Abstract
This study was conducted in order to know the colonization rate of MDR enterobacteria in neonates during their hospitalization in neonatal intensive care unit (NICU). Furthermore, we investigated risk factors for potential colonization and molecular epidemiology of isolated resistant bacteria. This prospective study was carried out in the neonatology and intensive care unit department of the University Hospital of Fez (Morocco) from February 2013 to July 2015. All consecutive admitted newborns were screened for intestinal and nasal carriage of MDR enterobacteria at admission of the babies and during the hospitalization. During the study period, a total of 641 Enterobacteriaceae were isolated and Klebsiella pneumoniae was the predominated bacteria. Bacterial identification and antibiograms were performed according to the international standards. On admission, 455 newborns were screened. A median age of these newborns was 1 day with an extended 147 days and their average weight was 2612 ± 1023 grams. 22.4% of neonates were found colonized by an ESBL producing Enterobacteriaceae (ESBL-E), 8.7% by a carbapenemases producing Enterobacteriaceae (CPE). During hospitalization, 207 of newborns were included in the acquisition study. 59.4% of newborns acquired an ESBL-E during their stay, 12.5% has acquired CPE. The blaCTXM-15 gene was the most frequently detected (81.2%) among ESBL-E. While, all CPE has expressed the blaOXA-48 gene exclusively. Two risk factors have been significantly associated with MDR enterobacteria colonization at admission which are newborns admission from maternity of the university hospital (95% CI, 1.859–5.129, P = 0.000) and neurological distress (95% CI, 1.038 to 4.694, P = 0.040). During hospitalization, the none risk factor was significantly associated with the carriage of MDR-E. The high rate of colonization, the MDR enterobacteria and the resistance genes found represent good indicator of cross-transmission in the NICU. An active strategy to control the spread of MDR enterobacteria should be applied.
Introduction
Multidrug-resistant Enterobacteriaceae (MDR-E) occurring in health care settings cause several healthcare associated infections (HAI), particularly bacteremia (45%), pneumonia (22%), gastrointestinal infection (8%), and urinary tract infection (5%) [1]. These infections remain a major cause of mortality and morbidity among hospitalized newborns, especially premature infants and babies with low birth weight [2–4]. According to the World Health Organization, HAI account from 4 to 56% of all causes of neonatal death among infants hospitalized in developing countries. It can attain 75% in Southeast Asia and Africa [5].
Newborns admitted to the neonatal intensive care unit (NICU) are frequently colonized by MDR bacteria. The risk factors related to this colonization has been widely reported and can start from anamnestic factors through to risk factors related to invasive procedures associated with health care. The acquisition of MDR-E in the NICU may result from the increasing use of antibiotics or transmission from one patient to another due to caregivers and the hospital environment [6]. Neonatal sepsis caused by multidrug-resistant gram-negative organisms, particularly Enterobacteriaceae is a concern in premature infants, and treatment options are limited [7]. In low- and middle-income countries (LMICs), these infections are associated with poor outcomes and high mortality rates [8].
Outbreaks occurring in NICUs are often caused by Enterobacteriaceae producing Extended-spectrum beta-lactamases (ESBLs) or carbapenemases (CPEs). This is a particular concern because these resistances are encoding by genes carried on mobile genetic elements, facilitating patient-to-patient transmission. Furthermore, these genes may carry resistance to other antimicrobial classes, which amplifies the risk of therapeutic failure [9]. Most previous reports regarding the molecular characterization of ESBLs concluded that the CTX-M variant encoding the ESBL were predominating worldwide. While, the prevalence of carbapenemases variants reported such as KPC, IMP, and VIM varies depending on the areas of the region. The OXA-48 remains the cabapenemases most identified in Mediterranean countries.
Although early screening of newborns colonized with ESBL or carbapenemase-producing bacteria can potentially contribute to the prevention and control of late-onset infections, few studies are addressing this issue in hospital children [10]. We established a systematic screening of newborn at admission to studied the acquisition of multidrug resistant bacteria. We reported, in previous work, the acquisition rate of Acinetobacter baumannii through hospitalized newborns. In the present study, we address the problem of MDR Enterobacteriaceae involved during newborn hospitalization. It aims to describe the colonization rate, risk factors and the molecular epidemiology of the bacteria involved during hospitalization.
Methods
Study
This prospective study was conducted at the neonatology and intensive care unit department of a University Hospital of Fez (Morocco) from February 2013 to July 2015. The setting was a medical and surgical NICU which has 18 beds divided into 2 sectors (9 beds were in each one); sector 1 corresponds to an intensive care unit and sector 2 corresponds to a pretermbaby unit. This NICU is the only one in Fez city (center of Morocco) with a population estimated approximately at 1.5 million inhabitants. Three seniors, 8 physicians, and 6 nursesare assigned to this ward daily.
Patients
During this period, all consecutive neonates admitted into the unit are included. Only the first NICU admission per neonate was included in the analysis. Neonates hospitalized during the week end, which have died or output before 48h of hospitalization were excluded. Babies were evaluated for Enterobacteriaceae intestinal (a) carriage at admission and (b) acquisition during hospitalization. Imported carriers and those without follow-up samples (due to death or discharge before the scheduled follow-up sampling) were excluded from acquisition analysis.
Ethics committee
Ethical approval was obtained from the local ethics committee in the University Hospital Center Hassan II in Fez- Morocco and all the parents’ babies were informed of the conditions related to the study and gave their written, informed consent.
Statistical analysis
Potential risk factors associated with Enterobacteriaceae colonization were studied. The sociodemographic and clinical characteristics of patients were collected prospectively by using a standard written questionnaire. The statistical analysis for our study was done using SPSS, version 20 (SPSS Inc., Chicago, IL, USA) software. It consisted primarily to describe the study population. Results for quantitative variables were presented as mean ± standard deviation and for qualitative variables; results were presented as number (percentage). Then, a univariate analysis was performed to establish all associations between gender, age, birth weight, prematurity, birthplace, admission route, delivery mode, date of hospital and NICU admission, and diagnosis on NICU admission. Antimicrobial therapy, breastfeeding, central or peripheral venous catheterization, and length of hospital stay were also recorded in the questionnaire. Chi-square test and fisher’s exact test were used to establish significant association as appropriate. The P < 0.05 was deemed statistically significant. The multivariate analysis was performed to identify potential risk factors associated with intestinal Enterobacteriaceae multi-drug-resistant colonization using simple logistic regression analysis. All variables with p < 0.2 in univariate analysis were included in a logistic regression model for multivariate analysis. Odds ratios were presented with the corresponding 95% confidence intervals (OR, CI 95%).
Sampling and screening
Two rectal swabs were collected from each newborn. The initial sample was performed up to 6 hours from admission to the NICUs and the second one after 5 days of hospitalization. Rectal swab specimens were enriched in nutrient broth BHI (Brain Heart infusion, Oxoid®) at 37°C for 24h. Then, they were inoculated on Mac Conkey agar plates and then incubated at 37° C for 24h. The identification of Enterobacteriaceae isolates was performed by classical bacteriological techniques and confirmed by using API20E galleries (Biomérieux, Marcy l’Etoile, France).
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined as recommended by the EUCAST 2013, the following antimicrobial agents (Oxoid®) were tested: amoxicillin (10 mg); amoxicillin/clavulanic acid (20/10mg); cefalotin (30mg); cefotaxime (30mg); céftazidime (30mg); ertapenem (10 mg); nalidixic acid (30 mg); ciprofloxacin (5 mg); norfloxacin (10 mg); gentamicin (10 mg); amikacin (30mg); fosfomycin(50mg); and trimethoprim/sulfamethoxazole (SXT) (1.25/23.75 mg). Enterobacteriaceae isolates resistant to three or more classes of antibiotics were considered as MDR. ESBL production was screened by the double-disk synergy test. The standard strains E. coli ATCC 25922 were used as negative control strains for ESBL production. The modified Hodge test was performed for screening carbapenemases production to all ertapenem resistant isolates.
Preparation of DNA template for PCR
Total DNA was extracted by suspending a few colonies of an overnight culture of Enterobacteriaceae isolates in 500μL of DNase- and RNase-free water (Invitrogen, Paisley, UK). The suspension was boiled at 100°C for 10min in a thermal block (Polystat 5, Bioblock Scientific, France), then centrifuged at 14000 x g for 10 min. An aliquot of 2 μL of the supernatant was used as a DNA template for PCR.
Detection of β-lactamase- and carbapenemase-encoding genes
All DNA from ESBLE and CPE isolates have been stored at -20°C for detection of subsequent resistance genes. Therefore, All ESBL-producing strains were screened by PCR as described previously for the following β-lactamase-encoding genes: blaCTX-M phylogenetic lineage groups 1, 2, and 9; blaTEM; and blaSHV. The blaOXA-48,blaKPC,blaNDM, blaIMP, and blaVIM genes were also detected to confirm the presence of carbapenemase-encoding genes. The known β-lactamase-producing strains, recuperated from Pasteur institute of Morocco, E. coli U2A1790(CTX-M-1), E. coli U2A1799 (CTX-M-9), Salmonellasp. U2A2145(CTX-M-2), Salmonella sp. U2A1446 (TEM-1 and SHV-12) were used as positive controls. The known carbapenemase-producing strains were used as positive controls. PCR products were detected on 1.5% agarose gels (FMC BioProducts, Rockland, ME) following ethidium bromide staining and ultraviolet illumination, and were photographed with an Olympus digitalcamera (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
Sequencing of resistance genes
All amplified products obtained were sequenced to validate their identities. Both strands of the purified amplicons were sequenced with the same primers used for PCR amplification. The nucleotide and deduced protein sequences were analysed with software available on the Internet at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov).
Results
Study population
During the study period, 455 neonates met the criteria for inclusion were swabbed at admission to the NICU. The average gestational age and mean birth weight were 35.2 (±3.2) weeks and 2612.1 g (±1023.2) respectively. 150 were carriers of MDR Enterobacteriaceae at admission. Of the 305 non-carriers, just 207 had follow-up rectal swabs discharge. The remaining 98 patients were excluded from acquisition analysis because they were discharged or died before the scheduled sampling (Fig 1).
Fig 1. Flowchart of the patients.
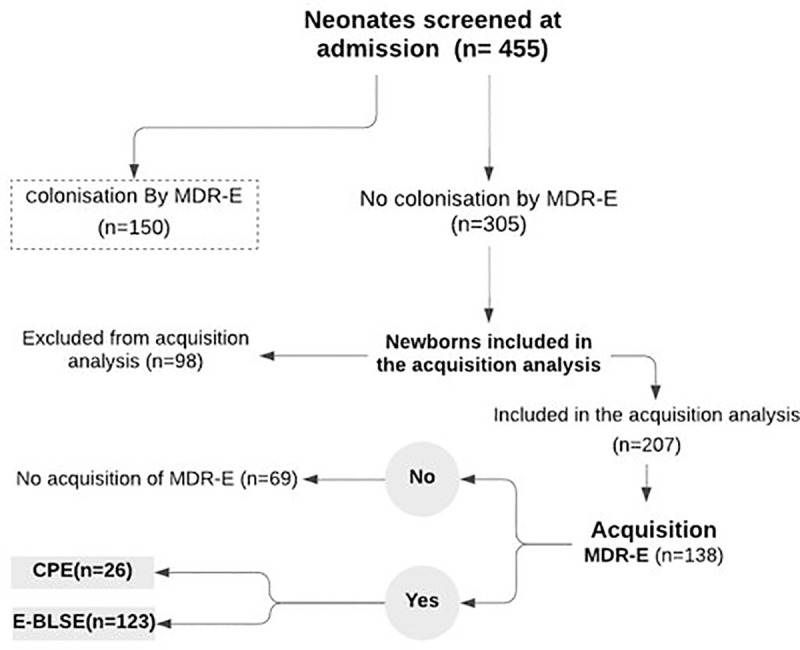
MDR Enterobacteriaceae intestinal carriage
During the study period, a total of 641 Enterobacteriaceae were isolated. Klebsiella pneumoniae was the predominated bacteria (n = 319), followed by Escherichia coli (n = 261). Furthermore, 17 strains of Enterobacter cloacae, 16 of Klebsiella oxytoca, 10 of Citrobacter freundii, 9 of Proteus mirabilis, 4 of Morganella morganii, 1 of Enterobacter aerogenes,1 of Proteus vulgaris, 1 of Citrobacter braakii, 1 of Providencia stuartii, and1 of Serratia marcescens were isolated.
In the current work, 33% of admitted newborns were carriers of MDR Enterobacteriaceae (150/455). We noted that 22.4% of all strains were producers of ESBL resistance and 8.7% were producers of carbapenemases resistance. Usually, MDR E. coli species was the most identified in this carriage (50.6%).
During NICU stay, the prevalence of MDR Enterobacteriaceae intestinal acquisition was 64.2% and 92.4% of them were ESBL producers. This time, MDR K. pneumoniae was the most predominant species acquired (88.7%). The prevalence of the different resistance types found in Enterobacteriaceae (ESBL and carbapenemases) were shown in Table 1.
Table 1. Prevalence of ESBL and CARBA carriage among newborns hospitalised in NICU.
| Enterobacteriaceae | Carriers at admission* N = 455[%(N)] | Colonised during NICU stay* N = 207[%(N)] |
|---|---|---|
| MDR-Enterobacteriaceae | 33 (150/455) | 64.2 (133/207) |
| ESBL-Enterobacteriaceae | 22.4 (102/455) | 59.4 (123/207) |
| ESBL-K. pneumoniae | 77.4 (79/102) | 94.3 (116/123) |
| ESBL-E.coli | 32.3 (33/102) | 26 (32/123) |
| ESBL-Other species | 8.8 (9/102) | 8.1 (10/123) |
| CARBA-Enterobacteriaceae | 8.7 (40) | 12.5 (26/207) |
| CARBA-K. pneumoniae | 42.5 (17/40) | 46.1 (12/26) |
| CARBA-E.coli | 45 (18/40) | 61.5 (16/26) |
| CARBA-Other species | 12.5 (5/40) | 7.6 (2/26) |
*Carriage of at least one MDR Enterobacteriaceae isolate.
Risk factors of MDR Enterobacteriaceae carriage
Univariate analysis performed between patient characteristics and intestinal carriage of MDR-Enterobacteriaceae on admission had shown that 57.3% of admitted newborns aged younger than 48h were carriers of MDR-E (p = 0.001). In addition, 54.7% of non-breastfed babies were found colonized at admission (p = 0.042) (Table 2).
Table 2. Association between patient’s characteristics and prevalence of multidrug-resistant Enterobacteriaceae carriage on the day of admission and during hospitalization at NICU.
| At admission | During NICU stay | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Category | Patients numbers (%) | MDR-EB - | MDR-EB +* | p-value | Patients numbers (%) | MDR-EB - | MDR-EB + | p-value | |
| n = 455 | [N (%)] | [N (%)] | n = 207 | [N (%)] | [N (%)] | ||||
| Gender | Male | 267 (58.7) | 180 (59) | 87 (58) | 0.457 | 124 (59.9) | 36 (65.5) | 88 (57.9) | 0.207 |
| Female | 188 (41.3) | 125 (41) | 63 (42) | 83 (40.1) | 19 (34.5) | 64 (42.1) | |||
| Age (days) | 0–2 | Mean±SD | 222 (72.8) | 86 (57.3) | 0.001 | Mean±SD | 39 (70.9) | 112 (73.7) | 0.408 |
| > 2 | 6.5±15.8 | 83 (27.2) | 64 (42.7) | 5.1±12.2 | 16 (29.1) | 40 (26.3) | |||
| Prematurity | Yes | 224 (49.2) | 156 (51.1) | 68 (45.3) | 0.140 | 113 (54.6) | 29 (52.7) | 84 (55.3) | 0.433 |
| No | 231 (50.8) | 149 (48.9) | 82 (54.7) | 94 (45.4) | 26 (47.3) | 68 (44.7) | |||
| Birth weight (g) | < 2500 | Mean±SD | 158 (51.8) | 68 (45.3) | 0.115 | Mean±SD | 31 (56.4) | 81 (53.3) | 0.408 |
| ≥ 2500 | 2612±1026g | 147 (48.2) | 82 (54.7) | 2466±975 | 24 (43.6) | 71 (46.7) | |||
| Pathology** | Respiratory distress | 259 (56.9) | 182 (59.7) | 77 (51.3) | 0.056 | 123 (59.4) | 32 (58.2) | 91 (59.9) | 0.475 |
| Icterus | 38 (8.4) | 21 (6.9) | 17 (11.3) | 0.078 | 14 (6.8) | 2 (3.6) | 12 (7.9) | 0.229 | |
| Surgical pathology | 31 (6.8) | 17 (5.6) | 14 (9.3) | 0.099 | 11 (5.3) | 4 (7.3) | 7 (4.6) | 0.328 | |
| Neonatal suffering | 34 (7.5) | 24 (7.9) | 10 (6.7) | 0.401 | 16 (7.7) | 4 (7.3) | 12 (7.9) | 0.573 | |
| Neonatal infections | 35 (7.7) | 20 (6.6) | 15 (10) | 0.134 | 16 (7.7) | 3 (5.5) | 13 (8.6) | 0.342 | |
| Neurological distress | 44 (9.7) | 34 (11.1) | 10 (6.7) | 0.086 | 22 (10.6) | 3 (5.5) | 19 (12.5) | 0.112 | |
| Congenital malformations | 11 (2.4) | 9 (3) | 2 (1.3) | 0.239 | 6 (2.9) | 2 (3.6) | 4 (2.6) | 0.505 | |
| Others | 49 (10.8) | 29 (9.5) | 20 (13.3) | 0.141 | 18 (8.7) | 7 (12.7) | 11 (7.2) | 0.168 | |
| Birthplace | Maternity of UH Fez | 265 (58.2) | 190 (41.8) | 75 (50) | 0.026 | 125 (60.4) | 33 (60) | 92 (60.5) | 0.703 |
| Other hospital | 164 (36) | 97 (31.8) | 67 (44.7) | 66 (31.9) | 19 (34.5) | 47 (30.9) | |||
| Home | 26 (5.7) | 18 (5.9) | 8 (5.3) | 16 (7.7) | 3 (5.5) | 13 (8.6) | |||
| Admission route | Maternity of UH Fez | 235 (51.6) | 178 (58.4) | 57 (38) | 0.000 | 117 (56.5) | 31 (56.4) | 86 (56.6) | 0.870 |
| Other hospital | 124 (27.3) | 77 (25.2) | 47 (31.3) | 56 (27.1) | 16 (29.1) | 40 (26.3) | |||
| Home | 96 (21.1) | 50 (16.4) | 46 (30.7) | 34 (16.4) | 8 (14.5) | 26 (17.1) | |||
| Delivery mode | vaginal | 310 (68.1) | 205 (67.2) | 105 (70) | 0.312 | 142 (68.6) | 34 (61.8) | 108 (71.1) | 0.137 |
| Caesarean section | 145 (31.9) | 100 (32.8) | 45 (30) | 65 (31.4) | 21 (38.2) | 44 (28.9) | |||
| Venous Catheterization | peripheral | -- | -- | -- | -- | 203 (98.1) | 55 (100) | 148 (97.4) | 0.288 |
| central | -- | -- | -- | 4 (1.9) | 0 (0) | 4 (2.6) | |||
| Breastfeeding | Breastfed newborn | 179 (39.3) | 111 (36.4) | 68 (45.3) | 0.042 | 69 (33.3) | 17 (30.9) | 52 (34.2) | 0.394 |
| Diet newborn | 276 (60.7) | 194 (63.6) | 82 (54.7) | 138 (66.7) | 38 (69.1) | 100 (65.8) | |||
| Antibiotherapy | Ceftriaxone+gentamicin | -- | -- | -- | -- | 116 (56) | 35 (63.6) | 81 (53.3) | 0.121 |
| Aximicin+gentamicin | -- | -- | -- | 74 (35.7) | 18 (32.7) | 56 (36.8) | 0.354 | ||
*Carriage of at least one MDR Enterobacteriaceae isolate
**Neonates may have more than one reason for hospitalization.
The same analysis was performed between patients’ characteristics and this time with the intestinal acquisition of MDR-E during hospitalization revealed that there is a significant association between the age < 48h (p = 0.001) and the acquisition of MDR-E during the stay. Any hospital stay ≥3 days (p = 0.007) was also linked to the acquisition of MDR-E. In other words, the results showed that 73.3% of newborns were younger than 48h at the time of admission and 96% of all babies which stayed at least 3 days in the ward were acquired an MDR-E (Table 2).
Multivariate analysis
On admission, the multivariate analysis showed that newborns imported from maternity of university hospital (95% CI, 1.859–5.129, P = 0.000) were 3 times more likely to be carriers of MDR-E than those imported from their houses. Also, neonates with neurological distress are 2 times more likely to be carriers of MDR-E (95% CI, 1.038 to 4.694, P = 0.040) than those who do not have this pathology. Table 3 shows all the risk factors for the carriage of MDR-E defined in this study.
Table 3. Multivariable analysis for MDR-Enterobacteriaceae carriage at NICU admission and acquisition during hospitalization.
| Variable | Multivariable analysis for MDR Enterobacteriaceae carriage at NICU admission | |
|---|---|---|
| OR (95%CI) | P value | |
| Neurological distress | 2.207 (1.038–4.694) | 0.040 |
| Imported from the maternity ward of the university hospital | 3.088 (1.859–5.129) | 0.000 |
| Imported from other wards or institutions | 2.010 (1.251–3.230) | 0.004 |
During hospitalization, the none risk factor was significantly associated with the carriage of MDR-E (Table 3).
Antibiotic resistance
On admission, the study of the resistance profiles of Enterobacteriaceae concerning the different antibiotics tested among newborns showed a high resistance rate for Ampicillin (75.3%) followed by Amoxicillin associated with Clavulanic acid (53.9%). Ceftazidime or cefotaxime resistance was also high with 47.6%. However, the resistance rate to Amikacin was very low (0.9%).
During the hospital stay, the study of resistance patterns of Enterobacteriaceae isolated showed that almost all isolates are resistant to ampicillin (97%) and amoxicillin/clavulanic acid (92%). The rate of resistance to ceftazidime/cefotaxime was 89.9% and gentamicin was 82.9%. Half of the isolates were resistant to nalidixic acid and ciprofloxacin. While only 3% of the isolates were resistant to amikacin. The different resistance profiles found according to the bacterial species identified are presented in Table 4.
Table 4. Resistance profiles of the different Enterobacteriaceae isolated on admission and during hospitalization.
| Enterobacteriaceae species (No.; %) n = 313 | No. (%) of resistant isolates at admission | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | FOX | CTX/CAZ | GN | AK | NA | NOR/CIP | SXT | ETP | |
| E. coli (n = 152; 48) | 88 | 65 | 36 | 56 | 51 | 2 | 53 | 46 | 38 | 18 |
| K. pneumoniae (n = 124; 39.6) | 124 | 80 | 3 | 79 | 72 | 0 | 39 | 39 | 49 | 17 |
| E. cloacae (n = 11; 3.5) | * | * | * | 9 | 7 | 0 | 7 | 5 | 6 | 4 |
| K. oxytoca (n = 10; 3.1) | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C. freundii (n = 5; 1.5) | * | * | * | 3 | 2 | 1 | 2 | 2 | 1 | 0 |
| P. mirabilis (n = 5; 1.5) | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. morganii (n = 2; 0.6) | * | * | * | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. braakii (n = 1; 0.3) | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P. vulgaris (n = 1; 0.3) | * | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P. stuartii (n = 1; 0.3) | * | * | * | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. marcescens (n = 1; 0.3) | * | * | * | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacteriaceae species (No.; %) n = 313 | 236 (75.3) | 169 (53.9) | 62 (19.8) | 149 (47.6) | 133 (42.4) | 3 (0.9) | 102 (32.5) | 93 (29.7) | 95 (30.3) | 40 (12.7) |
| No. (%) of resistant isolates during NICU stay | ||||||||||
| AMX | AMC | FOX | CTX/ CAZ | GN | AK | NA | NOR/CIP | SXT | ETP | |
| K. pneumoniae (n = 195; 59.4) | 195 | 184 | 10 | 184 | 169 | 0 | 95 | 90 | 99 | 18 |
| E. coli (n = 109; 33.2) | 100 | 96 | 63 | 93 | 89 | 9 | 79 | 77 | 28 | 26 |
| E. cloacae (n = 6; 1.8) | * | * | * | 6 | 3 | 1 | 3 | 3 | 3 | 2 |
| K. oxytoca (n = 6; 1.8) | * | 5 | 1 | 5 | 5 | 0 | 2 | 2 | 3 | 2 |
| C. freundii (n = 5; 1.5) | * | * | * | 4 | 3 | 0 | 1 | 1 | 0 | 1 |
| p. mirabilis (n = 4; 1.2) | 4 | 3 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 0 |
| M. morganii (n = 2; 0.6) | * | * | * | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| E. aerogenes (n = 1; 0.3) | * | * | * | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacteriaceae species (No.; %) n = 328 | 319 (97.2) | 302 (92) | 88 (26.8) | 295 (89.9) | 272 (82.9) | 10 (3) | 182 (55.4) | 173 (52.7) | 133 (40.5) | 49 (14.9) |
*Natural resistance
AMP: ampicillin; AMC: amoxicillin / clavulanic acid; FOX: cefoxitin; CTX: cefotaxim; CAZ: ceftazidim; GN: gentamicin; AK: amikacin; NA: nalidixic acid; NOR: norfloxacin; CIP: ciprofloxacin, SXT: cotrimoxazol; ETP: ertapenem.
Molecular analysis
All isolates found resistant to C3G and/or imipenem were tested for resistance genes. Phenotypic tests confirming the production of ESBL or CARBA enzymes have been carried out including the synergy test and the Hodge test.
Enterobacteriaceae producing ESBL
Of the 641enterobacterial isolates collected during this study, 330 were confirmed as ESBL producers by the synergy test (51.4%). K. pneumoniae was the predominant specie with a frequency of 76.9% (254/330), while 16.6% of the isolates were E. coli (55/330).
The blaCTX-M-1 gene was the most dominant with a frequency of 81.2% (268/330), followed by blaSHV with 69.6% (230/330). The blaTEM was present in 44.2% of isolates and the blaCTX-M-9 in 20.4% of isolates. However, blaCTX-M-2 was detected only in 5.7% of strains. Sequence analysis confirmed the presence of the five nucleotide sequences blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, blaSHV, blaTEM.
Enterobacteriaceae producing carbapenemases
The results of the antibiograms showed that 89 isolates are resistant to Imipenem and / or Ertapenem. E. coli.comes first with 44 isolates followed by K. pneumoniae with 35 strains. The Hodge test was used to select 42 positive isolates, which corresponds to 47%.
All Hodge positive isolates were screened for PCR production of carbapenemases. The results of the blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48 genes showed that all isolates k. pneumoniae, k. oxytoca and E. coli exclusively carried the blaOXA-48 gene. In contrast, isolates of E. cloacae and C. freundii did not harbor any of these genes. Sequence analysis confirmed the presence of the nucleotide sequence blaOXA-48.
Discussion
The study of colonization by MDR Enterobacteriaceae in hospitalized newborns showed a high prevalence of these bacteria, whether on admission (33%) or during hospitalization (59%). Several risk factors make acquisition almost inevitable (from postpartum to hospitalization risk factors) but the high rate only suggests that the application of universal and additional precautions is not optimal in-hospital services especially with regard to small children whose infectious state is bacteriologically documented., it can contribute to a cross-transmission given the charge of work and the invasive multitude acts in the unit.
Several studies have evaluated the prevalence of ESBL-E colonization among patients hospitalized in ICUs [11].
The predominant resistance mechanism in our study is the production of ESBL; we reported an acquisition rate of 59%, which means that 59% of the patients became colonized with ESBL during their hospital stay. Detsis et al. reported that the risk of subsequent ESBL-E infection in colonized patients is approximately 50 times higher than in non-colonized patients, and thus ESBL-E colonization of the gastrointestinal tract may be a useful tool for predicting ESBL-E infection [12]. Also, these colonized newborns can serve as reservoirs for other newborns as well as for potential epidemics [13, 14].
Of the ESBL-producing enterobacteria, K. pneumoniae has been widely implicated in numerous nosocomial and neonatal epidemics worldwide [15]. This specie has been responsible for 16–28% of bacteremia cases in different parts of the world [16]. In this study, 94% of the ESBL acquisition cases consisted of acquisition of ESBL-producing K. pneumoniae, which is consistent with another Italian report [17].
Also, the most recent challenge has been the spread of carbapenemase-producing Enterobacteriaceae (CPE) around the world [12]. We found that 8.7% of screened patients were colonized with CPE on admission and 12.5% during their stay. A recent study reported a CPE rate of 1.6% among newborns hospitalized in Algeria. This resistance was encoded exclusively by the blaOXA-48 gene [13].
Frequently colonizing of admitted newborns are K. pneumoniae and E. coli, which are common enterobacteria in the intestinal tract of hospitalized newborns. Both bacteria are naturally susceptible to many antibiotics but can develop resistance to many drugs, particularly beta-lactam antibiotics, the most commonly used antibiotics in NICUs. This resistance is a consequence of the selection pressure due to the excessive use of antibiotics and of their strong genetic determinism that gives them a high power of diffusion [19].
A high frequency of resistance to commonly used antibiotics such as cephalosporin 3G or gentamicin was observed in the isolates identified during our study. We noted that 90% of ESBL-E and 95% of EPCs, were resistant to gentamicin as well. More than 80% of these bacteria have co-resistance with quinolones and fluoroquinolones. Moreover, several authors have observed the same broad spectrum of resistance in enterobacterial strains isolated from hospitalized infants [14–16].
In recent decades, the predominant genotype in β-lactamase-producing enterobacteria has shifted from TEM and / or SHV to CTX-M (genes encoding ESBL). The prevalence of these genes varies widely. In Saudi Arabia, rates of 97.3% for SHV and 84.1% for TEM have been reported [17]. In contrast, in France, Birgy et al. reported lower rate: 6.3% and 9.4% respectively for TEM and SHV [18]. For the CTX-M β-lactamase, the blaCTX-M-1 group was the most detected in our work. An earlier study carried out in Morocco on isolates of ESBL-producing E. coli showed agreement (80%) with our results [19]. In South Asia, the CTX-M-1 and CTX-M-9 groups were identified, but none CTX-M-2 [17]. However, CTX-M-2 is more prevalent in most South American countries [20].
Subgroup blaCTX-M-15 is considered the most common type of ESBL worldwide [21–23]. In Moroccan hospitals, CTX-M-15 and CTX-M-28 were the two CTX-M enzymes identified in patients [19]. In the present study, blaCTX-M-15 was the only subclass found among all isolates harboring the blaCTX-M-1 gene.
On the other hand, the enzyme OXA-48 is a group D of β-lactamase, having the greatest catalytic property to imipenem, it also hydrolyzes penicillins and spares wide-spectrum cephalosporins. This enzyme, encoded by the blaOXA-48 gene, was initially identified from a strain of K. pneumoniae isolated in Turkey [24]. This gene is localized on a plasmid and has also been identified in E. coli and Citrobacter freundii, but not in A. baumannii [25]. Moreover, this corresponds to the results published in our previous manuscript which shows that all isolates of A. baumannii harbored blaOXA-23 and blaOXA-51 but no blaOXA-48 [26].
The identification of risk factors favoring the colonization of hospitalized newborns by MDR-E allows putting in place preventive strategies of transmission of MDR-E in health care structures. In SNRNs, the most commonly reported factors are birth weight; gestational age, use of invasive devices, previous treatment with antibiotics, and length of stay [27, 28].
The length of hospital stay as a risk factor has been associated with the acquisition of ESBL-E in neonates [29] as well as in adults hospitalized in ICUs [30]. On the other hand, inappropriate antibiotic therapy associated with ESBL-E colonization may result in prolonged hospital stays. Thus, the longer the hospital stay, the greater the risk of the onset of nosocomial bacteremia [31].
We also found that neurological distress and the origin from another hospital service presents risk factors associated with the carriage of an MDR-E on admission. Indeed, 38% of newborns coming from the maternity hospital of Fez University Hospital are carriers of MDR-E. These patients then constitute an important reservoir of MDR-E and will contribute, often through the hands of the medical staff, to the transmission of these bacteria to other hospitalized newborns. In the same context, Khiev and Veber reported that an intra- or extra-institutional transfer for hospitalization increases the risk of MDR-E colonization [32].
Cross-transmission of MDR-E between newborns through the environment and health care personnel remains the most likely route of newborn colonization. The prevention of colonization by MDR-E among hospitalized infants remains difficult. However, admission screening allows early detection of colonized patients and helps control the spread of these strains through the application of several appropriate control measures [33, 34].
The NICU of University Hospital Hassan II provides services to more than 1200 patients a year and drains a very important population that it is from the region of Fez than elsewhere. Building additional NICUs capacity in the region would help reduce local load and demand, thereby reducing the risk of transmission and infection.
Such as in many developing countries, low observance of hand hygiene practices among health professionals has been noted. Moreover, this situation amplifies the risk of transmission and spread of epidemic strains.
Any strategy for preventing MDR-E transmission and nosocomial infection is based primarily on respect for hand hygiene. Staff training is therefore an important element to succeed in this challenge.
In this context, we have developed and implemented a multimodal strategy focused on two different programs, the first is based on the implementation of hand hygiene and the optimization of standard and universal precautions. The second program is oriented towards the management of patients on the unit, starting with the systematic screening of newborns following childbirth at the hospital institutions and a reporting system for patients who have an infectious status that requires the initiation of the isolation or the cohorting of cases.
Our study has some limitations to consider. First, the moment of discharge screening can lead to biased estimates of the association between length of stay and MDR-E acquisition. Second, we did not evaluate for non-β-lactamase mechanisms of resistance such as those that relate to reduced outer membrane permeability, as we preferred to focus on resistance mechanisms with a high predilection for patient-to-patient transmission. Third, we were unable to carry out molecular analyses earlier, which led us to delay the publication of our results. at the same time, the inventory conducted very recently (in 2020) of the neonatal unit showed that the frequency of colonization is still high (around 60%). Last, since active surveillance for MDR-E was not consistent throughout the study period, all admitted newborns may not have been included in this study.
Conclusion
This study showed a high prevalence of MDR-E intestinal carriage, multiple antibiotic resistance profiles, and a diversity of encoding resistance genes in our NICU. This situation required the development of antimicrobial stewardship initiatives as well as the maintaining of antimicrobial resistance surveillance systems. In addition, the knowledge of risk factor profiles can lead to the development of strategies to prevent colonization at all levels, the proper management of newborns colonized by MDRs can lead to the prevention of bloodstream infections and the occurrence of sepsis among these fragile patients.
Supporting information
(XLSX)
Data Availability
All relevant data are within the manuscript and its S1 Dataset.
Funding Statement
This study was supported by University Hospital Hassan II (Fez, Morocco). The institution did not have any role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
References
- 1.Zingg W, Hopkins S, Gayet-Ageron A, Holmes A, Sharland M, Suetens C, et al. Health-care-associated infections in neonates, children, and adolescents: an analysis of paediatric data from the European Centre for Disease Prevention and Control point-prevalence survey. Lancet Infect Dis. 2017;17: 381–389. doi: 10.1016/S1473-3099(16)30517-5 [DOI] [PubMed] [Google Scholar]
- 2.Gortner L. Nosocomial infections in Very preterm neonates—Improvements by further scientific research or discussions in talk shows? Klin Pädiatrie. 2013;225: 55–56. doi: 10.1055/s-0033-1334959 [DOI] [PubMed] [Google Scholar]
- 3.Leistner R, Piening B, Gastmeier P, Geffers C, Schwab F. Nosocomial Infections in Very Low Birthweight Infants in Germany: Current Data from the National Surveillance System NEO-KISS. Klin Pädiatrie. 2013;225: 75–80. doi: 10.1055/s-0033-1334886 [DOI] [PubMed] [Google Scholar]
- 4.Bersani I, Speer C. Nosocomial Sepsis in Neonatal Intensive Care: Inevitable or Preventable? Z Geburtshilfe Neonatol. 2012;216: 186–190. doi: 10.1055/s-0032-1321837 [DOI] [PubMed] [Google Scholar]
- 5.World health Organization. Health care-associated infections. 2018.
- 6.Boyce JM. Environmental contamination makes an important contribution to hospital infection. J Hosp Infect. 2007;65: 50–54. doi: 10.1016/S0195-6701(07)60015-2 [DOI] [PubMed] [Google Scholar]
- 7.Yap PSX, Ahmad Kamar A, Chong CW, Yap IKS, Thong KL, Choo YM, et al. Intestinal carriage of multidrug-resistant gram-negative bacteria in preterm-infants during hospitalization in neonatal intensive care unit (NICU). Pathog Glob Health. 2016;110: 238–246. doi: 10.1080/20477724.2016.1229884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.BOO N, NG S, LIM V. A case-control study of risk factors associated with rectal colonization of extended-spectrum beta-lactamase producing sp. in newborn infants. J Hosp Infect. 2005;61: 68–74. doi: 10.1016/j.jhin.2005.01.025 [DOI] [PubMed] [Google Scholar]
- 9.Reygaert WC. Antimicrobial resistance mechanisms of Staphylococcus aureus. 2013. doi: 10.3389/fmicb.2013.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy SSS, Mello MJG, Gusmão-filho FAR, Correia JB. Colonisation by extended-spectrum β-lactamase-producing Klebsiella spp. in a paediatric intensive care unit. J Hosp Infect. 2010;76: 66–69. doi: 10.1016/j.jhin.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 11.Berezin EN, Solórzano F. Gram-negative infections in pediatric and neonatal intensive care units of Latin America. J Infect Dev Ctries. 2014;8: 942–953. doi: 10.3855/jidc.4590 [DOI] [PubMed] [Google Scholar]
- 12.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9: 228–236. doi: 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- 13.Mairi A, Touati A, Ait Bessai S, Boutabtoub Y, Khelifi F, Sotto A, et al. Carbapenemase-producing Enterobacteriaceae among pregnant women and newborns in Algeria: Prevalence, molecular characterization, maternal-neonatal transmission, and risk factors for carriage. Am J Infect Control. 2019;47: 105–108. doi: 10.1016/j.ajic.2018.07.009 [DOI] [PubMed] [Google Scholar]
- 14.Jett BD, Ritchie DJ, Reichley R, Bailey TC, Sahm DF. In vitro activities of various beta-lactam antimicrobial agents against clinical isolates of Escherichia coli and Klebsiella spp. resistant to oxyimino cephalosporins. Antimicrob Agents Chemother. 1995;39: 1187–90. doi: 10.1128/AAC.39.5.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villa L, Pezzella C, Tosini F, Visca P, Petrucca A, Carattoli A. Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended-spectrum beta-lactamase gene and a class 1 integron. Antimicrob Agents Chemother. 2000;44: 2911–4. doi: 10.1128/AAC.44.10.2911-2914.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended Spectrum Beta-lactamases Ghafourian et al. doi: 10.21775/cimb.017.011 [DOI] [PubMed] [Google Scholar]
- 17.Al-Agamy MHM, Shibl AM, Tawfik AF. Prevalence and molecular characterization of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in Riyadh, Saudi Arabia. Ann Saudi Med. 29: 253–7. doi: 10.4103/0256-4947.55306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.B A., M-K P., B P., D C., G N., C C., et al. Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: Prevalence of E. coli ST131. J Clin Microbiol. 2013;51: 1727–1732. doi: 10.1128/JCM.03255-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouchakour M, Zerouali K, Gros Claude JDP, Amarouch H, El Mdaghri N, Courvalin P, et al. Plasmid-mediated quinolone resistance in expanded spectrum beta lactamase producing enterobacteriaceae in Morocco. J Infect Dev Ctries. 2010;4: 779–803. doi: 10.3855/jidc.796 [DOI] [PubMed] [Google Scholar]
- 20.Gomes Chagas TP, Alves RM, Vallim DC, Seki LM, Campos LC, Asensi MD. Diversity of genotypes in CTX-M-producing Klebsiella pneumoniae isolated in different hospitals in Brazil. Brazilian J Infect Dis. 2011;15: 420–425. doi: 10.1016/S1413-8670(11)70222-7 [DOI] [PubMed] [Google Scholar]
- 21.Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006;9: 466–475. doi: 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 22.Rakotonirina HC, Garin B, Randrianirina F, Richard V, Talarmin A, Arlet G. Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol. 2013;13. doi: 10.1186/1471-2180-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: Diverse clones and clonal dissemination. Int J Antimicrob Agents. 2011;38: 160–163. doi: 10.1016/j.ijantimicag.2011.03.020 [DOI] [PubMed] [Google Scholar]
- 24.Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a University hospital in Morocco. Clin Microbiol Infect. 2014;20: 350–354. doi: 10.1111/1469-0691.12325 [DOI] [PubMed] [Google Scholar]
- 25.Matar GM, Dandache I, Carrër A, Khairallah M-T, Nordmann P, Sabra A, et al. Spread of OXA-48-mediated resistance to carbapenems in Lebanese Klebsiellapneumoniae and Escherichiacoli that produce extended spectrum β-lactamase. Ann Trop Med Parasitol. 2010;104: 271–274. doi: 10.1179/136485910X12647085215651 [DOI] [PubMed] [Google Scholar]
- 26.Arhoune B, Oumokhtar B, Hmami F, Fakir S El, Moutaouakkil K, Chami F, et al. Intestinal carriage of antibiotic resistant Acinetobacter baumannii among newborns hospitalized in Moroccan neonatal intensive care unit. PLoS One. 2019;14: 1–12. doi: 10.1371/journal.pone.0209425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crivaro V, Bagattini M, Salza MF, Raimondi F, Rossano F, Triassi M, et al. Risk factors for extended-spectrum beta-lactamase-producing Serratia marcescens and Klebsiella pneumoniae acquisition in a neonatal intensive care unit. J Hosp Infect. 2007;67: 135–41. doi: 10.1016/j.jhin.2007.07.026 [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Zhuang S, Du M. Risk Factors of Nosocomial Infection with Extended-Spectrum Beta-Lactamase-Producing Bacteria in a Neonatal Intensive Care Unit in China. Infection. 2007;35: 339–345. doi: 10.1007/s15010-007-6356-9 [DOI] [PubMed] [Google Scholar]
- 29.Nordberg V, Quizhpe Peralta A, Galindo T, Turlej-Rogacka A, Iversen A, Giske CG, et al. High Proportion of Intestinal Colonization with Successful Epidemic Clones of ESBL-Producing Enterobacteriaceae in a Neonatal Intensive Care Unit in Ecuador. PLoS One. 2013;8. doi: 10.1371/journal.pone.0076597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiddee A, Assawatheptawee K, Na-udom A, Treebupachatsakul P, Wangteeraprasert A, Walsh TR, et al. Risk factors for gastrointestinal colonization and acquisition of carbapenem-resistant gram-negative bacteria among patients in intensive care units in Thailand. Antimicrob Agents Chemother. 2018;62. doi: 10.1128/AAC.00341-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gayvallet-Montredon N, Sauvestre C, Bergeret M, Gendrel D, Raymond J. Archives de Pd’iatrie. /data/revues/0929693x/v0009i07/01009654/. Elsevier Science; [Google Scholar]
- 32.Khiev B, Veber B. Patient BMR +: risques de contamination et prévention en préhospitalier et aux urgences. 2010. [Google Scholar]
- 33.Schwaber MJ, Lev B, Israeli A, Solter E, Smollan G, Rubinovitch B, et al. Containment of a Country-wide Outbreak of Carbapenem-Resistant Klebsiella pneumoniae in Israeli Hospitals via a Nationally Implemented Intervention. Clin Infect Dis. 2011;52: 848–855. doi: 10.1093/cid/cir025 [DOI] [PubMed] [Google Scholar]
- 34.Kochar S, Sheard T, Sharma R, Hui A, Tolentino E, Allen G, et al. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect Control Hosp Epidemiol. 2009;30: 447–52. doi: 10.1086/596734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its S1 Dataset.


