Abstract
Histones serve to both package and organize DNA within the nucleus. In addition to histone post-translational modification and chromatin remodelling complexes, histone variants contribute to the complexity of epigenetic regulation of the genome. Histone variants are characterized by a distinct protein sequence and a selection of designated chaperone systems and chromatin remodelling complexes that regulate their localization in the genome. In addition, histone variants can be enriched with specific post-translational modifications, which in turn can provide a scaffold for recruitment of variant-specific interacting proteins to chromatin. Thus, through these properties, histone variants have the capacity to endow specific regions of chromatin with unique character and function in a regulated manner. In this Review, we provide an overview of recent advances in our understanding of the contribution of histone variants to chromatin function in mammalian systems. First, we discuss new molecular insights into chaperone-mediated histone variant deposition. Next, we discuss mechanisms by which histone variants influence chromatin properties such as nucleosome stability and the local chromatin environment both through histone variant sequence-specific effects and through their role in recruiting different chromatin-associated complexes. Finally, we focus on histone variant function in the context of both embryonic development and human disease, specifically developmental syndromes and cancer.
Organisms have evolved complex and elegant mechanisms of chromatin regulation to support cell function and specialization. The basic unit of chromatin — the nucleosome core particle — is composed of approximately 146 base pairs of DNA wrapped around a histone octamer, which contains two copies of each of the core histones H2A, H2B, H3 and H4 (REF.1). Wrapping of chromatin on nucleosomes is further stabilized by linker H1 histones bound at the entry and exit sites of DNA from each nucleosome, which is thought to contribute to chromatin compaction2.
Most histones present in chromatin are replication coupled: they are synthesized only during S phase and are deposited onto newly replicated DNA by dedicated histone chaperones that work in concert with the DNA polymerase3. Their genes are present as multiple, intronless copies in histone clusters within the genome, and their expression is tightly regulated at both the transcription level and the translation level. However, several histones are encoded by genes outside these canonical histone clusters. These genes are generally present in one or two copies within the genome and exhibit a more typical gene structure, with introns that can result in different splice isoforms in some cases. Their expression and chromatin incorporation at specific genomic regions is not coupled to cell cycle regulation. These histone variants are subject to dynamic exchange, and both their deposition and their selective eviction and/or recycling are regulated by specific chaperones or chromatin remodelling complexes (generally quite large in size) that recognize amino acid differences between these variants and their replication-coupled counterparts. Some histone variants are uniformly expressed, while others are tissue specific, with selective expression demonstrated in the male germ line (testes) and the brain. Some variants are highly conserved and appear to have arisen only once in evolution, while other variants seem to have diverged repeatedly in different lineages4. Histone variants also have highly similar isoforms called ‘subvariants’ (encoded mainly by pseudogenes), which are not conserved among species and might play a role in regulating tissue-specific gene expression5. All core histones in mammals exist as one or more such replication-independent variant counterparts, including the newly discovered Hominidae-specific histone H4 variant H4G6,7 (FIG. 1; TABLE 1).
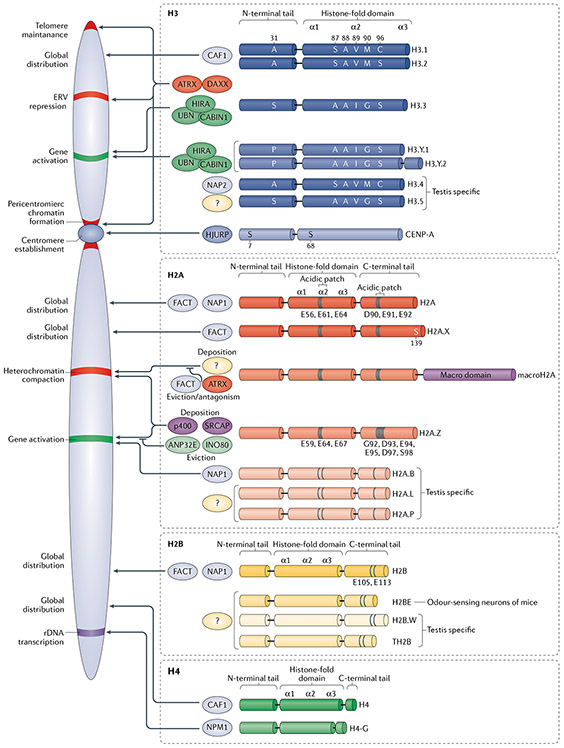
Fig. 1 ∣. Core histones, their variants and associated chaperone/remodeller machineries.
Replication-coupled and variant core histones with respective chaperones, genomic distribution and functional output are shown. Specific amino acid residues are illustrated at either key differences among members of a common histone protein family (for example, histone H3.3 Ser31 vs H3.1 Ala31) or at well-established histone variant-specific post-translational modifications (for example, centromeric protein A (CENP-A) Ser7). Different shades of colour in the histone’s structures are used to indicate differences within the domains compared with the replication-coupled histone. See the text and TABLE 1 for additional details and references. ANP32E, acidic leucine-rich nuclear phosphoprotein 32 family member E; ATRX, α-thalassaemia mental retardation syndrome X-linked; CABIN1, calcineurin-binding protein cabin 1; CAF1, chromatin assembly factor 1; DAXX, death domain-associated protein; ERV, endogenous retroviral element; FACT, facilitates chromatin transcription; HIRA, histone regulator A; HJURP, Holliday junction recognition protein; NAP, nucleosome assembly protein; NPM1, nucleophosmin; rDNA, ribosomal DNA; UBN, ubinuclein; SRCAP, Snf2-related CREBBP CBP activator protein.
Table 1 ∣.
Functions of core histone variants and their chaperones and their links to disease
| Core histone | Chaperones or chromatin remodelling complexes |
Tissue distribution |
General function |
Association with cancer |
Associated human genetic disorders |
Key references |
|
|---|---|---|---|---|---|---|---|
| Dysregulation in cancer |
Role in cancer | ||||||
| Histone variants with specific chaperones or remodelling complexes | |||||||
| H2A family | |||||||
| H2A.Z.1 | p400, SRCAP (deposition); INO80, ANP32E (eviction) | Global | Binding of regulatory complexes and chromatin dynamics | Amplification and missense mutations of unknown function | Oncogene expression, cell growth and epithelial–mesenchymal transition | FHS syndrome (SRCAP mutation) | 70,79,80,82,205,206,235 |
| H2A.Z.2 (occurring as 2 splice isoforms: H2A.Z.2.1 and H2A.Z.2.2) | p400, SRCAP (deposition); ANP32E (eviction) | Global | Binding of regulatory complexes and chromatin dynamics | Amplification and missense mutations of unknown function | Pro-oncogenic transcription and cell proliferation | FHS syndrome (SRCAP mutation) | 70,79,80,82,141,235 |
| H3 family | |||||||
| H3.3 | HIRA–UBN–CABIN1 | Global | Transcriptional activation and chromatin dynamics | Amplification and missense mutations (K27M, G34R/V/L and K36M) resulting in amino acid substitutions | Transcriptional response to pro-oncogenic signalling and dysregulation of global chromatin states | ND | 134,192,193,195,199,236-238 |
| ATRX–DAXX | Global | Heterochromatin formation and telomere stabilization | Missense (presumably loss of function) mutation in both ATRX and DAXX | Tumour suppressor through maintenance of telomere structure | α-Thalassaemia X-linked mental retardation syndrome (ATRX mutation) | 23,36,37,192,218,239 | |
| H3.Y.1 and H3.Y.2 (also known as H3.X)a | HIRA–UBN–CABIN1 | Testes and brain | Transcriptional activation | ND | ND | ND | 47 |
| CENP-A | HJURP | Global | Centromere identity and genome stability | Amplification or overexpression | Chromosome missegregation and chromosome instability | ND | 55,56,146,240 |
| Histone variants that, to date, rely on general histone deposition machinery | |||||||
| H2A family | |||||||
| macroH2A1 (occurring as 2 splice isoforms: macroH2A1.1 and macroH2A1.2) and macroH2A2 | FACT (eviction); ATRX (antagonizes deposition); ND (deposition) | Global | Gene silencing and higher-order chromatin compaction | Transcriptional repression and splicing defects and amplification | Tumour suppressor through heterochromatin and cell identity maintenance | ND | 95,201 |
| H2A.X | FACT | Global | DNA damage response and chromatin remodelling | Mutation or deletion | Tumour suppressor through prevention of genome instability and oncogenic translocations | Nijmegen breakage syndrome and ataxia–telangiectasia | 104,215,241 |
| H2A.B | NAP1 | Testes and brain | Nucleosome destabilization, and active transcription and mRNA splicing | ND | ND | ND | 61,242 |
| H3 family | |||||||
| H3.4 (also known as H3T) | NAP2 | Testes | Histone-to-protamine transition | ND | ND | ND | 45 |
| H4 family | |||||||
| H4G | Nucleophosmin | Global, mainly breast cancer | Upregulation of rDNA trascription | ND | Potentially oncogenic | ND | 6,7 |
| Histone variants with unknown or undetermined deposition machinery | |||||||
| H2A family | |||||||
| H2A.L (with several splice isoforms possible)b | ND | Testes | Histone-to-protamine transition shown for H2A.L2 | ND | ND | ND | 150 |
| H2B family | |||||||
| TH2B (also known as TS H2B.1) | ND | Testes | Histone-to-protamine transition | ND | ND | ND | 152 |
| H2B.W (also known as H2BFWT) | ND | Testes | ND | ND | ND | ND | |
| H3 family | |||||||
| H3.5 | ND | Testes | Histone-to-protamine transition | ND | ND | ND | 46 |
ANP32E, acidic leucine-rich nuclear phosphoprotein 32 family member E; ATRX, α-thalassaemia mental retardation syndrome X-linked; CABIN1, calcineurin-binding protein cabin 1; CENP-A, centromeric protein A; DAXX, death domain-associated protein; FACT, facilitates chromatin transcription; FHS, Floating-Harbor syndrome; HIRA, histone regulator A; HJURP, Holliday junction recognition protein; NAP, nucleosome assembly protein; ND, not determined; rDNA, ribosomal DNA; SRCAP, Snf2-related CREBBP activator protein; UBN, ubinuclein.
Encoded by two highly related genes.
Present in various mammals, including mice, but not humans.
Besides the widely appreciated mechanisms such as histone and DNA modification, chromatin remodelling and topological higher-order chromatin organization8, the incorporation of histone variants into chromatin contributes to the diversity of nucleosome structure and function9,10. The changes to chromatin associated with histone variant incorporation can be profound, resulting in nucleosomes that contain vastly different histone sequences and domains compared with their replicative counterparts, or relatively modest, with variant-containing nucleosomes differing by just a few amino acids yet displaying distinct genomic enrichment profiles and properties. Histone variants function in virtually all known DNA-templated processes. For example, the centromere-specific variant centromeric protein A (CENP-A) epigenetically marks the centromeric region of chromosomes, and H3.3 is highly associated with chromatin dynamics and nucleosome turnover. Furthermore, a number of distinct H2A variants influence DNA organization through participation in DNA repair, gene regulation and other processes, which are discussed in detail later. Accordingly, deregulation of histone variant expression and deposition has been implicated in both developmental syndromes and cancer (TABLE 1). As such, histone variants have been intensely studied, with many important insights having been made in recent years.
In this Review, we provide an overview of the core histone variants, describe recent insights into the molecular mechanisms that drive their deposition and discuss our current understanding of how they affect chromatin function. We further discuss the role that core histone variants play during mammalian development and end with recent advances in our understanding of the contribution of histone variants to developmental disorders and cancer. Of note, in addition to core histones, the linker histone H1 family consists of a large number of both somatic and germline-specific variants, which will not be covered here in detail (see REF.2 for a review).
Histone variants and their chaperones
The expression of histone variants throughout the cell cycle allows them to participate in genome regulation outside of replication, and their deposition is often referred to as replication independent, although some histone variants positively contribute to replication11-13. Studies of replication-coupled histones and their chaperones have provided important theoretical frameworks for our understanding of histone variant–chaperone interactions14. In the following subsections, we summarize our knowledge of the best-characterized H3 and H2A variants and their deposition complexes and provide a brief description of H2B and H4 variants, with a focus on histone variants found in mammals (FIG. 1).
H3 variants
The H3 variants demonstrate that small sequence differences can dramatically influence molecular outcomes of their incorporation into chromatin. In mammals, there are three main classes of H3 with tissue-wide expression: (1) replication-coupled H3.1 and H3.2, which differ by one Cys96-to-Ser96 amino acid substitution, whose significance is unknown; (2) the replacement variant H3.3 (described as such because of its role in histone replacement at active genes and promoters); and (3) CENP-A.
In mammals, H3.3 is encoded by two distinct genes, H3F3A and H3F3B (also known as H3-3A and H3-3B), whose translation results in an identical protein product that differs from H3.1 and H3.2 by only four or five amino acids (FIG. 1). Histone chaperones are able to distinguish between H3.1/H3.2 and H3.3 variants using an amino acid motif at the base of α-helix 2 in the core of the protein, wherein the Ser-Ala-Val-Met motif from amino acids 87–90 in H3.1 and H3.2 is replaced by the Ala-Ala-Ile-Gly motif in H3.3. Another difference between H3.1/H3.2 and H3.3 occurs in the highly post-translationally modified N-terminal tail with Ala at position 31 in H3.1 and H3.2 that is replaced in H3.3 by a highly conserved Ser. Phosphorylation of Ser31 of H3.3 has been implicated in several biological processes that are discussed in more detail below15-19.
Despite the observation that H3.3 nucleosomes are essentially structurally identical to H3.1 nucleosomes20, H3.3 deposition is generally an indication of dynamic chromatin, with enriched regions exhibiting high turnover rates on a genome-wide level21,22. Accordingly, H3.3 deposition is characteristic of euchromatin and specifically associated with active regulatory elements, including enhancers and promoters as well as gene bodies. H3.3 deposition is mediated by the histone regulator A (HIRA) chaperone complex, composed of HIRA, ubinuclein 1 or ubinuclein 2 (UBN1 or UBN2) and calcineurin-binding protein CABIN123-25 (FIG. 1). HIRA associates with RNA polymerase II and associated machinery26,27, transcription factors28 and replication protein A (RPA)-bound regions of nucleosome-free DNA29, all of which are proposed to be involved in recruitment of HIRA and subsequent H3.3 deposition at genes. HIRA does not make contacts with H3.3 and rather enhances interaction between H3.3 and its direct chaperone, UBN1 (REFS30,31). H3.3 incorporation at transcriptionally active regions can result either from de novo deposition of newly synthesized H3.3 or from recycling of H3.3 from existing nucleosomes. Recent biochemical and crystallographic analyses show that the HIRA subunit forms a homotrimer that binds two subunits of CABIN1, and that this stoichiometry and interaction with UBN1 are required for deposition of de novo synthesized H3.3 (REF.32). By contrast, transcription-mediated recycling of existing H3.3 (due to RNA polymerase II displacement of nucleosomes during elongation) does not require UBN1 and relies only on HIRA and the general H3 and H4 chaperone anti-silencing factor 1 (ASF1)33. This H3.3 recycling is linked to maintenance of histone H3 Lys36 trimethylation (H3K36me3)33 — a histone post-translational modification (PTM) found in gene bodies and associated with active transcription — suggesting that an important function of H3.3 recycling is to maintain chromatin PTM states at actively transcribed regions. Although it was initially surprising, it is well established that H3.3 is also deposited at repetitive heterochromatic regions such as telomeres and pericentromeric chromatin by a complex comprising α-thalassaemia mental retardation syndrome X-linked (ATRX) and death domain-associated protein (DAXX)23,34-36 (FIG. 1). Additionally, H3.3 is enriched at endogenous retroviral elements and imprinted regions in mouse embryonic stem cells (ES cells) in a DAXX-dependent manner37,38, although the contribution of ATRX at these regions is unclear, with studies providing evidence both for and against a role for ATRX37-40. Structural comparisons of DAXX and UBN1 bound to the H3.3–H4 dimer reveal striking similarities and suggest that, despite their lack of sequence conservation, these two chaperones have evolutionarily converged on recognizing the Ala-Ala-Ile-Gly motif as a mechanism to allow discrimination among H3 histones30,41,42. Protein folding of DAXX is coupled to H3.3–H4 substrate recognition and binding in vitro43, and loss of H3.3 results in reduced levels of nuclear DAXX37,40, suggesting a paradigm in which the substrate is required for chaperone stability. As such, DAXX and also HIRA44 require H3.3 to allow their recruitment to chromatin. While understanding the actual deposition process remains a major challenge, the presence of histone variant chaperones on chromatin provides support for the idea that one function of histone variants is to participate in the nucleation of chromatin-associated complexes at distinct regions of the genome.
In addition to H3.3, a number of additional replication-independent H3 variants exist. These variants may be expressed only in specific tissues, for example the testis-specific H3.4 (also known as H3T)45 and H3.5 (REF.46), and may be species specific, such as the primate-specific H3.Y.1 and H3.Y.2 (previously referred to as H3.Y and H3.X, respectively)47 and H3.5, which is specific to the hominid lineage (REF.46). Despite containing the Ala-Ala-Ile-Gly motif, H3.Y.1 interacts only with the HIRA complex and not with ATRX–DAXX, which was found to be dependent on a Val46-to-Leu46 substitution from H3.3 to H3.Y.1 (REF.48). This observation demonstrates the power of natural mutations in dissecting H3 variant–chaperone interactions and leaves open the question as to why higher organisms evolved an H3 variant that is a substrate for only one H3.3 deposition pathway. H3.Y.1–H3.3 heterotypic nucleosomes appear to be less compact (displaying higher susceptibility to micrococcal nuclease) than H3.3–H3.3 nucleosomes49. Furthermore, unpublished data demonstrate that DAXX-mediated H3.3 deposition actually helps to maintain a closed chromatin state at endogenous retroviral elements and ATRX-bound regions enriched with G-quadruplex DNA structures (personal communication with S. Elsässer and our own data), suggesting that H3.Y.1 deposition and nucleosome destabilization could be disruptive to the heterochromatin state found at these regions.
Evolution has also provided a highly specialized centromeric H3 variant, referred to as CenH3 or CENP-A in mammals, that is a key determinant of centromere identity and is required for genome stability (recently reviewed50) (FIG. 1). Given the critical importance of the centromere in chromosome segregation, one might expect CENP-A to be highly conserved. However, there is evidence that centromeric H3 variants are rapidly evolving and they are far less conserved than other H3 variants51. CENP-A exhibits 50–60% sequence similarity with most other H3 variants in the histone-fold domain and possesses a unique N-terminal tail. CENP-A-containing nucleosomes show transient unwrapping at the DNA entry–exit site, which may influence nucleosome assembly and stability. This has been attributed to the amino acid at position 49 in the protein sequence, which makes contacts with DNA: CENP-A contains a Lys residue at this position, whereas other H3 variants harbour an Arg residue52. CENP-A is chaperoned to the centromere by Holliday junction recognition protein (HJURP) and associates with at least 16 known centromere-associated network (CCAN) proteins53-56. The CCAN both recruits CENP-A to establish the kinetochore and serves as a structural core to directly recruit kinetochore proteins57. Despite divergent evolution, the structure of CENP-A with HJURP was used prospectively to aid determination of the H3.3–DAXX structure41, and there are many striking similarities between the two complexes41,58. Variations in stoichiometry of the CCAN complex components alter the physical properties of the nucleosome and the mode by which downstream CCAN and kinetochore components interact with CENP-A nucleosomes50. As with variations in the HIRA complex, these observations suggest that variant-specific chaperone complexes are flexible and allow distinct molecular outcomes based on the composition of the complex formed.
H2A variants
To date, the H2A family contains the greatest sequence diversity of identified variants. In humans, four classes of replication-independent H2A variants have been reported: H2A.X, H2A.Z, macroH2A and testis-specific short H2A variants (H2A.B, H2A.L, H2A.P and H2A.Q) (reviewed in REF.59) (FIG. 1). These variants span the spectrum of relatively modest amino acid differences — for example, the H2A.X variant contains an additional C-terminal Ser-Gln-Glu-Tyr motif, which is critical to its function — to large changes in protein composition — for instance, macroH2A contains an additional large C-terminal macro domain, whereas H2A.B lacks both the acidic patch (see further later) as well as the C-terminal α-helix and tail, both of which are fundamental for histone–histone interactions and nucleosome stabilization. While some H2A variants are known to partner with dedicated chaperones and ATP-dependent chromatin remodellers, the mechanisms that regulate localization of many H2A variants at specific genomic regions have been enigmatic. It is possible that general chaperone proteins such as nucleosome assembly protein 1 (NAP1) and facilitates chromatin transcription (FACT) perform this function60,61. In support of this notion, structural studies show that the FACT complex directly interfaces with H2B and is likely insensitive to the identity of H2A within the H2A–H2B dimer62,63. However, a general H2A–H2B chaperone cannot explain specificity of variant deposition, and thus additional undiscovered mechanisms must be at play to restrict H2A variants to specific regions of chromatin.
Histone H2A.Z is present in almost all organisms and is thought to have evolved early and only once64. H2A.Z shares extensive homology and structure with the core H2A in the histone-fold domain1,65, yet differs greatly in the sequence of its C-terminal tail (FIG. 1). In mammals, H2A.Z is encoded by two distinct paralogues, H2AFZ (also known as H2AZ1) and H2AFV (also known as H2AZ2), resulting in two different isoforms, H2A.Z.1 and H2A.Z.2, that differ by only three amino acids66-68. Additionally, H2AFV transcript can be alternatively spliced to give rise to two isoforms, H2A.Z.2.1 and H2A.Z.2.2, that differ in their C-terminal tails69. These three proteins are often collectively referred to as H2A.Z, although recent publications suggest that chromatin remodellers can distinguish among them, and these small differences have biological importance, which is discussed later70-72. Vertebrates contain two ATP-dependent chromatin remodellers responsible for incorporation of H2A.Z, p400 and Snf2-related CREBBP activator protein (SRCAP), both homologues of the Swi2/Snf2-related chromatin remodelling 1 (SWR1) complex73,74. The SWR1 complex mediates deposition of H2A.Z at regulatory elements and gene promoters in a stepwise fashion, generating first heterotypic H2A–H2A.Z nucleosomes and then homotypic H2A.Z nucleosomes75-77. Cells use distinct mechanisms to evict H2A.Z from nucleosomes, and the INO80 remodeller78 and acidic leucine-rich nuclear phosphoprotein 32 family member E (ANP32E) have been identified as H2A.Z-specific chaperones that are able to perform this function79,80. H2A.Z has been associated with both gene activation and gene silencing, potentially through regulating access of transcription factors and other chromatin-associated proteins to the underlying DNA64,81-83. H2A.Z is also necessary for the function of centromeres, and its loss leads to improper heterochromatin formation, loss of centromere cohesion, structural changes in pericentromeric heterochromatin and chromosome segregation defects84-87.
To date, no dedicated chaperones have been discovered for macroH2A, H2A.B or H2A.X, although FACT was recently identified as a factor promoting the ‘pruning’ of regions of macroH2A2 enrichment at transcribed chromatin88. Incorporation of macroH2A results in perhaps the most dramatic alteration to the nucleosome of any histone variant, as its C-terminal macro domain is roughly twice the size of its histone-fold domain89 (FIG. 1). In mammals, two non-allelic genes, H2AFY and H2AFY2, code for the two isoforms, macroH2A.1 and macroH2A.2. Additionally, H2AFY transcript can produce two splice variants, macroH2A.1.1 and macroH2A.1.2 (REF.90). The macro domain of macroH2A.1.1 is able to bind to ADP-ribose and poly(ADP-ribose)90-92, which is illustrative of the many links between chromatin regulation and metabolism93 (poly(ADP-ribose) is generated from the metabolite NAD+). The general view of macroH2A is as an epigenetic repressor of gene transcription due to its colocalization with Polycomb repressive complex 2 (PRC2)-mediated facultative heterochromatin, its relative depletion from active genes and its association with chromatin condensation94-96. The function of macroH2A in chromatin compaction was recently attributed not to the presence of the macro domain but to the short, unstructured linker region that separates this domain from the histone fold96, perhaps by promoting liquid–liquid phase separation of the protein and the formation of nuclear condensates. Of note, the plant histone variant H2A.W shows similarities in conserved motifs with macroH2A, suggesting that plants and animals share common mechanisms for heterochromatin condensation97 (see98 for a complete review of histone variants in plants). In contrast to macroH2A, H2A.B is the smallest H2A variant yet identified (FIG. 1). In mammals H2A.B is encoded by three intronless non-allelic genes, H2AFB1 (also known as H2AB1) H2AFB2 (also known as H2AB2) and H2AFB3 (also known as H2AB3), is strongly expressed in the testis and is thought to promote transcription through its destabilization of the nucleosome and through its ability to bind splicing factors and regulate mRNA processing99-101. Three additional short H2A variants exist in mammals (H2A.L, H2A.P and H2A.Q), although at least one type has been lost from all mammalian species102. For example, H2A.L is present in mammals, but not in humans, and its incorporation into chromatin impacts nucleosome structure and its remodelling99,100,103. Finally, H2A.X and its unique phosphorylation at Ser139 (resulting in γH2A.X; FIG. 1) has been historically associated with the DNA damage response, with deposition dependent on FACT and coupled to repair-associated DNA synthesis104 (see105 for a review of the role of histone variants in DNA damage).
H2B and H4 variants
Both H3 and H2A make homodimeric contacts in the octamer, whereas H4 and H2B contact only other histones1. Thus, probably, the different positions of the core histones within the nucleosome particle have subjected them to different evolutionary forces, leading to important diversifications of H2A and H3 but not as much for H2B and H4 (REF.4). Nevertheless, recently a new H4 variant, H4G, was found in a variety of human cell lines with localization primarily to the nucleoli6,7. Finally, minor sequence differences are observed in the four H2B variants identified so far (H2B, H2BE, TSH2B and H2B.W), which typically involve only a few amino acid substitutions (FIG. 1; TABLE 1). However, many of these substitutions occur in the histone-fold domain of these proteins, thereby destabilizing histone–histone interactions in a similar way to that of the H2A variants106.
The basis of histone variant functions
There are at least three important concepts when one is considering the functional role of histone variant deposition into chromatin. First, given the sequence differences between replication-coupled and histone variants, variants could endow nucleosomes with different inherent physical properties (FIG. 2). Second, through interaction with their unique domains and chaperone systems, histone variants can support the formation of distinct chromatin-associated complexes at specific regions of the genome (FIG. 3). Third, on the basis of both their specific genomic localization (that is, colocalization with specific chromatin-modifying enzymes) and their unique sequence, non-replicative histone variants are often enriched with specific PTMs or even can carry unique PTMs compared with their respective replication-deposited counterparts, which in turn can provide a scaffold for recruitment of variant-specific interacting proteins to chromatin (FIGS 1,3).
Fig. 2 ∣. Variant-containing nucleosomes influence nucleosome stability and chromatin properties.
The sequence-specific histone variant properties together with selective incorporation of multiple histone variants into chromatin can influence (positively or negatively) nucleosome stability and result in different functional output on chromatin organization and function. Histone H2A.B lacks the acidic patch and leads to decreased nucleosome stability. By contrast, H2A.Z contains an extended acidic patch that contributes to increased nucleosome stability in homotypic H2A.Z nucleosomes. MacroH2A contains an extra domain and is also associated with increased nucleosome stability. Centromeric protein A (CENP-A) confers a much more rigid subnucleosomal structure through the presence of two extra amino acid residues (Arg80 (R80) and Gly81 (G81)) in the loop 1 (L1) region, which provide the connection between two major helices in H3 and CENP-A. Incorporation of histone H3.3 together with H2A.Z at promoters and enhancers results in the loosening of histone–histone and histone–DNA interactions, leading to chromatin opening, transcription factor (TF) recruitment and promotion of transcription. H2A.Z nucleosomes also contribute to chromatin compaction. In this case, H2A.Z nucleosomes together with heterochromatin-associated H3K9me3 promote chromatin recruitment and stabilization of heterochromatin protein 1α (HPα), which coordinates chromatin compaction and transcriptional gene silencing. Furthermore, H2A.Z and macroH2A nucleosomes are both involved in the recruitment of Polycomb repressive complex 2 (PRC2), thus contributing to H3K27me3 and facultative heterochromatin establishment. Finally, incorporation of H3.3 into heterotypic CENP-A–H3.3 nucleosomes at centromeres during DNA replication can act as a placeholder for later replacement by CENP-A in G1 phase. RNAPII, RNA polymerase II.
Fig. 3 ∣. Variant-containing nucleosomes influence chromatin through various mechanisms.
a–c ∣ Histone variants can harbour post-translational modifications (PTMs) that have direct effects on chromatin structure and function through the recruitment of specific readers. Arabidopsis thaliana trithorax-related protein 5/6 (ATXR5/6) methylates only histone H3.1 and not H3.3 owing to steric hindrance by H3.3 Ser31 (part a). Variant-specific modification can promote chromatin-associated enzyme activity, specifically H3.3 Ser31 phosphorylation (ph) increases activity of the histone acetyltransferase p300 (part b). Additionally, H3.3S31ph promotes the histone methyltransferase SET domain-containing 2 (SETD2), resulting in H3.3K36me3 deposition — a mark of transcriptionally active chromatin. In turn, H3.3K36me3 is specifically recognized by zinc-finger MYND-type-containing 11 (ZMYND11), a reported repressor of transcription elongation, hence the role of H3.3S31ph in transcription regulation is likely multifaceted (part c). d ∣ Histone variants can also influence chromatin state through chaperone interaction with other chromatin-associated proteins to promote distinct chromatin states at regions of deposition. In the example shown, H3.3 incorporation leads to death domain-associated protein (DAXX)-dependent recruitment of Krüppel-associated box (KRAB)-associated protein 1 (KAP1) co-repressor and histone methyltransferase SET domain bifurcated 1 (SETDB1), all of which appear stabilized at chromatin in the presence of H3.3. e ∣ Another mechanism is combinatorial readout, such as the presence of acetylated histone H4 (H4ac) coupled with histone H2A.Z.1 which promotes bromodomain-containing 2 (BRD2) binding and transcription activation. f ∣ Histone variants can be modified in accordance with their site of deposition and thus modulate transcription. For example, H2A.Z can be acetylated at euchromatin or methylated and ubiquitylated at heterochromatin, contributing to gene activation or silencing, respectively. See TABLE 1 for additional details and references. PRC, Polycomb repressive complex.
Nucleosome structure and dynamics
One aspect of incorporation of histone variants into nucleosomes is changes to their structure and stability. For example, H2A.B and the related short H2A variants have truncated C termini and lack an acidic patch, which on other H2A family members is important for stabilizing histone–histone contacts with H4 in the nucleosome107. H2A.B nucleosomes accommodate a shorter stretch of ~118–130 bp of DNA compared with the normal 147 bp99,108, which inhibits chromatin folding109. Functionally, H2A.B deposition is correlated with active transcription and splicing both in vitro and in vivo101.
As another example, H2A.Z nucleosomes can be either homotypic (containing two H2A.Z molecules) or heterotypic (containing one H2A.Z molecule and one H2A molecule), and this composition translates to nucleosome stability: homotypic H2A.Z nucleosomes are more stable110 owing to their extended acidic patch111 and, because of their more stable state, stimulate PRC2 (REF.112) (FIG. 2). H2A.Z also contributes to heterochromatin formation by enhancing interaction between heterochromatin protein 1α and the nucleosome core in pericentromeric regions, which was required to promote kinetochore-driven microtubule formation and unperturbed chromosome segregation113-115. Heterotypic H2A.Z–H2A nucleosomes are reported to have reduced stability because of steric hindrance between their divergent L1 loops, sequences in the C-terminal tail that facilitate interaction between the two H2A proteins in the nucleosome65. Furthermore, primate-specific H2A.Z.2.2 results in nucleosome instability owing to changes in its C-terminal docking domain that reduce intranucleosomal contacts with H3 (REF.69). The observation that H2A.Z can destabilize the nucleosome is in agreement with the deposition profile of H2A.Z at enhancers and promoters, almost exclusively at the +1 nucleosome position116. These regions are more likely to be nucleosome-free than other regions of the genome117 and are further destabilized by the presence of nucleosomes containing both H2A.Z and H3.3 (see further later)118,119, perhaps allowing greater opportunity for transcription-factor binding to the underlying DNA82,111,120. While it is perhaps puzzling that H2A.Z can play both activating and inhibitory roles in transcription, we propose that, in general, H2A.Z is associated with dynamic regions undergoing changes in chromatin state, as seen for both hormone-induced gene activation and developmental formation of heterochromatin.
H3.3 is highly enriched in euchromatin, which is characterized by high levels of histone acetylation and is strongly correlated with active transcription. Studies have suggested that H3.3-containing nucleosomes are less stable than nucleosomes containing the replicative H3 (REF.118), which is consistent with an active role in transcription. However, ES cells lacking H3.3 do not show changes in the open state of chromatin at regulatory elements15, suggesting that H3.3 may not be causative in promoting nucleosome turnover at these regions. As such, the observation that H3.3-containing nucleosomes have physical stability properties different from those of H3-containing nucleosomes in cells might be attributed to phenomena such as the PTM state or the nature of chromatin-associated complexes established at sites of H3.3 enrichment. For example, loss of H3.3 results in reduced recruitment of the nucleosome remodelling deacetylase (NuRD) chromatin remodelling complex to active promoters121. A recently described subvariant of H3.3 called ‘H3mm7’, expressed in skeletal muscle stem cells (satellite cells), contains an amino acid substitution that destabilizes its contacts with H4 (REF.122), showing that H2A and H3 variants have both co-opted similar strategies to destabilize the nucleosome to promote access to underlying DNA. In addition to euchromatin, H3.3 has been shown to be deposited on centromeric regions in S phase, and it has been suggested that it acts as a placeholder for CENP-A on newly replicated chromatin, which is not deposited until much later in the cell cycle — telophase/G1 phase. In this case, the high turnover rates of H3.3 (REFS21,22) could support an efficient exchange of H3.3 for CENP-A after mitosis123.
CENP-A deposition also affects nucleosome structure and dynamics. Ectopic expression of CENP-A results in aberrant DAXX-dependent CENP-A deposition owing to disrupted stoichiometry between CENP-A and its chaperone, HJURP, and the formation of stabilized nucleosomes at regulatory elements of chromatin124. CENP-A influences nucleosome stability through the presence of two extra amino acid residues (Arg80 and Gly81) in the loop 1 region, which is solvent exposed and could potentially provide a binding site for trans-acting factors125 and confer a more rigid subnucleosomal structure when associated with H4 and centromeric proteins126. Findings from exogenous CENP-A expression perhaps become more relevant considering that CENP-A is amplified in a variety of cancers, including bladder, liver, lung and various gynaecological cancers127, and that heterotypic CENP-A–H3.3 nucleosomes have been observed in cancer cells124 (see also later). In these cases, increased deposition of CENP-A could impact chromatin structure outside centromeres (for example, in transcriptionally active regions). Ectopic CENP-A expression does not greatly influence gene expression in the steady state, but rather protects cells from DNA damage through unknown mechanisms124 and as such might play a role in mutagenesis and chemotherapy resistance.
Variant-specific chromatin states
Gene regulation by H3.3-specific Ser31 phosphorylation.
Generally, the PTMs that histone variants carry can be considered a product of their localization to specific regions, and it can be difficult to decouple this effect from functions that can be directly attributed to the histone variant itself. However, on the basis of sequence differences from replicative histones, it is possible for histone variant-specific PTMs to occur, and several striking examples demonstrate that such modifications can have profound impacts on the local chromatin environment (FIG. 3a-c).
Proteomics studies have shown that H3.3 is enriched with modifications associated with genome activation, such as H3K4me3, H3K36me3 and H3K27ac128. Although the residues modified are also found on H3.1 and H3.2, several recent studies show that expression of H3.3 mutants carrying substitutions at the site of modification alter global levels of that modification129,130. While it can be difficult to discriminate the importance of a modification being on a histone variant from its presence owing to genomic localization, there are cases in which the identity of the variant is critical. For example, in plants, the H3K27 methyltransferases ATXR5 and ATXR6 favour H3.1 over H3.3, which relies on Ala31 in H3.1 (FIG. 3a). Thereby, the repressive H3K27me1 mark is preferentially placed on H3.1, potentially protecting transcriptionally active, H3.3-enriched regions from H3K27me1 and Polycomb-mediated repression during DNA replication131,132.
Although most PTMs on histones are not restricted to particular variants, one example of a histone variant-specific PTM is the phosphorylation of a unique Ser31 on the H3.3 N-terminal tail (FIGS 1,3a). This phosphorylation was originally reported as a mitotic mark17, but several studies suggest that this serine and/or its phosphorylation are important developmentally and are linked to gene regulation. For example, a recent study demonstrated that a phosphomimetic H3.3 (with Asp31) but not a phosphomutant H3.3 (with Ala31) was able to rescue developmental defects caused by H3.3 deletion during Xenopus laevis gastrulation19. This dataset also identified factors that are either recruited (for example, members of the β-catenin signalling pathway during interphase) or repelled (for example, factors implicated in splicing such as small nuclear ribonucleoproteins during mitosis) by the phosphomimetic H3.3, suggesting that H3.3 phosphorylation at position 31 influences formation of chromatin effector complexes. Furthermore, the study found that phosphomimetic H3.3 promotes active chromatin states such as H3K27ac. In agreement, our work demonstrated that H3.3 Ser31 is required for activation of new transcriptional programmes through the acetylation of enhancers and promoters at developmental genes in differentiating ES cells, and that phosphorylated H3.3 stimulates p300 histone acetyltransferase activity, augmenting H3K27ac levels in vitro15 (FIG. 3b). Another recent study showed that in lipopolysaccharide-stimulated macrophages, H3.3 Ser31 phosphorylation recruits histone methyltransferase SET domain-containing 2 (SETD2) — a component of the active transcription machinery — to stimulation-responsive genes, resulting in H3.3 Lys36 trimethylation and rapid gene induction16. At the same time, there is evidence that H3.3 Lys36 trimethylation is read by a transcription repressor and tumour suppressor, zinc-finger MYND-type-containing 11 (ZMYND11)133,134 (FIG. 3c). As biochemical analysis shows that ZMYND11 binding is greatly reduced by H3.3 Ser31 phosphorylation133, it will be important to determine how the kinetics of H3.3 Ser31 phosphorylation influence both SETD2 activity and ZMYND11 function on chromatin. Of note, many studies show that H3.3 is not required to maintain ongoing transcription15,23,44,135. From the findings taken together, we suggest that H3.3 plays a more important role in rapid, signal-mediated gene activation than in the maintenance of existing gene expression programmes.
Histone variant-specific phosphorylation has also been observed at high-repeat genomic regions, and the presence of H3.3 Ser31 phosphorylation was reported during mitosis both at telomeres in ES cells and at centromeres in non-pluripotent cells17,136. In cancer cell lines that activate the alternative lengthening of telomeres (ALT) pathway (ALT cancers; see also the section entitled Relevance to human disease), loss of ATRX results in spreading of H3.3 Ser31 phosphorylation — which was attributed to the activity of checkpoint kinase 1 (CHK1) — from telomeres and centromeres to chromosome arms18. This result suggests that in the absence of ATRX, H3.3 may be more available to its other chaperone, the HIRA complex, for ectopic deposition. Notably, the appearance of H3.3 Ser31 phosphorylation at the centromere in non-pluripotent cells occurs in late prometaphase and metaphase17, suggesting that, unlike global histone phosphorylation at this stage137, H3.3 Ser31 phosphorylation does not play a role in early chromatin condensation. Perhaps it is important to limit heterochromatic states from spreading and/or it could serve as a mitotic bookmark for resuming euchromatin functions during G1/S phase. It will be important to test these ideas in future studies. The centromere-specific variant CENP-A is also subject to variant-specific modification, and phosphorylation, acetylation, methylation and ubiquitylation have all been connected to different roles in mitosis which are currently under investigation138.
Indirect effects of histone variant deposition on local chromatin states.
Histone variants can also influence chromatin state through a number of additional mechanisms (FIG. 3d-f). First, their chaperones can interact with other chromatin-associated proteins to promote distinct chromatin states at regions of deposition. For example, H3.3 contributes to H3K9me3 in ES cells through interaction of its chaperone, DAXX, with the histone methyltransferase SET domain bifurcated 1 (SETDB1) and the Krüppel-associated box (KRAB)-associated protein 1 (KAP1) co-repressor complex. Loss of H3.3 reduces KAP1 recruitment, and vice versa, suggesting positive feedback in maintenance of this complex on chromatin37,38,40,139 (FIG. 3d). Furthermore, variant-specific chaperones can influence the composition of the nucleosome; for example, ATRX prevents the deposition of macroH2A in a mechanism that is independent of ATRX association with DAXX140.
In addition to variant-specific PTMs discussed in the preceding subsection, universal PTMs can also contribute to functional diversification of variant histones. For instance, H4 acetylation in combination with incorporation of H2A.Z.1 into the nucleosome promotes the binding of bromodomain-containing protein 2 (BRD2) and transcription activation, indicating combinatorial readout of histone PTM and histone variant composition of the nucleosome72,141 (FIG. 3e). Histone variants can also be modified in accordance with their site of deposition and thus modulate transcription. For example, H2A.Z can be extensively acetylated at regulatory elements — which positively correlates with transcriptional output — favouring an open conformation that is more permissive to the transcription machinery72,81,82,142 (FIG. 3d). By contrast, H2A.Z at repressed regions promotes PRC2-mediated H3K27me3 deposition112 — a mark of facultative heterochromatin. Concomitantly, in regions of facultative heterochromatin, H2A.Z was shown to be monoubiquitylated by PRC1, and active deubiquitylation was required to activate expression of androgen receptor-regulated genes in prostate cancer cells143,144 (FIG. 3).
Functions in mammalian development
Given the high degree of sequence similarity of histone variants across phyla and their regulated developmental expression profiles (FIG. 4), it is not surprising that many histone variants are required for development135,145-147. Genetic analyses of the contributions of histone variants to development are possible because of their unique gene structure and position outside the histone clusters, and many of these knockout studies have been conducted in mouse models and other model systems over the past 20 years. In the following subsections, we describe our current understanding of histone variants in both the germline and during early development.
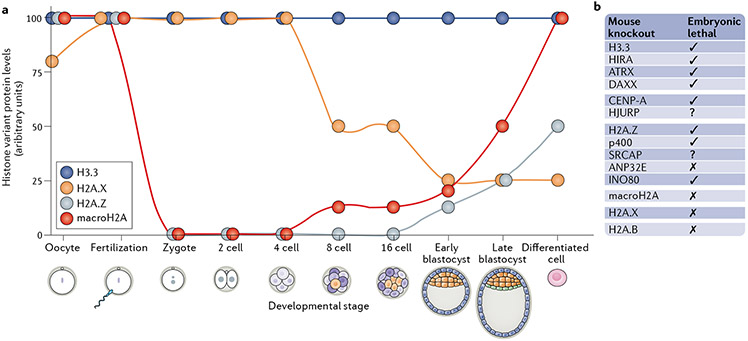
Fig. 4 ∣. Expression of histone variants during embryonic development.
a ∣ The expression levels of histone variants change during the early stages of embryonic development. Each dot represents the level of the histone variant at the specific developmental stage. Histones H3.3 and H2A.X are expressed already in the zygote and importantly contribute to all stages of development. H2A.Z is expressed only later in the blastocyst. MacroH2A is present in the oocyte but is excluded after fertilization, and its levels do not increase until the eight-cell stage, b ∣ Requirement of histone variants and associated chaperones169,170,243-245 for embryonic development in knockout mouse models. See the text for additional details and references. ANP32E, acidic leucine-rich nuclear phosphoprotein 32 family member E; ATRX, α-thalassaemia mental retardation syndrome X-linked; CENP-A, centromeric protein A; DAXX, death domain-associated protein; HIRA, histone regulator A; HJURP, Holliday junction recognition protein; SRCAP, Snf2-related CREBBP activator protein.
Roles in mammalian germ cells
The vast majority of known tissue-specific histone variants are specific to the germ line, of which the best studied are the testis-specific variants148. The eutherian testis-specific H2A variants fall into four clades, H2A.B, H2A.L, H2A.P and H2A.Q, and they all are subject to accelerated rates of protein evolution compared with other H2A proteins149. During spermatogenesis, haploid spermatids undergo extensive morphological changes, including compaction of their chromatin through the replacement of histones with protamines. Without exception, the testis-specific variants contain amino acid substitutions that destabilize the nucleosome102, facilitating the replacement of histones by protamines (histone-to-protamine transition) during sperm maturation150,151. For example, all short H2A.B variants have lost or contain a weakened acidic patch that would stabilize histone–histone interactions in the nucleosome (the original protein name for H2A.B was H2A.Lap1 for lack of acidic patch)100. The testis-specific H2B protein TH2B also contributes to nucleosome instability152, and the testis-specific human H3 protein H3T and mouse H3T both contain one or more amino acid substitutions that destabilize core histone contacts in the nucleosome or weaken interactions between H3 and DNA at the entry–exit region on the nucleosome153,154. Both mouse and human studies have revealed that a small percentage of H3.3-containing nucleosomes located at key regulatory regions are retained in the chromatin of mature spermatozoa155,156 and are removed rapidly on fertilization when the paternal genome is chromatinized using maternal stores of H3.3 (REFS157-159). It remains to be seen whether these paternal H3.3 nucleosomes dictate any function after fertilization. Much less is known about oocyte-specific core histone variants, although oocytes are known to express a cell-type specific H1 variant called H1oo160, and several of the variants already discussed have critical function in oocytes and fertilized embryos, as described in more detail next.
Roles in embryonic development
As for many chromatin-associated proteins, loss of H3.3, H2A.Z.1 and CENP-A results in embryonic lethality in mouse models before or around the time of gastrulation145-147. These variants have all been associated with formation and function of embryonic-specific pericentromeric heterochromatin and maintenance of genome stability, perhaps explaining their requirement for development. By contrast, macroH2A, H2A.X and H2A.B are dispensable for mammalian development161-163, although knockout of the genes encoding these variants in mice has been attributed to various phenotypes in adult mice. Until recently, technologies did not exist to allow genomic studies from pre-implantation embryos. Because of this, many laboratories have used ES cells and somatic cell reprogramming to infer the role in histone variants in cell fate transitions (BOX 1). Given that technologies such as low-input and single-cell genomics are now being developed, we expect that many concepts from these models will be tested in the context of early development in the coming years.
Box 1 ∣. Histone variants in embryonic stem cell biology.
Embryonic stem cells (ES cells) and induced pluripotent stem cells are popular model systems for studying the role of histone variants in both self-renewal and transitions between pluripotency and more differentiated states. Different histone variants have been implicated in different aspects of these processes. For example, ES cells lacking either histone H2A.Z or histone H3.3 maintain their self-renewal properties yet show defects in differentiation capacity15,44,75,82. While also associated with active genes, both H3.3 and H2A.Z have been associated with Polycomb repressive complex 2 (PRC2) activity, including at developmentally regulated promoters in ES cells and subsequent transcription of lineage-specific genes during differentiation15,44,75,112,248,249. H3.3 is also required for cell state transitions during reprogramming, where it is proposed to play a role in both erasing the transcription programme associated with terminal (in the reported case, fibroblast) differentiation, perhaps through histone exchange and removal of existing chromatin modification states, and in activating enhancers and gene expression to establish the pluripotency programme250. MacroH2A is also associated with PRC2-mediated heterochromatin formation94,95. While several studies show differentiation defects in ES cells lacking macroH2A, it has been reported that macroH2A-depleted ES cells are able to exit pluripotency180,251,252. However, there is consensus that macroH2A serves as a barrier to reprogramming towards induced pluripotent stem cells owing to its capacity to stabilize heterochromatin, thus preventing efficient reactivation of the pluripotency transcription programme253,254. One interesting observation from ES cell studies is that depletion of several variants, specifically H3.3 or H2A.X, leads to upregulation of genes that are transcribed specifically in the trophoblast, a developmental lineage from which ES cells are restricted, suggesting that histone variants may play a role in maintaining cell states once they are established44,255. of note, pluripotent chromatin is characterized by certain unique features, including high rates of histone exchange with relatively little traditional heterochromatin. It is therefore interesting to speculate that histone variants contribute to unique mechanisms of chromatin regulation that are specific to key developmental processes.
A developmental requirement for H3.3 is perhaps not surprising given the highly conserved nature of its two genes. Deletion of both genes encoding H3.3 in the mouse results in embryonic lethality around embryonic day 6.5 (E.6.5)135 (although it must be noted that flies lacking H3.3 develop normally and show only fertility defects, suggesting that, in some cases, H3 can compensate for loss of H3.3 (REFS164,165)). H3F3A and H3F3B can likely compensate for each other developmentally, although studies with individual gene deletion yielded contradictory phenotypes147,166-168 (probably owing to the use of different mouse strains and different targeting approaches that may affect not only protein translation but also, perhaps, transcription of non-coding regions such as the 3′ untranslated regions, which vastly differ between H3F3A and H3F3B). Because H3.3 is deposited at both euchromatic and heterochromatic regions23 (FIG. 1), it is difficult to attribute embryonic lethality to a particular function of H3.3. Assessment of chaperone gene knockouts does not provide further clarity, as deletion of HIRA, ATRX or DAXX results in embryonic lethality around the same time as H3.3 deletion or slightly after (as late as E11)169-171, and it is likely that each chaperone has a function outside of its role in H3.3 deposition. Relatively modest changes in transcription were reported in embryos lacking H3.3, and the embryonic lethality was attributed instead to chromosomal segregation defects135. In support of this notion, H3.3 is required to form pericentromeric heterochromatin in the paternal pronucleus after fertilization172-174, suggesting that an important function of H3.3 in early development is in heterochromatin formation, centromere definition and chromosome segregation. It is therefore tempting to speculate that H3.3 deposition at euchromatin is not important for regulating developmental transcription programmes. However, the pre-implantation embryo has access to maternal stores of H3.3 mRNA and protein that may provide sufficient levels of H3.3 to facilitate transcription required for early embryonic development175. Indeed, maternal H3.3 is necessary for activation of transcription from the paternal genome during zygotic genome activation159. Additionally, several studies suggest important roles for HIRA and H3.3 deposition in transcription during oogenesis and in the zygote, and for nascent transcription both in the steady state and during nuclear reprogramming29,176,177. While the mechanisms are likely many and varied, it will be important to determine exactly how H3.3 contributes to chromatin regulation of the early embryo in future studies.
Unlike H3.3 and H2A.Z, macroH2A is not essential for embryonic development, as both Macroh2a.1-knockout mice and Macroh2a.1 and Macroh2a.2-double-knockout mice are viable, although they do display defects in prenatal and postnatal growth and metabolism, increased perinatal death and defects in adult stem cell function161,178,179. Nevertheless, macroH2A.2 has been found to be essential in other developmental model systems such as zebrafish, and both macroH2A.1 and macroH2A.2 are required for unperturbed ES cell differentiation95,180. Several studies suggest that macroH2A may be a tumour suppressor (with the capacity to suppress cell proliferation and enforce/sustain a differentiated state), and in line with this function, macroH2A levels are generally low in the early embryo and in ES cells and increase during differentiation as cells specify their lineage180 (FIG. 4). Maternal macroH2A is excluded from the maternal genome shortly after fertilization at the late pronucleus stage and is not apparent again until the eight-cell stage181, incidentally around the point at which blastomeres become functionally distinct. While there is some disagreement in the field, many studies associate macroH2A with PRC2-mediated gene silencing182. Accordingly, macroH2A was originally implicated developmentally in X chromosome inactivation, although it is not required for the initiation of this process69. Overall, these data support a model in which macroH2A plays a role in maintaining differentiated cell states by stabilizing heterochromatin at silenced genes.
Relevance to human disease
As histone variants have such ubiquitous expression and pleiotropic roles, it is not surprising that they are involved in normal physiology as well as many diseases. For example, as histone variants presumably become the only source of new histone in postmitotic terminally differentiated tissues, such as the brain, it is possible that histone variants may critically contribute to brain-related diseases such as neurodevelopmental and neurodegenerative diseases. Indeed several studies suggest that H3.3 and H2A.Z variants critically contribute to brain function183-188. Moreover, considering that many histones variants are testis specific151,189, it follows that either the replacement or the modification of testis-specific histone variants might result in male infertility (recently reviewed190). Finally, many cancers and developmental syndromes are known to carry recurrent mutations in genes encoding chromatin-associated proteins, including histone variants and their deposition machineries127. In the following subsections we discuss important findings and recent advances pertaining to the involvement of histone variants in cancer and developmental diseases (TABLE 1, FIG. 5).
Fig. 5 ∣. ATRX-dependent mechanisms in physiology, cancer and human genetic disease.
Aa ∣ α-Thalassaemia mental retardation syndrome X-linked (ATRX) is recruited to heterochromatin regions marked by H3K9me3, where it deposits H3.3 (together with death domain-associated protein (DAXX))and contributes to chromatin compaction by binding to heterochromatin protein 1α (HP1α), promoting its stabilization on chromatin. Ab ∣ ATRX has also DAXX-independent function, whereby it antagonizes deposition of macroH2A1.1 and macroH2A1.2. Ac ∣ These activities are particularly important at telomeres, where ATRX functions to resolve G-quadruplex (G4) DNA during replication and is also important for the maintenance of telomeric heterochromatin (maintenance of high levels of H3K9me3). Ba ∣ ATRX is commonly mutated in cancers that activate a telomerase-independent mechanism of telomere maintenance via the alternative lengthening of telomeres (ALT) pathway. ATRX mutations in cancer are both missense and truncation mutations and are observed along the length of the gene246,247. These mutations in ATRX can cause loss of function of the protein and lead to the loss of telomeric heterochromatin, replication stress caused by G4 stabilization (see parts Aa and Ac) and subsequent DNA damage at telomeres as well as increased homologous recombination-dependent telomere sister chromatid exchange (T-SCE) downstream of macroH2A1 accumulation (see part Ab). Bb ∣ Large deletions of ATRX around the coding region are associated with in-frame fusion of the protein, which leads to the generation of truncated protein products that are redistributed to sites of active transcription — marked by H3K27ac — and display neomorphic function in activating gene expression. One of the targets of in-frame fusion ATRX is the transcription repressor REST, which on activation leads to repression of genes involved in neuronal differentiation, concomitant activation of neurogenesis programmes and increased cell proliferation228. This function was found to be DAXX independent. C ∣ ATRX mutations associated with α-thalassaemia X-linked mental retardation syndrome are generally missense mutations affecting key functional regions of the protein233. α-Thalassaemia X-linked mental retardation syndrome-related loss of ATRX activity was linked to increased deposition of macroH2A1 variants (see part Ab) at the α-globin locus, leading to heterochromatinization and silencing of the gene, thereby driving α-thalassaemia pathology. ATRX loss in the brain is linked to increased replicative damage and subsequent cell death, which was attributed to the role of ATRX in supporting replication through G4 (see part Ac). Of note, whether replication stress associated with α-thalassaemia X-linked mental retardation syndrome-causing mutations relies on aberrant H3.3 deposition is not clear. RNAPII RNA polymerase II.
Histone variants in cancer
Histone variant genes and their expression in cancer.
Many enzymes that modify chromatin are lost or rendered enzymatically inactive in cancers, but it can be difficult to attribute function to these mutations in part owing to the high mutational burden found in most adult cancers191. By contrast, paediatric tumours contain relatively few mutations, which are more likely to drive oncogenesis given the timing of disease progression relative to mutation load of the tumour. Mutations in H3.3 are highly recurrent in paediatric gliomas and chondroblastomas. These mutations tend to be at or near sites of post-translational histone modifications — including substitution of Met for Lys27 and substitution of Arg or Val for Gly34 (which lies in the vicinity of Lys36 and affects its methylation) in glioblastoma, and substitution of Met for Lys36 in chondroblastoma192,193 — and have been shown to function as endogenous inhibitors of Lys methylation194-196. These mutations have been the subject of intense study and have been extensively reviewed elsewhere197. We note two interesting observations in the context of this Review. First, the Lys27-to-Met substitution found on H3.3 in paediatric brain tumours is also found, albeit with lower frequency, on H3.1, and the two histone mutations co-occur with mutations in different tumour suppressors. As such, both the timing and the localization of deposition of each mutant results in distinct modes of oncogenic transformation198. Second, the Gly34 mutation occurs exclusively on H3.3 with high co-occurrence with ATRX mutation192, suggesting that amplification of HIRA-mediated H3.3 Gly34 mutant deposition at euchromatin may contribute to oncogenesis. In addition to neomorphic mutations, H3.3 is commonly amplified in a number of tumours, and a recent study found that HIRA-dependent H3.3 deposition is essential for tumour progression through response to extracellular signal-regulated kinase (ERK) activity199. It is tantalizing to hypothesize that H3.3 itself is phosphorylated at its unique serine (Ser31) by ERK or a downstream kinase to facilitate either enhancer activation or SETD2-mediated gene activity (FIG. 3a-c) required for tumour progression.
Given the role of macroH2A in preventing changes in cell state during reprogramming (see earlier and BOX 1), it is not surprising that macroH2A is considered to act as a tumour suppressor in several cancers200. Transcriptional repression of macroH2A in melanoma contributes to upregulation of oncoproteins that drive cell proliferation, such as cyclin-dependent kinase 8 (CDK8), and results in altered metabolism201,202. Splicing defects in macroH2A that result in reduced protein levels are also associated with tumour growth and invasion properties, including epithelial–mesenchymal transition203. Despite these observations, there are a number of cancers in which either macroH2A.1 or macroH2A.2 is upregulated rather than mutated or truncated. Specifically, macroH2A.1 is amplified in ~5% of clear renal cell carcinomas, and both macroH2A.1 and macroH2A.2 are amplified in cholangiocarcinoma127. These patient data demonstrate a complex and cell type-specific response to altered macroH2A levels that requires additional study.
The expression of H2A.Z is commonly upregulated in a host of tumours, ranging from breast and prostate cancer to lung cancer and metastatic melanoma200. H2A.Z and its acetylation are linked to active transcription and cell proliferation in hormone-dependent cancers, and high expression levels are indicative of poor prognosis142,204. H2A.Z has been both positively and negatively implicated in epithelial–mesenchymal transition, with one study reporting negative effects through activation of epithelial genes and repression of mesenchymal genes205, while another study reported a role for H2A.Z in activating mesenchymal genes during the transition state206. Functionally, knockdown of H2A.Z in various cancer cell lines reduces the proliferative capacity of those cells, and overexpression promotes growth207,208. Assessment of both genes encoding H2A.Z (H2AFZ and H2AFV) as well as their remodellers, SRCAP and EP400, shows that amplification of any one gene tends to be mutually exclusive of the other three, suggesting similar phenotypic outcomes for each gene127. The relationship between H2A.Z expression and tumorigenesis is not entirely clear and unidirectional, as many cancers carry missense mutations of unknown function (although one, resulting in H2A.Z.1 Arg80-to-Cys substitution, has been shown to disrupt nucleosome stability209) and p400, the remodeller responsible for H2A.Z deposition, has tumour suppressor function in lymphomas, carcinomas and colon cancer210,211. The patient data above imply that H2A.Z.1 and H2A.Z.2 might have redundant functions in promoting oncogenesis; however, several pieces of evidence suggest a more complicated scenario. First, while both H2A.Z.1 and H2A.Z.2 are upregulated in metastatic melanoma, only depletion of H2A.Z.2 reduced cell growth141. Furthermore, of the two genes, amplification of H2AFV is far more common across cancer types127.
Dysregulation of CENP-A is thought to be oncogenic in several cancers, and overexpression of centromere and kinetochore components (including both CENP-A and its chaperone HJURP) is associated with chromosome instability owing to chromosome missegregation and poor patient prognosis212,213. Finally, perhaps unsurprisingly, given its prominent role in DNA damage repair, H2A.X primarily acts as a tumour suppressor. Mutation or deletion of its encoding gene is observed in a number of cancers and is associated with genomic instability and oncogenic translocations162,214,215. The involvement of all the histone variants (and their chaperones) in tumour biology point to them as potentially important contributors to the establishment of tumour-specific gene programmes, and therefore finding ways to target them has great potential to be of therapeutic value.
ATRX deregulation in cancer.
Tumours rely on the extension of their telomeres for their growth, and some (10–15%) do so via a telomerase-independent mechanism known as ALT. Tumorigenic activation of ALT is strongly associated with inactivating mutations in ATRX and, less so, in DAXX216, although it is important to note that loss of ATRX is not sufficient to induce ALT. Approximately 80% of tumours with ALT harbour mutations in ATRX or DAXX (although mutations in ATRX and DAXX show a surprisingly high degree of tumour type specificity), and conversely 70–80% of cancers with ATRX mutations show ALT217. The oncogenic effect of ATRX mutation in tumours is generally attributed to telomere dysfunction (FIG. 5), with a number of groups demonstrating molecular phenotypes ranging from aberrant transcription of telomeric DNA to abnormal homologous recombination of telomeres that results in chromosomal rearrangements and genomic instability in the absence of ATRX. Mechanistically, ATRX was shown to bind G-rich repetitive elements characteristic of telomeres and pericentromeric regions23,218 (FIG. 5A). These elements are susceptible to the formation of G-quadruplexes, which has been widely implicated in replication stress, DNA damage and genome instability219. ATRX deficiency was shown to induce replication stress at telomeres220-222 (FIG. 5B), suggesting a role for ATRX in resolving G-quadruplexes. The role of ATRX in preventing replication stress in response to G-quadruplexes could be attributed to its function as an H3.3 chaperone, as ATRX-mediated suppression of ALT in U2OS cells was shown to depend on DAXX and H3.3 deposition at telomeres. In this context, telomeric H3.3 deposition could assist replication through G-quadruplexes223. Alternatively, loss of ATRX promotes deposition of the splice variants macroH2A1.1 and macroH2A1.2 at telomeres (FIG. 5A). Deposition of macroH2A1.1 prevents poly(ADP-ribosyl)ated tankyrase from localizing to telomeres and resolving cohesion, while macroH2A1.2 deposition facilitates homology-directed repair of damaged telomeres. Both result in increased homologous recombination-dependent telomere sister chromatid exchange, which may promote ALT140,224,225 (FIG. 5B). A recent report demonstrated that ATRX mutations found in chronic neuroblastoma result in production of truncated versions of ATRX (in-frame fusions) that lack the ADD domain — which interacts with the H3 tail and is responsible for its targeting to heterochromatin by binding to H3K9me3 (REFS226,227) — ultimately resulting in its redistribution from heterochromatin to active promoters (FIG. 5B). One of the targets of in-frame fusion ATRX was the transcription repressor REST, which was upregulated, leading to repression of genes involved in neuronal differentiation, concomitant activation of neurogenesis programmes and increased cell proliferation228. This study also showed that truncated ATRX can carry out its atypical function in the absence of its interaction with DAXX.
Implications for developmental diseases
Compared with cancer, histone variants and their chaperones have been implicated in relatively few developmental human syndromes, typically characterized by alterations in a variant-specific chaperone or chromatin remodeller and not the histone variant itself — perhaps unsurprising given the essential developmental roles of histone variants as discussed earlier229. For example, ATRX is named for its role in α-thalassaemia X-linked mental retardation syndrome, hallmarks of which are, as the name implies, mental retardation and α-thalassaemia — a loss of α-globin gene production230. Several studies link ATRX mutation to dysregulation of gene expression from the α-globin cluster (FIG. 5C). One study showed that ATRX promotes α-globin gene expression by protecting this region from the repressive effects of macroH2A deposition, mediated by DAXX-independent interaction between ATRX and macroH2A140. Additional studies demonstrated that α-thalassaemia X-linked mental retardation syndrome-relevant mutations could compromise its role in supporting replication at G-rich regions (of note, here the involvement of H3.3 downstream of ATRX mutations is unclear)12,218. Loss of ATRX specifically in the forebrain in mice led to increased replicative damage, cell death and physiological phenotypes such as reduced growth and decreased lifespan231. As shown in neural progenitor cells, DNA damage associated with the loss of ATRX was further exacerbated by the increase in the formation of G-quadruplexes. Altogether, these data suggest that ATRX protects neural progenitors from DNA damage by supporting replication through G-quadruplexes and preventing replication stress. In extension, this important role of ATRX in neural progenitors could explain the mental retardation associated with α-thalassaemia X-linked mental retardation syndrome. Given the high prevalence of acquired ATRX mutations in the broad spectrum of tumours that undergo ALT (see earlier), patients with α-thalassaemia X-linked mental retardation syndrome do not generally have an increased incidence of cancer (although limited cases of osteosarcomas were noted232). Mutations in patients with α-thalassaemia X-linked mental retardation syndrome tend to be missense mutations affecting the ADD domain (involved in targeting ATRX to heterochromatin) and the ATPase (SNF2)–helicase domain233 (which is important for translocation of ATRX on DNA) that may have rather subtle effects on ATRX function, whereas tumour mutations are less centred on these domains and are either missense or truncating alterations that are presumably loss-of-function mutations127. Additionally, loss of ATRX is not sufficient to induce ALT (see earlier)217, so it is likely that additional mutations are required for cancer progression. Coupled with the observation that, to our knowledge, DAXX mutations have not been reported in α-thalassaemia X-linked mental retardation syndrome and the fact that ATRX and DAXX mutations show a high degree of cancer-type specificity in tumours127, these observations add to the accumulating evidence that ATRX and DAXX have distinct biological roles in development and disease.
A recent study of the developmental disorder Floating-Harbor syndrome (FHS) implicated disease-associated deregulation of H2A.Z and provided new biological insight into the differences between H2A.Z.1 and H2A.Z.2 (REF.70). This rare disorder is characterized by heterozygous mutations in the H2A.Z remodeller gene SRCAP that result in reduced enrichment of H2A.Z.2 compared with H2A.Z.1 at enhancers and subsequent perturbations in the activation of neural crest gene programmes leading to craniofacial defects. With X. laevis as a developmental model system, the FHS could be phenocopied by knocking down H2A.Z.2 but not H2A.Z.1. The most surprising finding from this study was that the pathology could be attributed to one of the three amino acids that differ between H2A.Z.1 and H2A.Z.2. Specifically, H2A.Z.1 carrying an H2A.Z.2 substitution (Ser38 to Thr38) was able to rescue FHS developmental phenotypes. Studies suggest that this amino acid contributes to the conformation of the L1 loop in H2A.Z to influence nucleosome dynamics, with Thr38 nucleosomes displaying lower mobility234. These data imply that this single amino acid confers nucleosome properties required for regulating craniofacial development.
Conclusions and perspective
Over the past decade, considerable progress has been made in understanding the mechanisms by which histone variants are chaperoned to chromatin, the effect of their deposition on local chromatin landscapes and the function of histone variant and chaperone misexpression and mutations on human disease (TABLE 1). In our view, a crucial concept emerging from the past few years of study of histone variants is that a very small change in the amino acid sequence of the histone can drive comparatively large effects genome-wide on its incorporation into chromatin (for example, the biological consequences of differences between H2A.Z.1 and H2A.Z.2) and can allow these proteins to serve as mediators of signalling pathways that converge on chromatin (for example, H3.3 phosphorylation in gene activation).
We expect that ever-improving technologies will continue to drive impactful new discoveries in histone variant biology into the next decade. State-of-the-art quantitative mass spectrometry approaches will no doubt continue to elucidate undiscovered features of the histone variant interactome, consisting of nuclear and chromatin-bound proteins, which will be important to address the mechanisms by which histone variant chaperones are recruited to and promote deposition of histone variants at specific regions of chromatin. Improvements in structural biology approaches such as cryo-electron microscopy will provide unprecedented views of the relatively large complexes formed by the deposition machinery, as well as interactions with the nucleosome that allow both de novo deposition and histone exchange. New concepts such as that of the cellular roles of biomolecular condensates driven by liquid–liquid phase separation may provide new theoretical frameworks for our understanding of chaperone-mediated protein interactions, given that many of these proteins contain regions that are predicted to be unstructured (and hence have high propensity for phase separation) or have already been implicated in condensate biology (for example, DAXX and its association with promyelocytic leukaemia nuclear bodies). The ability to profile chromatin states and transcription programmes at the level of a single cell will allow us to determine how histone variants influence fertility and pre-implantation development, providing critical insights into cell fate decisions in a biologically relevant setting. From a biological perspective, we predict that one area ripe for discovery is that of ageing and neurodegenerative disorders. As histone variants presumably become the only source of new histone in postmitotic terminally differentiated tissues, it is possible that histone variants may take on many, so far unexplored functions in addition to those described herein, and it will be important to determine the effects of their loss on both healthy and diseased aged states. Combined, such novel approaches and concepts will ensure that the field of histone variant biology will continue to provide important contributions to studies of chromatin-mediated regulation of both development and disease for years to come.
Endogenous retroviral elements.
Subset of transposable elements (up to 8% of the total genome) with long terminal repeats, which can act as transcriptional elements.
Imprinted regions.
Regions of the genome that harbour genes that are expressed in a parental origin-specific manner.
Micrococcal nuclease.
Non-specific DNA and RNA endo–exonuclease that preferentially digests non-nucleosomal DNA.
G-quadruplex.
Guanine-rich DNA sequence that can fold into four-stranded, non-canonical secondary structures and is involved in genome functions such as replication and genome stability.
Centromere-associated network (CCAN) proteins.
Subcomplex in the kinetochore that binds centromeric chromatin and functions as a foundation for kinetochore formation.
Poly(ADP-ribose).
Reversible post-translational modification resulting in the covalent attachment of polymers of ADP-ribose units on target proteins.
Polycomb repressive complex.
Repressor family of protein complexes important for developmental gene regulation and embryo patterning through enzymatic activity on histone proteins.
Facultative heterochromatin.
Type of heterochromatin that has the ability to become transcriptionally active again (that is, the inactive X chromosome in mammals), typically marked by Polycomb group protein activity.
Liquid–liquid phase separation.
Physical process by which cells create non-membrane-bound compartments (biomolecular condensates) that have been implicated in various cellular processes from signalling to gene regulation.
Heterochromatin protein 1.
Family of chromodomain proteins that bind to H3K9me3 and are required for the formation of transcriptionally inactive heterochromatin.
+ 1 nucleosome.
The first (+1) nucleosome positioned after a promoter-associated nucleosome-free region.
Alternative lengthening of telomeres (ALT) pathway.
Telomerase-independent mechanism (homologous recombination-mediated DNA replication based) by which a number of human tumours maintain their telomeres.
Mitotic bookmark.
Binding by transcription and chromatin regulators at regulatory elements during mitosis to convey gene regulatory information to daughter cells.
Protamines.
Small proteins that replace histones during spermatogenesis, allowing denser packaging of DNA in the sperm than would be achieved with histones.
Pronucleus.
Cell nucleus characterized by a haploid set of chromosomes resulting from meiosis (female pronucleus) or just before fertilization (male pronucleus).
Zygotic genome activation.
Also known as maternal-to-zygotic transition, refers to the process that enables zygotic gene products to replace the maternal supply that initiated development.
Trophoblast.
Part of the outer trophectoderm layer. These cells contribute to extraembryonic tissues such as fetal placenta and to processes of early development.
X chromosome inactivation.
Transcriptional silencing of a random X chromosome in female mammalian cells that equalizes the dosage of gene products from the X chromosome between XX females and XY males.
Epithelial–mesenchymal transition.
Process that occurs during both development and cancer progression in which epithelial cells lose their adhesive properties to become invasive mesenchymal cells.
Homologous recombination-dependent telomere sister chromatid exchange.
Telomere extension events with crossover formation between sister chromatids, which are used as a template to repair homology-directed damaged telomeres.
α-Thalassaemia X-linked mental retardation syndrome.
Rare X-linked recessive syndrome caused by mutations in the ATRX gene, characterized by intellectual disability and reduced production of haemoglobin.
Floating-Harbor syndrome.
(FHS). Rare autosomal dominant disease caused by mutations in the SRCAP gene, characterized by a distinctive facial appearance, various skeletal malformations, delayed bone age and expressive and receptive language delays.
Neural crest.
Transient developmental structure unique to vertebrates that gives rise to diverse lineages, including melanocytes, craniofacial cartilage and bone, and most of the peripheral nervous system.
Promyelocytic leukaemia nuclear bodies.
Nuclear matrix-associated structures with characteristics of biomolecular condensates that contain promyelocytic leukaemia proteins, which are required for the assembly of a number of nuclear structures.
Acknowledgements
The authors thank E. Duncan and the reviewers for critical comments on this manuscript. L.A.B. is a Virginia Murchison Linthicum Scholar in Medical Research (University of Texas Southwester Medical Center Endowed Scholars Program in Medical Science) and a Peterson Investigator of the Neuroendocrine Research Foundation. This work was supported in part by grants form the Cancer Prevention and Research Institute of Texas (RR140042), the Welch Foundation (I-2025), the US Department of Defense (KCRP KC170230) and the NIH (R35 GM124958) to L.A.B., the American-Italian Cancer Foundation (S.M.) and the Green Center for Reproductive Biology Sciences.
Footnotes
Competing Interests
The authors declare no competing interests.
Peer review information
Nature Reviews Molecular Cell Biology thanks M. Buschbeck, F. Berger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Fyodorov DV, Zhou B-R, Skoultchi AI & Bai Y Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol 19, 192–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer PV et al. Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res. 46, 9907–9917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talbert PB & Henikoff S Histone variants — ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol 11, 264–275 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Maehara K et al. Tissue-specific expression of histone H3 variants diversified after species separation. Epigenetics Chromatin 8, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long M et al. A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic Acids Res. 47, 8399–8409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang MYH, Sun X, Ausió J & Ishibashi T Histone H4 variant, H4G, drives ribosomal RNA transcription and breast cancer cell proliferation by loosening nucleolar chromatin structure. J. Cell. Physiol 10.1002/jcp.29770 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Cavalli G & Heard E Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Venkatesh S & Workman JL Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol 16, 178–189 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Buschbeck M & Hake SB Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol 18, 299–314 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Long H et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 577, 576–581 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Clynes D & Gibbons RJ ATRX and the replication of structured DNA. Curr. Opin. Genet. Dev 23, 289–294 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Strobino M, Wenda JM & Steiner FA Loss of histone H3.3 results in DNA replication defects and altered origin dynamics in C. elegans. Preprint at bioRxiv 10.1101/854455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond CM, Strömme CB, Huang H, Patel DJ & Groth A Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martire S et al. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat. Genet 51,941–946 (2019).This work demonstrates that a single amino acid phosphorylation (H3.3 Ser 31 phosphorylation) can integrate signalling information and impact genome regulation globally and together with Sitbon et al. (2020) links H3.3 Ser 31 phosphorylation to H3 Lys27 acetylation.
- 16.Armache A et al. Phosphorylation of the ancestral histone variant H3.3 amplifies stimulation-induced transcription. Preprint at bioRxiv 10.1101/808048 (2019). [DOI] [Google Scholar]
- 17.Hake SB et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl Acad. Sci. USA 102, 6344–6349 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang FTM et al. CHK1-driven histone H3.3 serine 31 phosphorylation is important for chromatin maintenance and cell survival in human ALT cancer cells. Nucleic Acids Res. 43, 2603–2614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sitbon D, Boyarchuk E, Dingli F, Loew D & Almouzni G Histone variant H3.3 residue S31 is essential for Xenopus gastrulation regardless of the deposition pathway. Nat. Commun 11, 1256 (2020).This work demonstrates a critical developmental role for H3.3 Ser31, identifies interacting proteins that may depend on this amino acid, and together with Martire et al. (2019) links its phosphorylation to histone acetylation.
- 20.Tachiwana H et al. Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr. D. Biol. Crystallogr 67, 578–583 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Ha M, Kraushaar DC & Zhao K Genome-wide analysis of H3.3 dissociation reveals high nucleosome turnover at distal regulatory regions of embryonic stem cells. Epigenetics Chromatin 7, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deaton AM et al. Enhancer regions show high histone H3.3 turnover that changes during differentiation. eLife 5, e15316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg AD et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong C et al. UBN1/2 of HIRA complex is responsible for recognition and deposition of H3.3 at cis-regulatory elements of genes in mouse ES cells. BMC Biol. 16, 110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagami H, Ray-Gallet D, Almouzni G & Nakatani Y Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Sarai N et al. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 32, 2392–2406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray-Gallet D et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Soni S, Pchelintsev N, Adams PD & Bieker JJ Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for β-globin gene expression. Proc. Natl Acad. Sci. USA 111, 13337–13342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H et al. RPA interacts with HIRA and regulates H3.3 deposition at gene regulatory elements in mammalian cells. Mol. Cell 65, 272–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricketts MD et al. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun 6, 7711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banumathy G et al. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell. Biol 29, 758–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray-Gallet D et al. Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat. Commun 9, 3103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torné J et al. Two distinct HIRA-dependent pathways handle H3.3 de novo deposition and recycling during transcription. Preprint at bioRxiv 10.1101/2019.12.18.880716 (2019).This study demonstrates the flexibility of histone variant chaperone complexes and downstream functional consequences for maintenance of chromatin states.
- 34.Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC & Allis CD Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 107, 14075–14080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong LH et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drané P, Ouararhni K, Depaux A, Shuaib M & Hamiche A The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elsässer SJ, Noh K-M, Diaz N, Allis CD & Banaszynski LA Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Q et al. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell 17, 273–286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadic D et al. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 16, 836–850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoelper D, Huang H, Jain AY, Patel DJ & Lewis PW Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun 8, 1193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elsässer SJ et al. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature 491, 560–565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C-P et al. Structure of the variant histone H3.3–H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol 19, 1287–1292 (2012).This work and Elsässer et al. (2012) provide the structural basis for DAXX recognition of H3.3 over H3.1 or H3.2. See Ricketts et al. (2015) and Hu et al. (2011) for comparative structures of UBN1 bound to H3.3 and CENP-A bound to HJURP.
- 43.DeNizio JE, Elsässer SJ & Black BE DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res. 42, 4318–4331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banaszynski LA et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witt O, Albig W & Doenecke D Testis-specific expression of a novel human H3 histone gene. Exp. Cell Res 229, 301–306 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Schenk R, Jenke A, Zilbauer M, Wirth S & Postberg J H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120, 275–285 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Wiedemann SM et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol 190, 777–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zink L-M et al. H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements. Nucleic Acids Res. 45, 5691–5706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kujirai T et al. Structure and function of human histone H3.Y nucleosome. Nucleic Acids Res. 45, 3612–3612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gambogi CW & Black BE The nucleosomes that mark centromere location on chromosomes old and new. Essays Biochem. 63, 15–27 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Malik HS & Henikoff S Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Conde e Silva N et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol 370, 555–573 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Foltz DR et al. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol 8, 458–469 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Verdaasdonk JS & Bloom K Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol 12, 320–332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foltz DR et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunleavy EM et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Hori T, Shang W-H, Takeuchi K & Fukagawa T The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol 200, 45–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu H et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 (2011).This work together with Ricketts et al. (2015) and Liu et al. (2012) demonstrates the convergence of modes of H3 variant–chaperone structural interactions in the absence of sequence conservation.
- 59.Bönisch C & Hake SB Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 40, 10719–10741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belotserkovskaya R FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Okuwaki M, Kato K, Shimahara H, I. Tate S & Nagata K Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol 25, 10639–10651 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsunaka Y, Fujiwara Y, Oyama T, Hirose S & Morikawa K Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hondele M et al. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499, 111–114 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Giaimo BD, Ferrante F, Herchenröther A, Hake SB & Borggrefe T The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12, 37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suto RK, Clarkson MJ, Tremethick DJ & Luger K Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol 7, 1121–1124 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Eirín-López JM, González-Romero R, Dryhurst D, Ishibashi T & Ausió J The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol 9, 31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dryhurst D et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 7, 86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsuda R et al. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 38, 4263–4273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bönisch C et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 40, 5951–5964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greenberg RS, Long HK, Swigut T & Wysocka J Single amino acid change underlies distinct roles of H2A.Z subtypes in human syndrome. Cell 178, 1421–1436 (2019).This work demonstrates how the FHS pathology could be attributed to one of the three amino acids that differ between H2A.Z.1 and H2A.Z.2.
- 71.Dunn CJ et al. Histone hypervariants H2A.Z.1 and H2A.Z.2 play independent and context-specific roles in neuronal activity-induced transcription of Arc/Arg3.1 and other immediate early genes. eNeuro 10.1523/ENEURO.0040-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Semer M et al. DNA repair complex licenses acetylation of H2A.Z.1 by KAT2A during transcription. Nat. Chem. Biol 15, 992–1000 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Ruhl DD et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45, 5671–5677 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Liang X et al. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat. Struct. Mol. Biol 23, 317–YL323 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Creyghton MP et al. H2AZ is enriched at Polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong MM, Cox LK & Chrivia JC The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J. Biol. Chem 282, 26132–26139 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Luk E et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papamichos-Chronakis M, Watanabe S, Rando OJ & Peterson CL Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mao Z et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 4, 389–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Obri A et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014).Collectively, this work, Luk et al. (2010) and Papamichos-Chronakis et al. (2011) identify the remodellers and chaperones that both deposit and evict H2A.Z from nucleosomes.
- 81.Ku M et al. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 13, R85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu G et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180–192 (2013).This study provides support for the hypothesis that H2A.Z deposition at nucleosomes facilitates transcription factor and chromatin complex association with chromatin.
- 83.Weber CM, Ramachandran S & Henikoff S Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53, 819–830 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Greaves IK, Rangasamy D, Ridgway P & Tremethick DJ H2A.Z contributes to the unique 3D structure of the centromere. Proc. Natl Acad. Sci. USA 104,, 525–530 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hou H et al. Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J. Biol. Chem 285, 1909–1918 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krogan NJ et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl Acad. Sci. USA 101, 13513–13518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rangasamy D, Greaves I & Tremethick DJ RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol 11, 650–655 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Sun Z et al. Transcription-associated histone pruning demarcates macroH2A chromatin domains. Nat. Struct. Mol. Biol 25, 958–970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun Z & Bernstein E Histone variant macroH2A: from chromatin deposition to molecular function. Essays Biochem. 63, 59–74 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K & Ladurner AG Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol 12, 624–625 (2005). [DOI] [PubMed] [Google Scholar]
- 91.Karras GI et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Timinszky G et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol 16, 923–929 (2009). [DOI] [PubMed] [Google Scholar]
- 93.Li X, Egervari G, Wang Y, Berger SL & Lu Z Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol 19, 563–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Douet J et al. MacroH2A histone variants maintain nuclear organization and heterochromatin architecture. J. Cell Sci 130, 1570–1582 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Buschbeck M et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol 16, 1074–1079 (2009). [DOI] [PubMed] [Google Scholar]
- 96.Kozlowski M et al. MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 19, e44445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yelagandula R et al. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158, 98–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Probst AV, Desvoyes B & Gutierrez C Similar yet critically different: the distribution, dynamics and function of histone variants. J. Exp. Bot 10.1093/jxb/eraa230 (2020). [DOI] [PubMed] [Google Scholar]
- 99.Bao Y et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 23, 3314–3324 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Soboleva TA et al. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat. Struct. Mol. Biol 19, 25–30 (2011). [DOI] [PubMed] [Google Scholar]
- 101.Soboleva TA et al. A new link between transcriptional initiation and pre-mRNA splicing: The RNA binding histone variant H2A.B. PLoS Genet. 13, e1006633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang X, Soboleva TA & Tremethick DJ Short histone H2A variants: small in stature but not in function. Cells 9, 867 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Syed SH et al. The incorporation of the novel histone variant H2AL2 confers unusual structural and functional properties of the nucleosome. Nucleic Acids Res. 37, 4684–4695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Piquet S et al. The Histone Chaperone FACT Coordinates H2A.X-Dependent Signaling and Repair of DNA Damage. Mol. Cell 72, 888–901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hauer MH & Gasser SM Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 31, 2204–2221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maze I, Noh K-M, Soshnev AA & Allis CD Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet 15, 259–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K & Hansen JC The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 10, 20121022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arimura Y et al. Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin. Sci. Rep 3, 3510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou J, Fan JY, Rangasamy D & Tremethick DJ The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol 14, 1070–1076 (2007). [DOI] [PubMed] [Google Scholar]
- 110.Park Y-J, Dyer PN, Tremethick DJ & Luger K A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem 279, 24274–24282 (2004). [DOI] [PubMed] [Google Scholar]
- 111.Fan JY, Gordon F, Luger K, Hansen JC & Tremethick DJ The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol 9, 172–176 (2002). [DOI] [PubMed] [Google Scholar]
- 112.Wang Y et al. Histone variants H2A.Z and H3.3 coordinately regulate PRC2-dependent H3K27me3 deposition and gene expression regulation in mES cells. BMC Biol. 16, 107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ryan DP & Tremethick DJ The interplay between H2A.Z and H3K9 methylation in regulating HP1α binding to linker histone-containing chromatin. Nucleic Acids Res. 46, 9353–9366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fan JY, Rangasamy D, Luger K & Tremethick DJ H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16, 655–661 (2004). [DOI] [PubMed] [Google Scholar]
- 115.Vernì F & Cenci G The drosophila histone variant H2A.V works in concert with HP1 to promote kinetochore-driven microtubule formation. Cell Cycle 14, 577–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bagchi DN, Battenhouse AM, Park D & Iyer VR The histone variant H2A.Z in yeast is almost exclusively incorporated into the +1 nucleosome in the direction of transcription. Nucleic Acids Res. 48, 157–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nekrasov M et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol 19, 1076–1083 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Jin C & Felsenfeld G Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jin C et al. H3.3/H2A.Z double variant–containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet 41, 941–945 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Iwafuchi-Doi M et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kraushaar DC et al. The gene repressor complex NuRD interacts with the histone variant H3.3 at promoters of active genes. Genome Res. 28, 1646–1655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Harada A et al. Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration. Nat. Commun 9, 1400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dunleavy EM, Almouzni G & Karpen GH H3. 3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus 2, 146–157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lacoste N et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53, 631–644 (2014). [DOI] [PubMed] [Google Scholar]
- 125.Tachiwana H et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–235 (2011). [DOI] [PubMed] [Google Scholar]
- 126.Melters DP, Rakshit T, Grigoryev SA, Sturgill D & Dalal Y The ratio between centromeric proteins CENP-A and CENP-C maintains homeostasis of human centromeres. Preprint at bioRxiv 10.1101/604223 (2019). [DOI] [Google Scholar]
- 127.Cancer Genome Atlas Research Network. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McKittrick E, Gafken PR, Ahmad K & Henikoff S Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA 101, 1525–1530 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gehre M et al. Lysine 4 of histone H3.3 is required for embryonic stem cell differentiation, histone enrichment at regulatory regions and transcription accuracy. Nat. Genet 52, 273–282 (2020). [DOI] [PubMed] [Google Scholar]
- 130.Zhang T, Zhang Z, Dong Q, Xiong J & Zhu B Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 21, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jacob Y et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang D & Berger F DNA replication–coupled histone modification maintains Polycomb gene silencing in plants. Science 357, 1146–1149 (2017).This work shows that H3.3-enriched genes protect against heterochromatization during DNA replication in plants by inhibiting (through specific H3.3 Thr31) the methyltransferases ATXR5 and ATXR6.
- 133.Guo R et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 56, 298–310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wen H et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014).This work and Guo et al. (2014) identify ZMYND11 as an H3.3-specific reader of H3K36me3 that links histone variant-mediated transcription elongation control to tumour suppression.
- 135.Jang C-W, Shibata Y, Starmer J, Yee D & Magnuson T Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29, 1377–1392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wong LH et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sawicka A & Seiser C Sensing core histone phosphorylation — a matter of perfect timing. Biochim. Biophys. Acta 1839, 711–718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Srivastava S & Foltz DR Posttranslational modifications of CENP-A: marks of distinction. Chromosoma 127, 279–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Udugama M et al. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 43, 10227–10237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ratnakumar K et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 26, 433–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vardabasso C et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol. Cell 59, 75–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Valdes-Mora F et al. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 22, 307–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Draker R, Sarcinella E & Cheung P USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 39, 3529–3542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sarcinella E, Zuzarte PC, Lau PNI, Draker R & Cheung P Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol 27, 6457–6468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Faast R et al. Histone variant H2A.Z is required for early mammalian development. Curr. Biol 11, 1183–1187 (2001). [DOI] [PubMed] [Google Scholar]
- 146.Howman EV et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA 97, 1148–1153 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tang MCW et al. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet. 11, e1004964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hoghoughi N, Barral S, Vargas A, Rousseaux S & Khochbin S Histone variants: essential actors in male genome programming. J. Biochem 163, 97–103 (2018). [DOI] [PubMed] [Google Scholar]
- 149.Molaro A, Young JM & Malik HS Evolutionary origins and diversification of testis-specific short histone H2A variants in mammals. Genome Res. 28, 460–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Barral S et al. Histone variant H2A.L.2 guides transition protein-dependent protamine assembly in male germ cells. Mol. Cell 66, 89–101 (2017). [DOI] [PubMed] [Google Scholar]
- 151.Govin J et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol 176, 283–294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Montellier E et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 27, 1680–1692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ueda J et al. Testis-specific histone variant H3t gene is essential for entry into spermatogenesis. Cell Rep. 18, 593–600 (2017).Collectively, this work, Barral et al. (2017) and Montellier et al. (2013) demonstrate the importance of histone variants as ‘transitional’ proteins required for chromatin condensation during sperm maturation.
- 154.Tachiwana H et al. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl Acad. Sci. USA 107, 10454–10459 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hammoud SS et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Erkek S et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol 20, 868–875 (2013). [DOI] [PubMed] [Google Scholar]
- 157.Torres-Padilla M-E, Bannister AJ, Hurd PJ, Kouzarides T & Zernicka-Goetz M Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol 50, 455–461 (2006). [DOI] [PubMed] [Google Scholar]
- 158.Akiyama T, Suzuki O, Matsuda J & Aoki F Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 7, e1002279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kong Q et al. Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos. J. Biol. Chem 293, 3829–3838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tanaka M, Hennebold JD, Macfarlane J & Adashi EY A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 128, 655–664 (2001). [DOI] [PubMed] [Google Scholar]
- 161.Boulard M et al. Histone variant macroH2A1 deletion in mice causes female-specific steatosis. Epigenetics Chromatin 3, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Celeste A Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Anuar ND et al. Gene editing of the multi-copy H2A.B gene and its importance for fertility. Genome Biol. 20, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sakai A, Schwartz BE, Goldstein S & Ahmad K Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol 19, 1816–1820 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hödl M & Basler K Transcription in the absence of histone H3.3. Curr. Biol 19, 1221–1226 (2009). [DOI] [PubMed] [Google Scholar]
- 166.Couldrey C, Carlton MBL, Nolan PM, Colledge WH & Evans MJ A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet 8, 2489–2495 (1999). [DOI] [PubMed] [Google Scholar]
- 167.Tang MCW, Jacobs SA, Wong LH & Mann JR Conditional allelic replacement applied to genes encoding the histone variant H3.3 in the mouse. Genesis 51, 142–146 (2013). [DOI] [PubMed] [Google Scholar]
- 168.Bush KM et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin 6, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Roberts C et al. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol 22, 2318–2328 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Garrick D et al. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2, e58 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Michaelson JS, Bader D, Kuo F, Kozak C & Leder P Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 13, 1918–1923 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Loppin B et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 (2005). [DOI] [PubMed] [Google Scholar]
- 173.Santenard A et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol 12, 853–862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu Z et al. SUMOylated PRC1 controls histone H3.3 deposition and genome integrity of embryonic heterochromatin. EMBO J. 39, e103697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wen D et al. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl Acad. Sci. USA 111, 7325–7330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jullien J et al. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 5, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nashun B et al. Continuous histone replacement by Hira is essential for normal transcriptional regulation and De Novo DNA methylation during mouse oogenesis. Mol. Cell 60, 611–625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nashun B, Yukawa M, Liu H, Akiyama T & Aoki F Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 137, 3785–3794 (2010). [DOI] [PubMed] [Google Scholar]
- 179.Pehrson JR, Changolkar LN, Costanzi C & Leu NA Mice without macroH2A histone variants. Mol. Cell. Biol 34, 4523–4533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Creppe C et al. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol. Cell. Biol 32, 1442–1452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chang C-C et al. A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev. Biol 278, 367–380 (2005). [DOI] [PubMed] [Google Scholar]
- 182.Margueron R & Reinberg D The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xia W & Jiao J Histone variant H3.3 orchestrates neural stem cell differentiation in the developing brain. Cell Death Differ. 24, 1548–1563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Maze I et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Stefanelli G et al. Learning and age-related changes in genome-wide H2A.Z binding in the mouse hippocampus. Cell Rep. 22, 1124–1131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM & David Sweatt J Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515, 582–586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Piña B & Suau P Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol 123, 51–58 (1987). [DOI] [PubMed] [Google Scholar]
- 188.Michod D et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron 74, 122–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.McCARREY JR, Geyer CB & Yoshioka H Epigenetic regulation of testis-specific gene expression. Ann. N. Y. Acad. Sci 1061, 226–242 (2005). [DOI] [PubMed] [Google Scholar]
- 190.Wang T, Gao H, Li W & Liu C Essential role of histone replacement and modifications in male fertility. Front. Genet 10, 962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Morgan MA & Shilatifard A Chromatin signatures of cancer. Genes Dev. 29, 238–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schwartzentruber J et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).This study identifies recurrent mutations in the histone H3.3 gene at or near sites of post-translational modification in paediatric and young adult patients with glioblastoma, providing evidence that such modifications have important implications for human health.
- 193.Behjati S et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet 45, 1479–1482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lu C et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lewis PW et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fang D et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang X & Zhang Z Oncohistone mutations in diffuse intrinsic pontine glioma. Trends Cancer Res. 5, 799–808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nagaraja S et al. Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol. Cell 76, 965–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gomes AP et al. Dynamic incorporation of histone H3 variants into chromatin is essential for acquisition of aggressive traits and metastatic colonization. Cancer Cell 36, 402–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Corujo D & Buschbeck M Post-translational modifications of H2A histone variants and their role in cancer. Cancers 10, 59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kapoor A et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Longhitano L et al. MacroH2A variant suppress uveal melanoma progression and rewires cancer metabolic phenotype. FASEB J. 33, 790.8 (2019). [Google Scholar]
- 203.Dardenne E et al. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nat. Struct. Mol. Biol 19, 1139–1146 (2012). [DOI] [PubMed] [Google Scholar]
- 204.Valdés-Mora F et al. Acetylated histone variant H2A.Z is involved in the activation of neo-enhancers in prostate cancer. Nat. Commun 8, 1346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Domaschenz R, Kurscheid S, Nekrasov M, Han S & Tremethick DJ The histone variant H2A.Z is a master regulator of the epithelial-mesenchymal transition. Cell Rep. 21, 943–952 (2017). [DOI] [PubMed] [Google Scholar]
- 206.Yang HD et al. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget 7, 11412–11423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rispal J et al. The H2A.Z histone variant integrates Wnt signaling in intestinal epithelial homeostasis. Nat. Commun 10, 1827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Svotelis A, Gévry N, Grondin G & Gaudreau L H2A.Z overexpression promotes cellular proliferation of breast cancer cells. Cell Cycle 9, 364–370 (2010). [DOI] [PubMed] [Google Scholar]
- 209.Arimura Y et al. Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic Acids Res. 46, 10007–10018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mattera L et al. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene 28, 1506–1517 (2009). [DOI] [PubMed] [Google Scholar]
- 211.Gorrini C et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448, 1063–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 212.Sharma AB, Dimitrov S, Hamiche A & Van Dyck E Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Res. 47, 1051–1069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhang W et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun 7, 12619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Turinetto V & Giachino C Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 43, 2489–2498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bassing CH et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114, 359–370 (2003). [DOI] [PubMed] [Google Scholar]
- 216.Heaphy CM et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lovejoy CA et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Law MJ et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143, 367–378 (2010). [DOI] [PubMed] [Google Scholar]
- 219.Varshney D, Spiegel J, Zyner K, Tannahill D & Balasubramanian S The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol 10.1038/s41580-020-0236-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang Y et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun 10, 943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Huh MS et al. Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis. 7, e2220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Clynes D et al. ATRX dysfunction induces replication defects in primary mouse cells. PLoS One 9, e92915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Clynes D et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun 6, 7538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kim J et al. The macroH2A1.2 histone variant links ATRX loss to alternative telomere lengthening. Nat. Struct. Mol. Biol 26, 213–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ramamoorthy M & Smith S Loss of ATRX suppresses resolution of telomere cohesion to control recombination in ALT cancer cells. Cancer Cell 28, 357–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Iwase S et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol 18, 769–776 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Eustermann S et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol 18, 777–782 (2011). [DOI] [PubMed] [Google Scholar]
- 228.Qadeer ZA et al. ATRX in-frame fusion neuroblastoma is sensitive to EZH2 inhibition via modulation of neuronal gene signatures. Cancer Cell 36, 512–527 (2019).This study identifies DAXX-independent ATRX truncations with neomorphic function in neuroblastoma, adding to evidence that ATRX and DAXX have functions that are independent of each other and may be independent of their role in H3.3 deposition.
- 229.Quénet D Histone variants and disease. Int. Rev. Cell Mol. Biol 335, 1–39 (2018). [DOI] [PubMed] [Google Scholar]
- 230.Gibbons RJ, Picketts DJ, Villard L & Higgs DR Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-Xsyndrome). Cell 80, 837–845 (1995). [DOI] [PubMed] [Google Scholar]
- 231.Watson LA et al. Atrx deficiency induces telomere dysfunction, endocrine defects, and reduced life span. J. Clin. Invest 123, 2049–2063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chen X et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 7, 104–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gibbons RJ et al. Mutations in the chromatin-associated protein ATRX. Hum. Mutat 29, 796–802 (2008). [DOI] [PubMed] [Google Scholar]
- 234.Horikoshi N et al. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Crystallogr. Sect. D. Biol. Crystallogr 69, 2431–2439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shin H et al. Transcriptional regulation mediated by H2A.Z via ANP32e-dependent inhibition of protein phosphatase 2A. Biochim. Biophys. Acta 1861, 481–496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ray-Gallet D et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002). [DOI] [PubMed] [Google Scholar]
- 237.Gessi M et al. H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J. Neurooncol 112, 67–72 (2013). [DOI] [PubMed] [Google Scholar]
- 238.Wu G et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet 44, 251–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Forbes SA et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Shrestha RL et al. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8, 46781–46800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rogakou EP, Pilch DR, Orr AH, Ivanova VS & Bonner WM DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem 273, 5858–5868 (1998). [DOI] [PubMed] [Google Scholar]
- 242.Tolstorukov MY et al. Histone variant H2A.Bbd is associated with active transcription and mRNA processing in human cells. Mol. Cell 47, 596–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ueda T et al. Critical role of the p400/mDomino chromatin-remodeling ATPase in embryonic hematopoiesis. Genes Cell 12, 581–592 (2007). [DOI] [PubMed] [Google Scholar]
- 244.Reilly PT et al. Generation and characterization of the Anp32e-deficient mouse. PLoS One 5, e13597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Qiu Z et al. Ino80 is essential for proximal-distal axis asymmetry in part by regulating Bmp4 expression. BMC Biol. 14, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, l1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Li Z et al. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151, 1608–1616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Leatham-Jensen M et al. Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLoS Genet. 15, e1007932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Fang H-T et al. Global H3.3 dynamic deposition defines its bimodal role in cell fate transition. Nat. Commun 9, 1537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tanasijevic B & Rasmussen TP X chromosome inactivation and differentiation occur readily in ES cells doubly-deficient for macroH2A1 and macroH2A2. PLoS One 6, e21512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Barrero MJ, Sese B, Martí M & Izpisua Belmonte JC Macro histone variants are critical for the differentiation of human pluripotent cells. J. Biol. Chem 288, 16110–16116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pasque V, Gillich A, Garrett N & Gurdon JB Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 30, 2373–2387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gaspar-Maia A et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun 4, 1565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wu T et al. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell 15, 281–294 (2014). [DOI] [PubMed] [Google Scholar]