Abstract
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), which is an ongoing pandemic disease. SARS-CoV-2-specific CD4+ and CD8+ T-cell responses have been detected and characterized not only in COVID-19 patients and convalescents, but also unexposed individuals. Here, we review the phenotypes and functions of SARS-CoV-2-specific T cells in COVID-19 patients and the relationships between SARS-CoV-2-specific T-cell responses and COVID-19 severity. In addition, we describe the phenotypes and functions of SARS-CoV-2-specific memory T cells after recovery from COVID-19 and discuss the presence of SARS-CoV-2-reactive T cells in unexposed individuals and SARS-CoV-2-specific T-cell responses elicited by COVID-19 vaccines. A better understanding of T-cell responses is important for effective control of the current COVID-19 pandemic.
Keywords: COVID-19, SARS-CoV-2, T cell, vaccine
INTRODUCTION
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), an ongoing pandemic disease. As of April 3, 2021, more than 130 million cases of COVID-19 and more than 2.8 million deaths have been confirmed worldwide. SARS-CoV-2 infection results in a broad spectrum of clinical manifestations, from asymptomatic or mild disease to severe disease associated with exaggerated inflammatory responses (Huang et al., 2020; Merad and Martin, 2020). For effective control of the current COVID-19 pandemic, a better understanding of the immune responses against SARS-CoV-2 is urgently needed.
During viral infection, CD4+ and CD8+ T cells contribute to viral control by producing effector cytokines and exerting cytotoxic activity. After the emergence of COVID-19, many studies reported T-cell phenotypes in patients with COVID-19 and a relationship between the T-cell phenotype and COVID-19 severity (De Biasi et al., 2020; Kuri-Cervantes et al., 2020; Mathew et al., 2020; Song et al., 2020). In addition, several studies have investigated the phenotypes and functions of various subtypes of immune cells, including CD4+ and CD8+ T cells, from COVID-19 patients using single-cell RNA sequencing (scRNA-seq) (Lee et al., 2020; Liao et al., 2020; Wilk et al., 2020). However, these studies analyzed total CD4+ or CD8+ T cells without information on the SARS-CoV-2-specificity.
Other studies have detected and characterized SARS-CoV-2-specific CD4+ and CD8+ T cells in patients with COVID-19. SARS-CoV-2-specific T cells have been examined by ex vivo stimulation-based functional assays, including interferon (IFN)-γ ELISpot assays, intracellular cytokine staining, and activation-induced marker (AIM) assays. However, the phenotypes of T cells can change during ex vivo stimulation, and non-functioning T cells cannot be detected by stimulation-based functional assays. These limitations can be overcome by major histocompatibility complex (MHC) multimer techniques. Recently, the phenotypes and functions of SARS-CoV-2-specific T cells, particularly CD8+ T cells, were reported using MHC class I (MHC-I) multimers.
Here, we briefly review recent information on the phenotypes and functions of SARS-CoV-2-specific CD4+ and CD8+ T cells in COVID-19 patients and convalescents. In addition, we discuss the SARS-CoV-2-reactive CD4+ and CD8+ T-cell responses in unexposed individuals and T-cell responses elicited by COVID-19 vaccines.
T-CELL RESPONSES IN PATIENTS WITH COVID-19
Early after the emergence of COVID-19, several studies reported an exhausted phenotype of CD8+ T cells in patients with the disease and up-regulation of immune checkpoint inhibitory receptors, including PD-1 (De Biasi et al., 2020; Diao et al., 2020; Zheng et al., 2020a; 2020b). In addition, a recent scRNA-seq study reported an exhaustion cluster among SARS-CoV-2-reactive CD8+ T cells in patients with COVID-19 (Kusnadi et al., 2021). In this study, SARS-CoV-2-reactive CD8+ T cells were isolated from the peripheral blood mononuclear cells (PBMCs) of COVID-19 patients or healthy donors via modified antigen-reactive T-cell enrichment (ARTE). In modified ARTE, PBMCs were stimulated ex vivo with SARS-CoV-2 antigens, and responding CD8+ T cells were isolated based on the expression of CD137 and CD69. Next, they performed scRNA-seq analysis of SARS-CoV-2-reactive CD8+ T cells. The SARS-CoV-2-reactive CD8+ T cells exhibited exhausted phenotypes with a decreased capacity to produce cytokines.
However, our group recently examined SARS-CoV-2-specific CD8+ T cells using MHC-I multimers and demonstrated that IFN-γ is produced by SARS-CoV-2-specific CD8+ T cells in acute and convalescent COVID-19 patients regardless of PD-1 expression (Rha et al., 2021) (Fig. 1). Thus, SARS-CoV-2-specific PD-1+CD8+ T cells are functional, not exhausted. Given that T-cell inhibitory receptors, such as PD-1, can be upregulated by T-cell receptor-induced activation (Singer et al., 2016; Wherry and Kurachi, 2015), PD-1 expression on CD8+ T cells is likely to reflect activation, rather than functional exhaustion, in patients with COVID-19.
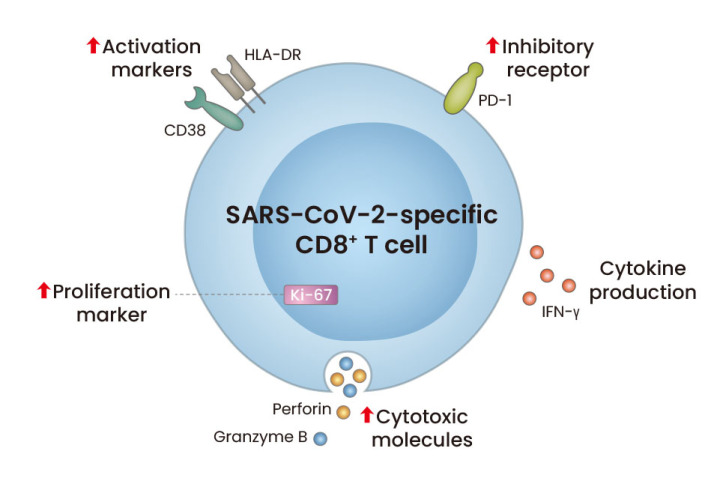
Fig. 1. Phenotypes and functions of SARS-CoV-2-specific CD8+ T cells in patients with acute COVID-19.
During acute COVID-19, SARS-CoV-2-specific CD8+ T cells express not only activation markers (CD38 and HLA-DR), a proliferation marker (Ki-67), and cytotoxic molecules (perforin and granzyme B), but also immune checkpoint inhibitory receptors (PD-1 and TIM-3). However, SARS-CoV-2-specific CD8+ T cells produce IFN-γ regardless of PD-1 expression, indicating that SARS-CoV-2-specific PD-1+CD8+ T cells are functional, not exhausted.
Some data demonstrate that SARS-CoV-2-specific T cells are fully activated during COVID-19. In patients with moderate/severe COVID-19, SARS-CoV-2-specific CD4+ and CD8+ T cells express activation and proliferation markers, including CD38, HLA-DR, and Ki-67 (Sekine et al., 2020). Analysis using MHC-I multimers has also shown that SARS-CoV-2-specific CD8+ T cells express activation markers (CD38 and HLA-DR), a proliferation marker (Ki-67), inhibitory receptors (PD-1 and TIM-3), and cytotoxic molecules (perforin and granzyme B) during acute COVID-19 (Sekine et al., 2020) (Fig. 1). Our group also reported that SARS-CoV-2-specific CD8+ T cells from acute COVID-19 patients exhibit an activated phenotype with high expression of CD38, HLA-DR, PD-1, perforin, and granzyme B (Rha et al., 2021). During the acute phase, the relative frequency of Ki-67+ proliferating cells and CD38+HLA-DR+ activated cells among MHC-I multimer+ cells decreases, with a decrease in the nasopharyngeal viral titer. However, the relative frequency of perforin+granzyme B+ cells and PD-1+ cells among MHC-I multimer+ cells is sustained during the course of COVID-19.
Among patients with COVID-19, SARS-CoV-2-specific T-cell responses have been analyzed in relation to disease severity. However, contradictory results have been reported (Peng et al., 2020; Sattler et al., 2020; Sekine et al., 2020; Tan et al., 2021). A recent study comprehensively evaluated all three arms of adaptive immunity, including CD4+ and CD8+ T-cell and humoral responses, in acute and convalescent COVID-19 patients (Rydyznski et al., 2020). The coordination in SARS-CoV-2-specific adaptive immune responses was found to be associated with mild disease. Interestingly, as a single parameter, the relative frequency of SARS-CoV-2-specific IFN-γ-producing CD8+ T cells inversely correlated with peak disease severity in acute COVID-19 patients, indicating a role of CD8+ T cells in protective immunity against SARS-CoV-2 infection.
Cytokine-induced activation of pre-existing bystander memory CD8+ T cells has been reported in many viral infections (Kim et al., 2018; Kim and Shin, 2019), and bystander activation has also been investigated in patients with COVID-19. A recent study described the activation of cytomegalovirus (CMV)-specific CD8+ T cells in a single patient who developed ventilator-associated Pseudomonas pneumonia during the post-acute phase of COVID-19 (Gregorova et al., 2020). However, the activity of CMV-specific CD8+ T cells was examined following ex vivo stimulation with CMV peptides, and this study could not show bona fide bystander activation. Another study using MHC-I multimers observed upregulation of CD38 on CD8+ T cells specific to SARS-CoV-2-unrelated viruses, including CMV and Epstein–Barr virus, in patients with severe COVID-19 (Sekine et al., 2020). However, this activation was not accompanied by upregulation of HLA-DR and Ki-67. Whether bystander memory CD8+ T cells are truly activated in patients with COVID-19 remains to be elucidated.
MEMORY T CELLS AFTER RECOVERY FROM COVID-19
Several studies have reported SARS-CoV-2-specific memory T-cell responses in the early convalescent phase of COVID-19 up to 3 months after symptom onset (Grifoni et al., 2020; Le Bert et al., 2020; Peng et al., 2020; Rodda et al., 2021). SARS-CoV-2-specific CD4+ and CD8+ T-cell responses were detected in 100% and approximately 70% of COVID-19 convalescents, respectively (Grifoni et al., 2020). T-cell responses targeted not only the spike (S) protein, but also the membrane, nucleocapsid (N), and other open reading frames. Interestingly, although SARS-CoV-2-specific T-cell responses significantly correlate with SARS-CoV-2 S-specific antibody titers (Grifoni et al., 2020; Ni et al., 2020; Peng et al., 2020; Zhou et al., 2020), memory T-cell responses have been detected in the absence of SARS-CoV-2-specific antibodies (Sekine et al., 2020). Considering that T-cell responses to SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) are long-lasting (Le Bert et al., 2020; Ng et al., 2016; Tang et al., 2011; Zhao et al., 2017), SARS-CoV-2-specific memory T cells are expected to be maintained long-term. Recent studies have shown that SARS-CoV-2-specific T-cell responses are maintained up to 6-8 months following infection (Jung et al., 2021; Zuo et al., 2021), though another study reported that SARS-CoV-2-specific T-cell responses decline with a half-life of 3-5 months (Dan et al., 2021).
MHC-I multimer staining has revealed that SARS-CoV-2-specific CD8+ T cells exhibit effector memory (CCR7−CD45RA−) or central memory (CCR7+CD45RA−) phenotypes with early (CD27+CD28+) or intermediate (CD27+CD28−) differentiation in COVID-19 convalescents (Peng et al., 2020). In another study, SARS-CoV-2-specific MHC-I multimer+CD8+ T cells exhibited an early differentiated memory phenotype (CCR7+CD127+CD45RA−/+TCF1+) in convalescents (Sekine et al., 2020). Our group also examined the phenotypes and functions of SARS-CoV-2-specific CD8+ T cells using MHC-I multimers (Rha et al., 2021) and found that SARS-CoV-2-specific MHC-I multimer+CD8+ T cells exhibit early differentiated effector memory phenotypes with a high proliferative capacity in the early convalescent phase.
Stem cell-like memory T (TSCM) cells have the capacity for self-renewal and multipotency to repopulate the broad spectrum of memory and effector T-cell subsets, and the generation of TSCM cells is required for long-term T-cell memory (Gattinoni et al., 2011; 2017). For example, long-lived memory T cells following live-attenuated yellow fever vaccination exhibit stem cell-like properties and contribute to life-long protection (Akondy et al., 2017; Fuertes Marraco et al., 2015). Our group initially demonstrated the generation of SARS-CoV-2-specific TSCM cells in the late convalescent phase of COVID-19 on the basis of the expression of CCR7 and CD45RA (Rha et al., 2021) and clearly defined TSCM cells as CCR7+CD45RA+CD95+ T cells in a subsequent study (Jung et al., 2021) (Fig. 2). Thus, our group showed that TSCM cells successfully develop after recovery from COVID-19 (Jung et al., 2021), indicating that SARS-CoV-2-specific T-cell memory is long-lasting. These findings were supported by the sustained polyfunctionality and proliferation capacity of SARS-CoV-2-specific T cells during an 8-month follow-up period in COVID-19 convalescents (Jung et al., 2021).
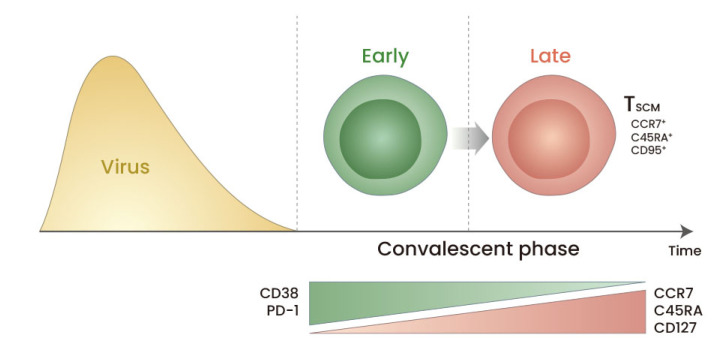
Fig. 2. Differentiation of SARS-CoV-2-specific CD8+ T cells during the convalescent period of COVID-19.
From the early convalescent to the late convalescent phases, the frequency of SARS-CoV-2-specific CD8+ T cells expressing CD38 and PD-1 decreases gradually, whereas the frequency of SARS-CoV-2-specific CD8+ T cells expressing CCR7, CD45RA, and CD127 increases. In particular, among SARS-CoV-2-specific CD8+ T cells, the frequency of CCR7+CD45RA+CD95+ stem cell-like memory T (TSCM) cells that have the capacity for self-renewal and multipotency increases in the late convalescent phase. The generation of TSCM cells is also observed among SARS-CoV-2-specific CD4+ T cells.
Recently, memory T cells were shown to contribute to host protection against SARS-CoV-2 challenge in animal models. In a rhesus macaque model, CD8-depleted convalescent animals exhibited limited viral clearance upon SARS-CoV-2 re-challenge, suggesting that memory CD8+ T cells contribute to viral clearance during SARS-CoV-2 re-infection (McMahan et al., 2021). Furthermore, T-cell vaccination partially protected hosts from severe disease in a mouse model of SARS-CoV-2 infection (Zhuang et al., 2021).
SARS-CoV-2-REACTIVE T-CELL RESPONSES IN UNEXPOSED INDIVIDUALS
SARS-CoV-2-reactive T cells have also been detected among unexposed individuals (Bacher et al., 2020; Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Nelde et al., 2021; Sekine et al., 2020; Sette and Crotty, 2020). SARS-CoV-2 epitopes recognized by these T cells exhibit high homology to endemic common cold coronaviruses (CCCoVs), including OC43, HKU1, 229E, and NL63 (Braun et al., 2020; Mateus et al., 2020; Nelde et al., 2021). Thus, memory T cells primed by previous CCCoV infections may be cross-reactive to SARS-CoV-2 antigens (Box 1). SARS-CoV-2-reactive T-cell responses in unexposed individuals were first detected by AIM assays (Grifoni et al., 2020). In this study, SARS-CoV-2-reactive CD4+ and CD8+ T-cell responses were detected in 50% and 20% of unexposed individuals, respectively. In another study, SARS-CoV-2 S-reactive CD4+ T-cell responses were detected in 35% of unexposed individuals (Braun et al., 2020). SARS-CoV-2-reactive CD4+ T-cell responses have been mapped at the epitope level across the SARS-CoV-2 proteome (Mateus et al., 2020). In this study, several epitopes of memory CD4+ T cells detected in unexposed individuals were reactive to both SARS-CoV-2 and CCCoVs with comparable affinity.
| Box 1. Unsolved issues regarding SARS-CoV-2-reactive T cells in unexposed individuals. |
|---|
| SARS-CoV-2-reactive T-cell responses are observed among a considerable proportion of individuals who have not been exposed to SARS-CoV-2. It remains to be elucidated whether these T cells have been originally primed by endemic common cold coronaviruses (CCCoVs), such as OC43, HKU1, 229E, and NL63, or other animal coronaviruses. In addition, it remains to be elucidated whether pre-existing memory T cells that are cross-reactive to SARS-CoV-2 play a protective or detrimental role in patients with COVID-19. |
Le Bert et al. (2020) examined individuals who recovered from severe acute respiratory syndrome 17 years ago and detected long-lasting T-cell responses that are cross-reactive to SARS-CoV-2. In this study, T cells reactive to the N, NSP7, and NSP13 proteins of SARS-CoV-2 were detected in unexposed individuals. Intriguingly, an NSP7-specific T-cell response among unexposed individuals showed reactivity to regions conserved among animal β-coronaviruses rather than human CCCoVs (Le Bert et al., 2020), suggesting that exposure to unknown β-coronaviruses of animal origin may induce SARS-CoV-2-reactive T-cell responses (Box 1).
Heterologous immunity is the phenomenon by which pre-existing memory T cells primed by an earlier infection are activated during a second unrelated infection. A major mechanism underlying heterologous immunity is the cross-reactivity of T-cell epitopes between unrelated viruses (Rehermann and Shin, 2005; Welsh et al., 2010). A similar phenomenon, heterosubtypic immunity, occurs in the case of repeated infections by related viruses (Bodewes et al., 2009; Rothman, 2011), and a beneficial role of heterosubtypic immunity has been reported (Ge et al., 2010; Sridhar et al., 2013; Wilkinson et al., 2012). In influenza A virus infection, the presence of cross-reactive T cells is significantly associated with better clinical outcomes (Sridhar et al., 2013; Wilkinson et al., 2012).
Heterologous/heterosubtypic immunity can play a protective or detrimental role depending on the relationship between the priming and subsequent viruses (Bodewes et al., 2009; Rehermann and Shin, 2005; Rothman, 2011; Welsh et al., 2010). Pre-existing memory T cells may have a low affinity for cross-reactive epitopes and exert suboptimal functions. However, pre-existing memory T cells can immediately respond to viruses harboring cross-reactive epitopes, proposing that heterologous/heterosubtypic immunity may contribute to the rapid elimination of viruses.
The clinical implication of pre-existing T cells that are cross-reactive to SARS-CoV-2 is not known. Interestingly, a recent study showed that COVID-19 patients with a recent history of laboratory-confirmed CCCoV infection had significantly milder disease than those without a recent history of CCCoV infection (Sagar et al., 2021). This suggests that pre-existing SARS-CoV-2-reactive memory T cells primed by recent CCCoV infections may alleviate the severity of COVID-19. Further studies are required to clarify the clinical implications of SARS-CoV-2-reactive memory T cells primed by previous CCCoV infections.
T-CELL RESPONSES ELICITED BY COVID-19 VACCINES
Currently, two COVID-19 mRNA vaccines (BNT162b2 by Pfizer-BioNTech and mRNA-1273 by Moderna) and two viral vector vaccines (ChAdOx1 nCoV-19 by AstraZeneca and Ad26.COV2.S by Johnson & Johnson/Janssen) have been authorized by the U.S. Food and Drug Administration (FDA) and/or European Medicines Agency (EMA). In clinical studies, COVID-19 vaccines were shown to successfully elicit not only neutralizing antibodies, but also SARS-CoV-2-specific T-cell responses (Anderson et al., 2020; Ewer et al., 2021; Keech et al., 2020; Sahin et al., 2020).
A study using a rhesus macaque model examined the contribution of antibodies and T cells to protection against SARS-CoV-2 infection (McMahan et al., 2021). They showed a critical role of antibodies, with adoptive transfer of purified IgG from convalescent animals to naïve animals, protecting recipient animals against SARS-CoV-2 challenge. They also demonstrated an importance role of CD8+ T cells in a depletion study. Depletion of CD8+ T cells in convalescent animals partially abrogated the protective immunity against SARS-CoV-2 re-challenge. They suggested that memory T-cell responses are crucial, especially when antibodies work sub-optimally. These convalescent animals might have memory T cells targeting multiple proteins of SARS-CoV-2 whereas the current COVID-19 vaccines elicit memory T cells targeting the S protein. However, according to the results from the rhesus macaque study, we can anticipate that vaccine-induced memory T cells play a critical role in host protection, particularly when exposed to SARS-CoV-2 variants escaping from vaccine-induced neutralizing antibodies.
The emergence and propagation of SARS-CoV-2 variants has raised concerns that some variants may escape from neutralizing antibodies elicited by COVID-19 vaccination or natural infection. SARS-CoV-2 variants harboring multiple mutations in the S protein have emerged in the United Kingdom, South Africa, and Brazil. Recent studies have demonstrated reduced neutralizing activity of COVID-19 vaccine-elicited antibodies against some SARS-CoV-2 variants (Chen et al., 2021; Wang et al., 2021a; 2021b). Therefore, COVID-19 vaccine-induced neutralizing antibodies may be less effective against emerging SARS-CoV-2 variants. However, vaccine-induced CD4+ and CD8+ T-cell immunity may be helpful in attenuating disease severity and decreasing mortality. Although SARS-CoV-2 mutations that abrogate binding to MHC have been reported (Agerer et al., 2021), a recent study reported insignificant impacts of variants on SARS-CoV-2-specific CD4+ and CD8+ T-cell responses (Tarke et al., 2021). Most T-cell epitopes were conserved among emerging SARS-CoV-2 variants.
CONCLUSION
During viral infection, both the humoral and cellular arms of the adaptive immune system contribute to eliminating virus from the host. Virus-specific CD4+ and CD8+ T cells produce effector cytokines and exert cytotoxic activity, whereas neutralizing antibodies directly interfere with viral entry of host cells. However, the roles of SARS-CoV-2-specific CD4+ and CD8+ T cells have been highlighted less often than the roles of neutralizing antibodies amid the current COVID-19 pandemic.
COVID-19 vaccines have been approved since December 2020 and have started to be administered to populations worldwide. However, there is a concern that the emergence of SARS-CoV-2 variants escaping vaccine-induced neutralizing antibodies may nullify the effects of the current COVID-19 vaccines. COVID-19 vaccines elicit not only neutralizing antibodies, but also SARS-CoV-2-specific memory T-cell responses. Vaccine-induced T-cell immunity may play a role in reducing the disease burden of COVID-19 by attenuating disease severity and decreasing mortality. To end the current COVID-19 pandemic, we need to better understand SARS-CoV-2-specific CD4+ and CD8+ T cells, as well as neutralizing antibodies.
ACKNOWLEDGMENTS
This research was supported by the 2020 Joint Research Project of Institutes of Science and Technology.
Footnotes
AUTHOR CONTRIBUTIONS
M.K.J. and E.C.S. conceptualized and wrote the manuscript. E.C.S. acquired funding and supervised.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Agerer B., Koblischke M., Gudipati V., Montano-Gutierrez L.F., Smyth M., Popa A., Genger J.W., Endler L., Florian D.M., Muhlgrabner V., et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8(+) T cell responses. Sci. Immunol. 2021;6:eabg6461. doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schroder I., Wieters I., Khodamoradi Y., et al. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Kreijtz J.H., Rimmelzwaan G.F. Yearly influenza vaccinations: a double-edged sword? Lancet Infect. Dis. 2009;9:784–788. doi: 10.1016/S1473-3099(09)70263-4. [DOI] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M., et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., Sharpe H., Makinson R., Morter R., Flaxman A., Wright D., Bellamy D., Bittaye M., et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- Fuertes Marraco S.A., Soneson C., Cagnon L., Gannon P.O., Allard M., Abed Maillard S., Montandon N., Rufer N., Waldvogel S., Delorenzi M., et al. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015;7:282ra248. doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- Gattinoni L., Lugli E., Ji Y., Pos Z., Paulos C.M., Quigley M.F., Almeida J.R., Gostick E., Yu Z., Carpenito C., et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L., Speiser D.E., Lichterfeld M., Bonini C. T memory stem cells in health and disease. Nat. Med. 2017;23:18–27. doi: 10.1038/nm.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Tan V., Bollyky P.L., Standifer N.E., James E.A., Kwok W.W. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J. Virol. 2010;84:3312–3319. doi: 10.1128/JVI.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorova M., Morse D., Brignoli T., Steventon J., Hamilton F., Albur M., Arnold D., Thomas M., Halliday A., Baum H., et al. Post-acute COVID-19 associated with evidence of bystander T-cell activation and a recurring antibiotic-resistant bacterial pneumonia. Elife. 2020;9:e63430. doi: 10.7554/eLife.63430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Rha M.S., Sa M.A., Choi H.K., Jeon J.H., Seok H.R., Park D.W., Park S.H., Jeong H.W., Choi W.S., et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescents for 8 months with successful development of stem cell-like memory T cells. MedRxiv. 2021 doi: 10.1101/2021.03.04.21252658. https://doi.org/10.1101/2021.03.04.21252658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chang D.Y., Lee H.W., Lee H., Kim J.H., Sung P.S., Kim K.H., Hong S.H., Kang W., Lee J., et al. Innate-like cytotoxic function of bystander-activated CD8(+) T cells is associated with liver injury in acute hepatitis A. Immunity. 2018;48:161–173.e5. doi: 10.1016/j.immuni.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Kim T.S., Shin E.C. The activation of bystander CD8(+) T cells and their roles in viral infection. Exp. Mol. Med. 2019;51:1–9. doi: 10.1038/s12276-019-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnadi A., Ramirez-Suastegui C., Fajardo V., Chee S.J., Meckiff B.J., Simon H., Pelosi E., Seumois G., Ay F., Vijayanand P., et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021;6:eabe4782. doi: 10.1126/sciimmunol.abe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D'Andrea K., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lubke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B., Shin E.C. Private aspects of heterologous immunity. J. Exp. Med. 2005;201:667–670. doi: 10.1084/jem.20050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rha M.S., Jeong H.W., Ko J.H., Choi S.J., Seo I.H., Lee J.S., Sa M., Kim A.R., Joo E.J., Ahn J.Y., et al. PD-1-expressing SARS-CoV-2-specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54:44–52.e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A.L. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011;11:532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sattler A., Angermair S., Stockmann H., Heim K.M., Khadzhynov D., Treskatsch S., Halleck F., Kreis M.E., Kotsch K. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J. Clin. Invest. 2020;130:6477–6489. doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Wang C., Cong L., Marjanovic N.D., Kowalczyk M.S., Zhang H., Nyman J., Sakuishi K., Kurtulus S., Gennert D., et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2016;166:1500–1511.e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., Cao W.J., Yang T., Dai X.P., Wang S.Y., et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., da Silva Antunes R., Frazier A., et al. Negligible impact of SARS-CoV-2 variants on CD4 (+) and CD8 (+) T cell reactivity in COVID-19 exposed donors and vaccinees. BioRxiv. 2021 doi: 10.1101/2021.02.27.433180. https://doi.org/10.1101/2021.02.27.433180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021a;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021b;592:616–622. doi: 10.1101/2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.M., Che J.W., Brehm M.A., Selin L.K. Heterologous immunity between viruses. Immunol. Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson T.M., Li C.K., Chui C.S., Huang A.K., Perkins M., Liebner J.C., Lambkin-Williams R., Gilbert A., Oxford J., Nicholas B., et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., Layqah L.A., Alghamdi M.G., Al Gethamy M.M., Dada A.M., et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017;2:eaan5393. doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020a;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020b;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K., Wong Y.C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.Y., Lau T.T., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Lai X., Sun J., Chen Z., Zhang Z., Dai J., Liu D., Li Y., Li F., Wang Y., et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J. Exp. Med. 2021;218:e20202187. doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Dowell A.C., Pearce H., Verma K., Long H.M., Begum J., Aiano F., Amin-Chowdhury Z., Hallis B., Stapley L., et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021;22:620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]