Abstract
The human zinc transporter ZnT8 (SLC30A8) is expressed primarily in pancreatic β-cells and plays a key function in maintaining the concentration of blood glucose through its role in insulin storage, maturation and secretion. ZnT8 is an autoantigen for Type 1 diabetes (T1D) and is associated with Type 2 diabetes (T2D) through its risk allele that encodes a major non-synonymous single nucleotide polymorphism (SNP) at Arg325. Loss of function mutations improve insulin secretion and are protective against diabetes. Despite its role in diabetes and concomitant potential as a drug target, little is known about the structure or mechanism of ZnT8. To this end, we expressed ZnT8 in Pichia pastoris yeast and Sf9 insect cells. Guided by a rational screen of 96 detergents, we developed a method to solubilize and purify recombinant ZnT8. An in vivo transport assay in Pichia and a liposome-based uptake assay for insect-cell derived ZnT8 showed that the protein is functionally active in both systems. No significant difference in activity was observed between full-length ZnT8 (ZnT8A) and the amino-terminally truncated ZnT8B isoform. A fluorescence-based in vitro transport assay using proteoliposomes indicated that human ZnT8 functions as a Zn2+/H+ antiporter. We also purified E. coli-expressed amino- and carboxy-terminal cytoplasmic domains of ZnT8A. Circular dichroism spectrometry suggested that the amino-terminal domain contains predominantly α-helical structure, and indicated that the carboxy-terminal domain has a mixed α/β structure. Negative-stain electron microscopy and single-particle image analysis yielded a density map of ZnT8B at 20 Å resolution, which revealed that ZnT8 forms a dimer in detergent micelles. Two prominent lobes are ascribed to the transmembrane domains, and the molecular envelope recapitulates that of the bacterial zinc transporter YiiP. These results provide a foundation for higher resolution structural studies and screening experiments to identify compounds that modulate ZnT8 activity.
Keywords: Circular dichroism spectrometry, Electron microscopy, Membrane protein expression, Diabetes mellitus, Zinc transporter
Graphical abstract
Highlights
-
•
An in vivo viability assay and an in vitro proteoliposome transport assay show that recombinant human ZnT8 functions as a zinc antiporter.
-
•
Structural analyses reveal that the transmembrane and carboxy-terminal domains of ZnT8 recapitulate the structure of the bacterial homolog YiiP.
Abbreviations
- A280
absorbance at 280 nm
- AA
amino acids
- β-ME
β-mercaptoethanol
- CMC
critical micellar concentration
- ctZnT8
carboxy-terminal domain of ZnT8
- cv
column volume
- DFA
differential filtration assay
- DM
n-decyl-β-D-maltopyranoside
- E. coli
Escherichia coli
- EM
electron microscopy
- FC-12
fos-choline 12
- FSC
Fourier shell correlation
- IMAC
immobilized metal-affinity chromatography
- MALDI-MS
matrix-assisted laser desorption/ionization mass spectrometry
- MOI
multiplicity of infection
- ntZnT8
amino-terminal domain of ZnT8
- OD
optical density
- OG
n-octyl-β-D-glucopyranoside
- PCR
polymerase chain reaction
- PMSF
phenylmethylsulfonyl fluoride
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- Sf9
Spodoptera frugiperda
- SNP
single nucleotide polymorphism
- TEA
triethanolamine
- TEV
tobacco etch virus
- T1D
Type 1 diabetes
- T2D
Type 2 diabetes
- Tm
melting temperature
- TM
transmembrane
- TRX
thioredoxin
- ZnT8A
full-length, 40 kDa ZnT8
- ZnT8B
amino-terminal truncated, 35 kDa ZnT8
1. Introduction
The human pancreatic β-cell integral membrane protein ZnT8 (also known as SLC30A8) is a major autoantigen for Type I diabetes mellitus (T1D) (Wenzlau et al., 2008; Howson et al., 2012). ZnT8 autoantibody levels are predictive biomarkers for T1D (Wenzlau et al., 2007, 2011). The importance of ZnT8 in the etiology of Type II diabetes mellitus (T2D) became clear when the ZnT8 gene SLC30A8 was first associated with a novel risk locus for T2D, in which the C-allele encodes a major nonsynonymous SNP at Arg325 (Saxena et al., 2007; Scott et al., 2007; Sladek et al., 2007; Zeggini et al., 2007). One study showed a 24% increase in disease frequency with this risk allele (Sladek et al., 2007), which was present at a very high level (55–75%) in the populations studied. The estimated population risk of T2D attributable to this polymorphism is ~9% (Cauchi et al., 2010). In particular, this risk allele is associated with (i) an increase in the ratio of circulating proinsulin-to-insulin, (ii) a decrease in the response to insulin (Lichten and Cousins, 2009) and (iii) an imbalance of zinc between secretory vesicles and the cytoplasm (Mocchegiani et al., 2008). Point mutations in the carboxy-terminal domain can increase the risk of T1D and T2D (Wenzlau et al., 2008). Paradoxically, frameshift or premature termination mutations cause a loss of function and decrease the risk of T2D (Flannick et al., 2014). The rare p.Arg138∗ nonsense allele is enriched in western Finland, and a large scale study of this population showed that carriers reproducibly exhibited increased glucose-stimulated insulin secretion (Dwivedi et al., 2019). Taken together, these results suggest that ZnT8 could be an important drug target in the treatment of diabetes (Flannick et al., 2014).
Zinc has a significant role in all processes of insulin trafficking - synthesis, storage and secretion - and may also be important in communication between α- and β-cells in the Islets of Langerhans (Wenzlau et al., 2008; Chimienti et al., 2005). ZnT8 is expressed almost exclusively in pancreatic islets, mainly restricted to membranes of insulin secretion granules in β-cells (Murgia et al., 2009). ZnT8 regulates transport of Zn2+ ions into the lumen of secretory granules in exchange for protons that are translocated by the vesicular ATPase proton pump (Chabosseau and Rutter, 2016). Within the lumen of insulin secretory granules, Zn2+ ions are chelated by hexamers of insulin to facilitate storage (Chimienti et al., 2005, 2006). Fluorescent dye transfer studies in proteoliposomes suggested that the activity of ZnT8 is dependent on the lipid environment, in particular by anionic phospholipids (Merriman et al., 2016). Interestingly, this work also showed that the wild-type Arg325 variant of ZnT8, which confers susceptibility to T2D, displayed higher Zn2+ transport activity than the Trp325 isoform.
Studies using transgenic mice have shown that ZnT8 plays a role in β-cell survival and the physiologic response to glucose (Petersen et al., 2011). As expected, ZnT8 knockout mice do not display zinc-insulin crystals within β-cell secretory granules; however, the animals displayed normal glucose homeostasis (Lemaire et al., 2009). Nevertheless, mice with a specific knock down of ZnT8 in islet cells display a more drastic phenotype, with abnormal β-cell morphology and impaired glucose homeostasis, which resembles that shown by human carriers of the ZnT8 risk allele (Wijesekara et al., 2010).
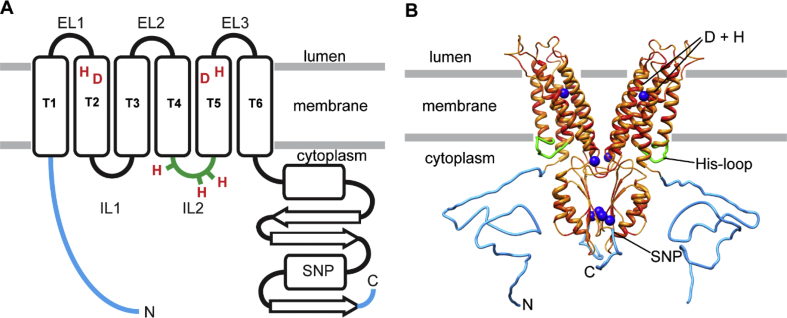
Hydropathy analysis suggests that the 40 kDa ZnT8 protein (369 amino acids) has a transmembrane (TM) bundle of six α-helices (Fig. 1 and Fig. S1) (Chimienti et al., 2004, 2006). It is notable that the ~8 kDa, acidic amino-terminal domain shows no similarity to any other protein sequence, and its function is unknown. A ~10 kDa acidic carboxy-terminal domain is predicted to have a mixed α-β fold (Fig. 1). An amino-terminal, truncated isoform of 35 kDa, designated as ZnT8B, has been found in vivo and is thought to be the result of translational initiation at a second Met codon at the end of the N-terminal domain (Davidson et al., 2014).
Fig. 1.
ZnT8 topology and structural models based on YiiP, a bacterial homolog. (A) Topology model of ZnT8 indicating positions of the R325W SNP, zinc-sensing His-rich loop (IL2), and pair of His (H) and Asp (D) residues in TM 2 and 5 that are conserved in mammalian ZnT proteins. Also shown are the 49 amino-terminal and ~12 carboxy-terminal residues not found in YiiP. (B) Homology model of ZnT8 showing identical residues in YiiP (red), conserved substitutions (dark orange), extended N- and C-termini of ZnT8 (light blue), and zinc atoms observed in YiiP (dark blue spheres). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The bacterial homolog, YiiP, has 17% sequence identity over 290 of 369 amino acids in ZnT8, and has provided the only available structures of a zinc transporter (Lemaire et al., 2009; Fu and Lu, 2007; Coudray et al., 2013; Lu et al., 2009). YiiP crystallized as a dimer in a detergent mixture of n-undecyl-β-D-maltoside (UDM) and fos-choline 12 (FC-12). The monomers are non-covalently associated via a charge ring of two Lys-Asp pairs, which are thought to form a pivot for transmitting structural changes from the cytoplasmic to the TM domains (Lu and Fu, 2007). Whereas the X-ray structure represents an outward-facing conformation (Lu et al., 2009), the structures determined by electron microscopy (EM) of YiiP incorporated into helical, lipid nanotubes were consistent with an inward-facing conformation (Coudray et al., 2013; Lopez-Redondo et al., 2018). The moderate 13 Å resolution cryoEM map helped explain the proposed alternating access mechanism for Zn2+ transport (Coudray et al., 2013). A subsequent structure at 4.1 Å resolution identified the interface between membrane domains as a strong determinant of dimerization. This could provide a stable scaffold for “rocking and twisting movements” of the four-helix bundle that is required for alternating access of Zn2+ to opposite sides of the membrane. The study also found that the characteristic splaying of TM domains observed in the X-ray structure was not necessary for transport activity, thus questioning the physiological relevance of that structure (Lopez-Redondo et al., 2018).
It is important to note that ZnT8 has several unique features not present in YiiP: (i) An additional 49 amino-terminal residues of unknown fold; (ii) A carboxy-tail extension of ~12 residues, which contains three cysteines; (iii) A cytoplasmic His-rich loop; and (iv) A predicted Zn2+ binding site composed of two paired His and Asp residues in TM domains 2 and 5. Regarding this last feature, YiiP has an Asp/Asp-Asp/His motif for binding Zn2+ ions, whereas the homologous site in ZnT8 bears a His/Asp-His/Asp motif (Fig. 1). These two pairs of amino acids form one of the zinc binding sites, mediating Zn2+ transport and selecting against Cd2+ (Hoch et al., 2012). The His-rich loop is essential for Zn2+ transport (Suzuki et al., 2005; Kawachi et al., 2008), and a working model suggests that this loop cooperates with the TM and the cytoplasmic C-terminal domains to function as a zinc sensor and regulate Zn2+ transport activity (Fukada and Kambe, 2011).
In addition to YiiP, a cryoEM structure is available for bacterial ZntB, which belongs to the CorA metal ion transporter (MIT) family (Gati et al., 2017). ZntB transports Zn2+, Cd2+ and Ni2+ across the membrane in a pH gradient-dependent manner, indicating that this protein is an importer of Zn2+, whereas YiiP is a Zn2+ antiporter. In addition, the oligomeric state and architecture of ZntB are radically different from YiiP, which is a homodimer, with each monomer comprised of a bundle of six TM α-helices and a smaller cytoplasmic domain. In contrast, ZntB assembles as a homopentamer, and each monomer is comprised of two TM α-helices and a larger, elongated cytoplasmic domain.
To our knowledge, no structure of a human zinc transporter has been determined. We therefore overexpressed and purified recombinant ZnT8 from the yeast Pichia pastoris and Sf9 insect cells, which are cost-efficient alternatives to mammalian expression systems. An in vivo assay in yeast cells showed that recombinant ZnT8 enhances zinc efflux from yeast cells. In addition, liposomes reconstituted with purified ZnT8 recapitulated the Zn2+/H+ antiport activity seen for YiiP and other ZnT proteins (Kambe, 2015). CD spectroscopy suggested that the amino-terminal domain contains predominantly α-helical structure, while the carboxy-terminal domain has a mixed α-helical and β-sheet structure. Biochemical analysis indicated that purified ZnT8 is dimeric when solubilized in detergent micelles, which was confirmed by EM. Single-particle image analysis showed that the overall molecular boundary of ZnT8 was similar to that of YiiP.
2. Materials and methods
2.1. Gene synthesis and cloning
2.1.1. Escherichia coli
The amino- and carboxy-terminal domains of ZnT8A (accession number NM_173851.3) were fused to the carboxy terminus of bacterial thioredoxin (TRX) using the pET32 Escherichia coli expression vector (Novagen, Madison, WI). Synthetic genes corresponding to the amino-terminal 73 amino acids or the carboxy-terminal 95 amino acids of full-length ZnT8 were generated (GenScript) and then subcloned into the vector between the NcoI and NotI restriction sites. A construct of TRX alone was generated using QuickChange PCR mutagenesis (Agilent, Santa Clara, CA) to insert a transcriptional stop codon sequence at the NcoI restriction site of pET32. These constructs were then transformed into BL21(DE3) E. coli for protein overexpression. All constructs were verified by DNA sequencing (GENEWIZ, Frederick, MD) from both the 5′ and 3’ ends of the insert.
2.1.2. Pichia pastoris
ZnT8 gene constructs (Fig. S1) were synthesized (GenScript) either as the full-length protein ZnT8A or based on the transcriptional variant ZnT8B in which an internal Met codon (Met50) forms the translational start site (Davidson et al., 2014). PpZnT8Bopt1 was constructed to have a carboxy-terminal G3H6 sequence with a proprietary Pichia pastoris-optimized codon bias (GenScript, Piscataway, NJ). PpZnT8Bopt2 was constructed with a proprietary P. pastoris-optimized codon bias (GenScript) and was designed to include a carboxy-terminal G3-tev-G3-H10-G3-FLAG sequence adding a tobacco etch virus (TEV) protease-cleavable decahistidine and FLAG tag. PpZnT8Bopt2 was also designed with a membrane protein-optimized P. pastoris codon bias (Bai et al., 2011). These genes were separately cloned into the pPICZ vector (Invitrogen) and transformed into P. pastoris strain KM71H (Invitrogen) by electroporation (Lin-Cereghino et al., 2005).
2.1.3. Spodoptera frugiperda (Sf9)
A ZnT8A gene construct was designed with a carboxy-terminal G3-TEV-H10 sequence adding a TEV protease-cleavable decahistidine tag. A ZnT8B gene construct was designed with a carboxy-terminal G3-H6 tag. Both genes were codon-optimized for expression in Sf9 insect cells (GenScript). These genes were cloned separately into pVL1393 donor plasmids (AB Vector), which were then used to generate virus expressing recombinant protein via a commercial baculovirus expression system (BD BaculoGold, BD Biosciences). Constructs were verified by DNA sequencing from both the 5′ and 3’ ends of the insert.
2.2. Protein expression
2.2.1. Escherichia coli
Expression of TRX and fusions with the amino- and carboxy-terminal domains of ZnT8A in E. coli was performed using an auto-induction method (Studier, 2005). Cells were initially grown in 2× Luria Broth media (2% tryptone, 1% yeast extract, 1% NaCl (pH 7.2)) with shaking at 37 °C to an optical density at 600 nm (OD600nm) of ~2, then diluted 1/100 into 1 L of ZYM-5052 media in 2.8 L baffled Fernbach flasks. The cell culture was shaken continuously, first at 37 °C for 2 h and then at 25 °C for 20–24 h. Cells were recovered by centrifugation at 5000×g for 10 min at 4 °C. The resulting cell pellet was frozen and stored at −80 °C.
2.2.2. Pichia pastoris
A fresh culture was started by incubation of an isolated yeast colony in 10 ml of BMGY medium (1% yeast extract, 2% peptone, 1.34% yeast nitrogen base, 100 mM potassium phosphate (pH 6.0), 4 × 10−5 % biotin, and 1% glycerol) overnight at 30 °C with shaking at 275 rpm. A larger volume of BMGY medium (100–200 ml) was inoculated with a 1/100 volume of the starting culture. The second culture was incubated for ~12 h at 30 °C with shaking at 250 rpm. Cells were harvested at an OD600nm of 1–4 and pelleted by centrifugation for 5 min at 1500×g. Induction of protein expression was initiated by resuspending the yeast cells in BMMY medium (1% yeast extract, 2% peptone, 1.34% yeast nitrogen base, 100 mM potassium phosphate (pH 6.0), 4 × 10−5 % biotin, and 1% methanol) to an OD600nm of 1–2 in a volume of 0.1–1.0 L. Incubation was continued at 28 °C with shaking at 175 rpm in baffled culture flasks for 24 h. Following induction, cells to be used in assays of protein function were stored at 4 °C and used within four days. Cells for protein purification were pelleted by centrifugation at 4000×g for 15 min at 4 °C. The cell pellet was then frozen and stored at −80 °C.
2.2.3. Spodoptera frugiperda
Insect cells were infected with baculovirus at a multiplicity of infection (MOI) of 5 and incubated at 27 °C with shaking at 150 rpm in ESF921 media (Expression Systems, Davis, CA). Cells were harvested 48–58 h post infection and pelleted by centrifugation for 15 min at 4000×g The cell pellet was washed with phosphate-buffered saline (PBS), and the washed cells were pelleted by centrifugation at 3000×g for 10 min. The cell pellet was then frozen and stored at −80 °C.
2.3. Protein purification
2.3.1. Amino- and carboxy-terminal ZnT8A domains from E. coli
Cells were resuspended in Lysis Buffer E (50 mM HEPES pH 7.5, 250 mM NaCl, 5 mM benzamidine, 20 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF)) and then disrupted by passage 3–4 times through an 87 μm interaction chamber at 25,000 PSI using a model M-110P microfluidizer (Microfluidics, Westwood, MA). Cell debris was removed by centrifugation of the lysate at 25,000×g for 15 min at 4 °C. The clarified solution was mixed with Talon metal affinity resin (Clontech, Mountain View, CA) by gentle stirring for 1–2 h at 4 °C. The resin was pelleted by centrifugation at 500×g for 10 min at 4 °C and transferred into a chromatography column. The column was washed six times with 10 column volumes (cv) of Lysis Buffer E supplemented with 5 mM β-mercaptoethanol (β-ME) and washed three times with 10 cv of Wash Buffer E (50 mM HEPES pH 7.5, 250 mM NaCl, 20 mM imidazole, 5 mM β-ME). The column was then eluted with four 1-cv aliquots of Elution Buffer E (25 mM HEPES pH 7.2, 100 mM NaCl, 250 mM imidazole, 5 mM β-ME). The eluates were pooled and transferred into a 10,000 MWCO Slide-A-Lyser (ThermoScientific) and dialyzed against Dialysis Buffer E (25 mM Bis-Tris pH 6.5, 100 mM K2SO4, 3 mM sodium azide, 10 mM β-ME) overnight at 4 °C. The dialysis buffer was then replaced with the same volume of fresh buffer, and dialysis was continued for another day. The dialysate was recovered and concentrated to 1 mL using a 10,000 MWCO VivaSpin15R concentrator (Vivaproducts, Littleton, MA) by centrifugation at 1000×g at 4 °C. This concentrate was applied to a HiLoad 16/600 Superdex 75 chromatography column (GE Healthcare, Marlborough, MA). Following equilibration, chromatography was performed at a flow rate of 1.0 mL/min with Dialysis Buffer E at 4 °C, and 2.0 mL fractions were collected. Fractions containing purified protein were identified by SDS-PAGE and stored at 4 °C.
To cleave TRX from the fusion proteins, aliquots of TRX-ntZnT8 or TRX-ctZnT8 were supplemented with CaSO4 to 2 mM. Recombinant bovine enterokinase (GenScript) was added following the manufacturer's recommendations and incubated for 20 h at room temperature. The samples were then applied to a 7.8 × 300 mm Zenix SRT-150 chromatography column (Sepax Technologies, Newark, DE) and equilibrated. Chromatography was performed at 0.5 mL/min with Dialysis Buffer E at 4 °C, and 0.25 mL fractions were collected. The ntZnT8A moiety co-migrated with TRX under these conditions. Therefore, we tested a Superdex 75 Increase column (GE Heathcare) with and without pretreatment using 10 mM Tris(2-carboxyethyl)phosphine hydrochloride, which yielded the same results. Consequently, full-length TRX-ntZnT8A was used for our studies. Fractions containing purified TRX-ntZnT8 or ctZnT8 were concentrated using a 5000 MWCO VivaSpin500 concentrator (VivaProducts) by centrifugation at 10,000×g at 4 °C. For use as an experimental control, TRX was purified, cleaved, and isolated following the same method.
2.3.2. ZnT8B from P. pastoris
Yeast cells were resuspended in Lysis Buffer (50 mM Na-PO4 pH 7.0, 500 mM NaCl, 10% glycerol (v/v), 1 mM PMSF) and then disrupted by injecting 7–10 times at 30,000 PSI through an 87 μm interaction chamber, using a model M-110P microfluidizer (Microfluidics). Cell debris was removed by centrifugation of the lysate at 4500×g for 15 min at 4 °C. Membranes were then harvested by centrifugation of the supernatant at 185,000×g for 45 min at 4 °C. The membrane pellet was resuspended in Lysis Buffer using a glass Dounce homogenizer, and centrifugation was repeated as before. The washed membranes were then resuspended in Extraction Buffer (50 mM Na-PO4 pH 7.0, 500 mM NaCl, 10% glycerol (v/v), 1 mM PMSF, 20 mM imidazole) using 15 pestle strokes in a glass Dounce homogenizer, and Triton X-100 was added to obtain a final detergent concentration of 4%. Four detergents were initially tested for their ability to solubilize yeast membrane proteins (Table S1). Triton X-100 was selected based on efficiency of solubilization and cost. The membranes were incubated in Extraction Buffer containing 4% Triton X-100 for 3 h at 4 °C. Insoluble material was then removed by centrifugation at 185,000×g for 45 min at 4 °C. The clarified solution was mixed with Talon IMAC resin (Clontech) by gentle stirring for 3 h at 4 °C. The resin was pelleted by centrifugation at 1000×g for 10 min at 4 °C, and the pellet was transferred into a chromatography column. The column was washed seven times with 10 cv of Wash Buffer (50 mM Na-PO4 pH 7.0, 500 mM NaCl, 10% glycerol (v/v), 1 mM PMSF, 20 mM imidazole, 0.14% fos-choline 12 (FC-12)) followed by elution with three cv of Elution Buffer (50 mM Na-PO4 pH 7.0, 500 mM NaCl, 10% glycerol (v/v), 1 mM PMSF, 500 mM imidazole, 0.14% FC-12). The eluates were pooled and injected into a 10,000 MWCO Slide-A-Lyser (ThermoScientific) for overnight dialysis at 4 °C against SEC/Dialysis Buffer (25 mM triethanolamine (TEA) pH 7.0, 500 mM NaCl, 5% glycerol (v/v), 1 mM PMSF, 0.14% FC-12). The dialysate was recovered and concentrated to ~1 ml by centrifugation at 225×g at 4 °C using a 50,000 MWCO VivaSpin4 concentrator (Vivaproducts). The concentrated solution was injected into a 7.8 × 300 mm SRT-C SEC300 SEC column (Sepax Technologies) that had been equilibrated at 4 °C in SEC/Dialysis Buffer. Chromatography was performed on an AKTA Explorer 10 (GE Healthcare) at 4 °C using a flow rate of 0.3 ml/min. Fractions (0.16 ml) containing purified ZnT8B were identified by SDS-PAGE and Western immunoblot analysis, and pooled fractions were stored at 4 °C. Blue native non-denaturing electrophoresis was performed as previously described (Heuberger et al., 2002). Protein concentrations were determined using the MicroBCA assay (Pierce Biotechnology, Rockford, IL).
2.3.3. ZnT8A and ZnT8B from Spodoptera frugiperda
Cell pellets were resuspended in 100 mL Low-Salt Buffer (50 mM TEA pH 7, 100 mM NaCl, 5% glycerol (v/v), 1 Complete protease inhibitor cocktail tablet without EDTA per 50 ml of lysate (Roche)). Cells were disrupted with 20–30 pestle strokes in a glass Dounce homogenizer. Nucleic acids were digested by adding MgCl2 to 2.5 mM and adding 12.5 units/mL of Benzonase (EMD Millipore), followed by 15 min of gentle stirring at 4 °C. Membranes were then harvested by centrifugation of the supernatant at 125,000×g for 30 min at 4 °C. The membrane pellet was washed twice with High-Salt Buffer (Low-Salt Buffer with NaCl increased to 500 mM). Membrane aliquots were flash frozen in liquid N2 and stored at -80 °C. When needed, membranes were thawed on ice and resuspended in Extraction Buffer (50 mM TEA pH 7, 500 mM NaCl, 5% glycerol (v/v), 20 mM imidazole) with 10–15 pestle strokes in a glass Dounce homogenizer. Membranes were solubilized by addition of n-decyl-β-D-maltopyranoside (DM) to a final detergent concentration of 1% (w/v) and gentle stirring for 3 h at 4 °C. Insoluble material was removed by centrifugation at 125,000×g for 45 min at 4 °C. His-tagged ZnT8 was purified from the clarified, solubilized membranes by Talon resin IMAC (Clontech). One ml of affinity resin was added per 50 ml detergent extract, and the suspension was stirred gently for 1 h at 4 °C. The resin was pelleted by centrifugation at 500×g for 5 min at 4 °C, and the resin was transferred into a 10 × 100 mm glass chromatography column. DM was exchanged for FC-12 on the column by washing with five column volumes (CVs) of Wash Buffer (50 mM TEA pH 7, 500 mM NaCl, 5% glycerol (v/v), 20 mM imidazole, 0.14% FC-12). During the fourth wash, the column was capped, kept stationary and equilibrated with the new detergent for 15 min at 4 °C. ZnT8 was eluted from the column with 2 CV of Elution buffer (50 mM TEA pH 7, 500 mM NaCl, 5% glycerol (v/v), 500 mM imidazole, 0.14% FC-12). The eluate was injected into a 10,000 MWCO Slide-A-Lyser (ThermoScientific) for overnight dialysis at 4 °C against SEC/Dialysis Buffer (25 mM TEA pH 7.0, 500 mM NaCl, 5% glycerol (v/v), 2 mM dithiothreitol (DTT), 0.14% FC-12). The dialysate was collected and concentrated to ~1 ml by centrifugation at 225×g at 4 °C using a 50,000 MWCO VivaSpin4 concentrator (VivaProducts). The concentrate was injected into a 7.8 × 300 mm SRT-C SEC300 SEC column (Sepax Technologies) that had been equilibrated at 4 °C in SEC/Dialysis buffer. Chromatography was performed with SEC/Dialysis Buffer on an AKTA Explorer 10 (GE Healthcare) at 4 °C using a flow rate of 0.3 ml/min. Fractions (0.16 ml) containing purified ZnT8B were identified by SDS-PAGE and Western immunoblot analysis, and pooled fractions were stored at 4 °C. Protein concentrations were determined by their molar absorptivity using a NanoDrop 1000 spectrophotometer (Thermo Scientific), assuming a molar absorption coefficient of 43,000 M−1cm−1 for ZnT8A and 40,600 M−1cm−1 for ZnT8B.
2.4. SDS-PAGE and Western immunoblot analysis
Protein aliquots were resuspended in SDS sample buffer (2% SDS, 50 mM Tris-HCl pH 7.5, 10% glycerol, 100 mM DTT), and incubated at 50 °C for 30 min. SDS-PAGE was performed using a 4–20% acrylamide gradient Tris-glycine gel (Bio-Rad) with Precision Plus (Bio-Rad) prestained protein molecular weight standards. Gel-separated proteins were then electroblotted onto a nitrocellulose membrane. Western immunoblot analysis (Harlow and Lane, 1988) was performed with ZnT8A polyclonal antibodies diluted 10,000-fold in Blocking Buffer (LI-COR, Lincoln, NE). Donkey anti-rabbit immunoglobulin G antiserum coupled to IRDye800 (LI-COR) was diluted 20,000-fold and used to label the primary antibodies. Detection of the bound fluorescent antibodies at a wavelength of 800 nm was accomplished using an Odyssey infrared scanner (LI-COR). Digitized images of the immunoblot results were corrected for background and contrast using Photoshop (Adobe Systems, San Jose, CA). Densitometry to quantify antibody labeling was performed using NIH-ImageJ (Rasband, 1997).
2.5. Detergent screening
An array of 96 non- and zwitter-ionic detergents was screened using a differential filtration assay (DFA; Selector Kit, Anatrace, Maumee, OH; Fig. S2) (Vergis et al., 2010). The estimated mass of the ZnT8B protein-detergent complex was 100–300 kDa, so that complexes that did not pass through the 300 kDa filter were an indication of protein aggregation. Western immuno-dot-blot analysis, performed as described above, identified 17 detergents in which the ZnT8B protein-detergent complexes passed through the 300 kDa cut-off filter (Table S2).
Solutions of each detergent were prepared in Wash Buffer with detergent concentrations 5–10 × the published critical micellar concentration (CMC) and in Elution Buffer with 1.5–3.0 × the CMC. (Bhairi, 2007; Anatrace, 2014). ZnT8B was bound to a Talon IMAC column in Wash Buffer with Triton X-100 as described above. The resin was washed with Wash Buffer, and the protein was eluted with Elution Buffer, in which both buffers contained each of the 17 detergents. The eluate was dialyzed as before, but with the new detergent at a concentration of 1.5–3.0 × CMC in SEC/Dialysis Buffer. ZnT8B samples adjusted to the same concentration in each of the 17 detergents were then examined by analytical size exclusion chromatography at 4 °C using a 5 × 150 mm Superdex 200 GL chromatography column (GE Healthcare) that had been equilibrated with SEC/Dialysis Buffer containing the same detergent used for elution from the IMAC column. Fractions collected at 0.3 ml/min were examined by Western immunoblot analysis to detect ZnT8B in the chromatogram. FC-12 and APX305 were the optimal detergents for solubilization, on the basis of having the highest OD280 nm ratio of the protein peak to the void peak in the SEC profile (Table S2, Fig. S2). Of these two, FC-12 was selected because of its lower cost, ready availability, and chemical homogeneity.
2.6. Fluorescent thermal stability assay
Thermal stability of ZnT8B in FC-12 was measured by the fluorescence of the cysteine-reactive dye 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM, Invitrogen) over a temperature gradient (10–95 °C). The ZnT8B construct has eight potentially free cysteines in the TM domain and five in the aqueous ectodomains. (As in the homologous YiiP X-ray structure, our homology model suggests that no cysteines are bonded.) With increasing temperature, ZnT8B unfolds, and the dye binds to the thiols and emits fluorescence.
An aliquot of CPM dye at 1 mM in dimethylformamide was diluted 1:75 into either a sample of protein at ~50 μg/ml or a buffer-only control. For these experiments, we used the SEC/Dialysis Buffer (25 mM TEA pH 7.0, 500 mM NaCl, 5% glycerol (v/v), 1 mM PMSF, 0.14% FC-12). Following a 15 min incubation on ice, the sample was transferred into a 1.5 mm pathlength quartz spectrofluorometer cuvette (Hellma Analytics, Müllheim, Germany), which was inserted into a FluoroMax-3 spectrofluorometer (Horiba Jobin-Yvon, Edison, NJ) equipped with a thermoelectric, temperature-controlled cuvette holder. The excitation and emission wavelengths were set to 387 nm and 463 nm, respectively. The sample was heated from 5 to 95 °C at a rate of 2 °C/min, and the fluorescence was recorded every 1 min. The fluorescence versus temperature profile was analyzed by non-linear regression using a Boltzmann sigmoidal equation. The sample melting temperature (Tm) was calculated from the inflection point of the resulting melting curve as previously described (Alexandrov et al., 2008).
2.7. Mass spectrometry
An aliquot of protein was diluted 1:10 with 0.1% trifluoroacetic acid and spotted on a stainless-steel target plate with an equal volume of 15 mg/ml sinapinic acid dissolved in 50% acetonitrile, 0.1% trifluoroacetic acid in water. MALDI-MS was performed using a MicroFlex LT (Bruker, Billerica, MA) calibrated with high molecular weight protein standards. The sample was shot at least 100 times in positive linear mode.
2.8. Circular dichroism spectrometry
Samples were prepared in 0.1 and 1.0 mm strain-free quartz cuvettes (New Era, Vineland, NJ) with a buffer of 2.5 mM HEPES-H2SO4 pH 6.5, 10 mM K2SO4, 1.0 mM β-ME and 0.3 mM sodium azide. Data were recorded at 10 °C on a Model 410 circular dichroism spectrometer (Aviv Biomedical, Lakewood, NJ) using a spectral bandwidth of 1.0 nm, a scanning step of 0.5 nm, and 4–8 s data averaging per step. Data requiring a photomultiplier voltage greater than 600 VDC was discarded since the signal is typically poor when collected at high voltages (Greenfield, 2006). Three to five scans were averaged to generate the final spectra, after which the buffer-only spectra were subtracted from the protein solution spectra. Protein concentrations were determined by UV–Vis spectrometry, using the predicted molar extinction coefficients (Pace et al., 1995) of 26,510 M−1cm−1 for TRX-ntZnT8, 7155 M−1cm−1 for ctZnT8, and 13,980 M−1cm−1 for thioredoxin. To compensate for cuvette path-length errors, data collected using a 0.1 mm cuvette were scaled to fit the data collected using a 1.0 mm cuvette before merging the two spectra. Spectrometer calibration and structural analyses were performed as previously described (Daniels and Yeager, 2019).
2.9. Electron microscopy
For EM of negatively-stained samples, a 3.0 μl drop of the FC-12 buffer containing ZnT8 at ~1 μg/ml was incubated on a glow-discharged, carbon-coated copper grid for 60 s. The grid was then washed with three drops of deionized water and then stained with a drop of freshly prepared 0.75% uranyl formate for 60 s (Ohi et al., 2004). Sample, wash drops and the stain were blotted with filter paper. EM was performed using a Tecnai G2 F20 electron microscope (FEI, Hillsboro, OR) equipped with a field-emission electron source, operating at 120 keV. Low electron dose images (~15 e−/Å2) were recorded on a 4 K × 4 K pixel CCD camera (Gatan, Pleasanton, CA) at 62,000× magnification (corresponding to 1.8 Å/pixel) with a 1 s exposure time and an underfocus value of ~1.5 μm.
2.10. Single-particle image analysis
The initial steps of image processing were performed using EMAN v2.05 software (Tang et al., 2007). To facilitate particle picking, the contrast of the micrographs was enhanced by high- and low-pass Gaussian filtering at 100 and 10 Å, respectively. A total of 3850 particles were picked manually using a square mask with an edge dimension of 168 Å. The image and particle coordinate files in .mrc and .box format, respectively, were imported into RELION v1.3 (Scheres, 2012). Corrections for effects of the contrast transfer function were performed using CTFFIND4 (Mindell and Grigorieff, 2003). All picked particles were screened visually using the Z-score, and ‘bad’ particles were removed interactively. The remaining particles were then sorted by reference-free 2D classification into 25 classes. We removed particles that belonged to outlier classes with either very few particles per class or signs of protein aggregation, giving a final dataset of ~3500 particles in 23 classes. Details of the 3D reconstruction are described in section 3.2.3. The resolutions of the maps in (Fig. S9B, C, E and F) were calculated using the FSC method in which the data sets were divided into two halves and refined independently.
2.11. Molecular modeling
To further interpret the 3D map of ZnT8, we generated a homology model of ZnT8 based on the X-ray structure of YiiP (Lemaire et al., 2009). Several excellent applications exist for this purpose as outlined in Dolan et al. (2012), and in line with their recommendations we chose the Modeller software package (Yang et al., 2012) for its user-friendliness. This homology model was then energy minimized and docked into the 3D EM reconstruction using the rigid fit tool in UCSF Chimera (Yang et al., 2012). Protein disorder calculations were performed using the MetaDisorder software package (Kozlowski and Bujnicki, 2012).
2.12. Functional assays
2.12.1. In vivo zinc uptake assay
P. pastoris transformed with pPICZ alone, ZnT8Bopt1/pPICZ, or ZnT8Bopt2/pPICZ were grown at 30 °C in BMGY media (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base without amino acids, 0.4 μg/ml biotin and 1% glycerol) to an OD600nm of 1–2, then diluted 1/100 in BMMY media (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base without amino acids, 0.4 μg/ml biotin and 1% methanol) and grown at 28 °C for 40–42 h with vigorous aeration in baffled flasks. An aliquot of each culture was then diluted to give an OD600nm of 10 in 25 ml BMMY media and transferred into 250 ml baffled flasks, which were then incubated either on ice or at 23 °C with gentle shaking. 65ZnCl2 (PerkinElmer, Boston, MA) was added to a concentration of 10 μM, after which 1.0 ml aliquots were collected at 15–30 min intervals. For each aliquot, cells were pelleted by centrifugation at 2700×g, and the supernatant was discarded. Each cell pellet was washed with 1.0 ml BMMY media and re-pelleted as above. Each cell pellet was then resuspended in 150 μL BMMY media and transferred into a 25 mL glass scintillation vial. A 20 ml aliquot of Ultima Gold liquid scintillation cocktail (PerkinElmer) was added to each vial, and 65Zn decay was measured using a model LS 6000SC scintillation counter (Beckman Instruments, Fullerton, CA). The experiments were replicated four times from three unique preparations of cells. The counting results for the four replicates were averaged and scaled so that the maximum signal from pPICZ-only control cells at the final time point was equal in each experiment.
2.12.2. In vitro proteoliposome-based transport assay
The lipid for protein reconstitution was prepared by mixing chloroform stocks of phosphatidylcholine (POPC) and phosphatidylserine (POPS) at a 2:1 mass ratio (Avanti Polar Lipids, Alabaster, AL). The lipid solutions were lyophilized for 2 h using a stream of dry argon gas followed by vacuum desiccation for 2 h. The dried lipids were rehydrated at room temperature in Lipid Buffer (25 mM HEPES pH 7.5, 100 mM KCl and 2.3% (w/v) n-octyl-β-D-glucopyranoside (OG)) to a final concentration of 10 mg/ml.
Aliquots of detergent-solubilized lipid were mixed with detergent-solubilized, IMAC-purified protein in a 1:3000 protein:lipid molar ratio, and the fluorescent Zn2+ indicator dye fluozin-1 (Life Technologies, Grand Island, NY) was added to a final concentration of 20 μM. The sample was incubated in the dark for 1 h at room temperature. Control, protein-free liposomes were prepared using the same procedure. An Econo-Pac 10DG desalting column (Bio-Rad) was equilibrated with Sample Buffer containing either 25 mM HEPES pH 7.5 and 100 mM KCl or 25 mM MES pH 6.0 and 100 mM KCl. Shortly before purification on the column, the liposome sample was diluted with Sample Buffer sufficient to produce an OG concentration of 0.4%, thus bringing the detergent concentration below its critical micellar concentration (0.53%). A 200 μl aliquot of 20 μM Fluozin-1 in Sample Buffer was applied to the desalting column, followed by 1 ml of liposome sample and 2 ml of Sample Buffer. Elution of the resulting liposomes was performed by applying an additional 1.5 ml of Sample Buffer to the column and collecting 500 μl fractions. The first fraction was discarded, and the two subsequent fractions were pooled and stored in the dark at room temperature.
Purified liposomes were mixed with 3 vol of Sample Buffer at either pH 7.5 or 6.0. The sample was transferred into a quartz cuvette (Hellma Analytics, Mullheim, Germany), which was then inserted into the temperature-controlled holder of a spectrofluorometer (Fluorolog-3, Horiba Scientific, Edison, NJ), with excitation and emission wavelengths set to 480 nm and 520 nm, respectively. The sample was constantly mixed using a magnetic stir bar, and after 1 min the extra-liposomal concentration of free Zn2+ was increased to 100 μM by the addition of ZnCl2. After 5 min the liposomes were solubilized by the addition of OG to 1% (w/v). Spectra were recorded and analyzed using the DataMax software package. All experiments were performed at 22 °C.
3. Results and discussion
To gain insight into the structure and transport mechanism of ZnT8, we developed in vivo and in vitro transport assays, performed physicochemical studies and determined a low-resolution EM structure that provide a foundation for higher-resolution structural studies and potential drug discovery efforts. We initially focused our efforts on the amino-terminal truncated isoform ZnT8B since the amino-terminal domain was predicted to be disordered (Fig. S3).
3.1. Functional studies: an in vivo assay in P. pastoris and an in vitro proteoliposome transport assay show that recombinant human ZnT8 functions as a zinc antiporter
3.1.1. In vivo transport assay shows that ZnT8 from P. pastoris is functionally active
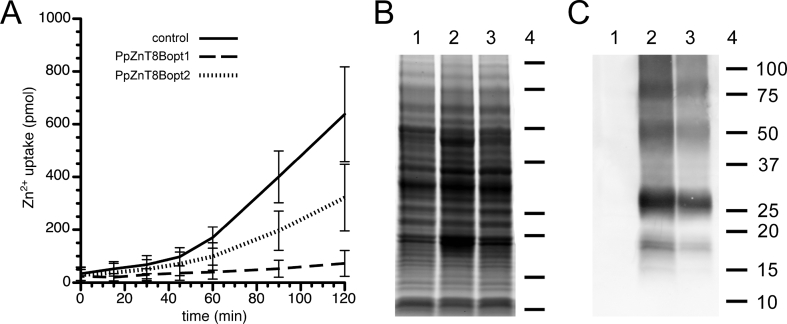
A zinc uptake assay in P. pastoris showed that yeast overexpressing ZnT8B constructs with different codon bias (designated PpZnT8Bopt1 and PpZnT8Bopt2) exhibited a significant reduction in zinc uptake compared with control yeast (Fig. 2A). The cell line producing the most ZnT8 (PpZnT8Bopt1) almost completely excluded labeled zinc (Fig. 2A, dashed line), while the cell line producing ~40% less ZnT8 (PpZnT8Bopt2) (Fig. 2C) accumulated half as much 65Zn2+ as mock-transformed control yeast (Fig. 2A, dotted line versus solid line). Although the opt2 construct was generated with a membrane protein-optimized P. pastoris codon bias (Bai et al., 2011), it showed no significant improvement on protein expression levels.
Fig. 2.
P. pastoris expressing ZnT8B exclude extracellular Zn2+in proportion to expression level. (A) Yeast overexpressing PpZnT8Bopt1 (dashed line), PpZnT8Bopt2 (dotted line), or expression vector alone (solid line) were supplemented with 10 μM 65ZnCl2, and aliquots were taken at time points thereafter. Uptake of 65Zn2+ into the cells was measured using a scintillation counter. Results show the average of seven replicates, and error bars indicate ±1 SD. SDS-PAGE of membrane-enriched protein fractions shown as a Coomassie stained gel (B) or Western immunoblot using ZnT8 antibodies (C). Extracts are from yeast expressing the expression vector alone (lane 1), PpZnT8Bopt1 (lane 2), or PpZnT8Bopt2 (lane 3). Similarity in protein staining profiles (B) indicates comparable loading of protein, while the corresponding immunoblot (C) shows a 40% reduction of ZnT8B in PpZnT8Bopt2 compared to PpZnT8Bopt1. Molecular weight markers indicated in lane 4, with weight in kiloDaltons (kDa) shown to the right of lane 4 in panel C.
Zinc uptake was unidirectional: once accumulated, cell-associated 65Zn2+ was not observed to decrease following incubation in media lacking Zn2+. Since P. stipitis and Saccharomyces cerevisiae yeast generally have a cytoplasmic pH that is one pH unit higher than the media (Lohmeier-Vogel et al., 1996), the resulting proton gradient would greatly favor zinc export from cells overexpressing a Zn2+/H+ antiporter such as ZnT8. Supporting this idea, zinc uptake decreased in more acidic media, as the proton gradient disfavored zinc uptake (Table 1). We also noted that cadmium competed with zinc for uptake through the ZnT8 transporter (Table 1). Yeast expressing PpZnT8Bopt1 showed less zinc uptake than yeast expressing PpZnT8Bopt2, which is likely because of a ~40% reduction in ZnT8 accumulation by PpZnT8Bopt2-expressing cells (Fig. 2C). Cell densities of the three cultures remained comparable during the experiment, with the difference less than 20% at the end of the 2-hr assay period.
Table 1.
Rates of zinc ion uptake in transformed P. pastoris cell cultures. Rates derived from linear domain of zinc uptake assay, from 1 h timepoint on. Units are in pmol/minute. Values are the average of at least three replicates and are shown with the standard error.
| MMY (unbuffered) |
BMMY |
||||
|---|---|---|---|---|---|
| pH 4.0 | pH 5.0 | pH 6.0 | pH 6.0 + Cd2+ | pH 6.0 | |
| vector only | 3.9 ± 0.7 | 6.2 ± 1.5 | 5.6 ± 0.1 | 2.7 ± 0.3 | 7.3 ± 0.6 |
| PpZnT8Bopt1 | 0.3 ± 0.2 | 0.6 ± 0.3 | 1.6 ± 0.4 | 0.4 ± 0.1 | 0.5 ± 0.2 |
| PpZnT8Bopt2 | 2.8 ± 0.5 | 4.3 ± 1.2 | 4.4 ± 1.1 | 1.4 ± 0.2 | 3.4 ± 0.5 |
The yeast cell wall can adsorb zinc (Ross et al., 1994), which could result in spuriously elevated levels of zinc uptake. To address this, we repeated our Zn2+ uptake assay with yeast cultures incubated on ice. A slight amount of zinc immediately became associated with the cells, but this amount did not change over time and was similar for the three cell lines tested (Fig. S4). Since this background signal did not change over time, its effect on our assay results is negligible. Taken together, these results suggest that any growth-related increase in the amount of zinc-adsorbing cell wall was insignificant compared to the effects of ZnT8 expression. Although we did not characterize the membrane(s) in which overexpressed ZnT8 is localized, sufficient protein should have been trafficked to the plasma membrane for our assay to show that ZnT8 is active. For instance, we found that even vacuole-specific membrane protein overexpressed in P. pastoris will be found in the plasma membrane (Daniels et al., 2006).
3.1.2. In vitro transport assay shows that ZnT8 from S. frugiperda is functionally active
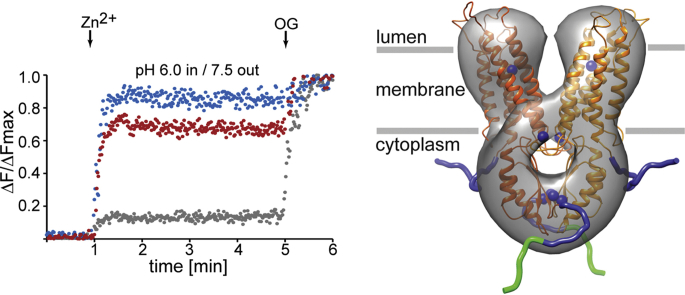
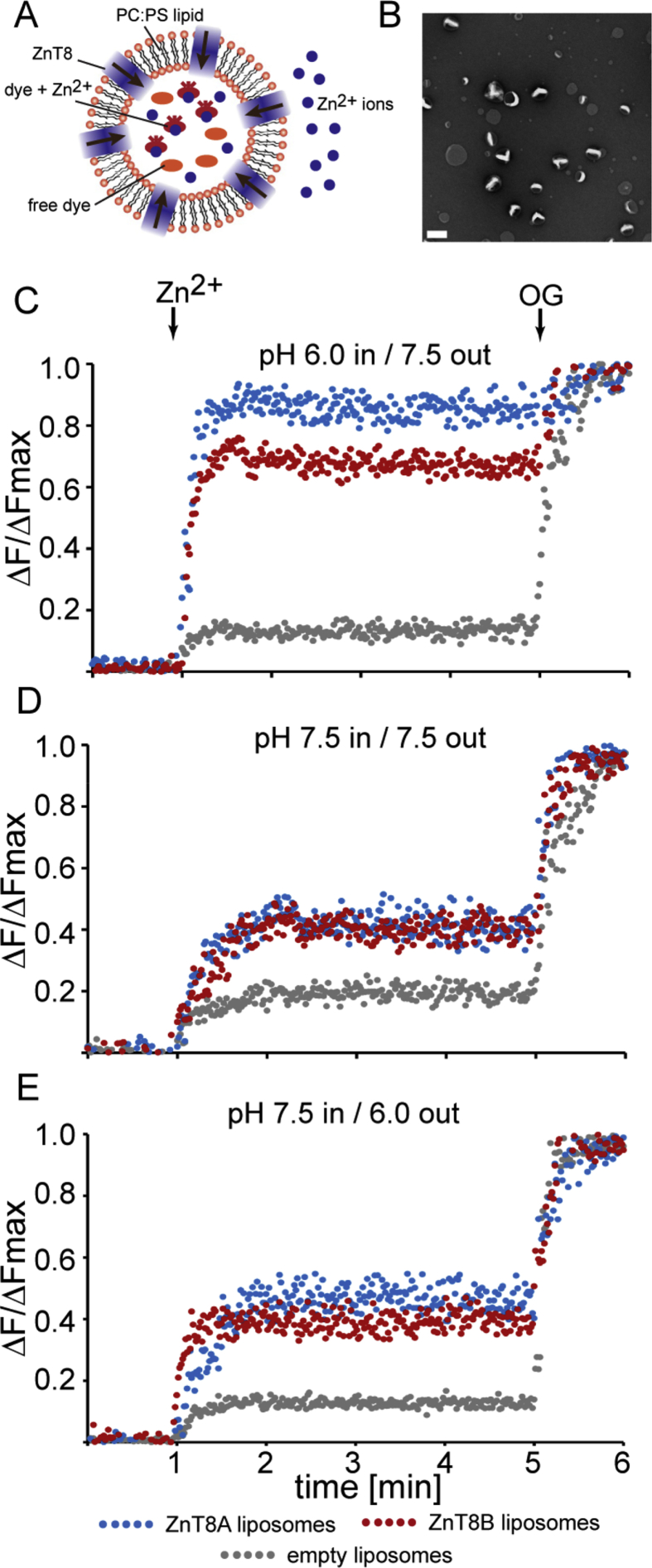
Proteoliposomes were formed by using size-exclusion chromatography to rapidly remove the solubilizing detergent from a ZnT8-lipid-detergent solution. The chromatography buffer included the zinc fluorophore fluozin-1, which was retained within the proteoliposomes as they formed (Fig. 3A). EM showed that negatively-stained proteoliposomes were primarily unilamellar and uniform in appearance with a diameter ranging from 70 to 120 nm (Fig. 3B). To measure zinc uptake, the liposomes were maintained with a TM proton gradient in which the internal pH was 6.0 and the external buffer pH was 7.5. ZnCl2 was added to 100 μM, and the change in fluozin-1 fluorescence over time was measured. At the conclusion of the assay, OG was added to a final concentration of 1%, which solubilized the liposomes and exposed the fluozin-1 dye to the zinc in solution, which enabled measurement of a reference maximum fluorescence. Compared with control liposomes, ZnT8-containing liposomes displayed a significant increase in fluorescence in the presence of Zn2+ (Fig. 3C). When the pH gradient was eliminated, the fluorescence change was reduced (Fig. 3D), which was also observed when the pH gradient was reversed (Fig. 3E), supporting our observation that Zn2+ uptake via ZnT8 was driven by H+ efflux resulting from the pH gradient across the liposome membrane. Since ionic metals do not have a membrane-permeable neutral form, any increase in liposome fluorescence results from ZnT8-mediated zinc transport. The increase in fluorescence observed for control, protein-free liposomes is possibly due to either adsorption of fluozin-1 to the lipid layer or residual dye in the buffer.
Fig. 3.
In vitro liposome assay indicates that purified ZnT8A and ZnT8B transport zinc. (A) A cartoon model representing the principle of the assay. (B) An electron micrograph of negatively-stained ZnT8 proteoliposomes; white bar denotes 200 nm. (C) Zinc transport with an internal liposome pH of 6.0 and an external buffer pH of 7.5. (D) Transport of Zn2+ by reconstituted ZnT8 with both liposomal and external pH of 7.5, (E) Transport of Zn2+ with an internal liposome pH of 7.5 and an external buffer pH of 6.0. Changes in fluozin-1 fluorescence upon Zn2+ and subsequent β-octyl glucoside (OG) addition (arrows), using either ZnT8A (blue trace) or ZnT8B proteoliposomes (red trace), or protein-free liposomes (grey trace). ΔF/ΔFmax is the relative change in fluorescence normalized to the maximum fluorescence following proteoliposome dissolution by OG. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
These in vitro results are in agreement with the prevailing notion that ZnT8 operates as a Zn2+/H+ antiporter, similar to the bacterial homolog YiiP. In addition, the results are consistent with the inward proton gradient between the insulin secretory vesicle lumen and the β-cell cytoplasm, which is a necessary component for ZnT8-mediated zinc loading of these vesicles (Chabosseau and Rutter, 2016).
3.1.3. Implications for the mechanism for zinc transport
A necessary component for zinc/proton antiport is a proton gradient across the cell membrane. We have not characterized the driving force for zinc ion export from the cell cytoplasm by ZnT8 in the P. pastoris plasma membrane, but we hypothesize that this yeast has the requisite proton gradient. Aerobically cultured yeast have a cytoplasmic pH of ~7 when grown in media buffered to pH 6 (Lohmeier-Vogel et al., 1996; Haworth and Fliegel, 1993), which would suggest that a favorable proton gradient is present. Rapid zinc uptake by P. pastoris is likely since S. cerevisiae and other yeast have a zinc import system comprised primarily of the Zrt1 high-affinity and the Zrt2 low-affinity zinc transporters (Zhao and Eide, 1996), which are both members of the taxonomically widespread ZIP protein family (Kambe et al., 2006). The mechanism of Zn2+ transport in this family is unknown, but may be via HCO3+ symport as described for human ZIP2 (Gaither and Eide, 2001).
3.2. Structural studies: ZnT8 is a homodimer, with transmembrane and carboxy-terminal domains that recapitulate the structure of the bacterial homolog YiiP
3.2.1. ZnT8B forms a stable dimer when purified in fos-choline 12
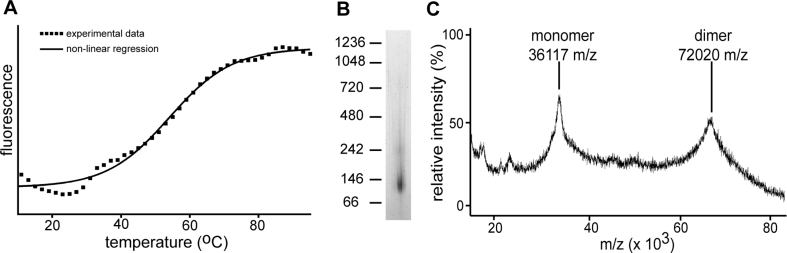
Full-length ZnT8A was expressed in P. pastoris, but low expression levels precluded further study of this isoform. In contrast, ZnT8B, the amino-terminal truncated form of ZnT8, expressed well in P. pastoris, and this protein was amenable to purification. Guided by previous protocols for transporter solubilization from P. pastoris and cost effectiveness (Aller et al., 2009; Roberts et al., 2011), Triton X-100 (TX100) was used to solubilize membranes. However, ZnT8B was unstable in this detergent, and we therefore used a differential filtration assay (DFA) of 96 detergents (Fig. S2) (Vergis et al., 2010) to determine that ZnT8B displayed minimal aggregation when solubilized in the zwitterionic detergent fos-choline 12 (FC-12; Table S2). TX100 was therefore exchanged to FC-12 during immobilized metal-affinity chromatography (IMAC). The protein was then further purified by size-exclusion chromatography (SEC) (Fig. S5). A thermal denaturation assay measuring the accessibility of a thiol-reactive fluorescent dye to free cysteine residues showed that the melting temperature (Tm) of the ZnT8B/FC-12 complex is ~57 °C (Fig. 4A), which is within the range observed for stably folded membrane proteins (Alexandrov et al., 2008). The addition of Zn2+ did not change the observed Tm (data not shown).
Fig. 4.
Purified ZnT8B forms a highly stable dimer. (A) Fluorescent thermal stability assay. Plot shows dye fluorescence per oC increase in temperature from which the calculated Tm of ZnT8B is 57 °C. (B) Non-denaturing PAGE of ZnT8B-G3H6, showing the protein-detergent micelle complex with an apparent molecular weight of ~140 kDa, which corresponds to an actual molecular weight of ~80 kDa. Positions of molecular weight markers in kDa (left). (C) MALDI-MS of purified ZnT8B-G3H6 showing number of sample ions detected and their corresponding mass-to-charge (m/z) ratio. The ZnT8B construct appears as both a monomer of 36.117 kDa and a dimer of 72.020 kDa.
Native gel electrophoresis suggested an apparent molecular mass of 140 kDa (Fig. 4B), which when the added mass of the Coomassie blue dye is accounted for (Heuberger et al., 2002) suggests a protein component of 70–80 kDa size. Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) of the purified ZnT8B in FC-12 showed a monomeric species of 36.1 kDa, as well as a dimer of 72.0 kDa (Fig. 4C), which corresponds to the predicted 36.05 kDa size of ZnT8B-G3H6. The presence of the monomeric species was expected, since with MALDI-MS non-covalent interactions between the two subunits together are expected to be lost. These results indicate that ZnT8B expressed in P. pastoris exists as a dimer following purification.
Purification of both ZnT8A and ZnT8B from S. frugiperda showed high protein purity, lower levels of protein aggregation (as compared with protein purified from P. pastoris), and good long-term stability (Fig. S6).
3.2.2. The ZnT8A amino- and carboxy-terminal ectodomains display ordered secondary structure
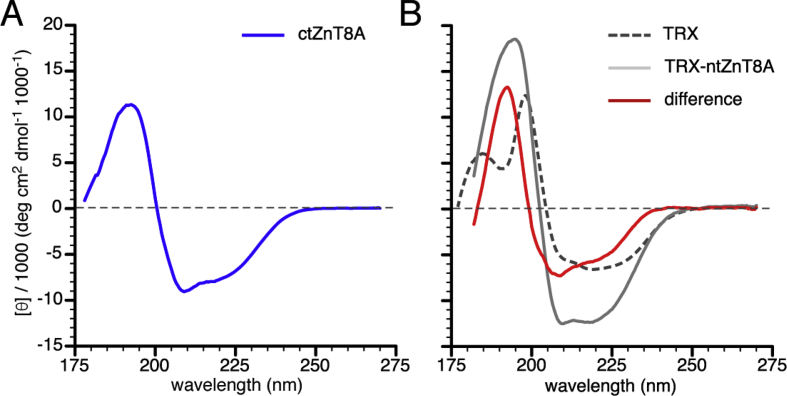
Circular dichroism (CD) spectroscopy was used to discern the secondary structure of the amino- and carboxy-terminal domains of ZnT8A. When examined in low salt buffer, all our protein constructs exhibited distinctive secondary structures (Fig. 5 and Table 2), which were deconvoluted using the CDSSTR algorithm (Sreerama and Woody, 2004) and the SMP180 reference dataset of 128 soluble and membrane proteins (Abdul-Gader et al., 2011) (Table 2). In this analysis it is important to keep in mind that most protein CD calculations use static structures as a reference, determined by X-ray diffraction, while our experimental data correspond to the dynamic solution-state structure and will thus show a greater amount of disorder (whereas crystal packing will reduce the disorder in X-ray structures).
Fig. 5.
CD spectroscopy shows that the carboxy- and amino-terminal domains of ZnT8 show a mixed α-β fold. Far-ultraviolet CD spectrometry of ZnT8 termini and thioredoxin (TRX) constructs, showing molar ellipticity per residue versus wavelength. (A) Spectrum for the carboxy-terminal domain (ctZnT8). (B) Spectra for the amino-terminal domain (ntZnT8) constructs. ntZnT8-TRX and TRX spectra are shown in grey and stippled grey, respectively. The ntZnT8 difference spectrum (red line) was derived by subtracting the TRX spectrum from that of the ntZnT8-TRX fusion protein. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Secondary structure fractions calculated by deconvolution of circular dichroism spectra. Shown are results for the bacterial thioredoxin-ZnT8A amino terminal domain fusion (TRX-ntZnT8A), the ZnT8A carboxy-terminal domain (ctZnT8A), the carboxy-terminal domain of YiiP (ctYiiP), and bacterial thioredoxin (TRX). Deconvolution solutions produced by the CDSSTR method, using a reference dataset of 128 membrane and soluble protein structures (Abdul-Gader et al., 2011). Secondary structure fractions noted are regular α-helix (αR), distorted α-helix (αD), regular β-strand (βR), distorted β-strand (βD), β-turn (T), and unordered (U), as defined by (Sreerama et al., 1999). The normalized root mean square deviation (NRMSD) is shown for each solution.
| protein | αR | αD | βR | βD | T | U | NRMSD |
|---|---|---|---|---|---|---|---|
| TRX-ntZnT8A | 0.23 | 0.16 | 0.11 | 0.07 | 0.12 | 0.32 | 0.016 |
| ctZnT8A | 0.12 | 0.11 | 0.18 | 0.10 | 0.12 | 0.36 | 0.039 |
| ctYiiP (X-ray)a | 0.33 | 0.26 | 0.19 | 0.23b | – | ||
| TRX | 0.13 | 0.10 | 0.23 | 0.07 | 0.10 | 0.36 | 0.029 |
| TRX (X-ray)a | 0.39 | 0.30 | 0.15 | 0.16 | – | ||
Secondary structure assignment using STRIDE (Kabsch and Sander, 1983).
12% unassigned content and 11% disordered.
As a control for correct folding of overexpressed protein and our CD methods, we examined E. coli-expressed and purified bacterial thioredoxin (TRX) (Fig. S7). Previous CD spectra of TRX showed a broad negative CD signal with a minimum of −6800 deg.cm2.dmol−1 at 219 nm and a sharp positive CD band with a peak of 17,300 deg.cm2.dmol−1 at 196 nm (Reutimann et al., 1981). Our results (Fig. 5B) are similar, with a broad negative CD signal with a minimum of −6800 deg.cm2.dmol−1 at 219 nm, a sharp positive CD signal with a peak of 11,000 deg.cm2.dmol−1 at 198 nm, and an additional peak of 5100 deg.cm2.dmol−1 at 185 nm with a trough at 191 nm between the two positive peaks. Our CD spectrum of bacterial TRX thus recapitulates previous results (Reutimann et al., 1981), supporting the fidelity of our protein overexpression, purification and CD spectrometry results.
Carboxy-terminal domain
The carboxy-terminal domain of ZnT8 was expressed in E. coli as a fusion with TRX. Following proteolytic digestion with enterokinase the carboxy-terminal domain was purified by SEC (Fig. S7), and MALDI-MS verified that the purified fragment showed the expected mass of ~10.5 kDa (data not shown).
Our homology model of ZnT8A shows the cytoplasmic carboxy-terminal domain to have a mixed α1-β1-β2-α2-β3 fold, as does an earlier homology model based on the E. coli homolog YiiP (Weijers, 2010). Correspondingly, our CD spectra of this 85 amino-acid domain showed a strong α-helical signal (Fig. 5A), with a negative CD minimum of −9100 deg. cm2.dmol−1 at 209 nm and a positive CD peak of 11,300 deg.cm2.dmol−1 at 193 nm. Spectral deconvolution by the CDSSTR algorithm (Sreerama and Woody, 2004) indicated a mixed α-β fold (28% α-helical, 25% β-sheet, 20% β-turn and 30% disordered; Table 2). This agrees well with the most recent X-ray structure of the E. coli homolog YiiP (Lu et al., 2009), in which the carboxy-terminal domain displays 33% α-helical, 26% β-strand, 19% β-turn and 12% unassigned content, as determined by STRIDE (Kabsch and Sander, 1983). Another 11% appears to be disordered, as 10 of the 95 residues of the carboxy-terminus were not resolved in the initial structure (Fu and Lu, 2007) and were removed in the later crystallographic study. Recent CD analysis of the carboxy-terminal domain of ZnT8 over a reduced wavelength range (195-260 nm) suggested that there is no significant difference beween the structures of the R325 and W325 SNP variants in this domain (Parsons et al., 2018).
Amino-terminal domain
Similar to the carboxy-terminal domain, the amino-terminal domain was expressed in E. coli as a fusion with TRX. Unfortunately, the cleaved amino-terminal domain co-migrated with TRX, and the two species could not be separated using SEC. Therefore, to discern the secondary structure of the amino-acid domain (ntZnT8A), the CD spectrum of TRX was subtracted from the spectrum of the purified fusion protein of TRX-ntZnT8A.
ZnT8 possesses an amino-terminal extension not found in the bacterial homolog YiiP so there is no structural model for this domain. Sequence analysis initially suggested that the 73 amino-acid amino-terminus is primarily disordered, particularly in the region between Gln28 and Gly63 (Fig. S3). However, the difference CD spectrum displayed a strong positive CD band at 192 nm and a minor negative CD band at 209 nm (Fig. 5B), indicating that the ZnT8A amino-terminal domain has significant α-helical content. The prominence of the 209 nm band suggests that some polyproline II helical content may also be present, since this structure is characterized by a strong negative band near 206 nm (Sreerama and Woody, 2004).
3.2.3. The molecular boundary of the ZnT8B dimer recapitulates that of YiiP, a bacterial homolog
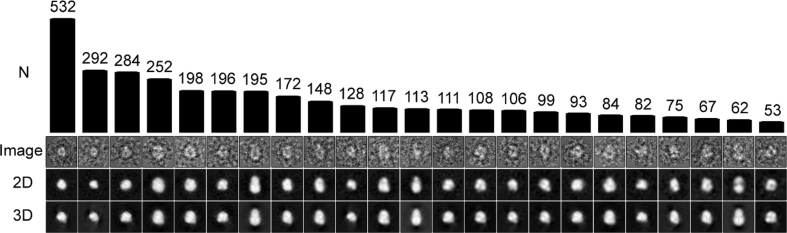
EM images of negatively stained ZnT8B derived from P. pastoris displayed a field of discrete particles showing minimal aggregation (Fig. S8) and were thus suitable for single-particle image analysis. Individual complexes were masked using the EMAN2 (Tang et al., 2007) software package. By use of the RELION software package (Scheres, 2012), 23 2D class averages were derived for ~3500 particles (Fig. 6).
Fig. 6.
EM and single particle analysis using RELION. Shown are the 23 final particle classes used in determining the ZnT8B structure. Top row indicates number of particles in each class average (N), second row shows image of a representative raw particle belonging to class, third row shows 2D class average, bottom row displays back projections of 3D map with view corresponding to class average shown in third row. Note the close correspondence of the 2D class averages (in which symmetry was not applied) with the back projections of the 3D map (in which C2 symmetry was imposed), suggesting that ZnT8 assembles as a symmetric dimer.
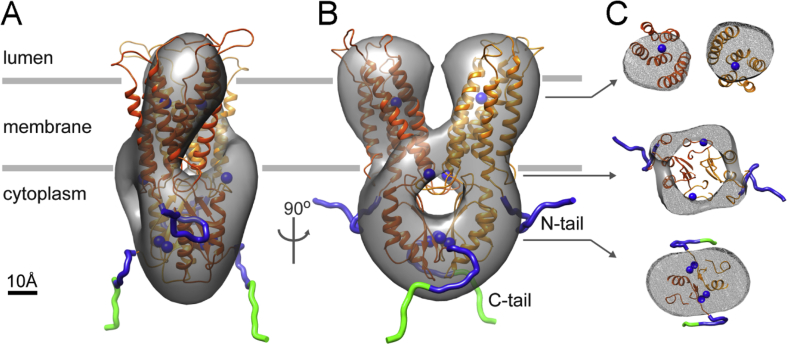
A 3D map without applied symmetry was obtained by reference-free methods using EMAN2. The map was truncated at 50 Å resolution (Fig. S9A), which served as a starting model for further refinement using RELION. The resulting non-symmetrized map (Fig. S9B) displayed two elongated lobes of density with apparent two-fold symmetry, consistent with our biochemical and MS results (Fig. 4B and C) that ZnT8B is a dimer in FC-12. We therefore applied C2 symmetry, and the resulting map had a calculated resolution of 20 Å using a conservative Fourier shell correlation (FSC) cut-off value of 0.5 (Fig. S10). Interpretation of the low-resolution map was aided by docking of a homology model using the structure of the bacterial YiiP homolog as a template. There was a good fit to the 105 Å × 70 Å × 55 Å molecular boundary and suggests an assignment of the transmembrane and cytoplasmic domains (Fig. S9C). To extend the analysis, we therefore used a 50 Å resolution map of YiiP as a reference (Fig. S9D) for refinement by RELION. The resulting non-symmetrized map of ZnT8 again demonstrated a bilobed appearance (Fig. S9E), and consequently we applied C2 symmetry during refinement of the final map (Fig. 7 and Fig. S9F), which had a calculated resolution of 20 Å using a conservative FSC cut-off value of 0.5 (Fig. S11). As expected, the subunits within the ZnT8 dimer have improved definition compared with the initial, reference-free map and slightly more compact dimensions of 90 Å × 70 Å × 50 Å (Fig. 7). At this resolution we do not expect to distinguish ZnT8 conformational states as was possible with YiiP at higher resolutions (Coudray et al., 2013; Lu et al., 2009). Nevertheless we predict our model to present an inward-facing aspect similar to Coudray et al. (2013) since the protein was prepared in the absence of zinc.
Fig. 7.
ZnT8 EM map displays a2-fold symmetricdimeric structure that recapitulates the architecture of the bacterial homolog YiiP. Front (A) and side (B) views of the ZnT8 homology model of YiiP fitted into the EM density map calculated at 20 Å resolution. The complex has a height of 90 Å and a girth of 70 Å × 50 Å. Axial sections (C) of density map corresponding to transverse planes in B as indicated. Subunits (orange and yellow), zinc ions (blue spheres), amino-terminal fragment (blue), His6 tag (green).
3.3. Future prospects: our results provide a foundation for higher resolution structural studies and screening experiments to identify compounds that modulate ZnT8 activity
We are hopeful that the in vivo transport assay in P. pastoris and the in vitro proteoliposome-based assay can be adapted for high-throughput screening to identify small molecule compounds modulating ZnT8 activity. Given the observed low expression levels of ZnT8 in both yeast and insect cells, the current protocol would require substantial scale-up to yield the milligram quantities required for X-ray crystallography. Even though the ZnT8B dimer has a mass of only ~72 kDa, it should nevertheless be amenable to cryoEM and single-particle analysis (Renaud et al., 2018; Wu et al., 2012). The use of contemporary cryoEM technologies (Cheng, 2015; Khoshouei et al., 2017), the dimeric symmetry and the addition of fusion proteins (Chun et al., 2012) and/or Fab fragments (Wu et al., 2012; Alam et al., 2018; Kim et al., 2015; Dutka et al., 2019; Mukherjee et al., 2020) will not only aid the identification of domains within the complex but will also add mass and identifiable structural features to facilitate the determination of particle orientations. Recent work showing that the loss of ZnT8 function is protective in diabetes (Dwivedi et al., 2019) highlights the importance of further structure-function work on this clinically relevant protein.
Funding
Supported in part by American Diabetes Association grant #7-13-BS-038 (M.J.D.) and National Institutes of Health R01 grants HL48908 (M.Y.) and GM138532 (M.Y.).
CRediT authorship contribution statement
Mark J. Daniels: Conceptualization, Methodology, Investigation, Data interpretation, Supervision, Visualization, Funding acquisition, Writing - original draft, Writing - review & editing. Maciej Jagielnicki: Methodology, Investigation, Data interpretation, Software, Formal analysis, Visualization, Writing - original draft. Mark Yeager: Conceptualization, Supervision, Funding acquisition, Methodology, Data interpretation, Writing - review and editing.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgments
The authors thank Janet Wenzlau for ZnT8 antisera, John Shannon and the University of Virginia Keck Mass Spectrometry Facility for MALDI-MS, Kelly Dryden for assistance with EM, Michael Purdy for advice on single-particle image processing, and Michael Wiener for use of reagents for detergent screening. EM was performed in the UVa Molecular Electron Microscopy Core, which has been supported by NIH grants G20 RR031199, S10 RR031199, S10 OD018149 and U24 GM116790.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2020.06.001.
Contributor Information
Mark J. Daniels, Email: markjd@virginia.edu.
Mark Yeager, Email: yeager@virginia.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdul-Gader A., Miles A.J., Wallace B.A. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinformatics. 2011;27(12):1630–1636. doi: 10.1093/bioinformatics/btr234. [DOI] [PubMed] [Google Scholar]
- Alam A. Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc. Natl. Acad. Sci. U. S. A. 2018;115(9):E1973–E1982. doi: 10.1073/pnas.1717044115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A.I. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Aller S.G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323(5922):1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anatrace . Anatrace; 2014. Detergent Properties Booklet. [Google Scholar]
- Bai J. A gene optimization strategy that enhances production of fully functional P-glycoprotein in Pichia pastoris. PloS One. 2011;6(8) doi: 10.1371/journal.pone.0022577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhairi S.M. EMD Biosciences; San Diego, CA: 2007. Detergents: A Guide to the Properties and Uses of Detergents in Biological Systems, Calbiochem. [Google Scholar]
- Cauchi S. Meta-analysis and functional effects of the SLC30A8 rs13266634 polymorphism on isolated human pancreatic islets. Mol. Genet. Metabol. 2010;100:77–82. doi: 10.1016/j.ymgme.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Chabosseau P., Rutter G.A. Zinc and diabetes. Arch. Biochem. Biophys. 2016;611:79–85. doi: 10.1016/j.abb.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161(3):450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimienti F. Identification and cloning of a β-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53(9):2330–2337. doi: 10.2337/diabetes.53.9.2330. [DOI] [PubMed] [Google Scholar]
- Chimienti F. In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J. Cell Sci. 2006;119(20):4199–4206. doi: 10.1242/jcs.03164. [DOI] [PubMed] [Google Scholar]
- Chimienti F., Favier A., Seve M. ZnT-8, a pancreatic beta-cell-specific zinc transporter. Biometals. 2005;18(4):313–317. doi: 10.1007/s10534-005-3687-9. [DOI] [PubMed] [Google Scholar]
- Chun E. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20(6):967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray N. Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A. 2013;110(6):2140–2145. doi: 10.1073/pnas.1215455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M.J., Yeager M. Phosphorylation of TIP3 aquaporins during Phaseolus vulgaris embryo development. Cells. 2019;8(11) doi: 10.3390/cells8111362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M.J., Wood M.R., Yeager M. In vivo functional assay of a recombinant aquaporin in Pichia pastoris. Appl. Environ. Microbiol. 2006;72:1507–1514. doi: 10.1128/AEM.72.2.1507-1514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson H.W., Wenzlau J.M., O’Brien R.M. Zinc transporter 8 (ZnT8) and β cell function. Trends Endocrinol. Metabol. 2014;25(8):415–424. doi: 10.1016/j.tem.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan M.A., Noah J.W., Hurt D. Comparison of common homology modeling algorithms: application of user-defined alignments. Methods Mol. Biol. 2012;857:399–414. doi: 10.1007/978-1-61779-588-6_18. [DOI] [PubMed] [Google Scholar]
- Dutka P. Development of “Plug and Play” Fiducial Marks for Structural Studies of GPCR Signaling Complexes by Single-Particle Cryo-EM. Structure. 2019;27(12):1862–1874. doi: 10.1016/j.str.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi O.P. Loss of ZnT8 function protects against diabetes by enhanced insulin secretion. Nat. Genet. 2019;51(11):1596–1606. doi: 10.1038/s41588-019-0513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46(4):357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Lu M. The structural basis of water permeation and proton exclusion in aquaporins. Mol. Membr. Biol. 2007;24(5–6):366–374. doi: 10.1080/09687680701446965. [DOI] [PubMed] [Google Scholar]
- Fukada T., Kambe T. Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metall. 2011;3(7):662–674. doi: 10.1039/c1mt00011j. [DOI] [PubMed] [Google Scholar]
- Gaither L.A., Eide D.J. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14(3-4):251–270. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- Gati C. The structural basis of proton driven zinc transport by ZntB. Nat. Commun. 2017;8(1):1313. doi: 10.1038/s41467-017-01483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006;1(6):2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. Cold Spring Harbor Laboratory; , Cold Spring Harbor: 1988. Antibodies: a Laboratory Manual. [Google Scholar]
- Haworth R.S., Fliegel L. Intracellular pH in Schizosaccharomyces pombe - comparison with Saccharomyces cerevisiae. Mol. Cell. Biochem. 1993;124:131–140. doi: 10.1007/BF00929205. [DOI] [PubMed] [Google Scholar]
- Heuberger E.H.M.L. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 2002;317:591–600. doi: 10.1006/jmbi.2001.5416. [DOI] [PubMed] [Google Scholar]
- Hoch E. Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proc. Natl. Acad. Sci. U.S.A. 2012;109(19):7202–7207. doi: 10.1073/pnas.1200362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howson J.M.M. Genetic association of zinc transporter 8 (ZnT8) autoantibodies in type 1 diabetes cases. Diabetologia. 2012;55(7):1978–1984. doi: 10.1007/s00125-012-2540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kambe T. Sequence similarity and functional relationship among eukaryotic ZIP and CDF transporters. Genomics Proteomics Bioinformatics. 2006;4(1):1–9. doi: 10.1016/S1672-0229(06)60010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambe T. The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev. 2015;95(3):749–784. doi: 10.1152/physrev.00035.2014. [DOI] [PubMed] [Google Scholar]
- Kawachi M. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 2008;283(13):8374–8383. doi: 10.1074/jbc.M707646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshouei M. Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. Nature Comm. 2017;8 doi: 10.1038/ncomms16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature. 2015;517(7534):396–400. doi: 10.1038/nature13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski L.P., Bujnicki J.M. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinf. 2012;13:111. doi: 10.1186/1471-2105-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K. Insulin crystallization depends on zinc transporter ZnT8 expression, but is not required for normal glucose homeostasis in mice. Proc. Natl. Acad. Sci. U.S.A. 2009;106(35):14872–14877. doi: 10.1073/pnas.0906587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten L.A., Cousins R.J. Mammalian zinc transporters: nutritional and physiologic regulation. Annu. Rev. Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino J. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. Biotechniques. 2005;38(1):44–48. doi: 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmeier-Vogel E.M., McIntyre D.D., Vogel H.J. Phosphorus-31 and carbon-13 nuclear magnetic resonance studies of glucose and xylose metabolism in cell suspensions and agarose-immobilized cultures of Pichia stipitis and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1996;62(8):2832–2838. doi: 10.1128/aem.62.8.2832-2838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Redondo M.L. Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP. Proc. Natl. Acad. Sci. U. S. A. 2018;115(12):3042–3047. doi: 10.1073/pnas.1715051115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Chai J., Fu D. Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 2009;16(10):1063–1067. doi: 10.1038/nsmb.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Fu D. Structure of the zinc transporter YiiP. Science. 2007;317(5845):1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- Merriman C. Lipid-tuned zinc transport activity of human ZnT8 protein correlates with risk for type-2 diabetes. J. Biol. Chem. 2016;291(53):26950–26957. doi: 10.1074/jbc.M116.764605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142(3):334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Mocchegiani E., Giacconi R., Malavolta M. Zinc signalling and subcellular distribution: emerging targets in type 2 diabetes. Trends Mol. Med. 2008;14(10):419–428. doi: 10.1016/j.molmed.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Mukherjee S. Synthetic antibodies against BRIL as universal fiducial marks for single−particle cryoEM structure determination of membrane proteins. Nature Comm. 2020;11(1598) doi: 10.1038/s41467-020-15363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia C. Diabetes-linked zinc transporter ZnT8 is a homodimeric protein expressed by distinct rodent endocrine cell types in the pancreas and other glands. Nutr. Metabol. Cardiovasc. Dis. 2009;19(6):431–439. doi: 10.1016/j.numecd.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Ohi M. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6(1):23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C.N. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D.S. The C-terminal cytosolic domain of the human zinc transporter ZnT8 and its diabetes risk variant. FEBS J. 2018;285(7):1237–1250. doi: 10.1111/febs.14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A.B. siRNA-mediated knock-down of ZnT3 and ZnT8 affects production and secretion of insulin and apoptosis in INS-1E cells. APMIS. 2011;119:93–102. doi: 10.1111/j.1600-0463.2010.02698.x. [DOI] [PubMed] [Google Scholar]
- Rasband W.S. ImageJ. 1997-2015. http://imagej.nih.gov/ij/ Available from:
- Renaud J.P. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 2018;17(7):471–492. doi: 10.1038/nrd.2018.77. [DOI] [PubMed] [Google Scholar]
- Reutimann H. A conformational study of thioredoxin and its tryptic fragments. J. Biol. Chem. 1981;256(13):6796–6803. [PubMed] [Google Scholar]
- Roberts M.F., Taylor D.W., Unger V.M. Two modes of interaction between the membrane-embedded TARP stargazin’s C-terminal domain and the bilayer visualized by electron crystallography. J. Struct. Biol. 2011;174(3):542–551. doi: 10.1016/j.jsb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross I.S. Uptake of Zinc by Fungi. In: Winkelmann G., Winge D.R., editors. Metal Ions in Fungi. Marcel Dekker; New York: 1994. pp. 237–258. [Google Scholar]
- Saxena R. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.J. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N., Venyaminov S.Y., Woody R.W. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8(2):370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N., Woody R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004;383:318–351. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- Sladek R. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Studier F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Suzuki T. Two different zinc transport complexes of cation diffusion facilitator proteins localized in the secretory pathway operate to activate alkaline phosphatases in vertebrate cells. J. Biol. Chem. 2005;280(35):30956–30962. doi: 10.1074/jbc.M506902200. [DOI] [PubMed] [Google Scholar]
- Tang G. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Vergis J.M., Purdy M.D., Weiner M.C. A high-throughput differential filtration assay to screen and select detergents for membrane proteins. Anal. Biochem. 2010;407:1–11. doi: 10.1016/j.ab.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers R.N. Three-dimensional structure of β-cell-specific zinc transporter, ZnT-8, predicted from the type 2 diabetes-associated gene variant SLC30A8 R325W. Diabetol. Metab. Syndrome. 2010;2(1):33. doi: 10.1186/1758-5996-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzlau J.M. Mapping of conformational autoantibody epitopes in ZNT8. Diabetes Metabol. Res. Rev. 2011;27(8):883–886. doi: 10.1002/dmrr.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzlau J.M., Hutton J.C., Davidson H.W. New antigenic targets in type 1 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2008;15(4):315–320. doi: 10.1097/MED.0b013e328308192b. [DOI] [PubMed] [Google Scholar]
- Wenzlau J.M. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc. Natl. Acad. Sci. U.S.A. 2007;104(43):17040–17045. doi: 10.1073/pnas.0705894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesekara N. Beta cell specific ZnT8 deletion in mice causes marked defects in insulin processing, crystallisation and secretion. Diabetologia. 2010;53(8):1656–1668. doi: 10.1007/s00125-010-1733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Fabs enable single particle cryoEM studies of small proteins. Structure. 2012;20:582–592. doi: 10.1016/j.str.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. UCSF Chimera. MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. U. S. A. 1996;93(6):2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.