ABSTRACT
Gene expression involving RNA polymerase II is regulated by the concerted interplay between mRNA synthesis and degradation, crosstalk in which mRNA decay machinery and transcription machinery respectively impact transcription and mRNA stability. Rpb4, and likely dimer Rpb4/7, seem the central components of the RNA pol II governing these processes. In this work we unravel the molecular mechanisms participated by Rpb4 that mediate the posttranscriptional events regulating mRNA imprinting and stability. By RIP-Seq, we analysed genome-wide the association of Rpb4 with mRNAs and demonstrated that it targeted a large population of more than 1400 transcripts. A group of these mRNAs was also the target of the RNA binding protein, Puf3. We demonstrated that Rpb4 and Puf3 physically, genetically, and functionally interact and also affect mRNA stability, and likely the imprinting, of a common group of mRNAs. Furthermore, the Rpb4 and Puf3 association with mRNAs depends on one another. We also demonstrated, for the first time, that Puf3 associates with chromatin in an Rpb4-dependent manner. Our data also suggest that Rpb4 could be a key element of the RNA pol II that coordinates mRNA synthesis, imprinting and stability in cooperation with RBPs.
KEYWORDS: RNA polymerase II, Puf3, Rbp, mRNA imprinting, mRNA decay, Saccharomyces cerevisiae
Introduction
Gene expression is a highly regulated process that comprises several coordinated steps to ensure appropriate mRNA levels, and is determined by two different processes: mRNA synthesis and decay. Both processes occur mainly in different cell locations and need a high coordination. Changes in mRNA levels are determined by their transcription and degradation rates, and the parallel co-regulation of global mRNA synthesis and decay is important to maintain mRNA homeostasis [1–4].
mRNA imprinting, defined by M. Choder as the co-transcriptional association of a protein to an mRNA that lasts throughout the mRNA lifetime and is required for proper regulation of all major stages that the mRNA undergoes [5], is crucial for coordinating transcription in the nucleus and the decay that occurs mainly in the cytoplasm [1,5–11]. Two RNA polymerase II subunits, Rpb4 and Rpb7, correspond to these mRNA imprinting proteins interconnecting both processes.
Rpb4 and Rpb7 bind mRNA in the RNA pol II context during transcription and remain associated with mRNA throughout its life by regulating processes such as export, translation and decay [9,12–16]. Rpb4 has been proposed to interact with mRNA in an Rpb7 dependent manner [7,9,17,18]. The Rpb4/7 dimer participates in several transcription steps, such as transcription initiation [19] and elongation [20], and dissociates from the RNA pol II core by interacting with mRNA during transcription [7,10,13], although this statement is not unanimously accepted [7,9,17,18,21]. Furthermore Rpb4 participates in transcription termination and gene looping [22]. Rpb4-mediated mRNA decay probably occurs by interacting with mRNA decay machinery elements, such as Pat1-Lsm1/7 [14]. Under optimal growth conditions, Rpb4 serves as a key protein to globally modulate mRNA stability and to coordinate transcription and decay [13]. Furthermore, Rpb4-mediated post-transcriptional regulation also plays a major role in controlling the Environmental Stress Response (ESR) at both the transcription and mRNA decay levels [13]. A recent study has demonstrated that disturbing Rpb4 dissociation from RNA pol II compromises the posttranscriptional roles of Rpb4 and leads to translation and mRNA decay defects [17].
Rpb4 acts as an RNA binding protein (RBP) by regulating gene expression from mRNA synthesis to mRNA degradation [23]. Rpb4, and also Rpb7, are examples of RNA pol II components that act as RBPs [7,9,17,18,23]. However, other RBPs associated with transcription machinery have been described, such as transcription elongation factors Spt4/5 [23], elements of the decaysome complex involved in mRNA degradation like Xrn1, Ccr4 or Dhh1 [4,8,24], or the Cbc elements of mRNA capping machinery [25]. RBPs are elements that govern the post-transcriptional regulation of the mRNA life cycle in all living organisms that usually bind 3ʹ UTR elements of their targeted mRNAs, modulating their splicing, stability, translation and cellular location in eukaryotic organisms [23,26–33]. Different RBPs bind to specific and distinct sets of mRNAs that typically encode proteins with related biological functions or destined to similar subcellular localizations [27,32,34]. Many RBPs shuttle between nucleus and cytoplasm [32]. The distribution of RBPs between the nucleus and cytoplasm impacts gene expression through their interaction with their cytosolic or nuclear mRNAs [32].
Despite Rpb4–mRNA imprinting having been demonstrated [7,13], very little is known about the specific mRNAs bound by Rpb4 and the molecular mechanisms governing mRNA life cycle regulation via Rpb4.
In this work, we investigated the precise role of Rpb4 in mRNA imprinting and degradation. The genome-wide analysis by RIP-Seq identified a set of almost 1500 mRNAs bound to Rpb4. We found that the average degradation rate of the Rpb4-bound mRNAs was higher than the overall one of mRNAs, which falls in line with a role for Rpb4 in mRNA stability. Notably a fraction of these transcripts are also targets of RPB Puf3. Puf3 is a well-studied example of RBPs of the PUF (Pumilio and FBF) family, a conserved family of mRNA regulators that bind to the 3ʹ UTR of many mRNAs to regulate mRNA stability and/or translation [35–39], which binds mRNAs for nuclear-encoded mitochondrial proteins [40–42]. In line with this concerted Rpb4 and Puf3 association to a group of mRNA, our results demonstrated that Rpb4 and Puf3 show genetic and physical interactions. The integrity of the Puf3 protein depends on Rpb4. We also found that the Rpb4 and Puf3 association with mRNAs depends on each other and that Puf3 associates with chromatin in an Rpb4-dependent manner. Therefore, we conclude that Rpb4 and Puf3 cooperate to control their respective association with mRNAs and the mRNA stability of a specific group of mRNAs, which suggest a concerted role in mRNA imprinting. Our data also point out Rpb4 as a hub that cooperates with different RBPs in the mRNA synthesis and degradation interplay.
Materials and methods
Yeast strains, genetic manipulations, media and genetic analysis
Common yeast media, growth conditions and genetic techniques were used as described elsewhere [43].
Yeast strains and primers are listed in Supplementary Tables S1-S2, respectively.
Isoform-specific RNA immunoprecipitation (isRIP)
RNA immunoprecipitation was carried out as described in [44]. Briefly, two biological replicates of 1 l cell cultures of the exponentially grown Rpb4-TAP-tagged strain (OD600 0.8–0.9) were pelleted, washed twice with 25 ml of buffer A (20 mM Tris-HCl, pH 8, 140 mM KCl, 1.8 mM MgCl2, 0.1% NP-40, 0.02 mg/ml heparin) and resuspended in 2 ml of buffer B (20 mM Tris-HCl, pH 8, 140 mM KCl, 1.8 mM MgCl2, 0.1% NP-40, 0.2 mg/ml heparin, 0.5 mM DTT, 1 mM PMSF, 50 U/µl RNasin Plus, 1X protease inhibitor cocktail [Complete; Roche]). Then cells were broken in a FastPrep24 agitator for 30 seconds at speed 5.5 and 2x for 20 seconds at 6.0, followed by a 1-minute break between each step by adding glass beads (425–600 µm, Sigma). Extracts were clarified by centrifuging twice at 7000 x g for 5 min. A 100 µl cell lysate aliquot was used as the INPUT control. Rpb4 was immunopurified from cell lysates by using 400 µl of Dyna M280 sheep anti-rabbit IgG beads (Invitrogen) for 2 h at 4 ºC with gentle rotation in a rotation mixer. After four 15-minute washes with buffer C (20 mM Tris-HCl, pH 8, 140 mM KCl, 1.8 mM MgCl2, 0.1% NP-40, 10% glycerol, 0.5 mM DTT, 1 mM PMSF, 50 U/µl RNasin Plus, 1X protease inhibitor cocktail [Complete; Roche]), RNA-Rpb4 complexes were eluted from beads by TEV protease cleavage of TAP-tag using 10 µl of 10 U/µl acTEV protease for 1 h at 21 ºC in a heatblock by shaking at 700 rpm.
The eluent was collected and RNA was isolated by extracting twice with one volume of phenol: chloroform: isoamyl alcohol (25:24:1), pH 4.7, and centrifuging at 13,000 rpm for 1 min at room temperature. The upper phase was recovered and an equal volume of chloroform-isoamyl alcohol (24:1) pH 4.7 was added, gently mixed and centrifuged for 1 minute at 13,000 rpm and room temperature. RNA was then precipitated from the upper phase with two volumes of 100% ethanol, 1/10 volume of NaAc 3 M and 3 μl of linear acrylamide as a carrier by incubating for 20 minutes on ice. Then RNA was pelleted by centrifugation at 14,000 rpm for 20 minutes at 4 ºC and washed with 500 μl of 70% ethanol. After centrifugation at 13,000 rpm for 1 min at room temperature, the pellet was dried and resuspended in 15 μl of milliQ H2O.
INPUT RNA extraction was performed with the RNeasy Mini Kit (Quiagen) according to the manufacture’s indications.
3 T-fill sequencing and data analysis
Two biological replicates were used and the RNA from the total extracts (input) and immunoprecipitated samples (RIP) was sequenced. Constructions of sequencing libraries were carried out as previously described [45] from 1 μg RNA for inputs and 150 ng RNA for the RIP samples. Data were aligned to yeast reference genome version R64 for strain S288c using a custom computational pipeline, followed by the calculation of RIP enrichment using the DESeq2 R Bioconductor package as previously described [44,46].
mRNA extraction and reverse transcription
Total RNA from yeast cells was extracted and quantified as previously described from 50 ml of culture cells [47].
First-strand cDNA was synthesized using 1 μg of RNA with the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer’s protocol. As a negative control for genomic DNA contamination, each sample was subjected to the same reaction without reverse transcriptase.
Quantitative real-time PCR (RT-qPCR)
Real-time PCR was performed in a CFX-384 Real-Time PCR instrument (BioRad) with the EvaGreen detection system “SsoFast™ EvaGreen® Supermix“ (BioRad). Reactions were performed in a total volume of 5 µl that contained the cDNA corresponding to 0.1 ng of total RNA. Each PCR reaction was performed at least 3 times with three independent biological replicates to gain a representative average. The 18S rRNA gene was used as a normalizer. Supplementary Table S2 lists the employed oligonucleotides.
Isolation of the mRNA-associated proteins
mRNA crosslinking was carried out as described in [13] with some modifications. Briefly, 250 ml of cell cultures with an OD600 ~ 0.6–0.8 were exposed to 1,200 mJ/cm2 of 254 nm UV in a UV crosslinker (Biolink Shortwave 254 nm), resuspended in 350 µl of lysis buffer (20 mM Tris, pH 7.5, 0.5 M NaCl, 1 mM EDTA, 1x protease inhibitor cocktail [Complete; Roche]) and 200 µl of glass beads (425–600 µm, Sigma), and were broken by vortexing for 15 min at 4 ºC. A lysate aliquot was used as the INPUT control. The lysate was incubated with 150 µl of oligo (dT)25 cellulose beads (New England BioLabs, cat no. S1408S) for 15 min at room temperature, washed 5 times with loading buffer (20 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 1 mM EDTA) and once with low-salt buffer (10 mM Tris–HCl, pH 7.5, 0.1 M NaCl, 1 mM EDTA). The RNA-associated proteins were eluted by resuspending beads in 250 µl of elution buffer (20 mM Tris–HCl, pH 7.5) pre-warmed at 70 ºC by incubating 5 min at room temperature (x 2). The two eluate samples were mixed, lyophilized and resuspended in 20 μl of milliQ H2O. The mRNA-associated proteins were analysed by SDS-PAGE and western blot with the appropriate antibodies: anti-Rpb4 (Pol II RPB4 (2Y14); Biolegend), anti-Pgk1 (22 C5D8; Invitrogen), anti-H3 (ab1791; Abcam); PAP (Sigma).
Protein extraction and TAP purification
Protein whole cell extracts and TAP purifications were performed as described in [47]. Briefly, 150 ml of cells grown exponentially (OD600 0.6–0.8) were pelleted and resuspended in 0.3 ml of lysis buffer (50 mM HEPES [pH 7.5], 120 mM NaCl, 1 mM EDTA, 0.3% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)), supplemented with 1X protease inhibitor cocktail (Complete; Roche), 0.5 mM phenylmethylsulonyl fluoride (PMSF), 2 mM sodium orthovanadate and 1 mM sodium fluoride. Cells were broken by vortexing (3 cycles, 5 min each) using 0.2 ml of glass beads (425–600 µm; Sigma)). TAP purification was carried out as previously described [48] with 2 mg of protein extract and 50 µl of Dynabeads Pan Mouse IgG (Invitrogen) per sample. The affinity-purified proteins were released from the beads by boiling for 10 min and performing western blotting with different antibodies.
For the RNase treatment, the cell-crude extracts (60 μg proteins) were incubated with 15 μg of RNase A, at 37 ºC for 1 h, before proceeding with the yChEFs method, as previously described [49].
SDS-PAGE and western blot analyses
Protein electrophoresis and western blot were carried out as described in [48].
For the western blot analyses, the anti-Rpb4 (Pol II RPB4 (2Y14); Biolegend), anti-Pgk1 (22C5D8; Invitrogen), anti-H3 (ab1791; Abcam) and PAP (Sigma) antibodies were used.
Yeast chromatin-enriched fractions preparation
The chromatin-enriched fractions preparation was carried out by the yChEFs procedure [49,50] using 75 ml of exponentially grown YPD cultures (OD600 ~ 0.6–0.8). The final pellet obtained by this methodology was resuspended in 20 μl of 1x Tris-Glycine SDS sample buffer and incubated for 5 min at 100 ºC to obtain chromatin-bound proteins, which were analysed by western blot with the different antibodies indicated above. The anti-histone H3 (ab1791; Abcam) and anti-phosphoglycerate kinase Pgk1 (459,250; Invitrogen) antibodies were employed to detect H3 histone, used as a positive control, and Pgk1 as a negative control of cytoplasmic contamination.
Chromatin immunoprecipitation and sequencing (ChIP-Seq)
Chromatin immunoprecipitation was performed using three biological replicates for strains Puf3-TAP WT and Puf3-TAP rpb4Δ, and two for the WT strain. For each sample, 100 ml of cells were grown in YPD medium to OD600 ~ 0.7. The SNF21-TAP-natR S. pombe strain (spike-in control, SP641, h90 ura-r TetR-tup11 KanR-tetO-snf21-TAP-natR ura4-D18) was grown in YES medium to OD600 ~ 0.7. Cells were crosslinked by adding 1% formaldehyde (Thermo Fisher) directly to the culture and shaking for 15 minutes. Formaldehyde was quenched for 5 min by adding 0.125 M glycine. Cells were pelleted, washed 4 times with cold TBS and flash-frozen.
Pellets were thawed on ice, resuspended in 500 µl of zymolyase solution (1.2 M sorbitol, 10 mM Tris-HCl, pH 8, 10 mM CaCl2, 1% v/v beta-mercaptoethanol, 0.1 U/µl zymolyase (Zymo Research)) per pellet and incubated at 37 ºC for 30 minutes with shaking at 500 rpm to digest cell walls. Spheroplasts were isolated by centrifugation (5 min at 3,500xg) and. resuspended in 540 µl of lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% v/v Triton X-100, 0.1% w/v sodium deoxycholate, 1 mM PMSF, 1 mM benzamidine, 1:100 yeast protease inhibitor cocktail (Sigma Aldrich, P8215)). Lysates were sonicated in a Covaris ME220 instrument in 130 µl screw-cap microtubes for 3 min to 100–500 bp (PIP = 75, DF = 15%, 1,000 cycles per burst, 9 ºC). The sonicated lysates were centrifuged at 17,000xg for 10 minutes to pellet the insoluble material. For IP, an equal amount of chromatin from each sample was taken based on the estimated DNA concentration, and S. pombe sonicated chromatin was spiked to approximately 1% in relation to that amount (S. pombe chromatin was obtained as described above, except for using a 2-fold volume of zymolyase solution for twice the time).
Next 250 µl of Dynabeads Pan Mouse IgG (Thermo Fisher) per sample were washed 3 times with PBS/BSA (PBS with 5 mg/ml BSA (Sigma Aldrich)), resuspended in the sonicated chromatin solution and rotated overnight at +4 ºC. They were washed 3 times with lysis buffer, lysis buffer containing 500 mM NaCl and LiCl wash buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and once with elution buffer (50 mM Tris-HCl, pH 8, 1% SDS, 10 mM EDTA). Chromatin was eluted from beads in elution buffer at 65 ºC with shaking for 20 minutes. The eluted material was diluted 5-fold to lower the SDS concentration to 0.2%, then 1:100 RNase cocktail (Thermo Fisher, AM2286) was added and samples were incubated at 37 ºC for 45 minutes to digest RNA. SDS up to a total of 0.5% and Proteinase K up to 0.5 mg/ml (Thermo Fisher, AM2546) were added, and samples were shaken overnight at 65 ºC to reverse the crosslinked DNA. Input controls (10 µl sonicated chromatin) were subjected to RNA digestion and crosslinking reversion in the same way. DNA was purified with the QIAquick Gel Extraction kit (QIAGEN) and eluted in 50 µl of water.
Sequencing libraries were prepared using the NEBNext Ultra™ II DNA Library Prep Kit (NEB). Four PCR cycles were used for the input samples and 17 cycles for the IP samples. The resulting libraries were pooled and dual size-selected with 0.6X and 1.2X AMPure XP (Beckman Coulter) bead ratios. The pool was sequenced in NextSeq 500 with paired-end with a read length of 38 bases.
Fastaq files were mapped to a combined S. cerevisiae (R64) and S. pombe genome using bowtie2 with default settings. The mapped files were deduplicated with Picard and filtered by mapping quality (MAPQ>2). Peaks were called with MACS2 (–nomodel mode, – extsize 170, -q 0.05) using all the Puf3-TAP and Puf3-TAP Δrpb4 samples by taking the WT control as input. Metagene profiles were plotted with VAP [51] over all the S. cerevisiae genes and the genes whose mRNA was bound by Puf3.
Results
Rpb4 physically interacts with mRNAs
To investigate the association of the RNA pol II specific subunit Rpb4 with mRNAs in vivo, we performed RIP experiments using the previously described isRIP methodology (isoform-specific RNA immunoprecipitation) [44], which allows the quantification of mRNA isoforms that were immunoprecipitated with a RBP.
For that purpose we used an Rpb4 TAP-tagged yeast strain grown in YPD medium at 30 ºC, and immunoprecipitated the RNA natively bound to Rpb4 without cross-linking procedure. Two biological replicates were used and the RNA from total extracts and the Rpb4-immunoprecipitated samples were sequenced by genome-wide 3ʹ isoforms sequencing [45], measuring the enrichment of each mRNA in the immunoprecipitated (IP) fraction relative to the input sample. As expected, the biological replicates showed high reproducibility (Fig. S1). By using the ‘DEseq2’ software [52], the normalized fold change in the IP samples was calculated (Fig. 1A). To define the Rpb4-bound transcripts, a false discovery rate (FDR) of < 10% was applied, as previously described [44], along with a specific cut-off consisting of Log2 fold-change of RIP vs. input higher than 0 and padj<0.1. We identified 1458 mRNAs differentially enriched in the RIP samples over the total extract (Table S3). Notably, when binding stringency was increased to a 1.5 fold change (padj<0.1), enrichment persisted and then 777 Rpb4-bound mRNAs were identified. However, as Rpb4 was expected to bind a significant fraction of the transcriptome (unlike other RPB), we decided to adopt the more permissive criteria.
Figure 1.
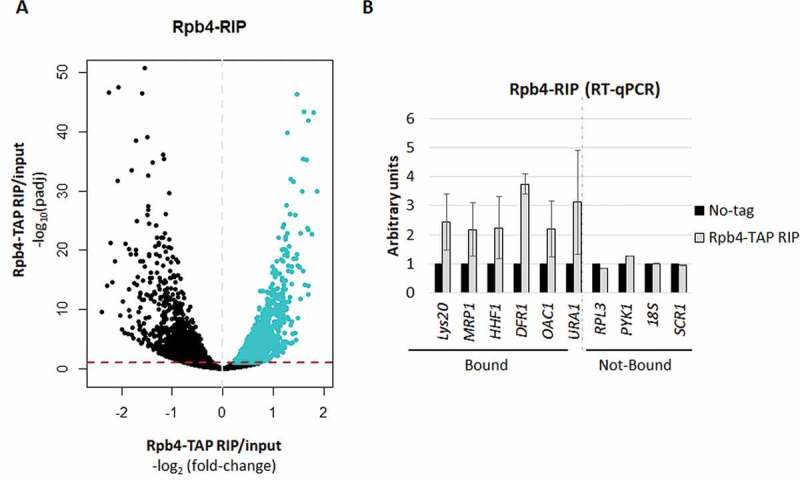
RNA immunoprecipitation (RIP) for Rpb4. (A) Volcano-plot generated to identify the significantly bound mRNAs by Rpb4 (blue), calculated by the DESeq2 software (RIP vs. input) [52]. A specific cut-off was applied to define the Rpb4-bounded transcripts consisting of Log2 Fold-Change of RIP vs. input higher than 0 (dashed grey line) and padj<0.1 (dashed red line). (B) RT-qPCR to analyse several Rpb4-bound and unbound mRNAs from the RIP experiments corresponding to the Rpb4-TAP and the no-TAP tag control strains. Data are shown as the average and standard deviation (SD) of at least three independent experiments (RIP vs. input). rRNA 18S was used as a normalizer
Furthermore, the RIP-Seq experiment results were corroborated by analysing the enrichment of some of the identified Rpb4-bound mRNAs, by RT-qPCR, in relation to both a no TAP-tag control strain and to some Rpb4 non-bound mRNAs, in the input and immunoprecipitated samples. As shown in Fig. 1B, all the analysed Rpb4-bound mRNAs were enriched in relation to the no TAP-tag control strain. As expected, the analysis of some Rpb4 non-bound mRNAs showed no enrichment either for the Rpb4 TAP-tagged yeast strain or the no TAP-tag control strain (Fig. 1B). In addition, the highly abundant 18S and SCR1 transcripts of RNA pol I and III, respectively, showed no enhanced signal in the RIP samples, which confirmed that Rpb4 was bound to specific RNA pol II transcripts, as expected from an RNA pol II specific subunit.
We performed a functional analysis of GO categories of the biological process for the Rpb4-bound mRNA identified in our genome-wide analysis, by using the String software [53]. We found, statistically significant, categories corresponding to mitochondrion, such as mitochondrial translation, mitochondrial gene expression or mitochondrial transport (Table S4). In addition, other statistically significant categories related to transmembrane transport and metabolic processes were detected.
Rpb4 and Puf3 associate with a common group of target mRNAs
As shown above, the Rpb4-bound mRNAs are enriched for the mitochondrion-related GO categories. In addition, the mRNA encoding mitochondrial proteins have been described to be bound by the member of the Pumilio and FBF family RBPs Puf3 at their 3ʹUTR region [40–42,44].
These results suggest a potential relation between Rpb4 and Puf3. Accordingly, we speculate that Rpb4-bound mRNAs could also be imprinted by Puf3. To explore it more in detail we compared our RIP data to different available datasets of Puf3 mRNA interactors identified by distinct methodologies. We firstly compared our data by the RIP-Seq procedure to data from two different Puf3 RIP-Seq analyses [44,54]. As shown in Fig. 2A and Figure S2, the Rpb4-bound transcripts significantly matched the Puf3-bound mRNAs compared to the dataset defined by Kershaw et al [54], and to a lesser extent compared to the more stringent dataset defined by Gupta et al [44], probably due to few identified Puf3-bound mRNAs as a consequence of the high fold-change used. To gain more insight, we also compared our data to other datasets of Puf3-bound transcripts identified by other methodologies, HITS-CLIP [55], RIP-ChIP [56] and PAR-CLIP [57]. As shown in Fig. 2B and Figure S2, again, a significant overlap was detected. To be more stringent, we also performed the same kind of analysis by selecting a 1.5 fold-change for our RIP data (RIP vs. input). In this case, the data from our analysis significantly matched the other used datasets, with similar or even higher percentage of overlapping (Figure S2).
Figure 2.
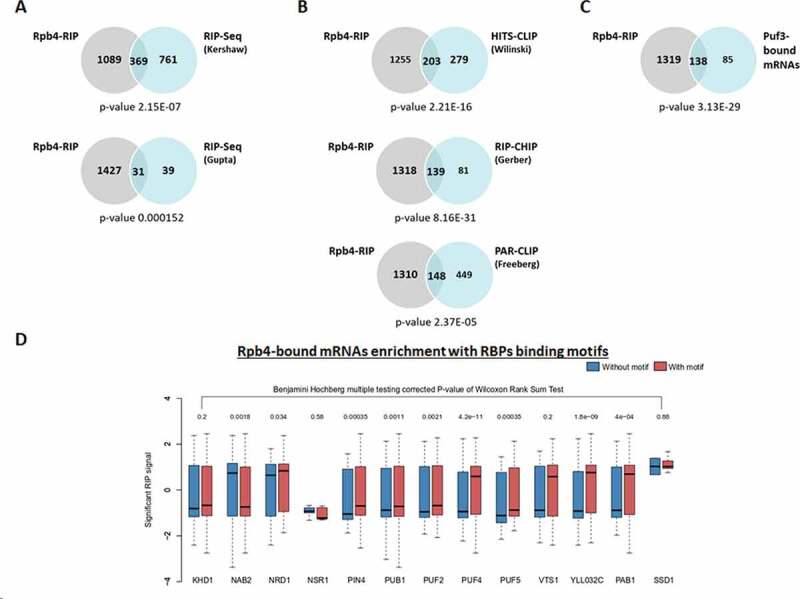
Rpb4-bound mRNAs correlates with Puf3-interactors. (A) Venn diagrams showing the overlapping found between the mRNAs bound by Rpb4 and Puf3 identified by RIP-Seq [44,54]. (B) Venn diagrams showing the coincidence between the Rpb4-RIP (grey circles) and Puf3-bound mRNAs (blue circles) identified by HITS-CLIP [55], RIP-CHIP [56] and PAR-CLIP [57]. (C) Venn diagrams depicting the common mRNAs bound by Rpb4 and Puf3 (for Puf3, they were selected as those in common in at least 3 of the 5 datasets indicated in B). (D) Boxplots showing the distribution of significant Rpb4 enrichment (Log2) over the two different categories of polyadenylation isoforms belonging to the genes with a particular predicted RBP binding motif (with motif or without motif). In the boxplots, the polyadenylation isoforms with no predicted motif are depicted in blue, and the isoforms with a predicted motif are denoted in red. The RBP motifs were taken from [58]. p-value <0.1 was considered significant
To gain more insight into these analyses, we then defined as Puf3-bound mRNAs all the Puf3 targets that were common in at least three of the five datasets used, which are presented in Fig. 2 [44,54–57]. Accordingly, we defined 223 mRNAs as Puf3-bound mRNAs (Table S5). Notably, most of Puf3-bound mRNAs, 138 (62%), matched the Rpb4-bound mRNAs identified by RIP-Seq in our analysis (Fig. 2C). Notably when a more stringent analysis was applied to identify the Rpb4-bound mRNAs (Log2 fold-change of RIP vs. input above 0.58 and padj<0.1), 104 (47%) of the Rpb4-bound mRNAs matched the Puf3-bound mRNAs. As expected, and in line with our above results and with previous results for Puf3 [40–42,44], the analysis of GO categories of Biological process, by using String software [53], of the Rpb4 and Puf3 common-bound mRNAs showed that the Biological process was related mainly to mitochondria and mitochondrial translation (Supplementary Table S6).
To extend our analysis beyond Puf3, and by taking into account that Rpb4 imprints almost 1,500 mRNAs (and only 138 in common with Puf3), we used the entire set of the Rpb4-bound mRNAs to analyse different RBPs with defined binding site motifs [58]. We compared the RIP signal (only the significantly enrichment signal with p-value <0.1) between the isoforms with and without the respective motif. In order to be well powered, we restricted it to only those RBPs with 200 or more predicted motif sites across 50 genes or more, limiting our analysis to the 13 RBPs shown in Fig. 2D. Notably, our results demonstrated that the Rpb4-bound mRNAs were enriched with binding motifs of other RBPs, such as Puf2, Puf4, Puf5, Pub1, Nab2, Nrd1, Pab1, Pin4 and YLL032C (Fig. 2D), which suggests that the interaction between Rpb4 and RBPs may extend beyond Puf3. This suggests that an interaction with specific RBPs is associated with the Rpb4 RIP signal, possibly through the imprinting mechanism. However, we cannot rule out that other interactions between Rpb4 bound mRNAs and other RBPs could exist.
Taken together, the above data indicate a functional relation between Rpb4 and Puf3 in the imprinting of at least a set of common mRNA targets, which is indicative of a coordinated role for these RPBs during this process.
Rpb4-bound mRNAs decay faster
The cells lacking Rpb4 or rpb1 mutants (rpo21-4 and rpb1-84) that affect the Rpb4 association with RNA pol II reduce Rpb4-mRNA imprinting and generally lead to increased mRNA stability. This demonstrates the role of Rpb4 in regulating mRNA stability [7,13,14], which probably depends on the association between Rpb4 and general mRNA decay machinery Pat1/Lsm1-7 [14].
Hence we wondered if a correlation between mRNA stability and its association with Rpb4 could exist. To explore this possibility, we compared our data from the Rpb4-TAP RIP experiments with previously published decay rate data obtained by MIST-Seq (Measurement of Isoform-Specific Turnover using Sequencing), which showed the half-life of polyadenylation or 3ʹ transcript isoform in yeast [44]. Notably as shown in Fig. 3A, the 3ʹ transcript isoforms that were enriched in the Rpb4 RIP experiments had lower half-lives than the overall mRNA. Consequently, these data suggest that the Rpb4-bound mRNAs degrade faster than global mRNA, which falls in line with previous data showing that decreasing mRNA imprinting by Rpb4 provokes increased mRNA stability [10,13].
Figure 3.
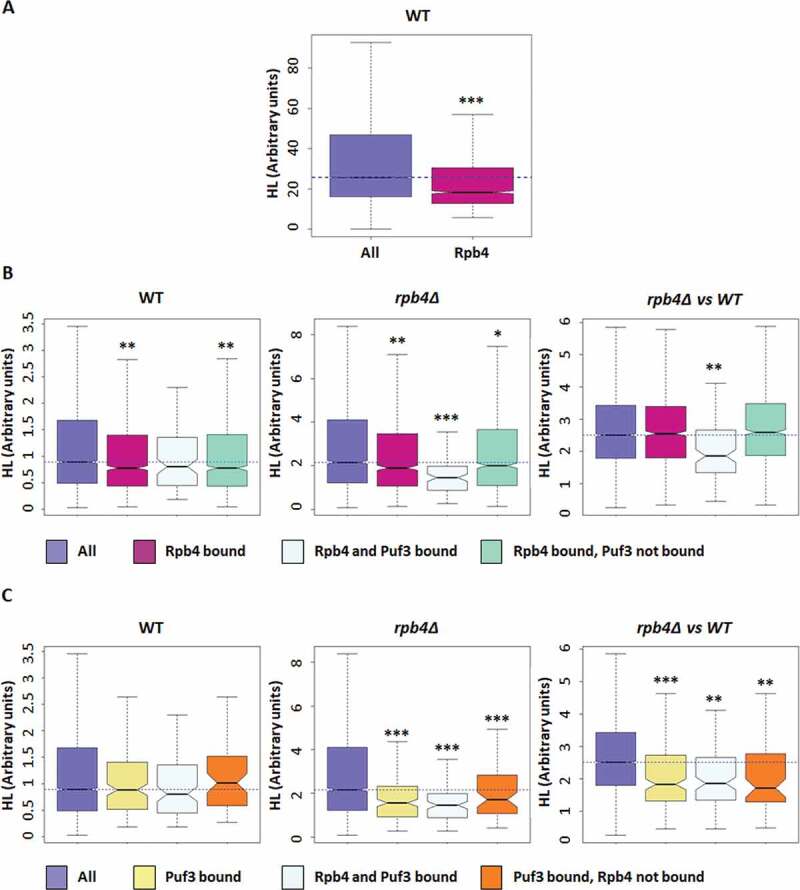
Rpb4-bound mRNAs show a higher decay rate. (A) Boxplots showing the half-lives of the general polyadenylation isoforms of a wild-type strain [44] for the Rpb4-bound mRNAs. (B) Boxplot depicting the half-lives of the Rpb4-bound mRNAs in a wild-type strain (left panel), the rpb4Δ mutant strain (middle panel), or the rpb4Δ/wt ratio (right panel). (C) Boxplot showing the half-lives for the Puf3-bound mRNAs in a wild-type strain (left panel), the rpb4Δ mutant strain (middle panel) or the rpb4Δ/wt ratio (right panel). Datasets used in B and C correspond to [13].* Wilcoxon-test p-value < 10−3; ** p-value < 10−5; *** p-value < 10−9 . Each group of analysed mRNA was compared to the group ‘all’
According to these results, we speculated that Rpb4-bound and Rpb4/Puf3 commonly-bound mRNAs could be differentially affected by lack of Rpb4 at the degradation/stability level in relation to the overall cellular mRNA. To test this hypothesis, we used the published wild-type and rpb4∆ mRNA half-life dataset, from the cells grown in YPD at the permissive temperature of 30 ºC [13], obtained by the GRO method [59]. As seen in Fig. 3B (left panel), corroborating the data in Fig. 3A, the Rpb4-bound mRNAs showed lower half-lives than the overall mRNA in the wild-type strain at the permissive temperature of 30 ºC.
When we analysed the mRNA half-lives in the rpb4Δ mutant strain, a significant general increase in mRNA stability was observed (Fig. 3B, middle panel) in agreement with previous reported data [13]. Strikingly, this increase was significantly less marked for the Rpb4-bound mRNAs that were also bound by Puf3 (Fig. 3B, middle panel). The less marked stability of the Rpb4-bound mRNAs versus the whole population was, however, identical when comparing the cells with (wt) or without (rpb4∆) this protein, except for the Rpb4/Puf3 commonly-bound mRNAs (Fig. 3B, right panel). These results indicate that Rpb4 had no specific effect on the stability of its mRNA targets, but a global effect on the whole transcriptome, except for the targets shared with Puf3. It is worth noting that Puf3 mRNA targets were destabilized in the cells grown exponentially in YPD medium [37,38]. Thus we conclude that the stability of Puf3 targets depends less on Rpb4 than the transcriptome average.
As a set of the Puf3 targets (as previously define) were not bound by Rpb4, we also explored the half-lives of the Puf3-bound mRNAs from the rpb4Δ mutant and its isogenic wild-type strain, using the same datasets. Unlike the Rpb4-bound mRNAs, the Puf3-bound mRNAs showed no significant differences in the half-lives in relation to the overall mRNAs in the wild-type strain (Fig. 3C, left panel). In addition, all the Puf3 targets (both those targeted and not by Rpb4) displayed a similar less marked increase in their stability than the overall mRNA population when Rpb4 lacks (Fig. 3C, middle and right panels). This finding could suggest that the effect of Rpb4 on the Puf3 targets does not require the simultaneous binding of both proteins to mRNAs.
Rpb4 physically and genetically interacts with Puf3
The above data and the demonstrated role of Rpb4 and Puf3 in mRNA stability [7,13,38] suggest a coordinated role of Rpb4 and Puf3 in mRNA imprinting.
To test this hypothesis, we first investigated whether Rpb4 and Puf3 physically interacted. To do so, we performed TAP pulldown from a strain containing a fully functional Puf3-TAP tagged protein and analysed the Rpb4 association using specific anti-Rpb4 antibodies. As shown in Fig. 4A, Rpb4 interacted with Puf3-TAP, although this interaction seemed weak or transitory compared to the positive control employed in the experiment. As a positive control in the experiment, a clear interaction between Rpb4 and Rpb3 (subunit of RNA pol II) was observed when performing Rpb3-TAP tagged protein pulldown. Conversely, and as a negative control, neither non-specific binding between Rpb4 and Imd2-TAP (inosine monophosphate dehydrogenase that catalyzes the rate-limiting step in GTP biosynthesis [60]) nor non-specific adsorption to the resin (no tag control) took place. We also investigated the association of Rpb4 with other PUF (Pumilio and FBF) family members that putatively interacted with the Rpb4-bound mRNAs, such as Puf2, Puf4 and Puf5 (see Fig. 2D). Additionally, we also investigated other RPBs suggested to either interact or not with the Rpb4 bound mRNAs (see Fig. 2D). To do so, we performed pulldowns with the TAP-tagged versions of the different selected RBPs (Fig. 4A). The results suggested a weak interaction between not only Rpb4 and Puf2, but also between Rpb4 and Nsr1, albeit to a lesser extent than with Puf3. Unlike what Fig. 2D suggest, no interaction among Rpb4 and Pub1, Puf4, Puf5, Nrd1 and Vts1 was observed. This suggests that unlike Puf3, other RBPs may not be physically associated with Rpb4, or may be transiently associated or at different times in the mRNA life cycle. It is noteworthy the possibility that these interactions depend on mRNA, at least partially, and/or that the transient association of Rpb4 and RBPs could account for the difficulty in detecting these interactions. Furthermore, in agreement with the Rpb4 and Puf3 physical interaction, a negative genetic interaction between mutants rpb4Δ and puf3Δ was observed as the puf3Δ mutation aggravated the slow growth phenotype of the single rpb4Δ mutant (Fig. 4B).
Figure 4.
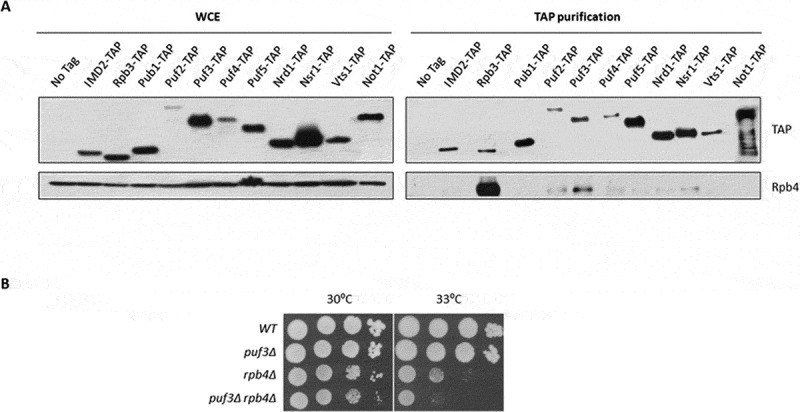
Rpb4 physically and genetically interacts with Puf3. (A). Whole-cell extract and TAP pulldown of the different RBPs TAP-tagged strains grown in YPD medium at 30 ºC. The resulting pulldowns were analysed by western blot with antibodies against Rpb4 and TAP-tag. No tag strain was used as a negative control and the Imd2-TAP strain was employed as a control of non-specific binding. The Rpb3-TAP strain was tested as a positive control for Rpb4 binding. (B) Growth assay of the wild-type and puf3Δ mutant strains in combination with RPB4 deletion in YPD medium at the indicated temperatures. All the experiments were performed at least in duplicate with independent biological replicates
All these data collectively indicate a functional association between Rpb4 and Puf3 that could mediate a common imprinting mRNA mechanism, and suggest that this could also account for other RBPs. Finally, as the Rpb4 and Puf3 interaction was the strongest between Rpb4 and RPBs, we decided to focus on dissecting this interaction.
Puf3 integrity depends on Rpb4
The above data prompted us to investigate the functional relation governing Rpb4 and Puf3 interaction. To explore it, we analysed whether lack of Rpb4 could influence the Puf3 protein level. Protein Puf3-TAP was analysed by western blot in crude extracts of rpb4Δ and its isogenic wild-type strains that contained a fully functional Puf3-TAP tagged protein, and were also grown in YPD at 30 ºC. Strikingly, a much faster migrating band (~92 KDa) appeared in the rpb4Δ strain (Fig. 5A and Supplemental Figure S3), while the full-length Puf3-TAP band (~120 KDa) decreased.
Figure 5.
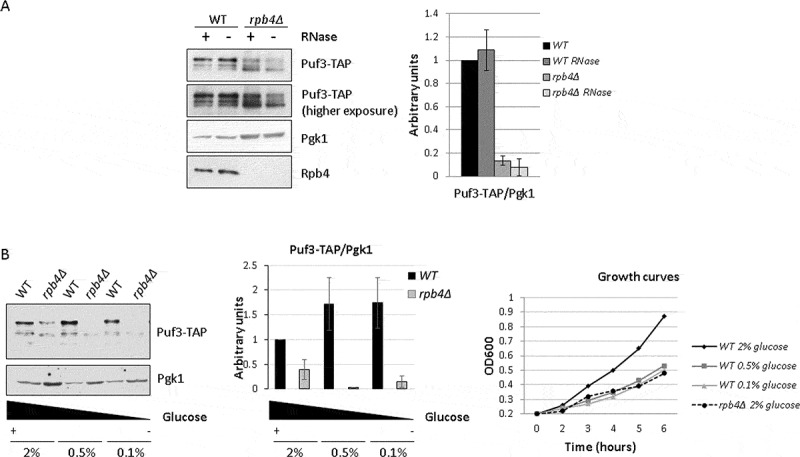
Puf3 integrity and PUF3 mRNA levels depend on Rpb4. (A) Whole-cell extract of the Puf3-TAP tagged wild-type and rpb4Δ mutant strains with and without RNase treatment for 1 h at 37 ºC. Extracts were analysed by western blot with antibodies against Rpb4, TAP-tag and Pgk1 used as a control. (B) Western blot showing the Puf3-TAP and Rpb4 proteins in whole-cell extracts of the Puf3-TAP tagged wild-type and rpb4Δ mutant strains grown in YPD medium at 30 ºC at different glucose concentrations (left panel). Growth curves of the wild-type and rpb4Δ mutant strains in YPD medium containing the indicated glucose concentrations at 30 ºC. Data are shown as the average and standard deviation (SD) of the results of at least three independent experiments with independent biological replicates. Growth curves are representative of two different experiments with independent biological replicates
It has been previously demonstrated that Puf3 is phosphorylated upon glucose depletion to modulate the fate of its target mRNAs from degradation to translation [37]. However, the faster migrating form of Puf3 that we observed did not seem to account for differences in Puf3 phosphorylation as no differences in Puf3 migration were observed upon glucose depletion (see below and Fig. 5B). Moreover, as the TAP tag of Puf3, which is recognized by the anti-TAP antibody, is located at the C-terminal part of the protein, the faster migrating Puf3 band should correspond to the proteolysis of the N-terminal domain, and would probably correspond to part of the Puf3 domain phosphorylated upon glucose depletion [37]. Nevertheless, the faster migration band did not seem to result from only a difference in Puf3 phosphorylation, because incubating the cell crude extracts of the wild-type cells (grown at 30 ºC in YPD) with phosphatase led to neither changes in the amount of the full-length Puf3-TAP band (~120 KDa) nor in the faster migrating band appearing (not shown).
In addition, and in order to rule out the notion that the appearance of the faster migrating band could be caused by the expression of a cryptic transcript leading to the production of a truncated protein [61], we confirmed by RT-qPCR that the production of full length transcript of PUF3 was similar in the WT and rpb4Δ strain (Supplemental Figure S4). These results indicated that the appearance of the ~92 kDa migrating band was not dependent on the internal cryptic promoter.
We also analysed whether the overall amount or Puf3 integrity depended on its association with RNA. To do so, we treated cell extracts with RNAse for 1 h and analysed tagged Puf3-TAP by western blot. As shown in Fig. 5A, RNAse treatment did not significantly affect the Puf3 levels in a wild-type strain. Similarly, the amount of the faster migrating Puf3 band observed when Rpb4 was lacking did not seem to be significantly altered.
The rpb4Δ mutant shows a slow growth phenotype [13,21]. In order to rule out whether the Puf3 faster migrating band could result from an indirect effect due to the slow growth of the rpb4Δ mutant cells, we analysed by western blot the Puf3-TAP protein in a wild-type and rpb4Δ strain growing at different glucose concentrations as the carbon source to modulate their doubling time (Fig. 5B). At 0.5% or 0.1% glucose, the wild-type strain showed a slow growth phenotype comparable to that of the rpb4Δ mutant at 2% glucose (Fig. 5B, right panel). However as we can see in Fig. 5B (left panel), increasing doubling time did not significantly alter Puf3 levels, nor the faster migrating Puf3 band appearing in a wild-type strain, unlike that observed upon RPB4 deletion (Fig. 5B, left panel).
Taken together, lack of Rpb4 led to a faster migrating Puf3 band of about 92 kDa appearing, which suggests Puf3 proteolysis. As this faster migrating band could also be observed in the wild-type strain under certain experimental conditions, we point out that Puf3 could be unstable in some situations that could lead to its proteolysis. According to our results, Rpb4 could act by stabilizing Puf3.
Puf3 integrity depends on Rpb4 associated with chromatin-bound RNA pol II and not on Rpb4-mRNA imprinting
The above data suggest that Rpb4 is necessary for Puf3 integrity. Then, in order to investigate whether the alteration in Puf3 migration could depend on either the Rpb4 subunit bound to chromatin-associated RNA pol II or free Rpb4, we took advantage of the rpb1-84 mutant strain, which affects RNA pol II assembly and partially dissociates Rpb4 from the rest of the enzyme [48]. This mutation leads to different chromatin-associated RNA pol II subcomplexes, some of which lack Rpb4, without altering Rpb4 cellular content [48]. Then the Puf3-TAP protein was analysed by western blot in crude extracts of the rpb1-84 mutant strain and its isogenic wild-type strain containing this TAP-tagged protein. As shown in Fig. 6A and Supplemental Figure S3, the Puf3-TAP faster migrating band (~92 kDa) was also observed in the rpb1-84 mutant strain. These results indicate that the appearance of the faster migrating Puf3 band depends on the Rpb4 association with chromatin-bound RNA pol II. Furthermore, bearing in mind that the rpb1-84 mutant and its wild-type strains have different genetic backgrounds than previously shown (above experiments), these data indicate that this phenomenon is independent of strain’s genetic background.
Figure 6.
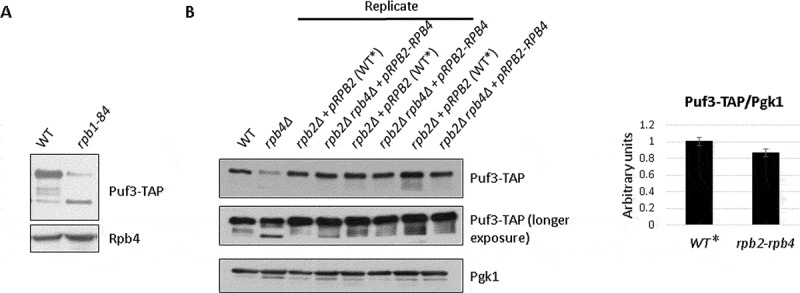
(A) Puf3 integrity is compromised by lack of Rpb4. Whole-cell extract of the Puf3-TAP tagged wild-type and rpb1-84 mutant strains analysed by western blot with antibodies against Rpb4 and TAP-tag. (B) Whole-cell extract of the Puf3-TAP tagged rpb2Δ strain expressing RPB2 from a plasmid (WT*), and the Puf3-TAP tagged rpb2Δ rpb4Δ strain expressing an RPB2-RPB4 fusion gene from the same plasmid analysed by western blot with antibodies against TAP-tag and Pgk1 as a control (left panel). An rpb4Δ mutant strain and its wild-type isogenic strain (WT) were used as a control. Protein quantification by gel densitometry (right panel). Data are shown as the average and standard deviation (SD) of the results of the three shown biological replicates
It has been demonstrated that Rpb4 fused to Rpb2 associates with RNA pol II and overcomes the transcriptional defect of the rpb4Δ mutant strain, [17,21], although only partially recovers its effect on the mRNA degradation rate [17]. Moreover, in the strain that expresses the Rpb2-Rpb4 fusion protein, a small fraction of Rpb2-Rpb4, but also of free Rpb4, are able to imprint mRNAs [17]. So by using this construction, we also explored whether Rpb4-dependent mRNA imprinting would be necessary for Puf3 integrity. To do so, we used two Puf3-TAP tagged stains, the rpb2Δ rpb4Δ strain expressing a Rpb2-Rpb4 fusion protein from a plasmid [17,21] and the isogenic rpb2Δ strain expressing the Rpb2 protein from the same plasmid and Rpb4 from the chromosomic RPB4 gene, considered to be wild type. As shown in Fig. 6B and Supplemental Figure S3, when cells expressed the Rpb2-Rpb4 fusion protein, no significant differences in the amount of the faster migrating Puf3 band in the whole cell crude extracts were observed versus the isogenic cells, as indicated above, considered to be the wild type (Fig. 6B, WT*), which indicates that Rpb2-Rpb4 suffices to maintain Puf3 integrity. As a control, an rpb4Δ strain showed the faster migrating band (~92 kDa), which was not observed in its isogenic wild-type strain (WT). By taking these data and those for the rpb1-84 mutant, we speculate that mRNA imprinting by Rpb4 is not necessary for maintaining Puf3 integrity.
Rpb4 has been proposed to interact with mRNA in the context of RNA pol II during transcription [7,10,13]. In order to decipher whether lack of Puf3 could alter the Rpb4 association with chromatin, we purified chromatin-enriched fractions by the yChEFs procedure [49,50] and analysed the Rpb4 association by western blot. Our results indicated that Rpb4 binding to chromatin did not seem dependent on Puf3 (Fig. 7A), as evidenced by similar Rpb4 levels associated with chromatin in the puf3Δ and wild-type strains. By employing specific antibodies against 3-phosphoglycerate kinase (Pgk1) as a control of cytoplasmic protein, and against histone-3 as a control of chromatin-associated proteins, we observed that chromatin was successfully isolated, and similar chromatin levels were purified and analysed in the wild-type and mutant cells.
Figure 7.
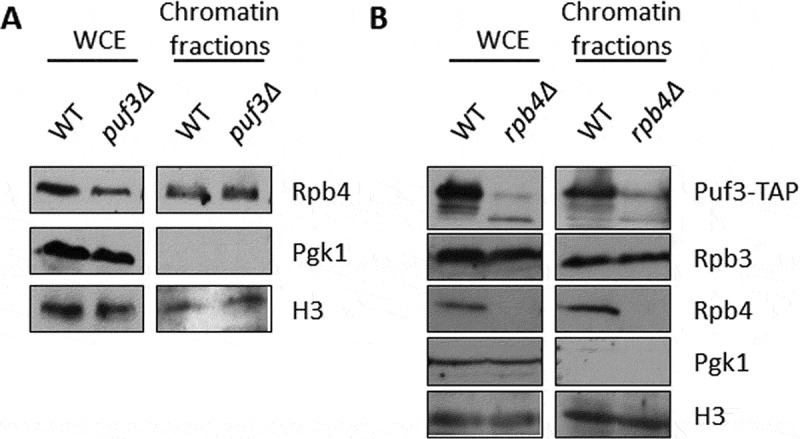
Puf3 and Rpb4 association with chromatin. (A) The whole-cell extract and chromatin-bound proteins isolated by the yChEFs procedure [49,50] from the wild-type and puf3Δ strains grown in YPD medium at 30 ºC were analysed by western blot using specific antibodies against Rpb4, H3 histone used as a positive control and Pgk1 as a negative control of cytoplasmic contamination. (B) Whole-cell extract and chromatin-bound proteins isolated by the yChEFs procedure [49,50] from the Puf3-TAP tagged wild-type and rpb4Δ strains grown in YPD medium at 30 ºC were analysed by western blot using specific antibodies against Rpb4, Rpb3 and TAP, and against H3 histone used as a positive control and Pgk1 employed as a negative control of cytoplasmic contamination. At least three independent experiments with independent biological replicates were performed
Although Puf3 has been described as a cytosolic protein located on the mitochondrial surface [40], we also explored if Puf3 could be associated with chromatin. After following the yChEFs procedure [49,50], Puf3-TAP was analysed by western blot in chromatin-enriched fractions of the rpb4Δ and wild-type strains expressing Puf3-TAP. Strikingly Puf3 was clearly detected in association with chromatin in a wild-type strain (Fig. 7B). Notably, the association of Puf3 with chromatin seemed to depend on Rpb4 as chromatin-associated Puf3 decreased in an rpb4Δ strain, and the lower migrating wild-type Puf3 band mainly appeared. In addition, the faster migrating band also appeared but to a lesser extent.
In order to go further into the chromatin association of Puf3, we performed ChIP-Seq experiment of Puf3-TAP in both a wild-type and an rpb4Δ strain using a no-tag strain as the negative control. Unfortunately, the analysis of three biological replicates showed a similar IP signal to that for the no-tag strain, which suggests that Puf3 did not bind likely chromatin tightly enough to be detected by chromatin immunoprecipitation (Supplemental Figure S5). Our results could also suggest a major indirect association of Puf3 with chromatin via the Rpb4-Puf3 interaction.
Taken together, these results indicated that Puf3 integrity depends on Rpb4-bound to the chromatin-associated RNA pol II and likely, not on the Rpb4-mRNA association. Furthermore, our results also demonstrated that Puf3 associates to chromatin in a similar way.
Rpb4 and Puf3 association with mRNAs depends on one another
As demonstrated above, Rpb4 and Puf3 genetically and physically interact. The Rpb4-RIP experiments showed that Rpb4 binds to Puf3 mRNAs, probably by modulating their stability. To decipher if Rpb4 also modulated the Puf3-mRNA interaction, we performed UV-crosslinking and protein-mRNA isolation experiments [13] to analyse the global association of Puf3 and Rpb4 to mRNAs by western blot in a wild-type strain and the rpb4Δ mutant strains expressing a TAP-tagged version of Puf3. As shown in Fig. 8A, only a very weak Puf3-mRNA association seemed to occur in the cells lacking Rpb4. We also investigated whether lack of Puf3 could influence the Rpb4-mRNA association in a wild-type strain and puf3Δ mutant strains, and demonstrated that the association of Rpb4 with mRNA clearly diminished when Puf3 was lacking (Fig. 8B).
Figure 8.
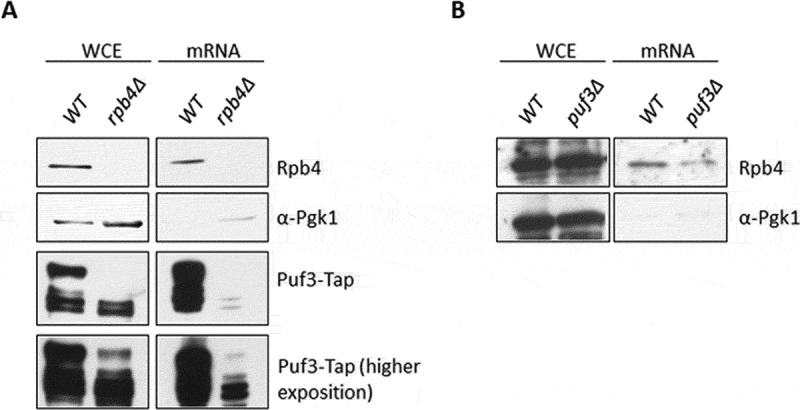
Rpb4 and Puf3 associations with mRNAs depend on each other. (A) Western blot of both whole-cell extracts and oligo-dT purified mRNAs after exposure to 1200 mJ/cm2 of 254 nm UV light to crosslink proteins to mRNAs in the Puf3-TAP tagged wild-type and rpb4Δ strains grown in SD medium at 30 ºC. Western blots were carried out with specific antibodies against Rpb4, Tap-tag and Pgk1 used as a control of cytoplasmic protein (unbound to mRNA). (B) Rpb4 association with mRNAs in a wild-type and puf3Δ strains analysed under similar conditions to those indicated in A. At least three independent experiments with independent biological replicates were performed
Those data indicate that the Rpb4 association with mRNA depends on Puf3, and the Puf3 association with mRNA depends on Rpb4, likely for a group of mRNAs.
Discussion
mRNA synthesis and decay are interconnected and coregulated converting gene expression in a circular system, which changes the classic linear flow view of the Central Dogma of Molecular Biology [8]. This mRNA crosstalk involves not only transcriptional machinery elements that impact mRNA decay, but also others from mRNA decay machinery that influence mRNA synthesis. Only Rpb4, a subunit of RNA pol II, has been clearly shown to be a transcriptional machinery element that participates in the entire mRNA life cycle [5,7,12–14,17]. This role by Rpb4 seems to be also participated by Rpb7, at least in part, as a dissociable dimer from the rest of the enzyme [15,18,62]. We herein demonstrate by RIP-Seq experiments that Rpb4 specifically associates with a large mRNA population. A group of these mRNAs are also target of RBP Puf3 and correspond to nuclear genes for mitochondrial proteins. Our results demonstrate that Rpb4 and Puf3 physically, genetically and functionally interact, and suggest that Rpb4 and Puf3 cooperate to regulate their mRNA association and mRNA stability. Finally, our data indicate that Rpb4 could be a central transcriptional machinery component in the interplay between mRNA synthesis and degradation.
Rpb4-mRNA binding has been previously reported for some specific mRNAs [7,14,17] or more generally [13]. Notably, our results demonstrate that Rpb4 associates with almost 1500 mRNAs, and point out that Rpb4 is a more general ‘imprinting element’ than previously shown [7,14], which falls in line with the proposed role of Rpb4 generally modulating mRNA imprinting and stability [13]. Accordingly, the functional analysis of the GO categories identified different biological processes, the most significantly related ones to mitochondria, but also others that had significant p-values and corresponded to transmembrane transport or metabolic processes. However, the RIP experiments did not detect the specific class of mRNAs that have been previously proposed to be the targets of Rpb4, namely those for RP and RiBi genes [14]. Although these data seem controversial, they could result from the fact that these targets have been established according to the role of Rpb4 in mRNA stability, and not directly based on its mRNA association [7,14]. However, we cannot rule out that Rpb4 imprints all or most mRNAs with different affinities, and the sensitivity of the applied technique was the main limitation to detect those weakly imprinted Rpb4-mRNA targets.
The most significantly identified GO categories establish a specific group of Rpb4-bound mRNA targets related to mitochondrion and mitochondrial translation, which correlate mainly with Pumilio family RPB Puf3-bound mRNAs [37,38,40–42,44,63]. Accordingly, these Rpb4/Puf3 common targets contain the previously described 3ʹUTR motifs for Puf3-mRNA binding [44]. In line with this, the physical and genetic interactions shown herein between Rpb4 and Puf3 corroborate these data. These results indicate that Rpb4 cooperates with RBPs, likely to modulate the mRNA turnover of a specific class of mRNAs, as our data also suggest by showing physical interactions between Rpb4 and Puf2 or Nsr1 [35,36,56,64,65]. Puf2 is also a Pumilio family RBP protein that participates in destabilizing ribosome biogenesis, RiBi, mRNAs [66], whereas Nsr1 is a RiBi component [65] whose mRNA turnover depends on Rpb4 [14]. Interestingly, these data point out the interplay between Rpb4-RPBs cooperation and the role of Rpb4 in mRNA decay of RiBi components, as previously demonstrated [7,14].
A role for Rpb4 as a key element in mRNA synthesis and decay has been clearly established [5,7,10,12–14,17]. Accordingly, the hypothesis that Rpb4 imprints mRNAs and decreases their stability [13,17] falls in line with the fact that Rpb4-bound mRNAs decay faster. However, the change in mRNA stability by lack of Rpb4 does not differ between Rpb4-bound mRNAs and other mRNAs, indicating a general effect of Rpb4 on mRNA degradation and not to specific effects on given groups of mRNAs. Our results also suggest that the simultaneous binding of Rpb4 and Puf3 to mRNAs is not necessary for the effect of Rpb4 on Puf3 mRNA targets.
Our results demonstrate for the first time, that Puf3 binds chromatin, albeit weakly or transiently, and likely imprints mRNAs co-transcriptionally. In addition, both processes seem to depend on Rpb4. In fact lack of Rpb4 affected the amount of both chromatin-associated Puf3 and, as expected, Puf3-mRNA binding. However, on the contrary, chromatin association of Rpb4 is independent of Puf3, since lack of Puf3 does not alter the amount of Rpb4 bound to chromatin.
Strikingly, Rpb4 is necessary to maintain Puf3 integrity as lack of Rpb4 affected Puf3 migration by leading to a faster-migrating band (~92 kDa) to accumulate (Fig. 5 and Supplementary Figure S3). This proteolysis seems to correspond to the amino terminal domain of the protein as the anti-PAP antibody used to detect Puf3-TAP recognizes the C-terminal domain. The amino terminal domain has been shown to be phosphorylated under glucose deprivation by activating the translation of pre-accumulated mRNAs, and quickly favouring mitochondria biogenesis under respiration conditions [38]. This function for the amino terminal domain of the protein accounts for Puf3 switching the fate of their mRNA targets from degradation to translation to facilitate cell acclimation to carbon-source availability and to fermentation/respiration conditions [37,38,41]. Moreover, Puf3 integrity depends on Rpb4 associated with chromatin-bound RNA pol II, and not on the Rpb4-mRNA association, as the Puf3 faster-migrating band does not appear when cells express a Rpb2-Rpb4 fusion protein that remains associated mainly with chromatin [17]. However, we were unable to rule out that the small fraction of Rpb2-Rpb4 or free Rpb4 that imprinted mRNAs in rpb4Δ cells expressing Rpb2-Rpb4 fusion protein could compensate, at least in part, the Puf3 integrity. It is worth noting that the faster-migrating Puf3 band bound chromatin less efficiently than the wild-type band (Fig. 7), and did not seem to bind mRNAs efficiently (Fig. 8). Accordingly, it is tempting to speculate that Rpb4 associates with Puf3 and stabilizes the protein to allow its efficient association to chromatin and its co-transcriptional mRNA imprinting. Furthermore, we also hypothesize that the low stability of Puf3 mRNA targets on the cells growing with glucose as a carbon source could be reached by the concerted action of Puf3 with Rpb4 because Rpb4-imprinted mRNAs decay faster and lack of Rpb4 increases mRNA stability.
Therefore, we speculated that Puf3-mRNA imprinting depended on Rpb4, probably as a consequence of Puf3 integrity. Similarly, Rpb4 mRNA imprinting would also be Puf3-dependent. We speculated that this dependence had to occur for commonly Rpb4- and Puf3- bound mRNAs, and could similarly take place for different classes of mRNAs with the concerted participation of Rpb4 and other RBPs based on the observed physical interaction between Rpb4 and other RPBs.
Taken together, our results showed that Rpb4 and Puf3 cooperated to regulate their mRNA association and mRNA degradation, and likely mRNA imprinting, at least for a common set of mRNAs associated with mitochondria. This could also occur for Rpb4 in concert with other RPBs.
Our results herein allowed us to hypothesize a functional model for Rpb4-Puf3 cooperation in mRNA stability. Puf3 bound to chromatin in an Rpb4-dependent manner, which also determined co-transcriptional Puf3-mRNA imprinting and influenced cytoplasmic mRNA degradation. Furthermore, Rpb4 imprinted a large set of mRNAs, part of which is common with Puf3. Notwithstanding, Rpb4 was able to modulate the cytoplasmic functions of mRNA degradation machinery. In line with this, the cooperation between Rpb4 and mRNA decay machinery, such as Pat1-Lsm1/7, has been proposed to mediate mRNA decay [14]. Rpb4-bound mRNAs have lower half-lives than overall mRNA and lack of Rpb4 increases mRNA stability. However, this increase did not seem to specifically occur for the set of Rpb4-bound mRNAs. This could be due to Rpb4’s very low level of imprinting in the rest of the transcriptome or because the effect of Rpb4 on global mRNA stability does not directly depend on mRNA imprinting. In any case, mRNA imprinting by Rpb4 seems related to mRNA stability if we consider that the mutants affecting the dissociation of Rpb4, or of dimer Rpb4/7 from RNA pol II, are also associated with lower mRNA imprinting and higher mRNA stability [13,17]. Notably however, Puf3-mRNA imprinting impacts mRNA stability as the stability of all Puf3-bound mRNAs increases upon Puf3 deletion [38], and also because the increase in mRNA stability provoked by lack of Rpb4 is attenuated for the Puf3-bound mRNAs. We speculated that the alteration of Puf3 integrity provoked by lack of Rpb4, which diminished Puf3-mRNA imprinting, negatively affected the increased Puf3-bound mRNA stability to a lower extent than for either other Rpb4-bound mRNAs or overall mRNAs, likely by the abnormal concerted action of Puf3 and mRNA degradation machinery.
Furthermore, by considering that the concerted action among Puf3, Pab1 and the Ccr4-Not complex has been shown to act on mRNA poly(A) tails to regulate eukaryotic mRNA stability and translation [11,67,68], we also speculated that Rpb4-Puf3 cooperation could serve to influence Ccr4-Not activity in mRNA degradation. Notably, the Puf3 binding motif is overrepresented in the most highly enriched Ccr4 targets [69]. Similarly to that occurs for Rpb4 over Puf3 integrity, Rpb4 could act on other elements of the mRNA degradation machinery and acting in concert on mRNA stability.
Finally, it would be interesting to analyse the possible mRNA targets of Rpb4 under different growth conditions to establish the most precise role of this protein as a central transcriptional machinery element that connects the mRNA degradation process.
Supplementary Material
Acknowledgments
We thank the “Servicios Centrales de Apoyo a la Investigación (SCAI)” of the University of Jaen for technical support. We thank Olga Calvo for sharing with us the S. pombe spike in.
Funding Statement
This work has been supported by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) and ERDF (BFU2016-77728-C3-2-P to F.N. and BFU2016-77728-C3-3-P to J.E.P-O), Spanish Ministry of Science and Innovation (MICINN) and ERDF (RED2018-102467-T to F. N. and J.E.P-O), the Junta de Andalucía (BIO258 to F. N.), Junta de Andalucía-Universidad de Jaén (FEDER-UJA 1260360 to F.N.), the Generalitat Valenciana (AICO/2019/088 to J.E.P-O) and DFG grant (STE 1422/4-1) to L.M.S. VP’s laboratory is funded by the Swedish Research Council (VR 2016-01842), a Wallenberg Academy Fellowship (KAW 2016.0123), the Swedish Foundations’ Starting Grant (Ragnar Söderberg Foundation), Karolinska Institutet (SciLifeLab Fellowship, SFO, KID and KI funds). IG is funded by a Seed funding grant provided by the Indian Institute of Technology, Delhi. A.I.G-G was financed by the University of Jaen, MINECO and ERDF funds (BFU2016-77728-C3-2-P to F.N.). F.G-S. is a recipient of a predoctoral fellowship from the Universidad de Jaén.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Pérez-Ortín JE, Alepuz P, Chávez S, et al. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425:3750–3775. [DOI] [PubMed] [Google Scholar]
- [2].Sun M, Schwalb B, Schulz D, et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Braun KA, Young ET.. Coupling mRNA synthesis and decay. Mol Cell Biol. 2014;34:4078–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Begley V, Corzo D, Jordán-Pla A, et al. The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4. Nucleic Acids Res. 2019;47:9524–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Choder M. mRNA imprinting: additional level in the regulation of gene expression. Cell Logist. 2011;1:37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dahan N, Choder M. The eukaryotic transcriptional machinery regulates mRNA translation and decay in the cytoplasm. Biochim Biophys Acta. 2013;1829:169–173. [DOI] [PubMed] [Google Scholar]
- [7].Goler-Baron V, Selitrennik M, Barkai O, et al. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008;22:2022–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Haimovich G, Medina DA, Causse SZ, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. [DOI] [PubMed] [Google Scholar]
- [9].Harel-Sharvit L, Eldad N, Haimovich G, et al. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2010;143:552–563. [DOI] [PubMed] [Google Scholar]
- [10].Shalem O, Groisman B, Choder M, et al. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet. 2011;7:e1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moqtaderi Z, Geisberg JV, Struhl K. Extensive structural differences of closely related 3ʹ mRNA isoforms: links to Pab1 binding and mRNA stability. Mol Cell. 2018;72:849–61e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Farago M, Nahari T, Hammel C, et al. Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol Biol Cell. 2003;14:2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garrido-Godino AI, García-López MC, García-Martínez J, et al. Rpb1 foot mutations demonstrate a major role of Rpb4 in mRNA stability during stress situations in yeast. Biochim Biophys Acta. 2016;1859:731–743. [DOI] [PubMed] [Google Scholar]
- [14].Lotan R, Bar-On VG, Harel-Sharvit L, et al. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 2005;19:3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lotan R, Goler-Baron V, Duek L, et al. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J Cell Biol. 2007;178:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kumar D, Varshney S, Sengupta S, et al. A comparative study of the proteome regulated by the Rpb4 and Rpb7 subunits of RNA polymerase II in fission yeast. J Proteomics. 2019;199:77–88. [DOI] [PubMed] [Google Scholar]
- [17].Duek L, Barkai O, Elran R, et al. Dissociation of Rpb4 from RNA polymerase II is important for yeast functionality. PLoS One. 2018;13:e0206161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Selitrennik M, Duek L, Lotan R, et al. Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot Cell. 2006;5:2092–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Edwards AM, Kane CM, Young RA, et al. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- [20].Sampath V, Balakrishnan B, Verma-Gaur J, et al. Unstructured N terminus of the RNA polymerase II subunit Rpb4 contributes to the interaction of Rpb4.Rpb7 subcomplex with the core RNA polymerase II of Saccharomyces cerevisiae. J Biol Chem. 2008;283:3923–3931. [DOI] [PubMed] [Google Scholar]
- [21].Schulz D, Pirkl N, Lehmann E, et al. Rpb4 subunit functions mainly in mRNA synthesis by RNA polymerase II. J Biol Chem. 2014;289:17446–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Allepuz-Fuster P, O’Brien MJ, González-Polo N, et al. RNA polymerase II plays an active role in the formation of gene loops through the Rpb4 subunit. Nucleic Acids Res. 2019;47:8975–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Forget A, Chartrand P. Cotranscriptional assembly of mRNP complexes that determine the cytoplasmic fate of mRNA. Transcription. 2011;2:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Villanyi Z, Ribaud V, Kassem S, et al. The Not5 subunit of the Ccr4-Not complex connects transcription and translation. PLoS Genet. 2014;10:e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li T, De Clercq N, Medina DA, et al. The mRNA cap-binding protein Cbc1 is required for high and timely expression of genes by promoting the accumulation of gene-specific activators at promoters. Biochim Biophys Acta. 2016;1859:405–419. [DOI] [PubMed] [Google Scholar]
- [26].Glisovic T, Bachorik JL, Yong J, et al. RNA‐binding proteins and post‐transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533. [DOI] [PubMed] [Google Scholar]
- [28].Halbeisen RE, Galgano A, Scherrer T, et al. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci. 2008;65:798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang H, Xu L, Wang Z, et al. Coordinating expression of RNA binding proteins with their mRNA targets. Sci Rep. 2014;4:7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Baltz AG, Munschauer M, Schwanhäusser B, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. [DOI] [PubMed] [Google Scholar]
- [31].Mitchell SF, Jain S, She M, et al. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Khong A, Parker R. The landscape of eukaryotic mRNPs. RNA. 2020;26:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Holmqvist E, Vogel J. RNA-binding proteins in bacteria. Nat Rev Microbiol. 2018;16:601–615. [DOI] [PubMed] [Google Scholar]
- [34].Klass DM, Scheibe M, Butter F, et al. Quantitative proteomic analysis reveals concurrent RNA-protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae. Genome Res. 2013;23:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. [DOI] [PubMed] [Google Scholar]
- [36].Wickens M, Bernstein DS, Kimble J, et al. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18:150–157. [DOI] [PubMed] [Google Scholar]
- [37].Lee CD, Tu BP. Glucose-Regulated phosphorylation of the PUF protein Puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep. 2015;11:1638–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Miller MA, Russo J, Fischer AD, et al. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Res. 2014;42:3954–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Voronina E. Diverse roles of PUF proteins in germline stem and progenitor cell development in C. elegans. Front Cell Dev Biol. 2020;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].García-Rodríguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lapointe CP, Stefely JA, Jochem A, et al. Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 2018;6:125–35e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Saint-Georges Y, Garcia M, Delaveau T, et al. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PloS One. 2008;3:e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].García-López MC, Mirón-García MC, Garrido-Godino AI, et al. Overexpression of SNG1 causes 6-azauracil resistance in Saccharomyces cerevisiae. Curr Genet. 2010;56:251–263. [DOI] [PubMed] [Google Scholar]
- [44].Gupta I, Clauder-Munster S, Klaus B, et al. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol Syst Biol. 2014;10:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wilkening S, Pelechano V, Jarvelin AI, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mirón-García MC, Garrido-Godino AI, García-Molinero V, et al. The prefoldin Bud27 mediates the assembly of the eukaryotic RNA polymerases in an Rpb5-dependent manner. PLoS Genet. 2013;9:e1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Garrido-Godino AI, García-López MC, Navarro F. Correct assembly of RNA polymerase II Depends on the foot domain and Is required for multiple steps of transcription in Saccharomyces cerevisiae. Mol Cell Biol. 2013;33:3611–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cuevas-Bermúdez A, Garrido-Godino A, Navarro F. A novel yeast chromatin-enriched fractions purification approach, yChEFs, for the chromatin-associated protein analysis used for chromatin-associated and RNA-dependent chromatin-associated proteome studies from Saccharomyces cerevisiae. Gene Rep. 2019;16:100450. [Google Scholar]
- [50].Cuevas-Bermúdez A, Garrido-Godino AI, Gutiérrez-Santiago F, et al. Fractions purification approach, yChEFs, from Saccharomyces cerevisiae. Bio-protocol . 2020;10:e3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Coulombe C, Poitras C, Nordell-Markovits A, et al. VAP: a versatile aggregate profiler for efficient genome-wide data representation and discovery. Nucleic Acids Res. 2014;42:W485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kershaw CJ, Costello JL, Talavera D, et al. Integrated multi-omics analyses reveal the pleiotropic nature of the control of gene expression by Puf3p. Sci Rep. 2015;5:15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wilinski D, Buter N, Klocko AD, et al. Recurrent rewiring and emergence of RNA regulatory networks. Proc Nat Acad Sci. 2017;114:E2816–E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Freeberg MA, Han T, Moresco JJ, et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013;14:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hogan DJ, Riordan DP, Gerber AP, et al. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell. 2004;15:303–313. [DOI] [PubMed] [Google Scholar]
- [60].Escobar-Henriques M, Daignan-Fornier B. Transcriptional regulation of the yeast gmp synthesis pathway by its end products. J Biol Chem. 2001;276:1523–1530. [DOI] [PubMed] [Google Scholar]
- [61].Wei W, Hennig BP, Wang J, et al. Chromatin-sensitive cryptic promoters putatively drive expression of alternative protein isoforms in yeast. Genome Res. 2019;29:1974–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem Sci. 2004;29:674–681. [DOI] [PubMed] [Google Scholar]
- [63].Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, et al. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011;17:1551–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hogan GJ, Brown PO, Herschlag D. Evolutionary conservation and diversification of Puf RNA binding proteins and their mRNA targets. PLoS Biol. 2015;13:e1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lee WC, Zabetakis D, Melese T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fischer AD, Olivas WM. Multiple Puf proteins regulate the stability of ribosome biogenesis transcripts. RNA Biol. 2018;15:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Webster MW, Stowell JA, Passmore LA. RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not. eLife. 2019;8. DOI: 10.7554/eLife.40670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. Embo J. 2000;19:6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Miller JE, Zhang L, Jiang H, et al. Genome-Wide Mapping of Decay Factor-mRNA Interactions in Yeast Identifies Nutrient-Responsive Transcripts as Targets of the Deadenylase Ccr4. G3 (Bethesda). 2018;8:315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


