Abstract
Previous studies suggest compounds such as sulforaphane (SFN) derived from cruciferous vegetables may prevent prostate cancer development and progression. This study evaluated the effect of broccoli sprout extract (BSE) supplementation on blood histone deacetylase (HDAC) activity, prostate RNA gene expression, and tissue biomarkers (histone H3 lysine 18 acetylation (H3K18ac), HDAC3, HDAC6, Ki67, and p21). A total of 98 men scheduled for prostate biopsy were allocated into either BSE (200 μmol daily) or a placebo in our double-blind, randomized controlled trial. We used nonparametric tests to evaluate the differences of blood HDAC activity and prostate tissue immunohistochemistry biomarkers between treatment groups. Further, we performed RNA-Seq analysis on the prostate biopsies and identified 40 differentially expressed genes correlated with BSE treatment, including downregulation of two genes previously implicated in prostate cancer development, AMACR and ARLNC1. Although urine and plasma SFN isothiocyanates and individual SFN metabolites were statistically higher in the treatment group, our results did not show a significant difference in HDAC activity or prostate tissue biomarkers. This study indicates BSE supplementation correlates with changes in gene expression but not with several other prostate cancer biomarkers. More research is required to fully understand the chemopreventive effects of BSE supplementation on prostate cancer.
Introduction
Prostate cancer (PCa) is the most frequently diagnosed noncutaneous cancer and is the second leading cause of cancer death in American men (1). Observational studies have shown that cruciferous vegetable intake is associated with decreased risk for many cancer types such as breast cancer (2) and lung cancer (3). However, recently pooled analyses of 15 prospective cohort studies showed cruciferous vegetable consumption was not associated with lower PCa risk (4). Part of the reason for the null association may be that the majority of observational studies assessed cruciferous vegetable intake through a self-report food frequency questionnaire, which does not accurately evaluate specific bioactive nutrients. Thus, clinical trials that directly evaluate bioavailable phytochemicals for association with PCa are important for developing effective PCa chemoprevention strategies.
Isothiocyanates (ITCs) are derived from cruciferous vegetables such as broccoli, Brussels sprouts, cauliflower, and cabbage. Sulforaphane (SFN) is an ITC derived from the glucosinolate precursor glucoraphanin, which is especially abundant in broccoli and broccoli sprouts (5, 6). When the plant is consumed, the enzyme myrosinase, released from plant and present in our gut, converts glucoraphanin to SFN. SFN is an effective chemoprotective agent in carcinogen-induced animal models (5, 7, 8), as well as in xenograft models of PCa (9).
Targeting the epigenome, including the use of histone deacetylase (HDAC) inhibitors, has shown promise in cancer clinical trials, making it an evolving strategy for chemoprevention and therapy. We have found that SFN inhibits HDAC activity in human colorectal and PCa cells (10, 11). There was a concomitant increase in accumulation of acetylated histones H3 and H4 and an upregulation of tumor suppressor genes p21 and Bax (12). In vivo, a dietary supplementation with broccoli sprouts or SFN inhibited prostate carcinogenesis and PCa progression in transgenic rodent models (13–17). In human intervention trials, broccoli supplementation decreased proliferation of Ki-67 positive cells in breast cancer patients with ductal carcinoma in situ (18). In this study, we completed a randomized, placebo-controlled clinical trial to examine the effects of short term broccoli sprout extract (BSE) supplementation on SFN metabolism, epigenetic biomarkers, and transcriptome profiles in men at risk for PCa.
Methods
Subjects
Subjects were recruited from the urology clinic at the VA Portland Health Care System (VAPORHCS), who were scheduled for prostate biopsy. We included men ≥21 years old who signed an informed consent. Exclusion criteria included 1) significant active medical illness that would preclude protocol treatment; 2) diagnosis of liver disease or abnormal baseline total bilirubin; 3) subject-reported allergy or sensitivity to cruciferous vegetables; 4) use of oral antibiotics (except doxycycline) within three months before randomization; 5) use of warfarin or need for therapeutic anticoagulation at time of biopsy or any time during the trial; 6) current oral steroid therapy; 7) current therapy with valproate or other pharmacological drugs associated with HDAC inhibition; 8) diagnosed dementia or other significant mental illness; 9) not being in another flagged study; 10) already taking BSE; and 11) any PCa-related treatment procedures.
The study sample size flowchart is depicted in Fig. 1 following CONsolidated Standards Of Reporting Trials (CONSORT) guidelines (19). After eligibility confirmation – 2 subjects had elevated bilirubin > 1.2 mg/dL and were excluded from the study – 98 consented subjects were randomized to one of the two treatment arms (BSE or placebo).
Figure 1.
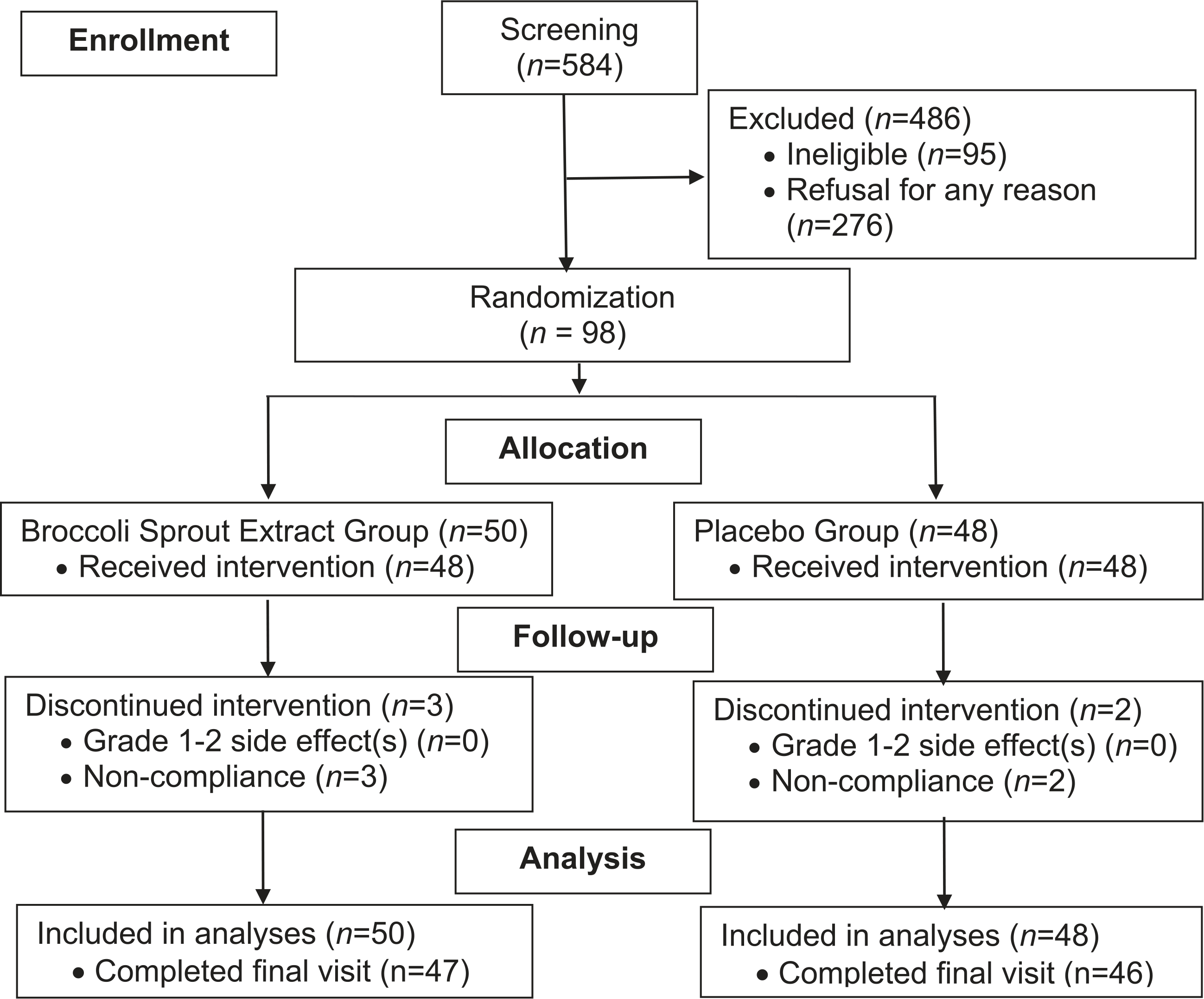
Randomized controlled trial sample size flowchart.
Study Design
After obtaining informed consent, 2 × 10 mL (one EDTA lavender-top tube and one red-top tube) and a 4 mL (assessing total bilirubin level) blood specimen were collected. A urine sample was collected at the same time. Before the intervention, the study coordinator explained the Diet History Questionnaire (20) and administered the risk factor, cruciferous vegetable, and adverse event questionnaires to obtain data on potential confounding variables and gain subjects’ baseline symptoms. Every two weeks during the study, the study coordinator administered three questionnaires: 1) a reporting form that included common adverse events for cruciferous vegetables, 2) changes to medications or supplement use over the past two weeks, and 3) a brief cruciferous vegetable intake checklist. Data were entered into a REDCap® database. After the BSE intervention and before biopsy, 2 × 10 mL blood (one EDTA lavender-top tube and one red-top tube) and urine samples were collected again for pre- to postintervention comparison. The original study protocol was approved by the Institutional Review Boards of Oregon Health & Science University (OHSU) and VAPORHCS.
The study biostatistician determined the treatment assignment based on the randomization protocol and provided this information directly to the research pharmacist. No other study team members or patients were aware of the study assignment. Once eligibility was confirmed, the Research Pharmacy dispensed enough BSE/Placebo capsules for the entire duration of the study, which would be considered complete at the time of the subject’s prostate biopsy (4–8 wk). Subjects returned any unused study drug to the study coordinator at the time of biopsy. The Research Pharmacy counted and recorded any remaining capsules. The study coordinator remained blinded to the subject’s study status throughout the intervention.
Treatment capsules consisted of myrosinase-treated BSE that provided 100 μmol SFN per capsule (21, 22). For quality control, SFN content was verified at Oregon State University (OSU). The matching placebo for the BSE consisted of a gelatin capsule containing microcrystalline cellulose. BSE supplements and placebos were obtained from John Hopkins University (Baltimore, MD). Subjects assigned to the treatment group took two BSE capsules daily (one capsule B.I.D.), providing 200 μmol of SFN in total. This dose is equivalent to the amount of SFN administered in our pilot study and other trials which achieved a significant increase in blood and urine SFN and SFN-derived metabolites and reduced HDAC activity (18).
Capsules were distributed in a container labeled only with dosing instructions, subject name, protocol number, subject identification number, and study drug name “broccoli sprout extract/placebo”. Should a subject’s prostate biopsy appointment be delayed due to nonstudy-related concerns, the subject would remain in the study for up to eight weeks. Subjects who took ≥80% of the prescribed pills were considered treatment compliant.
Adverse events questionnaires were completed at baseline, every two weeks during the study, at the biopsy visit, and at the 30-day follow-up after the trial. For any reported adverse event ≥ grade 3, according to the NCI Common Terminology Criteria for Adverse Events (Version.4.0), the responsible clinician was notified; the event was triaged and followed to resolution.
Biospecimen Sample Collection and Processing
For blood samples, plasma was collected, and peripheral blood mononuclear cell (PBMC) was isolated using a Ficoll Histopaque gradient per manufacture’s protocol. Plasma and urine samples were acidified with 10% trifluoroacetic acid (TFA) immediately after collection and were used for SFN metabolite evaluation. PBMC was cryopreserved in freezing media containing 10% DMSO and was used to determine HDAC activity. Prostate biopsies were obtained per clinical evaluation protocol with 10–20 cores taken for diagnosis and an additional four cores (three flash frozen and one formalin fixed) obtained solely for research purposes. Should the subject have cancer/prostatic intraepithelial neoplasia (PIN), research cores were collected away from the lesion in healthy tissue to determine the effect of treatment on similar tissue types. The research cores were embedded in optimal cutting temperature (OCT) compound and then suspended in methylbutane cooled with dry-ice. These flash-frozen tissues were then placed in cryotubes and stored in −80 °C freezer. Frozen prostate biopsy cores were used for SFN metabolite analyses and transcriptome analyses. All other clinical prostate biopsy specimens were immediately placed in 10% neutral buffered formalin for clinical pathological diagnosis. Formalin-fixed research specimens were stored and available for immunohistochemical (IHC) studies.
SFN Metabolites Analysis in Urine, Plasma, and Prostate Biopsy Samples
Plasma and urine samples were processed and analyzed for SFN metabolites as previously described (21). Acidified plasma and urine samples were briefly centrifuged at 12,000× g for 5 min at 4 °C to remove protein precipitates. Supernatants were collected and filtered using 0.22-μm Spin-X® filter columns (VWR). Filtered urine samples were further diluted 2-fold in 0.1% formic acid (v/v). For prostate biopsies, OCT-embedded prostate biopsies were thawed and rinsed on ice in prechilled 10% TFA to remove OCT compound. Tissues were blotted dry and tissue weights were recorded (average tissue weight = 4 mg). Prostate tissues were pulverized in liquid nitrogen using mortar and pestle, transferred to microcentrifuge tubes, and resuspended in 35 μL 10% TFA. Tissue homogenates were stored at −80 °C. Acidified prostate homogenates were thawed, vortexed vigorously, and centrifuged for 5 min at 4 °C, 12,000× g to remove protein precipitates and cellular debris. Tissue lysates were filtered using Spin-X columns (prewet with 20 μL 10% TFA to minimize sample loss). SFN metabolites in filtered samples were analyzed using MDS Sciex 4000 QTRAP LC-MS/MS instrument (Applied Biosystems) at the OSU Mass Spectrometry Center. The following precursor and product ions were used to detect SFN and its metabolites: SFN (178 > 114), SFN-glutathione (SFN-GSH, 485 > 179), SFN-cysteine (SFN-Cys, 299 > 114), SFN N-acetyl-L-cysteine (SFN-NAC, 341.1 > 114), and SFN-cysteinylglycine (SFN-CG, 356 > 114). Quantification was determined against known standards using 8-point linear standard curves (r2=0.99).
IHC Biomarkers
Deparaffinized slides were made of paraffin-embedded prostate tissues, and these specimens were rehydrated with graded alcohols, washed for 10 min in Tris-buffered saline (pH 7.2–7.6), heated for 10 min in a Russell-Hobbs programmable pressure cooker in 0.01 M citrate buffer (pH 6.0), and treated for 5 min with 3% aqueous H2O2 solution. After blocking for 1 h at 25 °C in 3% goat serum, slides were incubated for 1 h at 25 °C with primary antibodies for acetylated histone H3 lysine 18 (H3K18ac) (1:2000), p21 and Ki-67 (Abcam, Cambridge, MA), HDAC6 and HDAC3 (Santa Cruz Biotechnology, Inc., Dallas, TX), followed by mouse Envision (DAKO, Glostrup, Denmark) antibody, counterstained 1 min with Gill’s hematoxylin, rinsed, dehydrated, and coverslipped using Permount. Biomarkers were scored by our collaborating pathologist, Dr. George Thomas. A modified Histo-score (H-score) was recorded, which involved semiquantitative assessment of both staining intensity (graded as 1–3, with 1 representing weak staining, 2 moderate, and 3 strong) and percentage of positive cells.
PBMC HDAC Activity Analysis
The Cancer Prevention and Intervention Program’s Core Laboratory at OSU performed the analyses. PBMC samples were thawed on ice, followed by centrifugation to remove freezing media. Cell pellets were resuspended and washed once in cold PBS, and repelleted. PBMC cell lysates were prepared using IP lysis buffer, and protein concentrations were determined. Matched pre- and post-PBMC protein samples (15 μg protein/assay in triplicates) were evaluated for HDAC activities as previously described (13). Substrates and standards for the assay were custom synthesized by AAPPTec, LLC (Louisville, KY). HDAC activity was determined relative to deacetylated standards and was expressed as pmol/min/mg protein.
RNA Sequencing (RNA-Seq)
Prostate biopsy tissue was homogenized in Trizol (Thermo Fisher Scientific, Waltham, MA), and RNA was isolated and resuspended in 50 μl water. RNA quantity was measured using the Qubit RNA BR assay kit (Thermo Fisher Scientific). Total RNA samples were submitted to the OSU Center for Genome Research and Biocomputing core facility for library preparation and RNA sequencing. The PrepX PolyA mRNA Isolation kit was used for the mRNA enrichment, followed by the PrepX RNA-Seq for Illumina Library Kit (Wafergen Biosystems, Fremont, CA). Prepared libraries were quantified using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA). Library sizing was analyzed on the Agilent TapeStation 4200 using the HS-D5000 screen tape, followed by qPCR using the KAPA library quantification kit (KAPA Biosystems, Wilmington, MA). Samples were normalized, pooled, and sequenced (100-bp paired end) on an Illumina HiSeq 3000.
FASTQC was used to assess read quality and adapter contamination in the raw reads (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; Access Date: July-19, 2017). On average, samples had low rRNA and mtDNA contamination (mean values of <20% and ≤10%, respectively), and high-quality scores (mean Q scores ≥ 30). Reads were filtered by quality, trimmed, and aligned to the human genome (GRCh37/hg19) using the bcbio-nextgen pipeline (v 1. 06) (https://github.com/bcbio/bcbio-nextgen/, Access Date: December-20, 2017), with salmon (v. 0.8.1) as the specified alignment software, using default parameters (23).
Quantitative Real-Time PCR (qPCR)
cDNA was synthesized from prostate samples using 200 ng of total RNA and SuperScript III First-Strand Synthesis SuperMix (Thermo Fisher Scientific). qPCR was done using primers that amplify all known transcript isoforms of each human gene as a single product of expected size, between 110 and 175 bp. Primers were: AMACR, (forward) 5′-AGCATGGATGATT GGCCAGAA-3′ and (reverse) 5′-TGATAAACGAG CCCCGTTCC-3′; ACTB, (forward) 5′-TCTTCCA GCCTTCCTTCCTGGGCATG-3′and (reverse) 5′-GCTCAGGAGGAGCAATGATCTTGATC-3′; ARLNC1, (forward) 5′-GGTTGGTGGGTGATCTCAGG-3′ and (reverse) 5′-CGTTAGCCCTGGGGTTCATT-3′. Reactions were performed using Fast SYBR Green Mastermix (Thermo Fisher Scientific) on 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) as previously published (24).
Statistical Methods
The sample size and power analyses were performed for selected primary endpoints: ITC metabolite in urine, HDAC inhibition in plasma and tissue, and acetylated H3 & H4 in PBMC and tissue. Overall, an original sample size of 100 subjects (50 in each group) provided sufficient power to detect a biologically meaningful difference.
An intent-to-treat analysis was performed for the primary outcomes and included all randomized subjects using SAS 9.4 (SAS Institute Inc., Cary, NC, USA). Baseline characteristics were expressed as means and standard errors (SEs) for continuous variables, and counts (n) and percentages (%) for categorical variables, stratified by treatment group. The comparability of the two treatment groups for baseline characteristics was tested using t-tests for continuous variables and chi-squared tests for categorical variables. The key primary outcomes were PBMC HDAC level and prostate tissue biomarkers. Shapiro–Wilk normality tests were conducted for all continuous variables, and all of the primary outcomes did not follow normal distribution, so we used nonparametric tests for these outcomes. We used the Benjamini–Hochberg false discovery rate (FDR) method to adjust for multiple comparisons of the primary endpoints. Adverse events and compliance between the treatment groups were analyzed using Fisher’s exact tests as appropriate. The proportions of the compliant subjects were compared using a Chi-squared test. Tests of statistical significance were conducted using two-sided tests, and a P value 0.05 was considered statistically significant.
For RNA-seq data, significant differences in gene expression (Benjamin–Hochberg FDR corrected P< 0.10) were determined using the DESeq2 package (v. 1.18.1) implemented in R (v. 3.3.1) (25, 26). Several models were used (~phenotype, ~treatment, ~phenotype + treatment * phenotype:treatment) to compare the treatment effect (BSE vs placebo), cancer effect (the phenotype; cancer vs normal), and the interaction effect of BSE treatment on cancer.
For qPCR, data were normalized to the expression of the β-actin gene (ACTB) and analyzed using the standard 2-ΔΔCT method (24). Graphs were generated using GraphPad Prism software (La Jolla, CA), and significant differences between groups were calculated using two-way ANOVA.
Results
Patient Characteristics and Adverse Events
From July 2011 to December 2015, a total of 98 subjects aged 50–78 years (65.3 ± 5.2 years) were randomized into BSE group (n = 50) and placebo group (n = 48). Fig. 1 shows the flowchart of subjects’ enrolled in this study, with 47 (94%) completing the study in the treatment group and 46 (96%) completing in the placebo group. In addition, 84% of the subjects in the BSE group and 85% subjects in the placebo group had a compliance rate 80%. There was no statistically significant difference in compliance between the treatment groups (P = 0.44).
Table 1 describes the baseline characteristics of the subjects. There was no statistically significant difference in baseline characteristics including age, BMI, PSA at baseline, race, marital status, smoking, alcohol consumption, family history of PCa, or pathology diagnosis of PCa. In the BSE group, 20 (40.0%) subjects had confirmed diagnosis of PCa, while in the placebo group, 14 (29.2%) subjects were diagnosed with PCa. The only difference we observed was income status, with more subjects in the placebo group categorized in the lower income bracket.
Table 1.
| Subject characteristics | BSE (n= 50) Mean (SD) |
Placebo (n= 48) Mean (SD) |
P |
|---|---|---|---|
| Age, yr | 65.7 (5.4) | 64.9 (5.0) | 0.44 |
| BMI (kg/m2) at baseline | 28.9 (7.6) | 31.1 (6.4) | 0.12 |
| PSA at baseline | 8.4 (5.1) | 8.5 (6.4) | 0.90 |
| Cruciferous vegetable intake at baseline (g/day)5 | 91.3 (63.9) | 69.7 (83.8) | 0.15 |
| n (%4) | n (%4) | ||
| Race | 1.00 | ||
| White | 48 (96.0) | 46 (95.8) | |
| Non-White | 1 (2.0) | 1 (2.1) | |
| Unknown | 1 (2.0) | 1 (2.1) | |
| Family history of Prostate Cancer | 0.42 | ||
| Yes | 12 (24.0) | 15 (31.3) | |
| No | 38 (76.0) | 33 (68.8) | |
| Smoking | 0.78 | ||
| Current | 8 (16.0) | 10 (20.8) | |
| Former | 20 (40.0) | 17 (35.4) | |
| Never | 14 (28.0) | 12 (25.0) | |
| Missing | 8 (16.0) | 9 (18.8) | |
| Alcohol | 0.85 | ||
| Current | 13 (26.0) | 12 (25.0) | |
| Former | 13 (26.0) | 11 (22.9) | |
| Never | 8 (23.5) | 5 (10.4) | |
| Missing | 16 (32.0) | 20 (41.7) | |
| Income | 0.03 | ||
| ≤$25,000 | 8 (16.0) | 17 (35.4) | |
| $25,000–$50,000 | 18 (36.0) | 4 (8.33) | |
| >$50,000 | 7 (14.0) | 7 (14.6) | |
| Refuse/Don’t know/missing | 17 (34.0) | 20 (41.7) | |
| Education | 0.83 | ||
| ≤ 12 yr | 10 (20.0) | 13 (27.1) | |
| Some college/technical | 24 (48.0) | 22 (45.8) | |
| ≥ College graduate | 4 (8.0) | 4 (8.3) | |
| Missing | 12 (24.0) | 9 (18.8) | |
| Marital status | 0.11 | ||
| Single, Divorced, Widowed | 10 (23.8) | 16 (41.0) | |
| Married/partner | 31 (73.8) | 23 (59.0) | |
| Missing | 1 (2.4) | 0 (0.0) | |
| Pathology Biopsy Diagnosis | 0.34 | ||
| Benign | 30 (60.0) | 33 (68.7) | |
| Malignant | 20 (40.0) | 14 (29.2) | |
| Declined Biopsy | 0 (0.0) | 5 (2.1) | |
t-tests were conducted for continuous variables (age, BMI, PSA, and cruciferous vegetable intake); chi-square tests were conducted for categorical variables with expected cell frequencies ≥ 5; and Fisher’s exact tests were conducted for categorical variables with expected cell frequencies <5.
P value for t-tests or chi-square tests between supplement and placebo groups
P< 0.05.
For the categorical variables with missing values, chi-square tests or Fisher’s exact tests were conducted without including missing group.
Percentages may not add up to 100 due to rounding values.
Cruciferous vegetable intake includes sources from food lines, mixed dishes, and condiments.
There were no treatment group differences noted for each specific type of adverse event and the total number of adverse events (Table 2). No subjects experienced grade ≥ 3 adverse events.
Table 2.
Adverse events comparisons between BSE group and placebo group.
| Adverse Events (AE) | BSE (n= 50) Number (%) |
Placebo (n = 48) Number (%) |
|---|---|---|
| Bloating | 1 (0.02) | 0 (0.0) |
| Burping | 0 (0.0) | 0 (0.0) |
| Diarrhea | 0 (0.0) | 0 (0.0) |
| Nausea/vomiting | 0 (0.0) | 0 (0.0) |
| Bruising | 0 (0.0) | 0 (0.0) |
| Headache | 1 (0.0) | 0 (0.0) |
| Upset Stomach | 0 (0.0) | 0 (0.0) |
| Heartburn | 0 (0.0) | 0 (0.0) |
| Abdominal Pain | 0 (0.0) | 0 (0.0) |
| Muscle Pain | 0 (0.0) | 0 (0.0) |
| Taste Alteration | 0 (0.0) | 1 (0.02) |
| Other1 | 0 (0.0) | 0 (0.0) |
| All2,3 | 2 (0.04) | 1 (0.02) |
Other changes to health included back/neck pain and mental health issue.
Count of subjects who experienced at least one of the grade 2 adverse events.
P value for all adverse events comparing the treatment groups through Fisher’s exact test is 1.00.
Urinary, Plasma, and Prostate Tissue SFN Metabolites Level
Urinary and plasma SFN metabolite levels are presented in Table 3. Pre- to postintervention changes in total urinary SFN metabolites, and in individual SFN metabolites (SFN, SFN-NAC, and SFN-Cys), were statistically higher in the BSE versus the placebo group. SFN-GSH and SFN-CG changes in urine were greater in the BSE group but did not reach a significant level. In plasma, pre- to postintervention changes in total SFN ITCs and individual SFN metabolites (SFN-NAC, SFN-Cys, SFN-GSH, and SFN-CG) were statistically significant in the BSE group. No SFN metabolites were detected in plasma from the placebo group. Among all the subjects with prostate tissue available, three subjects in the BSE group had detectable SFN metabolites in the prostate tissue (data not shown due to small sample size).
Table 3.
Sulforaphane metabolite level changes in urine and plasma from pre- to post-treatment by treatment group among a random sample of subjects in the study (n = 98 randomized with n = 86 having urine/plasma).
| BSE Mean (SE) |
Placebo Mean (SE) |
p3 | |||||
| Variables from Urine | |||||||
| Preintervention n= 42 | Postintervention n= 42 | Changes2 n= 42 | Preintervention |
Postintervention |
Changes2 n= 44 | ||
| n= 44 | n= 44 | ||||||
| Total Urine Metabolites1 | 0.05 (0.02) | 4.80 (0.65) | 4.75 (0.64) | 0.04 (0.01) | 0.01 (0.003) | −0.02 (0.01) | <0.0001 |
| SFN | 0.002 (0.001) | 0.66 (0.16) | 0.66 (0.16) | 0.0007 (0.0003) | 0.0007 (0.0003) | 0.00005 (0.0003) | <0.0001 |
| SFN-NAC | 0.04 (0.01) | 2.90 (0.41) | 2.87 (0.40) | 0.03 (0.01) | 0.004 (0.002) | −0.02 (0.01) | <0.0001 |
| SFN-Cys | 0.008 (0.003) | 1.23 (0.19) | 1.22 (0.19) | 0.008 (0.002) | 0.005 (0.001) | −0.003 (0.002) | <0.0001 |
| SFN-GSH | 0.00 (0.00) | 0.0002 (0.0001) | 0.0002 (0.0001) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.08 |
| SFN-CG | 0.003 (0.001) | 0.005 (0.001) | 0.002 (0.002) | 0.004 (0.001) | 0.004 (0.001) | 0.0005 (0.002) | 0.73 |
| Variables from Plasma | |||||||
| Preintervention n = 42 | Postintervention n = 42 | Changes n = 42 | Preintervention n = 44 | Postintervention n = 44 | Changes n = 44 | ||
| Total Plasma Metabolites | 0.0002 (0.0002) | 0.12 (0.03) | 0.12 (0.03) | 0.0003 (0.0002) | 0.00 (0.00) | −0.0003 (0.0002) | <0.0001 |
| SFN | 0.00 (0.00) | 0.0001 (0.0001) | 0.0001 (0.0001) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.32 |
| SFN-NAC | 0.00 (0.00) | 0.03 (0.01) | 0.03 (0.01) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | <0.0001 |
| SFN-Cys | 0.00 (0.00) | 0.02 (0.01) | 0.02 (0.01) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | <0.0001 |
| SFN-GSH | 0.00 (0.00) | 0.03 (0.01) | 0.03 (0.01) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | <0.0001 |
| SFN-CG | 0.00 (0.00) | 0.04 (0.01) | 0.04 (0.01) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | <0.0001 |
Urinary SFN metabolite concentration units = micromolar (μM) concentrations of urinary SFN metabolite/millimolar (mM) concentrations of urinary creatinine. Plasma SFN metabolite concentrations are shown in micromolar (μM) concentrations.
Change = postintervention level minus preintervention level.
P values were calculated for difference of change in means across treatment groups. Mann–Whitney U-test was used for all the rest of the variables due to non-normal distribution.
HDAC Activity
Comparisons of PBMC HDAC activity levels (pre- to postintervention changes) between treatment groups are shown in Table 4. We also conducted stratified analyses by PCa diagnosis in order to identify potential differences between patients with and without cancer. Overall, there were no statistically significant differences of pre- to postintervention changes between BSE and placebo groups (Supplemental Figure S1). However, within the subgroup of subjects with confirmed PCa diagnosis, BSE supplement significantly increased HDAC activity (Supplemental Figure S2). Given that only three subjects had detectable SFN metabolites in prostate tissue, we can speculate that target tissue (prostate) HDAC activity levels also were unlikely to be changed by BSE versus placebo.
Table 4.
Comparison of PBMC HDAC activity among men scheduled for prostate biopsy.
| BSE Mean (SE) |
Placebo Mean (SE) |
P comparing changes | |||||
|---|---|---|---|---|---|---|---|
| All | Preintervention (n = 43) | Postintervention (n = 42) | Change (n = 42) | Preintervention (n = 44) | Postintervention (n = 44) | Change (n = 44) | |
| 849.7 (46.7) | 1080.0 (45.4) | 236.7 (59.0) | 937.1 (37.4) | 1080.9 (31.4) | 143.8 (39.9) | 0.14 | |
| Cancer | Preintervention (n = 15) | Postintervention (n = 15) | Change (n = 15) | Preintervention (n = 13) | Postintervention (n = 13) | Change (n = 13) | |
| 812.7 (63.5) | 1155.1 (61.9) | 342.3 (80.6) | 1007.3 (51.4) | 1053.8 (45.5) | 46.6 (65.8) | 0.01 | |
| NonCancer | Preintervention (n = 28) | Postintervention (n = 27) | Change (n = 27) | Preintervention (n = 31) | Postintervention (n = 31) | Change (n = 31) | |
| 869.5 (63.6) | 1038.2 (61.1) | 178.0 (78.3) | 907.6 (48.0) | 1092.2 (40.5) | 184.6 (48.2) | 0.95 | |
One postintervention sample in sulforaphane group had hemolysis.
IHC Biomarkers
In this study, 88 (90%) subjects had prostate tissue analyzed by IHC. Levels of H3K18ac, HDAC3, HDAC6, Ki-67, and p21 were evaluated by IHC from postintervention biopsy tissue (Table 5). There was no statistically significant difference between treatment groups for all the examined tissue biomarkers. Stratified by cancer and noncancer subgroup, no significant findings were found as well.
Table 5.
Comparisons of prostate tissue immunohistochemistry results between treatment groups.
|
All |
Cancer |
Noncancer |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue Biomarkers | Number of subjects with available information | BSE Mean (SE) |
Placebo Mean (SE) |
P comparing groups | FDR P value | Number of subjects with available information | BSE Mean (SE) |
Placebo Mean (SE) |
P comparing groups | FDR P value | Number of subjects with available information | BSE Mean (SE) |
Placebo Mean (SE) |
P comparing groups | FDR P value |
| H3K18ac H-score | 43/39 (Total = 82) | 185.8 (9.1) | 189.7 (8.0) | 0.83 | 0.91 | 17/13 (Total = 30) | 188.2 (14.6) | 184.6 (15.4) | 0.90 | 0.90 | 26/26 (Total = 52) | 184.2 (11.9) | 192.3 (9.5) | 0.71 | 0.89 |
| HDAC3 H-score | 44/43 (Total = 87) | 180.0 (15.0) | 172.3 (13.7) | 0.64 | 0.91 | 17/13 (Total = 30) | 167.6 (21.4) | 160.0 (24.5) | 0.75 | 0.90 | 27/30 (Total = 57) | 187.5 (20.5) | 177.7 (16.7) | 0.58 | 0.89 |
| HDAC6 H-score | 44/44 (Total = 88) | 187.0 (13.0) | 183.0 (12.5) | 0.91 | 0.91 | 17/14 (Total = 31) | 216.5 (20.1) | 182.1 (22.0) | 0.28 | 0.70 | 27/30 (Total = 57) | 168.5 (16.3) | 183.3 (15.4) | 0.43 | 0.89 |
| Ki67 % positivity | 41/42 (Total = 83) | 1.8 (0.2) | 1.9 (0.3) | 0.47 | 0.91 | 16/12 (Total = 28) | 1.7 (0.4) | 1.8 (0.5) | 0.84 | 0.90 | 25/30 (Total = 55) | 1.8 (0.3) | 1.9 (0.4) | 0.51 | 0.89 |
| P21 % positivity | 43/42 (Total = 85) | 2.4 (0.5) | 3.1 (0.6) | 0.41 | 0.91 | 17/13 (Total = 30) | 2.0 (0.8) | 3.8 (1.2) | 0.28 | 0.70 | 26/29 (Total = 55) | 2.7 (0.7) | 2.7 (0.6) | 0.94 | 0.94 |
Note: P values were calculated using Mann–Whitney U-test due to non-normal distribution.
Prostate Biopsy Gene Expression
Of the 98 patients within the study, we analyzed the transcriptomes from 55 prostate biopsy samples (30 BSE treated and 25 placebo) with good quality for RNA-seq analysis and 33% of the biopsies came from PCa patients. We obtained between 51.7 and 109 million reads per sample, totaling 4.5 billion reads with >80% of reads per sample mapped to the reference genome. rRNA contamination varied between samples, from 2.6% to 61.4%. Four samples with high duplication levels, rRNA contamination, and/or low read quality were filtered from the final analysis.
We identified only three significantly differentially expressed genes when examining the overall effect of BSE supplementation on gene expression (LINC00485, DYNC1I2P1, ADGRF2). These genes have no documented function in cancer progression, but are potentially interesting candidates for future studies to understand the effects of BSE supplementation on human physiology. Next, we hypothesized that the effects of BSE supplementation would be more pronounced in the samples from patients exhibiting signs of cancer. We next determined the interaction effect due to BSE on the cancer cells specifically and identified 40 genes (27 up, 13 down) that were significantly altered due to BSE treatment (Supplemental Table S1). When we examined the patterns of expression of these genes across all samples, we found that the changes in gene expression were not consistent between all samples within the same treatment (Fig. 2). We expected samples (columns within heat maps) to cluster first based on phenotype (tissue from patents with a cancer (tumor), or no cancer (normal)) and then by treatment (BSE/placebo), but instead we found samples clustered without respect to phenotype or treatment. Despite this variability, we identified a subset of the differentially expressed genes for more detailed examination (Supplemental Table S1, genes with bolded text). This gene list contained genes that were relevant to the etiology of cancer, including some related to the tumor microenvironment and cell migration. We also identified a cluster of six genes with similar patterns of expression in the heatmap (Fig. 2, box; Supplemental Table S1, bold genes) and noted that two of these genes were implicated in PCa development. These genes were alpha-methylacyl-CoA racemase (AMACR also known as P504S) and androgen receptor regulated long noncoding RNA 1 (ARLNC1 also called LINC02170, RP11–314O13.1, or PCa-Associated 47) (27, 28). We focused on these genes for confirmation work because C1orf64, SLIT1, RP11–627G23.1, and RP1–274L7.1 had less evidence of importance in PCa based on literature searches.
Figure 2. Expression patterns of significantly differentially expressed genes.
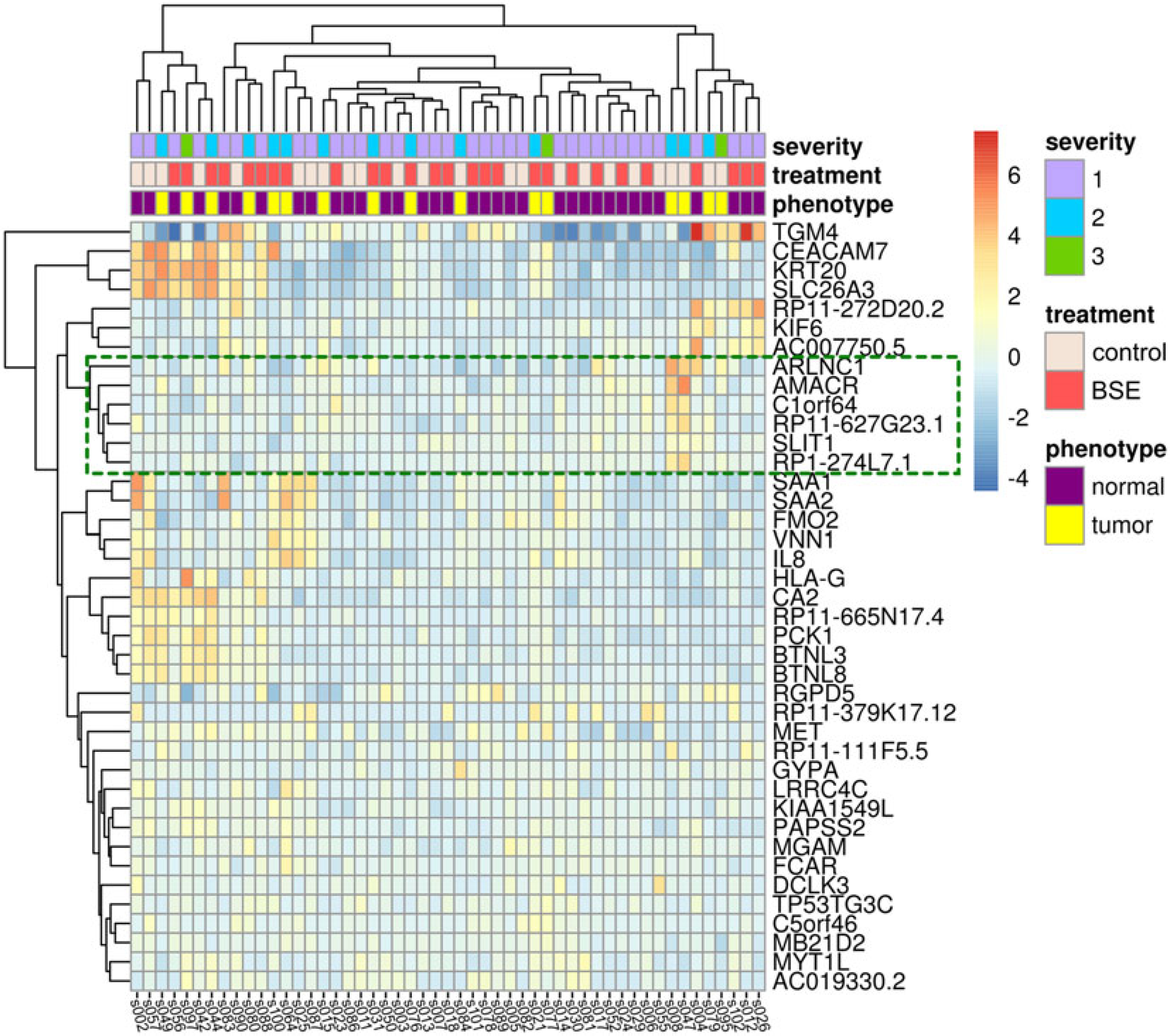
The heatmap shows the 40 significantly differentially expressed genes with respect to the BSE treatment on samples that came from subjects with evidence of prostate cancer. Columns represent expression of each sample, with sample labels at the bottom of the heatmap. Rows represent each differentially expressed gene, with the HUGO Gene Nomenclature Committee (HGNC) gene names listed along the right of the heatmap. Individual cells are colored relative to the row means of log2-fold changes. Each sample is annotated with level of cancer severity in the subject, treatment, and phenotype along the top of the heatmap. Dendrograms along the top and left side of the heatmap represent hierarchical clusters determined based on Euclidean distance calculations of sample-wide and gene-wide expression dissimilarities, respectively. The green box shows the cluster of six genes we identified, with two, AMACR and ARLNC1, having expression confirmed using qPCR (Figure 3).
ARLNC1 was upregulated in prostate tissue from cancer patients from the placebo group in the RNA-seq dataset, as compared to normal tissue (P = 0.0296), and a significant interaction between PCa and the effect of BSE supplementation was found (P = 0.0281) (Fig. 3). We verified this significant interaction by qPCR where ARLNC1 levels were 4.5-fold higher in samples from PCa patients that received the placebo than samples from patients with no cancer (Supplemental Figure S3). A 4.3-fold lower level of ARLNC1 was found among samples from cancer-positive patient treated with BSE as compared to placebo.
Figure 3. Effects of BSE supplements on ARLNC1 and AMACR expression.
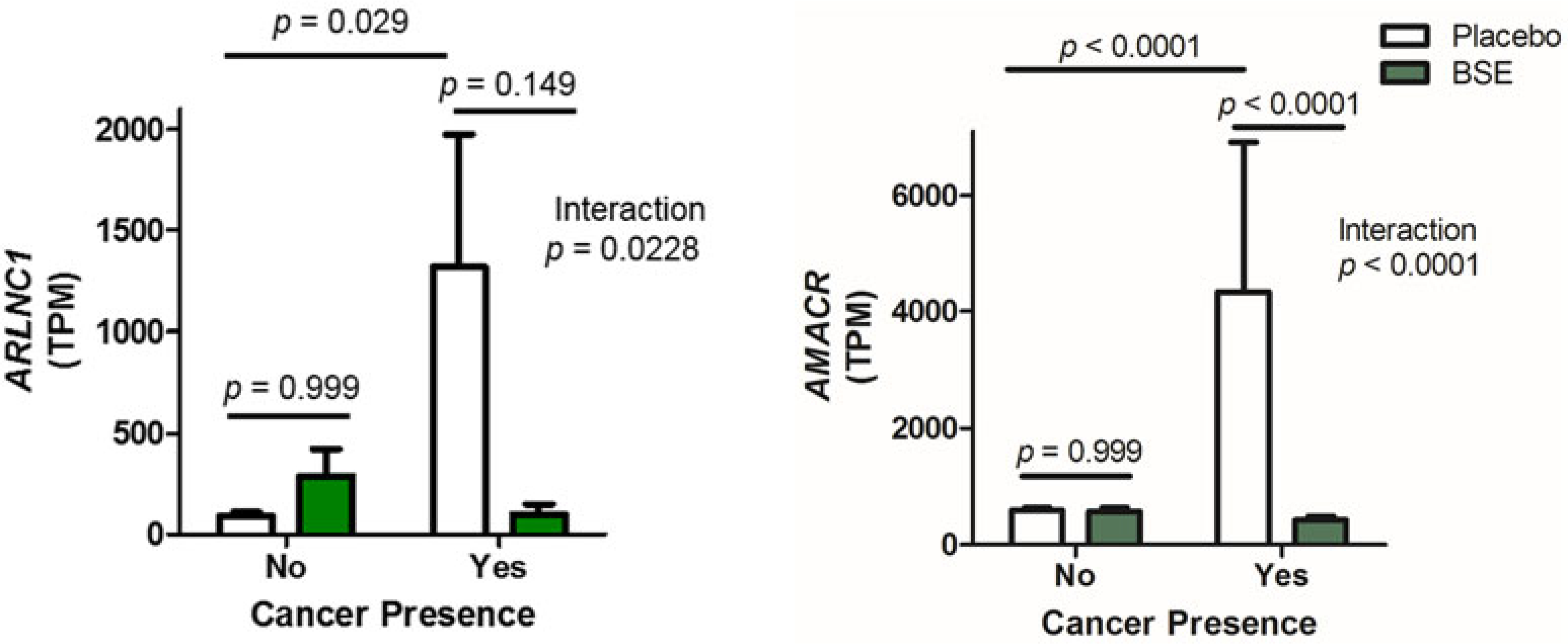
ARLNC1 and AMACR mRNA levels were detected in available prostate biopsy samples using RNA sequencing. Biopsies were designated no cancer if they were from subjects with benign tissue cores, while cancer positive samples came from subjects who had a severity score of 2–3. Subjects were given either a placebo or a BSE supplement. Bars indicate the mean expression level of the indicated gene with SEM. Statistics were calculated with DESeq2 software package (n = 7–18). TPM, transcripts per million.
AMACR mRNA levels were significantly increased in patients with PCa from the placebo group in the RNA-seq dataset (P < 0.0001), and AMACR mRNA levels were also significantly lower in biopsies of men that had cancer and took BSE supplements (P < 0.0001, compared to cancer positive placebo group) (Fig. 3). The qPCR results for AMACR generally confirmed the RNA-seq results, although the statistical significance was in the category of a trend (Supplemental Figure S3). More specifically, among the placebo-treated subjects, a 6.3-fold difference in AMACR mRNA was found between cancer positive and benign groups. Furthermore, a sevenfold decrease in the AMACR mRNA levels was found for cancer-positive subjects who took BSE supplements as compared to the cancer-positive placebo group. It was noted that the AMACR qPCR data were skewed in the biopsies from PCa patients, and when the data were log transformed, a significant overall effect of BSE supplements was found on AMACR mRNA levels (P = 0.0284).
Discussion
We conducted a randomized, placebo-controlled trial with BSE supplements in men and found significant increases in SFN metabolites in urine and plasma, as well as some significant interactions at the mRNA level with BSE treatment. In contrast, BSE supplementation was not associated with decreasing HDAC activity or significant changes in prostate tissue biomarkers among men, and few of the prostate biopsies had detectable levels of SFN metabolites. Indeed, we found only three subjects with detectable SFN levels in the prostate tissue, ranging from 23.0 to 62.3 pmole/g prostate tissue. We were unable to identify any associated metadata (e.g., patient demographic characteristics or urine/plasma metabolite levels) that correlated with the detectable SFN levels. Possible reasons for the null results may be that the intervention period was too short, the doses were too low, or the SFN metabolites were below detectable levels, including rapid elimination at the target site. It is also possible that SFN metabolites were lost for technical reasons, using OCT-embedded tissue processing methods. Previous studies have demonstrated that quick acidification is critical to maintain stability of SFN metabolites (29). While we attempted to thaw samples directly in acid to maintain the SFN levels, the OCT may have prevented the ability of acid to get into the tissue before it was thawed. Since none of the IHC biomarkers were significant, the semiquantitative H-score might not have been optimal. A more quantitative approach using high-content imaging and image analysis software might have identified pertinent changes.
The supplement we used was BSE treated with myrosinase, so SFN is directly delivered when consumed orally. In our study, the mean intervention period was 4.4 wk and the intervention doses were two 100-μmol SFN daily taken 12 h apart. The shorter duration or lower doses may restrict our ability to discern an effect of BSE supplementation, especially if the desired biological outcome caused by BSE supplementation requires cumulative intake. Additionally, not all of the subjects were PCa cases. Surprisingly, we found the BSE supplement significantly increased HDAC activity among PCa cases, contrary to our hypothesis. Our previous study using similar dosage among study volunteers showed that, although there were higher SFN metabolite levels in the plasma and urine of the BSE consumers, there was a transient decrease of HDAC activity 3 h after consuming either broccoli sprouts or BSE, followed by a significant increase of HDAC activity at the 12-h time point after consumption (21). The variation of HDAC changes between cancer and noncancer subjects may indicate that PCa patients have different SFN metabolizing mechanisms, or other effects on specific HDAC isoforms not influenced by SFN, compared to noncancer patients. In particular, the targeting of HDAC3 and HDAC6, versus other HDACs, may be worthy of further investigation (11–13, 30).
Our study results are applicable to men with a high risk for PCa identified at clinical examination. As we know, the majority of men with elevated PSA who undergo prostate biopsy will have no diagnosed cancer, although this differs by race (31). More research targeted toward this population will contribute to reducing the literature gap.
Clinical trials evaluating SFN effects in PCa were reviewed by Amjad et al., and our study was listed as one of the ongoing investigations (32). One earlier study was an open label clinical trial administering 400 g/wk broccoli and 400 g/wk pea to 22 patients with high-grade prostate intraepithelial neoplasia for 12 months. This study showed significant changes in TGFβ, insulin signaling, and EGF receptor pathways from broccoli dietary intervention, and broccoli consumption interacted with GSTM1 genotype (33). Two clinical trials have evaluated the effect of SFN in men with biochemical recurrence after radical prostatectomy, with both studies showing promising effect of decreasing the rate of PSA progression (34, 35). More specifically, the Alumkal et al.’s study was a single arm study among 20 patients who had recurrent PCa and were administered 200 μmol/day SFN for up to 20 wk, similar dosage to our study but with a longer treatment period. Results showed that PSA declined by ≥ 50% in most patients (35). In the Cipolla et al.’s study, 60 mg of stabilized free SFN was administered among 78 subjects for six months followed by two months without treatment. The effects on PSA were more pronounced from 3–6 months during the intervention and remained the same during the first 2-month postintervention (34). Our study showed no PSA difference between the two treatment groups, and the shorter duration may be the main reason. As far as we are aware, there are no additional updated publications from other ongoing clinical trials reviewed by Amjad et al. (32).
Also as far as we are aware, our study is the first to use RNA-seq to assess the effect of BSE on RNA expression in human prostate tissue. We were able to detect a significant interaction between BSE treatment and PCa, yet the patterns of gene expression associated with BSE treatment were variable within both treatment type and patient type. Despite this variability, we identified a cluster of six genes, AMACR, ARLNC1, C1orf64, SLIT1, RP11–627G23.1, and RP1–274L7.1, that would be of interest for future studies on gene expression. Expression patterns of these six genes are similar across samples, based on a distance-based clustering method, and AMACR and ARLNC1 have already been implicated as markers of PCa development (27, 28, 32, 36–38). We hypothesize that the four other genes (C1orf64, SLIT1, RP11–627G23.1, and RP1–274L7.1) could therefore also play a role in PCa development and are promising targets for future studies. In general, the qPCR experiments validated RNA-seq studies, although we were limited in the number of samples that came from patients positive for PCa with sufficient amounts of RNA for both assays. More samples would have increased the statistical power which would have been advantageous. Nevertheless, the data are consistent with the significant interactions identified in the RNA-seq dataset between cancer presence and BSE treatment. It is possible that these genes have a longer period of time that they are affected in response to BSE treatment, as compared to the period of time in which SFN metabolites are present. For example, our previously published RNA-seq dataset also found AMACR mRNA levels significantly downregulated in LNCaP cells at 6 and 24 h after a single SFN treatment (24). We first hypothesized that SFN could restore nuclear factor (erythroid-derived 2)-like 2 (Nrf2) expression; however, we did not identify Nrf2 response genes, consistent with another study that showed no effect of Nrf2 genes with SFN supplementation and may also be a timing issue (39). It could also be region specific, similar to a finding in our previous mouse study, which showed increases in an Nrf2 target gene only in the dorsolateral lobe of the prostate and not in the anterior or ventral lobes (13).
While the biological role of AMACR in cancer is complex, it is thought to link lipid metabolism with activity of nuclear receptors such as FXR and PPAR, and thus regulate the expression of enzymes such as cyclooxygenase-2 (27). It is exciting that SFN may inhibit its expression in PCa because efforts are currently underway to create chemical inhibitors or target AMACR expression with immunotherapy to treat PCa (36–38, 40). AMACR is also implicated in colon cancer carcinogenesis, and SFN treatment has been previously shown to significantly decrease AMACR expression in colon cancer Caco-2 cells (41).
The increasing abundance of ARLNC1 RNA we observed in samples from PCa patients is consistent with a recent study where the noncoding RNA was shown to directly interact with the androgen receptor and maintain a positive feedback loop that potentiated AR signaling during prostate cancer progression. ARLNC1 was also proposed to be an effective therapeutic target in androgen receptor positive prostate cancer because antisense oligos targeting this RNA decreased tumor growth in a mouse xenograft model (28). Given this potential as a target for cancer treatment, the lower abundance of ARLNC1 RNA in samples from prostate cancer patients that received BSE suggests that compounds from the diet, such as SFN, may also effectively decrease this RNA expression in cancer cells, although this would need to be further tested. Interestingly, we also identified that ARLNC1 was differentially expressed in multiple independent transcriptomics studies including esophageal, triple-negative breast cancer, and nonsmall-cell lung cancer, but any possible role this RNA plays in these cancers would need to be clarified as ARLNC1 was suggested to be a prostate lineage-specific lncRNA (42–48). Summarized differential gene expression for ARLNC1 from the Gene Expression Atlas is provided in Supplemental Table S2. The exploration of the role of ARLNC1 and other noncoding RNAs with particular attention to cell senescence (a key factor in cancer development) and causation is an important area of future research (49).
In conclusion, the BSE supplement was associated with significant interactions in gene expression among some genes that are related to PCa development, but did not significantly alter the expected PCa biomarkers. Small sample size of aggressive cancer diagnoses prohibits further examination of potential patterns in this trial. Future trials with longer treatment durations among a more homogenous study population should be considered.
Supplementary Material
Acknowledgments
We would like to thank all of the men who contributed their time and effort to join our study. We are grateful to OHSU pathology residents who prepared prostate tissues for research. Ms. Laura Peters (VAPORHCS) assisted with subject retention and specimen collection. We acknowledge staff at the OSU Mass Spectrometry Center and the Center for Genome Research and Biocomputing at Oregon State University. We also appreciate Dr. Shannon’s research coordinator, Mrs. Amy Palma, who spent time with study subjects and worked with surgical urologists.
Statements: Our study has not been published elsewhere nor has it been submitted simultaneously for publication elsewhere.
Funding
This study was supported by the National Institute of Health (P01 CA090890 and S10RR02787801) and Oregon Agricultural Experimental Station, Hatch Funds. Clinical Trial Registration: clinicaltrials.gov identifier: NCT01265953. Protocol ID = Portland VA-09-0607.
Abbreviations:
- AMACR
alpha-methylacyl-CoA racemase
- ARLNC1
androgen receptor-regulated long noncoding RNA
- BMI
body mass index
- BSE
broccoli sprout extract
- FDR
false discovery rate
- HDAC
histone deacetylase
- IHC
immunohistochemistry
- ITCs
isothiocyanates
- LS MEANS
least-squaresmeans
- NSAIDs
nonsteroidal anti-inflammatory drug
- OHSU
Oregon Health & Science University
- OCT
Optimal Cutting Temperature
- OSU
Oregon State University
- PBMC
peripheral blood mononuclear cells
- PCa
prostate cancer
- PIN
prostatic intraepithelial neoplasia
- PSA
prostate specific antigen
- qPCR
quantitative Real-Time PCR
- SFN
sulforaphane
- SE
standard error
- TFA
trifluoroacetic acid
- TPM
transcripts per million
- VAPORHCS
Veterans Affairs Portland Health Care System
References
- 1.Siegel RL, Miller KD, and Jemal A: Cancer statistics, 2018. CA Cancer J Clin 68, 7–30, 2018. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Farvid MS, Chen WY, Rosner BA, Tamimi RM, Willett WC, et al. : Fruit and vegetable consumption and breast cancer incidence: Repeated measures over 30 years of follow-up. Int J Cancer 144, 1496–1510, 2018. doi: 10.1002/ijc.31653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Bergan R, Shannon J, Slatore CG, Bobe G, et al. : The role of cruciferous vegetables and isothiocyanates for lung cancer prevention: Current status, challenges, and future research directions. Mol Nutr Food Res 62, 1700936, 2018. doi: 10.1002/mnfr.201700936 [DOI] [PubMed] [Google Scholar]
- 4.Petimar J, Wilson KM, Wu K, Wang M, Albanes D, et al. : A pooled analysis of 15 prospective cohort studies on the association between fruit, vegetable, and mature bean consumption and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev 26, 1276–1287, 2017. doi: 10.1158/1055-9965.EPI-16-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahey JW, Zhang Y, and Talalay P: Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci U S A 94, 10367–10372, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Talalay P, Cho CG, and Posner GH: A major inducer of anticarcinogenic protective enzymes from broccoli: Isolation and elucidation of structure. Proc Natl Acad Sci U S A 89, 2399–2403, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung FL, Conaway CC, Rao CV, and Reddy BS: Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis 21, 2287–2291, 2000. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Kensler TW, Cho CG, Posner GH, and Talalay P: Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci U S A 91, 3147–3150, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh AV, Xiao D, Lew KL, Dhir R, and Singh SV: Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis 25, 83–90, 2003. doi: 10.1093/carcin/bgg178 [DOI] [PubMed] [Google Scholar]
- 10.Myzak MC, Hardin K, Wang R, Dashwood RH, and Ho E: Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 27, 811–819, 2006. doi: 10.1093/carcin/bgi265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myzak MC, Karplus PA, Chung FL, and Dashwood RH: A novel mechanism of chemoprotection by sulforaphane: Inhibition of histone deacetylase. Cancer Res 64, 5767–5774, 2004. doi: 10.1158/0008-5472.CAN-04-1326 [DOI] [PubMed] [Google Scholar]
- 12.Ho E, Clarke JD, and Dashwood RH: Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr 139, 2393–2396, 2009. doi: 10.3945/jn.109.113332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaver LM, Löhr CV, Clarke JD, Glasser ST, Watson GW, et al. : Broccoli sprouts delay prostate cancer formation and decrease prostate cancer severity with a concurrent decrease in HDAC3 protein expression in TRAMP mice. Current Developments in Nutrition 2, nzy002, 2017. doi: 10.3945/cdn.117.002378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keum YS, Khor TO, Lin W, Shen G, Kwon KH, et al. : Pharmacokinetics and pharmacodynamics of broccoli sprouts on the suppression of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: Implication of induction of Nrf2, HO-1 and apoptosis and the suppression of Akt-dependent kinase pathway. Pharm Res 26, 2324–2331, 2009. doi: 10.1007/s11095-009-9948-5 [DOI] [PubMed] [Google Scholar]
- 15.Sakao K, Vyas AR, Chinni SR, Amjad AI, Parikh R, et al. : CXCR4 is a novel target of cancer chemopreventative isothiocyanates in prostate cancer cells. Cancer Prev Res (Phila) 8, 365–374, 2015. doi: 10.1158/1940-6207.CAPR-14-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SV, Warin R, Xiao D, Powolny AA, Stan SD, et al. : Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res 69, 2117–2125, 2009. doi: 10.1158/0008-5472.CAN-08-3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vyas AR, Hahm ER, Arlotti JA, Watkins S, Stolz DB, et al. : Chemoprevention of prostate cancer by d,l-sulforaphane is augmented by pharmacological inhibition of autophagy. Cancer Res 73, 5985–5995, 2013. doi: 10.1158/0008-5472.CAN-13-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atwell LL, Zhang Z, Mori M, Farris P, Vetto JT, et al. : Sulforaphane bioavailability and chemopreventive activity in women scheduled for breast biopsy. Cancer Prev Res (Phila) 8, 1184–1191, 2015. doi: 10.1158/1940-6207.CAPR-15-0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolignano D, Mattace-Raso F, Torino C, D’Arrigo G, Abd ElHafeez S, et al. : The quality of reporting in clinical research: The CONSORT and STROBE initiatives. Aging Clin Exp Res 25, 9–15, 2013. doi: 10.1007/s40520-013-0007-z [DOI] [PubMed] [Google Scholar]
- 20.Diet History Questionnaire, Version 1.0. Bethesda, MD: Applied Research Program, National Cancer Institute, National Institute of Health, 2002. [Google Scholar]
- 21.Atwell LL, Hsu A, Wong CP, Stevens JF, Bella D, et al. : Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol Nutr Food Res 59, 424–433, 2015. doi: 10.1002/mnfr.201400674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahey JW, Holtzclaw WD, Wehage SL, Wade KL, Stephenson KK, et al. : Sulforaphane bioavailability from glucoraphanin-rich broccoli: Control by active endogenous myrosinase. PLoS One 10, e0140963, 2015. doi: 10.1371/journal.pone.0140963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C: Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419, 2017. doi: 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaver LM, Buchanan A, Sokolowski EI, Riscoe AN, Wong CP, et al. : Transcriptome analysis reveals a dynamic and differential transcriptional response to sulforaphane in normal and prostate cancer cells and suggests a role for Sp1 in chemoprevention. Mol Nutr Food Res 58, 2001–2013, 2014. doi: 10.1002/mnfr.201400269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, and Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 5502014. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2016. [Google Scholar]
- 27.Lloyd MD, Yevglevskis M, Lee GL, Wood PJ, Threadgill MD, et al. : alpha-Methylacyl-CoA racemase (AMACR): metabolic enzyme, drug metabolizer and cancer marker P504S. Prog Lipid Res 52, 220–230, 2013. doi: 10.1016/j.plipres.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Pitchiaya S, Ciéslik M, Niknafs YS, Tien JC-Y, et al. : Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet 50, 814–824, 2018. doi: 10.1038/s41588-018-0120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al Janobi AA, Mithen RF, Gasper AV, Shaw PN, Middleton RJ, et al. : Quantitative measurement of sulforaphane, iberin and their mercapturic acid pathway metabolites in human plasma and urine using liquid chromatography-tandem electrospray ionisation mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 844, 223–234, 2006. doi: 10.1016/j.jchromb.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 30.Myzak MC, Tong P, Dashwood WM, Dashwood RH, and Ho E: Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 232, 227–234, 2007. [PMC free article] [PubMed] [Google Scholar]
- 31.Mahal BA, Chen YW, Muralidhar V, Mahal AR, Choueiri TK, et al. : Racial disparities in prostate cancer outcome among prostate-specific antigen screening eligible populations in the United States. Ann Oncol 28, 1098–1104, 2017. doi: 10.1093/annonc/mdx041 [DOI] [PubMed] [Google Scholar]
- 32.Amjad AI, Parikh RA, Appleman LJ, Hahm ER, Singh K, et al. : Broccoli-derived sulforaphane and chemoprevention of prostate cancer: From bench to bedside. Curr Pharmacol Rep 1, 382–390, 2015. doi: 10.1007/s40495-015-0034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traka M, Gasper AV, Melchini A, Bacon JR, Needs PW, et al. : Broccoli consumption interacts with GSTM1 to perturb oncogenic signalling pathways in the prostate. PLoS One 3, e2568, 2008. doi: 10.1371/journal.pone.0002568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cipolla BG, Mandron E, Lefort JM, Coadou Y, Della Negra E, et al. : Effect of sulforaphane in men with biochemical recurrence after radical prostatectomy. Cancer Prev Res (Phila) 8, 712–719, 2015. doi: 10.1158/1940-6207.CAPR-14-0459 [DOI] [PubMed] [Google Scholar]
- 35.Alumkal JJ, Slottke R, Schwartzman J, Cherala G, Munar M, et al. : A phase II study of sulforaphane-rich broccoli sprout extracts in men with recurrent prostate cancer. Invest New Drugs 33, 480–489, 2015. doi: 10.1007/s10637-014-0189-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honma I, Torigoe T, Hirohashi Y, Kitamura H, Sato E, et al. : Aberrant expression and potency as a cancer immunotherapy target of alpha-methylacyl-coenzyme A racemase in prostate cancer. J Transl Med 7, 103, 2009. doi: 10.1186/1479-5876-7-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgenroth A, Urusova EA, Dinger C, Al-Momani E, Kull T, et al. : New molecular markers for prostate tumor imaging: A study on 2-methylene substituted fatty acids as new AMACR inhibitors. Chem Eur J 17, 10144–10150, 2011. doi: 10.1002/chem.201003176 [DOI] [PubMed] [Google Scholar]
- 38.Wilson BA, Wang H, Nacev BA, Mease RC, Liu JO, et al. : High-throughput screen identifies novel inhibitors of cancer biomarker alpha-methylacyl coenzyme A racemase (AMACR/P504S). Mol Cancer Ther 10, 825–838, 2011. doi: 10.1158/1535-7163.MCT-10-0902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wise RA, Holbrook JT, Criner G, Sethi S, Rayapudi S, et al. : Lack of effect of oral sulforaphane administration on Nrf2 Expression in COPD: A randomized, double-blind, placebo controlled trial. PLoS One 11, e0163716, 2016. doi: 10.1371/journal.pone.0163716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carnell AJ, Hale I, Denis S, Wanders RJ, Isaacs WB, et al. : Design, synthesis, and in vitro testing of alpha-methylacyl-CoA racemase inhibitors. J Med Chem 50, 2700–2707, 2007. doi: 10.1021/jm0702377 [DOI] [PubMed] [Google Scholar]
- 41.Traka M, Gasper AV, Smith JA, Hawkey CJ, Bao Y, et al. : Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J Nutr 135, 1865–1872, 2005. doi: 10.1093/jn/135.8.1865 [DOI] [PubMed] [Google Scholar]
- 42.Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, et al. : Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 44, D746–52, 2016. doi: 10.1093/nar/gkv1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajan P, Sudbery IM, Villasevil ME, Mui E, Fleming J, et al. : Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol 66, 32–39, 2014. doi: 10.1016/j.eururo.2013.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren S, Peng Z, Mao JH, Yu Y, Yin C, et al. : RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res 22, 806–821, 2012. doi: 10.1038/cr.2012.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maag JLV, Fisher OM, Levert-Mignon A, Kaczorowski DC, Thomas ML, et al. : Novel aberrations uncovered in Barrett’s esophagus and esophageal adenocarcinoma using whole transcriptome sequencing. Mol Cancer Res 15, 1558–1569, 2017. doi: 10.1158/1541-7786.MCR-17-0332 [DOI] [PubMed] [Google Scholar]
- 46.Eswaran J, Cyanam D, Mudvari P, Reddy SD, Pakala SB, et al. : Transcriptomic landscape of breast cancers through mRNA sequencing. Sci Rep 2, 264, 2012. doi: 10.1038/srep00264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Djureinovic D, Hallstrom BM, Horie M, Mattsson JS, La Fleur L, et al. : Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight 1, e86837, 2016. doi: 10.1172/jci.insight.86837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, et al. : RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multi-class, and molecular pathway cancer diagnostics. Cancer Cell 28, 666–676, 2015. doi: 10.1016/j.ccell.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purcell M, Kruger A, and Tainsky MA: Gene expression profiling of replicative and induced senescence. Cell Cycle 13, 3927–3937, 2014. doi: 10.4161/15384101.2014.973327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


