Abstract
Seasonal influenza vaccination elicits a diminished adaptive immune response in the elderly, and the mechanisms of immunosenescence are not fully understood. Using Ig-Seq, we found a marked increase with age in the prevalence of cross-reactive (CR) serum antibodies that recognize both the H1N1 (vaccine-H1) and H3N2 (vaccine-H3) components of an egg-produced split influenza vaccine. CR antibodies accounted for 73% ± 18% of the serum vaccine responses in a cohort of elderly donors, 65% ± 15% in late middle-aged donors, and only 13% ± 5% in persons under 35 years of age. The antibody response to non-HA antigens was boosted by vaccination. Recombinant expression of 19 vaccine-H1+H3 CR serum monoclonal antibodies (s-mAbs) revealed that they predominantly bound to non-HA influenza proteins. A sizable fraction of vaccine-H1+H3 CR s-mAbs recognized with high affinity the sulfated glycans, in particular sulfated type 2 N-acetyllactosamine (Galβ1-4GalNAcβ), which is found on egg-produced proteins and thus unlikely to contribute to protection against influenza infection in humans. Antibodies against sulfated glycans in egg-produced vaccine had been identified in animals but were not previously characterized in humans. Collectively, our results provide a quantitative basis for how repeated exposure to split influenza vaccine correlates with unintended focusing of serum antibody responses to non-HA antigens that may result in suboptimal immunity against influenza.
Keywords: Infectious disease, Vaccines
Keywords: Adaptive immunity, Influenza
Introduction
Influenza is a serious global health threat causing nearly 5 million severe cases of illness and 650,000 deaths annually and particularly affecting individuals aged 65 years or older (1, 2). Among different types of influenza viruses, types A and B currently cause seasonal outbreaks in humans, yet influenza A viruses (IAVs) pose a greater threat of global pandemic, given their zoonotic potential (3, 4). IAVs consist of 8 negative-strand RNA segments that encode more than 10 proteins including nucleoprotein (NP), matrix protein 1 (M1), and matrix protein 2 (M2) in addition to the highly variable surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) (5, 6). The antigenic properties of HA and NA, which constitute the most antigenically variable and immunogenic proteins in the virus, further classify IAV into group 1 (e.g., H1N1) and group 2 (e.g., H3N2) (7). Antibodies against HA, and to a lesser extent against NA (8–10), are the most widely used correlate of protective influenza immunity (11–14). In addition to HA and NA, the 23-aa ectodomain of M2 (M2e) is the third and least abundant surface-accessible polypeptide of the virus, and animal studies as well as limited clinical data with an anti-M2e mAb indicated that antibodies against M2e can confer protection against influenza challenge, most likely in an Fc-dependent manner (15–17). Internal influenza virus proteins, specifically NP (18, 19) and M1 (20), may also have some contribution to protective adaptive immunity primarily via T cell–dependent mechanisms (21–23), and for this reason, whether or not they should be included in vaccine candidates continues to be debated (24, 25). Currently, HA serves as the antigenic component of seasonal influenza vaccines (26).
The majority of influenza deaths occur within the 65-and-older demographic despite higher rates of vaccination (~60%) (27). It is well established that the induction of protective immunity to influenza, as well as to other vaccines, is lower among older adults (28, 29). At the serological level, the poorer efficacy of influenza vaccination manifests as a decreased abundance of vaccine-specific antibodies and lower neutralization titers against the vaccine strains (30–32), which are often concomitant with a greater binding breadth toward divergent HAs (33, 34). Several of the molecular underpinnings of immunosenescence in the context of influenza vaccination have been reported, including: (a) contraction of the naive repertoire; (b) a decline in activation-induced cytidine deaminase and also in somatic hypermutation (SHM) of the entire peripheral B cell repertoire; (c) a decline in the number of germinal centers (at least in animal models); and (d) increased frequencies of anergic or dysfunctional B cells including age-associated B cells (33, 35–39). In a recent study by Wilson and colleagues (40), analysis of approximately 1000 antibody sequences encoded by peak plasmablasts revealed a significantly higher rate of intraclonal diversification in young donors. In contrast, antibodies secreted by plasmablasts from the elderly appeared to arise predominantly through a recall response of memory B cells targeting highly conserved epitopes. Of note, our analysis of the molecular composition of the serological repertoire over 5 years of repeated vaccinations was also consistent with an increased prevalence or enhanced “imprinting” of antibodies against the more conserved stem region of HA (41).
Here, we compared the IgG serological repertoire in a cohort of young, late middle-aged, and elderly participants following vaccination with a split, inactivated egg-based vaccine (Fluzone). We found that the IgG response to the vaccine became increasingly dominated by highly expanded antibody clonotypes that were cross-reactive (CR) between the group 1 and group 2 vaccine components, which, on average, accounted for more than 70% of the responses in the elderly donors. Analysis of CR serum monoclonal antibodies (s-mAbs) revealed that they overwhelmingly recognized conserved, non-HA influenza vaccine antigens that included internal viral proteins (NP and M1) and, surprisingly, a significant fraction of high-affinity s-mAbs bound to sulfated glycans that are prevalent on egg-produced proteins.
Results
Age-dependent features of the influenza A virus vaccine–reactive serum IgG repertoire after vaccination.
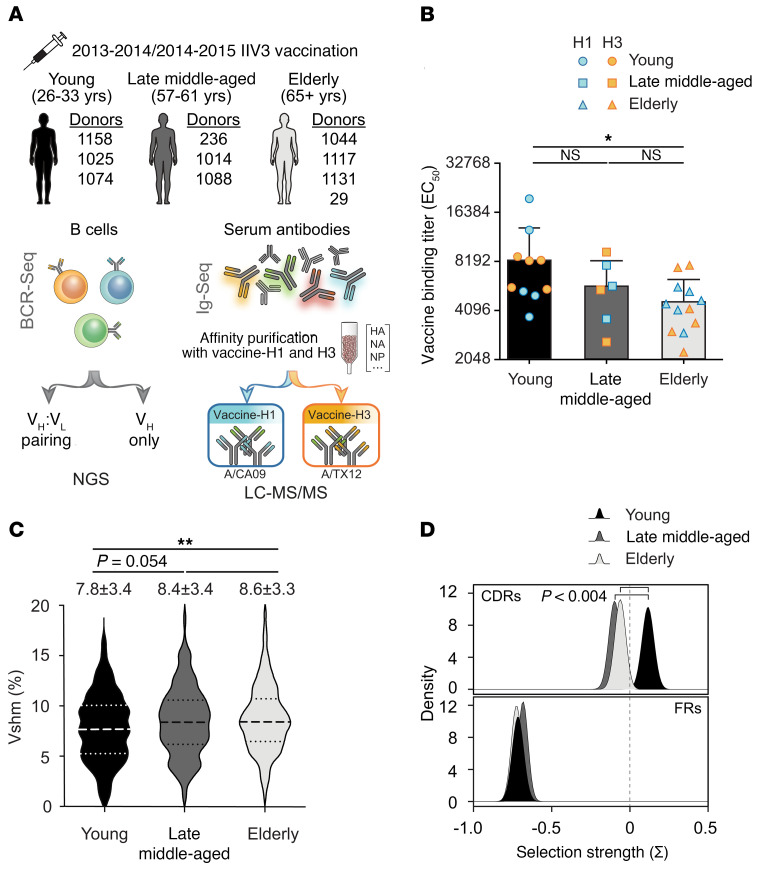
We used the Ig-Seq experimental pipeline to delineate the molecular features of the serum IgG repertoire reactive to each of the 2 inactivated influenza A virus (IAV) vaccine components as a function of age. A cohort of 10 donors belonging to 3 age groups (young: 26–33 years old; late middle-aged: 57–61 years old; and the elderly: ≥65 years of age) received trivalent inactivated influenza vaccine (IIV3; Fluzone) based on the CDC’s age stratification (18–49, 50–64, ≥65 years old) (42). The relatively limited number of participants studied here was due to the labor- and time-intensive nature of Ig-Seq experiments, which require repertoire analysis with manual curation of protein spectral matches together with B cell receptor sequencing (BCR-Seq) (43). The donors were selected from a cohort of 502 individuals who received either the 2013–2014 or 2014–2015 vaccine from the Nuñez et al. study (44), based on the availability of post-vaccination PBMCs and sufficient amounts of sera for Ig-Seq analyses. Four of the donors were vaccinated in both the 2013–2014 and 2014–2015 seasons. All 3 components of the IIV3, namely H1N1 A/California/7/2009 (vaccine-H1), H3N2 A/Texas/50/2012 (vaccine-H3), and B/Massachusetts/2/2012, were identical in both seasons (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI148763DS1). Blood samples were collected 7–9 days (designated D7) and 21–28 days (designated D21) after vaccination to capture the peak peripheral plasmablasts and peak serum titers, respectively. Consistent with earlier observations (30, 31), we observed a statistically significant decrease in D21 serum IgG binding titers against vaccine-H1 and vaccine-H3 when comparing the elderly group with the young group (Figure 1B). With 1 exception (donor 1131, 69 years of age), all other donors were seropositive (hemagglutination inhibition [HAI] titer ≥40) for either the H1N1 or H3N2 strain included in the vaccine or for both, while showing a different breadth for historical strains (Supplemental Figure 1A; donor 1131, although seronegative for the vaccine strains, was seropositive for historical strains, especially those of H3N2.
Figure 1. Age-dependent features of serological repertoire to the influenza A vaccine components.
(A) Experimental design. Ten donors were vaccinated in the 2013–2014 and/or 2014–2015 seasons with Fluzone trivalent inactivated influenza vaccines (IIV3). Peripheral blood samples were collected on D7 and D21 after vaccination. Serum IgG was split and affinity purified against vaccine-H1 and the vaccine-H3 components independently, processed into peptides, and then run using LC-MS/MS. (B) Post-vaccination (D21) serum binding titer (EC50) against vaccine-H1 and vaccine-H3 by age group. The mean ± SD is shown for each group (n = 10, n = 6, n = 12), with each data point representing the mean EC50. *P ≤ 0.05, by Kruskal-Wallis test followed by Dunn’s post-hoc test. (C) Vshm rates of all identified vaccine-reactive clonotypes. Dotted lines represent the 25th and 75th percentiles, and dashed lines indicate the median. The mean ± SD for each group is shown (n = 416, n = 242, n = 411). **P ≤ 0.01, by Kruskal-Wallis test followed by Dunn’s post-hoc test. (D) Estimated selection pressure imposed on complementary determining regions (CDRs) and framework regions (FRs) of the identified vaccine-reactive antibody sequences in different age groups using BASELINe. P < 0.004 by binomial test.
We determined the clonotypic composition of the serum antibodies against vaccine-H1 and vaccine-H3 by performing 2 separate affinity pull-down assays with the respective immobilized vaccine component and analysis via liquid chromatography with tandem mass spectrometry (LC-MS/MS) (41, 45). Mass spectra were assigned by searching against donor-specific databases obtained by high-throughput sequencing, and single-cell sequencing was performed in parallel to identify heavy- and light-chain pairing (VH:VL pair) for recombinant antibody expression (see Methods). We first analyzed the molecular features of the IAV vaccine–reactive serum antibodies. Recent reports indicate that the variable heavy-chain (VH) SHM (Vshm) rates of total plasmablasts (i.e., not antigen sorted) do not change with age, whereas those of influenza-reactive plasmablasts decrease (33, 40). Interestingly, we observed lower Vshm of the IAV vaccine–reactive serum antibodies in the young group (7.8% ± 3.4%) relative to the elderly 65 group (8.6% ± 3.3%, P = 0.002) (Figure 1C). BASELINe, a Bayesian-based framework for quantifying selection from somatic mutation patterns (46), suggested higher antigen-driven selection in the young group among the vaccine-reactive serum antibodies (Figure 1D). This model calculates the likelihood of the observed versus the expected, normalized frequency of nucleotide mutations in VH sequences, allowing the estimation of selection pressure among groups of VH sequences. In other words, the BASELINe analysis revealed that, while vaccine-induced serum antibodies accumulated a higher number of mutations in older donors, antigen-driven selection appeared to be stronger among individuals in the young group.
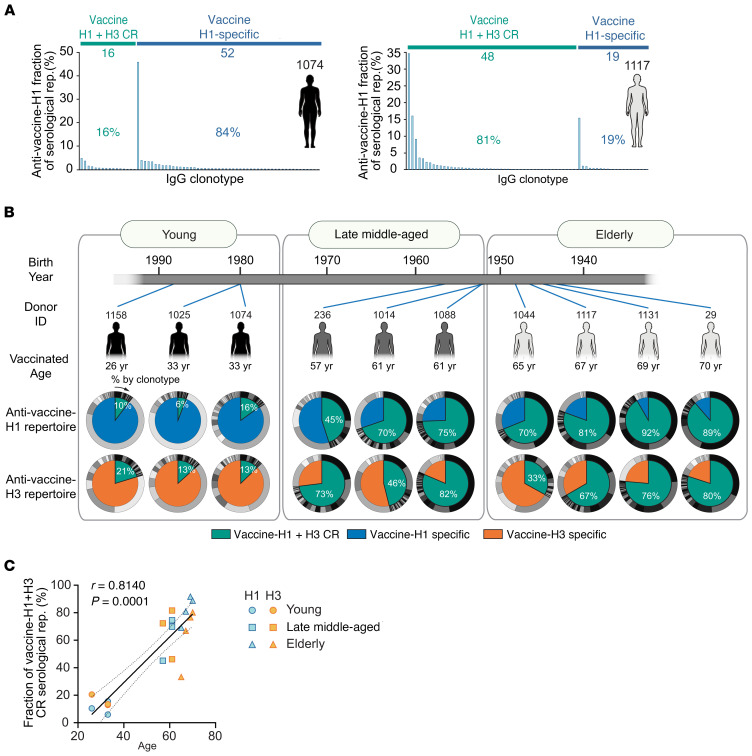
Clonal composition histograms for 2 representative donors, 33 and 67 years of age, are shown in Figure 2A. In concordance with previous observations, the serum repertoire following vaccination (41, 45, 47, 48) was dominated by a small number of clonotypes, with the most prevalent clonotype accounting for greater than 18% of the observed serum responses to the vaccine. We defined IgG clonotypes detected in both eluates from vaccine-H1 and vaccine-H3 affinity pull-down assays as vaccine-H1+H3 CR antibodies (i.e., they bound to both vaccine-H1 and vaccine-H3). Those enriched only in vaccine-H1 or vaccine-H3 eluates were defined as vaccine-H1 or vaccine-H3 specific (Supplemental Data File 1). In the older donors, the serum response to the IAV vaccine was dominated by vaccine-H1+H3 CR antibodies (Figure 2B). Overall, vaccine-H1+H3 CR antibodies accounted for 13.1% ± 4.9% (mean ± SD) in the young group, 65.0% ± 15.4% (mean ± SD) in the late middle-aged group, and 73.4% ± 18.3% (mean ± SD) in the elderly group. The similar levels of CR serum antibodies between the late middle-aged and elderly participants, as contrasted to the young participants, is not surprising, given the proximity of the ages of individuals in the late middle-aged and elderly groups. (Figure 2C). The increased prevalence of vaccine-H1+H3 CR antibodies in the serum repertoire of the older donors correlated with a higher frequency of CR clonotypes (i.e., an increase in the diversity of vaccine-H1+H3 CR antibodies) (Supplemental Figure 1, B and C). Vaccination with the same IIV3 in the following season was accompanied by an increase in the fraction of vaccine-H1+H3 CR antibodies, with young donors showing a more pronounced antibody expansion (Supplemental Figure 1D).
Figure 2. Age-dependent increase in the vaccine-H1+H3 CR serological repertoire.
(A) Clonal composition of the anti–vaccine-H1 serum antibody repertoire (rep.) in a representative young donor and a representative elderly donor. Vaccine-H1+H3 CR: antibody clonotypes were detected in both the anti–vaccine-H1 and anti–vaccine-H3 repertoires. Vaccine-H1 specific: antibody clonotypes were detected only in the anti–vaccine-H1 repertoire. (B) Vaccine-H1+H3 CR and monovalent vaccine-specific serum response in all donors. Inner wheel: percentage of the serum repertoire of vaccine-H1+H3 CR (green) and vaccine-H1 specific-antibody clonotypes (blue) or vaccine-H3 specific-antibody clonotypes (orange). Outer wheel: relative abundance of each clonotype estimated from the LC-MS/MS peak area. (C) Scatter plot showing Spearman’s correlation coefficient (and 95% CI of the regression line) between the fraction of the vaccine-H1+H3 CR serum repertoire and age.
Biochemical and functional characterization of anti-HA s-mAbs.
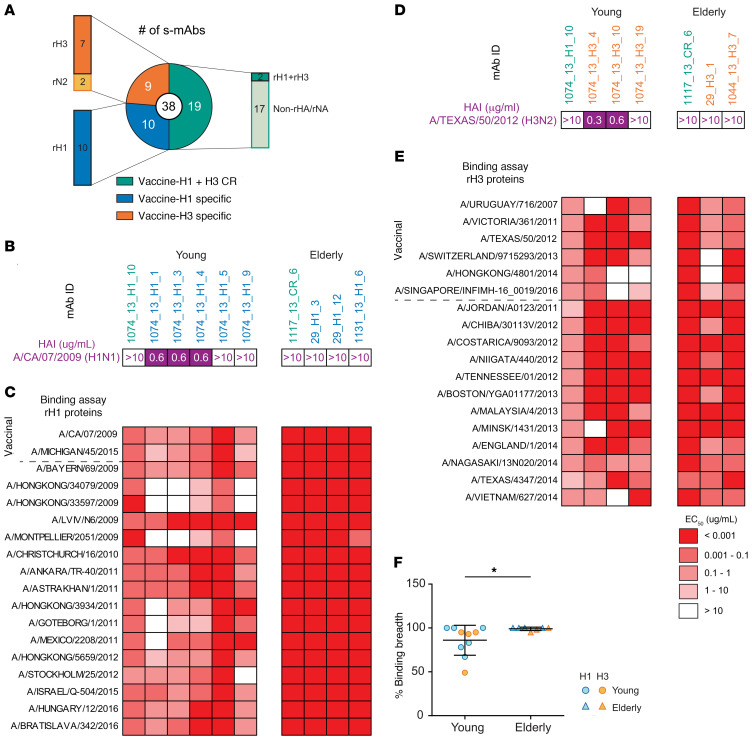
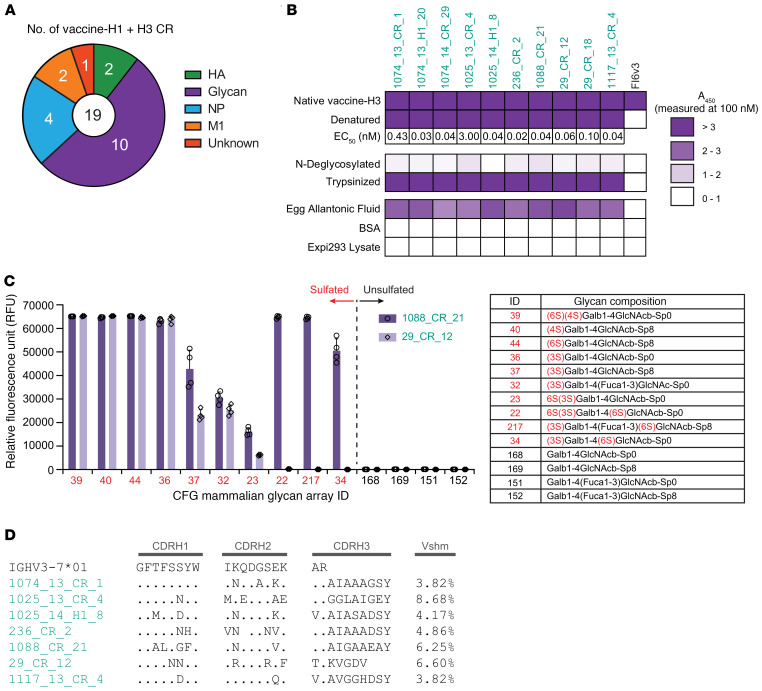
We recombinantly expressed a total of 38 s-mAbs identified from 8 participants to validate their binding specificities and characterize in greater depth (Supplemental Table 2). These were prioritized on the basis of their abundance in serum and availability of VH:VL pairs. Of the 19 verified vaccine-H1– or vaccine-H3–specific s-mAbs, 17 of 19 s-mAbs were further shown to bind to the respective recombinant HA (rHA), and 2 of 19 bound to recombinant NA. Among the vaccine-H1+H3 CR s-mAbs, 2 of 19 recognized both group 1 and group 2 rHAs from the vaccine strains, whereas 17 of 19 s-mAbs bound to other components in the IAV vaccine, which are further described below. In summary, 19 of the 38 s-mAbs were HA specific, while the other 19 bound to other components in the vaccine (Figure 3A).
Figure 3. Characterization of rHA-binding s-mAbs identified from different donors.
(A) Pie chart summarizing the binding specificity of recombinant s-mAbs. (B–E) HAI titer (B and D) and binding specificity (C and E) of anti-HA s-mAbs against rHAs from contemporary H1 strains or H3 strains are shown. (F) Comparison of the binding breadth of s-mAbs from the young and elderly cohorts. *P ≤ 0.05, by 2-tailed Mann-Whitney U test. The breadth is defined as the number of strains with detectable binding (EC50 ≤10 μg/mL) divided by the total number of tested strains. The mean ± SD is shown for each group (n = 10, n = 7).
We examined the binding breadth of anti-rHA s-mAbs from the young and elderly groups (9 and 6 s-mAbs, respectively) using a large panel of rHAs from contemporaneous strains (H1: 18 strains, H3: 41 strains) (Figure 3, B–E, and Supplemental Figure 2A). In agreement with earlier findings (33, 34, 40), serum antibodies from the elderly group had greater HA binding breadth than did those from the young group, with multiple s-mAbs from the elderly group recognizing the entire panel of the rHAs tested (Figure 3F).
Of note, 2 recombinant s-mAbs, 236_H1_1 and 236_H1_3 (s-mAb nomenclature: donor_vaccination year (if applicable)_Ig-Seq specificity_rank-ordered abundance in serum repertoire), from late middle-aged donor 236 were HA specific and corresponded to the donor’s top first- and third-most abundant vaccine-H1–specific serum antibodies (accounting for 26% and 2% of the serum responses to vaccine-H1, respectively) (Supplemental Table 2). Both s-mAbs showed substantial breadth of HA binding to HA from historical strains dating back to 1947, before the donor was born (Supplemental Figure 2B). Although neither antibody had HAI activity, both showed neutralization activity against the vaccinal H1 strain A/CA/7/2009 in microneutralization assays. Interestingly, 236_H1_3 neutralized all the strains tested, including A/Fort Monmouth/1/1947 (Supplemental Figure 2C). These 2 antibodies, along with 2 other antibodies isolated from 2 elderly donors, shared the characteristic IGHV1-69 anti-stem signature reported previously (Supplemental Figure 2D) (49). The 2 antibodies from the elderly donors competed for binding with a well-characterized stem binding FI6 (50) (Supplemental Figure 2E).
A substantial fraction of vaccine-H1+H3 CR s-mAbs recognize rare sulfated glycans.
Only 2 of the 19 vaccine-H1+H3 CR s-mAbs — 1074_13_H1_10 and 1117_13_CR_6 — bound to both group 1 and group 2 HAs, as discussed above. Of the remaining 17 CR s-mAbs, 4 bound to NP and 2 to M1 (Figure 4A). The elicitation of non-HA antibodies in response to the vaccine was not unexpected, as NP and other conserved antigens are well known to be present in licensed split, inactivated influenza vaccines including Fluzone (51). Vaccinal influenza strains are made by inserting HA and NA genes of the identified strains using reverse genetics or reassortment technology into egg-adapted A/Puerto Rico/8/34 for a high production yield (52). In other words, internal proteins from the A/Puerto Rico/8/34 strain are included in both vaccine-H1 and vaccine-H3. The anti-NP antibodies showed nanomolar KD by biolayer interferometry (BLI), whereas the 2 M1-specific antibodies exhibited weak affinity (Supplemental Figure 3, A and B). We were unable to confirm the binding target of 29_CR_2, as it did not bind the recombinant HA, NP, or M1 proteins tested, however, we noticed that it shares 84.3% aa sequence identity with the previously reported human anti-M2e antibody 3136_G05 (15) (Supplemental Figure 3C).
Figure 4. Characterization of s-mAbs that bind to sulfated glycans from egg components.
(A) Analysis showed that 17 of 19 vaccine-H1+H3 CR s-mAbs bound to components other than HA. (B) Binding profile of putative avian glycan–specific antibodies. ELISA assays were performed using 100 nM recombinant s-mAbs with their respective antigens. Denaturation was performed by boiling with SDS, and N-deglycosylation was from treatment with PNGase F after denaturation. Trypsinization was performed after reduction and alkylation. The EC50 was determined with denatured vaccine-H3 as the antigen. A450, absorbance at 450 nm. (C) Glycan array binding specificity of 2 s-mAbs with the glycan-targeting IGHV3–7 signature. RFU values for the top 10 glycans (red) and unsulfated versions (black) are shown with the mean ± SD for 4 replicates. (D) Sequence alignments of IGHV3–7 putative avian glycan–specific antibodies from different donors.
The remaining 10 of the 19 vaccine-H1+H3 CR s-mAbs did not recognize rHA or any of the internal influenza proteins tested but bound tightly to denatured (boiled and reduced) vaccine-H3 with a low- to sub-nanomolar EC50 by ELISA (Figure 4B). We subjected the denatured vaccine-H3 to trypsin treatment and then purified the resulting peptides; this treatment did not reduce binding for any of the 10 s-mAbs. However, this binding was abrogated following vaccine-H3 N-deglycosylated with peptide:N-glycosidase F (PNGase F), indicating that these antibodies recognize glycans or glycopeptides. We found no evidence of polyspecificity with BSA or with human cell lysates. Additionally, we detected no binding by ELISA with rHAs expressed in either Sf9 or HEK293 cells, suggesting that these s-mAbs recognize antigens that are more abundant in egg-prepared vaccine and not present in rHA expressed in insect or human cells. In support of this hypothesis, these 10 s-mAbs were found to bind to specific pathogen–free (SPF) and chemically deactivated chicken allantoic fluid. Allantoic fluid is the physiological equivalent of the mammalian amniotic fluid in birds and contains a high concentration of egg-derived glycoproteins but no influenza proteins (Figure 4B). For precise identification of the glycan structures recognized by these antibodies, we used a glycan array containing 585 synthetic glycans made available by the Consortium of Functional Glycomics (CFG) (53) and found that they targeted sulfated type Galβ1-4GalNAcβ and its variants including (6S) (4S), (4S), (6S), and (3S) (Figure 4C and Supplemental Table 3). This glycoform modification was reported to be present on HA and NA from egg-produced influenza vaccines (54–58). Although antibodies targeting this glycoform on egg-produced influenza virus and in allantoic fluid had been generated in animals, to our knowledge, our study together with a study by Guthmiller. et al that appeared while this work was in press are the first reports of human mAbs against sulfated type Galβ1-4GalNAcβ (56, 57, 59, 60). Seven s-mAbs isolated from 6 different individuals that follow the same glycan binding pattern utilize the IGHV3–7 germline with a very short CDR-H3 (≤10 aa) and show relatively low-to-moderate Vshm with evidence of convergent aa substitutions occurring in CDR-H1 or CDR-H2 (Figure 4D), suggesting that they constitute a public clonotype.
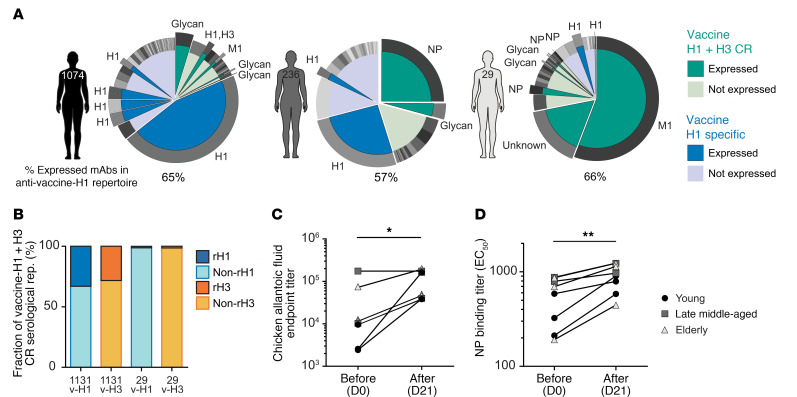
In total, we determined the binding specificity of the set of vaccine-H1+H3 CR s-mAbs analyzed and found that approximately 45% recognized influenza antigens (NP, M1, or conserved epitopes on HA), while the remainder bound to the sulfated glycans. Subsequently, we were able to assign the binding specificities for a significant portion of the overall antibody response in select donors. In donor 1074 (33 years of age), antibodies with known specificity comprised 65% of the observed serum repertoire to the vaccine-H1, and the response was dominated by anti-H1 HA antibodies with a small contribution from anti-glycan s-mAbs (Figure 5A, left). In a 57-year-old donor (donor 236), antibodies with known specificity accounted for 57% of the serum repertoire, with an anti-NP clone accounting for 25% of the total response (Figure 5A, middle). Finally, the repertoire of a 70-year-old donor (donor 29; 61% serum IgG specificity determined) was dominated by an anti-M1 antibody, which accounted for 56% of the response, with only a few anti-H1 HA clonotypes (Figure 5A, right), consistent with the data presented in Supplemental Figure 1, B and C.
Figure 5. Estimated contribution of non-HA response in the vaccine-induced serological repertoire.
(A) Representative examples of antigen specificities for expressed serum antibody clonotypes in the repertoire of a young donor (donor 1074), a late middle-aged donor (donor 236), and an elderly donor (donor 29). (B) Overall fraction of the estimated non-rHA repertoire within the vaccine-H1+H3 CR repertoire in 2 elderly donors (donors 1131 and 29). v-H1, anti–vaccine-H1 repertoire; v-H3, anti–vaccine-H3 repertoire. (C) Endpoint binding titer in 8 donors before and after vaccination against the SPF, chemically inactivated chicken embryo allantoic fluid (n = 8). *P ≤ 0.05, by 2-tailed Wilcoxon signed-rank test. (D) Pre- and post-vaccination serum binding titer (EC50) against NP (n = 8). The endpoint titer was determined as the maximum serum dilution that showed a higher signal than the average + (3 × SD) of 4 blank wells by ELISA. **P ≤ 0.01, by 2-tailed Wilcoxon signed-rank test.
To further support the finding that the serum response to the vaccine in the elderly was dominated by antibodies that react to non-HA proteins and to sulfated glycan antigens, we compared the serum antibody repertoire with vaccine-H1 or vaccine-H3 and determined the repertoire following affinity purification using rHA produced in insect cells by performing Ig-Seq on 2 donors from the elderly group. Antibody clonotypes that bound to the vaccine but not to insect cell–expressed rHA (i.e., antibodies that target non-HA components) accounted for between 67% (donor 1131) and an overwhelming 91% of the vaccine-H1+H3 CR response (donor 29) (Figure 5B). It is important to note, however, that even though in donor 29 the serum response to the vaccine was dominated by antibodies against proteins other than HA, the donor was still seropositive for H3 and had significant HAI breadth. This finding implies that the small fraction of the serum response to the vaccine that is HA specific may nonetheless be sufficient to confer significant HAI activity. Vaccination also boosted the serum titer against both glycans (i.e., allantoic fluid) and against NP (Figure 5, C and D).
Discussion
In an earlier repertoire profiling study of young adults following 2011–2012 Fluzone vaccination, we reported that more than 75% of the serum response was rHA specific, whereas the rest targeted non-HA antigens present in the vaccine (45). Here, we confirm an HA-focused response in a separate cohort of young donors who received the 2013–2014 Fluzone vaccine (Figure 2, A and B). Importantly, however, we found that the serum response to the vaccine in older adults was dominated by antibodies that bound to both the H1N1 and H3N2 vaccine components and primarily recognized conserved internal influenza proteins present in both vaccine components (Figures 4 and 5). Of note, the fraction of non-HA or NA in the serum response to IIV3 was higher than the fraction of such antibodies reported in an earlier analysis of peak plasmablasts following vaccination of an elderly cohort (40) (Figure 2A and Figure 5, A and B). Such disparity is not surprising, given the complex kinetics of plasmablast expansion following immunization and the fact that the day-21 post-vaccination serum response comprises antibodies contributed by both circulating plasmablasts and long-lived plasma cells (43, 47). The provenance of serum antibodies from long-lived plasma cells might also account for our observation of a higher rate of Vshm in the elderly. In contrast, earlier reports have suggested that antibodies encoded by day-7 post-vaccination plasmablasts from that age group have the same or lower Vshms than those from younger donors (33, 40). Nonetheless, in agreement with analyses of antibodies encoded by peripheral B cells isolated by single-cell cloning (40), we found evidence for higher antigen-driven selection in serum antibodies in the young group (Figure 1D).
Although the antibody response to internal influenza proteins such as NP, M1, and M2 is routinely observed following infection or vaccination (19, 61–63), to our knowledge, the dramatic increase in the fraction of the serum response to such antigens with age has not been previously reported. We now show that in the serum response, many such antibodies recognized with high affinity sulfated glycans, such as sulfated type Galβ1-4GalNAcβ, that are present on egg-produced HA (a component of the Fluzone and other egg-derived vaccines), but not on rHA expressed either in insect or human cells. These antibodies bound sulfated glycans with high avidity, and, unlike many anti-glycan antibodies, they were not polyspecific, as they showed no reactivity to mammalian cell lysates or BSA (Figure 4B). Additionally, they used predominantly IGHV3–7, had short CDR-H3 regions, and displayed relatively low Vshm, indicating that these sulfated glycan–binding antibodies probably belonged to public clonotypes. Along similar lines, it is noteworthy that within the serum response, we observed evidence for both public antibodies and antibodies with stereotypic features, including, in addition to the aforementioned anti-glycan antibodies (Figure 4D), stereotypic antibodies against HA stem (Supplemental Figure 2, B–D). Joyce et al. (64) identified HA-neutralizing antibodies from multiple donors that had convergent sequence motifs, and, separately, Boyd and coworkers (65) noted convergent VDJ rearrangements in the peripheral B cell repertoire following influenza vaccination. Our data now suggest that convergent antibodies against various influenza vaccine antigens are also readily detectable in the circulation and therefore underscore the notion that such antibodies are physiologically relevant. It should be noted that the B cells that encode public and stereotypical serum antibodies must not only derive from naive cells with the same or very similar rearrangements (which recent studies indicate occur at a small but significant frequency within the peripheral repertoire; refs. 66, 67), but they must also be selected for significant expansion and differentiation into antibody-secreting cells, as required for the production of detectable levels of circulating antibodies. Although we had noted the presence of 1 putative public serum antibody clonotype against tetanus in an earlier study (47), the results presented here suggest that public antibodies may in fact be relatively common in the serological repertoire.
There is some evidence that antibodies against the less antigenically variable influenza proteins (i.e., NP and M2e) may be able to contribute to the protection via Fc effector mechanisms (i.e., ADCP and ADCC) in the mouse model of lethal influenza challenge (68, 69), but whether such antibodies are physiologically relevant in humans remains to be determined. Of note, non–surface-exposed influenza proteins elicit strong CD4+ and CD8+ T cell responses that may be critical to protection (28, 51, 70). Therefore, more studies will be required to determine the impact of the increased serological response to non-HA/NA targets in the elderly on protection from infection. Natural infection has been reported to elicit stronger responses to non-HA influenza proteins compared with vaccination (34, 71, 72). Interestingly, peak plasmablasts with specificities for non-HA proteins are observed following vaccination either with an egg-based split vaccine (Fluzone) or an egg-based subunit vaccine (Fluvirin) (40). The expansion of non–HA-reactive plasmablasts in the latter case may either be in response to small amounts of residual internal influenza proteins (51) or a consequence of bystander B cell stimulation, as reported recently (73). It needs to be determined whether antibodies against sulfated glycans are boosted following vaccination with non–egg-based vaccines such as Flublok (insect) and FLUCELVAX (Madin-Darby canine kidney [MDCK]), acknowledging that MDCK cells that have also been reported to express sulfated type Galβ1-4GalNAcβ glycans (74). Although the immunological roles of the anti-glycan antibodies are to be determined, it is highly unlikely that they contribute to protection against human influenza viruses, which of course lack avian glycans. Overall, our results provide quantitative information on the degree to which the serological repertoire to the seasonal influenza vaccine in older adults is dominated by antibodies against conserved non-HA/NA antigens, including sulfated glycans that are prevalent in eggs. The focusing of a larger fraction of the repertoire in the older adults to non-HA antigens following a boost immunization may be contributing to the diminished adaptive immune response in the elderly. Along those lines, it would be of interest to compare and contrast in detail the breadth and quality of HA antibodies in older adults following a subunit vaccine (Flublok), which should not boost non–HA-reactive antibodies relative to inactivated vaccines. Findings from such studies will inform the design of more effective vaccine strategies for the elderly.
Methods
Study volunteers.
Volunteers who were vaccinated in the 2013–2014 (year 1) and/or 2014–2015 (year 2) seasons were included in the study during which the composition of vaccine remained identical (trivalent formulation of A/California/7/2009 [H1N1], A/Texas/50/2012 (H3N2), B/Massachusetts/2/2012 [Yamagata lineage]). Volunteers between the ages of 18 and 64 years received the standard dose (15 μg) or intradermal dose (9 μg) of inactivated influenza Fluzone (Sanofi Pasteur) vaccine (IIV-SD or IIV-ID), and participants over 65 years of age were offered either IIV-SD or a high dose (60 μg) of Fluzone IIV (IIV-HD) (Supplemental Table 1). For this study, 10 volunteers of different ages were grouped into 3 age cohorts: young (26–33 years old), late middle-aged (57–61 years old), and elderly (65 years or older). Two participants from the young cohort and 2 from the elderly cohort were vaccinated in both seasons. Blood was collected from the participants before vaccination (D0), 7–9 days after vaccination (D7: peak plasmablast), and 21–28 days after vaccination (D21: peak serum titer), and then aliquoted and stored at –150°C for future analysis. Blood samples were processed into PBMCs, and sera were processed as described below.
Library preparation and analysis for high-throughput sequencing of VH.
Total RNA was isolated from D7 (D21 for donor 236) post-vaccination PBMCs using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. To produce cDNA, 1 μg total RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific) and oligo-dT primer (Thermo Fisher Scientific) according to the manufacturer’s instructions. IgG VH transcripts were amplified using the FastStart High Fidelity PCR system (Roche) with gene-specific primers as previously described (75). V genes were sequenced using an Illumina MiSeq instrument (2 × 300 bp paired-end reads). Sequences were annotated using IMGT/HighV-QUEST, version 1.6.7 (76). IAV-reactive VH sequences were used to quantify the estimated selection pressure using the BASELINe framework, which statistically compares the estimated frequency of replacement mutations with the observed, normalizing intrinsic biases coming from germLine sequences (46). Sequences were first grouped into clonotypes using a Hamming distance of 0.1, and then the clonotypes were analyzed with calcBaseline, grouped by age with groupBaseline, and plotted with plotBaselineDensity from the SHazaM (version 0.1.11) package (77).
VH:VL-paired sequencing.
High-throughput paired heavy- and light-chain sequencing of D7 post-vaccination B cells was performed as previously described (78, 79). Briefly, single B cells were encapsulated in emulsion droplets using a custom flow-focusing apparatus. The droplets contained lysis buffer and poly(dT) conjugated magnetic beads to capture mRNA from single B cells. The magnetic beads with mRNA were collected and emulsified to undergo overlap extension reverse transcription PCR (RT-PCR), in which the VH and VL transcripts were physically linked and amplified by xenopolymerase (RTX) (80). VH:VL amplicons were extracted, amplified by nested PCR, and sequenced using the Illumina MiSeq instrument (2 × 300 bp paired-end reads). VH and VL regions were amplified separately for the recombinant expression of mAbs. For donor 236 and donor 1074 samples, VH:VL single-cell sequencing was additionally performed by Atreca Inc. utilizing FACS-based single-cell sorting into individual wells of microtiter plates (81).
Purification of total IgG from serum and subsequent digestion into F(ab′)2.
Each serum sample (1–2 mL) was diluted 1:1 with Dulbecco’s PBS (DPBS) (Gibco, Thermo Fisher Scientific) and passed through a 2 mL protein G agarose (Pierce, Thermo Fisher Scientific) affinity column 3 times in gravity mode. The column was washed with 10 column volume (cv) DPBS and eluted with 5 cv of 100 mM glycine-HCl (pH 2.7). The eluate was immediately neutralized with 1 mL of 1 M Tris-HCl (pH 8.0). Purified IgG was dialyzed into 20 mM sodium acetate (pH 4.5) overnight at 4°C and digested into F(ab′)2 with 1 mL immobilized pepsin resin (Pierce, Thermo Fisher Scientific) per 20 mg IgG for 6 hours on an inverter at 37°C. The digest was filtered with an Ultrafree-MC Centrifugal Filter Unit (MilliporeSigma) at 5000g for 2 minutes, and the filtrate was combined and adjusted to pH~7.
Antigen enrichment of F(ab′)2 and MS sample preparation.
IIVs or rHAs were immobilized on separate columns for the antigen-specific F(ab′)2 enrichment as previously described (45). Briefly, A/CA/07/2009 (H1N1) and A/TX/50/2012 (H3N2) vaccines were separately immobilized on N-hydroxysuccinimide–activated (NHS-activated) agarose resins (Pierce, Thermo Fisher Scientific) by overnight rotation at 4°C. For rHA columns, A/CA/07/2009 (rH1) and A/TX/50/2012 (rH3) expressed from the insect-baculovirus system (Hi-5) were provided by Stephen Harrison (Harvard Medical School, Boston, Massachusetts, USA) (82). The coupled agarose resins were washed with DPBS, and unreacted NHS groups were blocked with 1 M ethanolamine, pH 8.3, for 30 minutes at room temperature. The beads were further washed with DPBS and packed into 2 mL chromatography columns (Clontech). The columns were prewashed with 10 cv of 2 M urea and equilibrated with 20 cv DPBS.
The F(ab′)2 sample was divided into equal volumes and applied to the individual immobilized columns in gravity mode. Flow-throughs were collected, and the columns were washed with 10 cv DPBS and 10 cv diluted DPBS (1:2 in ddH2O). Antigen-enriched F(ab′)2 was eluted with 60 mM HCl (pH 1.7) in 0.75 mL fractions and neutralized with 1 M NaOH/Tris. Flow-through and elution fractions were assayed by indirect ELISA with the corresponding IIV, and the depletion of ELISA signal in each flow-through sample was evaluated. Elution fractions showing greater than 0.1 A450 were pooled and concentrated under vacuum to a volume of approximately 0.1 mL and desalted into double-distilled H2O (ddH2O) using a 0.5 mL Zeba spin column (Pierce, Thermo Fisher Scientific). The desalted eluate was further concentrated to 50 μL under vacuum.
For each enrichment, elution and flow-through samples were denatured in 50% (v/v) 2,2,2-trifluoroethanol (TFE), 50 mM ammonium bicarbonate, and 10 mM DTT at 60°C for 1 hour, and then alkylated by incubation with 32 mM iodoacetamide (MilliporeSigma) for 1 hour at room temperature in the dark. Alkylation was quenched by the addition of 20 mM DTT. Samples were diluted 10-fold with 50 mM ammonium bicarbonate and digested with trypsin (1:30 trypsin/protein) for 16 hours at 37°C. Formic acid was added to 1% (v/v) to quench the digestion, and the sample volume was reduced to approximately 100 μL under vacuum. Peptides were then bound to a C18 Hypersep SpinTip (Thermo Fisher Scientific), washed 3 times with 0.1% formic acid, and eluted with 60% acetonitrile and 0.1% formic acid. C18 eluate was dried under vacuum centrifugation and resuspended in 50 μL in 5% acetonitrile and 0.1% formic acid.
LC-MS/MS analysis.
Samples were analyzed by LC-MS/MS on a Dionex Ultimate 3000 RSLCnano uHPLC system (Thermo Fisher Scientific) coupled to an LTQ Orbitrap Fusion Tribrid or a Velos Pro mass spectrometer (Thermo Fisher Scientific). Peptides were first loaded onto an Acclaim PepMap RSLC NanoTrap column (Dionex, Thermo Fisher Scientific) prior to separation on a 75 μm × 15 cm Acclaim PepMap RSLC C18 column (Dionex, Thermo Fisher Scientific) using a 5%–40% (v/v) acetonitrile gradient over 120 or 245 minutes at 300 nL/min. Eluting peptides were injected directly into the mass spectrometer using a nano-electrospray source. The instruments were operated in a data-dependent mode with parent ion scans (MS1) collected at 120,000 or 60,000 resolution. Monoisotopic precursor selection and charge state screening were enabled. Ions with a charge of +2 or higher were selected for collision-induced dissociation fragmentation spectrum acquisition (MS2) in the ion trap. Dynamic exclusion was active with a 60 second or 45 second exclusion time for ions selected more than once or twice in a 60 second or 30 second window. Each sample was run 3 times to generate technical replicate data sets.
MS/MS data analysis.
Donor-specific protein sequence databases were constructed using the donor’s VH sequences with 2 or more reads and were then concatenated to a database of background proteins comprising non–donor-derived VL sequences (HD1) (47), a consensus human protein database (Ensembl 73, longest sequence per gene), and a list of common protein contaminants (MaxQuant). Donor-specific spectra were searched against the corresponding donor-specific databases using SEQUEST HT (Proteome Discoverer 1.4, Thermo Fisher Scientific). Searches considered fully tryptic peptides only, allowing up to 2 missed cleavages. A precursor mass tolerance of 5 ppm and a fragment-mass tolerance of 0.5 Da were used. Modifications of carbamidomethyl cysteine (static) and oxidized methionine (dynamic) were selected. High-confidence peptide-spectrum matches (PSMs) were filtered at a FDR below 1% as calculated by Percolator (q < 0.01, Proteome Discoverer 1.4, Thermo Fisher Scientific).
Iso/Leu sequence variants were collapsed into single peptide groups. For each scan, PSMs were ranked first by posterior error probability (PEP), then q value, and then finally based on XCorr. Only unambiguous top-ranked PSMs were kept; scans with multiple top-ranked PSMs (equivalent PEP, q value, and XCorr) were designated ambiguous identifications and removed. The average mass deviation (AMD) for each peptide was calculated as described before (83) using data from all injections and the peptides with an AMD of greater than 1.5 ppm were removed.
Peptide abundance was calculated from the extracted-ion chromatogram (XIC) peak area as described previously (47), using peak area values generated by the Precursor Ions Area Detector node in Proteome Discoverer. For each peptide, a total XIC area was calculated as the sum of all unique XIC areas of associated precursor ions, and the average XIC area across replicate injections was calculated for each sample. The peptide PSM was calculated as the sum of the aforementioned PSMs. For each antigen data set, the eluate and flow-through abundances were compared, and peptides with ≥10-fold higher signal in the elution sample were considered antigen specific.
Clonotype indexing and peptide-to-clonotype mapping.
VH sequences were grouped into clonotypes based on single-linkage hierarchical clustering as described previously (45, 47). Cluster membership required a 90% or higher identity across the CDR-H3 amino sequence as measured by Levenshtein distance. High-confidence peptides identified by MS/MS analysis were mapped to clonotype clusters, and peptides uniquely mapping to a single clonotype were considered “informative.” The abundance of each antibody clonotype was calculated by summing the XIC areas of the informative peptides mapping to 4 or more aa of the CDR-H3 region (or ≥3 aa mapping to the beginning of the CDR-H3, e.g., –CATK; i.e., CDR-H3 peptides). For each antibody clonotype, the most abundantly detected CDR-H3 peptide (in cases in which peptides from multiple somatic variants belonging to the same clonotype were detected) was used as a representative CDR-H3 sequence. Vshm rates for individual clonotypes were calculated by averaging the Vshm rates of all the VH sequences within each clonotype that contained the detected CDR-H3 sequences.
Clonotype abundance was calculated as the sum of the peptide abundances belonging to a clonotype. When multiple nonoverlapping tryptic peptides were available for a clonotype, 1 peptide was selected and used for the analysis to prevent double-counting. Clonotype abundances in elution samples were compared across antigens; clonotypes with ≥10-fold higher signal (abundance) for a single antigen were designated as specific (i.e., vaccine-H1 or vaccine-H3 specific), and clonotypes with less than a 10-fold difference or with PSMs of 3 or higher for both antigens were considered CR (e.g., vaccine-H1+H3 CR). Remaining clonotypes followed specificities of the antigen with a higher PSM. Binding specificities were further validated through recombinant expression and reclassified via ELISA. Among the 22 putative vaccine-H1– or vaccine-H3–specific clonotypes expressed as s-mAbs, 3 were found through ELISA to actually bind to both vaccine-H1 and vaccine-H3 and were reassigned as vaccine-H1+H3 CR. All 3 of these antibodies were present at a very low abundance (<2%) in the anti–vaccine-H1 repertoire, and their low abundance may have interfered with their detection in the vaccine-H3 affinity chromatography eluate because of the stochastic nature of data-dependent acquisition in LC-MS/MS (84).
Recombinant antibody synthesis, expression, and purification.
Variable heavy- and light-chain regions for select antibodies were codon optimized and synthesized as gBlocks (Integrated DNA Technologies) and cloned into a pcDNA3.4 vector (Invitrogen, Thermo Fisher Scientific) or a pTT5-based mammalian cell expression vector containing human IgG1 heavy- and κ or λ light-chain constant regions, respectively (41, 85). Heavy- and light-chain plasmids for each mAb were cotransfected into Expi293F or 293T cells (Thermo Fisher Scientific). After incubation for 5–6 days at 37°C with 8% CO2, the supernatant containing secreted antibodies was collected by centrifugation at 4000g for 15 minutes at 4°C. Supernatant was filtered with 0.22 μm syringe filters (Sartorius) and then passed over a column with 0.5 mL Protein G or A agarose resin (Pierce, Thermo Fisher Scientific) 3 times to ensure efficient capture. After washing the column with 20 cv DPBS, antibodies were eluted with 3 mL 100 mM glycine-HCl (pH 2.7) and immediately neutralized with 300 μL 1 M Tris-HCl (pH 8.0). Antibodies were buffer exchanged into DPBS using Amicon Ultra-30 centrifugal spin columns (MilliporeSigma).
ELISA.
Antigens for ELISA assays, in addition to IIVs (vaccine-H1, vaccine-H3) and rHAs, included recombinant NP, M1, NS1 (A/Puerto Rico/8/34/Mount Sinai H1N1 from Sino Biological), N1 (A/California/04/2009 H1N1pdm09 from BEI resources), N2 (A/Texas/50/2012 H3N2 from Sanofi-Pasteur), and allantoic fluid (SPF Chicken Embryo Allantoic Fluid, Chemically Inactivated, BEI Resources). N-deglycosylation of IIVs was performed with PNGase F (New England BioLabs [NEB]) following the manufacturer’s protocol. Briefly, IIVs (up to 20 μg) were reduced and denatured using glycoprotein denaturing buffer, and then GlycoBuffer 2 and NP-40 were added with 5 μL PNGase F (all from NEB). The reaction was incubated at 37°C overnight to ensure complete N-deglycosylation. Trypsinization was performed after denaturation and alkylation following the same protocol for F(ab′)2. Ninety-six-well EIA/RIA Assay Microplates (Corning) were coated with 4 μg/mL of the corresponding antigen at 4°C overnight. The plates were washed with PBST (DPBS plus 0.05% Tween-20) and blocked with 2% milk in DPBS for 2 hours at room temperature. After blocking, the plates were washed, and serially diluted recombinant antibodies or sera were added to the plates for 1 hour. The plates were washed again and 1:5000 diluted goat anti–human-IgG Fc HRP-conjugated secondary antibodies (Invitrogen, Thermo Fisher Scientific, catalog A18829) were added for 1 hour. After a final wash, 50 μL TMB-Ultra Substrate (Thermo Fisher Scientific) was added to each well to develop, and 50 μL 2 M H2SO4 was added to quench the reaction. Absorbance was measured at 450 nm (OD450) using a TECAN M200 plate reader. Data were analyzed and fitted for EC50 if necessary using a 4-parameter logistic, nonlinear regression model in GraphPad Prism (GraphPad Software).
HAI assay.
Influenza virus seed stocks used for HAI assays were obtained from the CDC. The HAI assay was performed following the standard methodology established by the WHO, which tests the quantity of serum or mAb required to interfere with the agglutinating activity of 4 agglutinating units of virus. Briefly, 2× serial dilutions of serum or mAb were mixed with 4 HA units of virus, followed by addition of 0.5% turkey red blood cells after a 1-hour incubation at room temperature. The HAI titer was determined as the lowest dilution of serum or concentration of mAb resulting in complete inhibition of HA.
Microneutralization assay.
The microneutralization assay was performed by Bioqual Inc., following standard operating procedure number BV-012 and the 2013 WHO Laboratory Procedures guidelines. Briefly, mAb was diluted to a starting concentration in assay diluent. Serial dilutions of mAb were incubated with influenza virus for 1 hour at 37°C before being transferred to 96-well plates with MDCK cells (American Type Culture Collection [ATCC]) with assay media containing 1 μg/mL trypsin. After 2 days of incubation, influenza virus was detected by ELISA to calculate the virus neutralization endpoint antibody titer. Reference serum, virus back-titration, and preimmune serum pools were included as controls.
Antibody forensics assay.
An antibody forensics assay was performed to determine the ability of antigen-specific antibodies to bind to rHA coupled to Bio-Plex Pro magnetic COOH beads (Bio-Rad) (85). Each rHA is coupled to a predetermined bead using a modification of a coupling procedure described by Bio-Rad (catalog 171406001). Prior to coupling, the carboxyl groups on the surface of the beads are activated with a carbodiimide derivative, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (Life Technologies, Thermo Fisher Scientific). The unstable intermediate is stabilized in aqueous solutions using sulfo-N-hydroxysulfosuccinimide (sNHS) (Life technologies, Thermo Fisher Scientific). These intermediates react with primary amines of rHAs to form amide bonds. Each rHA was coupled to a unique bead, and then rHA/bead complexes were combined into a multiplex mixture and used in antibody forensics assay. Briefly, beads were suspended in 100 mM MES buffer, and 10 mg/mL EDC and sNHS were added to the bead suspension and incubated on a rotating shaker at 600 rpm for 17 minutes in the dark. Then, the mixture was washed twice with ice-cold PBS (pH 7.4), and 2 μg rHA was added to approximately 6.25 × 104 activated beads. The beads and rHA mixtures were incubated for 48 hours at 4°C in a dark on a shaker at 600 rpm. Glycine (100 mM) was added to quench the reaction, followed by a 24-hour incubation at 4°C in a dark on a shaker at 600 rpm. Finally, after 2 washes with PBS containing 1% BSA, the mixture was resuspended in PBS containing 1% BSA and 10% v/v glycerol. Individual rHA-bead complexes were then combined into a single mixture and aliquoted into 2 mL vials and stored away from direct light at –20°C. mAbs were 10-fold serially diluted (starting from 1–10 μg/mL) and allowed to incubate with the beads with constant shaking (600 rpm) for 2 hours at 4°C. The plates were washed with PBST, and a biotinylated human-specific antibody against IgG (Jackson ImmunoResearch, catalog 109-065-008) was diluted in dilution buffer and added to the plates. The plates were incubated for 40 minutes at 4°C, washed with PBST, and incubated with streptavidin and R-phycoerythrin conjugate (SAPE) diluted in dilution buffer. The plates were washed with PBST and read using a BioPlex 100/200 bead array reader. The strength of antibody binding to the influenza-specific rHA was presented as a fluorescence intensity signal and then based on mAb titration converted to an EC50 titer (μg/mL). rHAs were expressed from either an insect-baculovirus system (Protein Sciences) or by HEK 293 cells in-house at Sanofi.
Glycan array experiment for glycan-binding mAbs.
The glycan-binding specificity of antibodies was determined with the CFG Printed Array, version 5.4 (585 glycans) at the National Center for Functional Glycomics (NCFG). Briefly, antibodies were diluted to 50 μg/mL in TSM binding buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween-20 and 1% BSA) and applied to the printed slide for 1 hour. The microarray slide was then rinsed 4 times in TSM wash buffer (TSM Binding buffer without BSA). Next, the fluorescently labeled anti–human IgG was diluted into TSM binding buffer and applied to the slide for 1 hour. After a series of final washing steps, the array was scanned with a GenePix 4300A microarray scanner (Molecular Devices). Analysis was carried out using GenePix Pro 7.0 analysis software (Molecular Devices). Glycans were printed on the array in replicates of 4, so relative fluorescence unit (RFU) values were determined by the average of the RFU values in each spot.
BLI.
Assays were performed on an Octet Red96e instrument (FortéBio) using anti–hIgG Fc Capture (AHC) biosensors (FortéBio). Sensors were hydrated with kinetics buffer (PBS plus 0.02% Tween-20, 0.1% BSA and 0.05% sodium azide) for 10 minutes, and then the antibodies were immobilized to a response level of approximately 0.6 nm. The sensors were dipped into wells containing different concentration of antigens (NP, M1, 2-fold diluted) for a 10-minute association including a reference well containing the kinetics buffer only. The dissociation was performed for 10 minutes in wells containing kinetics buffer. For duplicate measurements, regeneration was performed 5 times with 5 second injections into 10 mM glycine-HCl (pH 1.7). Data were subtracted by the reference and fit to a 1:1 binding model with Octet Data Analysis Software, version 11.1 (FortéBio) using the global fit. Average KD values and KD error of the duplicate measurements are shown in the figures. For the competition assay, the first antibody (FI6v3) (50) was immobilized at 40 μg/mL for 10 minutes and blocked with 50 μg/mL IgG isotype control for 5 minutes. Recombinant H1 from A/California/07/2009 (100 μg/mL; Sino Biological, catalog 11085-V08H) was associated for 5 minutes, and 40 μg/mL of the second antibody (29_H1_3, 1131_13_H1_6, 74_13_H1_1) was associated for 5 minutes to measure the degree of competition.
The proteomics data reported in this work are tabulated in Supplemental Data File 1, and the raw proteomics data with search result files are archived through MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under accession number MSV000086108.
Statistics.
Statistical analyses were performed using GraphPad Prism software, version 6.01 or 8.4.1. Comparisons of more than 2 groups were calculated using Kruskal-Wallis test followed by Dunn’s post-hoc test. Binomial test was used to compare the probability of selection between groups. Correlation analyses were performed using Spearman’s correlation. Significant differences between 2 paired groups were performed by the 2-tailed Wilcoxon signed-rank test. A P value of 0.05 or less was considered statistically significant.
Study approval.
The study procedures, informed consent, and data collection documents were reviewed and approved by the Western Institutional Review Board (Olympia, Washington, USA) and the IRBs of the University of Pittsburgh (Pittsburgh, Pennsylvania, USA) and the University of Georgia. The cohorts included in the study were recruited for a previous study published in 2017 (44). Briefly, eligible volunteers between the ages of 18 and 85 years were recruited from medical facilities in Pittsburgh, Pennsylvania, and Stuart, Florida, and provided written informed consent. Exclusion criteria included documented contraindications to Guillain-Barré syndrome, dementia, or Alzheimer’s disease, allergies to eggs or egg products, an estimated life expectancy of less than 2 years, medical treatment causing or diagnosis of an immunocompromising condition, or concurrent participation in another influenza vaccine research study.
Author contributions
JJ, STM, SD, HK, JL, and GG conceptualized the study. JJ, DP, GCI, JL, and GG designed the study methodology. JJ, STM, IVU, APH, DRB, SP, PP, JRM, GRK, DP, MDP, CY, BT, YT, JD, SD, TMR, JK, and JL collected specimens and experimental data. JJ, STM, IVU, APH, DRB, JRM, JD, SD, TMR, GCI, HK, JL, NCC, and GG analyzed data. JJ, JL, and GG wrote the original draft of the manuscript. All authors wrote, reviewed, and edited the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Stephen Harrison (Harvard Medical School, Boston, Massachusetts, USA) for insightful discussions and providing the rHA. We thank the Protein-Glycan Interaction Resource of the CFG and the NCFG at Beth Israel Deaconess Medical Center, Harvard Medical School (supporting grants P41 GM103694 and R24 GM137763) for glycan array resources and analysis. This work was supported by a master service agreement with Sanofi Pasteur Vaccines; by NIH grants 75N93019C00052 (to TDM and GG), P01AI089618 (to GG and JL), and P20GM113132 (to JL); and by the Clayton Foundation (to GG).
Version 1. 07/01/2021
Electronic publication
Funding Statement
For George Georgiou
Master service agreement
Footnotes
JJ’s present address is: Pandion Therapeutics, a wholly-owned subsidiary of Merck & Co., Inc., Kenilworth, New Jersey, USA.
PP’s present address is: Biologics Research, Sanofi, Framingham, Massachusetts, USA.
JRM’s present address is: Pfizer, Inc, Cambridge, Massachusetts, USA.
DP’s present address is: Department of Biological Sciences, Ajou University, Suwon, South Korea and Department of Molecular Science and Technology, Ajou University, Suwon, South Korea.
SD’s present address is: Ring Therapeutics, Cambridge, Massachusetts, USA.
HK’s present address is: Bill & Melinda Gates Foundation, Seattle, Washington, USA.
Conflict of interest: STM, IVU, SP, and JD are employees of Sanofi Pasteur; PP is an employee of Sanofi; SD is an employee of Ring Therapeutica; JRM is an employee of Pfizer; and JJ is an employee of Pandion Therapeutics, a wholly owned subsidiary of Merck & Co. Inc.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(13):e148763.https://doi.org/10.1172/JCI148763.
Contributor Information
Jiwon Jung, Email: jjung@utexas.edu.
Irina V. Ustyugova, Email: Irina.Ustyugova@sanofi.com.
Andrew P. Horton, Email: ahorton@utexas.edu.
Daniel R. Boutz, Email: dboutz@gmail.com.
Svetlana Pougatcheva, Email: Svetlana.Pougatcheva@sanofi.com.
Ponraj Prabakaran, Email: Prabakaran.Ponraj@sanofi.com.
Gregory R. King, Email: kinginator@gmail.com.
Daechan Park, Email: dpark@ajou.ac.kr.
Maria D. Person, Email: mperson@austin.utexas.edu.
Congxi Ye, Email: congxiye@gmail.com.
Bing Tan, Email: bingtan100@gmail.com.
Yuri Tanno, Email: takkiyuri0302@gmail.com.
Jin Eyun Kim, Email: jinjin2112@utexas.edu.
Joshua DiNapoli, Email: Joshua.Dinapoli@sanofi.com.
Simon Delagrave, Email: sdelagrave@biotechstudio.com.
Gregory C. Ippolito, Email: gregory.ippolito@gmail.com.
Harry Kleanthous, Email: hkleanthous@gmail.com.
Jiwon Lee, Email: jiwon.lee@dartmouth.edu.
George Georgiou, Email: gg@che.utexas.edu.
References
- 1.Iuliano AD, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med. 2010;363(21):2036–2044. doi: 10.1056/NEJMra1002842. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mostafa A, et al. Zoonotic potential of influenza A viruses: a comprehensive overview. Viruses. 2018;10(9):497. doi: 10.3390/v10090497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krammer F, et al. Influenza. Nat Rev Dis Primers. 2018;4(1):3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchinson EC, et al. Conserved and host-specific features of influenza virion architecture. Nat Commun. 2014;5:4816. doi: 10.1038/ncomms5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webster RG, et al. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y-Q, et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. 2018;173(2):417–429. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadlbauer D, et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science. 2019;366(6464):499–504. doi: 10.1126/science.aay0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilchuk IM, et al. Influenza H7N9 virus neuraminidase-specific human monoclonal antibodies inhibit viral egress and protect from lethal influenza infection in mice. Cell Host Microbe. 2019;26(6):715–728. doi: 10.1016/j.chom.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannoun C, et al. Immunogenicity and protective efficacy of influenza vaccination. Virus Res. 2004;103(1-2):133–138. doi: 10.1016/j.virusres.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 12.Coudeville L, et al. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med Res Methodol. 2010;10:18. doi: 10.1186/1471-2288-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JM, et al. Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin. Elife. 2019;8:1–28. doi: 10.7554/eLife.49324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krammer F. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol. 2019;19(6):383–397. doi: 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 15.Grandea AG, et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc Natl Acad Sci U S A. 2010;107(28):12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simhadri VR, et al. A human anti-M2 antibody mediates antibody-dependent cell-mediated cytotoxicity (ADCC) and cytokine secretion by resting and cytokine-preactivated natural killer (NK) Cells. PLoS One. 2015;10(4):e0124677. doi: 10.1371/journal.pone.0124677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos EL, et al. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J Infect Dis. 2014;211(7):1038–1044. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- 18.LaMere MW, et al. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J Virol. 2011;85(10):5027–5035. doi: 10.1128/JVI.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jegaskanda S, et al. Human seasonal influenza A viruses induce H7N9-cross-reactive antibody-dependent cellular cytotoxicity (ADCC) antibodies that are directed towards the nucleoprotein. J Infect Dis. 2016;215(5):818–823. doi: 10.1093/infdis/jiw629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanderven HA, et al. What lies beneath: antibody dependent natural killer cell activation by antibodies to internal influenza virus proteins. EBioMedicine. 2016;8:277–290. doi: 10.1016/j.ebiom.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaMere MW, et al. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J Immunol. 2011;186(7):4331–4339. doi: 10.4049/jimmunol.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yewdell JW, et al. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 1985;82(6):1785–1789. doi: 10.1073/pnas.82.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machkovech HM, et al. Positive selection in CD8+ T-cell epitopes of influenza virus nucleoprotein revealed by a comparative analysis of human and swine viral lineages. J Virol. 2015;89(22):11275–11283. doi: 10.1128/JVI.01571-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillie PJ, et al. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin Infect Dis. 2012;55(1):19–25. doi: 10.1093/cid/cis327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowell GH, et al. Back to the future: immunization with M-001 prior to trivalent influenza vaccine in 2011/12 enhanced protective immune responses against 2014/15 epidemic strain. Vaccine. 2017;35(5):713–715. doi: 10.1016/j.vaccine.2016.12.063. [DOI] [PubMed] [Google Scholar]
- 26.Doherty PC, et al. Influenza and the challenge for immunology. Nat Immunol. 2006;7(5):449–455. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 27. Center for Disease Control and Prevention (CDC). Estimates of Flu Vaccination Coverage Among Children. https://www.cdc.gov/flu/fluvaxview/coverage-1718estimates.htm. Accessed May 20, 2021.
- 28.Crooke SN, et al. Immunosenescence and human vaccine immune responses. Immun Ageing. 2019;16:1–16. doi: 10.1186/s12979-018-0141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciabattini A, et al. Vaccination in the elderly: the challenge of immune changes with aging. Semin Immunol. 2018;40:83–94. doi: 10.1016/j.smim.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki S, et al. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest. 2011;121(8):3109–3119. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frasca D, et al. Young and elderly patients with type 2 diabetes have optimal B cell responses to the seasonal influenza vaccine. Vaccine. 2013;31(35):3603–3610. doi: 10.1016/j.vaccine.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodwin K, et al. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24(8):1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 33.Ju C-H, et al. Plasmablast antibody repertoires in elderly influenza vaccine responders exhibit restricted diversity but increased breadth of binding across influenza strains. Clin Immunol. 2018;193:70–79. doi: 10.1016/j.clim.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moody MA, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6(10):e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dugan HL, et al. Aging and influenza vaccine-induced immunity. Cell Immunol. 2020;348:103998. doi: 10.1016/j.cellimm.2019.103998. [DOI] [PubMed] [Google Scholar]
- 36.de Bourcy CFA, et al. Phylogenetic analysis of the human antibody repertoire reveals quantitative signatures of immune senescence and aging. Proc Natl Acad Sci U S A. 2017;114(5):1105–1110. doi: 10.1073/pnas.1617959114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5(171):171ra19–ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frasca D, et al. Intrinsic defects in B cell response to seasonal influenza vaccination in elderly humans. Vaccine. 2010;28(51):8077–8084. doi: 10.1016/j.vaccine.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khurana S, et al. AID activity in B cells strongly correlates with polyclonal antibody affinity maturation in-vivo following pandemic 2009-H1N1 vaccination in humans. PLoS Pathog. 2012;8(9):e1002920. doi: 10.1371/journal.ppat.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry C, et al. Influenza virus vaccination elicits poorly adapted B cell responses in elderly individuals. Cell Host Microbe. 2019;25(3):357–366. doi: 10.1016/j.chom.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J, et al. Persistent antibody clonotypes dominate the serum response to influenza over multiple years and repeated vaccinations. Cell Host Microbe. 2019;25(3):367–376. doi: 10.1016/j.chom.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawood FS, et al. Interim estimates of 2019-20 seasonal influenza vaccine effectiveness - United States, February 2020. MMWR Morb Mortal Wkly Rep. 2020;69(7):177–182. doi: 10.15585/mmwr.mm6907a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavinder JJ, et al. Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires. Curr Opin Chem Biol. 2015;24:112–120. doi: 10.1016/j.cbpa.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Nuñez IA, et al. Impact of age and pre-existing influenza immune responses in humans receiving split inactivated influenza vaccine on the induction of the breadth of antibodies to influenza A strains. PLoS One. 2017;12(11):e0185666. doi: 10.1371/journal.pone.0185666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med. 2016;22(12):1456–1464. doi: 10.1038/nm.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaari G, et al. Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 2012;40(17):e134. doi: 10.1093/nar/gks457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lavinder JJ, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci U S A. 2014;111(6):2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindesmith LC, et al. Sera antibody repertoire analyses reveal mechanisms of broad and pandemic strain neutralizing responses after human norovirus vaccination. Immunity. 2019;50(6):1530–1541. doi: 10.1016/j.immuni.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pappas L, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516(7531):418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- 50.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 51.Koroleva M, et al. Heterologous viral protein interactions within licensed seasonal influenza virus vaccines. NPJ Vaccines. 2020;5(1):3. doi: 10.1038/s41541-019-0153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kilbourne ED. Future influenza vaccines and the use of genetic recombinants. Bull World Health Organ. 1969;41(3):643–645. [PMC free article] [PubMed] [Google Scholar]
- 53.Smith DF, et al. Use of glycan microarrays to explore specificity of glycan-bonding proteins. Methods Enzymol. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- 54.Sriwilaijaroen N, et al. Analysis of N-glycans in embryonated chicken egg chorioallantoic and amniotic cells responsible for binding and adaptation of human and avian influenza viruses. Glycoconj J. 2009;26(4):433–443. doi: 10.1007/s10719-008-9193-x. [DOI] [PubMed] [Google Scholar]
- 55.Downie JC. Host antigen as the sulphated moiety of influenza virus haemagglutinin. J Gen Virol. 1978;41(2):283–293. doi: 10.1099/0022-1317-41-2-283. [DOI] [PubMed] [Google Scholar]
- 56.Ward CW, et al. Antigenic determinants of influenza virus hemagglutinin. VII. The carbohydrate side chains of A/Memphis/102/72 hemagglutinin heavy chain which cross-react with host antigen. Virology. 1981;108(1):71–79. doi: 10.1016/0042-6822(81)90527-4. [DOI] [PubMed] [Google Scholar]
- 57.Ward CW, et al. Chemical and antigenic characterization of the carbohydrate side chains of an Asian (N2) influenza virus neuraminidases. Virology. 1983;126(1):370–375. doi: 10.1016/0042-6822(83)90486-5. [DOI] [PubMed] [Google Scholar]
- 58.She YM, et al. Remarkable structural diversity of N-glycan sulfation on influenza vaccines. Anal Chem. 2019;91(8):5083–5090. doi: 10.1021/acs.analchem.8b05372. [DOI] [PubMed] [Google Scholar]
- 59.Gerhard W, et al. The analysis of the monoclonal immune response to influenza virus. I. Production of monoclonal anti-viral antibodiesin vitro. Eur J Immunol. 1975;5(10):720–725. doi: 10.1002/eji.1830051013. [DOI] [PubMed] [Google Scholar]
- 60. Guthmiller JJ, et al. An egg-derived sulfated N-Acetyllactosamine glycan is an antigenic decoy of influenza virus vaccines. biorxiv. https://www.biorxiv.org/content/10.1101/2021.03.16.435673v1 Published March 16, 2021. Accessed June 9, 2021.
- 61.Khurana S, et al. Antigenic fingerprinting of antibody response in humans following exposure to highly pathogenic H7N7 avian influenza virus: evidence for anti-PA-X antibodies. J Virol. 2016;90(20):9383–9393. doi: 10.1128/JVI.01408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krejnusová I, et al. Antibodies to PB1-F2 protein are induced in response to influenza A virus infection. Arch Virol. 2009;154(10):1599–1604. doi: 10.1007/s00705-009-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong W, et al. Serum antibody response to matrix protein 2 following natural infection with 2009 pandemic influenza A(H1N1) virus in humans. J Infect Dis. 2014;209(7):986–994. doi: 10.1093/infdis/jit811. [DOI] [PubMed] [Google Scholar]
- 64.Joyce MG, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell. 2016;166(3):609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson KJL, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16(1):105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Briney B, et al. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566(7744):393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soto C, et al. High frequency of shared clonotypes in human B cell receptor repertoires. Nature. 2019;566(7744):398–402. doi: 10.1038/s41586-019-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Padilla-Quirarte HO, et al. Protective antibodies against influenza proteins. Front Immunol. 2019;10:1–13. doi: 10.3389/fimmu.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sedova ES, et al. Non-neutralizing antibodies directed at conservative influenza antigens. Acta Naturae. 2019;11(4):22–32. doi: 10.32607/20758251-2019-11-4-22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clemens E, et al. Harnessing the power of T cells: the promising hope for a universal influenza vaccine. Vaccines (Basel) 2018;6(2):18. doi: 10.3390/vaccines6020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He X-S, et al. Distinct patterns of B-cell activation and priming by natural influenza virus infection versus inactivated influenza vaccination. J Infect Dis. 2015;211(7):1051–1059. doi: 10.1093/infdis/jiu580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dugan HL, et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med. 2020;12(573):eabd3601. doi: 10.1126/scitranslmed.abd3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horns F, et al. Memory B cell activation, broad anti-influenza antibodies, and bystander activation revealed by single-cell transcriptomics. Cell Rep. 2020;30(3):905–913. doi: 10.1016/j.celrep.2019.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karaivanova VK, Spiro RG. Sulphation of N-linked oligosaccharides of vesicular stomatitis and influenza virus envelope glycoproteins: host cell specificity, subcellular localization and identification of substituted saccharides. Biochem J. 1998;329:511–518. doi: 10.1042/bj3290511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ippolito GC, et al. Antibody repertoires in humanized NOD-scid-IL2Rγnull mice and human B cells reveals human-like diversification and tolerance checkpoints in the mouse. PLoS One. 2012;7(4):e35497. doi: 10.1371/journal.pone.0035497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brochet X, et al. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta NT, et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31(20):3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeKosky BJ, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21(1):86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 79.McDaniel JR, et al. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat Protoc. 2016;11(3):429–442. doi: 10.1038/nprot.2016.024. [DOI] [PubMed] [Google Scholar]
- 80.Ellefson JW, et al. Synthetic evolutionary origin of a proofreading reverse transcriptase. Science. 2016;352(6293):1590–1593. doi: 10.1126/science.aaf5409. [DOI] [PubMed] [Google Scholar]
- 81.DeFalco J, et al. Non-progressing cancer patients have persistent B cell responses expressing shared antibody paratopes that target public tumor antigens. Clin Immunol. 2018;187:37–45. doi: 10.1016/j.clim.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 82.Raymond DD, et al. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat Med. 2016;22(12):1465–1469. doi: 10.1038/nm.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boutz DR, et al. Proteomic identification of monoclonal antibodies from serum. Anal Chem. 2014;86(10):4758–4766. doi: 10.1021/ac4037679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu A, et al. Technical advances in proteomics: new developments in data-independent acquisition. F1000Res. 2016;5:419. doi: 10.12688/f1000research.7042.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiu Y, et al. Mapping of a novel H3-specific broadly neutralizing monoclonal antibody targeting the hemagglutinin globular head isolated from an elite influenza virus-immunized donor exhibiting serological breadth. J Virol. 2019;94(6):e01035-19. doi: 10.1128/JVI.01035-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.