Genetic variation in gene regulation is an important source of phenotypic variation, contributing to human phenotypes and diseases [1,2] as well as evolution within and between species [3,4]. Expression variation between two individuals can be partitioned into diffusible/trans elements (e.g. transcription factors) or non-diffusible/cis elements (e.g. linked regulatory sequences like promoters or enhancers) [4]. By taking advantage of genetic crosses, we can gain insight into the mechanistic basis of expression variation that differentiates individuals [5,6]. Because parental genotypes share a single cellular compartment in F1 hybrids, they also share all diffusible regulatory factors. Thus, expression variation between alleles in an F1 hybrid reflects the portion of variation between the parents due to cis factors alone. The remaining portion of variation between parents not explained by variation in the F1 hybrids is due to variation in trans factors. Conceptually, this leads to the mechanistic perspective that allele specific expression (ASE) variation in F1 hybrids is equivalent to variation in cis elements whereas ASE variation in parents is a combination of variation in cis+trans factors [5]. By measuring the expression variation in both parents and their F1 hybrids, we can estimate the contribution of cis elements and trans factors to expression variation.
This ASE perspective facilitates estimation of important expression parameters on a genome scale [7,8], providing abundant fodder for making mechanistic inferences on the genetic basis of expression variation within and between species. However, an article in this issue of Trends in Genetics points out that, when cis and trans estimates share common F1 hybrid samples, they will be negatively correlated via error shared from the hybrid data [9]. One important consequence of this observation is that spurious inferences of compensatory evolution between cis and trans factors will occur when correlated error is not accounted for. This is because this type of compensatory evolution is defined as a negative relationship between cis and trans variation. As [9] points out, many studies continue to make precisely this error regarding compensatory evolution, and consequently, a solution is urgently needed. [9] argues that the simplest solution to this problem is to estimate cis and trans parameters from independent replicates of hybrid data so that error is no longer correlated. Indeed, an ASE inference framework formulated by [8] recommends correcting for error in just this way (cf Figures 2 and S2 from [8]). In order to demonstrate the utility of this approach, we investigate two ASE datasets. The first is an artificial dataset designed to be devoid of genetic variation in gene expression, and is constructed purely from biological replicates of the same strain from [10] (see Supplementary Materials for Methods). The second involves genetically distinct strains from [8] and therefore potentially exhibits compensatory variation in gene regulation.
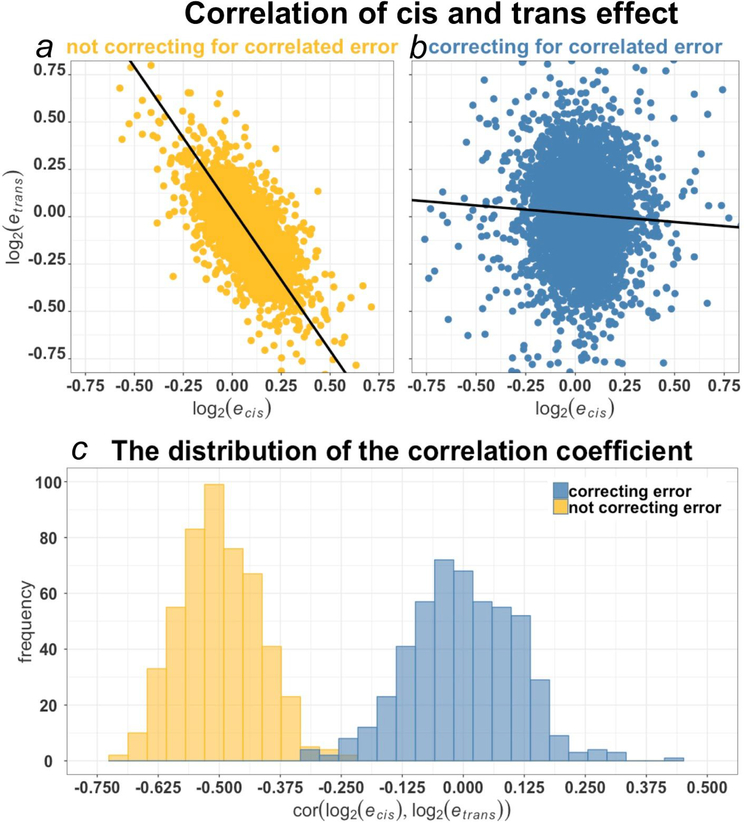
Figure 1a–b illustrates the estimation of cis and trans expression parameters both with and without correcting for correlated error in a representative random partition of the ASE dataset constructed to have no genetic variation between the parents (cf panel a to panel b). The negative correlation in panel a is large in magnitude and highly significant (r = −0.48, p < 0.0001) while that of panel b is small and marginally significant (r = −0.02, p = 0.02). Overall, when full biological replication is employed, correlations cluster around 0 (Figure 1c). Additionally, given that the approach in [8] places cis and trans expression parameters in a likelihood testing framework, it can address questions of compensatory evolution on a gene-by-gene basis in a way purely correlative approaches cannot. For example, in [8], the overall correlation between cis and trans was near zero in the independent estimates, offering no evidence for compensatory evolution (r = −0.028, p-value = 0.076), compensating for a spurious conclusion of rampant compensatory evolution suggested by the correlated estimates (r = −0.46, p-value < 10−15). However, by employing independent estimates of cis and trans, individual genes with evidence for differential expression can be identified. Of the 850 genes significant for cis and/or trans in the independent dataset of [8], 55% (466/850 with a 95% binomial confidence interval on the proportion 51%-58%) fall into the compensatory category at a significance threshold of 1%. Under a model of random expression variation, only 50% (425) are expected to fall in compensatory categories — quadrants II and IV — by chance (16 genes were excluded that have a cis estimate of 0 and cannot be classified as compensatory or reinforcing). Thus, while no evidence for a negative correlation between cis and trans is apparent at the genome level, the statistical evidence might support the action of compensatory evolution above the background expectation for at most a small number of genes (∼41). Alternatively, because of the nature of replication in [8] (replicate cultures were pooled before library preparation and subsequent replicates came from the same library), the variation associated with library preparation was not controlled, perhaps explaining the remaining small magnitude of excess compensatory evolution observed in the study. Clearly, however, a substantial proportion of the signal of compensatory variation was caused by correlated error arising from sequencing, as the method of [8] reduced the correlation from −0.46 to −0.028.
Figure 1.
The effect of correlated error on estimation of cis and trans expression variation ratios. The data considered in the figure was compiled from partitions of a highly replicated expression dataset in yeast [10]. a) Both cis and trans parameter estimates share a common sample of 11 hybrid individuals. b) cis parameters are estimated from one set of 7 hybrid individuals and trans parameters are estimated from a different set of 7 individuals. c) Summary of τ (Kendall rank correlation coefficient) for 500 randomly chosen partitions of both the correlated and independent estimation schemes. Panels a) and b) are representative instances of these random partitions.
This approach illustrates the utility of accounting for correlated error in a statistical inference framework. The ability to make inferences on individual genes is an important advantage in carefully measuring the extent of compensatory evolution. Indeed, any time estimates of cis and trans are considered jointly to make biological conclusions, correlated error should be considered, not just in cases of compensatory evolution. Modern datasets should be even better suited to addressing such questions, as lower sequencing costs allow us to achieve higher and higher replication, not only eliminating the correlated error problem, but also improving statistical power. Indeed, it would be irresponsible not to replicate parental and hybrid treatments in future ASE studies.
Supplementary Material
Acknowledgments
J.J.E. would like to thank Daniel Rokhsar for thoughtful comments. The work was supported by US National Institutes of Health (NIH) grant R01GM123303-1 (J.J.E.) and University of California, Irvine setup funds (J.J.E).
References
- 1.Lee TI and Young RA (2013) Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lappalainen T et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 [DOI] [PubMed] [Google Scholar]
- 4.Emerson JJ and Li W-H (2010) The genetic basis of evolutionary change in gene expression levels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2581–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittkopp PJ et al. (2004) Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88 [DOI] [PubMed] [Google Scholar]
- 6.Wittkopp PJ et al. (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40, 346–350 [DOI] [PubMed] [Google Scholar]
- 7.McManus CJ et al. (2010) Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20, 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerson JJ et al. (2010) Natural selection on cis and trans regulation in yeasts. Genome Res. 20, 826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser HB (2018) Improving Estimates of Compensatory cis-trans Regulatory Divergence. Trends Genet. DOI: 10.1016/j.tig.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gierlinski M et al. (2015) Statistical models for RNA-seq data derived from a two-condition 48-replicate experiment. Bioinformatics 31, 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.