Abstract
The last few decades have witnessed diversified in vitro models to recapitulate the architecture and function of living organs or tissues and contribute immensely to advances in life science. Two novel 3D cell culture models: 1) Organoid, promoted mainly by the developments of stem cell biology and 2) Organ-on-a-chip, enhanced primarily due to microfluidic technology, have emerged as two promising approaches to advance the understanding of basic biological principles and clinical treatments. This review describes the comparable distinct differences between these two models and provides more insights into their complementarity and integration to recognize their merits and limitations for applicable fields. The convergence of the two approaches to produce multi-organoid-on-a-chip or human organoid-on-a-chip is emerging as a new approach for building 3D models with higher physiological relevance. Furthermore, rapid advancements in 3D printing and numerical simulations, which facilitate the design, manufacture, and results-translation of 3D cell culture models, can also serve as novel tools to promote the development and propagation of organoid and organ-on-a-chip systems. Current technological challenges and limitations, as well as expert recommendations and future solutions to address the promising combinations by incorporating organoids, organ-on-a-chip, 3D printing, and numerical simulation, are also summarized.
Keywords: 3D printing, numerical simulations, organoids, organ-on-a-chip, synergistic engineering
1. Introduction
Drug discovery remains an unprecedented challenge and inadequate for treating many diseases that afflict humans because of enormous cost, time, and failure rates involved in clinical trials. Understanding disease mechanisms within sophisticated human organs needs more feasible models with lower costs, reduced time, and the greater simplicity of experiments.[1] Accurate human-representative models utilize many aspects of currently available technologies and hold promises for predicting the effectiveness and accuracy of drug responses to mimic key structural and functional properties of human organs from the subcellular to whole organ level.[2] Those developed systems have been committed to providing insights into basic biological principles, diseases, as well as potential treatments.[2] Animal models currently serve as gold standards for preclinical trials and exhibit comprehensive structural complexities and cell compositions like complex in vivo environments. They play a critical role in different application fields, such as experimental physiology and pathology, as well as drug screening).[3] Nevertheless, the complex physiology of animal models with specific signaling pathways and tissue-tissue or cell–cell interactions are difficult to be independent of other factors, and makes it challenging to distinguish and analyze the exact causal relationship, respectively. Moreover, animal models cannot precisely reflect human physiology, thus, have not been able to accurately predict in vivo toxicity responses upon drug treatment, may largely cause failure in drug development, particularly at the later stages.[3] For instance, animal model cannot assess toxicities of anticancer drug cisplatin because of its different species and impacts targeting to different membrane transporters that modulate drug accumulation between the human body and animals.[4] Furthermore, many animal models remain difficult to obtain and build for modeling numerous human immunological and neurological diseases (e.g., autoimmune diseases to cerebral cancer).[5] Besides, animal models always carry ethical issues that cannot be abused and discarded of the animals or employed for controversial experiments.[6] Therefore, in vitro models are urgently required to exhibit certain translation of their results to humans, and have attracted a lot of attention due to their lower costs, time saving, and the greater simplicity of experiments.
The survival and functionality of cells cultured in 2D and 3D models are noticeably dependent on the microenvironmental cues, such as spatiotemporal chemical treatments, physiological stimuli as well as cell–cell and cell–extracellular matrix (ECM) interactions.[7] 2D monolayer culture methods have been extensively and successfully adopted for their simplicity and cost-effectiveness. However, they usually exhibit insufficient physiological features for insufficient biophysical or biochemical stimuli as in vivo.[8] They are also inferior to the 3D culture that creates more defined and intricate microenvironment when recapitulating the living tissue or organ and modeling diseases.[9] The last decade witness the high-speed development of a novel microengineering device, also termed as organ-on-a-chip[10,11] or microphysiological system.[12] to recapitulate 3D tissue architectures, function, physiology, or pathology of living human organs in vitro. Existing organ-on-a-chip methods are primarily to combine proportional predifferentiated cells to imitate the composition of native tissues and perform their vital structural and functional features. Microengineering approaches are capable of precisely regulating the nutrient flow supply and shear stress stimulation, spatiotemporal chemical, and biological microenvironment, as well as the local electrical or mechanical behaviors of growing 3D tissues.[3] Furthermore, organ-on-a-chip technology can incorporate different 3D constructs into a dynamic circulation system that imitates the systematic interactions between a range of tissues and organs in the human body. They are also termed as human-on-a-chip[13] or body-on-a-chip,[14] being critical to drug discovery applications. Such multiorgans platform allows drugs and their metabolites to run through a range of organs before fulfilling its final action and subsequently allow them to perform in their final target sites in different parts of the organism.[15] Organ-on-a-chip acts as a promising model to ascertain the innermost human pathophysiology, as well as a suitable platform for disease modeling and drug discovery for its enhanced feasibility, productivity, and applicability.[16]
Unlike the rigorously controlled environments created by organs-on-chips, stem cell derived organoids developed from embryoid body cultures are largely self-organizing and similar to teratoma formation in vivo.[17,18] Organoids refer to 3D cell masses characterized by the presence of multiple organ-specific cell lineages, similar cellular organization to that of in vivo counterpart, as well as sophisticated 3D architecture and functional features.[19] Organoid models exhibiting higher physiological relevance than 2D models are more effectively benefit to introduce niche cues and narrow the gap between in vitro models and in vivo models. Pluripotent stem cells (PSCs) exhibit differentiation potential, which direct cell fate to specific tissue precursors and further down into organoids of targeted cell lineages through the supply of cues by activating or inhibiting signaling pathways.[20] Aggregates of PSCs process the processes of differentiation and morphogenesis after embedded in a hydrogel (often Matrigel) scaffolds, and imitate aspects of early embryonic development by improving appropriate exogenous factors.[21] Current organoids rely heavily on a high level of default robustness for the generation of a precisely organized tissue architecture of various shapes and sizes during spontaneous differentiation. Organoids are mostly superior in cellular heterogeneity, phenotype fidelity, and physiologically relevant complexity to organ-on-a-chip system which place predifferentiated cells at precise locations in an artificial manner. Accordingly, organoids can also address a gap in modeling pathology and diseases difficult or unlikely to study in animal models, and work as a promising personalized medicine approach.[22]
Due to recently evolving technological advancements in bioengineering and stem cell tissue engineering, organ-on-a-chip and organoid have emerged as two distinct approaches exhibiting their own merits and demerits for stem cell-derived 3D tissue preparation.[23,24] Accordingly, it is vital to identify the related pros and cons so as to enhance their applicability by correctly choosing cell sources according to research motivations and genetic backgrounds (e.g., cell lines or stem cells). Both of the two models uniquely summarize vital microstructures and functions concerning target tissues or organs and more effectively exploit structural fidelity in lineage specification, cell–cell interaction, and organ- or tissue-genesis.[25] This review systematically compares the physiological complexity, cellular diversity, and cellular genetic similarity, as well as microenvironmental control ability between self-renewing of organoids and well-defined manipulation of organ-on-a-chip to model disease etiology plus drug screening.
The overall goal of providing a path toward a superior, synergistic strategy of constructing tissues by integrating organ-on-a-chip to cultivate a range of organoids.[26] Each organoid covers different cell types requiring specific physiochemical cues and represents the structure and function of several organ systems.[27] Finding the right technological balance at the intersection of two promising approaches will be benefit to exploit an advanced integration strategy for high-fidelity stem cell-based human organ modeling, also known as collaborative engineering.[25] Strategic integration may address each approach’s limitations and figure out a large number of technical problems to some extent, and bring unprecedented bionic models suitable for various applications.[28] The strategy also increases structural and cellular fidelity varying from closed luminal cell spheroids to multilayers subtissue levels interfaces by recapitulating cell types and ratios of their counterpart in vivo.[29] The integration can enhance the spatial–temporal control of 3D tissue generation and microenvironment via biophysical stimuli, summarize various exogenous or endogenous cues as well as their concentration gradient, and further control the shape and size with built-in vascular system.[30] What is more exciting is achieving time-saving, labor-saving, and higher-throughput of drug screening and diversified applications by adding sensing systems and screenable readouts through biochemical and modular physical analysis as well as optical measurement in the integration models.[31–33]
Organoids or organs-on-chips have been more recently benefiting from the biocompatible BioMEMS (Biological Micro-Electro-Mechanical Systems)[34] and microfluidic chips.[35] The former enables precise control over cellular microenvironments under biological conditions, and the latter perfuses the system constructs by regulating fluid behavior and connections. Both of the mentioned systems have also been extensively adopted and utilized to develop 3D cell culture devices. However, the cumbersome user interfaces and time-consuming molding processes of lithography technology act as barriers to their clinical applications and commercial dissemination.[11] The novel 3D printing, also termed as rapid prototyping, is likely to bring unprecedented convenience and versatility and be adopted to produce microfluidic chips and 3D cell culture devices with precise shape, architecture, and structure of targeting tissues and organs.[36–38] The living systems on chips platform can also be reconstructed by the interaction of 3D printing-based biomimetic microfluidic chips and various microfluidics interface technologies and accessories (e.g., sensors, microvalves, and micropumps).[39] Researchers can acquire digital files of these molecules easily via the Internet, print them quickly by a 3D printer, and assemble the device manually, making it rapid and applicable to be employed in both clinics and research.[40,41] To be specific, 3D bioprinting can lay down biocompatible supporting materials and living cells simultaneously, and introduce the physiological relevant cues (e.g., well-defined cell arrangement) to more effectively simulate cellular diversity and microstructure exhibiting great consistency.[42–44] Various favorable merits are brought up and fully integrated, meanwhile, many drawbacks and difficulties in designing and optimizing culture devices are effectively avoided and complemented by integrating organoids/organs-on-chips and 3D printing.[45] In brief, 3D printing can simplify the fabrication of microfluidic devices and make 3D cell culture platforms cost-effective, time-saving, user-friendly, and less labor-intensive.
Numerical simulations using multiphysics software is a feasible tool that provides the necessary capabilities of modeling coupled fluid flow, mass transport, and biochemical, bioelectrical, and mechanical cues for designing and developing considerable proof-of-concept models.[46] Computational and mathematical modeling can contribute accurate and satisfactory results by simulating data gained from experiments, and avoid repetitive experimental measurements. This powerful tool can greatly reduce cost and time for analyzing, optimizing, and revising the design of 3D culture microfluidic chips, as well as shortening the design pipeline and boosting the development of elaborate 3D cell culture platforms.[47] For example, computational fluid dynamics (CFD) is more appropriate for analyzing flow patterns, pressure drops, wall shear stress profiles, and mechanical loads on microfluidic or membrane-based organ-on-a-chip devices. Multiphysics modeling can be employed as well to verify various speculations of cellular metabolic behaviors and activities (e.g., nutrient consumption, oxygen concentration, and distribution in microfluidic devices). Furthermore, it can imitate the effect of shear stress and flow field on cell migration, alignment, and phenotype, as well as the coupled effect between shear stress and submicrotopography.[48] Computational simulation of complex behavior arising in multicellular constructs, organoids, and multiorgan-on-a-chip or human-on-a-chip can provide critical insights for improving reproducibility or enhancing guidance. Nevertheless, incorporating numerical simulations into organoids or organ-on-a-chip models needs important advances in modeling and additional theoretical work in both simulation and experimental studies.
Organoid and organ-on-a-chip have been largely supported and increased funding from various funding agencies to build a flexible, reliable, affordable, and accessible in vitro microphysiological model to perform drug discovery, toxicity testing, and basic research.[49] In this review, we thoroughly compared their merits and differences between organoid and organ-on-a-chip models in cell sources, structural fidelity, cellular fidelity, and control ability from the application perspective, respectively. Then, we detailed the advantages and benefits by integrating organoid and organ-on-a-chip for enhancement in structural and cellular fidelity, increasing spatial–temporal control of 3D tissue generation and higher-throughput readouts. After-ward, the combinations of two 3D cell culture models with 3D printing or numerical simulation are separately illustrated for exploring their latest achievements, barriers, and future prospective. Furthermore, we provide the challenges and limitations facing organ-on-a-chip and organoids and 3D printing technologies and numerical simulation used for tissue models, and a conclusion with some personal insights to address the technologies which have their advantages, disadvantages, or scope of each model. The relatively elementary recommendations and future solutions are highly conducive to address the issue by integrating of organoid, organ-on-a-chip, 3D printing, and numerical simulation patient-specific disease-on-a-chip and human-on-a-chip.
2. Comparison Between Organ-on-a-Chip and Organoid Models
Organoid and organ-on-a-chip models are highly beneficial. Recognizing their merits and limitations of these approaches in terms of cell origin, structural fidelity, genetic stability, and environmental control capabilities and throughput will provide more insights into both approaches and their applicable environments. Moreover, such comparisons also reveal a future path to pursue in an integration approach integrating organoid, organ-on-a-chip, 3D printing, and numerical simulation toward patient-specific disease-on-a-chip and human-on-a-chip. In the present section, we first introduce the cell source of 3D cell culture which would be informative for comprehension, followed by comparing the above features and diversifications by presenting applications of both approaches in modeling biological processes of organ, cellular development, disease etiology plus drug screening. Subsequently, current technological challenges and future perspectives faced 3D cell culture models are also discussed.
2.1. Cell Source
Considerable cell sources from both human and animal origin are employed in existing organoids and organ-on-a-chip models. They fall into pre-existing well-differentiated cells (e.g., primary cells, immortalized cell lines, and tissue biopsies) and undifferentiated cells (e.g., embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and adult stem cells (ASCs)), each of which has distinct merits and demerits. Animal cell sources differ essentially from human physiological complexity and fail to summarize the inherent genetic mutations and variations of human cell sources for disease models and drug screening.[50] Pre-existing well-differentiated cells refer to well-established sources available extensively, as well as exhibiting genetically homogenous and slight phenotypic mismatches with actual tissues.[51] Undifferentiated cells are capable of differentiating successfully into specific lineages and genomes and encounter hurdles of arrested differentiation and inconclusive disease phenotypes, and potential mutagenesis and carcinogenicity by regulating mutations, chromosomal abnormalities, or epigenetic variations in DNA methylation patterns induced by viruses.[52] Thus, it is one of the critical procedures for both organoid models and organ-on-a-chip systems to correctly choose cell sources in line with research motivations and genetic background.[3] For instance, a liver-kidney-on-a-chip being seeded with Hep RG cells noticeably differs from that lined with HepaG2/C3A cells for drug screening duo to that only the former is capable of metabolizing ifosfamide into toxic chloroacetaldehyde.[53] Baudoin et al. clarified that tumor-derived hepatic cell lines exhibit inferiority to primary human hepatocytes in multiorgan-on-chips to summarize liver metabolism and functionality for predicting drug response application.[54] Furthermore, ESCs or iPSCs derived organoids also noticeably differ from primary tissue-based organoids and limit the presence of mesenchymal cell types and has no branding in the cultivating process when compared to the latter.[55,56]
Organ-on-a-chip systems are primarily dependent of the pre-existing well-differentiated cells.[16,57] The primary cell source refers to a type of mature cell covering a limited natural extracellular matrix and might vary from batch to batch. Given that primary cells derive from a specific patient, the primary cells based chips are capable of reestablishing disease models for individualized treatment and drug screening. Nevertheless, human primary cells are hard to acquire duo to the trauma of extraction and most primary cells adopted in the systems are of animal origin which significantly limit their translational ability and application areas.[58] Nevertheless, the primary cells based chips may also require the presence of cell lines in certain scenarios to regenerate the cell-or-organ level interactions.[59] Cell lines may be the most extensively used cell sources for organ-on-a-chip systems having been employed in research areas (e.g., kidney,[4] lung,[60] gut,[61] heart,[62] liver,[63] brain,[64] and as well as the integration of these organs on body-on-a-chip[65]). Consistent with primary cells, the inherent homogeneity of cell lines or mature cells within slightly natural extracellular matrix microenvironment leads to reproducible results and associated application in disease modeling. Nevertheless, this is at the expense of restricted patient specificity in contrast to other individual-specific cell sources.[3] Furthermore, immortalized cell lines would exhibit slight phenotypic diversifications from actual tissues in several cases (e.g., protein expression and metabolic pathways) and may not be the optimal choice.[66] Another common cell source for organ-on-a-chip is ex vivo tissue (e.g., intestinal and liver slices[67] or endocrine tissues).[68] ex vivo tissue biopsies originate from mature tissues and can more accurately reveal the biological information of living tissues consistent with primary cells.[69] Moreover, the biopsies may exhibit the additional benefit by incorporating some of the natural extracellular matrices and tissue structures and be capable of facilitating the recapitulation of organ tissues in the cultivating process. Nevertheless, current ex vivo tissues from human are not easily available except tumors for ethical issues and potentially cause rapid compromise of functions shortly after extraction, making them unsuitable for long-term culture and research.[70]
Organoid models are generally derived from primary tissue biopsies, or stem cells to summarize the structure and functionality of organ tissues.[71] Most common stem cell types consist of ASCs, ESCs, and iPSCs have the essential ability to be induced into one or clusters of specialized cells and be assembled into intricate structures resembling their counterparts in vivo.[72] ASCs can be derived from various sites of the human body of both juveniles and adults, thereby making them easy to access. ASC-derived organoids typically display a more mature phenotype and have been employed to model various organs. However, ASCs can only differentiate into certain types of cell lineages in accordance with derived-tissues and have various protocols that render heterogeneous phenotypes of organoid from batch to batch, making them less applicable to cell source of organoid in contrast to other stem cells.[73] ESCs originate from the inner cell mass (ICM) of an early-stage embryo named blastocyst. Both ESCs and ICM cells fall to totipotent or pluripotent cells.[74] Note that one type of the earliest entire organoid originates from mESCs (mouse embryonic stem cells) by culturing it under floating conditions within mixed culture media.[75] The most salient merit of the ESCs source is reflected by the unlimited differentiation potential and subsequent high phenotype fidelity. However, since researches associated with ESCs raise ethical difficulty and rigorous regulation, thus, it is extremely difficult nowadays to put this cell source into widespread practical research use.[76] In addition to ethical debates, there are still technical obstacles to guarantee creating various cell lines with genetic diversity, as well as exploring well-defined protocols to directional differentiation, and further hinder the application of ESCs.[77] iPSC (initially proposed in 2007 and awarded with Nobel Prize in 2012) represents the evident advancements in transfection protocols for stem cells and provide an ideal cell source for organoid models.[78] iPSCs can be conveniently generated from cells being harvested from specific tissues under defined factors by induced dedifferentiating process into the particular lineage and genomes.[78] Furthermore, iPSC can be easily harvested from donors with certain diseases and meanwhile without ethical concerns, making them overly applicable to studies on organ-level dysfunctions. The most advanced techniques (e.g., clustered regularly interspaced short palindromic repeats/CRISPR-associated nuclease9, CRISPR/Cas9 genome editing technology) may be employed to provide unique opportunities to scrupulously delve into more human diseases.[79] Few hurdles encountering the applications of iPSCs is also unavoidable to arrest their differentiation and vary indefinite disease’s phenotypes that be attributed to differences between the tissue of origin and genotype.[73] Therefore, future efforts are going forward to develop more reliable protocols for stem cell differentiation and to get rid of mutations, chromosomal abnormalities as well as epigenetic variations in DNA of stem cell sources.
2.2. Structural Fidelity
Self-organizing organoid and engineered organ-on-a-chip facilitate a thorough exploration of lineage specification, cell–cell interaction and organ-or tissue-genesis by adopting different strategies and possessing distinct merits. However, each model can also find its suitable applications concerning target tissues or organs because of their differences in structural fidelity. Organoid models can uniquely summarize important microstructures and functions of in vivo living tissues and provide ideal tools to explore human organs because of their high structural fidelity by rigorously applying timed chemical cues. Jo et al.[80] managed to induce self-organized multicellular 3D midbrain-like organoids by incorporating functional and electrically active midbrain dopaminergic (mDA), as well as specific layers of neuronal cells that could express characteristic human midbrain’s markers from human induced pluripotent stem cell (hiPSC) (Figure 1A). The midbrain organoids self-organized into neuro-melanin like granules whose structure was similar to the developing midbrain in vivo and performed function to produce cardinal midbrain dopaminergic neurons and dopamine after more than 2 months’ cultivation. For example, Vyas et al.[81] managed to employ hepatic organoid with high structural fidelity and self-organized ability to accurately imitate liver organogenesis and congenital diseases. The liver organoid originate from human fetal liver progenitor cells could re-establish parallel hepatobiliary organogenesis, high differentiated hepatocytes and biliary ductal structures after planted inside acellular ECM for cultivation. Moreover, they successfully extended their study and then developed a liver disease model resembling Alagille syndrome by inhibiting NOTCH signaling to interrupt duct morphogenesis. Furthermore, Zambrano et al.[82] induced human embryonic somatic cell (hESC) line AND-1 following formation protocol into typical finger-shaped structure of lung bud organoids for re-establishing a natural sequence of respiratory system differentiation from embryonic to alveolar stages (Figure 1B). They performed the trypsinization of the cells into small clumps covering 3–10 cells and subsequently seed and cultivate them on a low-attachment culture medium for around 25 days. The resultant organoids exhibited branching airway, early alveolar architecture as well as advanced paddle-racquet like structures, revealing that the organoids had reached the alveolar stage of lung differentiation and might become a versatile tool to model surfactant deficiency syndromes.
Figure 1.
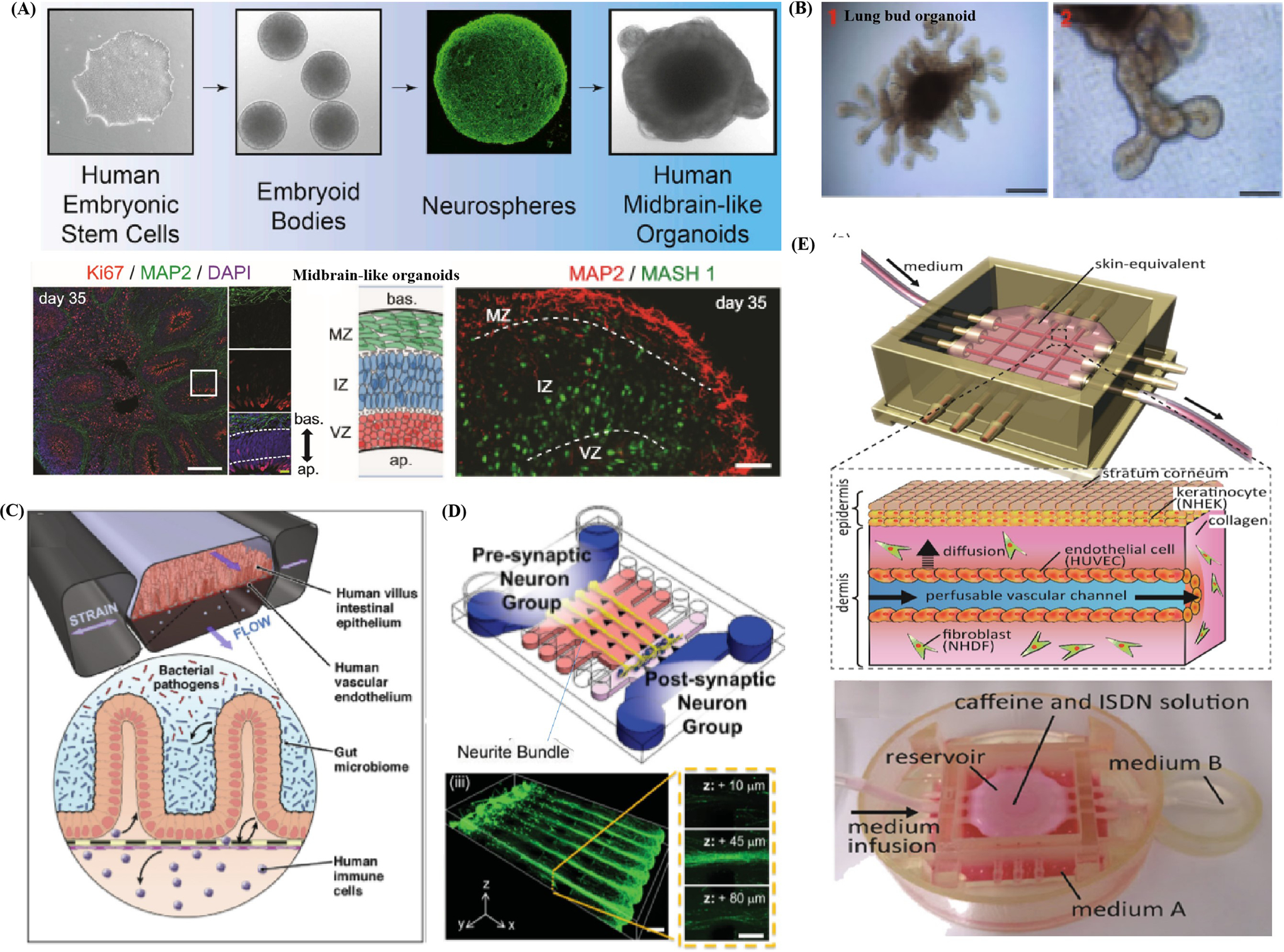
Illustration of structure fidelity of organoid models and organ-on-a-chip systems. A) Schematic demonstrates the human midbrain-like organoids (hMLOs) from hPSCs in 3D culture which contain distinct layers of neuronal cells expressing human midbrain markers. Reproduced with permission.[80] Copyright 2016, Elsevier. B) Illustration of typical finger-shaped extensions of human lung bud organoid. Reproduced with permission.[82] Copyright 2018, Springer Nature. C) Schematic represents a novel gut-on-a-chip system comprised of artificial human villus intestinal epithelium and vacuum chamber for applying cyclic strains. The system is able to mimic complex interaction between Caco-2, vascular epithelial cells, microbiome, bacterial and immune cells. Reproduced with permission.[83] Copyright 2018, Elsevier. D) Schematic shows the neurons that are aligned on the Matrigel and cultivated to form the 3D neural circuit. Reproduced with permission.[84] Copyright 2015, John Wiley & Sons, Inc. E) Schematic exhibits the microstructure of the microfluidic vascular channels embedded in the vascular-skin-equivalent-on-a-chip, which could provide perfusion flow. Reproduced with permission.[91] Copyright 2017, Elsevier.
In contrast to organoid models that demonstrate high structural fidelity for their spontaneous organization ability, engineering-based models can largely contribute to exploit microfabrication methods to imitate the structures of in vivo tissue in a controllable microenvironment. Bein et al.[83] exhibited a novel human intestine organoids-on-a-chip which represents the typical design principle of organ-on-a-chip systems (Figure 1C). Human intestine model have been engineered with increasing complexity that also include neighboring channels lined by microvascular endothelium, immune cells, commensal microbes, pathogenic bacteria, and some permit application of cyclic mechanical forces that mimic peristalsis-like deformations experienced by living intestine in vivo. Bang et al.[84] developed a simplified neural-on-a-chip with aligned 3D neuronal circuit in Matrigel which covered new micropillar arrays (Figure 1D). The neural-on-a-chip imitated the multilayered structure of neuronal circuit, axon fasciculation, as well as neural bundle by culturing primary rat cortical neurons with align ECM components. These researchers also facilitated the patterning of Matrigel cross-linking density distribution during gelation process by controlling under hydrostatic pressure and delivering continuous flow via the chip. Subsequently, they reported the neurite growth rate (an average speed of 250 μm per day), formation of axon bundle with fasciculation, and the evolution of neural network from presynaptic to postsynaptic neurons after seeding on one side of the Matrigel., Ho et al.[63] developed a lobule-mimetic liver-on-a-chip to imitate the liver tissue by covering concentric-stellate-tip microelectrode arrays to pattern hepatic and endothelial cells. They managed to guide, snare, and align the massive cells simultaneously inside the well-defined chamber, resembling the basic morphology of hepatic lobule by dielectrophoresis manipulation that created a delicate spatial electric field. The successful field-induced orientation of randomly scattered cells to desired stellate patterns was identified as revealed from the results of fluorescent assay to faithfully recapitulate lobule microstructure.
Structural fidelity of organ-on-a-chip system is inferior to that of organoid model in most cases and are difficult to recreate the elucidate tissues’ structure duo to current limitations of microfabrication techniques and knowledge concerning microstructure of tissues. It still faced with the lack of some necessary functions even if reproducing several basic architectures of tissues or organs. For instance, one of the cases of liver-on-a-chip cannot summarize the directional biliary ductal clearance or reestablish the secretion of metabolic enzymes, which requires long cultivation time.[43] Organoid models typically lack tissue–tissue interface and vascular compartment, and form enclosed luminal structure,[86] the organ-on-a-chip system can be extremely suitable and informative when modeling basic tissue architecture as barrier,[87,88] thin-film interface[89] as well as tubular structure. For instance, Booth et al.[88] fabricated a microfluidic blood–brain barrier (BBB) restrictive membrane recapitulating the sophisticated biological architecture of its counterpart in vivo. It is capable of imitating shear stress of the dynamic vascular environment, and hinder various exogenous compounds (e.g., amino acids, selected sugar, and electrolytes) in the blood from entering the central nervous system. The artificial multilayer BBB covered two glass substrates for support, four polydimethylsiloxane (PDMS) layers with a porous polycarbonate membrane within them to achieve endothelial cells and astrocytes culture, two channels for flow penetration and embedded electrodes for trans-endothelial electrical resistance (TEER) test. Note that in the experimental process, the tight junctions and high viability of cells in BBB were revealed from the optical imaging results. Besides, the TEER test of BBB expressed a distinct higher level in selective permeability than conventional models, verifying that such system could act as a useful platform for imitating and exploring barrier function.
Tavana et al.[90] created a lung-on-a-chip, recapitulating a thin film structure within a small airway, to delve into the underlying mechanism of surface tension-induced lung diseases. They imposed pulmonary pressures via micropumps and thoroughly observed the metabolic activities of human alveolar epithelial cells with or without Survanta (a type of clinical surfactant). The high mechanical stresses could lead to substantial cellular injury, and adding the surfactant could protect epithelium and down-regulate the death rate of these cells. Ingber et al.[61] fabricated a gut-on-a-chip to reproduce the convoluted architecture and associated barrier function of the intestine in vivo. The chip displayed two microfluidic channels split by a porous flexible thin film being coated with ECM and lined by Caco-2 cells and cultivated under dynamic conditions performing shear stress and cyclic strain. This bio-microfluidic chips are convenient to imitate the thin film structure within a small airway by quickly polarizing cells into a columnar epithelium and developing into intestinal villi-like architecture. The blood circulatory system is critical to the exchange of nutrients and maintaining the homeostasis across the whole body by linking all types of organs. Mori et al.[91] devised a vascular-skin-equivalent-on-a-chip with artificial vascular channels and perfusion systems to imitate the architecture of its living counterpart (Figure 1E). Two ends of microfluidic channels were coated with endothelial cells to test the ability of molecules to penetrate the epidermal layer into vascular systems. Moreover, the researchers fixed peristaltic pumps and silicone tubes to the channels’ ends to simulate the perfusion conditions and supply nutrition to artificial skin. Furthermore, histological analysis suggested the similarity of epidermal and vascular morphology also further verified the suitability of organ-on-a-chip to model vascular systems.
2.3. Genomic Stability
Organoid models also should be highlighted for their genomic stability duo to consist of diversified, genomic stable, self-renewing cells that can differentiate into fully developed mini-organs harboring all main cell types at a similar ratio to its living in vivo counterpart. Lancaster et al.[92] described a recently established protocol for generating 3D brain tissue, i.e. cerebral organoids, which closely mimics the endogenous developmental program give rise to developing cerebral cortex, ventral telencephalon, hippocampus, choroid plexus, and retinal identities within 1–2 months (Figure 2A). This straightforward protocol can easily be implemented in a standard tissue culture room and be applied to developmental studies, as well as to the study of a variety of human brain diseases. Cerebral organoids can also be maintained in long-term culture and be potential to model later events, such as neuronal maturation and survival. Qian et al. illustrated that forebrain organoids covered various cell types induced by two rounds of patterning factors; these brain-region-specific organoids exhibit high reproducibility, display a well-developed six cortical layers for human cortical development, and lead to the remarkable reduction of both tissue and temporal development heterogeneity.[21] Primary information obtained are concerning signaling pathways of solid organs (e.g., liver) and regarding their role in organogenesis remains a pivotal unanswered problem. Alagille syndrome is primarily attributed to mutation of the JAG1 gene and refers to a genetic disorder impairing liver by causing abnormalities in bile ducts. Guan et al. created iPSC-derived liver organoid[93] harbored endothelial cells, hepato-blasts, and mesenchymal cells in similar proportion to living tissue to imitate the elaborate intrahepatic bile ducts. They identified that C829X mutation leads to profound alteration, rather than G274D mutation of JAG1 for the ascertainment of these genetic disease-induced abnormalities. Furthermore, they could acquire valuable information concerned with the role of these critical signaling pathways and ascertain the role of JAG1 in epithelial cells assisted by the high cellular similarity.[93]
Figure 2.
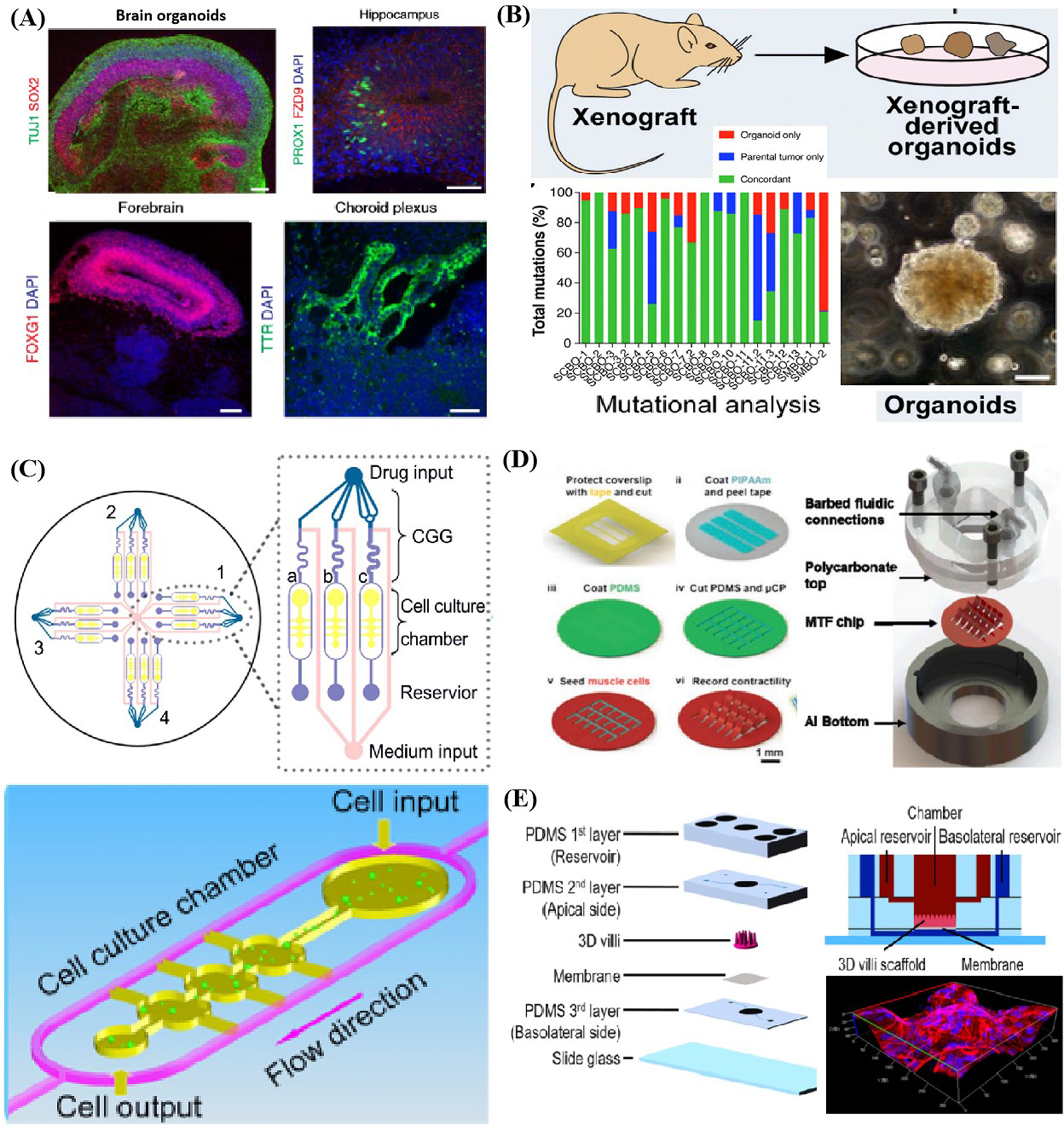
Examples of the genomic stability of organoid models, and displays of the environmental control ability and throughput of organ-a-chip systems. A) Established protocol for generating cerebral organoids which closely mimics the endogenous developmental program and can give rise to developing cerebral cortex, ventral telencephalon, choroid plexus and retinal identities. Reproduced with permission.[92] Copyright 2018, Springer Nature. B) Schematic represents the gene mutation rates identified in organoids compared with parental tumor and demonstrates gene fidelity during organoid cultivation. Reproduced with permission.[97] Copyright 2018, Elsevier. C) Microfluidic cell culture device comprised of four uniform units to create a series of concentration gradient for drug test. Reproduced with permission.[110] Copyright 2013, Elsevier. D) Illustration of microfluidic devices integrated with MTF chip for cyclic stretch application, barbed fitting for perfusion control, aluminum bottom for temperature maintenance and stimulator for electrical field loading. Reproduced with permission.[112] Copyright 2013, Royal Society of Chemistry. E) Schematic exhibited a gut-on-a-chip which fabricated 3D villi scaffold for cell arrangement and culture in dynamic environment. Reproduced with permission.[115] Copyright 2017, Springer Nature.
Research work published in Nature series demonstrated that the extent of the resultant genetic and transcriptional heterogeneity and its functional consequences of genetic variation within cell lines, and provides a framework for researchers to measure such variation in efforts to support maximally reproducible cancer research. Current cell lines used in 2D cell culture and organ-on-a-chip systems might also face the same problems in long-term cultivation, which makes somewhat untrustworthy and limits their applications in disease modeling and drug screening.[94–96] In contrast, the high genomic stability of organoid models also reflects in a conserved genomic landscape resembling their parental cells, thereby making organoid a feasible and expandable material source in search of genotype–phenotype relationship, drug response as well as functionality of certain tissue (e.g., tumor).[53] Lee et al.[97] fabricated and exhibited mutation rates of derived bladder cancer organoids originating from 22 patients compared with parental tumors (Figure 2B). The organoids maintained the heterogeneity of corresponding parental tumors, and kept histopathological and molecular diversity, and patterned a range of genomic variations resembling tumor evolution during cultivation. They could also gain insights into the partial relationship between the mutational profiles and verified certain responses when applying xenografts in vivo by phylogenetic analysis based on this useful and efficient organoids platform. Sylvia et al. employed pancreatic organoids derived normal and neoplastic tissues, which recapitulated the ductal and disease state features, to dissect the role of gene Kras in pancreatic neoplasia.[98] They managed to simulate the physiological spectrum of carcinomas generation from early-stage neoplasms to lesions by orthotopically transplanting the organoids. Furthermore, they reminisced of pancreatic intraepithelial neoplasms to invasive, metastatic tumor. They have identified the genes that drive the adenocarcinoma pathogenesis and critical molecular signaling pathways through inducing mutation of certain cancer genes followed by thoroughly proteomic profiles and transcriptional analyzing.
Additionally, the organoid regenerative methods can serve as an innovative tool to rescue those affected by specific mono-genic diseases by homologous engraftment of organoid to the right site after mutation correction. The most representative case is cystic fibrosis caused primarily by mutations in cystic fibrosis (CF) transmembrane conductance regulator (CFTR) gene. CFTR gene has been ascertained the functionality of an anion channel and is critical to electrolyte and fluid homeostasis of epithelium. Despite that various assays have been developed to assess the functionality of CFTR and explore efficient new drugs, existing reagents have poor performance to all for the mutation variability in different patients.[99] Ogawa et al.[100] fabricated bile organoid covered epithelialized cystic as well as ductal structures which could exhibit mature biliary markers from iPSC. They cocultured the organoid with OP9 mouse bone marrow stromal cells at the hepatoblast stage to achieve NOTCH signaling by performing serum-free protocol. Note that after CFTR modulators were added to stabilize protein and hinder misfolding, cysts originating from CF patients’ iPSC were ascertained to be recovered by forskolin-swelling tests. This verified the ability of organoid in regenerative correction of certain diseases or being engrafted in the right site in clinical perspective and even verified using murine test.[101] Moreover, Schwank et al.[102] developed intestinal organoids originating and expanded from both primary large(LI) and small(SI) intestinal stem cells to cure the identical disease. The cells originated from two homozygous CF patients having undergone common mutation at F508. They employed a range of sgRNAs with the plasmid, which encodes wild-type CFTR, to correct CFTR sequence based on CRISPR/Cas9 technology. Moreover, they introduced a silent mutation downstream the correction for polymerase chain reaction (PCR) testing and proved the successful repair of F508 and site-specific knock-in events in most cases. Similar examples cover the rectification of Kras gene mutation by Sylvia et al.,[55] correction of dyskerin gene (DKC1) in congenital dyskeratosis by Cas9-mediated intestinal organoid by Woo et al.,[103] reversion of certain functional loss attributed to RPGR gene mutation in retinitis pigmentosa by Deng et al.[104] In brief, researchers can construct matched diseased and healthy organoids with high cellular fidelity compositionally and genetically, and make them a regenerative clinical therapy and a feasible tool to access the disease progression, assess drug resistance, and toxicity.[94,97,99]
Indeed, engineering-based organ-on-a-chip systems are essential inferior to organoid models in physiological complexity, cellular diversity, and cellular genetic similarity for their over-simplified cellular composition and limited cell types.[4,16] However, it does not indicate that organ-on-a-chip models remain inferior to organoid in terms of applications as underlying biological exploration for organs, disease generation, as well as drug screening. Since organoid models rely on poorly defined Matrigel and essential organization ability of stem cells, which could lead to great variability in size, shape, and viability and hard dissection in the analysis of certain factors or signaling pathways.[99] Furthermore, deficiency of stromal components (e.g., immune cells) also hinders the use of organoids in modeling diseases or drug toxicity characterized by inflammatory responses.[38]
In contrast, organ-on-a-chip models can draw upon well-defined biochemical and physical modulators to determine the cell fate in regulated cell niches, thereby rendering simplicity to model monocue or several factors influences. For the features of organ-on-a-chip mentioned above, Sung et al.[105] developed a simple breast cancer-on-a-chip to summarize the transition of carcinoma, being critical to the breast cancer progression. The proposed breast cancer-on-a-chip, covering a compartmentalized culture system, could simulate the transition from ductal carcinoma in situ to invasive ductal carcinoma. They could conduct cell–cell signaling studies of a single cell without interference (human mammary fibroblasts) by coculture of epithelial cells with fibroblasts and exertion of spatial–temporal control. Furthermore, they managed to cast light on the fact that soluble factors just begin the transition, whereas only direct contact could lead to the transition of carcinoma. Their subsequent study identified the effects of specific critical ECM components during breast cancer progression with seven different combinations of three ECM proteins and by characterizing the proliferation and morphology of T47D clusters.[106] Moreover, the capability of organ-on-chip models to decipher the complex factors in microenvironment have been identified by numerous correlations of biochemical or physiological factors with biological process, including estrogen receptor protein with the proliferation of breast cancer cells,[107] transforming growth factor-β inhibitor with lung adenocarcinoma cell,[108] endothelin-1 and rho-associated kinase (ROCK) inhibitors with cardiomyocytes thin film,[109] etc.
2.4. Environmental Control Ability and Throughput
Organ-on-a-chip systems can control the delivery of different biochemicals or compounds and provide delicate spatial–temporal control over cell culture through manipulating input and output flow conditions (e.g., flow rates and associated shear stress) with syringe or micropump. Moreover, these microfluidic systems can also introduce a range of stimuli (e.g., concentration gradient,[110,111] electromechanical force,[112] mechanical force,[113,114] and shear stress[115]) in a high-throughput manner to simply combined with monitoring systems.
Organ-on-a-chip systems can manipulate the fluid flow spatial–temporally to re-establish the microniches for cell culture, which is one of the most prominent differences between organoid models and organ-on-a-chip systems. For instance, one drawback of tumor organoid is typically lacking vasculature, perfusion system, and many invasive cell types. Sobrino et al.[116] developed a vascularized organ-on-a-chip system with its unique ability to supply perfusion flow and was employed to imitate human microvessels and cultivate tumor cells to develop vascularized microtumors. The enormous benefits of controlled perfusion brought by microfluidic devices have been verified by tracking protein expression and status, and identified through considerable metabolic heterogeneity and correlation between tumor and vasculature, as well as their response to anticancer drugs.
The simple manipulation of reagent dosage by organ-on-a-chip systems makes them trustworthy and feasible tools to take efficient drugs, identify combination schemes and ascertain appropriate dosage according to toxicities and side effects. Xu et al.[110] developed a lung cancer-on-a-chip covering four microfluidic chips, each of which covered three culture chamber and a concentration gradient generator (Figure 2C). By regulating the concentration at drug inlet, reagents types as well as channel width, they could also produce a range of combinations and concentrations of drugs. Based on this efficient and high-throughput chip, they could provide a clear individualized prescription for eight patients in the meantime with a single or multidrug combination of appropriate dosage. Sara et al.[111] created spatial gradients of cellular composition and matrix in the hydrogel with U87 cells seeded in it. They could recreate the heterogeneous microenvironment of glioma and explore how gradations of biochemical or biophysical cues impact the malignant phenotype and treatment via the platform. In this study, a range of injecting precursors with spatial-gradated and well-defined chemical composition were produced using microfluidic mixing methods first, followed by patterning them into optically translucent hydrogel with certain structures. They could explicitly trace the effects of gradations in microenvironment by simultaneously performing polymerase chain reaction, enzyme-linked immunosorbent assay and metabolic activity assay of glioblastoma multiforme, thereby illustrating the great spatial control ability of the organ-on-a-chip model.
Furthermore, organ-on-a-chip systems can easily incorporate many microenvironmental stimuli (e.g., electromechanical force, mechanical force, and shear stress) for the essential convenience of integration with various engineering methods when comparison with organoid models. For instance, Agarwal et al.[112] advanced a high throughput heart-on-a-chip with orientated rat cardiomyocytes attached on thin films embedded in it, which could be employed to clinically alleviate translational barriers in the analysis of inotropic effects of β-adrenergic (Figure 2D). To be specific, they fabricated a submillimeter scaled PDMS cantilever and cultured the anisotropic rat cardiomyocytes cells and tissues on the substrate to imitate the laminar structure of the heart ventricle. Moreover, a metallic base was covered to stabilize the temperature. They also regulated the wash-in and wash-out fluid flow of drug by syringe pumps and exerted electrical field to induce contractile stresses. Likewise, Wang et al.[113] employed the identical heart-on-a-chip with patient-derived iPSC lined on the thin elastomer films to elucidate the efficacy of TAZ modRNA on Barth syndrome. They also employed fibronectin micropatterns to replicate the contractile pathophysiology in vitro under biophysical cues (e.g., mechanical force) to cultivate and manipulate the iPSC self-developed into laminar myocardium with immature phenotypes (e.g., aligned sarcomeres and metabolic abnormalities like in vivo model). Furthermore, Parker et al.[114] devised a stretchable heart-on-a-chip which could reestablish mechanical overload and failing myocardium of diseased heart in vitro. They employed cyclic stretch on engineered laminar ventricular tissue on a stretchable chip and ascertained the differences among animal models, clinical records and data harvested on-chip in various aspects (e.g., gene expression, myocyte architecture, calcium handling, as well as contractile function). The quantitative results demonstrated that the cyclic stretch could exert great influence on the gene expression profiles, myocyte shape as well as sarcomere alignment. Organ-on-a-chip could faithfully imitate the diseased heart in vitro on the basis of inference by replicating structural and mechanical cues. Shim et al.[115] created a gut-on-a-chip that re-established two features of its counterpart in vivo, the 3D villi structure and shear stress, which are conducive to the differentiation and phenotype fidelity of cell culture (Figure 2E). Subsequently, they designed a delicate collagen scaffold capable of recapitulating the structure of human intestinal villi, and combined it into the microfluidic chip to introduce shear stress by fluid perfusion.[116] They compared the morphology, representative enzymes’ activity and epithelium absorptive permeability of Caco-2 cells in three culture conditions, such as 2D culture on transwells, 2D culture on microfluidic chip and 3D culture on microfluidic chip, respectively. The results enabled them to identify noticeable enhancement in the Caco-2 cells’ metabolic activity and demonstrated the collective influence of 3D architecture and perfusion condition.
Besides spatial control ability, engineering-based organ-on-a-chip systems can also exert temporally control over culture processes. For example, Gretchen et al.[117] developed a liver-on-a-chip to prove the ADMET (e.g., absorption, distribution, metabolism, excretion, and toxicity) properties of acetaminophen by combining intestinal cells’ chambers and monolayers lined with HepG2/C3A cells. The device helping ascertain the dose-dependent hepatotoxicity induced by metabolic processes and glutathione depletion attributed to acetaminophen could serve as a good example to certify the temporal control ability of organ-on-a-chip systems. Researchers could regulate the residence time of drugs in the chamber to approximate to the identical durations that the drugs particularly stay in vivo, and study the potential toxicity of oral drugs or other chemicals that aided by microfluidic technology.
High throughput is another characteristic of organ-on-a-chip systems and conducive to clinical application which needing strict time requirement compared with organoid models. Multiwells organ-on-a-chip is able to simultaneously perform numerous experiments on the same chip promoted by engineering-based principles. For instance, Grosberg et al.[109] designed a muscle-on-a-chip covering 24-well plate to analyze reactions of muscular thin films (MTF) with ROCK inhibitor and endothelin-1 at different concentrations in the meantime. The MTFs seeded with muscular cells are patterned with ECM by microcontact printing and then placed in each well of the same chip. Subsequently, they tracked the projection and calculated the stress of MTF after treatment with reagents. The results certified the high throughput trait of multiwells organs-on-chips by adding high concentration of endothelin-1 to enhance contractility of MTFs while ROCK inhibited it. Wevers et al.[118] designed a high throughput 3D microfluidic platform to culture neuronal-glial networks, which is termed as OrganoPlate covering 384-well plate microtiter for 3D cell culture and coverslip-thickness glass for optical evaluation. Each independent culture unit of OrganoPlate contained four nearby wells, with two for providing medium and the other for cell/ECM mixture and monitoring, could be applied in the recapitulation of a miniaturized organ or tissue. In subsequent tests, the researchers implanted neurons and astrocytes into the chip and then subjected to immunofluorescence staining to classify the cells that came from a large number of cell sources. The responses of cells at different stages to various compound treatments with a range of concentrations are simultaneously ascertained by electrophysiological analysis.
3. Integration with Emerging Technologies
As previously discussed, neither organ-on-a-chip systems nor organoid models are perfect in the face of diverse and sophisticated studies or clinical purposes. Organ-on-a-chip systems may be inferior to organoid models in structural or cellular fidelity as they are sometimes not capable of replicating solid tissues or organs with elaborate microstructures as they rely on microfabrication methods and predifferentiated cell sources.[3,66] While organoids can narrow the gap between existing in vivo models and in vitro models by cultivating and manipulating cell sources long enough to self-organize and imitate various critical traits of target tissues, they still lack critical cell types, stromal components, and can recapitulate only the early phase of organogenesis.[81,85] Organoid models also lack tissue–tissue interfaces and generally form closed luminal structures which might entrap cells and render difficulties in analyzing luminal contents. Moreover, organoid models show variability in size and shape from batch-to-batch due to the poor definition of Matrigel components and the absence of microenvironmental control, making high-throughput analysis difficult.[83] In contrast, organ-on-a-chip systems are capable of providing cells with a consistent microenvironment and exerting great spatial–temporal control over 3D cell culture by incorporating biophysical or biochemical cues, spatial organization, well-defined cellular components, specific physiological functions, and cell–cell and cell–matrix interactions. Therefore, a strategic integration between these two 3D cell culture models may overcome the limitations of each approach and establish unprecedented and amenable biomimetic models for various applications.[83] Nevertheless, these 3D cell culture models are subject to numerous technical problems that hinder their utilization, propagation, and combination. For instance, the current fabrication methods—derived from conventional microelectro-mechanical systems (MEMS) manufacturing—are labor-intensive and require specific knowledge concerning micro- or nanomanufacturing, making it expensive, time-consuming, and difficult for researchers to create their own devices.[43,77] In addition, most 3D culture devices cannot faithfully reproduce the real 3D structure, reducing the merit of such platforms in providing topological clues for cell development due to the limitations of conventional manufacturing methods. These barriers can be overcome by integrating 3D printing, an emerging tool with convenient manufacturing processes that allows for the building of complex 3D architecture.[76] The combination of 3D printing technology and 3D cell culture technology can also enhance the integrity of biological microfluidic platforms, making it more user-friendly and easier to operate. Moreover, numerical simulation can be incorporated into the 3D cell culture procedures (e.g., device design and read-out), potentially enhancing the practicability and translational ability of biomicrofluidic chips. In Sections 3.1–3.3, the advantages of integrating organoid models, organ-on-a-chip systems, 3D printing, and numerical simulation are discussed. Furthermore, current limitations and prospects are illustrated to better clarify the roles of these novel technologies in biological research and clinical applications.
3.1. Integration Between Organoids and Organ-on-a-Chip
Organoids rely heavily on spontaneous self-assembly for the generation of a precisely organized tissue structure.[119] The formation process varies for each tissue type but generally follows the pattern of proliferation, differentiation, cell sorting, lineage commitment, and morphogenesis.[120] As 3D organoids increase in size and volume, growing cells in the organoid core become distant from the surface in contact with fresh medium; subsequently, simple diffusion becomes insufficient for providing oxygen and nutrients and limits waste removal, ultimately resulting in internal cell necrosis, which hinders the maximum size and extent of tissue maturation of the organoids.[121] There is also limited control over the size, shape, and relative arrangement of different cell types within 3D organoids, limiting their applications in reproducible quantitative studies, which are required for robust drug screening and testing. On the other hand, microfluidic organ-on-a-chip is an artificial bioengineering system composed of arranged cells that recreate the structural and functional features of human tissue/organ physiology, it has the advantage of a controlled environment that provides controlled fluid flow, cell–cell interaction, matrix properties, and biochemical and biomechanical cues.[122] Exposure of cells to physiological shear flow, mechanical stress, and substrate stiffness can have profound effects on cell and tissue physiology.[123,124] Moreover, sensors and actuators can be integrated into the microfluidic devices to enable precise monitoring and control.[125,126] Organoids or organ-on-a-chip alone have limited capacity to meet the broad range of needs that arise in the drug discovery process. The similarities of organoids to actual organs make them more attractive for target identification and validation early in the pipeline, whereas organ-on-a-chip as more reproducible and controllable engineered constructs are better suited for efficacy and safety screening.
Advances in microfluidic organ-on-a-chip approaches have allowed us to engineer organoids with essential structural and physiological features in a controlled manner and obtain microscale structures and parameters that approximate conditions in vivo.[127] By combining the strength of the two technologies, microfluidic organoid-on-a-chip can facilitate better nutrient and gas exchange to prevent cell death in the organoid core and recapitulate 3D tissue architecture and physiology.[128] This combination may also provide more versatile and predictive preclinical models that are broadly applicable to conventional and emerging drug discovery processes.[129] Notably, recent studies have demonstrated the proof-of-concept of engineering a perfused organoid-on-a-chip system by combining a 3D matrix, mechanical fluid flow, and in situ self-organization of multiple organoid types (e.g., brain, intestines, liver, pancreas, and lung) at a millimeter scale.[130] Organoid-on-a-chip may also enable the development of personalized disease models using patient-derived tissue specimens or by reprogramming iPSCs from skin cells as organoids.[131] Indeed, microengineered tumor organoid systems grown directly from patient biopsies may resolve some of the issues that often occur with unpredictable growth patterns and substantial heterogeneity.[132] The convergence of the two approaches to produce multiorganoid-on-a-chip or human organoid-on-a-chip is emerging as a new approach for building 3D models with higher physiological relevance.[133] Furthermore, the integration of chemically defined hydrogels with human organoid-on-a-chip may lead to the next generation of 3D models that show precise spatiotemporal control of niche factors. Additional bioengineering approaches, such as single-cell genomics, live imaging, and genome editing, may also be incorporated into organoid-on-a-chip systems to study human physiology, diseases, and organogenesis and achieve personalized medicine.
3.1.1. Enhancing Structural and Cellular Fidelity
Epithelial organoid models typically form closed luminal cell spheroids and lack tissue-tissue interfaces while organ-on-a-chip systems usually exhibit inferior structural fidelity at subtissue levels.[31] One recent advance in cerebral organoid technology was the adoption of a miniaturized multiwell spinning bioreactor (Spin Ω) as a cost-effective, simple-to-use system to facilitate nutrient and oxygen absorption, which enables formation of longer neuroepithelium-like zones and supports growth of large, complex organoids (Figure 3A).[21] This Spin Ω is fitting a standard 12-well tissue culture plate that dramatically reducing the required media volume and allowing to optimize protocols to generate forebrain organoids from human iPSCs with minimized heterogeneity and variability that enables quantitative analyses and better recapitulation of the developing human cortex. Above the cover of Spin Ω, spinning shafts are attached to a set of 13 interconnecting gears and driven by a single electric motor. They used computer-aided design software to design and 3D print each component. They also developed protocols for midbrain and hypothalamic organoids and employed the forebrain organoid platform to model Zika virus (ZIKV) exposure. These organoids recapitulate key features of human cortical development, including progenitor zone organization, neurogenesis, gene expression, and, notably, a distinct human-specific outer radial glia cell layer. This brain-region-specific organoids and Spin Ω provide an accessible and versatile platform for modeling human brain development and disease and for compound or drugs testing.
Figure 3.
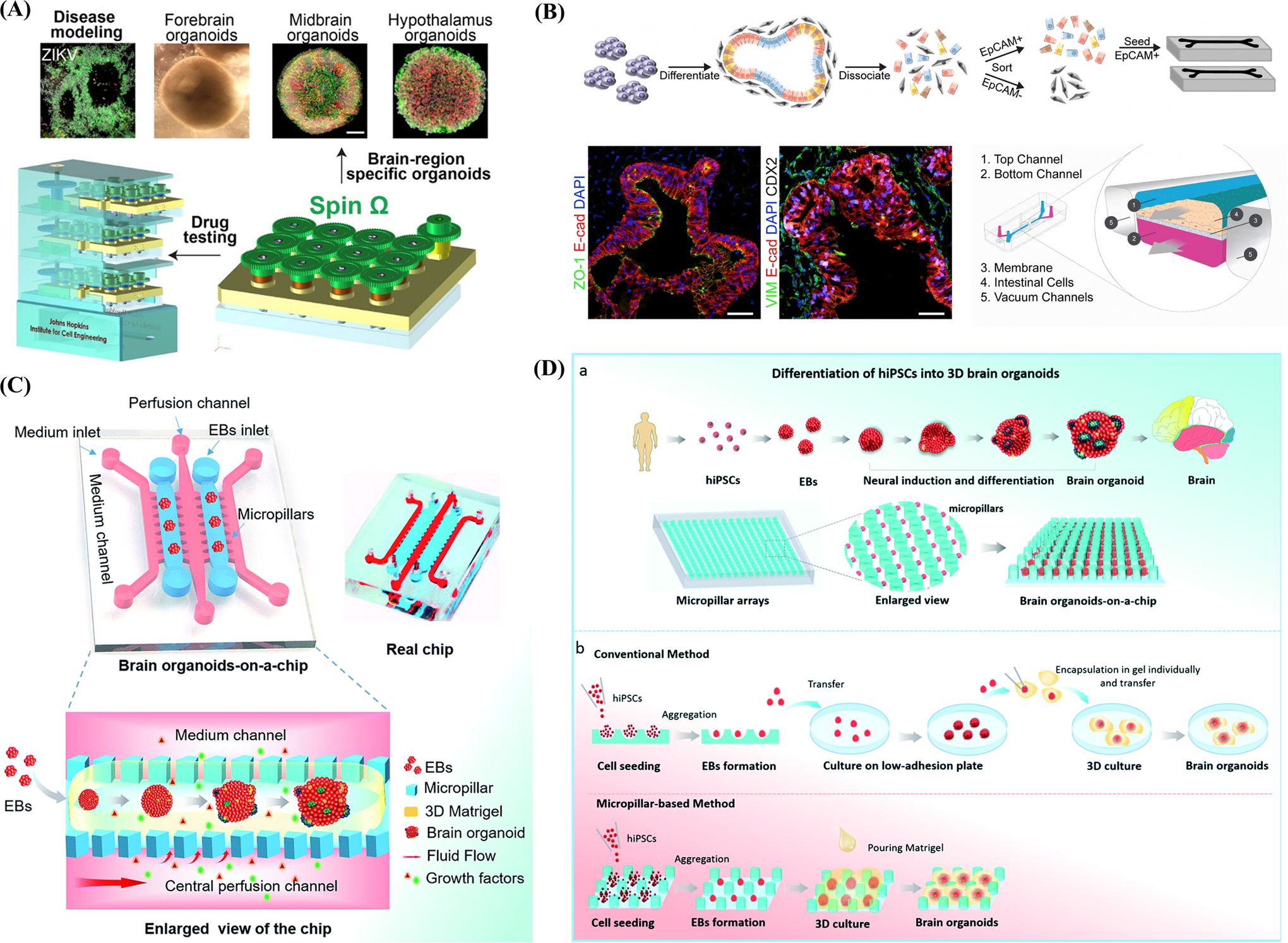
Schematic illustration of the benefits brought by the integration between organ-on-a-chip systems and organoid models and elucidates the concept of synergistic engineering among 3D cell culture, 3D printing and numerical simulation. A) Miniaturized spinning bioreactor (Spin Ω)-based brain-region-specific (forebrain, midbrain, and hypothalamic) organoids from human iPSCs culture system and modeling impact of Zika virus (ZIKV) exposure. Reproduced with permission.[21] Copyright 2016, Elsevier. B) The upper work-flow demonstrates the procedures of seeding epithelial cells derived from iPSC-based intestinal organoids into intestinal-on-a-chip. The lower immunofluorescence images represent the confluent monolayers of cells across the whole channel. Reproduced with permission.[135] Copyright 2018, Elsevier. C) Illustration of a biomimetic microfluidic chip comprised of micropillar arrays which allows direct formation from cell aggregates to brain organoids without much labor-intensive operation and reduces cultivation time. Reproduced with permission.[141] Copyright 2017, Royal Society of Chemistry. D) Schematic represents the brain organoid-on-a-chip devices which incorporated fluid flow in the EBs culture. Reproduced with permission.[147] Copyright 2017, Royal Society of Chemistry.
Furthermore, as each model has its suitable target organs or tissues, it is possible to combine organoid and organ-on-a-chip systems to overcome the disadvantages of each model. Kasendra et al.[85] combined intestine-on-a-chip and organoids to create a useful research tool that can emulate intestinal villus structure and functionality. The authors obtained epithelial cells from healthy intestinal biopsies and expanded them into intestinal organoids through culturing. Then, they dissociated and seeded these organoids on a porous membrane embedded in a microfluidic chip. In addition, the human intestinal microvascular endothelium was cultivated in another parallel microchannel within the same chip and exposed to perfusion flow and cyclic deformation. In this bio-microfluidic chip, the polarized epithelial cells differentiated into organoids and lined up to form villi-like projections. One side of intestinal organoids interacted with the endothelium, while the other side was exposed to an open lumen. Remarkably, transcriptomic analysis showed that the structural fidelity of this hybrid model in mimicking the human duodenum was greater than that of either organ-on-a-chip or organoid models alone. Another example that illustrates the utility of the hybrid model is the brain organoid-on-a-chip.
To test the early stage neurodevelopmental disorders caused by nicotine exposure, Wang et al.[134] integrated the organoid and organ-on-a-chip systems to fabricate a brain organoid-on-a-chip platform. Under controlled continuous perfusions, embryonic bodies could differentiate into brain organoids that re-established key brain-specific features, such as regional and cortical organization. Immunohistochemical and PCR analysis identified abnormal neuronal differentiation with enhanced TUJ1 expression and migration under nicotine exposure. Moreover, Lancaster et al.[92] successfully integrated organoid and organ-on-a-chip models to increase the surface area-to-volume ratio of artificial human forebrain tissue and maintain self-assembly during cultivation. Typically, organoid development requires nonadhesion cultivation conditions and cell–cell interactions, which are hard to attain in a microfluidic chip. By employing Poly(lactic-co-glycolic acid) fibers as floating scaffolds, the authors overcame the incompatibility between organ-on-a-chip systems and organoid protocols. Other than improved maturity of the neuroectoderm, this newly developed platform could also recapitulate characteristic cortical tissue, such as cortical plate and radial units, thanks to the basal membrane included in the chip. These studies prove that great advantages can be achieved by integrating different 3D culture models.
As far as genomic stability is concerned, organ-on-a-chip systems cannot fully recapitulate cell types and their ratios in vivo and may introduce unexpected mutations during cultivation. Organoid models also have a limited abundance of cell types, such as immune cells and stromal components, which may restrict their application. However, the combination of these models may, to some extent, overcome this drawback. Workman et al.[135] incorporated intestinal organoids derived from human iPSCs with a gut-on-a-chip to explore the normal and pathophysiologic reactions of the intestinal epithelium (Figure 3B). This platform can form polarized intestinal folds containing all intestinal epithelial subtypes and is biologically responsive to exogenous stimuli under a wide range of gastro-intestinal conditions. Flow cytometry was also employed for cell sorting to address the inhibition effect of mesenchymal cells on epithelial monolayer expansion, improving the efficiency of producing monolayers.
3.1.2. Increasing Spatial–Temporal Control of 3D Tissue Generation
Despite the fact that the organoid generation process (self-assembly, self-pattering, and self-morphogenesis[136]) depends heavily on the self-organizing ability and default robustness of stem cells, the recapitulation of various exogenous or endogenous cues, including the concentration gradient of biochemical cues,[137] growth factors,[138] and topological,[139] are extremely important in coaxing organized tissues in vitro. As mentioned in Section 3.1.1, engineering-based organ-on-a-chip systems can simultaneously incorporate a great number of temporal (resident time of specific factors or cultivation duration) and spatial controls.[137,140] Thus, the strategic integration of these two methods can compensate for the disadvantages of organoids, such as extremely long cultivation periods or undesirable scalability, and maximize the advantages of organ-on-a-chip by introducing biophysical stimuli as mechanical forces, electric or magnetic fields, and shear stress.[141]
Park et al.[140] incorporated a neuro-organoid induced from rat neural progenitor cells into a microfluidic chip that contains 50 cylindrical microwells to explore the effects of flow on neuro-organoids. Under slow cerebrospinal interstitial flow exerted by an osmotic micropump on the microwell arrays, the neuro-organoids exhibited more robust and elaborate neural architecture than those under a static culture environment. Furthermore, the authors extended their study to model Alzheimer’s disease (AD) by including amyloid-β, a major contributor to AD. They found that cell viability under dynamic conditions was lower than that of traditional devices, demonstrating the ability of the integrated chip to achieve more physiologically relevant results. Similarly, Wang et al.[141] employed an organ-on-a-chip comprised of 3D Matrigel to provide hiPSC-derived 3D brain organoids with multicellular tissue structure and fluid flow conditions (Figure 3C). The brain organoid under a perfused cultivation environment exhibited excellent recapitulation of key features of the development of human brain-like cortical architecture and neural circuit generation. Notably, organoids cultured in such a controlled manner displayed an improvement in expressing cortical layer makers. Taking these two cases into consideration, it is safe to conclude that controlled biophysical stimuli provided by organ-on-a-chip systems can pave the way for better recreation of living tissues by organoid models in vitro. Since organoid models rely heavily on spontaneous differentiation, it is difficult to envisage and control their shape and size in a favorable manner for experimental purposes or therapeutic applications. Moreover, without defined vascular systems to carry the blood and transport nutrients and waste, the mortality of cells inside the organoid models is extremely high, limiting the scale of the obtained organoid. To address this problem, Sakaguchi et al.[142] integrated thick tissues with built-in channels that function as surrogates of the vascular system. By seeding and lining endothelial cells within the channels as well as controlling the perfusion flow, cell-sustaining substances can reach the inner parts of the tissue. Similarly, Zhang et al.[143] proposed a hybrid strategy incorporating organ-on-a-chip, organoids, and 3D bioprinting to fabricate cardiovascular-organoid-on-a-chip with an endothelialized perfusion system. Through encapsulation and bioprinting directly into lattices, the endothelial cells can migrate to form the confluent endothelium. Furthermore, seeded cardiomyocytes can align to generate the myocardium with controlled anisotropy and are able to contract spontaneously and synchronously. Finally, the cardiovascular-organoid is embedded in a microfluidic bioreactor. Testing cardiovascular toxicity exhibited an impressive ability of this synthesized platform to recreate the human myocardium in vitro and screen pharmaceutical compounds.
3.1.3. Improving High-Throughput Readouts
Organoid models, which possess high structural and functional fidelity, usually lack a screenable readout, whereas organ-on-a-chip systems are convenient for integrating complicated sensing systems and are conducive for various analytical methods. Consequently, the integration of the two methods may improve high-throughput platforms for the exploration of basic biological principles or performing drug and toxicology measurements in the pharmaceutical industry.
Quadrato et al.[144] cultivated human whole-brain organoids over an extended period of time and then isolated cells from these organoids to investigate complicated cellular interactions. Notably, the authors stimulated photosensitive cells to control neuronal activity, and introduced a high-density microelectrode sensing system to analyze gene profiles of over 80 000 cells derived from 31 brain organoids. Facilitated by a high-throughput integrated platform, they managed to finish the largest-to-date molecular map of cell types and provided insights into the functions of neuronal circuits. Their study illustrates the high-throughput advantage achieved by integrating the two models. Zhang et al.[145] also integrated an automated, noninvasive, in situ, continual monitoring system with biomicrofluidic chips to assess various environmental parameters and biological responses over a long cultivation period. The platform enabled biochemical and modular physical analysis as well as optical measurement, and enhanced the throughput of drug screening. Furthermore, Devarasetty et al.[146] inserted a simple sensing system into a microfluidic chip. This integrated platform was used to capture the physiological activity of beating cardiac organoids and improve drug screening throughput. Additionally, the system was capable of translating captured files and creating beat kinetic plots for cardiac organoids. Using this system, the authors could distinguish differences in cardiac organoid beat rates under treatment with different agents, such as heart rate stimulating or decreasing agents, and at various concentrations. They were also able to verify the efficacy of certain drugs in a high-throughput manner.
Current protocols for inducing organoid generation are time-consuming, typically requiring several weeks or even months for stem cells and other cell sources to expand, differentiate, and form an assembly.[16,147] Research conducted in laboratories can bear long procedures; however it is not practical for clinical applications, such as elucidating personalized pathology or testing drug responses for specific patients. By exploiting the precise spatiotemporal control ability of organ-on-a-chip systems, we can shorten the generation time of organoids, leading to a more promising future of patient-specific organoid-on-a-chip technology.[16] Zhu et al.[147] combined organoids with an organ-on-a-chip system that contains micropillar arrays to form a novel platform for brain organoid production (Figure 3D). Under the control of well-designed micropillar microstructures in the microfluidic chip, embryoid bodies were able to differentiate into brain organoids directly and rapidly expand neuroepithelial cells. The authors were thus able to remarkably reduce labor-intensive procedures and form massive brain organoids within a relatively short period of time.
3.2. Integration with 3D Printing
Additive manufacturing, also termed 3D printing or rapid prototyping, is a layer-by-layer manufacturing technique that is widely utilized for its ability to rapidly fabricate versatile, customized objects using various materials.[148,149] A commercial bioprinter should include three essential elements, such as a robotic motion system, bioink dispensers, and computer-based software-enabled operational control to print bioink with satisfactory resolution. The design of computer-aided design (CAD) software-enabled blueprint to control mechanical motion trajectory of a robotic system as the preprocessing step, the movement of the motion system in x-, y-, and z-axes, and the dispensing system controls the accurate deposition of the print ink as the processing step. Finally, bioink is deposited, solidified, and stacked layer-by-layer in the 3D bioprinter as the postprocessing step (Figure 4A).[150] There has been a recent advancement in materials and the accuracy of 3D bioprinting, attracting an increasing number of researchers that have employed this promising fabrication technology in 3D cell culture.[151] One of the most exciting benefits of 3D printing is that printed objects can be of almost any shape or geometry. Moreover, the objects can be produced using digital model data from CAD models or other electronic data sources, such as computerized tomography (CT) scans. 3D printing also allows for simplified fabrication processes of convoluted devices free from the labor-intensive and time-consuming manual manipulations, leading to easy design revisions, manufacturing, testing, and iterations. Additionally, 3D printing is cost-effective and environmentally friendly as there is no need for any agents used in the etching process.[152] Furthermore, as 3D printing is inherently amenable to CAD and other computer-aided software, integration between 3D printing, and 3D cell culture can potentially allow for cloud manufacturing and commercialization of organoid- or organ-on-a-chip models. To summarize, it is possible to reconstruct living systems on chips with precise shape, architecture, and structure of target tissues and organs anywhere within a short time period with the help of 3D printing, bringing unprecedented versatility and convenience to the modeling process.[153,154]
Figure 4.
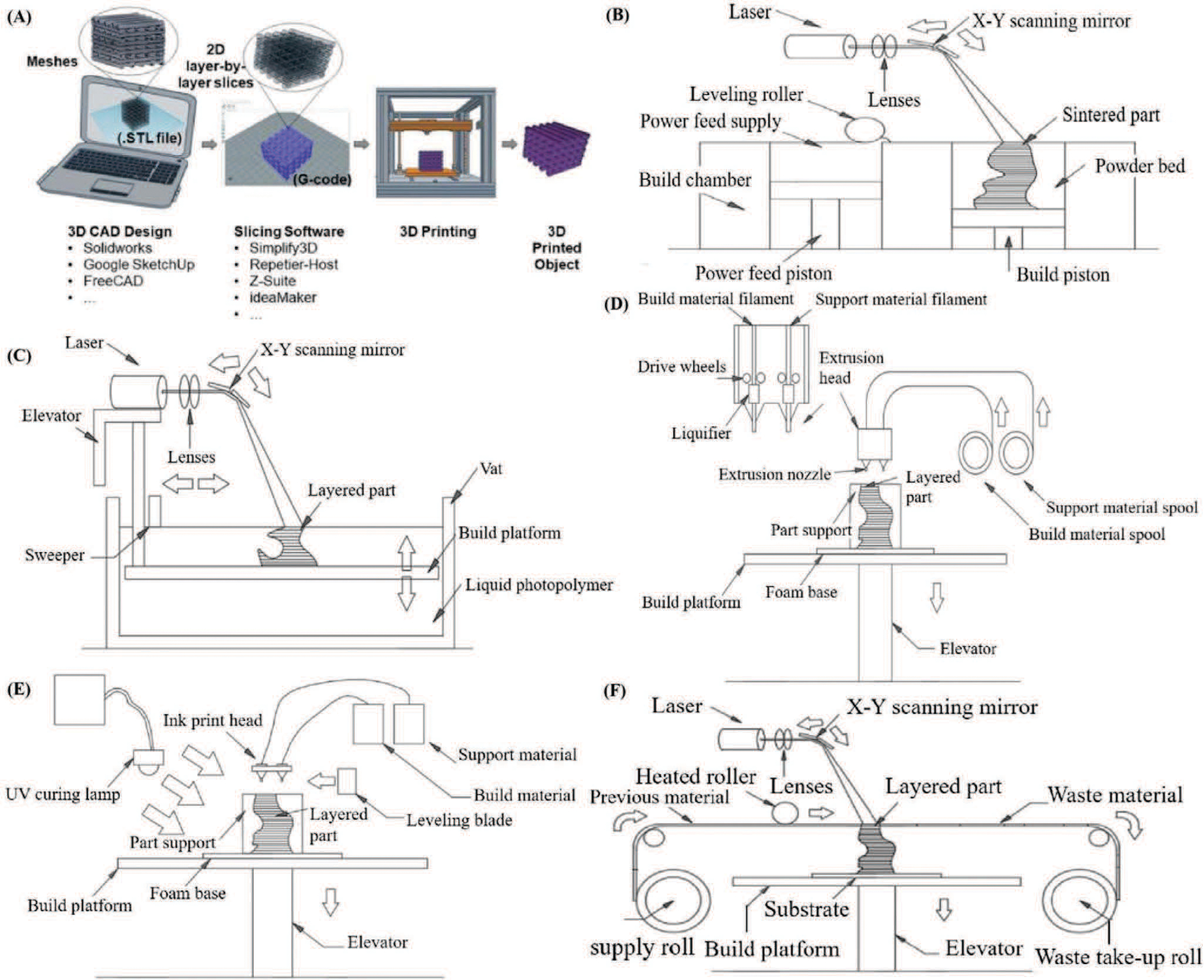
Schematic illustrations of the devices being made through different types of 3D printing processes. A). Illustration of 3D (bio)printing processes from the software designs of target tissue/organs to printed models. Reproduced with permission.[150] Copyright 2019, Elsevier. B). Schematic exhibits the printing system of selective laser sintering (SLS). The focused laser beam on the top of the platform scans the powder in the chamber to define each slice of target objects. C) Schematic displays the printing system of stereolithography (SLA). The system mainly comprises of two parts—a vat that contains liquid photopolymer resin and a laser light source that facilitates the formation of desired patterns. D) Schematic illustrates the printing system of fused deposition modeling (FDM). Thermoplastic filament or metal wire is extruded from the extrusion nozzle to fabricate desired structures. E) Schematic shows the printing system of photopolymer inkjet printing (IP). The system typically employs UV light source to cure the injected ink and fabricate complex structures. F) Schematic displays the printing system of laminated object manufacturing (LOM). The system utilizes a laser to cut plastic laminates, and employs glue or chemical bonding to assemble the object.
3.2.1. 3D Printing Technologies
Applications of 3D printing in the realm of biomimetic living systems can be roughly divided into two categories of organ-on-a-chip and tissue engineering. Essentially, not all 3D printing methods are suitable for the production of microfluidic chips. For instance, selective laser sintering (SLS), which utilizes focused laser beams to scan the powder layer-by-layer to define each slice of target objects, is more suitable for building anatomically correct scaffolds, such as bone tissues or surgical models of patients (Figure 4B). Although the application of 3D printing methods is currently more promising for tissue engineering than biomimetic microfluidic chips, there is still a great potential for integrating 3D printing with biomimetic microfluidic devices in the future.
The two most common techniques applied in the fabrication of 3D cell culture devices are stereolithography (SLA) and fused deposition modeling (FDM; also termed thermoplastic extrusion). SLA is a type of 3D printing technology that utilizes light to induce links between molecules and cure photopolymers (Figure 4C).[155] The SLA printing system is mainly comprised of two parts, a vat containing a liquid photopolymer resin and a laser light source that facilitates the formation of desired patterns in a layer-by-layer fashion. The laser can scan the liquid sequentially according to specific patterns controlled by the computer, which cures the resin and directly forms specific structures.[155,156] There are also alternative forms of SLA technologies, such as digital light procession (DLP)[157] and two-photon polymerization (2-PP),[158] which share the same principles. For DLP, the light source includes a controllable digital mirror that can either passively reflect or actively emit the laser light to cure target parts of the prepolymer; thus, DLP is capable of crosslinking the entire layer at once rather than scanning over the layer point-by-point in the traditional way.[159,160] The main difference between 2-PP and traditional single-photon polymerization (1-PP) is that polymerization occurs inside or near the surface of printing materials.[161] The polymerization process of 2-PP can start only when two photons are absorbed at the same time, which leads to the highest resolution among other printing approaches. Meanwhile, the FDM printing process relies on extruding heated thermoplastics, biocompatible and economical plastic filaments, or metal wires from a nozzle to fabricate the desired structures (Figure 4D). Once heated into a semiliquid form, the filaments are extruded at a defined location to form patterns. Then, layer after layer, the convoluted 3D architecture is achieved.[162] Nowadays, FDM, which is the cheapest method available, is used for home printers and is expected to promote the commercialization of organ-or organoid-on-a-chip models.[154,159] Similar to FDM, a newly developed noncontact printing method termed inkjet printing also relies on nozzles to extrude liquid photoresin or wax onto a substrate in a layer-by-layer manner. In contrast however, photopolymer inkjet printing often utilizes ultraviolet (UV) light to cure the ink and fabricate complex 3D structures (Figure 4E).
Other than the traditional methods mentioned above, there has been a recent boost in popularity and applicability of a variant of 3D printing termed 3D bioprinting, whose ink contains both scaffold material and living cells.[162,163] Bioprinting can simultaneously lay down biocompatible supporting materials and living cells in a single-step construction without requiring professional lithography skills or complex manipulation of cells, which greatly reduces the process time needed. Importantly, bioprinting can also introduce physiologically relevant cues, such as well-defined cell arrangements, to better simulate cellular diversity and microstructures with great consistency.[164] Last but not least, laminated object manufacturing (LOM) is another method that can be applied in manufacturing biomimetic microfluidic chips. LOM is a different kind of layer-by-layer technique that relies on a laser to cut plastic laminates and glue or chemical bonding to assemble the target 3D object (Figure 4F). This technique can be employed as a low-cost method to prototype complicated biomimetic microfluidic devices.[165]
3.2.2. 3D Printing Materials
Organ-on-a-chip platforms are microfluidic devices containing organized biological structures and microenvironments that emulate the physiological function, behavior, and response of their analogous organs in the human body.[166] Advances in biomaterials, engineering, and additive manufacturing in the realm of biomimetic living systems have led to novel opportunities for economic and rapid prototyping of printed microtissues or organ-on-chip systems in automated procedures for continuous production.[167,168] The synergistic application of 3D bioprinting to construct microtissues in organ-on-chip bioreactors has the potential to revolutionize in vitro organoid models through the inclusion of complex physiological structures in controlled extracellular environments.[169] Various biomaterials, such as natural, semisynthetic, or synthetic polymers have been widely used as bioink sources for construction of desired microtissues or organoids in organ-on-chip models using inexpensive desktop 3D bioprinter platforms.[170] Furthermore, the characteristics of bioinks need to be considered as important parameters of tissue/organ printing process, including printability, biocompatibility, biomimicry, and biodegradability, mechanical, and structural integrity.[171] The bioinks should be a filament-like structure when it is dispensed through the extrusion nozzle and retain its shape to support high printing fidelity. The rapidly crosslinkable bioinks are categorized into the physical (e.g., ionic, hydrophobic, supramolecular, hydrogen bonding), chemical (e.g., click-chemistry, Michael-type addition), photo-induced crosslinking (e.g., UV light-induced photopolymerization), and DNA hybridization methods.[172] The bioinks provide tissue-specific biochemical and physical stimuli to guide cellular behaviors, such as proliferation, differentiation, migration, and maturation. Synthetic bioinks (e.g., methacrylic acid (ma), polyethylene glycol (PEG)) have the characteristics of fast polymerization and high mechanical stability, they do not possess the cell-binding moieties required for cell adhesion, proliferation, spreading, and motility.[173] In contrast, natural polymers alone (e.g., hyaluronic acid (HA), gelatin, alginate, collagen, fibrin, decellularized extracellular matrix (dECM)) without additional crosslinking result in difficulties in supporting 3D structures.[174,175] Therefore, the subsequent several types of advanced hybrid bioinks by mixtures of natural, semisynthetic, or synthetic polymers with sequential crosslinking systems are largely adopted for in vitro microtissues or organoids in organ-on-chip models research.[176]
Bioprinting of gelatin-based hydrogel has been broadly reported in both fugitive and direct-write bioinks.[177] In particular, gelatin methacrylate (GelMA) has been used advanced bioinks as a single or customized material by mixture of GelMA macromere and lextrusion through exposure to UV light (360–480 nm in wavelength).[178] Unmodified HA has been widely used through blending with printable hydrogels, including GelMA, MeHA, norbornene-functionalized HA(norHA), and photocurable dextran, and via conjugation with thermoresponsive poly (N-isopropylacrylamide) (MeHA-pNIPAAM) and created stable 3D constructs.[179,180] Ma et al.[181] present the application of customized DLP-based 3D bioprinting system to the development of a 3D hydrogel-based triculture model that possesses the physiologically relevant cell combination and microarchitecture in a predefined biomimetic manner (Figure 5A). Specifically, 5% (wt/vol) GelMA with similar matrix stiffness in liver was chosen to support hiPSC-derived hepatic cells, and glycidal methacrylate-hyaluronic acid was mixed with GelMA for encapsulating the supporting endothelial and mesenchymal cells. Kolesky et al.[182] successfully fabricated an engineered vascular-embedded tissue constructs with aqueous fugitive ink composed of Pluronic F127 and cell-laden GelMA hydrogel inks by innovatively combined 3D bioprinting technology (Figure 5B). Pluronic F127 ink can be easily printed as a sacrifice template and removed under mild conditions because it undergoes thermally reversible gelation above a critical micelle concentration (CMC ≈ 21 wt%) and temperature (4 °C). They first coprinted two inks in a predefined sequential process and then deposited pure GelMA ink and photopolymerization, then the fugitive ink is liquefied and removed from the 3D construct yielding open channels. Based on the similar method of sacrificial template, Miller et al.[183] printed rigid 3D filament networks of an open, interconnected, self-supporting carbohydrate-glass lattice as a sacrifice template to supports convective and diffusive transport of blood under high-pressure pulsatile flow and sustains the metabolic function of 3D vascular liver architectures (Figure 5C). The process allows independent control of network geometry, endothelialization, and extravascular tissue to yield a monolithic tissue construct by covering carbohydrate-glass scaffold with a perivascular cell-containing hydrogel, and finally melted with cell media to form the hollowed cylindrical network.
Figure 5.
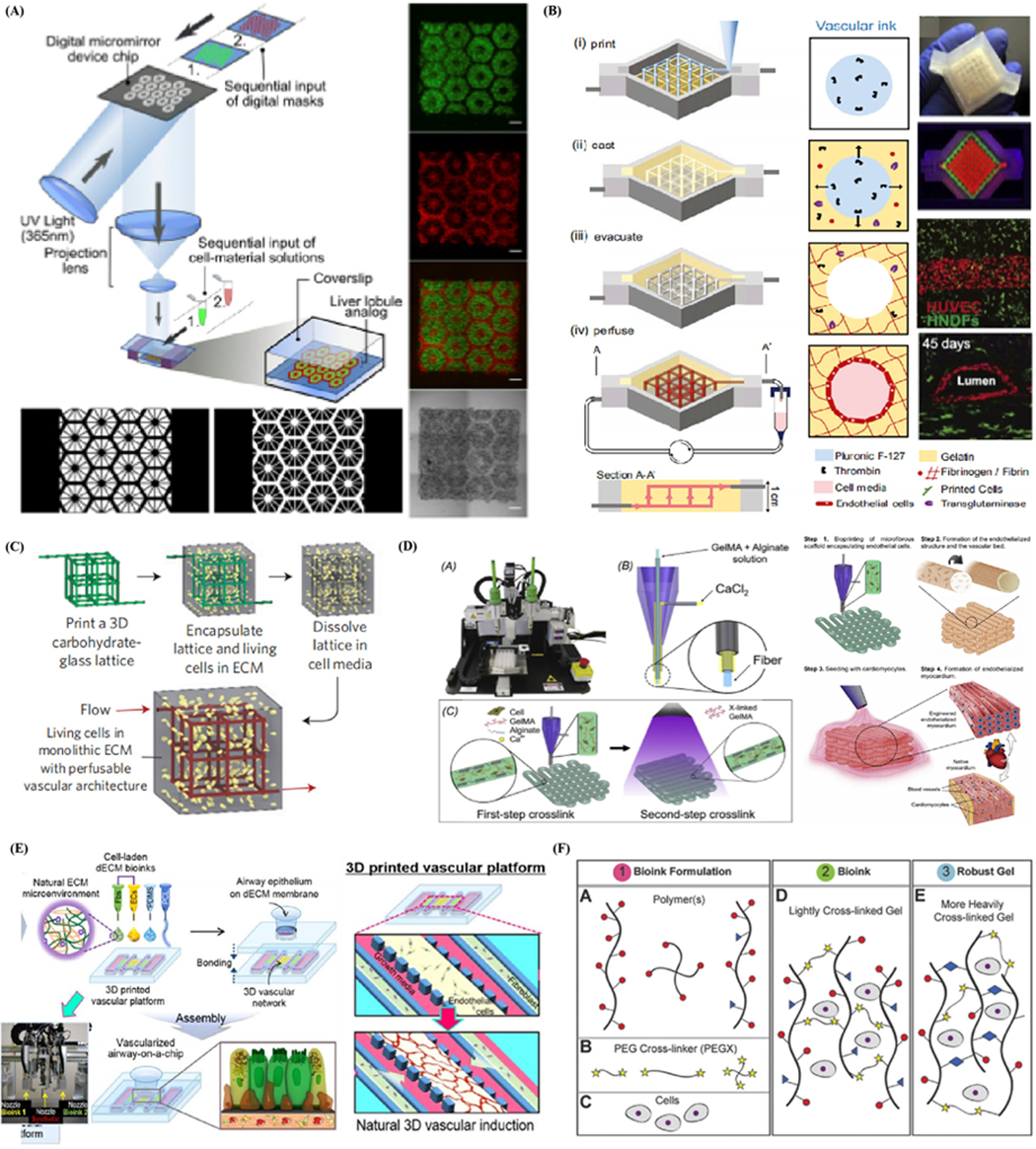
Various biomaterials such as natural, semisynthetic, or synthetic polymers have been widely used as bioink sources for construction of desired microtissues or organoids in organ-on-chip models using inexpensive desktop 3D bioprinter platforms. A) Schematic diagram of a two-step 3D bioprinting of hydrogel-based hepatic construct in which hiPSC-HPCs were patterned by the first digital mask with lobule structure (Left) followed by the patterning of supporting cells using a second digital mask with vascular structure. Reproduced with permission.[181] Copyright 2015, Macmillan Publishers Limited. B) Schematic illustration of the tissue manufacturing process of i) printing of fugitive (vascular) ink within a 3D perfusion chip; ii) casting of ECM material over the printed fugitive inks; iii) evacuating fugitive ink; and iv) perfusion via an external pump. Photographs and confocal microscopy image of bioprinted thick vascularized tissue. Reproduced with permission.[182] Copyright 2016, Macmillan Publishers Limited. C) Monolithic tissue construct containing patterned vascular architectures and living cells by printing carbohydrate-glass lattice serve as the sacrificial element and the formed vascular network with intervessel junction and vascular lumen. Reproduced with permission.[183] Copyright 2012, Springer Nature. D) Schematic of procedure of fabricating endothelialized myocardium by an Organovo Novogen MMX bioprinter and the coaxial needle where the bioink is delivered from the core and the ionic crosslinking CaCl2 solution is sheathed on the side by the two-step crosslinking process. Reproduced with permission.[143] Copyright 2016, Elsevier Ltd. E) Fabrication of vascularized airway-on-a-chip (VA-OC) by 3D cell printing with the bioink that mixture of tracheal mucosa-derived dECM (tmdECM) and Matrigel. Reproduced with permission.[188] Copyright 2019, IOP Publishing. F) Multimaterials bioinks optionally incorporated cells by d) mixing with polymers and polymer or polymer mixtures (PEGX) with linear (e.g., gelatin), branched (e.g., 4 arm PEG amine), or multifunctional (e.g., gelatin methacrylate) to form the bioink, and subsequence perform postprinting by secondary crosslinking to increase mechanical robustness. Reproduced with permission.[189] Copyright 2015, John Wiley & Sons, Inc.
Alginate-based hydrogel has been widely used as a bioink for extrusion bioprinting because of instantly polymerization by being mixed with multivalent cations (e.g., Ca 2+ or Ba 2+) during the printing process.[184] In general, Alginate-based bioinks require optimal printing condition of mixture of GelMA to increase controllable printability and cellular affifinity, and adding 4-arm poly (ethylene glycol)-tetra-acrylate to enhance the crosslinking density and increase the mechanical strength. Zhang et al.[143] utilized 3D bioprinting in a hybrid strategy to fabricate cardiovascular-organoid-on-a-chip with an endothelialized perfusion system (Figure 5D). The composition of the bioink enabled a dual-step crosslinking procedure duo to consist of a mixture of alginate, GelMA, and photoinitiator. During the bioprinting process, the ionic crosslinking of the alginate component of the bioink delivered through the core of the nozzle was first induced by exposing the extruded microfibers to a CaCl2 solution and carried by the sheath. When the scaffold was printed, a stable gelation was then achieved by crosslinking GelMA via UV exposure. Endothelial cells directly bioprinted within microfibrous hydrogel scaffolds gradually migrated toward the peripheries of the microfibers to form a layer of confluent endothelium, then seeded with cardiomyocytes to generate aligned myocardium capable of spontaneous and synchronous contraction. They further embedded the organoids into a specially designed microfluidic perfusion bioreactor to complete the endothelialized-myocardium-on-a-chip platform for cardiovascular toxicity evaluation.
Moreover, dECM isolated from various organs/tissues has been applied as bioink to simulate their complex ECM microenvironment.[185] dECM-based bioinks with inherent composition of various proteins, proteoglycans, and glycoproteins are potential to mimic native tissue-like ECM compositions.[186] Bioprinting with heart tissue-derived dECM bioink is a highly useful approach for providing a physiologically identical myocardium tissue microenvironment with similar mechanical stiffness properties by a two-step crosslinking method with sequential vitamin B2 (0.01% w/v)-induced UV crosslinking and thermal gelation to provide strength during the printing process.[187] Park et al.[188] also developed an in vitro airway epithelium model collectively reproduced a functional interface between the airway epithelium and the naturally-derived vascular network by direct 3D printing of cell-laden dECM bioinks (Figure 5E). dECM bioink derived from porcine tracheal mucosa (tmdECM) was used to encapsulate and print endothelial cells and fibroblasts within a designated polycarprolactone frame. tmdECM gradually drives endothelial reorientation and leads to the formation of a vessel network. This integrated platform was also applied to implement inflammatory responses induced by pathophysiological stimulation, and analysis the effects of functional blood vessels to the inflammatory responses of airway epithelium.
The lack of 3D-printable and cell-compatible bioinks as well as the limited ability to tune bioink material properties is cited as significant inhibitors to the growth of bioprinting. Many efforts are committed to establish a versatile method to create hydrogel bioinks of varying materials and permit the ability to tune the biological, physical, chemical, and mechanical properties of the resulting structures.[189] Developing a bioink synthesis technique compatible with low polymer fractions as well as many crosslinking chemistries could significantly expand the number of 3D-printable bioinks available.[190] Rutz et al.[189] developed a single crosslinker made of PEG that could be applied in amine-containing bioinks of both synthetic and natural bioinks (Figure 5F). Successful multimaterial bioinks were generated from the polymer solutions of natural proteins (fibrinogen and gelatin), and synthetic polymers (4 arm PEG amine), and synthetic-natural mixtures (4 arm PEG amine-gelatin), as well as modified proteins (GelMA) and protein mixtures (gelatin-fibrinogen). Single chemical crosslinker made of PEG can be applied in amine-containing bioinks of both synthetic and natural bioinks (such as gelatin, GelMA, and 4-arm PEG amine and their mixtures) and contains a homo-bifunctional polyethylene glycol ending in two reactive groups (PEGX). Its characteristic that tuning the concentration of PEGX to tailor the degree of crosslinking enables the customization of the printability as well as the changes of rheological properties of the bioinks. This selected bioinks can be potential used toward developing 3D tailorable platforms for studying cell–cell signaling and tissue morphogenesis, as well as creating more customized and biomimetic 3D-printed tissue constructs.
3.2.3. 3D Printing Advantages
Simplified Processes and Reduced Production Times:
Nowadays, clean-room lithography plays a leading role in fabricating biomimetic microfluidic chips whose manufacturing process generally includes the following steps: first, the masks are etched by soft lithography according to well-designed CAD files, after which elastomer precursors like PDMS are poured onto the masks to imprint the pattern. Then, the cured elastomer on the masks is carefully peeled off and subsequently activated via chemical solutions or oxygen plasma followed by bonding to glass substrates to seal the imprint and produce channels.[162,191] Despite the importance of lithography technology in the manufacturing of microfluidic chips, the cumbersome user interfaces and extremely slow molding processes act as barriers to the clinical application and commercial dissemination of 3D cell cultures on a chip.[192] However, integrating 3D printing technology with 3D cell culture may enable the easy fabrication of 3D cell culture devices without needing fabrication approaches, and promote cost-effective, time-saving, user-friendly, and less labor-intensive 3D cell culture platforms.
Generally speaking, the current 3D printing technologies used for generating biomimetic microfluidic chips can be divided into two categories of indirect and direct fabrication.[193] For indirect 3D printing, negative sacrificial molds are created first, followed by smoothing of the layered objects. After casting with a suitable bioink that will serve as the wall material, special types of reagents are employed to dissolve the sacrificial material, leaving behind the target model.[193,194] For example, sugar can be used as sacrificial material for printing 3D microfluidic chip using a modified low-cost desktop 3D printer (Figure 6A).[195] The process is that first print the sacrificial sugar lines on a platform and cast PDMS onto it, after which the printed chips were immersed in hot water to dissolve the solidified printed sugar lines. The whole procedure can be performed within a few hours and requires no professional experience.
Figure 6.
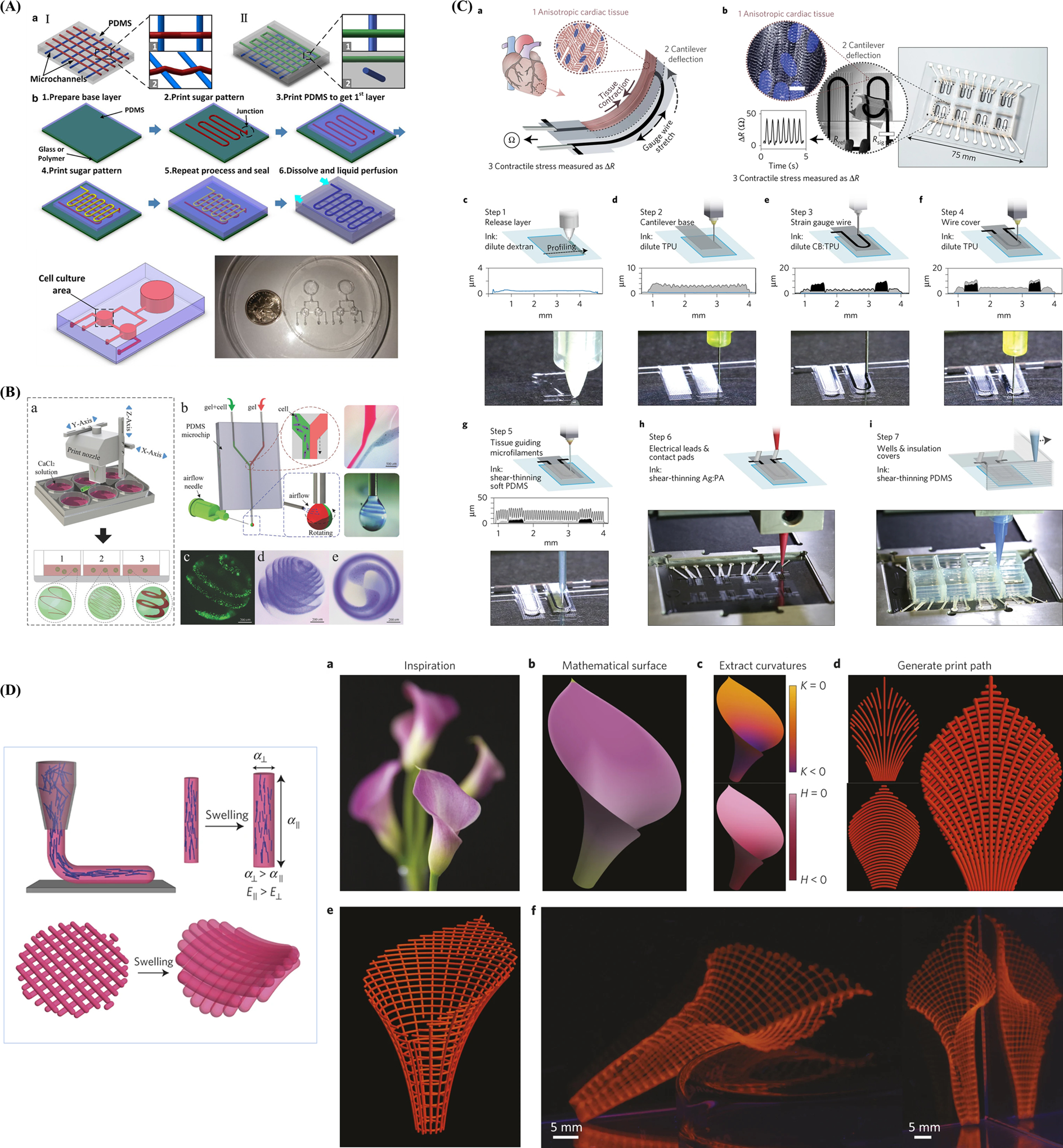
Enhanced structural fidelity, improved integrity and commercialization of 3D printing platforms and 4D biomimic printing. A) Schematic procedure of 3D microfluidic chip with supporting sugar structures microchannels with (I) and without (II) collapse (1 top view of adjacent microchannels; 2 3D view of adjacent microchannels). Reproduced with permission.[195] Copyright 2015, Springer Nature. B) 3D bioprinting spiral-based droplets and cell-laden microspheroids (spherical, rose-like, and tai chi-like) formation through extruding out of sodium alginate in PDMS microchip and from adjustable airflow-driven rotation within six-well plate. Reproduced with permission.[214] Copyright 2015, John Wiley & Sons, Inc. C) Instrumented cardiac microphysiological devices via multimaterial 3D printing principle, including contraction of an anisotropic engineered cardiac tissue, deflects a cantilever substrate, thereby stretching a soft strain gauge embedded in the cantilever, and automated 3D-printing procedure in 7 sequential steps. Reproduced with permission.[225] Copyright 2017, Springer Nature. D) Programming localized anisotropy via biomimetic 4D printing plant-inspired composite hydrogel architectures that are encoded with localized, anisotropic swelling behavior controlled by the alignment of cellulose fibrils along prescribed printing pathways. Reproduced with permission.[254] Copyright 2016, Springer Nature.
The direct 3D printing technique is as an even more promising approach for direct and efficient fabrication of microfluidic chips, wherein a variety of biomaterials can be deposited to create 3D architecture in a single step. Rogers et al.[196] constructed a microfluidic chip with an integrated membrane-based valve within an hour using a low-cost SLA printer and a customized resin formulation. The authors demonstrated that the printer can fabricate the desired rectangular cross-sections (350 μm in width and 250 μm in height) or cylindrical cross-sections (210 μm in diameter) with a 100% yield. Bhise et al.[197] also demonstrated the application of direct 3D printing for drug toxicity evaluation by encapsulating hepatic spheroids in GelMA hydrogels, which were then directly printed into a liver-on-a-chip platform via fused deposited direct-write bioprinting. In situ monitoring of secretion rates and immunostaining experiments confirmed the viability of the printed organ-on-a-chip for long-term 3D cell culture. Furthermore, it is possible to simultaneously print 3D tissue and directly fabricate the chip, which may greatly enhance the manufacturing of biomimetic microfluidic chips with faster design iteration, reduced costs, and shorter turnaround time.[198]
To summarize, the implementation of 3D printing techniques does not require researchers to be familiar with lithography protocols, disposal of silicon masters, or binding of elastomers on glass substrate surfaces. In other words, 3D printing simplifies the fabrication of microfluidic devices, achieving a cost-effective, time-saving, user-friendly, and less labor-intensive process.
Enhanced Structural Fidelity:
The great advantage of combining 3D printing and 3D cell culture is that it allows for true 3D structure modeling in contrast to conventional “2½D” chips, which simply elongate 2D designs into the third dimension or just pile up several layers.[199,200] Successful construction of repeatable and controllable 3D layered structures with great topographical flexibility and cellular fidelity can enable precise recapitulation of living tissues or organs in vivo. Bertsch et al.[201] used SLA to fabricate a microfluidic mixer which could not be accomplished by traditional MEMS fabrication methods; the printed mixer could mix the fluid efficiently within a short distance. Using the same technique, Wonjae et al.[202] printed a 3D immunomagnetic flow assay chip with immobilized antibody-functionalized magnetic nanoparticle clusters (AbMNCs) to capture Salmonella bacteria. Under a high flow rate inside the chip, which was induced by complex 3D cylindrical microstructures, Salmonella bacteria collides with AbMNCs and forms complexes instantly after injection into the microchannel, after which the complexes can be detected via ATP luminescence measurements. Huang et al.[203] also constructed a microchip and managed to illuminate the migration mechanisms and physical behaviors of HeLa cells. This microchip was constructed via DLP—an alternative form of stereolithography—and contained three channels of different widths (25, 40, 120 μm). Notably, the chip imitated the variation in blood vessel diameters and mimicked the honeycomb-like structure of vascular morphology. The authors were able to reveal the effects of channel size (representing vascular size) on the migration of cancer cells by analyzing cell migration rates and morphological changes. Furthermore, Liu et al.[204] employed DLP to print a variable height micromixer and internally fabricate complex, cell-laden scaffolds. First, the authors utilized digital micromirror devices to build labyrinthine rectangular columns with different heights, which served as efficient micromixers. After injecting a treated cell suspension and prepolymer mixture, the chambers were exposed to 365 nm light from a digital mask, allowing the prepolymers to form the desired patterns. With the same DLP technology, Spivey et al.[205] designed a device capable of capturing a single cell to monitor and study the cellular aging process. They could fabricate the desired curves, irregular top surfaces, or minute 3D structures as small as 4 μm that possessed unconventional geometries, which could not be achieved through traditional methods. These studies are excellent examples of how the design of biomimetic microfluidic chips limited by traditional approaches can be enhanced with the help of 3D printing.
Another advantage is that 3D printers can integrate multiple heads, which in turn can contain various materials during one printing process. This enables the fabrication of convoluted geometries that are not feasible via the traditional monomaterial methods.[206] Lozano et al.[207] employed extrusion-based 3D bioprinting and were able to create a novel brain-on-a-chip comprised of several separate layers of highly porous peptide-modified gellan gum. Thanks to the facile modeling ability of 3D bioprinting, this original device could maintain a stable nutrient and oxygen supply, which facilitated the proliferation of neuron cells and supported the formation of a neural network. Similarly, Lee et al.[208] utilized the same printing method and fabricated artificial neural tissues with murine neural stem cells, VEGF-releasing fibrin gel, and collagen hydrogel and managed to illustrate the migration and mechanisms of morphological change of murine neural stem cells.
Recently, 3D bioprinting has shown excellent potential for creating sophisticated microstructures by utilizing elaborately designed microfluidic devices, inspiring a new solution to the manufacturing of novel heterogeneous spheroids. The heterogeneous microspheroids have the advantages of multicomponent, controllable morphology and spatial organization, and ease of use to reconstructing microarchitecture of built-up tissue constructs. The Janus,[209] hybrid,[210] core–shell,[211] and multicompartment,[212] spheroids have been successfully used in bioapplications and undergo significant advances in tissue engineering for drug delivery, and regenerative medicine.[213] Zhao et al. report a novel airflow-assisted 3D bioprinting method which can print versatile spiral microarchitectures inside the microspheroids (Figure 6B).[214] A microfluidic nozzle is developed to improve the capability of intricate cell encapsulation with heterotypic contact and permit one-step and programmable bioprinting of fascinating hydrogel structures, such as the spherical helix, rose, saddle, and Tai chi pattern. A human multicellular organoid of spirally vascularized ossification is reconstructed by applying selected PDMS microfluidic chip as the body of dispensing nozzle, and several metal capillaries were inserted at the inlet/outlet orifices as an airflow spinning device. Cell-laden Na-alginate solutions can be precisely extruded into several jets with distinct patterns and boundaries by taking advantage of laminar flow. The heterogeneous structure of spiral-based spheroids is very convenient for building vascularized organoids in vitro by embedding multiple cells into the spheroid, contributing novel biomimetic asymmetrical prototypes for basic medical research and regenerative medicine.
When combined with 3D cell culture, 3D printing can further push the development of body-on-a-chip due to its capacity to recapitulate the architecture of intricate vascular systems in vivo. Body-on-a-chip is a type of biomimetic microfluidic chip integrated with various organ-like (key functionality or structure) chambers and a circulatory system. This platform can comprehensively resemble the physiology of the human body and allows for exploration of the dynamic processes of ADMET for drug screening in vitro.[3,215] Until now, many body-on-a-chip models such as multiple organ coculture,[216] integrated discrete multiple organ coculture,[217] microcell culture analog apparatus,[218] and 3D microfluidic cell culture system[219] have been established and demonstrate considerable applicability in several areas. However, most of them allow direct interstitial flow between different organ chambers, which does not occur in vivo because living organs are linked by vascular systems over different distances. Therefore, 3D printed vascular systems have the potential to better emulate the in vivo circulatory system by either printed microfluidic circuits or direct bioprinted vasculature. Bertassoni et al.[220] designed blood vessel-on-a-chip with cells directly printed into the microchannel. In addition, Costa et al.[221] fabricated a microfluidic chip-based vascular model with a resolution as small as 25 μm using SLA to replicate arterial thrombosis.
Improved Integrity of 3D Cell Culture Platforms:
Although various accessories like sensors, microvalves, and micropumps are widely applied in the analysis and control of biomimetic microfluidic chips, embedding these microdevices into the chip and making them compact is a practical problem for researchers. Therefore, another aspect of how the integration of 3D printing can benefit 3D cell culture is by easily enabling interactions between biomimetic microfluidic chips and various microfluidic interface technologies. To be more specific, applicable biosensors encompassed within nonconductive and noncorrosive materials can be directly inserted and integrated into microfluidic devices via 3D printing technology.[222] For example, an alternative form of FDM, which extrudes the liquid from a nozzle at room temperature, is able to economically incorporate many kinds of sensors and actuators into biomimetic microfluidic systems. Using such a fabrication method, Takenaga et al.[223] proposed a microfluidic unit that was printed and assembled on a light-addressable potentiometric sensor (LAPS) chip. After culturing and attaching Chinese hamster ovary (CHO) cells within the microfluidic chip, the authors could analysis metabolism process and elucidate the reaction of cells to different concentrations of fetal calf serum via the embedded LAPS. Erkal et al.[224] also managed to integrate an electrode into two commercially available, polymer-based chips; one chip was used for the detection of electrochemical cues, while the other detected the release of the stimulus and adenosine triphosphates (ATPs). Additionally, Lind et al.[225] integrated soft strain gauge in cardiac-on-a-chip via 3D printing to measure the contractile stress of laminar cardiac tissues (Figure 6C). The authors employed six customized functional inks, which were biocompatible and piezoresistive, to fabricate the instrumented cardiac microfluidic chips. This device is comprised of eight independent wells that contain strain sensors for monitoring, cantilevers for applying stress, a guiding layer for tissue growth, and an electrical interface for readout. In addition, thanks to its ability to monitor cellular activities continuously and noninvasively, this integrated biomimetic microfluidic chip was also utilized in drug response and human laminar cardiac tissue development studies. Notably, facilitated by the reproducible and transferable characteristics of 3D printing, the electrodes in the device can be transferred between locations many times, leading to more facile modifications and wider applications. Furthermore, Vittorio et al.[191] inserted various external components in a single chip via two-step FDM; they first extruded the ABS prepolymer to form the scaffold and immersed it in liquid PDMS. After curing at 75 °C for 2 h, the scaffold was removed by acetone. Through this method, the authors successfully inserted a 390 nm light-emitting diode in the microfluidic channel to detect optical density, electronic excitation, and nuclear magnetic resonance and perform molecular analysis. Further examples include Song et al.[139] who managed to integrate microvalves and optical windows in the microfluidic chip for controlling and observing, respectively, as well as Li et al.[226] who succeeded in fabricating a microchip capable of reverse transcription to perform RNA extraction and cDNA synthesis. Interestingly, there is another relatively simple method that suspends printing processes at appropriate time points to insert microdevices and then resumes the process. Shemelya et al.[227] embedded electronic interconnects through such a method to fabricate a microfluidic device comprised of controllers and capacitive sensors. Although 3D printing is as a suitable technique for integrating microdevices and 3D cell culture systems, it may be difficult for designers find a practical method to assemble and bind devices, indicating that further studies on optimal materials and sealing techniques are required.
Commercialization and Cyberization:
As mentioned above, traditional fabrication methods such as etching or molding of microfluidic chips are labor-intensive and time-consuming, making large-scale manufacturing of commercialized chips difficult. Moreover, the state-of-the-art devices used in laboratories often include complex microfluidic control systems that require professional knowledge. All these obstacles impede the dissemination of organoid or organ-on-a-chip systems in the market. Thus, 3D printing has gained widespread attention as a promising candidate for minimizing the manufacturing barriers of organoid or organ-on-a-chip systems to bring them closer to customers. Recently, several affordable 3D printers with sufficient resolution, such as Objet Connex (Stratasys) and ProJet 5500X (3D Systems), have emerged on the market. Moreover, there is an increasing number of commercially available printing materials ranging from traditional polylactic acid to products with proprietary formulations such as E-shell and ProBV-003 (summarized by Yazdi et al.[228]). Using the combinatorial single-step printing technique, clients would only need to export the already designed chips to the desktop printer to produce the desired customized organoid or organ-on-a-chip platforms, which can then be employed in the clinic within a short period of time. Dagmar et al.[229] successfully printed a microfluidic chip comprised of one reaction chamber, two microfluidic channels, and a dosing capillary via the commercial 3D printer Profi3D Marker in a single step to detect methicillin-resistant staphylococcus aureus bacteria. Notably, the authors enclosed environmental control systems, such as heating elements and temperature sensors, within the chip during the printing process to obtain a simple and user-friendly interface. This microfluidic chip can distinguish between different bacteria and serve as an early detection diagnostic tool through its utilization of gold nanoparticles (AuNP) probes that bind to DNA target sites (in this case, the mecA gene). Shallan et al.[230] also succeeded in fabricating a microfluidic chip for isotachophoresis analysis using the DLP-based, low-cost, and consumer-targeted 3D printer MiiCraft. The enclosed chip possessed a micromixer, droplet extractor, and gradient generator with small cross-sections, demonstrating that the resolution of commercial 3D printers can fulfill the requirements of customers.
Inspired by the analogy between microfluids and electricity, the principle of modular design can be used to facilitate the commercialization of 3D cell culture microfluidic chip systems.[231] Similar to electrical circuits, which rely on a combination of standard electronic components to fabricate the network, the production of microfluidic chips also depends on assembling predefined microfluidic devices, such as valves, pumps, and reservoirs.[41] Researchers can easily acquire digital files of these molecules via the Internet for printing using 3D printers, followed by manual assembly of the device, making this a rapid process that is applicable for both clinical and research purposes. Bhargava et al.[40] created a sample library of standard microfluidic elements and connectors using an SLA-based 3D printer and then verified the concept of modular design by assembling these components into a tunable concentration gradient generator. Notably, as 3D printing is aided by its intrinsic link to CAD/CAM, it can extend the modular concept from the physical field to the digital domain. Compared with the process of fabricating discrete elements and manual assembly, it is more efficient to download digital components from the Internet, construct and evaluate them in CAD software, and then print the chip in a single step.[192] Indeed, certain medical-level connectors are already available online and can be directly imported to CAD before printing.[232] Additionally, CAD designs can be shared with noncommercial licenses through a website created by MakerBot and designers can upload and sell their licensed designs on an online market launched by 3DSkema, demonstrating the promising future of printed 3D cell culture microfluidic chips on the market.[128,232]
Although there has been a rapid advancement in accuracy and materials of 3D printer, there are still limitations to the integration of 3D printing and biomicrofluidic chips. In terms of accuracy, there remains a resolution gap between conventional soft-lithography and 3D printing that hinders the creation of delicate and fine features,[233] even though the resolution of 3D printing (about 20 μm[197]) is comparable to that of the soft-lithography method. Moreover, as the scale of devices decreases to the extent where near-surface phenomena cannot be ignored, surface roughness achieved by 3D printing acts as a barrier to its application.[234] There are also other limitations associated with the materials used. For instance, although various figurative inks have been introduced for the production of biomicrofluidic chips, there are still many strict environmental and processing requirements, including high temperatures,[235] removal of supporting materials,[236] or mold fabrication,[237] that must be fulfilled, which outweigh the benefits of 3D printing. Moreover, some hydrophilic materials cannot be directly applied during the printing of microfluidic chips, while other materials fit for printing require pretreatment to improve hydrophilicity, making these processes rather complicated. Mechanical properties such as the ability to maintain multilayered structures over long cultivation periods under dynamic conditions are yet another concern. Finally, materials utilized in printed chips have to be biocompatible for cell attachment and differentiation, limiting the choice of materials.[198] To address these hurdles facing 3D printing, many researchers are currently exploring new materials feasible for manufacturing nano/microscale biochips and ways to enhance bioprinter nozzles, laser quality, and other critical components.[238] Therefore, we can still envision a promising future for the combination of 3D printing and 3D cell culture.
Organoids and organ-on-a-chip technology has the potential to disrupt drug development processes by replacing animal studies and/or significantly improving the outcomes of animal and clinical studies.[239] Several organizations in the USA and European Union have launched initiatives to promote organoid and organ-on-a-chip research.[240] Due to this international recognition, many companies in the private sector, such as GlaxoSmithKline, Sanofi, AbbVie, and Johnson & Johnson have collaborated with startup companies and institutes (e.g., Emulate and Wyss Institute at Harvard University) and made significant investments to commercialize this technology.[241] To make optimal use of this market opportunity and further this technological innovation, 3D printing has led to novel opportunities for the economic and rapid prototyping of printed microtissues or organoids in organ-on-a-chip systems, achieving continuous production via automated procedures.[242] There is no denying that entrepreneurs and intrapreneurs are also facing great challenges ahead of the commercialization of organ-on-a-chip technology and its alignment with 3D printing. Challenges include the printability, biocompatibility, and mechanical and structural integrity of 3D printing bioinks.[243] In addition, bioprinted organoids or iPSC-derived cell models within extracellular matrices or hydrogels are unable to achieve “adult-like” organ maturity due to restricted differentiation protocols, which complicates their translation to relevant clinical data.[244] Further challenges include making current commercial and laboratory-generated chips affordable, robust, and reproducible to easily configurable with standard assay detection platforms and workflows as well driving integration with automation and sophisticated detection technology. Moreover, the challenge of in vitro-in vivo (IVIV) translational models is clinical validation or mathematical surrogates that focus on their reliability and relevance in providing readily quantifiable results and relatability to in vivo data.[245] Achieving holistic, integrated physiological responses with human-on-a-chip currently remains a distant reality because of compounding technical, biological, and translational complexity across multiple organ functions, such as innervation, immune responses, gut-microbiome interactions, and the endocrine system.[246,247] Advanced label-free biosensing and real-time diagnostic technologies are currently being developed to significantly increase model viability, such as temperature, pH, oxygen level, and nutrient availability, in a dynamic and controlled environment.[247] Finally, commercialization challenges include the development of a standardized, reliable, and robust packaged test solution or the development of highly personalized chips using patient-derived stem cells according to the applied value in terms of end-user needs and demands.
3.2.4. 4D Bioprinting: Next-Generation Bioprinting
4D bioprinting is a cutting-edge additive manufacturing technology that integrates the fourth dimension of time into the 3D bioprinting process and relies on changes in various mechanical or physio-spatial aspects when subjected to predetermined stimuli or trigger sources.[248] 4D bioprinting can be used to place both living cells and growth factors in highly ordered, biomimetic motifs and promotes dynamic, structural, and cellular changes to achieve smart living tissue models.[249] Time-varying 4D bioprinting overcomes the static nature of 3D bioprinting by allowing physiologically relevant transformations that occur at more gradual, physiologically relevant timescales, such as tissue stretching, compression, or shifting of the biomaterial’s modulus, leading to enhanced mimicking of the developmental processes of native tissues/organs.[250] 4D bioprinting relies on shape deformation of conventional or smart materials and time-dependent maturation of the engineered construct.[251] 4D-bioprinted constructs are frequently established using stimuli-responsive materials or shape memory polymers and by varying conformation or physical characteristics that can be reversibly transformed between various temporary states as a function of selective triggering mechanisms.[252] 4D-bioprinted objects directly incorporate self-assembly features that arise from physically based information or modular cues into the construct’s design and printing material formulation, and guide dynamic transformation processes in response to external stimuli.[253] There are currently different approaches for achieving manual or spontaneous shape transformation of a material/biomaterial through stimuli-responsive and cell contraction actuating.
The use of stimuli-responsive (bio)materials offers several advantages including precise spatiotemporal control of shape transformation and folding of micrometer-sized objects as well as simultaneous folding of multiple objects made of different materials.[251] Moreover, dynamic and physiologically relevant transformations of 4D-bioprinted constructs can be modulated and fine-tuned by varying the functional chemistry of the biomaterials as well as the composition and ratio of the various chemical substituents of the bioink formulation. Typically, exogenous trigger mechanisms involve temperature responses, chemical or solvent immersion, electrical or magnetic stimulation, and light induction among others.[249] Frequently used stimuli are temperature and water, which have been used to induce shape transformation of bioprinted constructs by varying the temperature or water sorption and swelling. For example, thermos-responsive hydrogels swell and deform into capsule-like structures by folding star-shaped polymer bilayers at reduced temperatures, which then unfold and release the encapsulated cells after increasing the temperature. Lewis et al.[254] developed a biomimetic hydrogel composite ink composed of stiff cellulose fibrils embedded in a soft acrylamide matrix that can be 4D printed into programmable plant-inspired bilayer architectures patterned in space and time (Figure 6D). These hydrogels are endowed with localized swelling anisotropy that induces complex shape changes upon immersion in water, yielding complex 3D morphologies. Moreover, cell contraction exerts a traction force that induces self-folding-based shape transformation to fabricate the desired 3D cell-laden microstructure or complex cellular constructs by adhering to a substrate/biomaterial surface or within 3D tissues.
3.3. Integration with Numerical Simulation
Computational and mathematical modeling, also referred to as numerical simulation, is a powerful tool that can yield accurate solutions for many microfluidic devices before fabrication.[255] Numerical simulations can ensure whether a design is feasible and efficient, and provides a faster route to the desired outcomes by reducing experimental trial and error.[256] Recent advances in organ-on-a-chip technology can better represent the structural and functional complexity of living tissues/organs and reproduce the dynamic mechanical and biochemical microenvironment. These microphysiological systems are mostly focused on mimicking the physiology and pathology of human organs and diseases and describing experiments results rather than modeling and designing. Experiments alone cannot provide full insights into the biochemical, biophysical, and biomechanical processes that affect cell growth and behavior in organ-on-a-chip. Numerical simulation can provide additional theoretical information, predict multiple properties of different underlying processes, contribute accurate and satisfactory results, and avoid repetitive experimental measurements, reducing cost and time.[257] Nevertheless, incorporating numerical simulations into organ-on-a-chip models needs additional theoretical work in both simulation and experimental studies.
Microfluidic organ-on-a-chip devices should faithfully reproduce in vivo hydromechanical effects, such as blood, interstitial, and mucosal fluid mechanics, which influence multiple biochemical, physiological, and pharmacological processes.[258] Fluid flows directly affect cell morphology and cellular signaling as well as the vasculature structure and mucosal barriers in living organisms.[259] Advances in computer technology has made examining flow structure and drug distribution possible using simulation software such as CFD.[260] CFD provides a qualitative or quantitative prediction of fluid flows by means of mathematical modeling (partial differential equations), numerical methods (discretization and solution techniques), and software tools (solvers and pre- and postprocessing utilities).[261] Advanced models in CFD made feasible the simulation of complex transport phenomena in medicine and biology and created opportunities for solving problems in the clinic. Contemporary CFD tools, such as COMSOL, CoBi, and ANSYS Fluent CFD-ACE 1, provide the necessary capabilities of modeling coupled fluid flow, mass transport, and biochemistry for designing and developing microfluidic organ-on-a-chip devices.[257] High-fidelity models of single organ-on-a-chip are more appropriate for analyzing flow patterns, pressure drops, wall shear stress profiles, and mechanical loads on membranes.[262] Computational simulation of complex behavior arising in multicellular constructs can provide critical insights for improving reproducibility or guidance for achieving the desired form and function of self-assembled organoids from pluripotent precursors.[263] Multiscale models of multiorgan-on-a-chip or human-on-a-chip are more suitable for modeling long-term drug transport as well as pharmacokinetic (PK) and pharmacodynamic (PD) effects.[264,265] Sections 3.3.1–3.3.3 will present the most important advances in modeling and simulation as an integrated part of organoids and organ-on-a-chip research and development.
3.3.1. Numerical Simulation for Organoids
Advances in experimental tools for characterization, environmental manipulation, and live-cell imaging are needed to enhance spatiotemporal predictions of organoid properties. Organoid formation in terms of cell proliferation, lineage specification, and organ homeostasis involves complex biological phenomena, such as stem cell differentiation into mature cells, chemical diffusion, surface tension, cell–cell contact and signaling, and cell-substrate mechanical interactions.[266] Novel computational modeling and biophysical principles have yielded insights into the main structural and functional features of the organoid system as a function of spatiotemporal organization of the cell, which further aids analysis and optimization.[267]
Organoid development is a 3D process that includes cell proliferation, morphogenesis, and tissue expansion and exhibits its intrinsic complexity through multiple environmental cues.[268,269] Cell fate decisions are affected by proximal (direct neighbor) and distal interactions that arise from morphogen diffusion across tissues. Other drivers of differentiation, such as mechanosensing, membrane voltages, or gap junction communication, can be used in computational modeling and for testing the properties of spatial organization or symmetry breaking to uncouple fundamental mechanisms of cell–cell communication during organoid development.[270] Self-organization of multicellular structures is the use of cell–cell signaling networks to induce morphological changes.[271] Toda et al.[272] used the modular synNotch juxtacrine signaling platform to engineer artificial minimal intercellular genetic programs to yield assemblies with hallmarks of natural developmental systems: robust self-organization into multidomain structures, well-choreographed sequential assembly, cell type divergence, symmetry breaking, and the capacity for regeneration upon injury (Figure 7A). The ability of these networks to drive complex structure formation illustrates the power of interlinking cell signaling with cell sorting: Signal-induced spatial reorganization alters the local signals received by each cell, resulting in iterative cycles of cell fate branching. Various programing self-organizing synthetic structures with minimal logic of controlling cell adhesion (cadherin expression) through cell–cell communication (synNotch signaling). These results provide insights into the evolution of multicellularity and demonstrate the potential to engineer customized self-organizing organoids or materials.
Figure 7.
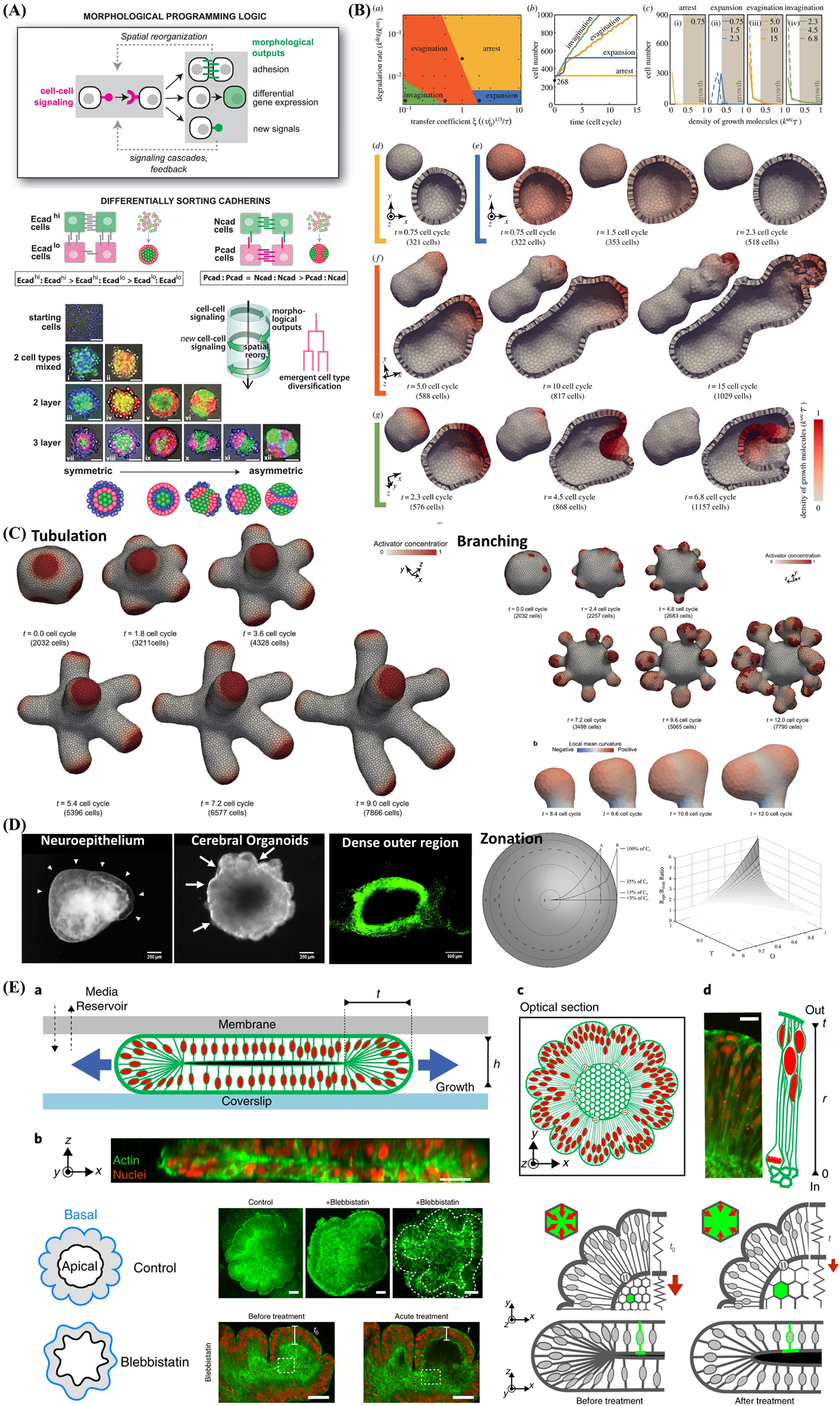
Computational simulation and mathematical modeling of complex behavior arising in multicellular constructs and self-assembled organoids from iPSC. A) Engineering cell–cell communication and signaling networks within self-organizing multicellular structures and organoids to program synthetic morphogenesis with spherically asymmetric structures by inducing differentially sorting adhesion molecules and using the simple synNotch→adhesion toolkit. Reproduced with permission.[272] Copyright 2021, American Association for the Advancement of Science. B) Computational simulations of signal-dependent epithelial growth and deformation during tissue morphogenesis that be categorized into four phases: arrest (yellow), expansion (blue), evagination (orange), and invagination (green). Reproduced with permission.[278] Copyright 2015, Royal Society. C) Combining Turing and 3D vertex models reproduces autonomous multicellular morphogenesis with undulation, tabulation (time series images of thin tube formation), and branching(time series images of whole tissue deformation and branch structure). Reproduced with permission,[279] Copyright 2018, Springer Nature. D) Analytic models of oxygen and nutrient diffusion, metabolism dynamics, and architecture optimization in a multicompartment spherical model for cerebral organoids with metabolically active region, intermediate region, hypoxic region, and ischemic region, respectively. Reproduced with permission.[280] Copyright 2016, Mary Ann Liebert, Inc. E) Brain organoid development and wrinkling occurs at a critical nuclear density and maximal strain. Nuclear motion and swelling during cell cycle lead to differential growth. Cytoskeletal forces maintain organoid core contraction and stiffness and adding cytoskeleton inhibition (blebbistatin) show the reduction in thickness and increase in inner surface area. Reproduced with permission.[283] Copyright 2018, Springer Nature.
Morphogenesis is a sequential process of multicellular deformations by signal-dependent cell activities, such as contraction, adhesion, migration, proliferation, and apoptosis in 3D space.[273] Cells have the characteristics of mechanically varied 3D structures such as apical, basal and lateral areas of epithelium, apical areas actively generate contractile forces by actomyosin activities,[6] and basal areas passively respond to extrinsic forces in a viscoelastic manner.[274,275] Shape deformation is critical for organoid morphogenesis to yield human-like organ structures.[276] Vertex models can successfully describe how spatial patterns of apical cell contractility induce deformation of epithelial shells, which is simulated by the formation of hexagonal prism-like cells with a fluid-filled cavity and a solid membrane.[277] Both 3D and 2D vertex models have demonstrated principles of evagination or invagination that correspond to the apical side, which comprises the inner shell surface or outer shell surface. To combine the mechanical properties and chemical interactions during morphogenesis of each cell, Okuda et al.[278] established a 3D vertex model that simulates intercellular signal-dependent epithelial morphogenesis, ultimately, the multicellular deformations, and biochemical patterns resulted in four types of 3D morphogenesis—arrest, expansion, invagination, and evagination (Figure 7B). In a later work, the same authors incorporated a 3D vertex model with a mathematical model of turing morphogen reaction-diffusion dynamics and demonstrated that diverse morphologies within the same tissue type depend on different time scales (Figure 7C).[279] Moreover, morphogenesis, including tubulation, branching, and undulation, was achieved from a 3D monolayer cellular sheet model. Other organoids established using these models include interstitial crypts, cortical polarized tissues, self-organizing epithelial acini, branching morphogenesis of mammary or salivary glands, and multilayered multilineage cysts.
An additional feature of 3D computational organoid modeling is the introduction of limitations of nutrient availability as a consequence of necrotic cores. Simulations with partial differential equations and experimentally engineering vascularized organoids are often appropriate for describing these properties of nutritional deficiency. Mcmurtrey et al.[280] developed an analytic models of oxygen and nutrient diffusion, metabolism dynamics, and architecture optimization in 3D tissue constructs with applications and insights in cerebral organoids. (Figure 7D). Oxygen, glucose, and other metabolic precursors necessary for anabolism are subjected to reaction-diffusion biotransport across a multicellular engineered tissue.[281] The lack of nutrient and gas exchange limits control over the size, shape, and relative cell arrangement of organoids and hinders the maximum size and extent of tissue maturation. The accumulation and diffusivity of secreted molecules (such as growth factors or chemokines) in intercellular regions/extracellular matrix represent a stable source of localized influence on cell fate decisions within organoids. Exposure of cells or organoids to physiological shear flow, mechanical stress, and substrate stiffness can have profound effects on their physiology and function.
Appropriate computational organoid modeling must reflect the high sensitivity to changes of biomechanics and biophysical forces that utilize surface tension and adhesive forces (or lack thereof) to encourage cell–cell interactions. When simulating 3D organoid growth, Hookean forces between cells dictate an outward “jostling” effect that maintains neighbor-to-neighbor distances and changes the degree of cellular packing when spheroid volumes expand as a result of cell division.[282] Human brain wrinkling emerges spontaneously due to compression forces arising during differential swelling of polymer gel. Karzbrun et al.[283] report the appearance of surface wrinkles during the in vitro development and self-organization of human brain organoids in a microfabricated compartment (Figure 7E). They observe the emergence of convolutions at a critical cell density and maximal nuclear strain. They identify two opposing forces contributing to differential growth, one is cytoskeletal contraction at the organoid core and the other is cell-cycle-dependent nuclear expansion at the organoid perimeter. It remarkably models well the physics of the folding brain with wrinkling wavelength exhibits linear scaling with tissue thickness, consistent with balanced bending, and stretching energies. Opposite evidence from smooth brain organoids display reduced convolutions, modified scaling, and a reduced elastic modulus. Taken together, organoid wrinkling is driven by a mechanical instability from differentially swelling materials. The emergent wrinkling pattern are ascribed to the increased growth in the organoid outer regions, and the actively contracting organoid inner surface. This human brain organoids on a chip approach successfully mimics the early developing cortex and reveal the physics of folding of brain.
The main goal of mathematical and computational oncology is to develop quantitative tools to determine the most effective therapies for each individual patient, otherwise known as precision medicine.[284] Mathematical modeling and computer simulations allow for relatively fast, efficient, and inexpensive simulations of innumerable treatment schedules to predict the most promising therapeutic regimen as well as the timing and dosage of administration.[285] Mathematical models explicitly take into account the spatial architecture of 3D tumor spheroids and patient-derived tumor organoid cultures to address tumor development, progression, and response to treatments.[286] The aim is to support the concept of virtual clinical trials and demonstrate the integration of mathematical, computational, and experimental approaches to unlock personalized treatment strategies.[287]
3.3.2. Numerical Simulation for Organ-on-a-Chip
Organ-on-a-chip can mimic the body’s multicellular architecture, tissue–tissue interfaces, physicochemical microenvironments, and vascular perfusion.[1] Modeling of the multiphysics behavior of microfluidic organ-on-a-chip is critical for their development and optimization. High-fidelity simulations of microfluidic organ-on-a-chip devices require a computational mesh generated from device geometry.[282] Typical organ-on-a-chip devices have relatively simple geometries and commonly feature two microchannels stacked on top of each other separated by a thin porous membrane for in vitro barrier modeling. The geometry/mesh models require specification of volume conditions (e.g., epi-channel, endo-channel, porous membrane, permeable sold, and cell layer) and boundary conditions (e.g., inlets, outlets, walls, and interfaces).[257] An essential component for adequately representing a subset of human organ or tissue functions in these microfluidic organ-on-a-chip systems is the concentration distribution of the bioactive compounds involved, especially the delicate balance between media mixing and cellular signaling for long-term maintenance of the multiple cell type coculture.[288] Experimental and numerical studies of the molecular concentration distributions resulting from convective-diffusive mass transport in both microchannels have been conducted by analyzing the effects of media flow rate and direction, separation membrane porosity, microchannel dimensions, and molecular size.[289]
Barrier-type computational models involve porous membranes functionalized by epithelial and endothelial cells and can be adapted to various barrier organs such as the gut, liver, lung, renal proximal tubule, blood–brain barrier, skin, and bone.[257,290] Hancock et al. developed a lung-on-a-chip consisting of auxiliary vacuum channels that enable stretching of a flexible membrane to simulate breathing and mimic the essential features of the blood-air barrier in human lungs (Figure 8A).[291] This lung-on-a-chip model makes use of the COMSOL Multiphysics software model for simulating fluid–structure interaction, laminar fluid flow, nonlinear structural materials, dilute species transport, and particle tracing capabilities. The authors first varied the vacuum pressure to produce a strain-pressure relationship for selecting a vacuum pressure waveform, which was then used in a time-dependent simulation involving membrane and channel deformation and air/liquid flow to simulate the breathing process. Additional features of lung model provide further insights, such as drug or nutrient transport within the culture medium and their uptake by cells or across the porous membrane, particle tracing to model bacteria or nanoparticle inhalation into lungs, and particulate flow and shear stress estimates on cells that attached on the porous membrane.
Figure 8.
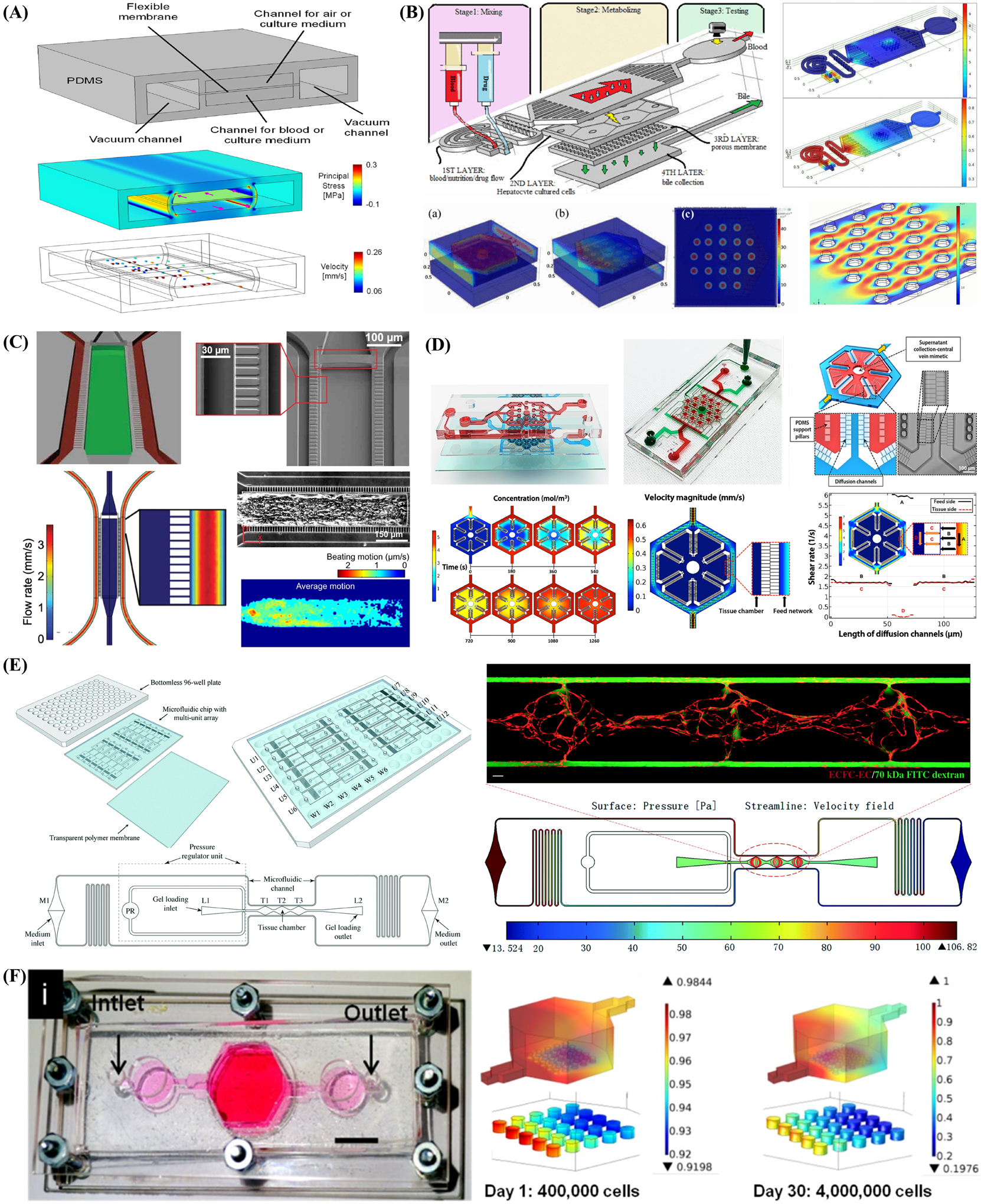
Summary of several applications of numerical simulations used for modeling organoids and organ-on-a-chip systems, especially computational and mathematical modeling yield accurate solutions for many microfluidic devices before fabrication. A) The Lung on a chip consists of a flexible membrane separating chambers for air and blood. Auxiliary vacuum channels enable stretching of the membrane to simulate breathing. The COMSOL software is used to simulate its fluid-structure interaction, nonlinear structural materials, laminar fluid flow, dilute species transport, and particle tracing capabilities.[291] B) Numerical simulations in COMSOL was performed and reported in a multilayered microfluidic liver-on-a-chip model that consists three processing steps: 1) mixing, 2) metabolizing, 3) and testing. Reproduced with permission.[292] Copyright 2017, IEEE Xplore. C) Schematic of the cardiac microphysiological system (MPS) nutrient channels (red) and cell-loading channel (green) by insetting the 2 mm endothelial-like barriers connecting the nutrient channel and the cell channel and imulated velocity profile of flow. Reproduced with permission.[294] Copyright 2014, Springer Nature. D) Schematic of liver-lobule-on-a-chip device incorporates 18 single lobule mimetic chambers and a separate seed-feed network, and numerical simulation of flow velocity and shear rate for diffusion-dominated nutrients mass transport in the device. Reproduced with permission.[295] Copyright 2016, IOP Publishing Ltd. E) A vascularized and perfused organ-on-a-chip platform and finite element simulation of interstitial flow with hydrostatic pressure and flow velocity of a whole tissue unit to induce vasculogenesis, which consists of 3 tissue chambers (T1–T3) connected to 2 adjacent microfluidic channels, 2 gel loading ports (L1–L2), 2 medium ports (M1 and M2), and one pressure regulator unit (PR). Reproduced with permission.[297] Copyright 2015, Royal Society of Chemistry. F) Schematic exhibited simulation of oxygen concentration variation within 3D printed liver-on-a-chip in Day 1 and Day 30 with consumption by hepatocytes taken into consideration. Reproduced with permission.[298] Copyright 2016, IOP Publishing Ltd.
Simulation proves the validity of the designed organ-on-a-chip device. Atiyat et al.[292] employed the fluid interaction module in COMSOL to evaluate the effectiveness and validity of the proposed multilayered live-on-a-chip device with theoretical and experimental results (Figure 8B). The proposed device is membrane-based culturing and the resulting designs with the ability to mimic the blood flow and bile excretion of actual cultured human liver cells. The system consists of three main parts: mixing, metabolizing, and testing. The mixing process takes place through a passive microfluidic micromixer by introducing blood or tested drug fluids through two inlets. Drugs (clopidogrel) can be introduced into the system with precultured liver cells over a porous membrane with the advantage of live observation to enzymatical activation. This research also demonstrated the COMSOL simulation results, such as surface diffusion flux and surface concentration of the mixing process of the tested drug and blood, flow of blood over the cultured cells, bile flow and excretion throw the membrane.
Other typical organ-on-a-chip devices exhibit relatively 3D axisymmetric geometries and commonly feature two lateral channels with a symmetrical chamber along the horizontal center line and evenly spaced micropillar arrays or endothelial-like barriers.[293] The balance between mass transport by convection and diffusion results in a concentration distribution that is specific to the design and optimization of the organ-on-a-chip by numerical simulation and experimental validation. The injection process of the mixture of 3D neural stem cells and hydrogel (collagen) can be simulated using the two-phase level set mode of COSMOL Multiphysics software. Moreover, the species transport mode of ANSYS Fluent software can simulate the diffusion process of nutrients (glucose and lactic acid) in the medium. The main parameters affecting the diffusion process include the concentration and diffusion coefficient of nutrients, the porosity and permeability of the polymerized hydrogel in the chamber, the flow rate of the medium, and the spacing between micropillars. Mathur et al.[294] reported a cardiac microphysiological system (MPS) contains a central cell chamber with self-organizing 3D cardiac microtissues, two adjacent channels for medium (30–40 μm wide) recapitulating the vasculature, and arrays of connecting microchannels (2 μm wide) for mimicking endothelial barriers (Figure 8C). They applied this MPS system to mimic many of the mass transport properties of functional ventricular myocardium, and tested the cardiac response with four model drugs. The narrow cross-section of these microchannels creates a total fluidic resistance into the cell culture area 10 times greater than through the media channel. Thus, the transport from the media channels to the cell chamber is purely diffusive. The cardiac MPS system is aligned with the 3D cardiac tissue structure, which provides consistent contractions to maintain the intracellular connections and electromechanical activity of the myocardium. They also demonstrated the heat map of the time-averaged beating motion and corresponding average beating kinetics in the MPS.
Banaeiyan et al.[295] demonstrated a microphysiological niche for hepatocytes in a very large-scale liver-lobule-on-a-chip device (Figure 8D). The chip consists of an integrated network of liver-lobule-like hexagonal tissue-culture chambers constructed in a hybrid layout with a separate seed-feed network. Each chamber contains a central outlet mimicking the central vein of a liver lobule. Separating chamber walls with 2 μm wide and 2 μm high diffusion channels located between the culture area and feed network protects cells from the shear force of the convective flow. Arrays of designated passages convey nutrients and xenobiotics into the tissue chambers by diffusion-dominated mass transport mimicking fenestrated endothelial cells in the liver-tissue microenvironment. The biorelevant geometry of the device enables coculturing of several cell types in a direct cell–cell contact, as well as in a separated manner by the diffusion barriers and lead to oxygen-gradient perfusion and the formation of metabolic zonation. They also used COMSOL software to simulate the flow velocity, shear stress, and diffusion of glucose molecules inside and outside the culture chambers under a continuous flow rate.
Frohlich et al.[296] utilized COMSOL to assess the shear stress field across the cell adhesion area, and designed a culture device named microscale tissue modeling device, which could provide a method to manipulate cell cultivation. They employed numerical simulation to emulate the influence of shear stress on cell migration, alignment, phenotype, and the coupled effect between shear stress and submicrotopography. Later, cell alignment and staining assays demonstrated the agreement between the results and predictions, proving that numerical simulation can be resorted to design a well-defined culture device rapidly and efficiently. Bhise et al.[297] demonstrated a vascularized and perfused organ-on-a-chip platform and undergone finite element simulation of interstitial flow with hydrostatic pressure and flow velocity of a whole tissue unit to induce vasculogenesis (Figure 8E). Each tissue unit consists of 3 tissue chambers (T1–T3) connected to 2 adjacent microfluidic channels, 2 gel loading ports (L1–L2), 2 medium ports (M1 and M2), and one pressure regulator unit (PR). Each vascularized micro-organ is independently-addressable and flow through the micro-organ is driven by hydrostatic pressure. They performed finite element simulations for interstitial flow through ECM embedded in the tissue chamber that confined for momentum transportation through a porous fibrin gel with low permeability. The hydrostatic pressure and interstitial flow velocity in both vertical and horizontal directions are within the optimal range (0.1–11 μm s−1) previously reported to continuously induce vasculogenesis.
Furthermore, the numerical simulations can also be employed to verify various speculations of cells’ metabolism and predict cellular behaviors. The results can also serve as a reference to account for experimental data. For instance, Schimek et al.[298] devised a full-thickness skin equivalent (ftSEs) in a 96-Well Insert Format, and utilized numerical simulation as an important method to test some of their assumptions. The simulation helped them to approximate the processes of the permeation of fluorescein sodium salt through the cultivated ftSE and predict results in the long run. Additionally, Loskill et al.[299] designed a microfluidic chip to separate transport effect, and utilized numerical simulation to show its efficiency. Via “Transport of Diluted Species,” which is a kind of inserted interface in COMSOL, concentration change within the chamber was assessed after the influx of liquid that contained diffusive solute. What is more, the restriction of the convective flow both in cell chambers and media channels is verified via a similar method. In this case, the numerical simulation technique served as a proof to demonstrate the effectiveness of the membrane to protect cells from shear stress. Here is another example to exhibit the benefits brought by the employment of numerical simulations.
Notwithstanding a wide variety of commercial sensors have been used to measure oxygen distribution and concentration, little is clear about the levels of oxygen inside microfluidic devices and the possible impacts on cell culture during the experiment. By numerical simulation technique, Funamoto et al.[300] successfully developed a numerical model to predict oxygen concentration and distribution in microfluidic devices during cell culture. Besides, they also forecast oxygen consumption by different cell types, such as ECs and hepatocytes. With the linear relationship between oxygen partial pressure and oxygen utilization rate taken into consideration, compatibility could be achieved between numerical simulation and experimental data, which showed that numerical simulation can be an effective and powerful tool for biomimic systems to predict some chemical-related cellular behaviors. For additional example, numerical simulation was also used to estimate the flow and oxygen fields in a liver-on-a-chip platform with bioprinted hepatic spheroids by Bhise et al.[113] Interestingly, they determined the flow rate that ensured sufficient oxygen concentration within the cell culture chamber especially in the vicinity of the hydrogel constructs (Figure 8F). Besides, computational simulation results of oxygen concentration with cellular consumption taken into account in different culture phase also supported the data gained from experiments.
For the design of 3D cell cultures’ microfluidic platforms, a potential issue for polymer-based platforms has been highlighted as absorption of hydrophobic drugs under certain assays, which might also cause the undesirable exchange between adjacent microfluidic channels. To tackle this problem, numerical simulation, which could take the processes of absorption into account, is employed in explaining the results of the experiment. For example, via COMSOL, Shirure et al.[301] took dissolution, convection, and diffusion into consideration to account for the loss of drugs within a polymer-based microfluidic device. They utilized four dimensionless numbers to characterize the unintended mixing of drugs caused by the absorption of polymer. Then, they modified the chip’s designs to acquire the desired experimental conditions that were conducive to cell culture. Furthermore, the results of the numerical simulation were validated by a subsequent experiment for three hydrophobic molecules (rhodamine B, cyanine NHS ester, and paclitaxel) in PDMS.
3.3.3. Numerical Simulation for Multiorganoid- or Multiorgan-on-a-Chip
Integration of multiple organ modules to construct multiorgan-on-a-chip—essentially a simplified and miniaturized version of the human body—has been demonstrated at a proof-of-concept level.[302] Human-on-a-chip or body-on-a-chip models were demonstrated to faithfully reproduce complex and dynamic interactions among tissues and organs and recapitulate human physiology and disease progression, and can aid (in vitro–in vivo) IVIV translation of drug response studies.[303,304] Multiorgan-on-a-chip require careful consideration of recreating tissue-like structures and functions as seen in single organ-on-a-chip, as well as special design considerations of scaling strategies that reflect human physiology factors in a quantitative sense, such as specification of organ sizes and operating conditions (e.g., flow rates in each organ module, cell numbers, ratios of cell types, and total volume of media in the system).[57,304] Current challenges in microphysiological systems is establishing appropriate scaling methods, which can be obtained by introducing additional engineering concepts or by applying mathematical/computational models to reduce unknown errors to achieve an adequate standard.[306,307] Robust mathematical modeling techniques that enable in vivo extrapolation will be essential elements for the design of multiorgan-on-a-chip and the interpretation of experimental results.[308]
Multiorgan interconnections are mainly achieved through a systemic fluid pool (common medium) that potentially transports nutrients, cell metabolites, pharmaceutical drugs, soluble ligands (e.g., cytokines, hormones, and growth factors) and cellular components (e.g., exosomes, nucleic acids, and proteins).[309] Developing an effective common medium that maintains the phenotypes and functions of all organs is a critical issue. Mixed organ-specific media in equal ratios aim to a certain degree to satisfy the special needs (such as essential growth factors) of specific organs. Ramme et al.[310] assumed that predifferentiated four organ on a chip models for the intestine, liver, brain, and kidney of iPSC origin could similarly maintain their phenotype during the 14 days of coculture in a common, growth factor-deprived medium (Figure 9A). Multiorgan MPSs are capable of emulating human biology in vitro at the smallest biologically acceptable scale, and further to mimic complex biological processes involving organ–organ interaction, system homeostasis and pharmacokinetics. A parallel, physiological-inspired flow scheme through the each organ compartment and a medium flow partitioning mimicking physiological ratios of blood flow were adapted for quantitative in vitro to in vivo extrapolation. The dynamic fluid flow is adjusted to enable physiological nutrition and oxygen supply of the tissues in a distinct percentage of the blood flow from the main channel with COMSOL Multiphysics simulation.
Figure 9.
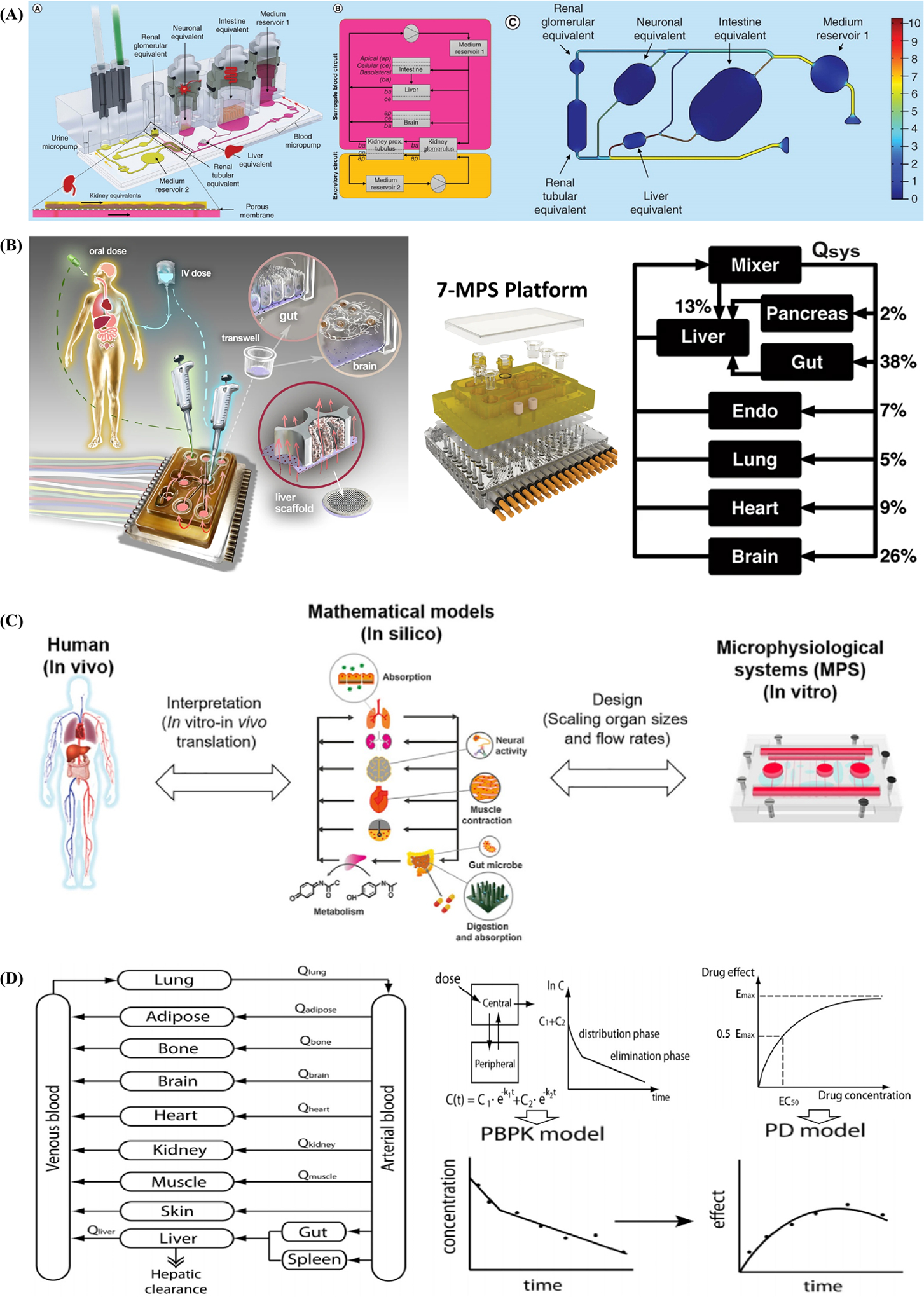
Computational and mathematical modeling of multiorganoids or multiorgans-on-a-chip require careful consideration of recreating tissue-like structures and functions as well as special design considerations of scaling strategies that reflect human physiology factors in a quantitative sense. A) Schematic of physiologically inspired model of four-organ-chip interconnecting miniaturized human intestine, liver, brain, and kidney equivalents or organoids from iPSCs and COMSOL simulation of its calculated velocity distribution and wall shear stress in the blood circuit. Reproduced with permission.[310] Copyright 2019, TissUse GmbH. B) Schematic overview of Physiome-on-a-chip devices that nurture many interconnected 3D MPS device representing specified functional behaviors of each organ of interest and corresponding flow partitioning for the 7-way platform. Reproduced with permission.[315] Copyright 2018, Springer Nature. C) Schematic diagram of pharmacokinetic–pharmacodynamic (PK–PD) models, physiologically based pharmacokinetic (PBPK) model, two-compartment PK model, and the relationship between the drug concentration and drug’s effect. Reproduced with permission.[307] Copyright 2015, AIP Publishing. D) A robust mathematical approach correctly reproduces human physiology, and how to interpret the MPS and translate experimental data to the in vivo case and the relationship between in vivo (human or animals), in silico (mathematical models), and in vitro (MPS) platforms. Reproduced with permission.[303] Copyright 2019, John Wiley & Sons, Inc.
Different fluidic interconnection platforms can greatly affect the efficacy of a common medium in supporting multiorgan functions and mediating interorgan communications. Compared to static fluidic integration, which relies on passive diffusion, dynamic microfluidic interconnections enable the establishment of controllable and reliable biochemical gradients that drive mass exchange between systemic fluid and local microenvironments.[311] The architecture of the interconnecting fluid networks can have a large impact on organ crosstalk in multiorgan systems. An open-loop, single-pass, multiorgan system usually involves perfusion through all organ modules in a sequential manner that is mostly unidirectional and lacks feedback loops from downstream organs to the upstream ones.[307] On the other hand, pumpless gravity-induced or pump-driven recirculating microfluidic systems provide a continuous unidirectional closed loop perfusion that better mimics blood circulation and facilitates reciprocal communications among organs.[312] Nevertheless, current multiorgan models require important design considerations, such as the arrangement of different organ modules to better mimic physiological processes and interconnection of organs using serial, parallel, or combined network architecture.[313]
Multiorgan-on-a-chip requires a well thought-out scaling strategy that successfully reproduces essential physiological functions of different organs and their response to drugs.[314] Physiologically unrealistic ratios of organs sizes and total medium (blood) volume will often distort the nature of their interactions and make it difficult to determine appropriate flow volume and rate among different organs within the device. Edington et al.[315] reported an approach to coculture multiple different MPSs with “4-way (liver, lung, gut, endometrium),” “7-way (adding brain, heart, and pancreas),” and “10-way (adding kidney, skin, and skeletal muscle)” linked together physiologically on reusable, open-system microfuidic platforms that are compatible with the quantitative study of a range of drugs (Figure 9B). They described the “physiome-on-a-chip” platform provides precise control over both intra- and inter-MPS flow partitioning and drug distribution by accommodating multiple different MPS flow configurations, each with internal recirculation to enhance exchange, and feature on-board pneumatically-driven pumps with independently programmable flow rates.[309]
Allometric scaling is widely used for estimating the key physiological parameters of multiorgan-on-a-chip.[314,315] Allometry is based on a governing law that dictates various physiological parameters dependent on organism size.[305] Allometric scaling laws can correlate the mass of organisms with physiological parameters, such as blood flow rate, metabolic rate, and heart rate. The multiorgan system requires that cell numbers and ratios enable appropriate physiological-like interactions and that flow rates do not cause shear stress-related damage to cells and allow adequate residence times for processing metabolic signals and convective oxygen transport.[257] So far, several scaling methods have been applied to multiorgan-on-a-chip, with each method suitable for a different set of objectives. The most straightforward and simple method for scaling down various organs is to directly scale down each organ proportionally according to easily accessible anatomical data.[307] However, directly scaling different organs using the same factors (e.g., proportional size of biomass, fluid-to-tissue ratio, and consumption/production rates) is likely to result in distortion of the appropriate relationships between differently scaled organs.[316] On the other hand, residence-time based scaling captures the essence of reaction kinetics that enable the generation and consumption of molecules in a quantitative sense within organ tissues and compartments.[317] The physiologically realistic constraints ensure that each organ is exposed to chemical cues (drugs) for the same amount of time and that generated chemical cues (metabolites) are diluted to an extent where they do not exert any observable effects.[318]
Ahluwalia et al. proposed two different allometric scaling laws terms as the cell number scaling method and the metabolic and surface scaling method, respectively, and applied for a two-organ model of hepatic-vascular crosstalk.[319,320] The allometric approach was then used to calculate the basal metabolic rate per cell hepatocyte and the vascular surface area from human parameters. Furthermore, organs can be scaled differently depending on whether their main function is a volume-mediated process (metabolic conversion by hepatocytes) or surface-mediated process (distribution through endothelium). Thus, the hepatic-vascular two-organ system was designed using the cell number scaling method to maintain the ratio of endothelial to hepatic mass after considering the fractions of total body weight for specific organs (6.28% for vascular endothelial tissues and 2.6% for hepatic tissues in human).[321]
Another allometric scaling approach termed “metabolically supported functional scaling” relies on the assumption that multiorgan systems can maintain in vivo cellular basal metabolic rates by ensuring the underlying prerequisite of limiting nutrient supply to cells.[308,321] An adipose-vascular two-organ system demonstrated that dispersed adipose cells (scaled with surface area) exhibit significantly higher glucose uptake than that of intact spheroids (scaled with volume) due to the difference in mass transport rates.[322] The “functional scaling” approach first defines the major function and specifies the functional parameter for each organ, such as heart (volume pumping), lungs (gas exchange), liver (metabolism), and kidneys (molecular filtering and transport).[307] However, some organs often carry out multiple functions, such as the brain, which does not just function as a neurovascular unit with a blood–brain-barrier; therefore, scaling a multiorgan system requires a multifunctional scaling approach or multivariate optimization. Gut-liver, liver-skin, and gut-liver-kidney multiorgan-on-a-chip devices successfully illustrated the use of a multifunctional scaling approach and mechanistic model in studying the PK of drugs after oral administration by specifying multiple objective parameters.[309,323] This multifunctional scaling algorithm defines the objective function as a weighted squared difference between a model outcome and the corresponding measurements.
The development of multiorgan-on-a-chip devices can bridge the gap between in vitro testing and animal or human models for drug screening applications. Sung et al.[307] demonstrated the relationship between in vivo (human or animals), in silico (mathematical models), and in vitro multiorgan-on-a-chip platforms (Figure 9C). Miniaturized organ-on-a-chip systems are ideal for reproducing microscale tissue structures of each organ, a strategy for correctly scaling between organ-on-a-chip systems and the human body is needed to build accurate multiorgan pharmacological models.[324] Interpretation of experimental data obtained from in vitro models and in vivo translation of the data requires appropriate mathematical modeling platforms to promote the development of pharmacology and pharmaceutical industry.[325] Multiorgan systems devices with interconnecting microchambers and microchannels enable precise manipulation of fluid flow to replicate blood circulation. The fluidic interconnections among organ models representing human responses depend on both single-organ functions reproduced in vitro and physiological relevance of organ–organ relationships embedded in the device design.[326] Multiorgan systems can mimic tissue–tissue interactions and provide a platform capable of simulating human metabolism with high authenticity, including the conversion of a prodrug to its effective metabolite as well as its subsequent therapeutic actions and toxic side effects.[327] Moreover, multiorganoid body-on-a-chip systems based on stem cells are positioned to be deployed for drug screening and are rapidly advancing as a gateway for individualized precision medicine.[328] As they can estimate efficacy and dose response, these multiorgan or multiorganoid systems have the potential to improve the drug development process before entering the expensive phase of clinical trials.[329]
In particular, the pharmacokinetic–pharmacodynamic (PK–PD) modeling technique and pharmacology approach for IVIV translation have been successfully applied as a mathematical modeling platform to analyze and predict the behavior and action of drugs in multiorgan-on-a-chip.[330,331] Lee et al.[303] has summarized the current status of organ-on-a-chip technology and microfluidic whole-body models for pharmacokinetic drug toxicity screening (Figure 9D). Drug bioavailability is governed by a complex process of absorption, distribution, metabolism, and excretion collectively known as ADME. PK is the study of time-dependent drug concentration, and PK modeling is based on the mass balance of drugs and their metabolites for optimizing drug formulation and dose as well as predicting toxicity and efficacy during drug development.[332] Physiologically based pharmacokinetic (PBPK) models are based on the actual physiology and anatomy of the human body and provide a mechanistic basis that better represents the drug mechanism of action and the effects elicited in multiorgan systems.[333] Meanwhile, PD modeling is based on empirical or mechanistic models that describe the action of drugs at the target site for verifying their effects.[334] Coupling PK, PBPK, PD, and PK–PD models has been frequently attempted to predict the pharmacological effect of a drug based on the administered dosage in multiorgan or multiorganoid systems. There are also several commercial PK–PD modeling tools, such as PK-Sim,[335] Simcyp,[336] and MATLAB/SimBiology,[337] that can be easily adapted to multiorgan systems. Michael L. Shuler[317] proposed a more refined approach based on the principles of PBPK modeling and derived parametric criteria to establish two different platforms that are involved in different stages of drug development; the μOrgan-on-a-chip platform allows only the extraction of PBPK parameters, while μHuman-on-a-chip allows direct simulation of drug concentration PK profiles in the human body.[307]
Multiscale modeling is particularly attractive for combining lumped (compartmental) and distributed (spatiotemporal) first principles-based mathematical models for capturing the intricate biophysical details of selected organs or tissues.[257] Multiscale computational modeling is also required for simulating organ-on-a-chip devices with complex geometries or studying cell/tissue-scale structures embedded in microdevices.[338] For example, detailed simulation of cellular spheroids or organoids integrated in a multiorgan-on-a-chip would simultaneously perform simulation of intraspheroid drug/metabolite transport and spheroid-medium exchange and requires several spheroidal objects embedded in the interconnecting microchamber medium pool.[339] Another example of multiscale mathematical modeling of liver-on-a-chip with complex geometry and zonation was shown to simulate fluid flow, oxygen transport and consumption, drug transport and intrinsic clearance, hepatocyte-specific metabolic pathways and fluxes, and enzyme kinetic mechanisms.[340,341] The recent progress in the development of multiorgan- or multiorganoid-on-a-chip appears promising and there is an active movement toward commercialization in collaboration with the pharmaceutical industry. Nevertheless, mathematical modeling platforms for multiorgan systems need further improvement and validation with increasing complexity. Quantitative systems pharmacology also relies on state-of-the-art computational methods and algorithms, such as machine learning, artificial intelligence, and cloud computing, and can be combined with multiorgan-on-a-chip technology in the future.[257,342] There are already cases of using deep learning for automated analysis of vascularization images or Bayesian algorithms for parameter estimation.[343] Development of hardware for organ-on-a-chip should be accompanied by a corresponding development of mathematical modeling techniques for designing and interpreting the systems.
4. Concluding Remarks
Organoids and organ-on-a-chip technology has the potential to disrupt the traditional drug development process by replacing animal models and/or significantly improving the outcomes of animal and clinical studies. Several organizations have made initiatives to promote organoid- and organ-on-a-chip related research. For instance, the Food and Drug Administration (FDA), National institutes of Health (NIH), and Defense Advanced Research Projects Agency (DARPA) of the USA are jointly supporting and funding the “Tissue Chip for Drug Screening” and “Tissue Chip in Space” programs that develop organ-on-a-chip devices for evaluating drug therapies and other national security purposes.[344,349] The European Union has also awarded funding to five organizations pursuing research on body-on-a-chip devices.[350] These massive investments in new tools with better predictive capabilities demonstrate the potential demand—and challenges—for reducing attrition rates, preclinical costs, and time-to-market.
Organ-on-a-chip microsystems represent a significant advancement but there remain technical and entrepreneurial challenges. The overarching consideration for organ-on-a-chip development is controlling the balance between complexity (to improve physiological relevance) and practicality (by focusing on practical operation and management). Focusing on the most important features of different diseases, simple albeit effective disease-on-a-chip models can be developed for studying microvascular diseases such as sickle cell disease,[351–352] microaneurysm in diabetic retinopathy,[342] etc. On the other hand, one of the critical technical challenges arises from the difficulty to generate and control physiologically relevant structural, biochemical, and mechanical cues that ensure more reliable and sustainable sources of human primary cells or stem cells. Major technical hurdles include the difficulty of developing downstream and online high-resolution biochemical analysis as they are incompatible with current measurement techniques. Furthermore, current laboratory prototyping PDMS-based fabrication techniques are not feasible for large-scale manufacturing of organ-on-a-chip for industrial applications.[244]
Organoids, self-organized organ-like cell aggregates that originate from multipotent stem cells, are emerging as a promising tool for understanding human development processes and disease progression. Organoids resemble small units of their organ of origin and accurately recapitulate tissue architecture and in vivo behavior. However, there are also limitations and major challenges to current fabrication technologies, such as uncontrollable size, lack of vascularization, poor reproductively, and inadequate complexity of organoids. Consequently, the architectural organization, maturation status, and functionality of organoids is not yet at in vivo levels due to limitations of insufficient nutrient and oxygen diffusion and a lack of controlled cell–cell and cell–matrix interactions. Advances in biomimetic hydrogels and 3D bioprinting will allow us to culture organoids or organoid-derived tissue constructs with well-distributed, interconnected vascular networks and highly defined spatial control. Organoid developmental processes coordinate multicellular communication, growth, and maturation to achieve physiological functionality and provide reliable, rapid, and cost-effective results for drug discovery and screening.
The strategic integration between organoids and organ-on-a-chip can address the limitations of each approach and provide a path toward a superior, synergistic strategy of constructing tissues. Advances in microfluidic organ-on-a-chip approaches allow us to engineer organoids with essential structural and physiological features in a controlled manner. The synergistic engineering of organoid-on-a-chip leads to more versatile and predictive preclinical models that may truly deliver on the promise of regenerative and precision medicine. Although many emerging opportunities lie ahead, organoid-on-a-chip is a nascent technology that also faces predictable challenges and limitations. Advanced engineering techniques (e.g., 3D printing) may hold the key for recapitulating 3D tissue architecture and physiology as well as facilitating better nutrient and gas exchange in a microengineered organoid platform. Typically, organoid-on-a-chip models have limited ability to recapitulate the dynamic environmental, structural, and functional changes that occur during organogenesis due to their predetermined design and construction manner. Addressing this limitation will require efforts to fully understand the spatiotemporal dynamics of organ development and achieve high-fidelity 3D stem cell-derived organoids or microtissues. Moreover, hydrogel materials (e.g., Matrigel) suffer from poorly defined compositions and exhibit batch-to-batch variability, which hinders environmental controllability and may be problematic for providing 3D structural support and proper morphogenesis. Addressing this problem will also require ongoing efforts to engineer new types of biomaterials with well-defined and tunable properties for organoid or microtissue culture. The convergence of the two approaches to produce multiorganoids-on-a-chip or human organoids-on-a-chip is emerging as a new direction for building 3D models with higher physiological relevance. Additional bioengineering approaches, such as live imaging, genome editing, and single-cell genomics, may also be incorporated into organoid-on-a-chip systems to study human physiology, diseases, and organogenesis and achieve personalized medicine.
3D printing is an ideal technology for building flexible, complex, monolithic devices, and creating organ-level biological architectures with precise 3D cell patterning and biomaterial heterogeneity. The convergence of 3D printing with microsystems aims to provide future strategies for more efficient, automated, modularly integrated, higher-throughput, and customizable organ-on-a-chip devices. The integration of 3D printing and human organoid-on-a-chip devices can lead to the next generation of 3D models with more precise self-organization and spatiotemporal control of the microenvironment. However, experiments alone cannot provide full insights into the biochemical, biophysical, and biomechanical processes that affect cell growth and behavior in microphysiological systems. Computational and mathematical modeling, also referred to as numerical simulation, can provide additional theoretical information and predict multiple properties of different underlying processes. Numerical simulation is also a powerful tool that can help obtain accurate and satisfactory results and reduce the costs and time associated with repetitive experimental measurements. Numerical simulation of the complex behavior of self-assembled organoids faces challenges arising from cell spatiotemporal organization, vascularization, shape deformation, morphogenesis, reproducibility, and guidance toward desired formation and function. Organ-on-a-chip combined with numerical simulation is suitable for simulating the optimization and validation of chip design, nutrient consumption and transduction, oxygen concentration and pressure distribution, shear stress, and flow field among others. Multiorganoids and multiorgan-on-a-chip or human-on-a-chip are more suitable for modeling multifunctional allometric scaling, interconnection via vascularization and innervation, long-term drug transport, and PK and PD effects. These specific advances can be leveraged to address major technical challenges by introducing modeling and simulation as an integrated part of 3D printing-assisted organoids and organ-on-a-chip platforms.
This massive investment in new research tools with better predictive capabilities demonstrates the potential demand and challenges for reducing attrition rates, preclinical costs, and time-to-market. Several companies have shown great interest in microphysiological system models as they offer an alternative to animal studies and reduce the ethical issues associated with drug testing. The adoption of organoids and organ-on-a-chip models has huge potential for a variety of commercial applications and can reduce failures in animal studies and clinical trials by identifying ineffective or unsafe drugs earlier. The synergistic combination of 3D bioprinting, organoids, and organ-on-a-chip has the potential to overcome the limitations and controversies of more traditional preclinical models and can offer an exciting new avenue for improving them. This integration has become more popular but still needs sophisticated bioprinting techniques capable of biofabrication in a scalable, accurate, rapid, and high-throughput manner to overcome challenges associated with resolution, bioink materials, and the limited coprint ability. In addition, integrated bioprinted tissues with organoids and organ-on-a-chip should exhibit proper tissue function, durability, and miniaturization to minimize the time and cost of fabrication. Coaxial bioprinting may also be used to better replicate vasculature and other vessels to study and understand complex organ systems. Moreover, advanced biosensing and diagnostic technologies can be used to significantly increase the prediction accuracy of capturing and measuring metabolites by providing a dynamic and controlled environment. Furthermore, iPSCs and their ability to differentiate into many cell types and self-assembly into organoids have opened up new avenues for achieving robust and reproducible personalized tissue constructs. Multiple miniaturized tissues or organoids can also be connected on a single organ-on-a-chip model to mimic the complexity of tissue function and responses with physiologically relevant flow rates and shear stress values. Establishing robust, patient-specific disease-on-a-chip and human-on-a-chip platforms, bioprinted organ-on-a-chip and human multiorganoids- or multiorgan-on-a-chip will lead the next generation of commercial devices for physiological research, diagnosis, drug screening, and personalized treatment.
Acknowledgements
F.Y.Z., Y.M.H.X. and H.L. contributed equally to this work. F.Y.Z. and Y.B.F. acknowledge support by the National Natural Science Foundation of China (NSFC) Grants (32001015), Beijing Science and Technology New Star Project (Z201100006820038), and Beihang Youth Top-notch Talent Support Program (YWF-20-BJ-J-1035 and YWF-21-BJ-J-1036), and Scientific Research Training Center for Chinese Astronauts Grants(SMFA15A02), and NSFC Grants (21635001, 81730087, 11827803 and 11421202) and Project supported by the National Key Scientific Instrument and Equipment Development Project (2013YQ190467). F.Y.Z. and M.D. acknowledge support by National Institutes of Health (NIH) Grants (U01HL114476 and R01HL121386). M.D. acknowledges support by National Institutes of Health (NIH) Grant (R01HL154150). The authors gratefully acknowledge Dr. Fang Kong and Megha Upadya for helpful discussions and suggestions.
Biography

Fuyin Zheng is currently Beijing Science and Technology New Star and an associate professor in Beijing Advanced Innovation Center for Biomedical Engineering at Beihang University from 2019. He received his Ph.D. degree in Biomedical Engineering at Southeast University in 2017. He also worked as a research scholar at the University of California, Berkeley, as a postdoctoral associate and fellow at Massachusetts Institute of Technology and Nanyang Technological University. He established the Bioinspired Organoids & Organ on a Chip (BOOC) lab, and his current scientific interests are focused on organoids and organ-on-a-chip, 3D and 4D bioprinting, structural color materials, and space medico-engineering.

Yuminghao Xiao is currently a Ph.D. student in the department of Naval Architecture and Marine Engineering at the University of Michigan, Ann Arbor. He obtained his bachelor’s degree in Mechanical Engineering from Huazhong University of Science and Technology (HUST). In 2018, he spent the summer working closely with Dr. Fuyin Zheng from the Nanomechanics Laboratory at Massachusetts Institute of Technology. His current scientific interests include Micro/Nano-fabrications of bio-mimetic microfluidic chips with additive manufacturing method and application of optimal control theory in path planning problems.

Hui Liu is currently a master student in the School of Biological Sciences and Medical Engineering and Beijing Advanced Innovation Center for Biomedical Engineering at Beihang University. She received her bachelor’s degree in Biomedical Engineering at Yanshan University in 2020. She participated in the research of brain regulation systems for mild brain cognitive impairment and face recognition system based on Python and OpenCV. She joined the Bioinspired Organoids & Organ on a Chip (BOOC) lab under Dr. Fuyin Zheng and Prof. Yubo Fan and focused on organoids and organ-on-a-chip (brain and heart), 3D & 4D bioprinting, and structural color materials.

Yubo Fan is currently the Cheung Kong Scholars distinguished professor in the School of Biological Sciences and Medical Engineering, Dean of the School of Engineering Medicine, Director of Beijing Advanced Innovation Center for Biomedical Engineering at Beihang University. He is also the Fellow of AIMBE, IAMBE, and FBSE. Professor Fan acquired his B.S. degree in Mechanics from Peking University in 1987 and Ph.D. in Biomechanics from Sichuan University of Science and Technology in 1992. His research areas include Biomaterials, Biomechanics, Mechanobiology, Rehabilitation Engineering and Organoids, etc. He has more than 300 peer-reviewed journal papers published in international academic journals.

Ming Dao Ming Dao is the Principal Investigator and Director of MIT’s Nanomechanics Laboratory, a Principal Research Scientist in the Department of Materials Science and Engineering at MIT. He is also a visiting professor in the School of Biological Sciences at Nanyang Technological University. He obtained his bachelor’s degree in engineering mechanics from Shanghai Jiao Tong University and his MS and Ph.D. degrees in materials science from the University of California at San Diego. His research interests include biomechanics and biophysics of the cell, microfluidic-based study of human diseases, nanomechanics of advanced materials, and machine learning for health and engineering applications.
Footnotes
Conflict of Interest
M.D. filed a patent application related to the technology reviewed in this paper.
Contributor Information
Fuyin Zheng, Key Laboratory for Biomechanics and Mechanobiology, Beijing Advanced Innovation Center for Biomedical Engineering, School of Biological Science and Medical Engineering, Beihang University, Beijing 100083, China; Department of Materials Science and Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; School of Biological Sciences, Nanyang Technological University, Singapore 639798, Singapore.
Yuminghao Xiao, Department of Mechanical Engineering, University of Michigan, Ann Arbor, MI 48109, USA.
Hui Liu, Key Laboratory for Biomechanics and Mechanobiology, Beijing Advanced Innovation Center for Biomedical Engineering, School of Biological Science and Medical Engineering, Beihang University, Beijing 100083, China.
Yubo Fan, Key Laboratory for Biomechanics and Mechanobiology, Beijing Advanced Innovation Center for Biomedical Engineering, School of Biological Science and Medical Engineering, Beihang University, Beijing 100083, China.
Ming Dao, Department of Materials Science and Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; School of Biological Sciences, Nanyang Technological University, Singapore 639798, Singapore.
References
- [1].Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE, Lab Chip 2012, 12, 2156. [DOI] [PubMed] [Google Scholar]
- [2].Karli G, Akhilesh G. Abhishek K,J, Biomaterials 2018, 14, 192. [Google Scholar]
- [3].Zheng F, Fu F, Cheng Y, Wang C, Zhao Y, Gu Z, Small 2016, 12, 2253. [DOI] [PubMed] [Google Scholar]
- [4].Jang KJ, Mehr AP, Hamilton GA, Mcpartlin LA, Chung S, Suh KY, Ingber DE, Integr. Biol 2013, 5, 1119. [DOI] [PubMed] [Google Scholar]
- [5].Huh D, Hamilton GA, Ingber DE, Trends. Cell. Biol 2011, 21, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu A, Zhang Q, Lambert G, Khin Z, Gatenby RA, Kim HJ, Pourmand N, Bussey K, Davies PC, Sturm JC, Proc. Natl. Acad. Sci. USA 2015, 112, 10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhuang P, Sun AX, An J, Chua CK, Chew SY, Biomaterials 2017, 154, 113. [DOI] [PubMed] [Google Scholar]
- [8].Hopkins AM, Desimone E, Chwalek K, Kaplan DL, Prog. Neurobiol 2015, 125, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatsura T, Oncol. Rep 2015, 33, 1837. [DOI] [PubMed] [Google Scholar]
- [10].Ahadian S, Civitarese R, Bannerman D, Mohammadi MH, Lu R, Wang E, Davenport-Huyer L, Lai B, Zhang B, Zhao Y, Adv. Healthcare Mater 2018, 7, 1700506. [DOI] [PubMed] [Google Scholar]
- [11].Zhang B, Korolj A, Lai BFL, Radisic M, Nat. Rev. Mater 2018, 3, 257. [Google Scholar]
- [12].Ashammakhi N, Darabi MA, Çelebi-Saltik B, Tutar R, Hartel MC, Lee J, Hussein SM, Goudie MJ, Cornelius MB, Dokmeci MR, Khademhosseini A, Small Methods 2020, 4, 1900589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Oleaga C, Lavado A, Riu A, Rothemund S, Carmona-Moran CA, Persaud K, Yurko A, Lear J, Narasimhan NS, Long CJ, Adv. Funct. Mater 2019, 29, 1970049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kratz SRA, Höll G, Schuller P, Ertl P, Rothbauer M, Biosensors 2019, 9, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao Y, Kankala RK, Wang S-B, Chen A-Z, Molecules 2019, 24, 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Takebe T, Zhang B, Radisic M, Cell Stem Cell 2017, 21, 297. [DOI] [PubMed] [Google Scholar]
- [17].Drost J, Clevers H, Nat. Rev. Cancer 2018, 18, 407. [DOI] [PubMed] [Google Scholar]
- [18].Kratochvil MJ, Seymour AJ, Li TL, Paşca SP, Kuo CJ, Heilshorn SC, Nat. Rev. Mater 2019, 4, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lou Y-R, Leung AW, Biotechnol. Adv 2018, 36, 132. [DOI] [PubMed] [Google Scholar]
- [20].Silva TP, Cotovio JP, Bekman E, Carmo-Fonseca M, Cabral J, Fernandes TG, Stem Cells Int 2019, 4508470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon K, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Tang H, Song H, Ming G, Cell 2016, 165, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rossi G, Manfrin A, Lutolf MP, Nat. Rev. Genet 2018, 19, 671. [DOI] [PubMed] [Google Scholar]
- [23].Akhtar AA, Sances S, Barrett R, Breunig JJ, Curr. Stem Cell Rep 2017, 3, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu H, Wang Y, Cui K, Guo Y, Zhang X, Qin J, Adv. Mater 2019, 31, 1902042. [DOI] [PubMed] [Google Scholar]
- [25].Takebe T, Zhang B, Radisic M, Cell Stem Cell 2017, 21, 297. [DOI] [PubMed] [Google Scholar]
- [26].Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J, Nikolova M, Cora V, Antkowiak L, Haq W, Elife 2019, 8, e46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jackson EL, Lu H, Integr. Biol 2016, 8, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang Y, Wang L, Guo Y, Zhu Y, Qin J, RSC Adv 2018, 8, 1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tao T, Wang Y, Chen W, Li Z, Su W, Guo Y, Deng P, Qin J, Lab Chip 2019,19, 948. [DOI] [PubMed] [Google Scholar]
- [30].Jin Y, Kim J, Lee JS, Min S, Kim S, Ahn DH, Kim YG, Cho SW, Adv. Funct. Mater 2018, 28, 1801954. [Google Scholar]
- [31].Dekkers JF, Alieva M, Wellens LM, Ariese HC, Jamieson PR, Vonk AM, Amatngalim GD, Hu H, Oost KC, Snippert HJ, Nat. Protoc 2019, 14, 1756. [DOI] [PubMed] [Google Scholar]
- [32].Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O, Cell Stem Cell 2016, 18, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, Massa S, Riahi R, Chae S, Hu N, Proc. Natl. Acad. Sci. USA 2017, 114, E2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bashir R, Adv. Drug Delivery Rev 2004, 56, 1565. [DOI] [PubMed] [Google Scholar]
- [35].Whitesides GM, Nature 2006, 442, 368. [DOI] [PubMed] [Google Scholar]
- [36].Yazdi AA, Popma A, Wong W, Nguyen T, Pan Y, Xu J, Microfluid. Nanofluid 2016, 20, 50. [Google Scholar]
- [37].Sochol RD, Sweet E, Glick CC, Wu S-Y, Yang C, Restaino M, Lin L, Microelectron. Eng 2018, 189, 52. [Google Scholar]
- [38].Liu X, Yuk H, Lin S, Parada GA, Tang TC, Tham E, de la Fuente-Nunez C, Lu TK, Zhao X, Adv. Mater 2018, 30, 1704821. [DOI] [PubMed] [Google Scholar]
- [39].Pourmand A, Shaegh SAM, Ghavifekr HB, Aghdam EN, Dokmeci MR, Khademhosseini A, Zhang YS, Sens. Actuators, B 2018, 262, 625. [Google Scholar]
- [40].Bhargava KC, Thompson B, Malmstadt N, Proc. Natl. Acad. Sci. USA 2014, 111, 15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee KG, Park KJ, Seok S, Shin S, Kim DH, Park JY, Yun SH, Lee SJ, Lee TJ, Rsc. Adv 2014, 4, 32876. [Google Scholar]
- [42].Elçin YM, Stem Cell Rev 2017, 13, 319. [DOI] [PubMed] [Google Scholar]
- [43].Jian H, Wang M, Wang S, Wang A, Bai S, Bio-Des. Manuf 2018, 1, 45. [Google Scholar]
- [44].Park JY, Jang J, Kang H-W, Microelectron. Eng 2018, 200, 1. [Google Scholar]
- [45].Yang Q, Lian Q, Xu F, Biomicrofluidics 2017, 11, 031301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Montes-Olivas S, Marucci L, Homer M, Front. Genet 2019, 10, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Barisam M, Saidi M, Kashaninejad N, Vadivelu R, Nguyen N-T, Micromachines 2017, 8, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang M, Zheng A, Zheng ZC, Wang MZ, Proc. Inst. Mech. Eng., Part H 2019, 233, 432. [DOI] [PubMed] [Google Scholar]
- [49].Jodat YA, Kang MG, Kiaee K, Kim GJ, Martinez AF, Rosenkranz A, Bae H, Shin SR, Curr. Pharm. Des 2018, 24, 5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hadley J, Animal Studies J 2020, 9, 216. [Google Scholar]
- [51].Yesil-Celiktas O, Hassan S, Miri AK, Maharjan S, Al-kharboosh R, Quiñones-Hinojosa A, Zhang YS, Adv. Biosyst 2018, 2, 1800109. [Google Scholar]
- [52].Jones JR, Zhang S-C, Curr. Opin. Biotech 2016, 40, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stefan W, Frank S, Augustinus B, Drug. Metab. Dispos 2003, 31, 1035.12867492 [Google Scholar]
- [54].Régis B, Matthieu PJ, Grégory N, Jessy B, Céline B, Cécile L, Henri B, Eric L, Xenobiotica 2013, 43, 140.22830982 [Google Scholar]
- [55].Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui BX, Gold JD, Wu JC, Nat. Methods 2014, 11, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Churko JM, Burridge PW, Wu JC, Methods Mol. Biol 2013, 1036, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hughes DJ, Kostrzewski T, Sceats EL, Exp. Biol. Med 2017, 242, 1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ad VDM, Van DBA, Integr. Biol 2012, 4, 461. [Google Scholar]
- [59].Chen MB, Suthan S, Wheeler AR, Simmons CA, Lab Chip 2013, 13, 2591. [DOI] [PubMed] [Google Scholar]
- [60].Huh D, Matthews BD, Mammoto A, Montoyazavala M, Hong YH, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kim HJ, Huh D, Hamilton G, Ingber DE, Lab Chip 2012, 12, 2165. [DOI] [PubMed] [Google Scholar]
- [62].Grosberg A, Alford PW, Mccain ML, Parker KK, Lab Chip 2011, 11, 4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ho CT, Lin RZ, Chen RJ, Chin CK, Gong SE, Chang HY, Peng HL, Hsu L, Yew TR, Chang SF, Lab Chip 2013, 13, 3578. [DOI] [PubMed] [Google Scholar]
- [64].Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, Biomicrofluidics 2015, 9, 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Miller PG, Shuler ML, Biotechnol. Bioeng 2016, 113, 2213. [DOI] [PubMed] [Google Scholar]
- [66].Pan C, Kumar C, Bohl S, Klingmueller U, Mann M, Mol. Cell. Proteomics 2009, 8, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Midwoud PM, Van MTM, Elisabeth V, Groothuis GMM, Lab Chip 2010, 10, 2778. [DOI] [PubMed] [Google Scholar]
- [68].Li X, Brooks JC, Hu J, Ford KI, Easley CJ, Lab Chip 2016, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Luni C, Serena E, Elvassore N, Curr. Opin. Biotechnol 2014, 25, 45. [DOI] [PubMed] [Google Scholar]
- [70].Verpoorte E, Oomen PE, Skolimowski MD, Mulder PPMFA, Midwoud PMV, Starokozhko V, Merema MT, Molema G, Groothuis GMM, presented at Transducers-Int. Conf. Solid-state Sensors, Anchorage, Alaska, June 2015. [Google Scholar]
- [71].Fatehullah A, Si HT, Barker N, Nat. Cell Biol 2016, 18, 246. [DOI] [PubMed] [Google Scholar]
- [72].Yin X, Mead B, Safaee H, Langer R, Karp J, Levy O, Cell Stem Cell 2016, 18, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wagner W, Ho AD, Stem. Cell Rev 2007, 3, 239. [DOI] [PubMed] [Google Scholar]
- [74].Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM, Science 1998, 282, 1145. [DOI] [PubMed] [Google Scholar]
- [75].Eiraku M, Sasai Y, Nature 2011, 472, 51.21475194 [Google Scholar]
- [76].Bredenoord AL, Clevers H, Knoblich JA, Science 2017, 355, eaaf9414. [DOI] [PubMed] [Google Scholar]
- [77].Marina B, Kriegstein AR, Cell 2013, 155, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Scott CW, Peters MF, Dragan YP, Toxicol. Lett 2013, 219, 49. [DOI] [PubMed] [Google Scholar]
- [79].Snyder JE, Hamid Q, Wang C, Chang R, Emami K, Wu H, Sun W, Biofabrication 2011, 3, 034112. [DOI] [PubMed] [Google Scholar]
- [80].Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, Saw TY, Tan CP, Lokman H, Cell Stem Cell 2016, 19, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vyas D, Baptista PM, Brovold M, Moran E, Gaston B, Booth C, Samuel M, Atala A, Soker S, Hepatology 2018, 67, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Magro-Lopez E, Palmer C, Manso J, Liste I, Zambrano A, Stem. Cell Res. Ther 2018, 9, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE, Cell Mol. Gastroenterol. Hepatol 2018, 5, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bang S, Na S, Jang JM, Kim J, Jeon NL, Adv. Healthcare Mater 2016, 5, 159. [DOI] [PubMed] [Google Scholar]
- [85].Bhatia SN, Ingber DE, Nat. Biotechnol 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- [86].Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Sci. Rep 2018, 8, 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jang KJ, Cho HS, Kang DH, Bae WG, Kwon TH, Suh KY, Integr. Biol 2011, 3, 134. [DOI] [PubMed] [Google Scholar]
- [88].Booth R, Kim H, Lab Chip 2012, 12, 1784. [DOI] [PubMed] [Google Scholar]
- [89].Huh D, Fujioka H, Tung YC, Futai N, Grotberg JB, Takayama S, Proc. Natl. Acad. Sci. USA 2007, 104, 18886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tavana H, Zamankhan P, Christensen PJ, Grotberg JB, Takayama S, Biomed. Microdevices 2011, 13, 731. [DOI] [PubMed] [Google Scholar]
- [91].Mori N, Morimoto Y, Takeuchi S, Biomaterials 2017, 116, 48. [DOI] [PubMed] [Google Scholar]
- [92].Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA, Nat. Biotechnol 2017, 35, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Guan Y, Xu D, Garfin PM, Ehmer U, Hurwitz M, Enns G, Michie S, Wu M, Zheng M, Nishimura T, JCI Insight 2017, 2, e94954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Marx V, Nat. Methods 2014, 11, 483. [DOI] [PubMed] [Google Scholar]
- [95].Masters JR, Stacey GN, Nat. Protoc 2007, 2, 2276. [DOI] [PubMed] [Google Scholar]
- [96].Artegiani B, Clevers H, Hum. Mol. Genet 2018, 27, R99. [DOI] [PubMed] [Google Scholar]
- [97].Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, Bergren SK, Pietzak EJ, Anderson CB, Benson MC, Coleman JA, Taylor BS, Abate-Shen C, McKiernan JM, Al-Ahmadie H, Solit DB, Shen MM, Cell 2018, 173, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Boj SF, Hwang CI, Baker LA, Chio I. I.n C., Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Öhlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FHM, Molenaar IQ, Rinkes IHB, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RGJ, Clevers H, Tuveson DA, Cell 2015, 160, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, Bronsveld I, van de Graaf EA, Nieuwenhuis EES, Houwen RHJ, Vleggaar FP, Escher JC, de Rijke YB, Majoor CJ, Heijerman HGM, de Winter-de Groot KM, Clevers H, van der Ent CK, Beekman JM, Sci. Transl. Med 2016, 8, 344ra84. [DOI] [PubMed] [Google Scholar]
- [100].Mina O, Shinichiro O, Bear CE, Saumel A, Stephanie C, Bin L, Markus G, Gordon K, Kamath BM, Anand G, Nat. Biotechnol 2015, 33, 853. [DOI] [PubMed] [Google Scholar]
- [101].Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, Nat. Med 2012, 18, 618. [DOI] [PubMed] [Google Scholar]
- [102].Schwank G, Koo B-K, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EES, Beekman JM, Clevers H, Cell Stem Cell 2013, 13, 653. [DOI] [PubMed] [Google Scholar]
- [103].Woo DH, Chen Q, Yang TB, Glineburg MR, Hoge C, Leu NA, Johnson FB, Lengner CJ, Cell Stem Cell 2016, 19, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Deng WL, Gao ML, Lei XL, Lv JN, Zhao H, He KW, Xia XX, Li LY, Chen YC, Li YP, Pan D, Xue T, Jin Z-B, Stem Cell Rep 2018, 10, P1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ, Integr. Biol 2011, 3, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Montanez-Sauri SI, Sung KE, Berthier E, Beebe DJ, Integr. Biol 2013, 5, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lang JD, Berry SM, Powers GL, Beebe DJ, Alarid ET, Integr. Biol 2013, 5, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li CY, Wood DK, Huang JH, Bhatia SN, Lab Chip 2013, 13, 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Grosberg A, Nesmith AP, Goss JA, Brigham MD, McCain ML, Parker KK, J. Pharmacol. Toxicol. Methods 2012, 65, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Xu Z, Gao Y, Hao Y, Li E, Wang Y, Zhang J, Wang W, Gao Z, Wang Q, Biomaterials 2013, 34, 4109. [DOI] [PubMed] [Google Scholar]
- [111].Sara P, Eftalda B, Harley BA, Adv. Mater 2015, 27, 1567. [DOI] [PubMed] [Google Scholar]
- [112].Agarwal A, Goss JA, Cho A, Mccain ML, Parker KK, Lab Chip 2013, 13, 3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wang G, Mccain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Nat. Med 2014, 20, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Megan M, Sean LS, Anna PG, Josue G, Kevin Kit AP, Proc. Natl. Acad. Sci. USA 2013, 110, 9770.23716679 [Google Scholar]
- [115].Shim KY, Lee D, Han J, Nguyen NT, Park S, Sung JH, Biomed. Microdevices 2017, 19, 37. [DOI] [PubMed] [Google Scholar]
- [116].Sobrino A, Phan DTT, Datta R, Wang X, Hachey SJ, Romerolópez M, Gratton E, Lee AP, George SC, Hughes CCW, Sci. Rep 2016, 6, 31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Mahler GJ, Esch MB, Glahn RP, Shuler ML, Biotechnol. Bioeng 2010, 104, 193. [DOI] [PubMed] [Google Scholar]
- [118].Wevers NR, Vught RV, Wilschut KJ, Nicolas A, Chiang C, Lanz HL, Trietsch SJ, Joore J, Vulto P, Sci. Rep 2016, 6, 38856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Park SE, Georgescu A, Huh D, Science 2019, 364, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Xu H, Jiao Y, Qin S, Zhao W, Chu Q, Wu K, Exp. Hematol. Oncol 2018, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Grebenyuk S, Ranga A, Front. Bioeng. Biotechnol 2019, 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Poznic M, Philos. Technol 2016, 29, 357. [Google Scholar]
- [123].Sateesh J, Guha K, Dutta A, Sengupta P, Rao KS, Microsyst. Technol 2019, 25, 2553. [Google Scholar]
- [124].Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE, Cell. Mol. Gastroenterol. Hepatol 2018, 5, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Young AT, Rivera KR, Erb PD, Daniele MA, ACS Sens 2019, 4, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Skardal A, Murphy SV, Devarasetty M, Mead I, Kang H-W, Seol Y-J, Zhang Y.u S., Shin S.u-R., Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Lee SJ, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, Atala A, Sci. Rep 2017, 7, 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Singh A, Nikkhah M, Annabi N, Biomaterials 2019, 198, 1. [DOI] [PubMed] [Google Scholar]
- [128].Yu F, Hunziker W, Choudhury D, Micromachines 2019, 10, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Haddrick M, Simpson PB, Drug Discovery Today 2019, 24, 1217. [DOI] [PubMed] [Google Scholar]
- [130].Ramme AP, Koenig L, Hasenberg T, Schwenk C, Magauer C, Faust D, Lorenz AK, Krebs A-C, Drewell C, Schirrmann K, Vladetic A, Lin G-C, Pabinger S, Neuhaus W, Bois F, Lauster R, Marx U, Dehne E-M, Future Sci. OA 2019, 5, FSO413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].van den Berg A, Mummery CL, Passier R, van der Meer AD, Lab Chip 2019, 19, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sontheimer-Phelps A, Hassell BA, Ingber DE, Nat. Rev. Cancer 2019, 19, 65. [DOI] [PubMed] [Google Scholar]
- [133].Skardal A, Shupe T, Atala A, Drug Discovery Today 2016, 21, 1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wang Y, Wang L, Zhu Y, Qin J, Lab Chip 2018, 18, 851. [DOI] [PubMed] [Google Scholar]
- [135].Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, Hamilton GA, Targan SR, Svendsen CN, Barrett RJ, Cell. Mol. Gastroenterol. Hepatol 2018, 5, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Yoshiki S, Nature 2013, 493, 318.23325214 [Google Scholar]
- [137].Dingal PCDP, Discher DE, Nat. Mater 2014, 13, 532. [DOI] [PubMed] [Google Scholar]
- [138].Lane SW, Williams DA, Watt FM, Nat. Biotechnol 2014, 32, 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Li W, Tang QY, Jadhav AD, Ankit N, Qian WX, Shi P, Pang SW, Sci. Rep 2015, 5, 8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ, Lee SH, Lab Chip 2014, 15, 141. [DOI] [PubMed] [Google Scholar]
- [141].Wang Y, Wang L, Guo Y, Zhu Y, Qin J, Rsc. Adv 2018, 8, 1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Sakaguchi K, Shimizu T, Horaguchi S, Sekine H, Yamato M, Umezu M, Okano T, Sci. Rep 2013, 3, 1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A, Biomaterials 2016, 110, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min YS, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Nature 2017, 545, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, Avci H, Zhang W, Silvestri A, Sanati Nezhad A, Manbohi A, De Ferrari F, Polini A, Calzone G, Shaikh N, Alerasool P, Budina E, Kang J, Bhise N, Ribas J, Pourmand A, Skardal A, Shupe T, Bishop CE, Dokmeci MR, Atala A, Khademhosseini A, Proc. Natl. Acad. Sci. USA 2017, 114, E2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Devarasetty M, Forsythe S, Shupe TD, Soker S, Bishop CE, Atala A, Skardal A, Biosensors 2017, 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Zhu Y, Wang L, Yu H, Yin F, Wang Y, Liu H, Jiang L, Qin J, Lab Chip 2017, 17, 2941. [DOI] [PubMed] [Google Scholar]
- [148].Javaid M, Haleem A, Clin. Epidemiol. Global Health 2020, 8, 586. [Google Scholar]
- [149].Wang X, Micromachines 2019, 10, 814. [Google Scholar]
- [150].Saygili E, Dogan-Gurbuz AA, Yesil-Celiktas O, Draz MS, Bioprinting 2020, 18, e00071. [Google Scholar]
- [151].Matai I, Kaur G, Seyedsalehi A, McClinton A, Laurencin CT, Biomaterials 2020, 226, 119536,. [DOI] [PubMed] [Google Scholar]
- [152].Au AK, Huynh W, Horowitz LF, Folch A, Angew. Chem., Int. Ed 2016, 55, 3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, Groll J, Hutmacher DW, Adv. Mater 2013, 25, 5011. [DOI] [PubMed] [Google Scholar]
- [154].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [155].Hwang HH, Zhu W, Victorine G, Lawrence N, Chen SC, Small Methods 2018, 2, 1700277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bertsch A, Bernhard P, Vogt C, Renaud P, Rapid. Prototyping. J 2000, 6, 259. [Google Scholar]
- [157].Bouge R, Assem. Autom 2013, 33, 307. [Google Scholar]
- [158].Ho CM, Ng SH, Li KH, Yoon YJ, Lab Chip 2015, 15, 3627. [DOI] [PubMed] [Google Scholar]
- [159].Yi L, Suhali MG, J. Biomed. Mater. Res. A 2010, 77A, 396. [DOI] [PubMed] [Google Scholar]
- [160].Melchels FPW, Feijen J, Grijpma DW, Biomaterials 2010, 31, 6121. [DOI] [PubMed] [Google Scholar]
- [161].Waldbaur A, Anal. Methods 2011, 3, 2681. [Google Scholar]
- [162].Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S, Trends. Biotechnol 2015, 33, 504. [DOI] [PubMed] [Google Scholar]
- [163].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [164].Kang HW, Sang JL, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [165].Frohlich EM, Zhang X, Charest JL, Integr. Biol 2012, 4, 75. [DOI] [PubMed] [Google Scholar]
- [166].Sun H, Jia Y, Dong H, Dong D, Zheng J, Curr. Opin. Chem. Eng 2019, 28, 1. [Google Scholar]
- [167].Hassan S, Heinrich M, Cecen B, Prakash J, Zhang YS, Biomaterials for Organ and Tissue Regeneration: New Technologies and Future Prospects, Elsevier, Amsterdam: 2020, p. 669. [Google Scholar]
- [168].Hou X, Zhang YS, Trujillo-de Santiago G, Alvarez MM, Ribas J, Jonas SJ, Weiss PS, Andrews AM, Aizenberg J, Khademhosseini A, Nat. Rev. Mater 2017, 2, 17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Miri AK, Mostafavi E, Khorsandi D, Hu S-K, Malpica M, Khademhosseini A, Biofabrication 2019, 11, 042002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Jang J, Park JY, Gao G, Cho D-W, Biomaterials 2018, 156, 88. [DOI] [PubMed] [Google Scholar]
- [171].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [172].Kesti M, Müller M, Becher J, Schnabelrauch M, D’Este M, Eglin D, Zenobi-Wong M, Acta Biomater 2015, 11, 162. [DOI] [PubMed] [Google Scholar]
- [173].Blondela D, Lutolf MP, Chimia 2019, 73, 81. [DOI] [PubMed] [Google Scholar]
- [174].Das S, Pati F, Choi Y-J, Rijal G, Shim J-H, Kim SW, Ray AR, Cho D-W, Ghosh S, Acta Biomater 2015, 11, 233. [DOI] [PubMed] [Google Scholar]
- [175].Pati F, Jang J, Ha D-H, Kim SW, Rhie J-W, Shim J-H, Kim D-H, Cho D-W, Nat. Commun 2014, 5, 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kratochvil MJ, Seymour AJ, Li TL, Pasca SP, Kuo CJ, Heilshorn SC, Nat. Rev. Mater 2019, 4, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR, Biofabrication 2014, 6, 024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Liu W, Heinrich MA, Zhou Y, Akpek A, Hu N, Liu X, Guan X, Zhong Z, Jin X, Khademhosseini A, Adv. Healthcare Mater 2017, 6, 1601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA, ACS Biomater. Sci. Eng 2016, 2, 1743. [DOI] [PubMed] [Google Scholar]
- [180].Ouyang L, Highley CB, Sun W, Burdick JA, Adv. Mater 2017, 29, 1604983. [DOI] [PubMed] [Google Scholar]
- [181].Ma X, Qu X, Zhu W, Li Y.i-S., Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chiena S, Chen S, Proc. Natl. Acad. Sci. USA 2016, 113, 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Kolesky DB, Homana KA, Skylar-Scotta MA, Lewis JA, Proc. Natl. Acad. Sci. USA 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS, Nat. Mater 2012, 11, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Lee KY, Rowley JA, Eiselt P, Moy EM, Bouhadir KH, Mooney DJ, Macromolecules 2000, 33, 4291. [Google Scholar]
- [185].Jang J, Kim TG, Kim BS, Kim S-W, Kwon S-M, Cho D-W, Acta Biomater 2016, 33, 88. [DOI] [PubMed] [Google Scholar]
- [186].Jang J, Park H-J, Kim S-W, Kim H, Park JY, Na SJ, Kim HJ, Park MN, Choi SH, Park SH, Biomaterials 2017,112, 264. [DOI] [PubMed] [Google Scholar]
- [187].Lee H, Han W, Kim H, Ha D-H, Jang J, Kim BS, Cho D-W, Biomacromolecules 2017, 18, 1229. [DOI] [PubMed] [Google Scholar]
- [188].Park JY, Ryu H, Lee B, Ha D-H, Ahn M, Kim S, Kim JY, Jeon NL, Cho D-W, Biofabrication 2019, 11, 015002. [DOI] [PubMed] [Google Scholar]
- [189].Rutz AL, Hyland KE, Jakus AE, Burghardt WR, Shah RN, Adv. Mater 2015, 27, 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Duan B, Kapetanovic E, Hockaday LA, Butcher JT, Acta Biomater 2014, 10, 1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Saggiomo V, Velders AH, Adv. Sci 2015, 2, 1500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Au AK, Huynh W, Horowitz LF, Folch A, Angew. Chem., Int. Ed 2016, 55, 3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Zhang Q, Austin RH, Bionanoscience 2012, 2, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Zhang YS, Yue K, Aleman J, Mollazadeh-Moghaddam K, Bakht SM, Yang J, Jia W, Dell’Erba V, Assawes P, Su RS, Ann. Biomed. Eng 2017, 45, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].He Y, Qiu J, Fu J, Zhang J, Ren Y, Liu A, Microfluidic. Nanofluidic 2015, 19, 447. [Google Scholar]
- [196].Rogers CI, Qaderi K, Woolley AT, Nordin GP, Biomicrofluidics 2015, 9, 016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Zhang YS, Su RS, Calzone G, Biofabrication 2016, 8, 014101. [DOI] [PubMed] [Google Scholar]
- [198].Knowlton S, Yenilmez B, Tasoglu S, Trends Biotechnol 2016, 34, 685. [DOI] [PubMed] [Google Scholar]
- [199].Butcher AL, Offeddu GS, Oyen ML, Trends Biotechnol 2014, 32, 564. [DOI] [PubMed] [Google Scholar]
- [200].Gross BC, Erkal JL, Lockwood SY, Chen C, Spence DM, Anal. Chem 2014, 86, 3240. [DOI] [PubMed] [Google Scholar]
- [201].Bertsch A, Heimgartner S, Cousseau P, Renaud P, Lab Chip 2001, 1, 56. [DOI] [PubMed] [Google Scholar]
- [202].Wonjae L, Donghoon K, Boram C, Gyoo Yeol J, Anthony A, Albert F, Sangmin J, Anal. Chem 2014, 86, 6683.24856003 [Google Scholar]
- [203].Huang TQ, Xin Q, Liu J, Chen S, Biomed. Microdevices 2014, 16, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Liu J, Hwang HH, Wang P, Whang G, Chen S, Lab Chip 2016, 16, 1430. [DOI] [PubMed] [Google Scholar]
- [205].Spivey EC, Blerta X, Shear JB, Finkelstein IJ, Anal. Chem 2014, 86, 7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Sung JH, Srinivasan B, Esch MB, Mclamb WT, Bernabini C, Shuler ML, Hickman JJ, Exp. Biol. Med 2014, 239, 1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Lozano R, Stevens L, Thompson BC, Gilmore KJ, Gorkin R, Stewart EM, in het Panhuis M, Romero-Ortega M, Wallace GG, Biomaterials 2015, 67, 264. [DOI] [PubMed] [Google Scholar]
- [208].Lee Y-B, Polio S, Lee W, Dai G, Menon L, Carroll RS, Yoo S-S, Exp. Neurol 2010, 223, 645. [DOI] [PubMed] [Google Scholar]
- [209].Yeo SJ, Park KJ, Guo K, Yoo PJ, Lee S, Adv. Mater 2016, 28, 5268. [DOI] [PubMed] [Google Scholar]
- [210].Maeda K, Onoe H, Takinoue M, Takeuchi S, Adv. Mater 2012, 24, 1340. [DOI] [PubMed] [Google Scholar]
- [211].Windbergs M, Zhao Y, Heyman J, Weitz DA, J. Am. Chem. Soc 2013, 135, 7933. [DOI] [PubMed] [Google Scholar]
- [212].Chen QL, Liu Z, Shum HC, Biomicrofluidics 2014, 8, 064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Fu F, Shang L, Zheng F, Chen Z, Wang H, Wang J, Gu Z, Zhao Y, ACS Appl. Mater. Interfaces 2016, 8, 13840. [DOI] [PubMed] [Google Scholar]
- [214].Zhao H, Chen Y, Shao L, Xie M, Nie J, Qiu J, Zhao P, Ramezani H, Fu J, Ouyang H, He Y, Small 2018, 14, 1802630. [DOI] [PubMed] [Google Scholar]
- [215].Esch MB, Smith AST, Prot JM, Oleaga C, Hickman JJ, Shuler ML, Adv. Drug. Delivery Rev 2014, 69–70, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Li AP, ALTEX 2008, 25, 43. [DOI] [PubMed] [Google Scholar]
- [217].Schimek K, Busek M, Brincker S, Groth B, Hoffmann S, Lauster R, Lindner G, Lorenz A, Menzel U, Sonntag F, Lab Chip 2013, 13, 3588. [DOI] [PubMed] [Google Scholar]
- [218].Sung JH, Shuler ML, Lab Chip 2009, 9, 1385. [DOI] [PubMed] [Google Scholar]
- [219].Zhang C, Zhao Z, Rahim NAA, Noort DV, Yu H, Lab Chip 2009, 9, 3185. [DOI] [PubMed] [Google Scholar]
- [220].Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Lab Chip 2014, 14, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Costa PF, Albers HJ, Jea L, Hht M, Van DHL, Passier R, Van DBA, Malda J, Ad VDM, Lab Chip 2017, 17, 2785. [DOI] [PubMed] [Google Scholar]
- [222].Jos M, Jetze V, Melchels FP, Tomasz J, Hennink WE, Dhert WJA, Jürgen G, Hutmacher DW, Adv. Mater 2013, 25, 5011. [DOI] [PubMed] [Google Scholar]
- [223].Takenaga S, Schneider B, Erbay E, Biselli M, Schnitzler T, Schöning MJ, Wagner T, Phys. Status Solidi A 2015, 212, 1347. [Google Scholar]
- [224].Erkal JL, Selimovic A, Gross BC, Lockwood SY, Walton EL, Mcnamara S, Martin RS, Spence DM, Lab Chip 2014, 14, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park SJ, Kotikian A, Nesmith AP, Campbell PH, Nat. Mater 2017, 16, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Li CG, Lee CY, Lee K, Jung H, Biomed. Microdevices 2013, 15, 17. [DOI] [PubMed] [Google Scholar]
- [227].Shemelya C, Cedillos F, Aguilera E, Maestas E, Ramos J, Espalin D, Muse D, Wicker R, Macdonald E, in Proc. 2013 IEEE SENSORS, IEEE, Piscataway, NJ: 2013, pp. 1–4. [Google Scholar]
- [228].Yazdi AA, Popma A, Wong W, Nguyen T, Pan Y, Xu J, Microfluidic. Nanofluidic 2016, 20, 50. [Google Scholar]
- [229].Dagmar C, Kristyna C, Sylvie S, Jan Z, Rodrigo MAM, Vedran M, David H, Pavel K, Radek V, Vojtech A, Electrophoresis 2015, 36, 457.25069433 [Google Scholar]
- [230].Shallan AI, Petr S, Monika C, Guijt RM, Breadmore MC, Anal. Chem 2014, 86, 3124. [DOI] [PubMed] [Google Scholar]
- [231].Oh KW, Lee K, Ahn B, Furlani EP, Lab Chip 2012, 12, 515. [DOI] [PubMed] [Google Scholar]
- [232].Au AK, Lee W, Folch A, Lab Chip 2014, 14, 1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].He Y, Wu Y, Fu JZ, Gao Q, Qiu JJ, Electroanalysis 2016, 28, 1658. [Google Scholar]
- [234].Jia ML, Meng Z, Yeong WY, Microfluidic. Nanofluidic 2016, 20, 1. [Google Scholar]
- [235].Lee J, Paek J, Kim J, Lab Chip 2012, 12, 2638. [DOI] [PubMed] [Google Scholar]
- [236].Song SH, Lee CK, Kim TJ, Shin IC, Jun SC, Jung HI, Microfluidic. Nanofluidic 2010, 9, 533. [Google Scholar]
- [237].Gelber MK, Bhargava R, Lab Chip 2015, 15, 1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Bishop GW, Satterwhite JE, Bhakta S, Kadimisetty K, Gillette KM, Chen E, Rusling JF, Anal. Chem 2015, 87, 5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].van de Burgwal LHM, Dorst PV, Viëtor H, Luttge R, Claassen E, PharmaNutrition 2018, 6, 55. [Google Scholar]
- [240].Junaida A, Mashaghia A, Hankemeiera T, Vulto P, Curr. Opin. Biomed. Eng 2017, 1, 15. [Google Scholar]
- [241].Zhang B, Radisic M, Lab Chip 2017, 17, 2395. [DOI] [PubMed] [Google Scholar]
- [242].Balijepalli A, Sivaramakrishan V, Drug Discovery Today 2017, 22, 397. [DOI] [PubMed] [Google Scholar]
- [243].Fetah K, Tebon P, Goudie MJ, Eichenbaum J, i Ren L, Barros N, Nasiri R, Ahadian S, Ashammakhi N, Dokmeci MR, Prog AK, Biomed. Eng 2019, 1, 012001. [Google Scholar]
- [244].Mccracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM, Nature 2014, 516, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Park DY, Lee J, Chung JJ, Jung Y, Kim SH, Trends Biotechnol 2020, 38, P99. [DOI] [PubMed] [Google Scholar]
- [246].Renggli K, Rousset N, Lohasz C, Nguyen OTP, Hierlemann A, Adv. Biosys 2019, 3, 1900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Skardal A, Murphy SV, Devarasetty M, Mead I, Kang H-W, Seol Y-J, Zhang YS, Shin S-R, Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Lee SJ, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, Atala A, Sci. Rep 2017, 7, 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Lapomarda A, Vozzi G, Biotechnol. Res. Asia 2016, 16, 15. [Google Scholar]
- [249].Khoo ZX, Teoh JEM, Liu Y, Chua CK, Yang S, An J, Leong KF, Yeong WY, Virtual Phys. Prototyping 2015, 10, 103. [Google Scholar]
- [250].Esworthy TJ, Miao S, Lee S.e-J., Zhou X, Cui H, Zuo YY, Zhang LG, Int. J. Smart Nano Mater 2019, 10, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Yang H, Leow WR, Wang T, Wang J, Yu J, He K, Qi D, Wan C, Chen X, Adv. Mater 2017, 29, 1701627. [DOI] [PubMed] [Google Scholar]
- [252].Raviv D, Zhao W, McKnelly C, Papadopoulou A, Kadambi A, Shi B, Hirsch S, Dikovsky D, Zyracki M, Olguin C, Sci. Rep 2014, 4, 7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].An J, Chua CK, Mironov V, Int. J. Bioprint 2016, 2, 02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Gladman AS, Matsumoto EA, Nuzzo RG, Mahadevan L, Lewis JA, Nat. Mater 2016, 15, 413. [DOI] [PubMed] [Google Scholar]
- [255].Madadelahi M, Acosta-Soto LF, Hosseini S, Martinez-Chapa SO, Madou MJ, Lab Chip 2020, 20, 1318. [DOI] [PubMed] [Google Scholar]
- [256].Huang M, Fan S, Xing W, Liu C, Math. Comput. Model 2010, 52, 2036. [Google Scholar]
- [257].Przekwas A, Somayaji MR, Organ-on-a-Chip, Academic Press, San Diego, CA: 2020, p. 311.. [Google Scholar]
- [258].Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, Khademhosseini A, J. Controlled Release 2014, 190, 82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Hagmeyer B, Schütte J, Böttger J, Gebhardt R, Stelzle M, Proc. SPIE 2015, 8615, 861509. [Google Scholar]
- [260].Shafa M, Panchalingam KM, Walsh T, Richardson T, Baghbaderani BA, Biotechnol. Bioeng 2019, 116, 3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Shafa M, Panchalingam KM, Walsh T, Richardson T, Baghbaderani BA, Biotechnol. Bioeng 2019, 116, 3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Savla U, Olson LE, Waters CM, J. Appl. Physiol 2004, 96, 566. [DOI] [PubMed] [Google Scholar]
- [263].Norfleet DA, Park E, Kemp ML, Curr. Opin. Biomed. Eng 2020, 13, 113. [Google Scholar]
- [264].McAleer CW, Pointon A, Long CJ, Brighton RL, Wilkin BD, Richard Bridges L, Sriram NN, Fabre K, McDougall R, Muse VP, Mettetal JT, Srivastava A, Williams D, Schnepper MT, Roles JL, Shuler ML, Hickman JJ, Ewart L, Sci. Rep 2019, 9, 9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Esch MB, Smith AST, Prot J-M, Oleaga C, Hickman JJ, Shuler ML, Adv. Drug Delivery Rev 2014, 69, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Lecuit L, Lenne PF, Nat. Rev. Mol. Cell Biol 2007, 8, 633. [DOI] [PubMed] [Google Scholar]
- [267].Poli D, Magliaro C, Ahluwalia A, Front. Neurosci 2018, 13, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Saglam-Metiner P, Gulce-Iz S, Biray-Avci C, Gene 2019, 686, 203. [DOI] [PubMed] [Google Scholar]
- [269].Clevers H, Cell 2016, 165, 1586. [DOI] [PubMed] [Google Scholar]
- [270].Kretzschmar K, Clevers H, Dev. Cell 2016, 38, 590. [DOI] [PubMed] [Google Scholar]
- [271].Maul TM, Chew DW, Nieponice A, Vorp DA, Biomech. Model. Mechanobiol 2011, 10, 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA, Science 2018, 361, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Okuda S, Inoue Y, Eiraku M, Sasai Y, Adachi T, Biomech. Model. Mechanobiol 2013, 12, 987. [DOI] [PubMed] [Google Scholar]
- [274].Kim HY, Varner VD, Nelson CM, Development 2013, 140, 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Osterfield M, Du X, Schüpbach T, Wieschaus E, Shvartsman SY, Dev. Cell 2013, 24, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Okuda S, Inoue Y, Eiraku M, Sasai Y, Adachi T, Biomech. Model. Mechanobiol 2013, 12, 627. [DOI] [PubMed] [Google Scholar]
- [277].Okuda S, Inoue Y, Eiraku M, Adachi T, Sasai Y, Biomech. Model. Mechanobiol 2015, 14, 413. [DOI] [PubMed] [Google Scholar]
- [278].Okuda S, Inoue Y, Watanabe T, Adachi T, Interface Focus 2015, 5, 20140095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Okuda S, Miura T, Inoue Y, Adachi T, Eiraku M, Sci. Rep 2018, 8, 2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].McMurtrey RJ, Tissue Eng., Part C 2016, 22, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Bánsági T, Vanag VK, Epstein IR, Science 2011, 331, 1309. [DOI] [PubMed] [Google Scholar]
- [282].Filipovic N, Nikolic M, Sustersic T, Biomaterials for Organ and Tissue Regeneration, Elsevier, Amsterdam, 2020, Ch. 28. [Google Scholar]
- [283].Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O, Nat. Phys 2018, 14, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Barbolosi D, Ciccolini J, Lacarelle B, Barlesi F, Andre N, Nat. Rev. Clin. Oncol 2016, 13, 242. [DOI] [PubMed] [Google Scholar]
- [285].Powathil GG, Swat M, Chaplain MAJ, Semin. Cancer Biol 2015, 30, 13. [DOI] [PubMed] [Google Scholar]
- [286].Montes-Olivas S, Marucci L, Homer M, Front. Genet 2019, 10, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Karolak A, Markov DA, McCawley LJ, Rejniak KA, Soc JR. Interface 2018, 15, 20170703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Frost TS, Estrada V, Jiang L, Zohar Y, Microfluid. Nanofluid 2019, 23, 114. [Google Scholar]
- [289].Ravetto A, Hoefer IE, den Toonder JMJ, Bouten CVC, Biomed. Microdevice 2016, 18, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Kulthong K, Duivenvoorde L, Sunb H, Confederat S, Wua J, Spenkelink B, de Haan L, Marin V, van der Zande M, Bouwmeester H, Toxicol. In Vitro 2020, 65, 104815. [DOI] [PubMed] [Google Scholar]
- [291].Hancock MJ, Elabbasi N, in Modeling a Lung-on-a-Chip Microdevice. COMSOL Conference Boston 2015, Boston, MA, USA, October 2015. [Google Scholar]
- [292].Atiyat SA, Karabsheh SM, in Proc. 2017 13th Iasted Int. Conf. Biomed. Eng. (Biomed), Innsbruck, Austria, February 2017. [Google Scholar]
- [293].Liu J, Zhang Y, Wang Z, Deng J, Ye X, Xue R, Ge D, Xu Z, Chin. J Anal. Chem 2017, 45, 1109. [Google Scholar]
- [294].Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks NC, Mandegar MA, Conklin BR, Lee LP, Healy KE, Sci. Rep 2015, 5, 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Banaeiyan AA, Theobald J, Paukštyte J, Wölfl S, Adiels CB, Goksör M, Biofabrication 2017, 9, 015014. [DOI] [PubMed] [Google Scholar]
- [296].Frohlich EM, Zhang X, Charest JL, Integr. Biol 2012, 4, 75. [DOI] [PubMed] [Google Scholar]
- [297].Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, i Lang Q, Zhang YS, Shin SR, Calzone G, Annabi N, Shupe TD, Bishop CE, Atala A, Dokmeci MR, Khademhosseini A, Biofabrication 2016, 8, 014101. [DOI] [PubMed] [Google Scholar]
- [298].Phan DTT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LYN, George SC, Lee AP, Hughes CCW, Lab Chip 2017, 17, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Schimek K, Hsu HH, Boehme M, Kornet JJ, Marx U, Lauster R, Pörtner R, Lindner G, Bioeng 2018, 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Loskill P, Sezhian T, Tharp KM, Lee-Montiel FT, Jeeawoody S, Reese WM, Zushin PH, Stahl A, Healy KE, Lab Chip 2017, 17, 1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Funamoto K, Zervantonakis IK, Liu Y, Ochs CJ, Kim C, Kamm RD, Lab Chip 2012, 12, 4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Sakolish CM, Esch MB, Hickman JJ, Shuler ML, Mahler GJ, EBioMed 2016, 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Lee JB, Sung JH, Biotechnol. J 2013, 8, 1258. [DOI] [PubMed] [Google Scholar]
- [304].Somayaji MR, Das D, Przekwas AJ, Drug Discovery Today 2018, 23, 1571. [DOI] [PubMed] [Google Scholar]
- [305].Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ, Lab Chip 2013, 13, 3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Christopher Somayaji MR, Das D, Przekwas A, Drug Discovery Today 2016, 21, 1859. [DOI] [PubMed] [Google Scholar]
- [307].Sung JH, Wang Y, Shuler ML, APL Bioeng 2019, 3, 021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Moraes C, Labuz JM, Leung BM, Inoue M, Chun TH, Takayama S, Integr. Biol 2013, 5, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Stokes CL, Cirit M, Lauffenburger DA, CPT: Pharmacometrics Syst. Pharmacol 2015, 4, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Patricia Ramme A, Koenig L, Hasenberg T, Schwenk C, Magauer C, Faust D, Lorenz AK, Krebs A-C, Drewell C, Schirrmann K, Vladetic A, Lin G-C, Pabinger S, Neuhaus W, Bois F, Lauster R, Marx U, Dehne E-M, Future Sci. OA 2019, 5, FSO413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Wikswo JP, Block FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, Samson PC, Schaffer DK, Seale KT, Sherrod SD, IEEE Trans. Biomed. Eng 2013, 60, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Esch MB, Ueno H, Applegatec DR, Shuler ML, Lab Chip 2016, 16, 2719. [DOI] [PubMed] [Google Scholar]
- [313].Wang YI, Carmona C, Hickman JJ, Shuler ML, Adv. Healthcare Mater 2018, 7, 1701000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].West GB, Brown JH, Enquist BJ, Science 1997, 276, 122. [DOI] [PubMed] [Google Scholar]
- [315].Edington CD, Chen W. L.i K., Geishecker E, Kassis T, Soenksen LR, Bhushan BM, Freake D, Kirschner J, Maass C, Tsamandouras N, Valdez J, Cook CD, Parent T, Snyder S, Yu J, Suter E, Shockley M, Velazquez J, Velazquez JJ, Stockdale L, Papps JP, Lee I, Vann N, Gamboa M, LaBarge ME, Zhong Z, Wang X, Boyer LA, Laufenburger DA, Carrier RL, Communal C, Tannenbaum SR, Stokes CL, Hughes DJ, Rohatgi G, Trumper DL, Cirit M, Grifth LG, Sci. Rep 2018, 8, 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Abaci HE, Shuler ML, Integr. Biol 2015, 7, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Sung JH, Wang YI, Kim JH, Lee JM, Shuler ML, AIChE J 2018, 64, 4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Ucciferri N, Sbrana T, Ahluwalia A, Front. Bioeng. Biotechnol 2014, 2, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Ahluwalia A, Sci. Rep 2017, 7, 42113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Vozzi F, Heinrich JM, Bader A, Ahluwalia AD, Tissue Eng., Part A 2009, 15, 1291. [DOI] [PubMed] [Google Scholar]
- [321].West GB, Woodruff WH, Brown JH, Proc. Natl. Acad. Sci. USA 2002, 99, 2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Guzzardi MA, Domenici C, Ahluwalia A, Tissue Eng., Part A 2011, 17, 1635. [DOI] [PubMed] [Google Scholar]
- [323].Wagner I, Materne E, Brincker S, Susbier U, Fradrich C, Busek M, Sonntag F, Sakharov DA, Trushkin EV, Tonevitsky AG, Lauster R, Marx U, Lab Chip 2013, 13, 3538. [DOI] [PubMed] [Google Scholar]
- [324].Maass C, Stokes CL, Griffith LG, Cirit M, Integr. Biol 2017, 9, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [325].Prot JM, Maciel L, Bricks T, Merlier F, Cotton J, Paullier P, Bois FY, Leclerc E, Biotechnol. Bioeng 2014, 111, 2027. [DOI] [PubMed] [Google Scholar]
- [326].Ribas J, Pawlikowska J, Rouwkema J, Microphysiol. Syst 2018, 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Kirouac DC, Onsum MD, CPT Pharmacometrics Syst. Pharmacol 2013, 2, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Sonntag F, Schilling N, Mader K, Gruchow M, Klotzbach U, Lindner G, Horland R, Wagner I, Lauster R, Howitz S, Hoffmann S, Marx U, J. Biotechnol 2010, 148, 70. [DOI] [PubMed] [Google Scholar]
- [329].Miranda CC, Fernandes TG, Diogo MM, Cabral JMS, Bioengineering 2018, 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Agoram BM, Martin SW, van der Graaf PH, Drug Discovery Today 2007, 12, 1018. [DOI] [PubMed] [Google Scholar]
- [331].Bugrim A, Nikolskaya T, Nikolsky Y, Drug Discovery Today 2004, 9, 127. [DOI] [PubMed] [Google Scholar]
- [332].Aarons L, Br. J. Clin. Pharmacol 2005, 60, 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Bois FY, Jamei M, Clewell HJ, Toxicology 2010, 278, 256. [DOI] [PubMed] [Google Scholar]
- [334].Dingemanse J, Appel-Dingemanse S, Clin. Pharmacokinet 2007, 46, 713. [DOI] [PubMed] [Google Scholar]
- [335].Willmann S, Thelen K, Lippert J, J. Pharm. Pharmacol 2012, 64, 997. [DOI] [PubMed] [Google Scholar]
- [336].Jamei M, Marciniak S, Feng K, Barnett A, Tucker GT, Rostamihodjegan A, Expert Opin. Drug Metab. Toxicol 2009, 5, 211. [DOI] [PubMed] [Google Scholar]
- [337].Hosseini I, Gajjala A, Yadav DB, Sukumaran S, Ramanujan S, Paxson RE, Gadkar K, J. Pharmacokinet. Pharmacodyn 2018, 45, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Kannan R, Chen ZJ, Singh N, Przekwas A, Delvadia R, Tian G, Walenga R, Int. J. Numer. Methods Biomed. Eng 2017, 33, e2838. [DOI] [PubMed] [Google Scholar]
- [339].Kannan RR, Singh N, Przekwas A, Int. J. Numer. Methods Biomed. Eng 2018, 34, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Sluka JP, Fu X, Swat M, Belmonte JM, Cosmanescu A, Clendenon SG, Wambaugh JF, Glazier JA, PLoS One 2016, 11, e0162428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Zuegge J, Schneider G, Coassolo P, Lave T, Clin. Pharmacokinet 2009, 40, 553563. [DOI] [PubMed] [Google Scholar]
- [342].Cai S, Li H, Zheng F, Kong F, Dao M, Karniadakis GE, Suresh S, Proc. Natl. Acad. Sci. USA 2021, 118, e2100697118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Urban G, Bache K, Phan DTT, Sobrino A, Shmakov A, Hachey SJ, Hughes CCW, Baldi P, IEEE/ACM Trans. Comput. Biol. Bioinform 2019, 16, 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Garrett-Bakelman FE, Darshi M, Green SJ, Gur RC, Lin L, Macias BR, McKenna MJ, Meydan C, Mishra T, Nasrini J, Science 2019, 364, e8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].Yeung CK, Koenig P, Countryman S, Thummel KE, Himmelfarb J, Kelly EJ, Clin. Transl. Sci 2020, 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Weber EJ, Lidberg KA, u Wang L, Bammler TK, MacDonald JW, Li MJ, Redhair M, Atkins WM, Tran C, Hines KM, Herron J, Xu L, Monteiro MB, Ramm S, Vaidya V, Vaara M, Vaara T, Himmelfarb J, Kelly EJ, JCI Insight 2018, 3, e123673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Wnorowski A, Sharma A, Chen H, Wu H, i Shao N-Y, Sayed N, Liu C, Countryman S, Stodieck LS, Rubins KH, Wu SM, Lee PHU, Wu JC, Stem Cell Rep 2019, 13, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [348].Low LA, Giulianotti MA, Pharm. Res 2020, 37, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Tagle DA, Curr. Opin. Pharmacol 2019, 48, 146. [DOI] [PubMed] [Google Scholar]
- [350].Mastrangeli M, Millet S, Mummery C, Loskill P, Braeken D, Eberle W, Cipriano M, Fernandez L, Graef M, Gidrol X, Picollet-D’Hahan N, Van Meer B, Ochoa I, Schutte M, Van den Eijnden-van Raaij J, ALTEX 2019, 36, 481. [DOI] [PubMed] [Google Scholar]
- [351].Du E, Diez-Silva M, Kato GJ, Dao M, Suresh S, Proc. Natl. Acad. Sci. USA 2015, 112, 1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Papageorgiou DP, Abidi SZ, Chang H-Y, Li XJ, Karniadakis GJ, Suresh S, Dao M, Proc. Natl. Acad. Sci. USA 2018, 115, 9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Du E, Dao M, Exp. Mech 2019, 59, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]


