Abstract
It has been more than a year since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first emerged. Many studies have provided insights into the various aspects of the immune response in coronavirus disease 2019 (COVID-19). Especially for antibody treatment and vaccine development, humoral immunity to SARS-CoV-2 has been studied extensively, though there is still much that is unknown and controversial. Here, we introduce key discoveries on the humoral immune responses in COVID-19, including the immune dynamics of antibody responses and correlations with disease severity, neutralizing antibodies and their cross-reactivity, how long the antibody and memory B-cell responses last, aberrant autoreactive antibodies generated in COVID-19 patients, and the efficacy of currently available therapeutic antibodies and vaccines against circulating SARS-CoV-2 variants, and highlight gaps in the current knowledge.
Keywords: COVID-19, humoral immunity, neutralizing antibody, SARS-CoV-2, SARS-CoV-2 variants
INTRODUCTION
Since the first emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, scientists have tried to reveal the characteristics of this virus that has had devastating consequences for human health and economies worldwide. Over the past year, not only the characteristics of this novel virus, but also the pathogenesis of coronavirus disease 2019 (COVID-19) and the immune responses induced in the human body have been abundantly studied, leading to the development and application of various therapeutic options and vaccines. Although humoral immunity is one important arm of protective immunity against viral infection, mainly by producing antibodies capable of neutralizing invading viruses, there has been much controversy over the role of humoral immune responses in COVID-19. In this mini-review, we introduce diverse aspects of humoral immunity that take part in protecting from or in the pathogenesis of COVID-19 and discuss the controversial findings observed in different groups, as well as future directions for ongoing issues.
HUMORAL IMMUNITY TO SARS-CoV-2 AND DISEASE SEVERITY
With their sterilizing immunity ability, antibodies are the first and most feasible target for vaccine development. Unlike the T-cell response, which is consistently impaired and the number of T cells dramatically reduced in severe COVID-19 patients, humoral responses to SARS-CoV-2 and their correlations with disease severity are diverse. Several early reports showed that higher antibody titers are associated with severe clinical manifestations (Garcia-Beltran et al., 2021a; Hashem et al., 2020; Tan et al., 2020; Zhao et al., 2020). A prospective study with 67 COVID-19 patients showed that anti-nucleocapsid protein (NCP) IgM and IgG started on day 7 and day 10 and peaked on day 28 and day 49, respectively. In addition, these antibodies appear earlier and their titers are significantly higher in severe patients than in non-severe patients. They also found that weak responders for IgG had a significantly higher viral clearance rate compared to stronger responders, indicating that a stronger antibody response is associated with delayed viral clearance and disease severity (Tan et al., 2020). Another study of 173 patients with SARS-CoV-2 infection also showed that a higher antibody titer is independently associated with a worse clinical classification (Zhao et al., 2020). These studies raise the possibility of the pathological role of antibody responses. A study of SARS-CoV infection in a macaque model showed aggravation of lung inflammation induced via antibody-dependent enhancement (ADE) (Liu et al., 2019). However, comprehensive studies are needed to explain the clinical correlates of antibody responses and define the involvement of ADE in severe cases of SARS-CoV-2 infection (Arvin et al., 2020).
With the accumulation of a comprehensive cohort of SARS-CoV-2 infections, the dynamics of the antibody response and the correlation with severity seem more complicated. Analysis of the functional humoral trajectories in hospitalized individuals with moderate to severe disease showed that SARS-CoV-2 antigen-specific IgM and IgA were nearly equivalently evolved across all groups (Zohar et al., 2020). However, S-specific IgG development occurred earlier, and higher IgG titers were observed in survivors of severe disease compared to non-survivors, indicating that rapid and potent IgG class switching is linked to survival (Zohar et al., 2020). Lucas et al. (2020) reported that deceased patients did not have higher overall humoral responses, including anti-spike IgG, anti-receptor-binding domain (RBD) IgG, and neutralizing antibody (NAb), and mounted a robust, yet delayed response compared to survivors. They also suggested that the generation of NAb before 14 days of disease onset is a key factor for recovery (Lucas et al., 2020). Interestingly, a study seeking early factors to predict later disease outcomes revealed that spike-specific humoral responses are enriched among convalescent individuals, whereas functional antibody responses to the nucleocapsid are elevated in deceased individuals (Atyeo et al., 2020). The spike-specific phagocytic and complement-fixing activities were enriched early in convalescents, indicating that these spike-specific humoral responses may be beneficial for the trajectory of SARS-CoV-2 infection. Collectively, antibody titers specific for SARS-CoV-2 do not simply correlate with the disease severity, but the various factors, including the kinetics of seroconversion, antibody isotype, and antigen specificity of antibodies, should be considered to determine the effect of the humoral response on disease severity.
NEUTRALIZING ANTIBODY AND CROSS-REACTIVITY
Neutralization is one of the most important functions of antibodies, inducing sterilizing immunity in viral infection. Most patients with a positive antibody response to SARS-CoV-2 have exhibited neutralizing activity measured by using pseudotyped or authentic SARS-CoV-2 virus. Several studies have identified potent NAbs against SARS-CoV-2 from convalescent COVID-19 patients (Brouwer et al., 2020; Ju et al., 2020; Shi et al., 2020; Zost et al., 2020). NAb can bind to the invading virus at the site of the RBD and other domains of the viral spike proteins, preventing the virus from docking onto its entry receptor, angiotension-converting enzyme 2 (ACE2). A study isolating RBD-specific monoclonal antibodies from eight patients with COVID-19 revealed a correlation between neutralizing activity and the competition with ACE2 for binding the RBD (Ju et al., 2020). Analysis of the crystal structure of RBD-bound antibody demonstrated that steric hindrance inhibits viral engagement with ACE2. This study also found that anti-SARS-CoV-2 antibodies and infected plasma do not cross-react with the RBDs of SARS-CoV or MERS-CoV, suggesting that anti-RBD antibodies are mostly viral-species-specific inhibitors (Ju et al., 2020). A similar study reported the isolation of two specific human monoclonal antibodies from a convalescent patient showing potent neutralizing activity against SARS-CoV-2 in vitro and in rhesus monkeys (Shi et al., 2020). Structural analysis revealed that monoclonal antibody binds to the epitopes corresponding to the part overlapping with ACE2-binding sites in the RBD, which interferes with virus-receptor interactions by both steric hindrance and direct competition. For the next step, many research groups have tried to isolate a great scale of human monoclonal antibodies capable of neutralizing SARS-CoV-2 and identify epitopes interacting with these monoclonal antibodies, as these antibodies will be promising candidates for COVID-19 therapeutics and prophylaxis. They also provide successful strategies for vaccine development against SARS-CoV-2 (Barnes et al., 2020; Cao et al., 2020; Liu et al., 2020).
Though a great number of studies investigating humoral immune responses in serum have been reported, a few studies have focused on the antibody response in respiratory mucosa where SARS-CoV-2 enters. Secretory IgA is the most abundant antibody isotype in the mucosal surfaces and is well known for having potent neutralizing activity. A recent study measured SARS-CoV-2-specific IgA antibodies in the serum, saliva, and bronchoalveolar fluid of 159 patients with COVID-19 and found that IgA antibodies are predominant in the early phase of SARS-CoV-2 infection (Sterlin et al., 2021). Moreover, IgA from serum and mucosal surfaces contributes to virus neutralization to a greater extent than IgG. Similarly, another study found that IgA dimers, the primary form of antibody in the mucosal surfaces, had almost 15-times more potent neutralization than IgA monomer, the dominant form in serum, and the monomer was 2-fold less potent than IgG equivalents (Wang et al., 2021a).
Another point that needed to be addressed was whether plasma or NAbs derived from other coronaviruses have neutralizing activity against SARS-CoV-2, as these can provide immediate protection for severe COVID-19 patients and are another possibility for the development of universal vaccines for highly virulent coronavirus (Pinto et al., 2020; Tian et al., 2020; Zhu et al., 2020). Several monoclonal antibodies identified from the memory B cells of an individual infected with SARS-CoV in 2003 target the spike protein of SARS-CoV-2, which shares 80% amino acid sequence identity with the spike protein of SARS-CoV (Pinto et al., 2020). Inversely, antibodies with neutralizing activity against SARS-CoV-2 isolated from COVID-19 convalescent individuals also cross-neutralize SARS-CoV and MERS-CoV (Zhang et al., 2021). The degree of cross-reactivity between human coronaviruses (HCoVs) and SARS-CoV-2 has been widely studied but resulted in controversial findings (Anderson et al., 2021; Ng et al., 2020; Nguyen-Contant et al., 2020; Poston et al., 2020; Song et al., 2020). Using a flow cytometry-based method, SARS-CoV-2 spike protein-reactive antibodies have been detected in SARS-CoV-2-uninfected individuals (Ng et al., 2020). SARS-CoV-2-uninfected donor sera including these S2-targeted IgG class antibodies showed specific neutralizing activity against SARS-CoV-2 and SARS-CoV-2 S pseudotypes. These pre-existing antibodies were more common in children and adolescents (Ng et al., 2020), suggesting a possibility that higher HCoV (common cold coronaviruses) infection rates in children than adults correlate with relatively less severe symptoms in children with COVID-19 (Castagnoli et al., 2020). In contrast, other studies have reported limited pre-existing cross-reactive antibodies against SARS-CoV-2 in unexposed individuals (Nguyen-Contant et al., 2020; Poston et al., 2020; Song et al., 2020). Specifically, only S2-targeted IgG antibody was detected and had some neutralizing activity, whereas anti-RBD IgG antibody was absent in unexposed individuals, though they found pre-existing cross-reactive memory B cells that were activated upon SARS-CoV-2 infection. Furthermore, some people in a pre-pandemic group had cross-reactive antibodies against SARS-CoV-2, but they are neither neutralizing nor protective against SARS-CoV-2 infection (Anderson et al., 2021). Further studies are needed to address how cross-reactivity between various coronaviruses affects the course and pathogenesis of infection with another coronavirus.
LONGEVITY OF THE ANTIBODY RESPONSE AND MEMORY B CELLS
The longevity of the immune memory response is critical for protection from pathogen reinfection. More data still needs to be accumulated over longer periods, but there are several controversial reports about how long antibody responses against SARS-CoV-2 could be sustained in patients with COVID-19 (Duysburgh et al., 2021; Gudbjartsson et al., 2020; Ibarrondo et al., 2020; Roltgen et al., 2020a; 2020b; Seow et al., 2020; Wajnberg et al., 2020). Some studies have shown that most antibodies against SARS-CoV-2 remain stable for several months after infection, whereas other studies have found a rapid decline in antibody titers within 3-4 months. Interestingly, the waning of anti-SARS-CoV-2 IgA antibodies seems to be affected less than other isotypes, including IgM and IgG antibodies (Gaebler et al., 2021). It is challenging to address the reason why these studies have shown different outcomes. In some studies, a rapid decline in antibody titers against SARS-CoV-2 was found in COVID-19 patients with mild symptoms or asymptomatic individuals, suggesting that the longevity of the antibody responses correlates with disease severity (Roltgen et al., 2020a; 2020b; Seow et al., 2020). These findings also suggest that boosting vaccine administration is required for long-lasting protection.
Although the longevity of antibodies against SARS-CoV-2 is still unclear, SARS-CoV-2-specific memory B cells have been found to persist for 3-6 months (Gaebler et al., 2021; Hartley et al., 2020; Rodda et al., 2021). In the study by Nussenzweig’s group, though IgM and IgG anti-SARS-CoV-2 RBD antibody titers significantly decreased and neutralizing capacity declined over time, the number of RBD-specific memory B cells was not changed up to 6 months after infection (Gaebler et al., 2021). Moreover, the humoral response in this cohort continued evolving as the antibodies from memory B cells underwent great somatic hypermutation in accordance with the persistence of SARS-CoV-2 in the small bowel (Gaebler et al., 2021). From 11 paired samples of COVID-19 patients between 4 and 242 days post-symptom onset, RBD- and NCP-specific memory B cells continued to increase until 150 days, and the number of RBD-specific IgG+ memory B cells significantly correlated with the number of circulating follicular helper T cells (Hartley et al., 2020). A more recent study investigated the phenotype of memory B cells in mild and severe COVID-19 patients up to 6 months after infection (Sokal et al., 2021). Using longitudinal single-cell and repertoire analysis, the B-cell response against SARS-CoV-2 showed a temporal switch from an extrafollicular reaction to a germinal center-dependent memory response generating anti-RBD NAbs. Notably, cross-reactive memory B cells against common cold coronaviruses contribute to the early extrafollicular antibody response against SARS-CoV-2. These findings suggest that the persistence and evolution of memory B cells contribute to the functional immunological memory that provides protection upon virus re-exposure and provides a basis for effective vaccination.
AUTOANTIBODY GENERATION IN COVID-19
Autoantibodies directed against host proteins can induce a perturbation in the host immune system. Severe inflammatory conditions, such as chronic viral infection, have been reported to increase the prevalence of autoantibodies. Several studies have shown that COVID-19 patients, especially individuals with severe symptoms, have a higher prevalence of autoantibodies against various host proteins (Bhadelia et al., 2021; Wang et al., 2020; Woodruff et al., 2020a; Zuniga et al., 2021). Higher levels of anti-Annexin A2 antibodies have been detected among hospitalized COVID-19 patients that died compared to non-critical hospitalized COVID-19 patients (Zuniga et al., 2021). Using the high-throughput autoantibody discovery technique, Wang et al. (2020) found that COVID-19 patients present with dramatic increases in autoantibody reactivities compared to uninfected controls. These autoantibodies are against immune-related proteins, including cytokines, chemokines, complement components, and cell surface proteins. They also showed that autoantibodies targeting tissue-associated antigens correlate with disease severity and the clinical characteristics of inflammation in COVID-19 patients. Another study identified activation of extrafollicular B cells in critically ill patients, and these B cells shared the B-cell repertoire features previously described in autoimmune settings, such as systemic lupus erythematosus (SLE) (Woodruff et al., 2020b). Autoantibodies against type I interferons (IFNs) were reported in at least 101 of 987 (10.2%) patients with life-threatening COVID-19 pneumonia, whereas none of the individuals with asymptomatic or mild SARS-CoV-2 infection and only 4 of 1,227 (0.33%) healthy individuals before the pandemic had autoantibodies against type I IFNs (Bastard et al., 2020). These autoantibodies had neutralizing activity against type I IFNs and their ability to block SARS-CoV-2 infection in vitro. Notably, 94% of patients with these autoantibodies were men. Collectively, it seems that the presence of autoantibodies against host proteins in SARS-CoV-2 infection correlates with the severity of COVID-19, but whether these autoantibodies will lead to autoimmune disease and how they affect the natural course of SARS-CoV-2 infection remains unknown.
EFFICACY OF CURRENT ANTIBODY-BASED THERAPEUTICS AND VACCINES AGAINST SARS-CoV-2 VARIANTS
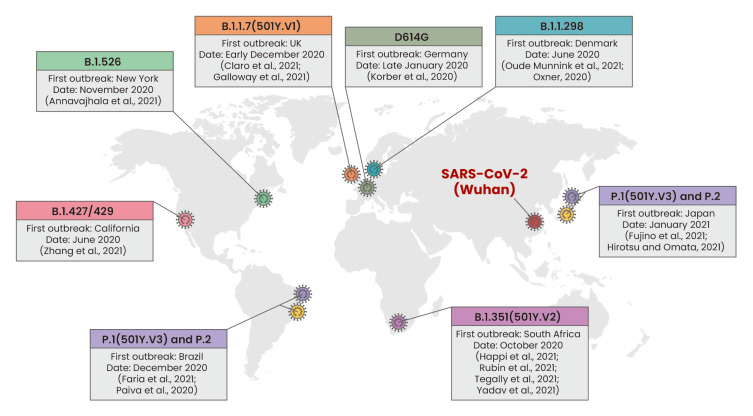
Strains of SARS-CoV-2 with a mutation in the spike protein were officially identified recently and are spreading rapidly worldwide (Fig. 1). Their altered transmissibility and impaired response to vaccination are increasing social anxiety. The first variant that emerged is D614G, which was first discovered in Germany at the end of February 2020. This has higher transmissibility than the wild-type virus (Wuhan) and became the world's most dominant virus at the end of March 2020 (Korber et al., 2020). In June 2020, B.1.1.298 was identified in Denmark, a SARS-CoV-2 variant that causes transmission between mink and human (Oude Munnink et al., 2021). Consequently, 17 million minks were killed to prevent interspecies transmission and the evolution of mutated viruses via mink (Oxner, 2020). Many variants with mutations in the RBD region have since been reported (Fig. 1, Table 1). Such a mutation is markedly resistant to certain spike protein monoclonal antibodies (Li et al., 2020) and has the potential to escape antibody recognition (Greaney et al., 2021). At the end of 2020, the B.1.1.7 variant harboring both the N501Y and D614G mutations in the RBD was reported in the UK (Claro et al., 2021; Galloway et al., 2021). This strain exhibits a greater transmission capacity than the D614G variant (Santos and Passos, 2020). To overcome this situation, there is an increasing need for research on whether current vaccines and therapeutics are effective against variants. The mRNA vaccines, including BNT162b2 (Pfizer) and mRNA-1273 (Moderna), which were manufactured based on SARS-CoV-2 isolated early in the epidemic from Wuhan, China, have demonstrated >94% efficacy at preventing COVID-19 in a phase 3 study and have been approved under Emergency Use Authorization (EUA) by the U.S. Food and Drug Administration (FDA) (Polack et al., 2020). After the emergence of variants, the efficacy of the vaccines against them was tested, revealing high efficacy against D614G (Europe) (Garcia-Beltran et al., 2021b; Mahase, 2021; Muik et al., 2021), B.1.1.7 (UK) (Garcia-Beltran et al., 2021b; Heath et al., 2021; Hoffmann et al., 2021; Muik et al., 2021; Wang et al., 2021b; Wu et al., 2021), B.1.1.298 (Denmark) (Garcia-Beltran et al., 2021b), and B.1.429 (US) (Garcia-Beltran et al., 2021b), but significantly reduced efficacy for variants harboring a mutation in the RBD, such as E484K (Garcia-Beltran et al., 2021b; Hoffmann et al., 2021; Wang et al., 2021b; Wu et al., 2021). Some monoclonal antibodies, including REGN-COV2 and Bamlanivimab, have had similar results in regards to efficacy (An EUA for bamlanivimab, 2020; Garcia-Beltran et al., 2021b; Hoffmann et al., 2021; Liu et al., 2021; Muik et al., 2021). Therefore, these results suggest that certain RBD mutations may have a core effect on the efficacy of neutralization. In recent studies, methods have been reported for screening the efficacy of monoclonal antibodies to minimize SARS-CoV-2 mutational escape. Baum et al. (2020b) demonstrated that antibody cocktail therapy can be an effective way of minimizing mutational escape by SARS-CoV-2. Greaney et al. (2021) and Starr et al. (2021) completely mapped RBD mutations that escape binding by certain neutralization antibodies by establishing a yeast-display system. These studies will strongly contribute to the discovery of vaccines and drugs that have broad neutralization potency against escape mutants. Outbreaks of SARS-CoV-2 variants will likely continue for a long time. Therefore, future research will require special efforts to continually track variants and find effective ways of preventing them. Furthermore, we should strive to build a system that can respond quickly to unexpected mutation of the virus.
Fig. 1. Global emergence of SARS-CoV-2 variants.
The location and date of the first outbreak of each SARS-CoV-2 variant, including D614G (Germany), B.1.1.298 (Denmark), B.1.427/429 (California), B.1.351 (501Y.V2, South Africa), B.1.526 (New York), B.1.1.7 (501Y.V1, UK), P.1 and P.2 (Brazil and Japan), are shown. The given references are the first reports of the emergence of the corresponding variant.
Table 1.
Efficacy of COVID-19 vaccines and neutralizing antibodies against SARS-CoV-2 variants
| Variants | Mutation region | Humoral immunity | Origin country |
|---|---|---|---|
| Wild type | X | Vaccination - High efficacy : BNT162b2 (Pfizer) (Polack et al., 2020), mRNA-1273 (Moderna) (Baden et al., 2021), NVX- CoV2373 (Mahase, 2021), AZD1222 (AstraZeneca) (Voysey et al., 2021), JNJ-7843673 (Johnson & Johnson) (Sadoff et al., 2021) Neutralization - High efficacy : REGN-COV2 (Baum et al., 2020a), Bamlanivimab (Chen et al., 2021; Gottlieb et al., 2021) |
Wuhan, China |
| D614G | D614G in spike | Vaccination - High efficacy (Weissman et al., 2021) : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b; Mahase, 2021; Zou et al., 2021), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b) |
Europe |
| B.1.1.7 (501Y.V1) | -Three amino acid deletions and seven missense mutations in spike -D614G as well as N501Y in the ACE2 receptor-binding domain (RBD) |
Vaccination - High efficacy : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b; Hoffmann et al., 2021; Muik et al., 2021; Wang et al., 2021b), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b; Wang et al., 2021b; Wu et al., 2021), NVX-CoV2373 (Heath et al., 2021; Muik et al., 2021) - Significantly decreased efficacy : BNT162b2 (Pfizer) (Collier et al., 2021) Neutralization - High efficacy : Imdevimab, Casirivimab, REGN-COV2, REGN10989, Bamlanivimab (Garcia-Beltran et al., 2021b; Muik et al., 2021) |
UK |
| B.1.1.298 | Two-amino acid deletion and four spike missense mutations including Y453F in RBD | Vaccination - High efficacy : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b) |
Denmark |
| B.1.427/429 | 4 spike missense mutations, L452R mutation in RBD | Vaccination - High efficacy : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b) - Significantly decreased efficacy : BNT162b2 (Pfizer) (Deng et al., 2021), mRNA-1273 (Moderna) (Deng et al., 2021) Neutralization - Significantly decreased efficacy (Li et al., 2020) : Bamlanivimab (An EUA for bamlanivimab, 2020) |
California, USA |
| P.2 (B.1.1.28 lineage) | 3 spike missense mutations E484K mutation in RBD |
Reinfection Cases of SARS-CoV-2 reinfection (Nonaka et al., 2021; Resende et al., 2021) Vaccination - Significantly decreased efficacy : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b) |
Brazil, Japan |
| P.1 (B.1.1.28 lineage, 501Y.V3) | 12 spike missense mutations E484K, K417T, N501Y mutation in RBD |
Reinfection Cases of SARS-CoV-2 reinfection (Naveca et al., 2021; Nonaka et al., 2021; Resende et al., 2021) Vaccination - Significantly decreased efficacy : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b; Nonaka et al., 2021), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b) Neutralization - High efficiency : Imdevimab, REGN-COV2 (Resende et al., 2021) - Significantly decreased efficacy : Casirivimab (partial), REGN10989, Bamlanivimab (An EUA for bamlanivimab, 2020; Hoffmann et al., 2021; Liu et al., 2021) |
Brazil, Japan |
| B.1.351 lineage (501Y.V2) | 3 RBD mutations, K417N, E484K, and N501Y, in addition to several mutations outside of RBD | Reinfection The risk of reinfection has been reported (Wibmer et al., 2021) Vaccination - High efficacy : JNJ-7843673 (Johnson & Johnson) (Sadoff et al., 2021), BNT162b2 (Pfizer) (Xie et al., 2021) - Significantly decreased efficacy : BNT162b2 (Pfizer) (Garcia-Beltran et al., 2021b; Wu et al., 2021), mRNA-1273 (Moderna) (Garcia-Beltran et al., 2021b; Wang et al., 2021b; Wu et al., 2021), NVX-CoV2373 (Heath et al., 2021; Mahase, 2021), AZD1222 (Voysey et al., 2021) Neutralization - High efficacy : Imdevimab, REGN-COV2, VIR-7831 (Hoffmann et al., 2021) - Significantly decreased efficacy : Casirivimab, REGN10989, Bamlanivimab (An EUA for bamlanivimab, 2020; Hoffmann et al., 2021; Liu et al., 2021) |
South Africa |
| B.1.526 | E484K, S477N mutation in RBD, L5F, T95I, D253G, D614G, and A701V in spike | Neutralization - Significantly decreased efficacy : Bamlanivimab (An EUA for bamlanivimab, 2020) |
New York, USA |
| Other variants (outcome prediction) | Mutations in RBD such as A475V, Q493R, R346S, N439K, 440K, etc. | Prediction of vaccines and therapeutics efficacy against various RBD mutants SARS-CoV-2 (Baum et al., 2020b; Greaney et al., 2021; Pinto et al., 2020; Starr et al., 2021) | - |
CONCLUDING REMARKS
Thus far, this devastating pandemic has resulted in 130 million cases and 2.84 million deaths from COVID-19 worldwide. During the past year, significant scientific knowledge has been generated on the features of novel SARS-CoV-2, as well as the host immune responses that are substantially involved in COVID-19 pathogenesis. We reviewed key discoveries regarding the humoral immune responses in COVID-19 that have formed the basis of currently available therapeutics, including monoclonal antibodies and vaccines. Like other viral infections, antibody responses against SARS-CoV-2 are important for neutralization and rapid clearance of the virus, but their impacts are more complicated than we expected like being involved in the pathogenesis and severity of COVID-19. The dynamics of the antibody responses, cross-reactivity of the NAbs, the longevity of antibodies and memory B cells, and the generation of autoantibodies collectively affect the pathogenesis and severity of COVID-19. Moreover, as the virus evolves, several variants of SARS-CoV-2 that are less responsive to current therapeutics are rapidly emerging. Therefore, additional studies are needed to address the gaps in current knowledge and find an effective way to prepare for emerging threats.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF-2021R1C1C1004546, NRF-2021M3A9H3015689).
Footnotes
AUTHOR CONTRIBUTIONS
E.L. and J.E.O. wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- An EUA for bamlanivimab - a monoclonal antibody for COVID-19. Med. Lett. Drugs Ther. 2020;62:185–186. [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1101/2020.11.06.20227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.C. A novel SARS-CoV-2 variant of concern, B. 1.526, identified in New York. MedRxiv. 2021 doi: 10.1101/2021.02.23.21252259. https://doi.org/10.1101/2021.02.23.21252259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020a;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020b;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadelia N., Belkina A.C., Olson A., Winters T., Urick P., Lin N., Rifkin I., Kataria Y., Yuen R.R., Sagar M., et al. Distinct autoimmune antibody signatures between hospitalized acute COVID-19 patients, SARS-CoV-2 convalescent individuals, and unexposed pre-pandemic controls. MedRxiv. 2021 doi: 10.1101/2021.01.21.21249176. https://doi.org/10.1101/2021.01.21.21249176. [DOI] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnoli R., Votto M., Licari A., Brambilla I., Bruno R., Perlini S., Rovida F., Baldanti F., Marseglia G.L. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA Pediatr. 2020;174:882–889. doi: 10.1001/jamapediatrics.2020.1467. [DOI] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claro I.M., da Silva Sales F.C., Ramundo M.S., Candido D.S., Silva C.A.M., de Jesus J.G., Manuli E.R., de Oliveira C.M., Scarpelli L., Campana G., et al. Local transmission of SARS-CoV-2 lineage B.1.1.7, Brazil, December 2020. Emerg. Infect. Dis. 2021;27:970–972. doi: 10.3201/eid2703.210038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I., Meng B., Datir R., Walls A.C., Kemp S.S., Bassi J., Pinto D., Fregni C.S., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. [2021 Mar 11 [Epub]];Nature. 2021 doi: 10.1038/s41586-021-03412-7. https://doi.org/10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-Gonzalez A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. MedRxiv. 2021 doi: 10.1101/2021.03.07.21252647. https://doi.org/10.1101/2021.03.07.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duysburgh E., Mortgat L., Barbezange C., Dierick K., Fischer N., Heyndrickx L., Hutse V., Thomas I., Van Gucht S., Vuylsteke B., et al. Persistence of IgG response to SARS-CoV-2. Lancet Infect. Dis. 2021;21:163–164. doi: 10.1016/S1473-3099(20)30943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. MedRxiv. 2021 doi: 10.1101/2021.02.26.21252554. https://doi.org/10.1101/2021.02.26.21252554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., Fujimoto T., Kuroda M., Wakita T., Ohmagari N. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg. Infect. Dis. 2021;27:1243–1245. doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L., et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021a;184:476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. [2021 Mar 12 [Epub]];Cell. 2021b doi: 10.1016/j.cell.2021.03.013. https://doi.org/10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., et al. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happi A.N., Ugwu C.A., Happi C.T. Tracking the emergence of new SARS-CoV-2 variants in South Africa. Nat. Med. 2021;27:372–373. doi: 10.1038/s41591-021-01265-1. [DOI] [PubMed] [Google Scholar]
- Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., Peleg A.Y., Boo I., Drummer H.E., Hogarth P.M., et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem A.M., Algaissi A., Almahboub S.A., Alfaleh M.A., Abujamel T.S., Alamri S.S., Alluhaybi K.A., Hobani H.I., AlHarbi R.H., Alsulaiman R.M., et al. Early humoral response correlates with disease severity and outcomes in COVID-19 patients. Viruses. 2020;12:1390. doi: 10.3390/v12121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath P.T., Galiza E.P., Baxter D., Boffito M., Browne D., Burns F., Chadwick D.R., Clark R., Cosgrove C., Galloway J., et al. Efficacy of the NVX-CoV2373 Covid-19 vaccine against the B. 1.1. 7 variant. MedRxiv. 2021 doi: 10.1101/2021.05.13.21256639. https://doi.org/10.1101/2021.05.13.21256639. [DOI] [Google Scholar]
- Hirotsu Y., Omata M. Discovery of a SARS-CoV-2 variant from the P. 1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J. Infect. 2021;82:276–316. doi: 10.1016/j.jinf.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. [2021 Mar 20 [Epub]];Cell. 2021 doi: 10.1016/j.cell.2021.03.036. https://doi.org/10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wei P., Zhang Q., Chen Z., Aviszus K., Downing W., Peterson S., Reynoso L., Downey G.P., Frankel S.K., et al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to Bamlanivimab in vitro. BioRxiv. 2021 doi: 10.1101/2021.02.16.431305. https://doi.org/10.1101/2021.02.16.431305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Klein J., Sundaram M., Liu F., Wong P., Silva J., Mao T., Oh J.E., Tokuyama M., Lu P., et al. Kinetics of antibody responses dictate COVID-19 outcome. MedRxiv. 2020 doi: 10.1101/2020.12.18.20248331. https://doi.org/10.1101/2020.12.18.20248331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- Muik A., Wallisch A.K., Sanger B., Swanson K.A., Muhl J., Chen W., Cai H., Maurus D., Sarkar R., Tureci O., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F., da Costa C., Nascimento V., Souza V., Corado A., Nascimento F., Costa Á., Duarte D., Silva G., Mejía M., et al. SARS-CoV-2 reinfection by the new Variant of Concern (VOC) P.1 in Amazonas, Brazil. virological 2021 [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. Bio. 2020;11:e01991–20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka C.K.V., Franco M.M., Graf T., de Lorenzo Barcia C.A., de Avila Mendonca R.N., de Sousa K.A.F., Neiva L.M.C., Fosenca V., Mendes A.V.A., de Aguiar R.S., et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg. Infect. Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxner R. Denmark to kill up to 17 million minks after discovering mutated coronavirus. [Retrieved March 24, 2021];2020 from https://www.npr.org/2020/11/05/931726205/denmark-to-kill-up-to-17-million-minks-after-discovering-mutated-coronavirus.
- Paiva M.H.S., Guedes D.R.D., Docena C., Bezerra M.F., Dezordi F.Z., Machado L.C., Krokovsky L., Helvecio E., da Silva A.F., Vasconcelos L., et al. Multiple introductions followed by ongoing community spread of SARS-CoV-2 at one of the largest metropolitan areas of Northeast Brazil. Viruses. 2020;12:1414. doi: 10.3390/v12121414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston D., Weisblum Y., Wise H., Templeton K., Jenks S., Hatziioannou T., Bieniasz P. Absence of SARS-CoV-2 neutralizing activity in pre-pandemic sera from individuals with recent seasonal coronavirus infection. [2020 Dec 3 [Epub]];Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1803. https://doi.org/10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Bezerra J.F., Vasconcelos R.H.T., Arantes I., Appolinario L., Mendonça A.C., Paixao A.C., Rodrigues A.C.D., Silva T., Rocha A.S., et al. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020. virological 2021 [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020a;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roltgen K., Wirz O.F., Stevens B.A., Powell A.E., Hogan C.A., Najeeb J., Hunter M., Sahoo M.K., Huang C., Yamamoto F., et al. SARS-CoV-2 antibody responses correlate with resolution of RNAemia but are short-lived in patients with mild illness. MedRxiv. 2020b doi: 10.1101/2020.08.15.20175794. https://doi.org/10.1101/2020.08.15.20175794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E.J., Baden L.R., Abdool Karim S.S., Morrissey S. Audio interview: Covid-19 in South Africa and a new SARS-CoV-2 variant. N. Engl. J. Med. 2021;384:e14. doi: 10.1056/NEJMe2100736. [DOI] [PubMed] [Google Scholar]
- Santos J.C., Passos G.A. The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. BioRxiv. 2020 https://doi.org/10.1101/2020.12.29.424708. [Google Scholar]
- Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., et al. Interim results of a phase 1-2a trial of Ad26.COV2.S Covid-19 vaccine. [2021 Jan 13 [Epub]];N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2034201. https://doi.org/10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O'Byrne A., Kouphou N., Galao R.P., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. BioRxiv. 2020 doi: 10.1038/s41467-021-23074-3. https://doi.org/10.1101/2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1101/2020.11.30.405472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claer L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Lu Y., Zhang J., Wang J., Dan Y., Tan Z., He X., Qian C., Sun Q., Hu Q., et al. Viral kinetics and antibody responses in patients with COVID-19. MedRxiv. 2020 doi: 10.1016/j.ebiom.2020.102999,. https://doi.org/10.1101/2020.03.24.20042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.Y., Mao T., Klein J., Dai Y., Huck J.D., Liu F., Zheng N.S., Zhou T., Israelow B., Wong P., et al. Diverse functional autoantibodies in patients with COVID-19. MedRxiv. 2020 doi: 10.1101/2020.12.10.20247205. https://doi.org/10.1101/2020.12.10.20247205. [DOI] [PubMed] [Google Scholar]
- Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021a;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021b;592:616–622. doi: 10.1101/2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Lee F.E.H., Sanz I. Clinically identifiable autoreactivity is common in severe SARS-CoV-2 Infection. MedRxiv. 2020a doi: 10.1101/2020.10.21.20216192. https://doi.org/10.1101/2020.10.21.20216192. [DOI] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020b;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. BioRxiv. 2021 doi: 10.1101/2021.01.25.427948. https://doi.org/10.1101/2021.01.25.427948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS- CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Yadav P.D., Gupta N., Nyayanit D.A., Sahay R.R., Shete A.M., Majumdar T., Patil S., Kaur H., Nikam C., Pethani J., et al. Imported SARS-CoV-2 V501Y. V2 variant (B. 1.351) detected in travelers from South Africa and Tanzania to India. Travel Med. Infect. Dis. 2021;41:102023. doi: 10.1016/j.tmaid.2021.102023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wu Q., Liu Z., Wang Q., Wu J., Hu Y., Bai T., Xie T., Huang M., Wu T., et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat. Microbiol. 2021;6:51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.J., Yuan Q., Wang H.Y., Liu W., Liao X.J., Su Y.Y., Wang X., Yuan J., Li T.D., Li J.X., et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 2020;6:eabc9999. doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schafer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Xie X., Fontes-Garfias C.R., Swanson K.A., Kanevsky I., Tompkins K., Cutler M., Cooper D., Dormitzer P.R., Shi P.Y. The effect of SARS-CoV-2 D614G mutation on BNT162b2 vaccine-elicited neutralization. NPJ Vaccines. 2021;6:44. doi: 10.1038/s41541-021-00313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga M., Gomes C., Carsons S.E., Bender M.T., Cotzia P., Miao Q.R., Lee D.C., Rodriguez A. Autoimmunity to the lung protective phospholipid-binding protein Annexin A2 predicts mortality among hospitalized COVID-19 patients. MedRxiv. 2021 doi: 10.1183/13993003.00918-2021. https://doi.org/10.1101/2020.12.28.20248807. [DOI] [PMC free article] [PubMed] [Google Scholar]