and stringent selection criteria (TI ≥6 and/or %Imax >70 at non-cytotoxic concentrations) (Fig. 4A). Among them, 15 compounds (1 early and 14 late HBV life cycle inhibitors) with a TI value of 2 to >500 were previously described HBV inhibitors (Table 2). The re-identification of known HBV inhibitors serves as another confirmation for the validity of the screening assay. The 15 re-identified HBV inhibitors interfere with retinoic acid receptor, microtubule, nucleic acid, and cholesterol synthesis. We selected 4 representative inhibitors for further MoA studies: 2 early HBV life cycle inhibitors, pranlukast (TI >12, %Imax = 92) and cytochalasin D (TI >20, %Imax = 86), and 2 late HBV life cycle inhibitors, fludarabine (TI >242, %Imax = 96) and dexmedetomidine (TI >8, %Imax = 82) (Fig. 4B,C and Table 2, Table 3).
Fig. 4.
HCS overview and representative inhibitor confirmation.
(A) Screening campaign summary of hit identification and confirmation. Number of hits, hit selection, and confirmation criteria are shown. Confirmation of selected early (B) (pranlukast and cytochalasin D) and late (C) (fludarabine and dexmedetomidine) HBV life cycle phase inhibitors by DRC analysis. %Imax, maximum inhibition; CC50, 50% cytotoxic concentration; dpi, days post-infection; DRC, dose–response curve; FDA, Food and Drug Administration; HCS, high-content screening; HTS, high-throughput screening; IFA, immunofluorescence analysis; TI, therapeutic index.
Table 2.
Antiviral efficacies and mechanisms of re-identified early and late HBV life cycle inhibitors.
| Name | %Imax | EC50 | CC50 | TI | Inhibitor published PMID | Mechanism published PMID | Target/proposed mechanism | |
|---|---|---|---|---|---|---|---|---|
| p1 inhibitor |
LE 135 |
94.5 |
11.68 |
>25 |
>2.2 |
25550158 |
RARβ |
|
| p2 inhibitor | Nocodazole | 91.8 | 1.06 | >25 | >23.7 | 28878350 | Microtubuli | |
| Vincristine | 96.9 | 1.27 | >25 | >19.6 | 28878350 | |||
| Floxuridine | 97.6 | 1.53 | >25 | >16.4 | 31247245 | Different steps in nucleic acid synthesis | ||
| Decitabine | 91.9 | 1.33 | >25 | >18.8 | 31247245 | |||
| GS-7340 | 99.4 | <0.05 | >25 | >500 | 16801428 | |||
| Entecavir | 96.7 | <0.05 | >25 | >500 | 12121928 | |||
| Foscarnet | 94.2 | <0.05 | >25 | >500 | 16280177 | |||
| Rimonabant | 99.4 | 0.23 | >25 | >109 | 31981244 | |||
| Raltitrexed | 93.7 | <0.05 | >25 | >500 | ||||
| Pralatrexate | 101.5 | <0.05 | >25 | >500 | ||||
| Ch 55 | 99.9 | 3.21 | >25 | >7.8 | 30224536 | RAR | ||
| Mevastatin | 100.8 | 2.02 | >25 | >12.4 | 19016777 | Cholesterol synthesis | ||
| Simvastatin | 99.9 | 2.24 | >25 | >11.2 | 19016777 | |||
| Posaconazole | 96.4 | 0.22 | >25 | >114.7 | 19016777 |
%Imax, percent maximum inhibition; CC50, 50% cytotoxic concentration; RAR, retinoic acid receptor; RARβ, retinoic acid receptor beta; TI, therapeutic index.
Table 3.
DRC analysis of reference inhibitors and confirmed novel early and late HBV life cycle inhibitors.
| Early HBV life cycle inhibitors |
Late HBV life cycle inhibitors |
|||||
|---|---|---|---|---|---|---|
| MyrB | Pranlukast | Cytochalasin D | Lamivudine | Fludarabine | Dexmedetomidine | |
| EC50 (μM) | 0.005 | 4.3 | 0.07 | 0.02 | 0.1 | 6.23 |
| CC50 (μM) | >0.8 | >50 | 1.4 | >10 | 13.4 | >50 |
| Imax (%) | 99 | 92 | 86 | 98 | 96 | 82 |
| TI | >10 | >12 | 20 | >333 | >117 | >8.0 |
| Published EC50 (μM) | 0.001–0.005 | No | No | 0.006–0.03 | No | No |
| Cellular target, drug use | HBV entry/NTCP inhibitor | Leukotriene receptor-1 antagonist, bronchospasm | Actin polymerisation inhibitor | Cytidine analogue, antiviral (HBV, HIV) | Purine analogue, blood cancer therapy | A2 adrenergic agonist |
| HBV proposed MoA | HBV–NTCP binding | HBV–NTCP binding | Actin-dependent HBV internalisation | HBV pol (RT) | HBV RT DNA synthesis | NR |
Calculated values of DRC regression analyses of EC50, CC50, %Imax, and TI for various classes of HBV inhibitors are presented. Information on EC50 values obtained in different published cell culture systems are provided. %Imax, percent maximum inhibition; CC50, 50% cytotoxic concentration; DRC, dose–response curve; MoA, mechanism of action; MyrB, myrcludex B; NR, not reported; NTCP, sodium taurocholate cotransporting polypeptide; RT, reverse transcription; TI, therapeutic index.
MoA studies with selected HBV inhibitors
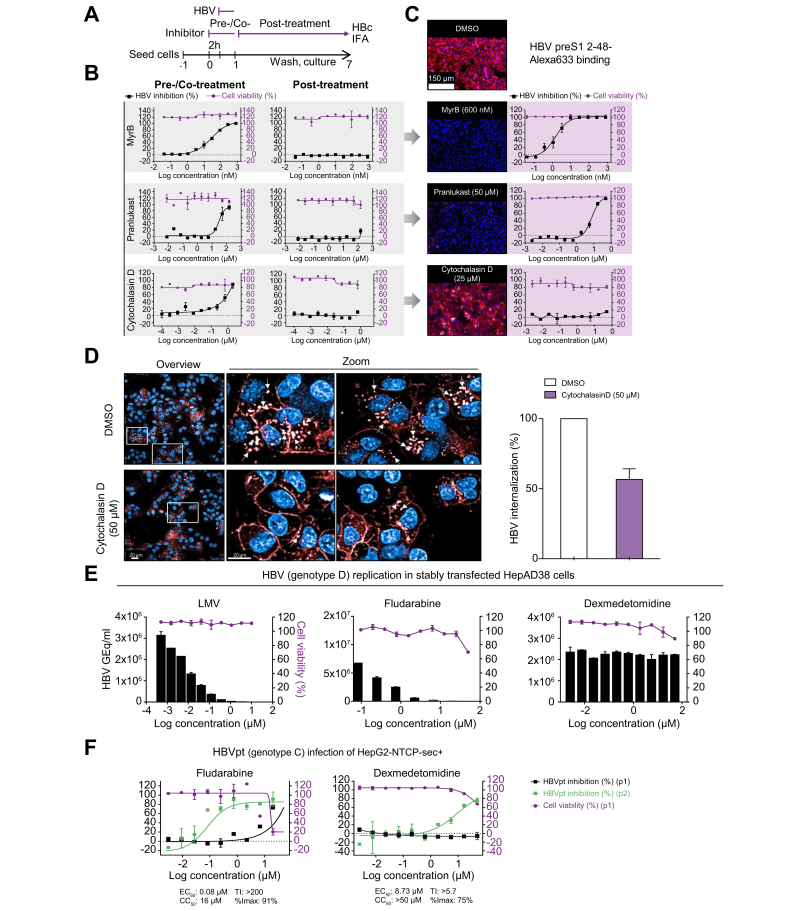
Pranlukast and cytochalasin D were chosen for additional MoA studies. Pranlukast is a leukotriene receptor-1 antagonist used for the treatment of bronchospasm, whereas cytochalasin D is an actin cytoskeleton disruptor. Time-of-addition experiments were conducted to characterise the MoA of pranlukast and cytochalasin D (Fig. 5A). Compounds were added to cells either 2 h before virus inoculation (pretreatment) and during HBV inoculation (cotreatment), or only added post-virus inoculation at 1−7 dpi. Similar to the reference entry inhibitor MyrB, pranlukast and cytochalasin D exhibited anti-HBV activity exclusively under the pretreatment/cotreatment conditions, thereby confirming antiviral activity during the early phase of the HBV life cycle (Fig. 5B). Identical to the reference entry inhibitor MyrB, pranlukast blocked the binding of fluorophore-conjugated, HBV surface protein-derived preS1 peptides to NTCP in a dose-dependent manner. These data confirm the cell membrane entry receptor NTCP as a potential drug target (Fig. 5C, middle panels). In contrast, cytochalasin D did not interfere with receptor–virus binding, as shown by constant membrane binding between HBV preS1 peptides and NTCP at concentrations up to 50 μM (Fig. 5C, lower panel). After cells were transferred to 37°C, internalisation of the preS1 probe into the cytosol of HepG2-NTSPsec+ cells was observed in high-magnification confocal microscopy live-cell imaging, as previously reported (Fig. 5D, upper panel).9 However, vesicular internalisation of the HBV preS1 peptide was partially reduced by cytochalasin D, suggesting that a functional actin cytoskeleton is a crucial factor in the unknown HBV internalisation mechanism (Fig. 5D, lower panel, and graph).
Fig. 5.
Mechanism of action studies with early and late HBV life cycle inhibitors.
(A) Time-of-addition experiment schedule. The reference inhibitors MyrB, pranlukast, and cytochalasin D were either pretreated and cotreated, or treated post-HBV inoculation in time-of-addition experiments. (B) DRC analysis of time-of-addition experiments. (C) Binding assay of HBV surface protein-derived Alexa633-labelled preS1 peptide under Mock (DMSO) or MyrB, pranlukast, cytochalasin D treatment. (D) Live-cell imaging of HBV surface protein-derived Alexa633-labelled preS1 peptide internalization (red) under cytochalasin D treatment. (E) Evaluation of HBV late step inhibitors in stably HBV replicating HepAD38 cells treated with serial dilutions of LMV, fludarabine, or dexmedetomidine. Supernatants were analysed by quantitative PCR at 7 days post-treatment. Cell viability was determined by automated nuclei counting. (F) Evaluation of late step inhibitors with HBVpt. HepG2-NTCPsec+ cells were pre-treated with 3-fold serially diluted fludarabine or dexmedetomidine (both starting from 50 μM) and infected with HBVpt genotype C. At 10 dpi, HBV inhibition of p1 cells (black squares) was analysed as described above, and supernatants were transferred to naïve p2 cells and analysed at 10 days post transfer (blue squares). Cell viability was determined as described before (red circles). Percentages of HBV inhibition and cell viability are indicated on the left and right y-axes, respectively. %Imax, percent maximum inhibition; CC50, 50% cytotoxic concentration; dpi, days post-infection; DRC, dose–response curve; GEq, genome equivalents; HBVpt, patient-derived HBV; IFA, immunofluorescence analysis; LMV, lamivudine; MyrB, myrcludex B; Ti, therapeutic index.
Next, utilizing stable HBV-replicating cells, we evaluated the MoA of the newly identified late HBV life cycle inhibitors fludarabine (Fludara; Selleckchem, Houston, TX, USA) and dexmedetomidine (Precedex; Selleckchem, Houston, TX, USA), which are an FDA-approved nucleic acid synthesis inhibitor used as a chemotherapy medication for the treatment of leukaemia and lymphoma and an anxiety-reducing sedative and pain medication, respectively. Similar to TDF or adefovir, fludarabine is a purine analogue fluorinated nucleotide/purine antimetabolite analogue of the antiviral agent vidarabine (ara-A), and all 3 drugs exhibit potent dose-dependent inhibition of HBV replication in HepAD38 cells, suggesting that fludarabine targets the viral polymerase (Fig. 5E). In contrast to LMV and fludarabine, dexmedetomidine mitigated the infectivity of secreted progeny HBV particles without reducing HBV replication or genome secretion in HepAD38 cells (Fig. 5E). To further investigate whether fludarabine and dexmedetomidine specifically interfere with HBV genome secretion, HCV and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected cells were evaluated. However, no inhibition was observed (Fig. S3A,B). Next, the specificity of cytochalasin D and pranlukast was evaluated using HIV-1- and SARS-CoV-2-infected cells. As a result, cytochalasin D had minor activity in both virus systems, whereas pranlukast only inhibited HIV-1, indicating that these drugs might have a broader antiviral activity for retroviruses and pararetroviruses (Fig. S3C,D).
Finally, to examine whether fludarabine and dexmedetomidine can interfere with the replication of patient-derived HBV (HBVpt) genotype C, we inoculated HepG2-NTCPsec+ cells with crude patient serum and analysed p1 and p2 cell infection as shown in Fig. 5F. The calculated EC50 values of fludarabine and dexmedetomidine were comparable with the values obtained using infectious cell culture-derived HBV genotype D (Fig. 4C and Table 2).
Discussion
Current therapeutic approaches utilizing direct-acting antivirals exclusively target 1 step of the HBV life cycle, resulting in viral load suppression in the blood of a patient with CHB. The lack of alternative treatments are a result of limitations of HBV-susceptible cell culture systems, their suitability for large-scale drug screening based on infectious HBV, and the inability to study the entire replicative cycle of the virus. The discovery of NTCP as a functional HBV receptor was a breakthrough for developing robust infectious cell culture models to study viral entry mechanisms.3 The authors of the study reported that infections of NTCP overexpressing HepG2 cells or primary human hepatocyte infections resulted in low levels of viral DNA in the media, despite significant amounts of HBeAg being secreted. As a consequence, no secondary infection was observed by passaging those supernatants onto other cells. It was speculated that differential expression of host factors in cell cultures might be disruptive for efficient formation or release of viral particles. Over the past years, HBV-susceptible cell culture systems have been constantly improved, yielding increased infection rates.10 However, none of the systems supported the efficient secretion of infectious HBV progeny upon infection that could be detected by IFA. Thereby, large- or medium-scaled target-based screening assays for the selective identification of best-in-class, single HBV life cycle step inhibitors, e.g. receptor binding or HBV RNaseH, were reported over the recent years.11,12 Here, we report on the first large-scale phenotypic assay that provides an empirical, target-unbiased screening approach of the entire HBV life cycle, suitable to identify novel first-in-class inhibitors. To reliably identify anti-HBV drugs, we carefully considered 3 key elements that must be implemented to develop any robust cell-based HCS assay (See Table 3).
First, we selected a cell line that recapitulates the biology of interest in vitro, conveys the disease's physiopathologic relevance, and holds true in the context of large-scale screening. This is especially important for drug discovery because if an irrelevant disease model is used, it can lead to invalid hit selection and delay the project. In terms of any infectious disease caused by an intracellular pathogen, cell-based phenotypic screening should support all relevant molecular host–pathogen interactions required for cell invasion, replication, morphogenesis, egress, and the spread of progeny. HepG2-NTCPsec+ cells were the first reported robust cell culture model supporting all molecular mechanisms of the entire HBV life cycle. Furthermore, HepG2-NTCPsec+ cells retain clinically relevant features of HBV-infected patients and animal models. For example, HBV has a reputation for being a stealth viral pathogen. This is highlighted by efficient virus amplification following de novo infection, release of highly infectious HBV large surface protein (LHB)-enveloped capsids, multiphasic kinetics of HBV amplification during infection establishment, spreading of secondary infection to naïve cells, and no significant host cell gene expression changes upon infection, all of which are characteristic of HepG2-NTCPsec+ cells.8 Here, we successfully utilised HepG2-NTCPsec+ cells to develop a HCS assay using 384-well plates, with minimal well-to-well and plate-to-plate variations in p1 and p2 infection. The system productively supports early and late HBV life cycle steps, as indicated by primary infection (p1) and secretion of infectious progeny virions leading to secondary infection (p2). Titrating the viral load for p1 cell inoculation resulted in a plateau at 80–95% in p1 cells using ≥625 HBV GEq/cell. However, after transferring the supernatants to p2 cells, infection rates ranged from 35% to 75%. This observation suggests that the viral load is less critical for the detection of HBV de novo infection in p1 cells but more important for the egress of infectious HBV particles from p1 cells. This could be as a result of multiple infection events of p1 cells when using higher viral loads, leading to manifold copies of cccDNA as templates for the propagation of progeny HBV.
We were able to prove that the HCS assay phenotypically discriminates reliably between 2 groups of inhibitors: (A) compounds interfering with the early HBV life cycle phase, comprising initial virus–receptor binding to the formation of persistent cccDNA genomes in the nucleus (establishment of infection), and (B) agents inhibiting in the late phase of the viral life cycle including all steps occurring after cccDNA formation from the cccDNA-driven transcription of PreS1 and PreS2/S mRNA to the secretion of virions (replication, morphogenesis, and egress). The exception to this distinction is compounds that specifically inhibit the transcription of the HBV pgRNA or the expression of HBc, which is the marker of p1 read-out. Such compounds would show the characteristic phenotype of an early life cycle inhibitor despite targeting a late life cycle step. Notably, a more selective target differentiation cannot be resolved using the assay, and compounds with multiple MoAs may show a mixed phenotype. In both cases, the implementation of secondary, target-based assays on a smaller scale is required for target identification and characterisation. As proof of concept, time of addition, as well as 3 mechanistic assays, provided deeper insights into the molecular targets/mechanisms of a small subset of the here identified compounds, including receptor binding (NTCP), HBV internalisation (p1 inhibitors), and viral replication or propagation inhibition (p2 inhibitors).
As a second key element for successful screening assay development, we identified a reliable biomarker to indicate cellular infection status in a phenotypic HCS approach. Combined Hoechst DNA and immunofluorescence HBcAg staining enabled robust simultaneous capture of the total number of nuclei (cells) and infected cells. These parameters enabled us to determine the HBV inhibitory and cytotoxic potency of reference and test compounds. A key advantage of using a cell-based HCS assay over cell-free, target-based assays is the ability to exclude cytotoxic and non-cell-permeable compounds at a very early phase in the drug discovery process. In addition to this, numerous other parameters can be extracted from images, and single cells can be analysed in a quantitative scoring manner. Information on the cellular or subcellular organelle morphology, size, volume, or cell-to-cell distance, as well as the subcellular distribution of HBcAg, can provide additional mechanistic insights regarding compounds of interest and be used to generate a phenotypic fingerprint of HBV inhibitors. For example, our high-content imaging assay enabled us to capture and quantify HBcAg translocation from the cytoplasm to the nucleus (data not shown). Interestingly, this phenotype may be correlated with viral replication status, as nuclear HBc reportedly reflects the level of viral replication in patients with CHB.13 Therefore, HBcAg translocation may serve as an interesting parameter for generating inhibitor fingerprints. As another example, we quantified the aggregated HBcAg phenotype in the cytoplasm or nucleus when the virus was challenged with class II core protein allosteric modifiers (CpAM) (i.e. GLS4 or Bay421-4109) (Fig. S4). This suggests the screening assay can indicate the MoA of test compounds that similarly promote HBcAg aggregation. In addition to HBcAg, the HCS assay can be easily adjusted for multiplex staining of other cellular or viral markers, i.e. HBsAg expression, as other attractive antiviral drug targets. Furthermore, the assay can be applied to studies of other members of the Hepadnaviridae family, because the conformational polyclonal anti-HBc antisera react against all human HBV genotypes and even more distantly related HBV genotypes from monkeys and bats.14
A third key element that must be considered to guarantee the reliability and robustness of the HBV HCS assay is the selection of proper controls to assess plate quality and normalise read-outs. Serial dilutions of 2 well-established early and late-phase reference inhibitors of the HBV life cycle functioned as suitable controls. These controls monitored the assay's robustness, reproducibility, and sensitivity during the screening campaign by enabling the calculation of values such as EC50, EC90, CC50, TI, signal-to-noise ratio, Z′, R2, %CV, and %Imax. Thereby, the new HBV HCS assay fulfils all important criteria for generating reliable and reproducible in vitro data sets for antiviral drug identifications. Interestingly, when examining high concentrations of the late-phase HBV life cycle reference inhibitors LMV, TDF, and HID, we found that 2–5% of p2 cells were still infected. As multiple washing steps were performed on p1 cells after removing the inoculum, these data indicate that none of these inhibitors are potent enough to prevent HBV propagation completely. These results are in accordance with a recent report demonstrating the presence of infectious virus in the serum of patients with CHB on nucleos(t)ide therapy, with detectable but not quantifiable HBV DNA.15
To evaluate whether the established HCS assay is suitable for discovering first-in-class HBV inhibitors, we performed a medium-scale pilot screen including 2,000 compounds and controls that span the whole range of the viral life cycle. Applying strict hit-selection criteria, we identified 103 candidates and confirmed 41 candidates in the primary assay. Importantly, we re-identified already known HBV inhibitors, e.g. entecavir and decitabine, and discovered new HBV inhibitors interfering with known proviral cellular factors such as microtubule polymerisation, different steps in nucleic acid and cholesterol synthesis, and retinoic acid receptor signalling (Table 2). These results suggest that our HCS assay authentically recapitulates the HBV biology.
We selected 2 representative novel inhibitors showing the characteristic phenotype of p1 (pranlukast and cytochalasin) or p2 inhibitors (fludarabine and dexmedetomidine) and conducted MoA studies to confirm inhibition in the early or late phase of HBV infection.
The FDA-approved drug pranlukast, a cysteinyl leukotriene receptor-1 antagonist, efficiently inhibited the binding of HBV preS1-derived peptide to its receptor, NTCP, with an EC50 of 7.6 μM. A previous study reported that pranlukast inhibits the bile acid transporter function in transiently NTCP-transfected U2OS cells.16 In the same study, a structurally related inhibitor, zafirlukast, significantly inhibited HBV and HDV infection at concentrations as low as 6 μM. Using infectious HBV and pranlukast, we obtained a comparable EC50 value of 4.3 μM, suggesting that zafirlukast and pranlukast have a similar MoA for inhibiting HBV entry in a comparable way to MyrB by targeting the initial step of the early HBV infection phase.
Cytochalasin D did not prevent the binding of NTCP to the HBV surface protein-derived myr-preS1–Alexa633 peptide. However, we showed that cytochalasin D caused partial retention of the peptide at the plasma membrane, thereby blocking the early HBV life cycle step of virus internalisation. These results demonstrate that a functional actin cytoskeleton is crucial for the currently unknown HBV entry mechanism. Cytochalasin D-sensitive actin- and myosin-dependent ‘surfing’ on cellular membranes has been described for different viruses, including retroviral murine leukaemia virus, human papillomavirus 16, and vaccinia virus, and seems particularly crucial for viral entry.17
Using infected and stable HBV-replicating cells, we demonstrated that fludarabine, a drug used in the treatment of leukaemia and lymphoma by slowing down the cellular DNA synthesis through the inhibition of the cellular ribonucleotide reductase and DNA polymerase, efficiently inhibited the production of infectious progeny of cell culture- and HBVpt genotypes C and D, respectively. Similar to fludarabine, all purine-based drugs target either cellular or viral enzymatic mechanisms of DNA synthesis and therefore are approved as anticancer or antiretroviral agents. In fact, fludarabine is used as an anticancer drug; however, structurally, it is the fluorinated purine antimetabolite analogue of the antiviral agent vidarabine, which was shown to be active against herpes viruses, poxviruses, rhabdoviruses, hepadnaviruses, etc. However, fludarabine can also induce severe and prolonged immunosuppression that was shown to be related to reactivation of overt and occult hepatitis B in patients.18 Despite a potent and direct antiviral effect against a late HBV life cycle step in vitro, the immunosuppressive effects of fludarabine make this drug unsuitable for the treatment of CHB.
In contrast, dexmedetomidine did not inhibit HBV DNA replication. Conversely, dexmedetomidine strongly suppresses the infectivity of secreted progeny HBV particles, irrespective of the virus source or the here tested different viral genotypes. Therefore, dexmedetomidine may represent a novel class of HBV propagation inhibitors that would not have been discovered using conventional assay systems to assess the read-out of secreted HBV DNA from stable HBV replicating cell lines.
Furthermore, to determine the specificity of the here described inhibitors, we proved that fludarabine and dexmedetomidine do not interfere in the production and spread of infectious HCV and SARS-CoV-2 (Fig. S3A,B). All 3 viruses are reported to be released via different secretory pathways (HBV = trans-Golgi network and multivesicular bodies (MVB), HCV = non-canonical route, SARS-CoV-2 = lysosomal exocytosis), and the results suggest that the identified inhibitors do not broadly inhibit cellular secretion pathways but rather have a certain specificity to prevent the formation of infectious HBV. Interestingly, both newly identified HBV p1 inhibitors partially inhibit entry of HIV-1, whereas only cytochalasin D (but not pranlukast) inhibits SARS-CoV-2 infection (Fig. S3C,D). It was previously shown that an actin-dependent receptor-HIV colocalisation is essential for viral entry, which is potentially sensitive to cytochalasin D.19 Furthermore, cytochalasin D was shown to prevent SARS-CoV-2 cell entry in a virus-free, live-cell assay.20 Importantly, all identified HBV inhibitors show antiviral activities at non-toxic compound concentrations, indicating that no essential cellular house-keeping functions are critically disturbed.
Next, we evaluated whether our novel screening assay is suitable to identify innate immune modulators. To answer this question, we treated p1 cells with IFNα or IFNλ and later transferred the supernatants to p2 cells. However, no or only insignificant antiviral effects of IFNα and IFNλ were observed on early and late HBV life cycle steps, respectively (Fig. S5A,B). Nonetheless, the cell line shows only a minor sensitivity to type I and III IFNs. Therefore, the HCS assay is unlikely to identify innate immune modulators because of the short-term experimental setup and strict selection criteria.
In conclusion, we developed an in vitro HBV HCS platform that enables target-free, empirical, robust, and convenient interrogation of the entire viral life cycle. Furthermore, we were able to prove that the assay reliably discriminates phenotypically between HBV early and late infection phase inhibitors. Unlike the plasmid-driven regulation of HBV replication in HepAD38 or HepG2.2.15 cells, the late phase of the HBV life cycle in our system is exclusively driven by transcriptional regulation of cccDNA, which originates from an authentic de novo HBV entry mechanism. As such, the entire HBV life cycle is covered by 1 in vitro system resembling the biological conditions during host infection. The platform described here should accelerate the discovery of novel antiviral interventions that will eradicate CHB and identify novel host drug targets.
Financial support
This work was financially supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2020R1A2C2009529, NRF-2019R1A2C1090515, and NRF-2017M3A9G6068246).
Authors' contributions
Performed the experiments: JY, AK, SP, EZ, DK, EJ. Conceived the project: DS, MPW. Provided crucial material: SKY, PSS.
Analysed the data: JY, AK, SP, DS, MPW. Wrote the manuscript: AK, MPW. Approved the manuscript: All authors
Data availability statement
The data supporting the findings of this study are available within the article and its supplementary materials.
Conflicts of interest
The authors declare no competing interests.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We would like to thank Dieter Glebe for scientific discussion, Diana Koo for editing the manuscript. We thank the National Research Foundation of Korea (MSIT and NRF) for financial support.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100296.
Supplementary data
The following are the supplementary data to this article:
References
- 1.World Health Organization . World Health Organization; 2017. Hepatitis B: fact sheet.http://www.who.int/mediacentre/factsheets/fs204/en/ [Google Scholar]
- 2.Gripon P., Rumin S., Urban S., Le Seyec J., Glaise D., Cannie I. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1 doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watashi K., Sluder A., Daito T., Matsunaga S., Ryo A., Nagamori S. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP) Hepatology. 2014;59:1726–1737. doi: 10.1002/hep.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai D., Mills C., Yu W., Yan R., Aldrich C.E., Saputelli J.R. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob Agents Chemother. 2012;56:4277–4288. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou T., Guo H., Guo J.T., Cuconati A., Mehta A., Block T.M. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antivir Res. 2006;72:116–124. doi: 10.1016/j.antiviral.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Shlomai A., Schwartz R.E., Ramanan V., Bhatta A., de Jong Y.P., Bhatia S.N. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.König A., Yang J., Jo E., Park K.H.P., Kim H., Than T.T. Efficient long-term amplification of hepatitis B virus isolates after infection of slow proliferating HepG2-NTCP cells. J Hepatol. 2019;71:289–300. doi: 10.1016/j.jhep.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 9.König A., Döring B., Mohr C., Geipel A., Geyer J., Glebe D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol. 2014;61:867–875. doi: 10.1016/j.jhep.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Hu J., Lin Y.Y., Chen P.J., Watashi K., Wakita T. Cell and animal models for studying hepatitis B virus infection and drug development. Gastroenterology. 2019;156:338–354. doi: 10.1053/j.gastro.2018.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saso W., Tsukuda S., Ohashi H., Fukano K., Morishita R., Matsunaga S. A new strategy to identify hepatitis B virus entry inhibitors by AlphaScreen technology targeting the envelope-receptor interaction. Biochem Biophys Res Commun. 2018;501:374–379. doi: 10.1016/j.bbrc.2018.04.187. [DOI] [PubMed] [Google Scholar]
- 12.Edwards T.C., Lomonosova E., Patel J.A., Li Q., Villa J.A., Gupta A.K. Inhibition of hepatitis B virus replication by N-hydroxyisoquinolinediones and related polyoxygenated heterocycles. Antivir Res. 2017;143:205–217. doi: 10.1016/j.antiviral.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu C.M., Yeh C.T., Chien R.N., Sheen I.S., Liaw Y.F. The degrees of hepatocyte nuclear but not cytoplasmic expression of hepatitis B core antigen reflect the level of viral replication in chronic hepatitis B virus infection. J Clin Microbiol. 1997;35:102–105. doi: 10.1128/jcm.35.1.102-105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Carvalho Dominguez Souza B., König A., Rasche A., de Oliveira Carneiro I., Stephan N., Corman V.M. A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J Hepatol. 2018;68:1114–1122. doi: 10.1016/j.jhep.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 15.Burdette D., Cathcart A., Shauf A., Win R., Zaboli S., Hedskog C. Evidence for the presence of infectious virus in the serum from chronic hepatitis B patients suppressed on nucleos(t)ide therapy with detectable but not quantifiable HBV DNA. J Hepatol. 2019;70:e95. [Google Scholar]
- 16.Donkers J.M., Zehnder B., van Westen G.J.P., Kwakkenbos M.J., IJzerman A.P., Oude Elferink R.P. Reduced hepatitis B and D viral entry using clinically applied drugs as novel inhibitors of the bile acid transporter NTCP. Sci Rep. 2017;7:15307. doi: 10.1038/s41598-017-15338-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann M.J., Sherer N.M., Marks C.B., Pypaert M., Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coppola N., Tonziello G., Pisaturo M., Messina V., Guastafierro S., Fiore M. Reactivation of overt and occult hepatitis B infection in various immunosuppressive settings. J Med Virol. 2011;83:1909–1916. doi: 10.1002/jmv.22199. [DOI] [PubMed] [Google Scholar]
- 19.Iyengar S., Hildreth J.E., Schwartz D.H. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol. 1998;72:5251–5255. doi: 10.1128/jvi.72.6.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Wang S., Wu Y., Hou W., Yuan L., Shen C. Virus-free and live-cell visualizing SARS-CoV-2 cell entry for studies of neutralizing antibodies and compound inhibitors. Small Methods. 2021;5:2001031. doi: 10.1002/smtd.202001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and its supplementary materials.