Abstract
Random encounters between proteins in crowded cells are by no means passive, but found to be under selective control. This control enables proteome solubility, helps to optimise the diffusive search for interaction partners, and allows for adaptation to environmental extremes. Interestingly, the residues that modulate the encounters act mesoscopically through protein surface hydrophobicity and net charge, meaning that their detailed signatures vary across organisms with different intracellular constraints. To examine such variations, we use in-cell NMR relaxation to compare the diffusive behaviour of bacterial and human proteins in both human and Escherichia coli cytosols. We find that proteins that ‘stick’ in E. coli are generally less restricted in mammalian cells. Furthermore, the rotational diffusion in the mammalian cytosol is less sensitive to surface-charge mutations. This implies that, in terms of protein motions, the mammalian cytosol is more forgiving to surface alterations than E. coli cells. The cellular differences seem not linked to the proteome properties per se, but rather to a 6-fold difference in protein concentrations. Our results outline a scenario in which the tolerant cytosol of mammalian cells, found in long-lived multicellular organisms, provides an enlarged evolutionary playground, where random protein-surface mutations are less deleterious than in short-generational bacteria.
Graphical abstract
Highlights
-
•
Random protein encounters and diffusibility in cells are controlled by surface charge.
-
•
Protein rotational diffusion is less restricted in human cells than in E. coli.
-
•
Human cells are less sensitive to alterations of protein charge than Escherichia coli cells.
1. Introduction
In the turbulent interior of live cells, soluble proteins constantly move and undergo countless encounters with their densely crowded surrounding. These diffusive “hand-shakes” with surrounding molecules are a prerequisite for life as they allow specific binding partners to be found and identified. The mechanism behind the ‘hand-shakes’ is the generic Brownian surface diffusion, allowing interaction partners to dynamically search one another's surfaces for a possible functional fit (Berg and von Hippel, 1985, Schreiber and Fersht, 1996, Camacho et al., 1999). However, this situation also comes with a trade-off: the life-times of the encounter complexes must be long enough to identify a putative binding partner, but short enough to avoid getting locally stuck by exaggerated close-range sampling of non-partners. Particularly so, when the partners are scarce and the search relies on covering large intracellular distances. Every formation of an encounter complex results in increased apparent size and slower rotational tumbling (Li et al., 2009, Nawrocki et al., 2017, Nawrocki et al., 2019), manifested as reduced rotational diffusion, Drot. The effect on rotational diffusion differs from the translational diffusion, Dt, in that spatial confinement per se does not have a direct impact on Drot: the cavities between larger structures and complexes enable relatively free rotation of small proteins, while sterically hindering translational movement, resulting in anomalous diffusion (Kozer et al., 2007). Nonetheless, all-atom MD simulations indicate that, taken over the whole ensemble, the crowded cytosol retards Dt and Drot to a similar extent, on timescales so short that anomalous nonlinear displacements are not yet detectable (Yu et al., 2016). Insight into the nature and biological optimisation of these diffusive events has recently been provided by NMR studies focusing on how Drot of various cytosolic proteins responds to surface mutations in live cells (Mu et al., 2017b, Ye et al., 2019, Wang et al., 2011, Cohen et al., 2015). Two results stand out: first, the diffusive protein interactions follow simplistic physicochemical rules, where the extent of in-cell retardation can be quantitatively predicted from surface net-charge density (Mu et al., 2017b, Ye et al., 2019), average surface hydrophobicity (Mu et al., 2017b, Majumder et al., 2015) and protein electric dipole moments (Mu et al., 2017). In particular, the repulsive negative charge stands out as a primary force in maintaining the cellular components soluble (Wennerstrom et al., 2020). Notably, a corresponding net charge effect on translational diffusion in bacterial cytoplasm was observed by fluorescence recovery after photo-bleaching (Schavemaker et al., 2017), qualitatively coupling Drot and Dt. Second, these mesoscopic properties are determined by the parts of the protein surfaces that are generally considered non-conserved since they show rapid divergence across organisms (Ma et al., 2003, Valdar and Thornton, 2001). Despite this, some degree of adaptation can be discerned through the variability of average physicochemical surface properties. A revealing example is given by proteins found in the cytosol of bacterial halophiles: while the structures of these proteins are generally conserved (Fedyukina et al., 2014), they possess an unusually high charge density (Mevarech et al., 2000).
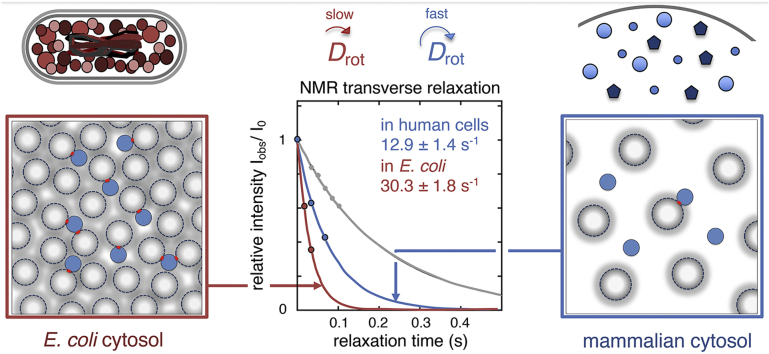
In accordance with this organism divergence, human proteins that are observed to tumble relatively unrestricted in the eukaryotic cytosol become arrested in Escherichia coli cells where they do not naturally belong (Mu et al., 2017). The human proteins can then be easily adjusted to tumble freely in the foreign E. coli environment by single point mutations, following the established relation between rotational diffusion and surface net-charge density (Mu et al., 2017). However, there is more to it: The difference between human and E. coli cells lies not only in their proteomes (Brocchieri and Karlin, 2005, Consortium, 2018) (Fig. 1), but also in their physiological status, macromolecular concentrations (Zimmerman and Trach, 1991, Milo, 2013, Theillet et al., 2014) and spatial compartmentalisation, all of which are expected to modulate the protein behaviour (Barbieri et al., 2015, Majumder et al., 2015, Danielsson et al., 2015, Ye et al., 2019) (Fig. 1). In E. coli, the maximum stability of SOD1 is significantly decreased despite being accompanied by an increase in thermal unfolding temperature, Tm (Danielsson et al., 2015).
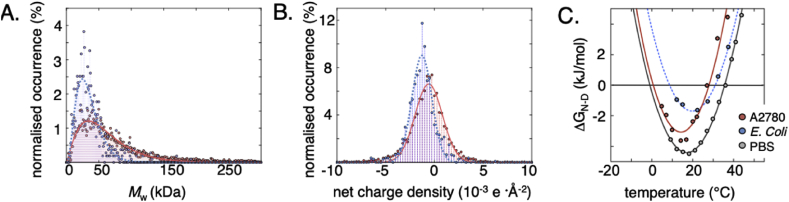
Fig. 1.
Properties of the E. coli and human cell cytosols, and their effect on protein stability. A. The cytosolic protein size distribution in E. coli (blue) and in human cells (red). The human proteome shows both a shift of the distribution peak towards larger protein size, as well as a significantly longer tail with larger proteins. Protein sequences of human (N = 5216) and E. coli cytosolic proteins (N = 1073) were collected from the Uniprot database (Consortium, 2018), in both cases the subset annotated as cytoplasmic was used for analysis. The distributions are fitted to a Γ-distribution. B. The surface net charge density of cytosolic proteins in E. coli (blue) and human cells (red). The charge density is somewhat higher in E. coli proteins, enabling stronger electrostatic repulsion. C. The temperature dependence of the folding free energy (ΔGN-D = −2.3RTlog [N]/[D], where [N] is the concentration of the folded and [D] of the unfolded state) of SOD1barrel shows the archetypical curvature of protein stability. SOD1barrel shows a significant destabilisation in A2780 cells (red) compared to in PBS-buffer (grey line). In the E. coli cytosol (blue line) the curve is shifted towards a higher melting temperature, while the maximum stability decreases significantly (Danielsson et al., 2015).
Efforts to further understand the differences in protein rotational diffusion between mammalian and bacterial cells have so far been challenged by difficulties in handling the intracellular protein concentration. In essence, any variation in the level of internalised or overexpressed target protein between experiments renders measurements of the NMR cross-peak heights alone insufficient for dynamic analysis. Such variations can be accounted for by normalisation to cell-lysate protein concentration, but with the intrinsic difficulty to reliably reproduce lysates and protein quantification therein (Barbieri et al., 2015, Mu et al., 2017b). In this study, we circumvent this problem by estimating Drot by NMR transverse relaxation, allowing in-cell analysis independent of protein concentration (Ye et al., 2019, Ye et al., 2018). The rate at which the NMR signal relaxes is dependent on the motional spectral density, which, in turn, is a function of the angular autocorrelation time, τc, of the randomly reoriented relaxation-inducing spin–spin interactions (Palmer 3rd, 1997). In protein NMR, τc is the harmonic mean of several correlations having various time scales, e.g. overall rotational correlation time, fast local correlation times and chemical exchange (Kay et al., 1989). For small globular proteins without dynamic disordered regions, the correlation time is completely dominated by the rotational correlation time, τr (Kay et al., 1989).
The results show that the human cells are overall less restrictive and more tolerant to surface-charge mutations than E. coli. Even so, the examined proteins show a similar relative change in apparent viscosity (ηapp), where = ()κ and κ = 1.96 ± 0.03 when transferred from mammalian to bacterial cells. The present findings are in excellent agreement with a recently published study where the net charge of a protein was modulated by a charged peptide tag (Ye et al., 2019). Together with (an extended analysis of) the accompanying changes of in-cell structural stability, the data indicate that the main difference between mammalian and E. coli cells lies not in the proteome properties per se, but more simply in the lower macromolecular concentration and collision frequency of the compartmentalised eukaryotic cytosol. The findings are discussed in the context of structural evolution, where the mammalian and bacterial cells seem to represent opposed trade-offs in functional optimisation, following their distinct fitness constraints.
2. Materials and methods
2.1. Protein production and purification
The model proteins are derived from the heavy metal binding protein TTHA1718, the human Atox1 homologue HAH1 and the human superoxide dismutase 1, SOD1. Design and mutagenesis of the model proteins TTHApwt, HAH1pwt and SOD1barrel have been extensively described in Mu et al. (Mu et al., 2017) and Danielsson et al. (Danielsson et al., 2011). The purification protocol (SI methods S1.2) described in Danielsson et al. (Danielsson et al., 2011) was used for SOD1barrel and HAH1pwt. The protocol described in Mu et al. (Mu et al., 2017) was used for TTHApwt.
2.2. In-cell NMR sample preparation and in-vitro experimental conditions
A2780 cell handling and electroporation protocol was mainly as in (Mu et al., 2017), the details are in (SI methods S1.3). In-vitro control NMR samples were in 10 mM MES pH 6.4 complemented by 150 mM NaCl and 10% D2O, unless otherwise stated.
2.3. NMR spectroscopy
All in-cell and lysate experiments (SI methods S1.4) were carried out on a Bruker Avance III 700 MHz spectrometer with a triple-resonance cryoprobe. All in-cell and in-vitro experiments (SI methods S1.4) were measured at 310 K. The data was processed using either the TopSpin (Bruker, Massachusetts, USA) or NMRPipe software (Delaglio et al., 1995). Signal intensities from one-dimensional relaxation data (SI methods S1.6) were fitted to a single exponential decay. The analysis was performed using in house Matlab (MathWorks, MA, USA) scripts. Signal intensities and relaxation rates from two-dimensional data (SI methods S1.6) were extracted using the CcpNmr software (Vranken et al., 2005).
2.4. NMR line width determination
Line widths were quantified by fitting a Gaussian function to a 1D slice in the 1H dimension of the in-cell and in-vitro control spectra for each well-resolved cross peak (SI methods S1.5). The fitting was performed using the line-shape tool in the NMRFAM-SPARKY software (Lee et al., 2015), and each fit was manually inspected and evaluated for fit quality.
3. Results
3.1. Setting the scene: the model proteins
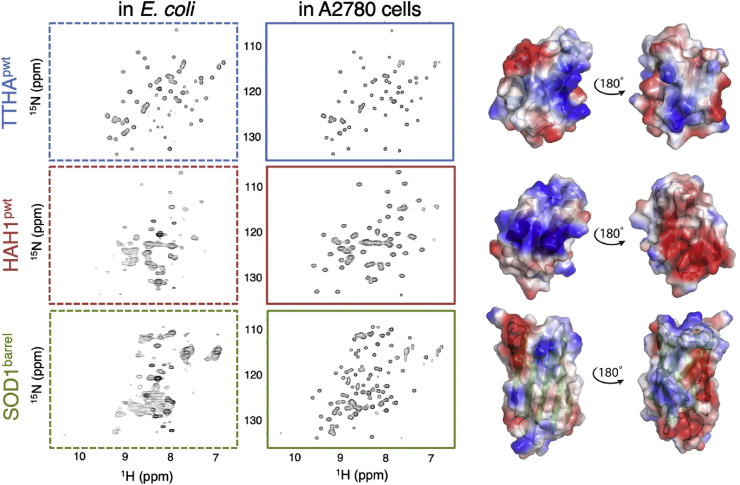
As probes for intracellular interactions, we use three well-characterized and similarly sized proteins. The putative heavy metal binding protein from Thermus Thermophilus, TTHA1718, the human antioxidant protein 1 homologue HAH1 (also known as Atox1), and the central barrel scaffold of human superoxide dismutase 1 (SOD1). All proteins were modified to be functionally inactive by removal of the metal coordinating cysteines in HAH1 and TTHA1718 and the long functional loops of SOD1 (SI methods S1.1, Table 1). This renders SOD1 monomeric and metal free, and abolishes the native interactions between HAH1 and its binding partners Wilson and Menkes proteins (Hamza et al., 1999). The inactive variants are hereafter referred to as: TTHApwt, HAH1pwt and SOD1barrel. All three proteins are thermodynamically stable and show no tendency to oligomerize or aggregate under the studied conditions (Mu et al., 2017). TTHApwt and HAH1pwt are structurally superimposable, while their surface properties differ. For example, both the charge distribution and the net charge density differs: HAH1pwt is net positive, while TTHApwt is net negative (Fig. 2). The three proteins differ also slightly in surface hydrophobicity, where TTHApwt is somewhat more hydrophilic than HAH1pwt and SOD1barrel. The distinct surface properties of the proteins render them suitable as probes of non-specific transient interactions in the crowded cell interior. By comparing NMR line broadening, we have previously demonstrated that the three proteins have distinct rotational motions in the E. coli cytosol (Mu et al., 2017), where TTHApwt tumbles more freely than HAH1pwt and SOD1barrel (Fig. 2). However, upon cell lysis and centrifugal removal of larger cellular components, all three proteins exhibit buffer-like HMQC spectra with similarly narrow cross peaks (SI controls S2.1, Fig. S1). The results suggest that the in-cell line broadening is not primarily a result of uniform steric crowding, which would have resulted in similar effects on especially the structurally very similar TTHApwt and HAH1pwt, but stems from transient interactions with the surrounding macromolecules that act differently on the three proteins. Consistently, the surface properties of TTHApwt, HAH1pwt and SOD1barrel differ in terms of net-charge density and hydrophobicity, accounting for their different response to the crowded E. coli interior (Mu et al., 2017) (Fig. 2, Table 1). Of particular interest is here that the prokaryotic TTHApwt tumbles relatively freely in the prokaryotic cytosol whereas the mammalian HAH1pwt and SOD1barrel become arrested, in line with that the proteins at some level may have adapted to the intracellular environment in which they have evolved (Mu et al., 2017) (Fig. 2). This raises the question how this situation changes when the evolutionary background is reversed, i.e. upon transfer of the very same proteins into eukaryotic cells? To investigate this, we opted again for in-cell NMR but now in a cultivated human cell line.
Table 1.
Collected physicochemical properties of the four probe proteins.
| protein | TTHApwt | HAH1pwt | SOD1barrel | HAH1K57E |
|---|---|---|---|---|
| sequence length | 66 | 68 | 110 | 68 |
| Mw (kDa)a | 7.0 | 7.4 | 11.0 | 7.4 |
| pI b | 5.4 | 7.8 | 6.2 | 5.6 |
| net charge c | −1.47 | 1.02 | −0.71 | −0.50 |
| net charge calc d | −1.47 | 0.94 | −0.70 | −1.08 |
| Rh (Å) e | 15.5 ± 0.2 | 15.7 ± 0.2 | 19.7 ± 0.2 | n.a. |
Molecular weight calculated from primary sequence.
pI calculated using propKa 3.0. (Li et al., 2005, Bas et al., 2008, Olsson et al., 2011).
Net charge determined from migration distance on native gel.
Net charge calculated using propKa 3.0.
Hydrodynamic radius from diffusion coefficients (Mu et al., 2017).
Fig. 2.
Improved in-cell NMR spectra in A2780 cells compared to E. coli. HMQC spectra of TTHApwt (blue frames), HAH1pwt (red frames) and SOD1barrel (green frames) in E. coli (dashed frames) and in live A2780 cells (solid frames). In E. coli, both HAH1pwt and SOD1barrel signals are severely broadened due to a high amount of interactions with the complex E. coli cytoplasm, i.e. the signals are barely visible in the HAH1pwt and SOD1barrel spectra even though the protein concentration is similar in all E. coli samples, as detected from the lysate signal intensity. In mammalian cells, all three proteins show significant improvement of the in-cell spectral properties, although still with varying line width. N.B. the contour levels differ in the E. coli dataset in order to visualise the broad low-intensity peaks of HAH1pwt and SOD1barrel. Right: the precited structures of TTHApwt (mutations templated on PDB id: 2ROE), HAH1pwt (mutations templated on PDB id: 1TL5) and the crystal structure of SOD1barrel (PDB id: 4BCZ) are shown. The calculated surface charge distribution is projected on the structure surfaces, with blue corresponding to positive charge density and red to negative, highlighting the differences in both charge distribution and charge clustering. Especially between the structural homologues TTHApwt and HAH1pwt severely broadened peaks. Even so, the line widths of the mammalian proteins narrow up in the cell lysates, showing that the Drot in the human cytosol is still reduced, albeit not as much as in E. coli (SI, Fig. S1). The results show thus that the mammalian cytosol affects the internalised proteins less than the E. coli cytosol, even to the extent that the protein-specific spectral characteristics are hard to distinguish at first sight.
3.2. The human cell cytosol is qualitatively very different from the E. coli cytosol
As a eukaryotic model, we use human ovary adenocarcinoma A2780 cells. This cell line has proven suitable for in-cell NMR experiments (Theillet et al., 2016, Danielsson et al., 2015), and is readily amenable to high-yield internalisation of isotope-labelled protein by electroporation (Danielsson et al., 2015, Theillet et al., 2016) (SI methods S1.3). In this study, we obtain intracellular concentrations of labelled protein between 20 and 40 μM, corresponding to 10–20 μM protein concentration in the detection volume (SI controls S2.2, Fig. S2), with negligible amount of leakage to the outside sample buffer (SI controls S2.1, Fig. S3). Although higher than typical endogenous protein concentrations, these low concentrations ensure that no self-interactions influence in-cell protein tumbling (Mu et al., 2017). It is immediately clear that all three proteins tumble relatively freely in the mammalian cytosol, indicated by only moderate line broadening (Fig. 2). The rotational diffusion in mammalian cells thus contrasts that in E. coli, where only the bacterial TTHApwt yields narrow cross peaks, while the spectra of HAH1pwt and SOD1barrel show severely broadened peaks (Fig. 2). Even so, the line widths of the mammalian proteins narrow up in the cell lysates, showing that the Drot in the human cytosol is still reduced, albeit not as much as in E. coli (Fig. 2, SI, Fig. S1). The results show thus that the mammalian cytosol affects the internalised proteins less than the E. coli cytosol, even to the extent that the protein-specific spectral characteristics are hard to distinguish at first sight (Fig. 2).
3.3. In-cell retardation inferred from NMR line-widths
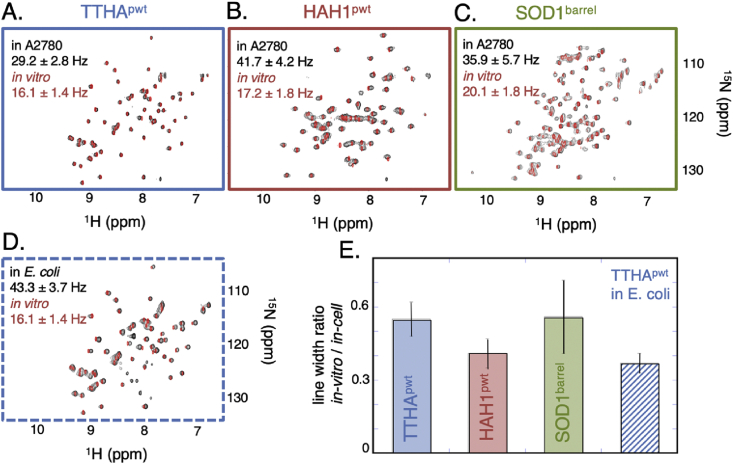
In-cell NMR line broadening reports not only on proteins’ global rotational diffusion (Drot), via τr, but also on local relaxation processes, such as chemical exchange broadening and/or changes in monomer conformational dynamics from local surface interactions with the crowded environment (Palmer 3rd, 1997). To deconvolute these contributions to the in-cell signal from A2780 cells, we determined first the line width (Δν1/2) of all well resolved cross peaks in the HMQC spectra, comprising 59–67% of the data (SI methods S1.5). The change of Δν1/2 upon cell internalisation thus gives a measure of the combined global and local changes induced by the cytosolic environment. The analysis shows that all three proteins undergo significant in-cell line broadening: Δν1/2 of TTHApwt increases on average from 16.1 ± 1.4 Hz (buffer) to 29.2 ± 2.8 Hz (cytosol), Δν1/2 of HAH1pwt from 17.2 ± 1.8 Hz (buffer) to 41.7 ± 4.2 (cytosol) Hz and Δν1/2 of SOD1barrel from 20.1 ± 1.8 Hz (buffer) to 35.9 ± 5.7 Hz (cytosol), where the ± -values represent one standard deviation in variability along the sequence (Fig. 3). On the whole, this variability is distributed relatively uniformly across the spectra of all three proteins, confirming that decreased Drot is the major contributor to the in-cell line broadening (SI, Fig. S4). Some notable exceptions are a few structurally clustered cross peaks of HAH1pwt and SOD1barrel that display small, but yet significant, deviations from the average in-cell broadening. In HAH1pwt, this clustered hotspot coincides with the edge strand β2, which is enriched in negative side-chain moieties that may play a role in locally modulating in-cell association (Otzen et al., 2000). In SOD1barrel, the hotspots comprise regions of loops 1, 3 and 7, which have previously been pin-pointed as sites for local structural motions (Danielsson et al., 2011, Danielsson et al., 2013a, Danielsson et al., 2013b) (SI, Fig. S4). Although these hotspots indicate that local dynamics to some minor extent influence the in-cell signals, their contributions in relation to the global Drot are negligible. In further support of this conclusion, the chemical-shift differences upon cell internalisation, which are expected to report on local structural adjustments, are overall small and mainly involve the dynamical regions of the proteins (SI controls S2.4, Figs. S5 and S6). The dominant influence of Drot on the line broadening therefore allows us to estimate the relative degree of in-cell retardation directly from the line-width ratios, i.e. (Fig. 3). From these ratios HAH1pwt stands out as the one that is most affected, while TTHApwt and SOD1barrel exhibit a smaller and similar degree of rotational retardation. Upon moving the analysis into E. coli, the line width of TTHApwt increases further from 29.2 ± 2.8 to 43.3 ± 3.7 Hz, while the cross-peaks of HAH1pwt and SOD1barrel are broadened beyond detection. The relative retardation of HAH1pwt in A2780 cells and TTHApwt in E. coli is 0.41 and 0.37 respectively, meaning that the protein that is most affected in mammalian cells (HAH1pwt) is still less affected than the least affected protein in bacteria (TTHApwt). Even so, the degree of rotational retardation in the mammalian cells remains coupled to protein surface properties: the structurally very similar TTHApwt and HAH1pwt show different rotational tumbling behaviour in mammalian cells, matching their divergent surface compositions (Fig. 2, Table 1).
Fig. 3.
First order quantification of the line broadening effect in A2780 cells. A-C. In-cell NMR spectra from A2780 cells in black overlaid by in-vitro spectra in dilute buffer in red, the frame colour code is as in Fig. 1. Inset in each spectrum is the average line width, determined from each well-separated cross peak. In D. the corresponding data for TTHApwt determined in E. coli is shown. E. The bars show the ratio between in-vitro and in-cell line widths.
3.4. Moving to in-cell NMR relaxation
Strictly speaking, the global rotational dynamics of a protein is characterised by the rotational correlation time (τr), describing how fast the rotational orientation of a molecule loses its correlation due to stochastic Brownian motions. Under ideal conditions, these diffusive motions are determined by protein size and the ‘drag’ of the solvent according to the Stoke–Einstein–Debye relationship,
| Eq. 1 |
where η is the effective viscosity, RH is the hydrodynamic radius, kB is Boltzmann's constant and T is the temperature. The rotational dynamics of a small globular protein, in conjunction with (in this case negligible) local dynamics and chemical exchange, is linked to the NMR line width (Δν1/2). The relation between τr and Δν1/2 is formally determined by the transverse relaxation (R2), where a large protein size and/or high solvent viscosity gives faster rates and broader lines. This is accompanied by slower longitudinal (R1) relaxation with consequently reduced sensitivity of the NMR experiment (Palmer 3rd, 1997, Kay et al., 1989). To obtain a more robust quantification of the intracellular motions, we opted for the direct determination of the transverse relaxation rate by in-cell NMR (SI methods S1.6).
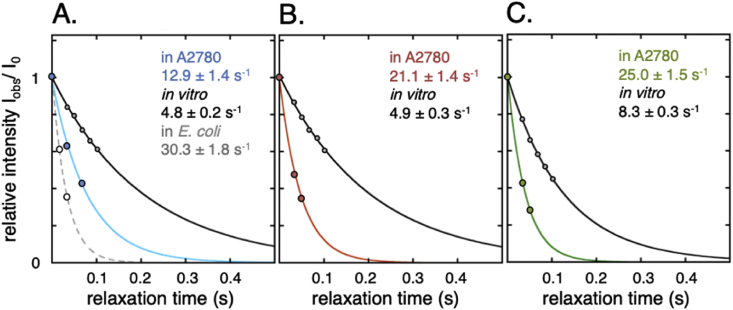
We focus here on 15N R2 as this is monotonically increasing with τr, i.e. an increase in R2 corresponds to an increase in τr for all relevant time regimes. The relative sensitivity of R2 to interactions and reduction in Drot also makes it especially suitable to low sensitivity in-cell NMR (Ye et al., 2018). Moreover, limited access to nutrients, oxygen and tight cell packing in the NMR tube results in a limited time frame for measurements on viable cells (Kubo et al., 2013, Inomata et al., 2017), typically less than 6 h before protein leakage starts to interfere with the in-cell signal (Danielsson et al., 2013a, Danielsson et al., 2013b, Theillet et al., 2016). Together with low signal-to-noise, this necessitates integration over the envelope of all signals to be able to measure 15N R2. In addition, we use interleaved data acquisition to reduce systematic, time-dependent errors from progressive cell packing in the NMR tube (SI controls S2.6, Fig. S7). For globular proteins, the determination of overall R2 from a series of 1D experiments yields the same information of τr as a 2D experiment, spare the sequential and structural variation details (SI controls S2.7, Fig. S8). Consistent with the line-width analysis, we find that R2 for all three proteins increases significantly upon cell internalisation (Fig. 4). Again, HAH1pwt stands out as the most affected protein in the mammalian cells with a more than 4-fold increase in R2, i.e. from 4.9 s−1 (buffer) to 21.1 s−1 (cytosol). The corresponding change for SOD1barrel is less than a factor of 3, even though the absolute in-cell relaxation of this protein is faster than for HAH1pwt due to its larger size. This illustrates nicely that the absolute values of relaxation rates cannot directly be translated into extent of in-cell rotational retardation if proteins of different sizes are to be compared (c.f. section 3.5). Finally, we measured R2 for TTHApwt in E. coli and find an additional increase in the relaxation rate, i.e. R2 changes from 12.9 s−1 (A2780 cytosol) to 30.3 s−1 (E. coli), indicating an even higher level of rotational restriction than observed in the mammalian cells (Fig. 4). Taken together, this is in good agreement with the direct measure of line broadening above, we find distinct relaxation differences in response to the cellular environment.
Fig. 4.
NMR relaxation data confirms the line broadening analysis. In-cell R2 relaxation rates in blue for TTHApwt (A.), red for HAH1pwt (B.) and in green SOD1barrel (C.), compared to the relaxation rates in dilute buffer (black solid lines). For all three proteins a significant increase in relaxation rate is observed, corresponding to a retardation of rotational diffusion. For TTHApwt, the R2 rate was determined also in E. coli, showing an even more pronounced retardation (grey dashed line in A.).
3.5. Accounting for protein size and shape
Even if the rotational-correlation times can be derived from R2 relaxation rates, the τr values themselves have an intrinsic shortcoming in more complex analyses. Assuming that two different proteins are tuned to identical surface properties to display identical in-cell interactions, these proteins will still yield different rotational-correlation times if they differ in shape and size (Ye et al., 2018, Guseman et al., 2018a, Guseman et al., 2018b). The reason is simply that a rod generally has a slower isotropic tumbling than a sphere, and a large sphere tumbles slower than a smaller one. To isolate the rotational retardation resulting from the diffusive in-cell interactions, we need therefore a measure that is independent of protein size and shape. One such measure is the apparent viscosity (ηapp), defined as the micro-viscosity in a pure solution that gives the same τr as observed in a more complex environment. In complex environments like the crowded and spatially heterogenous cytosol, the effective viscosity experienced by an immersed protein is the sum of the intrinsic microscopic solvent viscosity and all the restrictive interactions with the macromolecular environment (Yu et al., 2016). The apparent viscosity for molecule i is then given by:
| Eq. 2 |
where ηint is the intrinsic microscopic viscosity and the sum is over all interactions during the observation time, and where is the relative retardation of molecule i upon interaction with molecule j.
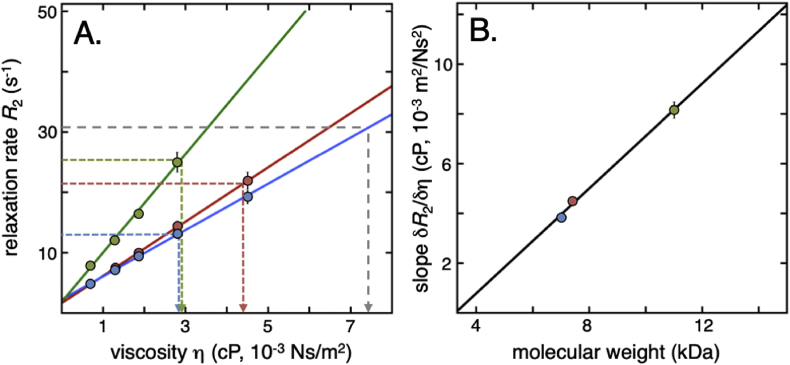
Since the coupling between ηint and the rotational-correlation time depends on size and shape (Ye et al., 2018, Guseman et al., 2018a, Guseman et al., 2018b), a reference curve must be determined for each studied protein. In this study, we use water/glycerol mixtures for benchmarking the viscosity dependence on R2, as the microscopic viscosity of these mixtures is well characterised (Cheng, 2008). We find that in the region 0–50% glycerol, the R2 relaxation rates increase linearly with viscosity for all three proteins (Fig. 5). Even so, the magnitude of the response in relaxation to increased viscosity (δR2/δη) differs between the proteins, accounting for the difference in size and shape (Fig. 5). From these reference curves, any in-cell relaxation rate can be translated to apparent viscosity (ηapp) to single out the contributions from the diffusive intracellular crosstalk.
Fig. 5.
Standard curves for determination of apparent viscosity. A. Reference curves for how the average R2 rates of the three probe proteins (shown in Fig. 4) change with increasing microscopic viscosity (corresponding to pure ηint in Eq. (2)). Colour coding is as in Fig. 1, and error bars are the translated error from signal to noise in signal intensity determination. The slope δR2/δη is directly linked to the size of the protein, as highlighted in the correlation plot in B.
3.6. Relation between ηapp and protein-net charge in mammalian cells
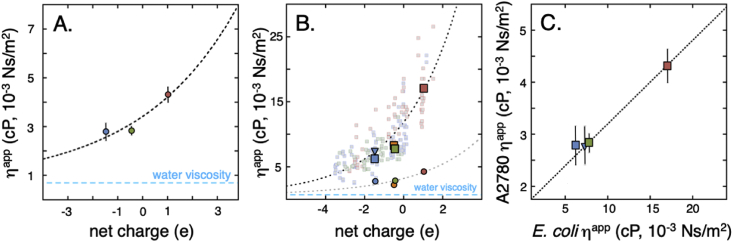
Resulting apparent viscosities for TTHApwt, HAH1pwt and SOD1barrel in A2780 cells are shown in Fig. 6. As expected, the apparent viscosity differs between the proteins, complying with their surface properties (Eq. (2)). The ηapp for TTHApwt and SOD1barrel are very similar at 2.7 ± 0.4 centiPoise (cP, 10−3 N/m2) and 2.8 ± 0.2 cP, respectively, whereas the less negatively charged HAH1pwt shows a larger retardation at 4.3 ± 0.3 cP, compared to 0.69 cP in pure buffer at 37 °C.
Fig. 6.
The apparent viscosity in A2780 human cells and in E. coli. A. The apparent viscosity, ηapp, shows a positive dependence on protein net charge, with increasing ηapp for positive protein surfaces. Colour codes are as in Fig. 1 and the dashed line is an exponential fit to the data points to guide the eye. The offset corresponds to water viscosity at infinite repulsion, while the exponentiality is adopted from the apparent exponential behaviour of the E. coli data. B. The same data as in the left panel compared to E. coli data (squares), N.B. the different axis scales. The large squares correspond to ηapp in E. coli for the three probe proteins, and the smaller faded squares to surface mutations, as determined from their in-cell NMR 1D-HMQC intensity. The blue triangle represents the apparent viscosity of TTHApwt in E. coli determined by the NMR relaxation rate R2, showing that the two methods are equivalent. Orange markers show the apparent viscosity for HAH1K57E. C. The observed ηapp in the two organisms correlate. The slope, however, is far from unity, indicating that the change in ηapp upon surface-charge mutation is 6 times stronger in E. coli.
Accordingly, ηapp shows a dependence on net charge (Fig. 6) where increased net-negative charge (SI methods S1.7) seems to speed up the in-cell tumbling by reducing the extent of arresting transient interactions. To further test this trend, we examined the effect of increasing the net-negative charge of HAH1pwt to that of SOD1barrel by the mutation HAH1K57E (SI methods S1.2, Table 1). The result shows that HAH1K57E falls almost precisely on top of SOD1barrel, accounting thus in a predictable manner for changes in charge, size and shape (Fig. 6). Plots of protein-charge dependence are not expected to be linear, as towards the limit of no in-cell interactions ηapp should approach the value of water. Conversely, increasing the interaction strength to the point where proteins fully stick will render ηapp excessively high. For simplicity, we model here this non-linear dependence between ηapp and protein net charge with a single-exponential function, including an offset to water viscosity (Fig. 6). This exponential-like property is also observed in the similar charge dependence found by 19F relaxation in U2OS cells (Ye et al., 2019). We also note that the effect on R2 in the dilute lysate controls differ slightly between the proteins (SI, Fig. S9), where the less interactive proteins TTHApwt and SOD1barrel show values similar to those in pure water. The higher R2 value of HAH1pwt, on the other hand, indicates that this protein partakes in transient interactions with residual cellular components in the dilute lysate (SI, Fig. S9). This raises once again the question to what extent these results compare with the behaviour in E. coli?
3.7. Comparison between the mammalian cytosol and E. coli
In the extensive bacterial dataset in Mu et al. (Mu et al., 2017), the in-cell retardation of molecular rotational tumbling (mobilityin-cell) was determined by 1D NMR, i.e. the signal intensity changes between the bacterial cytosol and dilute lysate conditions, keeping the protein concentration constant. To convert mobilityin-cell to ηapp values, we once again employ reference curves that relate the relative 1D intensity changes to a particular viscosity (SI methods S1.8, Fig. S10). The conversion shows that the apparent viscosity in E. coli is significantly higher than in mammalian cells: TTHApwt, HAH1pwt and SOD1barrel show rotational tumbling rates in E. coli that correspond to apparent viscosities of 6.8, 17.1 and 7.8 cP, respectively (Fig. 6), compared to 2.7, 4.3, and 2.8 cP in A2780 cells. This cross-species difference complies, in turn, with a simple power law independent of protein identity, where = ()κ and κ = 1.96 ± 0.03. From the full bacterial dataset in Mu et al. (2017), with >130 mutations specifically targeting surface properties of the three probe proteins, it is also clear that the dependence of ηapp on protein-net charge is significantly stronger in E. coli than in mammalian cells (Fig. 6). To confirm that the two datasets are indeed comparable, we used the R2 relaxation rate of TTHApwt in E. coli (Fig. 4) as an alternative measure on the ηapp. We find that the relaxation measurements yield a value of ηapp = 7.3 ± 0.5 cP, which matches the values of ηapp = 6.2 ± 0.3 cP obtained from line-broadening. Although not identical, the agreement between the values is reasonable considering that the R2 data reports on 15N relaxation as opposed to the 1H relaxation in the cross-peak line-broadening. The similar effect on nitrogen and proton relaxation also supports the interpretation that the rotational tumbling is the dominant contributor to the line width changes (Δν1/2) in these experiments, while exchange broadening and local dynamics play a minor role. The low effective protein concentration together with the limited sample lifetime makes a complete quantification of the exchange contribution to relaxation impossible. However, to further estimate the exchange contribution at μs - ms timescale, we measured 15N R2 relaxation rates without the refocusing CPMG pulse train (Loria et al., 1999) during the relaxation delay. The result shows that the obtained R2 values are similar with and without exchange suppression, emphasising that the contribution from exchange broadening is indeed small (SI controls S2.8, Fig. S11). Taken together, the data shows thus consistently that the mammalian cytosol comprises an environment that causes much less retardation on Drot than the bacterial cytosol. Moreover, the offset in ηapp between the organisms in Fig. 6B is not constant, but includes a real difference in the charge dependence: the correlation plot of the ηapp-values from the two organisms yields a slope of ≈ 6 (Fig. 6). Under the reasonable assumption that net charge is the main modulator of Drot in the cytosol (Mu et al., 2017b, Ye et al., 2019, Schavemaker et al., 2017), this suggests 6 times stronger surface net charge dependence on rotational motional freedom in E. coli.
3.8. First step towards a molecular picture
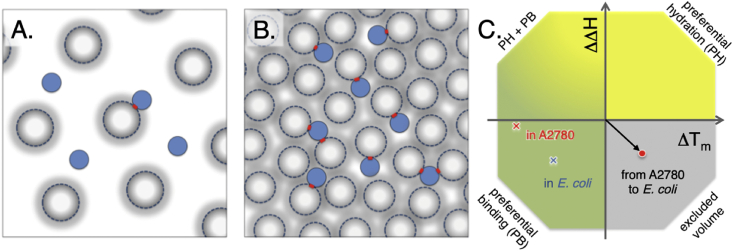
The question is now how the increased relaxation rates relate to the transient in-cell interactions, i.e. is it possible to derive a representative snapshot of the crowded intracellular environment? A reductionist way to approach this problem goes as follows: under fast-exchange conditions (on the NMR time scale), the observed transverse-relaxation rate is the population-weighted average of R2-values of the free probe protein and the ensemble average of the bound states. If we know the protein-mass distributions in mammalian and bacterial cells (Fig. 1, SI methods S1.9) (Brocchieri and Karlin, 2005, Consortium, 2018) and the transverse relaxation rate varies linearly with the particle mass (Farrow et al., 1995, Rossi et al., 2010), we can estimate the average fraction of probe proteins bound to other cellular proteins at any given time (SI methods S1.10). This comes with a set of assumptions. Even if proteins constitute the largest portion of the dry weight of the cytosol (Yu et al., 2016), the cytosol also comprises other interaction partners than proteins, e.g RNA, ribosomes, larger complexes, metabolites and organic molecules, all of which are involved in diffusive interactions with the probe proteins (Schavemaker et al., 2017, Majumder et al., 2015). First, including these additional macromolecules is not expected to significantly alter the size distribution. Second, they show a net negative surface charge distribution (Fedyukina et al., 2014) similar to that of proteins, implying that the model proteins will ‘feel’ them as part of the overall macromolecular distribution. Third, the estimate neglects any variation of abundance in the protein-size distributions, but this simplification is motivated by the observation that weighting for abundance have only moderate effects on the protein size and net-charge distributions (Geiger et al., 2012, Schmidt et al., 2016). Furthermore, as R2 varies approximately linearly with molecular mass, the relative populations of the three probe proteins relative to each other are not affected by a change in size distribution. Using this simplification, the population of bound TTHApwt in the relatively dilute human cells is estimated to pbound = 0.27, i.e. the majority of molecules are rotating freely at any given moment (Fig. 7). The corresponding numbers for HAH1pwt and SOD1barrel turn out slightly higher at pbound = 0.53 and pbound = 0.57, respectively. The populations are estimated under the assumption that, at any given time point, only one-to-one complexes are transiently formed. It is, however, reasonable to assume that larger complexes also are transiently formed (Nawrocki et al., 2017), but to a first order of approximation these larger transient complexes can be described by the same size distribution (Fig. 1). Notably, the apparent disagreement between these bound population values and the observation that HAH1pwt stands out as being more affected than SOD1barrel in terms of ηapp (Fig. 6), is explained by HAH1pwt being smaller. In essence, the relative mass change of HAH1pwt (Mw = 7.4 kDa) upon association with a human protein of effective size 34.5 kDa, from integration over the protein mass distribution (Fig. 1), is (34.5 + 7.4)/7.4 = 5.7 compared to 4.1 for SOD1barrel. This result is in good agreement with the observed difference in ηapp, even under assumption that mainly one-to-one complexes are formed. In contrast, in E. coli the relaxation rate of TTHApwt increases to such degree that even assigning full occupancy of binding to a cytosolic protein of effective mass 24.8 kDa (from the E. coli mass distribution in Fig. 1) is not enough (SI methods S1.9). To account for the observed in-cell retardation of rotational tumbling, we need to introduce an additional population of simultaneously bound cytosolic protein (Fig. 7). Our estimate of >1 contact at any given moment is further in good accord with a crowding corresponding to 3.5 near-neighbour partners for diffuse interactions, as previously observed in MD simulations of the bacterial interior (Yu et al., 2016, McGuffee and Elcock, 2010). Although this reductionist representation based on average parameters is bound to defy microscopic insight, it serves to show that our NMR results are fully consistent with these existing cellular data: the system behaves overall robustly and responds predictably to perturbation.
Fig. 7.
Snapshot representation of human and bacterial cytosol and thermodynamic classification of the effects from complex crowding solvents. A. A cartoon of the human cytosol with 60 mg/ml macromolecular concentration, here represented by ‘spherical’ proteins with the distribution of radii represented as soft edges. From relaxation data we can estimate that approximately 25% of the bacterial protein TTHApwt are involved in transient complexes at any given time point. B. The 6 times higher charge dependence on ηapp indicated 6 times higher macromolecular concentration in E. coli, and this results in that TTHApwt at any given time point is involved in a transient complex, and in more than 10% of the time it is involved in the formation of a transient trimer, with the slightly smaller proteins in the E. coli cytosol. C. The effects from altering the solvent can be classified from the effects on unfolding enthalpy (ΔH) and melting temperature (Tm), following the protocol by Ebbinghaus (Senske et al., 2014). The effects on SOD1barrel stability when comparing data from buffer to data from the A2780 cytosol is shown as a red ‘x’, and the corresponding effect when comparing to data from E. coli cytosol is marked as a blue ‘x’. Comparing human A2780 cell data to E. coli (red sphere) data results in a reduction in ΔH accompanied by an increase in Tm that can be classified as an excluded volume effect accompanied with increased transient binding, in line with an increased macromolecular concentration in the E. coli cytosol. The figure design is adapted from Senske et al. (Senske et al., 2014).
4. Discussion
4.1. Nature of diffusive in-cell interactions
When a protein interacts with another macromolecule, its rotational tumbling slows down. The reason for this retardation is primarily the increase in apparent mass experienced when free monomers become locked to the motion of larger complexes. Translated to the experiments performed here, this means that the observed retardation of Drot is the dynamic average of multiple transient mass increases, the size and duration of which are determined by the in-cell interactions. For a given distribution of macromolecules, the rotational tumbling of a particular protein will thus change if the particle–particle interactions are altered. In this study, we introduce such alterations by protein-surface changes to map out the physicochemical forces at play. To do this strictly, we first have to account for the differences of our reporter proteins TTHApwt, HAH1pwt and SOD1barrel in mass and shape (Fig. 5), as these differences alone will offset their intrinsic tumbling behaviour and obscure protein-to-protein comparison. In previous in-cell NMR studies, this ‘geometric’ problem has been circumvented by keeping the analysis to a single reporter protein (Barbieri et al., 2015, Majumder et al., 2015), or by accounting for size and shape differences by electrophoretic-mobility normalisation (Mu et al., 2017). Following the approach by Gierasch's (Wang et al., 2011) and Li's labs (Ye et al., 2013), we opt here for normalisation by converting data to apparent viscosity (ηapp). The principal advantage of this approach is that it not only accounts for differences in size and shape of the free monomers, but also for their different response to viscosity changes (Fig. 5). Conversion of data to ηapp reduces thus the initial size-related offsets between TTHApwt, HAH1pwt and SOD1barrel, and exposes a common dependence on net charge (Guseman et al., 2018a, Guseman et al., 2018b, Mu et al., 2017b, Majumder et al., 2015, Barbieri et al., 2015) (Fig. 6). It is also apparent that this net-charge dependence is different in mammalian and bacterial cells, where the latter stands out as overall more restrictive on the protein's rotational motions (Fig. 6). The conclusion from these results is that net-negative charge is the dominant modulator of diffusive in-cell interactions, in full accord with previous observations on other cellular systems (Mu et al., 2017b, Wang et al., 2011, Majumder et al., 2015). From a colloidal perspective, this suggests that the functional dispersion of the cellular components is, at a basal level, achieved by negative-charge repulsion, and in the case of cytosolic proteins this is maintained by the ‘non-conserved’ surface residues (Ma et al., 2003, Valdar and Thornton, 2001). In terms of describing the diffusive protein–protein interactions, however, there are clearly more factors at play. Most evidently, the considerable apparent random deviation from the fitted line in the E. coli data set (Fig. 6B) is partly accounted for by differences in the electric-dipole moment and surface hydrophobicity (Mu et al., 2017), both of which are well known to influence protein interactions at close range. Also, in the E. coli dataset there is a vast span of surface heterogeneity, explored in reference (Mu et al., 2017), that yields more specific flavours to the diffusive interplay. Acknowledging this complexity, the high–order interactions controlling the intracellular diffusion are often referred to as ‘quinary’ contacts (McConkey, 1982, Majumder et al., 2015, Mu et al., 2017b), following the seminal observation that certain proteins are diffusely attracted to one another, causing co-localization in the cytosol and co-elution during chromatographic separation (McConkey, 1982). Such biases of the diffusive interplay will naturally follow from the variation of net-charge across the human and E. coli proteins, where proteins on the negative side of the distribution will preferentially interact more strongly with the positive counterpart (Fig. 1). With respect to the surface detail, so called, ‘enhanced’ non-native interactions have been suggested to play a key role in directing proteins to their physiological targets (Luh et al., 2013, Cohen-Khait et al., 2017). The small folded protein Pin1 is an example where inactivation of the specific recognition site, using mutation, phosphorylation or competitive binding, increases the in-cell rotational motion also in absence of the binding target (Luh et al., 2013). In this study, we expect to distinguish such site-specific contributions from systematic offsets in the mesoscopic-charge dependence. As suitable candidate, we have the active-site truncated construct HAH1pwt where the removal of the copper coordinating cysteines abolishes specific binding. However, a positive charge cluster (R21, K25, K57) (Fig. 2) involved in the interaction interface with the native target MNK1 (Banci et al., 2009), and putatively involved in an enhanced non-native interaction, is left for mutational perturbation (PDB id: 2K1R, SI, Fig. S12). In E. coli, where the human MNK1 binding partner is naturally lacking, truncation of any of these positive charges yields still a larger than average gain in rotational motion, i.e. mobilityin-cell = 0.36 ± 0.03 compared to mobilityin-cell = 0.23 ± 0.05 for other positive-charge truncations (Mu et al., 2017). The pattern is even clearer with double mutations (SI controls S2.9, Fig. S12). Consistent with the protein Pin1 data, it is thus apparent that interfering with certain patches of surface charges yields larger effects on the rotational diffusion than the average charge replacement. With respect to the mutation HAH1K57E, this effect seems further enhanced in mammalian cells with the native MNK1 partner present: ηapp shows here 1.21 times the expected change, compared to 1.12 in E. coli, as estimated from the fitted average (Fig. 6). These deviations from the expected change can stem from the disruption of charge-patch patterns that by chance strengthen certain non- or semi-specific interactions, from cytosolic co-localization with specific subsets of proteins (Satori et al., 2013, Spitzer, 2011) and, in the case of mammalian cells, dynamic binding to the native MNK1 target. Although the validity of these interpretations remains to be tested, the very outline of the possibilities serves to emphasise that the targeted in-cell interactions are not simply rationalised as one-type-only. They rather represent a continuum stretching from ‘random encounters’ to ‘fully-specific binding’, all of which provide different levels of functional control under continuous optimisation of the amino-acid sequence.
4.2. The lower ηapp in mammalian cells complies with lower macromolecular concentration
Our most striking observation is that the apparent viscosity in the mammalian cytosol is considerably much lower than in the E. coli cells. The effect is consistent for all three proteins and complies phenomenologically with a power law = ()κ, where κ ≈ 2. Following our simplistic treatment of the in-cell interactions, the lower ηapp and weaker charge dependence in the mammalian cytosol can be due to (i) the surrounding molecules being more repulsive, decreasing the number of close encounters, (ii) the surrounding molecules being smaller leading to less rotational retardation upon association, or (iii) lower macromolecular concentration. The difference in repulsive net-charge density of the human and E. coli proteomes is obtained from sequence data (Consortium, 2018), indicating that the mammalian proteins carry a somewhat lower net-negative charge compared to their bacterial counterparts (−6.8·10−4 vs −12.3 eÅ−2, Fig. 1). As this is expected to increase rather than mitigate ηapp, we conclude that the protein–surface properties per se are not the cause of the difference. Similarly, the size of human proteins is overall larger than E. coli proteins, which, again, is inconsistent with the observed effect on ηapp (Consortium, 2018). This leaves us with the intracellular protein concentration as the most plausible factor to explain the data and, consistently, several reports show that the human cytosol is overall less crowded than E. coli cells (Theillet et al., 2014). That is, the lower protein concentration in mammalian cells decreases the collision frequency and, thereby, lowers the fraction of slowly tumbling encounter complexes at any given moment. To see if this scenario is reasonable, we estimated the difference in protein concentration between A2780 and E. coli cells from the relaxation data in Fig. 6 as follows. First, the collision frequency is proportional to the product of the concentrations of interacting molecules A and B: r = ξ [A][B], where ξ is a proportionality constant. The collision frequency per probe protein (the ensemble average observed in the NMR experiment is the same as the time average per molecule as Brownian motion is ergodic (Kallianpur and Robbins, 1953)) is then proportional to the concentration of surrounding molecules: r/[A] = ξ [B]. Second, the response of pAB to net-charge perturbation (ΔpB) is proportional to the observed values of Δηapp, based on the approximation that R2 is directly proportional to particle size in the dynamic range of these experiments (Fig. 5). Under the assumption that self-interaction of A in cells is negligible in the concentration range 0–1 mM, as previously shown to be the case for our probe proteins (24), the difference in [B] between mammalian and E. coli cells is given by [B]A2780/[B]E. coli = Δηapp, A2780/Δηapp, E. coli. Accordingly, we estimate the macromolecular concentration in A2780 cells to ~1/6 of that in E. coli, as judged from the slope of ηapp, A2780 vs. ηapp, E. coli (Fig. 6). Using 300–400 mg/ml as a benchmark for the protein concentration in E. coli (2), this yields 50–65 mg/ml for the mammalian cytosol. Despite the simplistic take, the result complies well with previous estimates of 50–200 mg/ml (35). Using finally the size distributions of the human and bacterial proteins to estimate the mass of the encounter complexes from relaxation data (SI methods S1.10), we arrive at a picture where our reporter proteins form on average 0.3 contacts with their neighbours in the mammalian cytosol, compared to > 1 in E. coli (Fig. 7). In line with this, one specific contributing factor can be assigned to diffuse transient interactions with ribosomes: recently proposed to be an important player in reduction of translational diffusion (Schavemaker et al., 2017). Interestingly, the ribosome concentration in E. coli is sometimes up-regulated to very high numbers, corresponding to 20% of the dry weight, compared to approximately 1% in human cells (Bremer and Dennis, 2008, Duncan and Hershey, 1983). This constitutes a minor but not negligible portion of the total interaction surface area. Transient interactions with the highly negative ribosomes (Knight et al., 2013) would be more pronounced in E. coli, and as such contribute to the stronger retardation in E. coli. This raises the question how far this mean-field representation of the protein–protein interactions will take us in terms of understanding the intracellular milieu in large. One example where this picture proves useful is in accounting for the in-vivo protein destabilization. Compared to dilute buffer conditions, the ALS-associated SOD1barrel conspicuously loses structural stability in mammalian cells. This structural destabilization is even further pronounced in E. coli. In essence, the unfolding temperature (Tm) is lowered in mammalian cells, while in E. coli it is in fact similar to that in buffer, despite its maximum stability showing a progressive decrease (Fig. 1). The comparison between mammalian and E. coli cells thus challenges the common belief that stability and Tm correlate and can be used interchangeably (Danielsson et al., 2015). Following the work by Ebbinghaus and co-workers (Senske et al., 2014), the contributions from transient interactions and excluded-volume effects when comparing different types of cells can be deconvoluted from the thermodynamic parameters of the protein-stability change itself. First, the comparison of thermodynamic stability of SOD1barrel in the A2780 cytosol as opposed to in-vitro buffer shows a decrease in Tm while ΔΔH is slightly negative (Danielsson et al., 2015) (Fig. 7). This is the signature of destabilisation due to preferential binding of solute components to the unfolded state, i.e. the folding equilibrium is shifted towards the unfolded state through mass action (Danielsson et al., 2015) (Fig. 7). In contrast, the Tm of SOD1barrel increases in E. coli accompanied by a decrease in ΔH when compared to data from A2780 (Fig. 7). This behaviour signals the involvement of two opposing terms, i.e. destabilisation from increased preferential binding to the unfolded state combined with stabilisation from increased excluded-volume effects.
Although the relative impact of binding and steric exclusion remains uncertain, a consistent mechanistic picture can be obtained from the mean-field results in Fig. 7. The indistinguishable contribution from steric exclusion when comparing dilute buffer conditions to the mammalian cytosol with a protein level of 60 mg/ml, indicates that transient binding at this point dominates the structural destabilisation (Senske et al., 2014, Danielsson et al., 2015). Upon increasing the protein concentration further to 350 mg/ml in E. coli, however, the stabilising contribution from steric crowding starts to become apparent, but remains superseded by the opposing term from increased transient binding. Accordingly, the observed cell-to-cell destabilisation does not need to be explained by different surface properties of the mammalian and E. coli proteomes, but are equally consistent with a situation where the cellular difference lays in a six-fold increase in protein concentration alone (Fig. 7).
4.3. Evolutionary implications of low cytosolic crowding
Here, the bacterial TTHApwt stands out as being the most freely tumbling protein in both mammalian and E. coli cells, possibly reflecting its natural adaptation to the densely crowded bacterial interior. Nevertheless, small changes of the protein's surface properties promote significant changes in diffusibility (Mu et al., 2017b, Majumder et al., 2015, Barbieri et al., 2015) (Fig. 6) and these changes manifest themselves most strongly in the bacterial cells. In other words, mutation of even a single surface group has pronounced impact on the intracellular interactions. Considering that many proteins are indeed found to evolve at the border of functional interactions and deleterious mis-interactions (Sikosek and Chan, 2014), such mutational sensitivity is clearly double edged. On the one hand, a random mutation can instantly promote beneficial new interactions; on the other hand, it can instantly compromise vital function. It is easy to envisage that this high-stake situation is advantageous for rapidly dividing and nutrient-limited bacteria, where the increased fitness of one cell typically out-weighs the loss of multiple others. For multicellular eukaryotes, however, the situation is quite different. The multicellular complexity and sexual reproduction of higher organisms require higher functional stability and cannot handle mutational drawbacks by cell sacrifice as freely as a bacterial colony. This raises the question to what extent does the eukaryote compartmentalisation promote such stability? Is it by simply mitigating the negative impact of random surface mutations? As indicated by our data, the separation of biomolecular processes and decreased cytosolic crowding does indeed render the system less sensitive to protein-surface perturbations. A corollary of this situation is that the mammalian cytosol allows for a larger chemical playground for the evolution of more complex surface functions. The decreased encounter frequencies from lower crowding not only bias the cytosol to specific interactions, but also promote a broader span of selectivity by moving the system further away from the glass transition, i.e. the point where the highly concentrated E. coli proteome undergoes a transition from a well-dispersed soluble state to a semi-solid unspecific collapsed state, rendering the bacterial cytosol rigid in terms of diffusion (Parry et al., 2014).
Although such optimisation at the same time loses the bacterial advantage of rapidly gaining new interactions, this may be an acceptable price to pay for increased evolutionary stability and more complex functional networks. Whatever the answer to these questions turns out to be, the present study shows that the diffusive quinary interactions in mammalian and E. coli cells are clearly distinct, and underlines that mechanistic and functional inferences between eukaryote and bacterial systems need to be done cautiously and with the different intracellular conditions in mind.
CRediT authorship contribution statement
Sarah Leeb: Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - review & editing. Therese Sörensen: Formal analysis, Validation, Investigation, Data curation, Writing - review & editing. Fan Yang: Investigation, Data curation, Writing - review & editing. Xin Mu: Investigation, Data curation. Mikael Oliveberg: Conceptualization, Validation, Formal analysis, Writing - review & editing, Supervision, Project administration, Funding acquisition. Jens Danielsson: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration.
Declaration of Competing Interest
We declare no conflict of interest.
Acknowledgements
The research was funded by the Knut and Alice Wallenberg Foundation (2017–0041), the Swedish Research Council (2017–01517) and the Magnus Bergvall Foundation (2017–02228).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2020.04.002.
Contributor Information
Therese Sörensen, Email: therese@dbb.su.se.
Mikael Oliveberg, Email: mikael@dbb.su.se.
Jens Danielsson, Email: jens.danielsson@dbb.su.se.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Berg O.G., von Hippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Calderone V., Della-Malva N., Felli I.C., Neri S., Pavelkova A., Rosato A. Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces. Biochem. J. 2009;422:37–42. doi: 10.1042/BJ20090422. [DOI] [PubMed] [Google Scholar]
- Barbieri L., Luchinat E., Banci L. Protein interaction patterns in different cellular environments are revealed by in-cell NMR. Sci. Rep. 2015;5:14456. doi: 10.1038/srep14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas D.C., Rogers D.M., Jensen J.H. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008;3 doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- Brocchieri L., Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33:3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C.J., Weng Z., Vajda S., DeLisi C. Free energy landscapes of encounter complexes in protein-protein association. Biophys. J. 1999;76:1166–1178. doi: 10.1016/S0006-3495(99)77281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.S. Formula for the viscosity of a Glycerol−Water mixture. Ind. Eng. Chem. Res. 2008;47:3285–3288. [Google Scholar]
- Cohen R.D., Guseman A.J., Pielak G.J. Intracellular pH modulates quinary structure. Protein Sci. 2015;24:1748–1755. doi: 10.1002/pro.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Khait R., Dym O., Hamer-Rogotner S., Schreiber G. Promiscuous protein binding as a function of protein stability. Structure. 2017;25:1867–1874 e1863. doi: 10.1016/j.str.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Consortium T.U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson J., Kurnik M., Lang L., Oliveberg M. Cutting off functional loops from homodimeric enzyme superoxide dismutase 1 (SOD1) leaves monomeric beta-barrels. J. Biol. Chem. 2011;286:33070–33083. doi: 10.1074/jbc.M111.251223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson J., Awad W., Saraboji K., Kurnik M., Lang L., Leinartaite L., Marklund S.L., Logan D.T., Oliveberg M. Global structural motions from the strain of a single hydrogen bond. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3829–3834. doi: 10.1073/pnas.1217306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson J., Inomata K., Murayama S., Tochio H., Lang L., Shirakawa M., Oliveberg M. Pruning the ALS-associated protein SOD1 for in-cell NMR. J. Am. Chem. Soc. 2013;135:10266–10269. doi: 10.1021/ja404425r. [DOI] [PubMed] [Google Scholar]
- Danielsson J., Mu X., Lang L., Wang H., Binolfi A., Theillet F.X., Bekei B., Logan D.T., Selenko P., Wennerstrom H. Thermodynamics of protein destabilization in live cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12402–12407. doi: 10.1073/pnas.1511308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Duncan R., Hershey J.W. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J. Biol. Chem. 1983;258:7228–7235. [PubMed] [Google Scholar]
- Farrow N.A., Zhang O., Szabo A., Torchia D.A., Kay L.E. spectral density function mapping using 15N relaxation data exclusively. J Biomol NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- Fedyukina D.V., Jennaro T.S., Cavagnero S. Charge segregation and low hydrophobicity are key features of ribosomal proteins from different organisms. J. Biol. Chem. 2014;289:6740–6750. doi: 10.1074/jbc.M113.507707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Wehner A., Schaab C., Cox J., Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. M111 014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman A.J., Perez Goncalves G.M., Speer S.L., Young G.B., Pielak G.J. Protein shape modulates crowding effects. Proc. Natl. Acad. Sci. U. S. A. 2018;115:10965–10970. doi: 10.1073/pnas.1810054115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman A.J., Speer S.L., Perez Goncalves G.M., Pielak G.J. Surface charge modulates protein-protein interactions in physiologically relevant environments. Biochemistry. 2018;57:1681–1684. doi: 10.1021/acs.biochem.8b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I., Schaefer M., Klomp L.W., Gitlin J.D. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K., Kamoshida H., Ikari M., Ito Y., Kigawa T. Impact of cellular health conditions on the protein folding state in mammalian cells. Chem. Commun. 2017;53:11245–11248. doi: 10.1039/c7cc06004a. [DOI] [PubMed] [Google Scholar]
- Kallianpur G., Robbins H. Ergodic property of the brownian motion process. Proc. Natl. Acad. Sci. U. S. A. 1953;39:525–533. doi: 10.1073/pnas.39.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L.E., Torchia D.A., Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Knight A.M., Culviner P.H., Kurt-Yilmaz N., Zou T., Ozkan S.B., Cavagnero S. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem. Biol. 2013;8:1195–1204. doi: 10.1021/cb400030n. [DOI] [PubMed] [Google Scholar]
- Kozer N., Kuttner Y.Y., Haran G., Schreiber G. Protein-protein association in polymer solutions: from dilute to semidilute to concentrated. Biophys. J. 2007;92:2139–2149. doi: 10.1529/biophysj.106.097717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo S., Nishida N., Udagawa Y., Takarada O., Ogino S., Shimada I. A gel-encapsulated bioreactor system for NMR studies of protein-protein interactions in living mammalian cells. Angew Chem. Int. Ed. Engl. 2013;52:1208–1211. doi: 10.1002/anie.201207243. [DOI] [PubMed] [Google Scholar]
- Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- Li C., Wang Y., Pielak G.J. Translational and rotational diffusion of a small globular protein under crowded conditions. J. Phys. Chem. B. 2009;113:13390–13392. doi: 10.1021/jp907744m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria J.P., Rance M., Palmer A.G., 3rd A relaxation-compensated Carr−Purcell−Meiboom−Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- Ma B., Elkayam T., Wolfson H., Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S., Xue J., DeMott C.M., Reverdatto S., Burz D.S., Shekhtman A. Probing protein quinary interactions by in-cell nuclear magnetic resonance spectroscopy. Biochemistry. 2015;54:2727–2738. doi: 10.1021/acs.biochem.5b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey E.H. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffee S.R., Elcock A.H. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevarech M., Frolow F., Gloss L.M. Halophilic enzymes: proteins with a grain of salt. Biophys. Chem. 2000;86:155–164. doi: 10.1016/s0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- Milo R. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays. 2013;35:1050–1055. doi: 10.1002/bies.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Choi S., Lang L., Mowray D., Dokholyan N.V., Danielsson J., Oliveberg M. Physicochemical code for quinary protein interactions in Escherichia coli. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:4556–4563. doi: 10.1073/pnas.1621227114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki G., Wang P.H., Yu I., Sugita Y., Feig M. Slow-down in diffusion in crowded protein solutions correlates with transient cluster formation. J. Phys. Chem. B. 2017;121:11072–11084. doi: 10.1021/acs.jpcb.7b08785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki G., Karaboga A., Sugita Y., Feig M. Effect of protein-protein interactions and solvent viscosity on the rotational diffusion of proteins in crowded environments. Phys. Chem. Chem. Phys. 2019;21:876–883. doi: 10.1039/c8cp06142d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M.H., Sondergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theor. Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Otzen D.E., Kristensen O., Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: a structural clue to amyloid assembly. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A.G., 3rd Probing molecular motion by NMR. Curr. Opin. Struct. Biol. 1997;7:732–737. doi: 10.1016/s0959-440x(97)80085-1. [DOI] [PubMed] [Google Scholar]
- Parry B.R., Surovtsev I.V., Cabeen M.T., O'Hern C.S., Dufresne E.R., Jacobs-Wagner C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P., Swapna G.V., Huang Y.J., Aramini J.M., Anklin C., Conover K., Hamilton K., Xiao R., Acton T.B., Ertekin A. A microscale protein NMR sample screening pipeline. J. Biomol. NMR. 2010;46:11–22. doi: 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satori C.P., Henderson M.M., Krautkramer E.A., Kostal V., Distefano M.D., Arriaga E.A. Bioanalysis of eukaryotic organelles. Chem. Rev. 2013;113:2733–2811. doi: 10.1021/cr300354g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schavemaker P.E., Smigiel W.M., Poolman B. Ribosome surface properties may impose limits on the nature of the cytoplasmic proteome. Elife. 2017;6 doi: 10.7554/eLife.30084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Kochanowski K., Vedelaar S., Ahrne E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., Heinemann M. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Fersht A.R. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- Senske M., Tork L., Born B., Havenith M., Herrmann C., Ebbinghaus S. Protein stabilization by macromolecular crowding through enthalpy rather than entropy. J. Am. Chem. Soc. 2014;136:9036–9041. doi: 10.1021/ja503205y. [DOI] [PubMed] [Google Scholar]
- Sikosek T., Chan H.S. Biophysics of protein evolution and evolutionary protein biophysics. J. R. Soc. Interface. 2014;11:20140419. doi: 10.1098/rsif.2014.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer J. From water and ions to crowded biomacromolecules: in vivo structuring of a prokaryotic cell. Microbiol. Mol. Biol. Rev. 2011;75:491–506. doi: 10.1128/MMBR.00010-11. second page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet F.X., Binolfi A., Frembgen-Kesner T., Hingorani K., Sarkar M., Kyne C., Li C., Crowley P.B., Gierasch L., Pielak G.J. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs) Chem. Rev. 2014;114:6661–6714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet F.X., Binolfi A., Bekei B., Martorana A., Rose H.M., Stuiver M., Verzini S., Lorenz D., van Rossum M., Goldfarb D. Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature. 2016;530:45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- Valdar W.S., Thornton J.M. Protein-protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001;42:108–124. [PubMed] [Google Scholar]
- Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhuravleva A., Gierasch L.M. Exploring weak, transient protein--protein interactions in crowded in vivo environments by in-cell nuclear magnetic resonance spectroscopy. Biochemistry. 2011;50:9225–9236. doi: 10.1021/bi201287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerstrom H. Colloidal stability of the living cell. Proc. Natl. Acad. Sci. U. S. A. 2020 doi: 10.1073/pnas.1914599117. www.pnas.org/cgi/doi/10.1073/pnas.1914599117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Liu X., Zhang Z., Wu Q., Jiang B., Jiang L., Zhang X., Liu M., Pielak G.J., Li C. (19) F NMR spectroscopy as a probe of cytoplasmic viscosity and weak protein interactions in living cells. Chemistry. 2013;19:12705–12710. doi: 10.1002/chem.201301657. [DOI] [PubMed] [Google Scholar]
- Ye Y., Wu Q., Zheng W., Jiang B., Pielak G.J., Liu M., Li C. Quantification of size effect on protein rotational mobility in cells by (19)F NMR spectroscopy. Anal. Bioanal. Chem. 2018;410:869–874. doi: 10.1007/s00216-017-0745-4. [DOI] [PubMed] [Google Scholar]
- Ye Y., Wu Q., Zheng W., Jiang B., Pielak G.J., Liu M., Li C. Positively charged tags impede protein mobility in cells as quantified by (19)F NMR. J. Phys. Chem. B. 2019;123:4527–4533. doi: 10.1021/acs.jpcb.9b02162. [DOI] [PubMed] [Google Scholar]
- Yu I., Mori T., Ando T., Harada R., Jung J., Sugita Y., Feig M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. Elife. 2016;5 doi: 10.7554/eLife.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S.B., Trach S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- Luh L.M., Hansel R., Lohr F., Kirchner D.K., Krauskopf K., Pitzius S., Schafer B., Tufar P., Corbeski I., Guntert P. Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition. J. Am. Chem. Soc. 2013;135:13796–13803. doi: 10.1021/ja405244v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.