Abstract
Background
Metabolic stress, as negative energy balance on one hand or obesity on the other hand can lead to increased levels of free fatty acids in the plasma and follicular fluid of animals and humans. In an earlier study, we showed that increased oleic acid (OA) concentrations affected the function of cultured bovine granulosa cells (GCs). Here, we focus on genome wide effects of increased OA concentrations.
Results
Our data showed that 413 genes were affected, of which 197 were down- and 216 up-regulated. Specifically, the expression of FSH-regulated functional key genes, CCND2, LHCGR, INHA and CYP19A1 and 17-β-estradiol (E2) production were reduced by OA treatment, whereas the expression of the fatty acid transporter CD36 was increased and the morphology of the cells was changed due to lipid droplet accumulation. Bioinformatic analysis revealed that associated pathways of the putative upstream regulators “FSH” and “Cg (choriogonadotropin)” were inhibited and activated, respectively. Down-regulated genes are over-represented in GO terms “reproductive structure/system development”, “ovulation cycle process”, and “(positive) regulation of gonadotropin secretion”, whereas up-regulated genes are involved in “circulatory system development”, “vasculature development”, “angiogenesis” or “extracellular matrix/structure organization”.
Conclusions
From these data we conclude that besides inhibiting GC functionality, increased OA levels seemingly promote angiogenesis and tissue remodelling, thus suggestively initiating a premature fulliculo-luteal transition. In vivo this may lead to impeded folliculogenesis and ovulation, and cause sub-fertility.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07817-6.
Keywords: Gene expression, Free fatty acids, Estradiol, Angiogenesis, Metabolic stress, Cell culture
Background
Dairy cows frequently suffer from negative energy balance (NEB) after parturition. Under these metabolic conditions, the serum levels of non-esterified fatty acids (NEFAs) become elevated and can negatively affect the reproductive performance [1–3]. Also in obese women, it has been reported that abnormal NEFA levels and lipid dysregulation are associated with fertility problems [4–6]. During NEB in lactating dairy cows, fat from adipose tissue is mobilized to meet the animal’s energy requirements. Consequently, levels of free fatty acids (FFAs) like palmitic acid (PA, 16:0), stearic acid (SA, 18:0), oleic acid (OA, 18:1) and of β-hydroxybutyric acid, increase in the plasma and follicular fluid [3, 7] and can negatively affect milk production and cause increased vulnerability to infections, various metabolic diseases and reduced fertility [8–10]. Also short-term fasting increases the levels of different fatty acids especially of OA in the follicular fluid [11]. Various studies showed that high levels of FFAs are indicators of abnormal lipid metabolism and can affect growth, differentiation and metabolism of cells by altering the gene expression levels [12–14]. Also in granulosa cells (GCs) it has been found that free fatty acids like PA, SA and OA affect cell survival in both humans and bovine [15, 16]. In our previous studies, we showed in a bovine GC culture model that OA affects the cell morphology and reduces expression of genes that are involved in 17-β-estradiol (E2) production and gonadotropin signalling as CYP19A1, FSHR and LHCGR [17, 18]. Also the GC identity marker FOXL2 was down-, whereas the marker of sertoli cells SOX9 was up-regulated [19]. Intriguingly, we could show in addition that OA elicited partly opposing effects compared to PA and SA if applied individually [18]. During the present genome-wide approach we selected to study OA effects because it was found at highest concentration in animals after fasting as compared to PA and SA [11] and it was shown to elicit strong and reproducible effects on cultured E2 producing GCs [17, 18].
To elucidate the pathways and upstream molecules that are involved in GC dysfunction under OA treatment we studied effects of OA on global gene expression and analyzed effects on signaling pathways, upstream regulators and biological processes.
Results
Oleic acid induced differentially expressed genes
Microarray analysis of OA vs. vehicle treated GCs after 8 days in culture resulted in 413 differentially expressed genes (DEGs, Supplementary Table 1). According to the statistical criteria used (FC > |1.5|, ANOVA p < 0.05, and FDR q < 0.05) OA treated and vehicle treated control samples (n = 4 in each group) were clearly separated by hierarchical cluster analysis (Fig. 1). Out of the 413 DEGs, 197 were down- and 216 genes up-regulated. ALDH1A1 (FC-6.15) was the most up-regulated gene, whereas CYP19A1 (FC − 4.68) was strongly down-regulated by OA (Table 1).
Fig. 1.
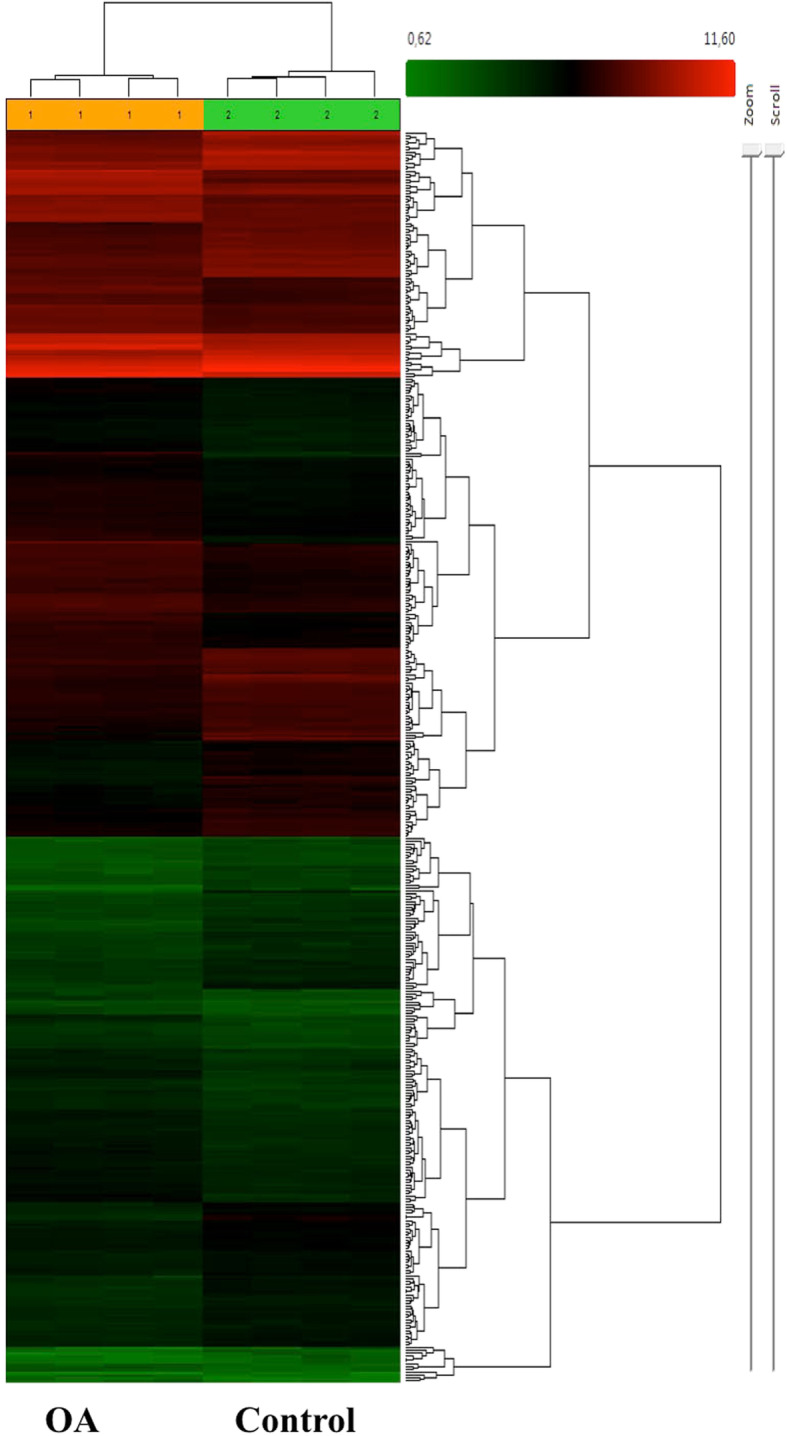
Microarray analysis. The figure shows a heat map analysis of DEGs. OA treated and untreated GCs were analyzed after 8 days in culture under stimulatory conditions with FSH and IGF-1 supplementation throughout the time of culture. The cells were exposed to OA or vehicle control from day 2. Data were obtained from four samples in each group
Table 1.
Top 24 DEGs affected by OA treatment of cultured GCs
| # | Gene Symbol | Fold change | p- value |
|---|---|---|---|
| 1 | ALDH1A1 | 6.15 | 0.000001 |
| 2 | ANGPTL4 | 5.69 | 2.10E-09 |
| 3 | PAG11 | 4.4 | 7.01E-08 |
| 4 | CD36 | 3.65 | 3.33E-08 |
| 5 | SLC38A4 | 3.59 | 0.000044 |
| 6 | PLA2R1 | 3.39 | 0.000014 |
| 7 | KIAA1199 | 3.3 | 1.98E-08 |
| 8 | DHRS9 | 3.08 | 8.76E-07 |
| 9 | LOC537017 | 2.77 | 0.000018 |
| 10 | ANKRD1 | 2.73 | 1.77E-09 |
| 11 | CD44 | 2.73 | 5.76E-07 |
| 12 | EFEMP1 | 2.69 | 3.33E-07 |
| 13 | CYP19A1 | -4.68 | 0.000004 |
| 14 | CNIH3 | −4.03 | 4.60E-08 |
| 15 | JAK3 | −3.38 | 2.94E-08 |
| 16 | TLL2 | −3.31 | 2.66E-07 |
| 17 | CDH20 | −3.07 | 0.000004 |
| 18 | SUSD4 | −2.91 | 0.000003 |
| 19 | NOS2 | −2.9 | 2.63E-07 |
| 20 | AFMID | −2.87 | 6.78E-07 |
| 21 | QRFPR | −2.77 | 2.00E-06 |
| 22 | SLC28A2 | −2.76 | 2.40E-05 |
| 23 | CMBL | −2.71 | 2.00E-06 |
| 24 | KCND2 | −2.68 | 1.40E-05 |
DEGs Differentially expressed genes, GCs Granulosa cells, OA Oleic acid
Real -time RT-PCR validation of differentially expressed genes
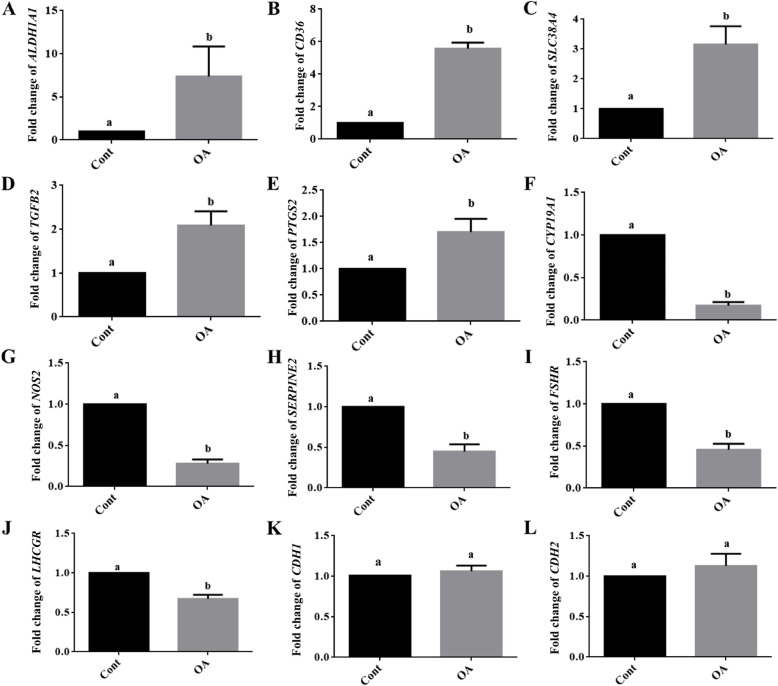
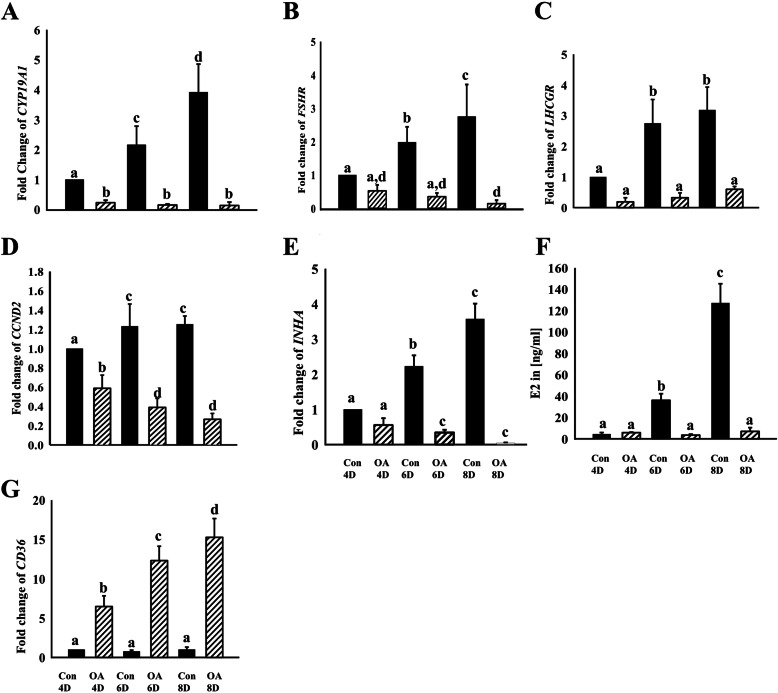
From the DEGs we selected functionally interesting up-regulated (ALDH1A1, CD36, SLC38A4, TGFB2 and PTGS2), down-regulated (CYP19A1, NOS2, SERPINE2, FSHR and LHCGR) and non-regulated genes (CDH1 and CDH2) for validation by real-time RT-PCR. The results clearly indicated similar OA effects after 8 days in culture thus confirming the microarray data (Fig. 2). An additional analysis also including earlier time points (4, 6 and 8 days in culture) indicated that the transcript abundance of several genes (CYP19A1, FSHR, LHCGR) remarkably increased under control conditions, but not in the presence of OA (Fig. 3). Instead, the abundance of some transcripts was even reduced by OA (CCND2, INHA). The E2 accumulated over time, but was nearly absent in OA treated samples. In contrast, CD36 was only expressed at very low levels under control conditions, but was largely stimulated by OA treatment.
Fig. 2.
Microarray data validation by real-time RT PCR. The abundance of different transcripts from OA affected and non-affected genes were analyzed by real-time RT-PCR. The results clearly showed that ALDH1A1, CD36, SLC38A4, TGFB2 and PTSG2 were up-regulated (A-E), CYP19A1, NOS2, SERPINE2, FSHR, LHCGR, (F-J) were down-regulated and CDH1 and CDH2 (K, L) were unaffected by OA treatment thus confirming the microarray data. GC samples were analyzed after 8 days in culture under stimulatory conditions with FSH and IGF-1 supplementation throughout the time of culture and OA or vehicle treatment from day 2. Different letters indicate significant differences relative to the respective controls, which were set to one (mean fold change ± standard deviation, P < 0.05, one-way ANOVA from four independent experiments). Cont denotes vehicle controls and OA denotes oleic acid treatment
Fig. 3.
Time dependent effects of OA on selected transcripts and E2 production. GCs were cultured under stimulatory conditions with FSH and IGF-1 supplementation throughout the time of culture. After 2 days, the cells were treated in addition with OA or vehicle control. Media with all respective supplements were replaced every other day. Transcript abundance of different genes (A, B, C, D, E, G) and levels of estradiol (F) were determined after 4, 6 and 8 days in culture in OA or vehicle treated GCs. OA treated samples showed reduced transcript abundance of CYP19A1, FSHR, LHCGR, CCND2, INHA FSHR, but increased levels of CD36. Different letters indicate significant differences of fold changes (mean fold change ± standard deviation, P < 0.05, one-way ANOVA from three independent experiments) relative to untreated, but FSH and IGF-1 stimulated cultures after day 4, which were set to one. Con denotes vehicle controls and OA denotes oleic acid treatment
Bioinformatics analysis of differentially expressed genes
IPA analysis of DEGs predicted that OA might modulate different Upstream Regulators, Canonical Signaling Pathways, and Functions and Diseases. 98 “Upstream Regulators” were affected with z-scores >|2|, out of which 74 were activated and 24 inhibited (Supplementary Table 2). OA activated the Upstream Regulators “TGFB1”, “Cg (choriogonadotropin)”, “Lipopolysaccharide”, “Rosiglitazone”, “Tgf beta”, “NFkB (complex)” and “HIF1A” etc., and inhibited “SB203580”, “FSH”, “PD98059”, “SMAD7”, “Y 27632” and “NR5A1” etc.
IPA analysis also predicted many affected Canonical Pathways. Top scores were indicated for “Hepatic Fibrosis / Hepatic Stellate Cell Activation”, “Ovarian Cancer Signaling”, “LPS/IL-1 Mediated Inhibition of RXR Function”, “Atherosclerosis Signaling” and “LXR/RXR Activation” (Supplementary Table 3).
Moreover IPA analysis of differentially expressed genes predicted 69 possibly affected diseases and functions. The most significant were “Cellular Movement”, “Cancer”, “Organismal Injury and Abnormalities”, “Cardiovascular System Development and Function” and “Organismal Development” (Supplementary Table 4).
Gene Ontology (GO) term analysis using the WebGestalt tool, showed that among others, down-regulated genes were associated with the terms “reproductive structure/system development”, “ovulation cycle process” and “regulation of follicle-stimulating hormone secretion” (Table 2 and Supplementary Figure 1). Simultaneously, the analysis of GO terms associated with up-regulated genes revealed effects on “circulatory system development”, “vasculature development”, “cardiovascular system development”, “extracellular matrix/structure organization/organization”, “angiogenesis”, “blood vessel morphogenesis” and “cell adhesion” (Table 2 and Supplementary Figure 2).
Table 2.
Affected Gene Ontology (GO) terms according to WebGestalt Over-Representation Analysis
| Regulation | GO | Name | gNum | FDR |
|---|---|---|---|---|
| Down | 0048608 | Reproductive structure development | 16 | 1.21E-02 |
| Down | 0061458 | Reproductive system development | 16 | 1.21E-02 |
| Down | 0048468 | Cell development | 39 | 1.21E-02 |
| Down | 0003006 | Developmental process involved in reproduction | 19 | 2.32E-02 |
| Down | 0022602 | Ovulation cycle process | 7 | 2.32E-02 |
| Down | 0046880 | Regulation of follicle-stimulating hormone secretion | 3 | 2.32E-02 |
| Down | 0046881 | Positive regulation of follicle-stimulating hormone | 3 | 2.32E-02 |
| Down | 0007167 | Enzyme linked receptor protein signaling pathway | 24 | 2.82E-02 |
| Down | 0032278 | Positive regulation of gonadotropin secretion | 3 | 2.82E-02 |
| Down | 0046884 | Follicle-stumulating hormone secretion | 3 | 2.82E-02 |
| Up | 0072359 | Circulatory system development | 42 | 4.61E-10 |
| Up | 0048646 | Anatomical structure formation involved in morphogenesis | 39 | 6.32E-08 |
| Up | 0001944 | Vasculature development | 30 | 9.44E-08 |
| Up | 0072358 | Cardiovascular system development | 30 | 9.82E-08 |
| Up | 0043062 | Extracellular structure organization | 22 | 1.24E-07 |
| Up | 0030198 | Extracellular matrix organization | 22 | 1.24E-07 |
| Up | 0009888 | Tissue development | 53 | 1.45E-07 |
| Up | 0001525 | Angiogenesis | 24 | 1.82E-07 |
| Up | 0048514 | Blood vessel morphogenesis | 26 | 2.48E-07 |
| Up | 0007155 | Cell adhesion | 50 | 7.64E-07 |
Time dependent effects of OA on GC morphology
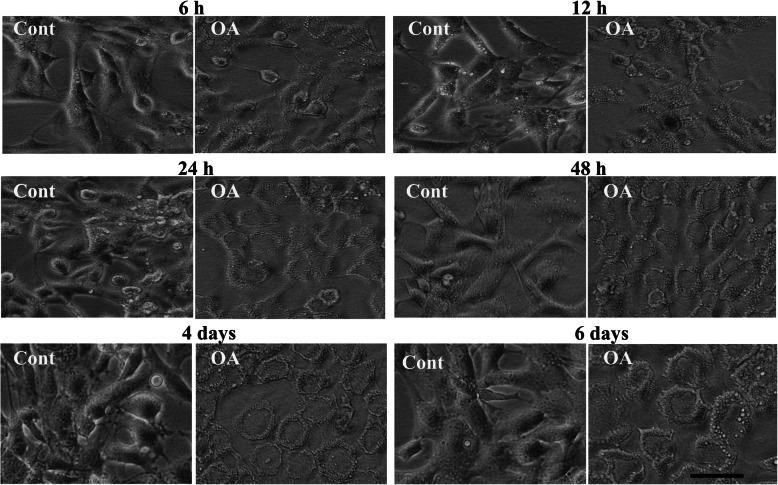
After treatment with OA, cultured GCs were evaluated under a microscope 6 h, 12 h, 24 h, 48 h, 4 days and 6 days after treatment. The cell morphology showed that OA induced morphological changed from a more fibroblast-like to a foam-cell-like structure in a time dependent manner (Fig. 4). In a previous study, we could show that this morphological change is mainly due to considerable lipid droplet accumulation [17].
Fig. 4.
Time dependent morphological changes of GCs after OA treatment. Following 2 days of stimulatory culture conditions with FSH and IGF-1 supplementation GCs were treated with OA or vehicle control in addition and photographed 6 h, 12 h, 24 h, 48 h, 4d and 6 later to study the OA induced changes of morphological features. A continuous change of the morphology from a fibroblast-like towards a flat, foam cell-like morphology can be seen. Cont denotes vehicle controls and OA denotes oleic acid treated cells. Scale bar = 50 μm
Discussion
Bioinformatic analysis of DEGs by IPA suggests that various upstream regulators were affected by OA treatment in cultured bovine GCs. Most interesting, FSH and NR5A1 (also known as steroidogenic factor 1, SF-1) were among the inhibited regulators. Both factors and associated pathways are known to play significant roles during folliculogenesis [20–23]. Down-regulation of FSH signaling is in line with our previously published data indicating that in particular the abundance of FSHR transcripts and thus the corresponding pathways and genes are down regulated by OA treatment [17]. This may result in the observed prevention of up-regulation of the functional key genes CYP19A1, CCND2, LHCGR and INHA and of E2 production from day 4 to the end of the experiment (see Fig. 3). In addition to this, also Over-Representation Analysis of down-regulated genes revealed that most of these are involved in female reproduction related processes such as “reproductive structure/system development”, “ovulation cycle process”, “(positive) regulation of follicle-stimulating hormone/gonadotropin (secretion)” (see Table 2). Thus, these data indicate that the presence of OA compromised GC functionality and suggest that OA may impair folliculogenesis and the ovarian cycle in cows. This is also supported by the observation of morphological changes in OA treated GCs showing lipid droplet accumulation. Also in human lipid droplet accumulation in GCs have been correlated with fertility problems [24]. Also recent in vivo data suggest that increased NEFA levels and in particular increased OA concentrations in the follicular fluid have detrimental effects on follicle growth and even prevent ovulation [18, 25]. However, the role of OA seems complex and is still controversial. Some studies also clearly demonstrated positive effects on oocytes and during early embryo development as well as protective actions against lipotoxicity [26, 27].
Among the activated upstream regulators the most interesting were TGFB1, Cg (choriogonadotropin), Tgf beta, NFkB, HIF1A, EDN1 and WNT3A (see Supplementary Table 2). The significant activation of the Cg pathway suggests that LH signaling was induced by increased OA concentrations via the luteinizing hormone/choriogonadotropin receptor (LHCGR) pathway. Together with the down-regulated GC functionality, this suggests that the cells are driven towards luteinization. In vivo the preovulatory LH surge induces a rapid and profound molecular transformation of follicular cells termed luteinization in particular in GCs [28–30]. These changes (down-regulation of E2 production and of follicular marker transcripts) seem to be at least partly mimicked by OA in cultured GCs. However, this transformation is certainly not complete. Luteinization of GCs is not only associated with down-regulation of E2 but in addition with up-regulation of progesterone (P4) production. This has been clearly shown in vivo after the preovulatory LH surge [28, 31]. However, we could show in our previous studies that OA treatment of cultured GCs is clearly associated with decreased E2 but not with increased P4 production. Contrary, P4 production was even down-regulated [17, 18].
Interestingly, also TGFB1, Tgf beta, NFkB, HIF1A and WNT3A were indicated as activated by OA treatment. According to previous studies these factors and associated pathways are involved in the regulation of the Epithelial-to-Mesenchymal Transition (EMT) in several cell types including ovarian cells [32]. Activation of these regulators thus suggests EMT up-regulation by OA treatment. Partial EMT is a characteristic feature of GCs entering the folliculo-luteal transformation [33]. It is also quite noticeable that EDN1 is among the activated upstream regulators. On one hand, endothelins are known to be involved in angiogenic processes [34–36] and on the other hand, the formation of new vasculature is an essential feature of the developing corpus luteum [37, 38]. EDN1, EDN2 and EDN3 as well as endothelin receptors type A and B (EDNRA, ENDRB) are present in bovine GCs [39] as well as in our cultured GCs whereby EDNRA is significantly down- and ENDRB significantly up-regulated (see Supplementary Table 2). Taken together, down-regulation of GC functionality and simultaneous activation of the endothelin pathway and of GO terms related to angiogenesis, vascularization and morphogenic processes (e.g. Anatomical structure formation, Extracellular structure/matrix organization, see Table 2) support our hypothesis that OA treatment induces a luteal-like transformation in cultured GCs. Not least, also the observed lipid droplet accumulation in OA treated cells suggests their more luteal-like character since lipid droplets are a major feature of steroidogenic luteal cells [40].
Intriguingly, IPA analysis of affected “Canonical Pathways” and “Cell functions and Diseases” provided also evidence that cancer-related signaling might be promoted by OA treatment (see Supplementary Tables 3 and 4). In addition, the up-regulation of genes involved in angiogenic and morphogenic processes can be interpreted as supportive of the alternative hypothesis that increased OA concentrations facilitate tumor initiation and progression. However, cancer studies are certainly overrepresented in databases used by IPA and other bioinformatic tools. This may lead to a biased output favoring pathway and regulators involved in cancerogenesis. In addition, the observed prevalence of GC cancer in cattle is very low (0.5–0.75%) according to several studies [41, 42]. Accordingly, this interpretation of our data must be treated with caution and certainly needs additional experimental evidence.
Conclusions
Based on our results and on studies from previous reports we propose that OA induces fatty acid uptake and lipid droplet accumulation in GCs and may inhibit FSH signaling and affect the expression of many genes, which are involved in GC functionality. On the other hand, genes and pathways involved in angiogenic and morphogenic processes are up-regulated thus suggesting that OA may activate pathways favoring folliculo-luteal transition.
Methods
Culture of granulosa cells
Isolation and culture of GCs were done as previously described [17, 43, 44]. Briefly, bovine ovaries were collected from a commercial slaughterhouse (Danish Crown Teterower Fleisch GmbH, Teterow). GCs were aspirated from small to medium follicles (2-6 mm diameter) that contain clear antral fluid with an 18-gauge needle in PBS with antibiotics. The percentage of viable cells was determined in a hemocytometer by using the trypan blue exclusion method. Routinely, 1.25 × 105 viable cells were seeded on 24-well collagen R (Serva, Heidelberg, Germany) coated plates per 0.5 ml of basal α-MEM (Merck/Biochrom, Berlin, Germany) without serum but supplemented with 25 ng/ml IGF-1, 20 ng/ml FSH and 2 μM androstenedione (Sigma Aldrich, Steinheim, Germany) to induce E2 production. Conditioned media were replaced with fresh media including all respective supplements every other day. Two days after seeding 400 μM of OA (Sigma Aldrich, Steinheim, Germany) dissolved in ethanol as described earlier [17] was added with the first change of media (i.e. after 2 days in culture). The concentration of 400 μM was selected, because similar levels have been found in vivo under NEB conditions and this concentration has been also shown to efficiently affect gene expression and hormone production in GCs [11, 17]. To allow the cells to recover from plating stress and to re-acquire GC-like morphological and physiological features (expression of marker genes and E2 production) the cells were cultured for 8 days [44] for microarray analysis. In addition, early alterations of the abundance of selected marker transcripts and of E2 concentrations in spent media were also studied after 4, 6 and 8 days. Early morphological changes were analyzed 6 h, 12 h, 24 h, 48 h, 4th day and 6th day after addition of OA by using a Nikon TMS-F inverted microscope.
RNA isolation and cDNA synthesis
RNA isolation and cDNA synthesis was done as previously described [19]. Briefly, RNA was isolated with the Nucleo Spin® RNA II Kit (Macherey-Nagel, Düren, Germany) and quantified with a NanoDrop1000 Spectrophotometer (Thermo Scientific, Bonn, Germany). The cDNA for real-time RT-PCR was prepared by using the SensiFAST cDNA Synthesis Kit (Bioline, Luckenwalde, Germany) from 200 ng RNA.
Microarray analysis
For mRNA microarray analysis RNA Integrity Numbers (RIN) were determined in a Bioanalyzer System (Agilent Technologies Inc.). Numbers were 9.7 on average in both experimental groups (OA treated and vehicle treated GCs, 4 samples per group) ranging from 9.6 to 10.0. Total RNA of the different samples was hybridized on Bovine Gene 1.0 ST Array (Affymetrix, St. Clara, CA, USA). Amplification, labelling and hybridization was performed with the “GeneChip Expression 3’ Amplification One-Cycle Target Labeling and Control Reagents” (Affymetrix) according to the manufacturer’s protocol. Hybridization was done overnight in the GeneChipR Hybridization Oven (Affymetrix) and visualized with the Affymetrix GeneChip Scanner 3000. Raw data were processed with the Expression Console (V1.4.1.46; Affymetrix). Normalization, background reduction and a gene-level summary was done using the RMA method (Robust Multichip Average). Results of the array have been submitted to the GEO database (GSE152307). Subsequent analysis was done with the Transcriptome Analysis Console 3.0 (TAC3.0, Affymetrix) to check for differentially expressed genes under different conditions. Analysis of Variance (ANOVA) was used to calculate the p-value and was additionally corrected for FDR (False Discovery Rate, Benjamini-Hochberg method) integrated in TAC3.0. Levels of significance for differentially expressed genes were set at fold change (FC) > |1.5|, ANOVA p < 0.05 and FDR < 0.05.
Real-time RT-PCR
Validation of microarray data was carried out by analysing the transcript abundance of four up-regulated (ALDH1A1, CD36, SLC38A4, TGFB2 and PTSG2), four down-regulated (CYP19A1, NOS2, SERPINE2, FSHR, LHCGR) and two unchanged genes (CDH1 and CDH2) by real time RT-PCR. For this, 0.25 and 0.5 μl cDNA of each sample were amplified using SensiFastTM SYBR No-ROX (Bioline, Luckenwalde, Germany) and gene-specific primers (Supplementary Table 5) in a LightCycler 96 instrument (Roche). To ensure amplification of the correct products melting points of products were routinely analysed. Initially, all amplicons were cloned and sequenced. These cloned plasmids were used as external standards. For preparation of an external standard curve, five fresh dilutions were prepared with concentrations from 5 × 1012 to 5 × 1016 g DNA/reaction. These were co-amplified with each run of samples. After amplification, values of 0.25 and 0.5 μl of cDNA were averaged considering different dilutions. The size of products was routinely controlled by agarose gel electrophoresis. For normalization, the abundance of target transcripts was divided by the corresponding abundance of TBP housekeeping control transcripts [45].
Bioinformatic analysis of differentially expressed genes
To identify “Upstream Regulators”, “Canonical Pathways”, and “Functions and Diseases” affected by OA treatment, differentially expressed genes (DEGs) were analyzed by the Ingenuity Pathway Analysis tool (IPA, QIAGEN, Hilden, Germany). In addition to this, gene enrichment and gene ontology analysis was done with the WEB-based GEneSeTAnaLysis Toolkit (WebGestalt).
Quantification of 17-β-estradiol
The levels of E2 in conditioned media collected at different days after OA treatment were estimated with an ultrasensitive 125I-RIA (DSL, Sinsheim, Germany) in 200-μL duplicates. The standard curve was established between 0.005 and 0.750 ngmL− 1. Radioactivity was measured with an automatic gamma counter with integrated RIA calculation (Wizard; Perkin Elmer, Rodgau, Germany). The detection limit of the method was found at 0.003 ngmL− 1. The intra- and inter-assay CVs were 8.8 and 9.7%, respectively. The analysis of media was done with 10 μl of the undiluted sample in duplicates as also described in previous studies [17, 46].
Statistical analysis
All experiments were carried out at least three times independently. Comparative data from OA effects on transcript abundance at different days were analysed by one-way analysis of variance (ANOVA) using the Holm-Sidak all pairwise multiple comparison procedure with the Sigma Plot 11.0 Statistical Analysis System (Jandel Scientific, San Rafael, CA, USA). Microarray re-evaluation data at the 8th day in culture were analysed by the paired t-test using the GraphPad prism 5.0 software. P values < 0.05 were considered significant.
Supplementary Information
Additional file 1: Figure S1. GO terms associated with OA down-regulated genes according to WebGestalt (significantly affected GO term in red).
Additional file 2: Figure S2. GO terms associated with OA up-regulated genes according to WebGestalt (significantly affected GO term in red).
Additional file 3: Table S1. List of OA regulated genes (DEGs).
Additional file 4: Table S2. OA regulated Upstream Regulators according to IPA analysis.
Additional file 5: Table S3. OA regulated Canonical Pathways according to IPA analysis.
Additional file 6: Table S4. OA regulated Diseases and Functions according to IPA analysis.
Additional file 7: Table S5. List of primers used for transcript quantification by real-time RT-PCR.
Acknowledgements
We thank Veronica Schreiter, Maren Anders and Ildiko Toth for their excellent technical assistance. We also thank and the Department of Animal Biology, School of Life Sciences, University of Hyderabad for providing technical facilities.
Abbreviations
- DEG
Differentially Expressed Gene
- E2
17-β-Estradiol
- EMT
Epithelial-to-Mesenchymal Transition
- FFA
Free Fatty Acid
- FSH
Follicle Stimulating Hormone
- GC
Granulosa Cell
- GO
Gene Ontology
- IGF-1
Insulin-like-Growth Factor 1
- IPA
Ingenuity Pathway Analysis
- LH
Luteinizing Hormone
- NEB
Negative Energy Balance
- NEFA
Non-Esterified Fatty Acid
- OA
Oleic Acid
- P4
Progesterone
- PA
Palmitic Acid
- SA
Stearic Acid
Authors’ contributions
VRY designed and conducted the experiments and wrote the manuscript. DK did the mRNA microarray analysis and JV contributed to the experimental design and preparation of the manuscript. All authors have read and approved the manuscript.
Funding
This study was funded by the core budget of the Leibniz Institute for Farm Animal Biology (FBN). VRY was supported by the DST-Inspire Faculty program (DST-India). Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152307
Primers for Real -time RT-PCR analysis were designed from published sequences with accession Nos: NM_174239.2, NM_001076372, NM_001278621.1, NM_001002763.1, NM_001166492.1, NM_174445, NM_174305, NM_174061, NM_174381, NM_174094.4, NM_001076799.1, NM_174669.2, NM_001205943.1, NM_001113252.1, NM_001075742.1.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Footnotes
The original version of this article was revised: affiliations 1 and 3 were mistakenly switched.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/24/2021
A Correction to this paper has been published: 10.1186/s12864-021-07933-3
Contributor Information
Vengala Rao Yenuganti, Email: vengal.ndri@gmail.com.
Jens Vanselow, Email: vanselow@fbn-dummerstorf.de.
References
- 1.Van Hoeck V, Leroy JL, Alvarez AM, Rizos D, Gutierrez-Adan A, Schnorbusch K, Bols PE, Leese HJ, Sturmey RG. Oocyte developmental failure in response to elevated nonesterified fatty acid concentrations: mechanistic insights. Reproduction. 2013;145(1):33–44. doi: 10.1530/REP-12-0174. [DOI] [PubMed] [Google Scholar]
- 2.Jorritsma R, Wensing T, Kruip TA, Vos PL, Noordhuizen JP. Metabolic changes in early lactation and impaired reproductive performance in dairy cows. Vet Res. 2003;34(1):11–26. doi: 10.1051/vetres:2002054. [DOI] [PubMed] [Google Scholar]
- 3.Leroy J, Vanholder T, Mateusen B, Christophe A, Opsomer G, de Kruif A, Genicot G, Van Soom A. Non-esterified fatty acids in follicular fluid of dairy cows and their effect on developmental capacity of bovine oocytes in vitro. Reproduction. 2005;130(4):485–495. doi: 10.1530/rep.1.00735. [DOI] [PubMed] [Google Scholar]
- 4.Holte J, Bergh T, Berne C, Lithell H. Serum lipoprotein lipid profile in women with the polycystic ovary syndrome: relation to anthropometric, endocrine and metabolic variables. Clin Endocrinol. 1994;41(4):463–471. doi: 10.1111/j.1365-2265.1994.tb02577.x. [DOI] [PubMed] [Google Scholar]
- 5.Niu Z, Lin N, Gu R, Sun Y, Feng Y. Associations between insulin resistance, free fatty acids, and oocyte quality in polycystic ovary syndrome during in vitro fertilization. J Clin Endocrinol Metab. 2014;99(11):E2269–E2276. doi: 10.1210/jc.2013-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calonge RN, Kireev R, Guijarro A, Cortes S, Andres C, Caballero P. Lipid dysregulation in seminal and follicular fluids could be related with male and female infertility. Endocrinol Metab Int J. 2018;6:7. [Google Scholar]
- 7.Rukkwamsuk T, Geelen MJ, Kruip TA, Wensing T. Interrelation of fatty acid composition in adipose tissue, serum, and liver of dairy cows during the development of fatty liver postpartum. J Dairy Sci. 2000;83(1):52–59. doi: 10.3168/jds.S0022-0302(00)74854-5. [DOI] [PubMed] [Google Scholar]
- 8.Esposito G, Irons PC, Webb EC, Chapwanya A. Interactions between negative energy balance, metabolic diseases, uterine health and immune response in transition dairy cows. Anim Reprod Sci. 2014;144(3–4):60–71. doi: 10.1016/j.anireprosci.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Moyes KM, Drackley JK, Salak-Johnson JL, Morin DE, Hope JC, Loor JJ. Dietary-induced negative energy balance has minimal effects on innate immunity during a streptococcus uberis mastitis challenge in dairy cows during midlactation. J Dairy Sci. 2009;92(9):4301–4316. doi: 10.3168/jds.2009-2170. [DOI] [PubMed] [Google Scholar]
- 10.Staples CR, Thatcher WW, Clark JH. Relationship between ovarian activity and energy status during the early postpartum period of high producing dairy cows. J Dairy Sci. 1990;73(4):938–947. doi: 10.3168/jds.S0022-0302(90)78750-4. [DOI] [PubMed] [Google Scholar]
- 11.Aardema H, Lolicato F, van de Lest CH, Brouwers JF, Vaandrager AB, van Tol HT, Roelen BA, Vos PL, Helms JB, Gadella BM. Bovine cumulus cells protect maturing oocytes from increased fatty acid levels by massive intracellular lipid storage. Biol Reprod. 2013;88(6):164. doi: 10.1095/biolreprod.112.106062. [DOI] [PubMed] [Google Scholar]
- 12.Duplus E, Forest C. Is there a single mechanism for fatty acid regulation of gene transcription? Biochem Pharmacol. 2002;64(5–6):893–901. doi: 10.1016/S0006-2952(02)01157-7. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura MT, Cheon Y, Li Y, Nara TY. Mechanisms of regulation of gene expression by fatty acids. Lipids. 2004;39(11):1077–1083. doi: 10.1007/s11745-004-1333-0. [DOI] [PubMed] [Google Scholar]
- 14.Georgiadi A, Kersten S. Mechanisms of gene regulation by fatty acids. Adv Nutr. 2012;3(2):127–134. doi: 10.3945/an.111.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu YM, Yanase T, Nishi Y, Tanaka A, Saito M, Jin CH, Mukasa C, Okabe T, Nomura M, Goto K, et al. Saturated FFAs, palmitic acid and stearic acid, induce apoptosis in human granulosa cells. Endocrinology. 2001;142(8):3590–3597. doi: 10.1210/endo.142.8.8293. [DOI] [PubMed] [Google Scholar]
- 16.Vanholder T, Leroy JL, Soom AV, Opsomer G, Maes D. Coryn M, de KA: effect of non-esterified fatty acids on bovine granulosa cell steroidogenesis and proliferation in vitro. Anim Reprod Sci. 2005;87(1–2):33–44. doi: 10.1016/j.anireprosci.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Yenuganti VR, Viergutz T, Vanselow J. Oleic acid induces specific alterations in the morphology, gene expression and steroid hormone production of cultured bovine granulosa cells. Gen Comp Endocrinol. 2016;232:134–144. doi: 10.1016/j.ygcen.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Sharma A, Baddela VS, Becker F, Dannenberger D, Viergutz T, Vanselow J. Elevated free fatty acids affect bovine granulosa cell function: a molecular cue for compromised reproduction during negative energy balance. Endocr Connect. 2019;8(5):493–505. doi: 10.1530/EC-19-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yenuganti VR, Vanselow J. Oleic acid induces down-regulation of the granulosa cell identity marker FOXL2, and up-regulation of the Sertoli cell marker SOX9 in bovine granulosa cells. Reprod Biol Endocrinol. 2017;15(1):57. doi: 10.1186/s12958-017-0276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelusi C, Ikeda Y, Zubair M, Parker KL. Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells. Biol Reprod. 2008;79(6):1074–1083. doi: 10.1095/biolreprod.108.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeyasuria P, Ikeda Y, Jamin SP, Zhao LP, De Rooij DG, Themmen APN, Behringer RR, Parker KL. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol. 2004;18(7):1610–1619. doi: 10.1210/me.2003-0404. [DOI] [PubMed] [Google Scholar]
- 22.Casarini L, Crépieux P. Molecular mechanisms of action of FSH. Front Endocrinol (Lausanne) 2019;10:305. doi: 10.3389/fendo.2019.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilha GF, Rovani MT, Gasperin BG, Antoniazzi AQ, Gonçalves PB, Bordignon V, Duggavathi R. Lack of FSH support enhances LIF-STAT3 signaling in granulosa cells of atretic follicles in cattle. Reproduction. 2015;150(4):395–403. doi: 10.1530/REP-15-0026. [DOI] [PubMed] [Google Scholar]
- 24.Raviv S, Hantisteanu S, Sharon SM, Atzmon Y, Michaeli M, Shalom-Paz E. Lipid droplets in granulosa cells are correlated with reduced pregnancy rates. J Ovarian Res. 2020;13(1):4. doi: 10.1186/s13048-019-0606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferst JG, Missio D, Bertolin K, Gasperin BG, Leivas FG, Bordignon V, Goncalves PB, Ferreira R. Intrafollicular injection of nonesterified fatty acids impaired dominant follicle growth in cattle. Anim Reprod Sci. 2020;219:11. doi: 10.1016/j.anireprosci.2020.106536. [DOI] [PubMed] [Google Scholar]
- 26.Aardema H, Vos PL, Lolicato F, Roelen BA, Knijn HM, Vaandrager AB, Helms JB, Gadella BM. Oleic acid prevents detrimental effects of saturated fatty acids on bovine oocyte developmental competence. Biol Reprod. 2011;85(1):62–69. doi: 10.1095/biolreprod.110.088815. [DOI] [PubMed] [Google Scholar]
- 27.Fayezi S, Leroy J, Novin MG, Darabi M. Oleic acid in the modulation of oocyte and preimplantation embryo development. Zygote. 2018;26(1):1–13. doi: 10.1017/S0967199417000582. [DOI] [PubMed] [Google Scholar]
- 28.Nimz M, Spitschak M, Schneider F, Fürbass R, Vanselow J. Down-regulation of genes encoding steroidogenic enzymes and hormone receptors in late preovulatory follicles of the cow coincides with an accumulation of intrafollicular steroids. Domest Anim Endocrinol. 2009;37:45–54. doi: 10.1016/j.domaniend.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert I, Robert C, Dieleman S, Blondin P, Sirard MA. Transcriptional effect of the LH surge in bovine granulosa cells during the peri-ovulation period. Reproduction. 2011;141(2):193–205. doi: 10.1530/REP-10-0381. [DOI] [PubMed] [Google Scholar]
- 30.Christenson LK, Gunewardena S, Hong X, Spitschak M, Baufeld A, Vanselow J. Research resource: preovulatory LH surge effects on follicular theca and granulosa transcriptomes. Mol Endocrinol. 2013;27(7):1153–1171. doi: 10.1210/me.2013-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berisha B, Steffl M, Amselgruber W, Schams D. Changes in fibroblast growth factor 2 and its receptors in bovine follicles before and after GnRH application and after ovulation. Reproduction. 2006;131(2):319–329. doi: 10.1530/rep.1.00798. [DOI] [PubMed] [Google Scholar]
- 32.Bilyk O, Coatham M, Jewer M, Postovit LM. Epithelial-to-mesenchymal transition in the female reproductive tract: from Normal functioning to disease pathology. Front Oncol. 2017;7:145. doi: 10.3389/fonc.2017.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Childs AJ, McNeilly AS. Epithelial-to-mesenchymal transition in granulosa cells: a key to activation of follicle growth? Biol Reprod. 2012;86(5):152. doi: 10.1095/biolreprod.112.100156. [DOI] [PubMed] [Google Scholar]
- 34.Rosanò L, Spinella F, Bagnato A. Endothelin 1 in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2013;13(9):637–651. doi: 10.1038/nrc3546. [DOI] [PubMed] [Google Scholar]
- 35.Wu MH, Huang CY, Lin JA, Wang SW, Peng CY, Cheng HC, Tang CH. Endothelin-1 promotes vascular endothelial growth factor-dependent angiogenesis in human chondrosarcoma cells. Oncogene. 2014;33(13):1725–1735. doi: 10.1038/onc.2013.109. [DOI] [PubMed] [Google Scholar]
- 36.Bagnato A, Spinella F. Emerging role of endothelin-1 in tumor angiogenesis. Trends Endocrinol Metab. 2003;14(1):44–50. doi: 10.1016/S1043-2760(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 37.Schams D, Berisha B. Angiogenic factors (VEGF, FGF and IGF) in the bovine corpus luteum. J Reprod Dev. 2002;48(3):233–242. doi: 10.1262/jrd.48.233. [DOI] [Google Scholar]
- 38.Zheng J, Redmer DA, Reynolds LP. Vascular development and heparin-binding growth-factors in the bovine corpus-luteum at several stages of the estrous-cycle. Biol Reprod. 1993;49(6):1177–1189. doi: 10.1095/biolreprod49.6.1177. [DOI] [PubMed] [Google Scholar]
- 39.Bridges PJ, Cho J, Ko C. Endothelins in regulating ovarian and oviductal function. Front Biosci (Schol Ed) 2011;3:145–155. doi: 10.2741/s140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talbott HA, Plewes MR, Krause C, Hou X, Zhang P, Rizzo WB, Wood JR, Cupp AS, Davis JS. Formation and characterization of lipid droplets of the bovine corpus luteum. Sci Rep. 2020;10(1):11287. doi: 10.1038/s41598-020-68091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-Martínez C, Durán-Navarrete AJ, García-Fernández RA, Espinosa-Alvarez J, Escudero Diez A, García-Iglesias MJ. Biological characterization of ovarian granulosa cell tumours of slaughtered cattle: assessment of cell proliferation and oestrogen receptors. J Comp Pathol. 2004;130(2–3):117–123. doi: 10.1016/j.jcpa.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Zemjanis R, Larson LL, Bhalla RP. Clinical incidence of genital abnormalities in the cow. J Am Vet Med Assoc. 1961;139:1015–1018. [PubMed] [Google Scholar]
- 43.Baufeld A, Vanselow J. A tissue culture model of estrogen-producing primary bovine granulosa cells. J Vis Exp. 2018;139:1. doi: 10.3791/58208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yenuganti VR, Vanselow J. Cultured bovine granulosa cells rapidly lose important features of their identity and functionality, but partially recover under long term culture conditions. Cell Tissue Res. 2017;368(2):397. doi: 10.1007/s00441-017-2571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baddela VS, Baufeld A, Yenuganti VR, Vanselow J, Singh D. Suitable housekeeping genes for normalization of transcript abundance analysis by real-time RT-PCR in cultured bovine granulosa cells during hypoxia and differential cell plating density. Reprod Biol Endocrinol. 2014;12(1):118. doi: 10.1186/1477-7827-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider F, Brüssow KP. Effects of a preovulatory administered depot gonadotrophin-releasing hormone agonist on reproductive hormone levels and pregnancy outcome in gilts. Reprod Fertil Dev. 2006;18(8):857–866. doi: 10.1071/RD06027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. GO terms associated with OA down-regulated genes according to WebGestalt (significantly affected GO term in red).
Additional file 2: Figure S2. GO terms associated with OA up-regulated genes according to WebGestalt (significantly affected GO term in red).
Additional file 3: Table S1. List of OA regulated genes (DEGs).
Additional file 4: Table S2. OA regulated Upstream Regulators according to IPA analysis.
Additional file 5: Table S3. OA regulated Canonical Pathways according to IPA analysis.
Additional file 6: Table S4. OA regulated Diseases and Functions according to IPA analysis.
Additional file 7: Table S5. List of primers used for transcript quantification by real-time RT-PCR.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152307
Primers for Real -time RT-PCR analysis were designed from published sequences with accession Nos: NM_174239.2, NM_001076372, NM_001278621.1, NM_001002763.1, NM_001166492.1, NM_174445, NM_174305, NM_174061, NM_174381, NM_174094.4, NM_001076799.1, NM_174669.2, NM_001205943.1, NM_001113252.1, NM_001075742.1.