Abstract
Objective
To estimate the proportion of presymptomatic transmission of SARS-CoV-2 infection that can occur, and the timing of transmission relative to symptom onset.
Setting/design
Secondary analysis of international published data.
Data sources
Meta-analysis of COVID-19 incubation period and a rapid review of serial interval and generation time, which are published separately.
Participants
Data from China, the Islamic Republic of Iran, Italy, Republic of Korea, Singapore and Vietnam from December 2019 to May 2020.
Methods
Simulations were generated of incubation period and of serial interval or generation time. From these, transmission times relative to symptom onset, and the proportion of presymptomatic transmission, were estimated.
Outcome measures
Transmission time of SARS-CoV-2 relative to symptom onset and proportion of presymptomatic transmission.
Results
Based on 18 serial interval/generation time estimates from 15 papers, mean transmission time relative to symptom onset ranged from −2.6 (95% CI −3.0 to –2.1) days before infector symptom onset to 1.4 (95% CI 1.0 to 1.8) days after symptom onset. The proportion of presymptomatic transmission ranged from 45.9% (95% CI 42.9% to 49.0%) to 69.1% (95% CI 66.2% to 71.9%).
Conclusions
There is substantial potential for presymptomatic transmission of SARS-CoV-2 across a range of different contexts. This highlights the need for rapid case detection, contact tracing and quarantine. The transmission patterns that we report reflect the combination of biological infectiousness and transmission opportunities which vary according to context.
Keywords: epidemiology, infection control, infectious diseases, virology, public health
Strengths and limitations of this study.
We generated estimates of presymptomatic transmission for different countries.
As this is a secondary analysis of published estimates, we did not analyse data at individual transmission-pair level.
As control measures such as rapid isolation of symptomatic people may increase the proportion of presymptomatic transmission, we generated estimates based on single locations and did not pool them.
Introduction
There is currently a pandemic of COVID-19, a recently emerged and rapidly spreading infectious disease that is caused by the novel coronavirus, SARS-CoV-2. There are large direct impacts of COVID-19 among known cases. As of 19 April 2021, the WHO has reported 140, 886 773 confirmed cases and 3 012 251 deaths due to COVID-19.1 In China, 14% and 5% of cases were classified as severe and critical, respectively.2 There are also major indirect impacts of COVID-19 and its control measures on other aspects of healthcare3–5 and on the economy.6 7
In addition to vaccination, primary control measures entail reducing transmission from infectious individuals. These include case isolation, contact tracing and quarantine, physical distancing, hygiene and ventilation measures.8 Infectious people are identified when they report symptoms, and are tested for SARS-CoV-2. Infectious people without symptoms may be identified when an active surveillance programme is in place.
In the absence of active surveillance, infectious people without symptoms may not be quarantined, and therefore may have more contacts with susceptible people resulting in increased SARS-CoV-2 transmission. Therefore, quantifying the transmission potential before or in the absence of symptoms will inform disease control measures and predictions of epidemic progression.
Characteristics of presymptomatic and asymptomatic transmission are potentially different, and separate approaches may be required to understand them. In this paper, we capitalise on the considerable information about presymptomatic transmission that can be inferred from contact tracing studies. Therefore, we focus on transmission from people before they develop symptoms rather than from people who never develop symptoms. This addresses the urgent need for more data on the extent of presymptomatic transmission which has been highlighted by those developing models to inform policies.9
Reports of presymptomatic transmission10–19 emerged as detailed contact tracing was conducted during early outbreaks of COVID-19. Further, both viral genome18 20–26 and live virus21 have been detected in upper respiratory samples prior to symptom onset. These findings are supported by quantitative studies based on contact tracing, with reports of serial intervals or generation times similar in duration or shorter than incubation periods in some situations,27–32 and even cases of symptoms manifesting in the infectee prior to the infector.24 30 33–37
Several studies have quantified the proportion27–30 38 and timing27 30 38 of presymptomatic transmission, using a variety of datasets and methodologies. Here, we compare presymptomatic transmission across a range of different contexts using a consistent methodology. We build on our rapid review of SARS-CoV-2 serial interval and generation time39 and rapid systematic review and meta-analysis of incubation period40 with a secondary analysis of published data to estimate the proportion and timing of presymptomatic transmission of COVID-19.
Methods
Principles of methodology
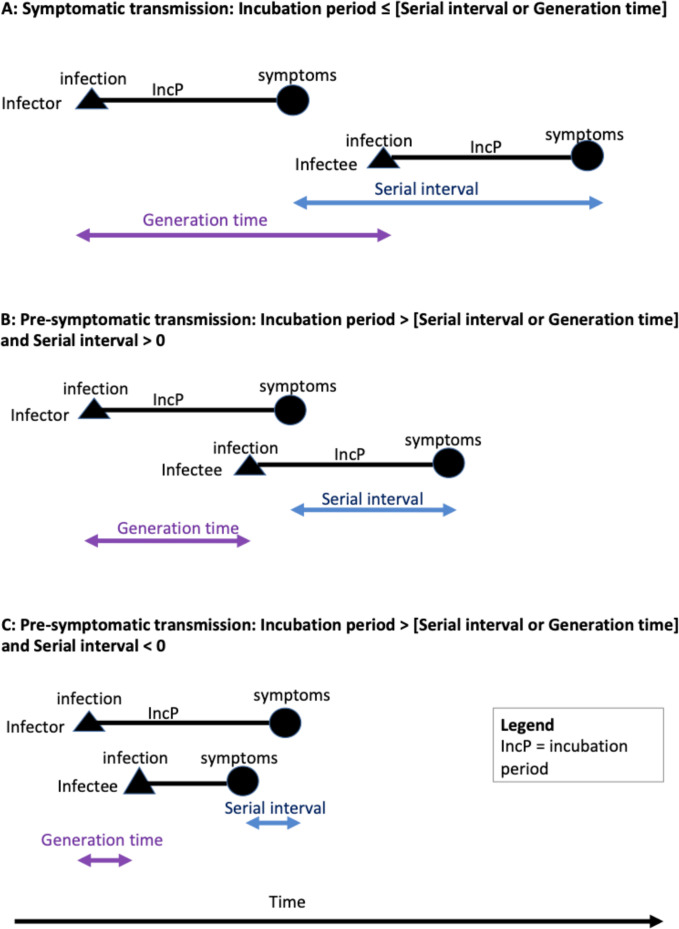
If transmission occurs after symptom onset, mean generation time, the duration in days between time of infection of a secondary case (infectee) and that of its primary case (infector), is longer than mean incubation period, the time between infection and symptom onset in the infector (scenario A in figure 1). If presymptomatic transmission occurs, mean generation time is shorter than mean incubation period (scenarios B and C in figure 1). If the incubation period of an infector and of an infectee are taken to be independent and identically distributed, serial interval, the time between infector and infectee symptom onset, can be taken as an approximation of generation time,41 42 although serial interval will have more variation.28 Our method entailed subtracting simulated values for incubation period from serial interval to estimate the timing and proportion of presymptomatic transmission in a range of different settings. Table 1 contains definitions relevant to our analysis.
Figure 1.
Schematic illustration of incubation period, generation time and serial interval at transmission pair level. Scenario A: if transmission occurs after symptom onset, mean generation time/serial interval is longer than mean incubation period. Scenario B: if presymptomatic transmission occurs, mean generation time/serial interval is shorter than mean incubation period. Scenario C: a negative serial interval is possible if symptoms manifest in the infectee before the infector. Relevant to all scenarios, if incubation period is assumed to be independent and identically distributed, mean serial interval will approximate mean generation time.
Table 1.
Definitions referred to in this review
| Asymptomatic | An infected person who never develops symptoms of the disease. |
| Presymptomatic | An infected person before they develop symptoms of the disease. |
| Duration of infectiousness | The time interval in days during which an infectious agent may be transferred directly or indirectly from an infected person to another person. |
| Incubation period | The time interval in days between invasion by an infectious agent and appearance of the first signs or symptoms of the disease in question. |
| Serial interval | The duration in days between symptom onset of a secondary case (infectee) and that of its primary case (infector). |
| Generation time or generation interval | The duration in days between time of infection of a secondary case (infectee) and that of its primary case (infector). |
| Transmission pair | An infected person (infector) and a person whom they transmit the pathogen to (infectee). |
| Latent period | The period from the point of infection to the beginning of the state of infectiousness. This period corresponds to the ‘exposed’ (E) compartment of a susceptible-exposed-infectious-recovered/removed model. |
| Transmission time relative to symptom onset | The time of transmission of an infectious agent from an infector to an infectee in days relative to the onset of symptoms in the infector. |
| Proportion of presymptomatic transmission | The proportion of all transmission events that occur before the onset of symptoms in the infector. |
Incubation period data
We used the incubation period estimate from our separately published rapid systematic review and meta-analysis40. That is, a lognormal distribution with meanlog and sdlog parameters of 1.63 (95% CI 1.51 to 1.75) and 0.50 (95% CI 0.46 to 0.55), respectively. The corresponding mean and median were 5.8 (95% CI 5.0 to 6.7) days and 5.1 (95% CI 4.5 to 5.8) days, respectively. As there is currently no evidence of country-specific drivers in variation of incubation period, we deemed it reasonable to use the estimate from this meta-analysis of incubation period40 to investigate presymptomatic transmission across a range of settings.
Serial interval and generation time data
We used serial interval estimates from our separately published rapid review of serial interval and generation time.39 In contrast to incubation period, interventions such as case isolation are reported to affect serial interval.39 43 44 Therefore, we analysed each serial interval or generation time estimate separately and excluded estimates based on data from a mixture of countries.
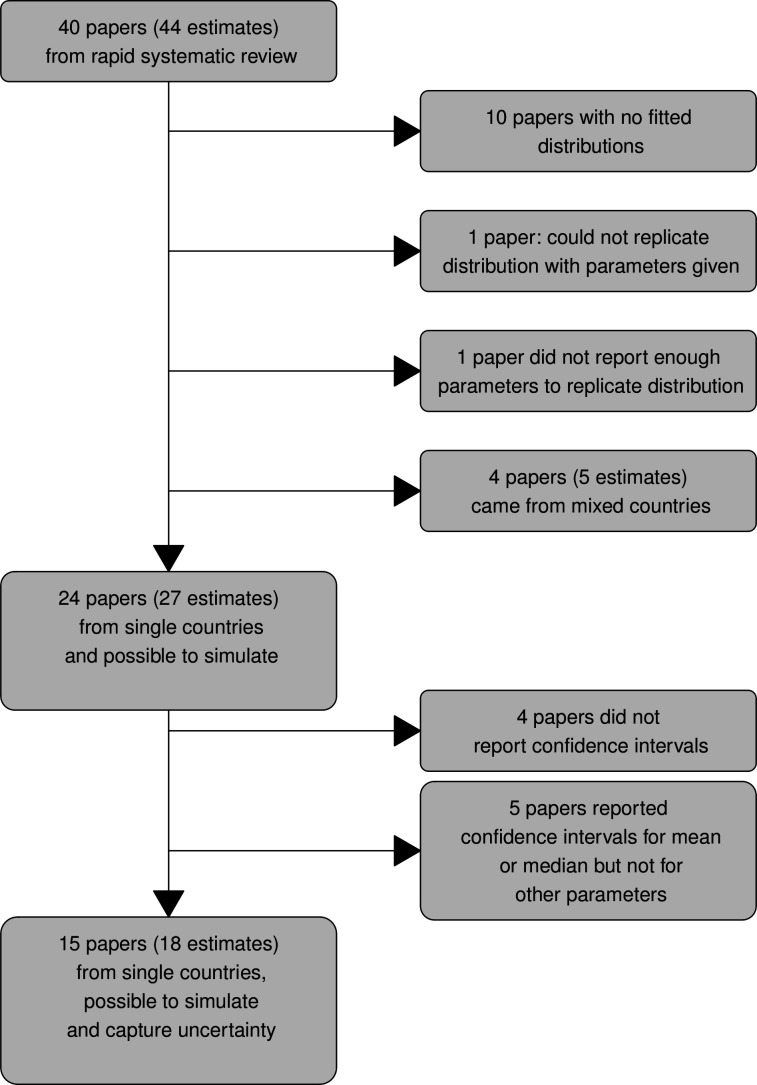
Figure 2 summarises how we selected serial interval or generation time estimates for inclusion in our analysis. From the 40 published papers included in the rapid review,39 we selected serial interval and generation time estimates based on data from single countries, for which statistical distributions were fitted, and which we could replicate (n=27 estimates from 24 papers). From this subset, we identified estimates for which enough information was provided, to allow us to simulate the uncertainty associated with their distributions (n=18 estimates from 15 papers).
Figure 2.
A summary of how serial interval and generation time estimates were selected for analyses.
Description of serial interval/generation time data
Building on initial data screening and assessment for quality and central estimates presented in our rapid review of serial interval and generation time,39 we highlighted country or region of origin, date-range for gathering of the data underlying the estimates, and sample-size.
Simulation
We subtracted samples from a simulated incubation period distribution from samples from simulated serial interval/generation time distributions to generate distributions of transmission time relative to symptom onset.
To calculate transmission time relative to symptom onset, we first replicated the reported serial interval/generation time distributions and the incubation period distribution from our meta-analysis.40 To achieve this, we sampled distribution parameters from their respective 95% CIs for each reported distribution (n=1000). We then simulated distributions using these parameters (n=1000). The incubation period sample was subtracted from each generation time or serial interval sample to give a resultant distribution indicating transmission time relative to onset of symptoms. The resultant 1 000 000 samples were resampled with replacement (n=1000 samples from each of 10 000 repeats) and 95% CIs from bootstrapping were calculated.
As we were conducting a secondary analysis based on published data, we did not incorporate potential correlations between serial interval and incubation period at transmission pair level. That is, we assumed that incubation period and generation time/serial interval were independent.
We presented the resultant simulated transmission time relative to symptom onset, and the proportion of presymptomatic transmission at the level of each underlying serial interval or generation time estimate, grouped by country or region.
In online supplemental figures 1–3 and table 1, we also present the result of simulations from the larger dataset of 27 estimates (defined in figure 2). These supplementary results include estimates based on serial intervals/generation times for which we could simulate distributions but not take the associated uncertainty into account. For this simulation, as only central estimates of serial interval/generation time parameters were used, we also used central parameter estimates of the incubation period (meanlog 1.63, sdlog 0.5).
bmjopen-2020-041240supp001.pdf (722.4KB, pdf)
bmjopen-2020-041240supp002.pdf (13.1KB, pdf)
bmjopen-2020-041240supp003.pdf (18.6KB, pdf)
bmjopen-2020-041240supp004.pdf (15.3KB, pdf)
bmjopen-2020-041240supp005.pdf (143KB, pdf)
All analyses were conducted in the R Statistical Environment.45 The extracted data and code that we used to generate our simulation is available through GitHub (https://github.com/miriamcasey/covid-19_presymptomatic_project).
Patient and public involvement statement
It was not appropriate or possible to involve patients or the public in the design, or conduct, or reporting, or dissemination plans of our research.
Results
Description of serial interval/ generation time data
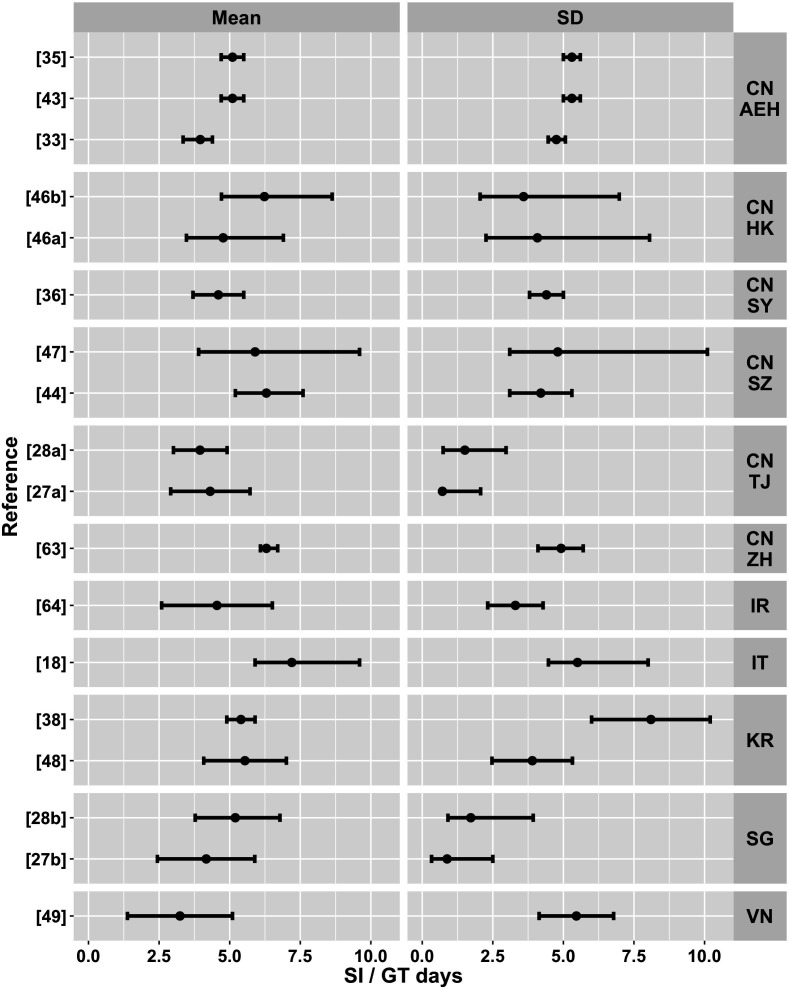
Building on the description of the serial interval and generation time estimates by Griffin et al,39 figure 3 summarises the country or region, collection date-range and sample size of the data underlying the serial interval and generation time that went into our simulation. Figure 4 summarises the mean and SD of each estimate. Of the 18 estimates from 15 papers for which we could incorporate uncertainty into our simulations, 11 came from China, 2 each came from the Republic of Korea and from Singapore and 1 each from the Islamic Republic of Iran, Italy and Vietnam. Sample sizes ranged from 1746 to 140735 transmission pairs.
Figure 3.
A summary of country or region and date ranges for the 18 serial interval and generation time estimates from 15 papers that were included in simulations to infer presymptomatic transmission. Line thickness is scaled to reflect sample size (ie, Kwok et al46 have the smallest and Xu et al35 the largest sample size). CN AEH, China: all regions excluding Hubei; CN HK, China: Hong Kong; CN SY, China: Shiyan (Hubei); CN SZ, China: Shenzhen; CN TJ, China Tianjin; CN ZH, China: Zhuhai; IR, Iran; IT, Italy; KR, The Republic of Korea; SG, Singapore; VN, Vietnam.
Figure 4.
A summary of the parameters from the serial interval and generation time estimates that were used in the simulation, by country or region and reference. Points indicate means and bars indicate 95% CIs. CN AEH, China: all regions excluding Hubei; CN HK, China: Hong Kong; CN SY, China: Shyan (Hubei); CN SZ, China: Shenzhen; CN TJ, China Tianjin; CN ZH=China: Zhuhai; IR, Iran; IT, Italy; KR, The Republic of Korea; SG, Singapore; VN, Vietnam.
Of the 11 estimates from China, 3 were based on datasets covering all of China excluding Hubei province. These three estimates were associated with the largest datasets in the study (n=1407,35 67743 and 46833 transmission pairs), and were associated with the same group of authors, who confirmed some overlap between the datasets underlying each paper. Xu et al35 and Ali et al43 both reported mean serial interval estimates of 5.1 days. It is also possible that there is some overlap between these general Chinese datasets and the smaller datasets associated with individual regions in China.
Both estimates from Hong Kong came from the same paper and dataset,46 but were based on samples of certain (n=17) and mixed certain and probable (n=26) transmission pairs. There is a difference of over a day in these two data subsets although they came from the same region and date range. The two estimates from Shenzhen44 47 had some overlap in date range but differed in sample size (48 transmission pairs,44 27 transmission pairs47). Ganyani et al28 and Tindale et al27 used the same datasets from Tianjin and Singapore. Son et al48 reported a serial interval estimate based on data from Busan in the Republic of Korea, whereas Chun et al38 used data from the whole country. Shiyan (Hubei province) and Zhuhai in China were associated with one estimate each, as were the remaining countries (figures 3 and 4).
Only Ganyani et al28 inferred generation time. The remainder of the estimates were based on serial intervals. Ten of the estimates were based on direct observation of transmission pairs. Eight serial interval estimates from six papers18 27 28 35 38 47 were based on inferences about transmission pairs from clusters of cases.
Many of the papers highlighted that serial interval was likely to be shorter if symptomatic cases were rapidly isolated. Bi et al44 quantified this as mean serial interval of 3.6 days if a case was isolated within less than 3 days of developing symptoms, increasing to 8·1 days if the infected individual was isolated on the third day after symptom onset or later, but with no further increase if isolation was delayed beyond 6 days after symptom onset. Ali et al43 quantified the contraction of serial interval over time, driven primarily by case isolation, and advocated for real-time estimation of serial intervals.
Simulation results
Figure 5 summarises the distributions of transmission time relative to symptom onset that were generated by the simulation. Table 2 provides summary statistics from the simulation output including the proportion of presymptomatic transmission. Mean transmission time relative to symptom onset ranged from −2.6 (95% CI −3.0 to –2.1) days before infector symptom onset in Vietnam49 to 1.4 (95% CI 1.0 to 1.8) days after symptom onset in Italy.18 The proportion of presymptomatic transmission was substantial in all contexts, ranging from 45.9% (95% CI 42.9% to 49.0%) in Italy18 to 69.1% (95% CI 66.2 to 71.9) in Tianjin.28 It was only possible to estimate the proportion of negative serial intervals, reflecting symptom onset in the infectee prior to the infector, from the five estimates that were fitted with distributions that allowed negative serial intervals. Simulations based on Chinese data ranged from 16.7% (95% CI 14.4 to 19.0) to 20.4% (95% CI 17.9 to 22.9), whereas the simulation using the data from Vietnam resulted in 30.9% (95% CI 28.0 to 33.8) negative serial intervals.
Figure 5.
A boxplot summarising simulation results showing transmission time in days relative to infector symptom onset. Purple triangles represent the mean of the simulation samples. CN AEH, China: all regions excluding Hubei; CN HK, China: Hong Kong; CN SY, China: Shiyan (Hubei); CN SZ, China: Shenzhen; CN TJ, China Tianjin; CN ZH, China: Zhuhai; IR, Iran; IT, Italy; KR, The Republic of Korea; SG, Singapore; VN, Vietnam.
Table 2.
A summary of simulation results
| Reference | Mean | SD | Median | PST |
| China—all excluding Hubei | ||||
| Xu et al35 | −0.7 (–1.1 to –0.3) | 6.2 (5.9 to 6.5) | −0.5 (–1 to –0.1) | 53.5 (50.4 to 56.6) |
| Ali et al43 | −0.7 (–1.1 to –0.3) | 6.2 (5.9 to 6.5) | −0.5 (–1 to 0) | 53.2 (50.1 to 56.3) |
| Du et al33 | −1.8 (–2.1 to –1.4) | 5.8 (5.5 to 6) | −1.6 (–2 to –1.1) | 61.2 (58.2 to 64.3) |
| China—Hong Kong | ||||
| Kwok et al46 | −1 (−1.4 to –0.7) | 5.3 (4.3 to 6.3) | −1.3 (–1.6 to –1.1) | 64.5 (61.5 to 67.4) |
| Kwok et al46 | 0.5 (0.2 to 0.8) | 5 (4.4 to 5.7) | 0.4 (0.1 to 0.7) | 46.3 (43.2 to 49.4) |
| China—Shiyan (Hubei) | ||||
| Yang et al36 | −1.2 (–1.5 to –0.8) | 5.7 (5.4 to 6) | −1 (−1.4 to –0.5) | 57.1 (54.1 to 60.2) |
| China—Shenzhen | ||||
| Wang et al47 | 0.1 (–0.2 to 0.5) | 6.2 (5.4 to 6.9) | −0.5 (–0.9 to –0.1) | 54.2 (51.1 to 57.2) |
| Bi et al44 | 0.5 (0.2 to 0.8) | 5.3 (5 to 5.6) | 0.1 (–0.2 to 0.5) | 48.6 (45.5 to 51.8) |
| China—Tianjin | ||||
| Ganyani et al28 | −1.8 (–2 to –1.6) | 3.5 (3.3 to 3.8) | −1.4 (–1.6 to –1.1) | 69.1 (66.2 to 71.9) |
| Tindale et al27 | −1.4 (–1.7 to –1.1) | 4.2 (3.9 to 4.5) | −1.1 (–1.4 to –0.8) | 61.1 (58.1 to 64.2) |
| China—Zhuhai | ||||
| Wu et al63 | 0.5 (0.2 to 0.9) | 5.8 (5.1 to 6.4) | 0 (–0.3 to 0.3) | 50.2 (47.1 to 53.3) |
| Iran—Qom | ||||
| Aghaali et al64 | −1.2 (–1.5 to –0.9) | 4.7 (4.4 to 5) | −1.4 (–1.7 to–1.1) | 63.5 (60.5 to 66.5) |
| Italy—Vo (village in Northern Italy) | ||||
| Lavezzo et al18 | 1.4 (1.0 to 1.8) | 6.4 (6.0 to 6.9) | 0.5 (0.1 to 0.9) | 45.9 (42.9 to 49.0) |
| Republic of Korea—all | ||||
| Chun et al38 | −0.4 (–1 to 0.1) | 8.8 (6.6 to 10.8) | −2 (−2.4 to –1.6) | 64.2 (61.2 to 67.2) |
| Republic of Korea—Busan | ||||
| Son et al48 | −0.3 (–0.6 to 0.1) | 5.1 (4.7 to 5.4) | −0.6 (–0.9 to –0.2) | 55.4 (52.3 to 58.4) |
| Singapore | ||||
| Ganyani et al28 | −0.6 (–0.8 to –0.4) | 3.7 (3.5 to 4) | −0.2 (–0.4 to 0) | 52.5 (49.4 to 55.6) |
| Tindale et al27 | −1.4 (–1.7 to –1.1) | 4.8 (4.5 to 5) | −1.1 (–1.5 to –0.8) | 60.0 (57.0 to 63.1) |
| Vietnam | ||||
| Pham et al49 | −2.6 (–3.0 to –2.1) | 7.2 (6.9 to 7.6) | −2.4 (–3 to –1.9) | 63.4 (60.5 to 66.4) |
The table shows the mean, standard deviation (SD) and median of transmission time relative to symptom onset in days as well as the proportion of presymptomatic transmission (PST).
For transmission time relative to symptom onset, negative values mean transmission before symptom onset and positive values mean transmission after symptom onset. The figures in parentheses represent the 95% confidence intervals from bootstrapping of simulation samples.
CN AEH, China: all regions excluding Hubei; CN HK, China: Hong Kong; CN SY, China: Shyan (Hubei); CN SZ, China: Shenzhen; CN TJ, China Tianjin; CN ZH, China: Zhuhai; IR, Iran; IT, Italy; KR, The Republic of Korea; SG, Singapore; VN, Vietnam.
Online supplemental figures 1–3 and table 1 show the results from simulations based on all 27 serial interval or generation time estimates from 24 papers, including the nine studies for which we could not incorporate uncertainty. The extra nine studies came from Brazil,34 Brunei Darussalam,24 China (all regions excluding Hubei),32 50 Tianjin,51 Wuhan,52 53 Iran54 and the Republic of Korea.22 Online supplemental table 1 also shows any estimates or comments relating to presymptomatic transmission that we found in the serial interval or generation time papers. Online supplemental table 2 compares the presymptomatic transmission time estimates of Ganyani et al,28 Tindale et al27 and this study which all refer to the same datasets from Singapore and Tianjin. Online supplemental tables 3 and 4 summarise virological studies and case reports of presymptomatic transmission which we refer to in our discussion.
Discussion
Our simulation study highlights the value of contact tracing data as a source of information about transmission dynamics of recently emerged diseases such as COVID-19. Using estimates of serial interval, generation time and incubation period from the published literature, our simulations highlight substantial potential for presymptomatic transmission of SARS-CoV-2.
Our estimation of mean transmission times ranged from 2.6 days before to 1.37 days after symptom onset. Virus transmission from an infector to an infectee requires both shedding of infectious virus from the infector and contact with a susceptible person under conditions that allow the virus to be transferred. Interventions such as rapid isolation of symptomatic people result in a greater proportion of transmission occurring earlier in the infectious period (shorter serial intervals and relatively more presymptomatic transmission).43 44 Well characterised infector–infectee data are required for serial interval estimation. It is possible that some of the cases associated with these data may be isolated more promptly than cases that were not detected by the public health authorities. Our transmission time estimates are therefore more likely to overlap with the earlier part of the infectious period. Consistently with this study, virological studies that show that viral load in upper respiratory samples peaks around symptom onset and rapidly declines towards undetectable levels about 2 weeks after symptom onset.30 55–58 Similarly, findings of detailed contact tracing in Shenzhen showed that isolation less than 3 days following symptom onset had a large effect in shortening serial interval whereas isolation at 6 days or later after symptom onset had no effect.44 This suggests reduced biological infectiousness beyond the first week of symptoms.
Our findings in support of transmission potential prior to symptom onset are consistent with multiple reports of both SARS-CoV-2 genome18 20 21 23–25 59 60 and live virus21 detection in upper respiratory samples prior to symptom onset. Bae et al22 reported viral genome detection up to 13 days prior to symptom onset and Arons et al21 isolated live virus from upper respiratory samples from nursing home residents 6 days prior to symptom onset. Of 48 residents testing positive for viral genome in upper respiratory tract samples, Arons et al21 reported that 24 of these residents tested positive a median of 4 (IQR 3–5) days in advance of symptom onset. Online supplemental table 3 provides a more detailed summary of the virological studies which we refer to. Case series with detailed descriptions of contact patterns and symptom onset10–19 (online supplemental table 4) further corroborate evidence from this study that transmission can occur well in advance of symptom onset.
In the majority of studies included in our simulation, there was commentary on the possibility of presymptomatic transmission, given reported serial intervals that were similar to, or shorter than, estimates for the incubation period of COVID-19 (online supplemental table 1). Another quantitative study investigating presymptomatic transmission30 used 77 transmission pairs from a mixture of countries to infer that infectiousness peaked at symptom onset (95% CI –0.9 to 0.9 days). The authors estimated that 44% (95% CI 30% to 57%) of transmission was presymptomatic. Ferretti et al29 also using data from a mixture of countries (40 transmission pairs), inferred that 37% (95% CI 27.5 to 45) of transmission was presymptomatic and that this accounted for almost enough transmission (0.9 of the effective reproduction number) to maintain an epidemic of its own.
Ganyani et al28 and Tindale et al27 used the same dataset to infer transmission pairs and estimate presymptomatic transmission. Their estimates were 48% (95% Credible interval (CrI) 32 to 67) and 74% for Singapore, and 62% (95% CrI 50 to 76) and 81% for Tianjin, respectively. This difference was likely to be due to different methods used to infer transmission pairs, different incubation periods and slightly different methods of estimating transmission time relative to symptom onset. Our estimates of presymptomatic transmission based on the generation times of Ganyani et al,28 and the serial intervals of Tindale et al27 also differ from the authors’ estimates (online supplemental table 2) due to using a different estimate for incubation period and a slightly different approach to transmission time calculation.
We estimate more presymptomatic transmission (64.2%) based on the serial interval of Chun et al38 than what is estimated in their paper (37%), as the incubation period used for our estimation of presymptomatic transmission (median 5.1 days) is much longer than that used in Chun et al’s calculations (median 2.9 days). This variation in estimates highlights the impact of inference method and also of incubation period on results. One of our motivations in this study was to facilitate comparisons between different countries or regions by removing some of the methodological variation due to different incubation period estimates and approaches to calculating transmission time.
The principle behind our analyses is that subtraction of incubation period from generation time allows us to estimate transmission time relative to symptom onset (figure 1). Generation time is difficult to observe directly and few papers estimate it. We included only a single estimate of generation time28 in our analyses. If the incubation period of an infector and of an infectee are taken to be independent and identically distributed, serial interval, the time between infector and infectee symptom onset, can be taken as an approximation of generation time,41 42 although serial interval will have more variation.28 The extra variation associated with serial interval should be borne in mind while interpreting our results.
There were further sources of variation that are challenging to address. Our description of the data sources underlying our simulation show large variation in sample size. With a relatively small sample size of 26, Kwok et al46 reported variation of more than a day in serial interval when certain and less certain subsets of transmission pairs were used, even though they were based on the same location and date range. The various methods (eg, Vink et al and te Beest et al41 61) for inferring transmission pairs from clusters of cases could also impact serial interval or generation time estimates. Griffin et al39 and Du et al33 highlight further variation associated with serial interval and generation time estimation, such as recall bias, resources for contact tracing and stage of epidemic, that could not be addressed with this current study.
We used published estimates rather than individual symptom onset data to inform our measures of presymptomatic transmission. Therefore, we could not investigate potential correlation between generation time/serial interval and incubation period. Using contact tracing data from Singapore and Tianjin, Tindale et al27 reported an intermediate signal for covariation between incubation period and serial interval. However, these authors showed that the degree of positive correlation did not greatly impact estimates of presymptomatic transmission. Liu et al62 simulated the effect of full correlation and anticorrelation between serial interval and incubation period on presymptomatic transmission estimates. However, the direction and magnitude of effects varied depending on which published estimates the simulations were based on. This highlights the need for ongoing investigations into SARS-CoV-2 transmission biology.
Despite the challenges associated with a highly variable international dataset, this study gives a clear signal that substantial presymptomatic transmission is occurring. This is consistent with evidence of virological studies, case reports and other quantitative studies. This means that extremely rapid and effective contact tracing, as well as isolation of contacts of cases before potential symptoms manifest, may be required to control disease spread.
Conclusion
Our study highlights substantial potential for presymptomatic transmission of COVID-19 in a range of different contexts. The proportion of presymptomatic transmission will vary by context, as this parameter is influenced by the contact rates between symptomatic infectious and susceptible people. These findings highlight the urgent need for extremely rapid and effective case detection, contact tracing and quarantine measures if the spread of SARS-CoV-2 is to be effectively controlled.
Supplementary Material
Footnotes
Twitter: @MiriamC51755360, @AndyByrneSci, @kieranwalshmpsi
Contributors: MC-B conceptualised the study, extracted parameter definitions from the literature, performed the analyses and drafted the manuscript. JG led the rapid review upon which the generation time and serial interval simulations are based. CM led the meta-analysis upon which the incubation period simulations are based upon. AC, KH, KOB and KW performed literature searches upon which the incubation period, generation time, serial interval and presymptomatic transmission information reported here are based upon. SM conceptualised, initiated and managed the overall project. MC-B, JG, CM, AByrne, JM, DME, AC, KH, ABarber, FB, EAL, KOB, PW, KW and SJM supplemented the literature review, discussed the study design, reviewed and edited the manuscript.
Funding: All investigators are full-time employees (or retired or former employees) of University College Dublin, the Irish Department of Agriculture, Food and the Marine (DAFM) or the Irish Health Information and Quality Authority (HIQA).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. All data and code used in the study are available through the following link: https://github.com/miriamcasey/covid-19_presymptomatic_project.
Ethics statements
Patient consent for publication
Not required.
References
- 1.WHO . COVID-19 situation reports, 2020. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [Accessed 19 Apr 2021].
- 2.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. JAMA 2020;323:1239. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 3.Roberts L. Polio, measles, other diseases set to surge as COVID-19 forces suspension of vaccination campaigns | Science | AAAS. Sci. Mag. Newsl, 2020. Available: https://www.sciencemag.org/news/2020/04/polio-measles-other-diseases-set-surge-covid-19-forces-suspension-vaccination-campaigns [Accessed 4 May 2020].
- 4.Saini KS, de Las Heras B, de Castro J, et al. Effect of the COVID-19 pandemic on cancer treatment and research. Lancet Haematol 2020;7:e432-e435. 10.1016/S2352-3026(20)30123-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawohl W, Nordt C. COVID-19, unemployment, and suicide. Lancet Psychiatry 2020;7:389–90. 10.1016/S2215-0366(20)30141-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes N. Economic effects of coronavirus outbreak (COVID-19) on the world economy. SSRN Journal 2020;8. 10.2139/ssrn.3557504 [DOI] [Google Scholar]
- 7.McKibbin WJ, Fernando R. The global Macroeconomic impacts of COVID-19: seven scenarios. SSRN Electron J. : 2020. [Google Scholar]
- 8.WHO . COVID-19 strategy update, 2020. Available: https://www.who.int/publications-detail/covid-19-strategy-update-14-april-2020
- 9.Peak CM, Kahn R, Grad YH, et al. Individual quarantine versus active monitoring of contacts for the mitigation of COVID-19: a modelling study. Lancet Infect Dis 2020;20:1025–33. 10.1016/S1473-3099(20)30361-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang R, Xia J, Chen Y, et al. A family cluster of SARS-CoV-2 infection involving 11 patients in Nanjing, China. Lancet Infect Dis 2020;20:534–5. 10.1016/S1473-3099(20)30147-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei WE, Li Z, Chiew CJ. Presymptomatic transmission of SARS-CoV-2 — Singapore, January 23–March 16, 2020. cent. Dis. control Morb. Mortal. Wkly. Rep;2020:411–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong Z-D, Tang A, Li K-F, et al. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis 2020;26:1052–4. 10.3201/eid2605.200198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian G, Yang N, Ma AHY, et al. COVID-19 transmission within a family cluster by presymptomatic carriers in China. Clin Infect Dis 2020;71:861–2. 10.1093/cid/ciaa316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y-C, Liao C-H, Chang C-F, et al. A locally transmitted case of SARS-CoV-2 infection in Taiwan. N Engl J Med 2020;382:1070–2. 10.1056/NEJMc2001573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu P, Zhu J, Zhang Z, et al. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J Infect Dis 2020;221:1757–61. 10.1093/infdis/jiaa077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med 2020;382:2019–20. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Na J, Zhou W. A familial cluster of COVID-19 indicating virus can be transmitted by asymptomatic carriers. Bull World Heal Organ 2020. [Google Scholar]
- 18.Lavezzo E, Franchin E, Ciavarella C, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of VO'. Nature 2020;584:425–9. 10.1038/s41586-020-2488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao J, Fan S, Chen J, et al. Epidemiological and clinical characteristics of COVID-19 in adolescents and young adults. Innovation 2020;1:100001. 10.1016/j.xinn.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 2020;63:706–11. 10.1007/s11427-020-1661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 2020;382:2081–90. 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae S, Kim H, Jung TY, et al. Epidemiological characteristics of COVID-19 outbreak at fitness centers in Cheonan, Korea. J Korean Med Sci 2020;35:e288. 10.3346/jkms.2020.35.e288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y, Zhang D, Yang P. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 2020;1:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong J, Chaw L, Koh WC, et al. Epidemiological investigation of the first 135 COVID-19 cases in Brunei: implications for surveillance, control, and travel restrictions. Am J Trop Med Hyg 2020;103:tpmd200771. 10.4269/ajtmh.20-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoehl S, Rabenau H, Berger A, Kortenbusch M, et al. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med 2020;382:26–8. 10.1056/NEJMc2001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimball A, Hatfield KM, Arons M, et al. Asymptomatic and Presymptomatic SARS-CoV-2 Infections in Residents of a Long-Term Care Skilled Nursing Facility - King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep 2020;69:377–81. 10.15585/mmwr.mm6913e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tindale LC, Stockdale JE, Coombe M, et al. Evidence for transmission of COVID-19 prior to symptom onset. Elife 2020;9. 10.7554/eLife.57149. [Epub ahead of print: 22 06 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganyani T, Kremer C, Chen D, et al. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Eurosurveillance 2020;25. 10.2807/1560-7917.ES.2020.25.17.2000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferretti L, Wymant C, Kendall M, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 2020;368:eabb6936. 10.1126/science.abb6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020;26:672–5. 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 31.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis 2020;93:284–6. 10.1016/j.ijid.2020.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren X, Li Y, Yang X. Evidence for pre‐symptomatic transmission of coronavirus disease 2019 (COVID‐19) in China. Influenza Other Respi Viruses 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, Xu X, Wu Y, et al. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg Infect Dis 2020;26:1341–3. 10.3201/eid2606.200357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prete CA, Buss L, Dighe A, et al. Serial interval distribution of SARS-CoV-2 infection in Brazil. J Travel Med 2021;28. 10.1093/jtm/taaa115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X-K, Liu XF, Wu Y, et al. Reconstruction of transmission pairs for novel coronavirus disease 2019 (COVID-19) in mainland China: estimation of Superspreading events, serial interval, and hazard of infection. Clin Infect Dis 2020;71:3163–7. 10.1093/cid/ciaa790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Dai J, Zhao J, et al. Estimation of incubation period and serial interval of COVID-19: analysis of 178 cases and 131 transmission chains in Hubei Province, China. Epidemiol Infect 2020;148:e117. 10.1017/S0950268820001338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan LT, Nguyen TV, Luong QC, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med 2020;382:872–4. 10.1056/NEJMc2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun JY, Baek G, Kim Y. Transmission onset distribution of COVID-19. Int J Infect Dis 2020;99:403–7. 10.1016/j.ijid.2020.07.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffin JM, Collins A, Hunt K. A rapid review of available evidence on the serial interval and generation time of COVID-19. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McAloon C, Collins Áine, Hunt K, et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open 2020;10:e039652. 10.1136/bmjopen-2020-039652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vink MA, Bootsma MCJ, Wallinga J. Serial intervals of respiratory infectious diseases: a systematic review and analysis. Am J Epidemiol 2014;180:865–75. 10.1093/aje/kwu209 [DOI] [PubMed] [Google Scholar]
- 42.Svensson A. A note on generation times in epidemic models. Math Biosci 2007;208:300–11. 10.1016/j.mbs.2006.10.010 [DOI] [PubMed] [Google Scholar]
- 43.Ali ST, Wang L, Lau EHY, et al. Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science 2020;369:1106–9. 10.1126/science.abc9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi Q, Wu Y, Mei S, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis 2020;20:911–9. 10.1016/S1473-3099(20)30287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R Core Team . R: a language and environment for statistical computing. R foundation for statistical computing., 2019. Available: http://www.r-project.org
- 46.Kwok KO, Wong VWY, Wei WI, et al. Epidemiological characteristics of the first 53 laboratory-confirmed cases of COVID-19 epidemic in Hong Kong, 13 February 2020. Eurosurveillance 2020;25:2000155. 10.2807/1560-7917.ES.2020.25.16.2000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Zhao S, Liao Y, et al. Estimating the serial interval of the novel coronavirus disease (COVID-19) based on the public surveillance data in Shenzhen, China, from 19 January to 22 February 2020. Transbound Emerg Dis 2020;67:2818–22. 10.1111/tbed.13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son H, Lee H, Lee M, et al. Epidemiological characteristics of and containment measures for COVID-19 in Busan, Korea. Epidemiol Health 2020;42:e2020035. 10.4178/epih.e2020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pham QT, Rabaa MA, Duong HL. The first 100 days of SARS-CoV-2 control in Vietnam. Clin Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Litvinova M, Wang W, et al. Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside Hubei Province, China: a descriptive and modelling study. Lancet Infect Dis 2020;20:793–802. 10.1016/S1473-3099(20)30230-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Teunis P. Strongly heterogeneous transmission of COVID-19 in mainland China: local and regional variation. Front Med 2020;7:329. 10.3389/fmed.2020.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382:1199–207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Zhou Q, He Y, et al. Nosocomial outbreak of COVID-19 pneumonia in Wuhan, China. Eur Respir J 2020;55:2000544. 10.1183/13993003.00544-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Najafi F, Izadi N, Hashemi-Nazari S-S, et al. Serial interval and time-varying reproduction number estimation for COVID-19 in Western Iran. New Microbes New Infect 2020;36:100715. 10.1016/j.nmni.2020.100715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walsh KA, Jordan K, Clyne B, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect 2020;81:357–71. 10.1016/j.jinf.2020.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrne AW, McEvoy D, Collins AB, et al. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open 2020;10:e039856. 10.1136/bmjopen-2020-039856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 58.Cevik M, Tate M, Lloyd O, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021;2:e13–22. 10.1016/S2666-5247(20)30172-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kam K-Q, Yung CF, Cui L, et al. A well infant with coronavirus disease 2019 with high viral load. Clin Infect Dis 2020;71:847–9. 10.1093/cid/ciaa201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J-Y, Chen T-J, Hwang S-J. Analysis of imported cases of covid-19 in Taiwan: a nationwide study. Int J Environ Res Public Health 2020;17:3311. 10.3390/ijerph17093311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.te Beest DE, Wallinga J, Donker T, et al. Estimating the generation interval of influenza A (H1N1) in a range of social settings. Epidemiology 2013;24:244–50. 10.1097/EDE.0b013e31827f50e8 [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Centre for Mathematical Modelling of Infectious Diseases nCoV Working Group, Funk S, et al. The contribution of pre-symptomatic infection to the transmission dynamics of COVID-2019. Wellcome Open Res 2020;5:58. 10.12688/wellcomeopenres.15788.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J, Huang Y, Tu C, et al. Household transmission of SARS-CoV-2, Zhuhai, China, 2020. Clin Infect Dis 2020;71:2099–108. 10.1093/cid/ciaa557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aghaali M, Kolifarhood G, Nikbakht R, et al. Estimation of the serial interval and basic reproduction number of COVID-19 in Qom, Iran, and three other countries: a data-driven analysis in the early phase of the outbreak. Transbound Emerg Dis 2020;67:2860–8. 10.1111/tbed.13656 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-041240supp001.pdf (722.4KB, pdf)
bmjopen-2020-041240supp002.pdf (13.1KB, pdf)
bmjopen-2020-041240supp003.pdf (18.6KB, pdf)
bmjopen-2020-041240supp004.pdf (15.3KB, pdf)
bmjopen-2020-041240supp005.pdf (143KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. All data and code used in the study are available through the following link: https://github.com/miriamcasey/covid-19_presymptomatic_project.