Abstract
Metastable glycosylated immunogens present challenges for GMP manufacturing. The HIV-1 envelope (Env) glycoprotein trimer is covered by N-linked glycan comprising half its mass and requires both trimer assembly and subunit cleavage to fold into a prefusion-closed conformation. This conformation, the vaccine-desired antigenic state, is both metastable to structural rearrangement and labile to subunit dissociation. Prior reported GMP manufacturing for a soluble trimer stabilized in a near-native state by disulfide (SOS) and Ile-to-Pro (IP) mutations has employed affinity methods based on antibody 2G12, which recognizes only ~30% of circulating HIV strains. Here, we develop a scalable manufacturing process based on commercially available, non-affinity resins, and we apply the process to current GMP (cGMP) production of trimers from clades A and C, which have been found to boost cross-clade neutralizing responses in vaccine-test species. The clade A trimer, which we named “BG505 DS-SOSIP.664”, contained an engineered disulfide (201C-433C; DS) within gp120, which further stabilized this trimer in a prefusion-closed conformation resistant to CD4-induced triggering. BG505 DS-SOSIP.664 was expressed in a CHO-DG44 stable cell line and purified with initial and final tangential flow filtration steps, three commercially available resin-based chromatography steps, and two orthogonal viral clearance steps. The non-affinity purification enabled efficient scale-up, with a 250 L-scale cGMP run yielding 9.57 g of purified BG505 DS-SOSIP.664. Antigenic analysis indicated retention of a prefusion-closed conformation, including recognition by apex-directed and fusion peptide-directed antibodies. The developed manufacturing process was suitable for 50 L-scale production of a second prefusion-stabilized Env trimer vaccine candidate, ConC-FP8v2 RnS-3mut-2G-SOSIP.664, yielding 7.8 g of this consensus clade C trimer. The successful process development and purification scale-up of HIV-1 Env trimers from different clades by using commercially available materials provide experimental demonstration for cGMP manufacturing of trimeric HIV-Env vaccine immunogens, in an antigenically desired conformation, without the use of costly affinity resins.
Keywords: GMP manufacturing, DS-SOSIP, envelope trimer, glycoprotein, HIV-1 vaccine, non-affinity chromatography, prefusion-closed conformation
1. Introduction
The HIV-1 envelope (Env) trimer is the only viral antigen to protrude through the virion-lipid bilayer and is thus the sole viral target of virus-neutralizing antibodies [reviewed in 1,2, 3]. Vaccine elicitation of such antibodies, however, has been hampered by the fragile nature of the trimer, which is metastable in conformation [4], labile to subunit dissociation [5], and covered by a dense shield of N-linked glycans comprising half of its mass [6–9]. The Env precursor gp160 is expressed as a single polypeptide, cleaved to gp120 and gp41 subunits in the Golgi, releasing at the newly cleaved N terminus of gp41 the fusion peptide that is critical for membrane fusion, and subsequently assembled via noncovalent interactions into a gp120-gp41 heterodimer. Three such moieties assemble to form the surface Env, a type 1 transmembrane quaternary homotrimer of heterodimers. Structural and functional studies have informed the development of soluble, native-like Env trimers, stabilized in a prefusion state that displays broadly neutralizing epitopes [10–24].
Vaccine studies with prefusion-stabilized Env trimers in multiple animal models have demonstrated the ability of these trimers to induce autologous neutralizing responses, capable of neutralzing only strains closely related to the vaccine immunogen [25, 26]. However, by using fusion peptide (FP)-carrier protein conjugates as priming immunogens to induce and expand immune responses targeting the FP-site of HIV-1 vulnerability [27, 28], Env trimer boosts could mature cross-clade neutralizing responses in mice, guinea pigs, and non-human primates [28], with FP-directed antibodies of up to almost 60% neutralization breadth being elicited in rhesus macaques [29]. Additional boosting with a heterologous trimer could increase the neutralization breadth and potency of fusion-peptide directed immune responses in guinea pigs [15, 30]. These findings suggest the potential utility of Env trimer immunogens in an effective HIV vaccine regimen, and several of these Env trimer immunogens are under development as vaccine candidates for phase I clinical trials.
To date, soluble HIV-1 Env trimers for research or clinical trials are predominantly produced in CHO- or HEK-based expression systems and are subsequently purified by methodologies including immunoaffinity, lectin, or Strep-Tactin affinity chromatography and size-exclusion chromatography (SEC) [30–38]. Commonly used immunoaffinity chromatography has utlilized antibodies like 2G12, PGT145, PGT151, VRC01, or a cocktail of V3-directed/CD4-induced antibodies to select for or against a specific conformational Env state. The cGMP production of trimeric HIV-1 Env immunogens has relied on the use of antibodies, such as the 2G12 antibody [39–42] used in the manufacture of the BG505 SOSIP.664 HIV-1 envelope glycoprotein trimer [34]. Immunoaffinity resins, while specific to well-formed, antigenically-pure trimer populations, require production or purchase of cGMP lots of each selector antibody as well as custom resin conjugation in order to use in a cGMP facility. Additionally, a Protein A flow-through step to remove any leached antibody (process-related impurity) is commonly employed, necessitating another costly affinity chromatography step. Size-exclusion chromatography, while commercially available, requires an ultrafiltration step prior to operation to meet load volume and process time restrictions. In-short, these existing strategies are complex, expensive, and difficult to scale-up for cGMP production.
Here we report a scalable, cost effective, cGMP-suitable manufacturing process using commercial, non-affinity purification methods to produce two HIV-1 Env trimer immunogens from different clades. We describe the cGMP production of the BG505 DS-SOSIP.664 (Trimer 4571) vaccine candidate, a prefusion-stabilized Env trimer derived from the clade A BG505 strain and developed through structure-based methods [12] to retain a prefusion-closed conformation even in the presence of the CD4 receptor, which is known to induce open, non-vaccine-preferred conformations of the Env trimer [4, 43]. The manufacturing process developed for BG505 DS-SOSIP.664 was applied to production of a second prefusion-stabilized Env trimer vaccine candidate, ConC-FP8v2 RnS-3mut-2G-SOSIP.664 (Trimer 6931). ConC-FP8v2 RnS-3mut-2G-SOSIP.664 was designed from a consensus clade C sequence [13] containing additional stabilizing mutations with a substitution in the fusion peptide epitope (FP8v2) [10, 15] and was developed to evaluate the utility of a heterologous HIV-1 Env trimer to boost immune responses directed to the fusion peptide in an epitope-focused vaccine regimen [15]. Both Env trimers were expressed in a CHO-DG44 stable cell line and purified by using two tangential flow filtration steps, three commercially available resin-based chromatography steps, and two orthogonal viral clearance steps. Non-affinity purification methods achieved high yields of good quality, pure Env trimers exhibiting the vaccine-desired antigenicity of a prefusion-closed state: recognition by broadly neutralizing antibodies (including antibodies PGT145 [44] and CAP256-VRC26.25 [45], which show strong preference for the recognition of the assembled, prefusion-closed Env trimer, with reduced or no recognition by poorly or non-neutralizing antibodies such as those that recognize CD4-induced states or regions of Env occluded on the prefusion-closed Env trimer). Our results provide proof-of-concept for a platform process suitable for the cGMP manufacture of trimeric HIV-Env vaccine immunogens, in their target antigenic state, through use of only commercial, non-affinity resins.
2. Materials and Methods
2.1. Upstream
The BG505 DS-SOSIP.664 Env trimer was expressed in stably transfected CHO-DG44 cells (obtained from Dr. Chasin, Columbia University) using VRC in-house vectors that were linearized by enzymatic digestion and transfected via electroporation (MaxCyte, Gaithersburg MD) as previously described [46]. Subsequently, a cGMP grade MCB was created from an isolated clone. A fed-batch bioprocess was used to produce the immunogen at various bioreactor scales, including a 50 L run for a toxicology study and 250 L for cGMP production. The ConC-FP8v2 RnS-3mut-2G-SOSIP.664 Env trimer was expressed in stably transfected CHO-DG44 cells using a double-gene VRC in-house vector which contained a second transgene expressing human furin. A fedbatch bioprocess at 50 L scale was performed using the optimized process parameters from the BG505 DS-SOSIP.664 process, as described accordingly: For both processes, the production media were ActiCHO P as basal with ActiCHO Feed A and ActiCHO Feed B as feed, all chemically defined from Cytiva (Piscataway, NJ). Both ActiCHO Feeds A/B were bolus added (3/0.3%, respectively, of initial bioreactor volume after inoculation) daily starting from day 2 until day 13. The bioreactors were controlled at pH 7.2 ± 0.2, dissolved oxygen 50%, and a biphasic temperature (37 °C from day 0–6, 32 °C from day 6 to harvest). On day 14, the BG505 DS-SOSIP.664 bioreactors were harvested for downstream clarification and purification, while a day 10 harvest was implemented for the 50 L ConC-FP8v2 RnS-3mut-2G-SOSIP.664 production run.
For downstream purification, the process described in Figure 1 was employed for three BG505 DS-SOSIP.664 50 L bioreactor consistency lots in-house and subsequently technology-transferred to the Vaccine Clinical Materials Program’s cGMP facility (Frederick, MD) and further scaled-up to produce a 250 L bioreactor of BG505 DS-SOSIP.664 material. After manufacturing, the same process with minor change in concentration and buffer exchange (II) step was applied in-house to a 50 L bioreactor of ConC-FP8v2 RnS-3mut-2G-SOSIP.664.
Fig. 1.
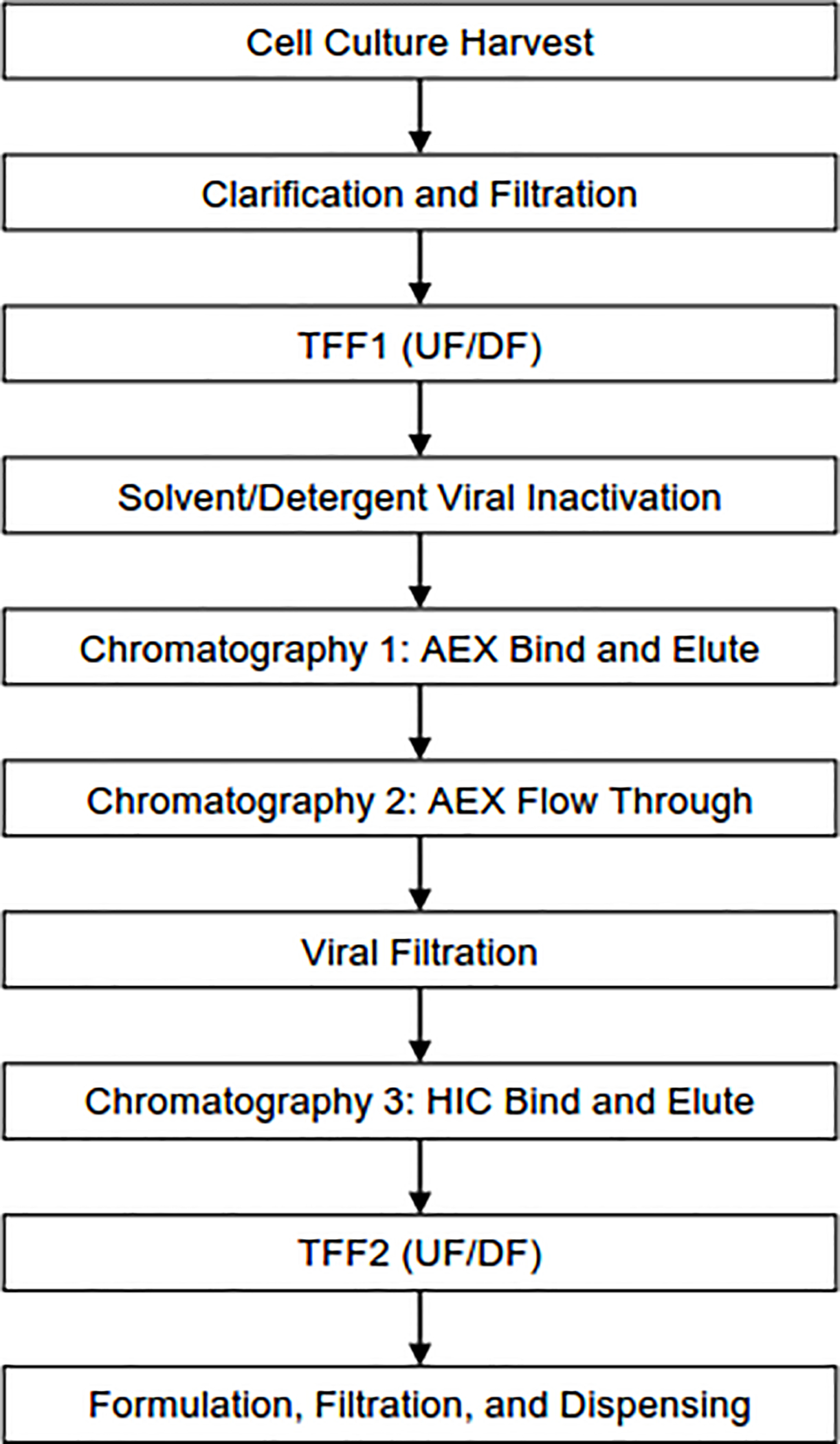
Downstream manufacturing process for purification of HIV-1 Env trimers. Process flow diagram outlines the chromatography and filtration steps used to purify BG505 DS-SOSIP.664 and ConC-FP8v2 RnS-3mut-2G-SOSIP.664 trimers from CHO-DG44 cell harvest. Abbreviations include AEX: Anion exchange, HIC: Hydrophobic Interaction Chromatography, TFF: Tangential flow filtration, UF/DF: Ultrafiltration/Diafiltration.
2.2. Clarification and Concentration/Buffer Exchange (I)
Removal of whole cells and debris from BG505 DS-SOSIP.664 harvest material was achieved by flowing harvest material through a Clarisolve 20MS depth filter followed by a Millistak+ F0HC filter (MilliporeSigma, Burlington, MA) at 60 LMH. An initial Tangential Flow Filtration (TFF1) step utilized a 500 kDa MWCO hollow-fiber polysulfone filter (Cytiva, Picastaway, NJ) to concentrate and buffer exchange clarified harvest five-fold into 20 mM MES, 25 mM NaCl, pH 6.5.
2.3. Solvent/Detergent (S/D) Treatment and Capture Chromatography
Post TFF1, the product-containing retentate was incubated with 1% polysorbate 80 and 0.3% tri-n-butyl phosphate for 1 h to perform S/D mediated viral inactivation. Subsequently, the material was loaded onto an anion exchange (AEX 1) capture column of TOYOPEARL NH2–750F resin (Tosoh Bioscience LLC, King of Prussia, PA). Bound material was eluted with five column volumes of 20 mM MES, 450 mM NaCl, pH 6.5.
2.4. Anion Exchange Chromatography in flow-through mode
Eluted material from capture chromatography was diluted with 50 mM Tris-HCl, pH 7.6 and subjected to a second anion exchange (AEX 2), performed in flow-through mode, using the weak anion exchange resin POROS 50 PI (ThermoFisher, Waltham, MA). The AEX 2 chromatography process step employed 20 mM Tris-HCl, 150 mM NaCl, pH 7.5 as an equilibration and chase buffer. The flow-through fraction was collected along with three column volumes of chase.
2.5. Nanofiltration
As an orthogonal viral removal step, the flow-through and chase pool from the previous step was passed through a filtration train consisting of a Viresolve prefilter followed by a 20 nm Viresolve Pro filter (MilliporeSigma, Burlington, MA).
2.6. Polishing Chromatography
The filtrate was spiked with 3.6 M ammonium sulfate, 50 mM Tris-HCl, pH 8.0 to attain conditioned load material in 2.0 M ammonium sulfate, 28 mM Tris-HCl, pH 8.0, which was then subjected to a final polishing chromatography step of TOYOPEARL PPG-600M resin (Tosoh Bioscience LLC, King of Prussia, PA). The hydrophobic interaction chromatography (HIC) process employs 50 mM Tris-HCl, 2 M ammonium sulfate, pH 8.0 for equilibration and wash, and 50 mM Tris-HCl, 1.45 M ammonium sulfate, pH 8.0 for elution.
2.7. Concentration/Buffer Exchange (II)
The elution fraction from the polishing chromatography was then concentrated two-fold and buffer exchanged fifteen-fold using a 100 kDa MWCO flat sheet PES membrane into a stabilizing buffer, 10 mM sodium phosphate, 10 mM NaCl, 7.5% sorbitol pH 7.2. After collection of the concentrated and diafiltered material, Pluronic F-68 surfactant was added to a final concentration of 0.01% (v/v) in accordance with the final formulation buffer specification. In case of ConC-FP8v2 RnS-3mut-2G-SOSIP.664, this step was executed using a 50 kDa MWCO flat sheet PES membrane to address lower step recoveries observed.
2.8. Antibody expression and purification
Antibody heavy and light chains were synthesized from published sequences, and subcloned into the pVRC8400 vector, as described previously [29]. Plasmids for both heavy and light chains were co-transfected into Expi293F cells (Thermo Fisher) with Turbo293 transfection reagent (Speed BioSystems). Cells were grown for six days, and antibodies were purified from the culture supernatant by protein A chromatography.
2.9. Reference Env trimer purified by affinity methods
For comparison, we used transient transfection of 293F-cells to express BG505 DS-SOSIP.664 and to purify in a research laboratory setting with VRC01-affinity resin and negative selection by V3-directed antibodies, as described previously [12]. Briefly, Env trimers were co-expressed with furin in transiently transfected 293 FreeStyle cells. Cells were cultured for six days, and the culture supernatants were harvested, filtered and loaded on a VRC01-affinity column. The column was washed and Env trimer was eluted with 3 M MgCl2, 30 mM Tris, pH 7.0. Eluted Env trimer was concentrated and subjected to size exclusion chromatography (Superdex 200 16/600; GE Healthcare). The Env trimer peak was pooled and further purified by negative selection first over a 447–52D column and then on a V3 antibody cocktail column (with six V3 antibodies 1006–15D, 2219, 2557, 2558, 3074, and 50.1) to remove aberrant trimer species.
2.10. Viral Clearance Study
Viral clearance and inactivation studies were performed at a partner Contract Research Organization (Texcell North America, Frederick, MD). As previously described [47], xenotrophic murine leukemia virus (XMuLV) and minute virus of mouse (MVM) were selected as model viruses to assess viral clearance in keeping with ICH Q5A viral clearance guidelines. The viruses were spiked at a known concentration to process intermediates and processed through a qualified scale down model. The presence of each model virus in the product fractions were detected by plaque-forming or TCID50 infectivity assay for XMuLV and MVM, respectively. The amount of reduction in virus is expressed as a Log Reduction Value (LRV).
Analytical methods
Several analytical methods were used to evaluate process performance and purified trimer quality. Concentration was primarily measured by absorbance at 280 nm, with an extinction coefficient of pure product defined as 1.57 mL/(mg*cm) for BG505 DS-SOSIP.664 and 1.56 mL/(mg*cm) for ConC-FP8v2 RnS-3mut-2G-SOSIP.664. Impurities were monitored using size-exclusion chromatography (SRT-C-500 Column, Sepax Technologies Inc.), CHO HCP ELISA (Immunoenzymetric CHO cell kit, Cygnus Technologies), and CHO host cell DNA MagMAX Extraction and QPCR (Thermo Fisher).
The size of the molecule was measured by SEC-MALS (SRT-C-500, Sepax Technologies Inc. and WYATT Dawn Heleos II MALS, Optilab TrEX Refractometer). Where applicable, the final material was also assessed for Endotoxin USP <85>, Bioburden USP<61>, pH USP<791>, and appearance USP<1>.
2.11. Antigenic analysis by Meso Scale Discovery (MSD)
Standard 96-well multi-array MSD plates (MSD, Cat #L15XA-3) were coated with HIV monoclonal antibodies at 4 μg/mL in 1x PBS, using 30 μL per well, and incubated overnight at 4°C. The panel of antibodies included HIV neutralizing antibodies (VRC01, b12, VRC13, PGT121, PGT128, 2G12, PGT145, CAP256-VRC26.25 [VRC26.25], 35O22, 8ANC195, PGT151 and VRC34.01), HIV non- or weakly-neutralizing antibodies (F105, 17b, 48D, 447–52D, 3074 and 2557) and a non-cognate influenza antibody CR9114. The following day, plates were washed three times with 1xPBS+0.05% Tween 20 (PBST), blocked with 150 μL of 5% MSD blocker A buffer (MSD, Cat #R93BA-4) and incubated on an Heidolph TITRAMAX 100 vibrational shaker at 650 rpm for 1 hour at room temperature. During incubation, HIV-1 Env trimers were diluted in 1% assay diluent (MSD Blocker A diluted in PBST) using 2-fold serial dilutions starting at 5 μg/mL, leaving the last well in each column as a blank. To evaluate CD4 induction, soluble CD4 (sCD4) was added to trimers at a constant molar concentration of 1 μM. Following incubation with blocking buffer, MSD plates were washed three times with PBST. Diluted trimers (+/−sCD4) were transferred to the MSD plates using 25 μL per well and plates were incubated with trimer on the plate shaker at 650 rpm for 2 hours at room temperature. MSD plates were washed as before prior to addition of SULFOTAG 2G12 antibody (2 μg/mL in 1% assay diluent) for detection, using 25 μL per well. Plates were incubated on the shaker at 650 rpm for 1 hour at room temperature and washed as described previously. 1x MSD Read Buffer T (MSD, Cat #R92TC-2) was added to the plates using 150 μL per well before plates were read on an MSD Sector 600 Imager.
2.12. Negative-stain electron microscopy
Samples were diluted to ~0.025 mg/ml with a buffer containing 10 mM HEPES, pH 7, and 150 mM NaCl. A 4.7-μl drop of the diluted sample was applied to a glow-discharged carbon-coated grid for 15 s. The drop was removed using blotting paper, and the grid was washed three times with 4.7-μl drops of the above buffer. Adsorbed proteins were negatively stained by applying consecutively three 4.7-μl drops of 0.75% uranyl formate. Micrographs were collected using SerialEM [48] on a FEI Tecnai T20 electron microscope operated at 200 kV and equipped with a 2k × 2k Eagle CCD camera or using EPU on a ThermoFisher Talos F200C electron microscope operated at 200 kV and equipped with a 4k × 4k Ceta CCD camera. The pixel size was 0.22 and 0.25 nm, respectively. Particles were picked automatically using in-house written software (Y.T., unpublished) and extracted using a box size of 128×128 and 100×100 pixels, respectively. Reference-free 2D classification was performed with Relion 1.4 [49].
3. Results
3.1. Downstream process development for purification of BG505 DS-SOSIP.664
To date, the purification of Env trimers has required expensive, difficult-to-scale methodologies such as custom-conjugated immunoaffinity chromatography [30, 33, 34] or other affinity methods including lectin and strep-tactin [31,35, 36, 38] and size-exclusion chromatography [30, 34, 35, 38]. In contrast with established trimer purification processes, a cost-effective, scalable method with several advantages for cGMP manufacturing is described herein.
An overview of the purification process employed for the 50 L bioreactor consistency runs and the 250 L cGMP production of BG505 DS-SOSIP.664 trimer is shown in Figure 1. The purification commenced with a depth filtration clarification step to remove cells and cell debris, followed by an ultrafiltration/diafiltration step to concentrate the clarified harvest, to remove spent cell culture media, and to buffer exchange the material into a suitable buffer for capture chromatography. The pool was subjected to solvent-detergent treatment by incubating with 1% Polysorbate 80, 0.3% Tri-n-butyl phosphate for 60–75 minutes. Following viral inactivation, Tosoh TOYOPEARL NH2–750F anion exchange resin was employed as a capture step. NH2–750F reduced levels of process-related impurities, such as residual solvent, detergent, HCP, and HCD, and increased purity to >70%.
Following NH2–750F, the material was conditioned and subjected to a flow-through anion exchange step by using POROS 50 PI resin with both a primary goal of viral clearance and a benefit in HCP reduction. The product pool was then filtered by using a 20 nm Viresolve Pro filter. Following nanofiltration, the product pool was conditioned and polished by using Tosoh Toyopearl PPG-600M resin, which decreased process- and product-related impurities, namely HCP and V3-exposed (open) conformations of Env, respectively, and increased purity to >99%. The pure trimer was then concentrated and buffer exchanged into a final formulation buffer of 10 mM NaPhosphate, 10 mM NaCl, 7.5% (w/v) Sorbitol, 0.01 % Pluronic® F68 (w/v), pH 7.2.
The optimized purification process was scaled-up from 3 L to 50 L (the cell culture harvest scale), and three 50 L consistency runs were produced in-house, including a lot for toxicological study requirements. Drug substance was manufactured at the 250 L scale upstream process under cGMP condition for Phase I clinical campaign, yielding 9.57 grams of purified trimer. The in-process HCP reduction and increase in purity by SEC for material produced by the cGMP manufacturing campaign are shown in Figure 2. Viral clearance studies performed with cGMP material and scale down models showed > 17.92 log reduction value (LRV) of XMulV and > 14.48 LRV of MVM (Table 1).
Fig. 2.
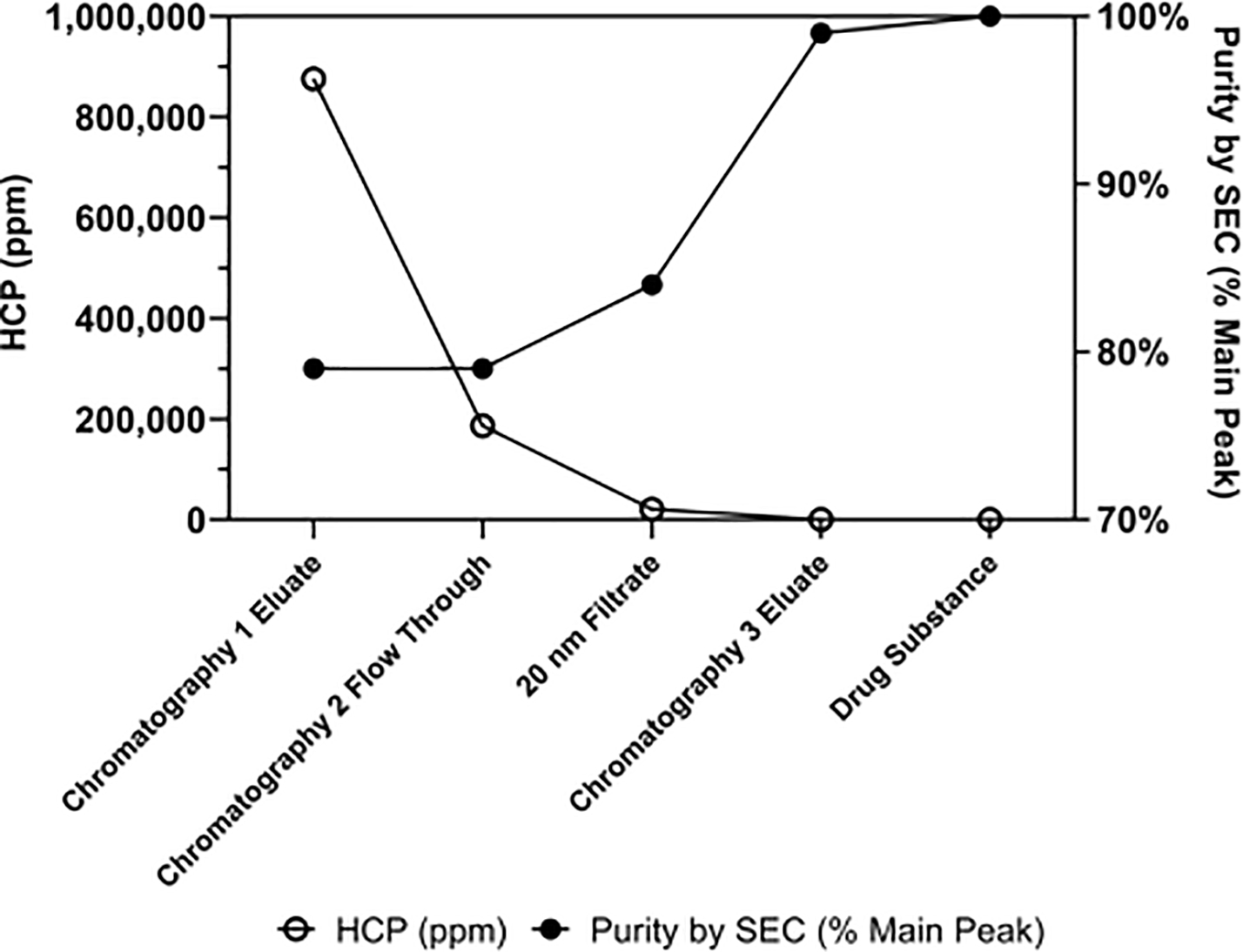
Process-related impurity (HCP) reduction and product quality improvement (Purity by SEC) across the purification process for BG505 DS-SOSIP.664 produced by the 250 L cGMP run. Abbreviations include HCP: Host Cell Protein, SEC: Size Exclusion Chromatography, ppm: parts per million.
Table 1.
Viral clearance/inactivation (LRV) for BG505 DS-SOSIP.664 downstream process unit operations*.
| XMuLV (LRV) | MVM (LRV) | |||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 1 | Experiment 2 | |
| S/D Treatment (60 min) | 2.57 | 2.81 | Not Applicable | |
| Chromatography 2: AEX | 5.17 | 4.97 | 3.13 | 2.97 |
| 20 nm Filtration | > 6.41 | > 6.52 | > 7.60 | > 7.52 |
| Chromatography 3: HIC | 3.76 | 3.63 | 3.83 | 3.91 |
| Total | > 17.92 | > 14.48 | ||
Abbreviations include S/D: solvent/detergent, AEX: anion exchange, HIC: hydrophobic interaction chromatography, XMulV: Xenotropic murine leukemia virus, MVM: minute virus of mice, LRV: log reduction value.
3.2. Quality assessment and characterization of purified BG505 DS-SOSIP.664
The quality metrics of the purified BG505 DS-SOSIP.664 material from 50 L (n=2) and 250 L bioreactors are shown in Table 2. A preceding 50 L run was also performed to evaluate scale-up process configurations (data not shown). Purity of BG505 DS-SOSIP.664 from both scales was found to be >99% as determined by SEC and process-related impurities such as HCP, HCDNA, PS80, and TnBP were all cleared to levels below LOQ. Thermostability of BG505 DS-SOSIP.664 purified from 50 L and 250 L runs was evaluated by differential scanning calorimetry (DSC), revealing consistent melting temperatures (Tm) of approximately 77 °C (Table 2).
Table 2.
Comparison of quality metrics assessed for purified BG505 DS-SOSIP.664 material from two 50 L GLP and one 250 L cGMP bioreactor runs*.
| Attribute | 50 L Run 1 (Tox Lot) | 50 L Run 2 | 250 L cGMP Run (Phase 1) |
|---|---|---|---|
| Yield (total amount of purified trimer, g) | 3.4 | 4.0 | 9.6† |
| Concentration | 0.57 mg/mL | 0.59 mg/mL | 0.506 mg/mL |
| Purity by SEC (% Main Peak) | 99.74% | 99.4% | 100% |
| Residual host cell protein | < 150 ng/mL (< 263 ppm) | < 30 ng/mL | < 30 ng/mL |
| Residual DNA | < 30 pg/mL | < 6.0 pg/mL | < 6 pg/mL |
| Endotoxin | < 8.8e-5 EU/μg | < 8.8e-5 EU/μg | < 0.0001 EU/μg |
| Bioburden | 0 CFU/10 mL | 0 CFU/10 mL | 0 CFU/10 mL |
| PH | 7.24 | 7.30 | 7.20 |
| Appearance | Clear, colorless, no visible particles | Clear, colorless, no visible particles | Clear, colorless, no turbidity; essentially free of particulates |
| Protein Mass (SEC-MALS) | 215 kDa | 213 kDa | 203 kDa |
| Glycan Mass (SEC-MALS) | 129 kDa | 128 kDa | 122 kDa |
| PS80 | < 15 ppm | < 15 ppm | < 15 ppm |
| TnBP | < 3 ug/mL | < 3 ug/mL | < 3 ug/mL |
| Melting temperature (Tm) by DSC | 77.0 | 76.7 | 77.3 |
Abbreviations include SEC: Size exclusion chromatography, SEC-MALS: Size exclusion chromatography-multi angle light scattering, PS80: polysorbate 80, TnBP: Tri-n-butyl phosphate, DSC: Differential scanning calorimetry, EU: endotoxin units, CFU: colony forming units, GLP: Good laboratory practices, cGMP: current good manufacturing practices.
Actual output, accounting for large volume in-process sampling to suffice IND-enabling studies.
The Meso Scale Discovery platform (MSD) was used to evaluate antigenic profiles of CHO-expressed BG505 DS-SOSIP.664 at the 50 L and 250 L scale (Figure 3A). For comparison, we included a 293F-expressed BG505 DS-SOSIP.664 reference that had been purified in a research laboratory setting by using VRC01-affinity resin and negative selection by V3-directed antibodies [12]. For all five panels of antibodies, MSD results showed consistent binding between different manufactured lots of BG505 DS-SOSIP.664. The antigenic behavior of CHO-expressed, non-affinity resin purified trimer was nearly indistinguishable from the 293-expressed, affinity-purified standard, with the only notable difference being an increased recognition by antibody 35O22 [50]. Two apex-directed antibodies, PGT145 and CAP256-VRC26.25 [44, 45], were well recognized, indicating proper conformation of the quaternary V1/V2-apex epitope. Notably, the gp120-gp41 interface antibodies all showed tight recognition – indicating trimers to hold both gp120 and gp41 in antibody-recognized conformations - including the VRC34.01 antibody [51], which recognizes the HIV-1 fusion peptide, a target site of vulnerability of vaccine importance. Negative-stain electron microscopy (NS-EM) images showed a predominant population of well-formed closed trimers with no visible aggregates and minimal monomer presence (Figure 3B), consistent with data reported for 293-expressed, affinity-purified BG505 DS-SOSIP.664 trimer [12, 16]. Overall, cGMP manufactured BG505 DS-SOSIP.664 displayed excellent antigenicity that was characteristic of the prefusion-closed state, with good recognition by broadly neutralizing antibodies (bNAbs) and little to no recognition by poorly or non-neutralizing antibodies, such as F105, 17b, or 48D, which bind the trimer in an open conformation.
Fig. 3.
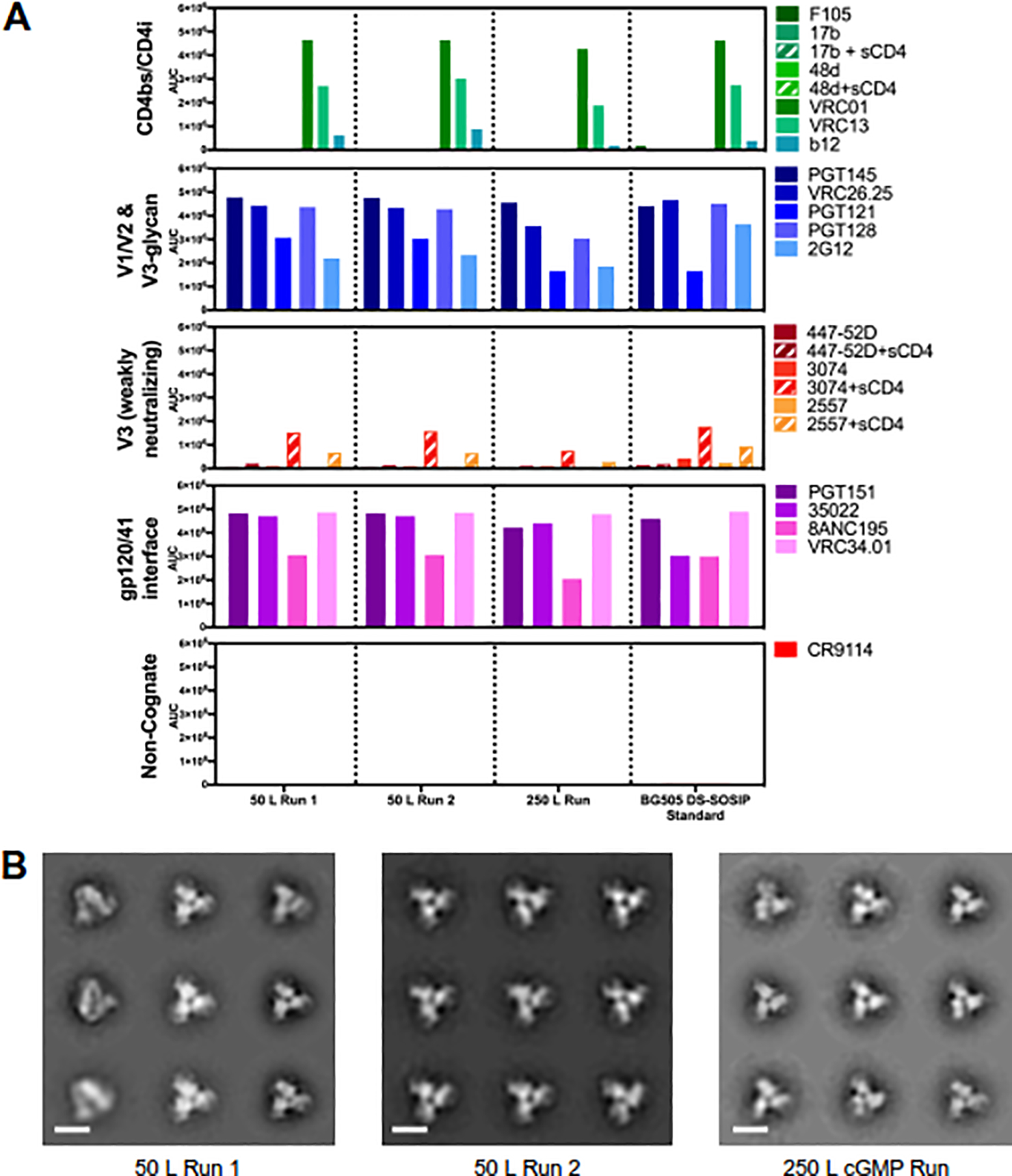
Antigenic and structural properties of BG505 DS-SOSIP.664 trimer purified from 50 L and 250 L bioreactor runs. (A) Antigenicity, as determined by MSD, is shown in AUC for CHO-expressed trimers purified using non-affinity chromatography and HEK293-expressed BG505 DS-SOSIP.664 purified using immunoaffinity chromatography (BG505 DS-SOSIP Standard, far right). Antigenicity was assessed on a diverse panel of antibodies, including broadly neutralizing antibodies directed against the CD4-binding site (VRC01, VRC13, b12), V1/V2-apex (PGT145, CAP256-VRC26.25), glycan-V3 site (PGT121, PGT128 and 2G12), gp120/gp41 interface (35022 and 8ANC195) and fusion peptide epitope (VRC34.01 and PGT151). The panel also includes non/weakly neutralizing CD4- induced antibodies (17b, 48d, F105) and V3-directed antibodies (447–52D, 3074 and 2557). (B) Negative stain electron microscopy class averages for BG505 DS-SOSIP.664 trimers purified from 50 L and 250 L runs. 2D classification produced classes indicative of correctly formed HIV-1 Env trimers in a prefusion-closed conformatioa Scale bars correspond to 10 nm.
3.3. Large-scale production of a stabilized consensus clade C Env trimer
We next evaluated whether the manufacturing processes developed for BG505 DS-SOSIP.664 could be applied to a clade C trimer vaccine candidate, ConC-FP8v2 RnS-3mut-2G-SOSIP.664, to enable its advancement into clinical development. The developed purification process (Figure 1) was found to be suitable for production of 7.8 g ConC-FP8v2 RnS-3mut-2G-SOSIP.664 trimer from a 50 L scale cell culture harvest, without the need for any additional product-specific optimization in the process. Assessment of key quality attributes showed the product pool to be 98.6% pure (by SEC), with process-related impurities (HCDNA, solvent and detergent) to be below detectable levels and 3972 ppm HCP (Table 3). Antigenic profiles, as assessed by MSD, were consistent between two different samples of purified ConC-FP8v2 RnS-3mut-2G-SOSIP.664 trimers and were indicative of the prefusion-closed state, as shown by strong binding of broadly neutralizing antibodies CAP256-VRC26.25, VRC01 and VRC34.01, but little to no binding of 447–52D and 17b antibodies, even in the presence of sCD4 (Figure 4A). NS-EM indicated homogenous samples of correctly formed Env trimers in a prefusion-closed conformation, as observed for the affinity-purified trimer [15], could be obtained by using the developed purification process for ConC-FP8v2 RnS-3mut-2G-SOSIP.664 (Figure 4B).
Table 3.
Summary of quality attributes for purified ConC-FP8v2 RnS-3mut-2G-SOSIP.664 trimer from 50 L run*.
| Attribute | 50 L Engineering Run |
|---|---|
| Yield (total amount of purified trimer, g) | 7.8† |
| Concentration | 0.564 mg/mL |
| Purity by SEC (% Main Peak) | 98.64% |
| Residual host cell protein | 3972 ppm |
| Residual DNA | < LLOQ (6 pg/mL = 10.6 ppb) |
| Endotoxin | 0.0016 EU/mcg Trimer |
| Bioburden | Not Performed |
| PH | 7.31 |
| Appearance | Clear, colorless, not turbid, not opalescene, two wispy visible particles |
| Protein Mass (SEC-MALS) | 206 kDa |
| Glycan Mass (SEC-MALS) | 108 kDa |
Abbreviations include LLOQ: lower limit of quantitation, ppb: parts per billion, EU: endotoxin units, SEC-MALS: size exclusion chromatography-multi angle light scattering, SEC: size exclusion chromatography.
Harvest volume from the 50 L reactor was 39 L.
Fig. 4.
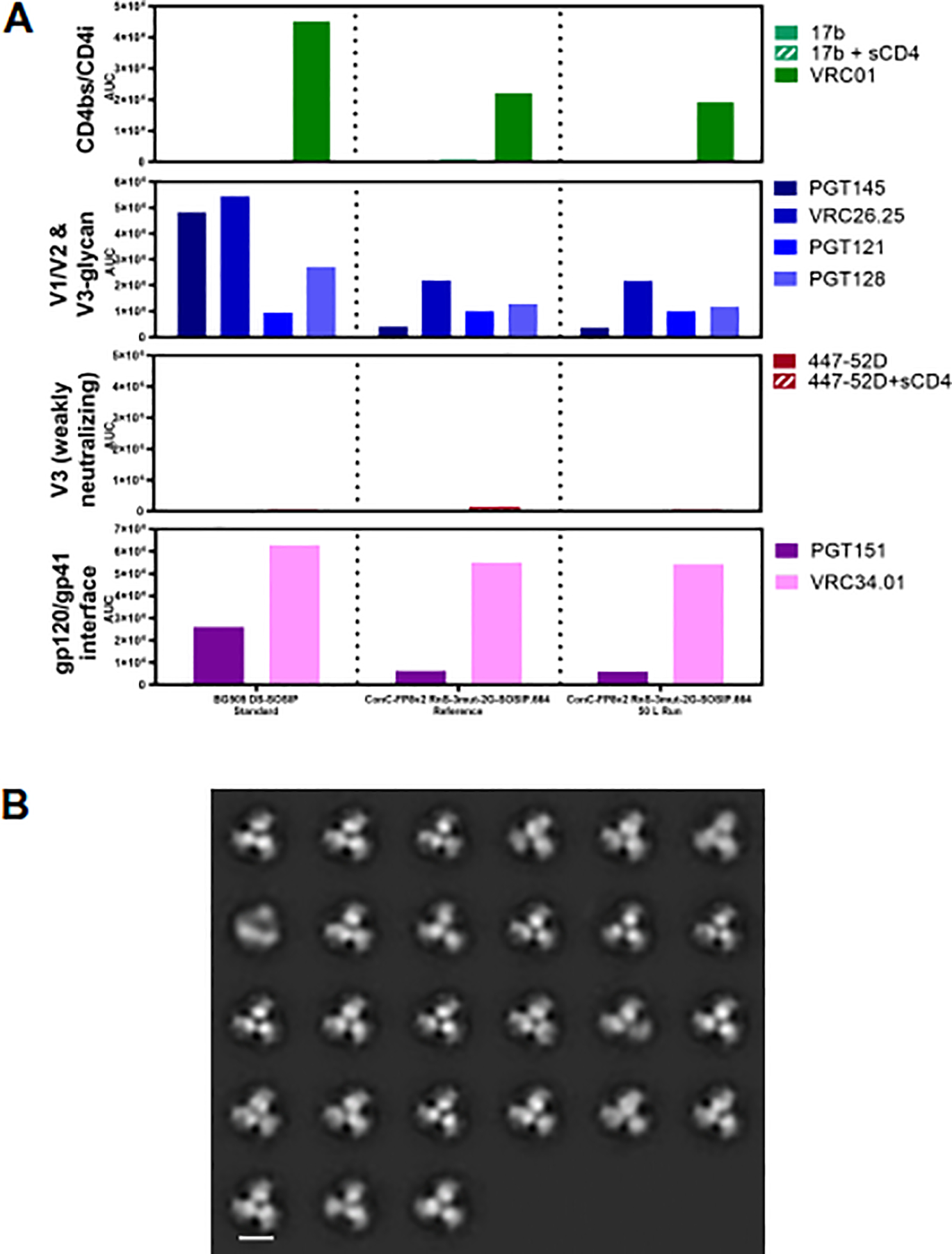
Antigenic and structural properties of ConC-FP8v2 RnS-3mut-2G-SOSlP.664 trimer. (A) Antigenicity, as determined by MSD, is shown in AUC for two development lots of ConC-FP8v2 RnS-3mut-2G-SOSlP.664 trimer and a BG505 DS-SOSIP standard. Antigenicity was assessed on a panel of antibodies, including broadly neutralizing antibodies directed against the CD4-binding site (VRC01), V1/V2-apex (PGT145 and CAP256-VRC2625), glycan-V3 site (PGT121 and PGT128)and fusion peptide epitope (VRC34.01 and PGT151). The panel also includes non/weakly neutralizing CD4-induced (17b) and V3-directed (447–52D) antibodies. (B) Negative stain electron microscopy class averages for purified ConC-FP8v2 RnS-3mut-2G-SOSIP.664 trimer from a 50 L scale production show correctly formed HIV-1 Env trimers in a prefusion-closed conformation. Scale bar corresponds to 10 nm.
4. Discussion
The successful application of the described conventional resin-based, cost-effective, scalable purification process to HIV-1 Env trimers from two different clades suggests a platform approach will be effective in future trimer purification. The method described herein is an improvement over recently published affinity chromatography-based cGMP manufacturing processes for Env trimers [34] by avoiding expensive custom mAb-conjugated resins and difficult-to-scale size-exclusion chromatography while producing Env trimers of high quality, thereby improving key attributes such as cost, complexity, and productivity. Affinity and size-exclusion chromatography steps were eliminated entirely; instead, resins that are inherently scalable and cost between $2,000 and $3,200 per liter were utilized. Additionally, the capture chromatography step showed robust clearance of solvent/detergent, so a dedicated detergent removal step could be avoided. The number of UF/DF steps was also decreased from four to two. The novel process achieved 9.57 g of purified trimer from a 250 L bioreactor, with a working volume of 173.4 L, compared to 3.52 g of purified trimer from a 200 L bioreactor using the traditional affinity chromatography-based process [34]. Though a direct comparison of quality is complicated by potential differences in analytical methods, each final material showed high purity (by an analytical SEC method), sufficient viral clearance, and low levels of impurities like HCP, HCDNA, solvent, and detergent.
The excellent antigenicity of the cGMP-manufactured BG505 DS-SOSIP.664 and development-grade ConC-FP8v2 RnS-3mut-2G-SOSIP.664 Env trimers provides experimental demonstration of the ability of commercial resins to separate open and/or V3-exposed forms of the Env trimer from those in a prefusion-closed conformation. This separation indicates the antigenic properties of the prefusion-closed conformation to be related to overall or global characteristics of the Env trimer, which are apparently detected by the specific recognition of each of the poorly or non-neutralizing antibodies. The fact that the manufacturing process reliably selected for native-like, well-formed trimers without the use of a conformationally-specific bNAb provides an avenue for further streamlining of HIV-1 Env immunogen manufacture, independent of clade. This advancement potentially eliminates the necessity of producing, obtaining, and screening broadly neutralizing antibodies for capture and negative selectors for polishing. Overall, the availability of a cGMP-suitable, inexpensive, platform-purification process should help to enable the manufacturing of candidate immunogens and to increase the catalog of available Env trimer immunogens for clinical evaluation.
Highlights.
Purification of Clade A and C HIV-1 Envelope trimers by a novel non-affinity method
Env trimers produced in pre-fusion closed conformation without antibody capture
Non-affinity purified trimers show minimal exposure of CD4-induced or V3 epitopes
Scalable process yields up to 0.2 g/L of purified prefusion-closed HIV-1-Env trimer
Acknowledgments
We thank Nga Tran, Lena Wang, Yoo-Jung Yang, and members of the downstream process development team at the Vaccine Production Program Laboratory for their assistance with the ConC-FP8v2 RnS-3mut-2G-SOSIP.664 program. We thank Jonathan Stuckey for assistance with figures, Shuishu Wang for assistance with manuscript preparation, and members of the Structural Biology Section for discussions and comments. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was also supported in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001E.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The VRC Production Program:
Nadia Amharref, Frank J. Arnold, Prasanthi Bandi, Nathan Barefoot, Christopher Barry, Elizabeth Carey, Ria Caringal, Kevin Carlton, Naga Chalamalsetty, Adam Charlton, Rajoshi Chaudhuri, Mingzhong Chen, Peifeng Chen, Hussain, Dahodwala, Gelu Dobrescu. Marianna Fleischman, Julia C. Frederick, Jason Gall, Isaac Godfroy, Deepika Gollapudi, Joe Horwitz, Althaf Hussain, Vera Ivleva, Lisa Kueltzo, Gabriella Lagos, Q. Paula Lei, David S. Lindsay, Robin Luedtke, Venkata Mangalampalli, Gabriel Moxey, Sarah O’Connell, Aakash Patel, Erwin Rosales-Zavala, Elizabeth Scheideman, Nicole A. Schneck, Richard M. Schwartz, William Shadrick, Shamitha Shetty, Alison Vinitsky, Lu Yang, and Yanhong Yang
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kwong PD, Mascola JR. HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity. 2018;48:855–71. [DOI] [PubMed] [Google Scholar]
- [2].Ward AB, Wilson IA. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev. 2017;275:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. [DOI] [PubMed] [Google Scholar]
- [4].Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–42. [DOI] [PubMed] [Google Scholar]
- [6].Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Myers G, Lenroot R. HIV glycosylation: what does it portend? AIDS Res Hum Retroviruses. 1992;8:1459–60. [DOI] [PubMed] [Google Scholar]
- [8].Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–12. [DOI] [PubMed] [Google Scholar]
- [10].Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, et al. Glycine Substitution at Helix-to-Coil Transitions Facilitates the Structural Determination of a Stabilized Subtype C HIV Envelope Glycoprotein. Immunity. 2017;46:792–803 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Joyce MG, Georgiev IS, Yang Y, Druz A, Geng H, Chuang GY, et al. Soluble Prefusion Closed DS-SOSIP.664-Env Trimers of Diverse HIV-1 Strains. Cell Rep. 2017;21:2992–3002. [DOI] [PubMed] [Google Scholar]
- [12].Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rutten L, Lai YT, Blokland S, Truan D, Bisschop IJM, Strokappe NM, et al. A Universal Approach to Optimize the Folding and Stability of Prefusion-Closed HIV-1 Envelope Trimers . Cell Rep. 2018;23:584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chuang GY, Lai YT, Boyington JC, Cheng C, Geng H, Narpala S, et al. Development of a 3Mut-Apex-Stabilized Envelope Trimer That Expands HIV-1 Neutralization Breadth When Used To Boost Fusion Peptide-Directed Vaccine-Elicited Responses. J Virol. 2020; 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, et al. Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity. J Virol. 2017,’91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, et al. Single-Chain Soluble BG505.SOSIP gp140 Trimers as Structural and Antigenic Mimics of Mature Closed HIV-1 Env. J Virol. 2015;89:5318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, et al. Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers. J Virol. 2015;90:2806–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun. 2016; 7:12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sliepen K, Han BW, Bontjer I, Mooij P, Garces F, Behrens AJ, et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat Commun. 2019;10:2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, et al. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity. 2016;45:483–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang P, Gorman J, Geng H, Liu Q, Lin Y, Tsybovsky Y, et al. Interdomain Stabilization Impairs CD4 Binding and Improves Immunogenicity of the HIV-1 Envelope Trimer. Cell Host Microbe. 2018;23:832–44 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ou L, Kong WP, Chuang GY, Ghosh M, Gulla K, O’Dell S, et al. Preclinical Development of a Fusion Peptide Conjugate as an HIV Vaccine Immunogen. Sci Rep. 2020; 10:3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med. 2018;24:857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kong R, Duan H, Sheng Z, Xu K, Acharya P, Chen X, et al. Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization. Cell. 2019;178:567–84 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cheng C, Xu K, Kong R, Chuang GY, Corrigan AR, Geng H, et al. Consistent elicitation of cross-clade HIV-neutralizing responses achieved in guinea pigs after fusion peptide priming by repetitive envelope trimer boosting. PLoS One. 2019;14:e0215163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].AlSalmi W, Mahalingam M, Ananthaswamy N, Hamlin C, Flores D, Gao G, et al. A New Approach to Produce HIV-1 Envelope Trimers: BOTH CLEAVAGE AND PROPER GLYCOSYLATION ARE ESSENTIAL TO GENERATE AUTHENTIC TRIMERS. J Biol Chem. 2015;290:19780–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bale S, Martine A, Wilson R, Behrens AJ, Le Fourn V, de Val N, et al. Cleavage-Independent HIV-1 Trimers From CHO Cell Lines Elicit Robust Autologous Tier 2 Neutralizing Antibodies. Front Immunol. 2018;9:1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cupo A, Cruz Portillo VM, Gelfand P, Yasmeen A, Klasse PJ, Moore JP. Optimizing the production and affinity purification of HIV-1 envelope glycoprotein SOSIP trimers from transiently transfected CHO cells. PLoS One. 2019;14:e0215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dey AK, Cupo A, Ozorowski G, Sharma VK, Behrens AJ, Go EP, et al. cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol Bioeng. 2018;115:885–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, et al. Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability. Immunity. 2019;51:915–29 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Pena A, de Taeye SW, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A. 2015;112:11947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89:3380–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Verkerke HP, Williams JA, Guttman M, Simonich CA, Liang Y, Filipavicius M, et al. Epitope-Independent Purification of Native-Like Envelope Trimers from Diverse HIV-1 Isolates. J Virol. 2016;90:9471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–71. [DOI] [PubMed] [Google Scholar]
- [40].Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, et al. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J Virol. 2016;90:76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen P, Chen M, Menon A, Hussain AI, Carey E, Lee C, et al. Development of a High Yielding Bioprocess for a Pre-fusion RSV Subunit Vaccine. J Biotechnol. 2021;325:261–70. [DOI] [PubMed] [Google Scholar]
- [47].Darling AJ. Design and Interpretation of Viral Clearance Studies for Biopharmaceutical Products. In: Desai MA, editor. Downstream Processing of Proteins: Methods and Protocols. Totowa, NJ: Humana Press; 2000. p. 195–209. [Google Scholar]
- [48].Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. [DOI] [PubMed] [Google Scholar]
- [49].Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]


