Abstract
Most members of basic leucine zipper (bZIP) transcription factor (TF) subgroup A play important roles as positive effectors in abscisic acid (ABA) signaling during germination and/or in vegetative stress responses. In multiple plant species, one member, ABA insensitive 5 (ABI5), is a major TF that promotes seed maturation and blocks early seeding growth in response to ABA. Other members, referred to as either ABRE‐binding factors (ABFs), ABRE‐binding proteins (AREBs), or D3 protein‐binding factors (DPBFs), are implicated as major players in stress responses during vegetative growth. Studies on the proteolytic regulation of ABI5, ABF1, and ABF3 in Arabidopsis thaliana have shown that the proteins have moderate degradation rates and accumulate in the presence of the proteasome inhibitor MG132. Exogenous ABA slows their degradation and the ubiquitin E3 ligase called KEEP ON GOING (KEG) is important for their degradation. However, there are some reported differences in degradation among subgroup A members. The conserved C‐terminal sequences (referred to as the C4 region) enhance degradation of ABI5 but stabilize ABF1 and ABF3. To better understand the proteolytic regulation of the ABI5/ABFs and determine whether there are differences between vegetative ABFs and ABI5, we studied the degradation of an additional family member, ABF2, and compared its in vitro degradation to that of ABI5. As previously seen for ABI5, ABF1, and ABF3, epitope‐tagged constitutively expressed ABF2 degrades in seedlings treated with cycloheximide and is stabilized following treatment with the proteasome inhibitor MG132. Tagged ABF2 protein accumulates when seedlings are treated with ABA, but its mRNA levels do not increase, suggesting that the protein is stabilized in the presence of ABA. ABF2 is also an in vitro ubiquitination substrate of the E3 ligase KEG and recombinant ABF2 is stable in keg lysates. ABF2 with a C4 deletion degrades more quickly in vitro than full‐length ABF2, as previously observed for ABF1 and ABF3, suggesting that the conserved C4 region contributes to its stability. In contrast to ABF2 and consistent with previously published work, ABI5 with C terminal deletions including an analogous C4 deletion is stabilized in vitro compared to full length ABI5. In vivo expression of an ABF1 C4 deletion protein appears to have reduced activity compared to equivalent levels of full length ABF1. Additional group A family members show similar proteolytic regulation by MG132 and ABA. Altogether, these results together with other work on ABI5 regulation suggest that the vegetative ABFs share proteolytic regulatory mechanisms that are not completely shared with ABI5.
Keywords: ABF, abscisic acid, bZIP, KEG, proteolysis, ubiquitin
1. INTRODUCTION
The Arabidopsis thaliana family of basic leucine zipper (bZIP) transcription factors consists of 67–78 members (Deppmann et al., 2004; Dröge‐Laser et al., 2018; Jakoby et al., 2002). bZIP proteins are characterized by the presence of a DNA‐binding basic region enriched in arginine or lysine residues and a leucine zipper dimerization motif that consists of a variable number of leucine heptad repeats. Arabidopsis bZIPs are divided into 13 groups (A–K, M, and S) defined by sequence similarity in the basic region and by shared conserved motifs outside of the bZIP domain (Dröge‐Laser et al., 2018). Group A, with thirteen members, can be further divided into four subgroups (Dröge‐Laser et al., 2018). Nine members in two subgroups share four additional regions called C(conserved)1–4 (Choi et al., 2000; Uno et al., 2000). The abscisic acid (ABA) insensitive 5 (ABI5) subgroup consists of ABA insensitive 5 (ABI5, also referred to as Dc3 protein‐binding factor or DPBF1) and three other DPBFs (DBPF2, DPBF3/AREB3 for ABA responsive element binding, and DPBF4/EEL for ENHANCED EM LEVEL). The next closest subgroup, with a diverged C4 region, is the ABRE‐binding factor (ABF) subgroup, consisting of ABF1, ABF2/AREB1, ABF3/DPBF5, ABF4/AREB2, and one member currently unnamed and uncharacterized (At5g42910) (Brocard et al., 2002; Dröge‐Laser et al., 2018). With the exception of At5g42910, which has not been tested, the other eight members show binding to a multimer of an ABA‐responsive element (Choi et al., 2000; Kim et al., 2002; Uno et al., 2000; Yamamoto et al., 2009). Multiple members of group A are broadly represented among land plants and are also present in the bryophytes Physcomitrella patens and Marchantia polymorpha. Two ABF/ABI5 members are identified in the charophyte Klesormidium nitens, which lacks an ABA‐dependent response but does have a desiccation tolerance response, suggesting the origins of land plant ABA‐dependent responses utilize group A bZIP proteins as an early component (Cuming, 2019; Hauser et al., 2011).
ABI5, the best characterized group A member, was first identified from a mutant screen in Arabidopsis for ABA‐insensitive germination (Finkelstein, 1994; Finkelstein & Lynch, 2000) and has emerged as a key player in ABA responses during germination and early seedling growth. ABI5 transcripts increase during seed development with highest levels in dry seeds but are also present in vegetative tissues at much lower levels (Brocard et al., 2002; Finkelstein & Lynch, 2000; Lopez‐Molina & Chua, 2000). There is a short window of time after seed stratification when exogenous ABA can strongly induce ABI5 mRNA and protein accumulation (Lopez‐Molina et al., 2001), which presumably inhibits germination and growth since abi5 loss‐of‐function (LOF) mutants germinate in normally inhibitory concentrations of ABA (Finkelstein, 1994; Lopez‐Molina & Chua, 2000). Although at levels ~100‐fold less than in 2‐day‐old seedlings, ABA can modestly increase ABI5 mRNA in 10‐day‐old plants (Brocard et al., 2002). ABI5 does play roles during nitrate‐mediated inhibition of lateral root growth (Signora et al., 2001), and in floral transition, the latter through positively modulating expression of FLOWERING LOCUS C (FLC) (Wang et al., 2013), a flowering repressing transcription factor (Michaels & Amasino, 1999).
Members of the ABF subgroup appear to have more significant roles during vegetative stress responses. Multiple ABF mRNAs increase in response to exogenous salt and ABA in mature plants (Choi et al., 2000; Uno et al., 2000). ABF1 (Yoshida et al., 2015), ABF2, ABF3, and ABF4 (Yoshida et al., 2010) all contribute to drought tolerance in mature plants. Overexpression of ABF2 (Kim et al., 2004), ABF3, or ABF4 (Kang et al., 2002) slows germination, indicating that they can modulate this process as well.
In addition to regulation at the transcript level, ABI5/ABF abundance is regulated at the protein level (Liu & Stone, 2014; Skubacz et al., 2016; Yu et al., 2015). Exogenous ABA slows the degradation of constitutively expressed ABI5 and ABF1 and ABF3 in transgenic seedlings (Chen et al., 2013; Lopez‐Molina et al., 2001), and the RING‐type E3 ligase KEEP ON GOING (KEG) plays an important role in ABI5, ABF1, and ABF3 degradation in vivo and ubiquitinates them in vitro (Chen et al., 2013; Stone et al., 2006). Additional ubiquitin ligases are implicated in promoting ABI5 degradation. DWD hypersensitive to ABA 1 (DWA1) and DWA2 are two substrate specificity factors for a Cullin4‐based ubiquitin E3 ligase (Lee et al., 2010) whose LOF mutants have ABA‐hypersensitive phenotypes. Both interact in vitro with ABI5 and dwa extracts degrade ABI5 more slowly. ABA‐hypersensitive DCAF 1 (ABD1), another CUL4 substrate receptor, interacts with ABI5 and plays a role in ABI5 degradation (Seo et al., 2014). ABD1 and ABI5 interact in a yeast two‐hybrid assay, and endogenous ABI5 pulled down with Myc‐ABD1 in a co‐IP from lysate of plants overexpressing Myc‐ABD1. When seeds were treated with CHX following ABA treatment, ABI5 protein degraded more slowly in abd1‐1 seeds than in wild‐type (WT) seeds (Seo et al., 2014). With the exception of KEG, the role of these ligases in the proteolytic control of other group A bZIPs is not known.
The earliest described Arabidopsis ABI5/ABF post‐translational modification is phosphorylation; using in‐gel kinase assays, a 42‐kDa kinase activity modifying peptide substrates corresponding to ABF2 or ABF4 C1, C2, or C3 regions is rapidly activated after in vivo ABA treatment (Furihata et al., 2006; Uno et al., 2000). Arabidopsis ABF in vivo phosphorylation at two sites increased after treatment of leaves with ABA; one corresponds to an AREB3 C3 serine, but the other phosphorylated C2‐Ser cannot be assigned to a specific ABF because the tryptic peptide containing the site is 100% conserved among seven ABF members (Kline et al., 2010). Interestingly, no net change in constitutively expressed Arabidopsis ABI5 phosphorylation is observed after ABA treatment of 8‐day‐old seedings, although migration on SDS‐PAGE is affected (Lopez‐Molina et al., 2001, 2002). Peptides that include the same C1‐3 regions, but not C4, were identified as singly phosphorylated in HA‐ABI5 by mass spectrometry from 4‐week‐old plants expressing ABI5 under the 35S promoter (Lopez‐Molina et al., 2002). SnRK2 kinases at ~42 to 44 kDa were identified as ABA‐inducible activities in a number of species (Johnson et al., 2002; Nakashima et al., 2009). At least two other kinases were observed in in‐gel kinase assays (Johnson et al., 2002; Nakashima et al., 2009), leading to identification of calcium‐dependent protein kinases (CDPKs), named CPKs in Arabidopsis. CPK32 interacts with ABFs 1–4 and phosphorylates an ABF4 peptide in vitro (Choi et al., 2005). CPK10 and 30 interact in yeast with ABF4 (Choi et al., 2005) and CPK4 and 11 phosphorylate full‐length ABF1 and ABF4 in in‐gel assays (Zhu et al., 2007). A calcinurin‐type kinase, CIPK26, interacts with ABI5 and phosphorylates it in vitro (Lyzenga et al., 2013). Recently, another CDPK, CPK6, was shown to interact with ABFs1–4 and ABI5 in planta, and phosphorylation in vitro of ABI5 and ABF3 was demonstrated (Zhang et al., 2020). These studies have revealed a complex network of phosphorylations, with additional kinases likely awaiting discovery and characterization. A phospho‐mimic form of AREB1/ABF2 with five Ser to Asp substitutions trans‐activates ABRE‐containing genes in an ABA‐independent manner, suggesting that ABA‐dependent phosphorylation activates the transcriptional activation function (Furihata et al., 2006). Recombinant ABFs, which are presumed to be unphosphorylated, bind to ABREs in vitro (Uno et al., 2000), in yeast one‐hybrid assays (Choi et al., 2000) and are active in yeast (Kim et al., 2002), so the precise activating mechanism remains unclear. Current evidence suggests that phosphorylation status does not affect protein longevity: ABI5 with C1–4 serine/threonine residues substituted either for alanine or aspartate were equivalently degraded in vitro (Liu & Stone, 2014).
Other post‐translational modifications of ABFs/ABI5 have been linked to protein longevity. ABI5 is S‐nitrosylated in an NO‐dependent manner, and this modification negatively affects ABI5 accumulation (Albertos et al., 2015). The SUMO‐modification pathway, another protein modification system, also modulates ABA responses. A reduction in sumoylation components increases ABA sensitivity in seed germination and root growth, and ABI5 mono‐sumoylation in vivo is dependent on SIZ1, the major plant SUMO E3 ligase (Miura et al., 2009). The genetic relationship between siz1 and abi5 mutants suggests that SIZ1 suppresses ABA responses in an ABI5‐dependent manner.
There are some differences and unknowns in the proteolytic regulation of ABFs. Of the seven group A members, only DPBF2 retains the cysteine required for S‐nitrosylation, and whether ABFs are sumoylated is relatively uncharacterized. ABFs lack the SUMO consensus sequence found in ABI5 (using sumoplot), and some members lack a similarly located lysine SUMO attachment site identified in ABI5. However, ABF3 was positive in a global screen for SUMO targets enriched after heat stress, although the attachment site was not reported (Rytz et al., 2018). Another example of a difference in proteolytic regulation among group A proteins is the role of the C4 region. These sequences enhance degradation of ABI5 (Liu & Stone, 2013) but slow ABF1 and ABF3 degradation in in vitro assays (Chen et al., 2013). ABA‐dependent phosphorylation at a conserved serine/threonine in the C4 region has been proposed to play a role in stabilizing ABF3 by promoting ABF3 interaction with a 14‐3‐3 protein (Sirichandra et al., 2010). This phosphorylation site is conserved in ABI5 and also plays a role in ABI5 interaction with 14‐3‐3 proteins. In barley, HvABI5 interacts with several 14‐3‐3 proteins, and introducing a phospho‐null substitution at T350 in the C4 region in HvABI5 eliminated interaction with 14‐3‐3 proteins in a yeast two‐hybrid assay (Schoonheim et al., 2007). Arabidopsis ABI5 also interacts with a 14‐3‐3 protein in a yeast two‐hybrid assay (Jaspert et al., 2011). While the C4 phosphorylation site appears to play similar roles in 14‐3‐3 binding for ABI5 and ABF3, sequence conservation between ABI5 and the ABFs is low in this region and could account for the different role of the C4 in regulating ABI5 compared to the other ABFs.
To better characterize ABF proteolytic regulation and to understand the proteolytic differences between ABF and ABI5 subgroups, we characterized the degradation of an additional ABF family member, ABF2. Like ABF1 and ABF3, ABF2 degrades in seedlings treated with cycloheximide (CHX), is stabilized following treatment with the proteasome inhibitor MG132, rapidly accumulates in seedlings treated with ABA, and is an in vitro KEG substrate. ABF2 with a C4 deletion degrades more quickly than full‐length ABF2 in vitro, while ABI5 degradation in the same assays is slowed by C‐terminal deletions, showing that the C4 region of ABF2 contributes to its stability and confirming differences between ABI5 and ABFs. We also characterized the in vivo activity of a C4 deletion form of ABF1. Seeds from lines overexpressing full‐length ABF1 germinated more slowly than seeds from lines overexpressing ABF1 with a C4 deletion, suggesting that the C4 region is important for ABF1 activity. A survey of other ABF/ABI5 subgroup members suggests that their regulation by ABA and the proteasome is similar to that of previously characterized ABFs. Altogether, these results suggest that the members of the ABF share proteolytic regulatory mechanisms that are not completely shared with ABI5.
2. MATERIALS AND METHODS
2.1. Plant growth conditions, materials, and genotyping
A. thaliana seeds were surface sterilized in a solution of 25% bleach and 0.02% Triton‐X100 for ten minutes, rinsed with sterile H2O, and stratified at 4°C for at least 24 hr before plating. For agar‐grown seedlings, seeds were plated on solid growth media (GM) consisting of 4.3 g/L of Murashige and Skoog basal salt mixture (Sigma), 1% sucrose, 0.5 g/L of MES, 1X B vitamins (0.5 µg/ml of nicotinic acid, 1 ug/ml of thiamine, 0.5 µg/ml of pyridoxine, and 0.1 µg/ml of myo‐inositol, Sigma), and 8 g/L of Bacto Agar (Becton Dickinson), pH 5.7. After 2 weeks at room temperature under constant light, seedlings were transplanted from agar GM plates to soil, and plants were grown at 20°C with 50% humidity and 16‐hr light/8‐hr dark. For liquid‐grown seedlings, approximately 100 seeds were added to 1‐ml liquid GM distributed around the periphery of a 60‐mm petri dish (Falcon #353002) and grown at room temperature under constant light.
A. thaliana ecotype Col‐0 (CS70000) and keg insertion alleles were obtained from the Arabidopsis Biological Resource Center in Columbus, Ohio (https://abrc.osu.edu/). The keg‐1 (At5g13530) T‐DNA line SALK_049542 and the keg‐2 T‐DNA line SALK_018105 (previously characterized in Stone et al., 2006) were maintained as heterozygotes because homozygous individuals do not progress beyond the seedling stage. Homozygous keg seedlings were identified by their phenotypic differences from WT siblings (Stone et al., 2006). The keg genotype was verified by PCR using primers listed in Table S1. For keg‐1 lines, primer 5‐253 was used with primer 5‐254 to produce a WT gene‐specific product, and primer 5‐254 was used with the T‐DNA left border primer 9‐001 for a T‐DNA junction product. For keg‐2 lines, primer 9‐113 was used with primer 9‐114 for a WT gene‐specific product, and primer 9‐114 was used with the T‐DNA left border primer 9‐001 for a T‐DNA junction product.
To generate transgenic A. thaliana lines expressing epitope‐tagged proteins, the plant expression constructs described below were transformed into Agrobacterium tumefaciens strain AGL1. The floral dip method (Clough & Bent, 1998) was used to transform the expression construct into Col‐0 plants. Transformants that segregated 3:1 on GM with 50 µg/ml of kanamycin were propagated and homozygous T3 or T4 seedlings were used in experiments (except for Figure 1b,c and Figure S3, Figure S4 and Figure S5, where T2 segregating seed was used). Transgenic line information is in Table S4.
FIGURE 1.

In seedlings, ABF2 protein decreases following cycloheximide treatment and increases following MG132 or ABA treatment. Seven‐day‐old liquid grown seedlings from independent lines expressing 10xMyc‐ABF2 were treated with (a) 0.2 mg/ml of CHX dissolved in GM or plain liquid GM as a solvent control for six hours, (b) 50‐μM MG132 in DMSO or 0.5% DMSO as a solvent control for six hours, or (c) with 50‐μM ABA in EtOH or 5% EtOH as a solvent control for 6 hr. Equal amounts of total protein per sample were loaded for SDS‐PAGE, and anti‐Myc western blotting was used to visualize the 10xMyc‐ABF2 protein in each sample (upper panels). For a loading control, anti‐actin western blotting was performed on the same membranes (lower panels). To measure the change in ABF2 protein for each line, all bands were quantified in Image Studio Lite, ABF2 values were normalized to actin values, and treatment value was divided by control. The treatment/control ratio is reported below each pair of samples. (d) Expression of Myc‐ABF2 and RD29A mRNAs in 7‐day‐old seedlings treated with ABA for 6 hr, relative to mRNAs in mock‐treated seedlings. Quantitative PCR (qPCR) results are shown as mean ± SD. N = three biological replicates with three technical replicates each. Data were normalized to ACT2. RD29A serves as a positive control for ABA treatment. (e) Seven‐day‐old seedlings expressing 10xMyc‐ABF2 were pretreated for 6 hr with 50‐µM ABA or 0.1% EtOH as a solvent control, followed by the addition of GM or 0.2 mg/ml of cycloheximide (CHX) for 6 hr. Myc‐ABF2 proteins were immunoprecipitated from each sample, visualized by western blot (Figure S2), and normalized to the – CHX sample for each treatment. Asterisks (**) indicate a significant difference (p < .01) according to an ANOVA. ns = not significant. Each data point represents an independent transgenic line. The seedlings used in A are from the homozygous T3 generation, the seedlings used in B and C are from the segregating T2 generation, and the seedlings used in D and E are from the homozygous T4 generation
2.2. CHX, MG132, and ABA treatments
Seedlings were grown in sterile liquid GM in 60‐mm petri dishes, and after 6 days, the liquid GM was replaced to allow seedlings to adjust to fresh media overnight. The following day, for CHX treatments, the liquid GM was replaced with liquid GM containing 0.2 mg/ml of CHX (Sigma) or plain liquid GM as a mock control. For MG132 treatments, the liquid GM was replaced with liquid GM containing 50‐µM MG132 (Peptides International #IZL‐3175‐v, or Selleck Chemical #S2619‐5MG) or 0.5% DMSO as a solvent control. For ABA treatments, the liquid GM was replaced with liquid GM containing 50‐μM ABA (Sigma) or 0.1% EtOH as a solvent control. For the ABA‐CHX chase assays, after 6 hr of 50‐μM ABA‐pretreatment, 0.2 mg/ml of CHX was added, and seedlings were incubated for an additional 6 hr. For all experiments, after the indicated times, seedlings were flash frozen in liquid nitrogen and ground in fresh buffer (50‐mM Tris pH 8.1, 250‐mM NaCl, 0.5% NP‐40, 1‐mM PMSF, 50‐µM MG132, and 1 cOmplete Mini EDTA‐free protease inhibitor tablet [Roche #11836170001] per 10‐ml buffer). Lysate was collected after centrifugation at 13,250 x g for 10 min at 4°C, and total protein concentration was measured using a Bradford assay (Protein Assay Dye Reagent Concentrate, Bio‐Rad). For the CHX, MG132, and ABA‐treated samples, equal amounts of total protein per sample were separated by 10% SDS‐PAGE, and 10xMyc‐ABF2 protein was visualized with anti‐Myc western blotting. Anti‐actin western blotting or Ponceau S staining of the membrane (Figure S3, Figure S4 and Figure S5) was used for a loading control. For the ABA pre‐treatment CHX experiments, equal total protein was immunoprecipitated using anti‐Myc immunobeads (EZview Red Anti‐c‐Myc Affinity Gel, Sigma). After incubation for 1 hr at 4°C, beads were washed three times in buffer and resuspended in 2.5X Laemmli sample buffer. Samples were heated and the supernatant was separated by 10% SDS‐PAGE. 10xMyc‐ABF2 protein was visualized by anti‐Myc western blotting as described above.
2.3. Cell‐free degradation assays
Cell‐free degradation assays were performed based on those described in Wang et al., (2009). Approximately 100 7‐day‐old liquid grown seedlings (0.15‐g fresh weight) were flash frozen in liquid nitrogen and stored at −80°C until use. Each 0.15‐g tube of frozen seedlings was ground in 350‐µl fresh buffer (25‐mM Tris, 10‐mM ATP, 10‐mM MgCl2, 10‐mM NaCl, and 1‐mM DTT). Lysate (approximately 2.5 µg/µl of total protein) was collected after centrifugation at 13,250 x g for 10 min at 4°C. Three hundred‐microliters of lysate were incubated at 22°C in a thermocycler, and approximately 2 μg of recombinant protein purified from Escherichia coli was added to the lysate. For assays comparing degradation of multiple proteins, one lysate was divided into 300‐µl aliquots. For assays with MG132, 1.5 µl of 10‐mM MG132 in DMSO, or 1.5 µl of DMSO as a solvent control, was added to the lysate prior to the addition of recombinant protein. Samples were removed at indicated time points, mixed with Laemmli sample buffer, and heated to 98°C for 7 min. Proteins were separated with 10% SDS‐PAGE and anti‐FLAG, anti‐HA, anti‐Myc, or anti‐GST western blotting was used to visualize the recombinant protein in each sample.
2.4. Germination experiments
Age matched seeds were sterilized and plated on GM as described above, stratified for 3 days at 4°C, then incubated at 20°C in the light. At specific time points, the plates were scored for germination (radicle emergence) and returned to the growth chamber. Time courses for each line were repeated three times with ~50 seeds per experiment.
2.5. RNA extraction and qPCR
Seedlings were grown in liquid GM under constant light for 7 days and treated for 6 hr with 50‐µM ABA or 0.1% ethanol as a solvent control. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, 74904) according to manufacturer's instructions. Two micrograms of total RNA was used in each 10‐µl reverse transcription reaction performed with the SuperScript III First‐Strand Synthesis System for RT‐PCR (real‐time polymerase chain reaction) (Invitrogen, 18080‐051) according to manufacturer's instructions. Real‐time PCR amplification was performed with 20‐µl reactions containing 2 µl of first‐strand cDNA (diluted 1/10 with H2O), 10 pmoles of each primer, and 10‐µl PowerUp SYBR Green Master Mix (Applied Biosystems, A25742). Relative transcript levels were obtained using the comparative Ct method, normalized to ACT2. The experiment was performed independently three times with three technical replicates each time. Primer sequences are listed in Table S5.
2.6. Cloning and constructs
Primer sequences for plant expression cloning are listed in Table S2. For the plant expression construct for 10xMyc‐ABF2 under control of the 35S promoter (p9186), plasmid pVM491 (ABF2 cDNA in pENTR223, stock # G85579) was obtained from the Arabidopsis Biological Resource Center (ABRC) in Columbus, Ohio (https://abrc.osu.edu/). A Gateway (Invitrogen) LR reaction was used to recombine the ABF2 CDS into pGWB21 (pVM259) (Nakagawa et al., 2007) following manufacturer protocols. Similarly, cDNAs for DPBF2 (G20103, pVM493) and DPBF4 (G83893, pVM492) were obtained from the ABRC and recombined into pGWB21. For ABF4 and AREB3, RNA was isolated from Arabidopsis seedlings, and cDNAs were synthesized as described above, amplified with primers 9‐363 and 9‐364 (ABF4) or 9‐365 and 9‐366 (AREB3), cloned into pDONR201 with Gateway BP reactions, sequence verified, and recombined into pGWB21 with Gateway LR reactions. For expression of His6‐HA3‐ABF1 (p9204) and His6‐HA3‐ABF1‐ΔC4 (p9205) under control of the UBQ10 promoter, cDNAs synthesized as described above were amplified with primers 9‐438 and 9‐439 (ABF1) or 9‐438 and 9‐452 (ABF1‐ΔC4). The PCR products were digested with AseI and BamHI, then ligated into p3756, a modified pGreenII plasmid (Gilkerson et al., 2015), that had been cut with NdeI and BamHI. The line expressing GFP under control of the UBQ10 promoter (line number 13240) was described in (Dreher, 2006).
Primer sequences for bacterial expression cloning are listed in Table S3. For the bacterial expression construct for full‐length His6‐FLAG‐ABF2 (p9193), a Gateway LR reaction was used to recombine the ABF2 cDNA from G85579 into the bacterial expression vector pEAK2 (Kraft, 2007).
For the bacterial expression construct for full‐length His6‐HA3‐ABF2 (p9213), p9208 (ABF2 cDNA cloned into a modified pGreenII plasmid using primers 9‐440 and 9‐441) was digested with NdeI and BamHI. The ABF2 sequence was ligated into NdeI and BamHI sites in the bacterial expression vector p3832, a modified pET3c (Novagen) plasmid containing a 5′ His6‐HA3 cassette.
For the bacterial expression construct for His6‐HA3‐ABF2‐ΔC4 (p6881), p9193 (ABF2 cDNA in pEAK2, described above) was used as a template, and primers 9‐440 and 6‐1072 were used to amplify the ABF2 sequence, introduce a stop codon after base 1245 (relative to A of translation start ATG), and add NdeI and BamHI restriction sites to the 5′ and 3′ ends, respectively. The sequence was ligated into NdeI and BamHI sites in the bacterial expression vector p3832.
For the bacterial expression constructs for His6‐HA3‐ABI5 (p6943), ‐ABI51‐343 (p6944), and ‐ABI5‐ΔC4 (p6945), ABI5 cDNA in pDONR (plasmid p9017, derived from the ABRC clone PYAt2g36270 described in Stone et al., 2006) was used as a template and primer 6‐1107 was used with 6–1109 (ABI5), 6‐1108 (ABI51‐343), or 6‐1110 (ABI5‐ΔC4) to amplify the ABI5 sequence, introduce stop codons after base 1029 for ABI51‐343 or base 1287 for ‐ΔC4, and add NdeI and BamHI restriction sites to the 5′ and 3′ ends, respectively. The ABI5, ABI51‐343, and ABI5‐ΔC4 sequences were ligated into NdeI and BamHI sites in the bacterial expression vector p3832.
The Myc‐TGA1 (p9109) and Myc‐GBF3 (p9107) constructs contain TGA1 or GBF3 cDNAs in p7296, a pEXP1‐DEST expression vector (Thermo) modified with a Myc9 tag. RNA was isolated from Arabidopsis seedlings, and cDNAs were synthesized as described above, amplified with primers 9‐367 and 9‐368 (TGA1) or 9‐361 and 9‐362 (GBF3), recombined into pDONR with Gateway BP reactions, then recombined with Gateway LR reactions into p7296. Sequences were verified. The GST‐EGL3‐FLAG construct (pVM567) contains EGL3 in the pGEX‐4T‐1 vector. It was a gift from Ling Yuan at the University of Kentucky. The GST‐FUS3 construct (pVM505) was obtained from Sonia Gazarrini as described in Chen et al. (2013). The GST‐KEG‐RKA construct was described in Stone et al. (2006). The UBC10 E2 construct was described in Kraft et al. (2005).
2.7. Recombinant protein expression
E. coli transformed with expression constructs was grown in 10‐ml cultures overnight at 37°C, then added to flasks containing 500‐ml LB (10 g/L of tryptone, 5 g/L of yeast extract, 5 g/L of NaCl, 15 g/L of agar, pH 7.5). The 500‐ml cultures were grown at 37°C for approximately 3 hr until the OD600 was around 0.4–0.6, at which point the cultures were moved to room temperature and protein expression was induced with the addition of 0.5‐mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) for 3–4 hr. Cells were collected by centrifugation and pellets were stored at −80°C. E. coli strains and induction compounds were as follows. His6‐HA3‐ABF2‐ΔC4, His6‐HA3‐ABI5, ‐ABI51‐343, and ‐ABI5‐ΔC4, Myc‐GBF3, Myc‐TGA1, and GST‐EGL3‐FLAG were expressed in BL21(DE3)pLysS cells and expression was induced with IPTG. His6‐HA3‐ABF2 and His6‐FLAG‐ABF2 were expressed in Lemo21(DE3) cells and induced with IPTG. GST‐FUS3 was expressed as described in Chen et al. (2013).
2.8. Protein purification/enrichment
Cell pellets were thawed on ice and resuspended in lysis buffer (25‐mM Tris pH 7.5, 500‐mM NaCl, 0.01% Triton‐X100, 30‐mM imidazole, and 1 cOmplete Mini EDTA‐free protease inhibitor tablet per 50‐ml buffer). Cells were lysed by sonication and supernatant was reserved after centrifugation. His‐ or GST‐tagged proteins were captured from the lysate by the addition of 100‐µl Ni Sepharose High Performance beads (GE Healthcare) or Glutathione Sepharose High Performance beads (GE Healthcare). Myc‐tagged proteins were captured by addition of 75‐μl EZview Red Anti‐c‐Myc Affinity Gel beads (Sigma). Beads were collected with centrifugation and rinsed at least three times with wash buffer (25‐mM Tris pH 7.5, 300‐mM NaCl, and 0.01% Triton‐X100). Proteins were eluted from the beads by addition of 300‐µl elution buffer (25‐mM Tris, 150‐mM NaCl, 0.01% Triton‐X100, and 300‐mM imidazole for His‐tagged proteins or 20‐mM reduced glutathione for GST‐tagged proteins) and shaking at 4°C for 30 min. Myc‐tagged proteins were eluted with 100 ug/ml of c‐Myc peptide in 250‐μl RIPA buffer. Supernatant was collected after centrifugation and mixed with glycerol for a final concentration of 20% glycerol, and frozen at −80°C.
2.9. In vitro ubiquitination assays
Substrate was incubated with E1, E2, E3, and ubiquitin in buffer (50‐mM Tris‐HCl pH 7.5, 10‐mM MgCl2, 1‐mM ATP, 200‐μM DTT, and 2.1 mg/ml of phosphocreatine) for 2 hr at 30°C. Each 30‐μl reaction included approximately 50 ng of yeast E1 (Boston Biochem), 250 ng of E2 (His‐AtUBC10, Kraft et al., 2005), 5–10 μl of bead‐bound E3 or 2–5 μl of soluble KEG E3, GST‐KEG‐RKA (Stone et al., 2006), 4‐μg ubiquitin (Sigma), and 2‐μl substrate (His6‐HA3‐ABF2). To stop the reaction, 10 μl of 5X LSB was added, and samples were boiled for 10 min.
2.10. SDS‐PAGE and western blotting
Proteins were separated on 10% polyacrylamide gels and transferred onto PVDF‐P membranes (Immobilon). Membranes were blocked in 5% nonfat powdered milk (Carnation) dissolved in TBS + 0.1% Tween 20 (Sigma). Proteins were detected with the following antibodies. FLAG‐tagged proteins were detected with Monoclonal ANTI‐FLAG M2‐Peroxidase (Sigma #A8592) at a dilution of 1:5,000. HA‐tagged proteins were detected with Anti‐HA‐Peroxidase (Roche #12013819001) at a dilution of 1:5,000. Myc‐tagged proteins were detected with Anti‐c‐myc‐Peroxidase (Roche #11814150001) at dilution of 1:5,000. GST‐tagged proteins were detected with rabbit polyclonal GST (Z‐5): sc‐459 (Santa Cruz Biotechnology) at a dilution of 1:5,000, followed by the secondary antibody peroxidase‐conjugated AffiniPure Goat Anti‐Rabbit IgG (H + L) (Jackson ImmunoResearch) at a dilution of 1:10,000. Actin was detected with Monoclonal Anti‐Actin (plant) antibody produced in mouse (Sigma #A0480‐200UL) at a dilution of 1:5,000, followed by peroxidase‐labeled Antibody to Mouse IgG (H + L) produced in goat (KPL #074‐1806) at a dilution of 1:10,000. SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific #34078) or ProSignal Dura (Prometheus #20‐301) was used as chemiluminescent substrates. Chemiluminescence was captured on X‐ray film (Phenix) or digitally imaged in the linear range of detection using the ImageQuant LAS400 imaging system (GE). Images were quantified using ImageStudio Lite. For quantitation of in vivo degradation or accumulation, ABF values were normalized to actin, and treatment values were divided by control values. Curve fitting, half‐life determinations, and t tests were performed using Prism (GraphPad) as described in each figure legend. In GraphPad, one‐phase decay is the term for first order decay.
3. RESULTS
3.1. ABF2 degrades in seedlings and increases following MG132 treatment
To examine the proteolytic regulation of ABF2 in plants, independent transgenic lines expressing Myc‐ABF2 under control of the 35S promoter were generated. Seven‐day‐old seedlings from three independent lines were treated with the protein synthesis inhibitor CHX. Once translation is inhibited by CHX, no new ABF2 protein is synthesized in the seedlings, and degradation of the existing ABF2 protein can be visualized by monitoring protein levels over time. In all three lines, Myc‐ABF2 protein was reduced over time following CHX treatment, indicating that Myc‐ABF2 is unstable in vivo (Figure 1a and Figure S1a–c). Proteasomal substrates typically accumulate after proteasome activity is inhibited by the cell‐permeable substrate analog MG132. Myc‐ABF2 protein increased when intact seedlings were treated with MG132 (Figure 1b and Figure S1d), indicating that Myc‐ABF2 degradation requires the proteasome.
3.1.1. ABF2 increases in seedlings following ABA treatment
Studies have shown that levels of constitutively expressed ABI5, ABF1, and ABF3 proteins increase in plants in response to exogenously supplied ABA (Chen et al., 2013; Lopez‐Molina et al., 2001). To test whether ABF2 behaves similarly, seedlings from four independent transgenic lines expressing Myc‐ABF2 under control of the 35S promoter were treated with ABA (in both the segregating T2 and homozygous T4 generations). Myc‐ABF2 protein increased in all four lines following ABA treatment (Figure 1c and Figure S1e,f). To support the hypothesis that this accumulation results from protein stabilization and not an increase in synthesis, the mRNA levels of the Myc‐ABF2 transgene were measured after 6 hr of ABA or after six hours of 0.1% EtOH as a solvent control. While mRNA for the positive control RD29A (Yamaguchi‐Shinozaki & Shinozaki, 1993) increased more than 100‐fold after ABA treatment, Myc‐ABF2 mRNAs did not increase as much as Myc‐ABF2 protein levels (Figure 1d). To further explore proteolytic control, seedlings were pre‐treated with ABA or solvent alone treated (mock), then Myc‐ABF2 degradation measured in a CHX‐chase assay. Myc‐ABF2 was more stable after ABA pre‐treatment (Figure 1e and Figure S2).
3.1.2. Survey of other group A bZIP proteins reveals regulation by MG132 and ABA
To investigate the proteolytic stability of other group A members, analogous epitope‐tagged ABF4, DPBF2, DPBF4, and AREB3 proteins were expressed from the same vector system described above, and three independent transgenic lines overexpressing each protein were tested for effects of MG132 and ABA on protein accumulation (Figure S3, Figure S4and Figure S5). ABF4, DPBF2, and DPBF4 accumulated in the presence of MG132 (AREB3 lines were not tested), and all four (ABF4, DPBF2, DPBF4, and AREB3) accumulated in the presence of ABA.
3.1.3. ABF2 degrades rapidly in vitro and is stabilized by MG132
To test whether ABF2 protein is unstable in vitro, we used a cell‐free degradation assay (as described in Wang et al., 2009) in which recombinant ABF2 protein expressed in and purified from E. coli was added to a lysate from WT Arabidopsis seedlings. Samples were removed from the reaction at various time points, and Western blotting was used to visualize the amount of tagged ABF2 protein remaining at each time point. We tested ABF2 with two different epitope tags (HA and FLAG) to show that ABF2 protein degradation in vitro does not depend on the nature of the tag. Both His6‐HA3‐ABF2 and His6‐FLAG‐ABF2 degraded quickly in the cell‐free degradation assay and were stabilized by the addition of MG132 (Figure 2). One‐phase decay curves predicted similar half‐lives of 2.3 and 2.8 min for His6‐HA3‐ABF2 and His6‐FLAG‐ABF2, respectively (Figure 2c,d).
FIGURE 2.
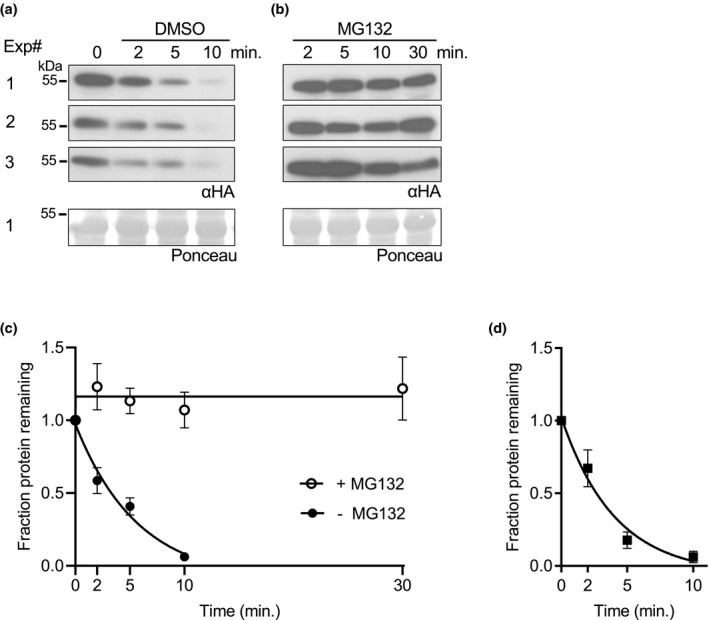
ABF2 degrades in vitro and is stabilized by MG132. In a cell‐free degradation assay, recombinant full‐length (a–c) His6‐HA3‐ABF2 or (d) His6‐FLAG‐ABF2 was added to lysate from Col seedlings, and the reaction was split into separate tubes with equal volumes of either (a) DMSO or (b) 200‐μM MG132 dissolved in DMSO and incubated at 22°C. Samples were removed at the indicated times, mixed with Laemmli sample buffer and heated to 98°C to stop the reaction. Three independent experiments were performed. (a, b) SDS‐PAGE followed by anti‐HA western blotting was used to visualize the ABF2 protein in each sample (upper panels). Ponceau staining was used to demonstrate equal loading (lower panels). Ponceaus for Experiment #1 are shown. (c) Western blots were quantified with Image Studio Lite and the fraction of protein remaining at each time point was graphed. For each reaction, GraphPad Prism nonlinear regression was used to fit a curve modeling one‐phase decay of ABF2. R 2 is .93. Bars are SE. N = 3 independent experiments. (d) Graph of identical time course with recombinant His6‐FLAG‐ABF2 (western blots not shown). R 2 is .91. Bars are SE. N = 3 independent experiments
3.1.4. ABF2 is ubiquitinated by KEG in vitro and KEG affects ABF2 degradation
The E3 ligase KEG ubiquitinates ABI5, ABF1, and ABF3 in vitro and promotes their degradation in vivo (Chen et al., 2013; Liu & Stone, 2013; Stone et al., 2006). Degradation of recombinant ABF1 and ABF3 is slowed in keg lysates (Chen et al., 2013), and endogenous ABI5 hyperaccumulates in keg seedlings (Stone et al., 2006). We used an in vitro ubiquitination assay to test whether ABF2 is also a potential in vivo KEG substrate. A C‐terminally truncated form of KEG, in fusion with GST (glutathione‐S‐transferase), called GST‐KEG‐RKA, was used in these assays. GST‐KEG‐RKA expresses higher quality and quantity of soluble protein in E. coli than the full‐length version facilitating these assays. KEG‐RKA (AAs 1‐829) contains the RING domain (required for E3 activity), a kinase domain and ankyrin repeat region, only lacking the C‐terminal HERC repeats (Stone et al., 2006). GST‐KEG‐RKA is an appropriate version to test in these assays because the KEG‐RKA truncation interacts with ABI5 in yeast‐2‐hybrid assays (Liu & Stone, 2013) and a smaller version of KEG, GST‐KEG‐RK, catalyzes ABI5 ubiquitination in vitro (Liu & Stone, 2010).
When recombinant His6‐HA3‐ABF2 was incubated with GST‐KEG‐RKA and other proteins required for ubiquitination (E1, E2, and free ubiquitin), higher migrating forms of His6‐HA3‐ABF2 were observed only in the complete reaction (Figure 3), indicating covalent ubiquitin attachment to His6‐HA3‐ABF2.
FIGURE 3.
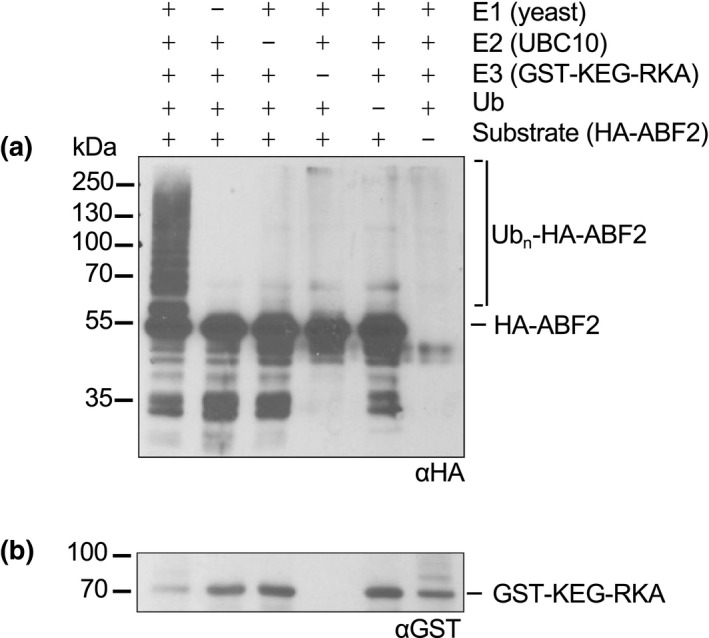
GST‐KEG‐RKA ubiquitinates ABF2 in vitro. In an in vitro ubiquitination assay, recombinant His6‐HA3‐ABF2 was incubated with E1 (yeast recombinant), E2 (recombinant AtUBC10), the E3 GST‐KEG‐RK, and ubiquitin (Ub). After 1 hr, reactions were separated by SDS‐PAGE. (a) His6‐HA3‐ABF2 was visualized with anti‐HA western blotting. Components in the reaction are indicated with a+. (b) GST‐KEG‐RKA was visualized with anti‐GST western blotting. Brackets indicate slower migrating forms of HA‐ABF2 present only in the complete reaction, indicating ubiquitination
To test whether KEG is important for ABF2 degradation, we used cell‐free degradation assays to compare the degradation of bacterially expressed ABF2 protein in lysates from WT seedlings to degradation in lysates from two different keg T‐DNA alleles (keg‐1 and keg‐2). keg‐1 contains a T‐DNA insertion in the second exon and produces little or no KEG transcript, while keg‐2 contains a T‐DNA insertion in the third intron and produces low levels of KEG transcript (Stone et al., 2006). Both lines exhibit similar seedling phenotypic differences from WT (Stone et al., 2006). His6‐HA3‐ABF2 protein degraded in lysates from WT Col seedlings but was stable in lysates from both keg‐1 and keg‐2 seedlings (Figure 4 and Figure S6, respectively) in parallel experiments with the same batch of recombinant protein. Less than 25% of His6‐HA3‐ABF2 protein remained after ten minutes in Col lysate, but there was no decrease in His6‐HA3‐ABF2 protein after ten minutes in either keg lysate.
FIGURE 4.
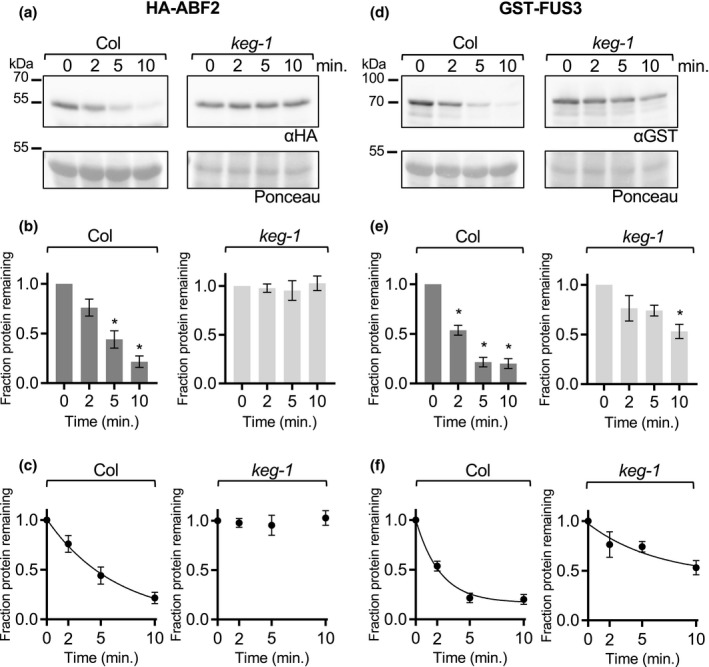
ABF2 protein is stabilized in keg‐1 lysate in vitro. In a cell‐free degradation assay, recombinant His6‐HA3‐ABF2 or GST‐FUS3 protein was added to lysate from Col or keg‐1 seedlings and the reaction was incubated at 22°C. Aliquots were removed at the indicated times, mixed with Laemmli sample buffer, and heated to 98°C to stop the reaction. (a and d) SDS‐PAGE followed by anti‐HA or anti‐GST western blotting was used to visualize the His6‐HA3‐ABF2 or GST‐FUS3 protein in each sample, respectively. Ponceau staining (below) was used to demonstrate equal loading. (b and e) Western blots were quantified with Image Studio Lite. Protein at time zero was normalized to 1 and the fraction of protein remaining at each time point was graphed. Asterisk (*) indicates the value differs significantly (p < .05) from time point zero based on ANOVA (GraphPad Prism). (c and f) For each reaction, GraphPad Prism nonlinear regression was used to fit a curve modeling one‐phase decay of His6‐HA3‐ABF2 or GST‐FUS3. The keg‐1 line in (c) is not a one‐phase decay curve because the data did not fit a decay curve. Bars are SE. N = 3 independent experiments (a and d show one representative experiment)
For the cell‐free degradation assays described above, Col lysates were prepared from 7‐day‐old seedlings. To obtain developmentally comparable keg lysates, keg seedlings were grown for 2 weeks before harvesting but were still smaller and paler than Col seedlings and rarely had true leaves. To demonstrate that the observed stability of ABF2 in keg lysates is not a property of the delayed development of keg seedlings and to further explore the substrate specificity of KEG, we tested whether keg‐1 lysates are capable of degrading other proteins. The cell‐free degradation assay was used to compare the degradation of four additional transcription factors in both Col and keg‐1 lysates: the B3 transcription factor FUCSA3 (FUS3), the bHLH transcription factor ENHANCER OF GLABROUS 3 (EGL3), the bZIP transcription factor TGACG sequence‐specific binding protein 1 (TGA1, in bZIP group D) and the bZIP transcription factor G‐box binding factor 3 (GBF3, in bZIP group G) (Figures 4, 5, 6). FUS3 was selected because it is reported to be a proteasomal substrate that degrades rapidly in cell‐free seedling degradation assays (Lu et al., 2010) and was previously used in our lab as a control for cell‐free degradation assays with ABF1 and ABF3 (Chen et al., 2013). EGL3 is also reported to be a proteasomal substrate that degrades in seedling lysates (Patra et al., 2013). TGA1 and GBF3 were selected because they are bZIP transcription factors but are in different groups than the ABFs and affect different physiological processes, suggesting that their proteolytic regulation might be different (Dröge‐Laser et al., 2018).
FIGURE 5.
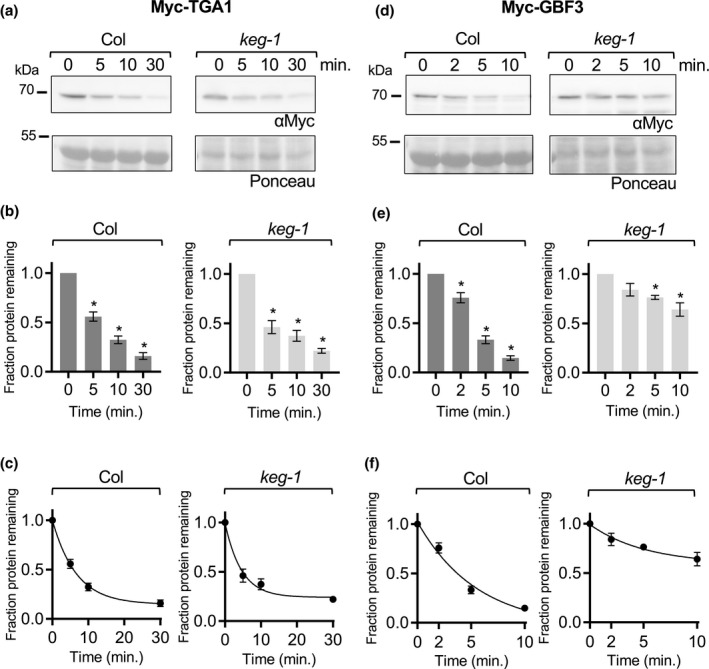
The bZIP proteins TGA1 and GBF3 degrade in keg‐1 lysate in vitro. In a cell‐free degradation assay, recombinant Myc‐TGA1 or Myc‐GBF3 protein was added to lysate from Col or keg‐1 seedlings, and the reaction was incubated at 22°C. Aliquots were removed at the indicated times, mixed with Laemmli sample buffer, and heated to 98°C to stop the reaction. (a and d) SDS‐PAGE followed by anti‐Myc western blotting was used to visualize the Myc‐TGA1 or Myc‐GBF3 protein in each sample. Ponceau staining was used to demonstrate equal loading. (b and e) Western blots were quantified with Image Studio Lite. Protein at time zero was normalized to 1 and the fraction of protein remaining at each time point was graphed. Asterisk (*) indicates the value differs significantly (p < .05) from time point zero based on ANOVA. (c and f) For each reaction, GraphPad Prism nonlinear regression was used to fit a curve modeling one‐phase decay of Myc‐TGA1 or Myc‐GBF3. Bars are SE. N = 3 independent experiments (a and d show one representative experiment)
FIGURE 6.
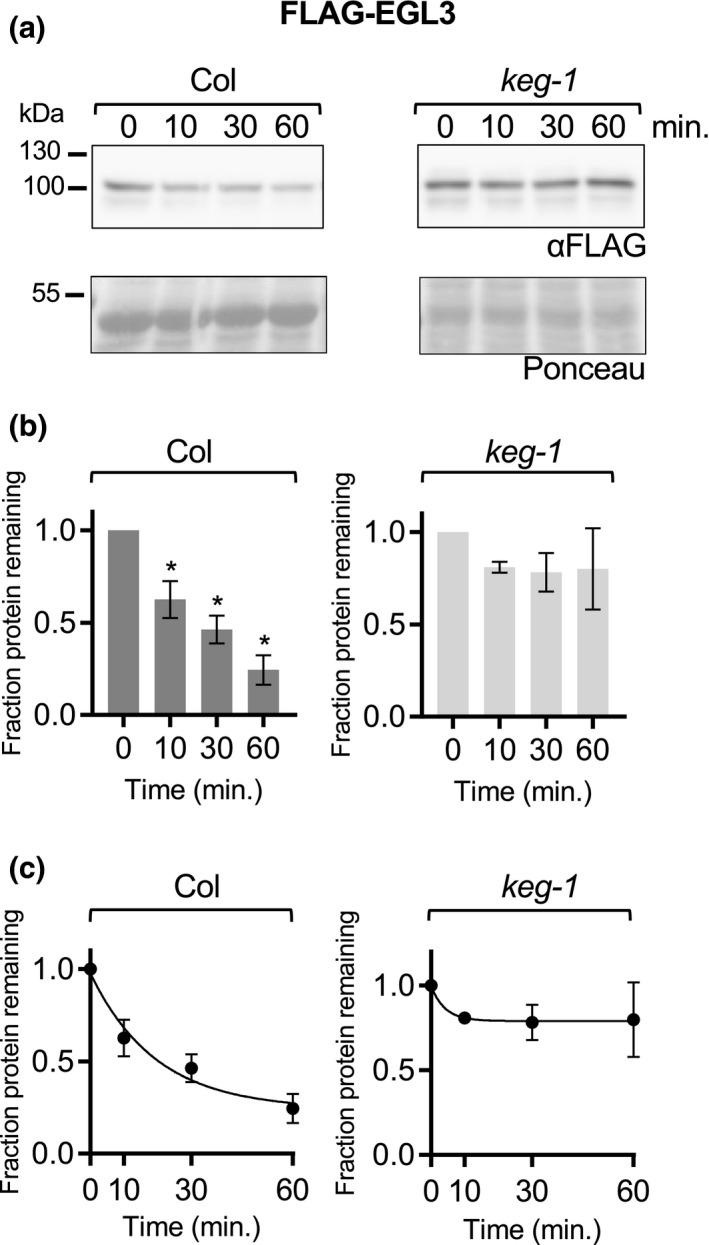
The bHLH protein EGL3 does not degrade significantly in keg‐1 lysate in vitro. In a cell‐free degradation assay, recombinant FLAG‐EGL3 protein was added to lysate from Col or keg‐1 seedlings, and the reaction was incubated at 22°C. Aliquots were removed at the indicated times, mixed with Laemmli sample buffer, and heated to 98°C to stop the reaction. (a) SDS‐PAGE followed by anti‐FLAG western blotting was used to visualize the FLAG‐EGL3 protein. Ponceau staining was used to demonstrate equal loading. (b) Western blots were quantified with Image Studio Lite. Protein at time zero was normalized to 1 and the fraction of protein remaining at each time point was graphed. Asterisk (*) indicates the value differs significantly (p < .05) from time point zero based on ANOVA. (c) For each reaction, GraphPad Prism nonlinear regression was used to fit a curve modeling one‐phase decay of FLAG‐EGL3. Bars are SE. N = 3 independent experiments (A shows one representative experiment)
For all proteins with observed degradation, the data fit a pseudo‐first order decay curve, as expected (graphs in C, Figures 4, 5, 6 and Figure S6). TGA1 degraded at similar rates in Col and keg‐1 lysates (Figure 5a–c). GBF3 and FUS3 also degraded in keg‐1 lysates, although their degradation was slower in keg‐1 lysates than in Col lysates (Figures 4, 5d–f, and 6d–f, respectively). Like ABF2, EGL3 was stabilized in keg‐1 lysates (Figure 6). Although the degradation of some proteins was slowed in keg‐1 lysates, these results show that protein degradation machinery is still functional in keg‐1 seedling lysates and that KEG is important for ABF2 degradation in vitro.
3.1.5. The C4 region affects ABF2 stability in vitro
It was previously reported that a truncated ABI5 protein (ABI51‐343) lacking 99 C‐terminal amino acids including the C4 region is more stable in vitro than full‐length ABI5 (Liu & Stone, 2013). To confirm that our cell‐free degradation assay setup could replicate this result and to directly compare truncated ABFs to comparably truncated ABI5 proteins, we cloned and recombinantly expressed ABI51‐343 and ABI5‐ΔC4, an ABI5 protein with a smaller deletion of just the C4 region (the 13 amino acids at the C‐terminus). ABI5‐ΔC4 is comparable to the ABF1 and ABF3 C4 deletions previously used in cell‐free degradation assays (Chen et al., 2013). In cell‐free degradation assays, His6‐HA3‐ABI51‐343 degraded more slowly than full‐length His6‐HA3‐ABI5, as previously reported (Liu & Stone, 2013) (Figure 7). There was significantly more ABI51‐343 protein than full‐length ABI5 protein at the 2‐, 5‐, and 10‐min time points, and the predicted half‐life of ABI51‐343 at 2.6 min was twice as long as the predicted half‐life of full‐length ABI5 at 1.2 min. His6‐HA3‐ABI5‐ΔC4 also degraded more slowly than full‐length His6‐HA3‐ABI5, but more quickly than His6‐HA3‐ABI51‐343 (Figure 7). The predicted half‐life of His6‐HA3‐ABI5‐ΔC4 was intermediate at 1.8 min, and there was significantly more ABI5‐ΔC4 protein than full‐length ABI5 protein at the 5‐ and 10‐min time points.
FIGURE 7.
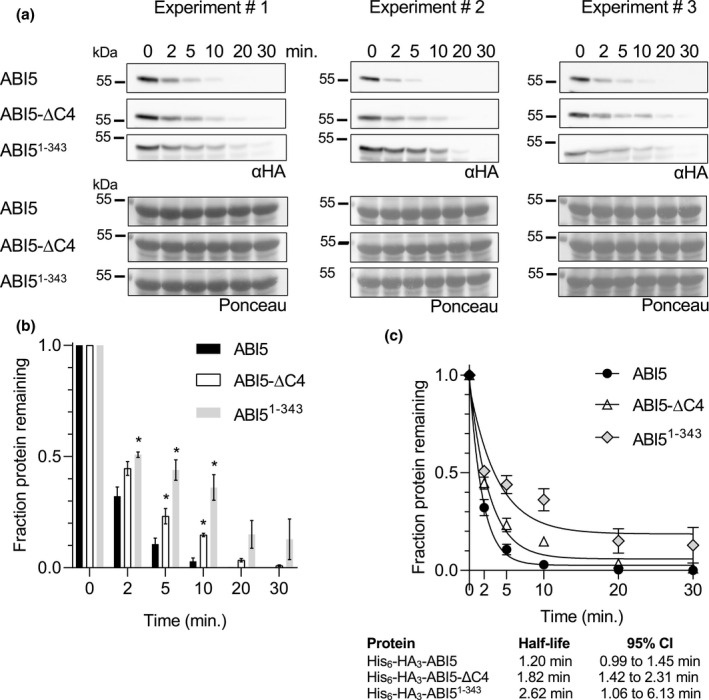
ABI51‐343 and ABI5‐ΔC4 degrade more slowly than full‐length ABI5 in vitro. In a cell‐free degradation assay, recombinant His6‐HA3‐ABI5, ‐ABI5‐ΔC4, or ‐ABI51‐343 protein was added to lysate from Col seedlings and the reactions were incubated at 22°C. Aliquots were removed at the indicated times, mixed with Laemmli sample buffer, and heated to 98°C to stop the reaction. (a) SDS‐PAGE followed by anti‐HA western blotting was used to visualize the HA‐tagged protein in each sample. Ponceau staining (lower panels) was used to demonstrate equal loading. (b) Western blots were quantified with Image Studio Lite. Protein at time zero was normalized to 1 and the fraction of protein remaining at each time point was graphed. Asterisk (*) indicates significant difference (p < .05) from fraction of full‐length ABI5 protein remaining at the same time point based on Student's t test. Significance not tested for 20‐ and 30‐min time points. (c) For each reaction, GraphPad Prism nonlinear regression was used to fit curves modeling one‐phase decay of each protein. R 2 values for His6‐HA3‐ABI5, ‐ABI5‐ΔC4, and ‐ABI51‐343 are 0.99, 0.98, and 0.86, respectively. Bars are SE. N = 3 independent experiments
In contrast to ABI5, ABF1 and ABF3 proteins are less stable in vitro when their C4 regions are deleted (Chen et al., 2013) (see Figure 8a for an alignment of the conserved regions in ABF proteins). To test whether ABF2 behaves similarly to ABI5 or ABF1 and ABF3, we cloned and recombinantly expressed a form of ABF2 lacking 12 amino acids at the C‐terminus (ABF2‐ΔC4). His6‐HA3‐ABF2‐ΔC4 protein degraded more quickly than full length ABF2 in a cell‐free degradation assay (Figure 8). The predicted half‐life of full‐length ABF2 at 3.8 min was longer than the predicted half‐life of ABF2‐ΔC4 at 2.1 min. Like ABF1 and ABF3, but in contrast to ABI5, ABF2 is stabilized by the C4 region.
FIGURE 8.
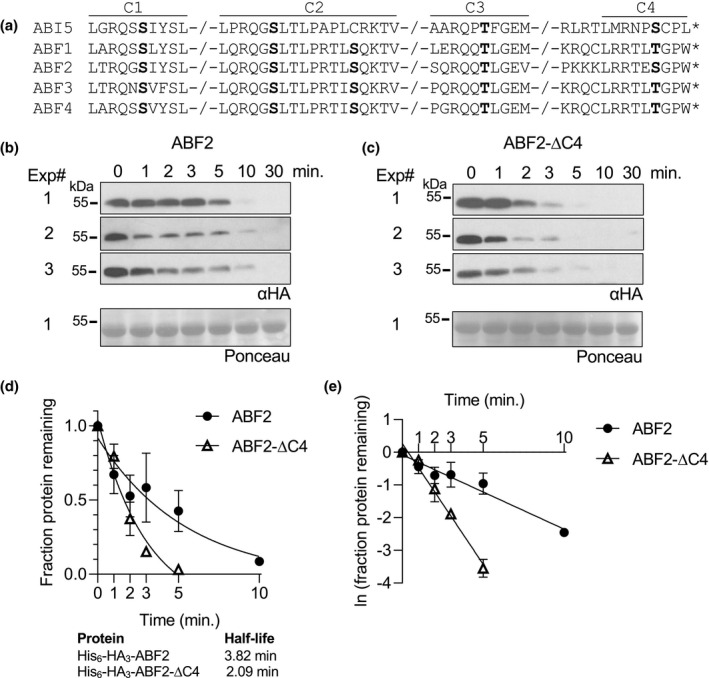
ABF2 lacking the C4 region degrades faster than full‐length ABF2 in vitro. (a) Alignment of the conserved regions of ABF proteins. Phosphorylation sites are in bold. In a cell‐free degradation assay, recombinant His6‐HA3‐ABF2 (b) or His6‐HA3‐ABF2‐∆C4 (c) was added to lysate from Col seedlings and the reaction was incubated at 22°C. Aliquots were removed at the indicated times, mixed with Laemmli sample buffer, and heated to 98°C to stop the reaction. Three independent experiments were performed. (b and c) SDS‐PAGE followed by anti‐HA western blotting was used to visualize the HA‐tagged protein in each sample. Ponceau staining was used to demonstrate equal loading (Ponceau for Experiment #1 is shown). (d) Western blots were quantified with Image Studio Lite. Protein at time zero was normalized to 1 and the fraction of protein remaining at each time point was graphed. GraphPad Prism nonlinear regression was used to fit a curve modeling one‐phase decay of each protein. Bars are SE. N = 3 independent experiments. (e) To compare slopes, a linear regression fit was performed in GraphPad Prism. Lines have significantly different slopes (p < .01)
3.1.6. Overexpression of full‐length ABF1 or ABF1‐ΔC4 causes delayed germination
Because overexpression of untagged ABF2 or ABF3 in Arabidopsis leads to delayed germination (Kang et al., 2002; Kim et al., 2004), we used this assay to determine whether the C4 region affects ABF function in vivo. We initially generated lines containing ABF2 FL and ABF2 delta C4 expressing transgenes, but rapid silencing in these lines prevented analyses. We were able to generate lines stably expressing versions of another ABF, ABF1, either full length (FL) His6‐HA3‐ABF1 or His6‐HA3‐ABF1‐ΔC4 (Figure 9a) and evaluated the germination of independent homozygous transgenic lines over time compared to age‐matched Col‐0 control. We observed that seeds from all three FL ABF1 OE lines germinated more slowly than Col seeds on the same GM plates (Figure 9b). After 32 hr, three lines overexpressing FL ABF1 reached 23.7%, 21.7%, and 0.6% germination, while Col (WT, age‐matched seeds) controls grown on the same plates reached 82.3%, 98%, and 94.8% germination, respectively. As a transformation control, germination of a transgenic line expressing GFP under the same UBQ10 promoter in the same plasmid backbone was analyzed, and its germination did not differ from that of untransformed Col (Figure S7).
FIGURE 9.
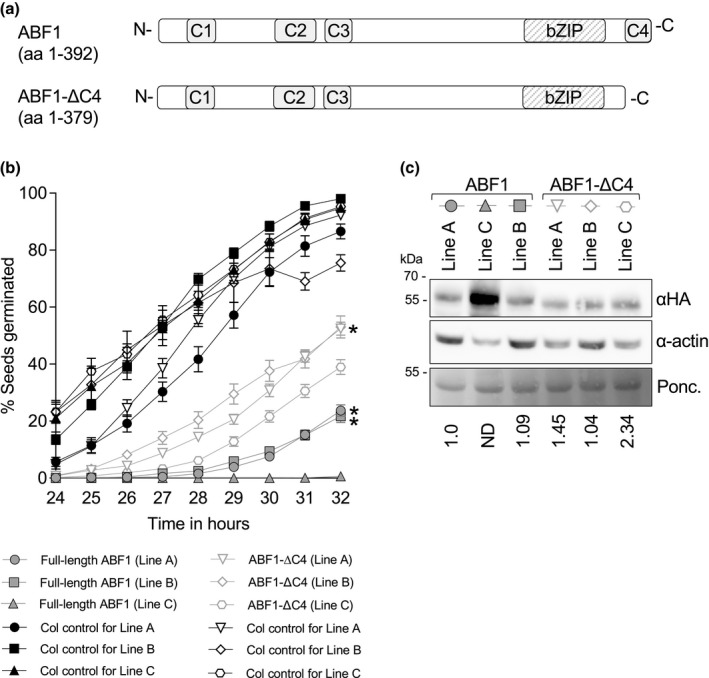
Overexpression of full‐length ABF1 or ABF1‐ΔC4 delays germination. (a) Diagram of full‐length ABF1 (upper) and ABF1‐ΔC4 (lower) proteins. (b) Seeds from plants overexpressing full‐length ABF1 or ABF1‐ΔC4 (or Col as a WT control on the same plate) were plated on GM, stratified at 4°C for 3 days, then incubated at 20°C under constant light. After plates were moved to 20°C, germination was scored hourly from 24 to 32 hr. Lines with similar protein expression levels are marked with an (*) (see Panel c). N = at least 7 independent experiments, with 50 seeds per genotype per experiment. Bars are SE. (c) Comparison of HA‐ABF1 protein level in each line. HA‐tagged proteins were visualized with anti‐HA western blotting (upper panel), and anti‐actin western blotting (middle panel) was used as a loading control. Ponceau staining (lower panel) is an additional loading control. The same membrane was used for the anti‐HA blot, anti‐actin blot, and Ponceau stain. HA bands were normalized to actin bands, then compared to the lowest expressing full length line (ABF1 line A). ND = not determined (band not in linear range)
To test whether the C4 region affects ABF1's ability to delay germination, we studied the germination of the ABF1‐ΔC4 expressing lines. Germination of seeds from all three ABF1‐ΔC4 lines was also slower than germination of the control Col seeds (Figure 9c, light gray lines and symbols), with 52.4%, 53%, and 39% germination at 32 hr, compared to 92.3%, 75.5%, and 95.3% for the Col controls. These results show that overexpression of the ABF1‐ΔC4 protein affects germination. To more directly assess in vivo activity, the ABF1 protein levels between lines were directly compared using the HA tag (Figure 9c). There seems to be a correlation between ABF1 protein and germination timing, with the higher expressing ABF1 FL line (Line C) exhibiting slower germination compared to ABF1 FL lines A and B. Similarly, among the ABF1‐ΔC4 lines, the line with twofold more expression (Line C) germinated more slowly than two other ABF1‐ΔC4 lines (A and B). When comparing lines with equivalent expression of ABF1 FL versus ABF1‐ΔC4, the ABF1‐ΔC4 lines (Line A and Line B) germinated faster than the FL lines (Line A and Line B), suggesting that the ΔC4 version is less effective in slowing germination than the FL protein.
4. DISCUSSION
ABF2 shares some aspects of proteolytic regulation with the other two previously analyzed ABF proteins in bZIP group A. These include instability both in vitro and in plants, accumulation regulated by the proteasome and ABA in vivo, in vitro ubiquitination by the E3 ligase KEG, KEG's importance for their cell‐free degradation, and the stabilizing effect of the C4 region in vitro. While most of these properties are consistent with proteolytic regulation of group A member ABI5, there are differences between the ABF proteins and ABI5. The C4 region destabilizes ABI5 in vitro but stabilizes ABF1, ABF2, and ABF3. ABF1‐ΔC4, ABF2‐ΔC4, and ABF3‐ΔC4 proteins degrade faster than full‐length proteins, while ABI5‐ΔC4 degrades more slowly in parallel experiments.
There are other aspects of ABI5 proteolytic regulation that are likely to differ. ABI5 stability is regulated by S‐nitrosylation, with modification of C‐153 conferring enhanced degradation in a CUL4‐ and KEG‐dependent manner (Albertos et al., 2015). ABFs1–4 lack a corresponding cysteine residue at this position, suggesting that they are not regulated by NO in an analogous manner. In addition, a lysine residue required for ABI5 degradation is not strictly conserved among ABF proteins (Liu & Stone, 2013). Further studies are needed to dissect the differences and similarities between ABF1–4 and ABI5 proteolytic regulation.
The differences in the rate of ABF degradation between in vivo and in vitro experiments are striking, with much slower degradation observed in vivo. While the reasons for these differences are not known, loss of in vivo compartmentalization when lysates are prepared or loss/absence of post‐translational modifications in the recombinant proteins that affect proteolysis are possible, non‐exclusive, hypotheses. It is known that KEG is cytoplasmic/trans‐Golgi/early endosome‐localized (Gu & Innes, 2011) and ABI5‐fluorescent fusions are largely nuclear localized (Lopez‐Molina et al., 2003). Because lysates lack separated compartments, they could expose added substrate to higher concentrations of E3 than found in vivo. Additionally, bacterially expressed substrates lack any eukaryotic post‐translational modifications, some of which might have regulatory roles in proteolysis in vivo.
Phosphorylation at a conserved site in the ABF3 C4 region (T451) is hypothesized to contribute to ABF3 stability by promoting ABF3 interaction with a 14‐3‐3 protein (Sirichandra et al., 2010). Whether C4 phosphorylation is important for other ABFs' interactions with a 14‐3‐3 protein has not been published. The C‐terminal amino acids of ABF1, 3, and 4 (LRRTLTGPW) and ABF2 (LRRTESGPW) contain both mode I (Rxx[S/T]xP) and mode II (Rxxx[S/T]xP) consensus 14‐3‐3 binding sites, while ABI5 (LMRNPSCPL), contains only a mode I site. However, in barley, HvABI5 interacts with several 14‐3‐3 proteins, and introducing a phospho‐null substitution at T350 in HvABI5 eliminated interaction with 14‐3‐3 proteins in a yeast two‐hybrid assay (Schoonheim et al., 2007), indicating that phosphorylation at the C4 site promotes 14‐3‐3 interaction with ABI5 as described for ABF3. Arabidopsis ABI5 also interacted with a 14‐3‐3 protein in a yeast two‐hybrid assay (Jaspert et al., 2011). Because the C4 phosphorylation site appears to play similar role in 14‐3‐3 binding to ABI5 and ABF3, there might be other factors in the C4 region that account for the different role of the C4 in regulating ABI5 protein compared to the other ABFs.
While we demonstrated that deletion of the C4 region has a small effect on proteolysis in vitro, we wanted to affirm that this region affects ABF function in plants to justify further studies. The lack of data on the in vivo role of ABF C4 prompted us to test whether its removal affected ABF biological activity. A delay in germination in plants overexpressing multiple ABF proteins has been observed (Kang et al., 2002; Kim et al., 2004) and is a sensitive and simple assay for function. Our in vivo germination assays suggest that the C4 region does affect ABF specific activity, indicating that further study of the role of these nine AAs will add important information on either ABF activity or longevity, or both.
In the cell‐free degradation assay, we explored the ability of keg extracts to degrade other types of transcription factors and tested extracts from two different keg LOF alleles. Both keg‐1 and keg‐2 lysates exhibited identical properties. TGA1 degraded similarly in keg‐1 and Col lysates, indicating that keg lysates are capable of degrading proteins. We also observed that degradation of several non‐ABF proteins was slower in keg‐1 lysates than in Col lysates. Degradation of GBF3 and FUS3 was slowed, and EGL3 was stable in keg‐1 lysates compared to Col lysates over the 10‐min time course. The FUS3 results were unexpected, because previous assays in our lab observed that FUS3 degraded equivalently in keg‐1 lysates (Chen et al., 2013). However, the time points used in Chen et al. (2013) extended from 30 to 90 min, and most FUS3 protein had degraded in both Col and keg‐1 lysate by the first time point at 30 min. Slower degradation at early time points might not have been observed in those experiments. Our FUS3 cell‐free degradation assay extended from 1 to 10 min, at which point just over 53% of FUS3 protein remained.
keg‐1 seedlings have a very high level of ABI5 protein (Stone et al., 2006). The slowed degradation of FUS3 and GBF3 in keg‐1 lysate might result from altered ABA signaling in keg‐1, since both FUS3 and GBF3 are involved in ABA responses and FUS3 degradation is ABA dependent. GFP‐tagged FUS3 protein increases following ABA treatment (Gazzarrini et al., 2004; Lu et al., 2010), and ABA inhibits FUS3 degradation in vitro (Chiu et al., 2016). Although the bZIP protein GBF3 is not in bZIP group A, it is also known to play a role in ABA response. Plants overexpressing GBF3 are more drought resistant than WT plants and exhibit less inhibition of root growth by ABA (Ramegowda et al., 2017). EGL3 is involved in trichome development and anthocyanin synthesis, and its degradation is mediated by the HECT E3 ligase ubiquitin protein ligase 3 (UPL3) (Patra et al., 2013). It is unclear why EGL3 is stabilized in keg‐1 lysate.
The reduced stability of additional proteins in keg extracts observed here is consistent with the pleotropic phenotype of keg mutants (Stone et al., 2006), and the observation that simultaneous removal of ABI5 and several ABF proteins in the keg background has only a minor effect on the keg phenotype (Chen et al., 2013). Identification of CIPK26 (Lyzenga et al., 2013), formate dehydrogenase (McNeilly et al., 2018), JAZ12 (Pauwels et al., 2015), and MKK4 and 5 (Gao et al., 2020) as KEG interactors/substrates further support the model that KEG has broad effects on plant growth and stress responses. The roles of KEG in immunity and vascular development implicate KEG in plant defense responses, intracellular trafficking (Gu & Innes, 2011, 2012), and cellular differentiation (Gandotra et al., 2013), in addition to its role in the regulation of group A bZIP proteins. These additional processes as well as downstream ABA responses affected in keg could indirectly modulate the proteolysis of other transcription factors.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
KL, Y‐TC, and JC designed the research; KL, Y‐TC, KK, and BS performed research; KL, Y‐TC, JC, and BS analyzed data; KL and JC wrote the paper.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Table S1‐S6
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation (IOS‐1557760 to J. Callis, PI) and funds from the Paul K. and Ruth R. Stumpf Endowed Professorship in Plant Biochemistry and Aggie Alumni Foundation Award to J.C. We thank Eli Nambara and Ruth Finkelstein for the gift of abi5‐7 seeds and Dr. Finkelstein for assistance with genotyping the abi5‐7 allele and helpful discussions. We thank Sonia Gazarrini (University of Toronto Scarborough) for the gift of the GST‐FUS3 construct, Ling Yuan (University of Kentucky) for the EGL3 in the pGEX‐4T‐1 vector, Sara Hotton for the synthesis of the modified pEXP1‐DEST Myc9 expression vector (p7296), and Jonathan Gilkerson for synthesis of the modified pET3c expression plasmid (p3832). We thank the University of California‐Davis Controlled Environment Facility (CEF) for assistance with the propagation of transgenic plants. We thank many past members of the Callis laboratory for helpful discussions, most recently John Riggs. We thank Jemina Cornejo, Eduards Norkvests, Anna Chen, and Stephanie Ng for assistance in propagation and analyses of transgenic lines.
Linden, K. J. , Chen, Y.‐T. , Kyaw, K. , Schultz, B. , & Callis, J. (2021). Factors that affect protein abundance of a positive regulator of abscisic acid signalling, the basic leucine zipper transcription factor ABRE‐binding factor 2 (ABF2). Plant Direct, 5, e00330. 10.1002/pld3.330
REFERENCES
- Albertos, P. , Romero‐Puertas, M. C. , Tatematsu, K. , Mateos, I. , Sánchez‐Vicente, I. , Nambara, E. , & Lorenzo, O. (2015). S‐nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nature Communications, 6, 8669. 10.1038/ncomms9669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard, I. M. , Lynch, T. J. , & Finkelstein, R. R. (2002). Regulation and role of the Arabidopsis Abscisic Acid‐Insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiology, 129, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.‐T. , Liu, H. , Stone, S. , & Callis, J. (2013). ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant Journal, 75, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, R. S. , Pan, S. , Zhao, R. , & Gazzarrini, S. (2016). ABA‐dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. Plant Journal, 88, 749–761. [DOI] [PubMed] [Google Scholar]
- Choi, H.‐I. , Hong, J.‐H. , Ha, J.‐O. , Kang, J.‐Y. , & Kim, S. Y. (2000). ABFs, a family of ABA‐responsive element binding factors. Journal of Biological Chemistry, 21, 1723–1730. 10.1074/jbc.275.3.1723 [DOI] [PubMed] [Google Scholar]
- Choi, H. I. , Park, H. J. , Park, J. H. , Kim, S. , Im, M. Y. , Seo, H. H. , Kim, Y. W. , Hwang, I. , & Kim, S. Y. (2005). Arabidopsis calcium‐dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid‐responsive gene expression, and modulates its activity. Plant Physiology, 139, 1750–1761. 10.1104/pp.105.069757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant Journal, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cuming, A. C. (2019). Evolution of ABA signaling pathways. In Seo M. & Marion‐Poll A. (Eds.), Advances in botanical research (Vol. 92, pp. 281–313). Academic Press. [Google Scholar]
- Deppmann, C. D. , Acharya, A. , Rishi, V. , Wobbes, B. , Smeekens, S. , Taparowsky, E. J. , & Vinson, C. (2004). Dimerization specificity of all 67 B‐ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B‐ZIP motifs. Nucleic Acids Research, 32, 3435–3445. 10.1093/nar/gkh653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, K. A. (2006). An investigation of the proteolytic profile and biological function of members of the Aux/IAA family of proteins in Arabidopsis thaliana. University of California. PhD Dissertation. [Google Scholar]
- Dröge‐Laser, W. , Snoek, B. L. , Snel, B. , & Weiste, C. (2018). The Arabidopsis bZIP transcription factor family‐an update. Current Opinion in Plant Biology, 45, 36–49. 10.1016/j.pbi.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Finkelstein, R. R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant Journal, 5, 765–771. 10.1046/j.1365-313X.1994.5060765.x [DOI] [Google Scholar]
- Finkelstein, R. R. , & Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell, 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata, T. , Maruyama, K. , Fujita, Y. , Umezawa, T. , Yoshida, R. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2006). Abscisic acid‐dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. PNAS, 103, 1988–1993. 10.1073/pnas.0505667103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra, N. , Coughlan, S. J. , & Nelson, T. (2013). The Arabidopsis leaf provascular cell transcriptome is enriched in genes with roles in vein patterning. Plant Journal, 74, 48–58. 10.1111/tpj.12100 [DOI] [PubMed] [Google Scholar]
- Gao, C. , Sun, P. , Wang, W. , & Tang, D. (2020). Arabidopsis E3 ligase KEG associates with and ubiquitinates MKK4 and MKK5 to regulate plant immunity. Journal of Integrative Plant Biology. 63, 327–339. 10.1111/jipb.13007 [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S. , Tsuchiya, Y. , Lumba, S. , Okamoto, M. , & McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Developmental Cell, 7, 373–385. 10.1016/j.devcel.2004.06.017 [DOI] [PubMed] [Google Scholar]
- Gilkerson, J. , Kelley, D. , Tam, R. , Estelle, M. , & Callis, J. (2015). Lysine residues are not required for proteasome‐mediated proteolysis of the auxin/indole acetic acid protein IAA1. Plant Physiology, 168, 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. , & Innes, R. W. (2011). The KEEP ON GOING protein of arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans‐golgi network/early endosome vesicles. Plant Physiology, 155, 1827. 10.1104/pp.110.171785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. , & Innes, R. W. (2012). The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. The Plant Cell, 24, 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, F. , Waadt, R. , & Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology, 21, R346–R355. 10.1016/j.cub.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M. , Weisshaar, B. , Droge‐Laser, W. , Vicente‐Carbajosa, J. , Tiedemann, J. , Kroj, T. , & Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends in Plant Science, 7, 106–111. 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Jaspert, N. , Throm, C. , & Oecking, C. (2011). Arabidopsis 14‐3‐3 proteins: Fascinating and less fascinating aspects. Frontiers in Plant Science, 2, 96. 10.3389/fpls.2011.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. R. , Wagner, R. L. , Verhey, S. D. , & Walker‐Simmons, M. K. (2002). The abscisic acid‐responsive kinase PKABA1 interacts with a seed‐specific abscisic acid response element‐binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiology, 130, 837–846. 10.1104/pp.001354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J. , Choi, H. , Im, M. , & Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress‐responsive abscisic acid signaling. The Plant Cell, 14, 343–357. 10.1105/tpc.010362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Kang, J. Y. , Cho, D. I. , Park, J. H. , & Kim, S. Y. (2004). ABF2, an ABRE‐binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant Journal, 40, 75–87. 10.1111/j.1365-313X.2004.02192.x [DOI] [PubMed] [Google Scholar]
- Kim, S. Y. , Ma, J. , Perret, P. , Li, Z. , & Thomas, T. L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA‐binding and transcriptional activities. Plant Physiology, 130, 688–697. 10.1104/pp.003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. Y. , Thomas, T. L. (1998). A family of novel basic leucine zipper proteins binds to seed‐specification elements in the Carrot Dc3 gene promoter. Journal of Plant Physiology, 152, 607–613. 10.1016/s0176-1617(98)80019-9. [DOI] [Google Scholar]
- Kline, K. , Barrett‐Wilt, G. , & Sussman, M. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. PNAS, 107, 15986–15991. 10.1073/pnas.1007879107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, E. (2007). An investigation of the ubiquitin conjugating enzymes and RING E3 ligases in Arabidopsis thaliana. University of California. PhD Dissertation. [Google Scholar]
- Kraft, E. , Stone, S. L. , Ma, L. , Su, N. , Gao, Y. , Lau, O.‐S. , Deng, X.‐W. , & Callis, J. (2005). Genome analysis and functional characterization of the E2 and RING domain E3 ligase ubiquitination enzymes of Arabidopsis thaliana . Plant Physiology, 139, 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Yoon, H.‐J. , Terzaghi, W. , Martinez, C. , Dai, M. , Li, M. , Byun, M.‐O. , & Deng, X.‐W. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4‐based E3 ligases, act together as negative regulators in ABA signal transduction. The Plant Cell, 22, 1716–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , & Stone, S. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting ligase self‐ubiquitination and proteasomal degradation. The Plant Cell, 22, 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , & Stone, S. L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor ABSCISIC ACID INSENSITIVE 5 is mediated by the RING‐type E3 ligase KEEP ON GOING. Journal of Biological Chemistry, 288, 20267–20279. 10.1074/jbc.M113.465369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , & Stone, S. L. (2014). Regulation of ABI5 turnover by reversible post‐translational modifications. Plant Signaling & Behavior, 9, e27577. 10.4161/psb.27577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , & Chua, N. H. (2000). A null mutation in a bZIP factor confers ABA‐insensitivity in Arabidopsis thaliana. Plant & Cell Physiology, 41, 541–547. 10.1093/pcp/41.5.541 [DOI] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , Mongrand, S. , & Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. PNAS, 98, 4782–4787. 10.1073/pnas.081594298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , Mongrand, S. , Kinoshita, N. , & Chua, N.‐H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes & Development, 17, 410–418. 10.1101/gad.1055803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , Mongrand, S. , McLachlin, D. T. , Chait, B. T. , & Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA‐dependent growth arrest during germination. Plant Journal, 32, 317–328. 10.1046/j.1365-313X.2002.01430.x [DOI] [PubMed] [Google Scholar]
- Lu, Q. S. , Paz, J. D. , Pathmanathan, A. , Chiu, R. S. , Tsai, A. Y. L. , & Gazzarrini, S. (2010). The C‐terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant Journal, 64, 100–113. [DOI] [PubMed] [Google Scholar]
- Lyzenga, W. J. , Liu, H. , Schofield, A. , Muise‐Hennessey, A. , & Stone, S. L. (2013). Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin‐proteasome system. Journal of Experimental Botany, 64, 2779–2791. 10.1093/jxb/ert123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeilly, D. , Schofield, A. , & Stone, S. L. (2018). Degradation of the stress‐responsive enzyme formate dehydrogenase by the RING‐type E3 ligase Keep on Going and the ubiquitin 26S proteasome system. Plant Molecular Biology, 96, 265–278. 10.1007/s11103-017-0691-8 [DOI] [PubMed] [Google Scholar]
- Michaels, S. D. , & Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell, 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Lee, J. , Jin, J. B. , Yoo, C. Y. , Miura, T. , & Hasegawa, P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. PNAS, 106, 5418–5423. 10.1073/pnas.0811088106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. , Kawai, T. , Tanaka, K. , Niwa, Y. , Watanabe, Y. , Nakamura, K. , Kimura, T. , & Ishiguro, S. (2007). Improved Gateway binary vectors: High‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology, and Biochemistry, 71, 2095–2100. 10.1271/bbb.70216 [DOI] [PubMed] [Google Scholar]
- Nakashima, K. , Fujita, Y. , Kanamori, N. , Katagiri, T. , Umezawa, T. , Kidokoro, S. , Maruyama, K. , Yoshida, T. , Ishiyama, K. , Kobayashi, M. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant & Cell Physiology, 50, 1345–1363. 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Patra, B. , Pattanaik, S. , & Yuan, L. (2013). Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant Journal, 74, 435–447. 10.1111/tpj.12132 [DOI] [PubMed] [Google Scholar]
- Pauwels, L. , Ritter, A. , Goossens, J. , Durand, A. N. , Liu, H. , Gu, Y. , Geerinck, J. , Boter, M. , Vanden Bossche, R. , De Clercq, R. , Van Leene, J. , Gevaert, K. , De Jaeger, G. , Solano, R. , Stone, S. , Innes, R. W. , Callis, J. , & Goossens, A. (2015). The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM‐DOMAIN12 stability. Plant Physiology, 169, 1405–1417. 10.1104/pp.15.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramegowda, V. , Gill, U. S. , Sivalingam, P. N. , Gupta, A. , Gupta, C. , Govind, G. , Nataraja, K. N. , Pereira, A. , Udayakumar, M. , Mysore, K. S. , & Senthil‐Kumar, M. (2017). GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana . Scientific Reports, 7, 9148. 10.1038/s41598-017-09542-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz, T. C. , Miller, M. J. , McLoughlin, F. , Augustine, R. C. , Marshall, R. S. , Juan, Y.‐T. , Charng, Y.‐Y. , Scalf, M. , Smith, L. M. , & Vierstra, R. D. (2018). SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. The Plant Cell, 30, 1077. 10.1105/tpc.17.00993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonheim, P. J. , Sinnige, M. P. , Casaretto, J. A. , Veiga, H. , Bunney, T. D. , Quatrano, R. S. , & de Boer, A. H. (2007). 14‐3‐3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant Journal, 49, 289–301. 10.1111/j.1365-313X.2006.02955.x [DOI] [PubMed] [Google Scholar]
- Seo, K. I. , Lee, J. H. , Nezames, C. D. , Zhong, S. , Song, E. , Byun, M. O. , & Deng, X. W. (2014). ABD1 is an Arabidopsis DCAF substrate receptor for CUL4‐DDB1‐based E3 ligases that acts as a negative regulator of abscisic acid signaling. The Plant Cell, 26, 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora, L. , De Smet, I. , Foyer, C. H. , & Zhang, H. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. The Plant Journal, 28, 655–662. 10.1046/j.1365-313x.2001.01185.x [DOI] [PubMed] [Google Scholar]
- Sirichandra, C. , Davanture, M. , Turk, B. E. , Zivy, M. , Valot, B. , Leung, J. , & Merlot, S. (2010). The Arabidopsis ABA‐activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14‐3‐3 binding site involved in its turnover. PLoS One, 5, e13935. 10.1371/journal.pone.0013935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubacz, A. , Daszkowska‐Golec, A. , & Szarejko, I. (2016). The role and regulation of ABI5 (ABA‐Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Frontiers in Plant Science, 7, 1884. 10.3389/fpls.2016.01884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S. L. , Williams, L. A. , Farmer, L. M. , VIerstra, R. D. & Callis, J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. The Plant Cell, 18, 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y. , Furihata, T. , Abe, H. , Yoshida, R. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid‐dependent signal transduction pathway under drought and high‐salinity conditions. PNAS, 97, 11632–11637. 10.1073/pnas.190309197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Zhu, D. M. , Huang, X. , Li, S. , Gong, Y. N. , Yao, Q. F. , Fu, X. D. , Fan, L. M. , & Deng, X. W. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell‐free assay system. The Plant Cell, 21, 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Li, L. , Ye, T. , Lu, Y. , Chen, X. , & Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. Journal of Experimental Botany, 64, 675–684. 10.1093/jxb/ers361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. , & Shinozaki, K. (1993). Characterization of the expression of a desiccation‐responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Molecular and General Genetics, 236, 331–340. 10.1007/BF00277130 [DOI] [PubMed] [Google Scholar]
- Yamamoto, A. , Kagaya, Y. , Toyoshima, R. , Kagaya, M. , Takeda, S. , Hattori, T. (2009). Arabidopsis NF‐YB subunits LEC1 and LEC1‐LIKE activate transcription by interacting with seed‐specific ABRE‐binding factors. The Plant Journal, 58, 843–856. 10.1111/j.1365-313x.2009.03817.x. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Fujita, M. , Sayama, H. , Kidohoro, S. , Maruyama, K. , Mizoi, J. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2010). AREB1, AREB2, ABF3 are master transcription factors that cooperatively regulate ABRE‐dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant Journal, 61, 672–685. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Fujita, Y. , Maruyama, K. , Mogami, J. , Todaka, D. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell and Environment, 38, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Wu, Y. , & Xie, Q. (2015). Precise protein post‐translational modifications modulate ABI5 activity. Trends in Plant Science, 20, 569–575. 10.1016/j.tplants.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Liu, D. , Yang, B. , Liu, W.‐Z. , Mu, B. , Song, H. , Chen, B. , Li, Y. , Ren, D. , Deng, H. , & Jiang, Y.‐Q. (2020). Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA‐responsive element‐binding factors. Journal of Experimental Botany, 71, 188–203. 10.1093/jxb/erz432 [DOI] [PubMed] [Google Scholar]
- Zhu, S.‐Y. , Yu, X.‐C. , Wang, X.‐J. , Rui Zhao, R. , Li, Y. , Fan, R.‐C. , Shang, Y. , Du, S.‐Y. , Wang, X.‐F. , Wu, F.‐Q. , Xu, Y.‐H. , Zhang, X.‐Y. , & Zhang, D.‐P. (2007). Two calcium‐dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. The Plant Cell, 19, 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Table S1‐S6


