ABSTRACT
Antibiotic resistant Klebsiella pneumoniae is a leading public health threat and gastrointestinal carriage is an established risk factor for subsequent infections during hospitalization. Our study contributes new knowledge of risk factors for gastrointestinal carriage and the genomic population structure of K. pneumoniae colonizing humans in a representative sample of a general population in a community setting. Altogether, 2,975 participants (54% women) >40 y in the population-based Tromsø Study: Tromsø7, Norway (2015–2016) were included. Fecal samples were screened for K. pneumoniae, which were characterized using whole-genome sequencing. Risk factors for carriage were analyzed using multivariable logistic regression on data from questionnaires and the Norwegian Prescription Database. Prevalence of K. pneumoniae gastrointestinal carriage was 16.3% (95% CI 15.0–17.7, no gender difference). Risk factors associated with carriage included age ≥60 y, travel to Greece or Asia past 12 months (adjusted odds ratio 1.49, 95% CI 1.11–2.00), Crohn’s disease/ulcerative colitis (2.26, 1.20–4.27), use of proton pump inhibitors (1.62, 1.18–2.22) and non-steroidal anti-inflammatory drugs past 6 months (1.38, 1.04–1.84), and antibiotic use the last month (1.73, 1.05–2.86). Prevalence was higher among those having used combinations of drug classes and decreased over time with respect to preceding antibiotic use. The K. pneumoniae population was diverse with 300 sequence types among 484 isolates distributed across four phylogroups. Only 5.2% of isolates harbored acquired resistance and 11.6% had virulence factors. Identification of risk factors for gastrointestinal carriage allows for identification of individuals that may have higher risk of extraintestinal infection during hospitalization. The findings that specific diseases and drugs used were associated with carriage show an impact of these possibly through modulating the human gut microbiota promoting colonization. The diverse population structure of carriage isolates reflects the ecologically adaptive capacity of the bacterium and challenges for vaccine prospects and the identification of reservoirs as a potential source for human colonization.
KEYWORDS: Klebsiella pneumoniae, carriage, risk factors, general population, bacterial genomics
Introduction
Klebsiella pneumoniae (Kp) is a key pathogen associated with nosocomial infections frequently accompanied by antibiotic resistance.1,2 As an opportunistic pathogen, Kp is particularly problematic among neonates, elderly, immunocompromised, and patients with underlying chronic diseases, and commonly causes pneumonia, urinary tract infections, and bacteremia.1 Additionally, the problem is exacerbated by the emergence and spread of community-acquired hypervirulent Kp causing infections in healthy individuals, usually presenting as pyogenic liver abscess occasionally accompanied with metastatic spread, but also as meningitis or endophthalmitis.3
Kp is known for its high prevalence and diversity of antibiotic resistance genes that challenge treatment options due to infections with multi-drug resistant variants. In Europe, antibiotic resistant Kp was responsible for more than 89,000 infections and 5,600 attributable deaths in 2015.4 Several new antibiotic resistance genes were first discovered within Kp before spreading to other pathogens.5 Consequently, The World Health Organization considers antibiotic-resistant Kp as a critical-priority bacterium in antibiotic research and development.6
Recent taxonomic updates show that Kp subdivides into five different species comprising seven phylogroups (Kp1-Kp7) referred to as the K. pneumoniae species complex (KpSC).7,8 The phylogroups include K. pneumoniae sensu stricto (Kp1), K. quasipneumoniae subsp. quasipneumoniae (Kp2), K. variicola subsp. variicola (Kp3), K. quasipneumoniae subsp. similipneumoniae (Kp4), K. variicola subsp. tropica (Kp5), ‘K. quasivariicola’ (Kp6) and K. africana (Kp7).7–12 Herein, we refer to “Kp” for all members of the KpSC unless otherwise specified. Kp has a broad environmental distribution, and transmission routes to humans are currently not well defined.8
Gastrointestinal carriage of Kp as a reservoir for healthcare-associated Kp infections was established in the early 1970s.13 Recent genomic studies show that gastrointestinal carriage is a risk factor for subsequent extraintestinal infections, and ~50% of Kp bloodstream infections are caused by the patient’s own gut isolates.14,15 Moreover, the relative abundance of Kp in the gastrointestinal tract is associated with an increased risk of Kp bacteremia.16
Cross-sectional studies have shown that Kp gastrointestinal carriage prevalence varies from 6% to 88% depending on geographical locations, detection methods and the populations investigated.14,15,17–19 However, we have a sparse understanding of risk factors for Kp gastrointestinal carriage and the population structure of Kp in the general human population. In a recent cross-sectional study of 911 pregnant women in low-income countries, Huynh et al. identified various country-specific environmental exposure factors linked to Kp gastrointestinal carriage and a diverse Kp population structure.19
Here, we investigated the prevalence of Kp carriage and associated risk factors among 2,975 participants in a cross-sectional study of a representative sample of a general adult population in Norway, a country with a low prevalence of antibiotic resistance and restricted antibiotic use. Additionally, we elucidated the Kp genomic population structure of carriage isolates.
Results
We analyzed data from 2,975 participants (1,615 women, 54.3%, Suppl. Table 1) 40 y and older. Median age of the participants was 65.0 y (Interquartile Range 59–71 y, no gender difference). Altogether, we identified 484 Kp carriers corresponding to a prevalence of 16.2% (95% CI 14.5–18.1) among women and 16.3% (14.4–18.4) among men.
Kp gastrointestinal carriage and associated factors
In analysis adjusting for all of the explanatory variables (Suppl. Figure 1), Kp gastrointestinal carriage was associated with age 60 y and older (compared to the reference group 40–49 y), self-reported travel to Greece or Asia during the preceding 12 months (AOR 1.49, 1.11–2.00) and Crohn’s disease/ulcerative colitis (2.26, 1.20–4.27) (Table 1). Furthermore, carriage was associated with the use of proton pump inhibitors (PPIs) (1.62, 1.18–2.22) and non-steroidal anti-inflammatory drugs (NSAIDs) (1.38, 1.04–1.84) in the past 6 months, and antibiotic use in the last month (1.73, 1.05–2.86).
Table 1.
K. pneumoniae (Kp) gastrointestinal carriage and associated factors among 2,975 participants in the Tromsø Study: Tromsø7 in crude and multivariable logistic regression analyses
| % (Kp) | n (Kp) | N | OR | 95% CI | p-Value | AOR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|---|---|---|
| Age (y) | 0.024 | 0.128 | |||||||
| 40–49 | 10.8 | 37 | 344 | 1.00 | 1.00 | ||||
| 50–59 | 15.4 | 67 | 435 | 1.51 | 0.98–2.32 | 1.34 | 0.85–2.11 | ||
| 60–69 | 17.3 | 222 | 1,286 | 1.73 | 1.20–2.51 | 1.56 | 1.06–2.30 | ||
| 70–84 | 17.4 | 158 | 910 | 1.74 | 1.19–2.55 | 1.56 | 1.03–2.36 | ||
| Current daily smoking | 0.798 | 0.717 | |||||||
| No | 16.2 | 421 | 2,598 | 1.00 | 1.00 | ||||
| Yes | 15.7 | 55 | 351 | 0.96 | 0.71–1.31 | 0.94 | 0.66–1.34 | ||
| Alcohol consumption frequency | 0.021 | 0.050 | |||||||
| Never to < monthly | 16.4 | 164 | 998 | 1.00 | 1.00 | ||||
| 2–4/month to 2–3/week | 16.9 | 298 | 1,765 | 1.03 | 0.84–1.27 | 1.13 | 0.88–1.46 | ||
| ≥4/week | 9.1 | 18 | 198 | 0.51 | 0.31–0.85 | 0.61 | 0.35–1.05 | ||
| Alcohol units/occasion | 0.501 | 0.365 | |||||||
| 1–4 | 16.2 | 412 | 2,540 | 1.00 | 1.00 | ||||
| ≥5 | 14.1 | 20 | 142 | 0.85 | 0.52–1.37 | 0.79 | 0.47–1.32 | ||
| Travel abroad past 12 monthsa | 0.024 | 0.009 | |||||||
| No | 15.5 | 196 | 1,276 | 1.00 | 1.00 | ||||
| Greece or Asia | 20.3 | 102 | 502 | 1.39 | 1.07–1.82 | 1.49 | 1.11–2.00 | ||
| All other countries | 15.3 | 179 | 1,171 | 0.99 | 0.79–1.23 | 0.97 | 0.76–1.26 | ||
| Hospitalization past 12 months | 0.145 | 0.844 | |||||||
| No | 15.9 | 412 | 2,588 | 1.00 | 1.00 | ||||
| Yes | 19.0 | 67 | 353 | 1.24 | 0.93–1.65 | 1.03 | 0.74–1.44 | ||
| Diabetes mellitusb | 0.011 | 0.056 | |||||||
| No | 16.0 | 427 | 2,674 | 1.00 | 1.00 | ||||
| Yes | 23.5 | 40 | 170 | 1.62 | 1.12–2.34 | 1.82 | 0.99–3.36 | ||
| Crohn’s disease/ulcerative colitis | 0.012 | 0.012 | |||||||
| No | 16.0 | 452 | 2,831 | 1.00 | 1.00 | ||||
| Yes | 28.3 | 17 | 60 | 2.08 | 1.18–3.68 | 2.26 | 1.20–4.27 | ||
| Proton pump inhibitors past 6 monthsc | <0.001 | 0.003 | |||||||
| No | 15.3 | 404 | 2,632 | 1.00 | 1.00 | ||||
| Yes | 23.3 | 80 | 343 | 1.68 | 1.28–2.20 | 1.62 | 1.18–2.22 | ||
| NSAIDs past 6 monthsd | 0.016 | 0.028 | |||||||
| No | 15.6 | 397 | 2,545 | 1.00 | 1.00 | ||||
| Yes | 20.2 | 87 | 430 | 1.37 | 1.06–1.78 | 1.38 | 1.04–1.84 | ||
| Antibiotic systemic use past 1 monthe | 0.001 | 0.032 | |||||||
| No | 15.8 | 453 | 2,866 | 1.00 | 1.00 | ||||
| Yes | 28.4 | 31 | 109 | 2.12 | 1.38–3.25 | 1.73 | 1.05–2.86 | ||
| Metformin past 6 monthsf | 0.090 | 0.764 | |||||||
| No | 16.0 | 460 | 2,867 | 1.00 | 1.00 | ||||
| Yes | 22.2 | 24 | 108 | 1.50 | 0.94–2.38 | 0.89 | 0.40–1.95 | ||
| Thyroid hormones past 6 monthsg | 0.065 | 0.333 | |||||||
| No | 15.9 | 431 | 2,714 | 1.00 | 1.00 | ||||
| Yes | 20.3 | 53 | 261 | 1.35 | 0.98–1.86 | 1.20 | 0.83–1.75 |
N, denominator; OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio; NSAIDs, non-steroidal anti-inflammatory drugs.
AOR adjusted for age, current daily smoking, alcohol consumption frequency, alcohol units/occasion, travel abroad past 12 months, hospitalization past 12 months, diabetes mellitus, Crohn’s disease/ulcerative colitis and drug use according to the Norwegian Prescription Database (A02BC, M01, J01, A07AA09, P01AB01, A10BA02, H03AA).
The multivariable model contains 2,446 participants with complete information on all variables.
aTraveled outside the Nordic countries >1 week duration in the past 12 months.
b20 participants who answered “Yes, previously” were excluded.
cA02BC, drugs used for peptic ulcer and gastro-esophageal reflux disease.
dM01, anti-inflammatory and anti-rheumatic products (non-steroids), anti-inflammatory/anti-rheumatic agents in combination and specific anti-rheumatic agents.
eJ01, A07AA09, P01AB01, antibacterials for systemic use, intestinal antiinfectives and nitroimidazole derivates used as antiprotozoals (metronidazole).
fA10BA02, blood glucose lowering drug used in diabetes.
gH03AA, natural and synthetic thyroid hormones.
Analysis of the significant variables in the multivariable model for the three most frequent species (Kp1, Kp2 and Kp3) is presented in Suppl. Table 2. In this descriptive analysis, we compared participants colonized by a single phylogroup to Kp non-carriers, excluding carriers of any other phylogroup. Kp1 was the dominating phylogroup responsible for the associations to selected variables, except for NSAID use which is solely significantly associated with Kp2. Kp3 is associated with PPI and antibiotic use.
The Kp prevalence of the three statistically significantly associated drug classes (usage in past 6 months) in relation to Kp carriage is shown in Figure 1. Kp prevalence was 16.1% among 286 antibiotic-only users, 17.4% among 305 NSAID-only users, and 21.5% among 209 PPI-only users. Kp prevalence increased in each overlapping area for two drug classes, and further increased to 30.8% in the overlapping area for all three.
Figure 1.
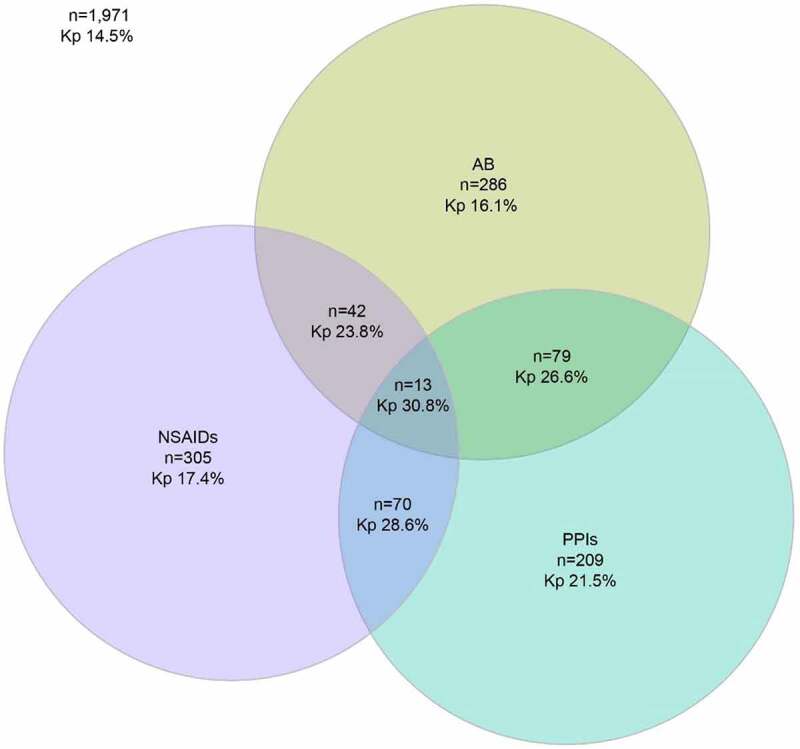
Proportional Venn diagram of Kp carriage prevalence related to statistically significantly associated drug classes (antibiotics (AB), nonsteroidal anti-inflammatory drugs (NSAIDs) and proton pump inhibitors (PPIs)) in the past 6 months among 2,975 study participants
Looking at the cumulative change in proportion of Kp carriers associated with antibiotic use during 1–12 months before fecal sampling, we found that Kp carriage prevalence was highest among those with antibiotic use in the past month (28.4%) and past 2 months (25.0%), decreasing to around 20.0% in the past 6–12 months (Figure 2). In contrast, prevalence of Kp carriage was significantly lower (15.2%) in the non-antibiotic using population.
Figure 2.
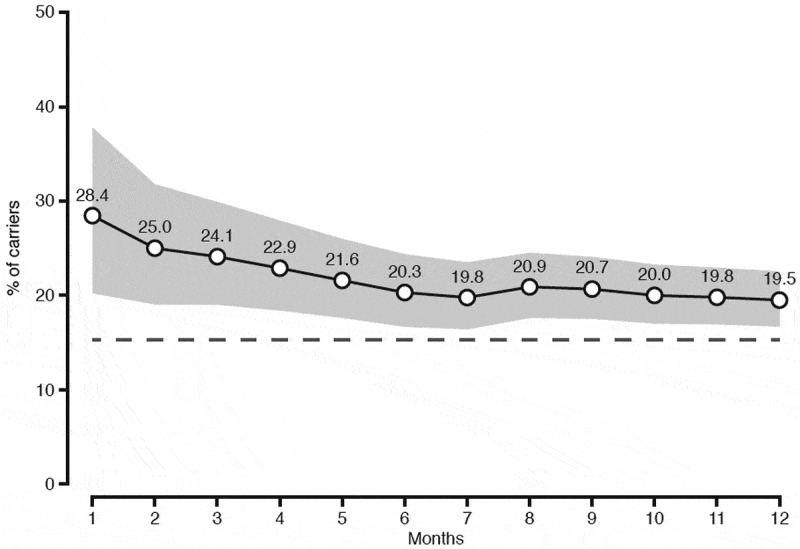
Cumulative change in the proportion of Kp carriers among those who had used antibiotics 1–12 months before the fecal sampling. Shaded gray area represents the 95% CI. The time period at each specified month includes data for the preceding months. The dashed black line indicates the prevalence of carriage in the non-antibiotic using population (15.2%)
Kp phylogeny and diversity
Phylogenetic analysis based on whole-genome sequencing of 484 K. pneumoniae isolates identified three species distributed in four phylogroups with a predominance of K. pneumoniae sensu stricto (Kp1, 62.6%), followed by K. variicola subsp. variicola (Kp3, 27.7%), K. quasipneumoniae subsp. quasipneumoniae (Kp2, 6.4%), and K. quasipneumoniae subsp. similipneumoniae (Kp4, 3.3%) (Figure 3, Suppl. Table 3). Phylogroups Kp5-Kp7 were not identified. Using multilocus sequence typing (MLST), we found a high degree of genotypic diversity (Simpson diversity index 99.5%) with a total of 300 different STs, including 96 (32%) novel STs (Suppl. Figure 2).20 The majority (79%) of the STs were represented by a single isolate. Only 17 STs (5.7%) were represented by more than five isolates. The most frequent were ST20 (n = 15, 3.1%), ST26 (n = 13, 2.7%), ST35 (n = 9, 1.9%), ST37 (n = 9, 1.9%), and ST2386 (n = 9, 1.9%). With regard to clonal relatedness, we found seven ST35 and three ST25 isolates with a range of zero to five SNPs (Suppl. Table 4), and seven to eight SNPs (Suppl. Table 5), respectively, indicating a recent clonal spread. No close clonal relatedness was identified among the remaining frequent STs (Suppl. Table 6).
Figure 3.
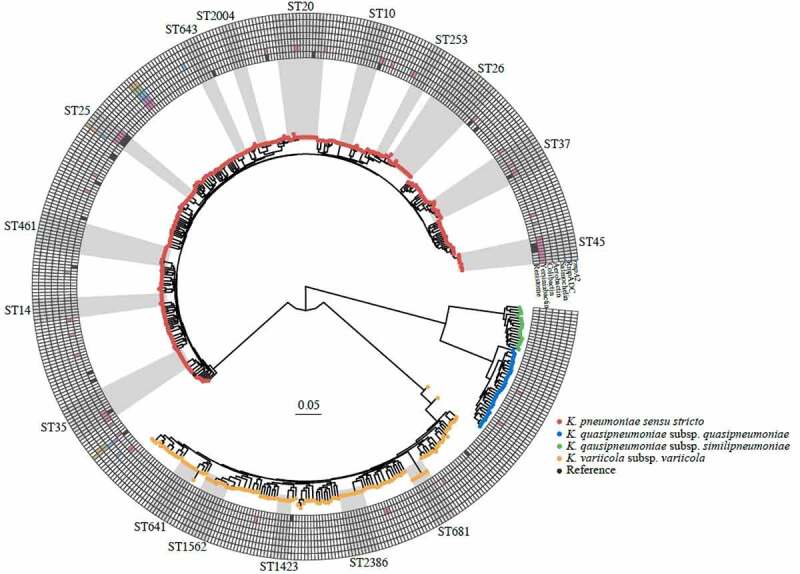
Core chromosomal maximum likelihood phylogeny of the 484 Kp genomes. The tips are colored by species. The heatmap shows presence (color) or absence (white) of acquired resistance genes (innermost ring) or virulence factors (remaining six rings). Clades corresponding to STs with five or more genomes are highlighted and labeled
Antimicrobial resistance and plasmid content
The prevalence of resistance was low and none of the isolates were resistant to cefotaxime, meropenem, aztreonam, or ciprofloxacin (Suppl. Figure 3). Resistance was observed against amoxicillin-clavulanic acid (0.6%), ceftazidime (0.2%), gentamicin (0.4%), cefuroxime (0.6%), trimethoprim-sulfamethoxazole (1.7%), piperacillin-tazobactam (4.8%), and mecillinam (5.0%). This was in concordance with the low number of intrinsic and acquired resistance genes (Figure 3, Suppl. Table 3). A high sequence diversity of species-specific intrinsic narrow-spectrum chromosomal β-lactamase gene alleles were identified (Suppl. Figures 4, 5, 6, and 7, Suppl. Table 3). Three ampicillin-susceptible isolates harbored either deleted blaSHV/blaLEN genes or a premature stop codon in blaSHV. Only 5.2% of the isolates harbored an acquired resistance gene (median number: 2, Suppl. Table 3). BlaSHV-51 and blaSHV-52 were assigned by Kleborate as extended-spectrum β-lactamase and inhibitor-resistant β-lactamase, respectively. However, both isolates were phenotypically susceptible to all tested β-lactams and β-lactam-inhibitor combinations. Thirty-two plasmid replicon types were identified among 393/484 (81.2%) isolates with IncFIB(K) (25.2%), Col (pHAD28) (15.0%), IncFIB(K) (pCAV1099-114) (11.4%), IncFIA (HI1) (10.5%), and IncFII (pKP91) (9.8%) the most frequent (Suppl. Table 3). The median number of replicon types per isolate was two.
Virulence factors and serotype prediction
The majority of isolates (n = 428, 88.4%) did not contain known acquired virulence determinants (Suppl. Table 3). Five K. pneumoniae sensu stricto isolates (1.0%) of different STs were defined as hypervirulent, including one isolate of the known hypervirulent ST23 clone.3 These five isolates harbored aerobactin (iuc1 or iuc2), salmochelin (iro1 or iro2) and rmpA (Suppl. Table 3); three additionally carried the yersiniabactin (ybt) locus. Three of the hypervirulent isolates harbored the capsular synthesis locus (KL) 1 or KL2, previously described as hypervirulence associated;3 the other two harbored KL23. Overall, the siderophore loci ybt, iuc, and iro were present in 10.7% (n = 52), 1.4% (n = 7), and 1.0% (n = 5) of the isolates, respectively (Suppl. Table 3). A total of 96 defined KL types were found (Suppl. Figure 8), with KL10 being the most frequent (n = 21; 4.7%) followed by KL28 (n = 18; 4.0%) and KL22 (n = 17; 3.8%). KL2 accounted for 2.9% (n = 13) and KL1 for only 0.9% (n = 4). We observed 11 different defined O-antigen types in 477 isolates (Suppl. Figure 9) with O1 (n = 133; 27.9%) being the most frequent followed by O3/3a (n = 113; 23.7%), O2 (n = 76; 15.9%), O5 (n = 58; 12.2%), and O3b (n = 47; 9.9%), accounting for 90% of the isolates. Five isolates harbored an O-locus with an unknown O-type.
Discussion
In this study of a large representative sample of a general population aged 40 y and older, we detected an overall Kp gastrointestinal carriage prevalence of 16.3% with no gender difference. We found that Kp carriage was associated with age 60 y and older, reported Crohn’s disease/ulcerative colitis, travel to Asia and Greece in the preceding 12 months, and recent use of PPIs, NSAIDs, and antibiotics. We showed that the Kp population among adults in the community setting in a high-income country with low antibiotic use was highly diverse and characterized by a low prevalence of acquired antibiotic resistance and virulence determinants.
The identified Kp carriage prevalence of 16.3% is lower than that observed among pregnant women in low-income countries (40–66%)19 and among healthy adults in Asian countries (19–88%),18 but in range with a hospital admission study in Australia (6–19%)14 and hospitalized patients in the USA (23%).15 This could be explained by the different populations investigated (e.g. healthy individuals, hospitalized patient groups, geographic setting, and/or ethnicity), but also by the sampling and detection strategies. Consistent with the culture-based Australian study of hospitalized patients on admission, we found that Kp carrier prevalence increased with age.14 The association of Kp gastrointestinal carriage with travel abroad, irrespective of resistance phenotype, has to our knowledge not been demonstrated before. The higher Kp prevalence associated with travel to Asia could be explained by the observed high carriage among people in this region.17–19
The increased prevalence of Kp carriage associated with Crohn’s disease/ulcerative colitis could be due to disease-specific gut microbiota alterations. The gut microbiota of patients with Crohn’s disease departs from the normal state as microbial diversity is significantly diminished including an increased abundance of Enterobacterales.21 A review by Kaur et al. emphasizes the possible role of Kp in the pathogenesis of lower intestinal tract diseases.22 This corresponds to our findings of a higher Kp carrier prevalence among participants with self-reported Crohn’s disease/ulcerative colitis. However, the pathogenesis of Crohn’s disease/ulcerative colitis is complex and involves an interplay between different factors which may include the microbiota composition.23
A large number of non-antibiotic drugs have been shown to inhibit growth of one or more representative bacterial species in the human gastrointestinal tract.24 The positive association of PPI and/or NSAID use with Kp carriage further underpins the influence of non-antibiotic drugs on fecal microbiota and the selection of specific bacterial species. PPI use is implicated in altered gut microbiota composition, bacterial colonization patterns, including multi-drug resistant microorganisms, and increased susceptibility to enteric bacterial infections.25–28 Although no direct evidence exists, it may be possible that Kp as a major etiological agent of liver abscess,29 particularly in Asian countries, could be linked to the observation that PPI therapy is associated with an increased risk of cryptogenic liver abscess.30 The role of NSAIDs as a risk factor is unclear, but NSAID use has been shown to influence the gastrointestinal microbiota toward a higher relative abundance of Enterobacteriaceae.31 Interestingly, in the phylogroup subanalysis, we found NSAID users solely significantly associated with Kp2, compared to the other significant variables which were dominantly associated with Kp1 (Suppl. Table 2). However, the data must be interpreted with caution due to the low number of cases in some subgroups.
Systemic antibiotics more obviously influence the microbiota composition. Kp is intrinsically resistant to aminopenicillins and carboxypenicillins, and thus has a selective advantage compared to other bacteria during treatment with penicillins, which constitutes approximately half of human antibiotic use in Norway.32 Antibiotics leave an imprint on the gastrointestinal bacterial community after treatment is removed, ranging from weeks to years in different studies.33 This is consistent with our finding of a significant increase in Kp prevalence related to antibiotic exposure even 12 months prior to fecal sampling. Notably, our data display a quantitative time–response relationship between antibiotic use and Kp carriage. The prevalence of Kp carriage decreased among individuals sampled from one to six-month post-exposure and reached a plateau at 5% above that of the non-antibiotic using population sampled at 6–12 months post-exposure. The latter indicates a potentially long-lasting effect on fecal Kp occurrence after antibiotic exposure.
The findings that specific diseases and drug treatments, often used in hospital settings, were associated with Kp gastrointestinal carriage merit further research to understand how these modulate the gastrointestinal tract microbiota promoting Kp colonization. Diseases,21 drug use,25,31 and diet profiles34 which are shown to be associated with an increased abundance of Proteobacteria may consequently also be associated with Kp carriage. In contrast, microbiome compositions favoring Firmicutes and Bacteroidetes were associated with a lower risk of Gram-negative intestinal domination and corresponding bloodstream infections.35 Further studies are required to investigate the potential relationship between microbial composition and Kp carriage.
Alcohol consumption is an established factor in altering the gastrointestinal microbiota,36 however, the effect of alcohol on the abundance of different taxonomic phyla is vague.37 Studies investigating oropharyngeal Kp colonization have found a higher prevalence among alcoholic patients compared to controls.38,39 In contrast, our data may indicate that more frequent alcohol intake might be associated with lower Kp gastrointestinal prevalence, but further studies including more cases are required.
Comparable to previous reports on Kp carriage isolates in pregnant women in the community-setting in low-income countries19 and hospitalized patients in high-income countries,14,15 we found a phylogenetically highly diverse Kp population with dominance of K. pneumoniae sensu stricto. This indicates individually adapted Kp populations with limited interconnection. The observed diversity in our study may also be an underestimation due to the selection of only one colony for sequencing. Considering the evidence that a high proportion of Kp extraintestinal infections originate from the patients’ own carriage isolates,14,15 we assume that the Kp carriage population structure among individuals in the community is partly mirrored in the clinical setting. This is in line with the finding that six of the ten most prevalent STs of the carriage isolates also are among the ten most frequently observed STs in an ongoing Kp bacteremia study in Norway (unpublished data; Fostervold et al.). The high bacterial diversity provides important data in terms of vaccine prospects and the identification of reservoirs as a potential source for human Kp colonization.5,40 The low level of antibiotic resistance in our study is consistent with national data in the clinical setting and reflects the relatively low antibiotic consumption in Norway.32
This is the first study investigating the prevalence of Kp gastrointestinal carriage and associated risk factors in a large representative sample of a general adult population in a community setting in a high-income country. An important methodological strength is the high study attendance rate. We used data from nearly 3,000 people aged >40 y, thereby avoiding the substantial selection bias related to convenience samples in healthcare settings. Major strengths also include the combined use of questionnaire data, drug use data from a national registry database, and both phenotypic and genomic laboratory results. The high-quality drug data enabled us to detect increasing Kp prevalence among participants having used two or three drug classes simultaneously, and additionally to assess the cumulative change over time in proportion of Kp carriers associated with antibiotic use.
The age restriction is a limitation as we only studied adults >40 y. It would have been interesting to analyze Kp prevalence among those younger than 40 whom use less antibiotics and have fewer chronic diseases.32 As we found that Kp prevalence increased with age, extrapolation may suggest younger adults to have a lower prevalence. Diet and oral hygiene are well-known factors effecting gastrointestinal microbial composition34,41; however, such data were not available in our study. Due to lack of resources, we did not screen all available fecal samples for Kp carriage which would have further increased the precision of the estimates. We used selective SCAI medium for detection of Kp, as this strategy has been shown to have a high Kp recovery supporting the growth of all Kp phylogroups.42 However, we acknowledge that positive culturing could reflect those with a relative high abundance of Kp and that molecular-based approaches, which are less dependent on abundance and phenotypic differentiation, may detect an even higher prevalence of Kp carriage. We are currently in a follow-up study performing whole metagenomic sequencing on a subset of the fecal samples and SCAI sweeps to investigate the abundance of Kp, microbial composition of carriers and non-carriers, and the phylogroup/ST diversity within single individuals. Home sampling, as conducted in our study, could be another biasing factor. However, we controlled for this by ensuring validity of the samples by assessing bacterial growth consistent with fecal flora on nonselective media and mean transport time from sampling to laboratory arrival was only 1.8 d.
In conclusion, our findings illustrate the association of non-antibiotic drugs and inflammatory bowel diseases with increased prevalence of Kp gastrointestinal carriage that warrants considerations with regard to risk stratification in the prevention of healthcare-associated infections and opens up for potential future gut microbiome modulation interventions. The highly diverse population structure of Kp colonizing humans illustrates the capacity for adaptive diversification of this species complex. This is challenging for vaccine prospects and complicates the identification of potential Kp cross-niche transmission, which will be important in detection of animal or environmental Kp reservoirs for clinically relevant human Kp carriage and infection from a One Health perspective.
Subjects and methods
Study population and design
The Tromsø Study is a population-based study with repeated cross-sectional health surveys in the municipality of Tromsø, Norway. Tromsø is considered as representative of a Northern European, urban population.43 The seventh survey of the Tromsø Study (Tromsø 7, https://uit.no/research/tromsostudy) was conducted from March 2015 to October 2016 and included two clinical visits. Unique national identity numbers from the official population-registry were used to invite all citizens >40 y (n = 33,423) (Figure 4). Sixty-five percent (n = 21,083, 11,074 women) attended the first clinical visit in the study. A total of 9,324 participants attending the first visit were invited for a second visit. These represent a random selection of 20% in age-group 40–59 y and 50% in age-group 60–84 y of the initially invited participants (n = 33,423). To enhance the proportion of participants in earlier Tromsø Studies, 3,154 participants aged 40–84 y who had attended clinical examinations in Tromsø 6 were also invited. From March 2015 to March 2016, 5,800 of the 9,324 participants invited for the second visit were consecutively offered a fecal self-sampling kit. In total, 87% (n = 5,042) returned a fecal sample either at the second visit, or by mail to the laboratory. Participants collected fecal material using nylon-flocked ESwab 490CE.A (Copan, Brescia, Italy). The first 3,009 of the 5,042 collected fecal samples were consecutively screened for the presence of Kp via selective culture. Due to resource limitations, the remaining 2,033 samples were not screened. All participants completed two self-administered structured questionnaires on socio-demographics, smoking, alcohol use, hospitalization, chronic diseases, and travel abroad. After excluding 12 participants with wrong or missing sample identification number and 22 with missing questionnaires, our final study population consisted of 2,975 participants.
Figure 4.
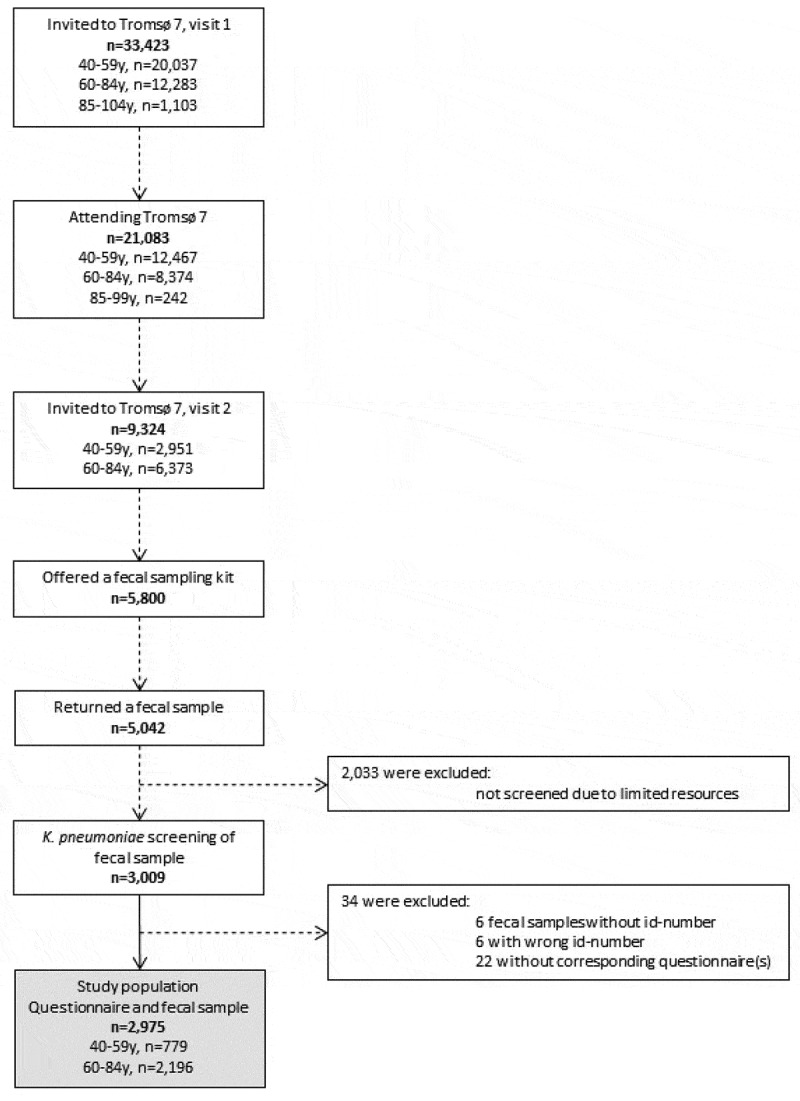
Flow diagram of study population
To assess the participants’ drug use during the preceding 12 months, data from Tromsø 7 were linked to the Norwegian Prescription Database (NorPD, http://www.norpd.no/).44 NorPD contains detailed information at the individual level on all dispensed prescription-drugs at all pharmacies in Norway. We defined drugs dispensed as drugs used and included the following groups in the Anatomical Therapeutic Chemical (ATC) classification system: A02 (acid-related disorders), A10 (diabetes), H03A (thyroid hormones), J01, A07AA09, P01AB01 (antibacterials for systemic use), and M01 (anti-inflammatory and anti-rheumatic drugs).
Klebsiella pneumoniae isolation
Upon arrival in the laboratory, 200 µl 85% glycerol was added to each ESwab tube and the samples were stored at −80°C. From the thawed media, 100 µl were plated onto Simmons citrate agar with inositol (SCAI) (both Sigma-Aldrich, Darmstadt, Germany) and incubated for 48 h at 37°C.42,45 Large, yellow, glossy colonies suspected of being Klebsiella spp. were identified using mass spectrometry (MALDI-TOF, Bruker Daltonics, Bremen, Germany). The first colony identified as either K. pneumoniae or K. variicola from each sample was kept and further analyzed. All samples were plated on cysteine lactose electrolyte deficient agar (MAST Group, Bootle, UK) to assess growth of fecal flora and validity of the samples.
Antimicrobial susceptibility testing
Susceptibility testing was performed according to the EUCAST disk diffusion method and interpreted using the EUCAST 2021 breakpoint table (https://eucast.org/).46
Genomic sequencing and bioinformatic analysis
DNA was extracted with the MagNA Pure 96 system (Roche Applied Science, Mannheim, Germany) and sequencing libraries were prepared according to the Nextera Flex sample preparation protocol (Illumina, San Diego, CA, USA). Sequencing was performed on the Illumina MiSeq platform to generate 300 bp paired-end reads. All reads were trimmed with TrimGalore v0.6.4 and assembled with Unicycler v0.4.8 including SPAdes v3.13.0.47–49 Kleborate v2.0.0 was used to determine sequence type (ST), species identification, and acquired genes encoding virulence or antibiotic resistance.50,51 Kaptive was used to identify capsule biosynthesis loci (KL), and LPS (O) antigen loci.52,53 Plasmid replicons were identified with PlasmidFinder v2020-07-13 using Abricate v0.9.9 (https://github.com/tseemann/abricate).54,55 Novel STs, blaSHV, blaLEN, blaOKP, and virulence alleles were assigned by the curators of the Institut Pasteur multilocus sequence type (MLST) and core genome MLST databases (https://bigsdb.pasteur.fr/klebsiella). To verify the absence of blaSHV/blaLEN, observed in two genomes, the raw reads were inspected with SRST2 v0.2.0.56
Phylogenetic analysis
To assess the phylogenetic relatedness, a core genome alignment of the 484 genomes against the K. pneumoniae ST23 NTUH-K2044 reference chromosome (GenBank accession: NC_012731.1) was generated using the RedDog pipeline v1beta.11 (https://github.com/katholt/RedDog), and inferred as described previously.57,58 To identify the number of single-nucleotide polymorphisms (SNPs) between any two genomes from the resulting alignment, snp-dists v0.7.0 (https://github.com/tseemann/snp-dists/) was used.
Definition of hypervirulent Kp
Hypervirulent Kp were defined here according to Huynh et al., as isolates harboring at least one of the genes rmpA and rmpA2, and/or at least one complete gene cluster among iucABCD-iutA (aerobactin) and iroBCDN (salmochelin).19
Statistical analysis
The primary analysis was a multivariable logistic regression model with outcome variable being culture-confirmed Kp gastrointestinal carriage using SPSS v26.0 (SPSS, Inc., Chicago, IL, USA). A priori known or assumed variables associated with Kp carriage were selected with the help of a directed acyclic graph constructed using DAGitty v3.0 (Suppl. Figure 1).59,60 All explanatory variables were kept in the fully adjusted model, independent of p-values. The strength of the associations was examined by calculating adjusted odds ratios (AORs) with 95% confidence intervals (CIs). Two-sided p-values <0.05 were considered statistically significant. Phylogroup subanalyses were performed using χ2 test. We used R v4.0.0 (Foundation for Statistical Computing, Vienna, Austria) to create a proportional Venn diagram for statistically significant drug groups associated with Kp carriage. STATA v16.1 (StataCorp LLC, Texas, USA) was used to analyze the cumulative change in proportion of Kp carriers associated with antibiotic use in the past 1–12 months.
Supplementary Material
Acknowledgments
We are grateful for technical assistance from Bjørg Haldorsen, Bettina Aasnæs and Ellen Josefsen in organizing the collection of fecal samples in the laboratory. Eva Bernhoff and Ragna-Johanne Bakksjø for performing whole-genome sequencing. Dorota Buczek for creating the Venn diagram. Rod Wolstenholme for figure editing. We thank the team of curators of the Institut Pasteur MLST and whole-genome MLST databases for curating the data and making them publicly available at https://bigsdb.pasteur.fr/.
Funding Statement
This study was supported by grants from the Northern Norway Regional Health Authority [HNF1415-18] and The Trond Mohn Foundation [TMF2019TMT03]. The funders of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Disclosure of potential conflicts of interest
All authors report no conflicts of interest.
Author contributions
ØS conceptualized and acquired funding for the study in collaboration with AS, KG, IHL, SB and KH. ØS was responsible for organizing the collection of fecal samples. LLEA did the screening of fecal samples and phenotypic testing. NR, KS, LS and KG conceptualized and conducted the epidemiological analysis of risk factors. IHL organized whole-genome sequencing of isolates. MAKH, NR and ØS did the analysis of whole genome sequence data. SB and KH provided tools for genomic analysis and data curation. NR, MAKH, ØS, and KG prepared first manuscript draft. All authors contributed to review and editing of the manuscript and approved the final version.
Data availability
Bacterial genome data (raw Illumina reads) are publicly available in NCBI under BioProject PRJEB42350. This study is based on data owned by a third party (The Tromsø Study, Department of Community Medicine, UiT The Arctic University of Norway). Confidentiality requirements according to Norwegian law prevent sharing of individual patient-level data in public repositories. Application of legal basis and exemption from professional secrecy requirements for the use of personal health data in research may be sent to a regional committee for medical and health research ethics (https://rekportalen.no/). The authors gained access to the data through the Tromsø Study’s application process. Guidelines on how to access the data are available at the website https://uit.no/research/tromsostudy. All enquiries about the Tromsø Study should be sent by e-mail to tromsous@ism.uit.no. All the questionnaire variables are published in the NESSTAR program system, and results can be viewed online: http://tromsoundersokelsen.uit.no/tromso/.
Ethics
The study, including the linking of data between Tromsø 7 and NorPD, was approved by the Regional Committee for Medical and Health Research Ethics, North Norway (REC North reference: 2016/1788 and 2014/940) and the Data Protection Officer at University Hospital of North Norway (reference: 2019/4264). The study complied with the Declaration of Helsinki. All participants in Tromsø 7 signed an informed consent form prior to participation.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Podschun R, Ullmann U.. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998. October;11(4):589–14. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017. May 1;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 3.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019. May 15;32(3):e00001–19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassini A, Hogberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019. January;19(1):56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018. October;45:131–139. doi: 10.1016/j.mib.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018. March;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues C, Passet V, Rakotondrasoa A, Diallo TA, Criscuolo A, Brisse S. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res Microbiol. 2019. Apr-May;170(3):165–170. doi: 10.1016/j.resmic.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020. June;18(6):344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 9.Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol. 2001. May;51(Pt 3):915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 10.Rosenblueth M, Martinez L, Silva J, Martinez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol. 2004. February;27(1):27–35. doi: 10.1078/0723-2020-00261. [DOI] [PubMed] [Google Scholar]
- 11.Brisse S, Passet V, Grimont PA. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol. 2014. September;64(Pt 9):3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues C, Passet V, Rakotondrasoa A, Diallo TA, Criscuolo A, Brisse S. Erratum to “Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov”. [Res Microbiol 170 (3) (2019) 165-170]. Res Microbiol. 2019. Sep-Oct;170(6–7):300. doi: 10.1016/j.resmic.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Selden R, Lee S, Wang WL, Bennett JV, Eickhoff TC. Nosocomial klebsiella infections: intestinal colonization as a reservoir. Ann Intern Med. 1971. May;74(5):657–664. doi: 10.7326/0003-4819-74-5-657. [DOI] [PubMed] [Google Scholar]
- 14.Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Pilcher DV, et al. Gastrointestinal carriage is a major reservoir of K. pneumoniae infection in intensive care patients. Clin Infect Dis. 2017. July 15;65(2):208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, Malani PN, Rao K, Bachman MA, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere. 2016. October 19;1(5):e00261–16. doi: 10.1128/mSphere.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimasaki T, Seekatz A, Bassis C, Rhee Y, Yelin RD, Fogg L, Dangana T, Cisneros EC, Weinstein RA, Okamoto K, et al. Increased relative abundance of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin Infect Dis. 2019. May 30;68(12):2053–2059. doi: 10.1093/cid/ciy796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung DR, Lee H, Park MH, Jung S-I, Chang -H-H, Kim Y-S, Son JS, Moon C, Kwon KT, Ryu SY, et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis. 2012. April;31(4):481–486. doi: 10.1007/s10096-011-1334-7. [DOI] [PubMed] [Google Scholar]
- 18.Lin YT, Siu LK, Lin JC, Chen T-L, Tseng C-P, Yeh K-M, Chang F-Y, Fung C-P. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 2012;12:13. doi: 10.1186/1471-2180-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh BT, Passet V, Rakotondrasoa A, Diallo T, Kerleguer A, Hennart M, Lauzanne AD, Herindrainy P, Seck A, Bercion R, et al. Klebsiella pneumoniae carriage in low-income countries: antimicrobial resistance, genomic diversity and risk factors. Gut Microbes. 2020. September 2;11(5):1287–1299. doi: 10.1080/19490976.2020.1748257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson EH. Measurement of diversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 21.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song S, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014. March 12;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur CP, Vadivelu J, Chandramathi S. Impact of Klebsiella pneumoniae in lower gastrointestinal tract diseases. J Dig Dis. 2018. May;19(5):262–271. doi: 10.1111/1751-2980.12595. [DOI] [PubMed] [Google Scholar]
- 23.Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018. February;11(1):1–10. doi: 10.1007/s12328-017-0813-5. [DOI] [PubMed] [Google Scholar]
- 24.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018. March 29;555(7698):623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJM, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016. May;65(5):740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reveles KR, Ryan CN, Chan L, Cosimi RA, Haynes WL. Proton pump inhibitor use associated with changes in gut microbiota composition. Gut. 2018;67(7):1369–1370. doi: 10.1136/gutjnl-2017-315306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willems RPJ, van Dijk K, Ket JCF, Vandenbroucke-Grauls C. Evaluation of the association between gastric acid suppression and risk of intestinal colonization with multidrug-resistant microorganisms: a systematic review and meta-analysis. JAMA Intern Med. 2020. April 1;180(4):561–571. doi: 10.1001/jamainternmed.2020.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bavishi C, Dupont HL. Systematic review: the use of proton pump inhibitors and increased susceptibility to enteric infection. Aliment Pharmacol Ther. 2011. December;34(11–12):1269–1281. doi: 10.1111/j.1365-2036.2011.04874.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang JH, Liu YC, Lee SS, Yen M-Y, Wang YSCH, Wann S-R, Lin -H-H. Primary Liver Abscess Due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 1998. June;26(6):1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 30.Wang YP, Liu CJ, Chen TJ, Lin YT, Fung CP. Proton pump inhibitor use significantly increases the risk of cryptogenic liver abscess: a population-based study. Aliment Pharmacol Ther. 2015. June;41(11):1175–1181. doi: 10.1111/apt.13203. [DOI] [PubMed] [Google Scholar]
- 31.Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016. February;22(2):178.e1–178.e9. doi: 10.1016/j.cmi.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NORM/NORM-VET 2019 . Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Tromsø/Oslo; 2020. 1502-2307 (print)/1890-9965 (electronic). [Google Scholar]
- 33.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011. April;9(4):233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 34.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019. January;16(1):35–56. [DOI] [PubMed] [Google Scholar]
- 35.Stoma I, Littmann ER, Peled JU, Giralt S, van den Brink MR, Pamer EG, Taur Y.. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clin Infect Dis. 2020. January 24;ciaa068. doi: 10.1093/cid/ciaa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorkhaug ST, Aanes H, Neupane SP, Bramness JG, Malvik S, Henriksen C, Skar V, Medhus AW, Valeur J. Characterization of gut microbiota composition and functions in patients with chronic alcohol overconsumption. Gut Microbes. 2019;10(6):663–675. doi: 10.1080/19490976.2019.1580097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee E, Lee JE. Impact of drinking alcohol on gut microbiota: recent perspectives on ethanol and alcoholic beverage. Curr Opin Food Sci. 2021;37:91–97. doi: 10.1016/j.cofs.2020.10.001. [DOI] [Google Scholar]
- 38.Fuxench-Lopez Z, Ramirez-Ronda CH. Pharyngeal flora in ambulatory alcoholic patients: prevalence of gram-negative bacilli. Arch Intern Med. 1978. December;138(12):1815–1816. doi: 10.1001/archinte.1978.03630370033017. [DOI] [PubMed] [Google Scholar]
- 39.Dao TT, Liebenthal D, Tran TK, Ngoc Thi Vu B, Ngoc Thi Nguyen D, Thi Tran HK, Thi Nguyen CK, Thi Vu HL, Fox A, Horby P, et al. Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PLoS One. 2014. March 25;9(3):e91999. doi: 10.1371/journal.pone.0091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipworth S, Vihta KD, Chau KK, Kavanagh J, Davies T, George S, Barker L, Vaughan A, Andersson M, Jeffery K, et al. Ten years of population-level genomic Escherichia coli and Klebsiella pneumoniae serotype surveillance informs vaccine development for invasive infections. Clin Infect Dis. 2021. January 7;ciab006. doi: 10.1093/cid/ciab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J Oral Microbiol. 2019. March 18;11(1):1586422. doi: 10.1080/20002297.2019.1586422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passet V, Brisse S. Association of tellurite resistance with hypervirulent clonal groups of Klebsiella pneumoniae. J Clin Microbiol. 2015. April;53(4):1380–1382. doi: 10.1128/JCM.03053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eggen AE, Mathiesen EB, Wilsgaard T, Jacobsen BK, Njolstad I. The sixth survey of the Tromso Study (Tromso 6) in 2007-08: collaborative research in the interface between clinical medicine and epidemiology: study objectives, design, data collection procedures, and attendance in a multipurpose population-based health survey. Scand J Public Health. 2013. February;41(1):65–80. doi: 10.1177/1403494812469851. [DOI] [PubMed] [Google Scholar]
- 44.Furu K. Establishment of the nationwide Norwegian Prescription Database (NorPD) – new opportunities for research in pharmacoepidemiology in Norway. Norsk Epidemiologi. 2009;18(2). doi: 10.5324/nje.v18i2.23. [DOI] [Google Scholar]
- 45.Van Kregten E, Westerdaal NA, Willers JM. New, simple medium for selective recovery of Klebsiella pneumoniae and Klebsiella oxytoca from human feces. J Clin Microbiol. 1984. November;20(5):936–941. doi: 10.1128/jcm.20.5.936-941.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matuschek E, Brown DF, Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect. 2014. April;20(4):O255–66. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
- 47.Krueger F. TrimGalore v0.6.4. August 2019. https://github.com/FelixKrueger/TrimGalore.
- 48.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017. June 8;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012. May;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam MM, Wick RR, Wyres KL, Holt KE. Genomic surveillance framework and global population structure for Klebsiella pneumoniae. bioRxiv. 2020. doi: 10.1101/2020.12.14.422303. [DOI] [Google Scholar]
- 51.Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018. October 29;10(1):77. doi: 10.1186/s13073-018-0587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE.. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016. December 12;2(12):e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wick RR, Heinz E, Holt KE, Wyres KL. Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol. 2018. May 25;56(6):e00197–18. doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. In silico detection and typing of plasmids using Plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014. July;58(7):3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seemann T. Abricate v0.9.9, Github. February 2020. https://github.com/tseemann/abricate.
- 56.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE.. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014. November 20;6(11):90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, Lam MMC, Duchêne S, Jenney A, Holt KE, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019. April 15;15(4):e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010. March 10;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Textor J, van der Zander B, Gilthorpe MS, Liskiewicz M, Ellison GT. Robust causal inference using directed acyclic graphs: the R package ‘dagitty’. Int J Epidemiol. 2016. December 1;45(6):1887–1894. doi: 10.1093/ije/dyw341. [DOI] [PubMed] [Google Scholar]
- 60.Williams TC, Bach CC, Matthiesen NB, Henriksen TB, Gagliardi L. Directed acyclic graphs: a tool for causal studies in paediatrics. Pediatr Res. 2018. October;84(4):487–493. doi: 10.1038/s41390-018-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bacterial genome data (raw Illumina reads) are publicly available in NCBI under BioProject PRJEB42350. This study is based on data owned by a third party (The Tromsø Study, Department of Community Medicine, UiT The Arctic University of Norway). Confidentiality requirements according to Norwegian law prevent sharing of individual patient-level data in public repositories. Application of legal basis and exemption from professional secrecy requirements for the use of personal health data in research may be sent to a regional committee for medical and health research ethics (https://rekportalen.no/). The authors gained access to the data through the Tromsø Study’s application process. Guidelines on how to access the data are available at the website https://uit.no/research/tromsostudy. All enquiries about the Tromsø Study should be sent by e-mail to tromsous@ism.uit.no. All the questionnaire variables are published in the NESSTAR program system, and results can be viewed online: http://tromsoundersokelsen.uit.no/tromso/.


