Abstract
The field of infancy research faces a difficult challenge: some questions require samples that are simply too large for any one lab to recruit and test. ManyBabies aims to address this problem by forming large-scale collaborations on key theoretical questions in developmental science, while promoting the uptake of Open Science practices. Here, we look back on the first project completed under the ManyBabies umbrella – ManyBabies 1 – which tested the development of infant-directed speech preference. Our goal is to share the lessons learned over the course of the project and to articulate our vision for the role of large-scale collaborations in the field. First, we consider the decisions made in scaling up experimental research for a collaboration involving 100+ researchers and 70+ labs. Next, we discuss successes and challenges over the course of the project, including: protocol design and implementation, data analysis, organizational structures and collaborative workflows, securing funding, and encouraging broad participation in the project. Finally, we discuss the benefits we see both in ongoing ManyBabies projects and in future large-scale collaborations in general, with a particular eye towards developing best practices and increasing growth and diversity in infancy research and psychological science in general. Throughout the paper, we include first-hand narrative experiences, in order to illustrate the perspectives of researchers playing different roles within the project. While this project focused on the unique challenges of infant research, many of the insights we gained can be applied to large-scale collaborations across the broader field of psychology.
Keywords: reproducibility, Open Science, infancy, infant-directed speech, collaboration
The seed of ManyBabies was planted over a lunch at Reading Terminal Market in Philadelphia, at the 2015 Biennial Meeting of the Society for Research in Child Development (SRCD). The Open Science Collaboration’s (2015) paper on failures to replicate high-profile results in psychology had not yet been released, but change was already in the air. Simmons, Nelson, and Simonsohn (2011) had reminded psychologists of the dramatic inflation of false positives caused by “questionable research practices,” and Button et al. (2013) had demonstrated the devastating consequences of running experiments with low statistical power. Further, Klein et al. (2014) had just reported the results of ManyLabs 1, in which a group of independent labs all ran the same set of replication protocols and pooled their data.
The topic of conversation that day at SRCD was that some hypotheses about infant development could perhaps never be adequately tested, simply because the necessary number of infants or conditions would exceed what a lab could complete in a feasible amount of time. Infant research is slow, and it can take several months (or even years with special populations) to complete data collection for a single condition. In many settings, research productivity is often measured by publication rate, placing pressure on researchers to publish at a rapid pace to secure promotion and/or research funding. This can limit motivation to conduct large-scale studies within an individual lab. For example, an experiment testing Hunter & Ames’s (1988) multi-factorial model of novelty and familiarity preferences in infancy would minimally require multiple age groups, fully crossed with exposure conditions and levels of stimulus complexity for somewhere between 8 – 27 carefully calibrated conditions. No infant lab would take on this project alone, given that this kind of high-risk investigation could likely consume all the lab’s recruitment resources for years! But we agreed that, in principle, such an investigation could be completed if labs worked together as the ManyLabs project had done.
By the end of 2015, this informal conversation led to a growing email thread, numerous lunch meetings and side conversations, and eventually a blog post (Frank, 2015). A vision began to emerge of a consortium of infancy labs, pooling data towards a single research question. In one memorable conversation, an infancy researcher declared “we cannot NOT do this,” and many of us agreed. Yet by the time we began planning the first study, it became clear that this large-scale collaborative model of research was very different from any process any of us had previously encountered in infancy research: Working together required, among many other things, for us to critically examine how to best coordinate our work, how to apportion credit and responsibility, and what standards we would require from participating researchers. Now four and a half years later, this paper describes some of the discussions that followed, the solutions we have found, and the challenges that remain.
The ManyBabies approach
What has emerged over the past several years, in what we refer to as “ManyBabies”, is a collaborative collective of infancy researchers committed to Open Science best practices and a large-scale collaborative research model. Our objective is to employ best research practices specifically within ManyBabies research, but also to model them for the larger research community with the hope of creating greater awareness and uptake of these practices. We cannot operate in a fully “Born Open” model described by Rouder (2015), where data are publicly available shortly after collection due to the privacy and ethics constraints on research with human infants. Nonetheless, we commit to fully open methods and stimuli, and data that are open to the greatest extent possible. Full datasets are made public once they have been scrubbed of identifying information and where consent has been obtained. Video records are shared within restricted repositories such as Databrary (2012). ManyBabies shares many core values with other large-scale collaborative efforts in psychology such as ManyLabs, the Psychological Science Accelerator, and ManyPrimates, although each network is distinct in its workflow and how it balances competing priorities.
ManyBabies operates within a framework that prioritizes collaboration in all aspects of the project - project selection, study design, stimulus creation, piloting, data collection, analysis, interpretation of the findings, and writing. We employ a consensus-based decision-making system, supported by leadership to keep things “moving along” and to bring decision-making to a close when necessary. Our relatively radical commitment to a consensus approach emerged in part out of our desire to increase the diversity of contributions to our science. We sought to do this by engaging with as broad a spectrum of laboratories and researcher backgrounds as possible. Early on, it became clear that one major barrier to developing best practices was the fact that our science took place within heavily siloed laboratories and that insights about methods typically stayed within those siloes. Building a community where open discussion about methodology and best practices could take place could only occur in an environment of trust and equality.
Crucially, ManyBabies does not engage in direct replication efforts, where a study is chosen from the literature and replicated exactly. Instead, the focus of ManyBabies is on testing key theoretical claims in the infancy literature by designing the best possible test of a claim -- whether or not it has been utilized before (i.e. conceptual replication). Further, we aim to examine sources of variation in effects across laboratories, methods, and populations. In other words, a central goal is to examine not only the reliability of a claim, but its generalizability and robustness across contexts. Typically and ideally, the main manipulations adopted for the “best test” of a phenomenon are developed based on a consensus of contributors, particularly those with different theoretical perspectives in order to ensure that the findings are accepted as conclusive by a wide variety of researchers. This means that in some cases, protocol development can take considerably longer than data collection itself, and may require significant discussion and debate. Although time-consuming and effortful, this method of group decision-making is crucial given the large number of laboratories contributing to (and resources committed to) a ManyBabies effort. Moreover, it is consistent with ManyBabies’ commitment to diversity as it pertains to researchers but also to populations and research questions.
By any reasonable metric, ManyBabies has thus far been a resounding success. We have now completed data collection on our first project – ManyBabies 1 – with many other projects in the works including ManyBabies 2–4, as well as numerous spin-off projects. Symposia and talks discussing ManyBabies at various conferences have seen a large, engaged, and supportive audience. Indeed, ManyBabies 1 was recently recognized with the Society for the Improvement of Psychological Science Mission Award. This progress has come with plenty of challenges, however, as we have had to learn how to work together in a new way. In the following sections, we outline many of the issues, insights, and processes that have emerged over the first few years of our endeavor. Our hope going forward is that other groups of researchers, in infancy and other fields, will embark on a similar journey, and that they can learn from our mistakes and benefit from our successes.
ManyBabies, Much Data: Scaling experimental design, data collection, and analysis
ManyBabies 1 was a large-scale project that investigated infants’ preference for infant-directed speech. In total, 69 labs from 16 countries tested 2845 infants, of which 2329 were included in the final analysis. Our main findings were that monolingual infants prefer infant-directed speech to adult-directed speech, and that the magnitude of their preference increases from 3 to 15 months. We observed a stronger preference in infants exposed to the dialect used in our stimuli, North American English. We also found that some methodologies were associated with larger observed effect sizes than others. These findings were published as a Registered Report in the journal Advances in Methods and Practices in Psychological Science (ManyBabies Consortium, 2019a). All stimuli, analysis scripts, and data are shared via the Open Science Framework (available at https://osf.io/re95x/), providing a rich set of resources for further experimental work, and secondary data analysis, as well as numerous spin-off projects (ManyBabies Consortium, 2019b). These spin-offs include projects analyzing factors that predict variability in rates of data exclusion due to infant fussiness (https://osf.io/ryzmb/, Kline et al., 2018); tracking later vocabulary outcomes for infants who participated in the ManyBabies 1 project (https://osf.io/2qamd/, Soderstrom et al., 2020); investigating the test-retest reliability of measuring an individual infant’s preference for infant-directed speech (https://osf.io/zeqka/, Schreiner et al., 2019); and examining bilingual infants’ preference for infant-directed speech (https://osf.io/zauhq/, Byers-Heinlein et al., 2020). But how did this large-scale collaboration actually happen, in the context of a field where most research is done within a single laboratory? In this section, we provide a behind-the-scenes view of the many steps along the way to ManyBabies 1.
Choosing a phenomenon
Since ManyBabies formed over a very broad focus across the research community – rather than emerging from a particular research question – specifying the first project was not easy, but it was done in a spirit of collegiality. Interested researchers started to discuss different project ideas informally via videoconference. There were about 20 different effects to be studied in the initial pool of nominations. We gathered and discussed the main arguments for and against each idea, for example, how certain we were of the effect, the ages that would have to be recruited and tested, etc. We ultimately decided that our main goal initially would be a “proof of concept” - to explore the feasibility of running such a large-scale collaboration and to examine lab-to-lab variability of an effect for which there was already a robust scientific consensus, rather than focusing on a more controversial phenomenon. Collaborators voted on their preferred effect, and the decision was made to test infant’s preference for infant-directed versus adult-directed speech. This became ManyBabies 1.
ManyBabies 1 and all of the subsequent main ManyBabies projects have been confirmatory (i.e., attempting to replicate an effect). However, the large and diverse samples we collect allow us to go beyond simple “confirmation”, to investigate important questions about whether a particular phenomenon, such as a characteristic presumed to be a developmental universal, truly applies across different populations. In a field fraught with small sample sizes and a strong bias towards North American participants (Nielson, Haun, Kärtner, & Legare, 2017), we argue that the field of infant research stands to benefit from large-scale confirmatory studies, which can reinforce existing findings and theories on which these findings are based. At the same time, there are crucial benefits to exploratory work. Our initial discussions and design decisions led us to realize that there were many interesting research questions that were related, yet distinct from our original research question. We couldn’t answer every question in a single project. In efforts to achieve a reasonable scope for ManyBabies 1, while simultaneously embracing these related questions, we used two approaches: secondary analyses, and spin-off projects.
We use the term “secondary analyses” to refer to additional analyses that can be done with the main dataset, for example, examining different moderators of our effect of interest. Demographic data (e.g., socio-economic status) will almost certainly be recorded as part of every ManyBabies project, and we are working on developing a minimal set of demographic questions that would be applied across ManyBabies studies to ensure comparability across, as well as within, different ManyBabies studies. We are also interested in what lab-level factors predict the magnitude of our effects, testing lore in the field. For example, in ManyBabies 1, labs could optionally collect data on the characteristics of the researcher testing the infant, to determine whether infants tested by graduate students show a larger effect than those tested by undergraduates, or whether infants tested by a bearded researcher complete fewer trials. Neither of these hypotheses has been supported in preliminary analyses (Kline et al., 2018), and a full-scale investigation is ongoing on these and other characteristics of the laboratories, researchers, and infant populations.
Other interesting questions require significant additional data to test, and we call these “spin-off projects”. As an example, many researchers in the original planning stages of ManyBabies 1 were interested in the question of whether bilingualism would affect infant’s preference for infant-directed speech. We decided to limit the main study to monolingual participants, but a subgroup of researchers decided simultaneously to launch a study to test bilinguals in the same paradigm, which we called ManyBabies 1 Bilingual. A Registered Report was drafted, and received in-principle acceptance (Byers-Heinlein et al., 2019). A total of 17 labs tested both monolingual and bilingual infants. Another spin-off project is examining whether individual differences in infants’ preference for infant-directed speech in ManyBabies 1 predict vocabulary growth longitudinally (https://osf.io/2qamd/, Soderstrom et al., 2020).
While the aim of our main projects is always replication of key phenomena, secondary and spin-off projects are typically exploratory, asking new and valuable questions about development and methodology. This combined approach balances control and hypothesis-led testing with exploratory research and post-hoc analysis.
Designing the study protocol
Once we decided that the research question for ManyBabies 1 would be to test infants’ preference for infant-directed over adult-directed speech, we needed to settle on a specific protocol that labs would use to conduct the experiment. Infants’ preferences for particular auditory stimuli are typically tested in looking-time paradigms: infants see an unrelated visual stimulus, and hear the target auditory stimuli across different trials. Their looking time towards the visual stimulus is taken as a measure of their interest in the auditory stimulus. In our case, longer looking during trials with infant-directed speech than during trials with adult-directed speech would indicate a preference.
However, infant looking-time paradigms come in many different flavours. As we quickly learned in discussions (and Google polls), different labs implement looking time paradigms in different ways, and there is no consensus amongst infant researchers about which paradigm is the best (see Eason, Hamlin, & Sommerville, 2017). Indeed, paradigms have different pros and cons related to ease of set-up, training required to run the paradigm, equipment needed, etc. There had been very little work testing the sensitivity of different paradigms to detect effects, and moreover, there is good reason to believe that this could differ based on developmental stage of participants and the research question being tested.
We soon realized that choosing any one paradigm would severely limit the number of labs who could participate. This was antithetical to ManyBabies’ objective to enable broad participation, while maintaining a consistent experimental protocol. We thus decided to allow flexibility in the specific looking time paradigm that labs could implement, letting labs choose between three common set-ups: the headturn-preference procedure, central fixation, and automatic eye-tracking. This decision had important advantages. First, it substantially increased the number of laboratories that could contribute data, thereby increasing the diversity of our sample. Second, it allowed us to compare the protocols directly, something that had never been done before in this kind of research. In the end, we found that one paradigm (the Headturn Preference Procedure) yielded a higher effect size than the other two. This has invited discussion and further research into understanding the origins of this effect, which could be related to general methodological differences, methodological differences that are specific to this research question, or correlated factors (i.e., different characteristics of labs that chose to use different procedures, given that this was not randomly assigned).
We also had to decide on a number of other key parameters of the experiment, such as the length and number of trials. Again, the different ways that labs implemented paradigms was surprisingly diverse, and sometimes inflexible: some key parameters of infant experimental design were mandatorily implemented in one way within certain software programs, while being impossible to implement in the same way in other software programs. For example, one issue was whether all experimental trials would have a fixed length (e.g., 20 s) or whether the length would be determined by infants’ attention during the trial. Because setting a strict standardization would likely exclude some labs from participating, we decided to define a standard protocol, but allowed labs to deviate and report the deviation if the standard protocol was not possible in their set-up. We are currently exploring different alternatives to this solution; for example, ManyBabies 4 is implementing a consistent experimental protocol in PyHab (Kominsky, 2019; based on PsychoPy, Peirce et al., 2019). This approach will increase standardization, but also places additional burdens on participating labs to implement a new procedure and to the central team to troubleshoot the many technical problems that emerge across platforms and hardware set-ups.
Stimulus design was another hurdle. Our research question pertained to language, but given the global nature of ManyBabies, infants in our study would be learning dozens of different native languages. In the end, we opted to create only one stimulus set using North American English. We made this decision for a number of interrelated reasons, which we elucidate here to illustrate some of the unique study design considerations that may arise in setting up large-scale collaborations.
First, we aimed to minimize the burden of time and resources for each laboratory. For this first ManyBabies project, a main goal was to generate interest from as many labs as possible, and we were certain that fewer labs would (or could) participate if they had to create their own stimuli. In addition to reducing our sample size, this would reduce the diversity of languages, labs, and nations represented in the sample.
Second, we wanted to create controlled, balanced, and natural infant-directed and adult-directed stimuli based on thorough input from the ManyBabies community (for details about stimuli, see ManyBabies Consortium, in press). For typical single-lab studies, researchers would create stimuli in their participants’ native language, but not all labs had the expertise necessary to generate such stimuli on their own (e.g., labs that focus on vision or social cognition). Third, the use of dozens of different sets of stimuli would produce an undesirable source of variation that would be confounded with other important variables of interest. We had theoretical reasons to expect that infants might perform differently based on the similarity of their native language to the stimulus language, and we had many long discussions that considered this issue. In the end, we decided by vote that the optimal route was to include language background as a moderating variable in our analyses.
Fourth, we chose North American English in part because of the robust research supporting preference for infant-directed speech in this language (as our goal was for ManyBabies 1 to be a “proof-of-concept” study for large-scale collaborations in our field), and in part because approximately half of labs would be testing infants learning North American English. This made it feasible to compare results for infants who were versus were not exposed to this language/dialect. Moreover, it would have been problematic to use a language that was non-native for the majority of infants, such as Dutch or Japanese, as this would complicate the interpretation of our findings (although this possibility was carefully considered).
It is worth noting that the decision to use just one stimulus set was, and remains, controversial within ManyBabies and the community of language acquisition researchers, due to the fact that it perpetuates an existing bias toward North American research (see ManyBabies Consortium, 2019a, for a broader discussion of this issue). We also had to make difficult decisions in other domains, e.g., inclusion/exclusion of infants born preterm or infants with sensory/developmental issues, which also differs across labs. The decisions we made as a group, most notably in using only North American English, were never intended to become norms for future studies. Instead, we deemed them reasonable first steps in building large-scale collaboration.
Another important part of the planning process was specifying our analysis plan. The dataset this project would generate would include trials nested within infants (something typical in our datasets) but also infants nested within labs (something we rarely if ever encounter). The novel structure of our data raised many important and difficult questions about model design and comparison that none of the collaborators had encountered before, much less received formal training in. Although we made reasonable compromises and were able to get some outside input at a few key junctures, the process would have been less time-consuming (and could have resulted in different choices in some cases) if we had obtained earlier and more consistent input from researchers and statisticians with expertise in analysing these types of data.
Finally, we had to determine the timeline for data collection. Given the slow pace of recruiting and testing infant participants, we decided that labs would be given a full year to gather their data. This would also allow for variation across labs in different regions that could affect the availability of research assistants to collect data (e.g., the cycle of undergraduate projects, graduate admissions, thesis deadlines, etc.).
Piloting
Given the large quantity of resources invested in the project, it was crucial to test the feasibility of our design, “work out the kinks”, and identify any failures before the protocols were distributed widely. Five labs piloted our procedure, and we collected data from 65 infants. In retrospect, these volunteer labs had quite a bit of experience with similar procedures and/or the kind of state-of-the-art best practices we were trying to implement, and it would have been beneficial to also include labs with less experience. Nonetheless, our pilot phase was important in testing the feasibility of the design, as well as data templates we had created. For example, we observed that many of the older infants tested in eye-tracking did not complete all 16 of the test trials. This solidified a design decision that we had made to set a very loose inclusion criterion, specifically that we would include babies for analysis if they contributed at least one pair of trials, and to evaluate post-hoc how the effect size would vary with different criteria.
Manuscript writing, pre-registration, and lab recruitment
Once the methodological details were in place, we began writing what would become the Phase 1 manuscript for a Registered Report, which was pre-registered on the Open Science Framework. Registered Reports are a recently-implemented publishing format used by an increasing number of journals, where the review process occurs in two stages (Nosek & Lakens, 2014). First, authors submit a “Stage 1” manuscript, that includes the introduction, and proposed methodology and analysis. This is peer reviewed for the appropriateness of the methods and planned analyses, and the potential of the paper to make a contribution to the literature. After the authors have incorporated feedback from the reviewers, the editor can accept the article “in principle”. At this point, the paper and its methods are pre-registered, and further methodological and analytic changes are not permitted except in rare situations. After data are collected and the analysis plan is carried out, the results and discussion of the paper are written, and the paper is submitted for “Stage 2” peer review. This stage of review checks that the research has been carried out as planned, and assesses the appropriateness of the discussion section. A particular benefit of the Registered Report format in the context of large-scale replications is that collaborators contributing data can be confident that efforts will be rewarded with publication. Additionally, the feedback received at Stage 1 can be invaluable in identifying potential confounds, missing controls, or errors before time and funds are invested in the data collection process.
The writing process for the ManyBabies Stage 1 Registered Report went relatively smoothly, and we refer readers to two recent papers that provide excellent tips for writing in the context of large-scale scientific collaborations (Moshontz, Ebersole, Weston, & Klein, 2019; Tennant et al., 2019). This writing process allowed us to pin down and codify all the details of our methodology, as well as our planned analyses. We submitted our Registered Report to the new Advances in Methods and Practices in Psychological Sciences in early 2017, and were one of the first to be accepted to this newly established journal.
We sent out a general call for participation on February 2, 2017, via list-servs and personal e-mails to researchers in the field. This occurred synchronously with piloting and manuscript writing. We knew that many of the researchers who had been involved in the planning would want to contribute data (although not all had access to infant populations), but it was unclear how many other labs would sign up.
Deciding whether or not to contribute data to ManyBabies 1 generated a number of novel considerations in the research community. An important consideration in laboratories’ decision-making was whether they supported the general approach: does the potential knowledge to be gained justify the field’s significant investment of resources? But even when fully supportive of the endeavor, labs faced very real practical considerations. Broadly, labs needed to weigh the benefits and costs to them of participating in ManyBabies, a topic we return to in a later section (see also Box 1 for narrative accounts from two labs).
Box 1: ManyBabies as kickstarter for a new lab.
The original ManyBabies 1 call came at a time when I was setting up a new developmental lab and the first of its kind at my institution. In preparation for a number of planned local studies, the basic infrastructure was in place: a bare-bones lab website, family-friendly testing rooms and equipment, and a commitment to open science, though no studies were in progress at the time. Being a small team of one PI with limited financial resources and only a couple of project students at the time meant that getting projects off the ground was a slow process.
The call for a multi-site, preregistered study provided a time-sensitive opportunity for us to actively recruit participants for a defined project, to train RAs, to systematise our testing protocols, and to run a well-designed study using easily-accessible funding, clear experimental protocols, and ready-made stimuli and analysis pipelines. More broadly, we benefited from high-quality training from a community of experts in how to run a lab effectively and under the principles of Open Science.
Since no existing study had to make way for ManyBabies 1, there was no opportunity cost. Two years on, the investment has been instrumental in setting the tone for our now busy and expanding lab. Contributing to the larger scientific enterprise for our inaugural study was a great way to launch the lab. Research opportunities continue via the rich and diverse international network and follow-up projects.
Catherine Davies, Leeds Child Development Unit
My perspective, coming from a relatively established lab, was that ManyBabies gave students in my lab a true taste of what Open Science means at a practical level. The approach adopted by ManyBabies is quite different to what we typically do and it provided a valuable educational opportunity for lab members. Assuming a general orientation towards Open Science in the future of psychological research, I felt ‘hands-on’ experience with this at a practical level was very valuable. An additional benefit came from the many group discussions between labs prior to commencing testing. We had many interesting conversations about our lab practices and protocols for which we would not normally have an opportunity. I found this aspect particularly educational and informative. I made some changes in my lab operations and protocols on account of these discussions, which I believe were definite improvements.
Leher Singh, Associate Professor, National University of Singapore Infant and Child Development Laboratory
In ManyBabies 1, we took several steps to support labs who were interested in participating. For example, to help offset some of the recruitment and personnel costs, ManyBabies 1 was thankfully able to secure a small grant from the Association for Psychological Science, from which we distributed funds to labs based on need and with the objective of increasing diversity in the sample. However, this was far from covering the full cost of ManyBabies 1, and finding sources of such funding continues to be challenging. To support labs in obtaining ethical approvals, we provided templates and an experienced point-person to answer questions, although in at least one case, lack of access to an appropriate ethics review board turned out to be a barrier to eventual participation. Moreover, as we detail below, we made decisions about our target populations and sample size requirements that would make it as easy as possible for labs to participate.
Against these potential costs, individual labs weighed the potential benefits of participation, including scientific insights and the opportunity to connect to a larger community of infant researchers focused on training and collaboration. As ManyBabies 1 was the first largescale collaboration that most labs had participated in, there was considerable uncertainty about the nature and extent of these benefits. For example, would the scientific discoveries warrant the large outlay of resources? What other benefits would come from being part of the network? Now that ManyBabies 1 is complete, these benefits have become considerably clearer, and we will return to them in a later section.
Study Implementation
We asked for labs to officially “sign up” to collect data via a detailed Qualtrics online questionnaire, and ultimately 69 labs contributed data to ManyBabies 1. We asked laboratories to commit to a particular sample size (or to an explicit stopping rule) and to provide detailed information about their lab characteristics and set-up. This form was also an opportunity to remind participating laboratories of their commitments around “data peeking”. For example, we expected that due to sampling error, some individual labs might observe null results or results that were just shy of conventional statistical significance. We wanted to avoid labs changing their data collection plans, or deciding not to submit data in these cases, as such practices would inadvertently affect Type 1 error and effect size estimates (see Schott et al., 2019). Additionally, having labs complete this questionnaire allowed the project leaders to assess and identify any missing information or potentially problematic deviations from protocol. We also asked labs to submit evidence of ethics approval, and contribute a laboratory “walk through” video. Each laboratory was asked to videotape their process from the time the participant arrives until they leave, providing comprehensive information about details such as size and colour of rooms, style of interaction with participants, etc. Because of ethical concerns regarding sharing videos of actual infant participants and their parents, some labs opted to use stand-ins, typically a doll and a research assistant. Although we have only begun to systematically examine and code these videos, they provide a rich and unprecedented peek behind the curtain of different infant lab setups and workflows.
Overall, we found coordinating so many laboratories to be very challenging, in part due to the novelty of the process both for the leadership team and the contributing laboratories. There was considerable confusion about lab sign-ups and one laboratory failed to complete the registration process until after data collection was complete. Having clear sign-up and approval processes in place is crucial. We gave labs a global “green light” to begin data collection once our Registered Report had a Stage 1 acceptance, although registration of new labs continued after this point in part to maximize the diversity of contributors.
To guide labs’ implementation of the protocols, we created a detailed manual with documentation about all aspects of the project. In retrospect, and given the many questions that we subsequently fielded from different labs, our original documentation was inadequate. Despite it being lengthy, some details of implementing the protocol were mistakenly omitted. It also did not address “corner cases”, for example, how labs might best adapt the protocol to the constraints of their own setup. Finally, although we were trying to be exhaustive in having a detailed documentation, in practice, there was so much documentation that researchers were less likely to fully read or/and remember everything (see also compliance with formatting instructions in the next section). Much of our documentation was created on an ‘as needed’ basis, and we could have benefitted from initially obtaining feedback on our drafts from different types of users who would eventually read the documentation (e.g. a PI considering joining the process, a researcher just about to plan data collection, an undergraduate research assistant testing infants, and a researcher returning to the manual as they prepared to send their final data in).
Our experience highlighted how aspects of the process were entirely new to many of the contributing laboratories. This was for several reasons, for example, because of differing siloed ideas about best practice or because participating laboratories did not utilize a particular paradigm in their own research, or because of the unique needs of such a large-scale project and our commitment to Open Science (e.g., strict adherence to data templates; submitting all data, etc.). Friction points often arose when our protocol differed from labs’ standard operating procedures (e.g. inclusion/exclusion criteria; data templates). We also had to modify some of our original policies to handle unanticipated issues. For example, we originally outlined strict rules asking labs not to perform interim analyses on their data or to present their own lab’s results. Ultimately, though, we had to adjust our policy to accommodate on-the-ground realities, e.g. for undergraduate students submitting a final year project prior to the completion of the data collection period. To help labs troubleshoot and address concerns as they arose, we provided clear points of contact for different types of questions, for example, about stimuli, implementing the method on particular hardware, general queries, etc.
Data Validation and Analysis
Once we had amassed the data from ManyBabies 1, we quickly came to realize that they posed a new analytic challenge from anything that any of us had ever faced. We had already written the analysis code for the specific statistical analyses we had planned using pilot data, so we assumed that much of the analysis would be easy to complete. However, some of the trickiest issues in ManyBabies 1 emerged around data validation and processing.
The process of labs uploading their contributed data seemed like it would be simple, but in fact, this process generated some unanticipated and challenging problems once we began processing and merging the datasets. Indeed, data validation and checking consumed the majority of the analysis teams’ efforts! Although we had provided data formatting templates with definitions of each variable, adherence to these templates was inconsistent. Problems were both numerous and sometimes difficult to diagnose. Each change that a lab made to the template was sensible on its own; for example, altering column names to add clarity, combining the two types of data (trial-level and participant-level) into a single workbook to make sure all the data was in one place, or reporting looking times in milliseconds (instead of seconds). These changes undoubtedly helped to ensure data entry quality *within* the lab; however, they made it extremely difficult to maintain that quality *across* labs. Moreover, there was the occasional typo that could only be corrected by going back to the original data (which was located in individual labs). Small data issues of this type are likely to occur frequently and to be relatively unproblematic in single-lab studies (because they can be quickly and easily corrected), but were made very salient because of our centralized data analysis process with data from 69 labs. Overall, the process of data validation highlighted the need for both automated data checking procedures (ideally, that labs could easily implement themselves before submitting their data) and hand checking of errors (see Box 2 for a narrative account from one lab when potential errors were detected in their data). Indeed, although automated procedures could, for example, identify errors such as column name mismatches, it is unlikely that this process will fully negate the need for some level of manual “sanity” checking given the creativity with which researchers (including those on the data processing team) unintentionally foiled the intended data protocols.
Box 2: When data don’t pass validation: The experience of one lab.
Data validation for ManyBabies 1 and spinoff projects was conducted centrally by the data analysis team. This included verifying the data format and values, and hand-checking and visualizing datasets to detect any anomalies. Our lab was one of several whose data had an issue that was “flagged” for follow-up communication. In our case this was for a spinoff project rather than the main ManyBabies 1 project. While for some labs, this flagging concerned only one or two values (mostly typos), our hand-coded data did not appear to match the automatic eye-tracking data retrieved from other labs. Thus, careful and rigidly controlled examination was needed on our part. First, we sought relevant ManyBabies collaborators who would form a rescue team, composed of experienced researchers who stepped forward to help. Our flagged data required in-depth verification, which was accomplished by comparing the original infant videos to the coded values we had submitted. As the rescue team members were located at other institutions, in other countries, we had to to refer back to ethics on data sharing policies and provide (limited) sharing under corresponding (regional) ethics guidance. After the backand-forth feedback from the rescue team members, we determined that the issue was related to incorrect initial coding of infants’ looking time, an issue which is not easily fixed. Our options were to either re-code the data from our video recordings and resubmit the corrected data, or drop the data, however painful it might feel. Ultimately, given timing and resource constraints, we decided to drop the data. This was not an easy decision, and we had to take a deep breath and keep in mind that this action was for the greater good - ultimately we were glad that the invalid data were not included in the analysis. The dropped data were nonetheless valuable in the lessons they taught us, and our authorship was not affected.
Connor Waddell, Undergraduate, Western Sydney University Liquan Liu, Lecturer, Western Sydney University
Throughout this process of data validation, and later our data analysis, we used a reproducible pipeline with distributed version control and project-management tools (Git and http://github.com). Since the eventual codebase contained thousands of lines of R code as well as contributions from 11 distinct contributors, it was critical to be able to share scripts that were run in multiple locations and to track issues with specific parts of the codebase.
We should also mention our experience with data blinding, which we had planned as part of our pre-registration to further reduce analytic bias. Our initial analysis was coded with condition labels randomized. This meant that we gave attention to the errors we encountered ‘fairly’, rather than potentially becoming most interested in some aspect of the data processing that seemed to be producing a surprising result. We only unblinded the data prior to presentation of our results as promised for the 2018 International Conference on Infant Studies. This workflow was effective, but time pressures meant that data validation was still being finalized when the unblinding occurred. This strongly illustrates the tension that can emerge between Open Science best practices and real-world constraints.
ManyBabies, Many Roles: Lessons for working with 100+ collaborators
For most or possibly all participants in ManyBabies, this was the single largest collaboration in which we had participated. Typical workflows with a smaller in-lab team, or even across two or three labs, may not work with 100+ collaborators. One of our biggest challenges was decision-making. Early on, the group committed to a consensus-based approach. This has been somewhat surprisingly successful, but it is not without pitfalls. In practice, “consensus” can sometimes mean that the loudest voice wins, and conversely can lead to stalemates where opposing views fail to reconcile.
While our approach is consensus-based, our organization is not without structure (see Figure 1). At our base are the many researchers and labs that are involved in ManyBabies projects. Individuals and labs can be involved in as many projects in whatever capacity they choose (for details see section Contribution), although the most common contribution was infant data. Leadership teams are formed around specific projects, which are named sequentially (MB1: ManyBabies 1). The leadership team plays a crucial role in ensuring that diverse views are heard and steering discussions toward a productive resolution. Additionally, the leadership team is important for pushing the project forward by assigning tasks, setting deadlines, and as a last resort, stepping in to make hard decisions once all the voices have been heard. As ManyBabies has grown, a higher level of leadership became necessary, known as the “Governing Board”, which is responsible for creating documentation and procedures to ensure that each project within the scope of ManyBabies conforms to its vision, and also that protocols (e.g. for data processing) and institutional memory about what works well are passed from one project to another.
Figure 1.
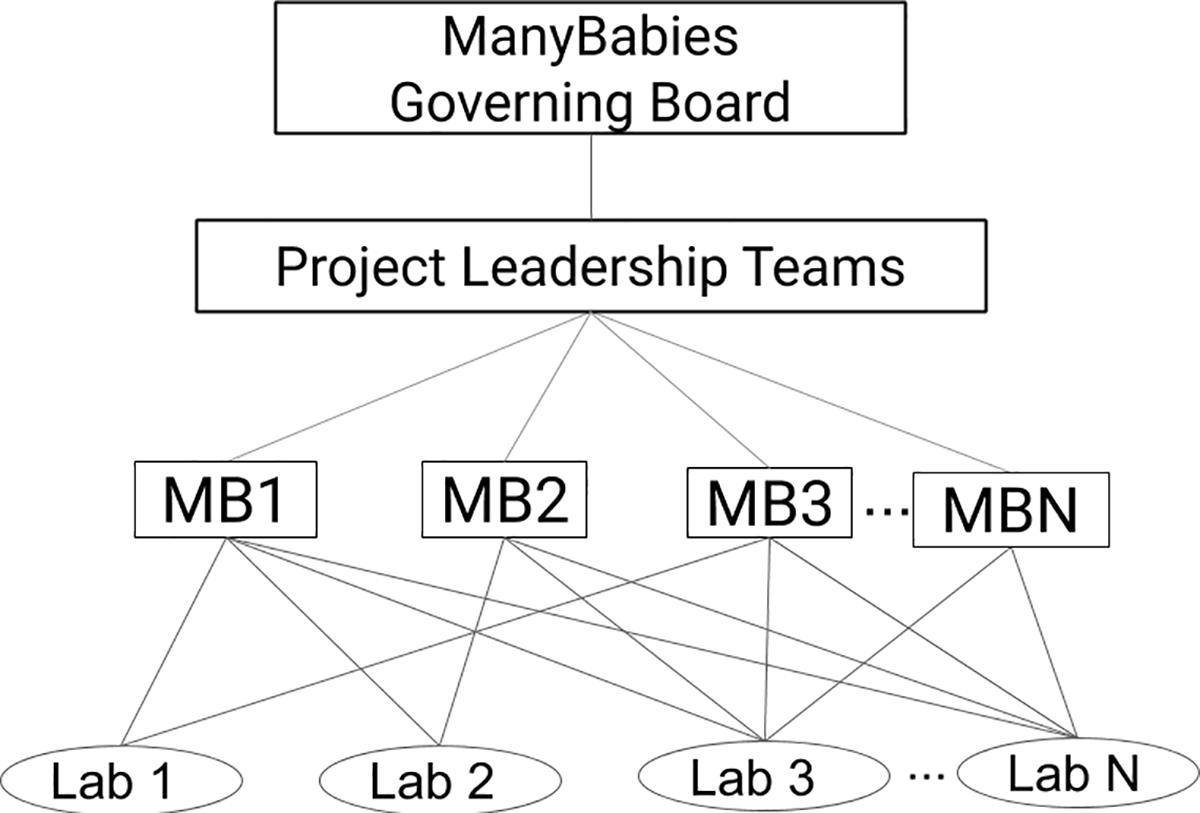
ManyBabies organizational structure
Over the course of ManyBabies 1, we experimented with a number of different communication approaches, such as video conference presentations and meetings, a Slack group, instruction manuals and wikis, collaborative writing within Google Docs, online surveys, Github issues, and email. Each of these channels had strengths and weaknesses, and worked better or worse depending on the objective. One global challenge was that each laboratory had its own internal workflows and experience (or lack thereof) with these different methods of communication. Slack in particular did not work as well as expected. The benefit of using a structured messaging system like Slack is that answers can be searched and shared across a larger group, more effectively ensuring a consensus answer to the various challenges and also making use of the collective knowledge store. However, Slack and similar message boards rely on active and ongoing use by a critical mass, which was never fully achieved. List-servs have been effective, but are used very sparingly, only for the most critical and often time-sensitive communications that require input from the larger group. Person-to-person email, the lowest common denominator, was used extensively to trouble-shoot individual problems, particularly during the “data cleaning” phase of the project. Some anticipated problems never materialized - for example, allowing edit access on documents and data to 100+ contributors could have generated chaos, but did not. We saw no evidence of scooping or data stealing (possibly in part due to our strict rules around the presentation of results associated with the project, and a general tendency of labs to err on the side of caution to ensure the success of the overall project), and manuscript writing emerged fairly organically from our highly structured outlines. When documents occasionally reached a point where the edits overwhelmed the text, self-appointed editors, usually from the leadership team, came in to resolve the issues and a coherent draft emerged within a relatively short time frame.
When only a few individuals are involved in a project, specific contributions can be easily recognized, and there is space for everyone to make a meaningful contribution to the manuscript itself. In a large-scale distributed project, this is not the case, and we therefore defined contributions and authorship broadly. We invited individuals to contribute in diverse ways including design, stimuli, data collection, presentation software, analysis code, and writing. This allowed collaborators to participate whether or not they had access to infants, although in practice there was still some confusion about this. While aiming to be inclusive, we also wanted to avoid the possibility of authorship padding. As a policy, we limited individual labs whose main role was data collection to 2 author-level contributors, the PI and a trainee. However, we were flexible with exceptions, such as for labs with multiple PIs, or situations where the trainee involved in the project unexpectedly had to be replaced.
As a result of our contribution model, the ManyBabies 1 paper had 149 authors. In terms of authorship order, we ultimately decided to have the two project leads be the first and last authors of the paper, with other authors listed alphabetically as middle authors (technically for publication purposes, the “author” of the manuscript is the ManyBabies Consortium, and the order is of the members within the consortium of authors). We created a contributions spreadsheet, where each author could indicate the parts of the research process they participated in, and the level of this contribution. This type of model has been described as a Contributorship rather than an Authorship model (https://psyarxiv.com/dt6e8/, Holcombe, 2019), which we discuss in further detail in the next section. In our case, this was done towards the end of the project, however, in the future it might be better to complete while projects are ongoing.
Ongoing challenges
While we are proud of the successes of our project, there are many challenges that remain. Below we describe some of the more significant ongoing challenges we face.
Funding
ManyBabies is unprecedented from the perspective of traditional consortium funding models. Rather than requiring a large amount of funding for a relatively small number of labs that are performing a single proposed project, the group has worked by disbursing a small amount of funding to a large number of labs who are collaborating to design new projects. While granting agencies are in the abstract supportive of our goals and objectives, traditional funding review processes have not looked kindly on the proposition that the consortium would select new projects during the funded project period. Sustainable funding that is tied to the consortium, rather than specific labs and projects, has been so far elusive. We are working with different funders around the globe to find solutions.
Technical infrastructure for large-scale collaborations
Planning, collecting data, and writing a paper with a group of 149 collaborators requires different kinds of tools and platforms than does working within a group of just 2–4 authors. ManyBabies benefitted immensely from software platforms including GitHub, the Open Science Framework, Google Documents, and Dropbox, but frequently faced tradeoffs to do with accessibility (who knows how to use this tool?), functionality (can this platform allow us to easily roll back errors?), and scaling up (you can ‘clean’ one or two datasets by hand and record the key processing steps, but this becomes unwieldy for 69 datasets). Different phases of ManyBabies 1 required different solutions to these problems, and have motivated, in some cases, the development of new standards and systems that can work for a project like ManyBabies. There is an ongoing need to further develop tools tailored to the unique needs of large-scale collaborations in the behavioural sciences.
Dedicated administrative support and personnel
In addition to funding constraints, each project needs dedicated personnel and administration to coordinate the project. In ManyBabies, this has to date been done by collaborating researchers (often Principal Investigators), who wedge this administrative work in between their many other commitments. The administrative load for this role can be significant. It includes (but is not limited to) coordination of web-based meetings, documenting key points from meetings, leading discussions between participating labs at each stage of project planning and data collection, data checking and quality checks after data collection, as well as analysis, leading manuscript writing and the revision process, and generally keeping the various stages of the project moving forward. An alternative model to having all aspects of the project administered by the respective leadership team would be to have two different types of project leads: a dedicated administrative lead (e.g. a research assistant) to help to facilitate meetings, coordinate sharing of documents and dissemination of information between participating labs, and to facilitate the implementation of the project and separate scientific leadership (typically a small group of leads) to focus on hypothesis formulation, experimental design, manuscript writing and revision, analyses and interpretation, which can be divided among sub-groups within the leadership team. This approach would reduce the logistical burden for researchers who assume scientific leadership for these large-scale research endeavors and it would at the same time provide valuable research experience for an administrative lead. However, having a dedicated administrative lead is challenging given the aforementioned barriers in funding this type of research.
Standards for lab participation
One of the promises of Open Science and initiatives such as ManyBabies is inclusivity and community-level collaboration. However, there may be differences between labs in levels of preparedness, enthusiasm, or resources associated with the planned experiments. One recommendation is to adopt a ‘buddy system’, where new labs are paired with more established ones (preferably geographically close by, to permit visits). This gives a new lab a ready resource person to consult in the event of uncertainty. Another safeguard (implemented in more recent ManyBabies projects) is to request a video of each lab’s testing protocol prior to data collection, which provides an independent check on whether lab set-ups and protocols are implemented in a consistent fashion. Note that this is similar to, but not identical to, the “walk through” video discussed above. The “walk through” video focused on documenting a lab’s practices after a study has been completed, whereas in this case, the goal is to review and identify areas of concern or deviation prior to implementing a study.
Balancing centralized standards vs. individual lab practices
A consistent challenge in implementing a large-scale collaboration is how to strike a balance between standardizing procedures across individual labs and minimizing the barriers to participation, given that individual lab practices may diverge from centralized standards. In general, the approach within ManyBabies projects has been to ensure that key experimental details are standardized across all labs – providing additional support to labs as necessary – while allowing labs as much freedom as possible to follow existing procedures and lab methods. However, there remain significant practical concerns in implementing this approach. For example, because infants are not model participants, each laboratory has standards and practices about when to exclude individual trials (trial-level exclusions), and when an infant participant’s data are fully discarded (session-level exclusions). Such exclusions occur due to concerns like infant fussiness, infant inattention, experimenter/technical errors, or parental interference, and decision-making regarding exclusions has long been a source of undisclosed variability in our field (Eason et al., 2017). In ManyBabies 1, trial-level exclusions were left up to individual labs while session-level exclusions were intended to be made centrally. In practice this created a number of challenges in implementation, because submitting data from nearly every infant (even those who became fussy after just a few trials) was at odds with most labs’ standard operating procedures, who routinely exclude these infants from datasets. Yet, this approach was powerful because it made visible methodological questions that are important for the field as a whole to consider. Indeed, we observed that effect sizes were larger when we excluded infants who completed fewer trials (ManyBabies Consortium, 2019). This illustrates a benefit of individual lab practices coming into tension with centralized standards: discussions that typically take place within a lab become broader conversations across all participating labs.
Interface of ManyBabies participation with traditional incentive structure
Time spent on ManyBabies projects may not directly result in outcomes that are traditionally valued for tenure, promotion, and funding, such as first/senior authored papers, grants awarded, citations, numbers of graduate students, and commercial, economic, or social applications of research findings. Authorship is one of the main currencies valued by hiring institutions, and it is currently unclear how employers, funders, and assessors will view authorship of large-scale collaborative projects like ManyBabies (although we note that an increasing number of job postings are specifically mentioning Open Science; cf. this project: https://osf.io/7jbnt/, Schönbrodt, et al., 2019).This issue is particularly acute within disciplines that have sole authorship as the dominant model of publishing. Some granting agencies, such as Canada’s Social Sciences and Humanities Research Council provide significant “white space” to describe roles in non-traditional authorship situations. We find it sensible for contributors to both list ManyBabies papers in the journal articles on their CVs, as well as under international collaborations, and to take advantage of opportunities to describe their contributions to large-scale collaborative projects. We hope that such issues can be resolved as the field embraces innovative contributorship models (e.g., the CRediT model, which provides a taxonomy of different contribution types ranging from data collection to project administration to securing funding; Holcombe, 2019). Additionally, ManyBabies provides opportunities for smaller groups of authors to launch spin-off projects culminating in traditionally-valued outcomes, for example, by probing ManyBabies datasets for evidence of a different hypothesis or running the same procedure on a distinct population of infants.
In light of the growing expectation to pursue international collaborations, ManyBabies 1 is a flagship example through its involvement of 149 collaborators from 16 countries. ManyBabies is firmly in line with funders’ increasing focus on interdisciplinary, challenge-led research. The consortium includes an interdisciplinary set of collaborators, and squarely addresses the challenge of replicability. We hope that these efforts mitigate some of the perceived risks of working on large scale replication studies, and that funders and institutions start to recognise not just excellence in the end products of research, but also excellence in the ways that research is done.
Technical skills barriers
Some parts of Open Scientific practices require significant technical skills. Not all researchers have been trained in these skills (or have managed to teach themselves), and moreover, applying these practices to large-scale collaborations tests the limits of even the most technically skilled researchers. Innovative platforms like the Open Science Framework and GitHub have made ManyBabies-style projects possible, but learning how to use them is nontrivial. These technical skills present barriers to the parts of a project where a lab or investigator can contribute. The analyses for ManyBabies 1 (and likely future ManyBabies projects) were conducted using R (R Core Team, 2018), in a series of data cleaning, analysis, and Markdown scripts that were more complex than those that might be developed for a typical single-lab infant project. R is transparent, open-source, and free. However, many researchers have been trained in other software such as SPSS, and may have limited knowledge of R as a statistical tool. Combining R with tools like Github and writing within R Markdown only add to the complication. This severely limited the pool of researchers who were able to contribute to the analysis stage of the project, even for seemingly trivial tasks like data-cleaning. More generally, projects like ManyBabies have demonstrated the need for tools that make Open Science easier to do. Some recent tools have tried to be both user-friendly and transparent (PsychoPy, JASP), and this is to be encouraged. However, we should also, as a field, emphasize the importance of training students, postdocs, and PIs to use tools that facilitate Open Science.
Benefits of participation and vision for the future
Benefits
Contributing to Open Science collaborations is often framed in terms of risks to participating researchers, particularly those in the early stages of their career. A number of us were, in fact, initially discouraged from participating by mentors and peers for this reason. Indeed, collaborative projects will naturally reduce some of the time researchers can devote to developing an independent program of research and requires balancing data collection time and resources that are often limited, particularly in infant labs. However, we think it is helpful to reframe these concerns by emphasizing the vast benefits researchers can derive from collaborative science. Below we discuss a number of benefits, many of which may be particularly attractive for early-career researchers.
Scientific benefits.
Given the considerable resources that a large-scale collaboration like ManyBabies requires, it is important that such work provides a clear benefit beyond traditional research models. One important benefit of ManyBabies is in providing a model of Open Science in action, implementing research practices such as pre-registration that buffer against the effects of publication bias. In ManyBabies, data are published regardless of the outcome of the data analysis (assuming sound implementation of the experiment). Traditionally, when experimenters see an experiment not producing the hypothesized set of results, they may terminate the experiment prior to completion to conserve lab resources, which can skew the scientific record, including meta-analyses (Schott et al., 2019). This is not an option in approaches such as ManyBabies, which may result in a dataset that better represents the full range of actual variation in the dependent variable. Pre-registration also buffers against flexibility in analytic choices, allowing for greater objectivity and reducing the chances of spurious analysis practices, such as p-hacking. The opportunity for independent analyses and complete consistency in data handling with other labs is typically not practiced across individual labs. Moreover, the rich dataset generated by ManyBabies projects allow for secondary analyses that might otherwise be unfeasible or too exploratory to warrant dedicated data collection efforts.
Mitigating risk for individual researchers.
One challenge for early-career researchers is that pursuing novel research questions in cognitive development will sometimes lead to null findings, dead ends, or findings that are difficult to explain in the context of past literature. There is always risk associated with research, because good questions do not always yield clear answers. Given that researchers face an incentive structure that focuses heavily on publishing positive findings, individual labs always face the risk that allocating resources to a given project is not guaranteed to yield tangible, positive outcomes (in the form of publications, conference presentations, grants, etc.). When contributing to a larger project, the risk that individual researchers take on in allocating resources to an experiment is shared across a larger whole. An analogy to farm shares is useful here. In farm shares, a community of consumers invests in a portion of a farmer’s crops before they are planted, providing farmers with the resources needed to grow crops and the certainty that these crops will reach the community. The risk of bad weather and a poor season (or alternatively, the benefit of a bumper crop) is shared across the community. Large-scale collaborative research projects function similarly. In our case, the ManyBabies community invests up-front, using both time and money, in a research question that will be valued by the scientific community. Thus, researchers “buy in” with the certainty of contributing to a tangible and visible scientific outcome. This outcome is, in fact, only possible with up-front, collective buy-in.
Training.
ManyBabies 1 collaborators ranged from senior scholars with decades of experience in infant research to undergraduates working on their first research project (for a narrative account from an undergraduate perspective, see Box 3). For many, this was their first exposure to Open Science practices such as pre-registration, the Registered Report journal format, and/or open data/analysis. Several labs reported implementing some of these practices as a result of participation in ManyBabies 1 (see Box 4). The documentation and manuals created for setting up the ManyBabies 1 can model a set of best practices for infancy research, e.g., how to document experiment setup and how to organize and process data to allow easy sharing and analysis. Trainees and more experienced researchers learned from one another as they puzzled out issues in experimental design and statistical analysis (see Box 5 for a graduate student’s perspective). Participating in a large-scale collaboration provides researchers with direct access to a community of researchers motivated to develop and share new techniques and best practices in the field.
Box 3: An undergraduate’s perspective on contributing to Manybabies.
I worked on ManyBabies 1 as part of my final-year undergraduate project. This was my first experience of doing research on the front lines. When I joined the project the foundation was already laid out: the design and manuscript were being created by the collaboration of researchers around the world. Although I had access to the manuscript, I did not personally participate in the writing process. My role was to understand every component of the manuscript and the experimental protocol. The visual and auditory stimuli were created by a few collaborators and then I combined them into videos and shared the files with participating labs. I taught the rest of my lab the procedure of the project, recruited infants for the study, and presented updates on the project at lab meetings. I was also able to present our lab’s preliminary results at undergraduate student conferences at our university.
It was a gratifying experience to be able to take on an important task of this project and to share my work with others. What was enlightening about this project was how helpful other research collaborators were; it was rewarding to be an undergraduate student in contact with so many accomplished researchers. I learned throughout this experience that there is so much more that happens behind the scenes, before the results are published in articles. Behind every sample size there should be a power calculation. Behind all stimuli and trials, there are countless hours of debate and fine-tuning. And behind every publication, there are researchers who were dedicated to investigating their research question. How many undergraduate students can say that they took part in an international collaborative study? It is one of the toughest, most challenging projects I have ever been a part of, but one of the most fulfilling. I learned about communicating and teaching others, as well as using different software programs. I never thought I would discover so much about research outside of the classroom, but I have found that engrossing myself in research first-hand and going beyond a textbook has been the most valuable learning experience.
Meghan Mastroberardino, Undergraduate Student, Concordia Infant Research Lab, Concordia University
Box 4: How did we do? A summary of the ManyBabies 1 lab exit questionnaire.
Following their participation in ManyBabies 1, labs were invited to complete an exit questionnaire to better understand their experiences. A total of 65 labs completed the questionnaire. Our first research question was whether participation in ManyBabies was effective in modeling and spreading Open Science practices. When asked whether lab practices changed after participating in ManyBabies, 13 (20%) labs reported yes, 21 (32%) labs reported maybe, and 30 (46%) labs reported no. It is not known how many labs who reported “no” had already implemented Open Science practices prior to participating in ManyBabies. The most common practices that were adopted/changed were open data (12), use of the Open Science Framework website (12), pre-registration (11), open materials (11), using Databrary, an online repository for video data (8), power analysis (6), and Registered Reports (3). In an open-ended question, labs also reported adding more language controls, and better parent controls/briefings. These results suggest that participation in large-scale collaborations that implement Open Science practices could be a powerful mechanism for uptake of new best practices amongst labs.
We also asked how satisfied labs were with their participation (n = 63). Labs had a high overall level of satisfaction, with 98% of labs reporting that their experience was “Excellent” or “Good”, and one lab reporting “Average”. In open-ended questions, there were many positive remarks including enthusiasm for ManyBabies, and gratitude for having been able to participate. Many respondents mentioned the importance of ManyBabies for building their network of collaborators, being a training opportunity for students and junior researchers, and increasing their proficiency in “best practices”. Negative remarks included specific concerns about the experimental design and stimuli, for example, that the stimuli were not natural enough, or that the questionnaires were too long or intrusive/inappropriate in some cultural contexts. In a question that specifically addressed documentation and communication, contributors (n = 63) were satisfied overall, with 93% giving one of the highest two ratings, although 4 labs rated their satisfaction as “poor”. Specific comments related to difficulty locating different materials across platforms, tracking different versions, and understanding where and how to upload data. These comments highlight the need for improved information architecture and infrastructure to support large-scale collaborations, but suggest a very high level of satisfaction overall with the experience.
The ManyBabies Governing Board
Box 5: A graduate student in the trenches with the data.
I initially became involved in ManyBabies 1 when the lab in which I was working as a PhD student at the University of Oregon signed on to participate in data collection. After data collection began, I received a list-serv posting advertising the opportunity to help with data analysis, and I was immediately interested in joining the data analysis team. However, I was apprehensive at first about whether my analysis and coding skills were up to par with others on the team and how much I would be able to contribute. In the end, working with the analysis team ended up being one of the best experiences of my graduate training. I was able to contribute to data analysis in a variety of ways, acquire new analysis and coding skills, and gain hands-on experience with a variety of issues unique to large-scale, collaborative research. For example, the data analysis team used Github to facilitate collaboration. While I had some basic experience with Github, I hadn’t yet used it to collaborate with multiple researchers. I quickly learned, with the support of other members of the analysis team, how to download and edit existing code and initiate “pull requests” (i.e., request that my edits be merged with existing code). Github additionally facilitated collaboration by allowing any contributor to post “issues” that any member(s) of the team could choose to address. In this way, I was able to select the issues that I was most interested in or felt I could best help with. These issues often involved unique problems related to merging data from more than sixty labs into one file for analysis. More generally, involvement in this project allowed me to observe and learn from others’ code and to take on new coding challenges and, in this way, improve my own skills.
Jessica Kosie, Postdoctoral Research Associate, Princeton University
Research networks.
Bringing researchers together to work on one problem naturally creates many opportunities to develop new ideas and projects. ManyBabies projects have created numerous new connections and collaborations between participating scientists. Early-career researchers and trainees – who may not have as many opportunities to expand their research networks – stand to benefit in particular from the connections formed organically as a consequence of collaborative work. Researchers at all levels interacted with each other both online, as well as through happy hour and lunch events that we organized at popular conferences in our field. Graduate students at these events were able to meet each other and to interact directly with potential postdoctoral advisors, and faculty members were able to learn about the research programs of a wider range of early-career researchers.
Facilitating broad participation and “contribute what you can” structure.
Finally, a guiding principle in many of the decisions made throughout ManyBabies was to create many different ways to contribute to the project and a variety of options in terms of the type and amount of resources any given researcher could commit to the project. For example, this principle led us to set the threshold for the minimum number of infants a lab needed to commit as part of the ManyBabies 1 project to a relatively small number (n = 16) and to place general calls to researchers to get involved at multiple stages over the course of the project. Some individuals focused almost exclusively on conceptual planning, some on data analysis, some on data collection, and some on writing. These focused contributions were essential for the success of ManyBabies 1, because they facilitated expertise in different aspects of the project. Our intuition is that the most effective path to advancing these goals is to get broad buy-in and participation from the field throughout the project. By keeping the initial commitments relatively modest and by practicing inclusivity when inviting new researchers and labs to join the effort, we hope to share the benefits of ManyBabies with an ever-growing group of developmental scientists.
Growth
ManyBabies continues to grow, despite the challenges we experienced during ManyBabies 1. We are now equipped with guidelines and best practices that will facilitate success for current, planned, and future collaborative projects. Key to the continued growth of ManyBabies is to enable minimal and time-intensive contributions by individual labs; some may only want to test a small sample of infants, and some may want to be a part of decision-making during all group conference calls for a particular project. Still others may want to take the lead in proposing and leading new collaborative projects. We are actively working to ensure that our core values of equity, diversity, and inclusion are at the forefront as we continue to develop our network.
ManyBabies and other large-scale collaborative efforts gain momentum from informal conversations that take place in the hallways of conferences, on Twitter, or within labs and departments. Our local colleagues have often been excited about ManyBabies, viewing it as a clearly important direction for the future of developmental science, both because it addresses problems related to replicability and because it encourages principles of Open Science. We encourage any interested graduate student or postdoc to raise the possibility of participating with their advisor, and we encourage every principal investigator to find out if somebody in their lab is interested in contributing. Even if data collection does not seem feasible, there are many additional ways to contribute to ManyBabies or to similar endeavors. We also encourage scientists to discuss the goals and ongoing ManyBabies projects with other scientists in their home labs and departments.
Diversity
A particularly important aspect of the ongoing development of ManyBabies is the involvement from labs in many different countries and cultures. Psychological science is plagued by reliance on what are known as WEIRD (western, educated, industrialized, rich, democratic) samples (Henrich, Heine, & Norenzayan, 2010). While these samples are often convenient, they do not represent the majority of human beings who live now or who have ever lived. This is especially important for infant research, as the environments in which infants are raised vary dramatically across the globe.
ManyBabies presents a special opportunity to collaborate with early child development researchers from a broad range of cultures and nations. However, the promise of this opportunity has yet to be fully realized. While ManyBabies 1 included dozens of labs from North America and Europe, there was minimal representation of labs in Australia and Southeast Asia, and there were no participating labs from Africa, Latin America, East Asia, South Asia, or the Middle East. That is, the Global South was particularly underrepresented, and developing countries were particularly underrepresented.
We have undertaken many efforts to broaden participation, and to date we have received one grant for doing so from the Jacobs Foundation to partner with labs in Africa. We found the names of professors, researchers, clinicians, and other professionals via word of mouth from colleagues, and we reached out to them to see if they would be interested in participating. Responses were almost uniformly positive, and we are currently in the process of training labs and initiating data collection for the ManyBabies 1 project on the infant-directed speech preference. This first attempt at collaboration with scientists in Africa will yield results that are either convergent or divergent from the findings reported in the initial ManyBabies 1 manuscript, and this will be useful for developmental science regardless of outcome. We are currently submitting grants with the goal of obtaining funding for interested scientists in both Latin America and Africa.
In the end, we hope to continue learning from each other about our research practices, and the value of this is likely to be high for labs regardless of their location, in Western regions or otherwise. Moreover, the sheer size of our total samples generate increased opportunities for studying hard-to-recruit populations that are too small to easily study within a single laboratory, such as bilingual infants (as in the ManyBabies 1 Bilingual project), as well as preterm infants and young children with developmental delays and disorders. We hope to collaborate with and learn from a global network of labs from six continents representing the diversity of human experience. Indeed, we would welcome contributions from Antarctica if a developmental lab is ever established there!
From Many Babies to Many Scientists
What makes a good scientist? If you simply ask people to name key characteristics of a successful scientist, “collaborative” would probably not rank highly on the list of frequently mentioned traits. The prototypical image of a scientist in the popular imagination is probably that of a solitary genius, toiling away at an idea or question, usually generated by them alone (or perhaps hitting them literally on the head, as in the apocryphal story of Isaac Newton’s apple tree and the theory of gravity). Individual, independent creativity and hard work will undoubtedly continue to be key to advancing developmental research, and psychological science in general. However, we think that large-scale collaborative projects like ManyBabies suggest a way to expand our sense of what makes a good scientist, moving collaborative work from the periphery to the center. This is of course in one sense nothing new - our science has always been a collaborative endeavor, with work shared across many individuals within and across labs. With projects such as ManyBabies, we hope to make large-scale collaborative science a more central part of how we approach the hardest problems in psychology - problems that no individual scientist alone can solve. Other fields have turned to large-scale collaboration to tackle key challenges. What springs to mind when envisioning cutting-edge work in physics today is not an individual pioneer like Newton - instead, it is the team of thousands of scientists working with the Large Hadron Collider to answer some of the most fundamental questions in physics. A number of initiatives, including – to name just a few – the Open Science Collaboration (Open Science Collaboration, 2015), the Psychological Science Accelerator (Moshontz et al., 2018), the ManyLabs initiative (Ebersole et al., 2016; Klein et al., 2014, 2018), ManyPrimates (Many Primates et al., 2019), ManyClasses (https://www.manyclasses.org/, ManyClasses, 2018), ManyNumbers (https://osf.io/e4xb7/, ManyNumbers, 2020), and the crowdsourcing platform StudySwap (https://osf.io/meetings/StudySwap/, StudySwap, 2019), are implementing variations on this approach, and we expect that a variety of collaboration models will be increasingly necessary to meet the fundamental challenges and open questions in psychology and related fields. In Table 1, we overview some considerations prior to launching a large-scale collaboration.
Table 1:
Some considerations prior to launching a large-scale collaboration.
| • Is the research question well-suited to a large-scale collaboration? Is the investment of resources commensurate with the anticipated contribution to knowledge? |
| • Is there sufficient interest from the research community to support the project? How many labs would be willing/able to contribute data? |
| • What will the leadership structure be? Is there sufficient conceptual knowledge, technical expertise, and diversity of perspectives? |
| • What steps will be taken to ensure an inclusive atmosphere and to support mentorship at all levels? |
| • Is the project feasible? Considerations include ethical approvals, access to relevant populations, methodological expertise, availability of research personnel and equipment, time, and monetary constraints. |
| • How will the project be administered? What mechanisms will be used to verify methodological standardization and data quality? |
In the initial ManyBabies projects, our primary goal was to address one of the most important problems we currently face - investigating the replicability of key findings in infant research while working to improve research methods. As our focus on testing a variety of theoretical questions in ManyBabies 1 makes clear, however, we do not think that large-scale collaborative projects are needed “just” for understanding replication issues. Instead, we hope that ManyBabies will help serve as a model of how to create collaborative projects to solve the hardest problems in our field – both methodological and theoretical.
Public significance statement.
ManyBabies is a large-scale collaboration across infant research labs, focusing on replicating important findings in infant research, as well as developing and modeling best practices, such as preregistration, open materials, and open data. ManyBabies further aims to increase growth and diversity both within infancy research and in psychological science in general. This paper shares “lessons learned” from our first project – ManyBabies 1 – which involved 100+ researchers and 70+ labs. Lessons emerged from different aspects of our collaboration, ranging from coordination challenges and operational practices to considerations for study design and data collection. The insights we gained can be applied to future collaborations in psychology at both small and large scales.
Acknowledgements:
We wish to thank the hundreds of researchers and thousands of families who contributed to the success of ManyBabies 1. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (2018–04390) and the Concordia Research Chairs program to KBH; the Eunice Kennedy Shriver National Institute of Child Health and Human Development (1R01HD095912–01A1) to CLW and KBH; the National Science Foundation Graduate Research Fellowship Program (DGE-1256259) to MZ; the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 798658 to LL.
Contributor Information
Krista Byers-Heinlein, Concordia University.
Christina Bergmann, Max Planck Institute for Psycholinguistics.
Catherine Davies, University of Leeds.
Michael C. Frank, Stanford University
J. Kiley Hamlin, University of British Columbia.
Melissa Kline, Center for Open Science.
Jonathan F. Kominsky, Rutgers University Newark
Jessica E. Kosie, Princeton University
Casey Lew-Williams, Princeton University.
Liquan Liu, University of Oslo; Western Sydney University.
Meghan Mastroberardino, Concordia University.
Leher Singh, National University of Singapore.
Connor P. G. Waddell, Western Sydney University
Martin Zettersten, University of Wisconsin-Madison.
Melanie Soderstrom, University of Manitoba.
References
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, & Munafò MR (2013). Power failure: why small sample size undermines the reliability of neuroscience. Nature Reviews Neuroscience, 14(5), 365–376. 10.1038/nrn3475 [DOI] [PubMed] [Google Scholar]
- Byers-Heinlein K, Bergmann C, Brown A, Carbajal MJ, Durrant S, Fennell CT, … & Wermelinger S (2019). A multi-lab study of bilingual infants: Exploring the preference for infant-directed speech. Stage 1 Registered Report Accepted in Advances in Methods and Practices in Psychological Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers-Heinlein et al. , (2020, February 5). ManyBabies1B - Preference for Infant-directed Speech in Bilingual Infants. Open Science Framework. Retrieved from osf.io/zauhq [Google Scholar]
- COPE Council. (2014). What constitutes authorship? COPE Discussion Document. Retrieved from https://publicationethics.org/files/Authorship_DiscussionDocument.pdf [Google Scholar]
- Databrary. (2012). The Databrary Project: A video data library for developmental science. New York: New York University. Retrieved from http://databrary.org [Google Scholar]
- Eason AE, Hamlin JK, & Sommerville JA (2017). A survey of common practices in infancy research: Description of policies, consistency across and within labs, and suggestions for improvements. Infancy, 22(4), 470–491. 10.1111/infa.12183 [DOI] [Google Scholar]
- Ebersole CR, Atherton OE, Belanger AL, Skulborstad HM, Allen JM, Banks JB, … Nosek BA (2016). Many Labs 3: Evaluating participant pool quality across the academic semester via replication. Journal of Experimental Social Psychology, 67, 68–82. 10.1016/j.jesp.2015.10.012 [DOI] [Google Scholar]
- Frank MC (2015, December 14). The ManyBabies Project [Blog post]. Retrieved from http://babieslearninglanguage.blogspot.com/2015/12/the-manybabies-project.html
- Henrich J, Heine SJ, & Norenzayan A (2010). Most people are not WEIRD. Nature, 466(7302), 29. 10.1038/466029a [DOI] [PubMed] [Google Scholar]
- Holcombe AO (2019). Contributorship, not authorship: Use credit to indicate who did what. MDPI Publications. 10.3390/publications7030048 [DOI] [Google Scholar]
- Hunter MA, & Ames EW (1988). A multifactor model of infant preferences for novel and familiar stimuli. Advances in Infancy Research, 5, 69–95. [Google Scholar]
- Kline ME (2018). Using the ManyBabies 1 dataset to understand variation in lab practices. Talk presented at the 21st Biennial International Conference on Infant Studies, Philadelphia, PA. [Google Scholar]
- Klein RA, Ratliff KA, Vianello M, Adams RB Jr, Bahník Š, Bernstein MJ, ... & Cemalcilar Z (2014). Investigating variation in replicability: A “many labs” replication project. Social Psychology, 45, 142–152. 10.1027/1864-9335/a000178 [DOI] [Google Scholar]
- Klein RA, Vianello M, Hasselman F, Adams BG, Adams RB, Alper S, … Nosek BA (2018). Many Labs 2: Investigating Variation in Replicability Across Samples and Settings. Advances in Methods and Practices in Psychological Science, 1(4), 443–490. 10.1177/2515245918810225 [DOI] [Google Scholar]
- Kline M et al. (2018, June 25). ManyBabies 1: Secondary Analysis of “Lab Factors” and Fuss-Out Rates. Open Science Framework. Retrieved from osf.io/ryzmb
- Kominsky JF (2019). PyHab: Open-source real time infant gaze coding and stimulus presentation software. Infant Behavior & Development, 54, 114–119. 10.1016/j.infbeh.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ManyBabies Consortium. (2019a). Quantifying sources of variability in infancy research using the infant-directed speech preference. Advances in Methods and Practices in Psychological Science. [Google Scholar]
- ManyBabies Consortium. (2019b, December 20). ManyBabies 1: Infant-Directed Speech Preference. Open Science Framework. Retrieved from osf.io/re95x [Google Scholar]
- ManyClasses. (2018, July 15). Retrieved from https://www.manyclasses.org/
- ManyNumbers. (2020, January 30). Open Science Framework. Retrieved from osf.io/e4xb7
- Many Primates, Altschul DM, Beran MJ, Bohn M, Call J, DeTroy S, … Watzek J (2019). Establishing an infrastructure for collaboration in primate cognition research. PLoS ONE, 14(10), e0223675. 10.1371/journal.pone.0223675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshontz H, Campbell L, Ebersole CR, IJzerman H, Urry HL, Forscher PS, Chartier CR (2018). The Psychological Science Accelerator: Advancing Psychology Through a Distributed Collaborative Network. Advances in Methods and Practices in Psychological Science, 1(4), 501–515. 10.1177/2515245918797607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshontz H, Ebersole CR, Weston SJ, & Klein RA (2019, August 14). A Guide for Many Authors: Writing Manuscripts in Large Collaborations. 10.31234/osf.io/92xhd [DOI]
- Nielsen M, Haun D, Kärtner J, & Legare CH (2017). The persistent sampling bias in developmental psychology: A call to action. Journal of Experimental Child Psychology, 162, 31–38. 10.1016/j.jecp.2017.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek BA, & Lakens D (2014). Registered reports: A method to increase the credibility of published results. Social Psychology, 45(3), 137–141. 10.1027/1864-9335/a000192 [DOI] [Google Scholar]
- Open Science Collaboration (2015). Estimating the reproducibility of psychological science. Science, 349(6251), aac4716. 10.1126/science.aac4716 [DOI] [PubMed] [Google Scholar]
- Peirce J, Gray JR, Simpson S, MacAskill M, Höchenberger R, Sogo H, ... & Lindeløv JK (2019). PsychoPy2: Experiments in behavior made easy. Behavior Research Methods, 51(1), 195–203. 10.3758/s13428-018-01193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- Rahal RM, & Open Science Collaboration. (2015). Estimating the reproducibility of psychological science. Science, 349(6251), aac4716. 10.1126/science.aac4716 [DOI] [PubMed] [Google Scholar]
- Rouder JN (2016). The what, why, and how of born-open data. Behavior Research Methods, 48(3), 1062–1069. 10.3758/s13428-015-0630-z [DOI] [PubMed] [Google Scholar]
- Schönbrodt FD, Mellor DT, Bergmann C, & Penfold N (2019, November 25). Academic job offers that mentioned open science. Open Science Framework. Retrieved from osf.io/7jbnt [Google Scholar]
- Schott E, Rhemtulla M, & Byers-Heinlein K (2019). Should I test more babies? Solutions for transparent data peeking. Infant Behavior and Development, 54, 166–176. 10.1016/j.infbeh.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Schreiner M, et al. (2019, June 6). Assessing test-retest reliability in infant-directed speech preference measures. Open Science Framework. https://osf.io/zeqka/ [DOI] [PubMed] [Google Scholar]
- Simmons JP, Nelson LD, & Simonsohn U (2011). False-positive psychology: Undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological Science, 22(11), 1359–1366. 10.1177/0956797611417632 [DOI] [PubMed] [Google Scholar]
- Soderstrom M, et al. (2020). ManyBabies 1 CDI Follow-up. Open Science Framework. Retrieved from osf.io/2qamd [Google Scholar]
- StudySwap. (2019, March 29). StudySwap: A platform for interlab replication, collaboration, and research resource exchange. Open Science Framework. Retrieved from osf.io/9aj5g [Google Scholar]
- Tennant J, Bielczyk NZ, Cheplygina V, Greshake Tzovaras B, Hartgerink CHJ, Havemann J, ... & Steiner T (2019, July 2). Ten simple rules for researchers collaborating on Massively Open Online Papers (MOOPs). 10.31222/osf.io/et8ak [DOI]


