Abstract
Aim:
To understand miRNA changes across gestation in healthy human placentae. This is essential before miRNAs can be used as biomarkers or prognostic indicators during pregnancy.
Materials & methods:
Using next-generation sequencing, we characterize the normative human placenta miRNome in first (n = 113) and third trimester (n = 47).
Results & conclusion:
There are 801 miRNAs expressed in both first and third trimester, including 182 with similar expression across gestation (p ≥ 0.05, fold change ≤2) and 180 significantly different (false discovery rate <0.05, fold change >2). Of placenta-specific miRNA clusters, chromosome 14 miRNA cluster decreases across gestation and chromosome 19 miRNA cluster is overall highly expressed. Chromosome 13 clusters are upregulated in first trimester. This work provides a rich atlas of healthy pregnancies to direct functional studies investigating the epigenetic differences in first and third trimester placentae.
Keywords: : chorionic villi, gestational differences, human transcriptome, miRNA, miRNA atlas, miRNome, placenta, pregnancy, stable miRNAs, uncomplicated pregnancies
Lay abstract
The human body produces miRNAs which affect the expression of genes and proteins. This study uses next-generation sequencing to identify the miRNA profile of first and third trimester human placentae using a large cohort (n = 113 first trimester; n = 47 third trimester). All pregnancies resulted in healthy babies. We identify miRNAs with significantly different expression between first and third trimester, as well as stably expressed miRNAs. This work provides a baseline for future studies which may use miRNAs to monitor maternal–fetal health throughout pregnancy.
The placenta plays a critical role in fetal development, forming the interface between the developing fetus and mother. It has multiple functions including provision of oxygen and nutrients to the fetus, removal of waste products, and constitutes an important barrier protecting the fetus from pathogens and environmental toxins throughout gestation. The formation and development of the placenta (placentation) that begins upon implantation and continues throughout the first trimester of pregnancy lays the foundation for a placenta leading to a healthy gestation. After implantation, the placenta produces growth factors, cytokines and hormones which target maternal physiological systems, facilitating the provision of additional blood flow and nutrient delivery to the fetus [1]. During the first trimester, cytotrophoblast cells differentiate into extravillous trophoblasts and syncytiotrophoblasts. Extravillous trophoblasts migrate by invading and remodeling the maternal decidual extracellular matrix and spiral arteries [2–5]. This occurs during a relative state of hypoxia, which promotes trophoblast invasion and angiogenesis [6–9]. It is also during this time that cytotrophoblasts fuse into syncytiotrophoblasts, creating a multinucleated epithelium that lines the intervillous space and also produces hormones including estrogen and progesterone for pregnancy maintenance and lactogen for fetal metabolism, growth and development [10–12]. The complexities of normal placental development are under the control of various epigenetic modifications that may be altered, ultimately leading to abnormal placentation and adverse outcomes [13,14].
miRNAs are noncoding, ssRNA molecules of about 22 nucleotides in length [15]. They are important post-transcriptional regulators of gene expression. They bind to RNA transcripts, causing RNA cleavage or mRNA translational repression, regulating 30–50% of all mammalian protein-coding genes [16,17]. While some miRNAs are universally expressed, others are expressed preferentially or exclusively in certain tissues [18]. Two large miRNA clusters are enriched in placenta, the chromosome 14 miRNA cluster (C14MC) and the chromosome 19 miRNA cluster (C19MC) [19,20]. C14MC is a large, imprinted, maternally-expressed miRNA cluster, with several members predominantly expressed in placenta and epithelial tissues [18]. C19MC is a large, imprinted, paternally-expressed miRNA cluster whose members have highest expression in placenta and cancer, with relatively weak expression in other tissue [18,20–24].
Widespread regulation of gene expression by miRNAs, the presence of placental-specific miRNAs and miRNA expression differences in various trophoblastic cell lines [25] suggest a role for miRNAs in trophoblast behavior and placental development and function. Furthermore, altered miRNA expression in the placenta may be involved in abnormal placentation and related pregnancy-associated diseases including preeclampsia [26–40] and intrauterine growth restriction [39–43]. Given that placental function changes from the first trimester during a critical state of placental development and continues to function for appropriate fetal development, epigenetic regulation through miRNAs likely plays a key role. In addition, due to their small size and stability, miRNAs are potential biomarkers of disease, particularly miRNAs from plasma exosomes [44–46]; however, it is essential to know how their expression varies across gestation. Previous studies comparing miRNA expression in first and third trimester placentae are limited to microarray analyses and have had conflicting results [25,47,48]. We performed next-generation sequencing (NGS) and expression analysis to identify and compare the normative miRNA signatures in first and third trimester placentae of healthy pregnancies resulting in delivery.
Materials & methods
Study population
The study population consisted of 157 singleton pregnancies with available first trimester placental tissue (n = 110), third trimester placental tissue (n = 44) or both (n = 3 in both groups), obtained between 2009 and 2018. Mothers with pre-existing diabetes or hypertension were excluded. All protocols were performed in accordance with the institutional review board’s guidelines at the Cedars-Sinai Medical Center. All subjects were enrolled under Institutional Review Board approved protocols (Pro00006806 and Pro00008600). All pregnancies had a normal karyotype and resulted in the delivery of a viable infant.
Analysis of demographic data
Demographic data included parental ages, races and ethnicities, maternal pre-pregnancy BMI, gestational age at chorionic villus sampling (CVS), fetal sex, maternal medical history and medication use, pregnancy complications, mode of delivery, gestational age at delivery, and birth weight. Means and standard deviations were reported for continuous variables. Proportions were reported as percentages. Demographics were compared between patients in the first and third trimester placenta, excluding three subjects sequenced in both groups to eliminate duplicate information. The t-test was used for normally distributed continuous variables, and the Wilcoxon rank-sum test was used for nonparametric data. Fisher’s exact test was used when appropriate. For comparison of categorical variables, the chi-square test was used.
Collection of first trimester placental samples
Samples from the first trimester of pregnancy were collected between 70–102 days gestation during CVS procedures done for prenatal diagnosis. Samples used for research consisted of extra tissue which is normally discarded after sending chorionic villi specimens for prenatal genetic diagnostic testing. Fetal-derived chorionic villi (first trimester placenta tissue) were cleaned and separated from any maternally derived decidua (nonplacenta tissue). Tissue samples (5–25 mg) were kept on ice and submerged in RNAlater RNA stabilization reagent (Qiagen, Hilden, Germany) within 30 min of collection and stored at -80°C.
Collection of third trimester placental samples
Samples from the third trimester of pregnancy were collected between 254–290 days gestation, after delivery of a viable neonate. Samples used for research consisted of tissue which would have otherwise been discarded. One centimeter cubed of placental tissue samples were obtained immediately after delivery from the fetal side of the placenta near the site of cord insertion beneath the amnion. The samples were cleaned and submerged in RNAlater RNA stabilization reagent (Qiagen) and stored at -80°C.
RNA extraction from first trimester placenta
RNA was extracted from leftover CVS tissue utilizing a method optimized for delicate tissue [49,50]. Briefly, tissue samples were thawed on ice with 600 μl of RLT Plus lysis buffer (Qiagen) containing 1% β-mercaptoethanol. Tissue was homogenized by passing at least ten-times through progressively thinner gauge needles (22-, 25- and 27-G) attached to an RNase-free syringe. Homogenates were loaded onto AllPrep spin columns and the remainder of sample processing was performed following manufacturer instructions using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen). RNA was eluted with 30–45 μl of RNase-free water at room temperature and the elution was passed through the column twice to improve yields, as previously described [49,50]. The average RNA integrity number for sequenced first trimester placenta samples was 8.87.
RNA extraction from third trimester placenta
Third trimester placenta tissue was thawed on ice, then a quarter of collected tissue was diced with RNase-free blades coated in RNAlater buffer, tissue was sonicated on ice in lysis buffer (600nμl RLT Plus lysis buffer with 1% β-mercaptoethanol) using 5 s pulses on a low setting (setting 2 on the Branson Sonifier 150D, CT, USA) until tissue fragments were small enough to complete homogenization with RNase-free needles. Further extraction was performed as described for first trimester tissue. The average RNA integrity number for sequenced third trimester placentae was 8.84.
Library preparation & miRNA sequencing
A miRNA sequencing library was prepared from total RNA using the QIASeq miRNA Library Kit (Qiagen). A pre-adenylated DNA adapter was ligated to the 3′ ends of miRNAs, followed by ligation of an RNA adapter to the 5′ end. A reverse-transcription primer containing an integrated unique molecular index (UMI) was used to convert the 3′/5′ ligated miRNAs into cDNA. After cDNA cleanup, indexed sequencing libraries were generated via sample indexing during library amplification, followed by library clean-up. Libraries were sequenced on a NextSeq 500 (Illumina, CA, USA) with a 1 × 75 bp read length and an average sequencing depth of 10.64 million reads per sample.
Differential expression analysis of miRNAs
The demultiplexed raw reads were uploaded to GeneGlobe Data Analysis Center (Qiagen) at https://www.qiagen.com/us/resources/geneglobe/ for quality control, alignment and expression quantification. Briefly, 3′ adapter and low quality bases were trimmed off from reads first using cutadapt v1.13 with default settings [51], then reads with <16 bp insert sequences or with <10 bp UMI sequences were discarded. The remaining reads were collapsed to UMI counts and aligned sequentially to miRBase release 21 mature and hairpin RNA databases using Bowtie v1.2 [52,53]. The UMI counts of each miRNA category were quantified, and then normalized by a size factor-based method in the R package DESeq2 v1.22.2 (Bioconductor) [54]. Data were averaged across all samples in each group (first and third trimester) for each respective miRNA and were reported as baseMean. The R package FactoMineR v1.41 was used to conduct principal components analysis (PCA), which was used to investigate clustering and potential outliers. Differential expression analysis was performed with DESeq2 to compare first versus third trimester expression, adjusting for fetal sex. Each miRNA was fitted into a negative binomial generalized linear model, and the Wald test was applied to assess the differential expressions between two sample groups (p-value). The Benjamini and Hochberg procedure was applied to adjust for multiple hypotheses testing, and those with false discovery rate (FDR) <0.05 were selected as significantly differentially (D) expressed (DE) miRNAs. The genome locations of miRNAs were identified by cross-referencing mature miRNA IDs with precursor miRNA accession IDs in miRBase release 21, then using R package biomaRt v2.45.8 and Ensembl release 91 (which contains miRBase release 21) to retrieve chromosome locations [55,56]. For miRNAs derived from >1 precursor, all precursors were counted in bar plots (e.g., hsa-miR-1184 is encoded by three precursors on chromosome X, thus was counted three-times) and each miRNA was plotted once per chromosome in scatter plots of genome distribution. Duplicate precursors were counted once to avoid copy number variants (e.g., hsa-miR-941-5 [57]), with the exception of Supplementary File 5.
Analysis of miRNA expression
Counts normalized for sequencing depth (baseMeans) were used as a measure of expression since miRNA lengths do not vary substantially. All expressed miRNAs were defined as any with baseMean >10 in first or third trimester placenta groups. The miRNAs common in both first and third trimester placentae, also known as similarly (S) expressed (SE), were defined by p ≥ 0.05, absolute fold change ≤2 and baseMean >10 in both trimesters. The p ≥ 0.05 threshold excludes all significantly different miRNAs (FDR <0.05) as well as other potentially different miRNAs (p < 0.05). DE miRNAs were defined as FDR <0.05, absolute fold change >2 and baseMean >10 in both trimesters. Higher expression thresholds were selected for target enrichment analysis when needed (next section). For miRNAs in C14MC and C19MC, these filters were applied: baseMean >1 and p ≥ 0.05 (SE) or baseMean >1 and FDR <0.05 (DE).
Enrichment analysis of predicted miRNA target genes
Ingenuity Pathways Analysis (IPA) software’s miRNA Target Filter application (Qiagen, CA, USA, http://www.qiagenbioinformatics.com/IPA) was used to generate a list of target RNAs based on sequence and experimental confirmation. Targets were included if biochemically confirmed using human tissue or nonspecies methods (sourced from Qiagen’s curated Ingenuity Knowledge Base, or the publicly available miRecords or TarBase). IPA’s core analysis function was used to test the hypothesis that the target genes are enriched in canonical biological pathways, as previously described [49,58,59]. Supplementary data also show core analysis results with additional targets predicted with high confidence according to IPA, based on the TargetScan algorithm previously described [60]. IPA designates high confidence as a cumulative weighted context score of ≤-0.4, predicting decreased expression by at least 25% due to a specific miRNA.
The input miRNAs were reduced with higher expression thresholds so that target gene numbers did not exceed IPA software limitations for core analysis. The following definitions were applied for highly expressed miRNAs (baseMean >10,000 in both trimesters), SE miRNAs (p ≥ 0.05, absolute fold change ≤2 and baseMean >1000 in both trimesters) and DE miRNAs (FDR <0.05, absolute fold change >2 and baseMean >1000 in both trimesters). Due to their smaller number, no additional miRNA filters were required for core analysis of C14MC and C19MC targets.
Heatmaps
Heatmap and dendrograms of samples versus miRNAs were created with a matrix of log2(baseMean) values scaled and centered by rows. The heatmaps and dendrograms were created with hierarchical clustering from R package gplots v3.1.1. Heatmaps of gene enrichment were created with R package pheatmap v1.0.12 with a matrix of -log10(P) output from IPA core analysis.
Validation with qRT-PCR
Expression of six selected miRNAs were reanalyzed with an independent cohort by quantitative real-time PCR (qRT-PCR) using the miRCURY LNA miRNA PCR system (Qiagen). The validation cohort was balanced for fetal sex and fetal race (all Caucasian). The six selected (hsa-miR-144-3p, hsa-miR-24-3p, hsa-miR-126-3p, hsa-miR-145-5p, hsa-miR-143-3p and hsa-miR-126-5p) had high expression to ensure signal from qRT-PCR (baseMean >1000), were significantly different in miRNA sequencing (FDR <10-13) between first and third trimester, and had good but relatively low fold changes (between 2–4). A highly expressed miRNA with stable expression in first and third trimester placentae was used as a reference gene (hsa-miR-130a-3p; baseMean >10,000 in both trimesters, p = 0.9693, FDR = 0.9889). RNAs from first trimester (n = 10) and third trimester (n = 6) placenta samples were extracted, then cDNA was synthesized using universal primers in the miRCURY LNA RT Kit (Qiagen). Expression was quantified by qRT-PCR using miRCURY LNA SYBR Green PCR Kit (Qiagen) and a BioRad MyIQ machine, and was analyzed using the ΔΔCt method [61], with hsa-miR-130a-3p as an internal reference. Statistics were performed using the Wilcoxon rank-sum test on ΔCt values.
Results
Cohort demographics & pregnancy outcomes
There were n = 113 first trimester placenta samples and n = 47 third trimester placenta samples studied, including three subjects with both first and third trimester placenta sequenced. PCA shows that first and third trimester placenta segregated into two distinct clusters divided by principal component 1 (Supplementary File 1). There were significantly more non-Hispanic, Caucasian parents and fetuses in the first trimester group (Table 1). However, race and ethnicity groups did not cluster in PCA analyses of the miRNA transcriptome (Supplementary File 1). Maternal pre-pregnancy BMI and thyroid disorders requiring thyroid replacement were significantly different among the groups (Table 1). Pregnancy outcome information was available for both groups, with no differences in gestational diabetes, placenta previa, mode of delivery, gestational age at delivery or birth weight (Table 1). There were more cases of pregnancy-induced hypertension requiring anti-hypertensives and/or magnesium in the third trimester placenta group, compared with none in the first trimester placenta group.
Table 1. . Demographics and birth outcomes.
| First trimester | Third trimester | p-value | |
|---|---|---|---|
| n | 110 | 44 | |
| Maternal age, years | 37.7 (3.0) | 37.3 (3.0) | 0.65 |
| Paternal age, years | 39.5 (4.8) | 38.6 (4.8) | 0.30 |
| Maternal race/ethnicity | |||
| Caucasian | 106 (96.4%) | 35 (79.6%) | 0.002 |
| Non-Hispanic | 107 (97.3%) | 35 (79.6%) | <0.001 |
| Paternal race/ethnicity | |||
| Caucasian | 104 (94.6%) | 37 (84.1%) | 0.04 |
| Non-Hispanic | 107 (97.3%) | 36 (81.8%) | 0.001 |
| Fetal race/ethnicity | |||
| Caucasian | 103 (93.6%) | 32 (72.7%) | <0.001 |
| Non-Hispanic | 106 (96.4%) | 33 (75.0%) | <0.001 |
| Fetal female sex | 57 (51.8%) | 18 (40.9%) | 0.22 |
| Maternal pre-pregnancy BMI, kg/m2 | 21.9 (3.4) | 24.0 (4.7) | 0.005 |
| Maternal pre-existing medical conditions | |||
| Hypertension | 0 (0%) | 0 (0%) | - |
| Diabetes | 0 (0%) | 0 (0%) | - |
| Thyroid disorder | 3 (2.7%) | 6 (13.6%) | 0.02 |
| Pregnancy complications | |||
| Hypertension (not pre-existing) | 0 (0%) | 8 (18.2%) | <0.001 |
| Gestational diabetes | 3 (2.7%) | 1 (2.3%) | 1.0 |
| Placenta previa | 0 (0%) | 1 (2.3%) | 0.49 |
| Placental abruption | 0 (0%) | 0 (0%) | - |
| Hypertension management in pregnancy | |||
| Anti-hypertensives | 0 (0%) | 3 (6.8%) | 0.022 |
| Any magnesium use (ante or postpartum) | 0 (0%) | 6 (13.6%) | <0.001 |
| Mode of delivery – cesarean section | 33 (30%) | 15 (34.1%) | 0.74 |
| Gestational age at delivery, days | 276.3 (7.0) | 276.5 (8.0) | 0.43 |
| Birth weight, g | 3435.4 (463.6) | 3473.1 (467.0) | 0.7 |
Values shown as mean (standard deviation) or n (%). P-values were adjusted for fetal sex.
All expressed miRNAs in placenta
We identified 2503 mature miRNAs with high-throughput sequencing of first and third trimester placentae, with 801 miRNAs reaching ten normalized counts in both first and third trimester placentae (baseMean >10). First trimester placenta expressed 872 mature miRNAs (baseMean >10), derived from 967 miRNA precursors from all chromosomes with annotated miRNAs (22 autosomes and the X chromosome) (Figure 1A & Supplementary File 2). By raw counts, expressed precursor miRNAs originate mostly from chromosomes 19 (12.7%), 14 (10.8%), X (9.6%) and 1 (6.8%). Third trimester placenta expressed 882 mature miRNAs (baseMean >10), derived from 985 miRNA precursors from all 22 autosomes and the X chromosome (Figure 1A & Supplementary File 2). The most represented chromosomes by raw counts were also 19 (12.4%), 14 (10.5%), X (9.7%) and 1 (7.1%). As a percentage of placenta-expressed miRNAs per all human miRNAs, the most represented chromosomes were 14 (64.6% first trimester; 64.0% third trimester), 19 (52.1; 51.7%), X (47.9; 49.5%) and 13 (47.4; 42.1%) (Figure 1A).
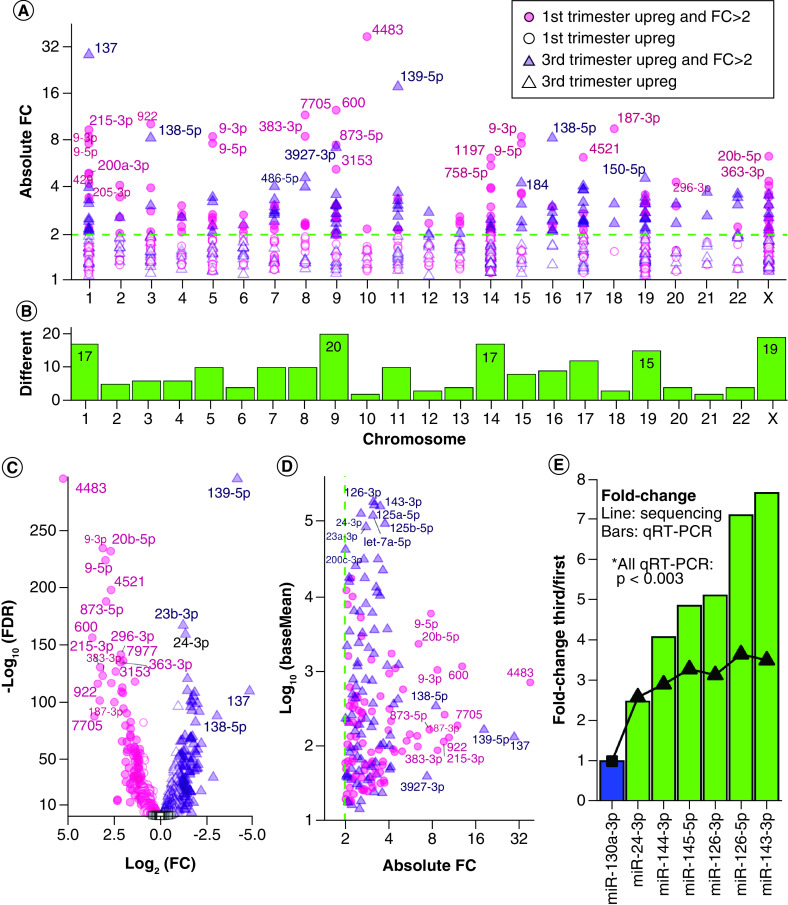
Figure 1. . Expressed miRNAs in first and third trimester placenta.
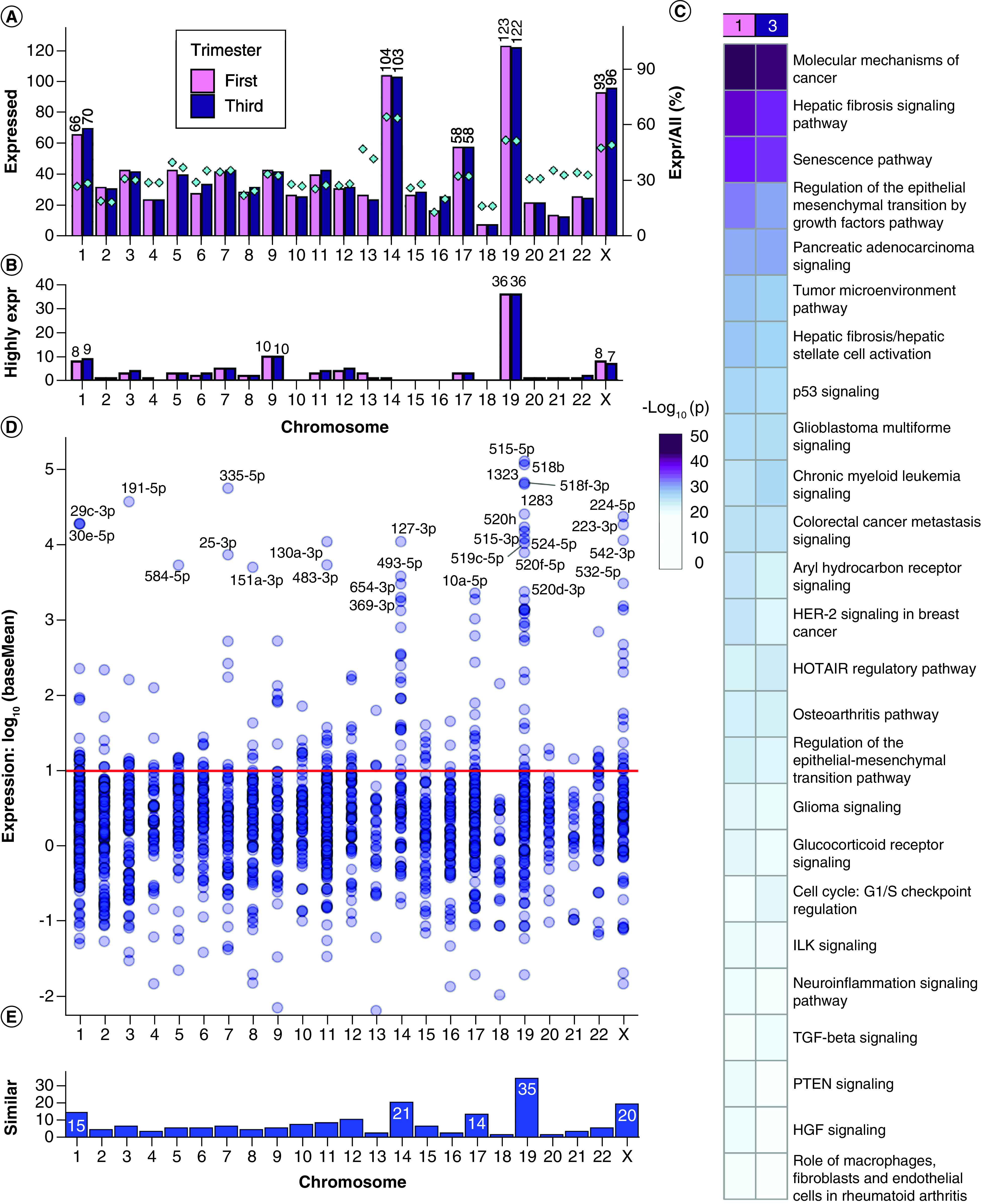
(A) Chromosome distribution of precursor miRNAs expressed at baseMean >10 in first or third trimester placenta. Barplot: frequency per chromosome. Diamond points and right y-axis: percentage of placenta-expressed miRNAs per total chromosome miRNAs. (B) Distribution of the most highly expressed precursor miRNAs at baseMean >10,000. (C) Pathway enrichment analysis with experimentally confirmed target genes of the most highly expressed miRNAs. (D) The expression distribution of miRNAs similarly expressed in first and third trimester at p ≥ 0.05 and fold change ≤2. The red line (baseMean = 10) is the threshold selected for stable expression. (E) Counts of similarly expressed miRNAs with p ≥ 0.05, fold change ≤2 and baseMean >10 in both trimesters.
Highly expressed miRNAs in placenta
Some miRNAs had expression values several orders of magnitude higher than most miRNAs, with the median at baseMean = 123.6 but the mean raised to baseMean = 5635 by these highly expressed miRNAs (Supplementary File 2). A threshold of baseMean >10,000 was selected for the ‘most highly expressed’ miRNAs. There were 75 mature miRNAs (derived from 96 precursors) in first trimester and 77 mature miRNAs (derived from 97 precursors) in third trimester placenta which reached this threshold (Figure 1B). The most highly expressed miRNA in first trimester was C19MC member miR-517b-3p (baseMean = 218,953). The most highly expressed miRNA in third trimester and overall most highly expressed was miR-126-3p (baseMean = 337,399). Chromosome 19 encoded 30 mature miRNAs (derived from 36 precursors) which reached baseMean >10,000 in both first and third trimester (Figure 1B), making chromosome 19 the source of over 37% of the most highly expressed precursor miRNAs in human placenta. Specifically, 28 of the 36 precursor miRNAs were C19MC members, and eight localized elsewhere on chromosome 19. The next chromosomes contributing the most highly expressed miRNAs were chromosome 9, chromosome 1 and chromosome X.
We performed pathway enrichment analysis on experimentally confirmed targets of the most highly expressed miRNAs (Figure 1C & Supplementary File 3 & Supplementary File 4Ai). The most significantly enriched canonical pathways in first and third trimester were ‘molecular mechanisms of cancer’, ‘hepatic fibrosis signaling’, ‘senescence’, ‘regulation of the epithelial mesenchymal transition by growth factors’ and ‘pancreatic adenocarcinoma signaling.’
Pathway enrichment analysis with both experimentally confirmed miRNA targets as well as targets predicted with high confidence demonstrated similar patterns, though third trimester showed relatively higher enrichment in ‘hepatic fibrosis/hepatic stellate cell activation’ and ‘regulation of the epithelial mesenchymal transition by growth factors’ compared with first trimester (Supplementary File 3 & Supplementary File 4Aii). Additional pathways were also enriched including inflammatory pathways such as ‘neuroinflammation’, ‘prolactin’, ‘systemic lupus erythematosus in B cell’ and ‘IL-6’ signaling (Supplementary File 4Aii).
SE miRNAs in first & third trimesters
There were 182 mature miRNAs similarly expressed (SE) in the first and third trimester placentae (p ≥ 0.05, fold change ≤2 and baseMean >10, Supplementary File 3), suggesting consistent expression throughout gestation (Figure 1D). These mature miRNAs are derived from 206 precursor miRNAs, with greatest representation from chromosomes 19 (17.0%), 14 (10.2%), X (9.7%) and 1 (7.3%) (Figure 1E). The most highly expressed SE miRNA was C19MC member hsa-miR-515-5p with first trimester baseMean = 129,659 and third trimester baseMean = 129,323, p = 0.902 between trimesters. This was followed closely by other C19MC members: hsa-miR-518b, hsa-miR-518f-3p, hsa-miR-1323 and hsa-miR-1283.
DE miRNAs between first & third trimesters
There were 781 mature miRNAs significantly DE between first and third trimester placentae at FDR <0.05 (31.2% of 2503 detected miRNAs), including 588 miRNAs with reliable expression at baseMean >10 (Figure 2A), further filtered to 180 miRNAs with fold change >2. Of these final 180 DE miRNAs (FDR <0.05, baseMean >10, fold change >2), 91 are upregulated in first trimester and 89 are upregulated in third trimester (Supplementary File 2). The 180 DE miRNAs were derived from 202 precursors with highest representation from chromosomes 9 (9.9%), X (9.4%), 1 and 14 (8.4% each), and 19 (7.4%) (Figure 2B). The end of chromosome X is enriched for first trimester upregulated miRNAs (Supplementary File 5). The most DE miRNA was hsa-miR-4483, with 38.2-fold higher expression in the first trimester placenta (FDR = 0) and a baseMean decrease from 984.1 to 25.5 from first to third trimester (Figure 2C & Supplementary File 2). The next most significantly DE miRNA was hsa-miR-139-5p with 18.1-fold higher expression in the third trimester (FDR = 3.69 × 10-298), baseMean increasing from 28.0 to 497.5 (Figure 2C). The DE miRNA with the highest overall expression was hsa-miR-126-3p, with a 3.13-fold higher expression (FDR = 4.52 × 10-97) in third trimester placenta (baseMean = 337,399) compared with first trimester placenta (baseMean = 107,787) (Figure 2D). Of the DE miRNAs, those with the greatest fold changes had lower to moderate expression around baseMean 100–1000 and were predominantly elevated in the first trimester, whereas those with the highest expression had lower fold changes and were predominantly elevated in the third trimester (Figure 2D). Zero DE miRNAs with baseMean >1000 reached fold change of 4 (Figure 2D & Supplementary File 2). Additionally, chromosome 13 miRNA distribution shows two small clusters enriched for first trimester upregulated miRNAs (Supplementary File 5).
Figure 2. . Differentially expressed miRNAs between first and third trimester placenta.
(A) Scatter plot of absolute fold change distribution across chromosomes for all DE miRNAs at FDR <0.05 and baseMean >10. The dotted line represents FC = 2. (B) Chromosome frequency of 180 DE miRNA precursors at FDR <0.05, FC >2, baseMean >10. (C) Volcano plot of all miRNAs with baseMean >10. Key as in (A), with addition of open black squares for nonsignificant miRNAs (FDR ≥0.05). (D) Expression versus absolute fold change for 180 DE miRNAs. (E) Six DE miRNAs (green) were selected for validation via quantitative real-time-PCR in an independent cohort. The bar plot shows qRT-PCR results normalized to an internal reference, hsa-miR-130a-3p (blue). The superimposed line shows fold changes in miRNA-seq. All six miRNAs were validated significantly different between first and third trimester with p < 0.003.
DE: Differentially expressed; FC: Fold change; FDR: False discovery rate.
Validation of DE miRNAs
Six DE miRNAs identified using NGS were selected for validation (Figure 2E). We performed qRT-PCR using an independent cohort of first (n = 10) and third trimester (n = 6) placenta samples. The miRNA hsa-miR-130a-3p was selected as an internal reference due to high and stable expression in first and third trimester placentae (p = 0.9693, fold change = 0.9984 first/third, baseMean = 11,097). All six validated miRNAs (hsa-miR-24-3p, hsa-miR-144-3p, hsa-miR-145-5p, hsa-miR-126-3p, hsa-miR-126-5p and hsa-miR-143-3p) were upregulated in third trimester placenta with 2.5- to 3.7-fold changes by sequencing, and all six were confirmed significant by qRT-PCR with p < 0.003 (Figure 2E).
Comparison of SE & DE miRNAs
A heatmap of the 182 SE miRNAs shows no clustering of the first and third trimester samples (Figure 3A). The heatmap of 180 DE miRNAs shows placenta sample clustering by trimester, and miRNAs clustering into two groups by direction of upregulation (Figure 3B). There was little subject variability within first and third trimester, but some miRNAs were not consistently expressed (baseMean = 0, red).
Figure 3. . Heatmaps showing sample miRNA variability.
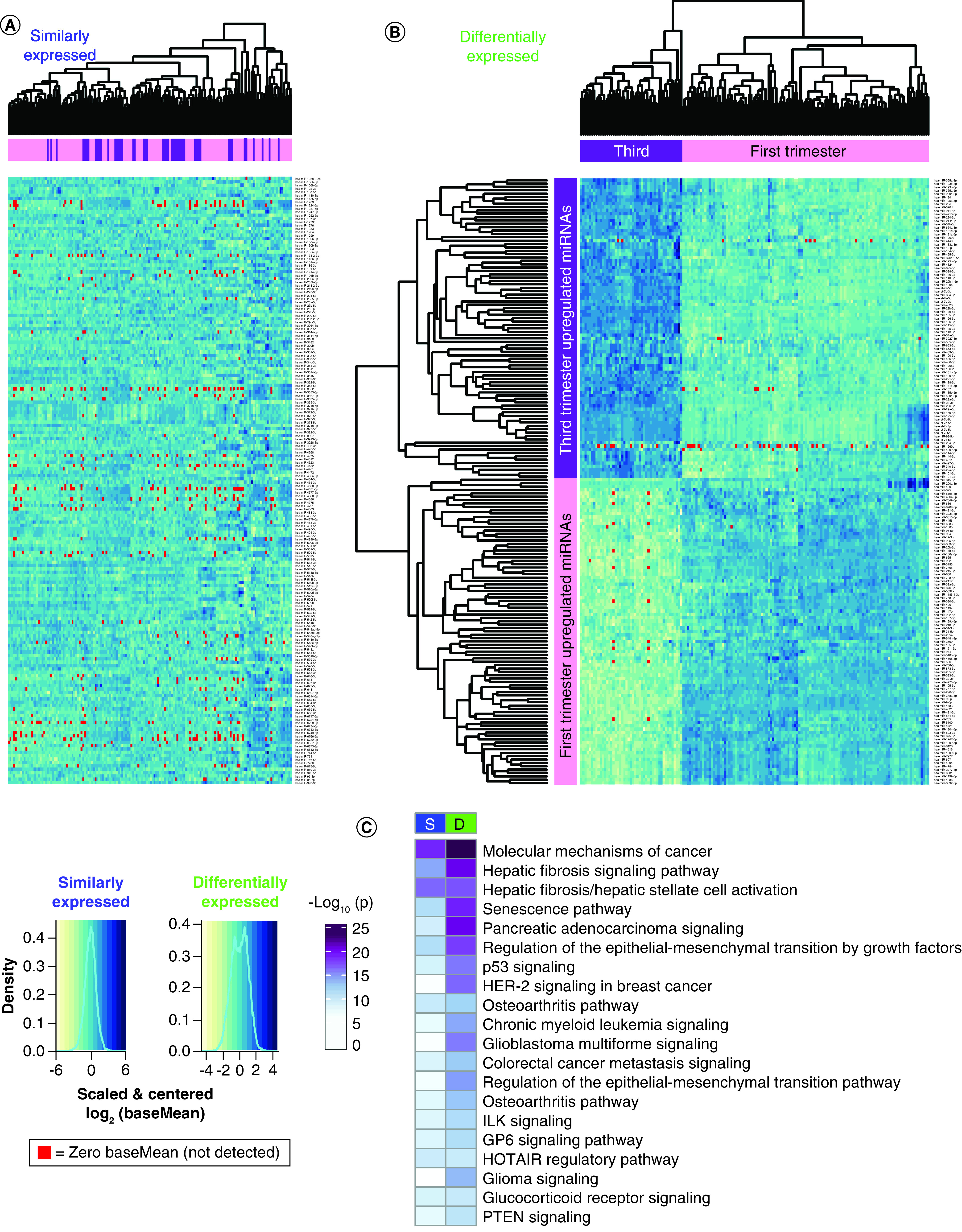
Heatmaps: rows = scaled and centered miRNA log2(baseMean), columns = hierarchically clustered samples. BaseMean = 0 samples are highlighted red. (A) One hundred eighty two similarly expressed miRNAs with p ≥ 0.05, fold change ≤2 and baseMean >10. The miRNAs are listed alphabetically. (B) One hundred eighty differentially expressed miRNAs with FDR <0.05, fold change >2 and baseMean >10. The miRNAs are hierarchically clustered. (C) Pathway enrichment analysis for experimentally confirmed targets of S and D expressed miRNAs.
D: Differentially; FDR: False discovery rate; S: Similarly.
Pathway enrichment analysis was performed for experimentally confirmed miRNA targets to identify potential regulatory roles of the miRNAs expressed in placenta (Figure 3C & Supplementary File 3 & Supplementary File 4B). The most significantly enriched pathways, targeted by both SE and DE miRNAs in first and third trimester placenta were ‘molecular mechanisms of cancer’ and ‘hepatic fibrosis signaling’. None of the top 20 pathways were more significantly targeted by SE miRNAs, suggesting high variability throughout gestation (Supplementary File 4Bi). DE miRNAs targeted more significantly by highly expressed miRNAs in the first trimester include ‘molecular mechanisms of cancer’, ‘hepatic fibrosis signaling’, ‘senescence’ and ‘regulation of the epithelial mesenchymal transition by growth factors’ pathways, suggesting these pathways are distinctly regulated by miRNAs in the first versus third trimester (Figure 3C & Supplementary File 4Bi).
When the pathway enrichment analysis was repeated with both experimentally confirmed miRNA targets as well as targets predicted with high confidence, additional patterns emerge. DE miRNAs target the ‘hepatic fibrosis/hepatic stellate cell activation’ pathway more heavily than SE miRNAs when predicted targets are included (Supplementary File 3 & Supplementary File 4Bii). Addition of predicted targets highlights specific cytokine and growth factor pathways including ‘IL-6’ and ‘IGF-1’ signaling, which are heavily targeted by SE miRNAs and less so by DE miRNAs (Supplementary File 4Bii).
Expression from C14MC & C19MC
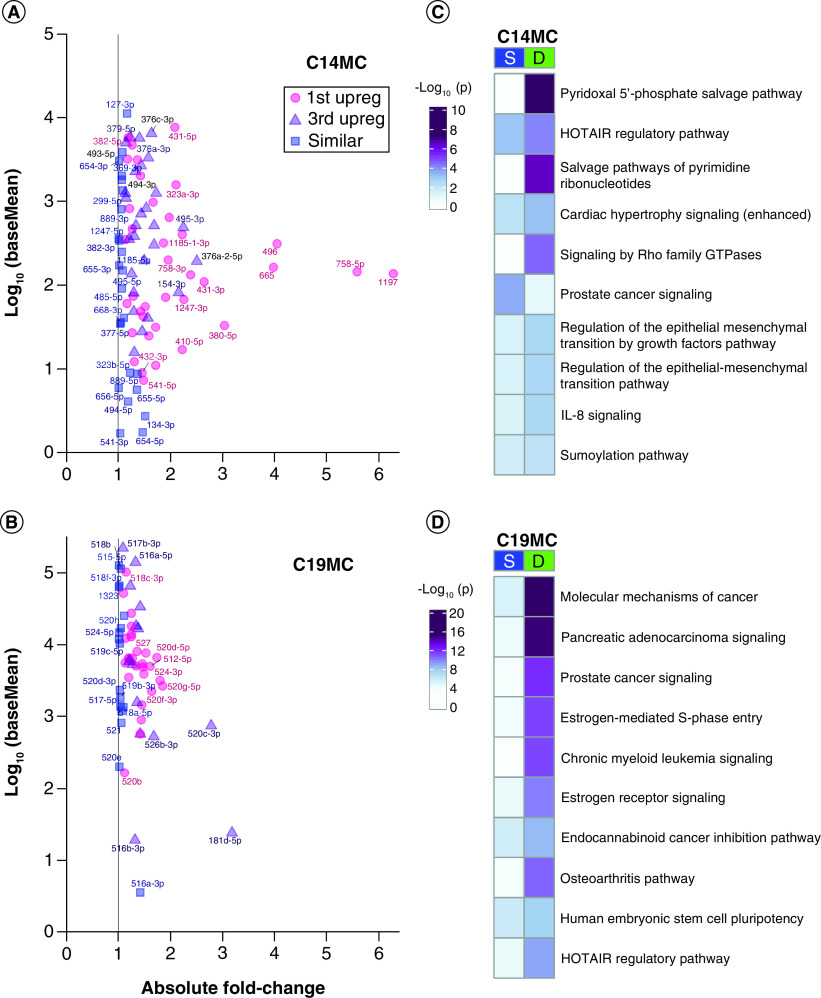
The placenta specific miRNA clusters expressed 42 mature SE miRNAs between first and third trimester placenta (p ≥ 0.05, baseMean >1), 24 from C14MC and 18 from C19MC (Figure 4A & B & Supplementary File 6). There were 105 mature DE miRNAs between first and third trimester (FDR <0.05 and baseMean >1), 64 from C14MC and 41 from C19MC (Figure 4A & B & Supplementary File 6). The cluster miRNAs with highest fold change came from C14MC: hsa-miR-1197 (6.28-fold, FDR = 4.83 × 10-118), hsa-miR-758-5p (5.59-fold, FDR = 5.07 × 10-101), hsa-miR-496 (4.05-fold higher in first, FDR = 3.92 × 10-109) and hsa-miR-665 (3.98-fold, FDR = 1.75 × 10-95), all higher in first trimester compared with third trimester placenta.
Figure 4. . Placenta-specific chromosome 14 miRNA cluster & chromosome 19 miRNA cluster.
(A & B) Expression versus absolute fold change plots for cluster miRNAs at baseMean >1. Pink = upregulated in first trimester at false discovery rate <0.05. Purple = upregulated in third trimester at FDR <0.05. Blue = similarly expressed with p ≥ 0.05. Point labels are the miRNA names minus the ‘hsa-miR-’ prefix. (A) C14MC miRNAs. (B) C19MC miRNAs. (C & D) Pathway enrichment analysis with experimentally confirmed targets of the similarly expressed or differentially expressed miRNAs in (C) C14MC or (D) C19MC.
C14MC: Chromosome 14 miRNA cluster; C19MC: Chromosome 19 miRNA cluster; D: Differentially expressed; S: Similarly expressed.
The most significantly upregulated third trimester miRNA was hsa-miR-520c-3p (2.78-fold higher, FDR = 2.91 × 10-111), followed by hsa-miR-181d-5p (3.18-fold higher, FDR = 6.38 × 10-70), both from C19MC. Overall, the C14MC contributed more DE miRNAs, reaching higher fold changes in first trimester than C19MC (Figure 4A). Although C19MC contributed fewer total miRNAs, and at lower fold change differences between trimesters, the C19MC baseMean distribution was an order of magnitude higher than C14MC distribution in both overall baseMean median (C19MC = 5696; C14MC = 198.7) and mean (C19MC = 21,278; C14MC = 1122) (Figure 4B & Supplementary File 6). This held true for both first and third trimester baseMeans expression values. Pathway enrichment analysis of experimentally confirmed target genes shows that distinct pathways are regulated by SE and DE miRNAs (Figure 4C). Metabolite salvage pathways and GTPase signaling were more significantly targeted by DE C14MC miRNAs, including ‘pyridoxal 5′-phosphate salvage pathway’, ‘salvage pathways of pyrimidine ribonucleotides’ and ‘signaling by Rho family GTPases’ suggesting these pathways are uniquely regulated by this cluster miRNAs between the first and third trimester. ‘Prostate cancer signaling’ was more significantly targeted by the SE C14MC miRNAs, suggesting this pathway is important throughout gestation (Supplementary File 4C). ‘Molecular mechanisms of cancer’, ‘pancreatic adenocarcinoma signaling’, ‘prostate cancer signaling’, ‘estrogen-mediated S-phase entry’ and ‘chronic myeloid leukemia signaling’ were more significantly targeted by DE C19MC miRNAs suggesting these pathways are uniquely regulated by this cluster miRNAs between the first and third trimester. Conversely, there were no pathways among the top 30 most enriched that were targeted by SE C19MC miRNAs, suggesting more variable regulation throughout gestation (Figure 4D & Supplementary File 4D). Pathway enrichment analysis with both experimentally confirmed miRNA targets as well as targets predicted with high confidence (Supplementary File 4D), demonstrated changes in regulated pathways, with greater representation of inflammatory pathways including the ‘systemic lupus erythematosus in B cell signaling’ and ‘coronavirus pathogenesis’ pathways, as well as several pathways more significantly targeted by SE miRNAs, including ‘role of PI3K/PKB signaling in the pathogenesis of influenza’.
Discussion
The placenta is a unique organ that changes function greatly throughout gestation, meeting different challenges and needs at different stages of pregnancy. Placentation in the first trimester sets the groundwork for its functions throughout gestation for fetal development. Placental function is in part epigenetically regulated through miRNAs including the placenta-specific miRNA clusters, C14MC and C19MC, that play critical roles in regulation of this vital organ. This is the first study to our knowledge to use high-throughput sequencing to compare miRNA expression between first and third trimester human placentae of healthy pregnancies resulting in delivery.
The miRNA expression profiles in first and third trimester have similar chromosome distributions, with expected peaks at chromosomes 14 and 19, as well as peaks at chromosome 1, the largest human chromosome and chromosome X, which has a higher density of miRNAs compared with autosomes [62,63]. These three chromosomes, as well as chromosome 13 which contains two small clusters [64], were overrepresented relative to their number of encoded miRNAs. The most highly expressed miRNA, hsa-miR-126-3p, was upregulated in third trimester, and was validated with qRT-PCR using an independent cohort. In a recent study comparing first and second trimester placenta, miR-126-3p was identified to be among the ten most highly expressed miRNAs and identified in maternal plasma [65]. Although variation among the first and second trimester was not different [65], our study identified hsa-miR-126-3p to be DE and highest in the third trimester, likely having a unique role in the third trimester compared with earlier in gestation. It is also highly abundant in fetal circulation and human umbilical vein endothelial cells [66], suggesting a role during parturition and fetal development and may become a potential biomarker for developmental origins of health and disease. The abundance of hsa-miR-126-3p in extracellular vesicles at third trimester is negatively correlated with gestational age at birth [67].
Among the most highly expressed miRNAs, over 37% were encoded in chromosome 19 (and 28/36 or 77.8% were specifically in C19MC), whereas none localized to chromosome 14. This supports an earlier miRNA-seq study which profiled 25 human placentae at delivery and identified higher expression from C19MC than C14MC miRNAs [68], and additionally we show the same pattern in first trimester. The C19MC miRNAs with high expression in the placenta may potentially be used for targets, as C19MC miRNAs have been identified in maternal circulation, as early as the first trimester, with elevations throughout gestation [69–71]. The most highly expressed miRNA that was SE was hsa-miR-515-5p, which is a member of the C19MC. Placental expression of hsa-miR-515-5p has been identified to play a key role in human trophoblast differentiation with aberrant upregulation contributing to pathogenesis of preeclampsia [35,72]. It has also been associated with preterm birth [73] and fetal growth restriction [43]. Although, it has been detected in maternal circulation, both in plasma and whole blood fractions, it has also been detected in whole blood fractions of healthy nonpregnant women [74], and may not be used solely as a biomarker of disease, but may be incorporated with other miRNAs with stable expression across gestation that change with disease using a bivariate biomarker disease approach described by Laurent [45]. Another highly and SE C19MC miRNA in this study, hsa-miR-518b, is elevated in the maternal plasma of pregnancies with preeclampsia (second and third trimester) [75] and pregnancies destined to develop gestational hypertension (first trimester) [76], compared with healthy pregnancies. Here, we show that placenta abundance of hsa-miR-518b is similar between first and third trimester in healthy pregnancies, suggesting hsa-miR-518b may be a good biomarker for preeclampsia at variable gestational ages.
This atlas identified 180 DE miRNAs which may be important for functional changes in the placenta throughout pregnancy. Among those, the most DE miRNAs were highest in the first trimester. The most significantly targeted pathways of experimentally confirmed targets by DE miRNAs and the most highly expressed in the first trimester was ‘molecular mechanisms of cancer’. Although identified in pathway enrichment analyses for tumor progression, many of the major signaling pathways involved in inter and intracellular communication of invasive phenotypes mimic those associated with migration and invasion of trophoblasts into the maternal decidua and spiral arteries. These essential placentation steps take place in an environment rich in hormones, cytokines and growth factors, and include responsible signaling pathways such as MAPK, PI3K/PKB, JAK/STATs, wingless and focal adhesion kinase pathways [2]. Of these miRNAs, the most DE with highest expression in first trimester, hsa-miR-4483, was also found to be strongly downregulated in the second trimester and hence likely plays a significant role in the first trimester placenta. It may function to regulate estradiol production early in gestation, as described in another hormone producing cell type and contribute to migration and invasion [77].
Differentiation of first trimester human placental cytotrophoblasts from an anchorage dependent epithelial phenotype into the mesenchymal-like invasive extravillous trophoblast is a crucial step for placentation. Illsley et al., previously demonstrated that an epithelial to mesenchymal transition takes place when first trimester cytotrophoblasts differentiate into extravillous trophoblasts [78,79]. miRNAs have been implicated in the epithelial to mesenchymal transition [80–85]. The most highly DE miRNA in first trimester placenta, hsa-miR-205, has been implicated in the epithelial to mesenchymal transition and the maintenance of the epithelial phenotype [80–84]. In human trophoblast cell lines it has been identified to silence MED1 under hypoxic conditions [80], suggesting it has a role in the first trimester regulating trophoblast differentiation during physiologic hypoxic conditions [80,86,87]. ‘Regulation of the epithelial mesenchymal transition by growth factors pathway’ was also one of the most significantly targeted pathways by DE miRNAs and most highly expressed in the first trimester, highlighting the differences between placentation when the placenta is invading maternal tissue and establishing itself in states of low oxygen tension versus time of delivery when the placenta has completed its purpose.
Two additional significantly targeted pathways by DE miRNAs and highly expressed in the first trimester included the ‘hepatic fibrosis signaling’ and the ‘senescence’ pathways. Hepatic fibrosis signaling is classically associated with extracellular matrix deposition [88], consistent with first trimester placental function when extravillous trophoblasts degrade and induce extracellular matrix remodeling to enable migration [2,5,89,90]. Cellular senescence is programmed cell-cycle arrest that restricts the propagation of cells, which is induced by various forms of cellular stress including oxidative stress. Cell fusion, has also been identified to trigger cellular senescence and has been described in the placenta, with the placental expressed fusogen, syncitin-1 (ERVWE1), which mediates cell fusion-induced senescence of the syncytiotrophoblast [91–93]. These senescent cells secrete inflammatory cytokines, chemokines and matrix metalloproteinases, known as the senescence associated secretory phenotype (SASP). SASP proteins promote an epithelial to mesenchymal transition and the degradation of basement membranes, increasing migration and invasion for appropriate placentation [94].
Our findings also support the importance of the placenta-specific miRNA clusters throughout gestation, with 42 miRNAs SE and 105 DE across first and third trimester. This indicates that while they are placenta-specific miRNAs, the majority have varying roles throughout pregnancy. DE canonical pathways targeted by the C14 and C19 clusters were more significant than those of all SE miRNAs suggesting these clusters have significantly different roles throughout gestation [69]. Similar to other studies using whole villous tissue and primary cytotrophoblasts [20,48,65], we identified a decrease in C14MC expression from first to third trimester. Another recent study did not identify a decrease throughout gestation, but their study only focused on the first and second trimester of presumably normal pregnancies [65]. Finally, we find that two small clusters in chromosome 13 are upregulated in first trimester. To our knowledge, this is the first time that the chromosome 13 miRNA clusters have been described in placenta, and future research is needed to understand their role across gestation.
The major strengths of this study are the use of first and third trimester tissue from healthy pregnancies resulting in delivery, the cohort size, the availability of detailed demographic information and birth outcomes, and the use of high-throughput sequencing. NGS, as opposed to other techniques such as array, allows for greater confidence in the conclusions regarding differential expression, since all known miRNA species previously annotated in the human genome are considered, and bias is not introduced by eliminating certain RNAs. Previous studies analyzing miRNA expression in first and/or third trimester placentae have used microarray technology and most examined very few samples (n = 2–6 in each group) [25,47,48,95]. There are currently few NGS miRNA profiles of the placenta, and our study is the first to profile both first and third trimester placentae with NGS and a large sample size. We successfully validated all six selected miRNAs using qRT-PCR and an independent cohort.
Our study has some limitations. Our cohort had advanced maternal age (>35 years matched in both groups) due to the demographics of women more likely to undergo CVS, the source of our first trimester tissue. There were some differences in the demographics between the groups from the first and third trimester placenta samples. This includes race, ethnicity, maternal BMI, thyroid disorders and pregnancy complications, specifically hypertension. However, the overall differences were small. In addition, PCA analysis did not demonstrate outliers. Furthermore, we performed pathway enrichment analysis using only experimentally confirmed targets. When performed using both experimentally confirmed and predicted with high confidence targets, although overall pathways and patterns remained consistent, when we only included experimentally confirmed targets, immune mediated pathways were not represented.
Conclusion
Overall, we identified and compared the normative miRNA signatures in the first and third trimester placentae. Our study shows many stably expressed miRNAs throughout gestation as well as significant differences between the miRNA signatures, including two novel clusters on chromosome 13. This work provides a rich atlas to direct functional studies investigating the epigenetic differences in first and third trimester placentae and development of disease related biomarkers or prognostic indicators that are gestational age specific.
Future perspective
As we improve our understanding of miRNA profiles in placenta and across gestation, miRNAs may be useful biomarkers for noninvasive prenatal diagnostic testing. Our knowledge of miRNA profiles is still in its infancy relative to our knowledge of the protein coding transcriptome. Until recently, most miRNA profiling papers of placenta used arrays with limited samples. However, protocols to capture small RNAs, synthesize cDNA and perform high-throughput NGS are improving rapidly. In 5–10 years’ time, we expect that the knowledge of human miRNA profiles in different tissue and extracellular locations will greatly improve as well. This will provide opportunities for biomarker discovery and diagnostic test development, since miRNAs are smaller, more stable RNAs than protein coding transcripts. Currently, the knowledge pool of miRNA targets has limited confirmed miRNA–RNA interactions, but this will improve as the miRNA field continues to evolve. Our work to profile miRNAs in first and third trimester provides a foundation for biomarker discovery during pregnancy and future advancements in maternal-fetal health.
Summary points.
This work creates an atlas of the miRNA expression profiles of first and third trimester human placenta from patients who delivered healthy babies, with pregnancy outcomes available from both groups.
Chromosome 19 contributes approximately 37% of the most highly expressed miRNAs in both first and third trimester placenta. Most of these miRNAs are localized to the pregnancy-associated miRNA cluster, chromosome 19 miRNA cluster (C19MC).
There are 182 miRNAs with similar expression across gestation. Other patient variables may affect the abundance of these miRNAs.
There are 781 mature miRNAs significantly different between first and third trimester placenta (false discovery rate <0.05), almost a third of all detected miRNAs (31.2% of 2503 miRNAs detected at any threshold).
After applying expression and fold change thresholds, 180 mature miRNAs remain significantly different between first and third trimester placenta (false discovery rate <0.05, baseMean >10, FC >2). These miRNAs may contribute to changes in placental function or be markers of different placental stresses throughout gestation.
Six miRNAs were successfully validated with qRT-PCR in an independent cohort.
The placenta-specific miRNA clusters (chromosome 14 miRNA cluster [C14MC] and C19MC) contain both similarly and differentially expressed miRNAs.
C14MC expressed miRNAs with greater fold change differences across gestation than C19MC miRNAs, though C14MC miRNAs are not among the most highly expressed miRNAs in placenta.
For both similarly and differentially expressed miRNAs, C19MC miRNA placenta expression was overall higher than C14MC expression.
Chromosome 13 has two miRNA clusters upregulated in first trimester placenta, never before described in placenta.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2021-0055
Financial & competing interests disclosure
This work was supported by the National Institute of Health grants: R01 HD091773, R01 HD074368, T32 DK007770 and U01 EB026421. The funding agency was not involved in the design, analysis or interpretation of the data reported. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest
- 1.Cross JC. Formation of the placenta and extraembryonic membranes. Ann. NY Acad. Sci. 857, 23–32 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Mendes S, Timóteo-Ferreira F, Almeida H, Silva E. New insights into the process of placentation and the role of oxidative uterine microenvironment. Oxid. Med. Cell. Longev. 2019, 9174521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu P, Wang Y-l, Piao Y-S et al. Effects of matrix proteins on the expression of matrix metalloproteinase-2, -9, and -14 and tissue inhibitors of metalloproteinases in human cytotrophoblast cells during the first trimester1. Biol. Reprod. 65(1), 240–246 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Staun-Ram E, Shalev E. Human trophoblast function during the implantation process. Reprod. Biol. Endocrinol. 3(1), 56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 3(12), a005058 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genbacev O, Zhou Y, Ludlow JW, Fisher SJ. Regulation of human placental development by oxygen tension. Science 277(5332), 1669–1672 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Rodesch F, Simon P, Donner C, Jauniaux E. Oxygen measurements in endometrial and trophoblastic tissues during early pregnancy. Obstet. Gynecol. 80(2), 283–285 (1992). [PubMed] [Google Scholar]
- 8.Genbacev O, Joslin R, Damsky CH, Polliotti BM, Fisher SJ. Hypoxia alters early gestation human cytotrophoblast differentiation/invasion in vitro and models the placental defects that occur in preeclampsia. J. Clin. Invest. 97(2), 540–550 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jauniaux E, Watson A, Burton G. Evaluation of respiratory gases and acid-base gradients in human fetal fluids and uteroplacental tissue between 7 and 16 weeks’ gestation. Am. J. Obstet. Gynecol. 184(5), 998–1003 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Yen and Jaffe's ReproductiveEndocrinology: Physiology, Pathophysiology, and Clinical Management. 7th ed. Strauss JF, Barbieri RL (Eds). Saunders; (2013). [Google Scholar]
- 11.Chuva de Sousa Lopes SM, Alexdottir MS, Valdimarsdottir G. The TGFβ family in human placental development at the fetal-maternal interface. Biomolecules 10(3), 453 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun T, Gonzalez TL, Deng N et al. Sexually dimorphic crosstalk at the maternal-fetal interface. J.Clin. Endocrinol.Metab. 105(12), e4831–e4847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman ML, Liang L, Valeri L et al. Regulation of birthweight by placenta-derived miRNAs: evidence from an arsenic-exposed birth cohort in Bangladesh. Epigenetics 13(6), 573–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducsay CA, Goyal R, Pearce WJ, Wilson S, Hu XQ, Zhang L. Gestational hypoxia and developmental plasticity. Physiol. Rev. 98(3), 1241–1334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11(9), 597–610 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1), 15–20 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentwich I, Avniel A, Karov Y et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37(7), 766–770 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J. Reprod. Immunol. 97(1), 51–61 (2013). [DOI] [PubMed] [Google Scholar]; • Chromosomal locations and backgrounds for chromosome 14 miRNA cluster and chromosome 19 miRNA cluster.
- 21.Jinesh GG, Flores ER, Brohl AS. Chromosome 19 miRNA cluster and CEBPB expression specifically mark and potentially drive triple negative breast cancers. PLoS ONE 13(10), e0206008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen PN, Huang CJ, Sugii S, Cheong SK, Choo KB. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J. Biomed. Sci. 24(1), 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radovich M, Solzak JP, Hancock BA et al. A large microRNA cluster on chromosome 19 is a transcriptional hallmark of WHO type A and AB thymomas. Br. J. Cancer 114(4), 477–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noguer-Dance M, Abu-Amero S, Al-Khtib M et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 19(18), 3566–3582 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S et al. MicroRNA expression profiles of trophoblastic cells. Placenta 33(9), 725–734 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Singh K, Williams J, Brown J et al. Up-regulation of microRNA-202-3p in first trimester placenta of pregnancies destined to develop severe preeclampsia, a pilot study. Pregnancy Hypertens. 10, 7–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am. J. Obstet. Gynecol. 204(2), 178 e112–e121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 200(6), 661 e661–e667 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Chen DB, Wang W. Human placental microRNAs and preeclampsia. Biol. Reprod. 88(5), 130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Li P, Hao S, Liu L, Zhao J, Hou Y. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin. Chem. Lab. Med. 47(8), 923–929 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Guo L, Tsai SQ, Hardison NE et al. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta 34(7), 599–605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lykoudi A, Kolialexi A, Lambrou GI et al. Dysregulated placental microRNAs in early and late onset preeclampsia. Placenta 61, 24–32 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Guo L, Yang Q, Lu J et al. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PLoS ONE 6(6), e21072 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod. Sci. 18(1), 46–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hromadnikova I, Kotlabova K, Ondrackova M et al. Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 34(6), 437–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy MS, Casselman RC, Tayade C, Smith GN. Differential expression of plasma microRNA in preeclamptic patients at delivery and 1 year postpartum. Am. J. Obstet. Gynecol. 213(3), 367 e361–e369 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Hemmatzadeh M, Shomali N, Yousefzadeh Y, Mohammadi H, Ghasemzadeh A, Yousefi M. MicroRNAs: small molecules with a large impact on pre-eclampsia. J. Cell. Physiol. 235(4), 3235–3248 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Soler-Botija C, Gálvez-Montón C, Bayés-Genís A. Epigenetic biomarkers in cardiovascular diseases. Front.Genet. 10, 950 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sõber S, Reiman M, Kikas T et al. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci. Rep. 5, 13336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hromadnikova I, Kotlabova K, Hympanova L, Krofta L. Cardiovascular and cerebrovascular disease associated microRNAs are dysregulated in placental tissues affected with gestational hypertension, preeclampsia and intrauterine growth restriction. PLoS ONE 10(9), e0138383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai PY, Li SH, Chen WN, Tsai HL, Su MT. Differential miR-346 and miR-582-3p expression in association with selected maternal and fetal complications. Int. J. Mol. Sci. 18(7), 1570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen H, Chen L, He J, Lin J. MicroRNA expression profiles and networks in placentas complicated with selective intrauterine growth restriction. Mol. Med. Rep. 16(5), 6650–6673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higashijima A, Miura K, Mishima H et al. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat. Diagn. 33(3), 214–222 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Li H, Ouyang Y, Sadovsky E, Parks WT, Chu T, Sadovsky Y. Unique microRNA signals in plasma exosomes from pregnancies complicated by preeclampsia. Hypertension 75(3), 762–771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Identifies miRNAs significantly different in preeclampsia patient extracellular vesicles and total plasma, compared to normal pregnancies.
- 45.Srinivasan S, Treacy R, Herrero T et al. Discovery and verification of extracellular miRNA biomarkers for non-invasive prediction of pre-eclampsia in asymptomatic women. Cell Rep. Med. 1(2), 100013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillet V, Ouellet A, Stepanov Y et al. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J. Clin. Endocrinol. Metab. 104(11), 5157–5169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farrokhnia F, Aplin JD, Westwood M, Forbes K. MicroRNA regulation of mitogenic signaling networks in the human placenta. J. Biol. Chem. 289(44), 30404–30416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu Y, Sun J, Groome LJ, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 304(8), E836–E843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Identifies first vs third trimester placenta miRNA differences in uncomplicated pregnancies, though first trimester pregnancies do not have birth outcomes.
- 49.Gonzalez TL, Sun T, Koeppel AF et al. Sex differences in the late first trimester human placenta transcriptome. Biol. Sex Differ. 9(1), 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pisarska MD, Akhlaghpour M, Lee B et al. Optimization of techniques for multiple platform testing in small, precious samples such as human chorionic villus sampling. Prenat. Diagn. 36(11), 1061–1070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1), 3 (2011). [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3), R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(D1), D68–D73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12), 550–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durinck S, Moreau Y, Kasprzyk A et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21(16), 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4(8), 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu HY, He L, Fominykh K et al. Evolution of the human-specific microRNA miR-941. Nat. Commun. 3, 1145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee B, Kroener LL, Xu N et al. Function and hormonal regulation of GATA3 in human first trimester placentation. Biol. Reprod. 95(5), 113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu N, Barlow GM, Cui J et al. Comparison of genome-wide and gene-specific DNA methylation profiling in first-trimester chorionic villi from pregnancies conceived with infertility treatments. Reprod. Sci. 24(7), 996–1004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 115(7), 787–798 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4), 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Di Palo A, Siniscalchi C, Salerno M, Russo A, Gravholt CH, Potenza N. What microRNAs could tell us about the human X chromosome. Cell. Mol. Life Sci. 77(20), 4069–4080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo X, Su B, Zhou Z, Sha J. Rapid evolution of mammalian X-linked testis microRNAs. BMC Genomics 10(1), 97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dunham A, Matthews LH, Burton J et al. The DNA sequence and analysis of human chromosome 13. Nature 428(6982), 522–528 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Detailed analysis of chromosome 13 identifies miRNA clusters.
- 65.Smith MD, Pillman K, Jankovic-Karasoulos T et al. Large-scale transcriptome-wide profiling of microRNAs in human placenta and maternal plasma at early to mid gestation. medRxiv (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah KB, Chernausek SD, Teague AM, Bard DE, Tryggestad JB. Maternal diabetes alters microRNA expression in fetal exosomes, human umbilical vein endothelial cells and placenta. Pediatr. Res. (2020) https://www.nature.com/articles/s41390-020-1060-x (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howe CG, Foley HB, Kennedy EM et al. Extracellular vesicle microRNA in early versus late pregnancy with birth outcomes in the MADRES study. Epigenetics 1–17 (2021). (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Extracellular vesicle miRNAs in first and third trimester maternal plasma.
- 68.Paquette AG, Chu T, Wu X, Wang K, Price ND, Sadovsky Y. Distinct communication patterns of trophoblastic miRNA among the maternal-placental-fetal compartments. Placenta 72–73, 28–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • A comparison of chromosome 14 miRNA cluster and chromosome 19 miRNA cluster expression at delivery from term placenta, fetal plasma and maternal plasma.
- 69.Dumont TMF, Mouillet JF, Bayer A et al. The expression level of C19MC miRNAs in early pregnancy and in response to viral infection. Placenta 53, 23–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miura K, Miura S, Yamasaki K et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 56(11), 1767–1771 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Hromadnikova I, Kotlabova K, Doucha J, Dlouha K, Krofta L. Absolute and relative quantification of placenta-specific micrornas in maternal circulation with placental insufficiency-related complications. J. Mol. Diagn. 14(2), 160–167 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Zhang M, Muralimanoharan S, Wortman AC, Mendelson CR. Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc. Natl Acad. Sci. USA 113(45), E7069–E7076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hromadnikova I, Kotlabova K, Ivankova K, Krofta L. Expression profile of C19MC microRNAs in placental tissue of patients with preterm prelabor rupture of membranes and spontaneous preterm birth. Mol. Med. Rep. 16(4), 3849–3862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation--identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 89(2), 185–191 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Jelena M, Sopić M, Joksić I et al. Placenta-specific plasma miR518b is a potential biomarker for preeclampsia. Clin. Biochem. 79, 28–33 (2020). [DOI] [PubMed] [Google Scholar]; • Identifies miR-518b as a promising biomarker for preeclampsia, significant with and without adjustment for gestational age.
- 76.Hromadnikova I, Kotlabova K, Hympanova L, Doucha J, Krofta L. First trimester screening of circulating C19MC microRNAs can predict subsequent onset of gestational hypertension. PLoS ONE 9(12), e113735–e113735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SY, Kang YJ, Kwon J et al. miR-4463 regulates aromatase expression and activity for 17β-estradiol synthesis in response to follicle-stimulating hormone. Clin. Exp. Reprod. Med. 47(3), 194–206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DaSilva-Arnold SC, Zamudio S, Al-Khan A et al. Human trophoblast epithelial-mesenchymal transition in abnormally invasive placenta. Biol. Reprod. 99(2), 409–421 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Illsley NP, DaSilva-Arnold SC, Zamudio S, Alvarez M, Al-Khan A. Trophoblast invasion: lessons from abnormally invasive placenta (placenta accreta). Placenta 102, 61–66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 24(6), 2030–2039 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 12, 1175–1184 (2006). [PubMed] [Google Scholar]
- 82.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl Acad. Sci. USA 105(49), 19300–19305 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gregory PA, Bert AG, Paterson EL et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10(5), 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7(20), 3112–3118 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Mong EF, Yang Y, Akat KM et al. Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition. Sci. Rep. 10(1), 3029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crawford SE, Qi C, Misra P et al. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 277(5), 3585–3592 (2002). [DOI] [PubMed] [Google Scholar]
- 87.Landles C, Chalk S, Steel JH et al. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol. Endocrinol. 17(12), 2418–2435 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Ying HZ, Chen Q, Zhang WY et al. PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics (Review). Mol. Med. Rep. 16(6), 7879–7889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu P, Wang Y, Piao Y et al. Effects of matrix proteins on the expression of matrix metalloproteinase-2, -9, and -14 and tissue inhibitors of metalloproteinases in human cytotrophoblast cells during the first trimester. Biol. Reprod. 65(1), 240–246 (2001). [DOI] [PubMed] [Google Scholar]
- 90.Staun-Ram E, Shalev E. Human trophoblast function during the implantation process. Reprod. Biol. Endocrinol. 3, 56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cox LS, Redman C. The role of cellular senescence in ageing of the placenta. Placenta 52, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Chuprin A, Gal H, Biron-Shental T et al. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev. 27(21), 2356–2366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gal H, Lysenko M, Stroganov S et al. Molecular pathways of senescence regulate placental structure and function. EMBO J. 39(15), e105972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Laberge RM, Awad P, Campisi J, Desprez PY. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 5(1), 39–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsamou M, Vrijens K, Wang C et al. Genome-wide microRNA expression analysis in human placenta reveals sex-specific patterns: an ENVIRONAGE birth cohort study. Epigenetics 16(4), 373–388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This is a large cohort microarray study of term placenta miRNAs.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.