Abstract
Background
Declining humoral immunity in coronavirus disease 2019 (COVID-19) patients and possible reinfection have raised concern. Mucosal immunity, particularly salivary antibodies, may be short lived although long-term studies are lacking.
Methods
Using a multiplex bead-based array platform, we investigated antibodies specific to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins in 256 saliva samples from convalescent patients 1–9 months after symptomatic COVID-19 (n = 74, cohort 1), undiagnosed individuals with self-reported questionnaires (n = 147, cohort 2), and individuals sampled prepandemic (n = 35, cohort 3).
Results
Salivary IgG antibody responses in cohort 1 (mainly mild COVID-19) were detectable up to 9 months postrecovery, with high correlations between spike and nucleocapsid specificity. At 9 months, IgG remained in blood and saliva in most patients. Salivary IgA was rarely detected at this time point. In cohort 2, salivary IgG and IgA responses were significantly associated with recent history of COVID-19–like symptoms. Salivary IgG tolerated temperature and detergent pretreatments.
Conclusions
Unlike SARS-CoV-2 salivary IgA that appeared short lived, specific saliva IgG appeared stable even after mild COVID-19, as for blood serology. This noninvasive saliva-based SARS-CoV-2 antibody test with home self-collection may be a complementary alternative to conventional blood serology.
Keywords: antibody, convalescence, COVID-19, immunoassay, saliva, serology
Mucosal defense plays an important role in COVID-19. Multiplex-based analysis of self-collected saliva demonstrates that broad-specific antibodies to SARS-COV-2 persist up to 9 months after recovery of mild COVID-19. Saliva is promising in monitoring and screening of COVID-19 immunity.
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) broke out in an abrupt fashion after its initial identification in Wuhan, China, in late December 2019 [1], and obligated the World Health Organization to declare a global health emergency, which escalated to a pandemic situation in March 2020. As of March 2021, SARS-CoV-2 has caused over 114 million cases of coronavirus disease (COVID-19) and up to 2.5 million deaths worldwide [2]. The human adaptive immune system plays a key role in eliminating and memorizing pathogens by launching a cascade of activities that activate B and T lymphocytes. B lymphocytes produce antibodies that recognize and neutralize SARS-CoV-2 and protect against reinfection [3–5]. Immunoglobulin G (IgG), IgA, and IgM antibodies are activated against SARS-CoV-2 and detected in the circulating blood of > 90% of infected individuals from 11 to 13 days post-symptom onset (PSO) [6–8]. A recent study showed that circulating antibodies after SARS-CoV-2 infection can persist for up to 8 months [9], while other studies have shown that this immunological memory persists for a certain period followed by a slight decline, especially in asymptomatic infected individuals [10–14].
Oral and nasal cavities are considered the main gateway for SARS-CoV-2 entry, and saliva secretory antibodies may be the first immunity arm to combat the infection through virus recognition. Salivary antibodies to SARS-CoV-2 can be detected early after symptom onset and persist for at least 3 months postinfection [8, 10, 12]. Hence, saliva sampling could be a suitable and noninvasive way to indicate SARS-CoV-2 exposure. Similar to the previous SARS-CoVs and Middle East respiratory syndrome coronavirus (MERS-CoV), the spike protein (S) of SARS-CoV-2 recognizes the angiotensin-converting enzyme 2 (ACE2) receptor and uses it to enter host cells [15–17]. Although antibodies play an important role in virus clearance [11, 18], differential features of anti-SARS-CoV-2 antibodies negatively impacting disease severity, especially those related to complement deposition and systemic inflammation, have also been described [19]. Understanding the dynamics and durability of antibody memory to SARS-CoV-2 is an instrumental step to manage the pandemic and may be useful in deploying vaccination strategies. As the mucosal immunity is known to be short-lived, the durability of SARS-CoV-2–specific antibodies in saliva could be limited. Whether they can be detected 3–4 months after infection [8, 10] is of great interest.
In this study, we exploited a highly sensitive and specific multiplex SARS-CoV-2 serology platform previously validated for seroprevalence studies [20] to investigate SARS-CoV-2 antibodies in saliva. Samples from individuals (1) with a diagnosis of mild COVID-19 in the convalescent phase, (2) 1–9 months after diagnosis of COVID-19, and (3) with or without a history of COVID-19 symptoms (undiagnosed) were analyzed and compared to prepandemic samples. Our data indicate that spike-specific IgG reactivity is detectable in saliva in the vast majority of patients at 1–9 months postinfection. This result was similar to those detected by blood serology performed in a clinical diagnostic laboratory. The IgA reactivity on the other hand was short-lived in saliva, detectable only during the first 3 months. Moreover, IgG and IgA reactivity to both spike and nucleocapsid significantly correlated with a history of COVID-19–like symptoms in undiagnosed individuals.
METHODS
Experimental Design
We applied a bead-based serology assay to detect IgG and IgA to SARS-CoV-2 proteins in saliva samples to evaluate its performance. The assay method is originally developed for detection of SARS-CoV-2–specific IgG in serum and plasma [20] where it showed 99.7% sensitivity and 100% specificity, and no cross-reaction when testing samples positive for other coronaviruses. Salivary antibody responses to 3 different SARS-CoV-2 antigens (2 spike and 1 nucleocapsid proteins) were first tested. The antigens’ performance in classifying positive and negative samples was evaluated for the single antigens as well as for antigens combined in panels. Best performing representations of spike and nucleocapsid were chosen in subsequent assessments.
Cohort Design
The study was approved by the Swedish Ethical Review Authority (Dnr 2020-01702 and Dnr 2020-06381) and complied with the Declaration of Helsinki. All participants were recruited after signing informed consent forms. Saliva samples (total n = 256) were collected and arranged in the following groups: cohort 1, convalescence COVID-19 samples (n = 74) from 72 patients (2 participants donated twice, 6 months apart) diagnosed with COVID-19 during March to April 2020 and collected from June to December 2020; cohort 2, samples from undiagnosed individuals donated during May to Nov 2020 (n = 147); cohort 3, anonymous saliva samples taken in 2018 before the COVID-19 outbreak (prepandemic, n = 35).
All convalescent patients (cohort 1) had COVID-19 diagnosis confirmed by SARS-CoV-2 reverse transcription polymerase chain reaction (RT-PCR), except 1 patient who had positive SARS-CoV-2 antibodies at 4 time points in the convalescence phase. Seroconversion was tested by clinical SARS-CoV-2 blood serology assays (described below). The patients were recruited from the Department of Infectious Diseases, Karolinska University Hospital (n = 65), and University Dental Clinic of Karolinska Institutet (n = 7). Clinical demographic data of convalescent patients was compiled from medical records or questionnaire. Among 72 patients, 95.8% had mild COVID-19 without hospitalization due to COVID-19 symptoms. Three were admitted to hospital for the purpose of isolation, and 3 were admitted due to COVID-19 symptoms. In the latter group, 2 were hospitalized without any required oxygen treatment and 1 received a maximum of 1.5 L oxygen treatment during hospitalization, indicating no severe disease outcome. The serum and saliva samples were grouped according to time of collection PSO, that is (1) less than 3 months PSO, (2) 3–8 months PSO, and (3) 9 months PSO. Cohort 2 comprised anonymous participants visiting the premises of University Dental Clinic of Karolinska Institutet or Eastman Institute, Stockholm during the study time, such as patients, staff, or their relatives. A questionnaire was used to collect COVID-19–related data of undiagnosed participants: (1) symptomatic, and (2) nonsymptomatic, based on their health in the 3 months prior to sampling.
Saliva Sample Collection
Expectorated unstimulated whole saliva was used throughout this study. All samples were self-collected using standardized instructions and sample tubes provided in this study. Samples were processed and stored at −80°C within 24 hours. Salivary stability tests were performed on sample subgroups to evaluate antibody reactivity following viral inactivation with either 1% Triton X-100 for 1 hour at room temperature (RT), or heat treatment at 56°C for 30 minutes [19]. Eighteen antibody-positive samples from cohort 1 and antibody-negative samples from cohort 2 were included in the comparison. Incubation at RT for 1 to 3 days was also tested in 5 samples to simulate the standard circumstances of the mailed-in saliva self-collection procedure. Saliva samples from convalescent patients (cohort 1) were collected on the same day as venous blood during a COVID-19 follow-up examination at the Department of Infectious Diseases, Karolinska University Hospital.
Clinical Serology Tests
Paired serum samples of all convalescent patients were tested at the Karolinska University Hospital Clinical Microbiology Laboratory. Three automated methods and 1 in-house diagnostic method were used for the included convalescent blood samples: SARS-CoV2-IgG test iFlash 1800 YHLO (CLIA), LIAISON SARS-CoV-2 S1/S2 IgG test DiaSorin (CLIA), and SARS CoV-2 IgG in-house enzyme-linked immunosorbent assay (ELISA) for samples taken prior to June 2020 (mainly early convalescent samples; <9 months); and the Elecsys Anti-SARS-CoV-2 antibody test Roche (ECLIA) for all late convalescent samples (9 months). YHLO determines antibodies against the SARS-CoV-2 nucleocapsid and spike protein, DiaSorin against spike protein, whilst Elecsys and the in-house ELISA determine antibodies against recombinant nucleocapsid protein. The tests use different techniques: chemiluminescence immunoassay (CLIA), electrochemiluminescence immunoassay (ECLIA), and ELISA.
Antigen Production
The proteins were produced as follows: (1) spike glycoprotein (spike-f) in a soluble trimeric form stabilized in its prefusion conformation was expressed in HEK293 cells and purified using a C-terminal Strep II tag; (2) spike S1 domain was expressed in CHO cells and purified using a C-terminal HPC4-tag; and (3) nucleocapsid C-terminal (NC-C) chain was expressed in Escherichia coli and purified using a C-terminal His-tag [21, 22].
SARS-CoV-2 Antibody Detection by a Bead-Based Assay
The analysis of salivary antibodies was performed as previously described [20] with some modifications. Briefly, each antigen was diluted to a final concentration of 80 µg/mL (100mM) with 2-(N-morpholino) ethanesulfonic acid buffer, pH 4.5 (SigmaAldrich) and immobilized on a uniquely color-coded bead type (bead ID) (MagPlex-C; Luminex). The antigen-immobilized beads were then pooled to form the bead array. Anti-human IgG (Jackson Immunoresearch), anti-human IgA (Bethyl), and the Epstein-Barr virus EBNA1 protein (Abcam) were also included as sample loading controls. Saliva samples were diluted 1:5 in assay buffer composed of 3% bovine serum albumin (w/v), 5% nonfat milk (w/v) in 1 × phosphate-buffered saline (PBS) supplemented with 0.05% (v/v) Tween20 (VWR), and incubated with the bead array for 1 hour at RT and 650 rpm. Afterwards, antigen-antibody complexes were cross-linked by adding 0.2% paraformaldehyde (AlfaAesar) in PBS 0.05% Tween 20 (PBST) for 10 minutes at RT. Detection was performed by applying R-phycoerythrin-conjugated anti-human IgG (Invitrogen) diluted to 0.4 µg/mL or R-phycoerithryne-conjugated anti-human IgA (Bethyl) diluted to 0.2 µg/mL in PBST for 30 minutes at RT. The read-out was performed by using a FlexMap3D system and xPONENT software (Luminex).
Statistical Analysis
Statistics and visualization of the multiplex bead array generated data were performed using R version 3.6.1 with RStudio version 1.2.1335, and additional packages heatmap version 1.0.10 and reshape2 version 1.4.3. In-house–developed functions were used for instrument file import and quality control. Bead array results were acquired as median fluorescent intensity per sample and bead identity. A cutoff for positivity was calculated per antigen as the mean plus 6 SD of 12 negative prepandemic reference samples carefully selected based on their signal intensity distribution. GraphPad Prism version 9.0.0 (86) was used for nonparametric comparisons: Mann-Whitney test and Spearman correlation analysis. Datasets also initially underwent normality distribution testing. The N − 1 χ 2 test was used for comparisons of binomial datasets in MedCal software calculator. Two-sided P values < .05 were considered significant.
RESULTS
Salivary Antibody Reactivity to SARS-CoV-2 Proteins
The assay performance was evaluated by comparing the ability of each of the 3 antigens included in the assay to classify convalescent samples (cohort 1, n = 74) and prepandemic samples (cohort 3, n = 35), of which 12 samples from cohort 3 were used to set the assay cutoffs. Among the 3 antigens, spike-f and NC-C showed the best performance in differentiating SARS-CoV-2 convalescent samples from the prepandemic samples. Spike-f showed 88% sensitivity and 100% specificity, with 1 negative control sample reaching intensity signal at the cutoff level. NC-C showed 66% sensitivity and 100% specificity (Table 1). We also evaluated the assay performance for all combined antigen panels of 2 and 3 antigens, considering a sample as positive when reactive to both antigens in a panel of 2 antigens and at least 2 in a panel of 3 antigens (Table 1). The best performance was reached by the spike-f, S1, NC-C triple combination, showing 72% sensitivity and 100% specificity. On the other hand, the IgA reactivities were identified only in a minority of cases, with higher prevalence of reactivity to spike-f (17%) in cohort 1 (Supplementary Table 1). It should be noted that larger sample sets are needed to establish and validate these sensitivity and specificity levels.
Table 1.
Specificity and Sensitivity of Single Antigen or Combination Antigen in Detecting SARS-CoV-2 IgG in Convalescent Saliva 1–9 Months After Symptom Onset and Prepandemic Saliva
| Antigen | Convalescent (n = 74) | Prepandemic (n = 23)a | ||||
|---|---|---|---|---|---|---|
| Sensitivity, % | No. Positive | No. Negative | Specificity, % | No. Positive | No. Negative | |
| Single antigen | ||||||
| Spike-f | 88 | 65 | 9 | 100 | 0b | 23 |
| S1 | 62 | 46 | 28 | 100 | 0 | 23 |
| NC-C | 66 | 49 | 25 | 100 | 0 | 23 |
| Combination antigen | ||||||
| Spike-f + S1 | 62 | 46 | 28 | 100 | 0 | 23 |
| Spike-f + NC-C | 66 | 49 | 25 | 100 | 0 | 23 |
| S1 + NC-C | 57 | 42 | 32 | 100 | 0 | 23 |
| Spike-f + S1 + NC-C | 72 | 53 | 21 | 100 | 0 | 23 |
Abbreviations: NC-C, nucleocapsid c-terminal chain; S1, spike S1 domain; spike-f, spike glycoprotein.
aAn additional 12 independent prepandemic saliva samples were used to establish assay cutoffs.
bOne sample shows intensity signal at cutoff level.
Serum and Salivary Antibody Reactivity Over Time After Covid-19
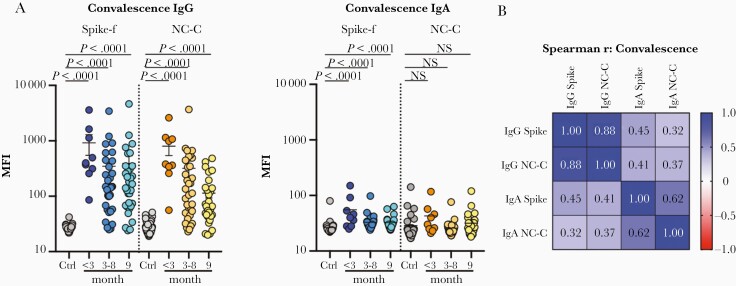
As shown in Table 2, cohort 1 mainly comprised patients who have had mild COVID-19 and were grouped according to duration after confirmed diagnosis. Some were hospitalized for isolation, but none received >1.5L oxygen treatment or required ventilation-related treatment. All individuals were free from respiratory symptoms at the 9-month follow-up but some residual symptoms were still noted in a minority of patients across all 3 groups (data not shown). As shown in Table 3, the vast majority of serum samples up to 9 months postinfection tested positive in clinical SARS-CoV-2 serology, with high seroprevalence across the whole time span of collection. Interestingly, paired saliva samples from cohort 1 tested with the multiplex bead array showed that the anti–spike-f IgG positivity rate in saliva remained remarkably high (100%–87.5%) and a similar range as was noted for serum antibodies (88.9%–96.9%) from early (<3 months) through to late convalescence (9 months) (Table 3, Figure 1A, and Supplementary Figure 1A). However, the NC-C–specific IgG in saliva dropped significantly after 3 months (from 88.9% to 69.7%–56.2%). As stated earlier, specific IgA responses to these antigens were detected only in a minority of the saliva samples, and were enriched in early convalescence (<3 months, 55.6% for spike-f and 22.2% for NC-C), while showing responses only in a minority of late convalescent samples (P < .01).
Table 2.
Demographic Characteristics of Convalescence Samples, Cohort 1, Grouped by Time of Sample Collection PSO
| Parameters | Time of Sample Collection PSO (n = 74) | ||
|---|---|---|---|
| <3 mo (n = 9) | 3–8 mo (n = 33) | 9 mo (n = 32) | |
| Sex, F:M | 8:1 | 23:10 | 6:26 |
| Age, y, median (range) | 59 (48–67) | 49 (20–63) | 57 (45–78) |
| Hospitalization status, % | |||
| Never hospitalized | 66.7 | 94 | 97 |
| Hospitalized only for isolation | 11 | 3 | 3 |
| Hospitalized due to COVID-19 symptoms | 22 | 3 | 0 |
| Days PSO, mean (SD) | 55 (20) | 120 (41) | 273 (11) |
Abbreviations: COVID-19, coronavirus disease 2019; PSO, post symptom onset.
Table 3.
Percent of Samples Positive Over Time for Saliva Antibodies to Spike-f or NC-C Compared to a Clinically Validated SARSCOV-2 Antibody Test of Serum
| Convalescence, mo | Serum Ab | Saliva IgG | Saliva IgA | ||
|---|---|---|---|---|---|
| SARS CoV-2 | Spike-f | NC-C | Spike-f | NC-C | |
| <3 | 88.9 | 100.0 | 88.9 | 55.6 | 22.2 |
| 3–8 | 90.9 | 84.8 | 69.7* | 12.5**** | 3.1** |
| 9 | 96.9 | 87.5 | 56.2**** | 9.7**** | 6.5**** |
Data are percent of samples positive.
* P < .05, ** P < .01, ****P < .0001 significance compared to clinical SARS-Cov-2 serum antibody diagnosis determined by N − 1 χ 2 test [23, 24].
Abbreviations: Ab, antibody; IgA, immunoglobulin A; IgG, immunoglobulin G; NC-C, nucleocapsid c-terminal chain; SARS CoV-2, severe acute respiratory syndrome coronavirus 2; Spike-f, spike glycoprotein.
Figure 1.
Measurement of IgG and IgA to spike-f (soluble trimeric form of the spike glycoprotein stabilized in the prefusion conformation) and NC-C (nucleocapsid C-terminal fragment) of SARS-CoV-2 in saliva of convalescent patients (cohort 1). A, Multiplex assay measured signal scores on indicated immunoglobulins to spike-f and NC-C in the pre–COVID-19 control samples (n = 35) and convalescent patient samples at indicated month postinfection (n = 74). The data are MFI and plotted using dot plots where each dot is 1 sample. Horizontal bars denote the mean and vertical lines represent standard error. Mann-Whitney U test for significance was performed. B, Spearman correlation analysis with coefficient indicated for respective antibody specificity pairs. Abbreviations: COVID-19, coronavirus disease 2019; Ctrl, control; IgG, immunoglobulin G; MFI, median fluorescence index; NS, not significant; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Moreover, salivary IgG to spike-f and NC-C were highly correlated in this cohort (r = 0.88, P < .0001, Spearman correlation test), with concordant serostatus in the majority of samples (Figure 1B). Significant, albeit moderate, correlations were also seen between IgA to spike-f and NC-C (r = 0.62, P < .001), and between spike-f–specific IgA and IgG (r = 0.45, P < .001; Figure 1B).
Salivary Antibody Reactivity to SARS-CoV-2 in Healthy Donors Is Associated With Recent History of COVID-19–Like Symptoms
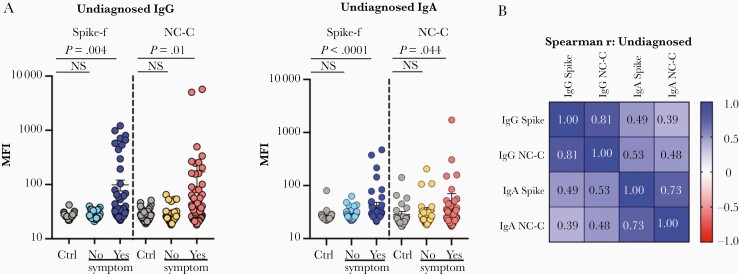
Next, we applied this assay platform to evaluate a second independent cohort, cohort 2. Participants here were self-reported symptom-free individuals visiting the University Dental Clinic’s premises of Karolinska Institutet and the Eastman Institute in Stockholm. A total of 147 individuals from May to November 2020 participated and donated saliva samples. Samples were collected and tested using the same standard operating protocol as for cohort 1. Shown in Figure 2A and based on antigen-specific cutoffs calculated on 12 negative controls, antibody reactivities to spike-f and NC-C in this cohort were as follows: IgG to spike-f was detected in 14% and to NC-C in 15%, while 11% had detectable IgG to both antigens; for IgA, 14% and 6% of the samples showed reactivity to spike-f and NC-C respectively, while only 6% showed reactivity to both. Salivary positivity was particularly enriched among participants with a self-reported recent history of COVID-19–like symptoms (14 days to 3 months prior to sampling time). Significant reactivities of IgG (P = .004 and P = .01) and IgA (P < .0001 and P = .044) to either spike-f or NC-C, respectively, was found to associate with a recent history of symptoms compared to prepandemic controls (Figure 2A). Adding risk factors together with presence of symptoms further enriched the salivary IgG positivity. The risk factors recent Covid-19 contact, travelling abroad, or clinical duties increased IgG positivity to 23%, 15%, and 13% for spike-f, and 23%, 19%, and 17% for NC-C, respectively (Supplementary Figure 1B).
Figure 2.
SARS-CoV-2–specific IgG and IgA in saliva of undiagnosed study participants (cohort 2) measured by the same method as in Figure 1. Samples were subgrouped by participant-reported COVID-19–like symptoms during 14 days to 3 months prior to the sampling. A, Multiplex assay measured signal scores on indicated immunoglobulins to spike-f and NC-C in pre–COVID-19 control samples (n = 35) and participants in cohort 2 reporting no or yes to recent COVID-19–like symptoms (n = 146). The data are MFI and plotted using dot plots where each dot is 1 sample. Horizontal bars denote the mean and vertical lines represent standard error. Mann-Whitney U test for significance was performed. B, Spearman correlation analysis with coefficient indicated for respective antibody specificity pairs. Abbreviations: COVID-19, coronavirus disease 2019; Ctrl, control; IgG, immunoglobulin G; MFI, median fluorescence index; NC-C, nucleocapsid C-terminal fragment; NS, not significant; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; spike-f, spike glycoprotein.
A correlation analysis (Figure 2B) gave a similar result to that observed for cohort 1, with the highest reported correlation between salivary IgG to spike-f and NC-C (r = 0.81, P < .0001, Spearman correlation test). Significant, albeit moderate, correlations were also seen between IgA to spike-f and NC-C (r = 0.73, P < .001), IgG and IgA to spike-f (r = 0.49, P < .001), and spike-f IgA to NC-C IgG (r = 0.53, P < .001).
Influence of Inactivation Pretreatment and Room Temperature on Saliva Antibody Stability
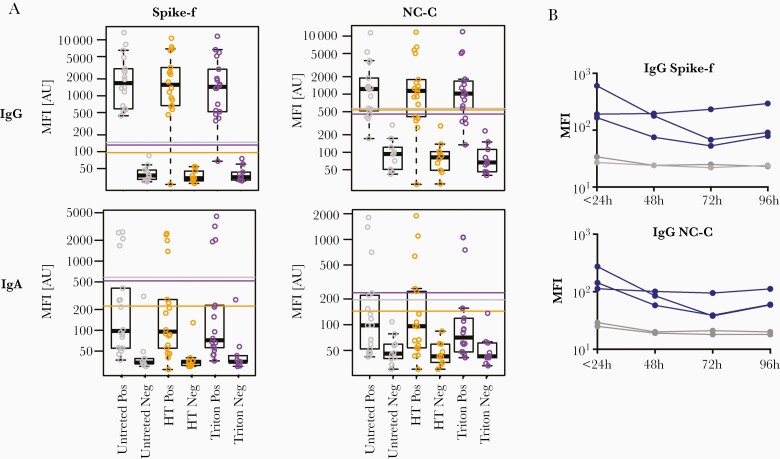
Next, the effects of virus inactivation by heat treatment at 56°C for 1 hour, 1% Triton X-100, and RT (identical aliquots stored at RT for 1-3 days) on the antibody results were determined (Figure 3). Both heat treatment and Triton X-100 showed little change in the cutoff (based on the 10 negative controls). A good correlation between treated and nontreated samples was noted (Figure 3 and Supplementary Figure 2), with a few exceptions of single samples that showed a drop in IgG reactivity. Simulation of RT storage (22°C) showed a slow decay in IgG signal intensity in positive samples over time, while the signal in negative samples remained low and stable. Based on these data, inactivation by heat treatment or Triton X-100 seems to have little effect on saliva samples. However, antibody decay variations showed slight IgG signal reduction by each day of RT storage.
Figure 3.
Stability tests of saliva samples subjected to HT, 1% Triton, and indicated time at room temperature. A, SARS-CoV-2–specific IgG and IgA reactivities in convalescent saliva samples (Pos) or prepandemic saliva samples (Neg) untreated, after HT at 56°C for 30 minutes, or after Triton inactivation for 60 minutes. Reactivities to spike-f and NC-C antigens are shown as box plots with each dot representing 1 sample. Each box represents the intensity signals interval included between the 25th and 75th interquartile, and the median is shown as a black line. Whiskers represent the interval including all intensity signals falling between the 5th and 95th percentile. For each condition (Untreated, HT and Triton), the cutoff for reactivity was calculated as the mean intensity of the negative samples included in the group plus 6x SD. The cutoff for each condition is represented by a colored line: grey = untreated, orange = HT, and purple = triton. B, Convalescent saliva samples (blue) or prepandemic saliva samples (gray) were aliquoted and placed at room temperature (22°C) for indicated times prior to freezing and subsequent measurement of SARS-CoV-2–specific IgG to spike or nucleocapsid. Abbreviations: HT, heat treatment; IgG, immunoglobulin G; MFI, median fluorescence index; NC-C, nucleocapsid C-terminal fragment; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; spike-f, spike glycoprotein; Triton, Triton-X-100.
Discussion
Comprehensive antibody testing and subsequent interventions are essential to monitor and control SARS-CoV-2 transmission. The present study demonstrates that salivary SARS-CoV-2–specific IgG after mild COVID-19 can serve as a complementary measure of exposure or immunity to SARS-CoV-2, particularly due to their frequent concurrence with serum IgG responses. Key findings included: (1) SARS-CoV-2–specific mucosal salivary antibodies coexisted with circulating blood antibodies for up to 9 months after natural infection in the majority of participants (88% in saliva vs 97% in blood); (2) natural infection induced salivary antibodies to recognize both viral spike and nucleocapsid proteins; (3) the response correlated significantly with recent COVID-19–like symptom history in undiagnosed individuals; and (4) salivary IgG is relatively stable, tolerating detergent and heat-based inactivation treatments. Taken together, these findings indicate saliva sampling is a noninvasive approach suitable for population-based immunity surveys. Ideally, if the saliva is sampled at home and mailed to the laboratory, it can help protect vulnerable persons at risk for severe COVID-19 by sparing them the need to visit laboratory units for blood sampling. It is therefore appealing, particularly during a pandemic, and can serve as a complementary test to conventional blood IgG assays. Our data also showed that sample inactivation by heat treatment or Triton X-100 were both viable options for biosafety handling procedures and caused minimal variation in assay performance. Options to combine with other point-of-care tests, such as lateral flow-based tests validated for blood, could be an interesting way forward [25].
Severe COVID-19 symptoms have been shown to induce strong antibody responses in 99% of convalescent individuals, but published data also show that these antibody responses tend to decline slower than in mild symptomatic cases [6, 9, 16, 19]. This may be attributed to the fact that tests developed earlier in the pandemic were based on detection of samples from severe COVID-19 cases resulting in suboptimal sensitivity to mild infections [26]. Furthermore, many of the initial test kits used the nucleocapsid as a target antigen and antibodies against it have been shown to decline more rapidly [27], as also demonstrated here. In this study, we deliberately used convalescent samples from mild COVID-19 patients to evaluate if the multiplex antibody platform was able to detect SARS-CoV-2–specific antibodies in saliva in such patients. In the present study, saliva reactivities were compared against blood serology using certified diagnostics (including anti-N pan-Ig ECLIA), which show high performance in detecting antibodies in late convalescent blood samples. Our result is in line with a South Korean group reporting recently that this diagnostic antibody assay is, among several others, effective in detecting SARS-CoV-2 antibodies in blood (90%) up to 8 months after either asymptomatic infection or mild symptomatic cases [28]. Here, the persistence of salivary IgG to structural viral proteins in the saliva samples 9 months after recovery from mild COVID-19 is intriguing, and possibly explained by a secondary exposure or spill-over from blood circulating responses. More studies are therefore warranted to clarify the mechanism underlying the magnitude of salivary responses with better matched study participants. It has been shown that the mucosal antibody response is triggered slightly earlier than the systemic response upon infection [10]. Information is still limited about the duration and kinetics of mucosal antibodies secreted into the mouth and nose, particularly in this patient group. A sensitive salivary antibody detection assay with the ability to identify infections with various severities would contribute to improving the current understanding of mucosal antibodies to SARS-CoV-2. For instance, such studies may compare low versus high avidity antibodies and their relation to neutralization or disease enhancement [10, 29, 30]. The advantage of multiplexed assays for antibody detection is that they minimize sample consumption and increase the throughput by maintaining high sensitivity and specificity. One example is the recent large-scale screenings of cross-reactivities to multiple pandemic or endemic coronaviruses [31], and the capacity to enable at-home testing for COVID-19 telemedicine diagnosis and monitoring, as proposed recently by Torrente-Rodríguez et al [32].
The hypothesis that antibodies towards previously known coronaviruses may block SARS-CoV-2 has raised questions about their functionality. However, such antibodies are known to be protective for only around 6 months after infection, and would therefore have disappeared by the time of emergence of SARS-CoV-2 [33, 34]. Clearly, further assessment of neutralizing capacity against SARS-Cov-2 virus and related coronavirus in human saliva is necessary. Other important applications for saliva immunoassays include evaluation of vaccine-induced mucosal immunity, which is ongoing in our laboratories for monitoring of local antibody recognition of virus mutations or vaccine-escape mutants. Because the mouth and nose are the first ports of entry for SARS-CoV-2, sensitive and accurate methods for quantitative measurements of local immunity will lead to better means to combat COVID-19.
One limitation of our study was the relatively small sample size and the predominantly male population. Another weakness was that blood samples were not analyzed in the same way as saliva and, as several diagnostic assays were used, only binary data were provided. Also, because of the cross-sectional design, we could not obtain baseline or longitudinal saliva samples. Moreover, we could not assess individual possibilities of reexposure or reinfection. However, it is unlikely that humoral immunity was boosted because in Stockholm, where the study took place, the period June to November 2020 (the second wave) showed an increase in the daily incidence rate of COVID-19 from 30 to 400 cases/100 000 population [35]. In conclusion, despite waning immunity concerns, the present study shows that our multiplex bead-based immunoassays can detect antibodies against SARS-CoV-2 in late convalescence saliva up to 9 months after mild COVID-19.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank all study participants who took an interest in this study.
Author contributions. E. P., A. M., P. N., and M. S. C. conceived and designed the study. H. A., S. B., A. M., and A. O. collected the material and performed the experiments. H. A., S. B., C. H., K. H., A. M., E. P., and M. S. C. analyzed the data. E. P., A. M., P. N., and M. S. C. supervised the work. K. L., S. A., G. B., and S. H. contributed material and data interpretations. H. A., S. B., E. P., and M. S. C. wrote the manuscript. K. H. and M. S. C. proof-read the revised submission. All authors reviewed and revised the manuscript critically.
Financial support. This work was supported by the Region Stockholm, Knut and Alice Wallenberg Foundation, Science for Life Laboratory, and the Erling-Persson Family Foundation (to S. H.). This project has received funding also from the European Union’s Horizon 2020 “HEalth data Linkage for ClinicAL benefit” training network, under the Marie Skłodowska-Curie grant agreement No. 813545.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Hassan Alkharaan, Department of Dental Medicine, Karolinska Institutet, Stockholm, Sweden; College of Dentistry, Prince Sattam bin Abdulaziz University, Al-Kharj, Saudi Arabia.
Shaghayegh Bayati, Department of Protein Science, Division of Affinity Proteomics, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, SciLifeLab, Stockholm, Sweden.
Cecilia Hellström, Department of Protein Science, Division of Affinity Proteomics, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, SciLifeLab, Stockholm, Sweden.
Soo Aleman, Department of Infectious Diseases, Karolinska University Hospital, Stockholm, Sweden; Department of Medicine, Karolinska Institutet, Stockholm, Sweden.
Annika Olsson, Department of Infectious Diseases, Karolinska University Hospital, Stockholm, Sweden.
Karin Lindahl, Department of Infectious Diseases, Karolinska University Hospital, Stockholm, Sweden; Department of Medicine, Karolinska Institutet, Stockholm, Sweden.
Gordana Bogdanovic, Department of Clinical Microbiology, Karolinska University Hospital, Stockholm, Sweden.
Katie Healy, Department of Dental Medicine, Karolinska Institutet, Stockholm, Sweden.
Georgios Tsilingaridis, Department of Dental Medicine, Karolinska Institutet, Stockholm, Sweden.
Patricia De Palma, Department of Dental Medicine, Karolinska Institutet, Stockholm, Sweden.
Sophia Hober, Department of Protein Science, Division of Protein Technology, KTH Royal Institute of Technology, Stockholm, Sweden.
Anna Månberg, Department of Protein Science, Division of Affinity Proteomics, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, SciLifeLab, Stockholm, Sweden.
Peter Nilsson, Department of Protein Science, Division of Affinity Proteomics, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, SciLifeLab, Stockholm, Sweden.
Elisa Pin, Department of Protein Science, Division of Affinity Proteomics, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, SciLifeLab, Stockholm, Sweden.
Margaret Sällberg Chen, Department of Dental Medicine, Karolinska Institutet, Stockholm, Sweden.
References
- 1. Zhu N, Zhang D, Wang W, et al. ; China Novel Coronavirus Investigating and Research Team . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johns Hopkins University. Coronavirus resource center. https://coronavirus.jhu.edu/map.html. Accessed 13 March 2021.
- 3. Krammer F. SARS-CoV-2 vaccines in development. Nature 2020; 586:516–27. [DOI] [PubMed] [Google Scholar]
- 4. Wang H, Zhang Y, Huang B, et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 2020; 182:713–21.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lumley SF, O’Donnell D, Stoesser NE, et al. ; Oxford University Hospitals Staff Testing Group . Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med 2021; 384:533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Premkumar L, Segovia-Chumbez B, Jadi R, et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 2020; 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 9. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021; 371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Isho B, Abe KT, Zuo M, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 2020; 5:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baumgarth N, Nikolich-Žugich J, Lee FE, Bhattacharya D. Antibody responses to SARS-CoV-2: let’s stick to known knowns. J Immunol 2020; 205:2342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pisanic N, Randad PR, Kruczynski K, et al. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. J Clin Microbiol 2020; 59:e02204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ripperger TJ, Uhrlaub JL, Watanabe M, et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity 2020; 53:925–33.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020; 26:1200–4. [DOI] [PubMed] [Google Scholar]
- 15. Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 2017; 27:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426:450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183:996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adeniji OS, Giron LB, Zilberstein NF, et al. COVID-19 severity is associated with differential antibody Fc-mediated innate immune functions. mBio 2021; 12:e00281–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rudberg AS, Havervall S, Månberg A, et al. SARS-CoV-2 exposure, symptoms and seroprevalence in healthcare workers in Sweden. Nat Commun 2020; 11:5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tegel H, Steen J, Konrad A, et al. High-throughput protein production—lessons from scaling up from 10 to 288 recombinant proteins per week. Biotechnol J 2009; 4:51–7. [DOI] [PubMed] [Google Scholar]
- 22. Kanje S, Enstedt H, Dannemeyer M, Uhlén M, Hober S, Tegel H. Improvements of a high-throughput protein purification process using a calcium-dependent setup. Protein Expr Purif 2020; 175:105698. [DOI] [PubMed] [Google Scholar]
- 23. Campbell I. Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat Med 2007; 26:3661–75. [DOI] [PubMed] [Google Scholar]
- 24. Richardson JT. The analysis of 2 × 2 contingency tables–yet again. Stat Med 2011; 30:890. [DOI] [PubMed] [Google Scholar]
- 25. Mulchandani R, Jones HE, Taylor-Phillips S, et al. ; EDSAB-HOME and COMPARE Investigators . Accuracy of UK rapid test consortium (UK-RTC) “AbC-19 Rapid Test” for detection of previous SARS-CoV-2 infection in key workers: test accuracy study. BMJ 2020; 371:m4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi S, Greenhouse B, Rodríguez-Barraquer I. Are seroprevalence estimates for severe acute respiratory syndrome coronavirus 2 biased? J Infect Dis 2020; 222:1772–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Havervall S, Jernbom Falk A, Klingström J, et al. SARS-CoV-2 induces a durable and antigen specific humoral immunity after asymptomatic to mild COVID-19 infection. medRxiv, doi: 10.1101/2021.01.03.21249162, 6. April 2021, preprint: not peer reviewed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choe PG, Kim KH, Kang CK, et al. Antibody responses 8 months after asymptomatic or mild SARS-CoV-2 infection. Emerg Infect Dis 2021; 27:928–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wen J, Cheng Y, Ling R, et al. Antibody-dependent enhancement of coronavirus. Int J Infect Dis 2020; 100:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol 2020; 5:1185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becker M, Strengert M, Junker D, et al. Exploring beyond clinical routine SARS-CoV-2 serology using MultiCoV-Ab to evaluate endemic coronavirus cross-reactivity. Nat Commun 2021; 12:1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torrente-Rodríguez RM, Lukas H, Tu J, et al. SARS-CoV-2 rapidplex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter 2020; 3:1981–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020; 583:290–5. [DOI] [PubMed] [Google Scholar]
- 34. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med 2020; 26:1691–3. [DOI] [PubMed] [Google Scholar]
- 35. Folkhälsomyndighetens. Folkhälsomyndighetens veckorapporter om covid-19, June–November 2020. https://www.folkhalsomyndigheten.se/folkhalsorappo. Accessed 13 March 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.