Abstract
We aimed to explore the mechanism of circular RNAs (circRNAs) and provide potential biomarkers for molecular therapy of diabetic foot ulcers (DFU). Gene expression profile of GSE114248, including five normal samples and five DFU samples, was downloaded from GEO database. Differentially expressed circRNAs (DEcircRNAs) between two groups were identified. Then, DEcircRNA‐miRNA and miRNA‐mRNA interaction was revealed, followed by the circRNA‐miRNA‐mRNA network construction. Moreover, functional and pathway analysis were performed based on mRNAs, followed by the DM‐related pathway exploration. Specific binding sites for key circRNAs and associated miRNAs were under investigation. Finally, RT‐qPCR was used to verify the candidate the relative expression level of circRNA between normal tissues and DFU. Totally, 65 DEcircRNAs were revealed between two groups, followed by 113 circRNA‐miRNA‐mRNA interactions explored. The mRNAs in these interactions were mainly assembled in functions like cell proliferation and pathways. Moreover, a total of 11 DM‐related pathways were revealed. Finally, circRNA‐miRNA specific binding‐site analysis revealed two key circRNAs, for example, circRNA_072697 and circRNA_405463, corresponding to their miRNAs. These two circRNAs were novel biomarkers for DFU. circRNA_072697 acted as a sponge of miR‐3150a‐3p in the progression of DFU via regulating KRAS. MAPK signaling pathway might contribute to the development of DFU.
Keywords: circRNA‐miRNA‐mRNA, circRNAs, diabetic foot ulcers, function and pathway enrichment analysis, miRNA‐mRNA binding sites
1. INTRODUCTION
Diabetic foot ulcers (DFUs) are the most common lower‐extremity complication in patients with diabetes mellitus (DM). 1 DFU and their complications contribute to the high morbidity and mortality of DM. 2 Despite common treatments, ulcers readily become chronic wounds. 3 Thus, it is necessary to understand the molecular and genetic basis of DFU to block chronic wounds and improve the efficiency of DFU treatment. 4
Molecular markers' exploration contributes to improvement of wound therapy effect and associated molecular mechanism investigation. 5 Circular RNAs (circRNAs) are a novel class of non‐coding RNA. 6 As potential novel biomarkers, differentially expressed circRNAs (DEcircRNAs) such as sha_circ_0084443 are proved to be important in the development of DFU via modulating keratinocyte migration and proliferation. 7 Actually, as competing endogenous RNAs (ceRNAs), cricRNAs can serve as microRNA (miRNA) sponges to regulate cell growth and suppress the progression of tumor cells. 8 Recently, the ceRNA network has been successfully used for exploring possible biomarkers of disease. 9 Based on an integrated microarray analysis, a previous study shows that the key circRNAs (such as has_circ_0078279) related networks are breakthrough for cancer mechanism investigation. 10 Importantly, the circRNA‐miRNA‐mRNA network can be used to reveal circRNAs in the progression of DM. 11 However, the important role of circRNAs and associated pathways/ceRNAs network in the progression of DFU has not been fully revealed yet.
In this study, the potential DEcircRNAs were investigated between normal samples and DFU samples based on circRNA expression profile. Then, DEcircRNA‐miRNA and miRNA‐mRNA interactions were revealed, followed by the ceRNA network construction. Moreover, functional and pathway analysis was performed based on mRNAs, followed by the DM‐related pathway exploration. Finally, specific binding sites for key circRNAs and associated miRNAs were investigation. The flow chart of experimental design in this study is listed in Supplementary Figure 1. This study hoped to reveal detail molecular mechanism and novel biomarkers for DFU.
2. MATERIALS AND METHODS
2.1. Data resource
Gene expression dataset GSE114248 that based on the platform of GPL21825074301 Arraystar Human CircRNA microarray V2 (update in February, 2020) was downloaded from GEO database. 12 The dataset was collected from human excisional skin wounds 7 days after injury samples (5 samples, normal group) and human DFU samples (5 samples, DFU group). The normal group included two men and three women donors with a mean age of 29 years (ranging 25‐43). The acute wounds (4 mm in diameter) were surgically created on the skin of healthy volunteers and the wound‐edge tissues (6 mm in diameter) were collected 7 days later. The DFU group enrolled five patients with a mean age of 74 years (ranging 54‐90), of which there were three men and two women. Chronic wound biopsies (4 mm in diameter) were collected from the non‐healing edges of DFUs.
2.2. DEcircRNAs investigation
Data normalisation were performed using robust multi‐array average (RMA) based on limma package in R (v.3.10.3) software. 13 The probe IDs were converted to circRNAs symbol based on the annotation files from the platform. Probes that did not correspond to the circRNAs symbol were excluded. For the circRNAs associated with different probes, the average value of the probes was used as the final expression value. Using the Bayesian method in limma package, 13 the DEcircRNAs between two groups were revealed. The DE‐circRNAs were required to be with adj. P Val < .001 and |log fold change (FC)| > 2. Then, the conversion of common name was performed on these DE‐circRNAs based on circBase, 14 followed by visualisation by heatmap and volcano plot.
2.3. DEcircRNA‐miRNA interaction prediction
Based on the FASTA format sequence files of DE‐circRNAs and all mature miRNAs, the DEcircRNA‐miRNA interactions were predicted using Miranda 15 (version: 3.3a). The score > 170 and energy < −30 were selected as cut‐off values for investigation of DEcircRNA‐miRNA regulatory relationship.
2.4. The miRNA‐mRNA interaction prediction
The target genes of up‐ and down‐regulated miRNAs were predicted using the miRWalk 3.0 software. 16 Then, with score > 0.95, a total of three databases including miRDB 17 Targetscan, 18 mirtarbase, 19 and Targetscan 20 were selected as default databases for miRNA‐mRNA interaction scanning. The miRNA‐mRNA interaction appeared in at least two databases were enrolled for the further analysis.
Moreover, genes directly related to DFU were explored based on Comparative Toxicogenomics Database (CTD) database (keyword: Diabetic Foot), 21 followed by miRNA regulatory network construction when compared DFU‐related genes with the target genes of miRNAs. Finally, DEcircRNA‐miRNA‐mRNA interactions were further obtained by integrated the DEcircRNA‐miRNA and miRNA‐mRNA pairs. The results were visualised using the Cytoscape software (version: 3.2.0). 22
2.5. The enrichment analysis and DM‐related pathway investigation
Gene Ontology (GO)‐biological process (BP) function enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis of mRNAs were performed using the DAVID software. 23 P value <.05 and count (the gene number) ≥ 2 were considered as cut‐off values of significant difference. Finally, the DM‐associated pathways provided by CTD database 21 and KEGG results were integrated to obtain the DM‐related pathways.
2.6. Specific binding‐site analysis for key circRNAs and associated miRNAs
Based on the DEcircRNA‐miRNA prediction results and DM‐related pathways obtained earlier, the specific binding sites of key circRNAs and associated miRNAs were investigated, followed by the specific binding site information visualised.
2.7. RT‐qPCR confirmation
Samples from eight normal excisional skin wounds 7 days after injury and nine DFUs were used in this study. For patients in the DFU group, there were six men and three women. The average age was 67 (ranging from 57 to 75) and the average duration of diabetes was 16 years. The normal group included four men and four women. The average age was 36 (ranging from 27 to 57). GADPH was used as the internal reference gene, and the primer sequences were as follows: F‐GGAGCGAGATCCCTCCAAAAT, R‐GGCTGTTGTCATACTTCTCATGG. The sequences of circRNA to be verified were as follows: hsa_circRNA_0072697‐F‐TTGCTGCTTCCGAAAAGAGT, hsa_circRNA_0072697‐R‐GAGGCCCAGTCCAGTATTCA. The annealing temperature is 60°C.
3. RESULTS
3.1. DEcircRNAs between normal group and DFU group
A total of 65 DEcircRNAs including 25 upregulated circRNAs and 40 downregulated circRNAs were identified between normal group and DFU group. The volcano plot showed that the upregulated genes and downregulated genes were significantly separated (Figure 1A), which was confirmed by the heatmap (Figure 1B).
FIGURE 1.
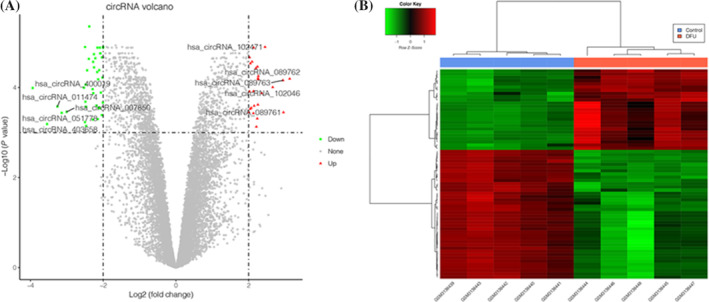
The volcano plots and heatmap for differentially expressed circular RNAs (DEcircRNAs)between diabetic foot ulcers (DFU)sample and normal sample. A, the volcano plots in current study; the X‐axis represents the value of log2 fold change, while the Y‐axis represents the value of ‐log10; the top5 up‐ and down‐regulated circRNAs were shown. B, the heatmap in current study; color from green to red indicated low to high representation value
3.2. The circRNA‐miRNA‐mRNA regulation network
A total of three miRNAs (miR‐3150a‐3p, miR‐370‐5p, and miR‐4649‐3p), four circRNAs (circRNA_072697, circRNA_406307, circRNA_066608, and circRNA_405463), and 4 edges were revealed based on DEcircRNA‐miRNA interaction prediction. Then, the target mRNAs of all three miRNAs were predicated, followed by DFU‐associated genes exploration. Finally, a total of 113 circRNA‐miRNA‐mRNA interactions such as circRNA_072697‐miR‐3150a‐3p‐KRAS were obtained in the current network. The result showed that there were 4 circRNA‐miRNA edges and 109 miRNA‐mRNA edges in current network. Meanwhile, a total of 3 miRNAs, 4 circRNAs, and 101 mRNAs were included in the current circRNA‐miRNA‐mRNA network (Figure 2).
FIGURE 2.
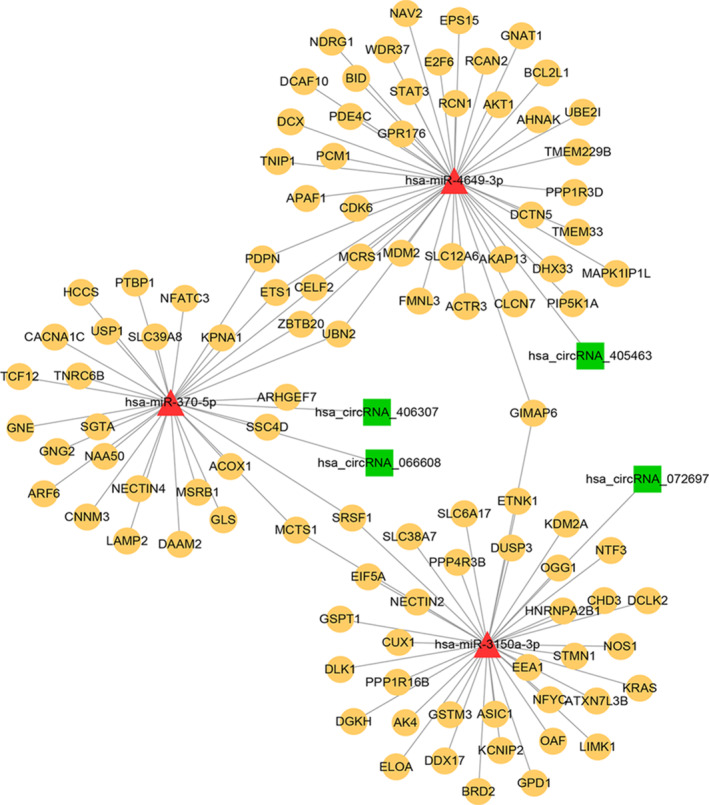
The circRNA‐miRNA‐mRNA interaction network. The red triangle represents miRNA; the green square represented circRNA; the yellow node represents the mRNA
3.3. GO function and KEGG pathways investigation
The KEGG pathway enrichment showed that target mRNAs of miR‐3150a‐3p, miR‐370‐5p, and miR‐4649‐3p were mainly enriched in 23 pathways like mitogen‐activated protein kinase (MAPK) signaling (hsa04010; genes: DUSP3, KRAS, NTF3, and STMN1), GABAergic synapse (hsa04727; genes: GLS, GNG2, and CACNA1C) and pathways in cancer (hsa05200; genes: BID, AKT1, ETS1, MDM2, CDK6, BCL2L1, and STAT3) (Figure 3A). Moreover, GOBP functional enrichment showed that the target mRNAs of miR‐3150a‐3p, miR‐370‐5p, and miR‐4649‐3p were mainly assembled in 26 functions including mRNA export from nucleus (GO:0006406; genes: SRSF1, HNRNPA2B1, and EIF5A), positive regulation of cellular component movement (GO:0051272; genes: PDPN and ETS1), and cell proliferation (GO:0008283; genes: EPS15, GNAT1, AKT1, PDPN, BCL2L1, and STAT3) (Figure 3B).
FIGURE 3.
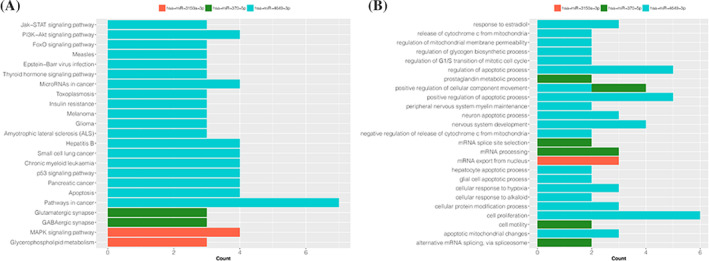
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. A, The result of KEGG pathway enrichment. B, The result of GO functional enrichment. The X‐axis represents the number of genes in certain item; the Y‐axis represents the different items of functions or pathways; different color indicated different miRNA
3.4. DM‐related pathway analysis
The CTD‐based analysis showed that there were 121 pathways associated with the development of DM. Integrated with all 23 pathways enriched by mRNAs, a total of 11 DM‐related pathways were revealed in the current study (Figure 4). CTD DM‐related pathways and circRNAs information are listed in Table 1.
FIGURE 4.
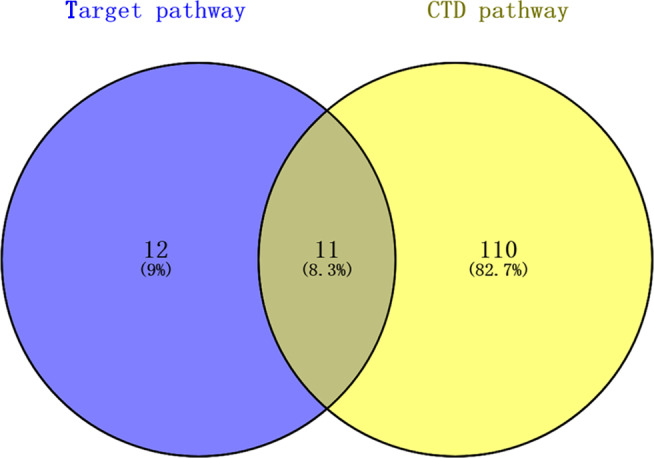
The Venn plot for pathways enriched by mRNAs and explored in Toxicogenomics Database (CTD) database. The purple circle represented the pathways enriched by target mRNAs of miRNAs. The yellow circle represented the DM‐associated pathways explored in CTD database
TABLE 1.
The detail information for DM‐related pathways in current study
| ID | circRNA | miRNA | Term | Count | P value |
|---|---|---|---|---|---|
| hsa04010 | hsa_circRNA_072697 | hsa‐miR‐3150a‐3p | MAPK signaling pathway | 4 | .02665 |
| hsa04068 | hsa_circRNA_405463 | hsa‐miR‐4649‐3p | FoxO signaling pathway | 3 | .037812 |
| hsa04151 | hsa_circRNA_405464 | hsa‐miR‐4649‐3p | PI3K‐Akt signaling pathway | 4 | .043071 |
| hsa04210 | hsa_circRNA_405465 | hsa‐miR‐4649‐3p | Apoptosis | 4 | .000359 |
| hsa04630 | hsa_circRNA_405466 | hsa‐miR‐4649‐3p | Jak–STAT signaling pathway | 3 | .043649 |
| hsa04931 | hsa_circRNA_405467 | hsa‐miR‐4649‐3p | Insulin resistance | 3 | .025392 |
| hsa05014 | hsa_circRNA_405468 | hsa‐miR‐4649‐3p | Amyotrophic lateral sclerosis (ALS) | 3 | .005822 |
| hsa05200 | hsa_circRNA_405469 | hsa‐miR‐4649‐3p | Pathways in cancer | 7 | .000164 |
| hsa05206 | hsa_circRNA_405470 | hsa‐miR‐4649‐3p | microRNAs in cancer | 4 | .02664 |
| hsa05212 | hsa_circRNA_405471 | hsa‐miR‐4649‐3p | Pancreatic cancer | 4 | .000413 |
| hsa05222 | hsa_circRNA_405472 | hsa‐miR‐4649‐3p | Small cell lung cancer | 4 | .000908 |
Note: circRNA, circular RNA; DM, diabetes mellitus.
3.4.1. Investigation of specific binding sites between candidate circRNAs and miRNAs
A total of four circRNA‐miRNA regulatory edges including three miRNAs and four circRNAs were enrolled in the current analysis. Based on the result of DM‐related pathways, two circRNA‐miRNA pairs including circRNA_072691‐miR‐3150a‐3p and circRNA_405463‐miR‐4649‐3p were enrolled for the analysis of current specific binding sites. The detail information of the binding sites for circRNA_072697‐miR‐3150a‐3p (score: 171.00; energy: −30.71) and circRNA_405463‐miR‐4649‐3p (score: 173.00; energy: −34.37) are shown in Figure 5.
FIGURE 5.

The detail information of the binding sites for circRNA_072697‐miR‐3150a‐3p and circRNA_405463‐miR‐4649‐3p. The “|” indicated perfect match of binding site; the “:” indicated incomplete match of the binding site
3.5. Hsa_circ_072697 showed a higher relative expression level in DFU
The relative expression of hsa_circ_072697 was detected in DFU and normal tissues. As shown in Figure 6, the result showed that the relative expression level of hsa_circ_072697 in DFU was significantly higher than in the normal tissues (P < .05).
FIGURE 6.
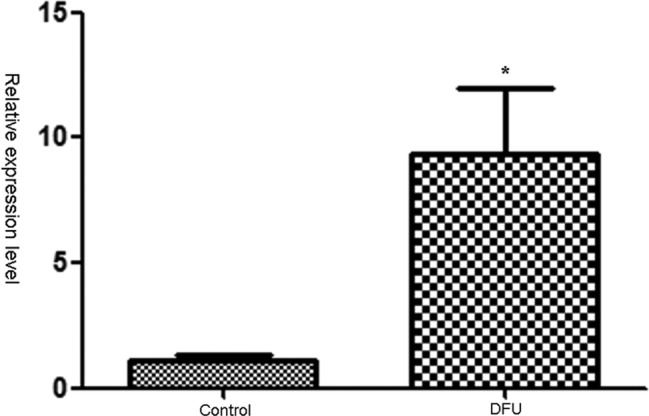
Relative expression level of circRNA_072697 in normal tissues (control) and DFU. DFU represented diabetic foot ulcers. *, P < .05. GAPDH was used as the reference gene
4. DISCUSSION
CircRNAs are proved to be vital for the development of various diseases including DM. However, the potential key circRNAs and associated detail mechanism during the process of DFU is still unclear. In current bioinformatics study, a total of 65DEcircRNAs were revealed between normal and DFU samples. Then, totally 113 circRNA‐miRNA‐mRNA edges (such as circRNA_072697‐miR‐3150a‐3p‐KRAS) were explored based on 4 circRNAs, 3 circRNA‐target miRNAs, and 101 miRNA‐target mRNAs. These mRNAs were mainly assembled in functions like cell proliferation and pathways like MAPK signaling. Moreover, a total of 11 DM‐related pathways including MAPK signaling were revealed in current study. Finally, circRNA_072697 and circRNA_405463 were two candidate genes revealed in the analysis of circRNA‐miRNA specific binding sites. The RT‐qPCR verification results of circRNA_072697 were also consistent with our bioinformatics analysis results.
Although epidermis of chronic wounds such as DFU has distinct morphology, the molecular mechanism and the associated biomarkers during this process were still unclear. 24 As novel focused biomarkers, circRNAs are widely been proved to participate in the development of DM‐related diseases. 25 , 26 A previous study indicates that the peripheral blood circRNA such as circRNA_0054633 can be used as a diagnostic biomarker for DM. 27 Wang et al showed that circRNA_0084443 was differentially expressed during the development of DFU and modulated the keratinocyte migration and proliferation. 7 As competing endogenous RNAs, circRNAs can serve as miRNA sponges to regulate cell growth in DM, and they can be used as a new class of biomarker. 28 , 29 A recent study shows that the dysregulated miRNAs like miR‐3150a‐3p contribute to the nephrogenic diabetes disease progression. 30 It is known to all that miRNAs are considered as novel therapeutic targets for DM wound healing. 31 Actually, the biological function of miRNA in promoting fibroblast migration is commonly realised by regulating target mRNA during the process of skin wound healing. 32 A previous study proves that novel identified circRNAs such as circRNA_010567 promotes fibrosis via suppressing miR‐141 by targeting mRNA TGF‐β1. 33 As a member of mRNA, KRAS Proto‐Oncogene, GTPase (KRAS) is being used as a research target for diabetes. 34 The mutation of KRAS has been observed in patients with rare neurocutaneous disorder categorised. 35 Although the role of circRNA‐miRNA‐mRNA has been gradually identified in different diseases such as gastric cancer and cardiovascular disease, 36 , 37 little is known about their associations with DFU. In the current study, circRNA_072697 and circRNA_405463 were two DEcircRNAs explored between normal samples and DFU samples. Meanwhile, circRNA_072697‐miR‐3150a‐3p‐KRAS was revealed as one of the most outstanding circRNA‐miRNA‐mRNA axis based on network analysis. Importantly, the binding site of circRNA_072697‐miR‐3150a‐3p had been proved in the analysis of circRNA‐miRNA specific binding sites. Thus, we speculated that the circRNA_072697 might act as a sponge of miR‐3150a‐3p in the progression of DFU via regulating KRAS. circRNAs including circRNA_072697 and circRNA_405463 might be utilised as novel biomarkers for DFU.
The high expression of mRNAs in MAPK signaling pathway is involved in the regulation of cascade reaction during the development of diabetic disease. 38 In diabetic rats, the effect of Curcumin in preventing diabetic cardiomyopathy is involved with the PKC‐MAPK signaling pathway. 39 By modulating MAPK signaling pathway in vitro and in vivo, the Aloesin from Aloe vera can be used to accelerate skin wound healing. 40 A previous study shows that MAPK signaling factors are upregulated during the wound therapy process in human diabetic foot wounds. 41 The negative pressure wound therapy can promote wound healing in diabetic foot patients via MAPK‐related pathway. 42 In the current study, KEGG pathway enrichment analysis showed that MAPK signaling pathway was the most significant pathway enriched by target genes of miR‐3150a‐3p. Meanwhile, the CTD‐based investigation showed that MAPK signaling pathway was one of the DM‐related pathways revealed in the current study. Thus, we speculated that MAPK signaling pathway might contribute to the development of DFU.
5. CONCLUSIONS
In conclusion, circRNA_072697 acted as a sponge of miR‐3150a‐3p in the progression of DFU via regulating KRAS. Moreover, circRNA_072697 and circRNA_405463 were novel biomarkers for DFU. Furthermore, MAPK signaling pathway played an important role in the development of DFU. In the future, studying the specific molecular mechanisms by which circRNAs function as miRNA sponges to regulate DFU occurrence and development will be a promising research field.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
Ming Tian, Huiying Jia: Conception and design of the research; Bo Yuan, Jiaoyun Dong: Acquisition of data; Bo Yuan, Jiaoyun Dong: Analysis and interpretation of data; Ming Tian: Statistical analysis; Huiying Jia, Ming Tian: Obtaining funding; Huiying Jia: Drafting the manuscript; Ming Tian: Revision of manuscript for important intellectual content. All authors read and approved the final manuscript.
Supporting information
Supplementary Figure 1 The flow chart of experimental design in this study
ACKNOWLEDGEMENTS
The authors wish to acknowledge SunteamBio Co. (Shanghai China) for its timely help on the experiments. This study is supported by the National Natural Science Foundation of China (No. 81401163 and No. 81670752).
Tian M, Dong J, Yuan B, Jia H. Identification of potential circRNAs and circRNA‐miRNA‐mRNA regulatory network in the development of diabetic foot ulcers by integrated bioinformatics analysis. Int Wound J. 2021;18:323–331. 10.1111/iwj.13535
Ming Tian and Jiaoyun Dong contributed equally to this work.
[Correction added on 21 January 2021, after first online publication: Bo Yuan was designated as corresponding author in this version.]
Funding information National Natural Science Foundation of China, Grant/Award Numbers: 81401163, 81670752
Contributor Information
Ming Tian, Email: tianming198@163.com.
Jiaoyun Dong, Email: dongjiaoyun@hotmail.com.
Bo Yuan, Email: hiyuanbo2002@163.com.
Huiying Jia, Email: jiawol380804@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Armstrong DG, Boulton AJ, Bus SA. Diabetic foot ulcers and their recurrence. New Engl J Med. 2017;376(24):2367‐2375. [DOI] [PubMed] [Google Scholar]
- 2. Chammas N, Hill R, Edmonds M. Increased mortality in diabetic foot ulcer patients: the significance of ulcer type. J Diabetes Res. 2016;2016:2879809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeffcoate WJ, Harding KG. Diabetic foot ulcers. The Lancet. 2003;361(9368):1545‐1551. [DOI] [PubMed] [Google Scholar]
- 4. Jhamb S, Vangaveti VN, Malabu UH. Genetic and molecular basis of diabetic foot ulcers: clinical review. J Tissue Viability. 2016;25(4):229‐236. [DOI] [PubMed] [Google Scholar]
- 5. Karam RA, Rezk NA, Rahman TMA, Al Saeed M. Effect of negative pressure wound therapy on molecular markers in diabetic foot ulcers. Gene. 2018;667:56‐61. [DOI] [PubMed] [Google Scholar]
- 6. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang A, Toma MA, Ma J, et al. Circular RNA hsa_circ_0084443 Is upregulated in diabetic foot ulcer and modulates keratinocyte migration and proliferation. Adv Wound Care. 2019;9(4):145‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151‐1164. [DOI] [PubMed] [Google Scholar]
- 9. Lu H‐c, J‐q Y, Yang X, et al. Identification of a potentially functional circRNA–miRNA–mRNA regulatory network for investigating pathogenesis and providing possible biomarkers of bladder cancer. Cancer Cell Int. 2020;20(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin X, Chen Y. Identification of potentially functional CircRNA‐miRNA‐mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. 2018;24:70‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang G, Ma Y, An T, et al. Relationships of circular RNA with diabetes and depression. Sci Rep. 2017;7(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barrett T, Suzek TO , Troup DB, et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33(suppl 1):D562‐D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smyth GK. limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2005:397‐420. [Google Scholar]
- 14. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA‐target interactions. Nat Methods. 2015;12(8):697. [DOI] [PubMed] [Google Scholar]
- 17. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146‐D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife Sci. 2015;4:e05005. 10.7554/eLife.05005.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheng‐Da H, Yu‐Ting T, Sirjana S, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA‐target interactions. Nucleic Acids Res. 2013;42(D1):D78‐D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi Y, Yang F, Wei S, Xu G. Identification of key genes affecting results of hyperthermia in osteosarcoma based on integrative ChIP‐Seq/TargetScan analysis. Int Med J Exp Clin Res. 2017;23:2042‐2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mattingly CJ, Colby GT, Forrest JN, Boyer JL. The comparative toxicogenomics database (CTD). Environ Health Perspect. 2003;111(6):793‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498‐2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dennis G Jr, Sherman BT, Hosack DA, et al. Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 24. Pastar I, Stojadinovic O, Yin NC, et al. Epithelialization in wound healing: a comprehensive review. Adv Wound Care. 2014;3(7):445‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li G, Liu H, Ma C, Chen Y, Wang J, Yang Y. Exosomes are the novel players involved in the beneficial effects of exercise on type 2 diabetes. J Cell Physiol. 2019;234(9):14896‐14905. [DOI] [PubMed] [Google Scholar]
- 26. L‐h J, D‐w S, J‐c H, Z‐l J. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1‐7. [DOI] [PubMed] [Google Scholar]
- 27. Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre‐diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan L, Feng J, Cheng F, et al. Circular RNA expression profiles in placental villi from women with gestational diabetes mellitus. Biochem Biophys Res Commun. 2018;498(4):743‐750. [DOI] [PubMed] [Google Scholar]
- 29. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42‐51. [DOI] [PubMed] [Google Scholar]
- 30. Shen Y, Chen G, Gao H, et al. miR‐939‐5p Contributes to the migration and invasion of pancreatic cancer by targeting ARHGAP4. OncoTargets Ther. 2020;13:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dangwal S, Foinquinos A, Thum T. MicroRNAs: novel therapeutic targets for diabetic wound healing. Aristidis Veves, The Diabetic Foot. New York, NY: Springer; 2018:237‐246. [Google Scholar]
- 32. Li X, Li D, Wikstrom JD, et al. MicroRNA‐132 promotes fibroblast migration via regulating RAS p21 protein activator 1 in skin wound healing. Sci Rep. 2017;7(1):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou B, Yu J‐W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR‐141 by targeting TGF‐β1. Biochem Biophys Res Commun. 2017;487(4):769‐775. [DOI] [PubMed] [Google Scholar]
- 34. Fujimoto T, Shirasawa S. KRAS‐induced actin‐interacting protein: a potent target for obesity, diabetes and cancer. Anticancer Res. 2011;31(7):2413‐2417. [PubMed] [Google Scholar]
- 35. Nagatsuma M, Takasawa K, Yamauchi T, et al. A postzygotic KRAS mutation in a patient with Schimmelpenning syndrome presenting with lipomatosis, renovascular hypertension, and diabetes mellitus. J Hum Genet. 2019;64(2):177‐181. [DOI] [PubMed] [Google Scholar]
- 36. Li M, Duan L, Li Y, Liu B. Long noncoding RNA/circular noncoding RNA–miRNA–mRNA axes in cardiovascular diseases. Life Sci. 2019;233(15):116440. [DOI] [PubMed] [Google Scholar]
- 37. Guan Y‐J, Ma J‐Y, Song W. Identification of circRNA–miRNA–mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toyoda M, Suzuki D, Honma M, et al. High expression of PKC‐MAPK pathway mRNAs correlates with glomerular lesions in human diabetic nephropathy. Kidney Int. 2004;66(3):1107‐1114. [DOI] [PubMed] [Google Scholar]
- 39. Karri V, Gowthamarajan K, Satish Kumar M, Rajkumar M. Multiple biological actions of curcumin in the management of diabetic foot ulcer complications: a systematic review. Trop Med Surg. 2015;3(179):2. [Google Scholar]
- 40. Wahedi HM, Jeong M, Chae JK, Do SG, Yoon H, Kim SY. Aloesin from Aloe vera accelerates skin wound healing by modulating MAPK/Rho and Smad signaling pathways in vitro and in vivo. Phytomedicine. 2017;28:19‐26. [DOI] [PubMed] [Google Scholar]
- 41. Yang SL, Han R, Liu Y, Hu LY, Li XL, Zhu LY. Negative pressure wound therapy is associated with up‐regulation of bFGF and ERK 1/2 in human diabetic foot wounds. Wound Repair Regen. 2014;22(4):548‐554. [DOI] [PubMed] [Google Scholar]
- 42. Wang T, Li X, Fan L, et al. Negative pressure wound therapy promoted wound healing by suppressing inflammation via down‐regulating MAPK‐JNK signaling pathway in diabetic foot patients. Diabetes Res Clin Pract. 2019;150:81‐89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 The flow chart of experimental design in this study
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


