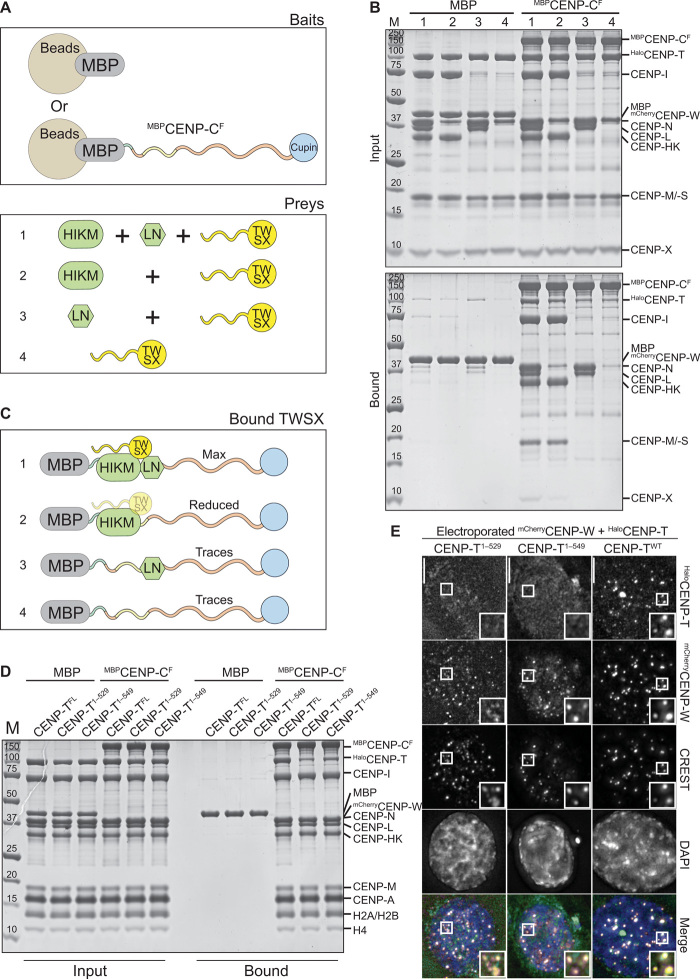
Biochemical reconstitution of human kinetochore delivers complexes with all subunits in the expected stoichiometry.
Abstract
Centromeres are epigenetically determined chromosomal loci that seed kinetochore assembly to promote chromosome segregation during cell division. CENP-A, a centromere-specific histone H3 variant, establishes the foundations for centromere epigenetic memory and kinetochore assembly. It recruits the constitutive centromere-associated network (CCAN), which in turn assembles the microtubule-binding interface. How the specific organization of centromeric chromatin relates to kinetochore assembly and to centromere identity through cell division remains conjectural. Here, we break new ground by reconstituting a functional full-length version of CENP-C, the largest human CCAN subunit and a blueprint of kinetochore assembly. We show that full-length CENP-C, a dimer, binds stably to two nucleosomes and permits further assembly of all other kinetochore subunits in vitro with relative ratios closely matching those of endogenous human kinetochores. Our results imply that human kinetochores emerge from clustering multiple copies of a fundamental module and may have important implications for transgenerational inheritance of centromeric chromatin.
INTRODUCTION
In addition to the hereditary information enshrined in the genetic code, chromosomes carry a wealth of epigenetic information that controls their structure and accessibility in time and space. Epigenetic information is usually implemented through covalent modifications of the DNA itself, or through modifications of the histones, proteins that pack DNA into nucleosomes, the fundamental organizational unit of eukaryotic genomes. In addition to these well-studied mechanisms, the most notable and unconventional example of epigenetic determination drives the establishment and maintenance of centromeres (1, 2). Centromeres are unique chromatin loci of the eukaryotic chromosome. They seed the assembly of kinetochores, large protein complexes that establish a point of contact between chromosomes and spindle microtubules during mitosis and meiosis (1, 2). The realization more than 20 years ago that centromere identity, with few exceptions, is established in a DNA sequence–independent manner was a crucial conceptual leap in centromere studies, as it redirected the spotlight onto the molecular basis of centromere persistence through cell division (1, 2).
The most characteristic and almost ubiquitous trait of centromeres is the enrichment of the histone H3-like centromeric protein A (CENP-A) (1, 2). Similar to H3, CENP-A forms a tight tetramer with H4, which additionally binds two H2A:H2B dimers to form an octameric nucleosome core particle (NCP) (3). The CENP-A nucleosome is surrounded by a group of 16 proteins collectively named the constitutive centromere-associated network (CCAN) and organized in different stable subcomplexes (4–8). Two CCAN subunits, CENP-N and CENP-C, interact directly with CENP-A (9–11). In addition, CENP-C, predicted to be predominantly intrinsically disordered, interacts with several other CCAN subunits to stabilize the overall globular assembly of CCAN (7, 8, 12–17). CENP-C and another CCAN subunit, CENP-T, also provide a platform for the assembly of the kinetochore’s outer layer (18, 19). The outer kinetochore comprises three further subcomplexes, known as the KNL1 (kinetochore null protein 1), MIS12 (minichromosome instability 12), and NDC80 (Nuclear division cycle protein 80) complexes, and collectively referred to as the KMN network (20). Besides important regulatory functions, the KMN network, through its NDC80 complex (NDC80C), provides a site for binding spindle microtubules, an interaction that promotes the alignment and segregation of chromosomes to the daughter cells (1, 2).
Kinetochores across eukaryotes display large differences in complexity. The Saccharomyces cerevisiae kinetochore assembles around ~125 base pairs (bp) of a defined centromeric DNA sequence (and therefore not epigenetically) that wraps around a single CENP-ACse4 nucleosome and binds a single microtubule, for which reasons it is identified as a “point” kinetochore. Conversely, kinetochores in most eukaryotes are identified as “regional” because they extend over much larger centromeric DNA segments (up to million base pairs in humans), expose multiple CENP-A nucleosomes, and bind multiple microtubules (2). This extreme range of complexity raises the question whether regional kinetochores are built by patching together multiple copies of a “unit module” related to the point kinetochore of S. cerevisiae. This idea remains untested, but it is plausible, and several observations support it. First, subunit composition and physical interactions appear to be largely conserved in point and regional kinetochores, as demonstrated by comprehensive biochemical reconstitutions and structural analyses (7, 8, 12, 13, 15, 21, 22). Second, the relative ratios of kinetochore subunits appear to be conserved in point and regional kinetochores, suggesting that regional kinetochores emerge from the multiplication of a fundamental point module (23–25). Last, the same relative subunit ratios observed at human kinetochores are also observed in partial reconstitutions of single, discrete particles of human kinetochores (15, 21, 23), once again suggesting that the larger assembly is created by convolution of individual modules.
From a reductionist perspective, this hypothesis raises the crucial question whether an individual module recapitulates some or even all fundamental properties of the regional structure, including promotion of biorientation and epigenetic specification and inheritance. Answering this question may have important technological implications, particularly toward the development of artificial chromosomes, but is technically and conceptually challenging. Despite very considerable progress in our understanding of the structural organization of the core protein complex surrounding the CENP-A nucleosome (7, 8, 21), there is persistent uncertainty on the organization of the continuous segment of centromeric chromatin underlying regional kinetochores. For example, even if CENP-A is greatly enriched within human centromeric chromatin in comparison to chromosome arms, it is surrounded and vastly outnumbered by histone H3 (26). Various models for the organization of regional centromeric chromatin have been discussed (1, 2), but conclusive evidence to support one or the other is missing. Furthermore, despite progress in the isolation of point kinetochores (27, 28), there are no robust procedures for the isolation of individual modules from natural regional kinetochores. To address these limitations, we applied bottom-up approaches based on the biochemical reconstitution of human kinetochore substructures of increasing complexity, followed by iterative assessments and validation of their functional and structural properties in comparison to those of real kinetochores, for instance, with regard to the role of KMN multivalency in kinetochore-microtubule attachment (29, 30).
Here, we engineered a full-length, dimeric form of human CENP-C. This allowed us to dissect how oligomerization and multivalency of CENP-C reflects on kinetochore assembly and the underlying chromatin. We demonstrate that full-length CENP-C can bind concomitantly to two individual nucleosomes or to two adjacent nucleosomes in a dinucleosome. We show that this organization supports the robust recruitment of all other kinetochore subunits with predicted stoichiometries, and we present a thorough dissection of the interactions that make this possible. Our studies represent an important step toward the reconstitution of a minimal regional kinetochore module.
RESULTS
Reconstitution of full-length CENP-C (CENP-CF) by protein fusion
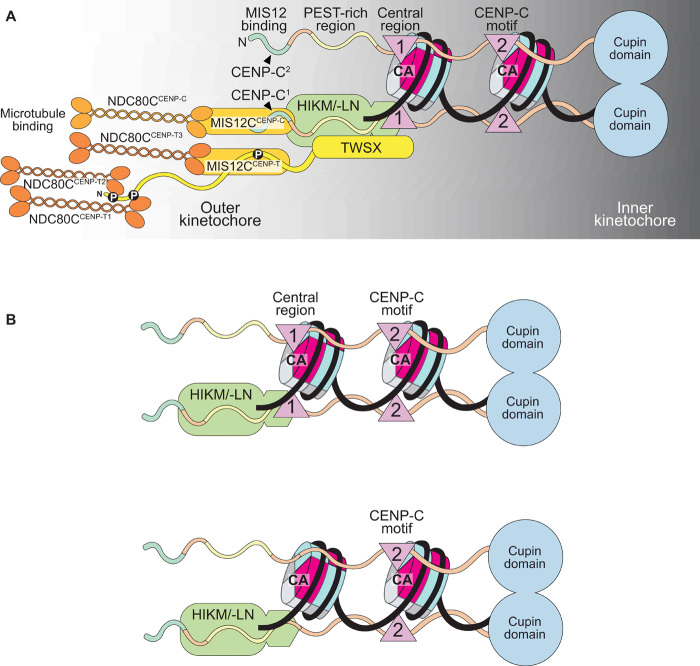
Human CENP-C is a 943-residue protein (Fig. 1A). Its N-terminal region interacts with the MIS12 complex (MIS12C) (18, 19, 31). The following PEST (proline–glutamic acid–serine-threonine)–rich region binds the CENP-HIKM and CENP-NL CCAN subcomplexes (12–14, 16). The central region encompasses a CENP-A–selective binding motif (indicated as “1” in Fig. 1A) that interacts with an exposed acidic patch on histones H2A and H2B as well as with the C-terminal tail of CENP-A (9, 11). It is required for robust kinetochore localization of CENP-C (9, 32, 33). The constellation of the N-terminal region, PEST, and central region spans the entire depth of the kinetochore from its outer domain until the centromere, and we have speculated that it provides spatial clues (a blueprint) for kinetochore assembly (12). Implicit in this analogy is that CENP-C promotes a multitude of interactions with kinetochore subunits that enhance the overall stability of the inner kinetochore complex. A fragment of CENP-C encompassing the MIS12C binding site, PEST, and central region (CENP-C1–544) was sufficient to reconstitute a recombinant 26-subunit kinetochore complex with a CENP-A NCP (CENP-ANCP) and most CCAN and KMN subunits (12, 15, 21).
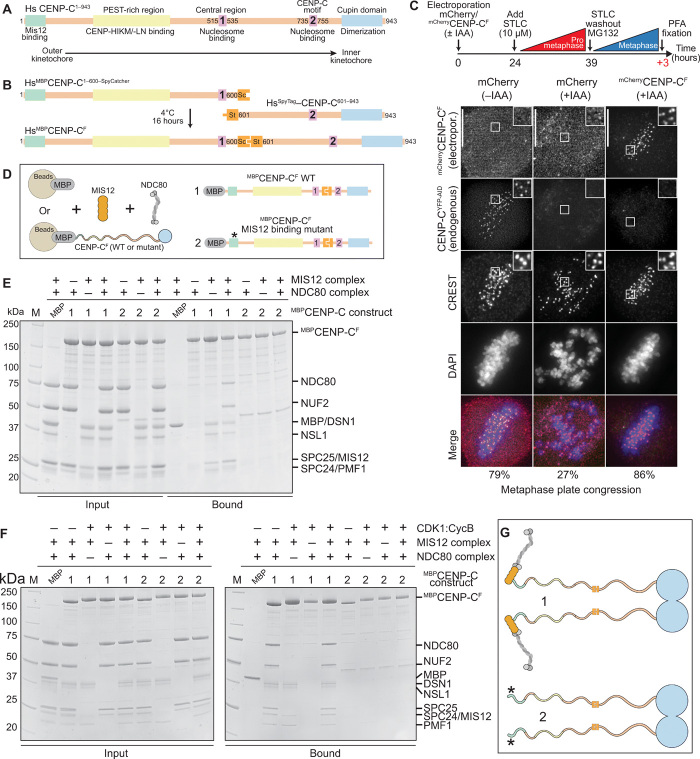
Fig. 1. Construction and validation of a CENP-C fusion protein.
(A) Organization of human CENP-C. The N-terminal MIS12 binding region is highlighted in green, and the CENP-HIKM/-LN binding region is highlighted in yellow. The central and C-terminal CENP-C motifs are highlighted in magenta. The C-terminal Cupin-like dimerization domain is highlighted in blue. The arrangement of the binding sites recapitulates the outer-to-inner kinetochore axis. (B) Strategy to purify full-length CENP-C using the SpyCatcher-SpyTag system. The two individual CENP-C fragments, CENP-C1–600–SpyCatcher and SpyTag–CENP-C601–943, were incubated together to form the full-length CENP-C ligation product. Sc, SpyCatcher; St, SpyTag. (C) Representative images showing fluorescence of electroporated mCherry or mCherryCENP-CF and endogenous CENP-CYFP-AID. Centromeres were visualized by CREST sera, and DNA was stained by 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm. Experimental regime: 24 hours after electroporation, cells were treated with 10 μM STLC for 15 hours. STLC (S-trityl-L-cysteine) was washed out three times, and 10 μM MG132 was added for 3 hours before paraformaldehyde (PFA) fixation. (D) Schematic of the performed amylose-resin pull-down assays. Maltose-binding protein (MBP) and two MBPCENP-CF variants, wild-type (WT) (1) and MIS12 binding mutant (2), were immobilized on amylose resin as bait. MIS12C and NDC80C were added as preys. (E and F) Result of the amylose-resin pull-down experiment. The MBPCENP-CF variant used as bait is indicated above each lane. MIS12C and NDC80C were added as indicated above each lane. MBPCENP-CF was additionally phosphorylated by CDK1:Cyclin B as indicated above each lane in (F). M, protein marker. (G) Graphical summary of the results shown in (E) and (F).
The functional properties of the C-terminal segment, CENP-C545–943, remain more enigmatic. The C-terminal Cupin domain promotes dimerization of CENP-C (34, 35). The other functionally relevant segment is the CENP-C motif (indicated as “2” in Fig. 1A and also known as the “conserved motif”), whose sequence is strongly related to that of the central region (fig. S1A). The CENP-C motif also binds CENP-ANCPs, but less robustly than the central region and less selectively, as it binds CENP-ANCPs and H3NCPs with similar binding affinities (11, 17, 36). The significance of the duplication of nucleosome-binding motifs in HsCENP-C (Homo sapiens Centromere Protein C) remains unclear but suggests that a CENP-C dimer may interact with more than one nucleosome.
To dissect this aspect of CENP-C organization and recapitulate in vitro expected increases in complex stability arising from dimerization, full-length CENP-C would be invaluable, but obtaining it proved difficult due to insolubility upon recombinant expression in different hosts. To overcome this technical hurdle, we sought to obtain full-length CENP-C from fusing two complementary segments of CENP-C with the SpyTag/SpyCatcher system, which allows the covalent fusion of two polypeptide chains through formation of an isopeptide bond (Fig. 1B) (37). We tested the efficiency of the fusion reaction with two complementary constructs collectively encompassing the entire CENP-C sequence, CENP-C1–600 and CENP-C601–943. These constructs, respectively, include the central region and the CENP-C motif and connect through a poorly conserved fragment, thus reducing the likelihood of functional inactivation. In isolation, CENP-C1–600 and CENP-C601–943 bound with CENP-ANCPs in size exclusion chromatography (SEC) experiments (fig. S1, B and C). We purified MBPCENP-C1–600–SpyCatcher (where MBP stands for maltose-binding protein, an affinity tag; fig. S2A) and SpyTag–CENP-C601–943, mixed them, and incubated them together for 16 hours to obtain the ligation product MBPCENP-C1–600-Spy-601–943 (MBPCENP-CF, where “F” indicates “fusion”; fig. S2B). The fusion product was separated from the excess of nonligated fragments using SEC, obtaining a product of sufficient purity for further biochemical analyses (fig. S2C).
We performed SEC experiments to assess whether MBPCENP-CF retains the ability to interact with CENP-ANCPs. MBPCENP-CF and CENP-ANCPs interacted robustly and eluted in an apparently stoichiometric complex from the SEC column (when mixed at a 1:2 molar ratio of MBPCENP-CF dimer and CENP-ANCPs, the significance of which will be discussed more thoroughly below) (fig. S2D, yellow trace). MBPCENP-CF bound only weakly to H3NCPs, as revealed by an only modest shift in the elution volume of H3NCPs, indicative of a low-affinity, rapidly dissociating interaction (fig. S2E, black trace).
CENP-CF is functional
As a stringent functional test, we asked whether CENP-CF localized to kinetochores and whether it complemented the loss of endogenous CENP-C. For this, we used a previously reported colorectal adenocarcinoma DLD-1 cell line in which both endogenous CENP-C alleles were C-terminally tagged with an auxin-inducible degron (AID) and enhanced yellow fluorescent protein (EYFP) (32, 38, 39). Upon addition of the auxin derivative indole acetic acid (IAA), endogenous CENP-C was efficiently depleted within 30 to 60 min (fig. S3, A and B), as described (32, 38).
Using the same fusion strategy that allowed us to obtain MBPCENP-CF, we generated mCherryCENP-CF and demonstrated that it also interacts with CENP-ANCPs and that it localizes to kinetochores of HeLa cells (figs. S2, F and G, and S3C). Next, we asked whether mCherryCENP-CF localized at kinetochores of DLD-1 cells depleted of the endogenous CENP-CAID-EYFP. We electroporated mCherryCENP-CF into DLD-1 cells undergoing a 12-hour IAA treatment to achieve complete removal of the endogenous CENP-CAID-EYFP. mCherryCENP-CF, but not an mCherry control, was readily identified at kinetochores, as indicated by overlapping mCherry and CREST (calcinosis, Raynaud phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia) (a centromere marker) signals, despite undetectable endogenous CENP-CAID-EYFP (fig. S3D). Thus, centromere localization of mCherryCENP-CF did not require endogenous CENP-C. After incorporation into chromatin, CENP-A remains stably associated with centromeres even after removal of CENP-C for at least several hours (9, 17, 40), suggesting that CENP-A directs re-recruitment of mCherryCENP-CF after electroporation in CENP-C–depleted cells.
Last, we assessed whether mCherryCENP-CF complemented the deleterious consequences on chromosome alignment caused by CENP-CAID-EYFP depletion. Seventy-nine percent of untreated cells reached metaphase alignment, against 20% in cells treated with IAA to deplete CENP-CAID-EYFP (Fig. 1C). Electroporated mCherryCENP-CF entirely rescued the effects of CENP-CAID-EYFP depletion, with >80% of mCherryCENP-CF–positive cells displaying metaphase alignment. Collectively, these results provide strong evidence that mCherryCENP-CF is functional.
CENP-CF binds the outer kinetochore
Mif2p, the S. cerevisiae ortholog of CENP-C, adopts an autoinhibited conformation that suppresses the ability of its N-terminal region to interact with the Mtw1CMIS12C complex before binding to Cse4CENP-A nucleosomes (41). To assess whether this was also true of human CENP-CF, we immobilized MBPCENP-CF and incubated it with MIS12C, NDC80C, or their combination. MIS12C bound in apparently stoichiometric amounts to CENP-C and attracted NDC80C when the complexes were combined (Fig. 1, D and E), arguing that the MIS12C binding motif of MBPCENP-CF is fully exposed. As explained below, the interaction of CENP-T with the MIS12C and NDC80C is regulated by cyclin-dependent kinase 1 (CDK1) phosphorylation. To assess whether phosphorylation created additional binding sites for the outer kinetochore also on MBPCENP-CF, we phosphorylated it with CDK1:Cyclin B. This caused a prominent electrophoretic migration shift of MBPCENP-CF, indicative of successful phosphorylation, but did not modify the stoichiometry of outer kinetochore subunits (Fig. 1F). CENP-C binding of outer kinetochore subunits was limited to the previously identified N-terminal MIS12C binding motif (19), because it was ablated when the motif was mutated (construct 2; Fig. 1, F and G). Thus, collectively, CENP-CF is not autoinhibited and is a target of CDK1 phosphorylation, but the latter does not generate additional binding sites for outer kinetochore subunits.
Role of CENP-C dimerization
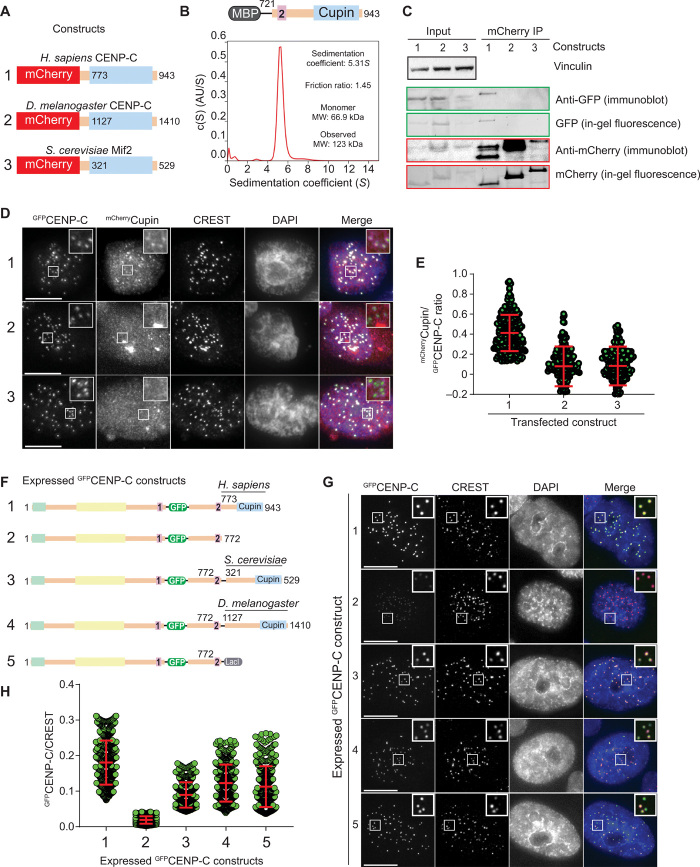
The C-terminal Cupin domains of the Drosophila melanogaster and S. cerevisiae CENP-Cs (DmCENP-C1127–1410 and Mif2321–529, respectively, depicted in Fig. 2A) form tight dimers (34, 35). Sedimentation velocity analytical ultracentrifugation (AUC) and SEC demonstrated that the human CENP-C Cupin domain also forms monodisperse, tight dimers (Fig. 2B and fig. S4A) [note that MBP is a monomer, as shown previously (42)]. When expressed in interphase HeLa cells depleted of endogenous CENP-C (fig. S4B), GFPCENP-C721–943 or GFPCENP-C760–943 failed to localize to kinetochores, whereas weak localization was observed in the presence of endogenous CENP-C (fig. S4, C to F), likely because of dimerization of the Cupin domain. When transfected in human cells together with full-length GFPCENP-C, only HsmCherryCENP-C773–943, but not DmmCherryCENP-C1127–1410 nor ScmCherryMif2p321–529, formed dimers with HsGFPCENP-C (Fig. 2C) and localized, albeit weakly, to kinetochores (Fig. 2, D and E). Thus, HsGFPCENP-C does not appreciably bind the fly or yeast CENP-C Cupin domains, revealing a strong preference for intraspecies homodimerization of the Cupin domain. When expressed ectopically in HeLa cells in the presence of endogenous CENP-C, a construct lacking the Cupin domain, HsCENP-C1–772, localized very weakly to kinetochores (Fig. 2, F to H, and fig. S4G). Conversely, chimeric constructs of HsCENP-C1–772 and the fly or yeast Cupin domains, or even with the LacI dimerization domain, restored very robust kinetochore localization. We conclude that dimerization through the Cupin domain stabilizes kinetochore CENP-C, likely through enforcement of multivalent interactions with other CCAN subunits.
Fig. 2. CENP-C dimerization promotes kinetochore localization efficiency.
(A) Schematic showing the three mCherry-tagged Cupin constructs from Homo sapiens, S. cerevisiae, and D. melanogaster that were coexpressed with human GFPCENP-C in HeLa cells. (B) Sedimentation coefficient distribution obtained from the sedimentation velocity AUC experiment using MBPCENP-C721–943. AU, arbitrary unit. The observed molecular weight (MW) of 123 kDa indicates that the sample forms a dimer. (C) Western blot showing the results of coimmunoprecipitation (IP) experiments using RFP-Trap beads. (D) Representative images showing the fluorescence of GFPCENP-C and three different mCherryCupin fragments as shown in (A). Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm. (E) Quantification of the fluorescence intensities showing the mCherry/green fluorescent protein (GFP) ratio. Centromeres were detected in the CREST channel using a script for semiautomated quantification. Shown are means and SD from three independent experiments. (F) Schematic showing the different GFPCENP-C variants that were expressed in HeLa cells. (G) Representative images showing the fluorescence of the different GFPCENP-C variants as shown in (F). Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm. (H) Quantification of the GFP fluorescence intensities. Shown are means and SD from three independent experiments.
CENP-CF binds two CENP-A nucleosomes
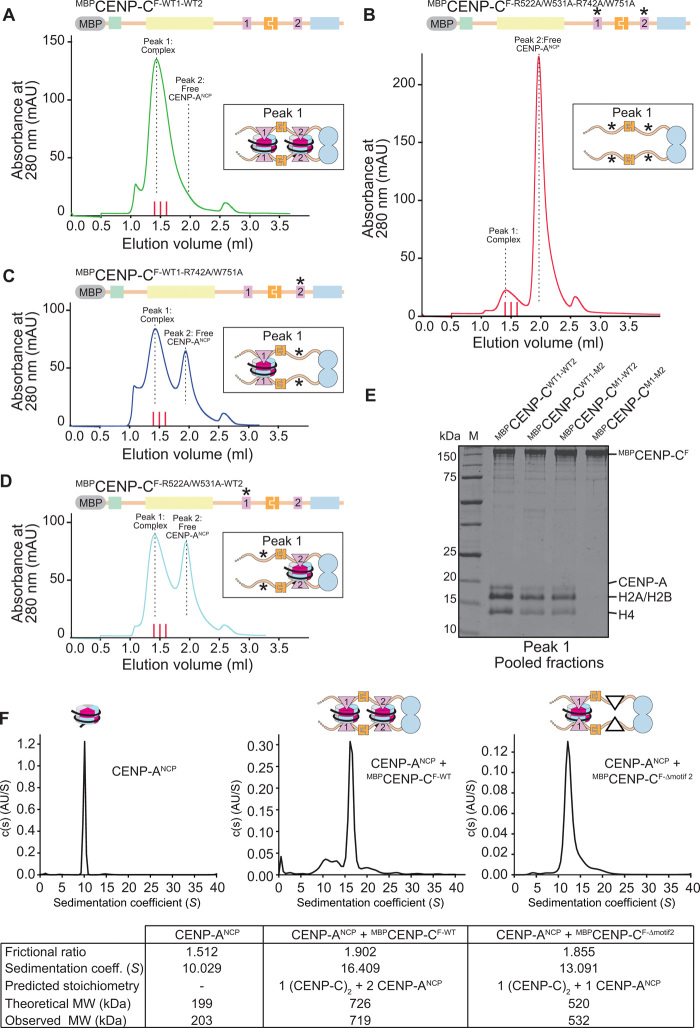
Because CENP-CF encompasses two nucleosome-binding motifs, its dimerization through the Cupin domain generates a dimer with four nucleosome-binding motifs. Because of its pseudo-twofold symmetric organization, an NCP may contain two binding sites for the CENP-A binding motifs of CENP-C (the central motif and the CENP-C motif), so that a CENP-C homodimer might be expected to interact with at least two NCPs, or more if further oligomerization came into play. To test this, we performed SEC experiments to assess how CENP-ANCPs interact with CENP-CF. As controls, we also included three CENP-CF variants in which the central region, the CENP-C motif, or both had been mutated to impair CENP-ANCP binding (11). Point mutations directed against conserved residues in the central region or in the CENP-C conserved motif (CENP-C1–600–R522A/W531A or CENP-C601–943–R742A/W751A, respectively) abolished CENP-A binding (fig. S1, B and C), indicating that each set of mutations is highly penetrant in isolation.
CENP-ANCPs combined with MBPCENP-CF-WT1-WT2 dimer at 2:1 molar ratio (5 μM CENP-ANCP and 2.5 μM CENP-C dimer) eluted from the SEC column almost entirely in one peak, with no residual free CENP-ANCPs (Fig. 3, A and E, and fig. S5A), implying that the two central regions and the two conserved motifs in the dimeric CENP-CF provide binding sites for at least two CENP-ANCPs. Conversely, CENP-C containing inactivating mutation both in the central region and in the CENP-C motif did not interact with CENP-A at all (Fig. 3, B and E, and fig. S5B). CENP-CF-R522A/W531A-WT2 and CENP-CF-WT1-R742A/W751A bound only one equivalent of CENP-ANCP, rather than two as CENP-CF (Fig. 3, C to E, and fig. S5, C and D; note that CENP-CF-R522A/W531A-WT2 and CENP-CF-WT1-R742A/W751A are, respectively, indicated as CENP-CM1-WT2 and CENP-CWT1-M2 in Fig. 3E). These stoichiometries were inferred both from the absorbance intensities of the peaks eluting from the SEC column and from the intensity of histone bands in Coomassie-stained SDS–polyacrylamide gel electrophoresis (PAGE) of the CENP-CF–bound fractions (Fig. 3E).
Fig. 3. CENP-C binds two CENP-A nucleosomes.
(A to D) Analytical SEC experiments were performed to demonstrate the interactions between different CENP-C variants and CENP-ANCPs. Red lines between 1.4 and 1.6 ml of elution volume indicate fractions collected for SDS-PAGE analysis shown in (E). The SEC chromatograms and SDS-PAGEs of all individual components are shown in fig. S5. (E) SDS-PAGE result showing the pooled fractions of the peak 1 complexes shown in the SEC chromatograms in (A) to (D). (F) Sedimentation coefficient distributions obtained from the sedimentation velocity AUC experiments using CENP-ANCP (left), CENP-ANCP + MBPCENP-CF-WT (middle), and CENP-ANCP + MBPCENP-CF-Δmotif2 (right). The table summarizes the results of the three experiments.
In an orthogonal approach, we used sedimentation velocity AUC to assess the molecular mass of the CENP-ANCP:CENP-CF dimer complex obtained at the 2:1 molar ratio. These experiments revealed an excellent agreement between the expected and predicted molecular mass for a complex containing a CENP-CF dimer and two CENP-ANCPs, with apparently little additional contamination from complexes of different stoichiometries (Fig. 3F and fig. S6, A to D). Furthermore, deletion of the CENP-C motif resulted in a complex containing a single CENP-ANCP, again with an excellent agreement between the theoretical and expected molecular masses. Collectively, these experiments indicate that a CENP-CF dimer has an intrinsic potential to bind two CENP-ANCPs, without forming substantial amounts of additional higher molecular weight species that might potentially arise when combining multivalent binding partners such as CENP-C and NCPs.
CENP-C interacts with CENP-A also through the CENP-HIKMLN complex
CENP-N interacts directly with CENP-ANCPs by recognizing the CENP-A L1 loop (14, 40, 43, 44). CENP-C, on the other hand, interacts with the acidic patch of H2A-H2B and with the CENP-A C-terminal tail (8, 9, 11, 15, 36, 40, 44, 45). These features may allow CENP-C and CENP-N to bind concomitantly to their cognate binding sites on the same CENP-A nucleosome, although the interaction may be further regulated by phosphorylation (44, 45). Furthermore, an alternative binding model where CENP-N occupies a noncanonical position has also been recently proposed (8). Because CENP-NL (and CENP-HIKM) interact tightly with CENP-C also through the PEST region (Fig. 1A) (12, 14), CENP-C may be expected to enable an additional contact of CCAN with the CENP-A nucleosome through CENP-N.
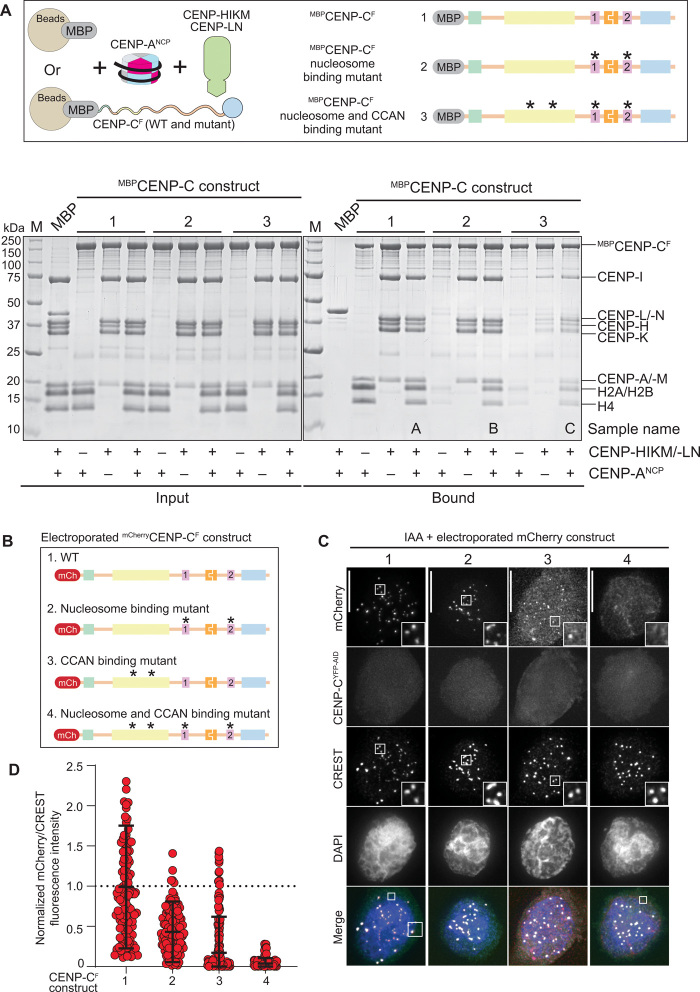
To investigate this possibility, we immobilized three MBP-tagged CENP-CF variants on amylose beads and used them as baits with CENP-ANCPs and CCAN subcomplexes (Fig. 4A). Specifically, we used CENP-CF, CENP-CF-R522A/W531A/R742A/W751A (indicated as “nucleosome binding mutant”), and a nucleosome binding mutant additionally impaired in the interaction with CENP-NL and CENP-HIKM through previously described point mutations (and indicated as “nucleosome and CCAN binding mutant”) (12, 14). We then incubated these baits with saturating concentrations of CENP-ANCPs, CENP-HIKMLN, or their combination. After washing, species retained on beads were resolved by SDS-PAGE and visualized by Coomassie staining. This revealed that CENP-CF binds the same levels of CENP-ANCPs and CENP-HIKMLN when presented individually or concomitantly (Fig. 4A and quantified in fig. S6E). Conversely, the nucleosome binding mutant of CENP-CF did not bind CENP-ANCPs but bound CENP-HIKMLN. When CENP-ANCPs and CENP-HIKMLN were coincubated with the nucleosome binding mutant, CENP-ANCP binding was restored, but at reduced levels. The residual binding of CENP-ANCP was through CENP-HIKMLN (and presumably through CENP-N in this complex), because the nucleosome and CCAN mutant of CENP-CF bound only residual levels of CENP-ANCPs or CENP-HIKMLN (Fig. 4A and quantified in fig. S6F). Thus, binding of CENP-CF to other CCAN subunits allows a robust point of contact with the CENP-A nucleosome through CENP-N (and possibly additional still undiscovered points of contact within the CENP-HIKMLN complex) even in the absence of functional CENP-A binding motifs.
Fig. 4. Relative contributions of CENP-C and CCAN to CENP-A binding.
(A) Schematic of the performed amylose-resin pull-down assays. MBP and three MBPCENP-CF variants, WT (1), nucleosome binding mutant (2), and nucleosome and CCAN binding mutant (3), were immobilized on amylose resin as bait. CENP-ANCP and the CCAN members CENP-HIKM/-LN were added as prey. The two SDS-PAGEs show the results of the amylose-resin pull-down experiment. The MBPCENP-CF variant used as bait is indicated above each lane. CENP-ANCP and CENP-HIKM/-LN was added as indicated below each lane. The left gel shows the input fractions, and the right gel shows the bound fractions. CCAN binding mutant: L265A, F266A, L267A, W317A, E302A, F303A, I305A, and D306A (12, 14). Nucleosome binding mutant: R522A, W531A, R742A, and W751A. (B) Schematic showing the different electroporated mCherryCENP-CF variants. (C) Representative images showing the fluorescence of endogenous CENP-CYFP-AID and the electroporated mCherryCENP-CF variants as shown in (B). Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm. (D) Quantification of the mCherry fluorescence intensities normalized to the centromere marker CREST and to mCherryCENP-CF-WT. Shown are means and SD.
Kinetochore localization of electroporated mCherryCENP-CF in cells depleted of endogenous CENP-CYFP-AID in DLD-1 cells demonstrated the main predictions from the in vitro assay (Fig. 4, B to D). Specifically, CENP-CF localized robustly to kinetochores, whereas the CENP-A binding mutant showed a partial localization impairment. Likely, this mutant is targeted to the centromere by CENP-HIKM or CENP-LN, because the CCAN binding mutant, already in isolation, showed very strongly impairment of kinetochore localization. This was compounded by additional mutation of the CENP-A binding motifs, which made the construct undetectable at the kinetochore. Collectively, these observations indicate that binding of CENP-C with centromeres depends simultaneously on multiple binding sites for CENP-A and other CCAN subunits.
The interaction of CENP-T with CCAN
The CCAN subunits CENP-S, CENP-T, CENP-W, and CENP-X are histone fold domain (HFD) proteins that associate in stable dimeric pairs (CENP-SX and CENP-TW) and as tetramer (46). We reconstituted the CENP-TWSX complex to identify conditions for its stable incorporation in reconstituted kinetochores. In solid-phase binding assays with immobilized MBPCENP-CF (Fig. 5, A and B), we observed binding of CENP-TWSX in the presence of CENP-HIKM and CENP-LN, slightly reduced binding upon omission of CENP-LN (fig. S6, G and H), and poor binding upon omission of CENP-HIKM or of both CENP-HIKM and CENP-LN (shown schematically in Fig. 5C). Thus, CENP-HIKM, as well as, marginally, CENP-LN, provides a binding platform for CENP-TWSX, in agreement with previous studies in S. cerevisiae (22, 28, 47–49). The HFD of CENP-T is followed by an extension, also containing the highly conserved C-terminal helical hairpin (α4 and α5 helices, predicted to encompass residues 532 to 546 of HsCENP-T), whose deletion does not affect CENP-W binding (50). This region has been shown to complete a binding interface for the CENP-HIK complex in S. cerevisiae (47, 48). In line with these previous results, deletions of this extension reduced or prevented binding of the CENP-TW complex to the MBPCENP-CF–CCAN complex in vitro and abrogated its kinetochore recruitment after electroporation (Fig. 5, D and E, and fig. S6, I and J).
Fig. 5. Determinants of CENP-T binding to CCAN.
(A) Schematic of the performed amylose-resin pull-down assays. MBP and MBPCENP-CF were immobilized on amylose resin as bait. CENP-TWSX was added as prey, either alone or in combination with CENP-HIKM and/or CENP-LN. (B) Result of the amylose-resin pull-down experiment. The used preys are indicated above each lane. The top gel shows the input fractions, and the bottom gel shows the bound fractions. (C) Graphical summary of the results obtained in (B). (D) SDS-PAGE result showing an amylose-resin pull-down experiment, in which three different Halo-tagged CENP-T variants were used, namely, CENP-T1–529, CENP-T1–549, and CENP-TFL. MBPCENP-C was used as bait, and MBP was used as negative control. (E) Representative images showing the fluorescence of mCherryCENP-W and three Halo-tagged CENP-T variants labeled with a green fluorescent ligand. Centromeres were visualized by CREST sera, and DNA was stained by DAPI. Scale bars, 10 μm.
Role of CENP-T in the outer kinetochore assembly
CENP-T, similar to CENP-C, is crucial for outer kinetochore assembly (18, 22, 23, 28, 29, 51–54). The flexible N-terminal region of human CENP-T binds two distinct NDC80 complexes through motifs encompassing the CDK1 targets Thr11 and Thr85 (29). A third CDK1 phosphorylation site, Ser201, possibly with a more modest additional contribution from Thr195, is further responsible for an interaction with an entire KMN network complex through a binding site on MIS12C (29, 55). Thus, each CENP-T recruits two NDC80Cs directly and one KMN (which contains an additional NDC80C), while CENP-C, as shown above, recruits a single KMN network through MIS12C for each of the subunits in the dimer.
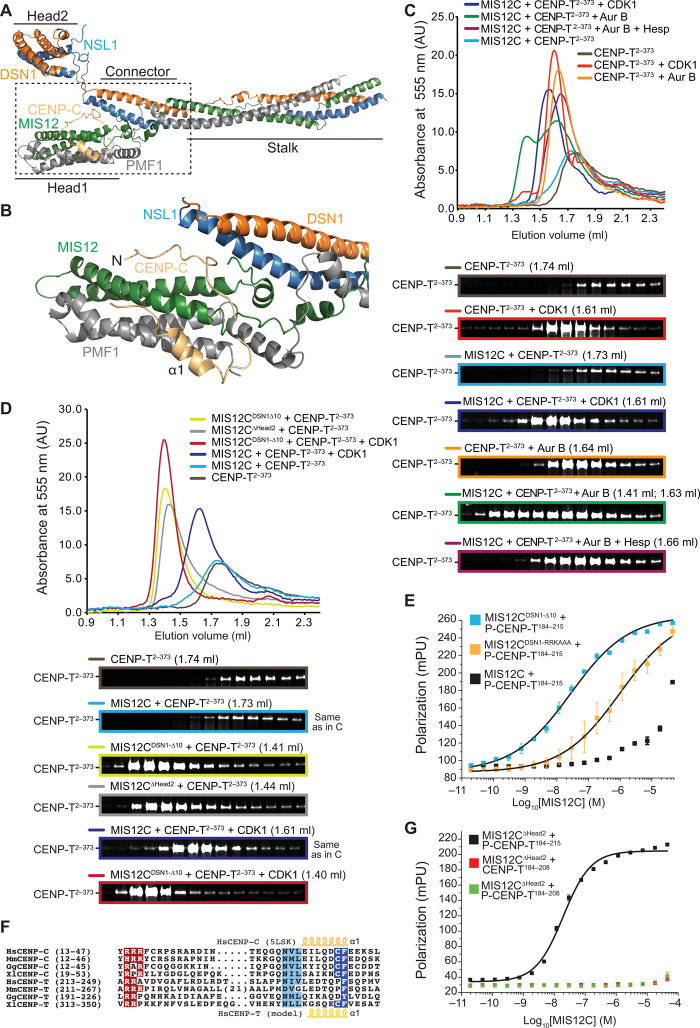
CENP-C binds MIS12C at the interface between the Head1 region [encompassing the N-terminal regions of the MIS12 and PMF1 (Polyamine Modulated Factor 1) subunits] and a “connector” encompassing the α3 helices of NSL1 and DSN1 (31). The regions of NSL1 (NNF1 Synthetic Lethal 1) and DSN1 (Dosage Suppressor of NNF1) preceding the connector form a globular domain named Head2 (Fig. 6, A and B). A disordered segment of DSN1 preceding Head2 contains at least two Aurora B target sites (Ser100DSN1 and Ser109DSN1) that control the access of CENP-C through an intramolecular regulatory mechanism (31, 55–58). A fragment encompassing the CENP-C N-terminal region binds MIS12C with ~150-fold higher affinity after Aurora B–mediated phosphorylation of Ser100DSN1 and Ser109DSN1 (31). Deletions of the entire N-terminal region of DSN1 and NSL1 including Head2 (indicated simply as MIS12C∆Head2) or of a 10-residue segment encompassing the two phosphorylation sites (MIS12CDSN1-∆10) bypass the requirement for phosphorylation and allow unhindered access to CENP-C (31, 55, 56). Previously, we showed that CENP-T and CENP-C cannot be simultaneously bound to the same MIS12C (29), suggesting partial or complete overlap of their binding sites on MIS12C. Whether access of CENP-T to MIS12C is also regulated by Aurora B activity, however, is unknown. More generally, because of lack of structural information on the CENP-T:MIS12C complex, as well as of obvious resemblances in the MIS12C-binding regions of CENP-C and CENP-T, how CENP-T binds MIS12C remains unclear.
Fig. 6. Molecular mechanism of the CENP-T:MIS12C interaction.
(A) Cartoon model of the human MIS12 complex in complex with the CENP-C N-terminal region [Protein Data Bank (PDB) ID 5LSK]. (B) Close-up of the MIS12C:CENP-C interaction interface. (C) SEC on a Superdex 200 column of the indicated species with the peak elution volume and resulting SDS-PAGE. (D) SEC on a Superdex 200 column of the indicated species with the peak elution volume and resulting SDS-PAGE. Note that the elution profiles and SDS-PAGE for the MIS12C + CENP-T sample (light blue) and for the MIS12C + CENP-T + CDK1 (dark blue) are the same as those displayed in (C) and are only displayed here for easier comparison. (E) Binding isotherms by fluorescence polarization with a fluorescent Ser201-phosphorylated CENP-T184–215 peptide and the indicated MIS12 complex variants. (F) Multiple sequence alignment of segments of the MIS12 complex binding regions of CENP-C and CENP-T from the indicated species. Hs, H. sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xl, Xenopus laevis. (G) Binding isotherms by fluorescence polarization obtained with the indicated fluorescent S201-phosphorylated or nonphosphorylated CENP-T peptides and the MIS12∆Head2 mutant complex. mpu, millipolarization unit.
To investigate this, we monitored the elution of fluorescently labeled CENP-T2–373 (29) from a SEC column upon addition of MIS12C. No binding of CENP-T2–373 to MIS12C was observed, as the elution profile of fluorescent CENP-T2–373 was essentially identical to that of CENP-T2–373 in isolation (Fig. 6C). In the absence of MIS12C, CDK1 or Aurora B phosphorylation caused CENP-T2–373 to elute at an earlier volume, possibly because phosphorylation causes decompaction of the N-terminal domain. CDK1 phosphorylation also enhanced CENP-T2–373 binding to MIS12C, as shown by a further decrease in the CENP-T2–373 elution volume. A more pronounced shift caused by addition of Aurora B was partially abrogated by the Aurora B inhibitor Hesperadin (Fig. 6C). Even if CENP-T2–373 appeared to be a robust Aurora B substrate in vitro, CENP-T2–373 binding to MIS12C was only observed upon their respective phosphorylation by CDK1 and Aurora B. Inverting the kinase-substrate pairs proved insufficient to promote robust binding (fig. S7A).
MIS12CDSN1-∆10 or MIS12C∆Head2, which lack the Aurora B phosphorylation motif, caused a more pronounced shift in the CENP-T2–373 elution volume, which was further reinforced after addition of CDK1 (Fig. 6D). Individually, MIS12C and MIS12CDSN1-∆10 eluted with the same elution volume (fig. S7B). Thus, phosphorylation of CENP-T and MIS12C by CDK1 and Aurora B, respectively, or deletion of the Aurora B target motif on MIS12C increases the binding affinity of the MIS12C:CENP-T complex, as previously observed with CENP-C (31, 55, 56). Accordingly, a fluorescent synthetic peptide encompassing residues 184 to 215 of CENP-T and phosphorylated on Ser201 (P-CENP-T184–215) bound to MIS12CDSN1-∆10 with a dissociation constant of ~27 nM in fluorescence anisotropy binding affinity measurements, approximately 1000-fold better than to control MIS12C (Fig. 6E and table S1).
Arginine residues in the Aurora B target motif of DSN1 are important for the autoinhibition mechanism, and their mutation to alanine (MIS12CDSN1-RRKAAA) is sufficient to enhance binding to CENP-C (31). We therefore speculated that these arginine residues compete directly with equivalent positively charged residues in the N-terminal region of CENP-C (Arg14-Arg15-Arg16; Fig. 6F) until Aurora B phosphorylation of DSN1 at neighboring serines neutralizes charge (31). Because MIS12CDSN1-RRKAAA also bound to the P-CENP-T184–215 peptide more tightly than control MIS12C (Fig. 6E), we speculated that a conserved short stretch of positive charges in CENP-T, Arg214, and Arg215 (Fig. 6F) plays a role equivalent to that of Arg14-Arg16 in CENP-C. In agreement with this idea, a peptide excluding this region (CENP-T184–208 instead of P-CENP-T184–215) did not bind MIS12C∆Head2, regardless of its phosphorylation status (Fig. 6G). CENP-C binds MIS12C also through the α1 helix downstream from the arginine-rich motif (Fig. 6B). A segment with helical propensity around residues 238 to 245 of CENP-T can be tentatively aligned to the CENP-C α1 helix (Fig. 6F). Collectively, these results suggest that residues 213 to 249 of CENP-T bind MIS12C similarly to residues 14 to 48 of CENP-C, as modeled in Fig. 7, A and B.
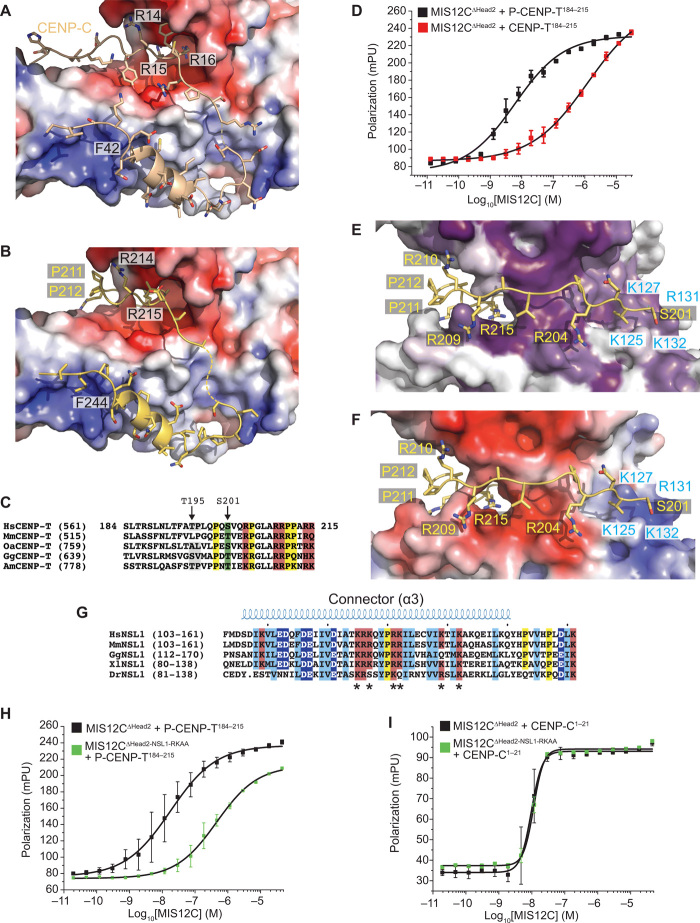
Fig. 7. Structural basis of CENP-T:MIS12C interaction.
(A) Surface electrostatics displayed on the MIS12 complex at the interface with CENP-C (wheat). (B) Structural model of the CENP-T peptide (residues 213 to 249) interaction with the MIS12C using the CENP-C N-terminal region as a template (PDB ID 5LSK) and the sequence alignment shown in Fig. 6F. CENP-T was modeled on the same interface. (C) Multiple sequence alignment of CENP-T in the unique region containing the Thr195 and Ser201 phosphorylation sites and encompassing the modeled residues (201 to 212) shown in (E) and (F), which precede the region aligned on CENP-C (Fig. 6F). (D) Binding isotherms by fluorescence polarization obtained with the indicated fluorescent S201-phosphorylated or nonphosphorylated CENP-T peptides and the MIS12C∆Head2 mutant complex. (E) Structural model of the CENP-T201–212:MIS12C interaction and the MIS12C surface conservation of residues lining the possible CENP-T binding site. (F) Charge distribution on the same interface discussed in (H). (G) Sequence analysis of the connector region of NSL1 pinpointing multiple conserved positively charged residues potentially involved in the phospho-dependent recognition of CENP-T. (H) Binding isotherms by fluorescence polarization obtained with the indicated fluorescent S201-phosphorylated peptide and the MIS12C∆Head2 control complex or the MIS12C∆Head2 with alanine mutations at Arg131NSL1 and Lys132NSL1. (I) As in (H) but with a fluorescent CENP-C1–21 peptide.
Phosphorylation of the CENP-T184–215 peptide on Ser201 (Fig. 7C) enhanced the binding affinity for MIS12C∆Head2 approximately 200-fold (Fig. 7D and table S1). Chemical cross-linking coupled with mass spectrometry on the MIS12C:P-CENP-T2–373 complex (fig. S7C and table S2) identified cross-links between Ser201CENP-T and Lys127NSL1 or Lys103MIS12 that allowed us to restrain the orientation of the CENP-T peptide on the MIS12C surface and model the ultimate path of the peptide (residues 201 to 212; Fig. 7, E and F). Lys127NSL1 and Lys103MIS12 are part of a positively charged, conserved surface ridge residing at the junction between the connector and Head1, opposite to the CENP-C binding site, and also comprising Arg131NSL1 and Lys132NSL1 (Fig. 7, E to G). Mutation of the latter two residues to alanine (MIS12CNSL1-RKAA) reduced the binding affinity for the P-CENP-T184–215 peptide approximately 25-fold (Fig. 7H and table S1), strongly implicating them in recognition of P-Ser201CENP-T. This model of the interaction of CENP-T with MIS12C predicts that two additional residues, Lys139NSL1 and Lys142NSL1, are well positioned to interact with P-Thr195CENP-T (29). In the structure of the MIS12C:CENP-C complex, this ridge is not involved in CENP-C binding (31). Accordingly, a CENP-C1–21 peptide bound MIS12CWT and MIS12CNSL1-RKAA with essentially identical affinity (Fig. 7I). Collectively, these data suggest that the CENP-T chain occupies a unique MIS12C ridge until it bends, around Pro211 and Pro212, to interact with the previously identified CENP-C binding interface. This model of the CENP-T–MIS12C interaction predicts the observed competitive binding to MIS12C of CENP-C and CENP-T.
Reconstitution of a complete kinetochore module
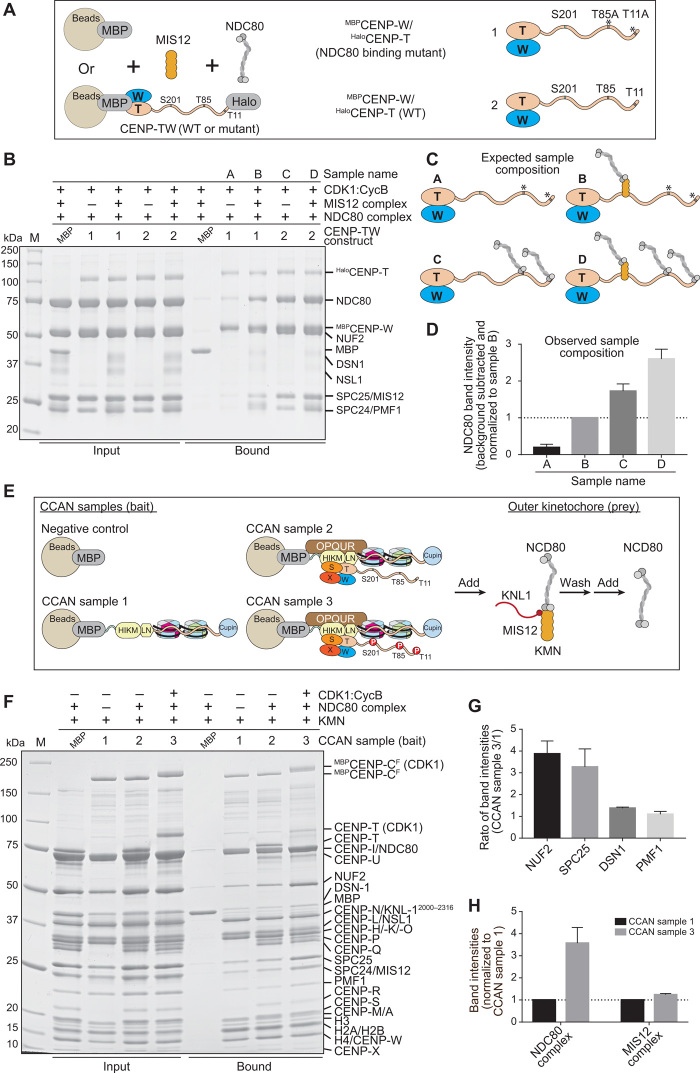
Last, we aimed to assemble a complete kinetochore module comprising all the inner and outer kinetochore subunits. To monitor the interaction of NDC80C with immobilized kinetochore proteins, we used wild-type (WT) CENP-T (Fig. 8A, construct 2) or a CENP-T mutant where the two direct NDC80C binding sites, Thr11 and Thr85, were mutated to alanine (construct 1). We then incubated these constructs as baits on solid phase with excess NDC80C and MIS12C and CDK1:Cyclin B (Fig. 8B; the outcome of this experiment is presented schematically in Fig. 8C and quantified in Fig. 8D and fig. S8A). Without MIS12C, construct 1 bound only background levels of NDC80C (sample A). Addition of MIS12C (sample B) resulted in the binding of one equivalent of NDC80C. Construct 2, on the other hand, bound approximately two equivalents of NDC80C in the absence of MIS12C (sample C) and three equivalents when MIS12C was also added (sample D). These stoichiometries suggest that phosphorylation of CENP-T on Thr11, Thr85, and Ser201 is complete and leads to the saturation of these binding sites.
Fig. 8. Reconstitution of a complete human kinetochore module.
(A) Schematic of pull-down experiments. MBPCENP-W/HaloCENP-TWT (1) and MBPCENP-W/HaloCENP-TT11A/T85A (2) immobilized on amylose beads as baits were incubated with NDC80C pray, alone, or with MIS12C. (B) SDS-PAGE result of pull-down experiment. MBPCENP-W/HaloCENP-T bait and added prey are indicated above each lane.( C) Expected composition of samples in (B). (D) Quantified NDC80 band intensities (normalized to sample B) and SDs (three repeats) of samples A to D indicated in bound fractions of (B). NDC80 band intensity of MBP control was subtracted as background. (E) Schematic of pull-down experiment. MBP and three CCAN samples assembled on MBPCENP-CF were immobilized on amylose resin as bait. KMN complex was added and incubated for 1 hour. Unbound KMN was washed, and NDC80C added for 1 hour. After washing unbound NDC80C, samples were analyzed by SDS-PAGE. (F) SDS-PAGE result of pull-down experiment. The CCAN bait and presence of CDK1:Cyclin B are indicated above each lane. (G) Quantified CCAN sample 3/sample 1 ratio and SD (three repeats) of indicated proteins shown in bound fractions of (F). (H) Quantified intensities and SD (three repeats) of the NDC80C (averaged over the NUF2 and SPC25 band intensities) and the MIS12C (averaged over the DSN1 and PMF1 band intensities) normalized to CCAN sample 1.
To obtain complete kinetochore reconstitutions, we reconstituted three MBPCENP-CF baits with dinucleosomes built on two consecutive human α-satellite repeats. We further decorated the dinucleosomes with a different complement of CCAN subunits (samples 1 to 3 in Fig. 8E; see also fig. S8, B to D, for an assessment of the influence of CDK1:Cyclin B on the interaction of CCAN subunits with CENP-CF). We settled on tandem CENP-ANCPs–H3NCPs dinucleosomes for these experiments because their propensity to interact with CCAN subunits was essentially indistinguishable from that of CENP-ANCPs–CENP-ANCPs dinucleosomes, but the resulting complexes appeared slightly more homogeneous in electrophoretic mobility shift assays (fig. S9, A and B).
Sample 3 contained all the CCAN subunits and was additionally phosphorylated by CDK1:Cyclin B. We first incubated the immobilized baits with the KMN network (generated with the MIS12∆10 mutant that binds maximally to CENP-C or CENP-T), and after washing the excess of unbound KMN, we added an excess of free NDC80C. After further washing, we analyzed bound proteins by SDS-PAGE and Coomassie staining (Fig. 8F). CDK1:Cyclin B phosphorylation was effective because it caused large shifts in the mobility of MBPCENP-CF and CENP-T. We quantified band intensities of scanned gels to determine the apparent stoichiometries of outer kinetochore subunits. The intensity of KMN subunits in sample 1, whose only linkage to the KMN is through the MBPCENP-CF baits, was taken as reference to assess the effects of integrating CENP-T and other CCAN subunits. We then examined the intensities of KMN subunits in sample 3 and compared them with those in sample 1. The sample 3/sample 1 ratio of MIS12C subunits (averaged over two subunits, DSN1 and PMF1) was approximately 1.5 (Fig. 8, G and H, and fig. S8, E to H), compatible with the addition of a single equivalent of CENP-T, if CENP-T remained fully saturated with MIS12C on the beads, as supported by the experiments in Fig. 8B. Alternatively, the observations are compatible with the addition of two equivalents if CENP-T underwent partial dissociation from MIS12C during the experiment. On the other hand, the sample 3/sample 1 ratio of NDC80C subunits [also averaged over two subunits, NUF2 (Nuclear Filament-containing protein 2) and SPC25 (Spindle Pole Component 25)] was higher than 3, compatible with the addition of two equivalents of CENP-T with full saturation of the Thr11 and Thr85 sites and partial saturation of the KMN binding site (Fig. 8, G and H, and fig. S8, E to H).
DISCUSSION
We have reconstituted on solid phase a sample incorporating all known core subunits of the human kinetochore, mostly in full-length form (Fig. 9A). The outer kinetochore subunits assemble on this sample with stoichiometries approaching the calibrated fluorescence intensity measurements performed on entire human kinetochores (23). CENP-TW, however, appeared to be generally substoichiometric in our reconstitutions, even in presence of nucleosomes, explaining why our observed stoichiometries do not yet reach the highest ratios of outer-to-inner kinetochore subunits observed at human kinetochores. Because the CENP-TW complex is one of the most deeply bound kinetochore components at the interface with centromeric chromatin (13, 18, 24, 59–61), failure to fully incorporate it in our reconstitutions suggests that an additional binding interface, not necessarily on DNA, may be missing. Characterization of the interaction of CENP-T with the MIS12C, on the other hand, brought to light notable similarities with the way that CENP-C interacts with this complex (31) and offered a glimpse on the potential regulation of this interaction by Aurora B kinase.
Fig. 9. Summary of main conclusions.
(A) The CENP-C dimer binds two nucleosomes (both being represented as CENP-A nucleosomes here). For clarity, binding interactions are only shown for one of the two protomers in the CENP-C dimer. CENP-HIKM/-LN binds near the PEST region, where it attracts CENP-TW. One full KMN network is recruited through the N-terminal region of CENP-C. Another one is recruited through CENP-TWSX. The latter recruits two additional NDC80 complexes. Therefore, each CENP-C protomer is associated with two KMN network complexes and a total of four NDC80 complexes. KNL1 was omitted for clarity. (B) CENP-C with (top) and without (bottom) the CENP-A binding motif in the central region could nevertheless interact with two nucleosomes through the CENP-C motif and the CENP-HIKM/-LN complex.
Recent single-particle cryo–electron microscopy (EM) structural analyses have revealed the overall organization of the CCANCtf19 complex of S. cerevisiae (7, 8), with insights into its point single kinetochore module. With the only known exception of kinetoplastids, eukaryotic kinetochores are evolutionarily related, despite considerable divergence in certain phyla, where loss of one or more CCAN subunits is not uncommon (62, 63). This seems to imply that regional kinetochores are built from multiple, adjacent repetitions of a module related to S. cerevisiae’s and that the complex we have reconstituted on solid phase is, or approaches, the human module. Structural analyses of kinetochores from additional species will have to clarify the extent to which CCAN complexes from other eukaryotes, including humans, resemble the yeast module.
CENP-C is found at kinetochores of most species, likely because its roles in connecting the CENP-A nucleosome to the outer kinetochore and as an organizer of the CCAN complex are indispensable. The apparent colinearity of binding motifs in CENP-C with the outer-inner kinetochore axis prompted us to define this protein as a blueprint for kinetochore assembly (12, 14, 15, 19). To gain deeper insights into how CENP-C shapes the kinetochore-centromere interface, we harnessed a protein ligation technique (37) to obtain a full-length version of human CENP-C. Electroporated in cells acutely depleted of endogenous CENP-C, the fusion protein was functional, showcasing the power of electroporation for targeted protein delivery and functional studies with recombinant proteins in cells (64).
Our observation that CENP-CF binds selectively to CENP-A nucleosomes over H3 nucleosomes even at micromolecular concentrations in vitro was unexpected, as previous studies had reported that at least the CENP-C motif, when isolated from the rest of CENP-C, binds relatively nonselectively to CENP-A and H3 NCPs (11, 36), although another study failed to detect an interaction of the CENP-C motif with H3.3 nucleosomes (65). Phosphorylation of CENP-C near the CENP-C motif has been recently shown to facilitate the interaction with CENP-ANCPs in vitro and CENP-C kinetochore localization (17, 45). The interaction of CENP-C with two nucleosomes, implied by the presence of CENP-ANCP binding motifs in the central region and the CENP-C motifs, had been postulated (2, 11) but remained undemonstrated. Here, we showed that CENP-C interacts with two disconnected CENP-A nucleosomes or with a reconstituted dinucleosome. In the absence of CCAN proteins, this ability reflects having two CENP-ANCP binding motifs, because mutation of both motifs abrogated binding of CENP-C to CENP-ANCPs in vitro, while mutation of either motif reduced the amount of bound CENP-A by half, suggesting that each binding site achieved saturation, at least at the low micromolar concentration of our experiments.
Many CENP-C orthologs have only the CENP-C motif and no evident CENP-A binding motif in the central motif (11). Even these CENP-C variants, however, may interact with two nucleosomes, because CENP-C interacts with CENP-ANCPs not only through its CENP-A binding motif(s) but also through CENP-N (Fig. 9B). Exemplifying this, addition of CENP-HIKM and CENP-LN complexes to CENP-ANCP binding assays performed with impairing mutants of the CENP-ANCP binding motifs of CENP-C rescued binding to CENP-ANCPs, while further mutation of the CENP-HIKM and CENP-LN binding sites largely abrogated binding in vitro and in cells. CENP-C localized quite robustly to centromeres after mutation or even deletion of the central region and CENP-C motif, as also reported in other studies (9, 17, 32). Thus, centromere recruitment of CENP-C relies not only on the CENP-ANCPs binding motifs but also equally, or even preferentially, on interactions with the CCAN. Studies of CENP-C localization in chicken DT40 cells have also demonstrated that recruitment of CENP-C requires CENP-HIKMLN and CENP-A nucleosome binding but that the dependence on these interactions varies depending on the cell cycle phase (16, 17). Rapid depletion of CENP-I or CENP-N in human DLD-1 cells demonstrated a partial dependence of CENP-C on these proteins for kinetochore localization in interphase but not in mitosis (13). Other studies in human cells showed partially reduced levels of CENP-C when single CCAN components were depleted by RNA interference (RNAi) (10, 15, 20). Collectively, these analyses identify CENP-C as an organizer of centromeric chromatin and of the inner kinetochore. They also pinpoint that the cell cycle, likely through phosphorylation, influences the localization requirements of CENP-C (17). Regretfully, structural analyses on yeast Ctf19CCAN have not yet visualized the Mif2pCENP-C polypeptide chain, either because it was not included in the reconstitution (7) or because it was, for the most part, invisible in density maps (8), a likely consequence of its predicted intrinsic disorder.
An interesting potential implication of the modular organization of regional kinetochores is that each module, in isolation, may encode its epigenetic specification and propagation. If this were true, then it might ultimately become possible to promote CENP-A loading in a reconstituted system provided that the loading machinery is offered the appropriate modular substrate. From a mechanistic perspective, however, how the organization of centromeric chromatin contributes to the recruitment of specialized CENP-A loading machinery—including the CENP-A chaperone HJURP (Holliday junction recognition protein), the MIS18:M18BP1 complex, and PLK1 (polo-like kinase 1)—and how CENP-A becomes loaded on chromatin by this machinery remain unclear (66).
Two features of CENP-A nucleosomes that must be considered in this context are (i) that once incorporated into centromeric chromatin, CENP-A is usually retained over extended, sometimes year-long periods of time, and (ii) that the levels of CENP-A at each chromosome appear to be maintained intergenerationally, implying that the amounts of newly loaded CENP-A are at least in first approximation identical to those of the centromere-residing CENP-A (66). Suppression by CDK limits loading to the early G1 phase of the cell cycle (42, 67, 68). The levels of CENP-A then halve during DNA replication, when CENP-A is equally distributed to the sister chromatids. New CENP-A loading is suppressed at this stage, and prereplication levels of CENP-A are only reestablished during the subsequent cell cycle. This contrasts with histone H3.1 deposition, which occurs along with DNA replication (69). Equal distribution of CENP-A to the sister chromatids during DNA replication engages HJURP, the same histone chaperone involved in CENP-A deposition, but the mechanistic basis of this process remains unclear (70). Vacancies generated by halving CENP-A may be temporarily filled with the “placeholder” histone H3.3, predicting that when CENP-A is loaded on chromatin in G1, histone H3.3 is concomitantly evicted (71).
Collectively, these features suggest a mechanism of CENP-A deposition in which a substrate of existing CENP-A nucleosomes determines how many new CENP-A nucleosomes are generated at every cell cycle. We previously adapted this idea to hypothesize that a minimal unit of two adjacent nucleosomes, with an appropriate complement of CCAN subunits, may satisfy the requirements for a substrate-based CENP-A loading reaction (2). For instance, the placeholder hypothesis (71) may be practically implemented on a dinucleosome scaffold, with a CENP-A nucleosome persisting through the cell cycle and an adjacent nucleosome alternating in composition, with an H3 nucleosome between S and G1 phases, and with a CENP-A nucleosome between G1 and S phases (2). While this remains merely conjectural, dimeric α-satellite units separated by a CENP-B box are a dominant feature of functional human centromeres; CENP-A is greatly enriched in adjacent peaks centered on each α-satellite unit in the dimer, while CENP-T associates more prominently with the linker (72, 73).
These considerations emphasize the importance of decoding the localization determinants of the CENP-A loading machinery that limits new CENP-A deposition to these sites, as the dissection of these determinants will reveal the molecular basis of epigenetic centromere specification. These determinants are probably complex and localized through the entire CCAN, because besides CENP-A itself, CENP-C and CENP-I also contribute to centromere recruitment of the Mis18 complex and M18BP1 (66).
All the work of in vitro reconstitution with CENP-A or Cse4 (the S. cerevisiae ortholog of CENP-A), histone H4, and the H2A/H2B dimer has so far uniformly converged on octameric, left-handed nucleosomes with somewhat looser DNA ends (3). Human kinetochores can be reconstituted on, and bind selectively to, the octameric CENP-ANCPs, as shown here and elsewhere (15), lending support to the idea that the left-handed octamer is the basic form of centromeric chromatin. CENP-A has been shown to form octameric nucleosomes also on centromeric chromatin in higher eukaryotes (3), but a possible caveat is that extensive nuclease digestions of centromeric chromatin lead to dissociation of CCAN subunits that might trigger postprocessing reorganization of CENP-A (72, 74).
In S. cerevisiae, the single kinetochore “module” is built on a specific, conserved centromere sequence and protects ~125 bp of DNA from nuclease digestion, of which approximately 85 bp, sufficient for a single gyre of DNA, wrapped around a CENP-A core proposed to be forming a tetrasome (75). In addition, the DNA of yeast centromeres is positively supercoiled (76, 77). The recent cryo-EM reconstruction of the Ctf19CCAN:Cse4NCPs complex (Cse4 is the S. cerevisiae ortholog of CENP-A), however, did not capture these features, as it was obtained with an octameric, left-handed Cse4NCP particle containing the 147-bp Widom 601 DNA sequence (8). In this structure, Chl4CENP-N occupies a different position from that previously observed with the isolated CENP-N protein on reconstituted CENP-ANCPs (14, 43) and that seems hardly compatible with the established function of human CENP-N in decoding the CENP-A L1 loop, where the sequences of CENP-A and H3 diverge (9, 78).
Thus, a fully convincing case for an octameric, left-handed nucleosome in S. cerevisiae has not yet been made, and the hypothesis that at least in this organism centromeric chromatin adopts a different organization remains open. In view of the possible modularity of regional kinetochores discussed above and of the considerable conservation of the CCAN subunits, and despite the considerable evidence supporting octameric CENP-A nucleosomes, presence of related noncanonical features in other eukaryotes remains a concrete possibility. How noncanonical features of centromeric chromatin could be built and stabilized, however, remains enigmatic. Biochemical reconstitution can contribute to solve this conundrum. A benchmark for successful reconstitution could be the reproduction of salient features of the original cellular object, including stoichiometries and successful binding interactions with the CENP-A loading machinery. Our future steps in biochemical reconstitutions will focus on this fundamental and still unresolved question.
MATERIALS AND METHODS
Plasmids
Plasmids for the production of Xenopus laevis H2A, H2B, H3, and H4 histones were a gift from D. Rhodes. Plasmids for the production of human CENP-A:H4 histone tetramer were a gift from A. F. Straight. Plasmids for the production of the “601” 145-bp DNA were a gift from C. A. Davey. The 165-bp CEN1 (centromere 1)–like DNA1 sequence (GTGGTAGAATAGGAAATATCTTCCTATAGAAACTAGACAGAATGATTCTCAGAAACTCCTTTGTGATGTGTGCGTTCAACTCACAGAGTTTAACCTTTCTTTTCATAGAGCAGTTAGGAAACACTCTGTTTGTAATGTCTGCAAGTGGATATTCAGACGCCCTTG) and the 183-bp CEN1-like DNA2 sequence (AGGCCTTCGTTGGAAACGGGATTTCTTCATATTCTGCTAGACAGAAGAATTCTCAGTAACTTCCTTGTGTTGTGTGTATTCAACTCACAGAGTTGAACGATCCTTTACACAGAGCAGACTTGAAACACTCTTTTTGTGGAATTTGCAGGCCTAGATTTCAGCCGCTTTGAGGTCAATCACCCC) were purchased from GeneArt. Eight tandem repeats of CEN1-like DNA fragment 1 or CEN1-like DNA fragment 2 were cloned into a pUC18 plasmid using restriction enzyme–based cloning and Gibson assembly. The plasmid pETDuet1-mChP-CENPW-HaloT-CENPT-8His was generated by insertion of the sequence of Halo-tagged CENP-T between Nco I and Xho I sites of pETDuet-8His (42). The plasmids pETDuet1-mChP-CENPW-HaloT-CENPT(1-549)-8His and pETDuet1-mChP-CENPW-HaloT-CENPT(1-529)-8His were generated by exchanging the coding sequence of CENP-T with CENP-T1–549 or CENP-T1–529 using restriction enzyme–based cloning. The plasmid pETDuet1-6His–tobacco etch virus (TEV)–MBP was generated by subcloning the coding sequence of Escherichia coli MBP with an N-terminal 6His-tag and a TEV protease cleavage site between Nco I and Sal I sites of pETDuet-1. A synthetic cDNA segment encoding human CENP-X isoform 1, codon-optimized for expression in bacteria, was subcloned in pET28a using Xba I and Sal I. A cDNA segment encoding human CENP-S isoform 1 was subcloned into the second cassette of the same plasmid using Sal I and Not I. Codon-optimized cDNA of human CENP-C, CENP-T, and CENP-W was purchased from GeneArt (Life Technologies). The plasmids containing the SpyCatcher and SpyTag sequences were ordered from Addgene. The plasmid pETDuet-MBP-8His was generated as previously described (42). The polymerase chain reaction (PCR)–amplified CDS (coding sequence) of CENP-C–601–943 or CENP-C–721–943 was inserted between the N-terminal MBP tag and the C-terminal 8His-tag using Bam HI and Xho I. The SpyTag sequence was inserted between the N-terminal MBP tag and the CENP-C–601–943 CDS using Bgl 2 and Bam HI. The PCR-amplified CDS of CENP-C–1–600 was inserted after the N-terminal MBP tag using Nhe I and Xho I. The PCR-amplified CDS of SpyCatcher was inserted using Kpn I and Sal I. In some constructs, MBP was exchanged against mCherry using Bgl 2 and Nhe I. All point mutations and sequence deletions were introduced by PCR-based site-directed mutagenesis.
For the coexpression of enhanced green fluorescent protein (EGFP)–CENP-C and different mCherry-tagged Cupin constructs, the CDS encoding full-length human CENP-C and three different CDS encoding Cupin domains from human, S. cerevisiae, and D. melanogaster were subcloned in pcDNA5-EGFP-NLS-P2AT2A-mCherry-PTS1 (42), a modified version of pcDNA5-FRT/TO (Thermo Fisher Scientific). The CDS of NLS was exchanged against the CDS of human CENP-C–1–943 using Bam HI and Xho I. The CDS of PTS1 (peroxisomal targeting signal 1) was exchanged against the CDS of human CENP-C–773–943, S. cerevisiae CENP-C–321–529, and D. melanogaster CENP-C–1127–1411 using Nhe I and Xma I. The CDS of S. cerevisiae CENP-C–321–529 was PCR-amplified from genomic yeast DNA. The CDS of D. melanogaster CENP-C was obtained from GeneArt. For the single expression of EGFP-tagged CENP-C constructs, the CDS of different CENP-C fragments were subcloned in a pcDNA5-EGFP-P2AT2A-2xStrepFLAG plasmid that was generated by inverse PCR. The PCR-amplified CDS of EGFP was inserted using Kpn I and Bam HI. The PCR-amplified CDS of CENP-C1-600 was inserted using Hind III and Kpn I. The PCR-amplified CDS of CENP-C–601–943 CENP-C–601–772, CENP-C–721–943, and CENP-C–760–943 were inserted using Bam HI and Xho I. The PCR-amplified CDS of S. cerevisiae CENP-C–321–529, D. melanogaster CENP-C–1127–1411, and the LacI dimerization domain (LacI-57-339) was inserted using Nhe I and Xma I.
Purification of CEN1-like DNA fragments for nucleosome reconstitution
The pUC18 plasmids were transformed into competent E. coli cells. Inoculated bacteria cultures were incubated overnight at 37°C in TB (Terrific Broth) media supplemented with ampicillin (100 μg/ml). Subsequently, the plasmid DNA was purified using the Giga Purification Kit (Macherey-Nagel). After Eco RV digestion, the insert DNA was separated from the backbone DNA by polyethylene glycol precipitation. The isolated DNA fragments were digested with Bst XI/Bgl I (DNA1) and Bgl I/Dra III (DNA2). The digested DNA fragments were loaded on a HiTrap Q FF anion exchange column (GE Healthcare) and eluted with a linear gradient from 0 to 2000 mM NaCl in 20 bed volumes. Fractions containing the DNA fragments were precipitated with ethanol and dissolved in 2 M NaCl.
Reconstitution of nucleosomes
CENP-A and H3 containing mononucleosomes assembled on 601 145-bp DNA were reconstituted precisely as described (15). For the assembly of dinucleosomes, CENP-A and H3 containing NCPs were first reconstituted using two different CEN1-like DNA fragments. One micromolar of NCPs reconstituted on CEN1-like DNA fragment 1 and CEN1-like DNA fragment 2 were ligated to each other for 16 hours at 4°C using 8xHis-tagged T4 DNA ligase. The ligated NCPs were incubated with cOmplete nickel beads (Roche) for 7 hours at 4°C to remove the T4 DNA ligase. The ligated dinucleosomes were concentrated, and glycerol was added at a final concentration of 2% (v/v). The mixture was loaded on a cylindrical gel containing 5% reduced bis-acrylamide. Native PAGE was carried out on a Mini Prep Cell apparatus (Bio-Rad) at 1 W of constant power. Dinucleosomes were eluted at a constant flow rate of 0.1 ml/min overnight at 4°C into a nucleosome buffer (50 mM NaCl, 10 mM triethanolamine, and 1 mM EDTA) and collected in 250 μl of fractions on 96-well plates. The OD260 (optical density at 260 nm) and OD280 of the individual fractions were measured using a CLARIOstar Plus plate reader (BMG Labtech). Fractions containing dinucleosomes were pooled, concentrated, and stored at 4°C.
Human cell lines
Parental Flp-In T-REx HeLa cells were a gift from S. Taylor (University of Manchester, Manchester, England, UK). Flp-In T-REx DLD-1–CENP-C–AID-EYFP cells were a generous gift from D. Fachinetti (Institut Curie, Paris, France) and D. C. Cleveland (University of California, San Diego, USA). These cells have both alleles of CENP-C tagged at the C terminus with an AID-EYFP fusion (38). Furthermore, a gene encoding the plant E3 ubiquitin ligase osTIR1 was stably integrated into the genome of the cells. To induce rapid depletion of the endogenous CENP-C, 500 μM of the synthetic auxin IAA was added to the cells.
Cell culture
In all cell culture experiments, cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; PAN-Biotech) supplemented with 10% tetracycline-free fetal bovine serum (Thermo Fisher Scientific), 2 mM penicillin/streptomycin (PAN-Biotech), and 2 mM l-glutamine (PAN-Biotech) at 37°C in a 5% CO2 atmosphere. Expression of CENP-C–GFP fusion proteins was induced by addition of doxycycline (50 ng/ml; Sigma-Aldrich) for at least 24 hours.
CENP-C RNAi
Gene expression of endogenous CENP-C was inhibited using a single small interfering RNA (siRNA) (target sequence: 5′-GGAUCAUCUCAGAAUAGAA-3′ obtained from Sigma-Aldrich), which targets the coding region of CENP-C mRNA. The expression of codon-optimized CENP-C rescue constructs was not affected by the siRNA treatment. Thirty nanomolars of CENP-C siRNA was transfected using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. To induce the expression of GFP-tagged CENP-C rescue constructs, doxycycline (50 ng/ml) (Sigma-Aldrich) was added to the cells at the time of siRNA transfection. Phenotypes were analyzed 60 hours after transfection by immunofluorescence microscopy or immunoblotting analysis, respectively.
Generation of stable cell lines
Stable Flp-In T-REx HeLa cell lines constitutively expressing various CENP-C constructs were generated by Flp/FRT recombination. Deletion mutants and point mutants of CENP-C were generated by PCR site-directed mutagenesis, and the sequences of all constructs were verified by Sanger sequencing (Microsynth Seqlab). CENP-C constructs were cloned into a pcDNA5/FRT plasmid and cotransfected with pOG44 (Invitrogen), a plasmid expressing the Flp recombinase, into cells using X-tremeGENE (Roche) according to the manufacturer’s instructions. Following 2 weeks of selection in hygromycin B (250 μg/ml; Thermo Fisher Scientific) and blasticidin (4 μg/ml; Thermo Fisher Scientific), single-cell colonies were collected and subsequently expanded. Expression of the transgenes was checked by immunofluorescence microscopy and Western blot analysis.
Immunofluorescence microscopy
The paraformaldehyde (PFA)–fixated cells were permeabilized with PBS-T [phosphate-buffered saline (PBS) buffer containing 0.1% Triton X-100] for 10 min and incubated with PBS-T containing 4% bovine serum albumin (BSA) for 40 min. Cells were incubated for 90 min at room temperature with CREST/anticentromere antibody (Antibodies Inc.; dilution, 1:200 2% BSA in PBS-T), washed three times with PBS-T, and were subsequently treated for 30 min with anti-human Alexa Fluor 647–conjugated secondary antibody (Jackson ImmunoResearch; dilution, 1:200 2% BSA in PBS-T) to immunostain the centromeres. To visualize DNA, 4′,6-diamidino-2-phenylindole (DAPI) (0.5 μg/ml; Serva) was added during the last washing step for 3 min. After drying, the coverslips were mounted with Mowiol mounting media (EMD Millipore) on glass slides and imaged using a ×60 oil immersion objective lens on a DeltaVision deconvolution microscope. The DeltaVision Elite System (GE Healthcare, UK) is equipped with an IX71 inverted microscope (Olympus, Japan), a PLAPON ×60/1.42 numerical aperture objective (Olympus) and a pco.edge sCMOS camera (PCO-TECH Inc., USA). Quantification of centromere signals was performed using the software Fiji with a script for semiautomated processing. Briefly, average projections were made from z-stacks of recorded images. Centromere spots were chosen on the basis of the parameters of shape, size, and intensity using the images of the reference channel obtained with CREST staining, and their positions were recorded. In the images of the data channels, the mean intensity value of adjacent pixels of a centromere spot was subtracted as background intensity from the mean intensity value of the centromere spot.
Immunoblotting
HeLa cells were harvested by trypsinization, and the cell pellet was washed with PBS. Cells were incubated in lysis buffer [75 mM Hepes (pH 7.5), 150 mM NaCl, 1.5 mM EGTA, 10 mM MgCl2, 10% glycerol, 0.1% NP-40, Benzonase (90 U/ml; Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor mix HP Plus (Serva)] for 15 min on ice and lastly subjected to sonication using Bioruptor Plus sonication device (Diagenode). The lysed cells were centrifuged at 16,000g for 30 min at 4°C, and SDS sample buffer was added to the supernatant. After Tricin–SDS-PAGE, the proteins were blotted on a nitrocellulose membrane and detected by Western blot analysis. The following primary antibodies were used: anti–α-tubulin (1:8000; Sigma-Aldrich, T9026), anti–CENP-C (1:1000; SI410, gift from S. Trazzi), anti-mCherry (1:2000; Novus Biologicals, NBP1-96752), anti-GFP (1:10,000; HRP-coupled; Abcam, 190584), and anti-vinculin (1:10,000; Sigma-Aldrich, V9131). As secondary antibodies, we used anti-mouse or anti-rabbit (1:10,000; Amersham, NXA931 and NA934) conjugated to horseradish peroxidase. After incubation with ECL Western blotting reagent (GE Healthcare), images were acquired with the ChemiDoc MP System (Bio-Rad) using Image Lab 5.1 software.
Electroporation of recombinant proteins into human cells
To electroporate recombinant mCherry or mCherry–CENP-C protein into DLD-1–CENP-C–AID-EYFP cells and recombinant mCherryCENP-W/Halo–CENP-T into HeLa cells, the Neon Transfection System Kit (Thermo Fisher Scientific) was used. Cells (3 × 106) were trypsinized, washed with PBS, and resuspended in electroporation buffer R (Thermo Fisher Scientific) to a final volume of 90 μl. Recombinant mCherry–CENP-C and mCherryCENP-W/Halo–CENP-T protein was diluted 1:2 in buffer R to 15 μM, and 30 μl of the mixture was added to the 90-μl cell suspension. After mixing the sample, 100 μl of the mixture was loaded into a Neon pipette tip (Thermo Fisher Scientific) and electroporated by applying two consecutive 35-ms pulses with an amplitude of 1005 V. The sample was subsequently added to 50 ml of prewarmed PBS, centrifuged at 500g for 3 min, and trypsinized for 7 min to remove noninternalized extracellular protein. After one additional PBS washing step and centrifugation, the cell pellet was resuspended in DMEM without antibiotics and transferred to a 12-well plate containing poly-l-lysine–coated coverslips. After the electroporation procedure, DLD-1–CENP-CAID-EYFP cells were additionally treated with 500 μM IAA (Sigma-Aldrich) to induce rapid depletion of endogenous CENP-C. Electroporated Halo–CENP-T was labeled using the Halo Tag Oregon Green fluorescent ligand (Promega) according to the manufacturer’s instructions.
Coimmunoprecipitation experiment
Purified cleared lysates from HeLa Flp-In T-REx cells coexpressing EGFP-tagged CENP-C and mCherry-tagged Cupin fragments from human, S. cerevisiae, and D. melanogaster were used for coimmunoprecipitation experiments. Four milligrams of lysate was incubated with 10 μl of RFP-Trap magnetic agarose beads (Chromotek) for 3 hours at 4°C. Subsequently, the beads were washed three times with 500 μl of washing buffer [75 mM Hepes (pH 7.5), 150 mM NaCl, 0.1% NP-40, and one-time protease inhibitor cocktail (Serva)]. Afterward, two times SDS-PAGE sample loading buffer was added to the dry beads. The samples were boiled for 5 min at 95°C and analyzed by SDS-PAGE and subsequent Western blotting analysis.
Protein expression and purification
The proteins 6xHis-TEV–MBP–CENP-C1–600–SpyCatcher, MBP-TEV–SpyTag–CENP-C601–943–8xHis, MBP-TEV–CENP-C721–943–8xHis, mCherry-PreSc–CENP-W/Halo-TEV–CENP-T–8xHis, 6xHis-PreSc–MBPCENP-W/Halo-TEV–CENP-T, 6xHis-TEV–CENP-W/Halo-TEV–CENP-T, and CENP-X/CENP-S were expressed under same conditions. E. coli BL21(DE3)-Codon-plus-RIL cells transfected with the pETDuet plasmid were grown in 2× YT (yeast extract tryptone) medium supplemented with ampicillin and chloramphenicol at 37°C to an OD600 of 0.8. Then, 0.2 mM ITPG (Isopropyl β-D-1-thiogalactopyranoside) was added to induce protein expression, and cells were cultured for 16 hours at 20°C. Bacterial cells were harvested and washed once with PBS, and the cell pellets were stored at −20°C. The proteins 6xHis-TEV–CENP-W/Halo-TEV–CENP-T and CENP-X/CENP-S were coexpressed by cotransformation of E. coli with two plasmids.
To purify the CENP-C constructs, the bacterial cell pellets were resuspended in lysis buffer [30 mM Hepes (pH 7.5), 500 mM NaCl, 10% glycerol, and 1 mM TCEP (Tris(2-carboxyethyl)phosphine)] supplemented with 1 mM PMSF and subsequently lysed by sonication. The crude lysate was cleared by centrifugation at 75,000g for 30 min at 4°C. The supernatant was passed through a 0.45-μm filter (Sarstedt) and incubated with cOmplete His-Tag purification resin (Roche) for 16 hours on a tube roller (StarLab) at 4°C. After incubation, the resin was washed with 100 ml of lysis buffer A supplemented with 10 mM imidazole. The bound protein was lastly eluted in 15 ml of lysis buffer supplemented with 400 mM imidazole. The samples were concentrated using centrifugal filters with a 30-kDa mass cutoff (Merck) and were subsequently applied to a Superose 6 10/300 size exclusion column (GE healthcare) equilibrated in SEC buffer [10 mM Hepes (pH 7.5), 300 mM NaCl, 2.5% glycerol, and 1 mM TCEP]. SEC was performed under isocratic conditions at a constant flow rate of 0.5 ml/min. Fractions (500 μl) were collected, and relevant fractions were analyzed by SDS-PAGE, pooled, concentrated, flash-frozen, and stored at −80°C until use for further experiments. Purification of mCherryCENP-W/Halo–CENP-T–8His and 6His–MBP–CENP-W/Halo–CENP-T was identical, but the lysis buffer contained 1 M NaCl and the SEC buffer contained 500 mM NaCl and 5% glycerol.
To purify the CENP-TWSX tetramer, the bacterial pellets were resuspended in lysis buffer [50 mM tris (pH 8.0), 1 M NaCl, 10% glycerol, 1 mM EDTA, 5 mM 2-mercaptoethanol, and 1 mM TCEP] supplemented with 1 mM PMSF, lysed by sonication, and cleared by centrifugation at 75,000g at 4°C for 1 hour. The cleared lysate was applied to cOmplete His-Tag purification resin (Roche), pre-equilibrated in lysis buffer, and incubated at 4°C for 6 hours on a tube roller. Subsequently, the beads were washed with lysis buffer supplemented with 10 mM imidazole. Subsequently, the CENP-TWSX complex was eluted in lysis buffer supplemented with 400 mM imidazole. His-TEV protease was added, and the CENP-TWSX complex was dialyzed three times against 2 liters of lysis buffer containing a reduced NaCl concentration of 300 mM. After dialysis, the protein complex was loaded on a HiTrap SP FF column, washed with 10 CV (calcium volumes) of 15% buffer B [50 mM tris (pH 7.4), 2 M NaCl, 5% glycerol, 1 mM EDTA, 5 mM 2-mercaptoethanol, and 1 mM TCEP], and eluted using a gradient of 300 to 2000 mM NaCl. The fractions containing CENP-TWSX were pooled, concentrated, and loaded onto a Superdex 200 10/300 SEC column (GE Healthcare) pre-equilibrated in SEC buffer [50 mM tris (pH 8.0), 500 mM NaCl, 5% glycerol, and 1 mM TCEP]. Fractions containing CENP-TWSX were pooled, concentrated, flash-frozen in liquid nitrogen, and stored at −80°C. All other proteins were purified as previously described (14, 15, 21, 29, 31).
Analytical SEC
Analytical SEC of samples containing CENP-C and NCPs was performed on a Superose 6 5/150 column (GE Healthcare) in SEC buffer containing 10 mM Hepes (pH 7.5), 300 mM NaCl, 2.5% glycerol, and 1 mM TCEP on an ÄKTAmicro system. Samples containing CENP-T2–373–TMR (Tetramethylrhodamine) and MIS12C were analyzed on a Superdex 200 5/150 column in SEC buffer containing 20 mM tris (pH 8), 150 mM NaCl, and 1 mM TCEP. All samples were eluted under isocratic conditions at 4°C in SEC buffer at a flow rate of 0.2 ml/min. Elution of proteins was monitored at 280 and 254 nm. Elution of mCherry–CENP-C was additionally monitored at 587 nm. Elution of CENP-T2–373–TMR was additionally monitored at 555 nm. Fractions (100 μl) were collected and analyzed by SDS-PAGE, Coomassie blue staining, and scans of in-gel fluorescence for samples containing CENP-T2–373–TMR. To detect complex formation of CENP-C and nucleosomes or CENP-T and MIS12C, the individual proteins were mixed at the indicated concentrations in a total volume of 50 μl, incubated for 30 min on ice, and lastly subjected to SEC.
SpyTag reaction for in vitro reconstitution of full-length CENP-C protein
To reconstitute full-length CENP-C, the SpyTag reaction was used (37). Briefly, MBP-TEV–SpyTag–CENP-C601–943–8xHis was incubated with TEV protease for 8 hours at 4°C to cleave off the N-terminal MBP tag. Subsequently, 6xHis-TEV-MBP–CENP-C1–600–SpyCatcher was added in threefold molar excess. After incubation at 4°C for 16 hours, the reconstituted MBP–CENP-C1–600–Spy–CENP-C601–943–8xHis protein was separated from unligated protein fragments on a Superose 6 10/300 size exclusion column (GE Healthcare) as described above. Relevant fractions were pooled and concentrated using 50-kDa cutoff Amicon filters. The concentrated protein was flash-frozen and stored at −80°C.
Analytical ultracentrifugation
Sedimentation velocity AUC was performed in a Beckman XL-A ultracentrifuge at 42,000 rpm at 20°C. CENP-A NCP (alone and together with MBP–CENP-CFLWT or MBP–CENP-C∆motif2) and MBP–CENP-C721–943 were diluted in AUC buffer [10 mM Hepes (pH 7.9), 100 mM NaCl, 2% glycerol, and 1 mM TCEP] and subsequently loaded into standard double-sector centerpieces. The cells were scanned at 260 or 280 nm. At least 150 scans were recorded for each sample. Data were analyzed using the program SEDFIT with the model of continuous c(s) distribution. The partial specific volumes of the proteins, buffer density, and buffer viscosity were estimated using the program SEDNTERP. For the DNA of CENP-A NCP, a partial specific volume of 0.55 ml/g was assumed. Figures were generated using the program GUSSI.
In vitro phosphorylation by CDK1:Cyclin B
When indicated, samples were phosphorylated during the amylose-resin pull-down experiments using in-house generated CDK1:Cyclin B:CKS1 (CCC). Phosphorylation reactions were set up in binding buffer [10 mM Hepes (pH 7.5), 300 mM NaCl, and 1 mM TCEP] containing 100 nM CCC, 3 μM CENP-T/CENP-C/CCAN substrate, 2 mM adenosine triphosphate, and 10 mM MgCl2. Reaction mixtures were incubated at 30°C for 120 min and then used in the pull-down experiments.
Amylose-resin pull-down assay
The amylose-resin pull-down assay was performed to check the binding of nucleosomes and kinetochore components to MBP-tagged CENP-C variants and MBP-tagged CENP-W/Halo–CENP-T variants. The proteins were diluted with binding buffer [10 mM Hepes (pH 7.5), 300 mM NaCl, 2.5% glycerol, and 1 mM TCEP] to 3 μM concentration in a total volume of 50 μl and mixed with 20 μl of amylose beads (New England Biolabs). After mixing the proteins and the beads, 20 μl were taken as input. The rest of the solution was incubated at 4°C for 1 hour on a thermomixer (Eppendorf) set to 1000 rpm. To separate the proteins bound to the amylose beads from the unbound proteins, the samples were centrifuged at 800g for 3 min at 4°C. The supernatant was removed, and the beads were washed four times with 500 μl of binding buffer. After the last washing step, 20 μl of 2× SDS-PAGE sample loading buffer was added to the dry beads. The samples were boiled for 5 min at 95°C and analyzed by SDS-PAGE and CBB (Coumassie Brilliant Blue) staining. In assays performed to investigate the interactions of CENP-T to CCAN, the buffer contained 150 mM NaCl, 15 mM Hepes (pH 7.5), 1 mM TCEP, and 0.01% Tween 20. The samples were incubated for 15 min at 1400 rpm and 4°C.
Binding measurements
Fluorescence polarization experiments were performed with a CLARIOstar plus plate reader using Corning 384-well low-volume black round-bottom polystyrene NBS (nonbinding surface) microplates. The experiment was performed in 20 μl of total reaction volume containing a fixed concentration (20 nM) of the indicated 5-FAM (5-Carboxyfluorescein) CENP-T or CENP-C peptide (obtained from GenScript). Indicated peptides were mixed in a titration series with an increasing concentration of the respective MIS12C variant in the binding buffer [20 mM tris (pH 8), 150 mM NaCl, and 1 mM TCEP] and equilibrated for 30 min before the measurement at approximately 25°C. Fluorescein (5-FAM) was exited with polarized light at 470 nm, and the emitted light was detected at 525 nm through both horizontal and vertical polarizers. Polarization values are shown as means ± SEM for at least three replicates. The values of the binding constants were derived using the Origin software (OriginLab, Northampton, MA) by nonlinear least-square fitting of the binding curve to quadratic equation
where [R]t is the total concentration of the labeled peptide, [P]t is the total concentration of the MIS12C added during the titration, and Kd is the dissociation constant.
Modeling
Multiple sequence alignments were carried out using MAFFT. The degree of sequence conservation was calculated using ConSurf and was mapped on the MIS12C structure. Human CENP-T residues 207 to 221 and 230 to 250 were modeled on the CENP-C peptide residues 6 to 22 and 28 to 48 according to the alignment shown in Fig. 6C by fitting the residues into the electron density of the Protein Data Bank (PDB) ID code dataset 5LSK structure (MIS12C with the CENP-C peptide) using Coot (79). The side chains with different or insufficient density were modeled according to geometrical constraints. Atomic coordinates of the resulting model are available upon request.
Cross-linking and mass spectrometry
Approximately 100 μg of MIS12/CENP-T complex in cross-linking buffer [50 mM Hepes (pH 7.5), 300 mM NaCl, and 1 mM TCEP] was incubated with 3 mM DSBU (Disuccinimidyl Dibutyric Urea) (diluted from 200 mM stock solution in dimethyl sulfoxide; Alinda Chemical Limited) at 25°C for 1 hour. The reaction was quenched by addition of 100 mM tris-HCl (pH 8) and incubated for 30 min. Diagnostic SDS-PAGE gels of samples before and after the cross-linking reaction were ran to ensure the completion of reaction. Samples were precipitated overnight with four volumes of cold acetone, spun down, and following the removal of the supernatant, the pellet was briefly air-dried before resuspension in urea. Samples were processed according to our previously described protocol (80) and analyzed using MeroX (81). Proximity maps were visualized in Circos plots and arranged using Adobe Illustrator.
Acknowledgments
We are grateful to D. Fachinetti and D. C. Cleveland for sharing reagents; to I. Hoffmann, C. Koerner, L. Schulze, I. Stender, A. Take, B. Voss, and S. Wohlgemuth for general technical assistance; and to F. Müller and P. Janning for help with mass spectrometry experiments. Funding: A.M. acknowledges funding by the Max Planck Society, the European Research Council (ERC) Advanced Investigator Grant RECEPIANCE (proposal 669686), and the DFG’s Collaborative Research Centre (CRC) 1093. Author contributions: K.W.: Conceptualization, investigation, project administration, visualization, and writing (original draft preparation). A.P.: Conceptualization, investigation, and visualization. D.P.: Conceptualization, investigation, and visualization. B.H.: Investigation, project administration, and visualization. D.V.: Investigation. I.V.: Conceptualization and visualization. A.M.: Conceptualization, funding acquisition, project administration, supervision, visualization, and writing (original draft preparation). Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabg1037/DC1
REFERENCES AND NOTES
- 1.McKinley K. L., Cheeseman I. M., The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musacchio A., Desai A., A molecular view of kinetochore assembly and function. Biology 6, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black B. E., Cleveland D. W., Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144, 471–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R. 3rd, Desai A., Fukagawa T., The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446–457 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R. III, Cleveland D. W., The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K., Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11, 673–684 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Hinshaw S. M., Harrison S. C., The structure of the Ctf19c/CCAN from budding yeast. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan K., Yang J., Zhang Z., McLaughlin S. H., Chang L., Fasci D., Ehrenhofer-Murray A. E., Heck A. J. R., Barford D., Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 574, 278–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll C. W., Milks K. J., Straight A. F., Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll C. W., Silva M. C. C., Godek K. M., Jansen L. E., Straight A. F., Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11, 896–902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H., Jiang J., Zhou B.-R., Rozendaal M., Feng H., Ghirlando R., Xiao T. S., Straight A. F., Bai Y., A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–1113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klare K., Weir J. R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., Musacchio A., CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinley K. L., Sekulic N., Guo L. Y., Tsinman T., Black B. E., Cheeseman I. M., The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol. Cell 60, 886–898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G. P., Weir J. R., Vetter I. R., Musacchio A., Luger K., Decoding the centromeric nucleosome through CENP-N. eLife 6, e33442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir J. R., Faesen A. C., Klare K., Petrovic A., Basilico F., Fischbock J., Pentakota S., Keller J., Pesenti M. E., Pan D., Vogt D., Wohlgemuth S., Herzog F., Musacchio A., Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 537, 249–253 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Nagpal H., Hori T., Furukawa A., Sugase K., Osakabe A., Kurumizaka H., Fukagawa T., Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell 26, 3768–3776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe R., Hara M., Okumura E. I., Herve S., Fachinetti D., Ariyoshi M., Fukagawa T., CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J. Cell Biol. 218, 4042–4062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gascoigne K. E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I. M., Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410–422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Screpanti E., De Antoni A., Alushin G. M., Petrovic A., Melis T., Nogales E., Musacchio A., Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391–398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheeseman I. M., Desai A., Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Pesenti M. E., Prumbaum D., Auckland P., Smith C. M., Faesen A. C., Petrovic A., Erent M., Maffini S., Pentakota S., Weir J. R., Lin Y. C., Raunser S., McAinsh A. D., Musacchio A., Reconstitution of a 26-subunit human kinetochore reveals cooperative microtubule binding by CENP-OPQUR and NDC80. Mol. Cell 71, 923–939.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pekgoz Altunkaya G., Malvezzi F., Demianova Z., Zimniak T., Litos G., Weissmann F., Mechtler K., Herzog F., Westermann S., CCAN assembly configures composite binding interfaces to promote cross-linking of Ndc80 complexes at the kinetochore. Curr. Biol. 26, 2370–2378 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Suzuki A., Badger B. L., Salmon E. D., A quantitative description of Ndc80 complex linkage to human kinetochores. Nat. Commun. 6, 8161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan X., O’Quinn R. P., Pierce H. L., Joglekar A. P., Gall W. E., DeLuca J. G., Carroll C. W., Liu S. T., Yen T. J., McEwen B. F., Stukenberg P. T., Desai A., Salmon E. D., Protein architecture of the human kinetochore microtubule attachment site. Cell 137, 672–684 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D., Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8, 581–585 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodor D. L., Mata J. F., Sergeev M., David A. F., Salimian K. J., Panchenko T., Cleveland D. W., Black B. E., Shah J. V., Jansen L. E., The quantitative architecture of centromeric chromatin. eLife 3, e02137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akiyoshi B., Sarangapani K. K., Powers A. F., Nelson C. R., Reichow S. L., Arellano-Santoyo H., Gonen T., Ranish J. A., Asbury C. L., Biggins S., Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468, 576–579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang J., Barber A., Biggins S., An assay for de novo kinetochore assembly reveals a key role for the CENP-T pathway in budding yeast. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huis in’t Veld P. J., Jeganathan S., Petrovic A., Singh P., John J., Krenn V., Weissmann F., Bange T., Musacchio A., Molecular basis of outer kinetochore assembly on CENP-T. eLife 5, e21007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkov V. A., Huis in’t Veld P. J., Dogterom M., Musacchio A., Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. eLife 7, e36764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovic A., Keller J., Liu Y., Overlack K., John J., Dimitrova Y. N., Jenni S., van Gerwen S., Stege P., Wohlgemuth S., Rombaut P., Herzog F., Harrison S. C., Vetter I. R., Musacchio A., Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell 167, 1028–1040.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L. Y., Allu P. K., Zandarashvili L., McKinley K. L., Sekulic N., Dawicki-McKenna J. M., Fachinetti D., Logsdon G. A., Jamiolkowski R. M., Cleveland D. W., Cheeseman I. M., Black B. E., Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun. 8, 15775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milks K. J., Moree B., Straight A. F., Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell 20, 4246–4255 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen R. L., Espelin C. W., De Wulf P., Sorger P. K., Harrison S. C., Simons K. T., Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell 19, 4480–4491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina-Pritchard B., Lazou V., Zou J., Byron O., Abad M. A., Rappsilber J., Heun P., Jeyaprakash A. A., Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1. EMBO J. 39, e103234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali-Ahmad A., Bilokapic S., Schafer I. B., Halic M., Sekulic N., CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail. EMBO Rep. 20, e48913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zakeri B., Fierer J. O., Celik E., Chittock E. C., Schwarz-Linek U., Moy V. T., Howarth M., Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fachinetti D., Han J. S., McMahon M. A., Ly P., Abdullah A., Wong A. J., Cleveland D. W., DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell 33, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Cao S., Zhou K., Zhang Z., Luger K., Straight A. F., Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Mol. Biol. Cell 29, 751–762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Killinger K., Böhm M., Steinbach P., Hagemann G., Bluggel M., Janen K., Hohoff S., Bayer P., Herzog F., Westermann S., Auto-inhibition of Mif2/CENP-C ensures centromere-dependent kinetochore assembly in budding yeast. EMBO J. 39, e102938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan D., Klare K., Petrovic A., Take A., Walstein K., Singh P., Rondelet A., Bird A. W., Musacchio A., CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. eLife 6, e23352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A. E., Bai Y., Subramaniam S., Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science 359, 339–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allu P. K., Dawicki-McKenna J. M., Van Eeuwen T., Slavin M., Braitbard M., Xu C., Kalisman N., Murakami K., Black B. E., Structure of the human core centromeric nucleosome complex. Curr. Biol. 29, 2625–2639.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ariyoshi M., Makino F., Watanabe R., Nakagawa R., Kato T., Namba K., Arimura Y., Fujita R., Kurumizaka H., Okumura E. I., Hara M., Fukagawa T., Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C. EMBO J. 40, e105671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hori T., Amano M., Suzuki A., Backer C. B., Welburn J. P., Dong Y., McEwen B. F., Shang W. H., Suzuki E., Okawa K., Cheeseman I. M., Fukagawa T., CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–1052 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Hinshaw S. M., Harrison S. C., The structural basis for kinetochore stabilization by Cnn1/CENP-T. Curr. Biol. 30, 3425–3431.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z., Bellini D., Barford D., Crystal structure of the Cenp-HIKHead-TW sub-module of the inner kinetochore CCAN complex. Nucleic Acids Res. 48, 11172–11184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basilico F., Maffini S., Weir J. R., Prumbaum D., Rojas A. M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., Krenn V., Massimiliano L., Valencia A., Vetter I. R., Herzog F., Raunser S., Pasqualato S., Musacchio A., The pseudo GTPase CENP-M drives human kinetochore assembly. eLife 3, e02978 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishino T., Takeuchi K., Gascoigne K. E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I. M., Fukagawa T., CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., Westermann S., CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14, 604–613 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Nishino T., Rago F., Hori T., Tomii K., Cheeseman I. M., Fukagawa T., CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32, 424–436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T., Westermann S., A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32, 409–423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara M., Ariyoshi M., Okumura E. I., Hori T., Fukagawa T., Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat. Cell Biol. 20, 1378–1388 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Rago F., Gascoigne K. E., Cheeseman I. M., Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25, 671–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S., Yu H., Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208, 181–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welburn J. P., Vleugel M., Liu D., Yates J. R. III, Lampson M. A., Fukagawa T., Cheeseman I. M., Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38, 383–392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akiyoshi B., Nelson C. R., Biggins S., The aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics 194, 785–789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fachinetti D., Folco H. D., Nechemia-Arbely Y., Valente L. P., Nguyen K., Wong A. J., Zhu Q., Holland A. J., Desai A., Jansen L. E., Cleveland D. W., A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki A., Badger B. L., Wan X., DeLuca J. G., Salmon E. D., The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev. Cell 30, 717–730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hori T., Shang W. H., Takeuchi K., Fukagawa T., The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200, 45–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drinnenberg I. A., Akiyoshi B., Evolutionary lessons from species with unique kinetochores. Prog. Mol. Subcell. Biol. 56, 111–138 (2017). [DOI] [PubMed] [Google Scholar]
- 63.van Hooff J. J., Tromer E., van Wijk L. M., Snel B., Kops G. J., Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, 1559–1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alex A., Piano V., Polley S., Stuiver M., Voss S., Ciossani G., Overlack K., Voss B., Wohlgemuth S., Petrovic A., Wu Y., Selenko P., Musacchio A., Maffini S., Electroporated recombinant proteins as tools for in vivo functional complementation, imaging and chemical biology. eLife 8, e48287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arimura Y., Shirayama K., Horikoshi N., Fujita R., Taguchi H., Kagawa W., Fukagawa T., Almouzni G., Kurumizaka H., Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 4, 7115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitra S., Srinivasan B., Jansen L. E. T., Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control. J. Cell Biol. 219, e202005099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jansen L. E., Black B. E., Foltz D. R., Cleveland D. W., Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuh M., Lehner C. F., Heidmann S., Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17, 237–243 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Mendiratta S., Gatto A., Almouzni G., Histone supply: Multitiered regulation ensures chromatin dynamics throughout the cell cycle. J. Cell Biol. 218, 39–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zasadzińska E., Huang J., Bailey A. O., Guo L. Y., Lee N. S., Srivastava S., Wong K. A., French B. T., Black B. E., Foltz D. R., Inheritance of CENP-A nucleosomes during DNA replication requires HJURP. Dev. Cell 47, 348–362.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunleavy E. M., Almouzni G., Karpen G. H., H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus 2, 146–157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thakur J., Henikoff S., CENPT bridges adjacent CENPA nucleosomes on young human α-satellite dimers. Genome Res. 26, 1178–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., Tachiwana H., Hori T., Kurumizaka H., Fukagawa T., The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 42, 1644–1655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ando S., Yang H., Nozaki N., Okazaki T., Yoda K., CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22, 2229–2241 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Henikoff S., Furuyama T., The unconventional structure of centromeric nucleosomes. Chromosoma 121, 341–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Furuyama T., Henikoff S., Centromeric nucleosomes induce positive DNA supercoils. Cell 138, 104–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diaz-Ingelmo O., Martinez-Garcia B., Segura J., Valdes A., Roca J., DNA topology and global architecture of point centromeres. Cell Rep. 13, 667–677 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Logsdon G. A., Barrey E. J., Bassett E. A., DeNizio J. E., Guo L. Y., Panchenko T., Dawicki-McKenna J. M., Heun P., Black B. E., Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208, 521–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Pan D., Brockmeyer A., Mueller F., Musacchio A., Bange T., Simplified protocol for cross-linking mass spectrometry using the MS-cleavable cross-linker DSBU with efficient cross-link identification. Anal. Chem. 90, 10990–10999 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Iacobucci C., Götze M., Ihling C. H., Piotrowski C., Arlt C., Schäfer M., Hage C., Schmidt R., Sinz A., A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein-protein interactions. Nat. Protoc. 13, 2864–2889 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabg1037/DC1