Abstract
SUMMARY
The TRPA1 ion channel is activated by electrophilic compounds through covalent modification of intracellular cysteine residues. How non-covalent agonists activate the channel, and whether covalent and non-covalent agonists elicit the same physiological responses, are not understood. Here, we report the discovery of a non-covalent agonist, GNE551, and determine a cryo-EM structure of the TRPA1-GNE551 complex, revealing a distinct binding pocket and ligand-interaction mechanism. Unlike the covalent agonist allyl isothiocyanate, which elicits channel desensitization, tachyphylaxis, and transient pain, GNE551 activates TRPA1 into a distinct conducting state without desensitization and induces persistent pain. Furthermore, GNE551-evoked pain is relatively insensitive to antagonist treatment. Thus, we demonstrate biased agonism of TRPA1, a finding that has important implications for the discovery of effective drugs tailored to different disease etiologies.
eTOC Blurb
Liu et al. determine the structural basis for the binding of a non-covalent agonist (GNE551) to the TRPA1 ion channel. Compared to the canonical covalent agonist AITC, GNE551 activates channels into distinct conducting states and induces different physiological responses, revealing a biased agonism of TRPA1 channel.
INTRODUCTION
Transient receptor potential ankyrin 1 (TRPA1) is an excitatory ion channel predominantly expressed in sensory neurons and co-localized with pain markers such as TRPV1 and CGRP (Jordt et al., 2004; Story et al., 2003). In rodents, knockout of the TRPA1 gene or treatment with channel antagonists produces analgesic and anti-inflammatory effects (Bandell et al., 2004; Bautista et al., 2006; Chen et al., 2011; Kwan et al., 2006); and in humans, a gain-of-function mutation of TRPA1 is linked to familial episodic pain syndrome (Kremeyer et al., 2010). TRPA1 has also been implicated in histamine-independent itch (Wilson et al., 2013), airway hypersensitivity (Caceres et al., 2009), and anti-apoptotic pathways (Takahashi et al., 2008), making TRPA1 an important therapeutic target (Bautista et al., 2013; Chen and Hackos, 2015).
The TRPA1 channel is a tetramer of four identical subunits, with each subunit containing six transmembrane helices (S1-S6), and intracellular N- and C-termini (Paulsen et al., 2015). The first four transmembrane helices (S1–S4) form the voltage-sensor like domain (VSLD) and the last two transmembrane helices (S5–S6) form the ion conduction pore module. One of the best characterized features of TRPA1 is its activation by electrophilic chemicals, including pungent ingredients of natural products (e.g., allyl isothiocyanate, or AITC), environmental irritants (e.g., acrolein), and reactive metabolites (e.g., 4-hydroxynonenal) (Bandell et al., 2004; Hinman et al., 2006; Jordt et al., 2004; Macpherson et al., 2007; Trevisani et al., 2007). These compounds covalently modify cysteine residues localized within the intracellular N-terminal domain of TRPA1 (Hinman et al., 2006; Macpherson et al., 2007). Recently, cryo-EM structures of TRPA1 in ligand-free form and in complex with covalent agonists (JT010 and BITC) have been determined (Suo et al., 2019). Open- and closed-state TRPA1 structures have also been reported by using an irreversible electrophilic agonist (iodoacetamide) and a small molecule antagonist (A-967079), respectively (Zhao et al., 2020). Together, these structures reveal that the opening of TRPA1 channel is mediated by rotation of the VSLDs around the pore module, concomitant with a straightening of the S4-S5 linker and S5 helix to open the intracellular S6 gate. Together, these studies have provided an unprecedented understanding of how electrophilic compounds modify cysteine residues and gate the TRPA1 channel.
Covalent activation by electrophiles represents a significant, albeit arguably narrow view of the multifaceted modulation of TRPA1, which is susceptible to a wide spectrum of alternative stimuli, including cold, heat, hypertonicity, Ca2+, trace metals, acids, and receptor-mediated pathways (Meents et al., 2019; Talavera et al., 2020). In particular, an increasing number of non-covalent agonists or modulators have been discovered, including natural products (e.g., carvacrol and menthol), marketed drugs (e.g., clotrimazole and propofol), and tool agents (e.g., piperidine carboxamides) (Chernov-Rogan et al., 2019; Jordt et al., 2004; Karashima et al., 2007; Ton et al., 2017; Xu et al., 2006). Additionally, several endogenous lipids or lipid products, including diacylglycerol, hepoxilin A3, cholesterol, and phosphatidylinositol 4,5-bisphosphate (PIP2), have been shown to modulate TRPA1 function (Bandell et al., 2004; Gregus et al., 2012; Karashima et al., 2008; Startek et al., 2019). In spite of these progresses, how non-covalent agonists interact with TRPA1 and, in particular, whether non-covalent and covalent agonists induce similar or different physiological responses, remain unknown.
In the current study, we identify and characterize GNE551, which belongs to a class of non-covalent agonists, and determine its mode of interaction with TRPA1 by phylogenic analysis and single particle cryogenic electron microscopy (cryo-EM). GNE551 binds to a transmembrane pocket conserved in several other TRP channels. Compared to the canonical covalent agonist AITC, GNE551 exhibits remarkable differences in channel modulation, pain response, and sensitivity to inhibitor treatment, hence demonstrating biased agonism of the TRPA1 channel.
RESULTS
GNE551 is a Potent, Selective, and Non-covalent Agonist of TRPA1 Channel
Using a Ca2+ influx assay of human TRPA1 channel, we screened 1.5 million compounds from our Genentech/Roche chemical library and found thousands of compounds with agonist activity. A vast majority of these compounds contained reactive moieties, similar to the electrophilic agonist AITC. However, a set of aminotriazole compounds, including GNE551, did not contain reactive moieties (Figures 1A and S1). In a liquid chromatography-mass spectrometry assay (Huang et al., 2015), GNE551 did not form an adduct with glutathione, a cysteine-containing antioxidant, confirming the lack of chemical reactivity of GNE551 (Figure S1A). In contrast, AITC formed a glutathione adduct due to its electrophilic isothiocyanate moiety (Figure S1B). Aminotriazole agonists showed overall similar structures and structure-function relationships (Figure S1C).
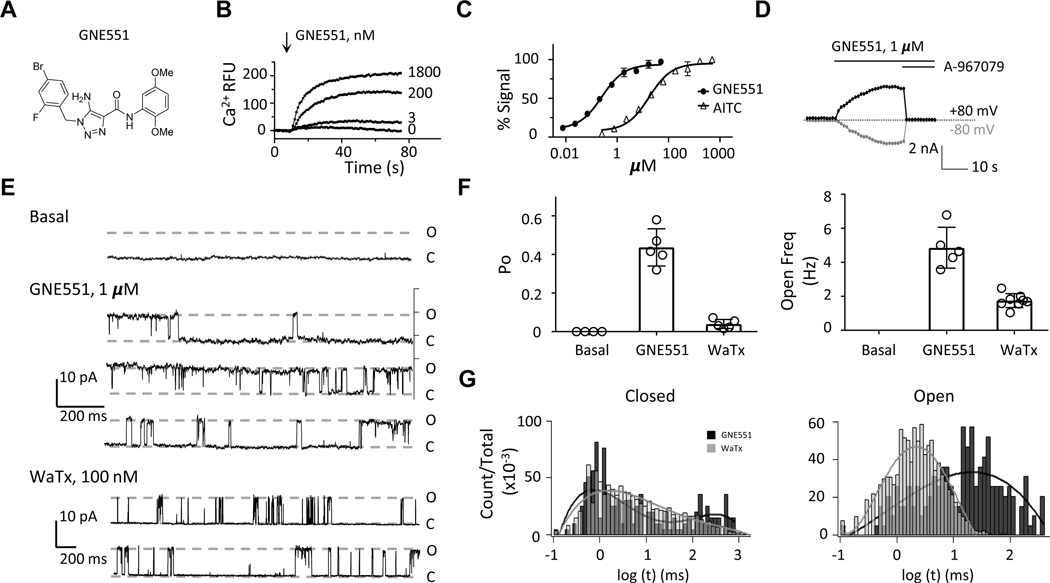
Figure 1. GNE551 Activates TRPA1 with Distinct Properties.
(A) Chemical structure of GNE551.
(B) In a Ca2+ influx assay using HEK293-F cells expressing human TRPA1, GNE551 activated the channel in a concentration-dependent manner. Representative traces of Ca2+ fluorescence (indicated as Ca2+ RFU, or relative fluorescence unit) are shown. Arrow indicates GNE551 addition.
(C) Concentration response of GNE551 and AITC in the Ca2+ influx assay. The EC50 was 254 nM for GNE551 (95% confidence interval, 207–312 nM, n = 4); in comparison, the EC50 for AITC was 17.5 μM (95% confidence interval, 13.7–22.5 μM, n = 4).
(D) In whole-cell recordings, GNE551 (1 μM) evoked outward currents at +80 mV (black trace) and inward currents at −80 mV (gray trace), which were blocked by 1 μM A-967079. Dotted line indicates zero current.
(E) Representative inside-out single channel current traces at the basal level and in the presence of GNE551 (1 μM) or WaTx (100 nM) recorded at +80 mV. O and C indicate open and closed state, respectively.
(F) Open probability (left) and open frequency (right) of TRPA1 channel under basal condition (n = 3), 1 μM GNE551 (n = 5) and 100 nM WaTX (n = 8).
(G) Closed and open dwell time histograms of a GNE551- or WaTx-activated channel. The open state dwell time was 20.9 milliseconds for GEN551 and 2.2 milliseconds for WaTx-activated channels, respectively.
In HEK293-F cells transiently expressing human TRPA1, GNE551 triggered calcium influx in a concentration-dependent manner (Figure 1B). GNE551 was more potent than AITC (EC50: 254 nM versus 17.5 μM, n = 4) and was equally efficacious (Figure 1C). GNE551 was further characterized in whole-cell voltage-clamp recordings. Nominally Ca2+-free intracellular and extracellular solutions were used to minimize channel desensitization, and a 0.2-second voltage ramp protocol (from −80 mV to +80 mV) was applied once per second. GNE551 evoked robust currents in TRPA1-expressing cells and the currents were blocked by A-967079, indicating a TRPA1-specific effect (Figure 1D). The EC50 of GNE551 determined from the currents at +80 mV was 141 nM (n = 3 – 6).
To assess its selectivity, we tested GNE551 on several ion channels. GNE551 (20 μM) did not activate or block human TRPV1, TRPM8, or TRPC6 (Table S1). Additionally, GNE551 (10 μM) did not inhibit human potassium channel hERG1 and sodium channels (hNav1.2., hNav1.5, or hNav1.7) (Table S2).
Among a CEREP panel of 35 receptors and ion channels, GNE551 (10 μM) exhibited robust binding inhibition (>75%) for Cav1.2 (L-dihydropyridine site) and NK1 receptor (Table S3). However, binding inhibition of GNE551 did not translate to significant agonistic or antagonistic effect on NK1 receptor, or inhibition of Cav1.2 at up to 30 μM (Table S4). These data suggest that GNE551 is likely to have good selectivity toward TRPA1.
GNE551 Promotes an Open State with Distinct Biophysical Properties
Electrophilic agonists, such as AITC, covalently modify intracellular cysteine residues on TRPA1 (Hinman et al., 2006; Macpherson et al., 2007; Suo et al., 2019; Zhao et al., 2020). Interestingly, the intracellular domain containing reactive cysteines also interacts with the cell-penetrating peptide toxin WaTx (Lin King et al., 2019). Hence, we characterized the single channel properties of TRPA1 channels in response to GNE551, WaTx, and AITC. In inside-out patch recordings, the single channel open probability (Po) of TRPA1 was extremely low, with no activity detectable under basal conditions (Figure 1E). GNE551 (1 μM) strongly activated the channel and produced a maximal Po of 0.41 ± 0.07 and an opening frequency of 4.6 ± 0.6 Hz (n = 5), whereas WaTx (100 nM) activated TRPA1 with a Po of 0.045 ± 0.012 and an opening frequency of 1.7 ± 0.5 Hz (n = 8, Figure 1F). The open state dwell time of GNE551-activated channels was 20.9 milliseconds, compared to an open dwell time of 2.2 milliseconds for WaTx (Figure 1G). Due to the rapid desensitization, we did not quantitate Po, opening frequency, or open dwell time for AITC-evoked single channels.
Next, we characterized the single channel conductance and ion permeability of TRPA1 activated by GNE551 and AITC. We kept the extracellular Na+ concentration constant (140 mM) while varying the cation species (Na+, Mg2+, or Ca2+) of the intracellular solutions. Consistent with previous reports (Lin King et al., 2019; Story et al., 2003), AITC-evoked currents displayed selectivity for divalent cations over Na+ (Figure S2A, Table 1). GNE551-activated currents also show selectivity for Mg2+ and Ca2+ (Figure S2A, Table 1). However, compared to AITC, GNE551 exhibited a lower single-channel conductance for all tested cations, and the GNE551-activated channels were less permeable to Mg2+ and more permeable to Ca2+ (Figure S2B, Table 1). Taken together, these data suggest that GNE551 and AITC promote TRPA1 into biophysically distinct conductive states.
Table 1.
Comparison of Biophysical Properties between GNE551- and AITC-activated Channels.
| Erev (mV) | Px/PNa | y (pS) | ||||
|---|---|---|---|---|---|---|
| Cation | AITC | GNESS1 | AITC | GNESS1 | AITC | GNESS1 |
| Na+ | - | - | - | - | 134.9 ± 11.6 | 101.4 ± 6.8 |
| Mg2+ | −19.1 ± 1.5 | −13.2± 2.7 | 10.2 ± 1.0 | 7.2 ± 1.3 | 30.1 ± 6.8 | 195 ± 1.9 |
| Ca2+ | −22.9. ± 2.4 | −25.6 ± 1.1 | 12.6 ± 0.81 | 15.7 ± 1.1 | 30.9 ± 6.2 | 21.6 ± 2.8 |
Erev, Px/PNa, and stand for reversal potential, ion selectivity against Na+, and single channel conductance, respectively. 1 μM GNE551 (n = 5) and 10 μM AITC (n = 4) were used.
GNE551 Activates TRPA1 without Desensitization or Tachyphylaxis
We observed drastic differences in desensitization and reversibility between GNE551- and AITC-activated channels. In nominally Ca2+-free solutions, AITC (10 μM)-activated currents desensitized within seconds (Figure 2A). After a wash off with external solution, a second application of AITC did not increase channel opening, indicating persistent channel desensitization. By contrast, GNE551 (1 μM) did not result in desensitization during a similar incubation time; and following a solution washout, a second application of GNE551 led to robust channel openings (Figure 2B). These striking differences are reflected by the time course of nPo (open probability times the number of channels in the macropatches) (Figure 2C) and all-point histograms of single channel events (Figure S2C). After an initial AITC application, GNE551 evoked robust opening, indicating the recovery of AITC-desensitized channels (Figure S2D). In whole-cell voltage clamp recordings, a second stimulation of AITC evoked much smaller currents compared to the first application (14.1 ± 6.3% at +80 mV, 19.4 ± 4.3% at −80 mV, n = 3, Figure 2D). In contrast, a second addition of GNE551 evoked relatively slow activating currents, but the peak currents were minimally reduced compared to the first application (93.5 ± 0.5% at +80 mV and 89.0 ± 6.3% at −80 mV, n = 3, Figure 2D). Hence, GNE551- and AITC-activated TRPA1 channels exhibit a spectrum of differing properties, including conductance, ion selectivity, desensitization, and reversibility of agonistic effects, indicating distinct activation mechanisms between the two ligands.
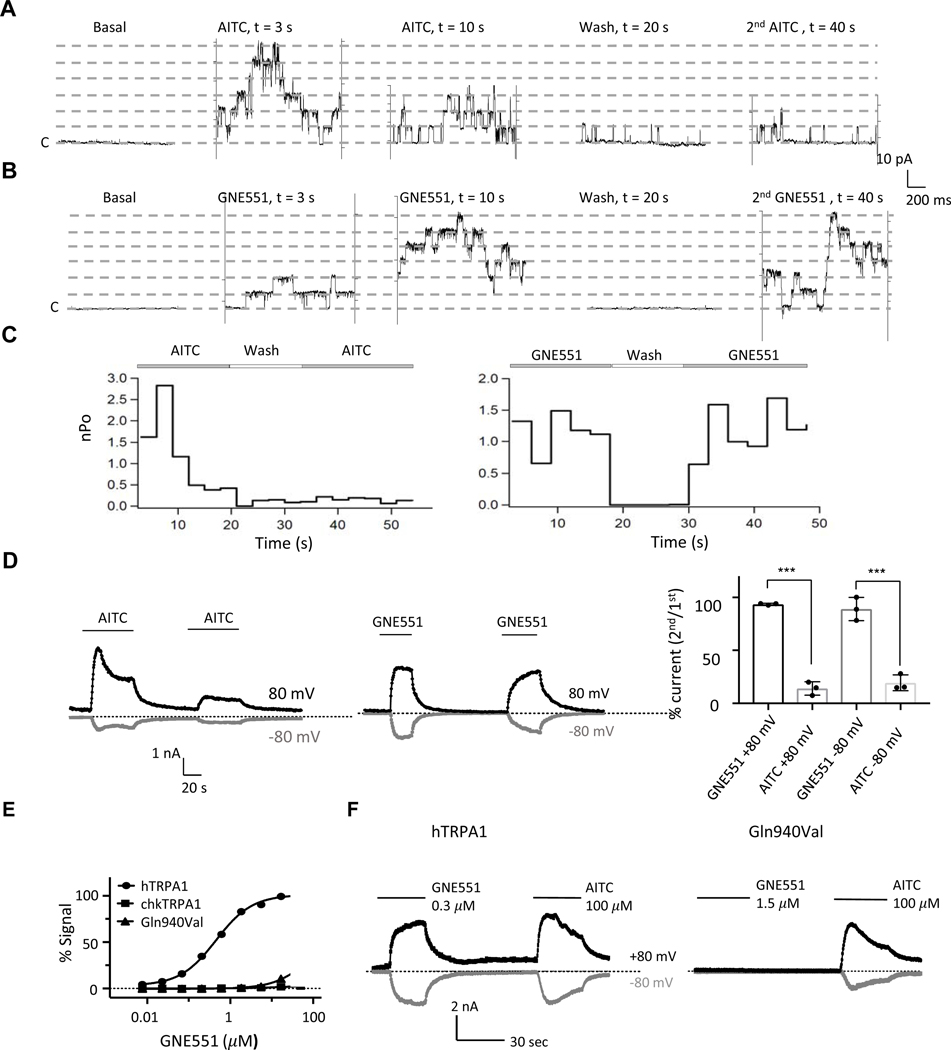
Figure 2. GNE551 does not Induce Desensitization and a Single Residue is Critical for Activation.
(A-B) Time course of single channel TRPA1 currents (inside-out macropatches, +80 mV) in response to 10 μM AITC and 1 μM GNE551.
(C) Plots of time course of nPo of TRPA1 channels activated by AITC (n = 4) and GNE551 (n = 3).
(D)Whole-cell currents in response to repeated stimulations by AITC (Left) and GNE551 (Middle), and percentage of currents of second compared to the first application (Right). The current during second application was reduced for AITC and mostly retained for GNE551. ***p<0.005.
(E) GNE551 concentration-response relationships in Ca2+ assay. GNE551 activated human channel (hTRPA1), but not chicken channel (chkTRPA1), or Gln940Val mutant human channel. n = 4.
(F) In whole-cell patch clamp recordings, GNE551 (300 nM) activated hTRPA1, but GNE551 (1.5 μM) failed to activate Gln940Val TRPA1 (n = 4). AITC was used as a positive control. Dotted lines indicate zero current.
Gln940 in Transmembrane Segment 6 is Critical for GNE551 Activation
Many TRPA1 modulators have species-specific effects (Chen et al., 2013; Chen et al., 2008; Chernov-Rogan et al., 2019; Xiao et al., 2008). Hence, we tested GNE551 on TRPA1 channels from several species. In addition to the human channel, GNE551 also activated TRPA1 from rhesus monkey, dog, guinea pig, and rat, but had no effect on chicken TRPA1 at a concentration as high as 50 μM (Figures S3A and S3B). To determine channel regions and residues underlying the species-specific effects, we systematically introduced regions of the chicken channel sequence into the human channel background and found that a chimera containing S1-S6 domains of chicken TRPA1 lost response to GNE551 (Figure S3B).
A sequence alignment of the S1-S6 segments between GNE551-insensitive (chicken TRPA1) and GNE551-sensitive channels (e.g., human TRPA1) revealed 39 amino acid residues unique to chicken TRPA1 (Figure S3C). Each of these residues was introduced into the human TRPA1 sequence, and the effects on GNE551 and AITC responses were tested in the Ca2+ assay. While most substitutions did not alter channel sensitivity to GNE551 to a large extent (Table S5), Gln940Val nearly abolished GNE551 activation (Figure 2E, Table S5). In whole-cell patch clamp recordings, GNE551 (1.5 μM) failed to activate Gln940Val channels despite a robust response to AITC (Figure 2F). Residue Gln940 is located on the S6 segment within the outer leaflet portion of the membrane (Figure S3C), and previously has not been implicated in interaction with any other known TRPA1 ligands. Conversely, several residues in the pore domain, including Ser873, Thr874, and Phe909, have been identified as critical for the effects of several agonists (i.e., menthol, isoflurane, and piperidine carboxamide) and the antagonist A-967079 (Chernov-Rogan et al., 2019; Paulsen et al., 2015; Ton et al., 2017; Xiao et al., 2008; Zhao et al., 2020). GNE551 activation was not affected by Ser873Val/Thr874Leu and Phe909Ala mutations (Table S5). Therefore, GNE551 activates TRPA1 by interacting with a novel binding site.
Structure of TRPA1 Channel in Complex with GNE551
We next sought to determine the ligand-binding mode of GNE551 to TRPA1 by cryo-EM. By using a truncated and non-functional construct (containing Ser448-Thr1078, Figure S4A) optimized for robust protein expression and biochemical stability, we generated a 3D reconstruction of human TRPA1 in complex with GNE551 at a resolution of ~3.0 Å (Figures 3A and S4, Table S6). Our structure conforms to the overall architecture of TRPA1 as described previously (Paulsen et al., 2015; Suo et al., 2019; Zhao et al., 2020). We observed unambiguous density for GNE551, but otherwise no major structural differences relative to a map of the unliganded full-length TRPA1 (PDB: 6PQQ) (Suo et al., 2019). In particular, the S6 gating constriction of GNE551-bound TRPA1 pore was in an identical conformation, and the S4-S5 linker and S5 helix were found in the typical kinked conformation seen in most structures to date (Figures S5A–S5C). While our current GNE551-bound structure is in a non-conducting state, it reveals the first structural view of the binding mode for a non-covalent TRPA1 agonist.
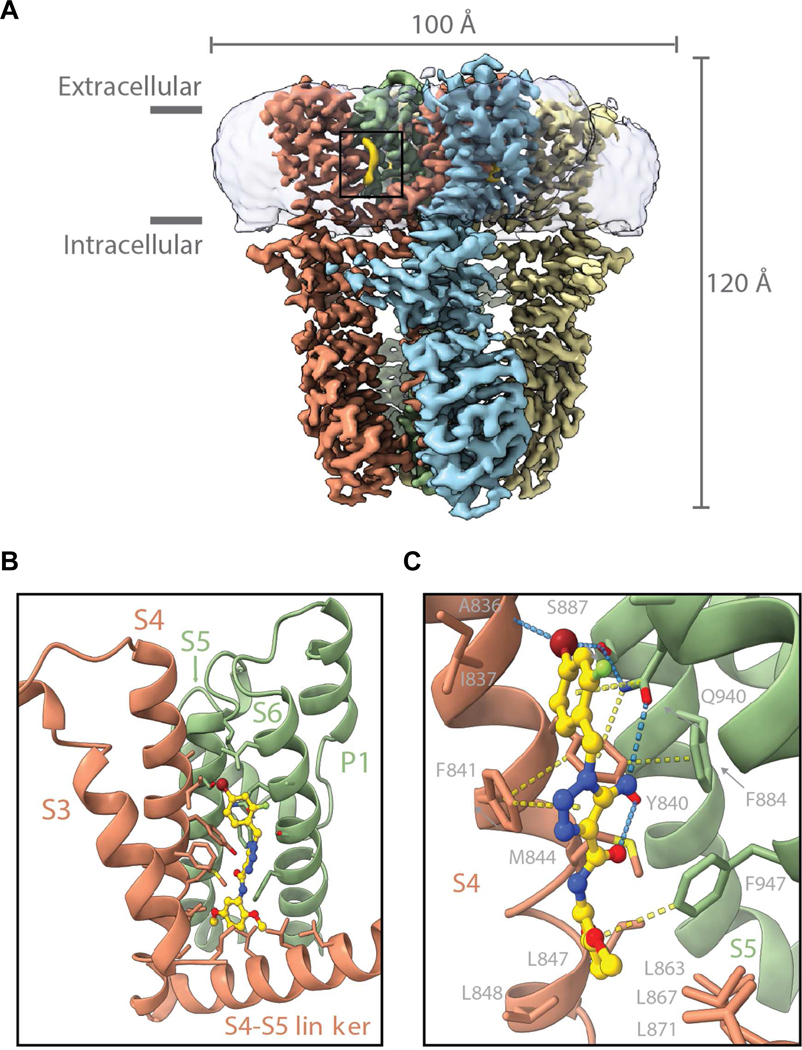
Figure 3. Structure of TRPA1 in Complex with GNE551 and Key Binding Pocket Interactions.
(A) Isosurface rendering of the cryo-EM map of human TRPA1 in complex with GNE551. TRPA1 protomers are colored in red, green, yellow, and blue; GNE551 in yellow and micelle density is shown in transparent.
(B)Close-up view of GNE551 binding site. GNE551 (shown in stick with carbon atoms colored in yellow, oxygen in red, and nitrogen in blue) binds in a pocket located at the interface between the S4 helix of one subunit (red) and pore module of an adjacent subunit (green). P1 = pore helix 1. Side chains for residues in close proximity of GNE551 are shown in stick.
(C)Molecular interactions between GNE551 and TRPA1. Hydrogen bonds are shown in blue. Favorable contacts involving aromatic side chains are in yellow. View is rotated 45 degrees relative to panel B.
GNE551 binds to a groove between the VSLD (S1-S4) of one subunit (in red, Figure 3) and the pore domain (S5-S6) of a neighboring subunit (in green, Figure 3), in a mostly hydrophobic pocket approximately halfway across the transmembrane region. Three notable polar interactions, involving Gln940, Tyr840, and Ser887, stand out in the receptor site otherwise dominated by non-polar, van der Waals contacts: Gln940 in the S6 domain forms a hydrogen bond with the primary amine of GNE551’s central triazole; Tyr840 of the S4 domain presents its terminal hydroxyl to form a hydrogen bond with the agonist’s amide carbonyl; and GNE551’s terminal bromine is positioned to interact with the Ser887 side chain and the backbone carbonyl of Ala836 (Figure 3C). To evaluate whether these polar interactions play a significant role in GNE551’s potency, we tested point mutations (Table S7) and studied structure-activity relationships among aminotriazole analogues (Figure S1C).
The conservative mutation of Tyr840 to Phe reduced the potency of GNE551 by six-fold, consistent with the hydrogen bonding between Tyr840 with GNE551, while more perturbing mutations at this position (Tyr840 to Trp, His, Leu, or Ala) completely abolished sensitivity to GNE551 (Table S7). In addition to its direct interaction with GNE551, residue Gln940 is involved in a set of stabilizing interactions with the neighboring S4 and S5 helices: its terminal amine group forms a favorable H-arene stack with Tyr840 from the S4 helix, and hydrogen-bonds with Ser887 from the S5 helix (Figure 3C). Consistent with this structural role in stabilizing the binding pocket, its hydrogen-bond interaction with the agonist, as well as a putative pi-stacking interaction with the benzene ring of GNE551, mutation of Gln940 to Asn, Val or Ala completely abolished sensitivity to GNE551 (Table S7). The notion that polar contacts between GNE551’s terminal bromine and the binding pocket potentiate agonism is supported by the loss of activation on mutation of Ser887 to Trp or Met (Table S7) and by structure-activity relationships observed among GNE551 analogues. Variations at the terminal bromine (R2 position as in Figure S1C) exhibited a pronounced impact on agonist potency, with highly polarizable bromo and chloro substituents being preferred to the smaller fluoro or larger methoxy groups at this position, consistent with favorable polar contacts occurring between GNE551’s bromine and the backbone of S4 at Ala836 and the side chain of Ser887 (Figures S1C and 3C).
In a recent cryo-EM structure of unliganded TRPA1 (PDB code: 6PQQ), a well-ordered phospholipid (designated as “lipid 5”) that occupies a binding pocket overlapping with that of GNE551 has been observed (Figures S5D–S5F and S6A) (Suo et al., 2019). Furthermore, comparison of unliganded versus covalent-agonist (BITC)-bound TRPA1 structures (PDB: 6PQQ versus 6PQP) indicated that another lipid (“lipid 7”) may be displaced upon channel opening (Suo et al., 2019). In these aspects, it is notable that TRPV1 and TRPA1 share a high degree of structural homology in the transmembrane region, and the GNE551/lipid pocket is equivalent to the vanilloid/lipid pocket in TRPV1 (Figure S6A) (Gao et al., 2016). In TRPV1, resiniferatoxin (RTX) binding leads to the displacement of a resident lipid and facilitate channel opening through a rearrangement of the S4-S5 linker and a stabilization of an interaction between residue Arg557 (of S4) and residue Glu570 (of the S4-S5 linker) (Gao et al., 2016). Furthermore, TRPV1 residue Glu570 forms polar interactions with the headgroup of the resident lipid in the closed state and is involved in activation upon capsaicin binding (Figure S6B) (Elokely et al., 2016; Yang et al., 2015). Since Glu570 of TRPV1 is conserved in TRPA1 as Glu864 (Figures S6A–S6C), we hypothesized that in TRPA1, Glu864 may similarly interact with the polar headgroup of lipid 5 and be involved in channel gating.
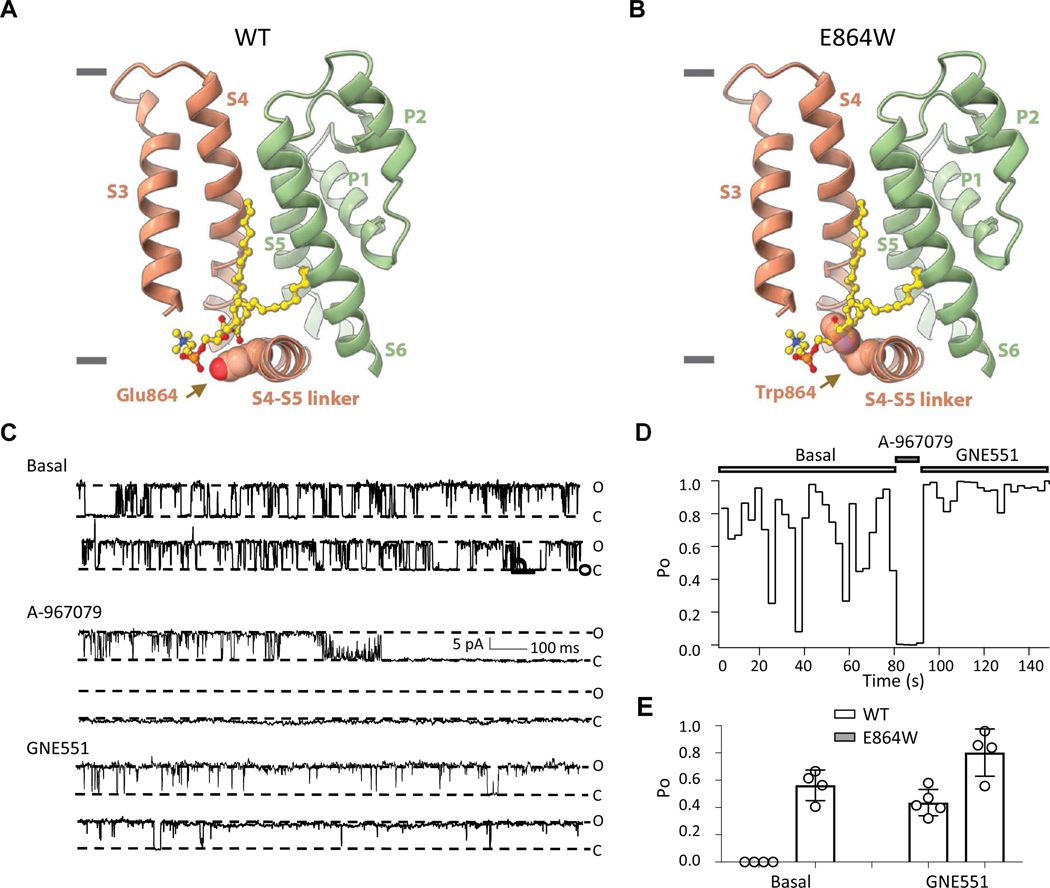
To test the above hypothesis, we introduced a Glu864Trp point mutation into TRPA1. Since the bulky side chain of tryptophan is well-known for its enrichment at the lipid-water interface (Yau et al., 1998), we reasoned that the Glu864Trp mutation might disrupt the interaction with lipid 5 and consequentially prevent the lipid from occupying its binding site (Figures 4A and 4B). Strikingly, in the Ca2+ assay, cells expressing TRPA1 Glu864Trp mutant had high background Ca2+ signals, and almost lost responses to GNE551 or AITC stimulation (Figure S6D). In single channel recordings, the Glu864Trp mutant had markedly increased basal activities in the absence of agonist (Po = 0.56 ± 0.05, n = 4, Figures 4C–4E, compared to a basal Po = 0 for wild type TRPA1, Figure 1F). The high basal activity of the Glu864Trp channel was blocked by A-967079 and could be further enhanced by application of GNE551 (Po = 0.80 ± 0.08, n = 4, Figures 4C–4E). These data suggest that the Glu864Trp mutation induces a mostly constitutively open channel by disrupting an endogenous lipid and enabling the S4-S5 linker rearrangements required for channel opening.
Figure 4. The Glu864Trp Mutation Renders TRPA1 Channel Constitutively Open.
(A) The structure of unliganded human TRPA1 (PDB: 6PQQ) revealed well-ordered lipid 5 (yellow), with its acyl chains occupying the GNE551-binding pocket, and its head group located in close proximity of Glu864 from the S4-S5 linker. TRPA1 is depicted in ribbon representation using the same colors as in Figure 3. The side chain of Glu864 is shown using a space-filling sphere representation. Gray bars indicate extracellular or intracellular boundaries.
(B) In the Glu864Trp mutant channel, the large hydrophobic side chain of the Trp would sterically clash with the lipid 5.
(C) Representative single-channel currents of Glu864Trp channel recorded at +80 mV under basal condition or in response to A-967079 (1 μM) or GNE551 (1 μM).
(D)Open probability (Po) over time of Glu864Trp at the basal level and in response to compounds.
(E) Glu864Trp increased the open probability at the basal level, and in response to GNE551.
AITC and GNE551 Evoke Distinct Pain Behaviors
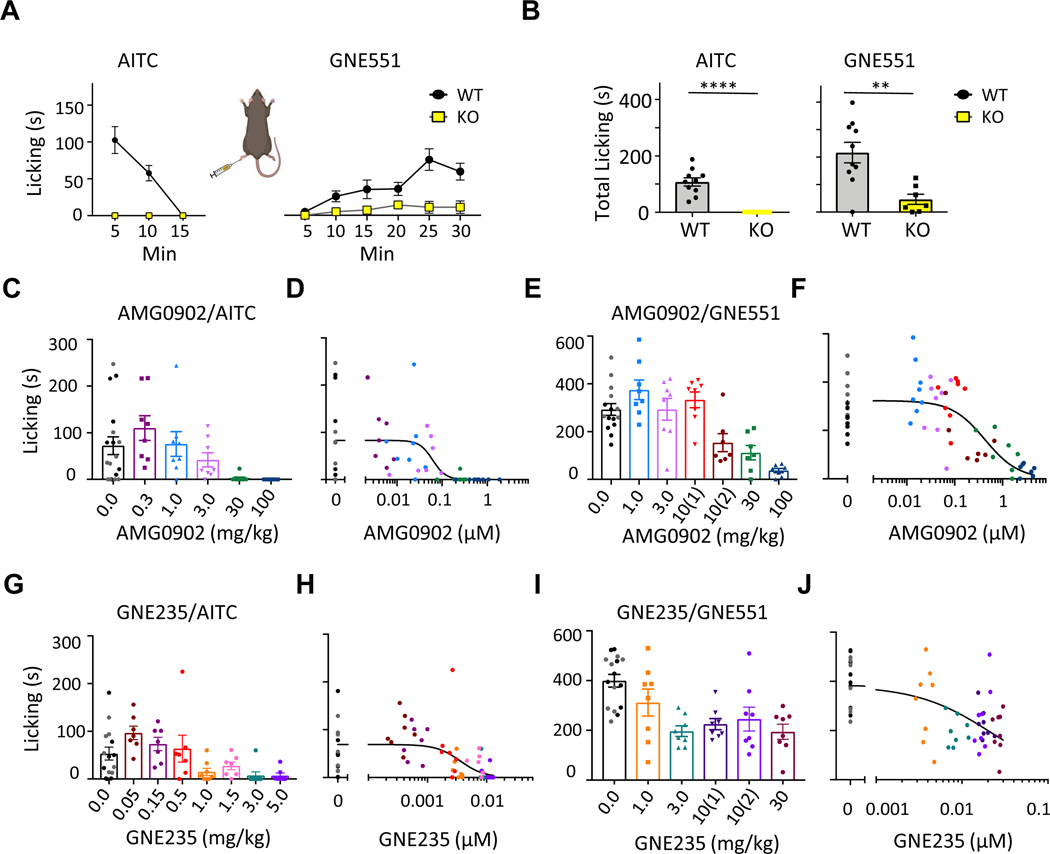
Due to their distinct binding modes and striking difference in modulating TRPA1 function, we wondered whether GNE551 and AITC induce different physiological responses in animal behavior studies. First, we confirmed the specificity of GNE551 by showing that GNE551 solely activated TRPA1 channels in rat dorsal root ganglion neurons (Figure S7A). An intraplantar injection of AITC (0.1%, 25 μl) to the hind paw elicited nocifensive behavior in wild type (WT) but not in knock out (KO) rats, consistent with previously reports in rats (Reese et al., 2020) and mice (Bautista et al., 2006; Kwan et al., 2006). The nocifensive responses to AITC, comprised of licking, flicking, and guarding of the injected paw, began immediately upon injection and subsided quickly (Figure 5A). For the first and second 5 min periods, AITC produced 102 ± 19 sec and 57.7 ± 11.1 sec of cumulative nocifensive time, respectively (n = 7), whereas the nocifensive response completely disappeared after 10 min (Figure 5A, Video S1). Intraplantar injection of GNE551 into WT rat paws also induced nocifensive response in a dose-dependent manner (1, 3, and 10 mM, 25 μl, Figures S7B and S7C), but the GNE511 responses had a strikingly different time course compared to that of AITC (Figure 5A, Video S1). During the first 5 min after injection of GNE551, there were virtually no paw flicking, licking, or guarding, but these behaviors gradually appeared, reaching a plateau approximately 15–20 min after injection and continuing through the end of the 30-min observation period (Video S1, Figures 5A and 5B). The slow onset of GNE551-response is consistent with its poor tissue permeability (Table S8), while the long duration of pain response is consistent with the lack of channel desensitization (Figure 2) and the lack of chemical reactivity of GNE551 (Figure S1A). A small residual response (13%) was present in TRPA1 KO rats, possibly due to a vehicle effect (Figure 5B). In WT rats, the latency to nocifensive response was also reduced by GNE551 injection (10 mM, 25 μl, Figure S7D).
Figure 5. GNE551 and AITC Elicit Pain with Distinct Kinetics and Sensitivity to Antagonists.
(A) Time course for time spent licking the ipsilateral paws after intraplantar injection of AITC (0.1%) or GNE551 (10 mM) in TRPA1 wild type rats (WT) and knockout rats (KO). n = 7 – 10.
(B) Quantification of total time licking in response to AITC (during the first 5 min) and GNE551 (during the first 30 min). ** p=0.0027, ****p<0.0001 (T-test). n = 7 – 10.
(C-F) Dose-dependent inhibition of AITC- or GNE551-evoked nocifensive behavior by TRPA1 inhibitor AMG0902 (C, E) and corresponding PK/PD plots (D, F). AMG0902 blocked the nocifensive responses evoked by AITC and GNE551 with EC50, (unbound free AMG0902 concentration at 50% efficacy) of 62 ± 26 nM (D) and 431 ± 145 nM (F), respectively. Data in E was collected over two sessions. First session: arms vehicle (black symbols), 1 mg/kg, 3 mg/kg, and 10 mg/kg (1); second session: arms vehicle (gray symbols), 10 mg/kg (2), 30 mg/kg, and 100 mg/kg. n = 7 – 14.
(G-J) Dose-dependent inhibition of AITC- or GNE551-evoked nocifensive responses by TRPA1 inhibitor GNE235 (G, I) and corresponding PK/PD plots (H, J). GNE235 blocked AITC- or GNE551-evoked responses with EC50 of 3.4 ± 1.1 nM (H) and 22 ± 6 nM (J), respectively. Data in I was collected over two sessions. First session: arms vehicle (black symbols), 1 mg/kg, 3 mg/kg, and 10 mg/kg (1); second session: arms vehicle (gray symbols), 10 mg/kg (2), and 30 mg/kg. n = 7 – 15. p<0.001 for 5C, 5E, 5G; and p=0.006 for 5I (Kruskall-Wallis non-parametric analysis).
AITC- and GNE551-evoked Pain Have Different Sensitivity to Antagonist Treatment
AITC-induced pain has been extensively utilized to evaluate the efficacy of TRPA1 antagonists, including A-967079 and AMG0902 (Chen et al., 2011; Lehto et al., 2016). Given their kinetic differences, we wondered whether AITC- or GNE551-evoked pain responses have different sensitivities to antagonist treatment. AMG0902 inhibited rat TRPA1 activation by AITC and GNE551 with EC50 values of 52 ± 4 nM and 84 ± 7 nM, respectively (n = 4), and its pharmacokinetic properties in rats were characterized (Tables S9 and S10). In the behavioral experiments, AMG0902 was administered first, and at the time of its peak plasma concentrations, AITC (0.1%, 25 μl) or GNE551 (10 mM, 25 μl) was injected into the rat paw to induce nocifensive responses. A PK (pharmacokinetic, free plasma concentration) versus PD (pharmacodynamic, licking time) plot was constructed based on data from each individual animal. AMG0902 blocked AITC-induced pain in a dose-dependent manner (Figure 5C), with an EC50 of 62 ± 26 nM (free plasma concentration, Figure 5D, Table S10). AMG0902 also inhibited GNE551-induced responses, but its potency was reduced (Figure 5E). For example, at 30 mg/kg dosing, AMG0902 blocked 86% of AITC-induced response, but only 38% of GNE551-induced response (Figures 5C and 5E). The EC50 for AMG0902 to block GNE551-induced pain was 431 ± 145 nM, or ~7-fold higher than its EC50 inhibiting AITC-evoked pain (Figures 5D and 5F, Table S10). In addition to the total pain responses, AMG0902 also lengthened the latency to nocifensive behavior with increasing doses (Figure S7E).
GNE235 is a TRPA1 antagonist belonging to a class of proline sulfonamides (Chen et al., 2018). It was more potent than AMG0902 (IC50: 3.3 ± 1.0 nM and 3.4 ± 0.9 nM on AITC and GNE551 activation, respectively, Tables S9), and had different PK properties compared to AMG0902 (Table S10). GNE235 inhibited AITC-evoked responses with a near maximal effect at 3 and 5 mg/kg (Figure 5G), and an EC50 of 3.4 ± 1.1 nM (Figure 5H). GNE235 significantly increased the latency of GNE551-induced pain responses (Figure S7F), but its efficacy plateaued at 51% at 30 mg/kg (Figure 5I). The calculated EC50 for GNE235 inhibiting GNE551-evoked pain was 22 ± 6 nM (Figure 5J), or ~ 7-fold higher than EC50 on AITC-evoked pain (Figure 5H). Therefore, compared to AITC-evoked pain, GNE551-evoked pain is relatively insensitive to treatment by two different classes of antagonists.
DISCUSSION
The discovery and characterization of GNE551 provide new insights into the mechanism of TRPA1 channel modulation and physiology. Our GNE551-bound TRPA1 cryo-EM structure reveals a hitherto unknown agonist binding pocket, which is localized in a cleft between the VSLD and pore module, and is completely embedded within the hydrophobic phase of the lipid bilayer. A recent cryo-EM structure of TRPA1 (PDB: 6PQQ) revealed the presence of a well-ordered phospholipid (“lipid 5”), with its aliphatic tail extending into the GNE551 pocket, and its head group positioned alongside the S4-S5 linker in the vicinity of Glu864 (Suo et al., 2019) (Figures 4, S5D–S5F, and S6A). Replacing Glu864 with a tryptophan produced a constitutively open channel, and molecular modeling suggests that the Trp864 side-chain impedes the binding of lipid 5. Based on these observations, we propose a scheme for TRPA1 gating: under basal conditions, a lipid occupies the transmembrane binding pocket and stabilizes the closed channel conformation; the displacement of this lipid by GNE551 binding, or by the large side chain of the introduced tryptophan residue, enables a conformational change in the S4-S5 linker and subsequent opening of the intracellular gate.
Consistent with our cryo-EM structure, phylogenic analysis independently identified Gln940 as critical for GNE551 activation (Figures 2E and S3, Table S5). The GNE551-binding pose was further corroborated by point mutations (Table S7) and by structure-function relationship for a series of aminotriazole agonists (Figure S1C). Of note, the GNE551-bound TRPA1 structure is in a non-conducting state; consequently, the exact structural mechanism of how GNE551 activates the channel remains uncertain. The observed non-conducting state could have resulted from the use of a truncated construct but may have occurred even with a fully-functional construct, given the suboptimal open probability of GNE551-bound TRPA1 (Po of 0.41). Previous TRPA1 structures containing electrophilic agonists AITC, JT010, and BITC were also non-conducting (Paulsen et al., 2015; Suo et al., 2019). In a recent report, HCN1 channel, expected to be in open state, has also yielded non-conductive state in structural analyses (Lee and MacKinnon, 2019). Nonetheless, an open state structure of TRPA1 has recently been obtained by the use of the irreversible electrophilic agonist iodoacetamide (Zhao et al., 2020), demonstrating that the closed to open transition involves large-scale rearrangements, most notably of the S4-S5 linker, which is located near the GNE551 binding pocket. To test whether the GNE551-TRPA1 interactions reported here are relevant in the open state, we overlaid our GNE551-TRPA1 structure with the recently resolved open state structure (Zhao et al., 2020) (Figure S8). As expected, the S4-S5 helix was dramatically displaced relative to the GNE551-binding pocket, suggesting that the “bottom” of the binding pocket is conformationally malleable and moves during openings and closings of the channel. In contrast, the fine structure of the remainder of GNE551-binding pocket is remarkably unchanged (Figure S8), indicating that molecular interactions between TRPA1 and all but the dimethoxy aniline moiety of GNE551 remain constant through TRPA1 activation.
The GNE551/lipid 5-binding pocket in TRPA1 is distinct from the cooling agent and antagonist binding site on TRPM8 channel, which is localized in the VSLD domain (Diver et al., 2019; Yin et al., 2019), but resembles the well-characterized vanilloid/lipid-pocket in TRPV1 channel (Gao et al., 2016). A phosphatidylinositol lipid binds to this TRPV1 pocket and interacts with the S4-S5 linker to stabilize the closed state of the channel. Resiniferatoxin (RTX) displaces the resident lipid and promotes an electrostatic interaction between residue Arg557 and Glu570, thereby pulling away the S4-S5 linker and opening the intracellular gate (Gao et al., 2016). Similar to RTX, binding of capsaicin and other agonists is also expected to displace the resident lipid in TRPV1. Likewise, the GNE551-binding pocket in TRPA1 may accommodate various ligands and lipids to modulate channel function. Besides TRPV1 and TRPA1, many other ion channels possess similar agonist/lipid binding pockets, including TRPV2, TRPV5, and Polycystin-2 (PC2) channels (Gao et al., 2016; Hughes et al., 2018; Wang et al., 2020; Zubcevic et al., 2019). Thus, channel activation via lipid displacement from vanilloid-like pockets may be a conserved ligand-recognition and activation mechanism.
As a polymodal receptor, the TRPA1 channel is modulated by a wide variety of stimuli. Menthol, propofol, and piperidine carboxamides interact with the pore domain, although the consequent activation mechanisms are not well understood (Chernov-Rogan et al., 2019; Ton et al., 2017; Xiao et al., 2008). Electrophilic compounds modify cysteine residues in a cytoplasmic structure (dubbed the “clam shell”, or “A-loop”) (Suo et al., 2019; Zhao et al., 2020). Cysteine modification leads to a conformational change in the upper part of the clam shell or A-loop, which propagates vertically to the TRP helix and S4-S5 linker, resulting in the straightening of S5 and the dilation of the intracellular gate. Concurrently, the covalent modification induces global conformational changes, such as the rotation of the VSLD and twisting of the pore domain. The same structure that recognizes electrophilic compounds also engages the peptide toxin WaTx, albeit through non-covalent ligand binding. In contrast to electrophiles and WaTx, GNE551 binds to a transmembrane pocket formed at the interface of the VSLD and the pore domain. Likely as a consequence of their different interaction mechanisms, WaTx, AITC, and GNE551 elicit distinct biophysical characteristics on TRPA1 channels. WaTx increases open time but not open probability, and reduces the Ca2+ permeability of open channels (Lin King et al., 2019). In contrast, GNE551 increases both open time and open probability, and retains Ca2+ permeability (Figures 1F and 1G, Table 1). Compared to AITC-activated channels, GNE551-activated channels have a smaller conductance and an altered Ca2+/Mg2+ permeability (Table 1). Furthermore, unlike the AITC-activated channels, which show rapid desensitization and tachyphylaxis, GNE551-activated channels do not desensitize and activation can be achieved in a reversible fashion (Figure 2).
The selectivity of GNE551 for TRPA1 is supported by several lines of evidence, including its lack of effect on 42 protein targets (Tables S1–S4), its ability of evoking Ca2+ influx in DRG neurons from WT, but not TRPA1 KO rats (Figure S7A), and the attenuation of GNE551 evoked-pain in TRPA1 KO rats (Figure 5B). In behavioral experiments, AITC and GNE551 evoke pain with fast onset and slow onset, respectively (Figures 5A and 5B, Video S1), while in in vitro experiments, they activate TRPA1 channel in the similar time frame (Figure 2D). This discrepancy may be resulted from their different ability in tissue penetration: GNE551 has a larger molecular weight, larger topical polar surface area, is more lipophilic, and predicted with poor tissue permeability (Table S8). Additionally, AITC evokes transient pain, whereas GNE551 induces a long-lasting pain response (Figures 5A and 5B). This remarkable difference is consistent with their different modes of channel modulation, namely, rapid desensitization and tachyphylaxis for AITC versus absence of desensitization and reversible activation for GNE551 (Figure 2). Other factors, such as pharmacokinetic difference, may also play a role. AITC is highly reactive and is not stable in a cytosolic environment containing high concentrations of nucleophiles (e.g., ~ 5 mM glutathione and ~ 0.2 mM cysteines). In contrast, GNE551 is non-reactive and expected to sustain its concentration for a much longer time, leading to more persistent activation of TRPA1. Regardless of the exact mechanisms, the finding that covalent and non-covalent TRPA1 agonists elicit different pain responses may have important ramifications. TRPA1 has been implicated in various pain pathologies (nociceptive, inflammatory, and neuropathic) and other disorders such as itch and asthma. These different pathological conditions may involve different sets of ligands with unique temporal, spatial, and channel-modulating characteristics. Currently, reactive species (e.g., 4-hydroxynonenal, methylglyoxal, 8-iso-prostaglandin A2, and H2O2) are considered as likely endogenous TRPA1 ligands (Bautista et al., 2013; Chen and Hackos, 2015). However, non-covalent ligands may regulate TRPA1 function under physiological and pathological conditions. For example, non-covalent lipids, including PIP2, cholesterol, hepoxilin A3, and diacylglycerol, are known to activate TRPA1 and likely play a role in mediating pain and inflammation (Bandell et al., 2004; Gregus et al., 2012; Karashima et al., 2008; Startek et al., 2019). In particular, PIP2 has been shown to modulate TRPV5 and PC2 via binding to sites equivalent to the GNE551 pocket (Hughes et al., 2018; Wang et al., 2020). It will be interesting to investigate whether PIP2 and other lipids also bind to the GNE551 pocket in TRPA1 underlying their physiological and pathological function.
As a noxious irritant, AITC has been used to induce pain and neurogenic inflammation for many decades (Landsteiner and Di Somma, 1938). Following the discovery of TRPA1 as its molecular target, AITC-based behavioral models have been widely used to assess TRPA1 antagonists and other potential analgesics (Chen et al., 2011; Lehto et al., 2016). Recently, experimental pain models in humans have also been developed using AITC, and JT010, also a covalent agonist (Andersen et al., 2017; Heber et al., 2019). Antagonists are tested in these models to assess target engagement, a critical step in advancing compounds in human clinical trials. Here, we show that for two different antagonists, GNE551-induced pain is ~7-fold less sensitive to antagonist treatment compared to AITC-induced pain (Figures 5C–5J). This phenomenon could be resulted from persistent channel activation by GNE551 due to its continued presence and the lack of channel desensitization. In addition to electrophilic ligands, TRPA1 can be modulated by endogenous, non-covalent ligands; and their underlying diseases can be less sensitive to antagonist treatment compared to AITC-evoked pain. Therefore, additional preclinical or human experimental models, using GNE551 or ligands with different modalities, should be explored for future drug discovery efforts.
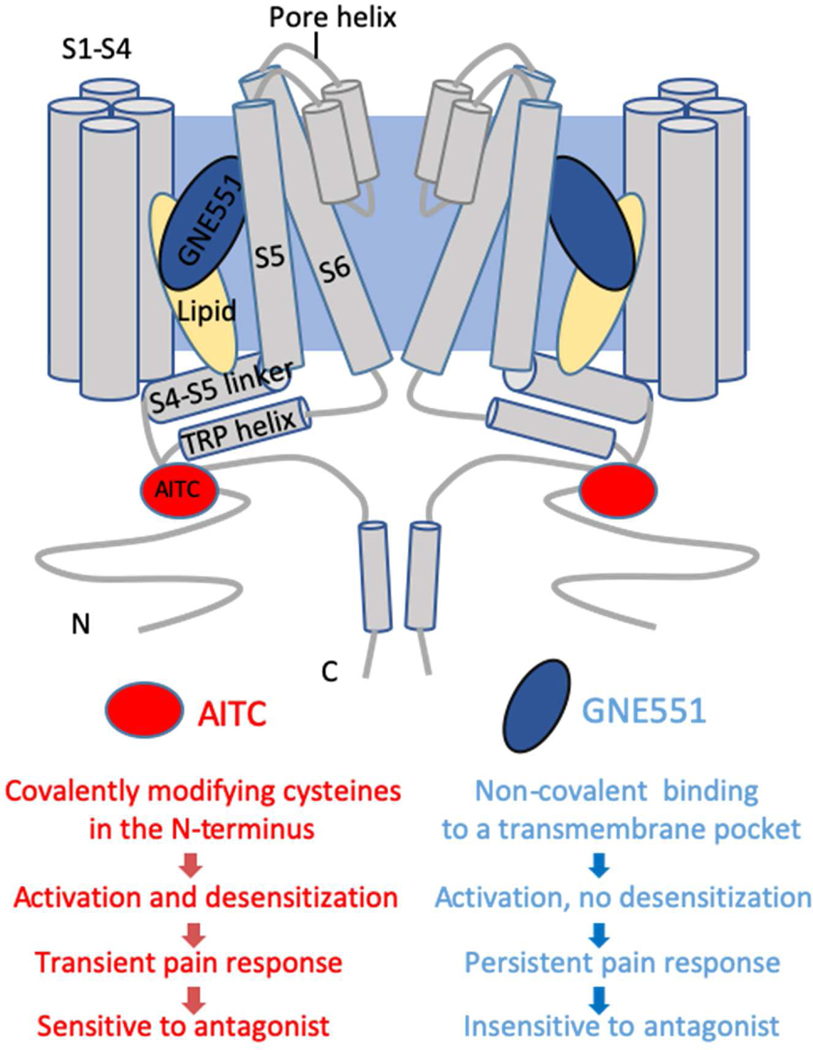
Our current work on TRPA1 invokes the notion of biased agonism, a concept that has profoundly changed the field of GPCRs (Jarpe et al., 1998). Biased signaling initially referred to ligands that target orthosteric binding sites (i.e., the sites of the endogenous ligands) and preferentially stimulate G protein signaling or arrestin pathways. Recently, the concept has been extended to allosteric ligands that bind to a site distinct from the orthosteric ligands, and to bitopic ligands that bind at both orthosteric and allosteric sites (Wisler et al., 2018), offering potential benefits such as new modes of receptor modulation, novel chemical scaffolds, enhanced subtype selectivity, and decreased side effects. Recent studies suggest that ion channel ligands may also exert biased agonism through different modes of modulation and alterations of biophysical properties (Perszyk et al., 2020). In TRPA1, binding of WaTx activates the channel with a reduced Ca2+ permeability compared to AITC, and elicits pain without neurogenic inflammation (Lin King et al., 2019). In the current study, we show that AITC and GNE551 interact with TRPA1 in different manners and produce distinct biophysical, physiological, and pharmacological consequences (Figure 6). In contrast to AITC, which covalently modify intracellular cysteine residues, GNE551 binds to a novel transmembrane pocket through non-covalent interactions, and activates channels with different ion selectivity and conductance. Most notably, AITC induces desensitization and tachyphylaxis, whereas GNE551 does not desensitize and activates the channel in a reversible manner. Furthermore, AITC evokes transient pain that is sensitive to antagonist treatment, whereas GNE551 evokes long-lasting pain with reduced antagonist sensitivity. Based on these findings, we postulate that different endogenous agonists of TRPA1 may underlie discrete physiology, disease etiology, and efficacy of clinical treatment. We also anticipate that biased agonism may play important roles in the physiology and pharmacology of many other ion channels. Since 15% of all marketed drugs target ion channels (Santos et al., 2017), understanding biased agonism in ion channels like TRPA1 could be essential to translational research and drug development.
Figure 6. Covalent and Non-covalent Agonists Exert Biased Agonism at the TRPA1 Channel.
AITC covalently modifies intracellular cysteines; GNE551 binds to a transmembrane pocket.
AITC activates and desensitizes TRPA1; GNE551 activates the channel without desensitization.
AITC induces transient pain; GNE551 induces persistent pain.
AITC-evoked pain is sensitive to antagonist treatment; GNE551-evoked pain is relatively insensitive to antagonist treatment.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contac, Jun Chen (chen.jun@gene.com).
Materials Availability
Plasmid constructs and reagents generated in this article are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The structure of GNE551-bound human TRPA1 is accessible with PDB code: 6X2J (atomic model) and EMDB code: EMD-22009 (map).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293-F cells (Thermo Fisher, R79007) were used for patch-clamp electrophysiology and Ca2+ assays. High-Five™ cells (Thermo Fisher, bti-tn-5b1–4) were used for protein production. Experimental procedures involving animals were approved by Genentech’s Institutional Animal Care and Use Committee and conducted in accordance with the recommendations of the International Association for the Study of Pain. Male Sprague-Dawley rats (Charles River, Hollister; strain code: 001) at 8 weeks of age or older were used. For experiments comparing TRPA1 KO rats and WT littermates, both males and females were used; animals were generated by crossing heterozygotes in our breeding facility. Rats had access to ad libitum food and water, and were housed in individually vented caging (Allentown IVC, 143 in2) on 36 slot racks. Cages were changed two times weekly and health checks were performed daily. The housing room was on a light/dark cycle of 14 hours on, 10 hours off (5:30am-7:30pm on) with all behavioral experiments taking place during the light cycle.
METHODS DETAILS
Chemical compounds
GNE551:5-amino-1-(4-bromo-2-fluorobenzyl)-N-(2,5-dimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, GNE551 analogs, WaTx, A-96079, AMG0902, and GNE235 were synthesized at Genentech. The following reagents were obtained commercially: allyl isothiocyanate (Sigma-Aldrich); Hank’s Balanced Salt Solution, FreeStyle Max Reagent, Calcium assay kit, and TCEP-HCl (Thermo Fisher); Facade®-EM (Avanti); Bio-Beads SM-2 (Bio-Rad); PMAL-C8 (Anatrace); and Amylose resin (New England Biolabs).
Glutathione reactivity assay
In each reaction, GNE551 or AITC (10 μM) was incubated with or without 1 mM glutathione in KPhosphate buffer (50 mM, pH7.4) at 37°C for 30 minutes. Reactions were stopped by chilling with ice. Samples were analyzed on the Thermo Fusion Lumos mass spectrometer equipped with a HESI source and coupled to an Ultimate 3000 UHPLC. Samples were separated using a Kinetex C18 (1.7 μM, 100Å, 50 × 2.1 mm) column at a flow rate of 0.4 mL/min and temperature at 40˚C. LC separation was as follows: Solvent A contained 0.1% formic acid in water and solvent B contained 0.1% formic acid in acetonitrile. A gradient program was run from 5% to 50% B over 15 minutes, followed by a wash at 95% solvent B.
Calcium influx assay
Calcium influx assay was performed as reported previously (Chen et al., 2013). Briefly, HEK-293F cells were transfected one day before the calcium imaging experiments. Transfected cells were loaded with calcium sensitive dye (Thermo Fisher, BD#640178) and incubated for one hour at 37˚C. Compound addition and fluorescent signal measurement were performed using the FDSS/μCell.
Whole-cell patch clamp recordings
Whole-cell currents were recorded one day after transient transfection of HEK293-F cells. Recording electrodes (3–5 M tip resistance) were fabricated with borosilicate glass capillaries (Harvard Apparatus). TRPA1 currents were triggered with a voltage ramp protocol composed of: 0 mV holding potential, a pre-step to −80 mV for 10 msec, a voltage ramp from −80 mV to +80 mV for 200 msec, and a post-step at +80 mV for 10 msec before stepping back to 0 mV holding potential. The protocol was applied once per second. Extracellular solution contained 140 mM NaCl, 2 mM KCl, 1 mM MgCl2, 10 mM HEPES, 0.1 mM EGTA, and 5 mM dextrose (pH 7.4). The intracellular solution contained 135 mM CsF, 10 mM NaCl, 10 mM HEPES, 1 mM MgCl2, and 10 mM EGTA (pH 7.2). Compounds were delivered using a multichannel fast solution exchange platform (Cellectricon).
Inside-out single channel recordings
Recordings were performed using a HEKA EPC10 amplifier paired with PatchMaster software (HEKA). Signals were filtered at 2.9 kHz and sampled at 10 kHz. Patch pipettes were prepared using borosilicate glass then heat polished to have a tip resistance of 6–10 Mι. Unless otherwise noted, bath and pipette solutions were symmetrical and consisted of 140 mM NaCl, 2 mM EGTA, and 5 mM HEPES with pH 7.2. Calcium-free solutions were achieved with the addition of 2 mM EGTA. Based on MAXCHELATOR program (http://maxchelator.stanford.edu), the total free calcium under this condition was < 500 pM. All recordings were performed at room temperature, and solutions were perfused through separate tubes onto the electrode tip containing the excised membrane patch using a rapid solution changer (RSC-200 Bio-Logic). For reversal potential measurements, 140 mM NaCl was replaced with 70 mM MgCl2 or 70 mM CaCl2 with EGTA excluded. Macropatch currents were recorded with a 500 msec voltage ramp protocol from −80 to +80 mV as described above. Single channel recordings were analyzed with Igor Pro 8 (WaveMetrics). Single channel amplitude and nPo were determined from all-point histograms while single channel open probability (Po) and dwell time measurements were determined using QuB software. Single channel conductance was determined by the slope conductance method, and permeability ratios were determined using the Goldman-Hodgkin-Katz equation.
Recombinant expression and purification of truncated hTRPA1
The coding sequence for human TRPA1 [Ser448-Thr1078] was synthesized (Genescript) and cloned into a modified pAcGP67A vector downstream of the polyhedron promoter. Recombinant baculovirus was generated using the Baculogold system (BD Biosciences) following standard protocols. Trichoplusia ni (High-Five) cells were infected for protein production and harvested 48 hours post-infection. Sequence of the construct for structural studies is shown below (the underlined sequence is the 6-His followed by a MBP affinity tag, TEV protease cleavage site is in bold and a (GGS)4 linker shown in italic was added in-between the TEV cleavage sequence and TRPA1 sequence):
MHHHHHHGKIEEGKLVIWINGDKGYNGLAEVGKKFEKDTGIKVTVEHPDKLEEKFPQVAATGDGPDIIFWAHDRFGGYAQSGLLAEITPDKAFQDKLYPFTWDAVRYNGKLIAYPIAVEALSLIYNKDLLPNPPKTWEEIPALDKELKAKGKSALMFNLQEPYFTWPLIAADGGYAFKYENGKYDIKDVGVDNAGAKAGLTFLVDLIKNKHMNADTDYSIAEAAFNKGETAMTINGPWAWSNIDTSKVNYGVTVLPTFKGQPSKPFVGVLSAGINAASPNKELAKEFLENYLLTDEGLEAVNKDKPLGAVALKSYEEELAKDPRIAATMENAQKGEIMPNIPQMSAFWYAVRTAVINAASGRQTVDEALKDAQTNSSSNNNNNNNNNNGENLYFQGSGGSGGSGGSGGSPLHFAASYGRINTCQRLLQDISDTRLLNEGDLHGMTPLHLAAKNGHDKVVQLLLKKGALFLSDHNGWTALHHASMGGYTQTMKVILDTNLKCTDRLDEDGNTALHFAAREGHAKAVALLLSHNADIVLNKQQASFLHLALHNKRKEVVLTIIRSKRWDECLKIFSHNSPGNKCPITEMIEYLPECMKVLLDFCMLHSTEDKSCRDYYIEYNFKYLQCPLEFTKKTPTQDVIYEPLTALNAMVQNNRIELLNHPVCKEYLLMKWLAYGFRAHMMNLGSYCLGLIPMTILVVNIKPGMAFNSTGIINETSDHSEILDTTNSYLIKTCMILVFLSSIFGYCKEAGQIFQQKRNYFMDISNVLEWIIYTTGIIFVLPLFVEIPAHLQWQCGAIAVYFYWMNFLLYLQRFENCGIFIVMLEVILKTLLRSTVVFIFLLLAFGLSFYILLNLQDPFSSPLLSIIQTFSMMLGDINYRESFLEPYLRNELAHPVLSFAQLVSFTIFVPIVLMNLLIGLAVGDIADVQKHASLKRIAMQVELHTSLEKKLPLWFLRKVDQKSTIVYPNKPRSGGMLFHIFCFLFCTGEIRQEIPNADKSLEMEILKQKYRLKDLTFLLEKQHELIKLIIQKMEIISET
250 grams of harvested cell pellet was resuspended in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 μg/mL benzonase, 0.5 mM TCEP, and Roche complete® protease inhibitor tablets). Cells were homogenized, microfluidized once and spun down at 8000 g for 20 min then spun down at 40000 g for 60 min. Membranes were solubilized in 25 mM Tris (pH 8), 150 mM NaCl, 10% glycerol, 0.5 mM TCEP, complete® protease inhibitor tablets, and 1% (w/v) FA-3 (Façade®-EM, Avanti) at 4°C for 2 hours. Insoluble debris was pelleted by ultracentrifugation at 40,000 rpm for 60 min, and the supernatant containing the solubilized protein was collected for affinity purification by batch binding to 3 mL of amylose resin for 1 hour at 4 °C. Solution was then poured onto a Bio-Rad column and re-packed by gravity flow. Column was washed with 5 CV of 25 mM Tris (pH 8), 150 mM NaCl, 0.5 mM TCEP with 0.06% FA-3 (Buffer A) and eluted with 3CV of Buffer A + 40 mM maltose. Elution fraction was concentrated down to 400 μL (~ 4 mg/ml) with a 100 kDa MWCO concentrators (Amicon Ultra-15 Centrifugal Filter Units Millipore Sigma). Three fold excess (w/w) of PMAL-C8 (Anatrace, 100 mg/mL stock solution in water) was added along to some TEV protease (prepared in-house) and EndoF1 and EndoH deglycosidases (prepared in-house). Sample was nutated overnight at 4°C. Biobeads were added to the sample and incubated at 4°C for 2 hours. Beads were filtered out and supernatant was loaded onto a Superose 6 3.2/300 column linked to an AKTA system pre-equilibrated with 25 mM Tris pH (8.2), 150 mM NaCl, 0.5 mM TCEP. Sample from the most concentrated fraction (around 1 mg/mL) was incubated with 100 μM of GNE551 (10 mM stock in 100% DMSO) at 4°C for 1 hour and applied to grids.
Cryo-EM sample preparation and data acquisition
Grids were prepared in the following manner. Holey carbon grids (Quantifoil, R0.6/1, 25nm, Au, 300 mesh) were plasma etched using the Solarus plasma cleaner (Gatan) in the hydrogen-oxygen setting for 3 seconds. 3 L of the peak fraction was applied to a glow-discharged holey Au grid, incubated for 30 seconds then hand blotted; this step was repeated 3 times. Another 3 L of the peak fraction was applied and grids were finally blotted in Vitribot Mark IV (Thermo Fisher) using 8 seconds blotting time with 100% humidity, and plunge-frozen in liquid ethane cooled by liquid nitrogen. Movie stacks were collected using SerialEM (Mastronarde, 2005) on a Titan Krios operated at 300 keV with bioquantum energy filter equipped with a K3 Summit direct electron detector camera (Gatan). Images were recorded at 165000x magnification corresponding to 0.83 Å/pixel. Each image stack contains 40 frames recorded every 0.25 seconds for an accumulated dose of ~40 e/Å2 and a total exposure time of 10 seconds. Images were recorded with a set defocus range of 0.8 to 2.5 m.
Cryo-EM data processing
Image processing from stacks of frames to refined 3D volumetric map was performed with cisTEM (Grant et al., 2018). Density modification was performed with Phenix, using all default parameters (Terwilliger, 2020). This density-modified map was used for model building and refinement. For atomic model building, PDB: 6PQQ (Suo et al., 2019) was rebuilt using interactive molecular dynamics in ISOLDE (Croll, 2018), and refined in real space using Phenix (Afonine et al., 2018) and Coot (Emsley et al., 2010). Map-model FSC was computed using cryo-EM validation job type in Phenix. The local resolution map was obtained following a re-implementation of the algorithm (Cardone et al., 2013). Molecular graphics were done with ChimeraX (Pettersen et al., 2020).
Animal behavioral studies
GNE551 was formulated in SEDDS (self-emulsifying drug delivery system; 50% Cremophor, 35% N-methyl-2-pyrrolidone, 15% miglyol). AITC (from Sigma) was formulated in mineral oil at 0.1% (10.2 mM). AMG0902 and GNE235 were formulated in 15% dimethyl sulfoxide (DMSO) / 85% MCT (0.5% methylcellulose / 0.2% Tween 80) and dosed by oral gavage in a volume of 5 mL/kg. AMG0902 or GNE235 was dosed first; at Tmax (time point of maximal plasma concentration, 1 hour for AMG0902 and 4 hours for GNE235), intraplantar injection of AITC or GNE551 was performed to elicit nocifensive responses. Animals were euthanized by CO2 inhalation immediately after the behavioral observation. For pharmacokinetic/pharmacodynamic (PK/PD) studies, blood samples were collected by cardiac puncture into K2EDTA-containing tubes and processed into plasma by centrifugation at 17,000 x g for 2 min and collection of supernatants. Plasma samples were frozen at −80°C until bioanalysis. Plasma concentrations of AMG0902 or GNE235 were determined using liquid chromatography tandem mass spectrometry.
Under light restraint, a unilateral intraplantar injection of AITC or GNE551 was made in a volume of 25 μL using a luer-tipped Hamilton syringe fitted with a 30-gauge disposable needle. The rat was immediately placed into a plastic observation chamber on an elevated glass surface with angled mirrors below, facilitating video recording of each animal from both side and bottom viewing angles. Observation chambers are of sufficient size that the rat can move around freely without restraint. Opaque side walls separated individual animals. Up to six rats at a time were video-recorded for 15 min for AITC injections or 30 min for GNE551 injections. Videos were later scored offline by an observer blinded to treatment group.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 7. Two-tailed Student’s t-test or Kruskall-Wallis non-parametric statistics were used to test for significance of genotype or drug effect. EC50 values were determined by nonlinear regression with the following constraints: EC50 > 0; Bottom = 0; 3 > Hillslope > 1. EC50 values were multiplied by fraction unbound (1-% PPB) to obtain free drug concentrations to allow comparison across assays. Results were considered statistically significant if p<0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| GNE551 | This paper | N/A |
| GNE551 analogs | This paper | N/A |
| Allyl isothiocyanate | Sigma-Aldrich | 377430 |
| WaTx | Lin-King, et al, 2019 | N/A |
| A-967079 | Chen et al., 2011 | N/A |
| AMG0902 | Lehto et al., 2016 | N/A |
| GNE235 | This paper | N/A |
| Hank’s Balanced Salt Solution | Thermo Fisher | 14025076 |
| FreeStyle Max Reagent | Thermo Fisher | 16447-100 |
| Calcium assay kit | Thermo Fisher | BD#640178 |
| Facade®-EM | Avanti | 850522 |
| Bio-Beads SM-2 | Bio-Rad | 1523920 |
| PMAL-C8 | Anatrace | P5008 |
| Amylose resin | New England Biolabs | E8021S |
| TCEP-HCl | Thermo Fisher | 20490 |
| Critical Commercial Assays | ||
| In vitro Safety Pharmacology | Eurofins | General Panel |
| NK1 Receptor Agonist Assay | Eurofins | Cat# 2190 |
| NK1 Receptor Antagonist Assay | Eurofins | Cat# 2192 |
| Cav1.2 Assay | Eurofins | IonChannelProfiler, GNT032916-1 |
| Deposited Data | ||
| Coordinates for TRPA1-GNE551 | This paper | PDB: 6X2J |
| Cryo-EM map for TRPA1-GNE551 | This paper | EMD-22009 |
| Experimental Models: Cell Lines | ||
| HEK293-F cells | Thermo Fisher | R79007 |
| High-Five™ | Thermo Fisher | bti-tn-5b1-4 |
| Experimental Models: Organisms/Strains | ||
| Sprague-Dawley Rat, Male | Charles River | Strain code: 001 |
| Sprague-Dawley Rat, Male, TRPA1 knockout | Reese et al., 2020 | N/A |
| Recombinant DNA | ||
| Plasmid: TRPA1 from human, rat, dog, guinea pig and chicken | Chernov-Rogan et al., 2018 | N/A |
| Plasmid: rhesus monkey TRPA1 | Chen et al., 2013 | N/A |
| Plasmid: human-chicken TRPA1 chimeras and point mutations | This paper | N/A |
| pAcGP67A | https://www.bdbiosciences.com/en-us/applications/research-applications/multicolor-flow-cytometry/vector-sequences-pacgp67a-vector | Addgene, 12130 |
| Software and Algorithms | ||
| FDSS software | Hamamatsu | N/A |
| pClamp10 | Molecular Devices | N/A |
| PatchMaster | HEKA | N/A |
| Igor Pro8 | WaveMetrics | https://www.wavemetrics.com/software/igor-pro-8 |
| UCSF ChimeraX | Godard et al., 2007 | https://www.rbvi.ucsf.edu/chimerax/ |
| serialEM | Mastronarde et al., 2005 | https://bio3d.colorado.edu/SerialEM/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| cisTEM | Grant et al., 2018 | https://cistem.org/ |
| Phenix | Afonine et al., 2018 | http://www.phenix-online.org/ |
| ISOLDE | Croll, 2018 | https://isolde.cimr.cam.ac.uk/ |
| Prism | GraphPad | https://www.graphpad.com |
| SwissAdme | SwissAdme | http://www.swissadme.ch/ |
| Other | ||
| Quantifoil R 0.6/1, 300 mesh, Gold | EMS | Q310AR-06 |
Highlights.
GNE551-TRPA1 co-structure reveals an agonist transmembrane binding pocket
Unlike AITC, GNE551 activates TRPA1 without channel desensitization
GNE551 induces persistent pain, whereas AITC induces transient pain
GNE551-evoked pain is less sensitive to TRPA1 antagonists
ACKNOWLEDGEMENTS
We would like to thank Kevin Ford, Michelle Dourado, Gloria Granados, and colleagues at BioMolecular Resources in Genentech for their assistance. We would also like to thank Craig Yoshioka (OHSU) for collecting the cryo-EM images and Dr. Michael Sanguinetti for his critical reading of the manuscript. The research is supported by Genentech, and by National Science Foundation (Grant NS103954 to J.Z.). A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC in OHSU through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
Footnotes
DECLARATION OF INTERESTS
All authors, except S.V. and J.Z., are current or former employees of Genentech, a member of the Roche group, and may hold Roche stock or stock options.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen HH, Lo Vecchio S, Gazerani P, and Arendt-Nielsen L. (2017). Dose-response study of topical allyl isothiocyanate (mustard oil) as a human surrogate model of pain, hyperalgesia, and neurogenic inflammation. Pain 158, 1723–1732. [DOI] [PubMed] [Google Scholar]
- Bandell M, Story GM, Hwang SW, Viswanath V, Eid SR, Petrus MJ, Earley TJ, and Patapoutian A. (2004). Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron 41, 849–857. [DOI] [PubMed] [Google Scholar]
- Bautista DM, Jordt SE, Nikai T, Tsuruda PR, Read AJ, Poblete J, Yamoah EN, Basbaum AI, and Julius D. (2006). TRPA1 Mediates the Inflammatory Actions of Environmental Irritants and Proalgesic Agents. Cell 124, 1269–1282. [DOI] [PubMed] [Google Scholar]
- Bautista DM, Pellegrino M, and Tsunozaki M. (2013). TRPA1: A gatekeeper for inflammation. Annu Rev Physiol 75, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres AI, Brackmann M, Elia MD, Bessac BF, del Camino D, D’Amours M, Witek JS, Fanger CM, Chong JA, Hayward NJ, et al. (2009). A sensory neuronal ion channel essential for airway inflammation and hyperreactivity in asthma. Proc Natl Acad Sci U S A 106, 9099–9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, and Steven AC (2013). One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol 184, 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Volgraf M, Do S, Kolesnikov A, Shore DG, Verma VA, Villemure E, Wang L, Chen Y, Hu B, et al. (2018). Discovery of a Potent (4 R,5 S)-4-Fluoro-5-methylproline Sulfonamide Transient Receptor Potential Ankyrin 1 Antagonist and Its Methylene Phosphate Prodrug Guided by Molecular Modeling. J Med Chem 61, 3641–3659. [DOI] [PubMed] [Google Scholar]
- Chen J, and Hackos DH (2015). TRPA1 as a drug target--promise and challenges. Naunyn Schmiedebergs Arch Pharmacol 388, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Joshi SK, DiDomenico S, Perner RJ, Mikusa JP, Gauvin DM, Segreti JA, Han P, Zhang XF, Niforatos W, et al. (2011). Selective blockade of TRPA1 channel attenuates pathological pain without altering noxious cold sensation or body temperature regulation. Pain 152, 1165–1172. [DOI] [PubMed] [Google Scholar]
- Chen J, Kang D, Xu J, Lake M, Hogan JO, Sun C, Walter K, Yao B, and Kim D. (2013). Species differences and molecular determinant of TRPA1 cold sensitivity. Nature communications 4, 2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang XF, Kort ME, Huth JR, Sun C, Miesbauer LJ, Cassar SC, Neelands T, Scott VE, Moreland RB, et al. (2008). Molecular determinants of species-specific activation or blockade of TRPA1 channels. J Neurosci 28, 5063–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov-Rogan T, Gianti E, Liu C, Villemure E, Cridland AP, Hu X, Ballini E, Lange W, Deisemann H, Li T, et al. (2019). TRPA1 modulation by piperidine carboxamides suggests an evolutionarily conserved binding site and gating mechanism. Proc Natl Acad Sci U S A 116, 26008–26019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diver MM, Cheng Y, and Julius D. (2019). Structural insights into TRPM8 inhibition and desensitization. Science 365, 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elokely K, Velisetty P, Delemotte L, Palovcak E, Klein ML, Rohacs T, and Carnevale V. (2016). Understanding TRPV1 activation by ligands: Insights from the binding modes of capsaicin and resiniferatoxin. Proc Natl Acad Sci U S A 113, E137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cao E, Julius D, and Cheng Y. (2016). TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N. (2018). cisTEM, user-friendly software for single-particle image processing. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregus AM, Doolen S, Dumlao DS, Buczynski MW, Takasusuki T, Fitzsimmons BL, Hua XY, Taylor BK, Dennis EA, and Yaksh TL (2012). Spinal 12-lipoxygenase-derived hepoxilin A3 contributes to inflammatory hyperalgesia via activation of TRPV1 and TRPA1 receptors. Proc Natl Acad Sci U S A 109, 6721–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber S, Gold-Binder M, Ciotu CI, Witek M, Ninidze N, Kress HG, and Fischer MJM (2019). A Human TRPA1-Specific Pain Model. J Neurosci 39, 3845–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman A, Chuang HH, Bautista DM, and Julius D. (2006). TRP channel activation by reversible covalent modification. Proc Natl Acad Sci U S A 103, 19564–19568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Huang L, and van Breemen RB (2015). Detection of reactive metabolites using isotope-labeled glutathione trapping and simultaneous neutral loss and precursor ion scanning with ultra-high-pressure liquid chromatography triple quadruple mass spectrometry. Anal Chem 87, 3646–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TET, Lodowski DT, Huynh KW, Yazici A, Del Rosario J, Kapoor A, Basak S, Samanta A, Han X, Chakrapani S, et al. (2018). Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nat Struct Mol Biol 25, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarpe MB, Knall C, Mitchell FM, Buhl AM, Duzic E, and Johnson GL (1998). [D-Arg1,D-Phe5,D-Trp7,9,Leu11]Substance P acts as a biased agonist toward neuropeptide and chemokine receptors. J Biol Chem 273, 3097–3104. [DOI] [PubMed] [Google Scholar]
- Jordt SE, Bautista DM, Chuang HH, McKemy DD, Zygmunt PM, Hogestatt ED, Meng ID, and Julius D. (2004). Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature 427, 260–265. [DOI] [PubMed] [Google Scholar]
- Karashima Y, Damann N, Prenen J, Talavera K, Segal A, Voets T, and Nilius B. (2007). Bimodal action of menthol on the transient receptor potential channel TRPA1. J Neurosci 27, 9874–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karashima Y, Prenen J, Meseguer V, Owsianik G, Voets T, and Nilius B. (2008). Modulation of the transient receptor potential channel TRPA1 by phosphatidylinositol 4,5-biphosphate manipulators. Pflugers Arch 457, 77–89. [DOI] [PubMed] [Google Scholar]
- Kremeyer B, Lopera F, Cox JJ, Momin A, Rugiero F, Marsh S, Woods CG, Jones NG, Paterson KJ, Fricker FR, et al. (2010). A gain-of-function mutation in TRPA1 causes familial episodic pain syndrome. Neuron 66, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Allchorne AJ, Vollrath MA, Christensen AP, Zhang DS, Woolf CJ, and Corey DP (2006). TRPA1 contributes to cold, mechanical, and chemical nociception but is not essential for hair-cell transduction. Neuron 50, 277–289. [DOI] [PubMed] [Google Scholar]
- Landsteiner K, and Di Somma AA (1938). Studies on the Sensitization of Animals with Simple Chemical Compounds : V. Sensitization to Diazomethane and Mustard Oil. J Exp Med 68, 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, and MacKinnon R. (2019). Voltage Sensor Movements during Hyperpolarization in the HCN Channel. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto SG, Weyer AD, Youngblood BD, Zhang M, Yin R, Wang W, Teffera Y, Cooke M, Stucky CL, Schenkel L, et al. (2016). Selective antagonism of TRPA1 produces limited efficacy in models of inflammatory- and neuropathic-induced mechanical hypersensitivity in rats. Mol Pain 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin King JV, Emrick JJ, Kelly MJS, Herzig V, King GF, Medzihradszky KF, and Julius D. (2019). A Cell-Penetrating Scorpion Toxin Enables Mode-Specific Modulation of TRPA1 and Pain. Cell 178, 1362–1374 e1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Dubin AE, Evans MJ, Marr F, Schultz PG, Cravatt BF, and Patapoutian A. (2007). Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature 445, 541–545. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Meents JE, Ciotu CI, and Fischer MJM (2019). TRPA1: a molecular view. J Neurophysiol 121, 427–443. [DOI] [PubMed] [Google Scholar]
- Paulsen CE, Armache JP, Gao Y, Cheng Y, and Julius D. (2015). Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 525, 552. [DOI] [PubMed] [Google Scholar]
- Perszyk RE, Swanger SA, Shelley C, Khatri A, Fernandez-Cuervo G, Epplin MP, Zhang J, Le P, Bulow P, Garnier-Amblard E, et al. (2020). Biased modulators of NMDA receptors control channel opening and ion selectivity. Nat Chem Biol 16, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2020). UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese RM, Dourado M, Anderson K, Warming S, Stark KL, Balestrini A, Suto E, Lee W, Riol-Blanco L, Shields SD, et al. (2020). Behavioral characterization of a CRISPR-generated TRPA1 knockout rat in models of pain, itch, and asthma. Sci Rep 10, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, et al. (2017). A comprehensive map of molecular drug targets. Nat Rev Drug Discov 16, 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Startek JB, Boonen B, Lopez-Requena A, Talavera A, Alpizar YA, Ghosh D, Van Ranst N, Nilius B, Voets T, and Talavera K. (2019). Mouse TRPA1 function and membrane localization are modulated by direct interactions with cholesterol. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story GM, Peier AM, Reeve AJ, Eid SR, Mosbacher J, Hricik TR, Earley TJ, Hergarden AC, Andersson DA, Hwang SW, et al. (2003). ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 112, 819–829. [DOI] [PubMed] [Google Scholar]
- Suo Y, Wang Z, Zubcevic L, Hsu AL, He Q, Borgnia MJ, Ji RR, and Lee SY (2019). Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Mizuno Y, Kozai D, Yamamoto S, Kiyonaka S, Shibata T, Uchida K, and Mori Y. (2008). Molecular characterization of TRPA1 channel activation by cysteine-reactive inflammatory mediators. Channels 2, 287–298. [DOI] [PubMed] [Google Scholar]
- Talavera K, Startek JB, Alvarez-Collazo J, Boonen B, Alpizar YA, Sanchez A, Naert R, and Nilius B. (2020). Mammalian Transient Receptor Potential TRPA1 Channels: From Structure to Disease. Physiol Rev 100, 725–803. [DOI] [PubMed] [Google Scholar]
- Terwilliger TCL, S J; Read RJ; and Adams PD (2020). Improvement of cryo-EM maps by deseity modification. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton HT, Phan TX, Abramyan AM, Shi L, and Ahern GP (2017). Identification of a putative binding site critical for general anesthetic activation of TRPA1. Proc Natl Acad Sci U S A 114, 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisani M, Siemens J, Materazzi S, Bautista DM, Nassini R, Campi B, Imamachi N, Andre E, Patacchini R, Cottrell GS, et al. (2007). 4-Hydroxynonenal, an endogenous aldehyde, causes pain and neurogenic inflammation through activation of the irritant receptor TRPA1. Proc Natl Acad Sci U S A 104, 13519–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Corey RA, Hedger G, Aryal P, Grieben M, Nasrallah C, Baronina A, Pike ACW, Shi J, Carpenter EP, et al. (2020). Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2. Structure 28, 169–184 e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SR, Nelson AM, Batia L, Morita T, Estandian D, Owens DM, Lumpkin EA, and Bautista DM (2013). The ion channel TRPA1 is required for chronic itch. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 9283–9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisler JW, Rockman HA, and Lefkowitz RJ (2018). Biased G Protein-Coupled Receptor Signaling: Changing the Paradigm of Drug Discovery. Circulation 137, 2315–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Dubin AE, Bursulaya B, Viswanath V, Jegla TJ, and Patapoutian A. (2008). Identification of transmembrane domain 5 as a critical molecular determinant of menthol sensitivity in mammalian TRPA1 channels. J Neurosci 28, 9640–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Delling M, Jun JC, and Clapham DE (2006). Oregano, thyme and clove-derived flavors and skin sensitizers activate specific TRP channels. Nat Neurosci 9, 628–635. [DOI] [PubMed] [Google Scholar]
- Yang F, Xiao X, Cheng W, Yang W, Yu P, Song Z, Yarov-Yarovoy V, and Zheng J. (2015). Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat Chem Biol 11, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau WM, Wimley WC, Gawrisch K, and White SH (1998). The preference of tryptophan for membrane interfaces. Biochemistry 37, 14713–14718. [DOI] [PubMed] [Google Scholar]
- Yin Y, Le SC, Hsu AL, Borgnia MJ, Yang H, and Lee SY (2019). Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel. Science 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lin King JV, Paulsen CE, Cheng Y, and Julius D. (2020). Irritant-evoked activation and calcium modulation of the TRPA1 receptor. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L, Hsu AL, Borgnia MJ, and Lee SY (2019). Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure of GNE551-bound human TRPA1 is accessible with PDB code: 6X2J (atomic model) and EMDB code: EMD-22009 (map).