Abstract
The noninvasive, fast acquisition of quantitative phase maps using digital holographic microscopy (DHM) allows tracking of rapid cellular motility on transparent substrates. On two-dimensional surfaces in vitro, MDA-MB-231 cancer cells assume several morphologies related to the mode of migration and substrate stiffness, relevant to mechanisms of cancer invasiveness in vivo. The quantitative phase information from DHM may accurately classify adhesive cancer cell subpopulations with clinical relevance. To test this, cells from the invasive breast cancer MDA-MB-231 cell line were cultured on glass, tissue-culture treated polystyrene, and collagen hydrogels, and imaged with DHM followed by epifluorescence microscopy after staining F-actin and nuclei. Trends in cell phase parameters were tracked on the different substrates, during cell division, and during matrix adhesion, relating them to F-actin features. Support vector machine learning algorithms were trained and tested using parameters from holographic phase reconstructions and cell geometric features from conventional phase images, and used to distinguish between elongated and rounded cell morphologies. DHM was able to distinguish between elongated and rounded morphologies of MDA-MB-231 cells with 94% accuracy, compared to 83% accuracy using cell geometric features from conventional brightfield microscopy. This finding indicates the potential of DHM to detect and monitor cancer cell morphologies relevant to cell cycle phase status, substrate adhesion, and motility.
Keywords: holography, cell motility, microscopy, support vector machine, breast cancer cell line, actins
Digital holographic microscopy (DHM) creates quantitative phase image maps from light transmitted through cells, and is useful for studying cell morphological alterations during division (1–3), differentiation (4), death (5,6), migration (7,8), and motility (9). Most cells have an index of refraction of n ~ 1.38, mismatched from the surrounding culture media at n ~ 1.33, and thickness in the range of 1–20 μm (10). These properties create an optical pathlength (OPL) difference that forms the basis for phase signal from cells in DHM. Despite sensitivity to alterations in OPL (of a few nanometers), DHM lacks specificity for cellular subcomponents, although the nucleus has a large phase signal and can be segmented from the rest of the cell (11,12). Multimodal DHM and epifluorescence microscopy systems (13) hold the promise of identifying subcellular determinants of phase signal by comparing optical phase and fluorescence label signals. Linking quantitative phase parameters to subcellular features would increase the utility of DHM to study in vitro cell behavior.
Cancer cells phase signatures from DHM reveal clinically relevant information about the imaged cells. Studies of cancer cells using DHM track cell response to toxic agents (14,15), distinguish the histological grade of the cancer (16), monitor substrate adhesion (14), and identify differences between cancer/non-cancer and metastatic/nonmetastatic cells (17). For example, the phase profile of adherent pancreatic tumor cells is lower following transfection of the cells with the tumor suppressor E-cadherin (14). Further, treatment of the tumor cells with an actin-depolymerizing toxin, latrunculin B, results in a mottled texture of the cell’s phase reconstruction, with a higher standard deviation (SD) across a lateral profile of the cell (14). Phase features from DHM images of cervical biopsies distinguished between grade 1 and 3 cervical intraepithelial neoplasias (16), and from cell suspensions distinguished between normal skin cells versus melanoma, and primary versus metastatic melanoma (17) and colon adenocarcinoma (11,17) cell lines. These studies suggest that DHM phase signatures are sensitive to the organization of the actin cytoskeleton, and that these quantitative metrics distinguish between cells of differing metastatic potential.
Highly invasive MDA-MB-231 breast cancer cells migrate on adherent substrates using several modes, which demonstrate adaptable extracellular matrix invasion strategies and possess distinct morphologies (18). Mesenchymal migration consists of elongated cells extending and retracting actin-rich protrusions, using cytoskeletal contractility to translate the cell by pushing against cell-matrix adhesions. Many viable MDA-MB-231 cells in vitro also possess rounded morphologies, some of which possess features of amoeboid migration, consisting of cytoplasmic streaming into pseudopodia, footlike extensions that allow the cell to translate by sliding along or squeezing through the local microenvironment (19). Cells which are mitotic, dead, or have temporarily lost substrate adhesions also appear round, preventing a clear assignment of phenotype related to morphology using single images. Time-lapse microscopy resolves this ambiguity and reveals aspects of both of these modes of migration and their control by microenvironmental factors such as pore size, extracellular matrix compliance, enzymatic degradation and microscale alignment (20,21). However, time-lapse microscopy experiments have low throughput since the same cells must be tracked for hours, limiting the amount of data that can be collected in a single experiment. Further, images of cancer cells using conventional phase contrast microscopy suffer from bright artefacts around the cell border that can obscure fine details of that region (22), while fluorescence microscopy requires exogenous labeling, or in the case of endogenous autofluorescence imaging, may induce photodamage caused by intense light fluence from the excitation source through the cell (23). In contrast, DHM delivers extremely low incident light power to imaged cells, and leaves the cell borders free of artefacts.
These previous studies plus the quantitative, noninvasive nature of DHM create the potential for experiments to determine the sensitivity of quantitative phase parameters to cell morphologies influenced by migration mode, cytoskeletal organization, substrate attachment, and the cell cycle. Such determinations, performed on cancer cells, would lead to quantitative understanding of processes relevant to cancer growth and invasiveness. Therefore, the hypothesis of the current study was that quantitative metrics from DHM phase maps of invasive breast cancer cells reflect cell morphological and cytoskeletal features, and distinguish between motile phenotypes characterized by different levels substrate attachment and modes of migration. Further, rounded/motile and dividing cells are distinguishable from single DHM phase reconstructions. To test this, MDA-MB-231 cells were plated on glass, tissue culture-treated polystyrene, and collagen hydrogels polymerized at two temperatures, and imaged using single-frame and time-lapse DHM and conventional phase contrast microscopy, followed by F-actin and nuclear staining, and epifluorescence microscopy. Phase maps of the adherent cells revealed morphology and motile phenotypes that depended on the type of substrate, the mode of migration, and the level of adhesion to the substrate. Some F-actin features and the nucleus were apparent in DHM maps of cells. A machine learning algorithm confirmed the high sensitivity and specificity of DHM phase signatures for distinguishing between rounded and elongated cells with different patterns of motility. These findings define the relationship between specific phase parameters and cancer cell behavior, and support the use of DHM to better understand cancer cell migration and substrate adhesion, in vitro behaviors with clinical relevance.
Materials and Methods
Brief Theory of Telecentric DHM
Figure 1 shows a bi-telecentric-DHM (BT-DHM) system in vertical transmission and afocal configuration, used for the quantitative analysis of biological samples (24–27), including this study. An extensive background of telecentric DHM theory is presented in Supporting Information, Section 5.1. Aspects of the DHM set-up and phase reconstruction have been previously published (24,25,27–29). In the afocal configuration (the back focal plane of the microscope objective (MO) coincides with the front focal plane of the tube lens (fo ≡ fTL)) the biological sample is placed at the front focal plane of the MO. This results in the optical mitigation of the bulk of the spherical phase curvature normally present in traditional nontelecentric DHM systems. A HeNe laser is used as a coherent optical source and is collimated using a combination of a spatial filter and a collimating lens to produce a plane wave beam. The collimated beam is split into a reference beam and an object beam. The object beam is focused on the biological sample using the afocal configuration. The two beams are then made to interfere with each other on a charge-coupled device (CCD) camera to generate an off-axis hologram. The resultant magnification of the BT-DHM system is . In this work the digital holograms are reconstructed by an angular spectrum technique (30,31). In a DHM system the pixel size in the image plane (the resolution with which the magnified image of the object is reconstructed), and scales according to: Δξmag = λd/(NΔxM), Δηmag = λd/(NΔyM), where Δx=Δy=L/N is the camera pixel size, L is the dimension of the CCD sensor, and N is the number of pixels in one dimension. In a DHM system, the spatial resolution is limited by the diffraction limit of the MO used in the optical setup which is: λ/(2nmedNA), where NA is the numerical aperture (NA) of the MO. For a transmissive biological phase object in a transmissive medium, the phase change is related to the sample thickness T and the change in index between the biological sample and the medium which can be expressed as: . In a regular DHM system the total phase of the object beam is expressed as: where R is the radius of curvature of the spherical curvature of the MO which superpose and obscure the phase due to the sample. The proposed bi-telecentric configuration optically compensates for the bulk of the parabolic curvature due to the MO, reducing the complexity of precisely measuring the optical setup parameters and of digital post-processing compensation.
Figure 1.
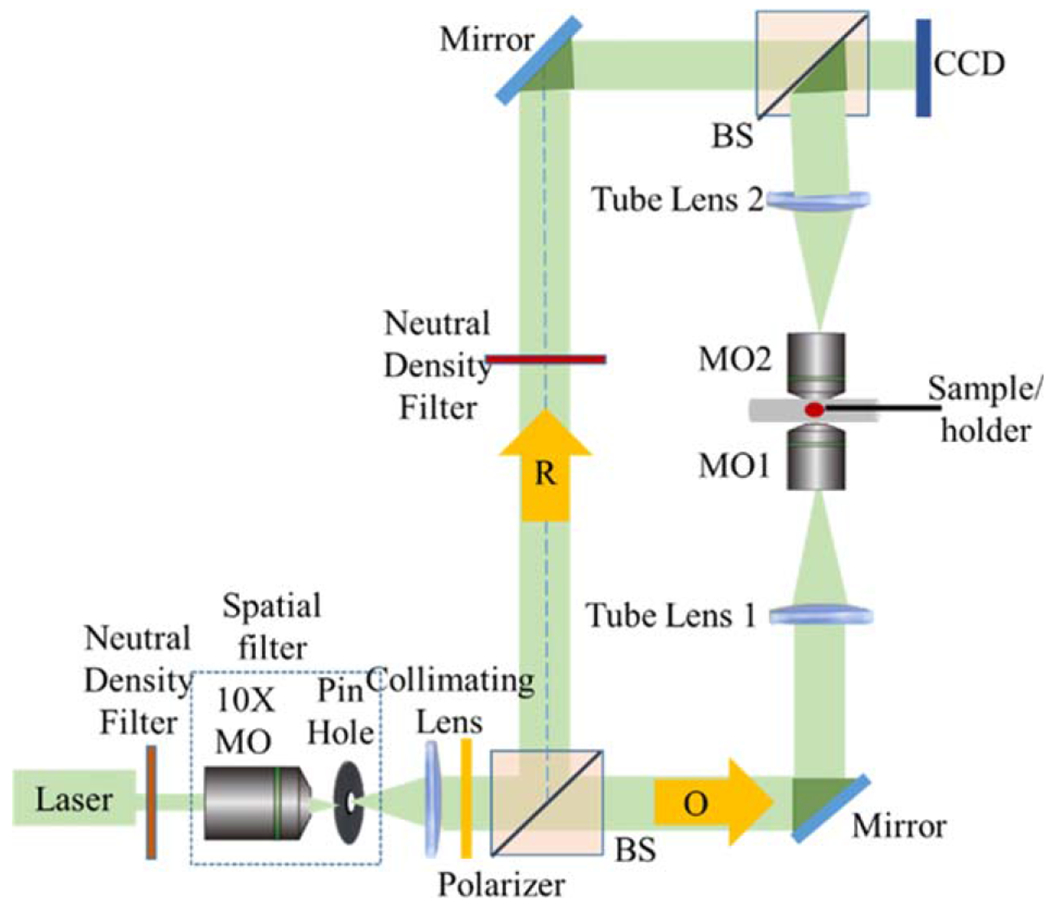
The BT-DHM system in transmission configuration. MO, beamsplitters (BS), object beam (O), reference beam (R), and CCD camera are labeled. [Color figure can be viewed at wileyonlinelibrary.com]
To obtain aberration-free reconstructed holograms, two post-processing techniques are employed sequentially. The first technique is fully automated and is based on principal component analysis (PCA) (31). This technique is only used to compensate for the remaining quadratic phase due to the MO and the tilt phase due to the angle of the reference beam: This phase aberration can be written in a matrix format as: , where * denotes the complex conjugate operator, and are two phase vectors. PCA is used to minimize the error orthogonal to the model. Since the phase of the biological sample φob (x, y) is small, it is considered to be a small perturbation obscured by the quadratic phase due to the MO and tilt due to the reference phase and hence can be written as: . In the PCA technique, aberration compensation reduces to computing the first principal component (PC) of the exponential term of the filtered hologram. The following steps summarize the algorithm: (a) Perform singular value decomposition (SVD) to obtain the first dominant PC, (b) obtain the linear and quadratic coefficients of the phase vectors from least square fitting of the two dominant singular vectors, (c) use these coefficients to compute φ(k, l), and (d) multiply the conjugate φ*(k, l) with the hologram to obtain which is the phase due to the biological sample without MO and tilt.
After canceling the main aberrations of the DHM system, higher order aberrations remain uncompensated for, optically or digitally. The second technique is semiautomatic and based on 2D polynomial model to perform surface fitting. To cancel the remaining phase aberration, the phase data was chosen manually at locations with a known, sample-free phase region. Then, 2D surface fitting is used to generate a phase mask based on the sample-free phase region. A 2D polynomial of Mth order has the following form:
| (1) |
where x and y represent pixel coordinates. To relate the regular polynomial fitting to the well-known Zernike polynomial fitting, two arrays are defined as: and , hold the polynomial model’s coefficients and the Zernike model’s coefficients, respectively, where l, k represent the polynomial order in x and y, respectively. The (2 + M)(1 + M)/2 coefficients of the P polynomial are used to calculate the coefficients of the Zernike polynomial model as shown in the following matrix format:
| (2) |
where zl.k,p is a matrix of size . Finally, the Zernike polynomials are used to construct the phase where Zk coefficients are expressed according to the Zemax® notation. In all the calculations discussed here M was set to 5. The refractive index of cells was assumed to be 1.381.
Preparation of Cancer Cell Cultures
As a first experiment, cancer cells from the highly invasive MDA-MB-231 breast cancer cell line were seeded in 35 mm diameter Petri dishes on glass, tissue-culture polystyrene (TCPS), and type I collagen hydrogels polymerized at 4 mg/ml and temperatures of 4 and 37°C. The gel polymerized at low temperature produced a coarse collagen network (CCN) with larger fiber and pore diameters than the fine collagen network (FCN) produced at 37°C. The MDA-MB-231 cell line was a generous gift from Dr. Zaver Bhujwalla (Johns Hopkins School of Medicine, Baltimore, MD). The cells on collagen were incubated for 24 hours in Dulbecco’s modified essential medium (DMEM, Sigma-Aldrich) containing 10% fetal bovine serum (FBS, Corning), 100 units/ml penicillin, and 100 μg/ml streptomycin (Corning) in standard tissue culture conditions of 37°C, 5% CO2, and 100% humidity.
DHM and Time-Lapse Brightfield Microscopy
Cells were taken from the incubator and imaged with the DHM setup described above. Image reconstruction was performed as described above to produce phase reconstruction maps depicting cells and collagen in a phase-cancelled background. For time-lapse DHM experiments, cells were imaged for 80–174 min at 2 min intervals. The lateral pixel size of the reconstruction was 0.19 μm, determined by imaging a resolution target, which oversampled the lateral resolution of 1.2 μm for the imaging system. The laser wavelength was 633 nm and imaging objective NA was 0.25. The surface power density at the specimen plane was measured to be < 0.1 mW/cm2. In separate time-lapse microscopy experiments, MDA-MB-231 cells cultured on glass, FCN, and CCN were placed on the CytoSMART® device (Lonza Group, Basel, Switzerland), an in-incubator brightfield imaging system, and imaged for 24 h at 5 min intervals.
Fluorescence Labeling and Epifluorescence Microscopy of Cancer Cells
After DHM imaging, MDA-MB-231 cells cultured on glass, polystyrene and collagen were fixed in 3.7% formaldehyde. Fixed cells were permeabilized in 0.1% Triton X-100 (Sigma) and incubated in PBS with 1% bovine serum albumen containing fluorescently labeled phalloidin (CytoPainter Phalloidin-iFluors 488, Abcam) and DAPI (Life Technologies) to stain F-actin and nuclei, respectively, according to the manufacturer’s protocol. Epifluorescence microscopy was performed on the cells using an Olympus BX60 microscope. Images were collected using 60×, NA 1.25 and 100×, NA 1.35 oil-immersion Plan Apo objectives, and a Photometrics CoolSNAP HQ2 high resolution camera with Meta-Morph software.
To co-register DHM, phase contrast, and epifluorescence microscopy images, two thin pieces of cellulose acetate tape were placed on the underside of a glass-bottomed petri dish, forming a long, ~2 mm narrow tape-free window in between. After 24 h of culturing, cells were imaged using DHM. The dish was placed so as to image cells between the tape strips. Immediately following imaging with DHM, cells were fixed with 4% paraformaldehyde, stained with phalloidin and DAPI, and imaged over a large region with a low-powered objective using phase contrast microscopy. Cells captured by DHM were noticed on the phase contrast images, after which higher resolution phase contrast and epifluorescence images of the cell were captured using the BX60 microscope and 60× objective. Digital images from DHM, epifluorescence, and conventional phase contrast microscopy were co-registered manually using ImageJ.
Image Analysis and Machine Learning from DHM Phase Maps
Cell segmentation, parameter quantification, and machine learning were all performed using custom-written codes and built-in functions and apps in MATLAB (2015a, Natick, MA). Other platforms such as Cell Profiler Analyst (Broad Institute, MA, http://cellprofiler.org/) are available and have been used for similar purposes (32). A MATLAB-based approach was chosen because of the ease of code construction and code segment testing in the MATLAB workspace, and extensive information available about built-in function libraries. Regardless of software used, image processing and machine learning results must be reproducible from methods presented (below), codes, and raw data (presented in the Supporting Information, Sections 5.3 and 5.4). One novel aspect of this study is in the application of texture analysis and PCA-SVM to classify the morphology of MDA-MB-231 cells.
To quantify cell shape and phase texture parameters from DHM phase maps, a custom-written Matlab code was used. Eleven shape and phase parameters were calculated from cells segmented by background subtraction (the mean from a background region of interest containing no cells), binary thresholding at a level of one SD above the mean background, followed by edge detection of the cell border and filling in of any low-signal holes in the cell object using morphological image processing techniques. The average cell phase height, first-order pixel statistics of SD, kurtosis, and skew, second order pixel texture statistics of contrast, correlation, homogeneity, and energy, and the shape parameters of area, eccentricity, and perimeter were calculated. The texture parameters calculated from the gray level co-occurrence matrix have been described elsewhere (33), and applied previously to DHM phase maps of cells (17). The Supporting Information Section 5.2 contains further explanation of first and second-order texture parameters, and texture analysis using the gray-level co-occurrence matrix. From brightfield microscopy images, the following cell geometric parameters were calculated, using built-in ImageJ measurements of background-subtracted, segmented images: cell area (A), perimeter (p), aspect ratio (AR = major axis/minor axis), circularity (4πA/p2), and solidity (area/convex area).
To distinguish between MDA-MB-231 cancer cells with elongated and rounded morphologies, cells were first classified as elongated, rounded/motile, rounded/dividing, and rounded/nonmotile (Fig. 2), by examining cells from the time-lapse brightfield microscopy data sets over 4 h. These cells were easily classified based on shape and movement, tracked over several hours (Fig. 3). Further, it was noticed that rounded/dividing and rounded/nonmotile cells combined to be < 15% of all cells cultured on the tested substrates (Fig. 2e), and that these cells always had smooth, unruffled borders, whereas rounded/motile cells usually had rough borders with a ruffled appearance (Fig. 2b–d). The appearance of MDA-MB-231 cells using these two modes of motility has been confirmed previously (34). Therefore, machine learning was performed on data sets containing only elongated or rounded/motile cells, determined either from time-lapse images (for conventional brightfield microscopy), or by visually comparing the cell to previously classified cells from time-lapse brightfield imaging (for single DHM phase maps). This visual comparison of cells from DHM phase maps to cells of known morphology and motility from time-lapse brightfield imaging was deemed sufficient for accurate assignment of cells from DHM images captured at single instants of time, especially considering the scarcity of dead and pre- or post-mitotic cells (Fig. 2e), which reduced the chances of visual misclassification. The data sets consisted of n = 150 cells for DHM, and n = 148 cells on glass imaged with conventional brightfield microscopy. For each data set, PCA extracted 2–8 features for learning, and the algorithm was evaluated using fivefold cross-validation. An initial cross-validation was used to determine how many principal components to use in the predictors from each cell. Then, the data sets were divided into roughly one-third for cross-validation, with two-thirds (n = 100 cells per data set) reserved as a data set naïve to the algorithm, and used for final testing of machine learning performance. This naïve testing data set was used to avoid overestimating the classification power of the machine learning algorithm.
Figure 2.
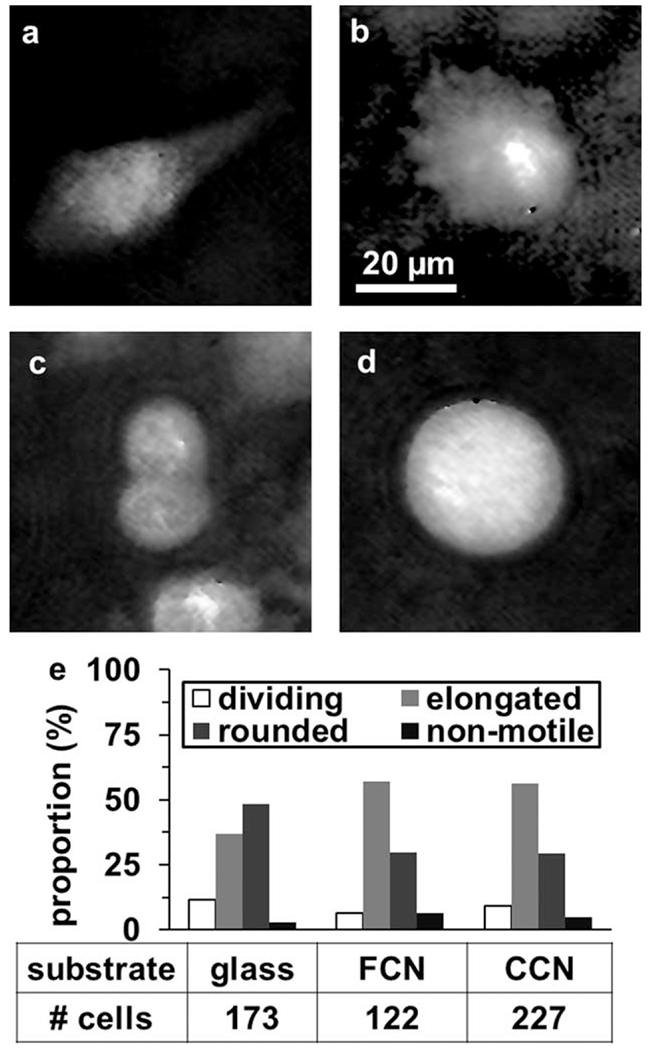
Phenotype of MDA-MB-231 cells on 2D substrates by DHM and time-lapse microscopy. DHM phase maps of typical (a) elongated motile, (b) rounded motile, (c) dividing, and (d) rounded/nonmotile cells cultured on glass. Scale bar is indicated. (e) The proportion of these four phenotypes in MDA-MB-231 cell populations cultured on glass, a FCN, and a CCN, visually assessed from in-incubator brightfield videomicroscopy, with the number of cells quantified in each condition listed.
Figure 3.
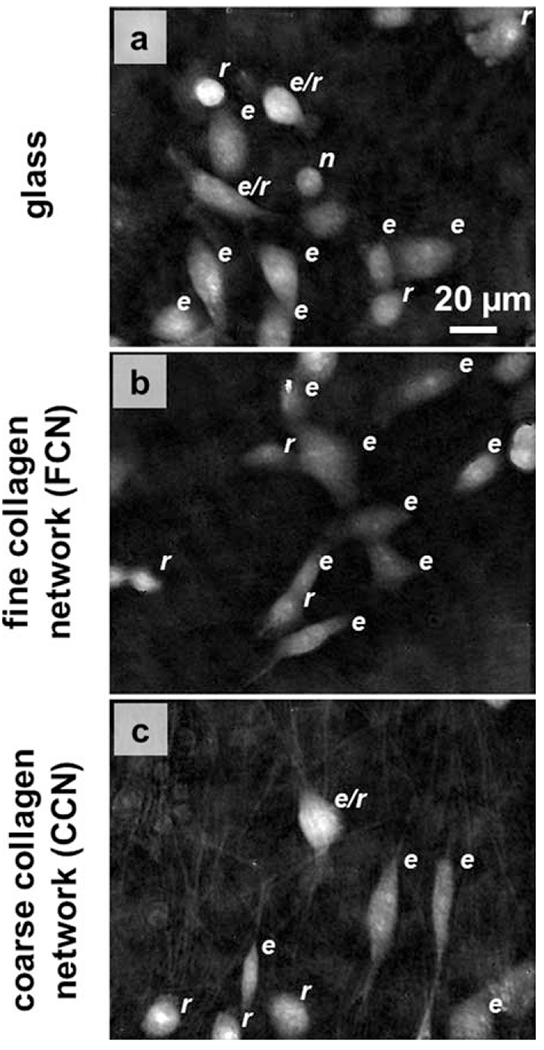
Phase maps from time-lapse DHM experiments of MDA-MB-231 cells on (a) glass (Video 1, MPEG, 1.8 MB), (b) a 4 mg/ml collagen hydrogel polymerized at 37°C (Video 2, MPEG, 1.6 MB), and (c) a 4 mg/ml collagen hydrogel polymerized at 4°C (Video 3, MPEG, 2.2 MB). Labels to the upper right of each cell indicate phenotype: elongated motile (e), rounded motile (r), rounded/dividing (d), and rounded/nonmotile (n). Scale is indicated.
A linear support vector machine learning algorithm was used based on excellent classification performance in previous studies using DHM data sets (17,35). Further, the method of PCA to reduce the dimensionality of features extracted for machine learning is well-established in the literature (36–39). The use of principal components reduced co-linearity of features used as predictors for machine learning, and avoided overfitting error. Further, only those parameters that were different between the two groups by t test were used to compute standardized feature scores from the principal components. The true positive rate was defined as the number of elongated cells identified over total number in the test set, and the false positive rate was the number of rounded cells identified as elongated. Receiver operating characteristic (ROC) curves were plotted from these tests, and the area under the curve was determined for each plot.
Results
Phase Signatures from DHM Assess Cell Morphology and Movement
Distinct phase features of typical elongated, rounded motile, rounded mitotic, and rounded nonmotile MDA-MB-231 cells appear in DHM phase maps on glass substrate (Fig. 2). Elongated cells possess a large central phase signal with tapering protrusions with smaller phase further from the cell body (Fig. 2a). Rounded motile cells also have a large central phase signal, but with an irregular and asymmetrical perimeter (Fig. 2b). Dividing cells are rounded but with smoother edges, and are larger before cytokinesis and smaller afterward (Fig. 2c). Rounded nonmotile cells also possess smooth phase borders, but do not divide or migrate in subsequent time-lapse image frames (Fig. 2d). Time-lapse microscopy revealed the proportion of each of these morphological subpopulations in MDA-MB-231 cells cultured on glass, a FCN and a CCN of equal collagen density (Fig. 2e). The proportion of dividing cells was 12%, 7%, and 9% for cells on glass, FCN, and CCN, respectively. The proportion of rounded motile cells was 48% on glass, dropping to 30% on both collagen substrates.
Time-Lapse DHM of Cancer Cells Reveal Aspects of Cell Motility and Substrate Adhesion
The appearance and motion of cancer cells is different on glass and collagen substrates as revealed by time-lapse DHM maps. The movement of MDA-MB-231 cells on glass and collagen hydrogels with two distinct microstructures was observed from DHM phase reconstructions acquired at two minute intervals (Fig. 3). Qualitative differences in cell morphology and movement on the substrates were apparent from the phase reconstructions. In particular, cells on glass had more prominent membrane ruffling at leading and trailing edges than on other substrates (Fig. 3a, Video 1). Cells were most motile on glass, less motile on collagen polymerized at 37°C (Fig. 3b, Video 2), and least motile on collagen polymerized at 4°C (Fig. 3c, Video 3). The background phase texture of the collagen hydrogel polymerized at 4°C, which possesses collagen fibers of larger diameter than those formed at 37°C, revealed a network of collagen fibers not apparent in phase maps from the other substrates. Cell morphologies varied from round (r) to elongated (e) on each substrate, with some cells undergoing shape transitions of round to elongated or elongated to round during the course of time-lapse DHM imaging. Other cells were dividing (d) or rounded and nonmotile (n).
Eleven geometric and texture parameters were measured from segmented cells in the phase reconstruction maps (Fig. 4). Using a semiautomated routine, isolated cancer cells were selected by tracing around the cell border (Fig. 4a), applying a threshold (Fig. 4b) to segment the cell (Fig. 4c). For each cell, an average phase height, SD of phase height, kurtosis, skew, area, perimeter, and eccentricity were calculated. Additionally, a gray-level co-occurrence matrix was calculated for the cell (Fig. 4d), clipping the first row and column that count neighboring pixels with values of 0 and x, where x > 1, to avoid edge effects, and the second-order texture parameters of contrast, correlation, energy, and homogeneity were calculated. All eleven phase signatures from the cell in Fig. 4c are listed in Fig. 4e.
Figure 4.
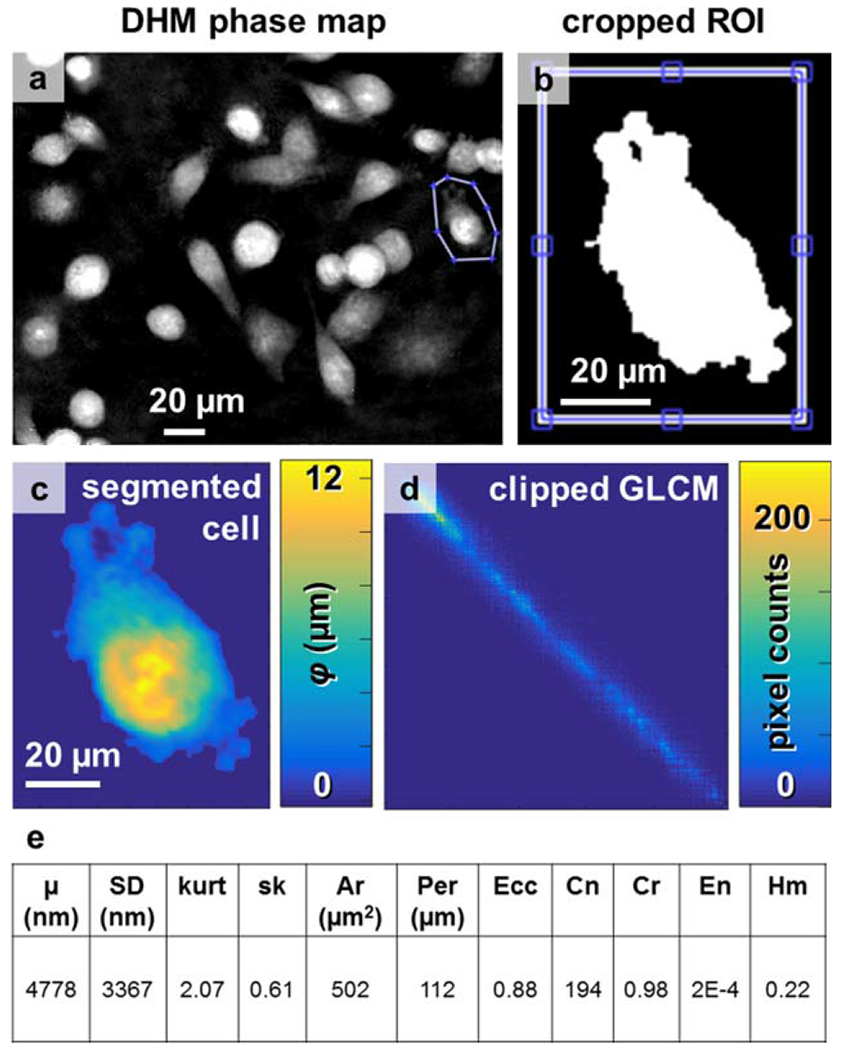
Cell segmentation and texture analysis. (a) A typical phase reconstruction of cultured MDA-MB-231 cells, with one cell outlined by a region of interest. (b) The binary threshold of the cell, used to create (c) the segmented cell. (d) The gray-level co-occurrence matrix with first row and column removed to avoid quantifying edge effects. (e) Eleven parameters were automatically recorded from each cell, including mean phase height (μ), SD, kurtosis (kurt), skew (sk), cell projected area (Ar), perimeter (Per), eccentricity (Ecc), and the texture parameters of contrast (Cn), correlation (Cr), energy (En), and homogeneity (Hm). [Color figure can be viewed at wileyonlinelibrary.com]
Cell phase signatures from segmented DHM phase maps had characteristic trends during matrix adhesion, deadhesion, and cell motility requiring generation of traction force. A dividing cancer cell was imaged with DHM and trends in phase signatures were identified (Fig. 5). The dividing cell was initially elongated and attached to the underlying glass substrate (Fig. 5a.i), but released attachments over the course of 52 min, becoming round (Fig. 5a.ii) and with 90% higher phase height and 41% smaller projected area than initially (Fig. 5b). Over the next 24 min the cell phase height and projected area remained fairly constant while cytokinesis occurred (Fig. 5a.iii,b,c). After cell division, over the remaining 94 min of DHM image acquisition (Fig. 5a.iv), the daughter cell average phase height decreased and projected area increased as substrate adhesion was re-established, 30% and 63% lower than the initial adherent parent cell and final premitotic parent cell phase height, respectively, and 51% and 18% lower than the initial adherent parent cell and final premitotic parent cell projected area, respectively. Three of the four second order contrast parameters were altered between the time periods predivision (0–52 min), during cytokinesis (52–76 min), post-division (78–138 min), and during re-adhesion to the substrate (138–172 min, Fig. 5d). The contrast parameter was 220 ± 19 predivision, 213 ± 20 during cytokinesis, and 330 ± 29 post-division. The energy parameter was 2.2 × 10−4 ± 3 × 1025 predivision, 2.6 × 10−4 ± 4 × 10−5 during cytokinesis, and 2.1 × 10−4 ± 4 × 10−5 post-division. The correlation parameter was 0.25 ± 0.01 predivision, 0.24 ± 0.01 during cytokinesis, and 0.21 ± 0.01 post-division. Over the final 34 min, as the cell was spreading on the substrate the contrast parameter decreased, while the energy and homogeneity parameters increased.
Figure 5.
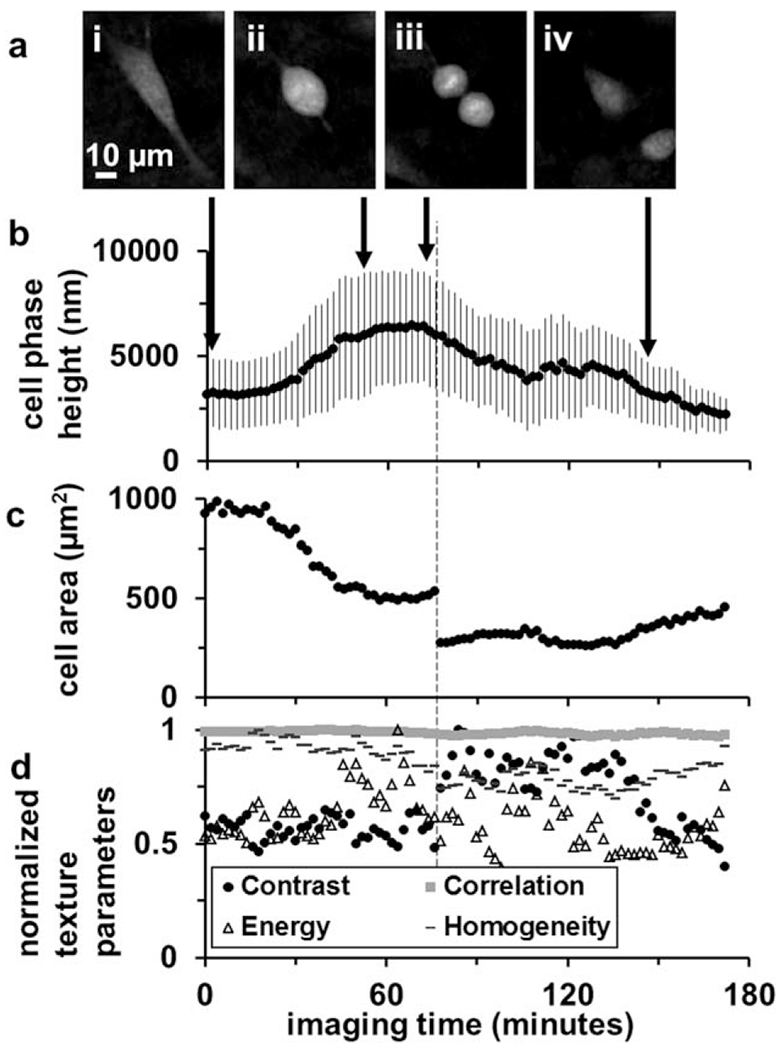
Quantitative phase parameters from a dividing cancer cell. (a) DHM phase reconstructions (i) before mitosis, (ii) during prophase-metaphase, (iii) anaphase, and (iv) post-mitosis. (b) The cell mean phase height and SD, (c) cell area, and (d) normalized texture parameters are plotted over the entire time-lapse acquisition duration, with arrows indicating timepoints of the frames in (i–iv).
Similar trends of phase signatures with substrate attachment and de-attachment were found in time-lapse DHM of a cancer cell undergoing morphological changes during migration. A partially elongated cell with prominent lamellipodia (Fig. 6a.i) becomes more elongated (Fig. 6a.ii) and then retracts, becoming round with a broad lamella (Fig. 6a.iii). The cell mean phase height becomes lower with greater elongation (i→ii), and following release of the cell edges (ii→iii), the average phase height becomes higher. Similar to the de-attaching cell precytokinesis, the second order contrast parameter decreased, while the energy and homogeneity parameters increased.
Figure 6.
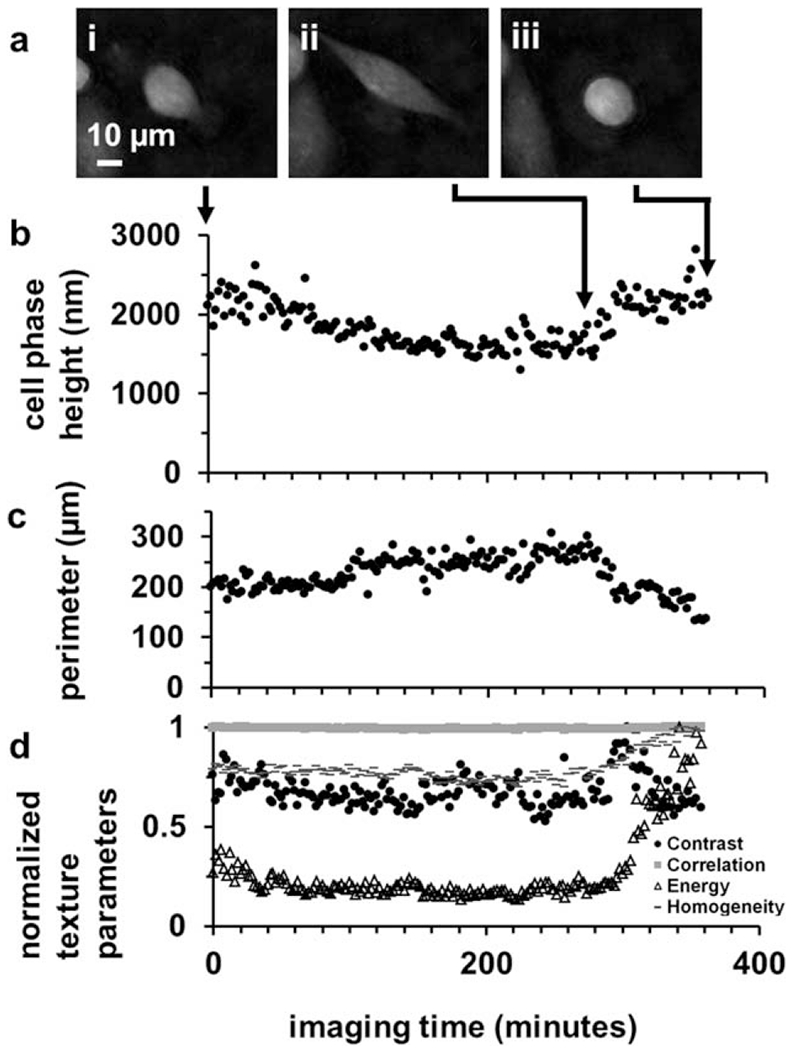
Quantitative phase parameters from a motile adherent cancer cell. (a) DHM phase reconstructions (i) at the beginning, (ii) end of an extension phase, and (iii) after retraction of membrane protrusions. (b) The cell mean phase height, (c) perimeter, and (d) normalized texture parameters are plotted over the entire time-lapse acquisition duration, with arrows indicating time-points of the frames in (i–iii).
Cancer Cell Phase Reconstructions Reflect Features of the F-Actin Cytoskeleton
Phase reconstructions from DHM of single cancer cells revealed several morphological variants associated with distinct F-actin features (Fig. 7). Similarly shaped cells from the same cultures on glass, TCPS, and the surfaces of type I collagen gels polymerized at 37°C and 4°C were imaged with DHM and epifluorescence microscopy. On glass, the selected cell is stellate in shape (Fig. 7a), and possesses several actin-rich lamellipodia containing multiple filopodia (Fig. 7b). On TCPS, the selected cell is oval-shaped (Fig. 7c), and possesses a ring of rich, cortical F-actin and multiple stress fibers (Fig. 7d). On collagen polymerized at 37°C, the selected cell is fan-shaped (Fig. 7e) and possesses a broad lamella with numerous filopodia and microspikes (Fig. 7f). Finally, on collagen polymerized at 4°C, the selected cell is spindle-shaped (Fig. 7g) and possesses stress fibers spanning substrate attachments at the leading and trailing edge (Fig. 7h). Additionally, punctate actin features indicative of smaller F-actin rich structures, such as podosomes and invadopodia, were located on cells in all conditions.
Figure 7.
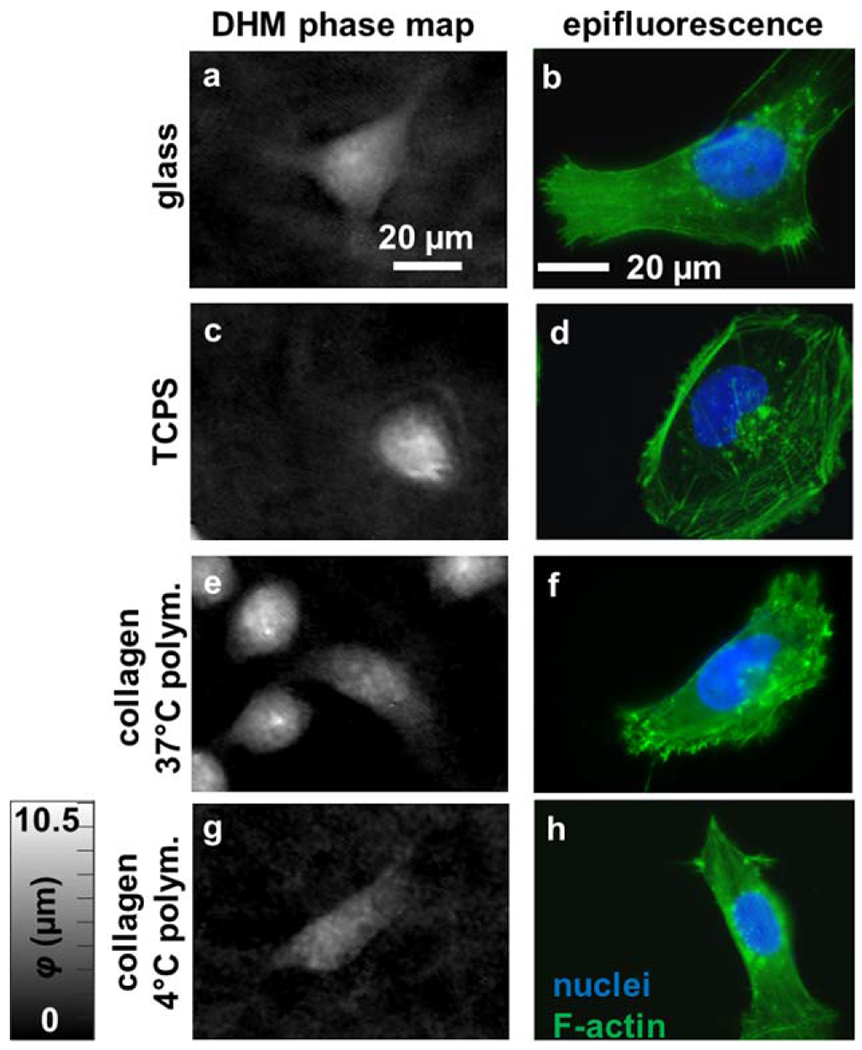
Comparing phase reconstructions to subcellular features. Cells with similar shape were imaged on (a,b) glass, (c,d) TCPS, and collagen gels polymerized at (e,f) 37°C and (g,h) 4°C, creating (a,c,e,g) DHM phase maps and (b,d,f,h) epifluorescence micrographs of nuclear (blue) and F-actin (green) fluorescence labels. The scale bar is indicated. [Color figure can be viewed at wileyonlinelibrary.com]
To confirm the relation of DHM phase features to the nucleus, actin cytoskeleton, and overall cell morphology, several DHM cell maps were co-registered with epifluorescence images of DAPI and phalloidin staining, and with conventional phase contrast microscopy (Fig. 8), for elongated (Fig. 8a,c,e,g) and rounded (Fig. 8b,d,f,h) cells. The DHM maps (Fig. 8e,f) noticeably lack artefacts present in conventional phase contrast (Fig. 8g,h), possess higher phase at positions containing nuclei (Fig. 8a,b), and possess textural features that overlap F-actin and phase contrast features (Fig. 8c–h).
Figure 8.
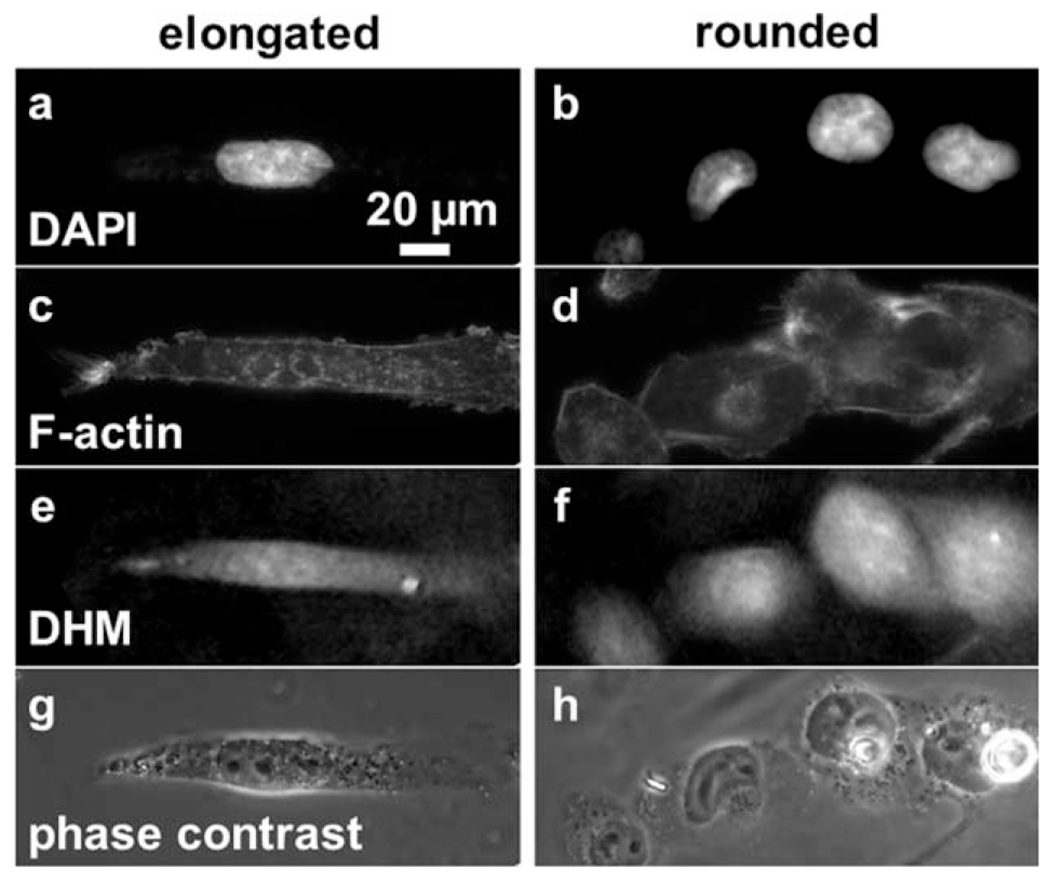
Co-registered DHM, epifluorescence, and phase contrast images of MDA-MB-231 cells. Epifluorescence images of (a,b) DAPI, and (c,d) phalloidin stains, as well as (e,f) DHM phase maps, and (g,h) conventional phase contrast microscopy images of the same cells were co-registered. Cells of (a,c,e,g) elongated, and (b,d,f,h) rounded morphologies cultured on glass were chosen. The scale bar is indicated.
Machine Learning Distinguishes between Elongated and Rounded Cell Morphologies
A support vector machine (SVM) algorithm distinguished between cells adherent to glass with elongated and rounded morphologies with higher accuracy based on DHM phase signatures than cell geometrical parameters (Table 1, Fig. 9). Using DHM phase signatures, an SVM algorithm was performed with fivefold cross-validation on two to six principal components derived from eight of the eleven phase signatures which were significantly different by t test. A plateau in predictive power of the classification was reached when using six principal components (Table 1, misclassifications indicated, Fig. 9a). In comparison, cell geometrical features from conventional brightfield microscopy reached a plateau in predictive power with four principal components, (Table 1, Fig. 9b). Classification accuracy and predictive power were higher using cell phase signatures from DHM, at 94% and AUC = 0.943, than using cell geometrical parameters from brightfield microscopy, at 83% and AUC = 0.838 (Table 1). These results were from testing of a naïve data set of n = 100 cells per imaging modality following fivefold cross-validation on n = 48–50 cells.
Table 1.
Results of linear support vector machine learning with fivefold cross-validation, elongated versus rounded motile cells
| PREDICTORS | AUC | ACCURACY | ERROR | R→E/ TOT. R | E→R/ TOT. E | N (# CELLS) |
|---|---|---|---|---|---|---|
| GI.BF.5 | 0.941 | 85.1% | 14.9% | 12/84 | 10/64 | 148 |
| GI.BF.4 | 0.940 | 83.8% | 16.2% | 12/84 | 12/64 | 148 |
| GI.BF.2 | 0.886 | 77% | 23% | 11/84 | 23/64 | 148 |
| GI.DHM.8 | 0.987 | 96% | 4% | 4/79 | 2/71 | 150 |
| GI.DHM.6 | 0.987 | 96% | 4% | 5/79 | 1/71 | 150 |
| GI.DHM 4 | 0 954 | 87.3% | 12.7% | 11/79 | 8/71 | 150 |
| GI.DHM.2 | 0.918 | 82% | 18% | 16/79 | 11/71 | 150 |
| GI.BF.4 fivefold crossval. | 0.934 | 81.3% | 18.8% | 4/24 | 5/24 | 48 |
| GI.BF.4 naive test | 0.838 | 83% | 17% | 12/60 | 5/40 | 100 |
| GI.DHM.6 fivefold crossval. | 0.982 | 94% | 6% | 2/25 | 1/25 | 50 |
| GI.DHM.6 naïve test | 0.943 | 94% | 6% | 5/54 | 1/46 | 100 |
GI, glass; AUC, area under the ROC; BF, morphological features selected from brightfield microscopy; DHM, morphological and phase features selected from DHM. The last two columns count the misclassifications of each type (E→R, e missclass. as r, R→E, r misclass. as e).
Figure 9.
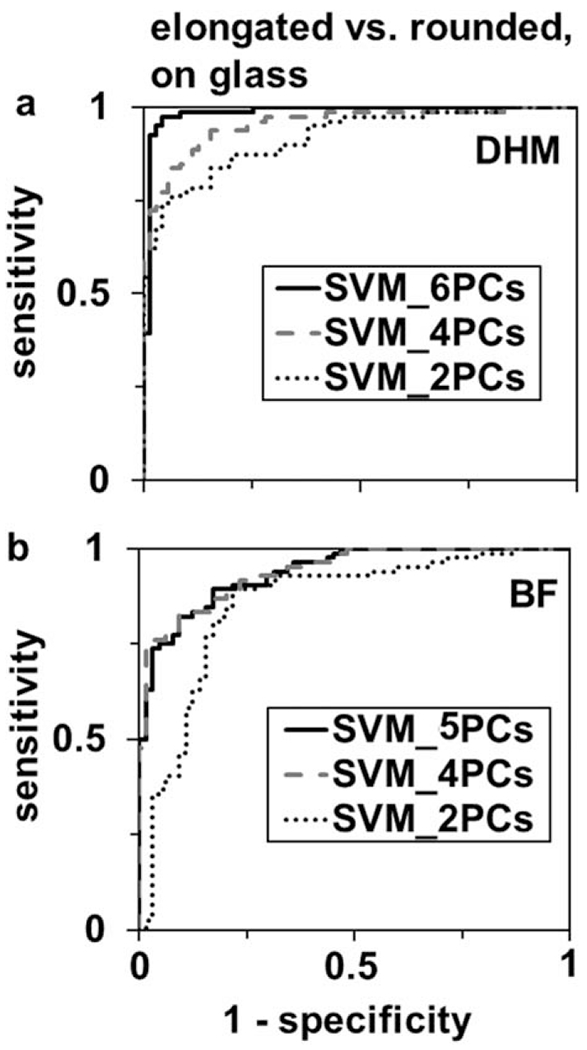
ROC curves for linear support vector machine learning classification of elongated versus rounded motile cells using PCA feature extraction of (a) cell morphological features and phase parameters from DHM and (b) cell morphological features from brightfield microscopy. Features derived from the first two (dotted line), four (dashed line) and five-six (solid line) principal components are used to generate the ROC curves.
Discussion
DHM is sensitive to cancer cell morphology and motility related to F-actin cytoskeletal features and substrate adhesion. Specifically, time-lapse sequences of DHM phase maps revealed membrane ruffling in lamellipodia of cells on glass substrates, higher motility on glass than on collagen substrates, and lamellipodia, lamellae, cortical actin, filopodia, and microspikes, discernable as features in DHM maps, as well as the underlying texture of a collagen network polymerized at 4°C to comprise fibers of ~3 pm diameter. Phase signatures quantitatively assess processes associated with cytoskeletal activity and altered substrate attachment, including cell division, elongation and retraction. The sensitivity of DHM to cell morphology and phase profile features permitted training of an SVM learning algorithm with high accuracy to distinguish between elongated and rounded/motile cancer cells.
The main strength of this study was in quantitative comparison of DHM phase maps of cancer cells to specific cytoskeletal features and substrate attachment patterns relevant to cancer cell migration and matrix invasion. The relation of phase map features to F-actin features associated with substrate adhesion (Figs. 7 and 8) suggests that DHM detects cancer cell cytoskeletal features responsive to the surrounding microenvironment. Further, quantitative phase parameters (Fig. 4e) help distinguish between elongated and rounded/motile cells with higher accuracy than cell geometric features only, from conventional brightfield microscopy (Table 1 and Fig. 9), suggesting the pixel phase values add additional information relevant to cell phenotypes, beyond cell geometry. The main study limitations were poor subcellular specificity of DHM phase signal to F-actin, and inadequate sample size of dividing and rounded/nonmotile cells to perform machine learning to distinguish between these subpopulations and rounded motile cells (Fig. 2). To work around the first limitation, serial DHM and epifluorescence microscopy was performed on the same specimens, so similar cells could be identified using phase and F-actin fluorescence signals. To mitigate the second limitation, dividing and nonmotile cells, comprising < 15% of the total, were excluded from the data sets used to cross-validate the machine learning classification algorithms, based on the smooth and even appearance of the borders of these cells. In the future, a multimodal instrument as described in Ref. 40 would facilitate comparison of fluorescently labeled subcellular components to their phase profiles. Collection of approximately 1,000 cells per culture condition would yield enough dividing cells (~ 100) for training and testing of machine learning algorithms to distinguish dividing from rounded/motile cells. Further time-dependent phase parameters could be determined from time-lapse DHM data sets which could improve the classification accuracy, although single-frame imaging and classification allows higher sample throughput and more cells to be imaged. An important aspect of DHM is the lack of phase artefacts, such as nonlinear shade-off near cell and organelle edges, compared to conventional phase contrast microscopy. Enhanced phase contrast of DHM would likely reveal more subtle morphological distinctions between cells cultured on different substrates and at different phases. This could be achieved in the future through improvements to reconstruction algorithms and DHM optical systems (41), including using super-resolution set-ups (42–47).
The DHM phase signal reveals information about the actin cytoskeleton and substrate adhesion status primarily through cell phase map morphology, profile, and texture (Figs. 4 and 7). The eleven quantitative parameters from this study overlap in part with morphological and phase parameters found to be useful in other DHM studies of cancer cells (17,35,48). Several other groups using holographic microscopy techniques have tracked migrating cancer cells (9), and identified phase textures of adherent cancer cells (11,14). Cancer cells from more advanced or metastatic disease have larger and more irregular nuclei (16), higher nuclear disorder parameters on the nanoscale, and higher micron-scale variation (11). Besides DHM, other label-free optical methods, including Raman spectroscopy (49) and Fourier transform light scattering (50) are useful for cell classification using machine learning techniques. Label-free, quantitative imaging methods such as DHM have several advantages in analyzing single cells, including the ease of spatial analysis of the optical signal from different regions of the cell, and the ability to directly compare with other imaging modalities, such as epifluorescence microscopy.
DHM and machine learning together form a powerful way to identify aspects of cancer cell behavior in transparent media, including morphologically distinct modes of motility. Numerous learning approaches are available and have been used on DHM data sets, including deep learning neural networks to distinguish SW480 colon cancer cells in suspension from white blood cells (48), anthrax from related microorganisms (51), and to detect and compensate for DHM background and phase aberrations (28). Machine learning paired with DHM has also recently been used to detect red blood cell infection by P. falciparum (52), discriminate between isogenic cell lines of differing metastatic stage (17), distinguish cancer grades from prostate biopsy microarrays (53), and distinguish healthy and nutrient-deprived cancer cells (35), and live and dead yeast cells (54). Recent commentaries and reviews highlight recent studies combining these two techniques to advance cancer research (55,56).
This study highlights that DHM is sensitive to cytoskeletal as well as nuclear and general cytoplasmic features, with special relevance to cancer cell motility. The MDA-MB-231 cell line is highly invasive, with switchable migration behavior depending on the microenvironment and protease activity (34). In the conditions studied, it is expected that about 50% of the cells are moving in a mesenchymal fashion, and the rest in an amoeboid mode (34), consistent with the proportions (Fig. 2) and appearance of elongated and rounded/motile morphologies observed in this study. Trends in DHM phase signatures during division (Fig. 5), motility (Fig. 6), and adhesion to different substrates relate to organization of the F-actin cytoskeleton (Figs. 7 and 8). Filopodia, lamellar structures, microspikes, and structures interface with substrate adhesion sites, are visible in phalloidin-labeled epifluorescence images and in DHM phase maps, indicating the mode of motility and quality of substrate attachment of each imaged cell. Relative to cell classification in a flow-based system (17), DHM of adherent cells on native biopolymers such as collagen captures functional aspects of cell behavior, such as division (Fig. 5) and motility (Fig. 2, 3, and 6). Given the ability of DHM to track cells migrating in three-dimensional (8), DHM of cells in a well-designed sample chamber could be used to develop a more quantitative matrix invasion assay than the conventional Boyden chamber assay (57). The relation of phase signatures to actin cytoskeletal features supports DHM as a tool to study mechanisms of cancer cell migration and invasiveness in vitro.
These data also indicate the possibility of tracking of cancer cell cycle status using DHM, relevant to studying mechanisms of cancer growth and response to therapeutic agents. Although only elongated and rounded/motile cells were classified from DHM phase parameters with machine learning algorithms, dividing, premitotic, mitotic, and post-mitotic cancer cells also appear to have distinct phase features. For example, the bright linear phase signal apparent in the middle of joined parent and daughter cells (Fig. 5.a.iii) could originate from centrosome-aligned chromosomes (1). Osmotic swelling of the cell during mitosis can also alter the phase signal (1). Indeed, DHM was found to monitor cell cycle phase distribution of etoposide-treated L929 cells with similar accuracy to flow cytometry, through measurement of cell phase volume (58). The use of DHM to distinguish stages of cell division is important for DHM-based cytometry, to segment cells in G1, G2, S, and M phases, or cells with subtly altered cell cycle induced by photodamage in time-lapse imaging experiments (23). With accurate DHM-based tracking of the cell cycle, the effects of kinase inhibitors targeting cell cycle regulation proteins as anti-cancer agents could be quantitatively determined (59).
In conclusion, DHM is an effective tool to assess cancer cell morphology and motility, especially in relation to substrate adhesion and de-adhesion. Machine learning applied to DHM data sets potentially separates cells based on behavior including cell division, mode of migration on substrates, and morphologies characteristic of distinct cancer phenotypes. These behaviors relate to the contractile and force-bearing actin cytoskeleton, which is most prominent in phase signal from the cell border, and with the nucleus contributes part of the total phase signal of the cell. Given a number of cytoskeletal proteins implicated as cancer biomarkers in vivo but with unknown effects on cancer cell behavior in vitro (60), DHM might reasonably provide a quantitative means to distinguish between the differential effects of these genetic markers on cancer cell behavior. Because DHM is nondamaging with very low power density at the sample (< 0.1 mW/cm2 in this study), is label-free, and has fast acquisition rates, it is an excellent tool for long-term time-lapse imaging of cancer cells to determine clinically relevant behavior.
Supplementary Material
Acknowledgments
Financial assistance from the School of Engineering at The Catholic University of America (VL, TN) is acknowledged. The authors would like to thank Dr. Zaver Bhujwalla (Johns Hopkins School of Medicine, Baltimore, MD) for the generous gift of the MDA-MB-231 cell line.
Footnotes
Additional Supporting Information may be found in the online version of this article.
The authors declare no conflicts of interest.
Literature Cited
- 1.Zlotek-Zlotkiewicz E, Monnier S, Cappello G, Le Berre M, Piel M. Optical volume and mass measurements show that mammalian cells swell during mitosis. J Cell Biol 2015;211:765–774. doi: 10.1083/jcb.201505056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappaz B, Cano E, Colomb T, Kühn J, Depeursinge C, Simanis V, Magistretti PJ, Marquet P. Noninvasive characterization of the fission yeast cell cycle by monitoring dry mass with digital holographic microscopy. J Biomed Opt 2009;14:034049. doi: 10.1117/1.3147385. [DOI] [PubMed] [Google Scholar]
- 3.Kemper B, Bauwens A, Vollmer A, Ketelhut S, Langehanenberg P, Müthing J, Karch H, von Bally G. Label-free quantitative cell division monitoring of endothelial cells by digital holographic microscopy. J Biomed Opt 2010;15:036009. doi: 10.1117/1.3431712. [DOI] [PubMed] [Google Scholar]
- 4.Chalut KJ, Ekpenyong AE, Clegg WL, Melhuish IC, Guck J. Quantifying cellular differentiation by physical phenotype using digital holographic microscopy. Integr Biol (Camb) 2012;4:280–284. doi: 10.1039/c2ib00129b. [DOI] [PubMed] [Google Scholar]
- 5.Pavillon N, Kuhn J, Moratal C, Jourdain P, Depeursinge C, Magistretti PJ, Marquet P. Early cell death detection with digital holographic microscopy. PLoS One 2012;7:e30912. doi: 10.1371/journal.pone.0030912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Schich Z, Molder A, Tassidis H, Harkonen P, Falck Miniotis M, Gjorloff Wingren A. Induction of morphological changes in death-induced cancer cells monitored by holographic microscopy. J Struct Biol 2015;189:207–212. doi: 10.1016/j.jsb.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Langehanenberg P, Ivanova L, Bernhardt I, Ketelhut S, Vollmer A, Dirksen D, Georgiev G, von Bally G, Kemper B. Automated three-dimensional tracking of living cells by digital holographic microscopy. J Biomed Opt 2009;14:014018. doi: 10.1117/1.3080133. [DOI] [PubMed] [Google Scholar]
- 8.Dubois F, Yourassowsky C, Monnom O, Legros JC, Debeir O, Van Ham P, Kiss R, Decaestecker C. Digital holographic microscopy for the three-dimensional dynamic analysis ofin vitro cancercell migration. J Biomed Opt 2006;11:054032. doi: 10.1117/1.2357174. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Hong J, Liu C, Cross M, Haynie DT, Kim MK. Four-dimensional motility tracking of biological cells by digital holographic microscopy. J Biomed Opt 2014;19:045001. doi: 10.1117/1.JBO.19.4.045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zangle TA, Teitell MA. Live-cell mass profiling: an emerging approach in quantitative biophysics. Nat Methods 2014;11:1221–1228. doi: 10.1038/nmeth.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damania D, Subramanian H, Backman V, Anderson EC, Wong MH, McCarty OJ, Phillips KG. Network signatures of nuclear and cytoplasmic density alterations in a model of pre and postmetastatic colorectal cancer. J Biomed Opt 2014;19:16016. doi: 10.1117/1.JBO.19.1.016016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zikmund T, Kvasnica L, Týčc M, Kříܞová A, Čolláková J, Chmelík R. Sequential processing of quantitative phase images for the study of cell behaviour in real-time digital holographic microscopy. J Microsc 2014;256:117–125. doi: 10.1111/jmi.12165. [DOI] [PubMed] [Google Scholar]
- 13.Quan X, Xia P, Matoba O, Nitta K, Awatsuji Y. Multi-Modal Digital Holographic Microscopy and Demonstration on Dual-Excitation Fluorescence. Heidelberg, Germany: The Optical Society (OSA); 2016. [Google Scholar]
- 14.Kemper B, Carl D, Schnekenburger J, Bredebusch I, Schaäfer M, Domschke W, von Bally G, Investigation of living pancreas tumor cells by digital holographic microscopy. J Biomed Opt 2006;11:34005. doi: 10.1117/1.2204609. [DOI] [PubMed] [Google Scholar]
- 15.Belashov AV, Zhikhoreva AA, Belyaeva TN, Kornilova ES, Petrov NV, Salova AV, Semenova IV, Vasyutinskii OS. Digital holographic microscopy in label-free analysis of cultured cells’ response to photodynamic treatment. Opt Lett 2016;41:5035–5038. doi: 10.1364/OL.41.005035. [DOI] [PubMed] [Google Scholar]
- 16.Benzerdjeb N, Garbar C, Camparo P, Sevestre H. Digital holographic microscopy as screening tool for cervical cancer preliminary study. Cancer Cytopathol 2016;124:573–580. doi: 10.1002/cncy.21727. [DOI] [PubMed] [Google Scholar]
- 17.Roitshtain D, Wolbromsky L, Bal E, Greenspan H, Satterwhite LL, Shaked NT. Quantitative phase microscopy spatial signatures of cancer cells. Cytometry A 2017;91A: 482–493. doi: 10.1002/cyto.a.23100. [DOI] [PubMed] [Google Scholar]
- 18.Clark AG, Vignjevic DM. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol 2015;36:13–22. doi: 10.1016/j.ceb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Lammermann T, Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr Opin Cell Biol 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: Three-dimensional amoeboid movement revisited. J Cell Biol 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen TH, Kandel M, Shakir HM, Best-Popescu C, Arikkath J, Do MN, Popescu G. Halo-free phase contrast microscopy. Sci Rep 2017;7:44034. doi: 10.1038/srep44034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magidson V, Khodjakov A. Circumventing photodamage in live-cell microscopy. Methods Cell Biol 2013;114:545–560. doi: 10.1016/B978-0-12-407761-4.00023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T, Nehmetallah G, Raub C, Mathews S, Aylo R. Accurate quantitative phase digital holographic microscopy with single- and multiple-wavelength telecentric and nontelecentric configurations. Appl Opt 2016;55:5666–5683. doi: 10.1364/AO.55.005666. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen T, Bui V, Tran D, Nehmetallah G. Deep learning bi-telecentric digital holographic microscopy for aberration compensation applied to cancer cells. Digital Holography and Three-Dimensional Imaging. Heidelberg, Germany: The Optical Society (OSA); 2017. p Tu2A.5. [Google Scholar]
- 26.Nguyen TC, Nehmetallah G, Lam V, Chung BM, Raub CB. Quantitative Assessment of Cancer Cell Morphology and Movement Using Telecentric Digital Holographic Microscopy.SPIE: San Francisco, CA; 2017. p 100740U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nehmetallah G, Nguyen TC. Optical and digital aberration compensation in DHM. Digital Holography and Three-Dimensional Imaging. Heidelberg, Germany: The Optical Society (OSA); 2016. p JW4A.5 [Google Scholar]
- 28.Nguyen T, Bui V, Lam V, Raub CB, Chang LC, Nehmetallah G. Automatic phase aberration compensation for digital holographic microscopy based on deep learning background detection. Opt Express 2017;25:15043–15057. doi: 10.1364/OE.25.015043. [DOI] [PubMed] [Google Scholar]
- 29.Nehmetallah G Multi-wavelength digital holographic microscopy using a telecentric reflection configuration. Digital Holography & 3-D Imaging Meeting. Germany: The Optical Society (OSA); 2015. p DM3A.7. [Google Scholar]
- 30.Nehmetallah G, Banerjee PP. Applications of digital and analog holography in 3D imaging. Adv Opt Photonics 2012;4:472–553. 10.1364/AOP.4.000472. [DOI] [Google Scholar]
- 31.Doblas A, Sanchez-Ortiga E, Martinez-Corral M, Saavedra G, Garcia-Sucerquia J. Accurate single-shot quantitative phase imaging of biological specimens with telecentric digital holographic microscopy. J Biomed Opt 2014;19:046022. doi: 10.1117/1.JBO.19.4.046022. [DOI] [PubMed] [Google Scholar]
- 32.Rappaz B, Breton B, Shaffer E, Turcatti G. Digital holographic microscopy: A quantitative label-free microscopy technique for phenotypic screening. Comb Chem High Throughput Screen 2014;17:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haralick RM. Statistical and structural approaches to texture. Proc IEEE 1979;67:786–804. 10.1109/PROC.1979.11328. [DOI] [Google Scholar]
- 34.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: Mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strbkova L, Zicha D, Vesely P, Chmelik R. Automated classification of cell morphology by coherence-controlled holographic microscopy. J Biomed Opt 2017;22:1–9. doi: 10.1117/1.JBO.22.8.086008. [DOI] [PubMed] [Google Scholar]
- 36.Dong P, Liu J. Hyperspectral image classification using support vector machines with an efficient principal component analysis scheme. In: Wang Y, Li T, editors. Foundations of Intelligent Systems: Proceedings of the Sixth International Conference on Intelligent Systems and Knowledge Engineering, Shanghai, China. Dec 2011 (ISKE2011). Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. p 131140. [Google Scholar]
- 37.Siuly S, Li Y. Designing a robust feature extraction method based on optimum allocation and principal component analysis for epileptic EEG signal classification. Comput Methods Programs Biomed 2015;119:29–42. doi: 10.1016/j.cmpb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Sahak R, Mansor W, Lee YK, Yassin AM, Zabidi A. Performance of combined support vector machine and principal component analysis in recognizing infant cry with asphyxia. Conf Proc IEEE Eng Med Biol Soc 2010;2010:6292–6295. doi: 10.1109/IEMBS.2010.5628084. [DOI] [PubMed] [Google Scholar]
- 39.Kriti Virmani J, Dey N, Kumar V. PCA-PNN and PCA-SVM Based CAD Systems for Breast Density Classification. In: Hassanien A-E, Grosan C, Fahmy Tolba M, editors. Applications of Intelligent Optimization in Biology and Medicine: Current Trends and Open Problems. Cham: Springer International Publishing; 2016. p 159180. [Google Scholar]
- 40.Quan X, Matoba O, Awatsuji Y. Image recovery from defocused 2D fluorescent images in multimodal digital holographic microscopy. Opt Lett 2017;42:1796–1799. doi: 10.1364/OL.42.001796. [DOI] [PubMed] [Google Scholar]
- 41.Bang LT, Wu HY, Zhao Y, Kim EG, Kim N. Depth enhancement of 3D microscopic living-cell image using incoherent fluorescent digital holography. J Microsc 2017;265:372–385. doi: 10.1111/jmi.12510. [DOI] [PubMed] [Google Scholar]
- 42.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 2000;198:82–87. [DOI] [PubMed] [Google Scholar]
- 43.Cotte Y, Toy F, Jourdain P, Pavillon N, Boss D, Magistretti P, Marquet P, Depeursinge C. Marker-free phase nanoscopy. Nature Photonics 2013;7:113–117. doi: 10.1038/nphoton.2012.329. [DOI] [Google Scholar]
- 44.Neumann A, Kuznetsova Y, Brueck SR. Optical resolution below lambda/4 using synthetic aperture microscopy and evanescent-wave illumination. Opt Express 2008;16:20477–20483. [DOI] [PubMed] [Google Scholar]
- 45.Kuznetsova Y, Neumann A, Brueck SR. Solid-immersion imaging interferometric nanoscopy to the limits of available frequency space. J Opt Soc Am A Opt Image Sci Vis 2012;29:772–781. doi: 10.1364/JOSAA.29.000772. [DOI] [PubMed] [Google Scholar]
- 46.Di J, Zhao J, Jiang H, Zhang P, Fan Q, Sun W. High resolution digital holographic microscopy with a wide field of view based on a synthetic aperture technique and use of linear CCD scanning. Appl Optics 2008;47:5654–5659. [DOI] [PubMed] [Google Scholar]
- 47.Paturzo M, Merola F, Grilli S, De Nicola S, Finizio A, Ferraro P. Super-resolution in digital holography by a two-dimensional dynamic phase grating. Opt Express 2008; 16:17107–17118. [DOI] [PubMed] [Google Scholar]
- 48.Chen CL, Mahjoubfar A, Tai LC, Blaby IK, Huang A, Niazi KR, Jalali B. Deep learning in label-free cell classification. Sci Rep 2016;6:21471. doi: 10.1038/srep21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beleites C, Neugebauer U, Bocklitz T, Krafft C, Popp J. Sample size planning for classification models. Anal Chim Acta 2013;760:25–33. doi: 10.1016/j.aca.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Jo Y, Jung J, Kim MH, Park H, Kang SJ, Park Y. Label-free identification of individual bacteria using Fourier transform light scattering. Opt Express 2015;23:15792–15805. doi: 10.1364/OE.23.015792. [DOI] [PubMed] [Google Scholar]
- 51.Jo Y, Park S, Jung J, Yoon J, Joo H, Kim MH, Kang SJ, Choi MC, Lee SY, Park Y. Holographic deep learning for rapid optical screening of anthrax spores. Sci Adv 2017;3:e1700606. doi: 10.1126/sciadv.1700606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park HS, Rinehart MT, Walzer KA, Chi J-TA, Wax A, Sullivan DJ. Automated detection of p. falciparum using machine learning algorithms with quantitative phase images of unstained cells. PLoS One 2016;11:e0163045. doi: 10.1371/journal.pone.0163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen TH, Sridharan S, Macias V, Kajdacsy-Balla A, Melamed J, Do MN, Popescu G. Automatic Gleason grading of prostate cancer using quantitative phase imaging and machine learning. J Biomed Opt 2017;22:36015. 10.1117/1.JBO.22.3.036015. [DOI] [PubMed] [Google Scholar]
- 54.Feizi A, Zhang Y, Greenbaum A, Guziak A, Luong M, Chan RYL, Berg B, Ozkan H, Luo W, Wu M, et al. Rapid, portable and cost-effective yeast cell viability and concentration analysis using lensfree on-chip microscopy and machine learning. Lab Chip 2016;16:4350–4358. doi: 10.1039/c6lc00976j. [DOI] [PubMed] [Google Scholar]
- 55.Raub CB, Nehmetallah G. Holography, machine learning, and cancer cells. Cytometry A 2017;91A:754–756. doi: 10.1002/cyto.a.23112. [DOI] [PubMed] [Google Scholar]
- 56.Wu Y, Ozcan A. Lensless digital holographic microscopy and its applications in biomedicine and environmental monitoring. Methods 2017; Available at: 10.1016/j.ymeth.2017.08.013 [DOI] [PubMed] [Google Scholar]
- 57.Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res 1987;47:3239–3245. [PubMed] [Google Scholar]
- 58.Falck Miniotis M, Mukwaya A, Gjörloff Wingren A, Kreplak L. Digital holographic microscopy for non-invasive monitoring of cell cycle arrest in L929 cells. PLoS One 2014;9:e106546. doi: 10.1371/journal.pone.0106546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickson MA, Schwartz GK. Development of cell-cycle inhibitors for cancer therapy. Curr Oncol 2009;16:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karantza V Keratins in health and cancer: more than mere epithelial cell markers. Oncogene 2011;30:127–138. 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


