Abstract
Glycogen and starch are the major carbon and energy reserve polysaccharides in nature, providing living organisms with a survival advantage. The evolution of the enzymatic machinery responsible for the biosynthesis and degradation of such polysaccharides, led the development of mechanisms to control the assembly and disassembly rate, to store and recover glucose according to cell energy demands. The tetrameric enzyme ADP-glucose pyrophosphorylase (AGPase) catalyzes and regulates the initial step in the biosynthesis of both α-polyglucans. AGPase displays cooperativity and allosteric regulation by sensing metabolites from the cell energy flux. The understanding of the allosteric signal transduction mechanisms in AGPase arises as a long-standing challenge. In this work, we disclose the cryoEM structures of the paradigmatic homotetrameric AGPase from Escherichia coli (EcAGPase), in complex with either positive or negative physiological allosteric regulators, fructose-1,6-bisphosphate (FBP) and AMP respectively, both at 3.0 Å resolution. Strikingly, the structures reveal that FBP binds deeply into the allosteric cleft and overlaps the AMP site. As a consequence, FBP promotes a concerted conformational switch of a regulatory loop, RL2, from a “locked” to a “free” state, modulating ATP binding and activating the enzyme. This notion is strongly supported by our complementary biophysical and bioinformatics evidence, and a careful analysis of vast enzyme kinetics data on single-point mutants of EcAGPase. The cryoEM structures uncover the residue interaction networks (RIN) between the allosteric and the catalytic components of the enzyme, providing unique details on how the signaling information is transmitted across the tetramer, from which cooperativity emerges. Altogether, the conformational states visualized by cryoEM reveal the regulatory mechanism of EcAGPase, laying the foundations to understand the allosteric control of bacterial glycogen biosynthesis at the molecular level of detail.
Keywords: Glycogen biosynthesis, Glycogen regulation, Nucleotide sugar biosynthesis, Enzyme allosterism
Abbreviations: AGPase, ADP-glucose pyrophosphorylase; EcAGPase, AGPase from E. coli; GS, glycogen synthase; GBE, glycogen branching enzyme; GP, glycogen phosphorylase; GDE, glycogen debranching enzyme; ATP, adenosine 5′-triphosphate; G1P, α-d-glucose-1-phosphate; PPi, pyrophosphate; GTA-like, glycosyltransferase-A like domain; LβH, left-handed β-helix domain; FBP, fructose 1,6-bisphosphate; AMP, adenosine 5′-monophosphate; SM, sensory motif; RIN, residue interaction network
Graphical abstract
Highlights
-
•
CryoEM structures of AGPase from E. coli were solved in complex with FBP and AMP.
-
•
FBP binds deeply into the allosteric cleft and overlaps the AMP site.
-
•
Allosteric regulators promote a conformational switch of the RL2 loop.
-
•
FBP modulates ATP binding and activates the enzyme.
-
•
A mechanism of allosteric regulation for bacterial AGPases is proposed.
1. Introduction
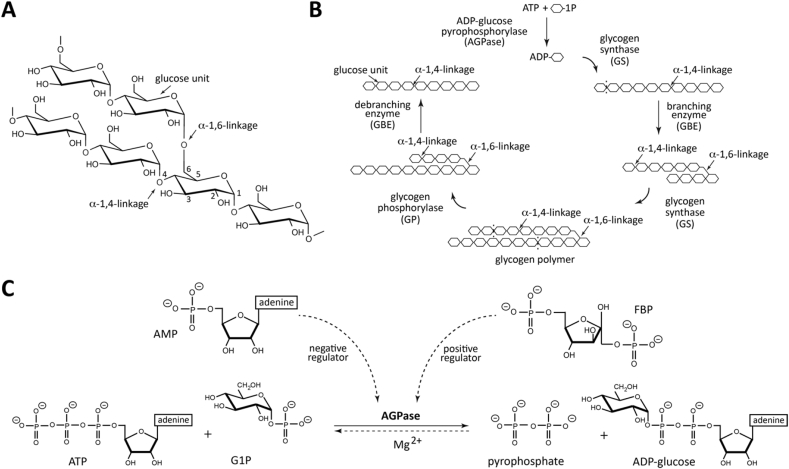
Glucose is a central carbon and energy currency in nature. The evolution led organisms to acquire specific pathways to store glucose in the form of α-glucan polysaccharides. The animal and bacterial glycogen, and the plant starch are the classical functional glucose storages in the cell (Ball & Morell, 2003). In addition, α-glucans were found as essential structural components of the bacterial capsule and biofilms playing an important role in pathogenesis (Bonafonte et al., 2000, Koliwer-Brandl et al., 2016, Kalscheuer et al., 2019). Glycogen and starch are large homopolymers composed by linear chains of α-(1 → 4)-glucose residues, containing α-(1 → 6)-linkages at branching points (Fig. 1A) (Ball and Morell, 2003, Cifuente et al., 2019). It is worth noting that the branch itself is composed of α-(1 → 4)-glucose residues. These chemical structures provide a high number of reactive-ends that facilitates rapid storage and recovery of glucose (Meléndez et al., 1999). Glucose activation is the first step required to overcome the energy barrier for subsequent polymerization (Fig. 1B). The classical pathway for bacterial glycogen biosynthesis involves the action of three enzymes: ADP-glucose pyrophosphorylase (AGPase), glycogen synthase (GS) and glycogen branching enzyme (GBE; Fig. 1B) (Cifuente et al., 2019, Espada, 1962, Recondo and Leloir, 1961, Trivelloni et al., 1962). AGPase catalyzes the biosynthesis of the activated-sugar ADP-glucose (Fig. 1B–C), whereas GS generates a linear α-(1 → 4)-linked glucose chain, and the GBE produces α-(1 → 6)-linked glucan branches in the polymer (Cifuente et al., 2019, Espada, 1962, Recondo and Leloir, 1961, Trivelloni et al., 1962). Glycogen degradation is carried out by glycogen phosphorylase (GP), which functions as a depolymerizing enzyme, and the glycogen debranching enzyme (GDE) that catalyzes the removal of α-(1 → 6)-linked ramifications (Fig. 1B) (Cifuente et al., 2019).
Fig. 1.
Classical bacterial glycogen structure, biosynthesis and regulation. (A) Glycogen is a very large-branched glucose homopolymer containing ~90% α-(1 → 4)-glucosidic linkages and 10% α-(1 → 6)-linkages. (B) The classical bacterial glycogen biosynthetic pathway involves the action of three enzymes: AGPase, GS; and GBE. Glycogen degradation is carried out by two enzymes: GP and GDE. (C) EcAGPase catalyzes the main regulatory step in bacterial glycogen. EcAGPase catalyzes the reaction between ATP and G1P in the presence of a divalent metal cation, Mg2+, to form ADP-Glc and PPi. ADP-Glc biosynthesis and hydrolysis directions are shown as a full and dotted lines, respectively. The two major positive and negative allosteric regulators, FBP and AMP, respectively, are shown.
Cell homeostasis requires precise and rapid mechanisms to sense the organism status to coordinate the metabolic network accordingly (Chubukov et al., 2014). To oversight the cell energy storage, glycogen and starch biosynthetic pathways are controlled by AGPase through cooperative and allosteric strategies (Perutz, 1989). The cooperativity emerges when the interaction of one molecule modulates the binding of another molecule of the same nature in other protomers, while allosterism arises when the association of a molecule modulates the binding of a different type of molecule to the oligomeric architecture of the enzyme (Perutz, 1989, Monod et al., 1963, Monod, 1971, Changeux, 2012). AGPase catalyzes the reversible condensation reaction between adenosine 5′-triphosphate (ATP) and glucose 1-phosphate (G1P) to produce ADP-glucose and pyrophosphate (PPi; Fig. 1C) (Cifuente et al., 2019, Preiss, 1978, Cifuente et al., 2016, Ballicora et al., 2003). Specifically, the oxygen on the phosphate group of G1P acts as a nucleophile attacking the α-PO4 group of the nucleoside triphosphate, leading to the liberation of PPi (Fig. S1) (Blankenfeldt et al., 2000). The reaction takes place in the presence of the divalent metal cation Mg2+, which minimizes the charge repulsion between phosphate groups, favoring nucleophile activation (Gentner and Preiss, 1968, Swift et al., 2012). Moreover, two positively charged residues polarize these groups, increasing the nucleophilic nature of the oxygen attacking the phosphor atom (Vithani et al., 2014, Führing et al., 2013). AGPase follows a sequential ordered bi–bi mechanism with ATP binding first, followed by G1P and by the ordered release of PPi and ADP-glucose (Paule & Preiss, 1971). The hydrolysis of PPi by inorganic pyrophosphatases results in a global irreversible and energetically expensive reaction in vivo (Kornberg, 1962, Lahti, 1983). Thus, evolution led AGPase to acquire an exquisite allosteric regulation mechanism to control its enzymatic activity by essential metabolites in the energetic flux within the cell (Cifuente et al., 2019, Preiss, 1978, Ballicora et al., 2003). AGPase activators are metabolites that reflect signals of high carbon and energy content of a particular bacteria or tissue, whereas inhibitors indicate low metabolic energy levels. As a consequence, AGPases have been classified into nine different classes taking into account the specific positive or negative allosteric regulators (Ballicora et al., 2003).
Bacterial AGPases are encoded by a single gene, giving rise to a native homotetrameric protein (α4) with a molecular mass of ca. 200 kDa (ca. 50 kDa for each protomer) (Cifuente et al., 2019, Ballicora et al., 2003). In contrast, plant AGPases are composed of two α and two β subunits, also referred to as ‘small’ and ‘large’ subunits, respectively, to form an α2β2 heterotetramer (Crevillén et al., 2003, Georgelis et al., 2007, Petreikov et al., 2010, Ventriglia et al., 2007). The two subunits have different functions; α is the catalytic subunit, whereas β is the regulatory subunit. To date, three crystal structures of AGPases have been reported, those of the bacterial homotetrameric AGPases from Escherichia coli (EcAGPase) (Cifuente et al., 2019, Cifuente et al., 2016, Comino et al., 2017) and Agrobacterium tumefaciens (AtAGPase) (Cupp-Vickery et al., 2008, Hill et al., 2015), and a recombinant homotetrameric version of the small subunit (α4) of the photosynthetic potato tuber AGPase (StAGPase) (Jin et al., 2005). The activity of the paradigmatic EcAGPase is enhanced by the high-energy glycolytic intermediate fructose-1,6-bisphosphate (FBP), whereas, the ubiquitous low-energy metabolite AMP inhibits the enzyme. Thus, the interplay of both regulators tune EcAGPase activity (Cifuente et al., 2019, Preiss, 1978, Gentner and Preiss, 1967). Moreover, EcAGPase shows positive cooperativity among ATP sites and a considerable S(ATP)0.5 (ATP concentration yielding half of the Vmax) reduction by FBP (Gentner and Preiss, 1967, Ghosh et al., 1992, Ballicora et al., 2007, Figueroa et al., 2011). We reported the first crystal structures of the paradigmatic EcAGPase in complex with FBP and AMP, respectively (Cifuente et al., 2016, Comino et al., 2017). These structures provided a first perspective of the regulatory sites and the identification of secondary structure elements likely participating into the allosteric regulation. However, the structural data prohibited the visualization of the architecture of EcAGPase in biologically relevant conformations and the comprehension on how the allosteric regulators act in a concerted manner to control the enzymatic activity. The structural framework that modulates these properties was historically provided by x-ray crystallography studies. However, the organization of the protein molecules inside the crystal packing prevents, in many cases, the visualization of native biologically relevant conformational states associated to the binding of substrates and regulatory molecules, impeding the construction of accurate models to explain allosteric regulation (Cifuente et al., 2016, Cupp-Vickery et al., 2008, Hill et al., 2015, Jin et al., 2005). In this work, we disclose the cryoEM structures of the homotetrameric EcAGPase (a ca. 200 kDa enzyme, with each protomer of 48.7 kDa), in complex with FBP and AMP, both at 3.0 Å resolution, respectively. In combination with biochemical, biophysical and bioinformatics data, the regulatory and cooperativity mechanisms of EcAGPase is unraveled, laying the foundations to understand the allosteric control of bacterial glycogen biosynthesis.
2. Results
2.1. The EcAGPase-FBP complex as visualized by cryoEM
To understand how the positive and negative signals are propagated across the EcAGPase structure in the native state in solution, we determined the single-particle cryoEM structures of EcAGPase in complex with either regulator, FBP or AMP (Fig. 2, Fig. 3, Fig. 4; Figs. S2 to S9; Table 1). EcAGPase is a homotetrameric enzyme that can be visualized as a dimer-of-dimers, with each protomer comprising an N-terminal glycosyltransferase A-like (GT-A-like) catalytic domain, and a C-terminal Left-Handed-β-helix (LβH) regulatory domain (Fig. 2, Fig. 3, Fig. 4; Fig. S8; Video S1: https://www.dropbox.com/s/fupmbncoxj9yo5k/vf_Video_S1.avi?dl=0) (Cifuente et al., 2016).
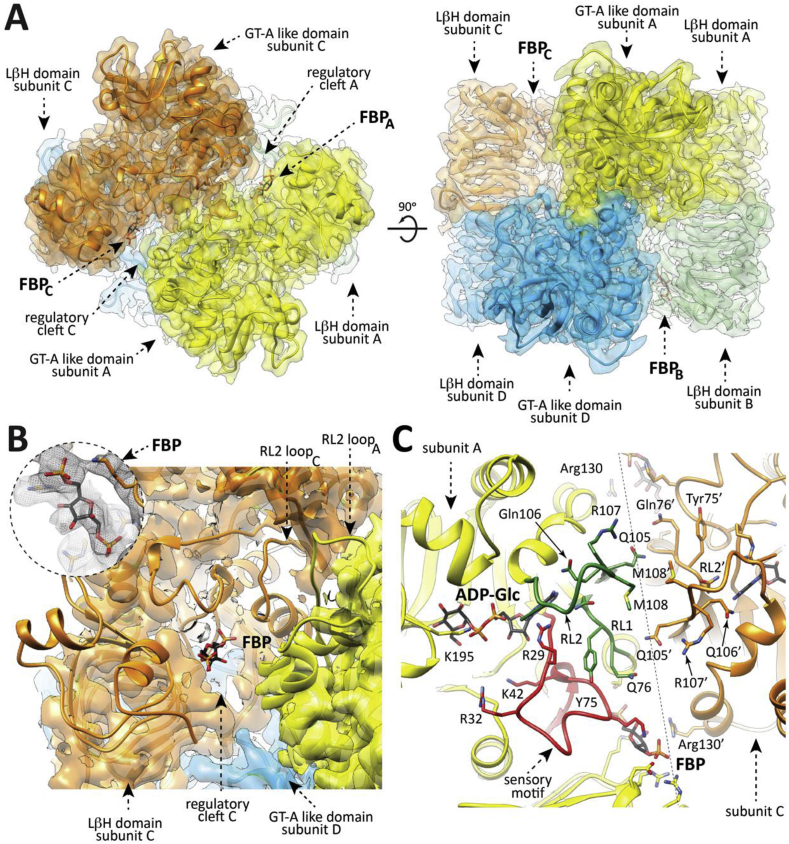
Fig. 2.
Overall structure of the EcAGPase-FBP complex as visualized by cryoEM. (A) The overall view of the electron density cryoEM map of the EcAGPase-FBPD2 complex, colored according to the four protomers (orange, yellow, blue and green) of the homotetramer. (B) A selected region corresponding to the neighborhood of the FBP binding site is masked to reveal the atomic model built into the electron density, with the protein shown as ribbons and the ligand FBP in sticks representation. Close-up of the EcAGPase-FBPD2 regulatory site showing the location and the electron density cryoEM map of the activator FBP. (C) Cartoon representation of the interface between the reference protomer (yellow) and the neighbor subunit (orange) of the EcAGPase-FBPD2 reconstruction. Note that the RL2 loop conformation as observed in the EcAGPase-FBPD2 reconstruction faces its own active site.
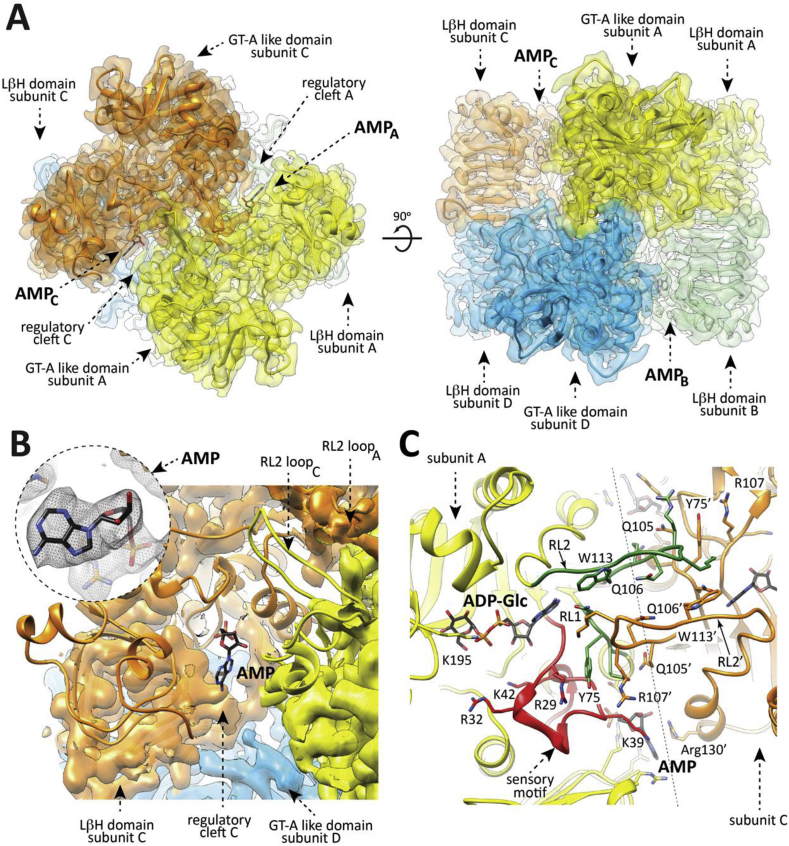
Fig. 3.
Overall structure of the EcAGPase-AMP complex as visualized by cryoEM. (A) The overall view of the electron density cryoEM map of the EcAGPase-AMPD2 complex, colored according to the four protomers (orange, yellow, blue and green) of the homotetramer. (B) A selected region corresponding to the neighborhood of the AMP binding site is masked to reveal the atomic model built into the electron density, with the protein shown as ribbons and the ligand AMP in sticks representation. Close-up of the EcAGPase-AMPD2 regulatory site showing the location and the electron density cryoEM map of the activator AMP. (C) Cartoon representation of the interface between the reference protomer (yellow) and the neighbor subunit (orange) of the EcAGPase-AMPD2 reconstruction. Note that the RL2 loop conformation, as observed in the EcAGPase-AMPD2 reconstruction, is oriented towards the neighbor active site.
Fig. 4.
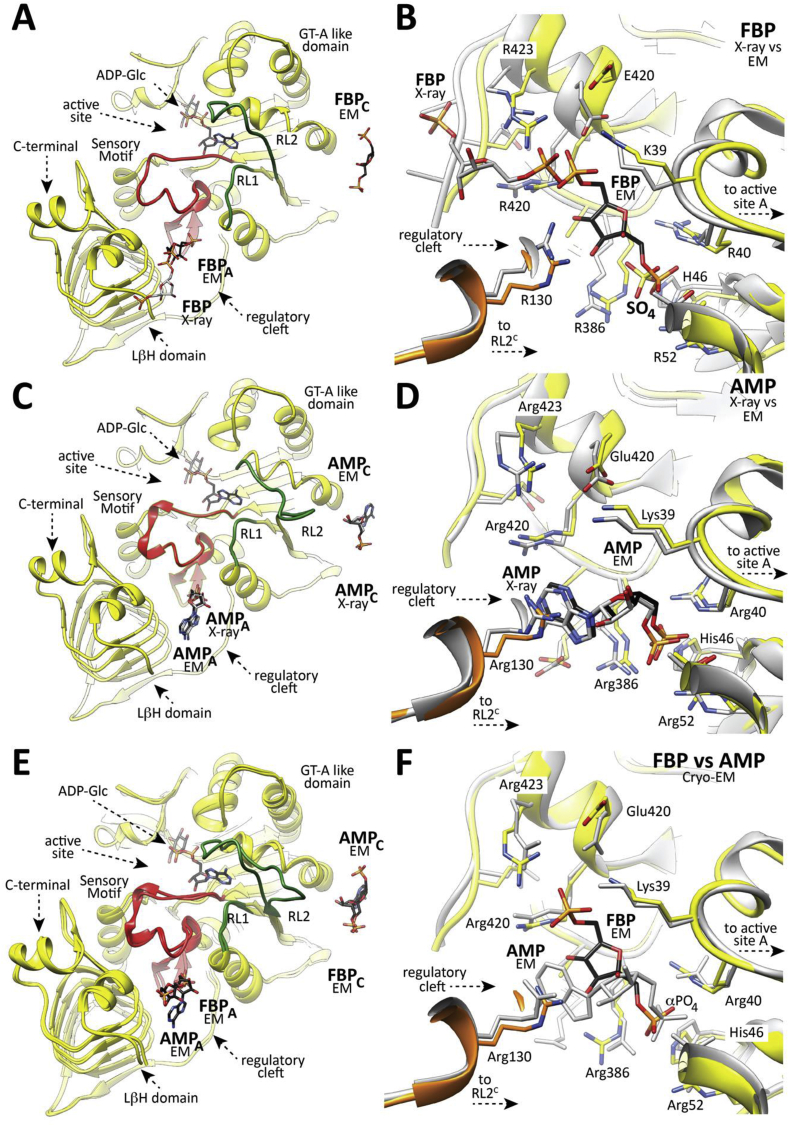
The location of FBP and AMP regulators into the allosteric site as observed by cryoEM. (A) Cartoon representation of one protomer of EcAGPase as observed in the EcAGPase-FBPD2 complex obtained by cryoEM. The secondary structure elements involved in the regulatory mechanism of EcAGPase are shown: (i) the SM (red) communicates the regulatory cleft and active sites of the same protomer, (ii) the RL1 (green) and (iii) RL2 loops (green). The FBP molecule is shown in black. The location of the FBP molecule as observed in the X-ray EcAGPase-FBP complex is shown in grey as a reference. (B) Closed view of the regulatory site as observed in the cryoEM EcAGPase-FBPD2 complex (yellow) and the EcAGPase-FBP complex (grey) obtained by X-ray crystallography. The FBP molecule is shown in black and grey, respectively. A molecule of ADP-Glc is shown in the active site as a reference. Note that the sulfate ion occupying the cleft cavity belongs to the structure of the EcAGPase-FBP complex solved by X-ray crystallography. (C) Cartoon representation of one protomer of EcAGPase as observed in the EcAGPase-AMPD2 complex obtained by cryoEM, and the location of AMP (black) into the regulatory site. The location of AMP in the EcAGPase-AMP complex solved by X-ray crystallography is depicted in grey. A molecule of ADP-Glc is shown in the active site as a reference. (D) Closed view of the regulatory site as observed in the EcAGPase-AMP complex obtained by cryoEM (EcAGPase-AMPD2; yellow) and X-ray crystallography (grey). The AMP molecule is shown in black. The location of the AMP molecule as observed in the X-ray EcAGPase-AMP complex is shown in grey as a reference. (E) Structural superposition of one protomer in the EcAGPase-FBPD2 and EcAGPase-AMPD2 complexes obtained by cryoEM. The FBP and AMP regulators are shown. A molecule of ADP-Glc is shown in the active site as a reference. (F) Closed view of the regulatory site as observed in the EcAGPase-FBPD2 (yellow) and EcAGPase-AMPD2 (grey) complexes.
Table 1.
CryoEM data collection, single-particle reconstruction maps, and model statistics.
| Data collection information | ||
|---|---|---|
| Data set | EcAGPase-FBP complex | EcAGPase-AMP complex |
| Nominal magnification | 47755x | 47755x |
| Voltage (kV) | 300 | 300 |
| Electron dose (e–/Å2) | 45.6 | 44.8 |
| Pixel size (Å) | 1.047 | 1.047 |
| Movie number | 2975 | 1681 |
| Defocus average and range (μm) | 1.4 (0.2–2.6) | 1.7 (0.3–5.0) |
| Frames | 40 | 40 |
|
Single-particle reconstruction information | ||||||
|---|---|---|---|---|---|---|
| Map | EcAGPase-FBP complex | EcAGPase-AMP complex | ||||
| Final number of particle images in map | 297275 | 94769 | ||||
| Symmetry imposed | D2 | C2 | C1 | D2 | C2 | C1 |
| EMDB code | EMD-4754 | EMD-10199 | EMD-10201 | EMD-4761 | EMD-10203 | EMD-10208 |
| Map resolution (Å) FSC=0.143 criterion |
3.05 | 3.16 | 3.24 | 2.95 | 3.09 | 3.26 |
| Map sharpening B factor (Å2) |
−148.1 |
−139.9 |
131.2 |
−116.8 |
−114.5 |
−99.9 |
| Model information | ||||||
| Model |
EcAGPase-FBP complex |
EcAGPase-AMP complex |
||||
| PDB ID | 6R8B | 6SHJ | 6SHN | 6R8U | 6SHQ | 6SI8 |
| Atoms (Non-H) | 26092 | 25974 | 25359 | 26760 | 26874 | 26590 |
| Protein residues | 1688 | 1664 | 1644 | 1700 | 1696 | 1700 |
| Ligands | 4 | 4 | 2 | 4 | 4 | 4 |
| Bonds (RMSD) | ||||||
| Length (Å) | 0.01 | 0.01 | 0.016 | 0.01 | 0.01 | 0.01 |
| Angles (°) | 1.34 | 1.01 | 1.34 | 1.35 | 1.38 | 1.40 |
| Mean B-factor | ||||||
| Protein | 153.64 | 163.46 | 94.72 | 134.96 | 136.07 | 88.79 |
| Ligand | 176.65 | 191.65 | 101.34 | 127.23 | 126.19 | 82.53 |
| MolProbity score | 2.11 | 2.02 | 1.89 | 2.30 | 2.04 | 1.79 |
| Clashscore | 7.93 | 3.54 | 2.21 | 7.17 | 5.36 | 2.14 |
| Rotamer outliers (%) | 3.12 | 3.58 | 4.81 | 3.32 | 2.28 | 2.45 |
| Ramachandran plot | ||||||
| Favored (%) | 95.71 | 92.90 | 94.47 | 91.43 | 92.06 | 91.31 |
| Allowed (%) | 4.05 | 6.98 | 5.41 | 8.57 | 7.94 | 8.33 |
| CC (volume) | 0.77 | 0.75 | 0.80 | 0.80 | 0.78 | 0.79 |
| Mean CC for ligands | 0.74 | 0.66 | 0.74 | 0.84 | 0.82 | 0.84 |
| CC (mask) | 0.77 | 0.75 | 0.81 | 0.80 | 0.79 | 0.79 |
Supplementary video related to this article can be found at https://doi.org/10.1016/j.crstbi.2020.04.005
The following is the supplementary data related to this article:
CryoEM single-particle analysis can profit from object symmetries for structural determination. In nature, object symmetry emerges when its configuration is preserved after following a combination of euclidean geometric transformations. As a consequence, diverse sets of operations define symmetry groups allowing the classification of structures. In the case of proteins, chirality disallows reflections. They belong to point groups only defined by a combination of rotation operations around intersecting axes. In the case of oligomeric proteins, multiple identical protomers may lead to different point groups labeled according to the Schönflies nomenclature in cyclic (Cn), dihedral (Dn), and platonic solids (T, O, and I) (Liboff, 2004). Specifically, Cn has an n-fold symmetry rotation axis (rotations of 360°/n). Dn has a main n-fold symmetry axis that intersects other n two-fold symmetry axis (rotation of 180°) contained in an orthogonal plane.
In cryoEM single-particle analysis of proteins, a suspected symmetry can be imposed in the reconstruction algorithm, benefiting from the redundancy of invariant parts that are averaged in the resulting density maps (Chiu et al., 2005). Nevertheless, due to flexibility, conformational changes, or ligand occupancy, symmetrical structures can undergo global or local functional breaking of symmetry (Blundell and Srinivasan, 1996, Goodsell and Olson, 2000). Some structural features can be hidden or blurred by the averaging (Chiu et al., 2005). Therefore, the analysis of all possible point groups may reveal details of the system of study not represented by only one symmetry. In that sense, the EcAGPase homotetramer architecture allows a maximum symmetry described by a D2. Here, all protomers are equivalent and can be superimposed to another, applying one 180° rotation around one of the three mutually orthogonal two-fold axes. However, the EcAGPase architecture may allow a C2 symmetry, conserving a single two-fold axis, resulting in two pairs of equivalent protomers. Finally, in the case of a complete lack of n-fold symmetry axes (C1), the homotetramer will not present equivalent protomers.
The three-dimensional reconstruction of the EcAGPase-FBP complex was carried out without imposing symmetry operators, C1 (EcAGPase-FBPC1; 3.24 Å resolution), enforcing C2 symmetry (EcAGPase-FBPC2; 3.16 Å resolution), and imposing D2 symmetry (EcAGPase-FBPD2; 3.05 Å resolution) (Figs. S2, S4 and S6 to S9). The similar resolution achieved by these reconstructions and the high correlation among them explain the highly symmetrical characteristic of this complex. The EcAGPase-FBPD2 reconstruction reveals a consensus density for FBP among the four regulatory clefts (Fig. S6). Strikingly, this binding mode shows a different location of the positive regulator compared to that observed in the EcAGPase-FBP complex obtained by X-ray crystallography (Fig. 2, Fig. 4B) (Cifuente et al., 2016, Bhayani et al., 2019). The FBP is found deeply buried into the cleft mainly defined by (i) the N-terminal β2-β3 hairpin (residues 46–52), α5, and the connecting loop α2-α3 (residues 37–42), which partially constitutes the so call ‘sensory motif’ (SM), communicating the regulatory and active sites of each subunit, and (ii) the C-terminal α15 (residues 419–425) and the connecting loops β28- β29 (residues 384–388) and β25-β26 (residues 367–371; Fig. 2A–C). The PO4 group at position 1 of FBP occupies a cavity rich in positively charged residues including the side chains of Arg40 (α3), His46 and Arg52 (β2-β3 hairpin) and Arg386 (LβH; Fig. 2A–C; Fig. 4A–B; Fig. S7). The O4 of the fructose ring makes a hydrogen bond with the guanidinium group of Arg419. The PO4 group at position 6 makes a critical interaction with the side chain of Lys39, essential for FBP binding and the activation of EcAGPase and with the guanidinium group of Arg423 (Fig. 4B) (Parsons and Preiss, 1978a, Parsons and Preiss, 1978b). The positive regulator was observed in the same location in both EcAGPase-FBPC1 and EcAGPase-FBPC2 reconstructions. Nevertheless, minor differences in the FBPC1 and FBPC2 conformation are revealed by the subtle movements of the PO4 group at position 1. Importantly, the EcAGPase-FBPC1 reconstruction exhibits clear electron density to model only two FBP molecules in two regulatory sites located in the same dimer, although residual electron density can be observed in the other two allosteric sites, suggesting that FBP may adopt distinct local conformations (Figs. S6 and S7).
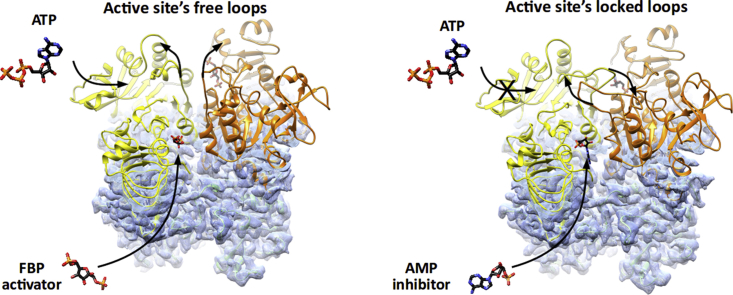
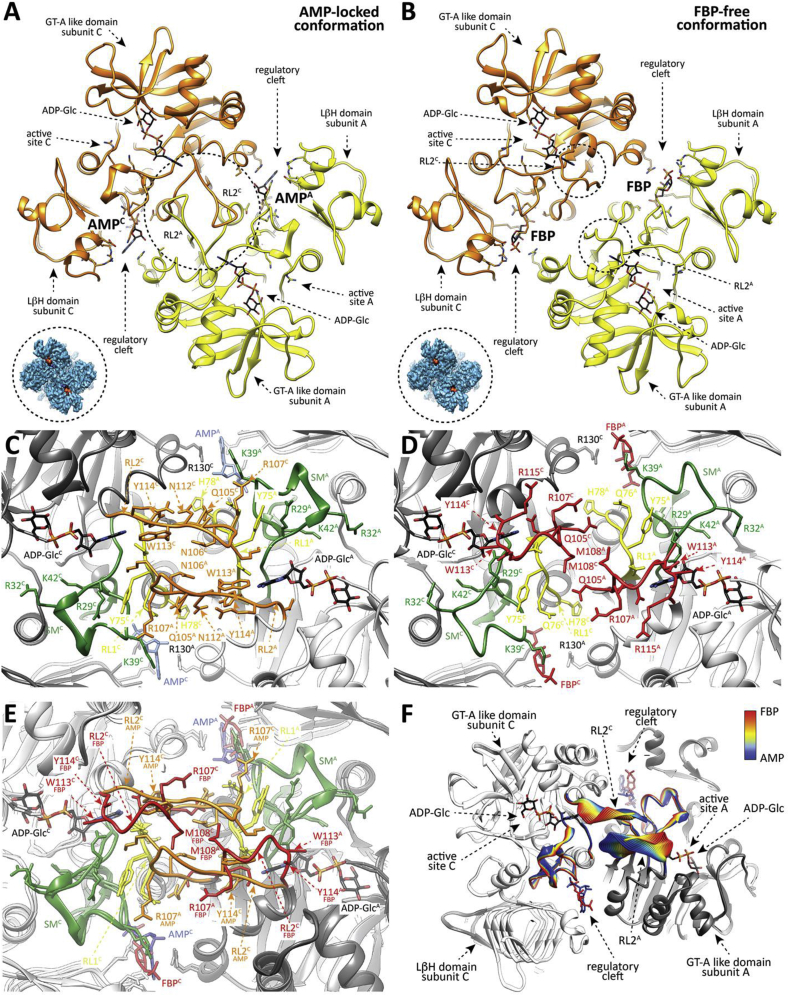
A detailed comparison of the overall EcAGPase-FBPC1, EcAGPase-FBPC2 and EcAGPase-FBPD2 reconstructions reveal remarkable structural differences in the conformation of two loops: one located at the center of the particle, hereafter called the protomer ‘core loop’ (CL; residues 88–97), and the long ‘regulatory loop’ RL2 (residues Ala104 to Gly116; Fig. 2C; Figs. S8 and S9) (Cifuente et al., 2016). Specifically, the side chains of residues Phe90 and Phe91, located in the CL loop, interact with each other in the EcAGPase-FBPC2 and EcAGPase-FBPD2 reconstructions. Nevertheless, in the EcAGPase-FBPC1 reconstruction, the aromatic ring of Phe91 moves away from the GT-A like core ~180° and interacts with Tyr309 of the neighbor dimer (Fig. S9). This structural rearrangement results in distinct pairs of CL conformations for each dimer in the EcAGPase-FBPC1 structure. The EcAGPase-FBPC1 and EcAGPase-FBPC2 reconstructions display bulky density impairing to model the position of the RL2 loop located in the ATP binding site (Cifuente et al., 2016). This electron density likely represents the RL2 loop in multiple conformational states, as suggested in our previous crystallographic studies on EcAGPase, as well as in other homologs, where the it could not be modeled except when participating in crystal contacts (Cifuente et al., 2016, Cupp-Vickery et al., 2008, Hill et al., 2015, Jin et al., 2005). Conversely, the EcAGPase-FBPD2 reconstruction reveals a consensus electron density for the RL2 loop in a relaxed “free” conformation (Fig. 2, Fig. 5). Moreover, the RL1 (residues Ile72 to His78) and the G-rich loops (residues 26 to 32; a constituent of the SM) appear in different conformations in these reconstructions, revealing the underlying dynamics of these loops. These findings suggest that the EcAGPase-FBP complex is a quasyD2 structure with local details that can be better recovered with C1 and C2 symmetries. Taken together, the conformations of the RL2 loop in an “free” state, and that of the SM G-rich loop and the RL1 loop, represent a configuration available for ATP binding.
Fig. 5.
The conformation of the RL2 loop as observed in the EcAGPase-FBPD2and EcAGPase-AMPD2complexes. (A) View of two neighboring protomers of different dimers (yellow and orange) in the EcAGPase-FBPD2 complex. FBP is observed in the regulatory cleft, whereas ADP-Glc is shown in the active site as a reference. The “free” conformation of the RL2 loops is indicated. A caption of the EcAGPase-FBPD2 complex map (blue) is shown with ADP-Glc placed in the active site. (B) As in panel (A), two neighboring protomers of different dimers in the EcAGPase-AMPD2 complex. AMP is observed in the regulatory cleft, whereas ADP-Glc is shown in the active site as a reference. The “locked” conformation of the RL2 loops is indicated. A caption of the EcAGPase-AMPD2 complex map (blue) is shown with ADP-Glc to observe map differences of the active shape compared with the EcAGPase-FBPD2 complex. (C) Closed view of the SM, RL1 and RL2 loops as observed in the EcAGPase-AMPD2 complex. (D) Closed view of the SM, RL1 and RL2 loops as observed in the EcAGPase-FBPD2 complex. (E) Structural comparison of the RL1 and RL2 loops as observed in the EcAGPase-FBPD2 and EcAGPase-AMPD2 complexes. (F) Cartoon representation showing the transition of the SM, RL1 and RL2 loops conformation as observed by cryoEM. Different positions of the main chain are colored according to the color scheme shown in the bar (FBP complex red, AMP complex blue).
2.2. The EcAGPase-AMP complex as visualized by cryoEM
The EcAGPase-AMP complex reconstructions also reach high resolution in all symmetries, C1 (EcAGPase-AMPC1; 3.26 Å resolution), C2 (EcAGPase-AMPC2; 3.09 Å resolution), and D2 (EcAGPase-AMPD2; 2.95 Å resolution), revealing a good correlation among them (Fig. 3, Fig. 4D; Figs. S3, S5, and S6 to S9; Table 1). These three reconstructions exhibit unambiguous electron density for the allosteric inhibitor in all four allosteric clefts, exhibiting an AMP-binding mode analogous to the one visualized in the EcAGPase-AMP X-ray crystal structure (Fig. 4D) (Cifuente et al., 2016). The AMP αPO4 group is located deep into the positively charged cavity and coordinated by the side chains of residues Arg40, His46, Arg53 (SM), Thr79 (α5), and Arg386 (LβH; Fig. 3A–C). The O2 of the ribose ring makes a strong interaction with the guanidinium group of Arg130 (Fig. 3C). The adenine heterocycle is also stabilized by a strong stacking interaction with Arg130 (α7) from the GT-A-like domain of a neighboring subunit. Strikingly, the EcAGPase-AMPD2 symmetry reconstruction reveals an extraordinary structural rearrangement of the RL2 loop, also observed in the lower symmetries (Fig. 2, Fig. 3B). The RL2 loop cross-over towards the active site of a neighbor protomer from a different dimer, adopting a “locked” state stabilized by important interactions with the RL2' loop, as well as with both RL1 and RL1’ loops. In addition, the CL loop is observed in the same conformation in all applied symmetries, indicating that AMP stabilizes a highly symmetrical quaternary conformation. We previously assigned to the AMP stacking interactions the main reason for a substantial stabilization, in more than 5°C of the Tm value followed by far-UV circular dichroism, of the enzyme (Cifuente et al., 2016, Comino et al., 2017). The cryoEM EcAGPase-AMP structure discloses now the contribution of the loops in the stabilization effect.
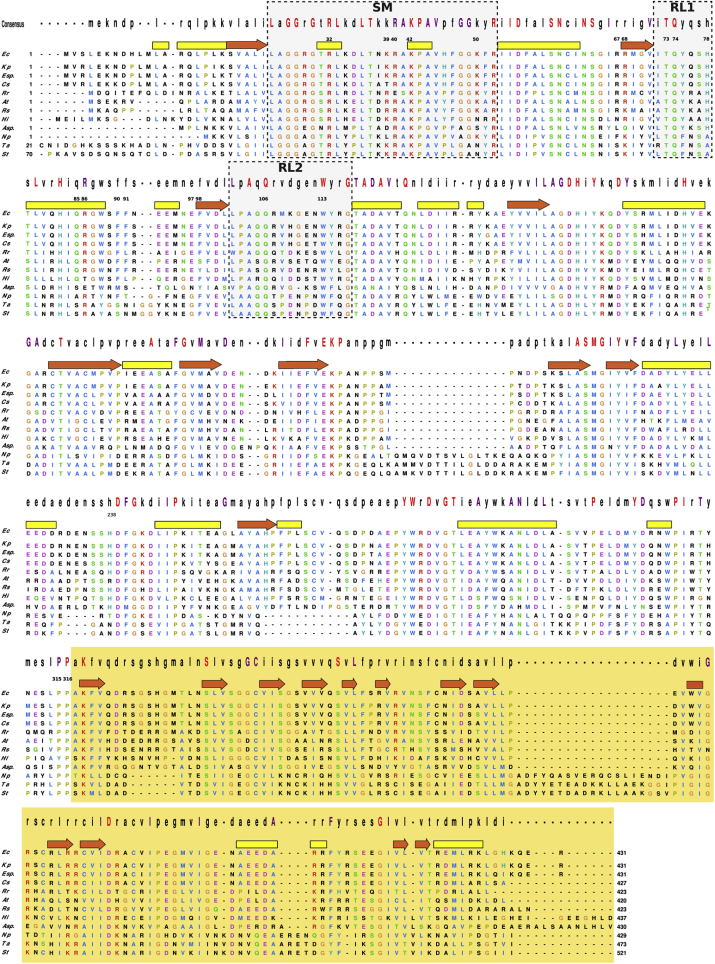
The structural comparison of the EcAGPase-AMPD2 and EcAGPase-FBPD2 reconstructions revealed important conformational changes (Fig. 4, Fig. 5). Tyr114 swings its side chain into the core of the protein, and makes a hydrogen bond with the side chain of Asn124, and interacts with the main chains of Leu102, Pro103 and Ala104, allowing the direct connection between the two ends of the RL2 loop (Fig. 5C–E). In addition, the main chain of Tyr114 also interacts with the side chain of Asn74 located in the RL1 loop of the same protomer. The replacement of Tyr114 by alanine displayed a drastic impact on the enzymatic activity, resulting in a reduced activation by FBP and a strong impairing effect in AMP-mediated inhibition (Kumar et al., 1988). Tyr114 and its equivalent residue Phe117 in StAGPase (Jin et al., 2005) lay in the proximity of the ATP adenine motif, suggesting that the observed effect is due to the impairment of ATP binding (Figueroa et al., 2011). The RL2 conformation is further stabilized by two hydrogen bonds between the main chain of Ala104 and the side chain of Asn112, and that of the side chain of Gln106 with the main chain of Glu111 (Fig. 2, Fig. 3, Fig. 5E). This structural arrangement is reinforced by the interaction between the main chain of Trp113 and the side chain of Gln74, located in the RL1 loop. Moreover, the side chain of Gln105 interacts with the side chain of Gln76, located in the RL1' loop of a neighbor subunit. Interestingly, the side chain of Arg107 makes a hydrogen bond with the main chain of Asn38, located in the SM motif (Fig. 5C–E). As a consequence, Arg29 swings its side chain out the ATP binding site and makes a strong electrostatic interaction with the side chain of Thr37. Similarly, the side chain of Trp113 comes out the catalytic pocket and adopt a completely different conformation. The replacement of Trp113 and Gln74 by alanine leads to a remarkable reduction in the activity and abolished the activation by FBP and inhibition by AMP (Figueroa et al., 2011, Hill et al., 2015). Furthermore, beyond their impact in the enzyme activity, the single point mutation to alanine of RL2 residues 105–111 reduce AMP inhibition (Hill et al., 2015). It is worth noting that RL2 residues are mainly conserved among AGPases from different sources, suggesting a common inhibition mechanism (Fig. 6).
Fig. 6.
Structural weighted alignment of EcAGPase with selected homologs. One-letter code residues are shown colored according to Clustal X scheme. The consensus sequence is displayed on top of the alignment where highly conserved residues in capitalized one-letter code (completely conserved residues in red, purple colored conservation ≥ 80%). The white background indicates the region corresponding to the N-terminal catalytic domain meanwhile the light orange background agrees with the C-terminal domain. Yellow and red bars indicate either α-helices and β-strand regions comprised in the EcAGPase structure, respectively. Regions corresponding to the sensory motif (SM), and the regulatory loops (RL1 and RL2) are enclosed in black boxes. The numbering of relevant EcAGPase residues is indicated. Shown are AGPases from E. coli (UniProt code P0A6V1), Klebsiella pneumoniae (B5XTQ9), Enterobacter sp. (A4WFL3), Cronobacter sakasakii (A7MGF4), Rodospirillum rubrum (Q2RS49), Agrobacterium tumefaciens (P39669), Rodobacter spheroids (A3PJX6), Haemofilus influenza (P43796), Arthrobacter sp. (A0JWV0), Nostoc punctiforme (B2IUY3), Triticum aestivum (P30523), and Solanum tuberosum (P23509).
2.3. A conformational switch modulates ATP binding
Our cryoEM studies on EcAGPase-FBP and EcAGPase-AMP complexes support the occurrence of two conformational states that operate in a concerted manner to allosterically modulate the enzymatic activity of the enzyme. A “locked” state where the binding of AMP promotes the close interaction between the RL2 and RL2′ from different protomers of different dimers, markedly reducing their availability for ATP binding (Fig. 5, Video S2: https://www.dropbox.com/s/6fu2cj8won6uu0i/vf_Video_S2.avi?dl=0); FBP displaces the negative regulator from the regulatory site and triggers the release of the RL2 and RL2′ loops adopting an “free” state (Fig. 4, Fig. 5). This conformational change makes the RL2 and RL2’ loops available for the interaction with ATP at the corresponding active sites. As a consequence, FBP breaks the symmetry of the particle, arguably from an EcAGPase-AMPD2 inhibited state to an EcAGPase-FBPquasyC2 active state, making the ATP site available. Interestingly, the C2 symmetric nature of the active enzyme can be assumed from early studies on EcAGPase (Haugen & Preiss, 1979). This mechanism accounts for the fact that sensitivity to AMP inhibition is modulated by the concentration of the activator FBP (Gentner and Preiss, 1968, Gentner and Preiss, 1967).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.crstbi.2020.04.005
The following is the supplementary data related to this article:
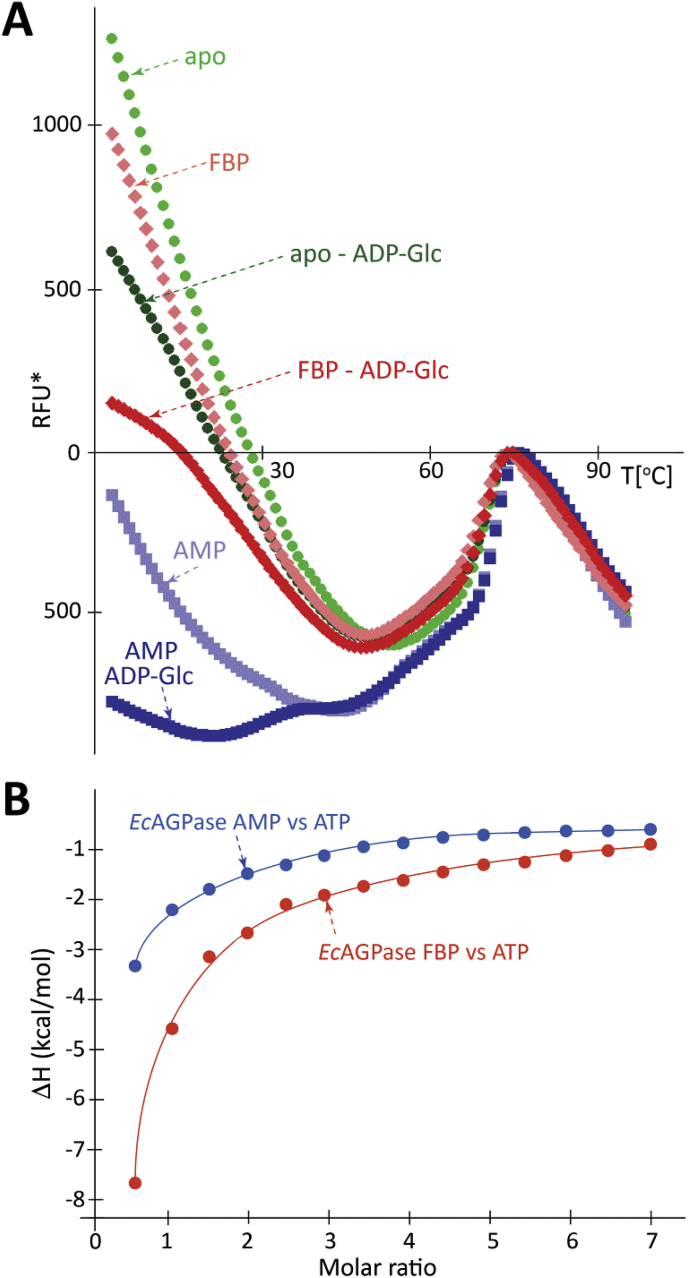
To determine whether the allosteric regulators modulate the recognition of ATP at the active site, we first determined the accessibility of the active site to substrates/products, in the presence/absence of the allosteric regulators by using a thermofluor assay (Fig. 7A; please see Materials and Methods in Supplementary Information). In the low temperature range (4–45°C), differences in fluorescence reflect the relative accessibility of the active and/or regulatory sites to the fluorophore SyPRO orange and therefore are attributable to the occupancy of the corresponding sites of EcAGPase in the native state. A reduction of the fluorescence signal in the presence of ADP-glucose is assigned to the occupancy of the active site. Conversely, differences in the fluorescence signal in the presence of allosteric regulators indicate the occupancy of the allosteric cleft. As depicted in Fig. 7A, the reduced fluorescence profile of EcAGPase-AMP compared with the EcAGPase-FBP complex strongly suggests greater accessibility of the active site to the fluorophore, and possibly the partial occupancy of the allosteric site by FBP. Moreover, in the high temperature denaturation range (45–95°C), the shift in the melting temperature clearly support the stabilization of the enzyme mediated by AMP association (Cifuente et al., 2016, Comino et al., 2017). Interestingly, AMP binding prevented the loss of enzymatic activity by chemical modification of the active site (Lee et al., 1986). Finally, to determine the binding of ATP to the EcAGPase-FBP and EcAGPase-AMP complexes, we performed ITC experiments. The titration of the EcAGPase-FBP with ATP discloses a very high affinity for the substrate, whereas, in contrast, the affinity towards ATP by the EcAGPase-AMP complex is markedly reduced (Fig. 7B; Fig. S10; Table S1), supporting our proposed allosteric model for EcAGPase (Fig. 8).
Fig. 7.
The allosteric regulators modulate the affinity for the substrate ATP. (A) Accessibility of ATP to the EcAGPase active site is modulated by the presence of the allosteric regulators FBP and AMP. (B) ITC measurements of EcAGPase-ligand interactions. The integrated heats of injections of the titrations corrected for the ligand heat of dilution and normalized to the ligand concentration. Solid lines correspond to the best fit of data.
Fig. 8.
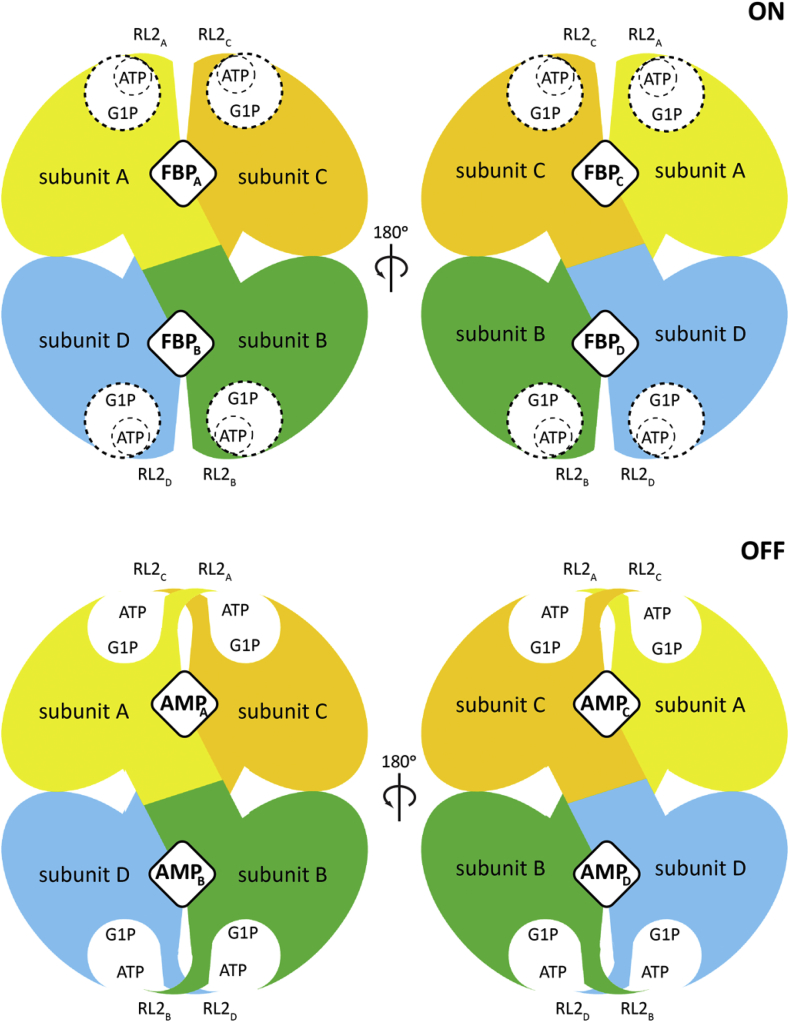
A molecular model for the allosteric regulation of EcAGPase. The EcAGPase tetramer is displayed in overlapping purple spheres, where the dotted lines are inter-protomer interphases. The active site is displayed as an open circle containing the substrates ATP and G1P. The allosteric clefts are indicated with rhombi at the interphases, that can alternatively be occupied by FBP or AMP, leading to the ON or OFF states, respectively. In the enzyme ON-state, the active site's RL2 loops (depicted in colors) are in a putative conformation allowing the interaction with ATP. In contrast, in the OFF inhibited AMP-state, the RL2 loops from neighbor protomers are engaged in the “locked” state, sequestering the loops for the interaction with ATP.
2.4. The allosteric network for signal transmission
To elucidate the pathways for the transmission of the positive and negative allosteric signals, we analyzed the residue interaction network (RIN) of (i) the FBP and AMP regulators, and (ii) the RL2 loop, towards the central core of the EcAGPase homotetramer, respectively (Figs. S11 and S12). As depicted in Fig. S12, there are important differences in the RIN profiles observed in the cryoEM EcAGPase-FBP and EcAGPase-AMP complexes. The negative regulator AMP interacts with Thr79 (α5), in close contact with His78 (α5), which in turn associates with Gln105 located in the RL2 loop adopting a “locked” conformation (Fig. 5). In contrast, in the presence of the positive regulator FBP, the interaction between Gln105 and His78 is missing, modifying the communication with the RL2 loop which adopts an “free” conformation (Fig. 5). These differences also impact the RIN associated with the CL loop. Notably, the CL loop in the EcAGPase-FBP complex encompasses a larger RIN (Figs. S11 and S12), compared to that observed in the EcAGPase-AMP complex, including the coordination between Arg67 and its counterpart in the GT-A-like domain of the opposite protomer. The replacement of Arg67 by alanine caused changes in the allosteric properties of EcAGPase (Ghosh et al., 1992). Furthermore, the CL interacts with a long loop that runs at the base of each protomer connecting the GTA-like domain and the LBH, here called the Inter-Domain Loops (IDL, residues 295–316). Interestingly, the CL interacts with the IDL from the same protomer and with both IDLs from the opposite dimer. The IDL anchors several regulatory motifs from the same and other protomers (SM, CL, IDL, LBH), changing its RIN in the FBP and AMP complexes. The IDL being at the core of the protein, therefore, represent a major element for allosteric signal transmission (Figs. S11 and S12).
3. Discussion
Bacterial glycogen and plant starch represent internal deposits of environmental carbon and energy surplus harvested by organisms (Ball and Morell, 2003, Cifuente et al., 2019). In the case of circumstantial scarcity, these storages are the source of energy permitting cell subsistence. From an evolutionary perspective, the rise of glycogen metabolism results in a competitive advantage to these organisms. This functionality requires the control of the energy flow towards and from the glycogen storage. Thus, signal-control systems to decouple glycogen synthesis and degradation have emerged to direct and adjust the energy flux between the glycogen compartment and the central energy metabolism. In bacteria, AGPase controls glycogen biosynthesis, whereas, GP regulates glycogen degradation (Cifuente et al., 2019). The coordination of these pathways is often evident, as in the case of E. coli, where AMP mediates the inhibition of AGPase and the activation of GP. In addition, ADP-glucose acts as a competitive inhibitor of GP in E. coli. This archetypal example demonstrates that allosteric enzymes are one of the finest products of evolution, providing rapid mechanisms of regulation and signal coordination of the whole metabolism.
3.1. The regulatory signaling mechanism of EcAGPase
Upon the addition of AMP to apo EcAGPase, the apparent melting temperature (Tm) value significantly increased, supporting the stabilization of the enzyme. In contrast, the addition of FBP to apo EcAGPase, did not affect the Tm, even at a high concentration of the positive regulator. The addition of FBP to the EcAGPase-AMP complex triggered a clear reduction in the Tm value, indicating that FBP competes with AMP and modifies the structural arrangement of the EcAGPase-AMP complex (Cifuente et al., 2016). Looking for differences in the secondary structure of EcAGPase in the presence or absence of allosteric regulators in solution, we assessed the circular dichroism spectrum of the enzyme in the far-UV region, from 200 to 250 nm (Fig. S13). Spectra were measured in a range of temperatures, from 10 to 40 °C. Overall, the spectral curves are similar, indicating that there are no large changes in the secondary structure content triggered by the regulators. Nevertheless, a close inspection of the 205–220 nm region shows slight differences supporting that EcAGPase resembles (i) more the AMP-state at lower temperatures, and (ii) more the FBP-state at high temperature. Interestingly, enzyme kinetics measurements revealed that the addition of AMP to the EcAGPase-FBP complex drastically reduced the enzymatic activity, compared to EcAGPase in the absence of the positive allosteric regulator (Haugen & Preiss, 1979). Altogether, the biochemical and biophysical observations suggest that, in the absence of allosteric regulators, the structural arrangement of the active site of EcAGPase is similar and/or visits more often the “locked” conformation observed in the EcAGPase-AMP complex. This view hints that the role of AMP is to stabilize a pre-existing low activity conformation already available in apo EcAGPase. In turn, FBP displaces AMP inducing a change in the RIN that ends with the activated “free” conformation of the RL2 loops in the active site (Fig. 5; Fig. S12).
Kinetic properties of EcAGPase single-point mutants have been previously reported (Hill et al., 2015, Figueroa et al., 2011, Bhayani et al., 2019). The harmonization of these results from different conditions and the interpretation of the kinetics parameters in the context of the structural data provided by X-ray crystallography and cryoEM represent a challenge. To reduce the complexity, we have performed a simplified meta-analysis of EcAGPase mutants restricted to the replacement of selected residues by alanine (Table 2; please see Materials and Methods in Supplementary Information). In the absence of AMP and FBP, the kinetics parameters Vmax and S0.5 (ATP concentration yielding Vmax/2; Table 2) did not significantly change in EcAGPase mutants of the active site. In contrast, the EcAGPase variations Arg40Ala, Arg52Ala, and Arg386Ala, localized in the allosteric cleft of the enzyme, showed a marked reduction in Vmax, suggesting that the positively charged pocket is required to maintain the functional structure of the SM (Fig. 2, Fig. 3, Fig. 4, Fig. 5; Fig. S7). Moreover, Arg40Ala exhibits an increased S(ATP)0.5, supporting a possible impact in the neighboring catalytic residue Lys42 (Fig. S1).
Table 2.
Qualitative meta-analysis of reported enzymatic properties of EcAGPase single point mutants relative to wild-type.
| Residue location | Element | Mutant | basal (apo) |
Activation(FBP) |
Inhibition(AMP and FBP) |
%Ident | Conserv score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vmaxratio | SATP(0.5)ratio | Vmax ratio | SATP(0.5)ratio | A(0.5)ratio | I(0.5)ratio | ||||||
| N-term domain (GTA-like) | Active site | RL2a | P103A | ~ | ↓ | ↓ | ↑ | ~ | ~ | 75 | 7 |
| Q105A | ~ | ~ | ~ | ↑ | ↑ | ↑ | 75 | 6 | |||
| Q106A | ~ | ~ | ↓ | ↑ | ↑ | ~ | 100 | 11 | |||
| R107A | ~ | ~ | ↓ | ↑ | ~ | ~ | 58 | 6 | |||
| M108A | ~ | ~ | ~ | ~ | ~ | ~ | 50 | 5 | |||
| K109A | ~ | ~ | ~ | ↑ | ~ | ~ | 33 | 2 | |||
| E111A | ~ | ~ | ~ | ~ | ~ | ~ | 41 | 2 | |||
| N112A | ~ | ~ | ~ | ↑ | ~ | ~ | 33 | 5 | |||
| W113Ad | ~ | ~ | ↓ | ↑ | ~ | ~ | 100 | 11 | |||
| Y114A | ~ | ~ | ↓ | ↑ | ~ | ~ | 66 | 9 | |||
| R115A | ~ | ~ | ↓ | ↑ | ↑ | ~ | 41 | 4 | |||
| RL1b | Q74A | n/a | ~ | n/a | ↑ | n/a | n/a | 100 | 11 | ||
| Regulator sitec | SM | R40A | ↓ | ↑ | ↓ | ↑ | n/a | n/a | 91 | 8 | |
| R52A | ↓ | ~ | ↓ | ↑ | n/a | n/a | 100 | 11 | |||
| α-7 | R130A | ↑ | ~ | ~ | ~ | ↑ | n/a | 33 | 1 | ||
| C-term domain (LBH) | LBH | R353A | ~ | ~ | ~ | ~ | ~ | n/a | 58 | 2 | |
| R386A | ↓ | ~ | ↓ | ↑ | ↑ | n/a | 50 | 5 | |||
| C-term α-helix | R419A | ~ | ~ | ~ | ~ | ~ | n/a | 33 | 2 | ||
| R423A | ~ | ~ | ~ | ~ | ↑ | n/a | 33 | 2 | |||
Mutants ad-hoc kinetic behavior grouping according to parameter ratios mutant/wild-type. The symbol “~” indicates values of the parameter ratio between 0.5 and 2.0. The symbol “↑” indicates values of the parameter ratio greater than 2.0. The symbol “↓” indicates ratios below 0.5. The conservation score of the alignment corresponds to that of Jalview (see Supplementary Materials and Methods).
Reference (Hill et al., 2015). ADP-synthesis assessed by a phosphatase-coupled malachite-green molybdate phosphate colorimetric end-point assay.
Reference (Figueroa et al., 2011). Radiolabeled ADP-synthesis assessed by a glycogen synthase-coupled glycogen radiometric end-point assay.
Reference (Bhayani et al., 2019). ADP-synthesis assessed as in reference 32.
Also reported in b.
In the FBP-activated state, mutants localized both in the active and regulatory sites of EcAGPase showed altered kinetics profiles (Table 2). In particular, mutants Pro103Ala, Gln106Ala, Arg107Ala, Trp113Ala, Tyr114Ala and Arg115Ala, located in the RL2 loop, are primarily characterized by a lack of response to FBP (Fig. 5C–E; Table 2) (Hill et al., 2015). Specifically, Pro103Ala displayed a reduced Vmax and reduced affinity for ATP. Pro103 is a strictly conserved residue that appears to play a fundamental role in the structural arrangement of the RL2 loop. Gln106Ala also displayed a reduced Vmax and affinity for ATP, and required a higher concentration of FBP to activate the enzyme. In the EcAGPase-FBPD2 complex, the side chain of Gln106 makes strong interactions with the main chain of Arg115, located in the RL2 loop, and Gln74, located in the RL1 loop (Fig. 5C–E; Table 2). In contrast, in the EcAGPase-AMPD2 complex, the side chain of Gln106 displays a completely different arrangement, making a hydrogen bond with the main chain of Gln112. The Arg107Ala mutant displays a reduced Vmax and affinity for ATP. The side chain of Arg107 makes an electrostatic interaction with the side chain of Gln123, stabilizing the “free” conformation in EcAGPase-FBPD2 complex. Arg107 is located in α7, a key secondary structure that mediates the signal transmission between the regulatory site and the active site through the RL2 loop. The Trp113Ala and Tyr114Ala mutants also showed a reduced Vmax and affinity for ATP. Although both residues have been suggested to participate in the binding of ATP, their side chains are not observed in the cryoEM structure of the EcAGPase-FBP complex, limiting the analysis. Arg115Ala showed a reduced Vmax and affinity for ATP, and required a higher concentration of FBP to activate EcAGPase. The side chain of Arg115 makes an electrostatic interaction with the side chain of His238, located in α7, contributing to the formation of the “free” conformation of the RL2 loop in the EcAGPase-FBPD2 complex. Finally, at the regulatory cleft, mutations Arg40Ala, Arg52Ala, and Arg386Ala displayed a reduced Vmax and S0.5 values, possibly by impairing the arrangement of the activator phosphate group in the corresponding pocket. Interestingly, although Arg130Ala and Arg423Ala display similar Vmax and S0.5 values; the mutants require higher concentrations of FBP to achieve half-full activation levels, an indication of their role in FBP recognition and the activation mechanism.
In the case the FBP-activated state was inhibited by AMP, all reported mutants located in the active site, including Pro103Ala, Gln106Ala, Arg107Ala, Met108Ala, Lys109Ala, Glu111Ala, Asn112Ala, Trp113Ala, Tyr114Ala, and Arg115Ala, did not display a substantial change in the I0.5, the AMP concentration required for half inhibition of the enzymatic activity of EcAGPase, possibly due to the preservation of the AMP site architecture (Fig. 5C–E). The Gln105Ala represents an exception, requiring a higher concentration of AMP for inhibition. In the AMP-inhibited state, Gln105 interacts with the side chain of Gln76, located in the RL1 loop; and the main chain of His78, located in the top of α5, where Thr79 participate in the interaction with the α phosphate group of AMP. Interestingly, mutants of the RL2 loop which exhibited the least sensitivity to FBP were also the least sensitive to AMP inhibition. EcAGPase was activated approximately 25-fold by FBP, compared to 1.5 fold for the mutants Pro103Ala, Trp113Ala, and Tyr114Ala. EcAGPase conserved just ca. 3% of its enzymatic activity with saturating AMP, whereas mutants Pro103Ala, Trp113Ala, and Tyr114Ala retained ca. 50–70%. Altogether, the structural and enzymatic data support the notion that these residues, located at both ends of the RL2 loop, play a major mechanistic role to facilitate the “free-to-locked” transitions, critical for the regulation of the enzyme (Fig. 5). We previously reported that mutants Lys39Ala, Arg40Ala, His46Ala, Arg52Ala, Arg386Ala, Arg419Ala and Arg423Ala, located in the regulatory site, failed to achieve the activation values mediated by FBP, compared to that obtained for and severely compromised the inhibition by AMP. Strikingly, we found that the Arg130Ala mutant deregulated AMP-mediated inhibition of the enzymatic activity, inducing the overproduction of glycogen in vivo. The interaction of Arg130 (α7) with the AMP nucleobase stabilizes the α7 helix in such a way facilitating the arrangement of a cavity for the accommodation of Tyr114 in the “locked” conformation.
3.2. The cooperativity signaling mechanism of EcAGPase
The enzyme cooperativity mechanism is disclosed by the EcAGPase-FBPD2 and EcAGPase-AMPD2 reconstructions, revealing clear pathways of communication between the active sites in the homotetrameric architecture. The signal transmission is mainly mediated by the interactions between the GT-A like domains (Fig. 8; Fig. S8). It occurs essentially in two manners: (i) side-by-side, where one GTA-like domain of a protomer interacts with a GTA-like domain of another protomer from a different dimer (Fig. 8); and (ii) top-to-bottom, where the interactions between GTA-like domains occur across the tetramerization interphase (Figs. S8 and S9). It has been extensively reported that FBP activation increases the positive cooperative effect displayed by EcAGPase. Our study clearly shows that FBP interaction with EcAGPase mediates the release of the RL2-to-RL2 lock, which means an immediate availability of two active sites in the active conformation. This event explains the drastic increment in the activity of the enzyme. Moreover, comparative RIN analysis between the AMP and FBP states reveal that the release of the “locked” conformation is accompanied by the reduction of the RL2 interactions. Simultaneously, there is an increment of the CL RIN, which is anchored to the α5, a central element in the architecture of AGPases. The α5 repositioning appears to contribute to the release of the RL1-RL2 interactions in the “locked” conformation. Finally, the changes in the CL conformation is disseminated across the tetramer by interactions with the IDL, which associates with several regulatory structural elements from different protomers.
Several residues involved in catalysis are conserved in the active site across all AGPase homologs, including (i) Arg32 involved in the anchoring of the γ phosphate group of ATP, and (ii) Lys42 and Lys195, involved in the interaction, respectively, with ATP and G1P, which polarize the phosphates groups to facilitate the nucleophilic reaction (G-rich loop; Fig. S1). Moreover, several residues implicated in substrate recognition are also preserved (Fig. 6). In contrast, most residues at the regulatory cleft implicated in the binding of FBP and AMP regulators in EcAGPase are not highly conserved, with the exception of Arg40 and Arg52 (Fig. 6). This observation agrees with the fact that AGPases from different sources use different allosteric regulators, providing a specific relationship to specific metabolic routes of relevance to the organism or tissue. The conservation of Arg40 and Arg52 can be explained because both residues participate in the interaction with the α phosphate group of ATP or a phosphate moiety of FBP, preserving the positively charged pocket shaped by the SM. This pocket might participate in the binding of other phosphorylated metabolites known to regulate other AGPases, as 3-phosphoglycerate (3PGA) in plants or fructose-6-phosphate (F6P) in Agrobacterium tumefaciens (Ballicora et al., 2003, Cupp-Vickery et al., 2008). The long RL2 loop appears to be an evolutionary feature of AGPases, not observed in other non-regulated Nucleotide-di-phosphate pyrophosphorylases (NDPases; Fig. S14). The structural characteristics, the close location to the ATP substrate, and the conservation of the RL2 loop strongly suggest this structural element as a major driving force in AGPase evolution (Fig. 6, Table 2). Altogether, these facts imply the evolution of the allosteric cleft is partially disentangled from the evolution of the active site, facilitating the divergence of regulatory cleft residues to acquire different enzymatic regulatory characteristics.
Considered the ‘second secret of life’ (Fenton, 2008), allosterism is a central biological phenomenon that provides coherence and harmony to the metabolism. An allosteric enzyme is a specialized product of evolutionary engineering enabling the control of catalysis by ligands that are chemically foreign to the chemical reaction; therefore, it is the base of metabolic coordination. In words of Monod, “the very gratuitousness of these systems, giving molecular evolution a practically limitless field for exploration and experiment, enabled it to elaborate the huge network of cybernetic interconnections which makes each organism an autonomous functional unit, whose performances appear to transcend the laws of chemistry” (Monod, 1971).
3.3. EcAGPase regulatory mechanism through the glass of different allosteric models
Different models have been proposed to contextualize the current understanding of the allosteric phenomena. The original Monod-Wyman-Changeux (MWC) concerted model presents a perspective of multimeric enzymes where all protomers are constrained to a single conformational state at a given moment (Monod et al., 1965). Two global conformational states exist in equilibrium, the so-called T (tense) and R (relaxed) states. The conformational transition between the two states occurs simultaneously, in a concerted manner, in all protomers. The ligands have different affinity for the two states, and the equilibrium is shifted thanks to the binding of an allosteric effector or substrate to a protomer. Since only one conformer can exist in a given enzyme molecule at the time, this model implies infinite coupling between protomers suppressing conformational intermediates (Hilser et al., 2012). The R-T shifting mediated by ligand binding represents the view of a conformational selection of the conformers. According to the MWC model view, the EcAGPase-AMP complex represents a T inhibited state since the “locked” mechanism requires all protomers to acquire the same conformation (Fig. 3, Fig. 4, Fig. 5; Fig. S8) (Cifuente et al., 2016, Comino et al., 2017). This approximation appears feasible since the inhibitor is active at very reduced concentrations, pointing to the high affinity of AMP, leading to an almost instant full occupancy of the allosteric cleft according to the reported intracellular AMP concentration, even if low amounts of FBP are present (Buchholz et al., 2001, Ishii et al., 2007, Bennett et al., 2009, Guo et al., 2013).
In the Koshland, Némethy and Filmer (KNF) sequential model, the protomers are not constrained to a global concerted conformational change of all protomers to the same conformation. Therefore, the KNF model contemplates the enzyme subunits to exist in different conformations at any given time (Koshland et al., 1966). The binding of a substrate or effector to a protomer influence in a different degree the conformation of other protomers and their affinity towards the ligands. The KNF model implies that ligands bind via induced fit phenomena, triggering conformational changes. In the case of AMP displacement by FBP, we observe that the activated form of EcAGPase shows protomers in different conformations, which concurs with this view. Moreover, our observations showing that FBP can have differences in the binding mode and differences in conformations are in agreement with the induced fit view.
Although these classical models of allosterism address some of the observations, the different conformations observed locally points towards other paradigms of allosterism. The EcAGPase model of regulation can be better reflected by the Ensemble Model (EM) in which the allosteric system is described as a population of conformational states statistically distributed according to their energy levels (Motlagh et al., 2014). In that sense, single-particle cryoEM reflects the view of the consensus of the particles. Other alternative paradigms present the allosteric phenomena without large motions of the enzyme but instead with changes in the dynamic and minimum global conformational changes. According to this view, the binding of the allosteric effector transfers the allosteric signals to the active site in the form of entropy-driven dynamics (Cooper & Dryden, 1984). Importantly, this model has been proposed for kinases having similar G-rich loops to AGPases (Kornev & Taylor, 2015). Specifically, the transmission of the signal can be defined by two mechanisms: (i) a ‘domino model’ propagating the signal through sequential local structural changes from the allosteric site via a single pathway to the active site and (ii) a ‘violin model' analogy mechanism, where the enzyme vibrational states, as the body of the violin, can change because of the binding of the effector, transferring the signal throughout the whole structure with no specific pathway (Kornev & Taylor, 2015).
4. Conclusions
Our work highlights how the advances of cryoEM can extraordinarily power the study of the cooperative and allosteric phenomena of enzymes. We observe novel biologically relevant conformations of EcAGPase that could not be visualized by other structural biology techniques – e.g. X-ray crystallography due to crystal constraints and crystallization artifacts. The single-particle reconstruction approach permits to build a model that represents the conformational consensus of the isolated enzyme. Strikingly, since the majority of allosteric and cooperative enzymes are multimeric proteins, the study of all allowed symmetries of the system reveal different views of the phenomena. In our case, although different symmetrized reconstructions result in a similar resolution, they allow different observations. In the activated state of EcAGPase, the high symmetry D2 allows to model a consensus for the RL2 loop in the “free” conformation, meanwhile, side chains conformation and the FBP occupancy of the allosteric cleft can be explored in the lower symmetries. On the other hand, all the reconstructions from the EcAGPase-AMP complex disclose the same conformation, indicating that the single-particle reconstruction represents a more homogeneous ensemble of particles. Many more allosteric enzymatic systems await for being explored or re-visited by cryoEM to reveal their mechanisms.
Author contributions
J.O.C. and M.E.G. conceived the project. A.M., N.C., and C.D. expressed and purified the protein. J.O.C and D.G.C. performed cryoEM grid optimization and cryoEM microscopy screening. J.O.C. and D.G.C. collected EM data. J.O.C. performed EM processing, built and refined the atomic models. J.O.C., D.A.J, N.C. and C.D. performed biophysical studies. J.O.C., C.D., N.C. and M.E.G. prepared figures and tables. J.O.C and M.E.G wrote the manuscript. All the authors critically discussed the interpretation of the data and the final manuscript.
Declaration of Competing Interest
The authors declare no financial conflict of interest.
Acknowledgments
This project was supported by grants from (i) the MINECO/FEDER EU contracts BFU2016-77427-C2-2-R, BFU2017-92223-EXP and Severo Ochoa Excellence Accreditation SEV-2016-0644, and (ii) the Basque Government contract KK-2019/00076 (to M.E.G.). We acknowledge Diamond Light Source for access and support of the cryo-EM facilities at the UK's national Electron Bio-imaging Centre (eBIC) under proposals EM6916 and EM17171, funded by the Wellcome Trust, MRC and BBRSC. We thank for valuable assistance in cryoEM data collection to the eBIC Diamond staff, especially to Alistair Siebert, Corey Hecksel and Kyle Dent.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2020.04.005.
Contributor Information
Javier O. Cifuente, Email: jcifuente@cicbiogune.es.
Marcelo E. Guerin, Email: mrcguerin@cicbiogune.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ball S.G., Morell M.K. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Ballicora M.A., Iglesias A.A., Preiss J. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol. Mol. Biol. Rev. 2003;67:213–225. doi: 10.1128/MMBR.67.2.213-225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballicora M.A., Erben E.D., Yazaki T., Bertolo A.L., Demonte A.M., Schmidt J.R., Aleanzi M., Bejar C.M., Figueroa C.M., Fusari C.M., Iglesias A.A., Preiss J. Identification of regions critically affecting kinetics and allosteric regulation of the Escherichia coli ADP-glucose pyrophosphorylase by modeling and pentapeptide-scanning mutagenesis. J. Bacteriol. 2007;189:5325–5333. doi: 10.1128/JB.00481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhayani J.A., Hill B.J., Sharma A., Iglesias A.A., Olsen K.W., Ballicora M.A. Mapping of a Regulatory site of the Escherichia coli ADP-Glucose pyrophosphorylase. Front. Mol. Biosci. 2019;6:89. doi: 10.3389/fmolb.2019.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W., Asuncion M., Lam J.S., Naismith J.H. The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA) EMBO J. 2000;19:6652–6663. doi: 10.1093/emboj/19.24.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell T.L., Srinivasan N. Symmetry, stability, and dynamics of multidomain and multicomponent protein systems. Proc. Natl. Acad. Sci. USA. 1996;93:14243–14248. doi: 10.1073/pnas.93.25.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafonte M.A., Solano C., Sesma B., Alvarez M., Montuenga L., García-Ros D., Gamazo C. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 2000;191:31–36. doi: 10.1111/j.1574-6968.2000.tb09315.x. [DOI] [PubMed] [Google Scholar]
- Buchholz A., Takors R., Wandrey C. Quantification of intracellular metabolites in Escherichia coli K12 using liquid chromatographic-electrospray ionization tandem mass spectrometric techniques. Anal. Biochem. 2001;295:129–137. doi: 10.1006/abio.2001.5183. [DOI] [PubMed] [Google Scholar]
- Changeux J.-P. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu. Rev. Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- Chiu W., Baker M.L., Jiang W., Dougherty M., Schmid M.F. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Chubukov V., Gerosa L., Kochanowski K., Sauer U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- Cifuente J.O., Comino N., Madariaga-Marcos J., López-Fernández S., García-Alija M., Agirre J., Albesa-Jové D., Guerin M.E. Structural basis of glycogen biosynthesis regulation in bacteria. Structure. 2016;24:1613–1622. doi: 10.1016/j.str.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Cifuente J.O., Comino N., Trastoy B., D'Angelo C., Guerin M.E. Structural basis of glycogen metabolism in bacteria. Biochem. J. 2019;476:2059–2092. doi: 10.1042/BCJ20170558. [DOI] [PubMed] [Google Scholar]
- Comino N., Cifuente J.O., Marina A., Orrantia A., Eguskiza A., Guerin M.E. Mechanistic insights into the allosteric regulation of bacterial ADP-glucose pyrophosphorylases. J. Biol. Chem. 2017;292:6255–6268. doi: 10.1074/jbc.M116.773408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A., Dryden D.T.F. Allostery without conformational change. Eur. Biophys. J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- Crevillén P., Ballicora M.A., Mérida A., Preiss J., Romero J.M. The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct kinetic and regulatory properties to the heterotetrameric enzyme. J. Biol. Chem. 2003;278:28508–28515. doi: 10.1074/jbc.M304280200. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery J.R., Igarashi R.Y., Perez M., Poland M., Meyer C.R. Structural analysis of ADP-glucose pyrophosphorylase from the bacterium Agrobacterium tumefaciens. Biochemistry. 2008;47:4439–4451. doi: 10.1021/bi701933q. [DOI] [PubMed] [Google Scholar]
- Espada J. Enzymic synthesis of adenosine diphosphate glucose from glucose 1-phosphate and adenosine triphosphate. J. Biol. Chem. 1962;237:3577–3581. [Google Scholar]
- Fenton A.W. Allostery: an illustrated definition for the “second secret of life”. Trends Biochem. Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa C.M., Esper M.C., Bertolo A., Demonte A.M., Aleanzi M., Iglesias A.A., Ballicora M.A. Understanding the allosteric trigger for the fructose-1,6-bisphosphate regulation of the ADP-glucose pyrophosphorylase from Escherichia coli. Biochimie. 2011;93:1816–1823. doi: 10.1016/j.biochi.2011.06.029. [DOI] [PubMed] [Google Scholar]
- Führing J., Cramer J.T., Routier F.H., Lamerz A.C., Baruch P., Gerardy-Schahn R., Fedorov R. Catalytic mechanism and allosteric regulation of UDP-glucose pyrophosphorylase from Leishmania major. ACS Catal. 2013;3:2976–2985. [Google Scholar]
- Gentner N., Preiss J. Activator-inhibitor interactions in the adenosine diphosphate glucose pyrophosphorylase of Escherichia coli B. Biochem. Biophys. Res. Commun. 1967;27:417–423. doi: 10.1016/s0006-291x(67)80116-5. [DOI] [PubMed] [Google Scholar]
- Gentner N., Preiss J. Biosynthesis of bacterial glycogen. VI. Differences in the kinetic properties of the Escherichia coli B adenosine diphosphate glucose pyrophosphorylase depending on whether Mg++ or Mn++ serves as divalent cation. J. Biol. Chem. 1968;243:5882–5891. [PubMed] [Google Scholar]
- Georgelis N., Braun E.L., Shaw J.R., Hannah L.C. The two AGPase subunits evolve at different rates in angiosperms, yet they are equally sensitive to activity-altering amino acid changes when expressed in bacteria. Plant Cell. 2007;19:1458–1472. doi: 10.1105/tpc.106.049676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Meyer C., Remy E., Peterson D., Preiss J. Cloning, expression, and nucleotide sequence of glgC gene from an allosteric mutant of Escherichia coli B. Arch. Biochem. Biophys. 1992;296:122–128. doi: 10.1016/0003-9861(92)90553-9. [DOI] [PubMed] [Google Scholar]
- Goodsell D.S., Olson A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biom. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- Guo A.C., Jewison T., Wilson M., Liu Y., Knox C., Djoumbou Y., Lo P., Mandal R., Krishnamurthy R., Wishart D.S. ECMDB: the Escherichia coli metabolome database. Nucleic Acids Res. 2013;41:D625–630. doi: 10.1093/nar/gks992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen T.H., Preiss J. Biosynthesis of bacterial glycogen. The nature of the binding of substrates and effectors to ADP-glucose synthase. J. Biol. Chem. 1979;254:127–136. [PubMed] [Google Scholar]
- Hill B.L., Wong J., May B.M., Huerta F.B., Manley T.E., Sullivan P.R.F., Olsen K.W., Ballicora M.A. Conserved residues of the Pro103-Arg115 loop are involved in triggering the allosteric response of the Escherichia coli ADP-glucose pyrophosphorylase. Protein Sci. 2015;24:714–728. doi: 10.1002/pro.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser V.J., Wrabl J.O., Motlagh H.N. Structural and energetic basis of allostery. Annu. Rev. Biophys. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N., Nakahigashi K., Baba T., Robert M., Soga T., Kanai A., Hirasawa T., Naba M., Hirai K., Hoque A., Ho P.Y., Kakazu Y., Sugawara K., Igarashi S., Harada S., Masuda T., Sugiyama N., Togashi T., Hasegawa M., Takai Y., Yugi K., Arakawa K., Iwata N., Toya Y., Nakayama Y., Nishioka T., Shimizu K., Mori H., Tomita M. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- Jin X., Ballicora M.A., Preiss J., Geiger J.H. Crystal structure of potato tuber ADP-glucose pyrophosphorylase. EMBO J. 2005;24:694–704. doi: 10.1038/sj.emboj.7600551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R., Palacios A., Anso I., Cifuente J.O., Anguita J., Jr Jacobs W.R., Guerin M.E., Prados-Rosales R. The Mycobacterium tuberculosis capsule: a cell structure with key implications in pathogenesis. Biochem. J. 2019;476:1995–2016. doi: 10.1042/BCJ20190324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliwer-Brandl H., Syson K., van de Weerd R., Chandra G., Appelmelk B., Alber M., Ioerger T.R., Jacobs W.R., Geurtsen J., Bornemann S., Kalscheuer R. Metabolic Network for the biosynthesis of intra- and extracellular α-glucans required for virulence of Mycobacterium tuberculosis. PLoS Pathog. 2016;12:1–26. doi: 10.1371/journal.ppat.1005768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. On the metabolic significance of phosphorolytic and pyrophosphorolytic reactions. In: Kasha H., Pullman P., editors. Horizons in Biochemistry. Academic Press; New York, NY: 1962. pp. 251–264. [Google Scholar]
- Kornev A.P., Taylor S.S. Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 2015;40:628–647. doi: 10.1016/j.tibs.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D.E., Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Kumar A., Tanaka T., Lee Y.M., Preiss J. Biosynthesis of bacterial glycogen. Use of site-directed mutagenesis to probe the role of tyrosine 114 in the catalytic mechanism of ADP-glucose synthetase from Escherichia coli. J. Biol. Chem. 1988;263:14634–14639. [PubMed] [Google Scholar]
- Lahti R. Microbial inorganic pyrophosphatases. Microbiol. Rev. 1983;47:169–178. doi: 10.1128/mr.47.2.169-178.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.M., Mukherjee S., Preiss J. Covalent modification of Escherichia coli ADPglucose synthetase with 8-azido substrate analogs. Arch. Biochem. Biophys. 1986;244:585–595. doi: 10.1016/0003-9861(86)90627-2. [DOI] [PubMed] [Google Scholar]
- Liboff R.L. Springer New York; New York, NY: 2004. Primer for Point and Space Groups. [Google Scholar]
- Meléndez R., Meléndez-Hevia E., Canela E.I. The fractal structure of glycogen: a clever solution to optimize cell metabolism. Biophys. J. 1999;77:1327–1332. doi: 10.1016/S0006-3495(99)76982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J., Changeaux J.P., Jacob F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- Monod J., Wyman J., Changeux J.P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Monod J. chance and necessity. Alfred A. Knopf, Inc.; New York, N.Y.: 1971. Microscopic cybernetics. [Google Scholar]
- Motlagh H.N., Wrabl J.O., Li J., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons T.F., Preiss J. Biosynthesis of bacterial glycogen. Incorporation of pyridoxal phosphate into the allosteric activator site and an ADP-glucose-protected pyridoxal phosphate binding site of Escherichia coli B ADP-glucose synthase. J. Biol. Chem. 1978;253:6197–6202. [PubMed] [Google Scholar]
- Parsons T.F., Preiss J. Biosynthesis of bacterial glycogen. Isolation and characterization of the pyridoxal-P allosteric activator site and the ADP-glucose-protected pyridoxal-P binding site of Escherichia coli B ADP-glucose synthase. J. Biol. Chem. 1978;253:7638–7645. [PubMed] [Google Scholar]
- Paule M.R., Preiss J. Biosynthesis of bacterial glycogen. X. The kinetic mechanism of adenosine diphosphoglucose pyrophosphorylase from Rhodospirillum rubrum. J. Biol. Chem. 1971;246:4602–4609. [Google Scholar]
- Perutz M.F. Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- Petreikov M., Eisenstein M., Yeselson Y., Preiss J., Schaffer A.A. Characterization of the AGPase large subunit isoforms from tomato indicates that the recombinant L3 subunit is active as a monomer. Biochem. J. 2010;428:201–212. doi: 10.1042/BJ20091777. [DOI] [PubMed] [Google Scholar]
- Preiss J. Regulation of adenosine diphosphate glucose pyrophosphorylase. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;46:317–381. doi: 10.1002/9780470122914.ch5. [DOI] [PubMed] [Google Scholar]
- Recondo E., Leloir L.F. Adenosine diphosphate glucose and starch synthesis. Biochem. Biophys. Res. Commun. 1961;6:85–88. doi: 10.1016/0006-291x(61)90389-8. [DOI] [PubMed] [Google Scholar]
- Swift R.V., Ong C.D., Amaro R.E. Magnesium-induced nucleophile activation in the guanylyltransferase mRNA capping enzyme. Biochemistry. 2012;51:10236–10243. doi: 10.1021/bi301224b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivelloni J.C., Recondo E., Cardini C.E. Adenosine diphosphate glucose and glucoside biosynthesis. Nature. 1962;195 doi: 10.1038/1951202a0. 1202–1202. [DOI] [PubMed] [Google Scholar]
- Ventriglia T., Ballicora M.A., Crevillén P., Preiss J., Romero J.M. Regulatory properties of potato-Arabidopsis hybrid ADP-glucose pyrophosphorylase. Plant Cell Physiol. 2007;48:875–880. doi: 10.1093/pcp/pcm047. [DOI] [PubMed] [Google Scholar]
- Vithani N., Bais V., Prakash B. GlmU (N-acetylglucosamine-1-phosphate uridyltransferase) bound to three magnesium ions and ATP at the active site. Acta Crystallogr. F Struct. Biol. Commun. 2014;70:703–708. doi: 10.1107/S2053230X14008279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.