Abstract
The adult olfactory epithelium (OE) regenerates sensory neurons and nonsensory supporting cells from resident stem cells after injury. How supporting cells contribute to OE regeneration remains largely unknown. In this study, we elucidated a novel role of Ym2 (also known as Chil4 or Chi3l4), a chitinase-like protein expressed in supporting cells, in regulating regeneration of the injured OE in vivo in both male and female mice and cell proliferation/differentiation in OE colonies in vitro. We found that Ym2 expression was enhanced in supporting cells after OE injury. Genetic knockdown of Ym2 in supporting cells attenuated recovery of the injured OE, while Ym2 overexpression by lentiviral infection accelerated OE regeneration. Similarly, Ym2 bidirectionally regulated cell proliferation and differentiation in OE colonies. Furthermore, anti-inflammatory treatment reduced Ym2 expression and delayed OE regeneration in vivo and cell proliferation/differentiation in vitro, which were counteracted by Ym2 overexpression. Collectively, this study revealed a novel role of Ym2 in OE regeneration and cell proliferation/differentiation of OE colonies via interaction with inflammatory responses, providing new clues to the function of supporting cells in these processes.
SIGNIFICANCE STATEMENT The mammalian olfactory epithelium (OE) is a unique neural tissue that regenerates sensory neurons and nonsensory supporting cells throughout life and postinjury. How supporting cells contribute to this process is not entirely understood. Here we report that OE injury causes upregulation of a chitinase-like protein, Ym2, in supporting cells, which facilitates OE regeneration. Moreover, anti-inflammatory treatment reduces Ym2 expression and delays OE regeneration, which are counteracted by Ym2 overexpression. This study reveals an important role of supporting cells in OE regeneration and provides a critical link between Ym2 and inflammation in this process.
Keywords: chitinase-like protein, inflammation, olfactory epithelium, regeneration, supporting cell, Ym2
Introduction
The olfactory epithelium (OE) is a pseudostratified epithelial structure mainly composed of supporting cells, olfactory sensory neurons (OSNs), and progenitors/basal cells (Schwob, 2002). Mitotically active globose basal cells (GBCs) continuously generate OSNs throughout life (Schwob et al., 2017), while dormant horizontal basal cells (HBCs) adherent to the basal lamina do not contribute to the maintenance of normal OE (Leung et al., 2007). After severe tissue injury, HBCs are activated and transited to GBCs, leading to the regeneration of various cell types constituting the OE (Schnittke et al., 2015; Gadye et al., 2017; Schwob et al., 2017). Selective loss of OSNs by olfactory bulbectomy is shown to recruit HBCs in some studies (Iwai et al., 2008) but not in others (Leung et al., 2007). Depletion of supporting cells in the OE activates HBCs to generate supporting cells, but rarely neurons (Lin et al., 2017). Although supporting cells have been implicated in OE regeneration, the underlying mechanisms are largely unknown.
Chitinases and chitinase-like proteins (CLPs) belong to the glycoside hydrolase family 18 (Shuhui et al., 2009; Sutherland et al., 2009). Chitinases are enzymatically active, but chitinase-like proteins do not have enzymatic activity (Bussink et al., 2007; Funkhouser and Aronson, 2007). There are eight subtypes of chitinase-like proteins, including Ym1(Chil3) and Ym2 (Chil4), which share ∼95% identity in mice (Ohno et al., 2014). Ym1/Ym2 serve multiple functions in regulating inflammation and infection (Falcone et al., 2001; Nair et al., 2005; Müller et al., 2007; Cai et al., 2009; Qureshi et al., 2011), immunity (Chupp et al., 2007; Ober et al., 2008), and progression of certain diseases (Furuhashi et al., 2010; Burman et al., 2016; Kumagai et al., 2016; Kzhyshkowska et al., 2016). A number of studies indicate that CLPs are involved in tissue injury and regeneration (Bonneh-Barkay et al., 2010; Pizano-Martínez et al., 2011; Maddens et al., 2012; Zhou et al., 2014; Puthumana et al., 2017; Zhang et al., 2018). Although Ym1/Ym2 expression has been reported in the OE (Giannetti et al., 2004; Heron et al., 2013), their roles in OE remain elusive.
Acute and chronic inflammation have different effects on neurogenesis (Chesnokova et al., 2016). Chronic inflammation has direct inhibitory effect on OE progenitor/stem cell proliferation, mediated by secreted tumor necrosis factor (TNF)-α (Lane et al., 2010). Suppression of inflammatory reaction promotes olfactory nerve recovery after injury (Kobayashi et al., 2018). Inflammation also impairs sensory neurogenesis in a chronic rhinosinusitis model (Rouyar et al., 2019). In olfactory tissue cultures, TNF-α and interleukin (IL)-5 inhibit olfactory regeneration through promoting apoptosis (Kim et al., 2019). These findings are consistent with a study in the CNS, which demonstrates that chronic inflammation impairs adult hippocampal neurogenesis (Monje et al., 2003). By contrast, acute inflammation exhibits the opposite effect on neurogenesis, leading to enhanced proliferation of neural progenitors (Kyritsis et al., 2012). In the OE, acute inflammation has been shown to facilitate regeneration (Pozharskaya et al., 2013; Chen et al., 2017). Since Ym1/Ym2 proteins regulate inflammatory responses in several tissues (Falcone et al., 2001; Nair et al., 2005; Müller et al., 2007; Cai et al., 2009), it is tempting to test the potential interaction between Ym1/Ym2 and inflammatory responses during OE regeneration.
Here we elucidated the potential role of Ym2 in regulating cellular dynamics during OE regeneration and in OE colonies. This protein was highly expressed in supporting cells of the open-side OE after unilateral naris occlusion and in the methimazole-lesioned OE. Genetic knockdown of Ym2 in supporting cells attenuated regeneration of the injured OE, while Ym2 overexpression accelerated OE recovery. Anti-inflammatory treatment inhibited Ym2 expression and delayed OE regeneration, counteracted by Ym2 overexpression, which increased inflammatory cells. Moreover, Ym2 downregulation inhibited cell proliferation/differentiation in cultured OE colonies. Anti-inflammatory treatment downregulated Ym2 and attenuated cell differentiation, reversed by Ym2 overexpression. This study reveals a novel function of supporting cells in OE regeneration via regulating Ym2 expression, providing a critical link between Ym2 and inflammation in OE regeneration.
Materials and Methods
Animals.
Transgenic Sox2-CreERT2 mice (stock #017593, The Jackson Laboratory; Arnold et al., 2011) in which expression of Cre recombinase is under the control of the Sox2 (SRY-box containing gene 2) promoter were purchased from The Jackson Laboratory. Transgenic Ym2fl/fl mice in which the Chil4 gene (encoding Chitinase-like four protein, Ym2) is flanked by LoxP sites were purchased from the Wellcome Trust Sanger Institute Mouse Genetics Project [Sanger MGP Allele:Chil4tm1a(EUCOMM)Hmgu (funding and associated primary phenotypic information may be found at www.sanger.ac.uk/mouseportal; White et al., 2013]. Sox2-CreERT2 mice were crossed with Ym2fl/fl mice to achieve inducible knockdown of Ym2 in Sox2+ cells. OMP (olfactory marker protein)-Cre mice (stock #006668, The Jackson Laboratory; Li et al., 2004) and Rosa26-TdTomato mice [stock #007909, The Jackson Laboratory (also known as Ai9 or Cre-dependent TdTomato reporter line); Madisen et al., 2010] were from The Jackson Laboratory. OMP-Cre mice were crossed with Rosa26-TdTomato mice to generate OMP-TdTomato mice from which OMP-TdTomato+ OE colonies were derived. Breeding pairs of wild-type C57BL/6J mice were purchased from Shanghai Model Organisms. The procedures of animal handling and tissue harvesting were approved by the Institutional Animal Care and Use Committees. Both male and female mice were used in this study, and the data were grouped because no sex difference was evident.
Chemicals.
Dexamethasone (Dex; Sigma-Aldrich) was dissolved in DMSO to make a 1000× stock solution. The concentrations of dexamethasone were 0.1 and 1 μm in cultured colonies, and 1 mg/kg in animals. Recombinant Ym2 protein (#CSB-YP842057MO, Cusabio) was dissolved in PBS. The concentrations of Ym2 protein were 0.5, 1, and 2 μg/ml in OE colonies, which were incubated for 10 d.
Unilateral naris occlusion.
On postnatal day 1 (P1), mice were anesthetized by hypothermia. Unilateral naris occlusion was achieved by a brief (<1 s) cauterization of one nostril (Small Vessel Cauterizer Kit, catalog #18000–00, Fine Science Tools). Pups were then placed on a heating plate until they recovered from anesthesia. Two weeks after the procedure, the cauterized nostril was examined under a stereo microscope. Only mice with complete occlusion were killed at P30 for tissue collection.
Ym2 identification by mass spectrometry.
Proteins extracted from the olfactory mucosa of the closed and open sides were precleared with 200 µl of Protein G Sepharose overnight and separated by SDS-PAGE. Gel was fixed for 3 h (10% acetic acid, 40% ethanol), stained with Coomassie Blue [10% (NH4)2SO4, 1.2% orthophosphoric acid, 0.1% Coomassie Blue], and destained in H2O. Bands ∼38 kDa in size were cut out from the gel and analyzed via mass spectrometry at Penn Quantitative Proteomics Resource Core.
Olfactory epithelium lesion.
Four- to 6-week-old animals were intraperitoneally injected with a single dose of methimazole (50 μg/g body weight) as previously reported (Leung et al., 2007). In lentiviral experiments, methimazole was administrated on day 10 after viral injection. In saline controls, the animals were injected with the same amount of PBS.
ELISA.
The ELISA was performed to measure the concentration of Ym2 in the nasal lavage, which was collected from nostrils of methimazole-treated mice (30 d postinjection) or age-matched control mice. The mice were placed horizontally on the operating table with their heads tilted up 45° to avoid choking. Sterile PBS (100 µl) was slowly dropped into the single side of nasal cavity, and the liquid was sucked out and collected after 10 s. A mouse Ym2 ELISA kit (catalog #EM0806, Fine BioTech) was used, and the kit protocol was followed. Briefly, 100 μl of standard and lavage samples were added to each well, and the plate was incubated for 90 min at 37 °C. After being rinsed with wash buffer, 100 μl of biotin-labeled antibody working solution was added, and the plate was incubated for 60 min at 37°C. Then, 100 μl of HRP-streptavidin conjugate working solution was added and incubated for 30 min at 37°C, and subsequently 90 μl of TMB (3,3',5,5'-tetramethylbenzidine) substrate solution was added per well and incubated for 20 min at 37°C. After adding 50 μl of stop solution, the plate was read at 450 nm immediately.
Genetic ablation of Ym2.
For genetic ablation of Ym2 in Sox2+ cells including supporting cells, Sox2-CreERT2 mice were crossed with Ym2fl/fl mice. Tamoxifen dissolved in sunflower oil at 0.22 mg/g body weight was injected every other day until the animals were killed. In the control group, the Sox2-CreERT2/Ym2fl/fl mice were injected with the same doses of sunflower oil.
OE colony culture.
The OE colony culture was followed by our previously reported protocol (Dai et al., 2018). Briefly, intact nasal epithelium of wide type C57BL/6J mice at 2–3 months of age was dissected and digested in 0.25% trypsin-EDTA, and a single-cell suspension was prepared. Cells were cultured in conditioned OE colony growth medium. The ingredients of growth medium is based on DMEM/F12 medium (Thermo Fisher Scientific) supplemented with R-spondin-1 (200 ng/ml; PeproTech), Noggin (100 ng/ml; PeproTech), Wnt3a (50 ng/ml; R&D Systems), Y27632 (10 μm; Sigma-Aldrich), epidermal growth factor (50 ng/ml; Thermo Fisher Scientific), N2 (1%; Thermo Fisher Scientific), B27 (2%; Thermo Fisher Scientific), HEPES (1 mm; Thermo Fisher Scientific), GlutaMAX (1%; Thermo Fisher Scientific), and Matrigel [4% (v/v); BD Biosciences]. Medium was changed every 3 d. Visible colonies were observed on day 3 after culturing. Colonies were passaged every 10 d.
Viral infection.
Lentiviral vectors expressing the full length of Ym2 cDNA (Lenti-Ym2) or three different short hairpin RNAs (shRNAs: Lenti-shYm2-1, Lenti-shYm2-2, and Lenti-shYm2-3, which target different parts of Ym2 sequence) were constructed. Lentivirus was collected and purified by Shanghai Tauto Biotech Co., Ltd. Lentivirus expressing the empty vector (Lenti-Ctrl) or scramble shRNA (Lenti-shCtrl) was used as a negative control. For the in vitro infection, the OE colonies were digested by 0.25% trypsin-EDTA to prepare a single-cell suspension and seeded in a low-attached 96-well plate with a density of 5000 cells/well; 2.5 × 105 transduction units (TU) of virus were added into each well, and apparent green fluorescence was observed on day 3 postinfection. For the in vivo injection, 5 μl of saline solution containing 1 × 106 TU of lentivirus was injected into each side of OE using a 1 μl microsyringe (Fine Science Tools) on a stereotaxic instrument (RWD).
OE colony cryosection preparation.
The protocol was described previously (Lancaster and Knoblich, 2014). Briefly, colonies were collected through centrifuging, washed with PBS, and fixed in 4% paraformaldehyde for 15 min. Then, colonies were dehydrated in 30% sucrose at 4°C overnight, followed by being wrapped into warm gelatin/sucrose solution and stored at 37°C for 15 min to equilibrate the colonies. Then, colonies in a small amount of gelatin/sucrose solution (∼20 μl) were solidified at room temperature and then were covered completely by warm gelatin/sucrose solution, which was polymerized at 4°C. The entire block of gelatin-containing colonies was immersed in a cold bath, and the sections at 20 μm thickness were prepared using a Cryostat (model CM1950, Leica).
Immunohistochemistry.
Mice were deeply anesthetized by intraperitoneal injection of ketamine-xylazine (200 and 15 mg/kg body weight, respectively) before decapitation. The heads were fixed in 4% paraformaldehyde (Sigma-Aldrich) overnight at 4°C and infiltrated with a series of sucrose solutions before being embedded in OCT (optimal cutting temperature) compound. The frozen tissues were cut into 20 μm coronal sections on a cryostat. After rinsing with PBS, the tissue sections were blocked for 60 min in 0.3% Triton X-100 in PBS with 5% bovine serum albumin, and then incubated at 4°C with the primary antibodies overnight. The primary antibodies included rabbit anti-TurboGFP (1:500; Thermo Fisher Scientific), goat anti-Sox2 (1:100; Santa Cruz Biotechnology), rabbit anti-Sox2 (1:100; Proteintech), goat anti-Sox2 (1:500; R&D Systems), rabbit anti-Ym2 (1:200; a gift from Shioko Kimura, National Cancer Institute, Bethesda, Maryland; Ward et al., 2001), mouse anti-Sus4 (1:200; provided by James Schwob, Tufts University, Boston, MA; Goldstein and Schwob, 1996), rabbit anti-PGP9.5 (1:200; Proteintech), rabbit anti-Krt14 (Proteintech; 1:200), rabbit anti-OMP (1:200; Abcam), chicken anti-OMP (1:500; provided by Qizhi Gong, University of California, Davis, Davis, CA; Chen et al., 2005), mouse anti Tuj1 (1:200; Abcam), rat anti-CD45 (1:200; Thermo Fisher Scientific), rat anti-F4/80 (1:500; Bio-Rad), or goat anti-ICAM1 (1:500 R&D Systems). All the antibodies and their specific targets are listed in Table 1. The secondary antibodies (Thermo Fisher Scientific) were diluted at 1:300 with which the sections were incubated for 1 h at room temperature. The nuclei were counterstained with DAPI (Thermo Fisher Scientific). Tissues were mounted in Vectashield (Vector Laboratories).
Table 1.
Primary antibodies used in this study
| Primary antibody | Source/vendor/catalog no. | Cell type |
|---|---|---|
| Gt@Sox2 | Santa Cruz Biotechnology, #sc-17320 | Supporting cell, basal cell |
| Gt@Sox2 | R&D Systems, #AF2018 | Supporting cell, basal cell |
| Rb@Sox2 | Proteintech, #11064–1-AP | Supporting cell, basal cell |
| Rb@Ym2 | Dr. Shioko Kimura (Ward et al., 2001) | Supporting cell |
| Mo@Sus4 | Dr. James Schwob (Goldstein and Schwob, 1996) | Supporting cell |
| Rb@PGP9.5 | Proteintech, #14730–1-AP | Olfactory sensory neuron |
| Rb@Krt14 | Proteintech, #10143–1-AP | Horizontal basal cell |
| Rb@OMP | Abcam, #ab183947 | Olfactory sensory neuron |
| Mo@Tuj1 | Abcam, #ab78078 | iOSN |
| Gt@ICAM1 | R&D Systems, #AF796 | Horizontal basal cell |
| Chk@OMP | Dr. Qizhi Gong (Chen et al., 2005) | Olfactory sensory neuron |
| Mo@Ki67 | BD Biosciences, #550609 | Proliferating cell |
| Mo@Krt18 | Abcam, #ab668 | Supporting cell |
| Rb@TurboGFP | Thermo Fisher Scientific, #PA5-22688 | Lentiviral infected GFP+ cell |
| Ra@CD45 | eBioscience, #14–0451-81 | Inflammatory cell |
| Ra@F4/80 | Bio-Rad, #MCA497GA | Macrophage |
For the immunostaining on cultured colonies, the cryosections of colonies were prepared as described above. Sections were washed by PBS, followed by incubation with blocking buffer containing SuperBlock (Thermo Fisher Scientific), 2% (v/v) donkey serum and 0.3% Triton X-100 at room temperature for 1 h. Primary antibody incubation was performed overnight at 4°C. After washing with PBS, appropriate secondary antibodies were used to visualize staining. The nuclei were counterstained with DAPI (Thermo Fisher Scientific). Sections were mounted in Vectashield (Vector Laboratories). The primary antibodies used were as follows: rabbit anti-Ym2 (1:200), rabbit anti-Sox2 (1:100; Proteintech), mouse anti-Ki67 (1:100; BD Biosciences), chicken anti-OMP (1:500; provided by Qizhi Gong at University of California, Davis; Chen et al., 2005), mouse anti-Tuj1 (1:200, Abcam), mouse anti-Krt18 (1:200; Abcam), and mouse anti-Sus4 (1:200, provided by James Schwob at Tufts University; Goldstein and Schwob, 1996). Fluorescent images were taken under a confocal microscope (model SP8, Leica) with LAS AF Lite software. The contrast and brightness of the images were set at an equal level.
Fluorescent in situ hybridization.
FITC-labeled riboprobes were synthesized using an FITC RNA labeling kit (catalog #11175025910, Roche). The template for Ym2 gene was amplified from mouse OE cDNA by PCR and subcloned into vector pGEM-T Easy (catalog #A1360, Promega). Primers used to amplify Ym2 cDNA were as follows: 5′-CATCTCTTCAGTGTTCTGGTGC-3′ and 5′-CCTAAATTGTTGTCCTTGAGCC-3′. fluorescence in situ hybridization (FISH) was performed as described previously with some modification (Fleming et al., 2012). Briefly, the sections were hybridized with 1–2 ng/μl FITC-labeled riboprobes diluted in hybridization buffer (containing 50% formamide, 5× SSC, 0.3 mg/ml yeast tRNA, 100 μg/ml heparin, 1× Denhardt's solution, 0.1% Tween 20, 0.1% CHAPS, and 5 mm EDTA in RNase-free H2O) overnight under Parafilm at 62°C. The sections were incubated in anti-FITC-POD (1:100 in 0.5% blocking reagent; catalog #11426346910, Roche) overnight at 4°C. FITC riboprobes were developed using the TSA Plus System (catalog #NEL741001KT, PerkinElmer). Slides were then rinsed in PBS and mounted with Vectashield (Vector Laboratories). Images were taken under a confocal microscope (model SP8, Leica) with LAS AF Lite software. The contrast and brightness of the images were set at equal levels.
Quantitative real-time PCR.
Total RNA was extracted by using the E.Z.N.A. Total RNA Kit I (catalog #R6834-02, Omega) according to the manufacturer manual. The extracted RNA was immediately dissolved in RNase-free water, and the purity and concentration were determined using a BioPhotometer (Metash). First-strand cDNA was synthesized using a PrimeScript RT Master Mix (catalog #RR036A, Takara). Primers used in this study were synthesized by Ruidibio. Quantitative real-time PCR was performed on an AnalytikJena Real-Time PCR System. The reaction mixtures included a cDNA template, 0.2 mm primers, SYBR qPCR SuperMix (catalog #E096-01B, Novoprotein), and double distilled H2O. Reaction conditions included an initial denaturation at 95°C for 1 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 30 s. The relative expression levels were calculated using the 2-ΔΔCt method. Primer sequences were as follows: IL-1β: AGCCCATCCTCTGTGACTCA and TGTCGTTGCTTGGTTCTCCT; TNF-α: CCCAGGCAGTCAGATCATCTTC and GGTTTGCTACAACATGGGCTACA; Ym2: TTGGAGGATGGAAGTTTGGACCT and TGACGGTTCTGAGGAGTAGAGACCA and GAPDH: TCAATGAAGGGGTCGTTGAT and CGTCCCGTAGACAAAATGGT.
Experimental design and statistical analysis.
Cell counting under any experimental condition was derived from three mice or three independent cultures. For each mouse, multiple sections along the anterior–posterior axis were used. The locations of sections and image capturing were kept consistent across animals to minimize counting bias. For each section, 10 nonoverlapping fluorescent images were captured to cover nearly the entire section, and the number of a particular cell type (per linear length) was quantified from ∼4 mm OE (∼0.4 mm/image). The only exception was Figure 1N, in which regions with the most densely labeled Ym2+ cells were compared because of patchy Ym2 expression in the open-side OE of naris-occluded mice. While images were not captured by someone blinded to experimental conditions, cell counting was performed by blinded individuals. For quantitative PCR and ELISA, data were derived from at least three independent experiments. Cell counts from the confocal images were performed using ImageJ and corrected using formula of Abercrombie (1946), as follows: corrected number = count × [section thickness/(section thickness + mean nuclei size)]. Fluorescence intensity data in immunohistochemistry and in situ hybridization were obtained in ImageJ using built-in functions. For all the fluorescence intensity, except for that seen in Figure 2M, we calculated the following average optical density = (fluorescence gray value – background gray value)/area. For Figure 2M, we calculated the integrated optical density (IntDen) as follows: IntDen = (fluorescence mean – background mean) × area. To make the statistical assessment as accurate as possible, all images from each OE section were included. Data were presented as the mean ± SEM. The statistical difference between two groups was determined by unpaired Student's t test and across multiple groups by one-way ANOVA or two-way ANOVA using GraphPad Prism 6.0 software.
Figure 1.
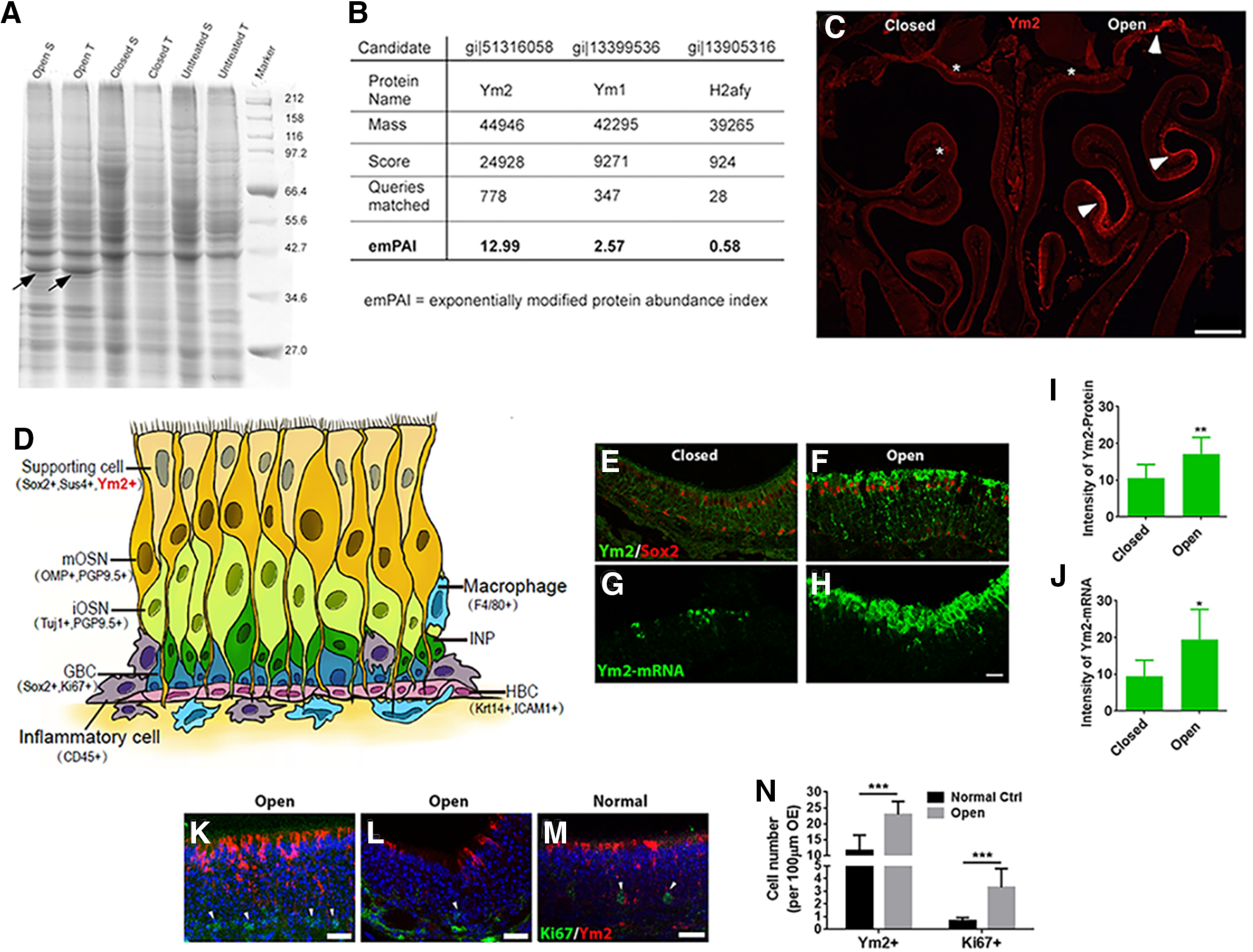
Ym2 is highly expressed on the open side of naris-occluded mice. A, Coomassie Brilliant Blue staining on an SDS-PAGE gel revealed a protein at ∼38 kDa (arrows) with the greatest difference between the closed and open sides. Tissues were collected from three mice that underwent unilateral naris closure from P1 to P30. S, Septum; T, turbinates. B, Mass spectrometric analysis revealed that the protein was most likely to be Ym2 with the highest emPAI (exponentially modified protein abundance index). Ym1 had the second highest emPAI (but much lower than Ym2), presumably because of its high sequence homology with Ym2. C, Higher Ym2 expression was observed in the open side. Arrowheads denote Ym2 antibody staining in the supporting cell layer and asterisks denote the lamina propria. D, Schematic of the OE containing multiple cell types with biomarkers in parentheses. E–H, Immunostaining and FISH indicated Ym2 was abundant in the supporting cell layer in the open side (F, H) compared with the closed side (E, G). I, J, Statistical analysis of Ym2 protein and mRNA intensity in the closed and open side (n = 9 and 10 sections, respectively, from three mice; arbitrary unit). K–M, Immunostaining against Ym2 and Ki67 in the OE of the open side of naris-occluded mice (K, L) and untreated control mice (M). Arrowheads mark Ki67+ cells. N, Statistical analysis of the number of Ym2+ and Ki67+ cells per 100 μm OE (n = 10 and 11 sections in control and open side from three mice in each condition). Because of the patchy expression of Ym2 in the open-side OE, the most densely labeled regions under the two conditions were captured and compared. Statistical significance was determined by unpaired Student's t test. I: **p = 0.009 (t(17) = 2.986); J: *p = 0.046 (t(17) = 2.524); N: ***p < 0.001; Ki67+: t(19) = 5.537; Ym2+: t(19) = 5.808. Scale bars: C, 0.5 mm; H, K–M, 20 μm.
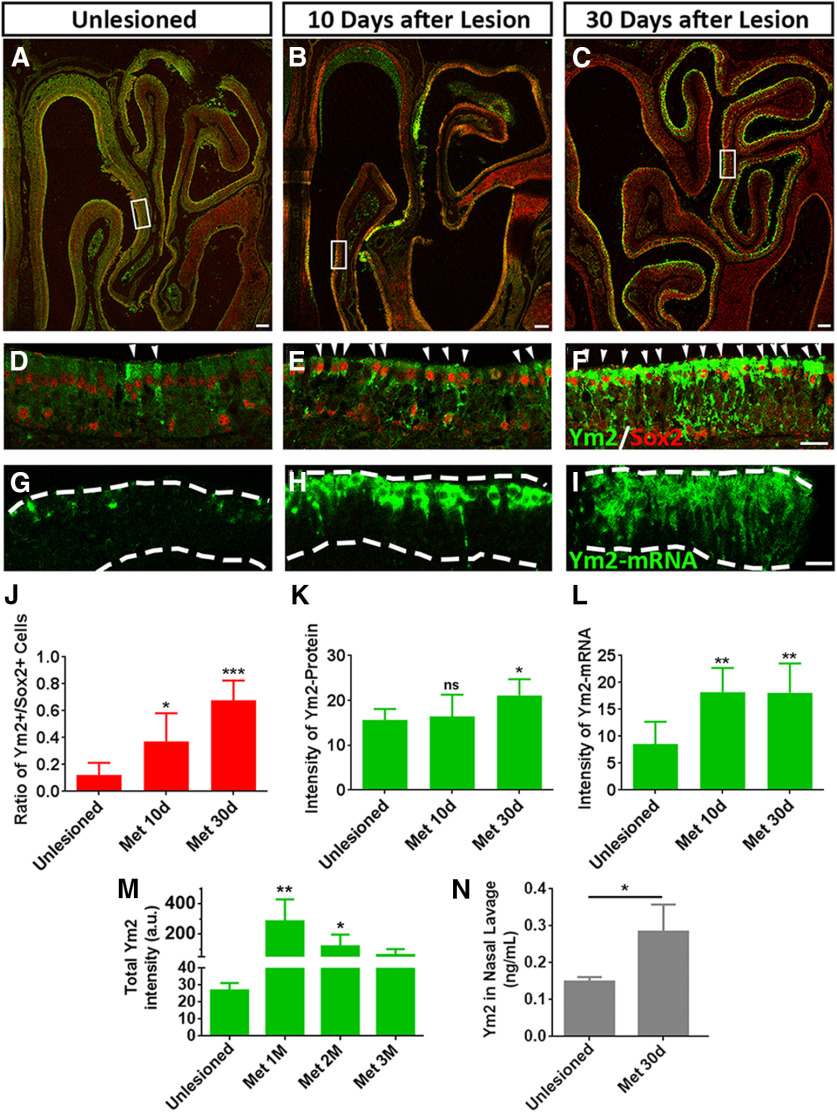
Figure 2.
Ym2 is highly abundant in the lesioned OE. A–F, Immunostaining revealed that Ym2 protein was upregulated in methimazole-lesioned OE on day 10 (B, E) and day 30 (C, F) compared with the unlesioned OE (saline injection; A, D). G–I, FISH analysis revealed higher Ym2-mRNA expression in the lesioned OE on day 10 (H) and day 30 (I) compared with the unlesioned control (G). J–L, Statistical analysis of the percentage of Ym2+/Sox2+ cells (J; n = 4, 10, and 9 sections from three mice in each group), immunostaining signal intensity (K; n = 5, 10, and 14 sections), and FISH signal intensity (L; n = 6, 12, and 20 sections). M, Statistical analysis of the total intensity of immunostaining against Ym2 in unlesioned and lesioned OE (months 1, 2, and 3 postinjury; n = 5, 6, 6, and 5 sections from three mice in each group). a.u., arbitrary unit. N, Concentration of Ym2 in the nasal lavage from methimazole-treated (30 d postinjection) and age-matched unlesioned mice (n = 3 mice in each condition). Statistical significance was determined by one-way ANOVA with Dunnett's multiple-comparisons test (J–M) and by unpaired t test (N). ns, Not significant. J: F(2,20) = 16.21, p < 0.0001; K: F(2,26) = 5.764, p = 0.0085; L: F(2,35) = 4.902, p = 0.0133; M: F(3,18) = 8.456, p = 0.0016; N: *p = 0.022 (t(4) = 2.268). Asterisks indicated the significance when compared with the unlesioned group. The rectangles in A–C are enlarged in D–F. Arrowheads in D–F point to Sox2+/Ym2+ cells. The unit of Ym2-protein and Ym2-mRNA intensity in K, L, and M is arbitrary. The dashed lines in G–I mark the OE borders. Met, methimazole. Scale bars: A–C, 100 μm; F, I, 20 μm.
Results
OE injury induces higher expression of Ym2 in supporting cells
To search for molecules potentially involved in neurogenesis of the OE, we initially used mice that underwent unilateral naris occlusion from P1 to P30 because the open-side OE has enhanced neurogenesis (among other changes) compared with the closed side (Coppola, 2012; Coppola et al., 2014), and this model provides within-subject comparison between the two sides. We compared the denatured proteins in the OE between the open and closed sides using SDS-PAGE electrophoresis. Coomassie Brilliant Blue staining revealed a protein at ∼38 kDa that displayed the most dramatic difference between sides (Fig. 1A). It was highly abundant in the open side, but faintly detectable in the closed side and in untreated mice. Using mass spectrometric analysis, this protein was identified as Ym2, a chitinase-like protein (Fig. 1B). In the naris-occluded mice, we observed patchy Ym2 expression via antibody staining in the open side (Fig. 1C). The OE is a pseudostratified epithelium mainly composed of supporting cells, immature OSNs (iOSNs), mature OSNs (mOSNs), immediate neuronal progenitors, GBCs, and HBCs (Fig. 1D). Biomarkers used in this study to label different OE cell types are shown in Figure 1D and Table 1. Double staining indicated enhanced Ym2 expression in Sox2+ supporting cells (Fig. 1E,F). The intensity of Ym2 staining increased by 62 ± 13% in the open side compared with the closed side (Fig. 1I). Since the antibody against Ym2 labels both Ym1 and Ym2, we designed an antisense RNA probe specific to Ym2 and detected Ym2-mRNA expression via FISH (Fig. 1G,H). The intensity of Ym2-mRNA in the open side was drastically enhanced by 106 ± 25% compared with the closed side (Fig. 1J). Thus, unilateral naris occlusion increased Ym2 expression in supporting cells of the open-side OE. Compared with untreated mice, the number of Ym2+ cells (per 100 µm OE length throughout this article) increased by 98 ± 11% on the open side of naris-occluded mice (Fig. 1K,M,N), and Ki67+ cells increased by 387 ± 65% (Fig. 1K,M,N). In the open-side OE, regions with fewer Ym2+ cells also contained fewer Ki67+ cells (Fig. 1L). Therefore, the abundance of Ym2+ cells seemed to be correlated with the proliferation of basal cells, suggesting a potential role of Ym2 in OE neurogenesis.
Given that the open-side OE experiences other changes during postnatal development (e.g., increased airflow and trauma), unilateral naris occlusion does not provide an optimal model for neurogenesis. We then turned our attention to a pharmacological OE injury regeneration model and asked whether Ym2 expression changed in this process. After the OE was ablated via methimazole injection, which spared basal cells, more Ym2+ cells were present in Sox2+ supporting cells on day 10 (Fig. 2B,E) and day 30 (Fig. 2C,F) compared with saline controls (Fig. 2A,D). The percentage of Ym2+/Sox2+ supporting cells increased by 201 ± 53% on day 10 and 449 ± 39% on day 30 after methimazole administration compared with the unlesioned OE (Fig. 2J). The intensity of Ym2 antibody staining increased in the lesioned OE on day 30 by 35 ± 6% compared with the unlesioned OE (Fig. 2K). Higher Ym2-mRNA expression in the lesioned OE was also confirmed by FISH. Compared with the unlesioned control (Fig. 2G), Ym2-mRNA signals were widely spread across the lesioned OE on both day 10 (Fig. 2H) and day 30 (Fig. 2I). The lack of Ym2-mRNA signals in the basal cell layer suggests that Ym2 is only expressed in supporting cells (Fig. 2H,I; see below). The intensity of Ym2-mRNA increased by 113 ± 15% on day 10 and 111 ± 13% on day 30 (Fig. 2L). The intensity of Ym2 antibody staining showed a decrease 2 and 3 months postinjury compared with 1 month (Fig. 2M). The concentration of secreted Ym2 in the nasal cavity was elevated in lesioned OE on day 30 (Fig. 2N; for potential technical limitations, see Discussion). Thus, methimazole-induced lesion enhanced Ym2 expression in supporting cells of the OE.
Ym2 regulates regeneration of the injured OE
We wished to test whether Ym2 contributes to OE regeneration by manipulating Ym2 expression in supporting cells. Sox2-CreERT2/Ym2fl/fl mice received intraperitoneal injections of tamoxifen to achieve inducible knockdown of Ym2 expression in Sox2+ cells (for details, see Materials and Methods). Since Sox2 labels both basal cells and supporting cells in the OE, we first confirmed Ym2 expression exclusively in supporting cells, not in basal cells by immunostaining against Sox2 protein and FISH against Ym2 mRNA in OE sections (Fig. 3A). Therefore, tamoxifen injection in Sox2-CreERT2/Ym2fl/fl mice provided a viable approach to downregulate Ym2 expression specifically in supporting cells (Fig. 3B). The tamoxifen-injected mice showed reduced Ym2 staining on day 7 post-methimazole lesion compared with controls receiving sunflower oil (solvent for tamoxifen) injections. The overall intensity of Ym2 decreased by 45 ± 7% with tamoxifen induction (Fig. 3C). The weaker Ym2 expression was found in all regions examined, anterior, middle, and posterior (Fig. 3D–I,D′–I′), confirming inducible knockdown of Ym2 expression in OE supporting cells of Sox2-CreERT2/Ym2fl/fl mice.
Figure 3.
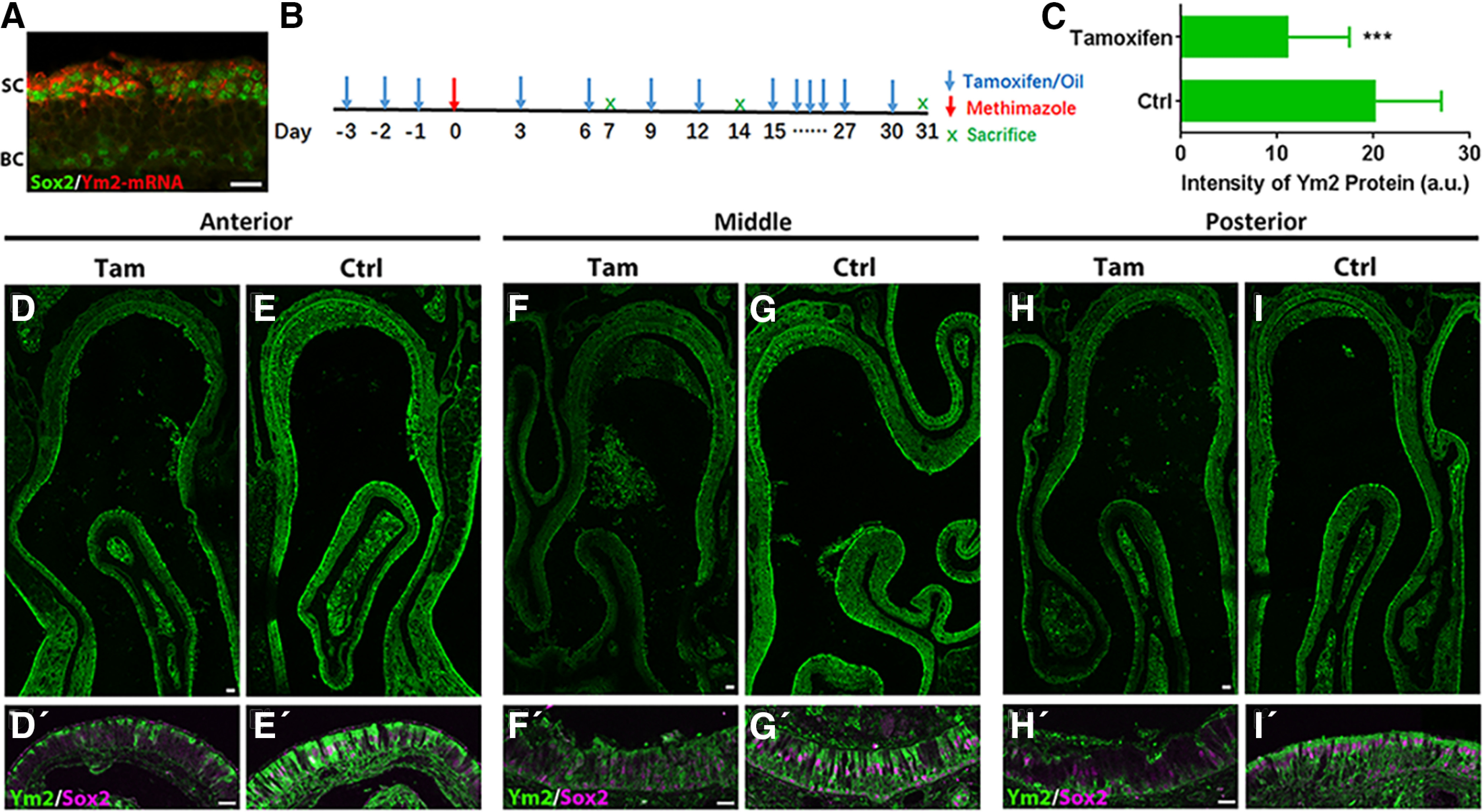
Ym2 knockdown in Sox2-CreERT2/Ym2fl/fl mice with tamoxifen induction. A, Confocal image of FISH against Ym2-mRNA and immunostaining against Sox2 on an OE section from a 3-week-old mouse. B, Scheme of tamoxifen-induced Ym2 knockdown in methimazole-lesioned OE. Repeated tamoxifen injections ensure Ym2 knockdown in continuously generated supporting cells. C, Statistical analysis on intensity of Ym2 antibody staining in the OE on day 7 post-methimazole injection (n = 20 and 17 sections from three control and tamoxifen-treated mice, respectively). a.u., Arbitrary units. D–I′, Confocal images of immunostaining against Ym2 and Sox2 in anterior (D, D′), middle (F, F′), and posterior (H, H′) regions with tamoxifen induction compared with the corresponding regions (anterior, E, E′; middle, G, G′; posterior, I, I′) in controls injected with sunflower oil (day 7 post-methimazole injection). Statistical significance was determined by unpaired Student's t test. C: ***p = 0.0002 (t(35) = 4.157). SC, Supporting cell; BC, basal cell; Tam, tamoxifen; Ctrl, control. Scale bars: D, F, H, 100 μm; A, D′, F′, H′, 20 μm.
We then determined whether Ym2 knockdown could affect OE regeneration. On day 7 post-methimazole injection, compared with control Sox2-CreERT2/Ym2fl/fl mice (Fig. 4A), there were 58 ± 8% fewer OMP+ cells in tamoxifen-injected mice (Fig. 4B,M), suggesting that Ym2 knockdown attenuated neuronal regeneration in the OE. Similarly, on day 31 post-OE lesion, OMP+ cells were reduced by 17 ± 7% in tamoxifen-treated mice compared with the controls (Fig. 4C,D,M). Since supporting cells play an important role in keeping HBCs in dormancy (Lin et al., 2017), we examined the effect of Ym2 knockdown on HBCs by immunostaining against ICAM1. On day 7 postlesion, the intensity of ICAM1 staining was significantly weakened by 52 ± 4% in the OE with Ym2 knockdown (Fig. 4E,F,N). Similar observation was present on day 31, with the ICAM1 intensity reduced by 37 ± 5% (Fig. 4G,H,N). This finding suggested that Ym2 knockdown impeded recruitment of HBCs in lesioned OE, which may lead to attenuation in OE recovery. Furthermore, genetic knockdown of Ym2 also decreased Sox2+ supporting cells by 42 ± 11% on day 7 (Fig. 4I,J,O) and by 29 ± 9% on day 31 (Fig. 4K,L,O). Collectively, these data indicated that Ym2 knockdown delayed recovery of the injured OE.
Figure 4.
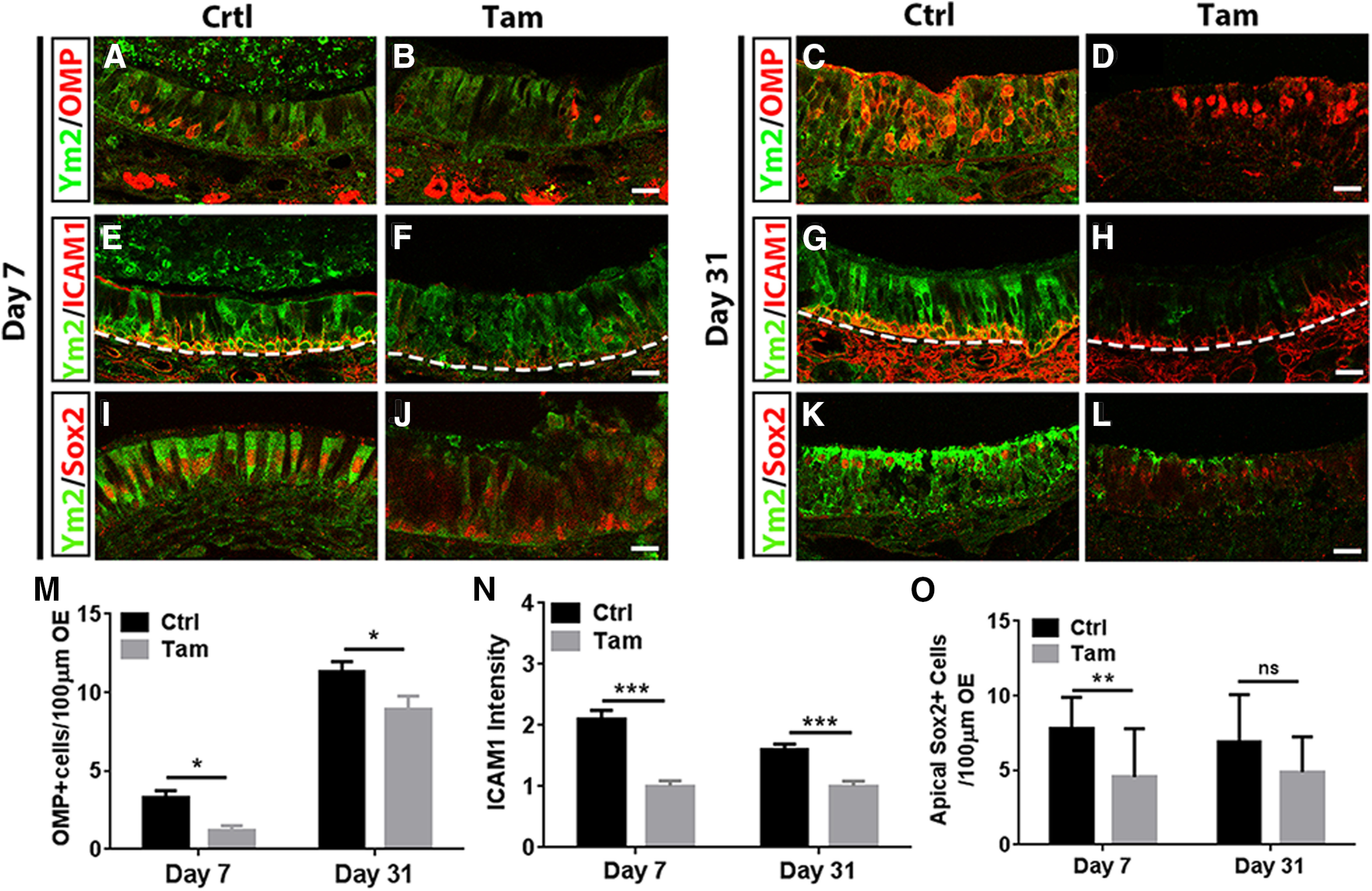
Genetic downregulation of Ym2 attenuates regeneration in the lesioned OE. A–L, Immunostaining showed that Ym2 downregulation through tamoxifen (Tam) induction in Sox2-CreERT2/Ym2fl/fl mice decreased the percentage of OMP+ cells (A–D), ICAM1+ cells (E–H), and Sox2+ supporting cells in the OE (I–L) on day 7 and day 31 postinjury compared with oil-injected controls (Ctrl). M, Statistical analysis on the number of OMP+ cells per 100 µm OE on day 7 and day 31 postinjury in control and Ym2 downregulation groups (n = 26 sections from three mice in each group). N, Statistical analysis of the ICAM1 expression intensity on day 7 and day 31 postinjury (n = 12 sections in each group). O, Statistical analysis on the number of apical Sox2+ cells/100 μm OE (n = 15 and 22 sections on day 7 and day 31 postinjury). The dashed line in E–H separated OE from lamina propria. Statistical significance was determined by two-way ANOVA. M: F(1,100) = 24.45, p < 0.0001; N: F(1,44) = 48.70, p < 0.0001; O: F(1,70) =15.16, p = 0.0002. ns, Not significant. Asterisks were determined by Sidak's multiple-comparisons test. Scale bars, 20 μm.
In addition to genetic knockdown of Ym2 expression, we also examined the effects of Ym2 downregulation and upregulation on methimazole-lesioned OE using viral infection. Wild-type C57BL/6J mice were injected with Lenti-shYm2 (lentivirus expressing shRNA targeting Ym2) or Lenti-Ym2. Initially, we injected lentivirus 1 d after OE injury, but OE regeneration was not affected under this condition. This is likely because OE regeneration actively occurs within a few days after lesioning, whereas lentiviral infection may take longer to effectively change Ym2 expression in enough cells. We then injected lentivirus 10 d before methimazole injection (Fig. 5A). This protocol allowed sufficient time for lentiviral infection to regulate Ym2 expression in the OE before and during the process of OE ablation. Multiple OE cell types, including supporting cells, OSNs, and basal cells, could be infected based on positive GFP signals on day 14 postinjury (Fig. 5B; for implications of this finding, see Discussion). On day 7 postinjury, OE regeneration was attenuated since the cell numbers in different layers were significantly decreased (Fig. 6A,B). Compared with the Lenti-shCtrl group, Ym2 downregulation via Lenti-shYm2 infection led to 88 ± 12% fewer PGP9.5+ neurons (Fig. 6A,F), 71 ± 19% fewer apical Sox2+ supporting cells (Fig. 6B,G), 71 ± 7% fewer OMP+ cells (Fig. 6B,E), 69 ± 25% fewer ICAM1+ cells (Fig. 6A,I), and 92 ± 7% fewer basal Sox2+ cells (Fig. 6H). By contrast, overexpression of Ym2 via Lenti-Ym2 infection led to increases in PGP9.5+, apical Sox2+, OMP+, ICAM1+, and basal Sox2+ cells by 236 ± 75% (Fig. 6A,F), 149 ± 63% (Fig. 6B,G), 300 ± 95% (Fig. 6B,G), 127 ± 22% (Fig. 6A,I) and 58 ± 14% (Fig. 6H), respectively.
Figure 5.
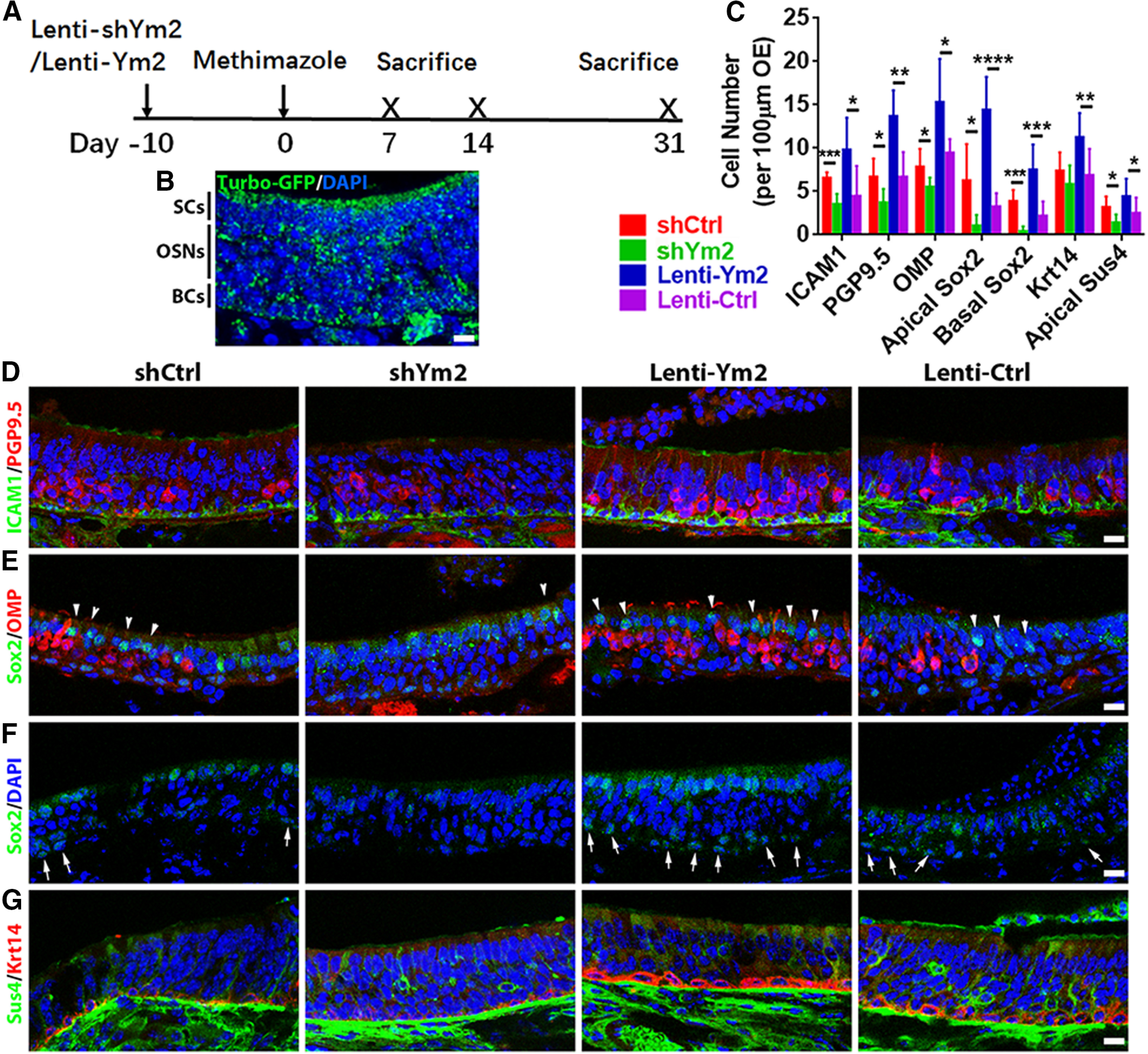
Ym2 regulates the recovery of the lesioned OE. A, The timeline of lentiviral and methimazole administration. B, Confocal image of Lenti-shYm2-infected OE immunostained with anti-TurboGFP. C, Statistical analysis of ICAM1+, PGP9.5+, OMP+, apical Sox2+, basal Sox2+, Krt14+, and Sus4+ cells/100 µm OE in Lenti-shCtrl, Lenti-shYm2, Lenti-Ym2, and Lenti-Ctrl groups on day 14 postlesion (n = 9 sections from three mice in each group). D–G, Confocal images of ICAM1+ and PGP9.5+ (D), apical Sox2+ and OMP+ (E), basal Sox2+ (F), and Sus4+ and Krt14+ (G) cells in the OE on day 14 post-OE lesion from mice injected with Lenti-shCtrl, Lenti-shYm2, Lenti-Ym2, or Lenti-Ctrl. Apical and basal Sox2+ cells are denoted by arrowheads and arrows in E and F. Statistical significance was determined by unpaired Student's t test. ICAM1+: ***p = 0.0003 (t = 5.47) and *p = 0.022 (t = 2.664); PGP9.5+: *p = 0.018 (t = 2.769) and **p = 0.0013 (t = 4.261); OMP+: *p = 0.041 (t = 2.372) and *p = 0.027 (t = 2.781); apical Sox2+: *p = 0.028 (t = 2.605) and ****p < 0.0001 (t = 7.109); basal Sox2+: ***p = 0.0002 (t = 5.55) and ***p = 0.0003 (t = 4.631); Krt14+: p = 0.237 (t = 1.251) and **p = 0.008 (t = 3.05); apical Sus4+: *p = 0.02 (t = 2.742) and *p = 0.023 (t = 2.065). The degree of freedom for all t tests is 16. Scale bars, 10 μm.
Figure 6.
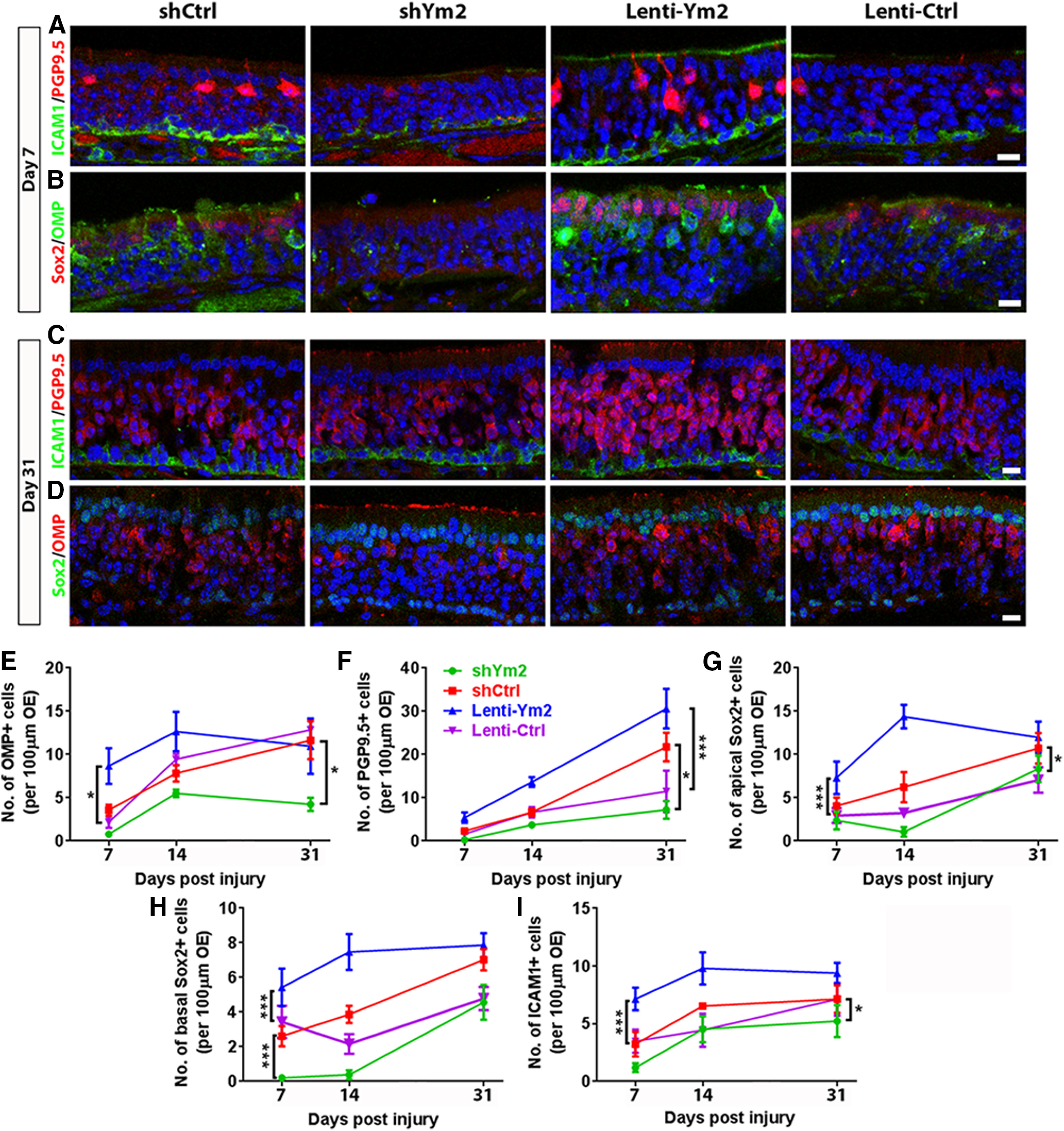
Ym2 regulates regeneration in the lesioned OE. A–D, Confocal images of ICAM1+, PGP9.5+, OMP+, and Sox2+ cells in the lesioned OE from mice injected with Lenti-shCtrl, Lenti-shYm2, Lenti-Ym2, or Lenti-Ctrl on day 7 and day 31 postinjury. E–I, Statistical analysis on the number of OMP+, PGP9.5+, apical Sox2+, basal Sox2+, and ICAM1+ cells/100 µm OE of mice injected with Lenti-shYm2, Lenti-shCtrl, Lenti-Ym2, or Lenti-Ctrl on day 7, day 14, and day 31 postinjury (OMP: n = 6 sections from three mice in each group; PGP9.5: n = 5–7 sections; apical Sox2+: n = 5–7 sections; basal Sox2+: n = 5–9 sections; ICAM1+: 5–7 sections). Statistical significance was determined by unpaired Student's t test between Lenti-shCtrl and Lenti-shYm2 or between Lenti-Ym2 and Lenti-Ctrl at the same time points, or by two-way ANOVA at all three time points together. Statistical analysis shown in E–I was based on two-way ANOVA. E: F(3,60) = 12.10, p < 0.0001; F: F(3,56) = 16.49, p < 0.0001; G: F(3,59) = 17.9, p < 0.0001; H: F(3,65) = 22.36, p < 0.0001; I: F(3,57) = 12.21, p < 0.0001. Asterisks was determined by Tukey's multiple-comparisons test. Scale bars, 10 μm.
Two weeks after OE injury, Lenti-shYm2 infection mice had fewer PGP9.5+ and OMP+ neurons (by 44 ± 9% and 77 ± 6%, respectively) compared with the Lenti-shCtrl group (Fig. 5C–E). Likewise, there were fewer apical Sox2+ and Sus4+ supporting cells by 83 ± 9% and 56 ± 12%, respectively (Fig. 5C,E,G). Furthermore, Ym2 downregulation reduced the generation of basal cells, evidenced by decreased Sox2+ basal cells by 90 ± 7% (Fig. 5C,F), ICAM1+ HBCs by 46 ± 8% (Fig. 5C,D), and Krt14+ HBCs by 21 ± 12% (Fig. 5C,G). By contrast, Ym2 overexpression significantly increased the cell numbers at this time point, as follows: PGP9.5+ neurons by 106 ± 17% (Fig. 5C,D), OMP+ OSNs by 62 ± 23% (Fig. 5C,E), apical Sox2+ supporting cells by 348 ± 43% (Fig. 5C,E), Sus4+ supporting cells by 95 ± 26% (Fig. 5C,G), Sox2+ basal cells by 246 ± 48% (Fig. 5C,F), ICAM1+ HBCs by 121 ± 31% (Fig. 5C,D), and Krt14+ HBCs by 64 ± 13% (Fig. 5C,G).
On day 31 postlesion, Ym2 downregulation led to 67 ± 9% fewer PGP9.5+ cells (Fig. 6C,F), whereas Ym2 overexpression caused an increase by 167 ± 40% (Fig. 6C,F). While Ym2 downregulation led to 53 ± 8% fewer OMP+ neurons (Fig. 6D,E), Ym2 overexpression did not significantly alter the number of OMP+ cells (Fig. 6D,E), suggesting a ceiling effect of the Ym2 level on OE regeneration. Changes in apical and basal Sox2+ cells as well as ICAM1+ cells are summarized in Figure 6G–I. Together, these findings support the idea that Ym2 regulates regeneration of the injured OE.
Ym2 regulates cell proliferation and differentiation in OE colonies
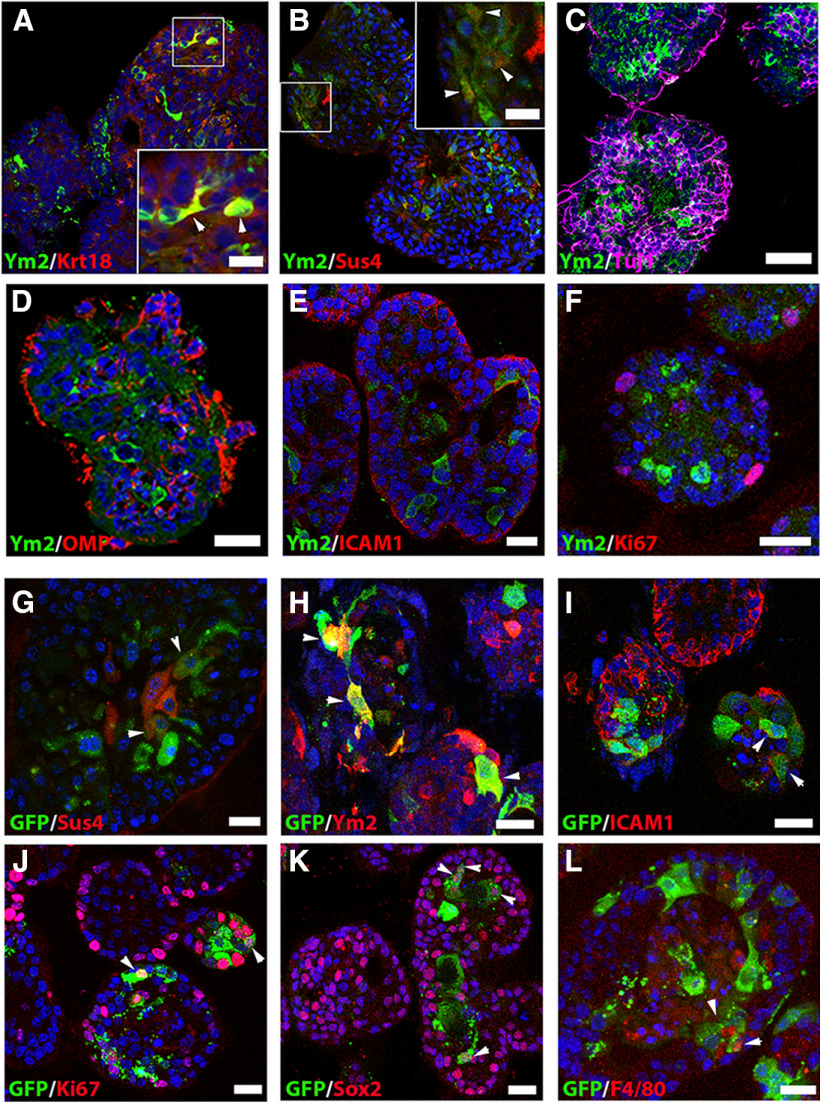
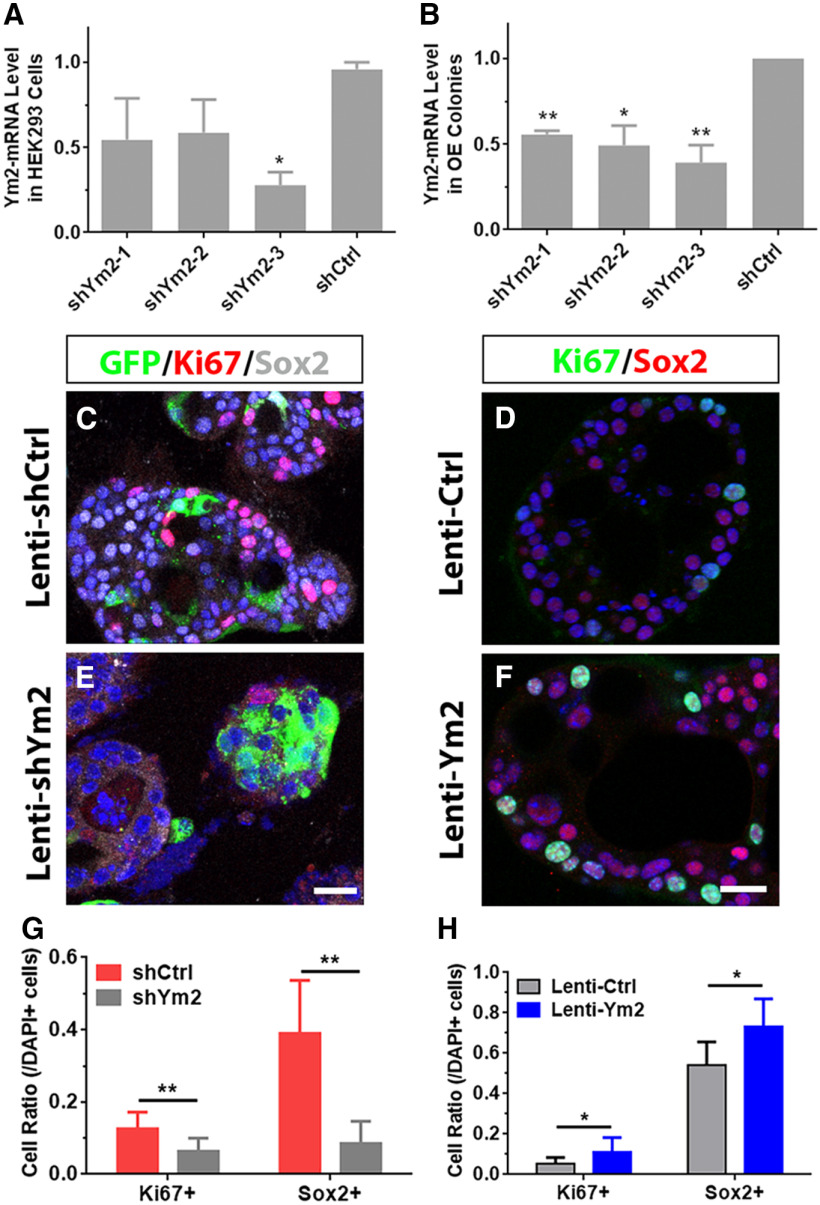
We then determined whether Ym2 is expressed in OE colonies. Ym2 was colocalized with Krt18+ and Sus4+ cells in vitro, but not in Tuj1+ and OMP+ sensory neurons, ICAM1+ HBCs, or Ki67+ proliferative cells Ym2 (Fig. 7A–F). This is consistent with the staining pattern in the OE, suggesting that Ym2 is exclusively expressed in presumptive supporting cells. OE colonies were then infected with lentivirus to regulate Ym2 expression to further verify its role. Similar to the OE (Fig. 5B), lentiviral infection in OE colonies was not cell type specific. Infected GFP+ cells included Sus4+ and Ym2+ supporting cells, ICAM+ HBCs, Ki67+ proliferative cells, Sox2+ progenitor or supporting cells, and even F4/80+ inflammatory cells (Fig. 7G–L; for implications of this finding, see Discussion). The efficiency of shYm2-3 lentiviral infection was confirmed in HEK293 cells overexpressing Ym2 (Fig. 8A) as well as in OE colonies (Fig. 8B). Thus, we used Lenti-shYm2-3 to infect OE colonies for further analysis. Ym2 downregulation decreased the percentage of Ki67+ (marker of proliferation) by 46 ± 9% and Sox2+ cells by 78 ± 7% in OE colonies compared with the control colonies infected with Lenti-shCtrl (Fig. 8C,E,G). By contrast, Ym2 overexpression led to an increase in the percentage of Ki67+ cells by 153 ± 30% and Sox2+ cells by 36 ± 10% (Fig. 8D,F,H). Ym2 downregulation reduced the percentage of Sus4+ supporting cells by 71 ± 14% and OMP+ mOSNs by 68 ± 5% in shYm2-infected colonies compared with the Lenti-shCtrl group (Fig. 9A,B,E,F,I). By contrast, Ym2 overexpression via Lenti-Ym2 infection led to an increase in the percentage of OMP+ cells by 30 ± 9% (Fig. 9C,D,I). However, we did not observe a significant change in the percentage of Sus4+ cells in OE colonies infected with Lenti-Ym2 (Fig. 9G–I). To confirm the effect of Ym2 overexpression on cell proliferation and differentiation, OE colonies were treated with recombinant Ym2 protein. The ratio of Ki67+ cells increased by 69 ± 11%, 52 ± 7%, and 35 ± 16% in the presence of 0.5, 1, and 2 μg/ml Ym2 protein, while the Sox2+ cell ratio was not significantly altered (Fig. 9J–M,R). Meanwhile, the ratio of Ki67+Sox2+ cells increased by 71 ± 10%, 47 ± 8%, and 42 ± 15%, and the ratio of OMP+ cells increased by 253 ± 66%, 292 ± 40%, and 226 ± 45% with treatment of 0.5, 1, and 2 μg/ml Ym2 protein (Fig. 9N–R). However, we did not find significant changes in the ratio of Krt14+ cells with Ym2 incubation (Fig. 9N–R). Collectively, these data support that Ym2 regulates cell proliferation and differentiation in OE colonies.
Figure 7.
Presumptive supporting cells in OE colonies express Ym2, and lentiviral infection in OE colonies is not cell type specific. A–F, Confocal images of Ym2+ and CK18+ (A), Sus4+ (B), Tuj1+ (C), OMP+ (D), ICAM1+ (E), and Ki67+ (F) cells in OE colonies. G–L, Lentiviral-infected (GFP+) OE colonies were immunostained with Sus4 (G), Ym2 (H), ICAM1 (I), Ki67 (J), Sox2 (K), and F4/80 (L). Lenti-Ctrl was used in H; Lenti-shYm2 was used in G, and I–L. Arrowheads mark Ym2+/CK18+, and Ym2+/Sus4+ cells in A and B, and positively stained GFP+ cells in G–L. Scale bars: C–L, 20 μm; A, B, 10 μm.
Figure 8.
Ym2 regulates cell proliferation of OE colonies. A, B, Quantitative PCR analysis showed the efficiency of Lenti-shYm2 of downregulating Ym2-mRNA expression in Ym2-overexpressed HEK293 cells or OE colonies (n = 3 independent experiments). shYm2-1, shYm2-2, and shYm2-3 were lentiviruses targeting three different parts of the Ym2 sequence. C–F, Confocal images of Ki67+ or Sox2+ cells in Lenti-shCtrl-, Lenti-shYm2-, Lenti-Ctrl-, or Lenti-Ym2-infected OE colonies. G, H, Statistical analysis of the percentage of Ki67+ and Sox2+ cells in OE colonies with Lenti-shCtrl, Lenti-shYm2, Lenti-Ctrl, or Lenti-Ym2 infection (n = 12, 12, 6, and 6 sections from three independent cultures, respectively). Statistical significance in A and B was determined by unpaired t test. A: *p = 0.0156, t(4) = 7.923; B: **p = 0.0013, t(4) = 27.93; *p = 0.0247, t(4) = 6.239; and **p = 0.0017, t(4) = 8.443. Statistical significance was determined by two-way ANOVA in G and H, and asterisks were determined by Sidak's multiple-comparisons test. shCrtl and shYm2: F(1,44) = 34.59, p < 0.001; Lenti-Ctrl and Lenti-Ym2: F(1,20) = 9.951, p = 0.005. Scale bars, 20 μm.
Figure 9.
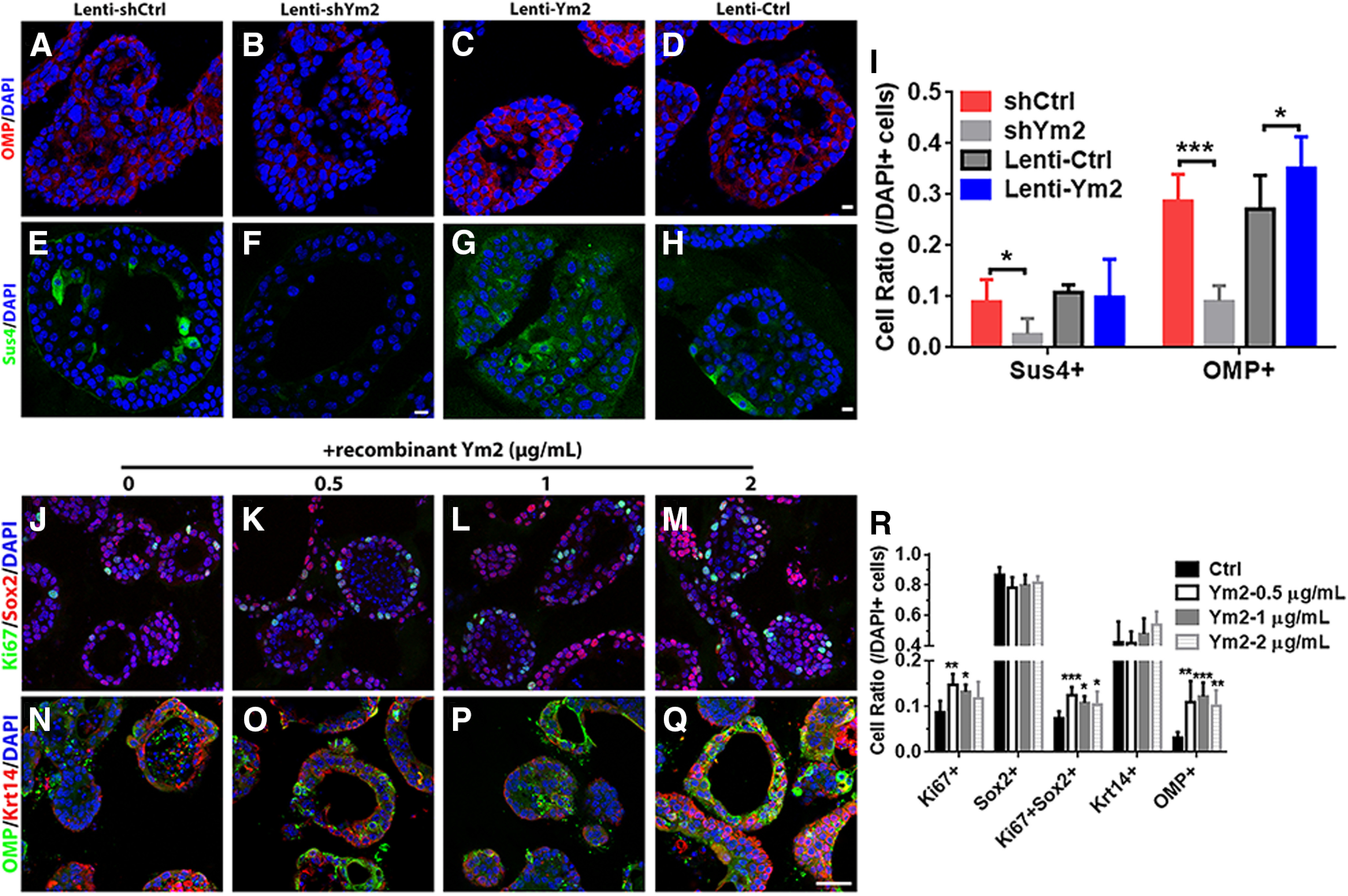
Ym2 regulates cell differentiation in OE colonies. A–D, Confocal images of OMP+ cells in colonies infected with Lenti-shCtrl, Lenti-shYm2, Lenti-Ctrl, or Lenti-Ym2. E–H, Immunostaining against Sus4 in Lenti-shCtrl-, Lenti-shYm2-, Lenti-Ctrl-, or Lenti-Ym2-infected colonies. I, Statistical analysis of Sus4+ and OMP+ cell ratios in colonies with Lenti-shCtrl, Lenti-shYm2, Lenti-Ctrl, or Lenti-Ym2 infection (Sus4+: n = 7, 6, 7, and 6 sections from three independent cultures in each group; OMP: n = 6 sections in each group). J–M, Confocal images of Sox2+ and Ki67+ cells in OE colonies treated with 0, 0.5, 1, and 2 μg/ml Ym2. N–Q, Confocal images of OMP+ and Krt14+ cells in OE colonies treated with 0, 0.5, 1, and 2 μg/ml Ym2. R, Statistical analysis of Ki67+, Sox2+, Ki67+Sox2+, Krt14+, and OMP+ cells in untreated and recombinant Ym2-treated colonies (Ki67+, Sox2+, Ki67+Sox2+: n = 6 sections from three independent cultures in each group; Krt14+, OMP+: n = 5 sections in Ctrl and 0.5 μg/ml Ym2-treated groups; n = 6 sections in 1 and 2 μg/ml Ym2-treated groups). Statistical significance in I was determined by two-way ANOVA: F(3,42) = 26.05, p < 0.0001. Asterisks in I were determined by Sidak's multiple-comparisons test. Statistical significance in R was determined by one-way ANOVA, and asterisks were measured by Dunnett's multiple comparisons test. Ki67+: F(3,20) = 5.691, p = 0.0055; Sox2+: F(3,20) = 2.389, p = 0.0991; Ki67+Sox2+: F(3,20) = 6.707, p = 0.0026; Krt14+: F(3,18) = 1.696, p = 0.2036; OMP+: F(3,18) = 7.838, p = 0.0015. Scale bars, 10 μm.
Attenuation of acute inflammatory responses inhibits Ym2 expression and delays OE regeneration
Previous studies suggest that OE regeneration is accompanied by acute inflammatory responses (Chen et al., 2017). Since Ym2 plays essential roles in inflammation in other tissues (Rouse et al., 2007; Cai et al., 2009; Lee et al., 2011; Ooi et al., 2012; Kurpińska et al., 2019), we asked whether Ym2 elevation in the injured OE was a prerequisite for the occurrence of inflammation or vice versa. We administrated methimazole to lesion the OE as well as Dex, a type of corticosteroid with anti-inflammatory effects (Fig. 10A). Dex treatment significantly decreased the Ym2-mRNA level in the injured OE compared with the untreated control (Fig. 10B). Inhibition of inflammatory responses after Dex treatment was confirmed by significantly reduced TNF-α and IL-1β levels (Fig. 10B). Interestingly, Dex-induced reduction in TNF-α and IL-1β levels was not evident on day 1 postinjury when a decrease of the Ym2 level was apparent (Fig. 10B). Given that injury enhanced Ym2 expression in the OE (Fig. 2), these data suggested that the modulation of Ym2 by Dex in the injured OE occurred before the inhibition of inflammatory responses. Dex administration in methimazole-lesioned mice drastically reduced the intensity of Ym2 expression in the supporting cell layer by 62 ± 8% compared with saline controls on day 14 postinjury (Fig. 10C,E,K), and led to 86 ± 10% fewer OMP+ neurons (Fig. 10C,E,L) and 75 ± 11% fewer PGP9.5+ neurons (Fig. 10D,F,L). On day 31, 36 ± 13% fewer OMP+ cells (Fig. 10G,I,M; not statistically significant) and 47 ± 13% fewer apical Sox2+ cells (Fig. 10H,J,M) were found in the OE with Dex administration. Thus, the inhibition of acute inflammatory responses downregulated Ym2 expression and attenuated regeneration in the injured OE.
Figure 10.
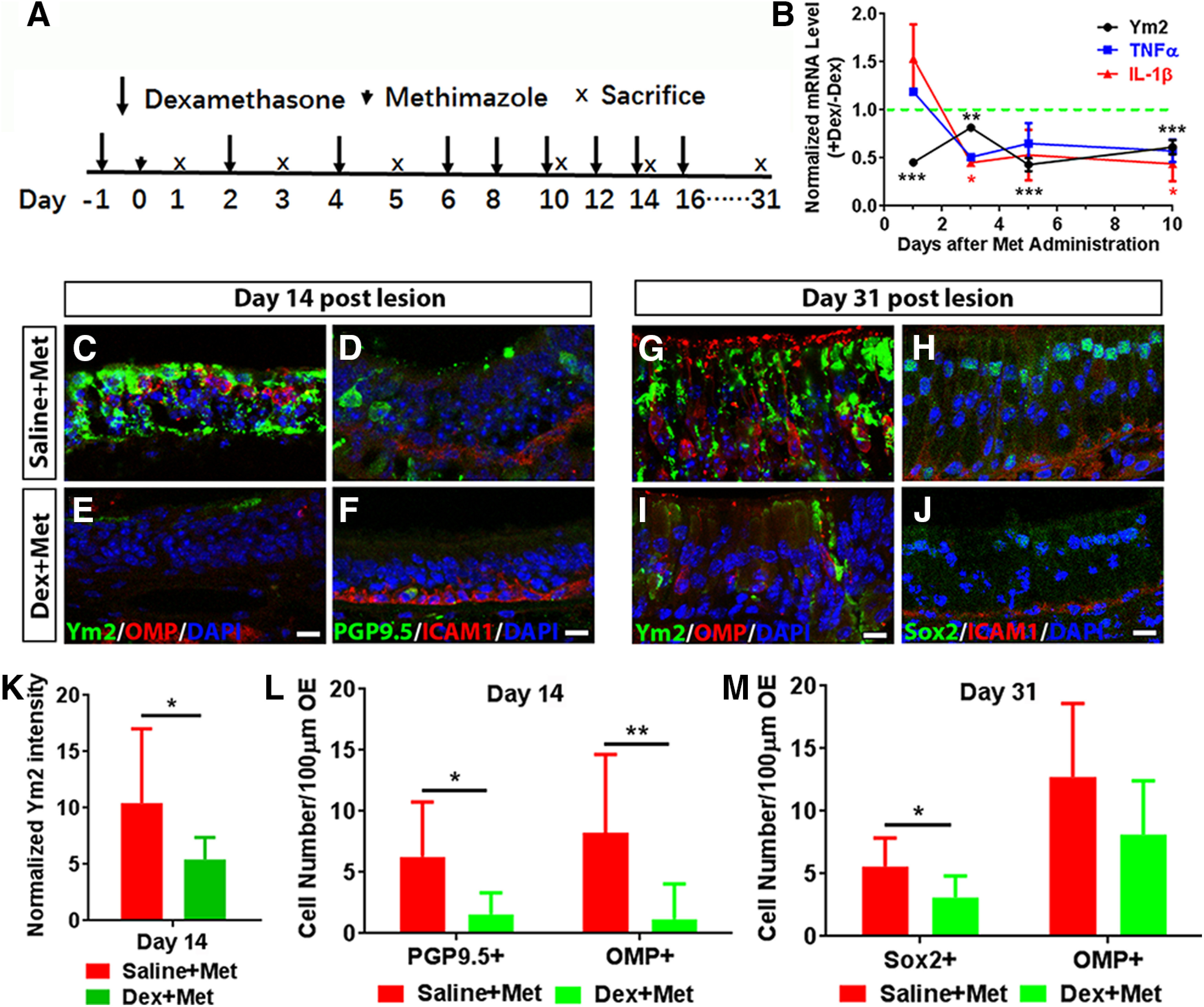
Dexamethasone (Dex) treatment reduces the Ym2 level and defers OE regeneration. A, Scheme showed Dex injection in mice with methimazole-induced OE lesion. B, Quantitative PCR analysis on Dex-induced alteration in the Ym2-, TNF-α-, and IL-1β-mRNA levels in lesioned OE (n = 3 independent experiments). C–J, Confocal images of immunostaining against Ym2, OMP, PGP9.5, Sox2, and ICAM1 in the OE of mice with saline or Dex injection on day 14 and day 31 postlesion. K, Analysis on Ym2 intensity in lesioned OE on day 14 with saline or Dex administration (n = 9 sections from three mice). L, M, Statistical analysis of PGP9.5+, OMP+, or apical Sox2+ cells in the OE of saline- or Dex-injected mice on day 14 (PGP9.5+: n = 6 sections; OMP+: n = 6 and 12 sections in Dex-treated and saline-treated groups) and day 31 postlesion (Sox2+: n = 6 sections; OMP+: n = 5 sections). Statistical significance was determined by one-way ANOVA (B), by unpaired Student's t test (K), and by two-way ANOVA (L, M). B: Ym2: F(4,10) = 74.57, p < 0.0001; TNF-α: F(4,10) = 16.00, p = 0.0004; IL-1β: F(4,10) = 14.69, p = 0.0003; K: *p = 0.0456 (t(16) = 2.18); L: F(1,26) = 14.95, p = 0.0007; M: F(1,18) = 4.609, p = 0.0457. Asterisks were determined by Dunnett's multiple-comparisons test (B) and by Sidak's multiple-comparisons test (L, M). Scale bars, 10 µm.
Ym2 overexpression counteracts the inhibitory effect of Dex on OE regeneration
To further corroborate the interaction between Ym2 and inflammatory responses, we induced Ym2 overexpression in the injured OE of Dex-injected mice (Fig. 11A). Compared with the Lenti-Ctrl group, Ym2 overexpression via Lenti-Ym2 infection increased various cell types in the OE on day 14 postlesion, as follows: PGP9.5+ neurons by 306 ± 59% (Fig. 11B–D,N), ICAM1+ HBCs by 197 ± 25% (Fig. 11B–D,N), Sox2+ supporting cells by 1385 ± 265% (Fig. 11E–G,N), and OMP+ mOSNs by 181 ± 59% (Fig. 11E–G,N). On day 31, the ICAM1+, PGP9.5+, and Sox2+ cells in the Ym2-overexpressed OE increased by 40 ± 7%, 70 ± 11%, and 154 ± 25%, respectively, compared with the Lenti-Ctrl group (Fig. 11H–M,O). However, there was no significant change in the number of OMP+ cells on day 31 (Fig. 11K–M,O). Ym2 overexpression in Dex-injected mice led to an increase in OE thickness by 39 ± 8% on day 14 and 9 ± 2% on day 31 postinjury (Fig. 11P). To further verify the potential connection between Ym2 and inflammation, we counted F4/80+ macrophages and CD45+ inflammatory cells in the OE. On day 14 postinjury, Ym2 overexpression led to significant increases of F4/80+ and CD45+ cells by 110 ± 16% and 78 ± 19%, respectively, compared with Lenti-Ctrl mice (Fig. 11Q–U). Collectively, these data showed that Ym2 overexpression could counteract the inhibitory effect of Dex on OE regeneration, implying that Ym2 plays a regulatory role in inflammation-mediated OE recovery from lesion.
Figure 11.
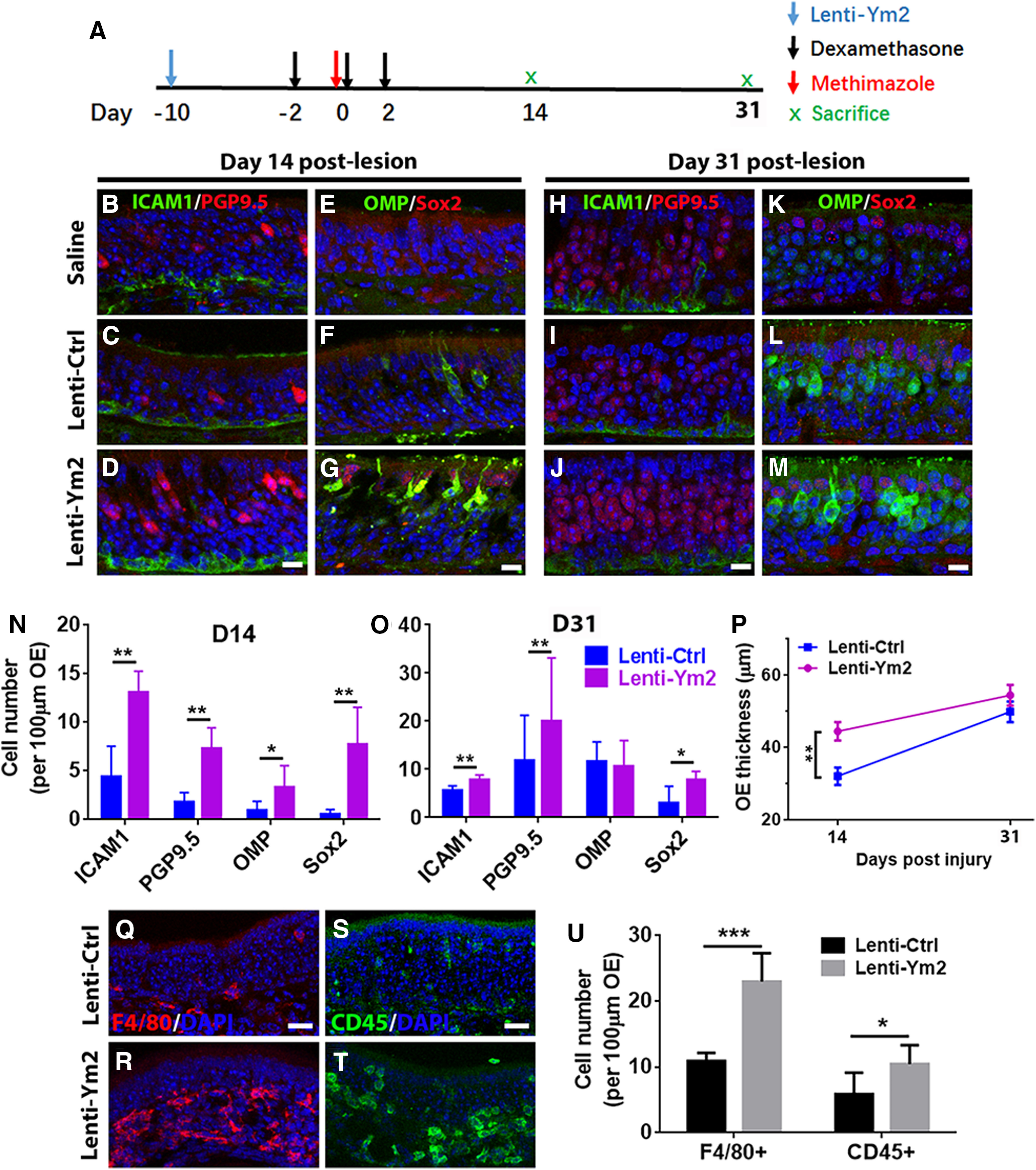
Ym2 overexpression counteracts Dex-induced attenuation in OE regeneration. A, The timeline of Lenti-Ym2, Dex, and methimazole administration. B–M, Immunostaining revealed alterations in the number (per 100 µm OE) of ICAM1+ and PGP9.5+ cells (B–D, H–J) as well as OMP+ and Sox2+ cells (E–G, K–M) by saline, Lenti-Ctrl, or Lenti-Ym2 injection in Dex-treated mice, killed on day 14 and day 31 post-OE lesion. N, O, Statistical analysis of the number of ICAM1+, PGP9.5+, OMP+, and apical Sox2+ cells per 100 µm OE in Lenti-Ym2/Dex-treated mice and Lenti-Ctrl/Dex-injected controls on day 14 and day 31 after OE injury (n = 5 sections from three mice). P, Statistical analysis of the OE thickness in Dex-treated mice injected with Lenti-Ctrl or Lenti-Ym2. OE thickness: 32.1 ± 7.1 and 44.5 ± 8.5 μm in mice receiving Lenti-Ctrl and Lenti-Ym2 on day 14; 49.9 ± 6.3 and 54.5 ± 7.0 μm on day 31; n = 10 sections on day 14 and n = 6 sections on day 31 from three mice. Q–T, Confocal images of F4/80+ and CD45+ cells in the OE of Lenti-Ctrl- and Lenti-Ym2-injected mice. U, Statistical analysis of the number of F4/80+ and CD45+ cells/100 µm OE (n = 6 sections from three mice). Statistical significance was determined by two-way ANOVA (N, O, P). N: F(1,32) = 58.79, p < 0.0001; O: F(1,32) = 6.650, p = 0.0013; P: F(1,28) = 8.868, p = 0.0061. Asterisks in N, O, and P were determined by Sidak's multiple-comparisons test. Statistical significance in U was determined by unpaired t test. *p = 0.0347, t(10) = 2.485; ***p < 0.001, t(10) = 6.544. Scale bars: D, G, J, M, 10 μm; Q, S, 20 μm.
Anti-inflammation downregulates Ym2 and inhibits cell differentiation in OE colonies
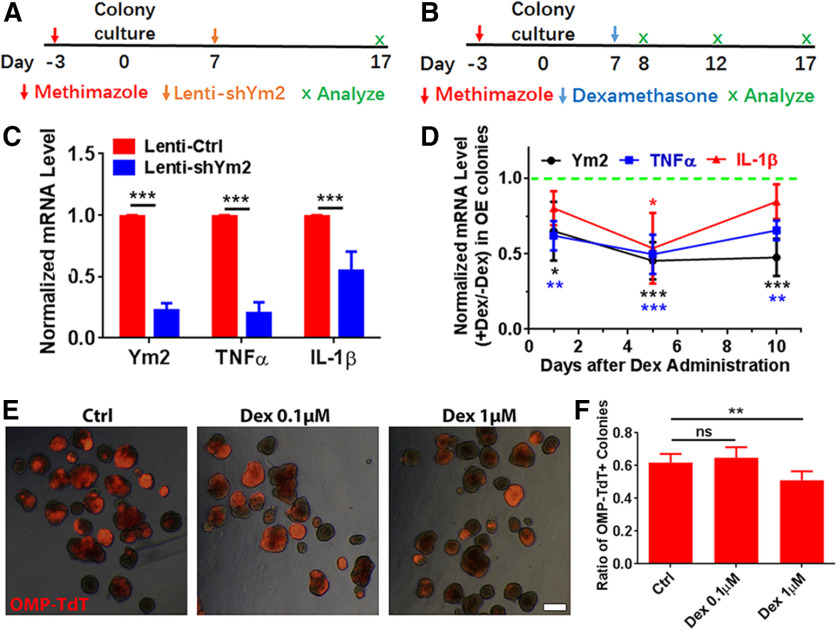
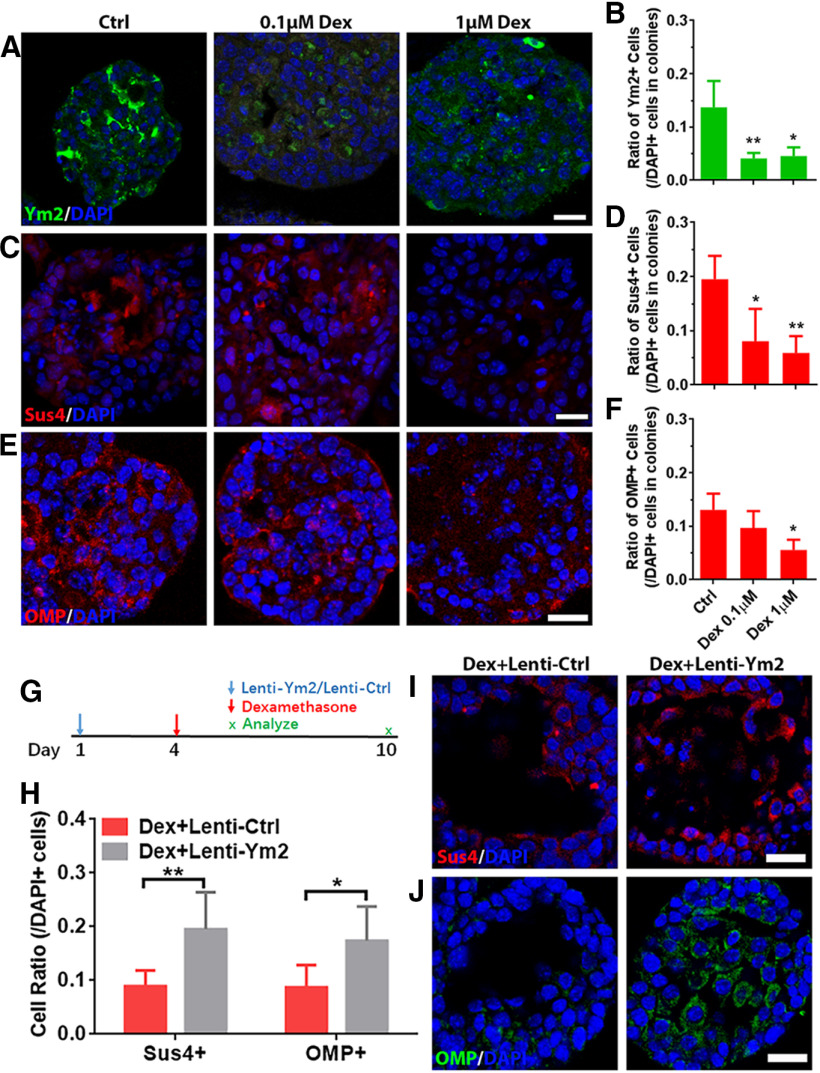
We then determined whether Dex affected Ym2 expression and cell differentiation in OE colonies. Ym2 downregulation in vitro (Fig. 12A) led to a drastic decrease in the TNF-α-mRNA level compared with the control (Fig. 12C). Compared with the colonies infected with Lenti-shCtrl, Ym2 downregulation also reduced the IL-1β-mRNA level (Fig. 12C). These data further support the notion that Ym2 may regulate inflammatory responses in the OE. Furthermore, OE colonies with 1 μm Dex treatment (Fig. 12B) showed a decreased Ym2-mRNA level compared with untreated controls on day 5 and day 10 (Fig. 12D), demonstrating that anti-inflammation in vitro also restrained Ym2 expression. In addition, treatment with 1 μm Dex decreased TNF-α-mRNA and IL-1β-mRNA expression (Fig. 12D). Immunostaining analysis on OE colonies showed 67 ± 8% or 63 ± 13% fewer Ym2+ cells with the treatment of 0.1 or 1 μm Dex, compared with untreated controls (Fig. 13A,B). Treatment of 0.1 or 1 μm Dex significantly reduced the percentage of Sus4+ cells by 50 ± 16% or 63 ± 8% in OE colonies (Fig. 13C,D). Furthermore, 1 μm Dex treatment reduced the percentage of OMP+ cells in OE colonies by 46 ± 7% (Fig. 13E,F). This was further supported by an 18 ± 3% decrease in OMP-TdT+ (TdTomato) colonies in the presence of 1 μm Dex (Fig. 12E,F), suggesting that the inhibition of inflammatory responses restrained neuronal differentiation in OE colonies. We then overexpressed Ym2 in Dex-treated OE colonies (Fig. 13G), which led to an increase in the percentage of Sus4+ cells by 120 ± 31% (Fig. 13H,I), and OMP+ cells by 102 ± 30% (Fig. 13H,J). These data indicated that anti-inflammatory treatment inhibited Ym2 expression and hindered cell differentiation in OE colonies, and this effect was counteracted by Ym2 overexpression.
Figure 12.
Anti-inflammatory administration interacts with Ym2 expression and affects cell differentiation in OE colonies. A, Schematic of methimazole and Lenti-shYm2 treatment in OE colonies. B, Schematic of methimazole and Dex administration in OE colonies. C, Quantitative PCR analysis showed decreases in the Ym2-, TNF-α-, and IL-1β-mRNA levels by Ym2 downregulation (n = 3 experiments). D, Quantitative PCR analysis on Ym2-, TNF-α-, and IL-1β-mRNA levels with Dex treatment in OE colonies (n = 3–5 experiments). E, Images of OMP-TdT+ (TdTomato+) colonies treated with saline, 0.1 μm Dex, or 1 μm Dex. F, Statistical analysis of the percentage of OMP-TdT+ colonies in the presence of saline, 0.1 μm Dex, or 1 μm Dex (n = 6, 6, and 7 sections from three independent cultures in each group). Statistical significance was determined by two-way ANOVA with Sidak's multiple-comparisons tests (C), and by one-way ANOVA with Dunnett's multiple-comparisons test (D, F). ns, Not significant. C: F(1,12) = 342.5, p < 0.0001; D: Ym2: F(3,12) = 12.34, p = 0.0004; TNF-α: F(3,10) = 18.15, p = 0.0002; IL-1β: F(3,8) = 5.203, p = 0.0335; F: F(2,16) = 10.83, p = 0.0011. Scale bars, 0.2 mm.
Figure 13.
Ym2 overexpression alleviates the inhibitory effect of Dex on cell differentiation of OE colonies. A, C, E, Confocal images of Ym2+, Sus4+, and OMP+ cells in saline-treated, 0.1 μm Dex-treated, or 1 μm Dex-treated OE colonies. B, D, F, Statistical analysis of the Ym2+, Sus4+, and OMP+ cell percentages in OE colonies treated with saline, 0.1 μm Dex, or 1 μm Dex (Ym2+: n = 3, 4, and 3 sections in each group; Sus4+: n = 4 sections; OMP+: n = 4, 4, and 3 sections). G, Scheme showing the Lenti-Ctrl or Lenti-Ym2 and Dex administration in OE colonies. H, Statistical analysis on the percentage of Sus4+ or OMP+ cells in Dex-treated colonies infected with Lenti-Ctrl or Lenti-Ym2 (Sus4+: n = 7 and 6 sections from three independent cultures in each group; OMP+: n = 6 sections). I, J, Confocal images of Sus4+ and OMP+ cells in Lenti-Ctrl- or Lenti-Ym2-infected colonies treated with Dex. Statistical significance was determined by one-way ANOVA with Dunnett's multiple-comparisons test (B, D, F), and by two-way ANOVA with Sidak's multiple-comparisons test (H). B: F(2,7) = 11.29, p = 0.0064; D: F(2,9) = 7.732, p = 0.0111; F: F(2,8) = 5.916, p = 0.0265; H: F(1,21) = 20.74, p = 0.0002. Scale bars, 20 μm.
Discussion
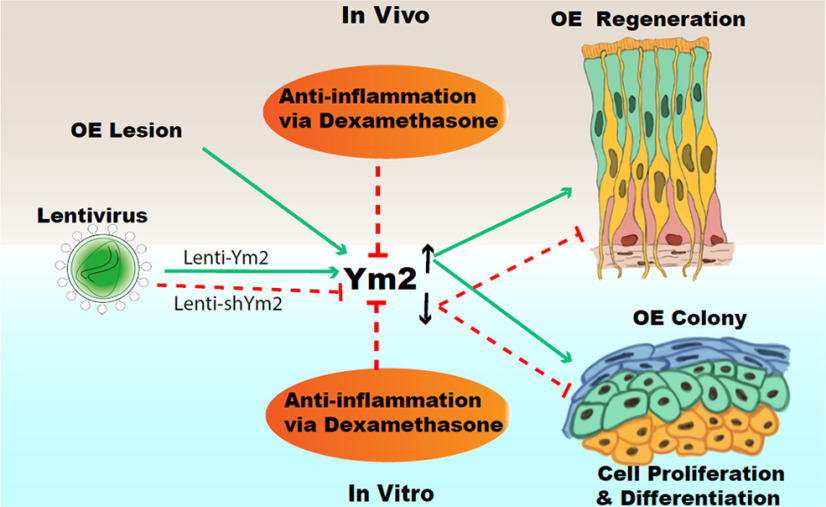
In this study, we discovered a novel role of Ym2 in regeneration of the injured OE and provided a critical link between Ym2 and inflammation in this process. As summarized in Figure 14, we found that (1) Ym2 expression in supporting cells is upregulated in the open-side OE of unilateral naris occlusion mice as well as in methimazole-lesioned OE; (2) Ym2 upregulation facilitates OE regeneration in vivo and cell proliferation/differentiation in OE colonies in vitro, whereas Ym2 downregulation has the opposite effects; and (3) anti-inflammatory treatment reduces Ym2 expression and delays OE regeneration in vivo and cell proliferation/differentiation in vitro, which is counteracted by Ym2 overexpression.
Figure 14.
Summary of the main findings in this study. Green lines denote “facilitate,” and red lines denote “impede.”
Basal cells play central roles in mediating OE regeneration (Schwob et al., 2017). Selective depletion of supporting cells leads to HBC activation (Lin et al., 2017), suggesting a potential role of supporting cells in HBC recruitment. However, it is not entirely clear how supporting cells contribute to this process. The accumulation of Ym1/2 protein in the apical part of the injured mouse OE was first reported in ZnSO4 irrigation and bulbectomy models (Giannetti et al., 2004). Five days after unilateral bulbectomy when basal cell proliferation is greatly increased, Chil4 (Chi3l4) mRNA is significantly higher on the lesion side, and remarkably, it is the mRNA with the greatest fold difference among all transcripts (Heron et al., 2013). Consistent with these studies, we showed increased Ym2 expression in the open-side OE of unilateral naris occlusion mice as well as in the methimazole-induced OE lesion model (Figs. 1, 2). In the naris occlusion model, the open side (the obligated breathing nostril) is subject to extra airflow, dryness, and environmental insults, and thus resembles a lesion model. Through genetic ablation and viral infection, we found that Ym2 regulates regeneration of OMP+ and PGP9.5+ sensory neurons; apical Sox2+ and Sus4+ supporting cells; and Sox2+, ICAM1+, and Krt14+ basal cells in the injured OE. Genetic or viral knockdown of Ym2 attenuates, while Ym2 overexpression accelerates recovery of the lesioned OE (Figs. 4–6), demonstrating that Ym2 serves as an important regulator in OE regeneration. Interestingly, Ym2 expression stays high 1–3 months postinjury when OE recovery is presumably complete (Fig. 2). We speculate that higher Ym2 expression may be necessary to maintain the turnover of the recovered OE and/or Ym2 expression increases with age, as reported previously (Giannetti et al., 2004).
Our ELISA data from the nasal lavage supports that Ym2 is a secreted protein (Fig. 2N), which is consistent with a previous report (Ward et al., 2001), and the presence of the secretory signal peptide in the Ym2 N terminus, as in Ym1 (Nio et al., 2004). Although we cannot completely rule out the possibility of contamination from nonsecreted Ym2 in the supporting cells during collection of the nasal lavage, the higher level of Ym2 in the lesioned OE compared with the control is consistent with the upregulation of Ym2 after injury. Although more studies are required to determine how Ym2 (presumably secreted from supporting cells) regulate OE regeneration, there are several possible scenarios, which are not mutually exclusive. First, Ym2 protein could act on supporting cells themselves in a cell-autonomous fashion to modulate their own functions, which in turn influence other cell types (e.g., basal cells and sensory neurons). Different cell types may mutually interact during OE regeneration. Cell–cell interactions between newly born neurons and progenitor cells control the ratio of supporting cells and neurons (Kam et al., 2016). Second, secreted Ym2 could act on other cell types such as basal cells in a cell-nonautonomous manner to regulate OE regeneration. Since Ym2 expression also affects the ICAM1 intensity and ICAM1+ cell percentage of HBCs in the OE (Figs. 4–6), Ym2+ supporting cells may regulate the basal cell activity through modifying ICAM1 expression and then affect cell turnover during OE recovery. The effectiveness of recombinant Ym2 in promoting cell proliferation and differentiation in vitro (Fig. 9) lends further support to this scenario. Third, Ym2 protein could exert its action via OE inflammatory cells (Fig. 11), which in turn modulate basal cell function (see below).
Lentiviral infection allows the manipulation of Ym2 expression in the OE (Figs. 5, 6). We applied Lenti-shYm2 infection 10 d before methimazole injection to allow sufficient time for lentiviral infection to regulate Ym2 expression in the OE. Upon methimazole injection, most OE cells are depleted within a couple of days. The effectiveness of this protocol suggests that manipulating Ym2 level before and during the process of OE ablation is sufficient to influence the function of the remaining HBCs and OE regeneration. Alternatively, viral infection may affect Ym2 level in the remaining basal cells (especially in overexpression experiments), which may lead to different Ym2 levels in regenerated supporting cells.
In genetic or viral downregulation of Ym2 expression experiments, the observed effects are likely because of the downregulation of Ym2 expression solely in supporting cells, since other cell types (e.g., basal cells) do not express detectable Ym2 in both the in vivo and in vitro systems (Figs. 1–3, 7). However, in Lenti-Ym2 viral overexpression experiments, infection can occur in multiple cell types (including supporting cells and basal cells; Figs. 5, 7), which may contribute to the observed effects of Ym2 overexpression.
Similar to Ym2, neuropeptide Y (NPY), a neuroproliferative factor, is synthesized in the postnatal OE by a subset of supporting cells (Hansel et al., 2001; Montani et al., 2006; Jia et al., 2013). Genetic knockout of NPY causes a decreased number of dividing olfactory neuronal precursor cells and results in significantly fewer OSNs, revealing a role of NPY in maintaining neuronal homeostasis in the OE (Hansel et al., 2001; Montani et al., 2006). Moreover, NPY is also reported to mediate injury-induced neuroregeneration in the OE (Jia and Hegg, 2012). Our study adds Ym2 to the list of critical factors synthesized by supporting cells that affect OE regeneration.
The specific mechanism regulating Ym2 expression in OE supporting cells requires further investigation. It has been reported that Ym2 expression depends on IL-4 and IL-13 signal transduction in remodeling of the airway wall in the allergic lung (Webb et al., 2001). Meanwhile, mouse CLPs including Ym2 can induce the accumulation of neutrophils through regulating IL-17 (Sutherland et al., 2014). It is likely that IL-4 and IL-13 are upstream, while IL-17 is downstream of Ym2 expression. Accordingly, it is plausible that the interaction with interleukin molecules may contribute to Ym2 function in regulating OE regeneration.
Inflammation exerts complex influences on regeneration and neural function of the OE. TNF-α-induced chronic inflammation leads to olfactory sensory neuron dysfunction revealed by electro-olfactogram (Sousa Garcia et al., 2017) and represses neurogenesis (Lane et al., 2010). However, TNF-α-mediated acute inflammation promotes stem cell proliferation and contributes to OE regeneration (Chen et al., 2017). These results suggest that TNF-α regulates OE stem cell behavior differentially in acute versus chronic inflammation. Murine Ym1/Ym2 is a critical regulator of inflammation in airway allergy (Zhao et al., 2005; Song et al., 2008) and pulmonary infection (Müller et al., 2007). Through genetic ablation and viral infection, we found that Ym2 regulates the regeneration of sensory neurons, supporting cells, and basal cells in the injured OE (Figs. 4–6) via its interaction with inflammatory responses. This is supported by the fact that anti-inflammatory treatment reduces Ym2 expression and delays OE regeneration, reversed by Ym2 overexpression, which increased inflammatory cells in the OE (Fig. 11). Similarly, Ym2 interacts with inflammatory responses and regulates cell proliferation and differentiation in OE colonies in vitro (Figs. 12, 13). Collectively, this is the first study to demonstrate the interaction of Ym2 and acute inflammatory responses in OE regeneration. Some steroids such as glucocorticoids are routinely used to treat chronic rhinosinusitis, preventing inflammation, relieving nasal obstruction, impeding OSN deaths, and facilitating the recovery of olfactory functions (Wolfensberger and Hummel, 2002; Kim et al., 2017). However, it is paradoxical that glucocorticoids cause impairment in OSN regeneration via downplaying protein synthesis (for review, see Chang and Glezer, 2018). Since Ym2 regulates OE recovery via interaction with inflammatory responses, it may act as an important mediator to balance the injury and regeneration induced by inflammation.
Footnotes
This work was supported by the National Natural Science Foundation of China (Grants 32070996 and 31771155 to Y.Y., 81970856 to H.Y., 31900714 to W.R.); the Shanghai Municipal Human Resources and Social Security Bureau, Shanghai Talent Development Fund (Grant 2018112 to Y.Y.); Eye, Ear, Nose and Throat Hospital, Fudan University, Excellent Doctors-Excellent Clinical Researchers Program (Grant SYB202002 to Y.Y.); the New Technologies of Endoscopic Surgery in Skull Base Tumor: CAMS Innovation Fund for Medical Sciences (Grant 2019-I2M-5-003 to H.Y.); and the National Institute of Deafness and Other Communication Disorders, National Institutes of Health (Grant DC-006213 to M.M.). We thank Dr. Shioko Kimura for providing the Ym1/Ym2 antibody, Dr. Qizhi Gong for the OMP antibody, and Dr. James Schwob for the SUS4 antibody.
The authors declare no competing financial interests.
References
- Abercrombie M (1946) Estimation of nuclear population from microtome sections. Anat Rec 94:239–247. 10.1002/ar.1090940210 [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K (2011) Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9:317–329. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneh-Barkay D, Zagadailov P, Zou H, Niyonkuru C, Figley M, Starkey A, Wang G, Bissel SJ, Wiley CA, Wagner AK (2010) YKL-40 expression in traumatic brain injury: an initial analysis. J Neurotrauma 27:1215–1223. 10.1089/neu.2010.1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman J, Raininko R, Blennow K, Zetterberg H, Axelsson M, Malmeström C (2016) YKL-40 is a CSF biomarker of intrathecal inflammation in secondary progressive multiple sclerosis. J Neuroimmunol 292:52–57. 10.1016/j.jneuroim.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Bussink AP, Speijer D, Aerts JM, Boot RG (2007) Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics 177:959–970. 10.1534/genetics.107.075846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Kumar RK, Zhou J, Foster PS, Webb DC (2009) Ym1/2 promotes Th2 cytokine expression by inhibiting 12/15(S)-lipoxygenase: identification of a novel pathway for regulating allergic inflammation. J Immunol 182:5393–5399. 10.4049/jimmunol.0803874 [DOI] [PubMed] [Google Scholar]
- Chang SY, Glezer I (2018) The balance between efficient anti-inflammatory treatment and neuronal regeneration in the olfactory epithelium. Neural Regen Res 13:1711–1714. 10.4103/1673-5374.238605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kohno K, Gong Q (2005) Conditional ablation of mature olfactory sensory neurons mediated by diphtheria toxin receptor. J Neurocytol 34:37–47. 10.1007/s11068-005-5046-8 [DOI] [PubMed] [Google Scholar]
- Chen M, Reed RR, Lane AP (2017) Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc Natl Acad Sci U S A 114:8089–8094. 10.1073/pnas.1620664114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova V, Pechnick RN, Wawrowsky K (2016) Chronic peripheral inflammation, hippocampal neurogenesis, and behavior. Brain Behav Immun 58:1–8. 10.1016/j.bbi.2016.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, Dziura JD, Reed J, Coyle AJ, Kiener P, Cullen M, Grandsaigne M, Dombret MC, Aubier M, Pretolani M, Elias JA (2007) A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med 357:2016–2027. 10.1056/NEJMoa073600 [DOI] [PubMed] [Google Scholar]
- Coppola DM (2012) Studies of olfactory system neural plasticity: the contribution of the unilateral naris occlusion technique. Neural Plast 2012:351752. 10.1155/2012/351752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola DM, Craven BA, Seeger J, Weiler E (2014) The effects of naris occlusion on mouse nasal turbinate development. J Exp Biol 217:2044–2052. 10.1242/jeb.092940 [DOI] [PubMed] [Google Scholar]
- Dai Q, Duan C, Ren W, Li F, Zheng Q, Wang L, Li W, Lu X, Ni W, Zhang Y, Chen Y, Wen T, Yu Y, Yu H (2018) Notch signaling regulates Lgr5(+) olfactory epithelium progenitor/stem cell turnover and mediates recovery of lesioned olfactory epithelium in mouse model. Stem Cells 36:1259–1272. 10.1002/stem.2837 [DOI] [PubMed] [Google Scholar]
- Falcone FH, Loke P, Zang X, MacDonald AS, Maizels RM, Allen JE (2001) A Brugia malayi homolog of macrophage migration inhibitory factor reveals an important link between macrophages and eosinophil recruitment during nematode infection. J Immunol 167:5348–5354. 10.4049/jimmunol.167.9.5348 [DOI] [PubMed] [Google Scholar]
- Fleming MS, Ramos D, Han SB, Zhao J, Son YJ, Luo W (2012) The majority of dorsal spinal cord gastrin releasing peptide is synthesized locally whereas neuromedin B is highly expressed in pain- and itch-sensing somatosensory neurons. Mol Pain 8:52. 10.1186/1744-8069-8-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkhouser JD, Aronson NN Jr (2007) Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 7:96. 10.1186/1471-2148-7-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi K, Suda T, Nakamura Y, Inui N, Hashimoto D, Miwa S, Hayakawa H, Kusagaya H, Nakano Y, Nakamura H, Chida K (2010) Increased expression of YKL-40, a chitinase-like protein, in serum and lung of patients with idiopathic pulmonary fibrosis. Respir Med 104:1204–1210. 10.1016/j.rmed.2010.02.026 [DOI] [PubMed] [Google Scholar]
- Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Fletcher RB (2017) Injury activates transient olfactory stem cell states with diverse lineage capacities. Cell Stem Cell 21:775–790.e9. 10.1016/j.stem.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannetti N, Moyse E, Ducray A, Bondier JR, Jourdan F, Propper A, Kastner A (2004) Accumulation of Ym1/2 protein in the mouse olfactory epithelium during regeneration and aging. Neuroscience 123:907–917. 10.1016/j.neuroscience.2003.09.024 [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Schwob JE (1996) Analysis of the globose basal cell compartment in rat olfactory epithelium using GBC-1, a new monoclonal antibody against globose basal cells. J Neurosci 16:4005–4016. 10.1523/JNEUROSCI.16-12-04005.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel DE, Eipper BA, Ronnett GV (2001) Neuropeptide Y functions as a neuroproliferative factor. Nature 410:940–944. 10.1038/35073601 [DOI] [PubMed] [Google Scholar]
- Heron PM, Stromberg AJ, Breheny P, McClintock TS (2013) Molecular events in the cell types of the olfactory epithelium during adult neurogenesis. Mol Brain 6:49. 10.1186/1756-6606-6-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai N, Zhou Z, Roop DR, Behringer RR (2008) Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells 26:1298–1306. 10.1634/stemcells.2007-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Hegg CC (2012) Neuropeptide Y and extracellular signal-regulated kinase mediate injury-induced neuroregeneration in mouse olfactory epithelium. Mol Cell Neurosci 49:158–170. 10.1016/j.mcn.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Hayoz S, Hutch CR, Iqbal TR, Pooley AE, Hegg CC (2013) An IP3R3- and NPY-expressing microvillous cell mediates tissue homeostasis and regeneration in the mouse olfactory epithelium. PLoS One 8:e58668. 10.1371/journal.pone.0058668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam JW, Dumontier E, Baim C, Brignall AC, Mendes da Silva D, Cowan M, Kennedy TE, Cloutier JF (2016) RGMB and neogenin control cell differentiation in the developing olfactory epithelium. Development 143:1534–1546. 10.1242/dev.118638 [DOI] [PubMed] [Google Scholar]
- Kim DK, Choi SA, Eun KM, Kim SK, Kim DW, Phi JH (2019) Tumour necrosis factor alpha and interleukin-5 inhibit olfactory regeneration via apoptosis of olfactory sphere cells in mice models of allergic rhinitis. Clin Exp Allergy 49:1139–1149. 10.1111/cea.13401 [DOI] [PubMed] [Google Scholar]
- Kim DH, Kim SW, Hwang SH, Kim BG, Kang JM, Cho JH, Park YJ, Kim SW (2017) Prognosis of olfactory dysfunction according to etiology and timing of treatment. Otolaryngol Head Neck Surg 156:371–377. 10.1177/0194599816679952 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Tamari K, Al Salihi MO, Nishida K, Takeuchi K (2018) Anti-high mobility group box 1 antibody suppresses local inflammatory reaction and facilitates olfactory nerve recovery following injury. J Neuroinflammation 15:124. 10.1186/s12974-018-1168-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai E, Mano Y, Yoshio S, Shoji H, Sugiyama M, Korenaga M, Ishida T, Arai T, Itokawa N, Atsukawa M, Hyogo H, Chayama K, Ohashi T, Ito K, Yoneda M, Kawaguchi T, Torimura T, Nozaki Y, Watanabe S, Mizokami M, et al. (2016) Serum YKL-40 as a marker of liver fibrosis in patients with non-alcoholic fatty liver disease. Sci Rep 6:35282. 10.1038/srep35282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurpińska A, Suraj J, Bonar E, Zakrzewska A, Stojak M, Sternak M, Jasztal A, Walczak M (2019) Proteomic characterization of early lung response to breast cancer metastasis in mice. Exp Mol Pathol 107:129–140. 10.1016/j.yexmp.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Kyritsis N, Kizil C, Zocher S, Kroehne V, Kaslin J, Freudenreich D, Iltzsche A, Brand M (2012) Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 338:1353–1356. 10.1126/science.1228773 [DOI] [PubMed] [Google Scholar]
- Kzhyshkowska J, Yin S, Liu T, Riabov V, Mitrofanova I (2016) Role of chitinase-like proteins in cancer. Biol Chem 397:231–247. 10.1515/hsz-2015-0269 [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA (2014) Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 9:2329–2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AP, Turner J, May L, Reed R (2010) A genetic model of chronic rhinosinusitis-associated olfactory inflammation reveals reversible functional impairment and dramatic neuroepithelial reorganization. J Neurosci 30:2324–2329. 10.1523/JNEUROSCI.4507-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA (2011) Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol 73:479–501. 10.1146/annurev-physiol-012110-142250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR (2007) Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 10:720–726. 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Li J, Ishii T, Feinstein P, Mombaerts P (2004) Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature 428:393–399. 10.1038/nature02433 [DOI] [PubMed] [Google Scholar]
- Lin B, Coleman JH, Peterson JN, Zunitch MJ, Jang W, Herrick DB, Schwob JE (2017) Injury induces endogenous reprogramming and dedifferentiation of neuronal progenitors to multipotency. Cell Stem Cell 21:761–774.e5. 10.1016/j.stem.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddens B, Ghesquière B, Vanholder R, Demon D, Vanmassenhove J, Gevaert K, Meyer E (2012) Chitinase-like proteins are candidate biomarkers for sepsis-induced acute kidney injury. Mol Cell Proteomics 11:M111.013094. 10.1074/mcp.M111.013094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje ML, Toda H, Palmer TD (2003) Inflammatory blockade restores adult hippocampal neurogenesis. Science 302:1760–1765. 10.1126/science.1088417 [DOI] [PubMed] [Google Scholar]
- Montani G, Tonelli S, Elsaesser R, Paysan J, Tirindelli R (2006) Neuropeptide Y in the olfactory microvillar cells. Eur J Neurosci 24:20–24. 10.1111/j.1460-9568.2006.04878.x [DOI] [PubMed] [Google Scholar]
- Müller U, Stenzel W, Köhler G, Werner C, Polte T, Hansen G, Schütze N, Straubinger RK, Blessing M, McKenzie AN, Brombacher F, Alber G (2007) IL-13 induces disease-promoting type 2 cytokines, alternatively activated macrophages and allergic inflammation during pulmonary infection of mice with Cryptococcus neoformans. J Immunol 179:5367–5377. 10.4049/jimmunol.179.8.5367 [DOI] [PubMed] [Google Scholar]
- Nair MG, Gallagher IJ, Taylor MD, Loke P, Coulson PS, Wilson RA, Maizels RM, Allen JE (2005) Chitinase and Fizz family members are a generalized feature of nematode infection with selective upregulation of Ym1 and Fizz1 by antigen-presenting cells. Infect Immun 73:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nio J, Fujimoto W, Konno A, Kon Y, Owhashi M, Iwanaga T (2004) Cellular expression of murine Ym1 and Ym2, chitinase family proteins, as revealed by in situ hybridization and immunohistochemistry. Histochem Cell Biol 121:473–482. 10.1007/s00418-004-0654-4 [DOI] [PubMed] [Google Scholar]
- Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, Lester LA, Gern JE, Lemanske RF Jr, Nicolae DL, Elias JA, Chupp GL (2008) Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 358:1682–1691. 10.1056/NEJMoa0708801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Kida Y, Sakaguchi M, Sugahara Y, Oyama F (2014) Establishment of a quantitative PCR system for discriminating chitinase-like proteins: catalytically inactive breast regression protein-39 and Ym1 are constitutive genes in mouse lung. BMC Mol Biol 15:23. 10.1186/1471-2199-15-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi AT, Ram S, Kuo A, Gilbert JL, Yan W, Pellegrini M, Nickerson DW, Chatila TA, Gomperts BN (2012) Identification of an interleukin 13-induced epigenetic signature in allergic airway inflammation. Am J Transl Res 4:219–228. [PMC free article] [PubMed] [Google Scholar]
- Pizano-Martínez O, Yañez-Sánchez I, Alatorre-Carranza P, Miranda-Diaz A, Ortiz-Lazareno PC, García-Iglesias T, Daneri-Navarro A, Vázquez-Del Mercado M, Fafutis-Morris M, Delgado-Rizo V (2011) YKL-40 expression in CD14(+) liver cells in acute and chronic injury. World J Gastroenterol 17:3830–3835. 10.3748/wjg.v17.i33.3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozharskaya T, Liang J, Lane AP (2013) Regulation of inflammation-associated olfactory neuronal death and regeneration by the type II tumor necrosis factor receptor. Int Forum Allergy Rhinol 3:740–747. 10.1002/alr.21187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthumana J, Hall IE, Reese PP, Schröppel B, Weng FL, Thiessen-Philbrook H, Doshi MD, Rao V, Lee CG, Elias JA, Cantley LG, Parikh CR (2017) YKL-40 associates with renal recovery in deceased donor kidney transplantation. J Am Soc Nephrol 28:661–670. 10.1681/ASN.2016010091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi AM, Hannigan A, Campbell D, Nixon C, Wilson JB (2011) Chitinase-like proteins are autoantigens in a model of inflammation-promoted incipient neoplasia. Genes Cancer 2:74–87. 10.1177/1947601911402681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse RL, Boudreaux MJ, Penn AL (2007) In utero environmental tobacco smoke exposure alters gene expression in lungs of adult BALB/c mice. Environ Health Perspect 115:1757–1766. 10.1289/ehp.10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyar A, Classe M, Gorski R, Bock MD, Le-Guern J, Roche S, Fourgous V, Remaury A, Paul P, Ponsolles C, Françon D, Rocheteau-Beaujouan L, Clément M, Haddad EB, Guillemot JC, Didier M, Biton B, Orsini C, Mikol V, Leonetti M (2019) Type 2/Th2-driven inflammation impairs olfactory sensory neurogenesis in mouse chronic rhinosinusitis model. Allergy 74:549–559. 10.1111/all.13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI, Jang W, Schwob JE (2015) Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc Natl Acad Sci U S A 112:E5068–E5077. 10.1073/pnas.1512272112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE (2002) Neural regeneration and the peripheral olfactory system. Anat Rec 269:33–49. 10.1002/ar.10047 [DOI] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Hewitt Coleman J (2017) Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J Comp Neurol 525:1034–1054. 10.1002/cne.24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuhui L, Mok YK, Wong WS (2009) Role of mammalian chitinases in asthma. Int Arch Allergy Immunol 149:369–377. 10.1159/000205583 [DOI] [PubMed] [Google Scholar]
- Song HM, Jang AS, Ahn MH, Takizawa H, Lee SH, Kwon JH, Lee YM, Rhim TY, Park CS (2008) Ym1 and Ym2 expression in a mouse model exposed to diesel exhaust particles. Environ Toxicol 23:110–116. 10.1002/tox.20319 [DOI] [PubMed] [Google Scholar]
- Sousa Garcia D, Chen M, Smith AK, Lazarini PR, Lane AP (2017) Role of the type I tumor necrosis factor receptor in inflammation-associated olfactory dysfunction. Int Forum Allergy Rhinol 7:160–168. 10.1002/alr.21855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland TE, Maizels RM, Allen JE (2009) Chitinases and chitinase-like proteins: potential therapeutic targets for the treatment of T-helper type 2 allergies. Clin Exp Allergy 39:943–955. 10.1111/j.1365-2222.2009.03243.x [DOI] [PubMed] [Google Scholar]
- Sutherland TE, Logan N, Rückerl D, Humbles AA, Allan SM, Papayannopoulos V, Stockinger B, Maizels RM, Allen JE (2014) Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nat Immunol 15:1116–1125. 10.1038/ni.3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Yoon M, Anver MR, Haines DC, Kudo G, Gonzalez FJ, Kimura S (2001) Hyalinosis and Ym1/Ym2 gene expression in the stomach and respiratory tract of 129S4/SvJae and wild-type and CYP1A2-null B6, 129 mice. Am J Pathol 158:323–332. 10.1016/S0002-9440(10)63972-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DC, McKenzie AN, Foster PS (2001) Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J Biol Chem 276:41969–41976. 10.1074/jbc.M106223200 [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin A-K, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC, Tannahill D, Logan DW, Macarthur DG, Flint J, et al. (2013) Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154:452–464. 10.1016/j.cell.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfensberger M, Hummel T (2002) Anti-inflammatory and surgical therapy of olfactory disorders related to sino-nasal disease. Chem Senses 27:617–622. 10.1093/chemse/27.7.617 [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhu Y, Wang S, Zachory Wei Z, Jiang MQ, Zhang Y, Pan Y, Tao S, Li J, Wei L (2018) Temporal gene expression profiles after focal cerebral ischemia in mice. Aging Dis 9:249–261. 10.14336/AD.2017.0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhu H, Wong CH, Leung KY, Wong WS (2005) Increased lungkine and chitinase levels in allergic airway inflammation: a proteomics approach. Proteomics 5:2799–2807. 10.1002/pmic.200401169 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Peng H, Sun H, Peng X, Tang C, Gan Y, Chen X, Mathur A, Hu B, Slade MD, Montgomery RR, Shaw AC, Homer RJ, White ES, Lee CM, Moore MW, Gulati M, Lee CG, Elias JA, Herzog EL (2014) Chitinase 3-like 1 suppresses injury and promotes fibroproliferative responses in mammalian lung fibrosis. Sci Transl Med 6:240–276. [DOI] [PMC free article] [PubMed] [Google Scholar]