Abstract
RNA viruses have developed specialized mechanisms to subvert host RNA‐binding proteins (RBPs) favoring their own gene expression. The Leader (L) protein of foot‐and‐mouth disease virus, a member of the Picornaviridae family, is a papain‐like cysteine protease that self‐cleaves from the polyprotein. Early in infection, the L protease cleaves the translation initiation factors eIF4GI and eIF4GII, inducing the shutdown of cap‐dependent translation. However, the cleavage sites on the viral polyprotein, eIF4GI, and eIF4GII differ in sequence, challenging the definition of a consensus site for L targets. Identification of Gemin5 and Daxx proteolytic products in infected cells unveiled a motif centered on the RKAR sequence. The RBP Gemin5 is a member of the survival of motor neurons complex, a ribosome interacting protein, and a translation downregulator. Likewise, the Fas‐ligand Daxx is a multifunctional adaptor that plays key roles in transcription control, apoptosis, and innate immune antiviral response. Remarkably, the cleavage site on the RNA helicases MDA5 and LGP2, two relevant immune sensors of the retinoic acid‐inducible gene‐I (RIG‐I)‐like receptors family, resembles the L target site of Gemin5 and Daxx, and similar cleavage sites have been reported in ISG15 and TBK1, two proteins involved in type I interferon response and signaling pathway, respectively. In this review we dissect the features of the L cleavage sites in essential RBPs, eventually helping in the discovery of novel L targets.
This article is categorized under:
RNA in Disease and Development > RNA in Disease
Translation > Translation Regulation
Keywords: Gemin5, Leader protease targets, LGP2, MDA5, RNA‐binding proteins
Overview of RNA‐binding proteins modifications operating in cells during RNA viral infections.
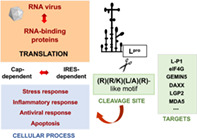
1. INTRODUCTION
RNA‐binding proteins (RBPs) are key actors in virtually all steps of gene expression regulation. These essential host factors are frequently the target of viral proteins. RNA viruses have very limited coding capacity. These pathogens, however, have developed different strategies to take advantage of host factors for viral multiplication in the infected cell (Walsh & Mohr, 2011). RNA viruses hijack host factors required for viral replication, and simultaneously, evade the interference with the abundant cellular mRNA competitors. Not surprisingly, distinct types of RBPs are targeted in viral infections (Flather & Semler, 2015), fully consistent with their prominent role in diverse RNA‐dependent pathways.
In the last decades, numerous studies carried out in cells infected with positive‐strand RNA viruses have documented the interference of cellular functions. Inactivation of specific, essential host factors is a fast process, which affects central cellular functions including antiviral immune response and often implies the inactivation of the normal function of RBPs by modification of the posttranslational state of the proteins involved, redistribution of the cellular compartment, or proteolysis, among other modifications (Figure 1). As typically observed in picornavirus infected cells, the cleavage of essential proteins governing mRNA polyadenylation, nucleus‐cytoplasmic transport, initiation of translation, or stress granules formation induces a strong shutdown of cellular mRNAs. Representative examples are the cleavage stimulation factor 64 (CstF‐64), the nucleoporin Nup98, the polyA‐binding protein (PABP), the translation initiation factors (eIF)‐4G, eIF4A, and eIF5B, and the stress granules proteins G3BP1–2 (Belsham et al., 2000; Castello et al., 2009; de Breyne et al., 2008; Galan et al., 2017; Visser et al., 2019; Weng et al., 2009; White et al., 2007; Zhang et al., 2007). However, proteolysis of host factors follows different kinetics, depending on the virus, the cell line, as well as the multiplicity of infection and the viral protease involved in the cleavage. In general, proteolysis of eIF4GI is the first cleavage event detected in all picornavirus infected cells, rapidly inhibiting cap‐dependent translation. Degradation and/or modification of other host factors follow this step. For instance, cleavage of Nup98, a member of the nuclear pore complex occurs very early in poliovirus infected cells, almost coincident with eIF4GI cleavage, whereas proteolysis of Nup153 is a late event (Castello et al., 2009). Similarly, relative to eIF4GI cleavage, proteolysis of PABP, or G3BP1 in infected cells, is also a late event.
FIGURE 1.
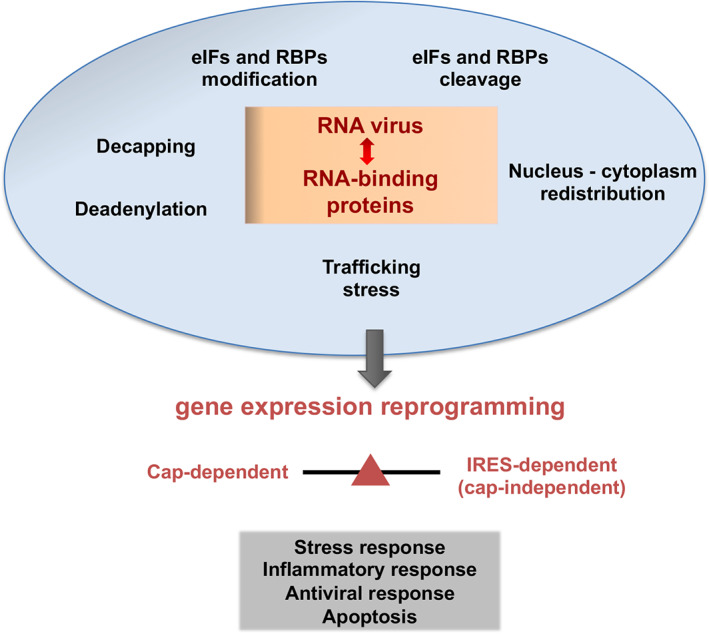
Overview of RNA‐binding proteins modifications operating in cells during RNA viral infections. Major alterations affecting RNA‐dependent pathways in cells infected with RNA viruses are indicated
Yet, despite the inactivation of the respective protein activity for cellular processes, the products remaining after host factor cleavage can still be active for RNA virus multiplication, including viral RNA translation, ultimately leading to extensive gene expression reprogramming. A paradigmatic example was provided by cleavage of the translation initiation factor eIF4G, which results in the separation of the PABP and eIF4E binding domains at the N‐terminus of the protein. Cleavage of eIF4G prevents cap‐dependent translation, in agreement with the general translation shutdown observed in foot‐and‐mouth disease virus (FMDV)‐infected cells (Gradi et al., 2004). However, the C‐terminal polypeptide carrying the eIF4A and mnk1‐binding moieties is sufficient to direct viral protein synthesis, which is driven by an internal ribosome entry site (IRES) element (Belsham & Brangwyn, 1990; Kuhn et al., 1990; Martinez‐Salas et al., 1993).
Picornaviruses contain a single‐stranded RNA genome of positive polarity that encodes a long open reading frame flanked by regulatory untranslated regions (Figure 2a). Immediately following virus entry into susceptible cells and uncoating, the viral genome is translated into a polyprotein that is cotranslationally processed by viral proteases. The vast majority of picornaviruses encode two cysteine proteases, designated 2A and 3C. However, the genome of FMDV encodes the Leader (L) protein at the N‐terminus of the polyprotein (Forss et al., 1984). The L gene product is a protease that can be produced in two forms, designated Lab and Lb, resulting from initiation of protein synthesis at two in frame functional initiation codons (AUG1, AUG2) separated by 84 nucleotides (Belsham, 1992). The inter‐AUG region folds as a hairpin (Andreev et al., 2007) and is involved in start codon recognition favoring selection of AUG2 as the main start codon (Cao et al., 1995; Lopez de Quinto & Martinez‐Salas, 1999). Both, the Lab and Lb proteins have protease activity and can cleave the polyprotein at the junction between the L protein and the structural protein precursor P1 (Strebel & Beck, 1986).
FIGURE 2.
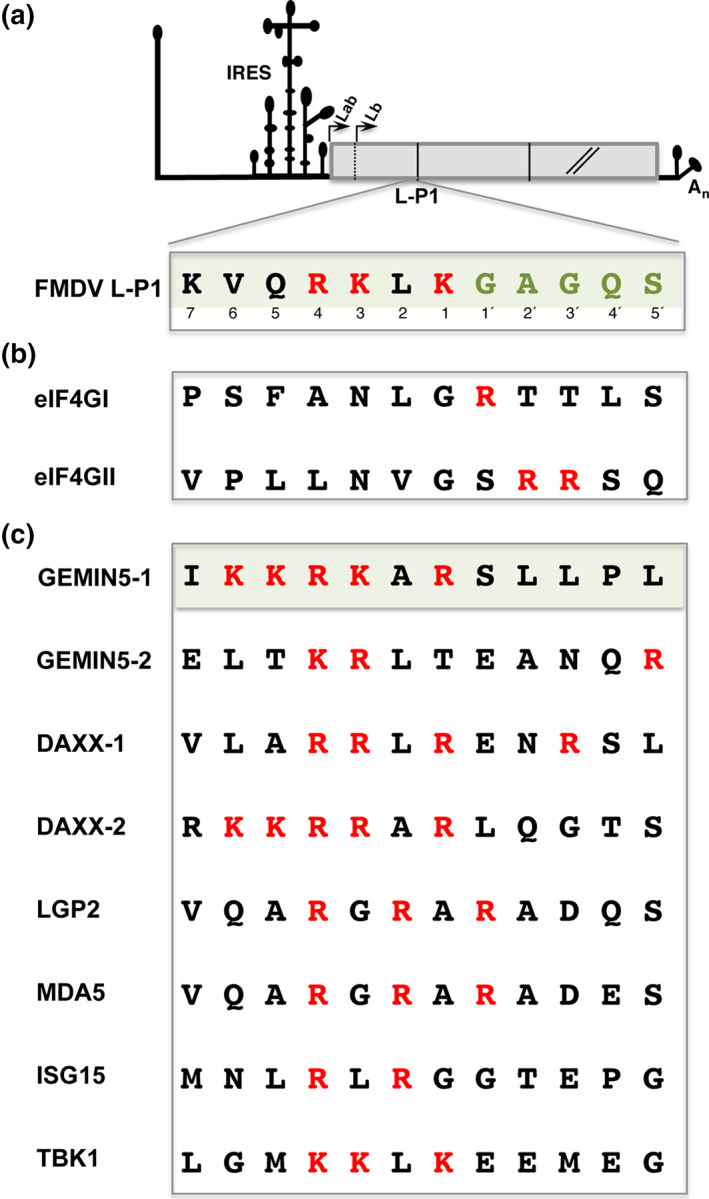
Amino acid sequences of the L protease target sites. (a) Diagram of the FMDV viral genome. For simplicity, only the IRES element and a truncated form of the open reading frame are depicted. Arrows indicate the initiation sites of the Lab and Lb proteins. The amino acid sequence cleaved by Lpro at the L‐P1 junction is indicated. Red letters denote basic residues on the cleavage site, while green letters depict the GAGQS sequence conserved among FMDV isolates. Numbers indicate the position of residues upstream (1–7) and downstream (1′–5′) of the scissile bond. (b) Amino acid sequence of eIF4GI and eIF4GII cleavage sites. (c) Sequence of the identified cleavage sites in Gemin5, Daxx, LGP2, MDA5, ISG15, and TBK1 proteins. Red letters depict the position of basic residues; the shaded box highlights the similarity of the type of amino acid between Gemin5‐1 and FMDV L‐P1
2. LPRO, A PAPAIN‐LIKE PROTEASE TARGETING HOST FACTORS INVOLVED IN CAP‐DEPENDENT TRANSLATION
The FMDV L gene product is a papain‐like cysteine proteinase (thereafter Lpro) that cleaves itself from the nascent viral polyprotein (Guarne et al., 1998). The cleavage site on the L‐P1 junction of the polyprotein is conserved in FMDV genomes (Figure 2a). Noteworthy, this site contains a conserved GAGQS motif at the N‐terminus of P1 and basic residues (K/R) at the C‐terminus. However, while the GAGQS is strongly conserved in a large number of FMDV isolates representing the seven serotypes of this highly variable virus (Carrillo et al., 2005), the tolerance for amino acids substitution toward the amino side of the scissile bond (RKLK) was significantly higher. Presumably, this marked difference in amino acid sequence variability led researchers to infer that the GAGQS was the critical part of the Lpro substrate. Later studies using purified Lpro and short synthetic peptides revealed that the protein possesses specific binding sites from positions 1 to 7 (Santos et al., 2009). When synthetic peptides containing variants of the viral polyprotein sequence were used in vitro, Lpro displayed specificity till position 5′ (Nogueira Santos et al., 2012). Moreover, presence of P instead of A at position 2′ was tolerated, revealing the enzyme flexibility for the GAGQS motif in this in vitro system.
Lpro comprises a globular catalytic domain reminiscent of cysteine proteases of the papain superfamily, and a flexible C‐terminal extension (Guarne et al., 1998). The sequence of the viral polyprotein (KLKGAG) cleaved during viral replication is recognized in a negatively charged groove traversing the active center. Solving the three‐dimensional structure of a protein mutant carrying a substitution of Cys‐133 to Ser defined the active center of the enzyme (Guarne et al., 2000). Similar to canonical papain‐like proteases, the catalytic center of Lpro contains Cys/His residues. However, in marked contrast with other members of this family, Lpro is sensitive to cation concentration increase and it is active only across a narrow pH range, due to three aspartate residues located near the active site which are absent in papain‐like enzymes. The canonical papain‐like proteinases are characterized by a Cys/His/Asn catalytic triad, and also possess conserved tryptophans that stabilize the hydrogen bond formed between His and Asn residues (Berti & Storer, 1995).
Beyond the Lpro proteolytic activity on the viral polyprotein, early studies conducted in FMDV‐infected cells documented the cleavage of the essential translation initiation factor eIF4G soon after infection, even before detectable viral protein expression (Devaney et al., 1988). Also, it was later shown that both forms of the L protease, Lab and Lb, were able to induce the cleavage of eIF4G (Medina et al., 1993). Cleavage of eIF4G in infected cells, as well as in L‐expressing cells prevents cap‐dependent translation, while viral protein synthesis driven by an IRES element remains fully active (Lozano & Martinez‐Salas, 2015).
Mutational studies conducted by different research groups aiming to determine the active site of Lpro indicated that Cys‐51 and His‐148 are required for both L‐P1 and eIF4G cleavages (Roberts & Belsham, 1995), and substitution of Cys‐23 by Ala yielded a protein unable to cleave at the L‐P1 and eIF4GI sites (Piccone et al., 1995). Moreover, Leu‐143 restricts the specificity of Lpro (Mayer et al., 2008). Leu‐143 forms part of the pocket, which was shown to interact with Leu in the polyprotein substrate (Steinberger et al., 2014). The residue 143 is generally occupied by Leu and Met on the viral polyprotein (Carrillo et al., 2005), suggesting that bulky side chains determine Lpro specificity.
3. FEATURES OF THE LPRO CLEAVAGE SITES ON THE HOST FACTORS eIF4GI AND eIF4GII
In mammals, the eIF4G translation initiation factor is expressed in two forms, eIF4GI and eIF4GII. These proteins, however, differ in sequence (Gradi, 1998), showing only 56% sequence similarity concentrated in the central and C‐terminal regions. in vitro Lpro cleavage assays identified the target site in eIF4GI between residues G674 and R675 (Kirchweger et al., 1994) (Figure 2b). This site resembled the enterovirus 2A protease (2Apro) cleavage (Lamphear et al., 1993), despite 2Apro is related to the family of cysteine proteases (Allaire et al., 1994). Likewise, cleavage of eIF4GII induced by Lpro generated cleavage products similar to those induced by enterovirus and rhinovirus 2Apro (Gradi et al., 2003). FMDV Lpro cleaves eIF4GII between residues G700 and S701 (Gradi et al., 2004), immediately adjacent to the site (V699/G700) cleaved by rhinovirus 2Apro.
The two host factors initially identified as Lpro substrates in vitro contained basic residues at positions 1′ or 2′ (Figure 2b). Taking into consideration the L‐P1 cleavage site, these observations suggested that proteins containing basic residues at both 1 and 1′ positions could be Lpro substrates. However, the Lpro cleavage sites on the viral polyprotein (VQRKLKGAGQSS) (Strebel & Beck, 1986), eIF4GI (S669FANLGRTTLST680) and eIF4GII (P695LLNVGSRRSQP706) (Gradi et al., 2004; Kirchweger et al., 1994) differ in sequence (Figure 2b), precluding the definition of a consensus motif for Lpro. Although it was proposed that the R residue at P2 provided the positive charge required for Lpro substrate binding (Guarne et al., 1998), a basic residue at either the 1 or 1′ position flanking the scissile bond was absent in eIF4GII, and an S residue was not observed at the polyprotein cleavage site. Furthermore, since the eIF4GII D683FGRQT688 sequence, partially resembling the cleavage site on eIF4GI, was not substrate of Lpro it was inferred that the presence of F at position 2 impaired Lpro activity (Kuehnel et al., 2004). Further puzzling this issue, another study showed that Lpro recognized a mengovirus polypeptide, the human cyclin A and poliovirus replicase‐related polypeptides (Ziegler et al., 1995), but again, sequence comparisons of the cleavage sites with eIF4GI and eIF4GII revealed no primary sequence similarities between any of these proteins.
Thus, in spite of the information available from all these works, the differences in the cleavage sites reported for eIF4GI, eIF4GII, and the L‐P1 viral polyprotein slowed down the identification of additional targets for this singular protease. In this respect, it is important to point out that target sequences defined by in vitro assays, using relatively large amounts of both enzyme and purified substrate in the absence of other cellular and viral factors, might not fully reflect the molecular interactions occurring under physiological conditions in infected cells.
4. IDENTIFICATION OF THE CLEAVAGE SITE IN GEMIN5 UNVEILED THE RKAR MOTIF AS A CORE RECOGNITION SITE FOR LPRO
Widespread definition of Lpro targets has been hampered by the lack of sufficient number of characterized substrates, hindering the definition of a sequence motif common to all Lpro substrates. As mentioned earlier, translation of the picornavirus RNA into a polyprotein is governed by the IRES element, and such translation regulatory elements are present in every picornavirus genome (Lozano & Martinez‐Salas, 2015; Yamamoto et al., 2017). Picornavirus IRES activity is resistant to the cleavage of eIF4G, which however, causes the shutoff of cap‐dependent protein synthesis affecting the vast majority of cellular mRNAs. In general, degradation of host factors in picornavirus infected cells reveals the viral interference of cellular pathways. All picornaviruses express the chymotrypsin‐like cysteine protease 3C; in addition, enteroviruses express the cysteine protease 2A (mentioned above) whereas aphthoviruses express an active L protease. These viral proteases recognize as substrates host factors related to RNA‐mediated pathways, including important proteins involved in translation control (Bonderoff et al., 2008; Perera et al., 2007; Rodriguez Pulido et al., 2007). Additionally, host proteins involved in antiviral immunity have been shown to be substrates of picornavirus proteases. Representative examples are RIG‐I (the retinoic acid‐inducible gene 1, a cytoplasmic RNA helicase that senses viral infection), the innate immune adaptor molecules MAVS and TRIF (mitochondrial antiviral signaling and the Tol/IL1 receptor domain‐containing adaptor inducing interferon‐beta proteins, respectively), and the p65‐RelA subunit of NF‐κB (Barral et al., 2009; Mukherjee et al., 2011; Neznanov et al., 2005).
Beyond canonical translation factors mentioned earlier, various IRES‐binding factors are targets of picornavirus proteases (Lee et al., 2017; Martinez‐Salas et al., 2015), coupling viral gene expression and inactivation or repurposing of the normal activity of specific RBPs. In the course of experiments looking for host factors interacting with the FMDV IRES, the study by Pineiro et al. (2012) discovered the proteolysis of Gemin5 in FMDV‐infected cells, at similar times than PABP and polypyrimidine tract‐binding protein cleavage, but later than eIF4GI. Immunoblots carried out with soluble lysates prepared from infected cells revealed that Gemin5 is hydrolyzed in FMDV infection but not in cells infected with other picornaviruses (swine vesicular disease virus or encephalomyocarditis virus) (Pineiro et al., 2012). In that regard, Gemin5 had been previously found associated with the FMDV and hepatitis C virus IRES in pull‐down experiments (Pacheco et al., 2008), and subsequently shown to be a downregulator of translation (Pacheco et al., 2009).
Gemin5 is a long multifunctional protein, initially reported as a member of the survival of motor neurons (SMN) complex (Gubitz et al., 2002). Since the SMN complex is responsible for the cytoplasmic delivery of Sm cores onto small nuclear RNAs (Battle et al., 2006), a critical step for small nuclear ribonucleoproteins (snRNPs) biogenesis, disruption of Gemin5 function would be expected to induce widespread defects in mRNA splicing. Moreover, given its role in cap‐dependent and IRES‐dependent translation (Francisco‐Velilla et al., 2016; Pacheco et al., 2009), lack of Gemin5 activity may also have consequences for gene expression in picornavirus infected cells. Gemin5 (1508 amino acids long) contains 14 tryptophan‐aspartic (WD) repeats at the N‐terminal end (Xu et al., 2016), a tetratricopeptide (TPR)‐like domain in the central region responsible for the dimerization of the protein (Moreno‐Morcillo et al., 2020), and a non‐canonical RNA‐binding site at the C‐terminal end (Pineiro et al., 2013) (Figure 3a). Proteolysis of Gemin5 was directly associated with expression of Lpro, yielding two C‐terminal products, p85 and p57 (Pineiro et al., 2015). Interestingly, while the full‐length protein behaves as a negative regulator of IRES‐dependent translation, the p85 fragment upregulates IRES activity (Figure 3a). Thus, cleavage of Gemin5 by FMDV Lpro causes a switch in the activity of this protein leading to opposite functional outcomes, such that the full‐length Gemin5 protein inhibits IRES‐dependent translation, likely acting as an antiviral factor, while the p85 fragment enhances IRES‐dependent translation, presumably acting as a proviral factor. Also consistent with this possibility, a C‐terminal fragment of about 21 kDa, released from a secondary cleavage on p85, which contains the IRES repressor activity (Fernandez‐Chamorro et al., 2014) could be detected in cells overexpressing Lpro but not in FMDV infected cells (Pineiro et al., 2012), presumably due to protein instability or degradation. This scenario partially resembles that of eIF4GI proteolysis in poliovirus infected cells, where cleavage of the full‐length protein inactivates its normal function in cap‐dependent translation, but stimulates IRES‐dependent activity (Hambidge & Sarnow, 1992).
FIGURE 3.
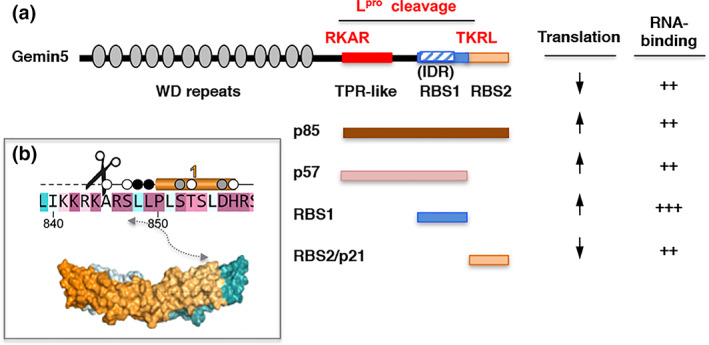
Outline of Gemin5 functional domains. (a) Diagram of the Lpro cleavage sites in Gemin5 protein. The structural domains of Gemin5 (170 kDa) are represented at the top. The WD repeats located at the N‐terminus are involved in snRNPs assembly and ribosome interaction. The TPR‐like domain in the central region mediates dimerization of the protein. The non‐canonical RNA binding site (RBS1) located at the C‐terminus harbors an IDR, and contributes to selective translation control counteracting the negative effect of Gemin5 in global translation. In contrast, the unstable RBS2/p21 moiety downregulates IRES‐dependent translation. The products p85 and p57 resulting from the sequential Lpro cleavage at the RKAR and TKRL sites are depicted. the role of the protein fragments in translation upregulation or downregulation, and RNA‐binding capacity is indicated. (b) Structural features of the RKAR motif. Residues are colored according to the conservation, from magenta (identity) to cyan (variable). The scissors mark the Lpro cleavage motif. The helix 1 is depicted above the sequence. Residues buried up to 40%, 80%, or 100% in the dimerization interface are denoted with a white, gray or black dot. The approximate position of the RKAR motif on the three‐dimensional structure of the TPR‐like dimer is depicted by a double arrow
A search in silico of putative Lpro target sequences within Gemin5 protein identified two potential sites partially homologous to the L‐P1 junction. However, rather than selecting conservation of the scissile‐bond GAGQS downstream motif (Figure 2a), we explored the importance of sequence conservation on the upstream residues. Briefly, this screening was performed selecting tracts of 12 mostly accessible amino acids that had at least two conserved basic residues (R, K) upstream of five residues, of which two were polar, flexible, or hydrophobic, but not aromatic (W, Y). Additionally, the putative sites arising from the search in silico were taken into consideration only if the estimated molecular weight of the resulting fragments matched the proteolysis products observed in lysates prepared from infected cells. Subsequent mutational analysis revealed that the primary cleavage site of Lpro in Gemin5 included the RKAR motif (Figure 2c). Replacement of the positively charged R or K amino acids in the IKKRKARSLLPL region, either individually, by pairs or in triplicate, by the negatively charged E residue was sufficient to inhibit Lpro‐induced cleavage (Pineiro et al., 2012). The second Gemin5 cleavage site for Lpro was the sequence ELTKRLTEANQR (Figure 2c), and as before, substitution of the consecutive KR residues of this motif by EE or PP abolished the cleavage of a tagged‐Gemin5 protein in Lpro‐expressing cells (Pineiro et al., 2012). Neither the RKAR nor the TKRL residues form part of the intrinsically disordered region (IDR) located within the non‐canonical RNA‐binding domain (RBS1) (Francisco‐Velilla et al., 2020). Interestingly, the Fas‐ligand Daxx was also identified as a putative Lpro substrate in the in silico analysis by carrying a VLARRLRENRSL motif (Figure 2c). Daxx is a multifunctional adaptor that plays key roles in transcriptional control, apoptosis, and the innate immune antiviral response (Yang et al., 1997). Consistently, Daxx was also proteolyzed in FMDV‐infected and Lpro‐expressing cells, and substitution of RRLR to EELR abrogated Lpro cleavage of the mutant protein. Therefore, disruption of the RKAR and RRLR motifs in Gemin5 and Daxx, respectively, abolished cleavage by the L protease.
Comparison of the Lpro target sequence within the viral L‐P1 junction with Gemin5 and Daxx shows strong homology in positions centered on the RKAR motif, but also extends beyond these four amino acids. Specifically, there is a notable enrichment in positively charged residues in positions 1 to 7, while hydrophobic residues are placed in positions 1′ to 5′ (Figure 2c). Concerning the structural organization of the Gemin5 cleavage site, the three‐dimensional structure of the amino terminus of p85 was recently resolved (Moreno‐Morcillo et al., 2020). This region adopts a TPR‐like structure organized in 17 helices of which the RKAR motif is located at the amino end of the TPR‐like moiety (Figure 3b). Importantly, this domain is responsible for the dimerization of the protein, both in vitro and in living cells. However, the residues around this motif form part of a loop and they are not buried in the dimer, indicating that they could be accessible both in the monomer and the dimer conformation of the protein. In addition, as mentioned above, the target sequence of Gemin5 identified by mutational analysis yielding p85, as well as the RRLR and RRAR motifs found in Daxx, shows higher resemblance with the L‐P1 cleavage site than to eIF4GI or eIF4GII sites. Therefore, the sequence (R)(R/K)(L/A)(R) defined a minimal core motif useful to identify putative targets of Lpro among host factors.
Identification of the motif recognized in Gemin5 provided the basis for the search in silico of novel substrates of this protease in mammalian cells (Pineiro et al., 2012). Among others, putative targets of Lpro found to contain variations of the (R)(R/K)(L/A)(R) motif were the nuclear fragile X mental retardation‐interacting protein 2 (NUFIP2) (KKRKARRN), the chromodomain helicase DNA‐binding protein 1 (CHD1) (KRRKARAK), the methyl CpG‐binding protein 2 (MECP2) (GWTRKLKQR), the melanoma‐associated antigen B1 (MAGEB1) (KRRKAREE), the melanoma‐associated antigen B2 (MAGEB2) (KRRKARDET), the porcupine‐homologue isoform CRA (PORCN) (KKRKARGT), the vang‐like protein 1 (VANGL1) (KKRKARLV), the testis specific Y encoded‐like protein 2 (TSPYL2) (KKRKTRGR), and Neuroguidin (NGDN) (AKRRALS), an eIF4E‐binding and cytoplasmic polyadenylation element‐binding (CPEB)‐protein (Pineiro et al., 2012). Furthermore, PABP1 that carries the RRSL motif was previously shown to be substrate of Lpro in transfected as well as FMDV‐infected cells (Rodriguez Pulido et al., 2007). Collectively, the list of proteins retrieved from databases carrying variants of this R/K‐rich motif includes nucleic acid‐binding proteins, and factors involved in cell cycle arrest, differentiation, and tumorigenesis. Not surprisingly, nucleic acid‐binding proteins, and in particular RBPs, are enriched in basic residues, suggesting that Lpro targets could be more abundant within proteins related to RNA‐mediated pathways. Although these proteins need to be tested for Lpro proteolysis, conserved motifs in RBPs are often related to the presence of functional domains that preserve the structure–function relationship. However, it is not expected that all computationally predicted targets will be substrates of the protease and experimental evidence should be provided. Differences in accessibility within the cellular environment, as well as three‐dimensional protein structure and/or interaction with other factors in macromolecular complexes may impair cleavage under physiological conditions. In summary, the finding that the Lpro target sequences within the viral L‐P1 junction and several host factors, namely Gemin5 and Daxx, aligned well in positions centered on the RKAR motif provided a useful tool to identify novel targets of this singular protease in mammalian cells.
The discovery of Gemin5 proteolysis in infected cells opened new avenues to investigate its organization in structural domains and the functional relevance in the context of normal cell physiology. Similarly, the study of additional substrates could help to understand the activity of the respective protein targets in response to different situations. For instance, identification of the Lpro cleavage at the RKAR motif led us to study the influence of the separated fragments of the protein in translation control. These studies indicated that while the full‐length protein behaves as a negative translation regulator, the p85 fragment resulting from Lpro cleavage stimulates IRES‐dependent translation. As a result of this unexpected observation, subsequent examination of the p85 fragment revealed the presence of a non‐canonical bipartite RNA‐binding domain at the C‐terminus, designated RBS1‐2 (Figure 3a). The unusual features of the RBS1 domain prompted the search of cellular RNAs interacting with this singular domain, underscoring its selective role in translation control (Francisco‐Velilla et al., 2016, 2018, 2019; Pineiro et al., 2013). Evidence accumulated from different laboratories has shown that this multifaceted protein is involved in regulation of cellular mRNA translation, transplicing, and gene expression reprogramming (Martinez‐Salas et al., 2020). Furthermore, cleavage of Gemin5 during FMDV infection adds to the proteolysis of the RNA helicase DDX20/Gemin3 by the enterovirus 2Apro (Almstead & Sarnow, 2007). Gemin3 is not fully cleaved by the enterovirus 2Apro in poliovirus infected cells and occurs late relative to eIF4GI. Notwithstanding, the resulting amino‐terminal proteolytic product retains intact the RNA helicase motifs (I to VI), suggesting that cleavage of Gemin3 has the potential to alter host activities unconnected to its effect on snRNP assembly. This possibility is reminiscent of the properties observed in the cleavage products of Gemin5 following proteolysis by FMDV Lpro (Francisco‐Velilla et al., 2019).
5. LPRO IMPACT IN RNA‐MEDIATED PATHWAYS INVOLVED IN ANTIVIRAL DEFENSE
Recognition of viral RNA by (RIG‐I)‐like receptors (RLRs) activates a signaling cascade that leads to transcription of type I interferon (IFN‐α/β) genes and the subsequent induction of expression of hundreds of antiviral proteins stimulated by IFN in infected and uninfected cells (Hur, 2019). RLRs primarily recognize viral RNAs, acting as sensors of pathogen associated molecular patterns, not found in host cells (Yoo et al., 2014). RLRs comprise a family of three members, RIG‐I, the melanoma differentiation associated gene 5 (MDA5), and the Laboratory of Genetics and Physiology 2 (LGP2). RIG‐I recognizes RNAs with a 5′‐triphosphate moiety, whereas MDA5 preferentially recognizes dsRNAs. Additionally, LGP2 synergizes with MDA5 to promote the antiviral response induced by picornavirus infection. RLRs direct signaling toward the downstream effector MAVS. Aggregation of MAVS protein activates the transcription factors IRF‐3, IRF‐7, and NF‐κB in the cytoplasm resulting in their translocation to the nucleus and triggering of the expression of IFNs and pro‐inflammatory cytokines (Mogensen, 2009). Interestingly, the RNA helicase LGP2 undergoes proteolysis during FMDV infection, resulting in a drastic decrease of full‐length LGP2 and accumulation of cleavage products (Rodriguez Pulido et al., 2018). The Lpro target site in LGP2 was mapped to the RGRAR sequence (Figure 2c) in a conserved helicase motif, strongly resembling the RKAR, RRLR, or RRAR cleavage sites in Gemin5 and Daxx (Pineiro et al., 2012). Replacement of the RGRAR sequence for EGEAE abrogated LGP2 cleavage by Lpro, defining the RGRAR sequence in the conserved helicase motif VI as the Lpro target site in LGP2. The expression of an inactive form of Lpro failed to cleave LGP2, strongly suggesting that LGP2 is a substrate of Lpro. Remarkably, cleavage of LGP2 removes part of the helicase domain and the CTD region, thereby inactivating critical functions of this protein. The CTD region determines RNA recognition specificity, and synergizes MDA5 activity (Deddouche et al., 2014; Uchikawa et al., 2016). In agreement with this, expression of catalytically active Lpro subverted the type I IFN antiviral response promoted by LGP2 within cells (Rodriguez Pulido et al., 2018). These findings suggested that LGP2 cleavage by Lpro was a new immune evasion mechanism exerted by RNA viruses. FMDV is highly sensitive to IFNs and has evolved different strategies to circumvent the antiviral response elicited in infected cells at different levels (Rodriguez Pulido & Saiz, 2017), many of them relying on the enzymatic activity of Lpro, which is considered as a key virulence factor. Cleavage of the essential initiation factor eIF4GI early during infection inhibits translation of cellular capped mRNAs, hence preventing the synthesis of IFN and other cytokines. The Lpro‐dependent degradation of the p65 subunit of NF‐κB for IFN suppression has been reported (de los Santos et al., 2007). Interestingly, Lpro can remove ubiquitin molecules from several proteins required for type I IFN expression and signaling pathway, such as RIG‐I, TBK1, TRAF3, and TRAF6 (Wang et al., 2011).
The cellular response against picornavirus infection largely relies on the early detection of the dsRNA intermediates generated during infection by MDA5 (Feng et al., 2012), while LGP2 promotes the viral RNA‐MDA5 interaction (Bruns & Horvath, 2015). Interestingly, MDA5 pulldown carried out with FMDV‐infected cell lysates retrieved FMDV viral RNA (Rodriguez Pulido et al., 2020). Given that the RGRAR sequence within motif VI in helicase domain 2 that provided the LGP2 cleavage site for Lpro (Rodriguez Pulido et al., 2018) is conserved in MDA5, it was plausible to hypothesize that MDA5 could be also a target for Lpro. Indeed, the expected size N‐ and C‐terminal fragments generated after Lpro cleavage were readily detected in FMDV‐infected cells. The cleavage pattern was confirmed by overexpression of MDA5 and the catalytically active Lpro. Also, a mutant of MDA5 bearing substitutions in the RGRAR motif was fully resistant to cleavage, unequivocally identifying MDA5 as a target for Lpro. Cleavage at the RGRAR sequence would remove part of the helicase motif VI and the CTD from both MDA5 and LGP2, yielding inactive proteins for antiviral signaling. In accordance with these results, cells expressing the non‐cleavable proteins retained antiviral activity. Hence, FMDV Lpro targets may share conserved motifs in functional domains playing a critical role on the antiviral response. These findings suggest that FMDV has developed an “all‐in‐one” solution to prevent the innate immune response induced by both RLRs, giving the virus a clear advantage for replication and spread in the host.
In response to viral infection, cells trigger an inflammatory response that at least in part relies on ubiquitin posttranslational modifications. Conversely, viruses have acquired specialized strategies to antagonize the host signaling processes reversing these modifications. Initiation of the IFN signaling pathway stimulates the expression of a large number of genes (designated ISGs) responsible for triggering the antiviral response (Ivashkiv & Donlin, 2014). One of these genes is the ubiquitin‐like modifier ISG15, which comprises two ubiquitin fold domains in tandem and is attached to Lys residues on target proteins via its C‐terminal Gly‐Gly motif. Cotranslational attachment of ISG15 to viral capsid proteins inhibits virus assembly (Durfee et al., 2010). Lpro cleaves a peptide bond in the C‐terminus of ISG15 (Figure 2c) resulting in removal of the modifier from substrate proteins by an irreversible inactivation mechanism during FMDV infection (Swatek et al., 2018).
Collectively, identification of Lpro substrates can contribute to our understanding of the mechanistic basis of the inhibitory activity of Lpro in IFN‐α/β induction. Relevant proteins of this pathway (RIG‐I, MAVS, TBK1, IRF‐3) are regulated by ubiquitin‐dependent modifications. In this respect, the TBKI kinase, responsible for phosphorylation of the transcription factor IRF3 is also cleaved by Lpro in FMDV‐infected cells (Visser et al., 2020) at a KKLK site, resembling the minimal motif defined in Daxx and Gemin5 (Figure 2c). Interestingly, the mutations in Lpro that impaired the reduction of IFN‐β mRNA levels displayed cleavage defects and/or degradation of the signaling proteins MAVS, TBK1, and NF‐κB p65, establishing a direct link between Lpro activity and IFN reduction. Thus, Lpro's ability to cleave RLR signaling proteins but not its deubiquitination/deISGylation activity correlates with suppressing IFN‐α/β gene transcription (Medina et al., 2020; Visser et al., 2020).
In summary, the cleavage by Lpro during FMDV infection of essential proteins, such as eIF4G, Gemin5, MDA5, or LGP2, indicates the evolutionary selection of a robust mechanism to interfere with the action of key host proteins, subverting their activity and favoring the expression of the viral RNA. Moreover, the identification of the cleavage site of Lpro in diverse host factors led us to compile the basic features that appear to characterize the substrates of this viral protease. Thus, searching for sequences resembling the (R)(R/K)(L/A)(R) motif could facilitate the identification of novel Lpro targets after experimental verification in infected or Lpro‐expressing cells.
6. CONCLUDING REMARKS AND PERSPECTIVES
The interference in RNA‐driven processes has become a widespread mechanism of evading the antiviral response elicited in infected cells (Girardi et al., 2020). However, both the cellular response to RNA virus infection and the subsequent viral‐induced antagonism depend on the execution of spatial and temporal ordered events. Early during infection, distinct types of RNA viruses induce the inactivation of specific RBPs, critical for RNA‐dependent pathways and therefore, required for host survival. This includes all steps of the RNA lifespan, from transcription to maturation, transport, translation, and degradation. Remarkably, many proteins involved in RNA‐dependent pathways display a modular structure, combining specific subsets of domains in distinct functional factors (Jarvelin et al., 2016). This is reflected in the presence of specific repeat motifs in the plethora of cellular RBPs. Conversely, this building‐block strategy allows the development of counter‐stratagems that could simultaneously disable the normal activity of a subset of host factors and subvert the cleavage products for viral‐related functions. Noteworthy, the strong shutoff observed in infected cells is driven by changes in posttranslation modification and/or proteolysis of dedicated essential translation factors (Jan et al., 2016; Kwan & Thompson, 2019; Mailliot & Martin, 2018). This is an ordered response that allows rapid silencing of cellular processes involving RNA activity.
Not surprisingly, RNA viruses have developed a sophisticated master plan including a pleyade of molecular strategies aimed to counteract the antiviral response elicited in infected cells. A paradigmatic example is illustrated by the translation initiation mechanism governed by IRES elements, a cap‐independent manner to initiate protein synthesis that is resistant to posttranslation modifications and/or proteolysis of host factors induced in infected cells (Lozano & Martinez‐Salas, 2015). Another paradigmatic example is epitomized by the L protease of FMDV, a singular papain‐like proteinase that cleaves itself from the nascent viral polyprotein chain, and also targets host factors involved in RNA‐mediated processes (Steinberger et al., 2014). Most remarkably, these combined strategies employed by FMDV synergize, as the proteolytic activity of Lpro on specific RBPs favors IRES‐dependent translation used by the viral RNA and also abrogates the antiviral immune response elicited in infected cells.
Beyond the cleavage site on the polyprotein, early works reported the cleavage sites in the scaffolding factors eIF4GI and eIF4GII (Devaney et al., 1988; Gradi et al., 2004; Kirchweger et al., 1994). However, additional targets of this unusual protease were hidden for a long time. Differences in the amino acid sequence of the three proven cleavage sites for this protease hampered the identification of additional substrates by computational and experimental approaches. More recently, a twist in the approach for searching putative Lpro cleavage sites allowed the identification of a target sequence centered on the RKAR motif in Gemin5, and subsequently predicted and experimentally mapped the cleavage site RRLR in Daxx (Pineiro et al., 2012). Despite these proteins share a similar (R)(R/K)(L/A)(R) motif, they are involved in different steps of RNA metabolism (snRNPs assembly, translation control, apoptosis and antiviral response). This observation suggests that combinatorial targeting of host factors sharing the Lpro cleavage motif may be a strategy to disable almost simultaneously families of proteins at early times during infection, thereby inactivating a whole set of cellular RNA‐driven events. Importantly, small variation of this motif (RGRAR) has been unequivocally identified in LGP2 and MDA5, two RNA helicases that sense the presence of viral RNAs in infected cells, which become cleaved and inactivated in cells infected with FMDV or expressing the active form of Lpro (Rodriguez Pulido et al., 2018, 2020). Additionally, two proteins related to the IFN‐pathway, ISG15 and TBK1 (Swatek et al., 2018; Visser et al., 2020), are cleaved by this protease at a very similar motif.
Therefore, the (R)(R/K)(L/A)(R)‐like motif has become a signature for Lpro targets, which may open new ways to search for novel substrates of this virulence factor. Inactivation of the IFN pathway is a common feature of many viral infections (Feng et al., 2014; Nelemans & Kikkert, 2019; Rodriguez Pulido & Saiz, 2017). In depth study of virulence factors and viral immunomodulatory proteins can lead us to their targets, which are often vital signaling nodes for cell homeostasis that play a key role during infection. Even small RNA viruses encode multifunctional proteins evolved to counteract crucial host factors. As discussed in this review, the FMDV L protease is an outstanding example. Undoubtedly, improving our knowledge on Lpro activity, specificity, and function in infected cells can take advantage of the recent development of global approaches to identify subtle changes in the cellular proteome (Garcia‐Moreno et al., 2019). Therefore, understanding how Lpro and other viral proteases recognize and cleave host targets may help to design and/or improve therapeutic tools to combat FMDV in particular, and RNA viruses in general. In addition, it may contribute to disentangle the intricacies of the antiviral immune response.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Margarita Saiz: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; validation; visualization; writing‐original draft; writing‐review and editing. Encarna Martinez‐Salas: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; validation; visualization; writing‐original draft; writing‐review and editing.
RELATED WIREs ARTICLE
TRIM25 and its emerging RNA‐binding roles in antiviral defense
ACKNOWLEDGMENTS
We thank Miguel Rodriguez‐Pulido, David Piñeiro, and lab members for their contribution to this work.
Saiz M, Martinez‐Salas E. Uncovering targets of the Leader protease: Linking RNA‐mediated pathways and antiviral defense. WIREs RNA. 2021;12:e1645. 10.1002/wrna.1645
Edited by: Jeff Wilusz, Editor‐in‐Chief
Funding information Consejería de Educación e Investigación, Grant/Award Numbers: B2017/BMD‐3770, S2018/BAA‐4370; Consejo Superior de Investigaciones Científicas, Grant/Award Number: 201820I019; Ministerio de Economia y Competitividad, Grant/Award Numbers: BFU2017‐84492‐R, AGL2014‐58675
REFERENCES
- Allaire, M. , Chernaia, M. M. , Malcolm, B. A. , & James, M. N. (1994). Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin‐like serine proteinases. Nature, 369(6475), 72–76. [DOI] [PubMed] [Google Scholar]
- Almstead, L. L. , & Sarnow, P. (2007). Inhibition of U snRNP assembly by a virus‐encoded proteinase. Genes & Development, 21(9), 1086–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev, D. E. , Fernandez‐Miragall, O. , Ramajo, J. , Dmitriev, S. E. , Terenin, I. M. , Martinez‐Salas, E. , & Shatsky, I. N. (2007). Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot‐and‐mouth disease virus RNA. RNA, 13(8), 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, P. M. , Sarkar, D. , Fisher, P. B. , & Racaniello, V. R. (2009). RIG‐I is cleaved during picornavirus infection. Virology, 391(2), 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle, D. J. , Lau, C. K. , Wan, L. , Deng, H. , Lotti, F. , & Dreyfuss, G. (2006). The Gemin5 protein of the SMN complex identifies snRNAs. Molecular Cell, 23(2), 273–279. [DOI] [PubMed] [Google Scholar]
- Belsham, G. J. (1992). Dual initiation sites of protein synthesis on foot‐and‐mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. The EMBO Journal, 11(3), 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G. J. , & Brangwyn, J. K. (1990). A region of the 5′ noncoding region of foot‐and‐mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: Involvement with the role of L protease in translational control. Journal of Virology, 64(11), 5389–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G. J. , McInerney, G. M. , & Ross‐Smith, N. (2000). Foot‐and‐mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. Journal of Virology, 74(1), 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti, P. J. , & Storer, A. C. (1995). Alignment/phylogeny of the papain superfamily of cysteine proteases. Journal of Molecular Biology, 246(2), 273–283. [DOI] [PubMed] [Google Scholar]
- Bonderoff, J. M. , Larey, J. L. , & Lloyd, R. E. (2008). Cleavage of poly(A)‐binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site‐mediated translation. Journal of Virology, 82(19), 9389–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns, A. M. , & Horvath, C. M. (2015). LGP2 synergy with MDA5 in RLR‐mediated RNA recognition and antiviral signaling. Cytokine, 74(2), 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X. , Bergmann, I. E. , Fullkrug, R. , & Beck, E. (1995). Functional analysis of the two alternative translation initiation sites of foot‐and‐mouth disease virus. Journal of Virology, 69(1), 560–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo, C. , Tulman, E. R. , Delhon, G. , Lu, Z. , Carreno, A. , Vagnozzi, A. , Kutish, G. F. , & Rock, D. L. (2005). Comparative genomics of foot‐and‐mouth disease virus. Journal of Virology, 79(10), 6487–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello, A. , Izquierdo, J. M. , Welnowska, E. , & Carrasco, L. (2009). RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. Journal of Cell Science, 122(Pt 20), 3799–3809. [DOI] [PubMed] [Google Scholar]
- de Breyne, S. , Bonderoff, J. M. , Chumakov, K. M. , Lloyd, R. E. , & Hellen, C. U. (2008). Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology, 378(1), 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T. , Diaz‐San Segundo, F. , & Grubman, M. J. (2007). Degradation of the nuclear factor kappa B during foot‐and‐mouth disease virus infection. Journal of Virology, 81, 12803–12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche, S. , Goubau, D. , Rehwinkel, J. , Chakravarty, P. , Begum, S. , Maillard, P. V. , Borg, A. , Matthews, N. , Feng, Q. , van Kuppeveld, F. J. M. , & Reis e Sousa, C. (2014). Identification of an LGP2‐associated MDA5 agonist in picornavirus‐infected cells. eLife, 3, e01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaney, M. A. , Vakharia, V. N. , Lloyd, R. E. , Ehrenfeld, E. , & Grubman, M. J. (1988). Leader protein of foot‐and‐mouth disease virus is required for cleavage of the p220 component of the cap‐binding protein complex. Journal of Virology, 62(11), 4407–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, L. A. , Lyon, N. , Seo, K. , & Huibregtse, J. M. (2010). The ISG15 conjugation system broadly targets newly synthesized proteins: Implications for the antiviral function of ISG15. Molecular Cell, 38(5), 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q. , Hato, S. V. , Langereis, M. A. , Zoll, J. , Virgen‐Slane, R. , Peisley, A. , Hur, S. , Semler, B. L. , van Rij, R. P. , & van Kuppeveld, F. J. (2012). MDA5 detects the double‐stranded RNA replicative form in picornavirus‐infected cells. Cell Reports, 2(5), 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q. , Langereis, M. A. , & van Kuppeveld, F. J. (2014). Induction and suppression of innate antiviral responses by picornavirus. Cytokine & Growth Factor Reviews, 25, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Chamorro, J. , Pineiro, D. , Gordon, J. M. , Ramajo, J. , Francisco‐Velilla, R. , Macias, M. J. , & Martinez‐Salas, E. (2014). Identification of novel non‐canonical RNA‐binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Research, 42(9), 5742–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flather, D. , & Semler, B. L. (2015). Picornaviruses and nuclear functions: Targeting a cellular compartment distinct from the replication site of a positive‐strand RNA virus. Frontiers in Microbiology, 6, 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forss, S. , Strebel, K. , Beck, E. , & Schaller, H. (1984). Nucleotide sequence and genome organization of foot‐and‐mouth disease virus. Nucleic Acids Research, 12(16), 6587–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco‐Velilla, R. , Azman, E. B. , & Martinez‐Salas, E. (2019). Impact of RNA‐protein interaction modes on translation control: The versatile multidomain protein Gemin5. BioEssays, 41(4), e1800241. [DOI] [PubMed] [Google Scholar]
- Francisco‐Velilla, R. , Embarc‐Buh, A. , Rangel‐Guerrero, S. , Basu, S. , Kundu, S. , & Martinez‐Salas, E. (2020). RNA‐protein coevolution study of Gemin5 uncovers the rple of the PXSS motif of RBS1 domain for RNA binding. RNA Biology, 17(9), 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco‐Velilla, R. , Fernandez‐Chamorro, J. , Dotu, I. , & Martinez‐Salas, E. (2018). The landscape of the non‐canonical RNA‐binding site of Gemin5 unveils a feedback loop counteracting the negative effect on translation. Nucleic Acids Research, 46(14), 7339–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco‐Velilla, R. , Fernandez‐Chamorro, J. , Ramajo, J. , & Martinez‐Salas, E. (2016). The RNA‐binding protein Gemin5 binds directly to the ribosome and regulates global translation. Nucleic Acids Research, 44(17), 8335–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, A. , Lozano, G. , Pineiro, D. , & Martinez‐Salas, E. (2017). G3BP1 interacts directly with the FMDV IRES and negatively regulates translation. The FEBS Journal, 284(19), 3202–3217. [DOI] [PubMed] [Google Scholar]
- Garcia‐Moreno, M. , Noerenberg, M. , Ni, S. , Järvelin, A. I. , González‐Almela, E. , Lenz, C. E. , Bach‐Pages, M. , Cox, V. , Avolio, R. , Davis, T. , Hester, S. , Sohier, T. J. M. , Li, B. , Heikel, G. , Michlewski, G. , Sanz, M. A. , Carrasco, L. , Ricci, E. P. , Pelechano, V. , … Castello, A. (2019). System wide profiling of RNA‐binding proteins uncovers key regulators of virus infection. Molecular Cell, 74(1), 196–211.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi, E. , Pfeffer, S. , Baumert, T. F. , & Majzoub, K. (2020). Roadblocks and fast tracks: How RNA‐binding proteins affect the viral RNA journey in the cell. Seminars in Cell and Developmental Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A. , Foeger, N. , Strong, R. , Svitkin, Y. V. , Sonenberg, N. , Skern, T. , & Belsham, G. J. (2004). Cleavage of eukaryotic translation initiation factor 4GII within foot‐and‐mouth disease virus‐infected cells: Identification of the L‐protease cleavage site in vitro. Journal of Virology, 78(7), 3271–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A. , Svitkin, Y. V. , Imataka, H. , & Sonenberg, N. (1998). Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci U S A, 95(19), 11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A. , Svitkin, Y. V. , Sommergruber, W. , Imataka, H. , Morino, S. , Skern, T. , & Sonenberg, N. (2003). Human rhinovirus 2A proteinase cleavage sites in eukaryotic initiation factors (eIF) 4GI and eIF4GII are different. Journal of Virology, 77(8), 5026–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarne, A. , Hampoelz, B. , Glaser, W. , Carpena, X. , Tormo, J. , Fita, I. , & Skern, T. (2000). Structural and biochemical features distinguish the foot‐and‐mouth disease virus leader proteinase from other papain‐like enzymes. Journal of Molecular Biology, 302(5), 1227–1240. [DOI] [PubMed] [Google Scholar]
- Guarne, A. , Tormo, J. , Kirchweger, R. , Pfistermueller, D. , Fita, I. , & Skern, T. (1998). Structure of the foot‐and‐mouth disease virus leader protease: A papain‐like fold adapted for self‐processing and eIF4G recognition. The EMBO Journal, 17(24), 7469–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz, A. K. , Mourelatos, Z. , Abel, L. , Rappsilber, J. , Mann, M. , & Dreyfuss, G. (2002). Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. The Journal of Biological Chemistry, 277(7), 5631–5636. [DOI] [PubMed] [Google Scholar]
- Hambidge, S. J. , & Sarnow, P. (1992). Translational enhancement of the poliovirus 5′ noncoding region mediated by virus‐encoded polypeptide 2A. Proceedings of the National Academy of Sciences of the United States of America, 89(21), 10272–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur, S. (2019). Double‐stranded RNA sensors and modulators in innate immunity. Annual Review of Immunology, 37, 349–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv, L. B. , & Donlin, L. T. (2014). Regulation of type I interferon responses. Nature Reviews. Immunology, 14(1), 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, E. , Mohr, I. , & Walsh, D. (2016). A cap‐to‐tail guide to mRNA translation strategies in virus infected cells. Annual Review of Virology, 3, 283–307. [DOI] [PubMed] [Google Scholar]
- Jarvelin, A. I. , Noerenberg, M. , Davis, I. , & Castello, A. (2016). The new (dis)order in RNA regulation. Cell Communication and Signaling: CCS, 14, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchweger, R. , Ziegler, E. , Lamphear, B. J. , Waters, D. , Liebig, H. D. , Sommergruber, W. , Sobrino, F. , Hohenadl, C. , Blaas, D. , & Rhoads, R. E. (1994). Foot‐and‐mouth disease virus leader proteinase: Purification of the Lb form and determination of its cleavage site on eIF‐4 gamma. Journal of Virology, 68(9), 5677–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnel, E. , Cencic, R. , Foeger, N. , & Skern, T. (2004). Foot‐and‐mouth disease virus leader proteinase: Specificity at the P2 and P3 positions and comparison with other papain‐like enzymes. Biochemistry, 43(36), 11482–11490. [DOI] [PubMed] [Google Scholar]
- Kuhn, R. , Luz, N. , & Beck, E. (1990). Functional analysis of the internal translation initiation site of foot‐and‐mouth disease virus. Journal of Virology, 64(10), 4625–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, T. , & Thompson, S. R. (2019). Noncanonical translation initiation in eukaryotes. Cold Spring Harbor Perspectives in Biology, 11, a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear, B. J. , Yan, R. , Yang, F. , Waters, D. , Liebig, H. D. , Klump, H. , Kuechler, E. , & Rhoads, R. E. (1993). Mapping the cleavage site in protein synthesis initiation factor eIF‐4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. The Journal of Biological Chemistry, 268(26), 19200–19203. [PubMed] [Google Scholar]
- Lee, K. M. , Chen, C. J. , & Shih, S. R. (2017). Regulation mechanisms of viral IRES‐driven translation [review]. Trends in Microbiology, 25(7), 546–561. [DOI] [PubMed] [Google Scholar]
- Lopez de Quinto, S. , & Martinez‐Salas, E. (1999). Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology, 255(2), 324–336. [DOI] [PubMed] [Google Scholar]
- Lozano, G. , & Martinez‐Salas, E. (2015). Structural insights into viral IRES‐dependent translation mechanisms. Current Opinion in Virology, 12, 113–120. [DOI] [PubMed] [Google Scholar]
- Mailliot, J. , & Martin, F. (2018). Viral internal ribosomal entry sites: Four classes for one goal. WIREs RNA, 9(2). 10.1002/wrna.1458. [DOI] [PubMed] [Google Scholar]
- Martinez‐Salas, E. , Embarc‐Buh, A. , & Francisco‐Velilla, R. (2020). Emerging roles of Gemin5: From snRNPs assembly to translation control. International Journal of Molecular Sciences, 21(11):3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Salas, E. , Francisco‐Velilla, R. , Fernandez‐Chamorro, J. , Lozano, G. , & Diaz‐Toledano, R. (2015). Picornavirus IRES elements: RNA structure and host protein interactions. Virus Research, 206, 62–73. [DOI] [PubMed] [Google Scholar]
- Martinez‐Salas, E. , Saiz, J. C. , Davila, M. , Belsham, G. J. , & Domingo, E. (1993). A single nucleotide substitution in the internal ribosome entry site of foot‐and‐mouth disease virus leads to enhanced cap‐independent translation in vivo. Journal of Virology, 67(7), 3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, C. , Neubauer, D. , Nchinda, A. T. , Cencic, R. , Trompf, K. , & Skern, T. (2008). Residue L143 of the foot‐and‐mouth disease virus leader proteinase is a determinant of cleavage specificity. Journal of Virology, 82(9), 4656–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, G. N. , Azzinaro, P. , Ramirez‐Medina, E. , Gutkoska, J. , Fang, Y. , Diaz‐San Segundo, F. , & de Los Santos, T. (2020). Impairment of the DeISGylation activity of foot‐and‐mouth disease virus lpro causes attenuation in vitro and in vivo. Journal of Virology, 94(13), e00341–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, M. , Domingo, E. , Brangwyn, J. K. , & Belsham, G. J. (1993). The two species of the foot‐and‐mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology, 194(1), 355–359. [DOI] [PubMed] [Google Scholar]
- Mogensen, T. H. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clinical Microbiology Review, 22(2), 240–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Morcillo, M. , Francisco‐Velilla, R. , Embarc‐Buh, A. , Fernandez‐Chamorro, J. , Ramon‐Maiques, S. , & Martinez‐Salas, E. (2020). Structural basis for the dimerization of Gemin5 and its role in protein recruitment and translation control. Nucleic Acids Research, 48(2), 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, A. , Morosky, S. A. , Delorme‐Axford, E. , Dybdahl‐Sissoko, N. , Oberste, M. S. , Wang, T. , & Coyne, C. B. (2011). The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathogens, 7(3), e1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans, T. , & Kikkert, M. (2019). Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses, 11(10):961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neznanov, N. , Chumakov, K. M. , Neznanova, L. , Almasan, A. , Banerjee, A. K. , & Gudkov, A. V. (2005). Proteolytic cleavage of the p65‐RelA subunit of NF‐kappaB during poliovirus infection. The Journal of Biological Chemistry, 280(25), 24153–24158. [DOI] [PubMed] [Google Scholar]
- Nogueira Santos, J. A. , Assis, D. M. , Gouvea, I. E. , Judice, W. A. , Izidoro, M. A. , Juliano, M. A. , Skern, T. , & Juliano, L. (2012). Foot and mouth disease leader protease (Lbpro): Investigation of prime side specificity allows the synthesis of a potent inhibitor. Biochimie, 94(3), 711–718. [DOI] [PubMed] [Google Scholar]
- Pacheco, A. , Lopez de Quinto, S. , Ramajo, J. , Fernandez, N. , & Martinez‐Salas, E. (2009). A novel role for Gemin5 in mRNA translation. Nucleic Acids Research, 37(2), 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco, A. , Reigadas, S. , & Martinez‐Salas, E. (2008). Riboproteomic analysis of polypeptides interacting with the internal ribosome‐entry site element of foot‐and‐mouth disease viral RNA. Proteomics, 8(22), 4782–4790. [DOI] [PubMed] [Google Scholar]
- Perera, R. , Daijogo, S. , Walter, B. L. , Nguyen, J. H. , & Semler, B. L. (2007). Cellular protein modification by poliovirus: The two faces of poly(rC)‐binding protein. Journal of Virology, 81(17), 8919–8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccone, M. E. , Rieder, E. , Mason, P. W. , & Grubman, M. J. (1995). The foot‐and‐mouth disease virus leader proteinase gene is not required for viral replication. Journal of Virology, 69(9), 5376–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro, D. , Fernandez, N. , Ramajo, J. , & Martinez‐Salas, E. (2013). Gemin5 promotes IRES interaction and translation control through its C‐terminal region. Nucleic Acids Research, 41(2), 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro, D. , Fernandez‐Chamorro, J. , Francisco‐Velilla, R. , & Martinez‐Salas, E. (2015). Gemin5: A multitasking RNA‐binding protein involved in translation control. Biomolecules, 5(2), 528–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro, D. , Ramajo, J. , Bradrick, S. S. , & Martinez‐Salas, E. (2012). Gemin5 proteolysis reveals a novel motif to identify L protease targets. Nucleic Acids Research, 40(11), 4942–4953 d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P. J. , & Belsham, G. J. (1995). Identification of critical amino acids within the foot‐and‐mouth disease virus leader protein, a cysteine protease. Virology, 213(1), 140–146. [DOI] [PubMed] [Google Scholar]
- Rodriguez Pulido, M. , & Saiz, M. (2017). Molecular mechanisms of foot‐and‐mouth disease virus targeting the host antiviral response. Frontiers in Cellular and Infection Microbiology, 7, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Pulido, M. , Sanchez‐Aparicio, M. T. , Martinez‐Salas, E. , Garcia‐Sastre, A. , Sobrino, F. , & Saiz, M. (2018). Innate immune sensor LGP2 is cleaved by the Leader protease of foot‐and‐mouth disease virus. PLoS Pathogens, 14(6), e1007135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Pulido, M. , Serrano, P. , Saiz, M. , & Martinez‐Salas, E. (2007). Foot‐and‐mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA‐binding proteins. Virology, 213, 140–146. [DOI] [PubMed] [Google Scholar]
- Rodriguez Pulido, M. , Martínez‐Salas, E. , Sobrino, F. , & Sáiz, M. (2020). MDA5 cleavage by the Leader protease of foot‐and‐mouth disease virus reveals its pleiotropic effect against the host antiviral response. Cell Death & Disease, 11(8), 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. A. , Gouvea, I. E. , Judice, W. A. , Izidoro, M. A. , Alves, F. M. , Melo, R. L. , Juliano, M. A. , Skern, T. , & Juliano, L. (2009). Hydrolytic properties and substrate specificity of the foot‐and‐mouth disease leader protease. Biochemistry, 48(33), 7948–7958. [DOI] [PubMed] [Google Scholar]
- Steinberger, J. , Grishkovskaya, I. , Cencic, R. , Juliano, L. , Juliano, M. A. , & Skern, T. (2014). Foot‐and‐mouth disease virus leader proteinase: Structural insights into the mechanism of intermolecular cleavage. Virology, 468–470C, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel, K. , & Beck, E. (1986). A second protease of foot‐and‐mouth disease virus. Journal of Virology, 58(3), 893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek, K. N. , Aumayr, M. , Pruneda, J. N. , Visser, L. J. , Berryman, S. , Kueck, A. F. , Geurink, P. P. , Ovaa, H. , van Kuppeveld, F. J. M. , Tuthill, T. J. , Skern, T. , & Komander, D. (2018). Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies. Proceedings of the National Academy of Sciences of the United States of America, 115(10), 2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa, E. , Lethier, M. , Malet, H. , Brunel, J. , Gerlier, D. , & Cusack, S. (2016). Structural analysis of dsRNA binding to anti‐viral pattern recognition receptors LGP2 and MDA5. Molecular Cell, 62(4), 586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, L. J. , Aloise, C. , Swatek, K. N. , Medina, G. N. , Olek, K. M. , Rabouw, H. H. , de Groot, R. J. , Langereis, M. A. , de Los Santos, T. , Komander, D. , Skern, T. , & van Kuppeveld, F. J. M. (2020). Dissecting distinct proteolytic activities of FMDV Lpro implicates cleavage and degradation of RLR signaling proteins, not its deISGylase/DUB activity, in type I interferon suppression. PLoS Pathogens, 16(7), e1008702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, L. J. , Medina, G. N. , Rabouw, H. H. , de Groot, R. J. , Langereis, M. A. , de Los Santos, T. , & van Kuppeveld, F. J. M. (2019). Foot‐and‐mouth disease virus Leader protease cleaves G3BP1 and G3BP2 and inhibits stress granule formation. Journal of Virology, 93(2), e00922‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, D. , & Mohr, I. (2011). Viral subversion of the host protein synthesis machinery. Nature Reviews. Microbiology, 9(12), 860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Fang, L. , Liu, L. , Zhong, H. , Chen, Q. , Luo, R. , Liu, X. , Zhang, Z. , Chen, H. , & Xiao, S. (2011). Foot‐and‐mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon‐lambda1 pathway. Molecular Immunology, 49(1–2), 407–412. [DOI] [PubMed] [Google Scholar]
- Weng, K. F. , Li, M. L. , Hung, C. T. , & Shih, S. R. (2009). Enterovirus 71 3C protease cleaves a novel target CstF‐64 and inhibits cellular polyadenylation. PLoS Pathogens, 5(9), e1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. P. , Cardenas, A. M. , Marissen, W. E. , & Lloyd, R. E. (2007). Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host & Microbe, 2(5), 295–305. [DOI] [PubMed] [Google Scholar]
- Xu, C. , Ishikawa, H. , Izumikawa, K. , Li, L. , He, H. , Nobe, Y. , Yamauchi, Y. , Shahjee, H. M. , Wu, X.‐H. , Yu, Y.‐t. , Isobe, T. , Takahashi, N. , & Min, J. (2016). Structural insights into Gemin5‐guided selection of pre‐snRNAs for snRNP assembly. Genes & Development, 30(21), 2376–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H. , Unbehaun, A. , & Spahn, C. M. T. (2017). Ribosomal chamber music: Toward an understanding of IRES mechanisms. Trends in Biochemical Sciences, 42(8), 655–668. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Khosravi‐Far, R. , Chang, H. Y. , & Baltimore, D. (1997). Daxx, a novel Fas‐binding protein that activates JNK and apoptosis. Cell, 89(7), 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J. S. , Kato, H. , & Fujita, T. (2014). Sensing viral invasion by RIG‐I like receptors. Current Opinion in Microbiology, 20, 131–138. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Morace, G. , Gauss‐Muller, V. , & Kusov, Y. (2007). Poly(A) binding protein, C‐terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Research, 35(17), 5975–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, E. , Borman, A. M. , Deliat, F. G. , Liebig, H. D. , Jugovic, D. , Kean, K. M. , Skern, T. , & Kuechler, E. (1995). Picornavirus 2A proteinase‐mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology, 213(2), 549–557. [DOI] [PubMed] [Google Scholar]


