Abstract
Problem:
Various etiological factors, such as infection and inflammation, may induce the adverse outcomes of pregnancy of miscarriage, stillbirth, or preterm birth. The pathogenic mechanisms associated with these adverse pregnancies are yet unclear. We hypothesized that a common pathogenic mechanism may underlie variant adverse outcomes of pregnancy, which are induced by genetic-environmental factors. The specific objective of the current study is to uncover the common molecular mechanism(s) by identifying the specific transcripts that are present in variant subtypes of pregnancy loss and preterm birth.
Method of Study:
Transcriptomic profiling was performed with RNA expression microarray or RNA sequencing of placentas derived from pregnancy loss (which includes spontaneous miscarriage, recurrent miscarriage, and stillbirth) and spontaneous preterm birth, followed by bioinformatic analysis of multi-omic integration to identify pathogenic molecules and pathways involved in pathological pregnancies.
Results:
The enrichment of common differentially expressed genes between full-term birth and preterm birth and pregnancy loss of miscarriage and stillbirth revealed different pathophysiological pathway(s), including cytokine signaling dysregulated in spontaneous preterm birth, defense response, graft-versus-host disease, antigen processing and presentation, and T help cell differentiation in spontaneous miscarriage. Thirty-three genes shared between spontaneous preterm birth and spontaneous miscarriage were engaged in pathways of interferon gamma–mediated signaling and of antigen processing and presentation. For spontaneous miscarriage, immune response was enriched in the fetal tissue of chorionic villi and in the maternal facet of the placental sac. The transcript of nerve growth factor receptor was identified as the common molecule that is differentially expressed in all adverse pregnancies: spontaneous preterm birth, stillbirth, spontaneous miscarriage, and recurrent miscarriage. Superoxide dismutase 2 was up-regulated in all adverse outcomes of pregnancy except for recurrent miscarriage. Cytokine-cytokine receptor interaction was the common pathway in spontaneous preterm birth and spontaneous miscarriage. Defense response was enriched in the fetal tissue of miscarriage and in the maternal tissue in spontaneous miscarriage.
Conclusions:
Our results indicated that the chemokine-cytokine pathway may play important roles in and function as a common pathogenic mechanism associated with, the different adverse outcomes of pregnancy, which demonstrated that differentially expressed transcripts could result from a common pathogenic mechanism associated with pregnancy loss and spontaneous preterm birth, although individual pregnancy outcomes may differ from each other phenotypically.
Keywords: infection-inflammation, mechanism, pathogenesis, pregnancy loss, Preterm birth
1 |. INTRODUCTION
Pregnancy loss and preterm birth are the most common adverse outcomes of pregnancies. Pregnancy loss usually includes spontaneous miscarriages, which occur for the first time before the 20th gestational week (GW),1 and stillbirth if the fetus dies after GW 20.2 Spontaneous miscarriage that occurs two or more times consecutively is considered recurrent miscarriage.3 Preterm birth is defined as premature birth delivered at GW <37, which is the leading cause of neonatal death and the second largest direct cause of pediatric deaths in children younger than 5 years of age.4
1.1 |. Etiology
Etiologically, it is estimated that about 50–75% of total pregnancies are miscarried. Among most of the aborted embryos, genetic defects (usually chromosomal abnormalities) can be found as the cause for cessation of early intrauterine development of the embryo soon after implantation, appearing as menorrhagia or delayed menstruation that escapes from notice of pregnancy.5,6 These adverse outcomes could be triggered by various risk factors at different periods of gestation. Maternal causes of spontaneous pregnancy loss include medical conditions such as diabetes or thyroid disease, hormone imbalance, maternal physical disabilities, infection, immune system responses, or uterine abnormalities. These maternal-originated etiological factors, such as infection or inflammation, comprise the intrauterine micro-environmental conditions that could result in abnormal placentation in miscarriage at early gestation.7,8
The risk factors for stillbirth include maternal factors (demographic- and fertility-related, infection, nutrition, lifestyle, non-communicable diseases, and environmental factors) and fetal factors (male sex, post-term pregnancy, small for gestational age, congenital abnormalities, Rhesus disease).9 Problems with the placenta, genetic defects with the fetus, fetal infections, and other physical problems in the fetus may cause stillbirth.9,10 Spontaneous preterm birth presents as preterm premature rupture of fetal membrane or preterm labor, with an estimated 15 million babies born too soon annually.4 Common causes of spontaneous preterm birth include multiple pregnancies, infections, and pregnancy complications (diabetes and preeclampsia). Intrauterine and systemic infection and inflammation may induce preterm premature rupture of fetal membrane or spontaneous preterm labor, in which the premature activation of the pro-inflammatory pathway may lead to a breakdown of feto-maternal tolerance and/or activate premature contraction of uterine muscle to induce parturition, resulting in preterm birth.
1.2 |. Pathogenic mechanism
Pathologically, spontaneous miscarriage, including recurrent miscarriage, stillbirth, and spontaneous preterm birth, have been found to share many key overlapping features and risk factors. Maternal diabetes, cigarette smoking, hypertension, and infection during pregnancy are independently dominant risk factors for miscarriage, stillbirth, and prematurity, in which an estimated 15% of premature births are caused by preeclampsia-eclampsia.11–14 Much evidence supports that abnormal inflammation responses are involved in the development of most reproductive system disorders15–21 and has confirmed the involvement of non-coding RNA as an epigenetic regulator in reproductive disorders,22–24 especially in relation to activation of chemokine-cytokine pathways by infection and inflammation.25 Our earlier studies have demonstrated that transcripts including coding and long non-coding RNAs involved in the pathogenic pathway of cytokine and chemokine receptor interactions were differentially expressed and associated with early miscarriages and late preterm birth.26–30 As a result, we proposed that the epigenetic regulation of the chemokine-cytokine pathway could present a common epi-pathogenic mechanism associated with variant adverse outcomes of pregnancy.31
1.3 |. Rationale of this study
In one-dimension study on miscarriage, stillbirth, and preterm birth, the focus is traditionally on searching for a specific gene or gene product as a disease-specific marker. However, human pregnancy is a longitudinally dynamic course along with intrauterine development of the placenta, which could be under the influence of environmental etiologic factors, such as infection and inflammation. Various environmental factor(s) could play an etiologic role to trigger a similar pathogenic pathway in different pathological phenotypes, although the factor may act at various time points during the course of pregnancy and results in variant adverse outcomes, such as miscarriage, intrauterine growth retardation, or preterm birth. Therefore, the variant outcomes of pregnancy may share a similar pathogenic mechanism.
1.4 |. Hypothesis and objective
Given the fact that miscarriage, stillbirth, and preterm birth occur at different time points of pregnancy, arising from different pathogeneses, and present as variant subtypes of phenotypic outcome, infection/inflammation has been documented as an etiological trigger in many studies with a traditional one-dimensional approach, in which cytokine/chemokine receptor interactions pathway can be documented. We hypothesize that variant pathogenic molecule(s) involved in this pathway may be present among variant adverse outcomes of pregnancy. The specific objective of this study was to identify the specific transcripts that are present in variant subtypes of pregnancy loss and preterm birth.
2 |. MATERIALS AND METHOD
2.1 |. Ethics statement
This study was approved by the Lianyungang Municipal Hospital for Maternal and Children’s Health (Hospital Ethics Committee Approval # SH1337, October 2013) to utilize their prebanked clinical specimens along with clinical information from an existing pregnancy cohort. Any information related to personal identification during the biobanking of the placental specimens has been encoded and was anonymous to the authors of this study. Written informed consent was obtained from the pregnant women who participated in the biobanking project for use of the prebanked tissues in the research study.
2.2 |. Pregnancy cohort
All placental specimens used in this current transcriptomic study of stillbirth and of previously genotyped miscarriages and preterm births26–30 were selected retrospectively from our pregnancy cohort that has been biobanked in the Chinese Birth Cohort Network for Preterm Birth and Pregnancies we developed in 2008, supported by the March of Dimes Global Network for Maternal and Infant Health.31–34 Criteria used to select cases from our prebanked pregnancy cohort included: (i) age of the pregnant woman 18–45 years old, (ii) no clinically recognized infection/inflammation, which was determined by phenotypically notable fever; no increased counts of peripheral white blood cells; and/or no increased IL6 and/or TNFα before and/or during pregnancy, (iii) primipara and singleton without medically induced abortion, (iv) vaginal delivery without premature rupture of chorioamniotic membrane, (v) no vaginal bleeding during the pregnancy, (vi) no other pregnancy-related complication(s) and no clinical intervention with antibiotics, steroids, or tocolytics during the pregnancy, (vii) no family history of birth defects, (viii) no chromosomal abnormality reported, and (ix) no consanguinity. Any cases not meeting the above criteria were excluded. Placentas from age-matched pregnant women were divided into five groups, with 10 placentas per group, which were spontaneous miscarriage, recurrent miscarriage, stillbirth birth, spontaneous preterm birth, and full-term birth. Spontaneous miscarriage was further subgrouped as spontaneous miscarriage of fetal tissue, referring to chorionic villi that are fetus-originated, and spontaneous miscarriage of maternal tissue, referring to the maternal portion in the miscarried embryo sac. As a control group, 40 cases of full-term birth were selected from the physiological pregnancies delivered at GW 39–40+6 without premature rupture of membrane and were subjected in variant comparison studies.
2.3 |. Tissue process
Although the placental tissue used in this study was selected from tissue that had been prebanked and stored in our biobank, a standardized and harmonized procedure for banking placental tissues was performed. Briefly, immediately following delivery, the placentas were flushed with 200 ml cold distilled water until no blood was visible, dried with clean paper towels, sliced with a sterile scalpel into 1 × 1 cm2 cubes at a site 2 cm from the edge of the placenta, juxtaposed from the maternal decidua through all layers of placenta reaching the chorioamniotic membrane (including the membrane), quickly frozen in liquid nitrogen for 30 min, and stored at −80°C until use. The entire procedure was by necessity completed within 30 min after the placenta was delivered.
2.4 |. RNA microarray hybridization and bioinformatics analysis
The microarray hybridization procedure used in this study has been described elsewhere.26,27 Briefly, total RNA was isolated from the biobanked placental tissues of variant adverse pregnancies with the standard procedure we described.26,27 mRNA was purified from total RNA after removal of ribosomal RNAs with the mRNA-ONLY™ Eukaryotic mRNA Isolation Kit (Epicentre). Each sample was amplified and transcribed into fluorescent complementary RNA (cRNA) along the entire length of the transcript without a 30 bias by utilizing the random priming method. The labeled cRNAs were purified by using a RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured with NanoDrop ND-1000. One microgram of each labeled cRNA was fragmented by adding 5 μl 10× blocking agent and 1 μl of 25× fragmentation buffer and then heating the mixture to 60°C for 30 min. Finally, 25 μl 2×GE of hybridization buffer was added to dilute the labeled cRNA. Fifty μl of hybridization solution was dispensed onto the gasket slide and assembled with the expression microarray slide. The Arraystar microchip slides (www.arraystar.com) were hybridized with the labeled cRNA, incubated for 17 h at 65°C in an Agilent hybridization oven, washed, fixed, and scanned by using the Agilent DNA Microarray Scanner (Agilent Technologies). Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed by using the GeneSpring GX v12.1 software package (Agilent Technologies). p value <0.05 and fold change threshold (absolute ratio) >2 were considered as significantly different. The gene GO and pathway enrichment was analyzed by using g:profiler online tools (https://biit.cs.ut.ee/gprofiler/gost database updated on 02/10/2019) and DAVID 6.8 online analysis software (https://david.ncifcrf.gov/), as we reported.26–30 Online STRING (https://string-db.org) was applied to study gene associations.
2.5 |. Statistical analysis
The transcriptomic data were measured for placental tissues from variant groups and were compared with each other in a one-way ANOVA. All data were examined for normality and homogeneity of variance among all groups, as we have described elsewhere.26–30 The significance of cutoff for data generated from each individual intragroup of transcriptomic differential expression profiles (DEGs) was set as p < 0.05, false discovery rate (FDR) < 0.05, and log (fold change) > 2. If variances were unacceptably unequal, adjustments for unequal variances were performed. If these measures were insufficient, bootstrapped or non-parametric methods were used. A significance criterion of 0.01 was used throughout. Our principal concern, however, was to establish effect sizes and their confidence bounds.35
3 |. RESULTS
3.1 |. Differential expression genes in spontaneous miscarriage, recurrent miscarriage, stillbirth, and spontaneous preterm birth
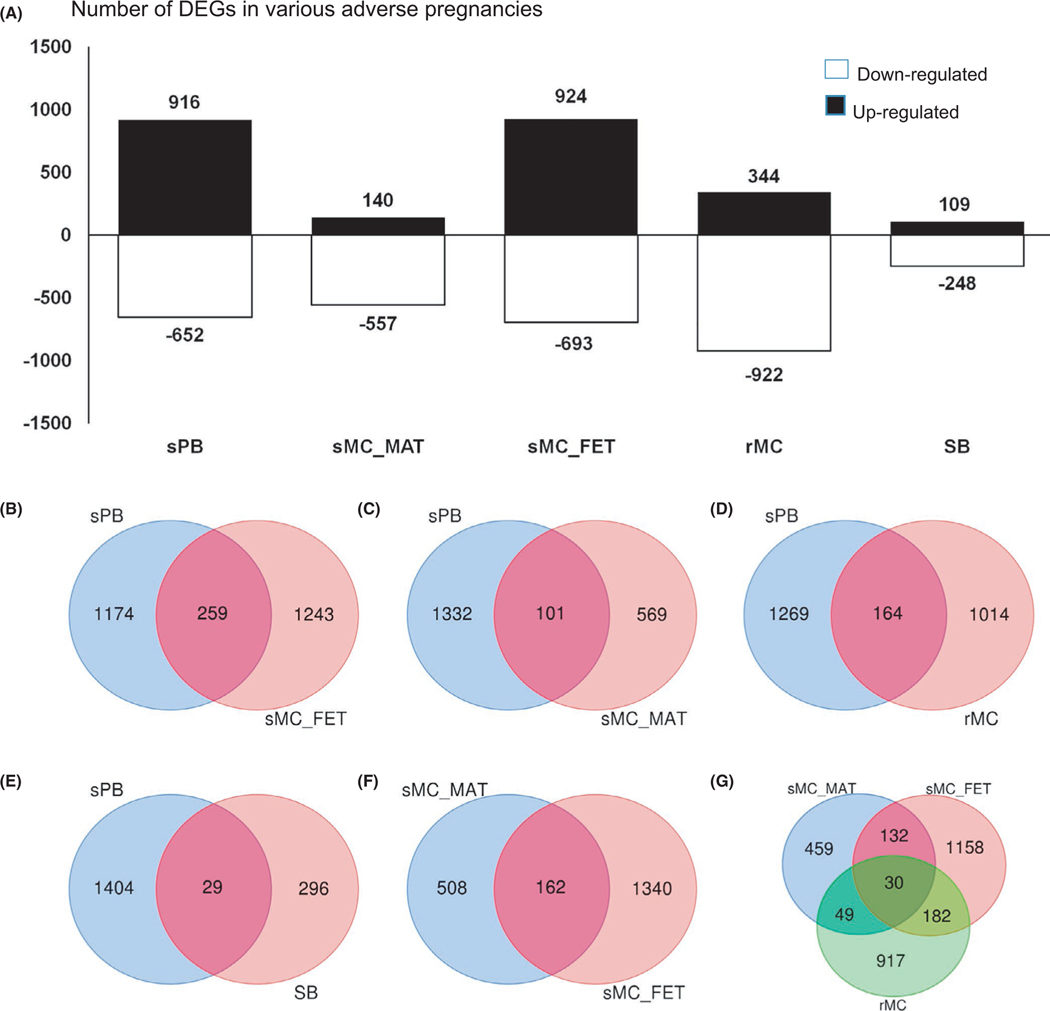
The DEGs of transcripts were generated from five groups (Figure 1A). Variant comparisons were performed with FTB vs. X, where X represents variant outcome of pregnancies. In total, 1568 DEGs in spontaneous preterm birth; 697 in the maternal tissues of sMC-MAT; 1617 in the fetal tissue of sMC-FET; 1266 in recurrent miscarriage; and 357 in stillbirth were identified (Figure 1A). Due to the fact that different variants or isoforms may display the same gene, duplication of gene symbols could be presented in the same groups. After the duplicated gene symbols were removed, there were 1433 DEGs in spontaneous preterm birth; 259 being present in sMC_FET (Figure 1B), 101 in sMC_MAT (Figure 1C), 164 in recurrent miscarriage (Figure 1D), and 29 in stillbirth (Figure 1E). Details of the overlapped DEGs among these groups are presented in Table 1. We also found that the fetal facet and the maternal facet of placenta from spontaneous miscarriage show very different DEG profiles and that 162 common DEGs were shared in both groups (Figure 1F). Within these 162 DEGs, 30 transcripts were overlapped with recurrent miscarriage (Figure 1G, Table 1). Interestingly, nine transcripts (FAM129C, FAM167A, MSLN, TSHB, FGFR1, CACNA1G, DERL3, MATK, PLAC4) were overlapped between miscarriage (spontaneous miscarriage and recurrent miscarriage recurrent miscarriage) and spontaneous preterm birth, two (HBG2, ACOT7) between miscarriage and stillbirth, and one nerve growth factor receptor (NGFR) among miscarriage, stillbirth, and spontaneous preterm birth (Table 1), among which the differentially expressed transcript of NGFR was documented as down-regulated when compared to the control group.
FIGURE 1.
Differential expression genes (DEGs) and Venn diagram of interactions. A, DEGs in spontaneous miscarriage (sMC), maternal (sMC_MAT) and fetal (sMC_FET); recurrent miscarriage (rMC); stillbirth (SB); and spontaneous preterm birth (sPB). B-G, Venn diagram of interactions between sPB and sMC_FET (B), sPB and sMC_MAT (C), sPB and rMC (D), sPB and SB (E), sMC_MAT and sMC_FET (F), and sMC_MAT/sMC_FET, and rMC (G).
TABLE 1.
DEGs shared among adverse pregnancies
| Adverse pregnancy* | Number of DEGs shared | DEGs shared between/among pregnancies |
|---|---|---|
| sMC_MAT & sPB | 101 | PTN MATK CHRDL1 MBNL2 BTLA FGFR1 SOX4 LRRN1 AK3L1 FAM129C ELANE SMAD3 MRVI1 IL17RC ISM1 BDNF STEAP3 TIPARP PCSK6 NM_006896 C2orf63 DYRK2 ZNF709 MORF4L2 CAMK2B NGFR FAM22G LDHA FAM167A CPT1B PPP2R5C LOR TMEM146 BCAR1 PODXL KCNH6 KCNMB3 ANG GART ANO7 NM_014212 C13orf33 GAL C17orf62 TMPRSS2 FBXO15 TEF BIN1 KIAA0040 NUDT2 NM_002144 SLC1A6 RARA BDH1 NEIL2 CD59 PRR21 SH3BP5L PCDH11Y MOV10L1 SPOP PLEKHN1 CLEC7A CMPK1 DBNDD1 NM_014620 RAD54L TAZ IL1F8 PANX2 REG1A FAM3A CMTM5 GABRB3 ZNF185 POLR2J3 FGFBP2 MSLN NM_153715 NOG HBS1L LSR RASA1 KCNB1 SSX2IP MT1H PLAC4 FLJ36031 RPN2 KCNIP2 CD69 TSHB NRG1 TGFBI TEX11 GIT2 NCOA4 CACNA1G DUSP13 RFX2 DERL3 |
| sMC_FET & sPB | 259 | CXCR4 CXCL3 HOXD11 ZG16 SOD2 FGFR1 HLA-DRB5 AK3L1 TSC22D1 LHB FAM129C CD38 LAIR1 BTN3A1 KLHL30 C12orf75 ERBB2IP HLA-DRA CHST6 DAPP1 GPR171 ENHO CARS PAEP OMD OAS1 TNF DIXDC1 NGFR IL8 RCAN1 UPK1B MYST3 TEX9 CASP1 GALNTL2 CFHR1 GLIPR2 MSR1 CCL4 SST RHOV TRPC4 EPSTI1 KCNMB3 PSMB8 NM_014212 EOMES TNFRSF8 C13orf33 MAPK8IP2 MKKS RGS1 PPARD FAM63A PDZK1IP1 VASH2 EIF2AK2 PAM NEIL2 NFKBIL1 POSTN MOV10L1 SPOP PRKD2 IGFBP6 MXRA7 CFL2 PDLIM4 CGB ERP27 TTC8 CCL3L1 CD74 F2R GZMB BMP1 DPF1 HLA-G MTDH SLC25A20 ADAR PTPRN C6orf127 ABCA9 CAMTA2 GAS1 GABRB3 CXCL10 IQSEC3 NM_153715 RASA1 CTNND1 KMO CLDN1 UPF3B HPSE FLJ36031 NM_021192 NLE1 DCN CNNM2 CGB8 SERPINA3 IRF9 NDP CD3E DDX20 RELL2 CGB1 RNASE4 HTRA4 CH25H CACNA1G VGLL4 AS3MT SCARF1 RFX2 HOPX DERL3 UGCG TSPAN11 ADRA2C ZNF837 L3MBTL MATK FAM43B FAM109A HCP5 HOMER3 PRL ACACA COG6 CCL3L3 SHISA3 TMEM45A NM_014621 LAIR2 BTN3A2 BDNF FGG CLEC2B PPAP2A TSPAN8 PPP1R8 DUSP2 ZNF692 IGFALS ECE2 TP73 ANTXR2 GZMK KLRC2 KLF6NKG7 ABCA8 LEPROT FBLN1 HLA-DPA1 MPO S100A14 CD27 FAM167A KYNU CAPG TBXAS1 NM_018952 CRLF1 ABI3BP CGB2 LRRC15 DIRAS2 CAMP CRYBG3 KCNH6 CD9 CCL4L1 NDOR1 RGR ANG G0S2 ANO7 REG3G RXFP1 TMEM63B CCL2 NUDT3 FAM26F SIGLEC6 ISYNA1 ACE IRF4 NUDT2 HLA-F ASGR2 CGB7 C7orf10 ZNF580 LAYN MGLL SCARA5 MAOB BTNL8 DMKN IFI6 GZMA CDH16 FGF7 PION CYS1 FOXL2 DOCK10 SPOCK2 CA10 ARAP2 PRR15 GJB2 ALDH1A1 CHIC2 TMCC1 ANK1 HIF1A ACSF3 C5orf20 LILRB5 COL17A1 DMAP1 ZSWIM5 PRKAR1B SERPINE1 ARC CDH23 MSLN HLA-DRB1 KLK10 NM_019102 PIP FABP4 KCNB1 PCP4 GTF2H2D ZBED2 CLDN8 CITED4 CCL25 PLAC4 MAGEB4 CD69 KLF10 BCAT1 AHNAK2 TSHB CCL3 FBXO32 FAM123C GATA4 CENPF NUSAP1 PSMB9 |
| rMC & sPB | 164 | HSD17B1 IFT122 FGFR1 CSF3R LHB FAM129C ITM2B ZNF83 PDCD4 NM_002145 FBXO11 KLHL30 CAPN12 PAEP AHR SLC35E2 COQ10B MUC15 RBBP6 PRND SOX9 CYHR1 COQ10A ASCL2 SH2B1 CASP1 LYPD3 KCNAB1 SNCA HRAS HN1 CSHL1 KIF9 UXT SNRNP48 LAMA3 GAL GNGT1 OR5H14 AGA LRRFIP1 VASH2 PAM FLT1 NUCKS1 WDR70 NFKBIL1 POSTN CGB APOLD1 FBLN2 TRA2A BMP1 MTDH REG1A CAMTA2 PDE4DIP FBXL16 IMPDH1 KMO AGBL5 KCNIP2 DEK NLE1 IFNAR2 CGB8 PSG11 HIST1H1C CYFIP2 SRRM2 HNRNPA1KTN1 C1orf104 CACNA1G HOPX OAT DERL3 KCTD14 MATK SOX4 NANOG PSG8 SMPD4 EXT2 PTPN12 CD55 SYNGR3 CMTM1 C20orf201 OSMR ARID3A ZNF117 ZNF692 PPAP2C HIST1H2BC IGFALS ERAP1 TBXA2R SMAGP RCC2 PDCD7 KLF6 KIAA1217 FBLN1 PPHLN1 GSN MPO GH2 FAM167A CPT1B KYNU CAPG PSG5 TPM1 HEXB TMEM169 CGB2 CLCN7 JPH4 SFN RND3 IL1RAP TMEM106A TMED2 C16orf88 KCNK12 TMEM63B ALDOA HNRNPH3 HMGB2 LPAR2 ACE DEFA3 PRKCSH HLA-F ASGR2 CGB7 CD59 SCARA5 PCDH11Y LILRB1 PIH1D2 DOK3 ISM2 C21orf91 PANX2 RAD17 CDKN1C DMAP1 PRKAR1B SERPINE1 MSLN KIAA0319 PATE3 CYB561D1 TMEM150A PLAC4 TSHB NRG1 FAM123C WAC BAX SFRS11 GATA4 |
| sPB & SB | 29 | MAP4 SOD2 MAX TPM3 FYB SMAGP PERP RDX SVIL NGFR LDHA TBXAS1 MCM10 UBE2Q2 UXT NR2F2 LMNB1 RAP1B MARK2 CSAD ALDH1A1 RAD54L COL17A1 PDE4DIP TFEB CALD1 LTBP3 CENPF NUSAP1 |
| rMC & sMC_FET & sMC_MAT | 30 | KCNK16 CYP11A1 FGFR1 BBC3 FAM129C HBZ ACOT7 ARAP1 HBG2 PGM1 SLCO1C1 TRIM34 CLSTN3 LGALS13 CACNA1G DERL3 MATK GPR162 LRTOMT SCOC MAMSTR FAM167A LGALS14 C11orf21 CTBP2 MSLN PEMT PSG6 PLAC4 TSHB |
| rMC & sMC_FET & sMC_MAT & sPB | 9 | FAM129C FAM167A MSLN TSHB FGFR1 CACNA1G DERL3 MATK PLAC4 |
| sMC_FET & sMC_MAT & sPB & SB | 1 | NGFR |
| rMC & sMC_FET & sMC_MAT & SB | 2 | HBG2 ACOT7 |
sMC, spontaneous miscarriage; sMC_MAT, spontaneous miscarriage maternal; sMC_FET, spontaneous miscarriage fetal; rMC, recurrent miscarriage; SB, stillbirth; sPB, spontaneous preterm birth
3.2 |. Functional enrichment of differential expression genes in spontaneous miscarriage, recurrent miscarriage, stillbirth, and spontaneous preterm birth
Functional enrichment analysis (including GO and pathways enrichment) of DEGs in each type of adverse pregnancy is described in this article (Figure 2). For spontaneous preterm birth vs. full-term birth (Figure 2A), the most enriched biological pathways were cellular response to organic substance and positive regulation of biological process, and the greatest molecular function was cell adhesion molecule-binding and signaling receptor binding. The only reactome pathway that was enriched was interferon gamma signaling, in which 13 genes (IFNG, IRF9, GBP3, IFNGR1, OAS1, GBP4, HLA-G, SUMO1, IRF2, JAK1, HLA-DPA1, HLA-DRB5, and HLA-DRA) were involved. For stillbirth vs. full-term birth (Figure 2B), the top two enriched biological processes were DNA package and chromosome organization, the molecule functions were protein heterodimerization activity and DNA binding, and the cell components were DNA packaging complex and nucleosome. The KEGG pathways were systemic lupus erythematosus, alcoholism, and hypertrophic cardiomyopathy.
FIGURE 2.
Enrichment GO functions or pathways of DEGs using g:Profiler. A, sPB vs. FTB; B, sPB vs. FTB; C, sPB vs. FTB; and D, rMC vs. FTB; E, Comparisons of the functional enrichment of DEGs in sMC_MAT. F, Comparisons of the functional enrichment of DEGs in MC_FET.
For spontaneous miscarriage vs. full-term birth (Figure 2C), in which spontaneous miscarriage includes both fetal and maternal portions of the placenta, the top three enriched biological processes were monocyte chemotaxis, leukocyte migration, and the molecular functions receptor ligand activity, signaling receptor binding, and chemokine receptor binding; the cell components were the extra-cellular region and the MHC protein complex. The top three KEGG pathways were graft-versus-host disease, type I diabetes mellitus, and allograft rejection.
The functional enrichment of DEGs in recurrent miscarriage is shown in Figure 2D. Seven genes were involved in pregnancy, which was consistent with clinical features. At the molecular function level, hormone activity and oxygen carrier activity were the main functions. The reactome pathway clustering showed that cell surface interactions at the vascular wall (PSG8, PSG1, PSG5, PSG4, PSG3, PSG7, PSG11, PSG9) and glycoprotein hormone pathways (TSHB, CGB8, INHA, LHB, CGA) are involved in recurrent miscarriage.
As indicated in Figure 2E,F, the immune response was enriched in fetal tissue of chorionic villi and in maternal-originated tissue of placental sac. For molecular function, chemokine receptor binding and receptor ligand activity as well as graft-versus-host disease and allograft rejection pathways were mainly enriched in the fetal villi. However, fibroblast growth factor-activated receptor activity and potassium channel activity were the top two enrichment functions in sMC_MAT. Of the seven biological processes involved in both the fetal and the maternal tissues, regulation of cell population proliferation and cell-cell signaling were the top two processes (Table 2).
TABLE 2.
The common pathways involved in both sMC_MAT and sMC_FET
| Term name | Term_ID | Adjusted p value sMC_MAT | Intersection size sMC_MAT | Adjusted p value sMC_FET | Intersection size sMC_FET |
|---|---|---|---|---|---|
| Regulation of cell population proliferation | GO:0042127 | 0.014424 | 86 | 2.50E-06 | 107 |
| Cell-cell signaling | GO:0007267 | 0.005249 | 82 | 2.08E-05 | 119 |
| System development | GO:0048731 | 0.016813 | 205 | 2.28E-05 | 228 |
| Developmental process | GO:0032502 | 0.038194 | 260 | 2.63E-05 | 281 |
| Anatomical structure development | GO:0048856 | 0.044655 | 235 | 4.68E-05 | 265 |
| Animal organ development | GO:0048513 | 0.000135 | 105 | 0.000712 | 256 |
| Skeletal system development | GO:0001501 | 0.003388 | 38 | 0.001446 | 69 |
| Anatomical structure morphogenesis | GO:0009653 | 0.005859 | 129 | 0.008479 | 120 |
| Fibroblast growth factor-activated receptor activity | GO:0005007 | 0.001934 | 4 | 1 | 1 |
| Potassium channel activity | GO:0005267 | 0.002006 | 16 | 1 | 11 |
| Animal organ formation | GO:0048645 | 0.000152 | 13 | 1 | 7 |
| Pattern specification process | GO:0007389 | 0.000384 | 36 | 1 | 19 |
| Chemokine receptor binding | GO:0042379 | 1 | −1 | 2.93E-10 | 18 |
| Defense response | GO:0006952 | 1 | 49 | 2.72E-20 | 138 |
| Graft-versus-host disease | KEGG:05332 | 1 | −1 | 1.95E-10 | 13 |
| Interferon gamma signaling | REAC:R-HSA-877300 | 1 | 1 | 2.01E-05 | 13 |
| Interleukin-10 signaling | REAC:R-HSA-6783783 | 1 | 1 | 2.56E-05 | 14 |
Abbreviations: sMC_FET, spontaneous miscarriage fetalsMC_MAT, spontaneous miscarriage maternal.
Although the expression pattern was quite different between spontaneous miscarriage and recurrent miscarriage, enrichment analysis indicated that heme binding was the common molecular function, with three down-regulated genes (CYP11A1, HBZ, and HBG2) involved, and positive regulation of the T cell apoptotic process was the common pathway, in which three down-regulated DEGs (BBC3, LGALS13, and LGALS14) were involved. The apoptosis pathway (including the leukocyte apoptotic process) and the Kennedy pathway from the pathway of sphingolipids were enriched (Table 3).
TABLE 3.
Functional enrichment of common DEGs in sMC_FET and sMC_MAT
| Term_name | Term_ID | Adjusted p value | Intersection_size | Intersections |
|---|---|---|---|---|
| Leukocyte apoptotic process | GO:0071887 | 0.0047 | 8 | BBC3, BIRC7, SLC7A11, LGALS14, BCL2L11, LGALS13, FCER1G, CRKL |
| Lymphocyte apoptotic process | GO:0070227 | 0.042673 | 6 | BBC3, BIRC7, LGALS14, BCL2L11, LGALS13, CRKL |
| Apoptosis - multiple species | KEGG:04215 | 0.020838 | 4 | BBC3, BIRC7, NGFR, BCL2L11 |
| Kennedy pathway from sphingolipids | WP:WP3933 | 0.039604 | 3 | ETNK2, SGPL1, PEMT |
Abbreviations: DEGs, differential expression genes; sMC_FET, spontaneous miscarriage fetal; sMC_MAT, spontaneous miscarriage maternal.
3.3 |. The specific and common pathways shared among spontaneous miscarriage, recurrent miscarriage, stillbirth, and spontaneous preterm birth
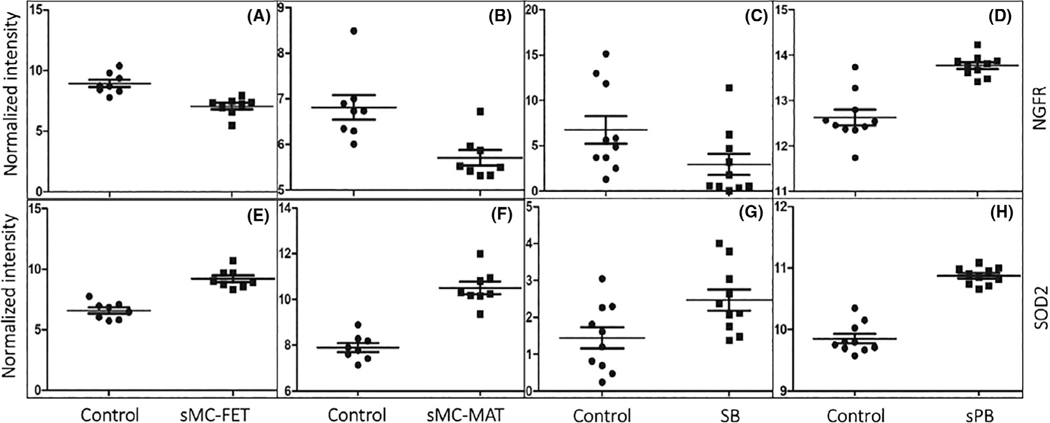
Many functional pathways were found to be involved in one of the spontaneous pregnancy losses or in spontaneous preterm birth (Figure 3A). For example, response to lipopolysaccharide and cytokine signaling was dysregulated in spontaneous preterm birth; DNA packaging and metabolism was the most enriched in stillbirth; defense response, graft-versus-host disease, antigen processing and presentation, and T help cell differentiation were involved in spontaneous miscarriage; and the oxygen transport, cell surface interactions at the vascular wall, and hemostasis were mainly disturbed in recurrent miscarriage. These specific functional characteristics reflected the difference among these abnormal reproductive outcomes, whereas some pathways, as indicated in Figure 3B, were identified to be dysregulated in three or more adverse pregnancies. For example, spontaneous preterm birth, spontaneous miscarriage, and recurrent miscarriage all responded significantly to cytokine signaling, and the haptoglobin binding pathway was the common pathway that was observed in stillbirth, spontaneous miscarriage, and recurrent miscarriage. The detailed molecules involved in either a unique pathway or in a pathway shared by variant adverse pregnancies can be documented. For example, SOD2 was the only gene up-regulated in spontaneous preterm birth, stillbirth, and fetal facet from spontaneous miscarriage, as validated by qRT-PCR (Figure 4). In which, NGFR that was the only transcript presented in all adverse outcome of pregnancies was shown to be up-regulated in sPB but down-regulated in sMC-FET, sMC-Mat, and SB. Thirty-three DEGs were determined to be dysregulated in both spontaneous preterm birth and spontaneous miscarriage. In addition, LMNB1 and RAP1B were differentially expressed in stillbirth and spontaneous preterm birth, and MUC1 and HLA-A were up-regulated in spontaneous miscarriage and spontaneous preterm birth. These results suggested that spontaneous preterm birth may have shared common pathogenic mechanism(s) with spontaneous miscarriage, recurrent miscarriages, or stillbirth.
FIGURE 3.
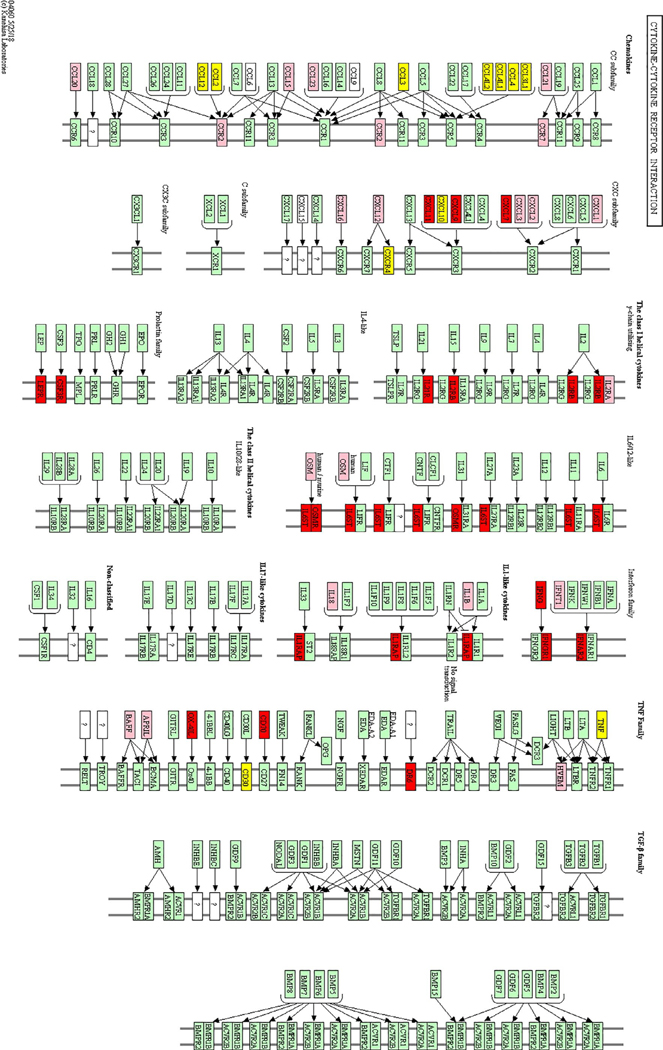
Functional pathways analysis. A, Specific functional pathways among sPB, SB, sMC, and rMC. 1. Reproductive structure development; 2. Placenta development; 3. Cellular response to lipopolysaccharide; 4. Type II interferon signaling (IFNG); 5. DNA packaging; 6. Systemic lupus erythematosus; 7. Alcoholism; 8. Hypertrophic cardiomyopathy (HCM); 9. Cytokine activity; 10. Th17 cell differentiation; 11. G protein-coupled receptor binding; 12. Defense response; 13. Interleukin-10 signaling; 14. Monocyte chemotaxis; 15. Immune response; 16. Response to external stimulus; 17. Leukocyte migration; 18. Graft-versus-host disease; 19. Allograft rejection; 20. Antigen processing and presentation; 21. Th1 and Th2 cell differentiation; 22. Oxygen transport; 23. Cell surface interactions at the vascular wall; 24. Hemostasis. B, Common functional pathways among sPB, SB, sMC, and rMC. 1. Cellular response to chemical stimulus; 2. Response to organic substance; 3. Cellular response to interferon gamma; 4. Response to interferon gamma; 5. Response to cytokine; 6. Multi-multicellular organism process; 7. Receptor ligand activity; 8. Signaling receptor activator activity; 9. Response to endogenous stimulus; 10. Response to chemical; 11. Cellular response to cytokine stimulus; 12. Negative regulation of cellular process; 13. Protein dimerization activity; 14. Receptor regulator activity; 15. Hormone activity; 16. Female pregnancy; 17. Haptoglobin binding. C, STRING analysis of 10 common DEGs in cytokine-cytokine receptor interaction. Red indicates eight genes involved in the chemokine signaling pathway, and purple indicates six genes involved in interleukin-10 signaling.
FIGURE 4.
Verification of differential expression of NGFR and SOD2. The differentially expressed NGFR and SOD2 identified through transcriptomic studies were verified in sMC, SB, and sPB. With qRT-PCR, transcript of NGFR was shown to be significantly down-regulated in sMC-FET (p < 0.01), sMC-MAT (p < 0.05), SB (p < 0.05), but up-regulated in sPB (p < 0.01). The transcript of SOD2 was up-regulated in all sMC-FET (p < 0.01), sMC-MAT (p < 0.01), SB (p < 0.05), and sPB (p < 0.01).
The KEGG pathway indicated that cytokine-cytokine receptor interaction was the most enriched pathway in spontaneous preterm birth and spontaneous miscarriage (Table 4). To gain additional understanding of the response to cytokines in the biological processes involved in spontaneous preterm birth, stillbirth, and spontaneous miscarriage, the DEGs in response to cytokines were analyzed further. Table 5 shows the DEGs involved in response to cytokines in spontaneous preterm birth, stillbirth, and spontaneous miscarriage. Twenty-seven DEGs in spontaneous preterm birth and 26 DEGs in spontaneous miscarriage were mapped to the KEGG pathway of cytokine-cytokine receptor interaction (Figures 3C and 5). Ten common DEGs (CCL3, TNF, CCL2, CXCL3, TNFRSF8, CCL4, CXCL10, CXCR4, CCL3L3, and CCL4L1), which are up-regulated in both spontaneous preterm birth and spontaneous miscarriage, were mainly focused on chemokines (including CC subfamily and CXC subfamily), TNF, and TNFRSF8 (CD30). For spontaneous preterm birth, the DEGs focused on the CXC subfamily, class I helical cytokines, and interferon family. The DEGs in stillbirth included chemokines, IL2RA, OSM, and the interferon family.
TABLE 4.
KEGG pathway of DEGs involved in response to cytokines in SB, sMC, and sPB
| Adverse pregnancy | KEGG pathway | p value | Count | DEG |
|---|---|---|---|---|
| SB | hsa05322:Systemic lupus erythematosus | 8.21E−15 | 14 | |
| hsa05034:Alcoholism | 3.17E−13 | 14 | ||
| hsa05202:Transcriptional misregulation in cancer | 4.18E−12 | 13 | ||
| hsa05130:Pathogenic Escherichia coli infection | 0.023197714 | 3 | ||
| sMC | hsa04060:Cytokine-cytokine receptor interaction | 6.28E−20 | 26 | CXCL1, CCL3, TNF, CCL2, CXCL3, IL18, TNFRSF8, TNFSF13, CXCL12, CCL4, CXCL10, CCL23, CCL20, IFNE, CCL21, CXCR4, CCL3L3, IL1B, IL2RA, CCL4L1, TNFRSF14, OSM, CCR7, TNFSF13B, CXCL16, CCR2 |
| hsa05323:Rheumatoid arthritis | 2.39E−15 | 16 | ||
| hsa05168:Herpes simplex infection | 8.41E−13 | 18 | ||
| hsa04062:Chemokine signaling pathway | 1.10E−12 | 18 | ||
| hsa04940:Type I diabetes mellitus | 4.41E−12 | 11 | ||
| sPB | hsa04060:Cytokine-cytokine receptor interaction | 1.31E−16 | 27 | TNFRSF21, CCL3, CCL2, TNF, OSMR, IL6ST, LEPR, CXCL3, IL21R, CXCL9, TNFRSF8, CD70, CXCL11, CCL4, CXCL10, CXCR4, CCL3L3, IFNG, IL1RAP, CSF3R, CD27, IFNGR1, IL2RB, TNFSF4, CCL4L1, IFNAR2, PPBP |
| hsa05164:Influenza A | 2.21E−11 | 19 | ||
| hsa04612:Antigen processing and presentation | 4.56E−10 | 13 | ||
| hsa05168:Herpes simplex infection | 4.70E−10 | 18 | ||
| hsa04630:Jak-STAT signaling pathway | 1.23E−09 | 16 |
Abbreviations: DEGs, differential expression genes; SB, stillbirth; sMC, spontaneous miscarriage; sPB, spontaneous preterm birth.
TABLE 5.
DEGs involved in response to cytokine in sPB, SB, and sMC
| Adverse pregnancy | Common DEGs number | Gene name |
|---|---|---|
| SB & sMC & sPB | 1 | SOD2 |
| SB & sPB | 2 | LMNB1 RAP1B |
| sMC & sPB | 33 | IRF4 CXCL10 FGG CAMP CXCR4 HLA-F SPOCK2 CCL4L1 EIF2AK2 CXCL3 OAS1 CD74 TNF HLA-DRA HLA-DPA1 HLA-DRB1 CCL3 HLA-DRB5 TNFRSF8 IL8 HLA-G FABP4 IFI6 KYNU CCL2 CCL3L3 POSTN SMAD3 KMO PTPRN PSMB9 CRLF1 CCL4 |
| SB & sMC | 2 | MUC1 HLA-A |
| sPB | 98 | NFAT5 HCK OSMR LEPR UGCG IFNG ABCG4 CARD14 GBP3 RARG IL1RAP SNCA CDC5L VCAM1 SOX9 RBM15 SUMO1 IFNAR2 HLA-C NPNT REL IFNGR1 VRK2 CAMK2B PSMB8 HK2 IL33 RARA GSN PRTN3 COMMD7 AGPAT1 CXCL9 ITGB1 AVPR2 BCL2L1 LGALS9 IRF9 CRK TNFRSF21 PPBP NANOG HSPA1B JAK1 CSF3R IRF7 HNRNPU IL6ST TNFSF4 HSP90AB1 SMPD4 CISH ARID5B ETS1 PTPN12 CD27 EPRS HSP90B1 NCL IL2RB SFRP1 CASP1 GBP4 XAF1 HIF1A ADAM9 PIAS1 PARP16 CXCL11 GGT5 SOCS5 PDCD4 KARS ADAR MAP4K3 BCLAF1 CLDN1 MKKS PARP14 CD70 SAA1 IL21R ELF1 REG1A CD38 FN1 FBXW11 IRF2 ADAMTS12 IL17RC RFX2 HSP90AA1 TWISTNB PIM1 RFFL DOCK8 DHX9 CD14 |
| SB | 39 | HIST1H3B TYMS PLVAP NDUFA13 IRAK1 SBNO2 HIST1H3A RPLP0 HIST2H3D HIST1H3H HELLS PCOLCE2 HIST1H3J HIST1H3D VIM eACTG1 HIST1H3F DDOST KRT8 PSMA3 HIST1H3E NR1H2 ACP5 HIST1H3C HIST1H2BJ HIST2H3A EED STAT1 MYBL2 PDCD10 B2M BIRC5 HIST1H3I TUBA1B HIST2H3C HCLS1 ACTR2 PF4 HIST1H3G |
| sMC | 54 | LCN2 GIPC1 F2RL1 CD44 CCL21 GPR17 MTF2 TNFSF13B HAMP CEACAM1 SELPLG SMARCA4 CCL23 HLA-DPB1 CXCL12 OAS2 CRHBP IFNE RELB CCR2 JUN STAT4 SOCS3 CXCL16 CD300LF IL2RA TIMP1 CRKL FGB NCAM1 MAP3K5 PIK3R1 CXCL1 CHI3L1 FOXO1 CCL20 TRIM34 TNFRSF14 IL1RL1 OSM TREM2 ALDH1A2 TNFSF13 CD58 CCR7 TRIM6 IL1B IL18 CCRL2 MMP9 FCER1G TFPI IFI27 ADIPOQ |
Abbreviations: DEGs, differential expression genes; SB, stillbirth; sMC, spontaneous miscarriage; sPB, spontaneous preterm birth.
FIGURE 5.
KEGG pathway of cytokine-cytokine receptor interaction involved in sPB and sMC. Yellow: molecules shared in sPB and sMC; red: DEGs in sPB; and pink: DEGs in sMC.
4 |. DISCUSSION
4.1 |. Research design
Many pregnancies undergo subclinical infection that triggers intrauterine inflammation. Longitudinally, the unrecognized infection and inflammation may induce miscarriage, stillbirth, or preterm birth, depending on when the induction occurs or which molecule(s) have been impacted by the epigenetic regulation that is involved in variant differentially expressed pathways. To identify the molecules that may be involved in the pathogenic mechanism that associates with infection and inflammation, we applied a novel approach to integrate the transcript molecules that had been found to be involved in the chemokine-cytokine pathway in individual phenotypes of pregnancy loss and in preterm birth. It was hoped that with this approach, we could identify the transcripts that are common among adverse pregnancies, regardless of whether or not these pregnancies may present at different stages of gestation through multiple etiological factors. This approach could facilitate possible functional analysis for the purpose of better understanding the molecular mechanism and the pathogenic role of common (or unique) pathways/transcripts in various phenotypes of pregnancy.
Applying an integrative approach to merge all transcriptomic data from variant adverse pregnancies, which may have a high variability in gestational age, causality, mothers’ co-morbidities, and pathologies with separate phenotypic outcomes, to study the common shared pathogenic pathways and molecules could create the impression that the study was designed illogically, if our vision appeared to be only to examine individual phenotypes and outcomes at the end point of pregnancy. Therefore, with a broader vision looking from embryo to the later gestational stage of fetus, we hypothesized that variant adverse outcomes of pregnancy could share common pathogenic pathway(s) and initiated this current investigation with an integrative transcriptomic approach.
Pregnancy is a longitudinal and dynamic process, progressing from a fertilized single cell to the human body consisting of multiple organs and systems. The placenta, as the maternal-fetal interface, plays a key function during the entire pregnancy period, particularly at the early stage of gestation. The early development and function of the placenta have a critical role in physiological pregnancy and intrauterine fetal development. Should they be attacked by pathogenic factor(s), the normal pregnancy would be affected, and pregnancy loss as well as preterm birth could occur. Many genetic and environmental risk factors including gene-environmental interaction have been known to interfere with placental development and function, which could result in the phenotypes of adverse pregnancy. These risk factors may attack the placenta at different gestational timepoints or target different placental cells, resulting in variant subtypes of pregnant outcomes. If interference with the placenta’s development and function was the only risk factor associated with the outcome of pregnancy, it would be much easier to understand how the adverse outcomes would occur. Unfortunately, other maternal and/or fetal conditions or factors may also impact the pregnancy. Among such heterogenous and complicated conditions and factors, infection and inflammation that are associated with the chemokine-cytokine pathway present a good indication that clinical infection and inflammation can occur at any time during gestation to trigger maternal and fetal immunoresponses. Such immunoresponses may include activation of the cytokine and chemokine molecules through epigenetic regulation and transcription. In this case, it is logical to believe that the transcribed molecule may be present in the placenta since the placenta is the maternal-fetal interface. Therefore, it would not be surprising to identify the common pathway(s)/molecule(s) among variant outcomes of adverse pregnancies. The common pathways and molecules identified and documented in this study provide solid evidence to demonstrate our success.
4.2 |. Identification of cytokine pathway among variant pregnancies
To find the common pathways or common molecules involved in variant adverse outcomes, function/pathway enrichment was analyzed for all DEGs in each disorder. Our results showed that the top-scoring pathway in variant groups of adverse pregnancies differed from each other such that cytokine signaling was dysregulated in spontaneous preterm birth; DNA packaging and metabolism in stillbirth; defense response, graft-versus-host disease, antigen processing and presentation, and T help cell differentiation involved in spontaneous miscarriage; and oxygen transport in recurrent miscarriage. However, the chemokine-cytokine pathway is present in all groups. Interestingly, SOD2, a key molecule involved in the cytokine pathway, was identified as the common gene shared by spontaneous miscarriage, stillbirth, and spontaneous preterm birth. SOD2 is a mitochondrial antioxidant enzyme that plays a crucial role in controlling reactive oxygen species production, being responsible for detoxification of superoxide anion in human reproduction.36,37 Oxidative stress with inflammation is known to be linked to various pregnancy complications, but it has not been studied extensively in preterm labor or preterm premature rupture of fetal membrane,38,39 nor have been the magnitude of oxidative stress generated by the environmental risk factor, antioxidant capacity of feto-maternal tissues, oxidative stress-induced cellular damage, and signaling pathways activated by cellular elemental damage.40 SOD2 has been determined to be associated with recurrent miscarriage.41 SOD2 level in serum of miscarriage was reported to be reduced, and activity of SOD2 was decreased in plasma and placenta tissues in recurrent miscarriage.42 In chorioamniotic membrane samples, SOD2 was over-expressed in term labor compared to term with no labor,43 which could restrain the ability of cytotrophoblasts to fuse and differentiate into syncytiotrophoblasts.44 A few studies have reported the pathogenic function of SOD2 in stillbirth. However, overexpressing SOD2 reversed the maternal diabetes-induced increase of miR-27a, and the suppression of Nrf2 and Nrf2-controlled antioxidant enzymes suggested that dysregulated SOD2 may have a critical role in the early development of embryos or fetuses that may be involved in stillbirth.45 Our finding that SOD2 was differentially expressed in the placentas of stillbirth, in addition to miscarriages and preterm birth, provided evidence that SOD2 may function as an irregulated antioxidant enzyme in the development of stillbirth. Furthermore, our data showed that SOD2 expression was dysregulated only in the fetal facet of placenta, but not in the maternal tissue of the aborted tissues, in spontaneous miscarriage. Whether this finding indicates that the SOD2 up-regulation involved in spontaneous miscarriage is derived from the fetus rather than the pregnant mother is an open question.
4.3 |. Cytokine-chemokine receptor interactions
Chemokines are a family of small-molecular weight cytokines that are involved in leukocyte stimulation and chemotactic gradient determining. The increase in chemokine concentration could be associated not only with infection but also with the mechanism of labor.46 It has been suggested that pro-inflammatory cytokines, but hardly any anti-inflammatory cytokines, were present at term labor.37 Successful pregnancy requires carefully coordinated communications between the mother and fetus. Immune cells and cytokine signaling pathways participate as mediators of these communications to promote healthy pregnancy. The balance between pro-inflammatory and anti-inflammatory cytokines is crucial for implantation of the fetus, preparation of placenta, and pregnancy outcome. Chemokines and their receptors are key factors in embryo implantation and placenta vascularization. In fact, the cytokine-cytokine receptor interaction was the common one shared by spontaneous preterm birth and spontaneous miscarriage, in which 27 DEGs in spontaneous preterm birth and 26 DEGs in spontaneous miscarriage engaged. Ten common DEGs, including CXCR4, mainly focusing on CC, the CXC chemokine subfamily, were at the core position in the gene interaction network (Figure 3C).47–49 Most DEG expression was found to be elevated, except for CCR7, which was decreased in spontaneous miscarriage. OSMR, LEPR, IL21R, CSF3R, and TNFSF4 were down-regulated in spontaneous preterm birth, and CCR7 was down-regulated in recurrent miscarriage.50 OSMR, LEPR, IL21R, and CSF3R were involved in the Jak-STAT signaling pathway, which transduces a multitude of signals for development and homeostasis. Ligands of CXCR3, CXCL9, CXCL10, and CXCL11 were all up-regulated in spontaneous preterm birth, and CXCL10 was elevated in spontaneous miscarriage. An isolated elevation in amniotic fluid CXCL10 concentration was reported to be associated with the delivery of a placenta with histologic chronic chorioamnionitis or lesions consistent with maternal anti-fetal rejection,48 and another study indicated that CXCR3 blockade protected against Listeria monocytogenes infection-induced fetal wastage.51
4.4 |. NGFR is shared by all adverse pregnancies
The NGFR (OMIM: 162010), also referred to as p75(NTR), is the only transcript identified to be present in spontaneous miscarriage, recurrent miscarriage, stillbirth, and spontaneous preterm birth (including preterm labor and preterm premature rupture of membrane), through integrative omic-analyses (Table 1). It functions through binding at low affinity of NGF, but also through other neurotrophins, including brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NTF3), and neurotrophin-4 (NTF4).52 It can mediate cell survival as well as cell death of neural cells through inactivation of RHOA and plays a role in the regulation of the translocation of GLUT4 to the cell surface in adipocytes and skeletal muscle cells in response to insulin, probably by regulating RAB31 activity, and thereby contributes to the regulation of insulin-dependent glucose uptake,53 which is necessary for the circadian oscillation of the clock genes ARNTL/BMAL1, PER1, PER2, and NR1D1 in the suprachiasmatic nucleus of embryo/fetal developmental process.54 Although originally NGFR was studied in neuronal cells, it has now become evident that as the receptor of NGF, NGFR has functions in the balance of normal pregnancy progression, through the NGF-NGFR pathway.55 Additionally, in vitro findings revealed that NGF is able to promote a Th2 cytokine shift (ie, decreased secretion of TNF and increased secretion of IL10) in isolated uterine lymphocytes. These findings are consistent with the results of previous studies demonstrating a selective expression of NGF and NTRK1 in Th2 cells, supporting Th2 cells and suppressing Th1-cell function at the maternal-fetal interface, which modulates a cytokine environment compatible with the maintenance of pregnancy.56,57 Further analysis of the expression profiles of NGF-NGFR at the maternal-fetal interface pointed to a pivotal role of this pathway in the establishment of balanced immune-endocrine interactions during pregnancy in mice study, in which the expression of NGF occurs mainly in decidual tissue.58 Interestingly, it was found that an increased percentage of NTRK1+ decidual NK cells and a decreased percentage of NGF-expressing CD11c+ DCs follow NGF neutralization, which indicates that the functions of the innate immune cell subsets might also be deregulated in the absence of NGF signaling at the maternal-fetal interface of placenta.59–61 Epigenetic regulation via differential methylation may result in imbalanced NGFR, which could consequently disrupt the NGF pathway during early post-implantation stages. Study of 536 infants born less than 30 weeks post-menstrual age and participating in the neonatal neurobehavior and outcomes in very preterm infants showed that NGFR was differentially methylated with a p value 0.0086 and FDR < 0.05.62 It has been documented that during the early post-implantation period, if pregnant mice were exposed to supraphysiological levels of NGF, which functions through NGFR, spontaneous abortion syndrome may be induced. Typical signs of neurogenic inflammation were observed in such NGF-treated mice, including increased infiltration of NGF-expressing CD4+ and CD8+ T cells, increased innervation with TAC nerve fibers, enhanced mast cell degranulation, and a Th1 cytokine shift in decidual lymphocytes characterized by increased secretion of IL6 and decreased secretion of IL10, thus, in a manner similar to stress-triggered abortions.63
5 |. IN SUMMARY
Many studies have focused on the chemokine-cytokine pathway involved in adverse pregnancies. However, information about the transcribed molecule(s) commonly shared between or among variant adverse outcomes of pregnancy is lacking. Through integration of transcriptomic data generated from pregnancy loss—spontaneous miscarriage, recurrent miscarriage, stillbirth, and spontaneous preterm birth including preterm labor and preterm premature rupture of fetal membrane—we have identified pathogenic pathways associated with pregnancy loss and preterm birth. We hope to pursue further intensive investigations to uncover the pathogenic functions of the transcripts SOD2, CXCR4, and NGFR in the development of adverse pregnancies.
ACKNOWLEDGMENTS
This study is supported in part by the National Institutes of Health (R01HD094381) and the New York State Office for People with Developmental Disabilities (914-3280).
Funding information
National Institutes of Health, Grant/Award Number: R01HD094381; New York State Office for People With Developmental Disabilities, Grant/Award Number: 914-3280
Footnotes
DATA AVAILABLE STATEMENT
Our data are available and comply with the Expects Data Policy.
REFERENCES
- 1.Alves C, Rapp A. Spontaneous Abortion (Miscarriage). Treasure Island, FL: StatPearls; 2020. [Google Scholar]
- 2.Cousens S, Blencowe H, Stanton C, et al. National, regional, and worldwide estimates of stillbirth rates in 2009 with trends since 1995: a systematic analysis. Lancet. 2011;377:1319–1330. [DOI] [PubMed] [Google Scholar]
- 3.Jeve YB, Davies W. Evidence-based management of recurrent miscarriages. J Hum Reprod Sci. 2014;7:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencowe H, Cousens S, Chou D, et al. Born too soon: the global epidemiology of 15 million preterm births. Reprod Health. 2013;10(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen EC, Christiansen OB, Kolte AM, Macklon N. New insights into mechanisms behind miscarriage. BMC Med. 2013;11:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinar MH, Gibbins K, He M, Kostadinov S, Silver R. Early pregnancy losses: review of nomenclature, histopathology, and possible etiologies. Fetal Pediatr Pathol. 2018;37:191–209. [DOI] [PubMed] [Google Scholar]
- 7.WHO: recommended definitions, terminology and format for statistical tables related to the perinatal period and use of a new certificate for cause of perinatal deaths. Modifications recommended by FIGO as amended October 14, 1976. Acta Obstet Gynecol Scand. 1977;56:247–253. PMID: 560099. [PubMed] [Google Scholar]
- 8.Bender Atik R, Christiansen OB, Elson J, et al. ESHRE guideline: recurrent pregnancy loss. Hum Reprod Open. 2018;2018:hoy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawn JE, Blencowe H, Waiswa P, et al. Stillbirths: rates, risk factors, and acceleration towards 2030. Lancet. 2016;387:587–603. [DOI] [PubMed] [Google Scholar]
- 10.Group SCRNW. Association between stillbirth and risk factors known at pregnancy confirmation. JAMA. 2011;306:2469–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton JR, Barton LA, Istwan NB, et al. Elective delivery at 340(/)⁷ to 36⁶(/)⁷ weeks’ gestation and its impact on neonatal outcomes in women with stable mild gestational hypertension. Am J Obstet Gynecol. 2011;204(44):e41–45. [DOI] [PubMed] [Google Scholar]
- 12.Smulian JC, Ananth CV, Vintzileos AM, Scorza WE, Knuppel RA. Fetal deaths in the United States. Influence of high-risk conditions and implications for management. Obstet Gynecol. 2002;100:1183–1189. [DOI] [PubMed] [Google Scholar]
- 13.Pineles BL, Park E, Samet JM. Systematic review and meta-analysis of miscarriage and maternal exposure to tobacco smoke during pregnancy. Am J Epidemiol. 2014;179:807–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui H, Gong TT, Liu CX, Wu QJ. Associations between passive maternal smoking during pregnancy and preterm birth: evidence from a meta-analysis of observational studies. PLoS One. 2016;11:e0147848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vannuccini S, Clifton VL, Fraser IS, et al. Infertility and reproductive disorders: impact of hormonal and inflammatory mechanisms on pregnancy outcome. Hum Reprod Update. 2016;22:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedger M, Hodyl NA, Moss TJ. Inflammation in reproduction, pregnancy and development. J Reprod Immunol. 2018;130:23–24. [DOI] [PubMed] [Google Scholar]
- 17.Weiss G, Goldsmith LT, Taylor RN, Bellet D, Taylor HS. Inflammation in reproductive disorders. Reprod Sci. 2009;16:216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilman-Sachs A, Dambaeva S, Salazar Garcia MD, et al. Inflammation induced preterm labor and birth. J Reprod Immunol. 2018;129:53–58. [DOI] [PubMed] [Google Scholar]
- 19.Maduro MR. Inflammatory pathways and parturition. Reprod Sci. 2018;25:165. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nigro G, Mazzocco M, Mattia E, et al. Role of the infections in recurrent spontaneous abortion. J Matern Fetal Neonatal Med. 2011;24:983–989. [DOI] [PubMed] [Google Scholar]
- 22.Tran AM, Chalbatani GM, Berland L, et al. A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular RNAs. Front Cell Dev Biol. 2020;8:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y, Ji H, Liu H, et al. Pro-inflammatory cytokine-induced microRNA-212–3p expression promotes myocyte contraction via methyl-CpG-binding protein 2: a novel mechanism for infection-related preterm parturition. Mol Hum Reprod. 2019;25:274–282. [DOI] [PubMed] [Google Scholar]
- 24.Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–165. [DOI] [PubMed] [Google Scholar]
- 25.Liu KS, Li TP, Ton H, Mao XD, Chen YJ. Advances of long noncoding RNAs-mediated regulation in reproduction. Chin Med J (Engl). 2018;131:226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo X, Shi Q, Gu Y, et al. LncRNA pathway involved in premature preterm rupture of membrane (PPROM): an epigenomic approach to study the pathogenesis of reproductive disorders. PLoS One. 2013;8:e79897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Cao Q, Ge J, et al. LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lncRNAs in early spontaneous abortion. Am J Reprod Immunol. 2014;72:359–375. [DOI] [PubMed] [Google Scholar]
- 28.Luo X, Pan J, Wang L, et al. Epigenetic regulation of lncRNA connects ubiquitin-proteasome system with infection-inflammation in preterm births and preterm premature rupture of membranes. BMC Pregnancy Childbirth. 2015;15:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan J, Mor G, Ju W, et al. Viral Infection-induced differential expression of lncRNAs associated with collagen in mouse placentas and amniotic sacs. Am J Reprod Immunol. 2015;74:237–257. [DOI] [PubMed] [Google Scholar]
- 30.Zhao X, Dong X, Luo X, et al. Ubiquitin-proteasome-collagen (CUP) pathway in preterm premature rupture of fetal membranes. Front Pharmacol. 2017;8:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen C, Zhong N. Long non-coding RNAs: the epigenetic regulators involved in the pathogenesis of reproductive disorder. Am J Reprod Immunol. 2015;73:95–108. [DOI] [PubMed] [Google Scholar]
- 32.Howson CP, Zhong N, Padilla C, Yunis K, Giugliani R. The March of Dimes Global Network for Maternal and Infant Health: harnessing the power of experts in lower-income countries to improve the health of women, mothers, newborns and babies. Beijing Da Xue Xue Bao Yi Xue Ban. 2009;41:392–394. [PubMed] [Google Scholar]
- 33.Reeve ME, Charafeddine L, Zhong N, et al. Preconception health assessment in China, Lebanon and the Philippines: applicability to other countries. Matern Child Health J. 2014;18:1066–1074. [DOI] [PubMed] [Google Scholar]
- 34.Howson CP, Cedergren B, Giugliani R, et al. Universal newborn screening: a roadmap for action. Mol Genet Metab. 2018;124:177–183. [DOI] [PubMed] [Google Scholar]
- 35.StataCorp. Stata: Release 13. Statistical Software. College Station, TX: StataCorp LP. 2013. [Google Scholar]
- 36.Liu X, Zhang L, Wang P, et al. Sirt3-dependent deacetylation of SOD2 plays a protective role against oxidative stress in oocytes from diabetic mice. Cell Cycle. 2017;16:1302–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadley EE, Richardson LS, Torloni MR, Menon R. Gestational tissue inflammatory biomarkers at term labor: a systematic review of literature. Am J Reprod Immunol. 2018;79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myatt L, Cui X. Oxidative stress in the placenta. Histochem Cell Biol. 2004;122:369–382. [DOI] [PubMed] [Google Scholar]
- 39.Agarwal A, Gupta S, Sharma RK. Role of oxidative stress in female reproduction. Reprod Biol Endocrinol. 2005;3:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menon R, Boldogh I, Hawkins HK, et al. Histological evidence of oxidative stress and premature senescence in preterm premature rupture of the human fetal membranes recapitulated in vitro. Am J Pathol. 2014;184:1740–1751. [DOI] [PubMed] [Google Scholar]
- 41.Ghneim HK, Al-Sheikh YA, Alshebly MM, Aboul-Soud MA. Superoxide dismutase activity and gene expression levels in Saudi women with recurrent miscarriage. Mol Med Rep. 2016;13:2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fried M, Kurtis JD, Swihart B, et al. Systemic inflammatory response to malaria during pregnancy is associated with pregnancy loss and preterm delivery. Clin Infect Dis. 2017;65:1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burton GJ, Jauniaux E. Placental oxidative stress: from miscarriage to preeclampsia. J Soc Gynecol Investig. 2004;11:342–352. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Dong D, Reece EA, Wang AR, Yang P. Oxidative stress-induced miR-27a targets the redox gene nuclear factor erythroid 2-related factor 2 in diabetic embryopathy. Am J Obstet Gynecol. 2018;218:136.e131–136.e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haddad R, Tromp G, Kuivaniemi H, et al. Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am J Obstet Gynecol. 2006;195:394.e1–394.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaturvedi V, Ertelt JM, Jiang TT, et al. CXCR3 blockade protects against Listeria monocytogenes infection-induced fetal wastage. J Clin Invest. 2015;125:1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krow-Lucal ER, Kim CC, Burt TD, McCune JM. Distinct functional programming of human fetal and adult monocytes. Blood. 2014;123:1897–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giakoumelou S, Wheelhouse N, Brown J, et al. Chlamydia trachomatis infection of human endometrial stromal cells induces defective decidualisation and chemokine release. Sci Rep. 2017;7:2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ao D, Li DJ, Li MQ. CXCL12 in normal and pathological pregnancies: a review. Am J Reprod Immunol. 2020;84:e13280. [DOI] [PubMed] [Google Scholar]
- 51.El Hachem H, Crepaux V, May-Panloup P, et al. Recurrent pregnancy loss: current perspectives. Int J Womens Health. 2017;9:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. [DOI] [PubMed] [Google Scholar]
- 53.Lin Z, Tann JY, Goh ET, et al. Structural basis of death domain signaling in the p75 neurotrophin receptor. Elife. 2015;4:e11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baeza-Raja B, Eckel-Mahan K, Zhang L, et al. p75 neurotrophin receptor is a clock gene that regulates oscillatory components of circadian and metabolic networks. J Neurosci. 2013;33:10221–10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank P, Barrientos G, Tirado-González I, et al. Balanced levels of nerve growth factor are required for normal pregnancy progression. Reproduction. 2014;148:179–189. [DOI] [PubMed] [Google Scholar]
- 56.Arredondo LR, Deng C, Ratts RB, et al. Role of nerve growth factor in experimental autoimmune encephalomyelitis. Eur J Immunol. 2001;31:625–633. [DOI] [PubMed] [Google Scholar]
- 57.Sekimoto M, Tsuji T, Matsuzaki J, et al. Functional expression of the TrkC gene, encoding a high affinity receptor for NT-3, in antigen-specific T helper type 2 (Th2) cells. Immunol Lett. 2003;88:221–226. [DOI] [PubMed] [Google Scholar]
- 58.Kanai-Azuma M, Kanai Y, Matsuda H, et al. Nerve growth factor promotes giant-cell transformation of mouse trophoblast cells in vitro. Biochem Biophys Res Commun. 1997;231:309–315. [DOI] [PubMed] [Google Scholar]
- 59.Ashkar AA, Di Santo JP, Croy BA. Interferon gamma contributes to initiation of uterine vascular modification, decidual integrity, and uterine natural killer cell maturation during normal murine pregnancy. J Exp Med. 2000;192:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu B, Li X, Sun R, et al. Natural killer cells promote immune tolerance by regulating inflammatory TH17 cells at the human maternal-fetal interface. Proc Natl Acad Sci U S A. 2013;110:E231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tirado-González I, Barrientos G, Freitag N, et al. Uterine NK cells are critical in shaping DC immunogenic functions compatible with pregnancy progression. PLoS One. 2012;7:e46755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Everson TM, Marsit CJ, Michael O’Shea T, et al. Epigenome-wide analysis identifies genes and pathways linked to neurobehavioral variation in preterm infants. Sci Rep. 2019;9:6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tometten M, Blois S, Kuhlmei A, et al. Nerve growth factor translates stress response and subsequent murine abortion via adhesion molecule-dependent pathways. Biol Reprod. 2006;74:674–683. [DOI] [PubMed] [Google Scholar]