Summary
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) has become the preferred method for mapping chromatin accessibility, due to its time and input material efficiency. However, it can be difficult to evaluate data quality and identify sources of technical bias across samples. Here, we present ataqv, a computational toolkit for efficiently measuring, visualizing, and comparing quality control (QC) results across samples and experiments. We use ataqv to analyze 2,009 public ATAC-seq datasets; their QC metrics display a ten-fold range. Tn5 dosage experiments and statistical modeling show that technical variation in the ratio of Tn5 transposase to nuclei and sequencing flowcell density induces systematic bias in ATAC-seq data by changing the enrichment of reads across functional genomic annotations including promoters, enhancers, and transcription factor bound regions, with the notable exception of CTCF. Ataqv can be integrated into existing computational pipelines and is freely available at https://github.com/ParkerLab/ataqv/.
Introduction
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) is the current preferred method for mapping chromatin accessibility due to its simplicity, speed, and low input material requirements (Buenrostro et al., 2013). In ATAC-seq, intact nuclei are exposed to Tn5 transposase, which preferentially cuts protein-free unprotected DNA to ligate sequencing adapters to the cleaved ends. After sequencing, the reads are aligned to a reference genome and peak calling is performed to determine the regions of the genome enriched for transposase-accessible DNA. This information can be used to inform the prediction of active regulatory regions (Buenrostro et al., 2013), nucleosome positioning (Schep et al., 2015), and transcription factor binding (Quach and Furey, 2016; Schmidt et al., 2017).
The number of publicly-available ATAC-seq datasets is rapidly growing, but the quality of these datasets can vary widely. ATAC-seq libraries may differ in PCR amplification bias, fragment length distribution, transcription start site (TSS) enrichment, nuclei prep quality, proportion of mitochondrial reads, and other variables (Benjamini and Speed, 2012). ATAC-seq involves a number of experimental and computational steps which may introduce such heterogeneity. Some of these confounders are shared with many other high-throughput sequencing-based assays (e.g., PCR amplification bias), while others are more ATAC-seq specific (e.g., potentially high proportions of mitochondrial reads and variable nuclei prep quality). Identifying these confounders and adjusting for them in downstream analyses is an important part of reproducible and rigorous ATAC-seq analyses.
Few computational quality control (QC) tools exist for ATAC-seq, and each of the existing tools have notable limitations. The ENCODE ATAC-seq processing pipeline includes a script (ATAqC; https://github.com/kundajelab/ataqc) that produces a QC report, but this script is difficult to utilize as a standalone tool. Considerable effort is required to integrate it into a custom pipeline as one must install a complete conda environment, and it supports only the human and mouse reference genomes. It produces one report per sample (rather than a unified report for multiple samples), complicating cross-sample comparisons. A second tool, ATACseqQC (Ou et al., 2018), exists as an R Bioconductor package. This package provides R functions for QC of BAM files and preprocessing for common downstream analyses. Because it provides functions rather than generating a single report, it is a flexible framework but places additional work on the end user and renders the package inaccessible to those unfamiliar with R. Like ATAqC, it generates separate plots for each bam file, making it less practical for cross-sample comparisons. Alfred (Rausch et al., 2019) is a third tool with ATAC-seq QC functionality. It is run on the command line and a web server is available for visualizing the results. It is quick to set up and run, but does not have an option to visualize several libraries simultaneously, and can handle only three read groups per bam file in the case that the user wishes read groups to be analyzed separately.
Several additional software packages built to assist in ATAC-seq data processing and analysis exist, and these each include QC steps; however, they are not meant to provide comprehensive QC on bam files. These packages include ATAC-pipe (Zuo et al., 2019), which supports only two reference genomes (hg19 and mm9) and does not perform QC on a user-provided bam file (the primary QC function accepts raw fastq files, tying read mapping and QC together); and esATAC (Wei et al., 2018), which similarly provides few read mapping statistics when starting from a bam file (rather than a fastq file) and produces individual QC plots (e.g., fragment length distribution and TSS coverage) for each sample/replicate, complicating cross-sample comparisons.
In order to address these shortcomings, and to facilitate the unified analysis of thousands of ATAC-seq datasets, we developed a new ATAC-seq QC and visualization software package, ataqv (Fig. S1). Ataqv overcomes the primary limitations of existing packages. It eases cross-sample and cross-experiment comparisons, can easily be integrated into existing data processing pipelines, and produces interactive reports that are easy to share. We apply ataqv to thousands of publicly-available libraries and observed a broad range of results across diverse QC metrics. We therefore carefully constructed Tn5 dosage experiments to explore the influence of technical variation on ATAC-seq profiles and find that experimental conditions that influence the ATAC-seq fragment length distribution, such as sequencing lane cluster density and Tn5:nuclei ratio, robustly skew QC metrics and alter the biological interpretation of ATAC-seq results. QC reports and metrics from the ataqv package can help identify these technical biases and adjust for them in downstream analyses.
Results
Ataqv is a modular and accessible tool for ATAC-seq quality control and visualization
Ataqv allows quick visualization and comparison of 35 metrics and potential confounders across samples (Table S1; Fig. S2). It produces both machine-readable (JSON format) metrics and an interactive HTML report (Fig. S1b) that is accessible to experimental scientists and easy to share. It is simple to integrate into existing ATAC-seq pipelines and can handle thousands of samples, an important consideration as single-cell analyses come of age and sample sizes grow. The only inputs are a BAM file of aligned reads, an optional BED file of peaks, and the name of the organism to which they were aligned (Fig. S1a). Human, mouse, rat, worm, fly, and yeast reference metadata is built in; metadata for other organisms (autosomal and mitochondrial chromosome names) can be easily supplied. If desired, metrics can be calculated separately for each read group in a BAM file (facilitating the processing of BAM files that may contain many libraries, as is often the case for single-cell data). A demonstration of the interactive ataqv HTML report is at https://parkerlab.github.io/ataqv/demo/ and the ataqv source code is freely available under the GPL3 license at https://github.com/ParkerLab/ataqv/.
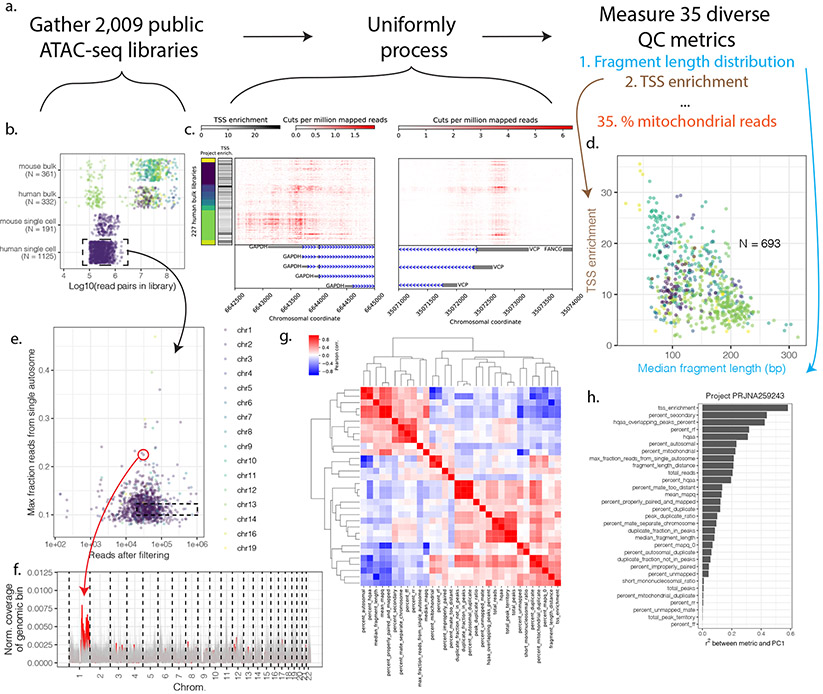
To demonstrate the utility of ataqv and assess the heterogeneity of publicly-available datasets, we downloaded and uniformly processed 2,009 human and mouse ATAC-seq libraries (Table S2 and Fig. 1a-c). The fragment length distributions (FLDs) and TSS enrichment for these libraries display over ten-fold variability (Fig. 1d), and considerable heterogeneity exists even between libraries from the same study (Fig. 1c,d, S3). Links to the interactive ataqv sessions for these uniformly processed data sets are available in the Methods section. These sessions make clear the heterogeneity in public ATAC-seq data and may be helpful as a point of reference to compare new ATAC-seq datasets.
Figure 1. Survey of public ATAC-seq data.
(a) 2,009 public ATAC-seq libraries representing 23.4 billion read pairs were downloaded and uniformly processed. (b) Number of libraries and total read pairs per species and project (colors represent different projects). (c) ATAC-seq signal at promoters of two housekeeping genes (GAPDH and VCP) across human bulk libraries with at least 5M reads post-filtering. Colors along the y-axis represent project. (d) TSS enrichment and median fragment length for the 693 processed bulk (not single cell) datasets. (e) Maximum fraction of autosomal reads derived from a single autosome for public human single-cell ATAC-seq data. (f) Normalized read coverage in 2Mb windows (with 1Mb steps between them) across chromosomes for the outlier circled in red from (e) and for a set of 90 non-outlier cells from the same cell type (GM12878; all lying within the dotted box in (e)). The outlier’s read coverage is represented by the red line; non-outliers are shown in gray. One arm of chromosome 1 shows abnormally high coverage in the outlier cell. (g). Correlation between ataqv metrics across public bulk ATAC-seq datasets. Metric abbreviations are listed in Table S1 (h). Correlation between PC1 and ataqv metrics in project PRJNA259243.
QC of single-cell ATAC-seq (scATAC-seq) data is especially critical to ensure meaningful and reproducible results, as a portion of the sequencing reads produced in scATAC-seq experiments may be derived from background DNA released by non-viable cells. Per-cell scATAC-seq fragment counts, sometimes in combination with TSS enrichment, is commonly used to filter the data to those reads derived from high-quality cells (Rai et al., 2020; Satpathy et al., 2019). Ataqv introduces another metric that we believe will be useful in filtering single-cell data in particular: the maximum fraction of autosomal sequencing reads derived from a single autosome. Most scATAC-seq data is produced using microfluidics platforms, and in such systems free DNA from dead or dying cells may end up being transposed and barcoded, perhaps in close proximity to (and with the same barcodes as) healthy cells. As a result some cellular barcodes that appear to represent healthy cells and could pass common QC thresholds may contain reads from this free DNA. If large, free-floating chromosomal segments are represented in such barcodes, the observed distribution of reads across chromosomes would not match the expected distribution. To demonstrate this, we examined the maximum fraction of autosomal sequencing reads derived from a single chromosome for all cells in public scATAC-seq data (Fig. 1e). This metric illuminated extreme chromosomal read imbalance in some of the cells. Plotting the read coverage in genomic bins across each chromosome for some of these cells frequently showed that large contiguous segments of the chromosomes have increased coverage relative to the rest of the chromosomes (Fig. 1f), consistent with a scenario in which broken chromosome(s) derived from another cell received the same barcode as the reads from a potentially healthy cell. Such cases should be filtered out during QC of scATAC-seq data. We additionally examined this metric in the public bulk ATAC-seq libraries, where outliers may reflect abnormal karyotypes (Fig. S4). Consistent with this notion, the outliers we observed (e.g., K562 and mESCs) tended to be cell lines with known abnormal karyotypes (Naumann et al., 2001; Rebuzzini et al., 2008; Sugawara et al., 2006).
To explore the relationship between QC metrics, we calculated the correlation between all QC metric pairs across the public bulk libraries analyzed (Fig. 1g). We find that TSS enrichment positively correlates with % of reads in peaks, and negatively correlates with median fragment length. Read count positively correlates with the number of peaks called, likely because greater read count increases the statistical power to call peaks (Landt et al., 2012). A few metrics show such high correlation that they may be considered somewhat redundant for the purposes of standard QC; e.g., the number of peaks unsurprisingly shows very high correlation with peak territory (the amount of the genome covered by peaks). The ataqv software includes an option to output a reduced set of QC metrics by pruning out several metrics that tend to show very high correlation with other metrics. Ataqv metrics can be correlated with principal component (PC) scores in order to determine which characteristics of the libraries may be contributing most to the variance in the data across libraries. To demonstrate this, we performed a principal component analysis on the project with the most bulk ATAC-seq libraries from a single cell type and correlated the PC1 scores against ataqv metrics (Fig. 1h; we selected data from a single project and cell type because in a cross-project or cross-cell-type analysis PC1 would capture project or cell type). TSS enrichment showed the highest correlation with PC1 scores (Figs. 1h, S5), indicating that TSS enrichment may be a particularly important variable to examine during QC.
Ataqv metrics may be useful as covariates in downstream analysis, in part because they may reflect latent variables. For example, while examining a subset of ATAC-seq libraries from one study, we noticed that half of the libraries displayed a considerably different fragment length distribution than the other half. Through inspection of the sequencing read names we inferred the sequencing run and flowcell that each library was sequenced on and found that the median fragment lengths of each library covaried with the sequencing flowcell (Fig. S6a), suggesting that the QC metric was capturing a batch effect that otherwise may not have been apparent from the metadata (public metadata is frequently difficult to parse or missing altogether). Running a differential peak analysis with and without the QC metric median fragment length as a covariate, we found a robust shift towards more extreme p-values from the analysis when the covariate was included (Fig. S6b), which indicates increased statistical power after controlling for the batch effect.
To further demonstrate the utility of ataqv in identifying problematic variance, we used it to systematically explore two potential sources of bias. ATAC-seq experiments produce a stereotypical FLD, distinguished by many short (< 100 bp) fragments and a tail of longer (> 147 bp) fragments in multiples of the nucleosomal unit size. Because chromatin structure differs across classes of regulatory elements, different regulatory elements produce different local FLDs (Buenrostro et al., 2013). We therefore hypothesized that variables perturbing the FLD will systematically change ATAC-seq results. We therefore designed experiments to test the influence of two technical variables: Tn5:nuclei ratio and sequencing lane cluster density. As noted in (Buenrostro et al., 2015), the ratio of Tn5 enzyme to nuclei number is a determining factor in the experiment FLD. Increasing this variable should shift the FLD toward shorter fragments. Sequencing lane cluster density also affects the length distribution of sequenced fragments, with high cluster density generally favoring shorter fragments (Bronner et al., 2014; Gohl et al., 2019). Importantly, while both of these variables affect the FLD, they do so in different ways. In the case that the Tn5:nuclei ratio changes, both the global (genome-wide) as well as local (locus-specific) FLDs should shift. When the sequencing lane cluster density changes, the true underlying global and local FLDs do not change; however, they are subsampled in different manners between the sequencing runs (high cluster density runs should sample more from the left-most part of the FLD than do low cluster density runs).
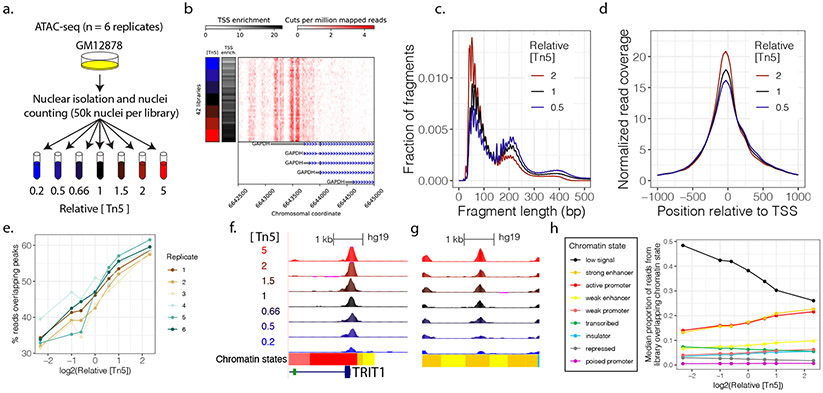
To quantify the influence of cluster density and Tn5:nuclei ratio, we performed two sets of ATAC-seq experiments. In one, we performed ATAC-seq on GM12878 using seven different Tn5 concentrations all using 50k nuclei as input, and sequenced each library on two separate sequencing runs, one run having 124% the cluster density of the other (411M vs 508M clusters passing filtering; Fig. S7a; n = 3 independent nuclear isolations, producing a total of 21 libraries). Importantly, during this experiment we observed that the number of PCR cycles required for each library strongly covaried with Tn5 concentration (Fig. S8). Because PCR amplification can influence the fragment length distribution (Frohman et al., 1988) and introduce other biases, we designed a second experiment in which we again performed ATAC-seq on GM12878 nuclei using seven different concentrations of Tn5 while holding the number of nuclei constant at 50k (n = 6 independent nuclear isolations, producing a total of 42 libraries; Fig. 2a, b) but additionally held the number of PCR cycles constant across libraries (Fig. S9). We refer to these experiments as the ‘PCR-variable’ and ‘PCR-constant’ experiments, respectively. The interactive ataqv reports for both of these experiments are available online (see Methods).
Figure 2. Tn5 concentration systematically alters ATAC-seq results.
(a) ATAC-seq was performed on GM12878 cells, using seven different concentrations of Tn5 transposase while keeping the number of nuclei constant. Six replicates were performed. (b) GAPDH locus coverage. (c) Increasing Tn5 concentration shifts the fragment length distribution towards shorter fragments. (d) Increasing Tn5 concentration increases TSS enrichment. (e) Increasing Tn5 concentration increases the percentage of high-quality, autosomal reads overlapping peaks. (f) UCSC genome browser screenshot displaying a Tn5-sensitive promoter peak (http://genome.ucsc.edu/)(Casper et al., 2018; Kent et al., 2002). (g) UCSC genome browser screenshot displaying a Tn5-sensitive enhancer peak. (h). The percentage of ATAC-seq reads falling into enhancer and active TSS chromatin states increases with increasing Tn5, while the percentage of reads falling into low signal regions decreases. Values shown represent the median values across replicates in the PCR-constant experiment.
Sequencing lane cluster density biases ATAC-seq library fragment length metrics and TSS enrichment
First, we examined the effect of sequencing lane cluster density on ATAC-seq results. As expected, despite the fact that the same libraries were sequenced in both runs, the fragment length distributions from the high cluster density run were consistently shifted toward shorter fragments relative to the low cluster density run (Fig. S7b). The average difference in median fragment length between sequencing runs was 12 bps. Interestingly, TSS enrichment was consistently higher in the high cluster density sequencing run (average difference of 1.83; Fig. S7c). Other QC metrics differed consistently but to a lesser degree (see ataqv HTML report). We conclude that sequencing run cluster density has a systematic effect on ATAC-seq QC metrics, likely because different cluster densities effectively ‘subsample’ the actual library fragment length distribution in a biased manner and this changes the representation of different functional regions (like the TSS) across the genome.
ATAC-seq results are sensitive to Tn5:nuclei ratio
Next, we examined the effect of the Tn5:nuclei ratio, using the results of the PCR-constant experiment and of the high cluster density sequencing run of the PCR-variable experiment. As expected, when the number of PCR cycles was held constant, the fragment length distribution shifted toward a greater proportion of shorter fragments as Tn5 concentration increased (Fig. 2c, S10a). This correlation was attenuated when PCR cycles were allowed to vary, likely reflecting the influence of PCR cycles on FLDs (Fig. S11a). Furthermore, in both experiments we found that increasing Tn5 concentration negatively correlated with the percent of mitochondrial sequencing reads (Fig. S10b, S11b). We speculate that as Tn5 concentrations increase, an increasing proportion of mitochondrial DNA (which competes with nuclear DNA for the pool of Tn5) (Montefiori et al., 2017) is digested to the extent that it is no longer effectively sequenced. Alternatively, it may be that an increasing proportion of nuclear genomic DNA is digested sufficiently to be effectively sequenced. As mitochondrial reads are typically filtered out during standard ATAC-seq data processing, reducing mitochondrial reads increases the amount of sequence available for downstream analysis. Read duplication rate negatively correlated with Tn5 in both experiments (Fig. S10d, S11e). Overall, increasing the amount of Tn5 resulted in a considerably greater proportion (approximately four-fold higher comparing the extremes of Tn5 concentration) of reads surviving filtering in both experiments (Fig. S10e, S11f). Additionally, we found Tn5 concentration positively correlated with the enrichment of fragments around TSSs (Fig. 2d) in the PCR-constant experiment but not in the PCR-variable experiment (Fig. S11c). The enrichment of reads in ATAC-seq peaks increased approximately 1.75-fold from the lowest Tn5 concentration to the highest (Fig. 2e, S11d). Examining the peaks called for each library, we found that the number of peaks increases with Tn5 concentration, and that this relationship is not solely due to differences in the number of reads surviving bioinformatic filtering at each Tn5 concentration (Fig. S12). Furthermore, we found that as Tn5 concentration increases, peak calls become more reproducible, such that the mean Jaccard index between peak calls from two replicates increases as Tn5 concentration increases (Fig. S13). We performed a principal component analysis and found that the first principal component correlated with Tn5 concentration (Fig. S14), confirming that this technical variable has a systematic effect on ATAC-seq results.
In order to determine whether there is a subset of peaks that are Tn5 sensitive or whether Tn5 sensitivity is a shared property of all peaks, we used a negative binomial generalized linear model (GLM) to model the number of reads in an ATAC-seq peak as a function of the Tn5 concentration, controlling for replicate and using the PCR-constant experiment. At a false discovery rate (FDR) of 5%, we identified 49,989 Tn5 sensitive peaks (of 70,658 total and 62,576 for which the model converged; Fig. S15, top panel; Fig. 2f,g, Fig. S16-18), of which the overwhelming majority (99%) displayed a positive relationship between Tn5 concentration and peak signal (49,443 of the 49,989 5% FDR peaks). Similar results were obtained in the PCR-variable experiment (29,355 peaks significant of 43,447 that converged; Fig. S19). This massive shift (79.9% of converged peaks in PCR-constant experiment) indicates that Tn5 sensitivity is a common quantitative trait of peaks across the genome. Adding a covariate summarizing the fragment length distribution of each library to the GLM reduced the number of Tn5 sensitive peaks detected in the PCR-constant experiment (Fig. S15; of the 62,576 peaks that converged in all models, 79.9% were Tn5 sensitive at 5% FDR when using no covariate and 8.3% were sensitive after adding median fragment length to the model as a covariate). Such covariates had no effect in the PCR-variable experiment (Fig. S19).
Our results suggest that higher Tn5 concentration increases the ATAC-seq signal-to-noise ratio (at least over the tested range of Tn5 concentrations). In order to determine if this holds for both promoter and enhancer regions, we calculated the proportion of reads that overlapped with chromHMM-derived GM12878 chromatin states (Fig. 2f,g,h, S20). (Ernst and Kellis, 2012; Parker et al., 2013). We found that the percentage of reads falling in strong enhancer and active promoter chromatin states increases with increasing Tn5 (Bonferroni-adjusted p-values of 6.34e-22 and 1.2e-16, respectively, in the PCR-constant experiment; 2.61e-9 and 3.87e-7 in the PCR-variable experiment), and that this is accompanied by a decrease in the proportion of reads falling in the low signal state (Bonferroni-adjusted p = 1.04e-21 and p = 1.2e-8 in the PCR-constant and PCR-variable experiments, respectively). This increase in signal-to-noise due to a technical variable is therefore observed for both TSS-proximal and TSS-distal regulatory elements.
To determine if the binding of certain transcription factors (TFs) might influence the change in ATAC-seq signal, we examined ATAC-seq reads and peaks in relation to ENCODE GM12878 reproducible ChIP-seq peaks (ChIP-seq experiments on 85 TFs; Table S3) (ENCODE Project Consortium, 2012; Sloan et al., 2016). For all TFs, the proportion of ATAC-seq reads overlapping with ChIP-seq peaks increased as the Tn5 concentration increased (Fig. S21). This is consistent with our chromatin state findings (Fig. 2h), given that TF binding will commonly overlap with enhancers and promoters which themselves show increased signal with increasing Tn5 concentration. In order to determine if the binding of certain TFs correlates with Tn5 sensitivity, we examined the probability that a peak is Tn5 sensitive given that it is bound by a certain TF, controlling for peak size (Fig. S22). We performed logistic regression and discovered that binding of nearly all TFs (82 out of 85) are significantly (Bonferroni adjusted p < 0.05) associated with increased Tn5 sensitivity. The only exceptions are CTCF, RAD21, and REST. These factors are commonly associated with strongly phased nucleosomes (Fu et al., 2008; Harwood et al., 2019; Sadeh and Allis, 2011; Wiechens et al., 2016); we speculate that this may render regions bound by them less sensitive to variability in Tn5 concentration. Overall, these results show that technical variation in ATAC-seq data is associated with selectively biased profiling of functional genomic regions.
Discussion
We conclude that ATAC-seq experiments performed for the purpose of identifying enhancers and promoters will likely achieve better signal-to-noise with increased Tn5 concentration. We note that while this relationship holds over the 25-fold range of Tn5 concentrations we have tested, it is likely that continuing to increase Tn5 concentration beyond a certain point will begin to reduce data quality as highly accessible regions are digested to an extent that they can no longer be effectively sequenced. We have not, however, reached this concentration in the data presented here. Another important caveat is that these relationships may change when nuclei numbers are limiting. The ATAC-seq protocol published in (Buenrostro et al., 2015) states that when “too few” cells are used, the proportion of reads derived from inaccessible regions of the genome increases. Our data was generated using a large enough number of cells that Tn5, rather than cell number, appears to be the limiting factor in library complexity. When this is the case, we find that increasing the Tn5 concentration increases the proportion of reads in peaks, in enhancer and promoter chromatin states, and in most TF bound regions. These findings are generally consistent with another recent publication, which adjusted several experimental variables in cell lines and generally found that increasing Tn5 concentration yielded more peaks and greater enrichment of reads around TSS (Fujiwara et al., 2019); however another publication, utilizing mouse embryonic stem cells, concluded that changing Tn5 concentration had little effect on ATAC-seq results (Corces et al., 2017).
We note that, although we generated our data under a large range (25X) of Tn5:nuclei ratios, considerable differences are apparent even over lower ranges that are likely to be encountered in real-world lab settings. Differences between samples in the number of input cells and the efficiency of nuclear isolation can easily generate two-fold or greater differences in Tn5:nuclei ratio (Fig. S23). We expect that these differences may be especially extreme in cases of variable sample quality, or when working with tissues for which nuclear isolation is especially difficult (e.g., adipose tissue). Accordingly, nuclei counting should be a standard step in the ATAC-seq protocol to ensure consistent results.
The observed relationship between Tn5:nuclei ratio and PCR cycles is also an important finding. When the ratio is low, additional PCR cycles may be necessary in order to further enrich for short fragments in the library. This is another reason to control the Tn5:nuclei ratio, as failure to do so may lead one to differentially amplify libraries later in the protocol, which may introduce additional PCR-related biases.
Another recent publication (Gohl et al., 2019) found considerable differences in the fragment length biases of different Illumina sequencing machines, and flagged this as a point of concern for those performing ATAC-seq. Our results build on and extend these published results, as we find that cluster density differences across runs on the same type of sequencing machine systematically perturb fragment length as well. Therefore, both the type of sequencing machine and the loading concentration of sequencing libraries should be taken into account when planning and analyzing ATAC-seq experiments.
When performing QC before proceeding with data analysis, a common question involves which QC metrics to focus on and which thresholds to select. We urge caution in following such hard- and-fast rules for several reasons. First, QC metrics may differ systematically according to factors like cell type. For example, embryonic stem cells are thought to have greater genome-wide chromatin accessibility than more differentiated cells (Aughey et al., 2018; Le Gros et al., 2016) which could result in a significantly different distribution of ATAC-seq reads and therefore differences in TSS enrichment, number of peaks, percent of reads in peaks, etc. Similarly, cell types vary in their mitochondrial DNA copy numbers (Kelly et al., 2012; Sun and St John, 2018) which may lead to different levels of mitochondrial reads in different cell types, and sample heterogeneity (e.g., a homogenous cell line vs a tissue sample composed of several different cell types) likely affects many of these metrics. Second, ideal QC metrics may depend on analysis goals. For example, if one wishes to map precise nucleosome positions adjacent to open chromatin using a method such as NucleoATAC (Schep et al., 2015), a mix of shorter and longer reads are favorable, and therefore a library with very high TSS enrichment but short median fragment length (few reads longer than 150 bps) might be considered a “poor” library for this purpose. These study-specific goals are therefore different, and the associated QC metrics that indicate ‘good’ may not be shared. Third, one’s threshold for “acceptable” data will realistically vary depending on sample availability and analysis needs. If working with valuable clinical samples or very rare, hard-to-obtain cell types, the amount of material per sample or the number of samples available may be a limiting factor. In this case, one may settle for relatively lower-quality data than one would accept if one were creating abundant cell line ATAC-seq data (for which sample availability is not likely to be an issue). Lastly, we have found that the details of the calculation of a metric can make a significant difference in the resulting QC values. The calculation of TSS enrichment is a prime example. A variety of methods for calculating TSS enrichment exist among the QC packages and pipelines available. Ataqv calculates coverage around the TSS using entire ATAC-seq fragments, while other packages calculate coverage using only the cutsite or by shifting and extending individual sequencing reads such that the reads are centered on the cutsite. We have found that different methods can result in considerably different TSS enrichment values for the same library (Fig. S24). Unsurprisingly, the TSS list used for calculation of TSS enrichment can change results as well (Fig. S25) (Corces et al., 2017). Given all of the above factors, we believe that when selecting QC thresholds researchers should look at the distribution of many QC metrics calculated uniformly across libraries, and use those distributions to determine reasonable thresholds. To demonstrate this and provide one point of reference to users, we have plotted the distributions of several QC metrics in the different cell types from the analyzed public bulk ATAC-seq data (Fig. S26). Similarly, if researchers wish to compare the characteristics of their ATAC-seq libraries to previously-generated libraries, a suitable reference library is probably one that is species- and cell type-matched, was processed using the same genome annotations, and that has already been shown to give quality results in the downstream analyses that the author(s) plan to utilize the newer libraries for. The ataqv packages facilitates easy implementation of all these considerations.
The systematic relationships between technical variance and change in ATAC-seq signal highlighted here demonstrate the importance of identifying and adjusting for heterogeneity in ATAC-seq data. The heterogeneity of the data may also inform one’s choice of downstream methods. For example, several existing methods leverage the characteristic ATAC-seq fragment length distribution to call peaks (Tarbell and Liu, 2019), predict TF binding (Li et al., 2019), or determine nucleosome positioning (Schep et al., 2015). Cross-sample heterogeneity in FLDs may confound such analyses.
It has become increasingly clear that rigorous analysis of quantitative chromatin signatures will be critical for understanding complex human traits and diseases (Alasoo et al., 2018; Khetan et al., 2018; Kumasaka et al., 2019; Varshney et al., 2019). We expect ataqv to be useful for scrutinizing confounding heterogeneity and it will therefore be an important tool in dissecting biological mechanisms.
STAR Methods
Lead contact and materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stephen C.J. Parker (scjp@umich.edu). This study did not generate new unique reagents.
Experimental model and subject details
We cultured GM12878 cells following the ENCODE GM12878 cell culture protocol (https://www.encodeproject.org/documents/1bb75b62-ac29-4368-9855-68d410e1963a/), except that we added plasmocin (Invivogen, San Diego, CA; 50 ug/mL) to the growth media to prevent mycoplasma contamination. The cell line was not authenticated.
C2C12 cells were proliferated at 37 degrees Celsius in growth media (DMEM + 20% FBS + 1% penicillin streptomycin) in a CO2 incubator (5% CO2). The cell line was not authenticated.
Method details
ATAC-seq experiments
We conducted ATAC-seq as described in Buenrostro et al. (2015) using a home-made Tn5 that we synthesized as described in (Picelli et al., 2014). We isolated nuclei from three independent cultures (“replicates”) for the ‘PCR-variable’ experiment and six additional cultures for the ‘PCR-constant’ experiment. For each replicate we incubated 50,000 nuclei with various concentrations of enzyme (⅕X, ½X, ⅔X, 1X, 1.5X, 2X, 5X; 1X corresponds to 2.5 uL of 1:1 Tn5-A/B mix) at 37°C for 30 minutes in a 50 uL reaction. We column-purified the tagmented DNA using the Zymo DNA Clean & Concentrator-5 kit (Zymo Research, Irvine, CA). In the PCR-variable experiment, we PCR-amplified the entire eluate until amplification curve reached its mid-log phase (⅓ to ½ of max signal; the number of PCR cycles required to reach this phase differed among groups, see Results section); whereas in the PCR-constant experiment, we amplified the entire eluate with a fixed number of PCR cycles (16) for all samples. We purified the products using SPRI beads prepared as in (Rohland and Reich, 2012) and eluted in 20 uL of TE buffer with Tween-20 (10 mM Tris-HCl, 0.1 mM EDTA, 0.05% Tween-20, pH 8). Libraries were multiplexed and sequenced on an Illumina NextSeq 500 instrument.
GM12878 ATAC-seq data processing
All reads were trimmed to 36 bps using fastx_trimmer (from fastx-toolkit v 0.0.14). Adapters were trimmed using cta (v. 0.1.2; https://github.com/ParkerLab/cta). Reads were aligned to hg19 (Lander et al., 2001) using bwa mem (v. 0.7.15; flags: -M) (Li and Durbin, 2009). For the ATAC-seq experiments that were used to observe the effect of Tn5 concentration, each library was sequenced on two sequencing runs; bam files from the two sequencing runs were merged using samtools merge. Picard MarkDuplicates (v. 2.18.27; http://broadinstitute.github.io/picard) was used for duplicate removal (options: VALIDATION_STRINGENCY=LENIENT) and samtools (v. 1.7) (Li et al., 2009) was used to filter for autosomal, properly-paired and mapped read pairs with mapping quality >= 30 (samtools view -b -h -f 3 -F 4 -F 8 -F 256 -F 1024 -F 2048 -q 30). Peak calling was performed using MACS2 callpeak (v. 2.1.1.20160309; options: --nomodel --broad --shift −100 --extsize 200 --keep-dup all) (Zhang et al., 2008). Peaks were filtered against ENCODE blacklists (ENCODE Project Consortium, 2012) (downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/wgEncodeDacMapabilityConsensusExcludable.bed.gz and http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/wgEncodeDukeMapabilityRegionsExcludable.bed.gz) using bedtools intersect (option -v; v. 2.27.1) (Quinlan, 2014). Ataqv (v. 1.1.0) was run on the bam files with duplicates marked, and the blacklists were passed as excluded regions. For the TSS file, we took the hg19.tss.refseq.housekeeping.ortho.bed.gz TSS file packaged with ataqv (representing TSS for genes with 1:1:1 human:mouse:rat orthologues where the human gene is a housekeeping gene (Eisenberg and Levanon, 2013); GitHub commit f4b655) and further filtered the list to remove genes that had more than one TSS in human, mouse, or rat. One library from the PCR-variable experiment had very few reads (~0.5M in one sequencing run and ~0.25M in the second) and was excluded from downstream analysis. For figures displaying ATAC-seq coverage, we normalized the signal to account for differences in library size and all signal track plots show the same range. Normalization was performed on the MACS2-created *treat_pileup.bdg bedgraph files. The script used for normalization is available on GitHub (https://github.com/porchard/normalize_bedgraph; commit 82ab906; run with parameters ‘--to-number-reads 10000000’). The normalized bedgraph files were then converted to bigwig format using bedGraphToBigWig (v. 4) (Kent et al., 2010).
Public ATAC-seq data (mouse and human) processing
The processed read groups/libraries are listed in Table S2. Libraries were processed in the same manner as the GM12878 ATAC-seq libraries (mapping to mm9 or hg19 as appropriate) (Lander et al., 2001; Mouse Genome Sequencing Consortium et al., 2002). For mm9 we used the blacklist available at http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm9-mouse/mm9-blacklist.bed.gz (downloaded on Jan. 25, 2013) (ENCODE Project Consortium, 2012) and the mm9.tss.refseq.housekeeping.ortho.bed.gz TSS file packaged with ataqv, further filtered as described above for hg19.tss.refseq.housekeeping.ortho.bed.gz.
Determination of high-confidence peaks
To generate the list of peaks used in downstream analyses, we used bedtools merge to calculate the union of the FDR 1%, blacklist-filtered peaks from libraries created using the 1X Tn5 concentration for each of the two experiments. We then kept, as master peaks, those intervals that overlapped with FDR 1%, blacklist-filtered peak calls from at least two of the 1X Tn5 libraries from that experiment.
Ataqv metrics
Ataqv collects many common measurements of ATAC-seq results, as well as several new metrics that are illuminating when comparing experiments. These metrics are listed in Table S1.
One of the metrics, fragment length distribution (FLD) distance, quantifies the similarity between each experiment’s FLD and a reference FLD (“distance to reference distribution”; Fig. S2). This provides a quantitative indicator of over- or under-transposition of samples and may be used as a covariate in downstream analyses. The distance to reference distribution metric is similar to a signed Kolmogorov-Smirnov statistic, with the magnitude representing the maximum vertical difference between the empirical distribution functions of the reference distribution and the experiment’s distribution. It is calculated as:
Where S is the statistic, x represents a fragment length, and Fe and Fr represent the empirical distribution functions of the experiment’s fragment length distribution and the reference fragment length distribution, respectively. The greater the magnitude of this metric, the less similar the experiment’s FLD is to the reference FLD. A positive value indicates over-transposition relative to the reference FLD (a greater proportion of short fragments in the distribution relative to nucleosomal fragments), while a negative value indicates under-transposition relative to the reference. The interactive ataqv report includes plots of these FLD metrics, allowing for the quick visual identification of outliers.
Ataqv calculates TSS enrichment using fragments. Fragment coverage over the TSS +/− 1kb is computed, and the enrichment for each position is calculated by dividing this coverage by the average coverage over the outermost 200 bps in the 2kb interval (100 bp upstream, 100 bp downstream).
Overlap of reads with chromatin states
Chromatin states were downloaded from https://research.nhgri.nih.gov/manuscripts/Collins/islet_chromatin/hg19/ChromHMM/GM12878_chromHMM.bb (Parker et al., 2013). The bigBed file was converted to bed format using bigBedToBed (v. 1) (Kent et al., 2010). Reads were filtered against ENCODE blacklist regions using bedtools intersect prior to the analysis. Each read was assigned to the chromatin state with which it showed the most overlap (according to bedtools intersect). To determine the statistical significance of the relationship between Tn5 concentration and the percentage of reads falling in each chromatin state, we ran one linear model per chromatin state, modeling proportion_of_reads_in_chromatin_state ~ replicate + log2(relative Tn5 concentration). P-values for the Tn5 concentration coefficient were Bonferroni adjusted.
Overlap of reads with ChIP-seq peaks
IDR ChIP-seq peaks were downloaded from ENCODE (Table S3) (ENCODE Project Consortium, 2012; Li et al., 2011; Sloan et al., 2016). Reads were filtered against ENCODE blacklist regions prior to the analysis. Bedtools intersect was used for the overlap; a single base pair was considered sufficient to call a read overlapping with a peak.
Estimating the efficiency of nuclear isolation
10 nuclear isolations were performed using C2C12 cells in order to characterize the variability in nuclear isolation efficiency. Cells were trypsinized and washed, and 250K cells were used for each nuclear isolation. Nuclear isolation was performed as in Supplementary Protocol 1 of (Corces et al., 2017). For each of the 10 replicates, nuclei were counted twice using trypan blue dye in a Countess II FL instrument, and the average of the two counts used to determine the number of final nuclei.
Quantification and statistical analysis
Modeling Tn5-sensitive peaks
To detect Tn5-sensitive peaks, we used the glm.nb function in the MASS R package (v. 7.3-50) (Venables and Ripley, 2002). We used the following model: Reads in peak ~ replicate + log2(relative Tn5 concentration) + offset(log(size_factor)) Where replicate represents the nuclear isolation (of which there were 6), relative Tn5 concentration is one of (0.2, 0.5, 0.66, 1, 1.5, 2, 5), and size_factor is the total number of reads after filtering the bam file (the ‘offset’ term adjusts for the variable number of reads in each library after filtering). The ‘reads in peak’ value was determined by passing the bed file of high-confidence peaks and each filtered bam file to bedtools’ coverageBed (‘-counts’ option). In the case that the model did not converge, we excluded the peak from the downstream analysis. For the PCR-constant experiment, all 42 libraries were used. For the PCR-variable experiment, the 20 libraries that passed QC were used.
Logistic regression to estimate TF ChIP-seq peak sensitivity
To determine if binding of each TF is associated with increased Tn5 sensitivity, we modeled (peak_is_tn5_sensitive ~ median_atac_peak_signal + overlaps_TF_chipseq_peak) using R’s glm function. The median_atac_peak_signal term controls for differences in NB GLM power as peak size increases. To calculate this term, we first gathered read counts for all Tn5 = 1X libraries in all peaks, and normalized these counts by the median count within each library to get a peak signal score for each peak in each library. We then took the median signal score across libraries for each peak. P-values for each TF were Bonferroni adjusted. Peak signal and whether or not the peak was Tn5 sensitive was derived from the PCR-constant experiment.
Data and code availability
All raw and processed data generated during this study have been deposited to GEO under the accession: GSE130450. We have created a GitHub repo containing the code necessary to reproduce the analyses in this work (https://github.com/ParkerLab/ataqv-2019). Interactive QC reports for previously-published ATAC-seq libraries are available at https://theparkerlab.med.umich.edu/data/porchard/ataqv-public-survey/. Interactive QC reports for our original data are available at at https://theparkerlab.med.umich.edu/data/porchard/ataqv-tn5-series-pcr-controlled (PCR-constant experiment, with six replicates per Tn5 concentration) and https://theparkerlab.med.umich.edu/data/porchard/ataqv-tn5-series-not-pcr-controlled (PCR-variable experiment, and sequencing at high and low cluster density).
Supplementary Material
IDR ChIP-seq peaks downloaded from ENCODE, Related to STAR Methods.
List of public ATAC-seq datasets analyzed, Related to STAR Methods and Figure 1.
KEY RESOURCES TABLE
Acknowledgements
We thank members of the Parker laboratory for testing ataqv and recommending improvements to the design and functionality of the software. We thank Minjun Jin and Nandini Manickam for providing the nuclear isolation efficiency data. This work was supported by National Institutes of Health (N.I.H.) grant R01 DK-117960 (to S.C.J.P.), the American Diabetes Association Pathway to Stop Diabetes Grant 1-14-INI-07 (to S.C.J.P.), grant T32 HG00040 from the National Human Genome Research Institute of the N.I.H. (to P.O.), grant T32 DK101357 from the N.I.H. (to Y.K.), and a University of Michigan Rackham Predoctoral Fellowship (to P.O.)
Footnotes
Declaration of interests
The authors declare no competing interests.
References
- Alasoo K, Rodrigues J, Mukhopadhyay S, Knights AJ, Mann AL, Kundu K, Hale C, Dougan G, and Gaffney DJ (2018). Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet 50, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey GN, Estacio Gomez A, Thomson J, Yin H, and Southall TD (2018). CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. ELife 7, e32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Speed TP (2012). Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 40, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner IF, Quail MA, Turner DJ, and Swerdlow H (2014). Improved Protocols for Illumina Sequencing. Curr. Protoc. Hum. Genet 80, 18.2.1–42. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol 109, 21.29.1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. (2018). The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46, D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, and Levanon EY (2013). Human housekeeping genes, revisited. Trends Genet. 29, 569–574. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, and Kellis M (2012). ChromHMM: automating chromatin-state discovery and characterization. Nat. Publ. Group 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, and Martin GR (1988). Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, and Weng Z (2008). The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome. PLOS Genet. 4, e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Baek S, Varticovski L, Kim S, and Hager GL (2019). High Quality ATAC-Seq Data Recovered from Cryopreserved Breast Cell Lines and Tissue. Sci. Rep 9, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Magli A, Garbe J, Becker A, Johnson DM, Anderson S, Auch B, Billstein B, Froehling E, McDevitt SL, et al. (2019). Measuring sequencer size bias using REcount: a novel method for highly accurate Illumina sequencing-based quantification. Genome Biol. 20, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood JC, Kent NA, Allen ND, and Harwood AJ (2019). Nucleosome dynamics of human iPSC during neural differentiation. EMBO Rep. 20, e46960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RDW, Mahmud A, McKenzie M, Trounce IA, and St John JC (2012). Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNa polymerase gamma A. Nucleic Acids Res. 40, 10124–10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetan S, Kursawe R, Youn A, Lawlor N, Jillette A, Marquez EJ, Ucar D, and Stitzel ML (2018). Type 2 Diabetes–Associated Genetic Variants Regulate Chromatin Accessibility in Human Islets. Diabetes 67, 2466–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka N, Knights AJ, and Gaffney DJ (2019). High-resolution genetic mapping of putative causal interactions between regions of open chromatin. Nat. Genet 51, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros MA, Clowney EJ, Magklara A, Yen A, Markenscoff-Papadimitriou E, Colquitt B, Myllys M, Kellis M, Lomvardas S, and Larabell CA (2016). Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. Cell Rep. 17, 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, and Bickel PJ (2011). Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat 5, 1752–1779. [Google Scholar]
- Li Z, Schulz MH, Look T, Begemann M, Zenke M, and Costa IG (2019). Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 20, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori L, Hernandez L, Zhang Z, Gilad Y, Ober C, Crawford G, Nobrega M, and Jo Sakabe N (2017). Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Sci. Rep 7, 2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Naumann S, Reutzel D, Speicher M, and Decker H-J (2001). Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk. Res 25, 313–322. [DOI] [PubMed] [Google Scholar]
- Ou J, Liu H, Yu J, Kelliher MA, Castilla LH, Lawson ND, and Zhu LJ (2018). ATACseqQC: a Bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics 19, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, NISC Comparative Sequencing Program, et al. (2013). Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci 110, 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund ÅK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach B, and Furey TS (2016). DeFCoM: analysis and modeling of transcription factor binding sites using a motif-centric genomic footprinter. Bioinformatics 33, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR (2014). BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinforma 47, 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai V, Quang DX, Erdos MR, Cusanovich DA, Daza RM, Narisu N, Zou LS, Didion JP, Guan Y, Shendure J, et al. (2020). Single-cell ATAC-Seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures. Mol. Metab 32, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Hsi-Yang Fritz M, Korbel JO, and Benes V (2019). Alfred: interactive multi-sample BAM alignment statistics, feature counting and feature annotation for long- and short-read sequencing. Bioinformatics 35, 2489–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebuzzini P, Neri T, Mazzini G, Zuccotti M, Redi CA, and Garagna S (2008). Karyotype analysis of the euploid cell population of a mouse embryonic stem cell line revealed a high incidence of chromosome abnormalities that varied during culture. Cytogenet. Genome Res 121, 18–24. [DOI] [PubMed] [Google Scholar]
- Rohland N, and Reich D (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh R, and Allis CD (2011). Genome-wide “Re”-Modeling of Nucleosome Positions. Cell 147, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, Corces MR, et al. (2019). Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol 37, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, and Greenleaf WJ (2015). Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F, Gasparoni N, Gasparoni G, Gianmoena K, Cadenas C, Polansky JK, Ebert P, Nordström K, Barann M, Sinha A, et al. (2017). Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res. 45, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, et al. (2016). ENCODE data at the ENCODE portal. Nucleic Acids Res. 44, D726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara A, Goto K, Sotomaru Y, Sofuni T, and Ito T (2006). Current status of chromosomal abnormalities in mouse embryonic stem cell lines used in Japan. Comp. Med 56, 31–34. [PubMed] [Google Scholar]
- Sun X, and St John JC (2018). Modulation of mitochondrial DNA copy number in a model of glioblastoma induces changes to DNA methylation and gene expression of the nuclear genome in tumours. Epigenetics Chromatin 11, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbell ED, and Liu T (2019). HMMRATAC: a Hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 47, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney A, VanRenterghem H, Orchard P, Boyle AP, Stitzel ML, Ucar D, and Parker SCJ (2019). Cell Specificity of Human Regulatory Annotations and Their Genetic Effects on Gene Expression. Genetics 211, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, and Ripley BD (2002). Modern Applied Statistics with S (Springer-Verlag; ). [Google Scholar]
- Wei Z, Zhang W, Fang H, Li Y, and Wang X (2018). esATAC: an easy-to-use systematic pipeline for ATAC-seq data analysis. Bioinformatics 34, 2664–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechens N, Singh V, Gkikopoulos T, Schofield P, Rocha S, and Owen-Hughes T (2016). The Chromatin Remodelling Enzymes SNF2H and SNF2L Position Nucleosomes adjacent to CTCF and Other Transcription Factors. PLOS Genet. 12, e1005940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Jin Y, Zhang W, Lu Y, Li B, and Qu K (2019). ATAC-pipe: general analysis of genome-wide chromatin accessibility. Brief. Bioinform 20, 1934–1943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IDR ChIP-seq peaks downloaded from ENCODE, Related to STAR Methods.
List of public ATAC-seq datasets analyzed, Related to STAR Methods and Figure 1.
Data Availability Statement
All raw and processed data generated during this study have been deposited to GEO under the accession: GSE130450. We have created a GitHub repo containing the code necessary to reproduce the analyses in this work (https://github.com/ParkerLab/ataqv-2019). Interactive QC reports for previously-published ATAC-seq libraries are available at https://theparkerlab.med.umich.edu/data/porchard/ataqv-public-survey/. Interactive QC reports for our original data are available at at https://theparkerlab.med.umich.edu/data/porchard/ataqv-tn5-series-pcr-controlled (PCR-constant experiment, with six replicates per Tn5 concentration) and https://theparkerlab.med.umich.edu/data/porchard/ataqv-tn5-series-not-pcr-controlled (PCR-variable experiment, and sequencing at high and low cluster density).