Abstract
Background
Congenital heart disease (CHD) is the leading cause of mortality from birth defects. In adult CHD patients with successful surgical repair, cardiac complications including heart failure develop at late stage, likely due to genetic causes. To date, many mutations in cardiac developmental genes have been associated with CHD. Recently, regulatory variants in genes have been linked to many human diseases. Although mutations and splicing variants in GATA4 gene have been reported in CHD patients, few regulatory variants of GATA4 gene are identified in CHD patients.
Methods
GATA4 gene regulatory region was investigated in the patients with atrial septal defects (ASD) (n = 332) and ethnic-matched controls (n = 336).
Results
Five heterozygous regulatory variants including four SNPs [g.31360 T>C (rs372004083), g.31436G>A, g.31437C>A (rs769262495), g.31487C>G (rs1053351749) and g.31856C>T (rs1385460518)] were only identified in ASD patients. Functional analysis indicated that the regulatory variants significantly affected the transcriptional activity of GATA4 gene promoter. Furthermore, two of the five regulatory variants have evidently effected on transcription factor binding sites.
Conclusions
Our data suggested that GATA4 gene regulatory variants may confer ASD susceptibility by decreasing GATA4 levels.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12872-021-02136-w.
Keywords: Atrial septal defects, Genetics, GATA4, Promoter, Regulatory variants
Introduction
Congenital heart disease (CHD) is the leading cause of mortality from birth defects. CHD prevalence is about 1% of live births [1]. Genetic factors play a critical role in the CHD development. Although hundreds of gene mutations and variants are implicated in CHD, precise genetic basis for sporadic CHD is largely unclear [2, 3]. In adult CHD patients with successful surgical repair, cardiac complications (heart failure, arrhythmia and cardiac sudden death) develop at late stage, likely due to genetic causes [4, 5]. Thus, understanding the genetic etiology of CHD are required for potential precision medicine and genetic counseling.
The human heart formation is a complicated morphogenetic process, including cell specification, differentiation, proliferation and migration, heart tube formation, looping and chamber separation. Cardiac morphogenesis is spatiotemporally controlled by transcription factors, cofactors, epigenetic regulators, cell signaling molecules and non-coding RNAs [6, 7]. Disruption in integrity and function of cardiac gene regulatory networks causes CHD. Accumulating evidence has demonstrated that mutations, copy number variation and regulatory variants in cardiac developmental genes cause different types of CHD, such as ASD (atrial septal defects), VSD (ventricular septal defect) and TOF (tetrology of Fallot) [2, 3].
Transcription factor GATA4 is required for cardiac specification, differentiation, proliferation and morphogenesis [8, 9]. GATA4 gene is expressed in all types of cardiac cells. GATA4 regulates many cardiac genes in the process of cardiac morphogenesis [10, 11]. During the heart development, GATA4 plays an essential role for proepicardium generation, heart tube formation, separation and outflow tract development. GATA4-null mouse embryos die early, displaying various heart defects derived from disrupted looping morphogenesis and septation [12–15]. Conditional deletion of GATA4 in the myocardium reveals that GATA4 regulates cardiomyocyte proliferation, right ventricle and atrioventricular canal formation [16]. GATA4 also regulates myocardial angiogenesis and development of cardiac conduction system [17, 18].
Mutations in GATA4 gene cause diverse types of CHD, including VSD, ASD, TOF and PS [8, 9, 19]. Specifically, mutations (non-synonymous and synonymous) and variants (noncoding and intronic) in GATA4 gene have been found in sporadic ASD cases [20–23]. However, regulatory variants in the GATA4 gene promoter have not been reported in ASD patients. Since GATA4 is a dosage-dependent transcription factor in the heart development [24], we postulated that GATA4 gene regulatory variants may contribute to the CHD development. In previous studies, a few GATA4 gene regulatory variants have been identified in VSD [25]. In this study, we studied the human GATA4 gene regulatory regions in ASD patients and ethnic-matched controls.
Materials and methods
Study participants
ASD patients (n = 332) were recruited from Affiliated Hospital of Jining Medical University (Jining, Shandong, China), including male 125 and female 207. The age range was from one month to 38 years and the mean age was 9.00 years. All ASD patients had no familial history of CHD. Diagnosis was further confirmed with echocardiography and following surgical procedures. Ethnically-matched controls (n = 336) were from Division of Pediatric Surgery in the same hospital, including male 159 and female 177. The age range was from four months to 13 years and the mean age for controls was 5.07 years. Controls with familial history of heart diseases and other inherited disorders were ruled out. This work was conducted according to the principles of the Declaration of Helsinki. The research protocol was approved by the hospital Human Ethic Committee. Written consents were informed and signed by the parents of participants. For participants older than 16, written and informed consents were obtained from the participants themselves.
Direct DNA sequencing
Genomic DNA preparation was prepared. Direct sequencing of the GATA4 gene promoter region was carried out as previously reported [25]. Two overlapped DNA fragments of GATA4 gene promoter, 510 bp ( − 961 bp to − 451 bp) and 569 bp (− 502 bp to + 67 bp), were amplified by PCR and directly sequenced by Shanghai Sangon Biotech Company (Shanghai, China). GATA4 gene regulatory variants were identified by comparing to human GATA4 gene (NG_008177.2).
Dual-luciferase reporter assay
GATA4 gene promoter (971 bp, from − 932 bp to + 39 bp) was generated by PCR and subcloned into the SacI and Hind III sites of pGL3-basic, a luciferase reporter vector. The rat cardiomyocyte line H9c2 cells (CRL-1446, ATCC, Manassas, VA, USA) were transfected with designated expression constructs according to the transient transfection procedure previously reported [25]. In brief, H9c2 cells were cultured in 6-well plates. Expression constructs (1.0 µg) and Lipofectamine (3.0 µl) were used for each well. The vector pRL-TK expressing Renilla luciferase (25 ng) was used as an internal control for transfection efficiency. Forty-eight hours later, the transfected cells were collected and luciferase activity was examined with the Promega dual-luciferase reporter assay system. The transcriptional activity was expressed as ratios of luciferase activity over Renilla luciferase activity. Wild type GATA4 gene promoter activity was set as 100%. Relative activity of variant GATA4 gene promoter was calculated. All transfection experiments were performed three times independently, in triplicate.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed with the LightShift® Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the procedure. H9c2 cell nuclear extracts were prepared with NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). Biotinylated double-stranded oligonucleotides (30 bp) with or without the variants were used as probes. The DNA–protein reactions were incubated for 20 min at room temperature. The reaction mixtures were separated on a 6% polyacrylamide gel, and subsequently transferred onto a nylon membrane (Thermo Fisher Scientific). Oligonucleotides were cross-linked using the UV Stratalinker 1800 (Agilent Technologies, Inc., Santa Clara, CA, USA). Signals were examined by chemiluminescence.
Statistical analysis
ANOVA was used to analyze quantitative data, which was represented as mean ± SEM. SPSS 23.0 was used to compare frequencies of regulatory variants between two groups. P < 0.05 was taken as statistically significant.
Results
Regulatory variants of GATA4 gene
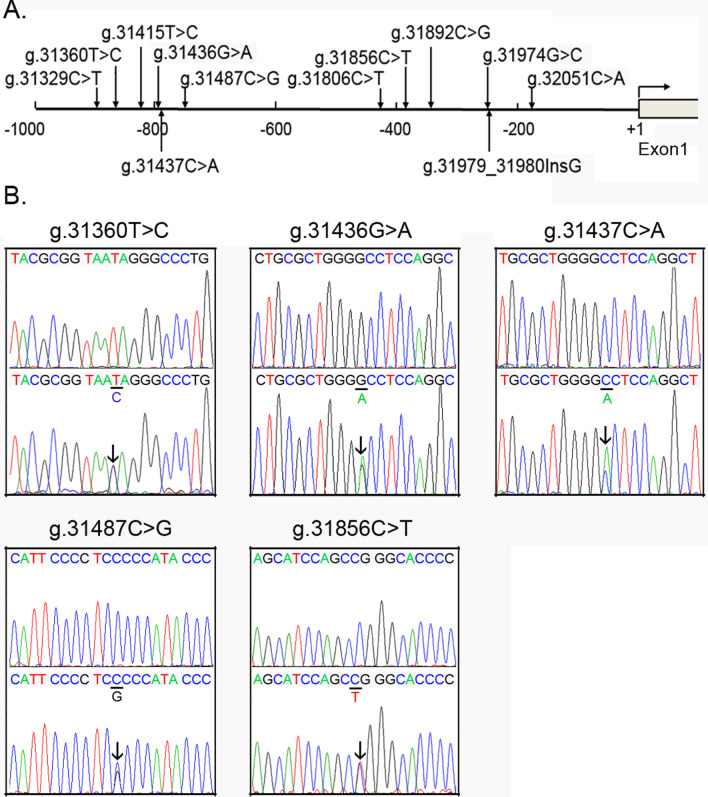
In this study population, twelve regulatory variants, including eight SNPs, were identified (Fig. 1a and Table 1) by compared to dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) and gnomAD database (http://gnomad-sg.org/). Five heterozygous variants including four SNPs [g.31360 T>C (rs372004083), g.31436G>A, g.31437C>A (rs769262495), g.31487C>G (rs1053351749) and g.31856C>T (rs1385460518)] were found in six ASD patients (Fig. 1b). All these six ASD patients suffered from ostium secundum ASD (type II). Six heterozygous variants including four SNPs [g.31329C>T (rs1429952472), g.31415 T>C, g.31806C>T (rs1204517110), g.31892C>G (rs1204747694), g.31974G>C (rs560860578) and g.32051C>A) were only found in controls. An insertion variant (g.31979_31980InsG) was found in both ASD patient and controls.
Fig. 1.
Regulatory variants of GATA4 gene. a Locations of regulatory variants upstream to the transcription start site of human GATA4 gene (NG_008177.2). b Sequencing chromatograms of GATA4 gene regulatory variants identified in ASD patients. Top panels are wild type and bottom heterozygous variants (marked with arrows). Sequencing orientations are forward
Table 1.
GATA4 gene regulatory variants in ASD patients and controls
| Regulatory variants | Genotypes | Locationa | Controls (n = 336) | ASD (n = 332) | P value | Frequencies (gnoMAD) |
|---|---|---|---|---|---|---|
| g.31329C>T(rs1429952472) | CT | − 904 bp | 1 | 0 | – | Not found |
| g.31360 T>C(rs372004083) | TC | − 873 bp | 0 | 1 | – | 0.000287 |
| g.31415 T>C | TC | − 818 bp | 1 | 0 | – | – |
| g.31436G>A | GA | − 797 bp | 0 | 1 | – | – |
| g.31437C>A(rs769262495) | CA | − 796 bp | 0 | 2 | – | 0.000287 |
| g.31487C>G(rs1053351749) | CG | − 746 bp | 0 | 1 | – | 0.000064 |
| g.31806C>T(rs1204517110) | CT | − 427 bp | 1 | 0 | – | Not found |
| g.31856C>T(rs1385460518) | CT | − 377 bp | 0 | 1 | – | Not found |
| g.31892C>G(rs1204747694) | CG | − 341 bp | 1 | 0 | – | 0.000032 |
| g.31974G>C(rs560860578) | GC | − 259 bp | 1 | 0 | – | 0.003761 |
| g.31979_80InsG | –/G | − 256 bp | 5 | 1 | 0.104 | – |
| g.32051C>A | CA | − 182 bp | 1 | 0 | – | – |
aRegulatory variants are located upstream (–) to the transcription start site of GATA4 gene at 32233 of NG_008177.2
Regulatory variant-affected binding of transcription factors
We analyzed the human GATA4 gene promoter with JASPAR to predict binding sites for transcription factors. The GATA4 gene regulatory variants identified in ASD patients were analyzed in details. The variant [g.31360 T>C (rs372004083)] may abolish the binding site of DLX6 (distal-less homeobox 6) and LHX1 (LIM homeobox 1). The variant (g.31436G>A) may abolish a TFAP2A (transcription factor AP-2 alpha) site. The variant [g.31437C>A (rs769262495)] may abolish a TFAP2A site and create a MZF1 (myeloid zinc finger 1) site. The variant [g.31487C>G (rs1053351749)] may abolish a SP1 (SP1 transcription factor) binding site, create a THAP1 (THAP domain containing 1) site and modify the sites of KLF5 (Kruppel like factor 5) and ZNF148 (zinc finger protein 148). The variant [g.31856C>T (rs1385460518)] may create a BHLHE22 (basic helix-loop-helix family member E22) binding site, and modify the site for HIC1 (hypermethylated in cancer 1).
Effects of regulatory variants on GATA4 gene promoter activity
Expression constructs including pGL3-WT (wild type), pGL3-31360C, pGL3-31436A, pGL3-31437A, pGL3-31487G, pGL3-31856 T and pGL3-31979_80InsG were transfected into H9c2 cells. Transfection results showed that variants [g.31436G>A, g.31437C>A (rs769262495), g.31487C>G (rs1053351749) and g.31856C>T (rs1385460518)] significantly decreased GATA4 gene promoter activity (P < 0.01). The variant [g.31360 T>C (rs372004083)] had minimal effect (P>0.05). In contrast, the variant (g.31979_80InsG) had no effects (P>0.05) (Fig. 2).
Fig. 2.
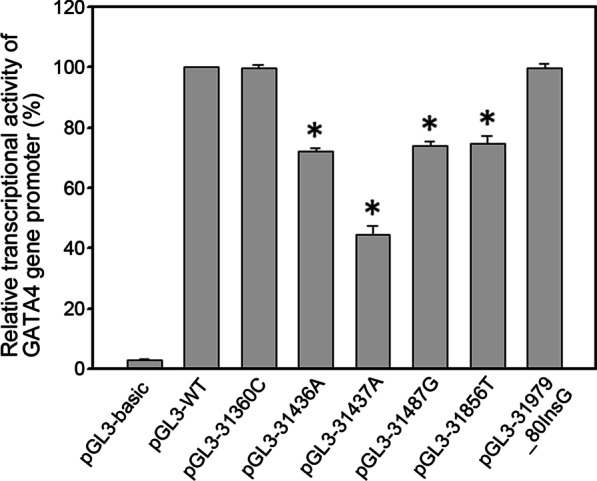
Relative activities of GATA4 gene promoters with or without regulatory variants in H9c2 cells. All transfection experiments were performed three times independently, in triplicate. The results were represented as means ± SEM. Bar graph represented the mean and error bar indicated SEM. Empty pGL3-basic was used as a negative control. Activity of wild type GATA4 gene promoter was set as 100%. WT, wild type. *P < 0.01
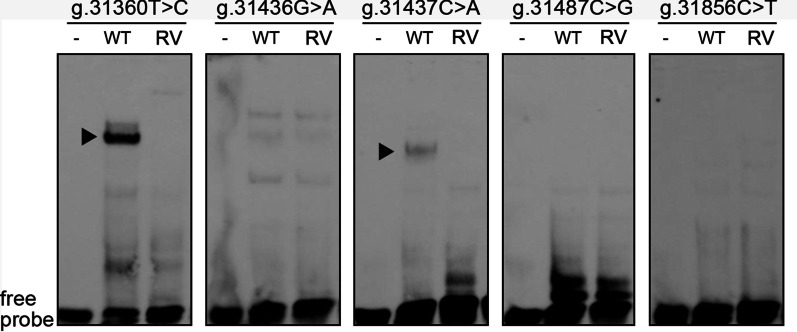
Binding of transcription factors determined by EMSA
EMSA was performed to determine whether the regulatory variants effected on the binding of transcription factors. The oligonucleotides (30 bp) were biotin-labelled as probes (Table 2). EMSA showed that variants [g.31360 T>C (rs372004083) and g.31437C>A (rs769262495)] significantly weakened or abolished the binding ability of unknown transcription factors (Fig. 3). The affected transcription factors probably functioned as activators, requiring further investigation. In addition, EMSA did not detect the effects of other three variants on transcription factor binding (data not shown), likely due to the sensitivity limitation.
Table 2.
The double-stranded biotinylated oligonucleotides for EMSA
| Regulatory variants | Oligonucleotide sequences | Locations |
|---|---|---|
| g.31360 T>C (rs372004083) | 5′-TTTTTACACGGTAA(T/C)AGGGGCCCTGTGATTG-3′ | 31346–31375 |
| g.31436G>A | 5′-CCCGCTGCGCTGGG(G/A)CCTCCAGGCTCTGAC-3′ | 31422–31451 |
| g.31437C>A (rs769262495) | 5′-CCGCTGCGCTGGGG(C/A)CTCCAGGCTCTGACG-3′ | 31423–31452 |
| g.31487C>G | 5′-GACACATTCCCCTC(C/G)CCCATACCCTGGAAG-3′ | 31473–31502 |
| g.31856C>T | 5′-AACTAGCATCCAGC(C/T)GGGCACCCCGGGTGA-3′ | 31842–31871 |
EMSA was performed with biotin-labeled oligonucleotide and H9c2 cell nuclear extracts. Free probe was indicated at the bottom. Solid arrows indicated the affected binding for unknown transcription factors
WT wild type, RV regulatory variant
Fig. 3.
Effects of regulatory variants on transcription factor binding. EMSA was performed with biotin-labeled oligonucleotide and H9c2 cell nuclear extracts. Free probe was indicated at the bottom. Solid arrows indicated the affected binding for unknown transcription factors. WT, wild type. RV, regulatory variant.
Discussion
Misregulation of gene expression have been implicated in many human diseases [26]. The clinical significance of de novo variation has been recently highlighted in sporadic CHD [27]. Rare inherited variants and de novo variations within regulatory regions of heart developmental genes have been demonstrated to cause CHD [28–31]. In this study, we focused on the GATA4 gene promoter, and found five functional regulatory variants in six ASD patients. Collectively, frequency of GATA4 gene regulatory variants in ASD patients was 1.81% (6/332). As shown in Table 1, the variants g.31360 T>C (rs372004083), g.31437C>A (rs769262495) and g.31487C>G (rs1053351749) were more frequent compared to dbSNP database and gnoMAD database. The variant g.31856C>T (rs1385460518) was more frequent compared to dbSNP database, and was not found in gnoMAD database. The variant g.31436G>A was not found in dbSNP database and gnoMAD database.
The human GATA4 gene, located to chromosome 8p23.1-p22, is expressed in all cardiac cells [32–34]. There are conserved GC-boxes, E-box and GATA motif within the GATA4 gene promoter [35]. GATA4 exhibits cell-specific DNA-binding ability and tissue-specific function [36]. During development of the human heart, GATA4 gene expression is regulated by NKX2-5, F-actin binding protein NEXN, BMP signaling and other GATA factors [37–39]. It has been reported that GATA4 gene is significantly upregulated in coronary artery disease [40]. However, the human GATA4 gene expression and regulation remains to be further investigated [41]. In this study, the GATA4 gene regulatory variants identified in ASD patients did not affect the conserved motifs.
More importantly, two regulatory variants, g.31360 T>C (rs372004083) and g.31437C>A (rs769262495), evidently affected the transcription factor binding in the EMSA assay. As predicted, the variant [g.31360 T>C (rs372004083)] may abolish the binding of DLX6 and LHX1, and the variant [g.31437C>A (rs769262495)] may abolish a TFAP2A site. The abolished binding of the potential transcription factors was consisted to the repressive effect of the two variants on the GATA4 gene promoter, suggesting that the potential transcription factors were probably transcriptional activators. When appropriate antibodies were available, further EMSA experiments will be conducted to identify these transcription factors.
GATA4 is implicated in a cardiac gene regulatory network integrating cardiac transcription factors, co-factors, epigenetic regulators and microRNAs [42–44]. During the heart development, GATA4 regulates expression of downstream target genes, including atrial natriuretic factor, brain natriuretic protein, connexin 40 and myosin heavy chain genes [8–11, 45]. Decreased GATA4 level may affect its interaction with other factors in cardiac gene regulatory networks, disrupting the atrial septum. Therefore, GATA4 gene expression may be upregulated with genetic approaches or small molecules in further studies. Correction of deficient GATA4 gene expression may provide a potential way to prevent cardiac complications in the adult CHD patients carrying these variants.
Conclusions
In this study, functional regulatory variants of GATA4 gene were identified in ASD patients. These GATA4 gene regulatory variants may confer susceptibility to ASD development by decreasing GATA4 levels.
Supplementary Information
Additional file 1: Original EMSA images for Fig. 3 were included in this document file.
Acknowledgements
Not applicable.
Abbreviations
- ASD
Atrial septal defects
- BHLHE22
Basic helix-loop-helix family member E22
- CHD
Congenital heart disease
- DLX6
Distal-less homeobox 6
- EMSA
Electrophoretic mobility shift assay
- GATA4
Transcription factor GATA4
- HIC1
Hypermethylated in cancer 1
- KLF5
Kruppel like factor 5
- LHX1
LIM homeobox 1
- MZF1
Myeloid zinc finger 1
- SP1
SP1 transcription factor
- TFAP2A
Transcription factor AP-2 alpha
- THAP1
THAP domain containing 1
- TOF
Tetrology of Fallot
- VSD
Ventricular septal defects
Authors' contributions
DF, JS and QC diagnosed the patients. DF, SP and JC performed the experiments and interpreted the data. JS and QC collected the clinical data and samples. DF, QC and BY designed the study and wrote the manuscript. BY reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by Shandong Taishan Scholar Project, China (tshw201502063) and National Natural Science Foundation of China (81370271, 81870279).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study protocol was approved by the hospital Human Ethic Committee. Written consents were signed by the parents of participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qianjin Cheng, Email: 18678766720@163.com.
Bo Yan, Email: yanbo@mail.jnmc.edu.cn.
References
- 1.Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, Mital S, Priest JR, Pu WT, Roberts A, Ware SM, Gelb BD, Russell MW; American Heart Association Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; and Council on Genomic and Precision Medicine. Genetic basis for congenital heart disease: revisited: a scientific statement from the American Heart Association. Circulation 2018;138:e653–e711. [DOI] [PMC free article] [PubMed]
- 2.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112:707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res. 2017;120:923–940. doi: 10.1161/CIRCRESAHA.116.309140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, Chapman KA, Kisling M, John AS. Genetic considerations for adults with congenital heart disease. Am J Med Genet C Semin Med Genet. 2020;184:149–153. doi: 10.1002/ajmg.c.31777. [DOI] [PubMed] [Google Scholar]
- 5.Verheugt CL, Uiterwaal CS, van der Velde ET, Meijboom FJ, Pieper PG, van Dijk AP, Vliegen HW, Grobbee DE, Mulder BJ. Mortality in adult congenital heart disease. Eur Heart J. 2010;31:1220–1229. doi: 10.1093/eurheartj/ehq032. [DOI] [PubMed] [Google Scholar]
- 6.Buijtendijk MFJ, Barnett P, van den Hoff MJB. Development of the human heart. Am J Med Genet C Semin Med Genet. 2020;184:7–22. doi: 10.1002/ajmg.c.31778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore-Morris T, van Vliet PP, Andelfinger G, Puceat M. Role of epigenetics in cardiac development and congenital diseases. Physiol Rev. 2018;98:2453–2475. doi: 10.1152/physrev.00048.2017. [DOI] [PubMed] [Google Scholar]
- 8.Lentjes MH, Niessen HE, Akiyama Y, de Bruïne AP, Melotte V, van Engeland M. The emerging role of GATA transcription factors in development and disease. Expert Rev Mol Med. 2016;18:e3. doi: 10.1017/erm.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development. 2018;145:dev164384. doi: 10.1242/dev.164384. [DOI] [PubMed] [Google Scholar]
- 10.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 11.Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 13.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 14.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L, Liu J, Xiang M, Olson P, Guzzetta A, Zhang K, Moskowitz IP, Xie L. Gata4 potentiates second heart field proliferation and Hedgehog signaling for cardiac septation. Proc Natl Acad Sci U S A. 2017;114:E1422–E1431. doi: 10.1073/pnas.1605137114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heineke J, Auger-Messier M, Xu J, Oka T, Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ, Robbins J, Crombleholme TM, Molkentin JD. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J Clin Invest. 2007;117:3198–3210. doi: 10.1172/JCI32573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malek Mohammadi M, Kattih B, Grund A, Froese N, Korf-Klingebiel M, Gigina A, Schrameck U, Rudat C, Liang Q, Kispert A, Wollert KC, Bauersachs J, Heineke J. The transcription factor GATA4 promotes myocardial regeneration in neonatal mice. EMBO Mol Med. 2017;9:265–279. doi: 10.15252/emmm.201606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 20.Dixit R, Kumar A, Mohapatra B. Implication of GATA4 synonymous variants in congenital heart disease: a comprehensive in-silico approach. Mutat Res. 2019;813:31–38. doi: 10.1016/j.mrfmmm.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Posch MG, Perrot A, Berger F, Ozcelik C. Molecular genetics of congenital atrial septal defects. Clin Res Cardiol. 2010;99:137–147. doi: 10.1007/s00392-009-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, Goldmuntz E, Broman KW, Benson DW, Smoot LB, Pu WT. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43:677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet. 2007;44:779–783. doi: 10.1136/jmg.2007.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Wu G, Shan J, Pang S, Wei X, Zhang H, Yan B. Genetic analysis of the promoter region of the GATA4 gene in patients with ventricular septal defects. Transl Res. 2012;159:376–382. doi: 10.1016/j.trsl.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blue GM, Kirk EP, Giannoulatou E, Sholler GF, Dunwoodie SL, Harvey RP, Winlaw DS. Advances in the genetics of congenital heart disease: a clinician's guide. J Am Coll Cardiol. 2017;69:859–870. doi: 10.1016/j.jacc.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 28.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, Hung WC, Haider S, Zhang J, Knight J, Bjornson RD, Castaldi C, Tikhonoa IR, Bilguvar K, Mane SM, Sanders SJ, Mital S, Russell MW, Gaynor JW, Deanfield J, Giardini A, Porter GA, Jr, Srivastava D, Lo CW, Shen Y, Watkins WS, Yandell M, Yost HJ, Tristani-Firouzi M, Newburger JW, Roberts AE, Kim R, Zhao H, Kaltman JR, Goldmuntz E, Chung WK, Seidman JG, Gelb BD, Seidman CE, Lifton RP, Brueckner M. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, Prigmore E, Rajan D, Abdul-Khaliq H, Banka S, Bauer UM, Bentham J, Berger F, Bhattacharya S, Bu'Lock F, Canham N, Colgiu IG, Cosgrove C, Cox H, Daehnert I, Daly A, Danesh J, Fryer A, Gewillig M, Hobson E, Hoff K, Homfray T; INTERVAL Study, Kahlert AK, Ketley A, Kramer HH, Lachlan K, Lampe AK, Louw JJ, Manickara AK, Manase D, McCarthy KP, Metcalfe K, Moore C, Newbury-Ecob R, Omer SO, Ouwehand WH, Park SM, Parker MJ, Pickardt T, Pollard MO, Robert L, Roberts DJ, Sambrook J, Setchfield K, Stiller B, Thornborough C, Toka O, Watkins H, Williams D, Wright M, Mital S, Daubeney PE, Keavney B, Goodship J; UK10K Consortium, Abu-Sulaiman RM, Klaassen S, Wright CF, Firth HV, Barrett JC, Devriendt K, FitzPatrick DR, Brook JD; Deciphering Developmental Disorders Study, Hurles ME. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet 2016;48:1060–5. [DOI] [PMC free article] [PubMed]
- 30.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, DePalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe'er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, Cosgrove C, Blue G, Granados-Riveron J, Setchfield K, Thornborough C, Breckpot J, Soemedi R, Martin R, Rahman TJ, Hall D, van Engelen K, Moorman AF, Zwinderman AH, Barnett P, Koopmann TT, Adriaens ME, Varro A, George AL, Jr, dos Remedios C, Bishopric NH, Bezzina CR, O'Sullivan J, Gewillig M, Bu'Lock FA, Winlaw D, Bhattacharya S, Devriendt K, Brook JD, Mulder BJ, Mital S, Postma AV, Lathrop GM, Farrall M, Goodship JA, Keavney BD. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013;45:822–824. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang WY, Heng HH, Liew CC. Assignment of the human GATA4 gene to 8p23.1-->p22 using fluorescence in situ hybridization analysis. Cytogenet Cell Genet. 1996;72:217–218. doi: 10.1159/000134194. [DOI] [PubMed] [Google Scholar]
- 33.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development [published correction appears in Genome Biol. 2016 Dec 19;17 (1):263] Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohara Y, Atarashi T, Ishibashi T, Ohashi-Kobayashi A, Maeda M. GATA-4 gene organization and analysis of its promoter. Biol Pharm Bull. 2006;29:410–419. doi: 10.1248/bpb.29.410. [DOI] [PubMed] [Google Scholar]
- 36.Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, Kelley DR, Gifford CA, Cacchiarelli D, Rinn JL, Gnirke A, Ziller MJ, Meissner A. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat Genet. 2018;50:250–258. doi: 10.1038/s41588-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riazi AM, Takeuchi JK, Hornberger LK, Zaidi SH, Amini F, Coles J, Bruneau BG, Van Arsdell GS. NKX2-5 regulates the expression of beta-catenin and GATA4 in ventricular myocytes. PLoS ONE. 2009;4:e5698. doi: 10.1371/journal.pone.0005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Zhou L, Wang Q, You X, Li Y, Zhao Y, Han X, Chang Z, He X, Cheng C, Wu C, Wang WJ, Hu FY, Zhao T, Li Y, Zhao M, Zheng GY, Dong J, Fan C, Yang J, Meng X, Zhang Y, Zhu X, Xiong J, Tian XL, Cao H. NEXN inhibits GATA4 and leads to atrial septal defects in mice and humans. Cardiovasc Res. 2014;103:228–237. doi: 10.1093/cvr/cvu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- 40.Kontaraki JE, Kochiadakis GE, Marketou ME, Chlouverakis G, Igoumenidis NE, Saloustros IG, Vardas PE. Early cardiac gene transcript levels in peripheral blood mononuclear cells reflect severity in stable coronary artery disease. Hellenic J Cardiol. 2014;55:119–125. [PubMed] [Google Scholar]
- 41.Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- 42.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA, Pu WT. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun. 2014;5:4907. doi: 10.1038/ncomms5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7:1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linhares VL, Almeida NA, Menezes DC, Elliott DA, Lai D, Beyer EC, Campos de carvalho AC, Costa MW. Transcriptional regulation of the murine. Cardiovasc Res. 2004;64:402–411. doi: 10.1016/j.cardiores.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Original EMSA images for Fig. 3 were included in this document file.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.