Abstract
Severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) is a novel virus that causes coronavirus disease 2019 (COVID-19). To understand the identity, functional characteristics and therapeutic targets of the virus and the diseases, appropriate infection models that recapitulate the in vivo pathophysiology of the viral infection are necessary. This article reviews the various infection models, including Vero cells, human cell lines, organoids, and animal models, and discusses their advantages and disadvantages. This knowledge will be helpful for establishing an efficient system for defense against emerging infectious diseases.
Keywords: coronavirus, COVID-19, infection model, organoid, SARS-CoV-2
INTRODUCTION
Currently, coronavirus disease 2019 (COVID-19), resulting from severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), is an ongoing global pandemic. As of the 16th of June 2021, more than 177 million cases have been diagnosed with ~3.8 million deaths resulting in one of the most deadly pandemics in human history. SARS-CoV-2 was first identified in December 2019 (Zhu et al., 2020). Since then, scientific communities have investigated the characteristics of the virus to develop strategies for efficient prevention, diagnosis, and antiviral treatment.
At the early phase of the global pandemic, many of the non-functional characteristics, such as genomics, phylogeny, and taxonomy, were explored by metagenomic RNA-sequencing of the virus isolated from bronchoalveolar lavage fluid samples from patients with severe pneumonia (Wu et al., 2020; Zhou et al., 2020; Zhu et al., 2020). Its genomic sequence revealed that the virus is a novel betacoronavirus, with ~80% and ~96% sequence similarity to SARS-CoV and a bat coronavirus, respectively. The genome sequence of the virus suggested bats and pangolins as probable reservoir hosts (Wu et al., 2020; Zhou et al., 2020). Most obviously, the sequence of the S protein of the SARS-CoV-2 is distinct from that of its relative viruses, sharing only ~73% amino-acid similarity with the SARS-CoV. Interestingly, four amino acid residues are specifically inserted in the S protein, generating a polybasic cleavage site that can be more efficiently cleaved by furin and other proteases (Walls et al., 2020).
As the number of infected cases increases, epidemiological and observational studies have identified the characteristics of SARS-CoV-2 and SARS-CoV-2 infection, such as its clinical courses, potential therapeutics, emergence of mutant viruses, and global transmission routes. For example, patient demographics indicated that the infection rate is much lower in young children (O'Driscoll et al., 2021). Remdesivir was suggested to be an effective therapeutic regimen (Beigel et al., 2020). A more infectious mutant virus has been observed harboring the D614G spike protein mutation (Korber et al., 2020), and a viral quasispecies is also being monitored (Jary et al., 2020). However, these studies are unable to answer the functional characteristics of the novel virus, such as its mechanisms of cell entry and transmission, kinetics in replication and transcription, host responses, and therapeutic targets. To this end, efficient laboratory models are necessary, which enable viral infection and subsequent examination of the infected cells in controlled and reproducible environments. Most importantly, screening novel therapeutics is impossible without competent infection models.
Functional studies of infectious diseases are sometimes challenging due to the specificity of the host species and target tissues, known as species and cell-type tropisms. For example, a wild-type mouse is not susceptible to SARS-CoV-2 infection due to the difference in its angiotensin-converting enzyme 2 (ACE2) receptor, the molecule for viral entry (Munoz-Fontela et al., 2020). Other animal models, such as golden hamsters or ferrets, are susceptible (Kim et al., 2020d; Sia et al., 2020), but they sometimes fail to reproduce the pathophysiology found in humans. Ordinary cell lines, mostly immortalized from normal tissues and/or derived from neoplastic cells, may not always be susceptible to the virus, and infected cells do not reflect the responses of the human target tissues (Chu et al., 2020). The recent development of organoid cultures, an organotypic culture of adult normal stem cells, offers a new innovative possibility for viral studies, but they are in their infancy. In this mini-review, we examine and compare the many infection models that have been used to understand the SARS-CoV-2 pathophysiology which can be extended to other respiratory viruses (Figs. 1 and 2).
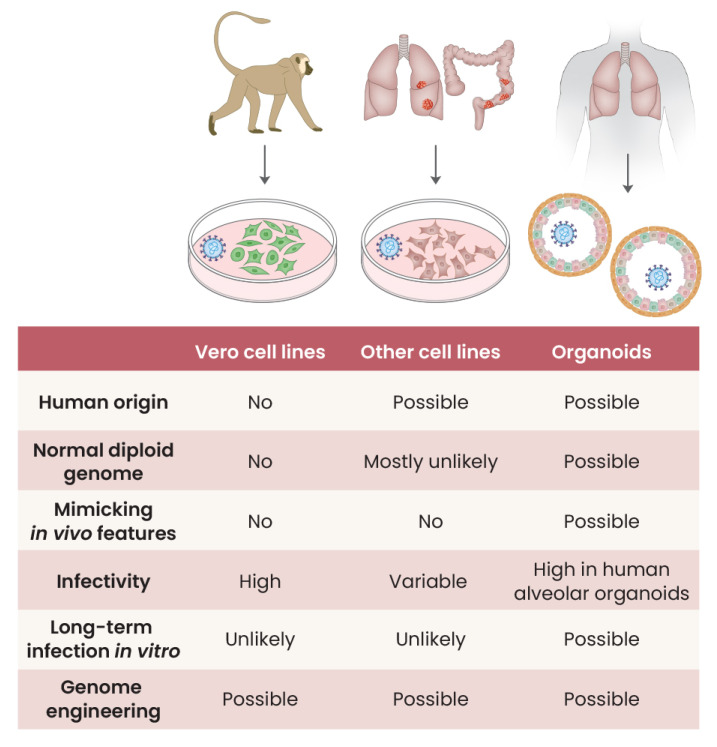
Fig. 1. In vitro models that have been used in SARS-CoV-2 infection studies and their features.
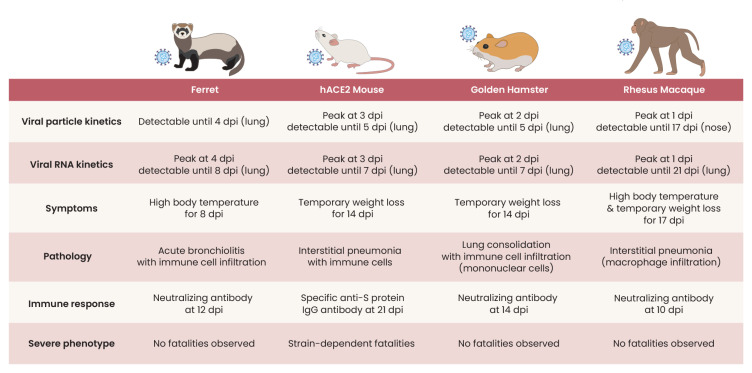
Fig. 2. In vivo models that have been used in SARS-CoV-2 infection studies and their features.
dpi, days post-infection.
IN VITRO INFECTION MODELS
Vero cells have been widely used for infection studies (Osada et al., 2014) (Fig. 1). For example, verotoxin, also known as Shiga toxin, was first identified in 1976 with an observation of cytopathies when a cell line was exposed to extracts of specific Escherichia coli strains (Konowalchuk et al., 1977). Vero cells have also been frequently used as host cells for many growing viruses, such as simian polyomavirus SV-40, measles virus, rubella virus, arbovirus, and adenovirus, to name a few (Osada et al., 2014). Vero cells were established in 1962 from the kidney tissue of an African green monkey (Chlorocebus sabaeus) (Ammerman et al., 2008). Several sublines, such as Vero 76 and Vero E6, were then obtained. Vero cells are non-tumorigenic with pseudo-diploid karyotypes and have been found to be susceptible to various viruses (Osada et al., 2014). Of note, Vero cells have a homozygous 9 Mb-long deletion on chromosome 12, resulting in the complete loss of the type I interferon genes which have particular importance as an early line of innate immune defense (Osada et al., 2014). Due to the endogenous lack of interferon genes, viruses can be grown into much higher titers in Vero cells.
Vero cells have also been used in SARS-CoV-2 studies. In one of the earliest studies of SARS-CoV-2 (Zhou et al., 2020), Vero cells were exposed to bronchoalveolar lavage fluid specimens obtained from a COVID-19 patient. At 3 days post-infection, infected cells showed clear cytopathies, which could be visualized by immunofluorescence and electron microscopy. Using the Vero cell system, viral particles of SARS-CoV-2 can be prepared for more sophisticated infection studies (Kim et al., 2020c). The landscapes of the SARS-CoV-2 transcriptome, including expressed genomic and subgenomic RNAs with the patterns of splicing and gene fusion, deletion and frameshift, were also revealed from the nanopore direct, long-read sequencing of RNAs extracted from the infected Vero cells (Kim et al., 2020a). In addition, vaccines against SARS-CoV-2 were also developed by inactivation of the SARS-CoV-2 from inoculated Vero cells (Zhang et al., 2021). Of note, Vero cells have been used for polio and rabies vaccine production for over 30 years (Montagnon et al., 1999) and now are the most widely accepted cell lines by regulatory authorities in vaccine development.
Although Vero cells have numerous advantages for amplifying many viruses, the cell lines are not suitable for investigating the interaction between viruses and host cells. Most importantly, Vero cells are not of human origin and are not normal cells as they lack type I interferon genes. The molecular changes of the cell lines after viral infection may not recapitulate the responses of human cells against the virus. In the case of SARS-CoV-2, cell-type tropism is another issue; that is, Vero cells are of kidney origin thus are not a physiological model for respiratory viruses.
To avoid species tropism and to investigate target organ-specific responses to virus infections, using human cell lines is a reasonable option. Viruses typically show different susceptibilities, infection kinetics, and cellular damages/responses according to the species and cell type of the host cells. Therefore, finding an optimal cell line that recapitulates physiological infection phenotypes is an important step for studying the infection of an emerging virus.
For the SARS-CoV-2 virus, Chu et al. (2020) conducted a screening study with a systematic comparison of 25 cell lines derived from different tissues/organs of a human and non-human species. Their cell lines included ones from the human respiratory tract (A549, Calu3, HFL), human gastrointestinal tract (Caco2), human liver (Huh7), human cervix (HeLa), human kidney (293T), human brain (U251), and human muscle (RD) and from four organs of a bat, the kidney of a porcupine, the kidney of a non-human primate (Vero E6, FRhK4, LLCMK2), the kidney of a Dog, Cat, Pig, Rabbit, and Hamster, and the fibroblast of a mouse and chicken. Of the nine human cell lines tested, Calu3 (lung adenocarcinoma) and Caco2 (colorectal adenocarcinoma) showed the most robust SARS-CoV-2 replication. Although hepatic, renal, and neuronal cell lines showed evidence of viral replication, cell lines of the pulmonary origin (A549, lung adenocarcinoma; HFL, embryonic lung fibroblasts), cervix origin (HeLa, cervical adenocarcinoma), and muscle (RD, rhabdomyosarcoma) were not robustly susceptible to the virus. Of the sixteen non-human cells, SARS-CoV-2 replicated in the renal lines of non-human primates, cat, rabbit, and pig. Cell lines from bat, porcupine, and mouse tissues were not susceptible to SARS-CoV-2.
Cell lines were used to specify the cellular receptor for viral entry during the early days of the SARS-CoV-2 pandemic. SARS-CoV-2 was used to infect the HeLa cell line, which does not express the human ACE2 receptor, but transiently expresses the human, Chinese horseshoe bat, civet, or pig ACE2 receptor (except for the mouse ACE2). It suggests that the ACE2 receptor is the cellular entry receptor for SARS-CoV-2, similar to SARS-CoV (Zhou et al., 2020). Of note, transient expression of ACE2 enabled intracellular SARS-CoV-2 entry into a cell line (BHK-21 cells), which otherwise is non-susceptible to the virus, suggesting that the virus uses ACE2 for cellular entry (Hoffmann et al., 2020). Furthermore, viral entry to the cell can be enhanced by increasing the expression levels of the bicistronic vector of ACE2 and TMPRSS2 (Wang et al., 2021).
Despite these advantages, cell lines do not recapitulate the processes of the host defense that are operative in normal human tissues. Most of the cell lines were established from human cancers, and their genomes are extensively altered from the normal diploid status (Barretina et al., 2012; Salawu et al., 2016). The phenotypes of cell lines are usually well adapted to the two-dimensional in vitro culture conditions, substantially shifted from its normal three-dimensional in vivo characteristics (Kapalczynska et al., 2018). For instance, some lung cancer cell lines (i.e., A549) are not susceptible to the SARS-CoV-2 (Chu et al., 2020). Cell lines can also show irregular responses after infection. For example, Caco-2 (colorectal adenocarcinoma) does not show substantial transcriptome changes, although the cells are readily infected by SARS-CoV-2 (Wyler et al., 2021; Youk et al., 2020). In addition, long-term impacts of viral infection are difficult to observe in cell line models because infected cell lines typically lose their viability in two-dimensional cell culture conditions. Therefore, although cell lines are classical and accessible infection models, observations and findings from these models should be interpreted cautiously.
Although cell lines are easily accessible and represent some of the organ’s responses to virus infections, characterization of a virus should be investigated by a model reflecting a normal specific cell type physiology. To achieve this, organoids are the ideal method. The organoid technique is a special cell culture system that can expand and differentiate tissue stem cells ex vivo. Typically, stem cells are growing and self-organizing into three-dimensional multicellular structures, reflecting the organotypic cell-type heterogeneity in the in vivo tissue architecture (Fujii et al., 2018). Applying the organoid system, robust long-term cultures of normal human cells are possible.
Recent advances in organoids have provided a powerful platform in infection studies by enabling long-term cultures for various types of normal human cells (Kim et al., 2020b; Ramani et al., 2018). Compared with the above-mentioned typical cell lines, cells in organoids established from normal tissues have diploid genomes (Huch et al., 2015), and their gene expression profiles are more congruent to that of normal tissues compared to cell lines. Indeed, organoids are in vivo-like mini-organs that can be managed in ex vivo conditions. By modifying the basic culture condition that was initially developed for intestinal organoids (Sato et al., 2009), multiple types of organoids from >10 human tissues, including the lung, liver, and kidney, have been generated (Kim et al., 2020b). Practically, organoids can be established from tissue stem cells directly extracted from primary adult tissues (Broutier et al., 2016). Alternatively, induced pluripotent stem cells can be used for deriving organoids by differentiating the cells with specific developmental cues (Lancaster et al., 2013).
A number of studies have used organoid models in viral diseases. For example, brain organoid models were applied to the Zika virus infection (Garcez et al., 2016), which revealed the damaging impact of the infection in human brain development. In addition, more virulent strains (Cugola et al., 2016), viral proteins underlying the brain damage (Yoon et al., 2017), and potential therapeutics (Xu et al., 2016) have also been revealed using organoid models. Other viruses, such as hepatitis B virus (Crignis et al., 2020), noroviruses (Ettayebi et al., 2016), respiratory syncytial viruses (Sachs et al., 2019), and influenza viruses (Zhou et al., 2018), also have been studied using organoid models.
The infection mechanisms of SARS-CoV-2 have also been explored using organoid models. However, studies of SARS-CoV-2 infection in human lung normal cells were challenging in the early phase of the pandemic because organoid techniques for human alveolar cells were not fully defined. Instead, renal (Monteil et al., 2020) and intestinal (Lamers et al., 2020) organoids were initially applied for the infection studies. A series of more advanced organoid models for SARS-CoV-2 infection then followed using robust human alveolar organoids established in chemically defined conditions from alveolar and induced pluripotent stem cells (Huang et al., 2020; Jacob et al., 2020; Katsura et al., 2020; Pellegrini et al., 2020; Yang et al., 2020; Youk et al., 2020). These models enabled the real-time monitoring of the very early phase of SARS-CoV-2 infection in human alveolar cells that are otherwise impossible. Diverse techniques, such as classical assays in virology, immunostaining and ultrastructural imaging, and genomic approaches, can be implemented on the organoid infection models, providing deep insights into the infection mechanism and the responses of human alveolar cells at real-time and single-cell resolutions. By screening therapeutics on the infection models, inhibitors against SARS-CoV-2 were also identified (Han et al., 2021).
Despite the many advantages, organoid models also have a few limitations. Organoid techniques are yet to be generally accessible to many researchers because these techniques have been recently developed. In typical organoids, immune cells are lacking. Thus, cellular and molecular interactions among pathogens, host cells, and immune cells could not be investigated.
IN VIVO INFECTION MODELS
The most widely used animal model for biological investigation, the mouse (Mus musculus), has no appropriate receptors to SARS-CoV-2 (Munoz-Fontela et al., 2020). The mouse ACE2 receptor does not effectively bind to the spike protein of SARS-CoV-2. Adaptation processes, such as modification of the viral sequence of the receptor-binding domain to enhance binding to the mouse ACE2 (Bao et al., 2020) or modification of the mouse to express human ACE2 (Winkler et al., 2020), are required to use mice as animal models for SARS-CoV-2 infection (Fig. 2). Compared to most mouse models with mild and reversible immunopathology of SARS-CoV-2 infection, K18-hACE2 transgenic mice expressing human ACE2 recapitulate severe COVID-19 with immunological features of severe inflammation (Winkler et al., 2020). Another human ACE2 transgenic mouse, HFH4-hACE2 in a C3B6 mouse, presented with typical interstitial pneumonia after SARS-CoV-2 infection, and pre-exposure to SARS-CoV-2 protected this model mouse from a lethal challenge (Jiang et al., 2020). Other human ACE2 transgenic mice based on the BALB/c strain or C57BL/6 were also infected by SARS-CoV-2 and resulted in active viral replication and histologic inflammatory changes (Hassan et al., 2020; Sun et al., 2020). Improvement to mimic the whole spectrum of COVID-19 using a mouse model is still ongoing.
Golden Syrian hamster (Mesocricetus auratus) has been suggested to be a useful animal model of SARS-CoV-2 infection due to its structural similarity with human ACE2 (Sia et al., 2020). Indeed, SARS-CoV-2-inoculated hamsters show clinical and histological features of COVID-19. Nevertheless, SARS-CoV-2 infected golden hamsters recovered spontaneously in 2 weeks (Sia et al., 2020). Hence, hamster models are considered as an animal model of mild COVID-19. Due to their small size and cost-effectiveness, antiviral agents and other potential therapeutic agents were tested in hamsters (Driouich et al., 2021; Kaptein et al., 2020; Rogers et al., 2020).
The ferret (Mustela putorius furo) has been used as an animal model for studying the pathogenesis of respiratory virus infection since 1933, when the natural susceptibility of the ferret to influenza virus was discovered (Maines et al., 2006; van Riel et al., 2007). There are advantages that make the ferret an ideal animal model for respiratory virus infection in human patients. Its long and narrow thorax is suitable for compartmentalization to study the differential effects of the virus in the upper and lower airway (Maher and DeStefano, 2004). In addition, the expression and glycomic status of receptors lining the bronchial mucosa of ferret are similar to that of humans, which provide a superior susceptibility to respiratory viruses than that of the mouse models (Enkirch and von Messling, 2015; Richard et al., 2020). Infection and rapid transmission of SARS-CoV-2 in the ferret model were well-demonstrated by a co-housing experiment (Kim et al., 2020d). The immune response to SARS-CoV-2 was also similar to human COVID-19 during the early phase of infection (Lee et al., 2020). Therefore, ferrets were used for evaluating the efficacy of newly developed therapeutic agents and vaccines (Cox et al., 2021; Kim et al., 2021). One of the cautions to observe when using the ferret model is that SARS-CoV-2-infected ferrets manifest the infection as an upper respiratory infection rather than severe or fatal pneumonia. Ferret models are considered as an animal model of mild COVID-19.
Non-human primates are the animal model genetically close to humans and useful for evaluating the efficacy of COVID-19 vaccines. SARS-CoV-2 infected rhesus macaque showed pulmonary infiltrates which are similar to those in chest radiographs and recapitulate the pathologic features of COVID-19 patients with moderate disease activity (Munster et al., 2020). The protective efficacies of the adenovirus vector-based vaccine ChAdOx1 nCoV-19 and mRNA vaccine against SARS-CoV-2 were tested in rhesus macaques (Corbett et al., 2020; van Doremalen et al., 2020). African green monkeys were also studied as an animal model of SARS-CoV-2 infection and its immunopathogenesis (Woolsey et al., 2021).
Unlike cell lines and organoid models, animal models recapitulate the clinical and immunological features of COVID-19. However, these animal models do not fully encompass the phenotype of severe inflammation frequently observed in human patients. Animal models showing severe inflammation with lethality need to be developed to understand the pathogenesis of severe COVID-19 patients and their potential therapeutic targets.
CONCLUDING REMARK
For the last 18 months, we have witnessed the catastrophic impact of emerging viruses in our society. Unfortunately, additional pandemic events are also likely in the near future due to the higher population density, more frequent traveling, and more opportunities to contact wild animals. To reduce the negative impact of these novel infectious diseases, we need to build up efficient and scalable experimental systems that can be used for the identification of the characteristics of viruses and their diseases, screening of therapeutics and development of vaccines and prevention strategies. Our understanding of the pros and cons of many infection models will be helpful for establishing an efficient system for defense against emerging infectious diseases.
ACKNOWLEDGMENTS
This work was supported by the Suh Kyungbae Foundation (SUHF-18010082), the National Research Foundation of Korea (Leading Researcher Program NRF-2020R1A3B2078973), and the Human Frontier Science Program Research Grant (RGY0071/2018).
Footnotes
AUTHOR CONTRIBUTIONS
T.K. and J.S.L. wrote the manuscript. Y.S.J. conceived and supervised the overall process.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ammerman N.C., Beier-Sexton M., Azad A.F. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. 2008;Appendix 4:Appendix 4E. doi: 10.1002/9780471729259.mca04es11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.K., Huch M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crignis E.D., Romal S., Carofiglio F., Moulos P., Verstegen M.M.A., Khalid M.M., Pourfarzad F., Rao S., Bazrafshan A., Koutsothanassis C., et al. Human liver organoids; a patient-derived primary model for HBV infection and related hepatocellular carcinoma. BioRxiv. 2020 doi: 10.7554/eLife.60747. https://doi.org/10.1101/568147. [DOI] [PMC free article] [PubMed]
- Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L., Guimaraes K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S., et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich J.S., Cochin M., Lingas G., Moureau G., Touret F., Petit P.R., Piorkowski G., Barthelemy K., Laprie C., Coutard B., et al. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021;12:1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkirch T., von Messling V. Ferret models of viral pathogenesis. Virology. 2015;479-480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.L., Qu L., et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Matano M., Toshimitsu K., Takano A., Mikami Y., Nishikori S., Sugimoto S., Sato T. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 2018;23:787–793.e6. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Garcez P.P., Loiola E.C., Madeiro, da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., Rehen S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J., et al. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell. 2020;27:962–973.e7. doi: 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S.R., Huang W.K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M., et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27:937–950.e9. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jary A., Leducq V., Malet I., Marot S., Klement-Frutos E., Teyssou E., Soulie C., Abdi B., Wirden M., Pourcher V., et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020;26:1560.e1–1560.e4. doi: 10.1016/j.cmi.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapalczynska M., Kolenda T., Przybyla W., Zajaczkowska M., Teresiak A., Filas V., Ibbs M., Blizniak R., Luczewski L., Lamperska K. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018;14:910–919. doi: 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S.J.F., Jacobs S., Langendries L., Seldeslachts L., Ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Barthelemy K., et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. U. S. A. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J., et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020a;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020b;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Chung Y.S., Jo H.J., Lee N.J., Kim M.S., Woo S.H., Park S., Kim J.W., Kim H.M., Han M.G. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020c;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Kim D., Yu K.M., Seo H.D., Lee S.A., Casel M.A.B., Jang S.G., Kim S., Jung W., Lai C.J., et al. Development of spike receptor-binding domain nanoparticles as a vaccine candidate against SARS-CoV-2 infection in ferrets. Bio. 2021;12:e00230–21. doi: 10.1128/mBio.00230-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020d;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konowalchuk J., Speirs J.I., Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul, van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Koh J.Y., Yi K., Kim Y.I., Park S.J., Kim E.H., Kim S.M., Park S.H., Ju Y.S., Choi Y.K., et al. Single-cell transcriptome of bronchoalveolar lavage fluid reveals dynamic change of macrophages during SARS-CoV-2 infection in ferrets. BioRxiv. 2020 doi: 10.1038/s41467-021-24807-0. https://doi.org/10.1101/2020.11.18.388280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J.A., DeStefano J. The ferret: an animal model to study influenza virus. Lab Anim. (N.Y.) 2004;33:50–53. doi: 10.1038/laban1004-50. [DOI] [PubMed] [Google Scholar]
- Maines T.R., Chen L.M., Matsuoka Y., Chen H., Rowe T., Ortin J., Falcon A., Nguyen T.H., Mai le Q., Sedyaningsih E.R., et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnon B.J., Vincent-Falquet J.C., Saluzzo J.F. Experience with vero cells at Pasteur Merieux Connaught. Dev. Biol. Stand. 1999;98:137–140. discussion 167. [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Perez-Perez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., Fontanet A., Cauchemez S., Salje H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T., Kuroda M., Hanada K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 2020;27:951–961.e5. doi: 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani S., Crawford S.E., Blutt S.E., Estes M.K. Human organoid cultures: transformative new tools for human virus studies. Curr. Opin. Virol. 2018;29:79–86. doi: 10.1016/j.coviro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., van den Brand J.M.A., Bestebroer T.M., Lexmond P., de Meulder D., Fouchier R.A.M., Lowen A.C., Herfst S. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat. Commun. 2020;11:766. doi: 10.1038/s41467-020-14626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Bottinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salawu A., Fernando M., Hughes D., Reed M.W., Woll P., Greaves C., Day C., Alhajimohammed M., Sisley K. Establishment and molecular characterisation of seven novel soft-tissue sarcoma cell lines. Br. J. Cancer. 2016;115:1058–1068. doi: 10.1038/bjc.2016.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., Liu S.S., Zhang N.N., Li X.F., Xiong R., et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Munster V.J., de Wit E., Rimmelzwaan G.F., Fouchier R.A., Osterhaus A.D., Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C., Borisevich V., Prasad A.N., Agans K.N., Deer D.J., Dobias N.S., Heymann J.C., Foster S.L., Levine C.B., Medina L., et al. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat. Immunol. 2021;22:86–98. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E., Mosbauer K., Franke V., Diag A., Gottula L.T., Arsie R., Klironomos F., Koppstein D., Honzke K., Ayoub S., et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. Science. 2021;24:102151. doi: 10.1016/j.isci.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.K., Qian X., Tcw J., Kouznetsova J., Ogden S.C., Hammack C., et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.J., Song G., Qian X., Pan J., Xu D., Rho H.S., Kim N.S., Habela C., Zheng L., Jacob F., et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell. 2017;21:349–358.e6. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J., Kim T., Evans K.V., Jeong Y.I., Hur Y., Hong S.P., Kim J.H., Yi K., Kim S.Y., Na K.J., et al. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–919.e10. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Sachs N., Chiu M.C., Wong B.H., Chu H., Poon V.K., Wang D., Zhao X., Wen L., et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6822–6827. doi: 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]