Abstract
The outbreak of coronavirus disease 2019 (COVID-19) has not only affected human health but also diverted the focus of research and derailed the world economy over the past year. Recently, vaccination against COVID-19 has begun, but further studies on effective therapeutic agents are still needed. The severity of COVID-19 is attributable to several factors such as the dysfunctional host immune response manifested by uncontrolled viral replication, type I interferon suppression, and release of impaired cytokines by the infected resident and recruited cells. Due to the evolving pathophysiology and direct involvement of the host immune system in COVID-19, the use of immune-modulating drugs is still challenging. For the use of immune-modulating drugs in severe COVID-19, it is important to balance the fight between the aggravated immune system and suppression of immune defense against the virus that causes secondary infection. In addition, the interplaying events that occur during virus–host interactions, such as activation of the host immune system, immune evasion mechanism of the virus, and manifestation of different stages of COVID-19, are disjunctive and require thorough streamlining. This review provides an update on the immunotherapeutic interventions implemented to combat COVID-19 along with the understanding of molecular aspects of the immune evasion of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which may provide opportunities to develop more effective and promising therapeutics.
Keywords: COVID-19, immune escape, pathology, SARS-CoV-2, therapeutics
INTRODUCTION
Coronaviruses (CoVs) infect a wide range of hosts, including mammals and avian species. They are, therefore, a hectic challenge not only to human health but also to livestock and the world economy. Species of human CoVs, including HCoV-OC43 and HCoV-229E, have long been known to cause minor respiratory infections such as common cold (Hamre and Procknow, 1966; McIntosh et al., 1967). Other species of CoVs, such as HCoV-HKU1 and HCoV-NL63, which cause similar seasonal infections, have recently been identified (van der Hoek et al., 2004). Moreover, the emergence of the highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 over the past 20 years has made the CoVs even more challenging.
The outbreak of SARS-CoV and MERS-CoV infections resulted in the development of prophylactic and therapeutic interventions. However, since they were not pandemic, enough attention was not paid to eradicate them and effective treatments were not devised to cope with further emergencies. This, at least in part, led to the catastrophic outbreak of coronavirus disease 2019 (COVID-19) last year, which still awaits effective intervention. The whole genome sequence of SARS-CoV-2 was publicly available after the initial assessments, which accelerated the development of vaccines and therapeutics. CoV-related research has been conducted since the emergence of the avian infectious bronchitis virus (the first CoV), and later, HCoV-OC43 and HCoV-229E, leading to a better understanding of the replication of CoVs and their interactions with the host (Schalk and Hawn, 1931). The emergence of SARS-CoV and MERS-CoV further accelerated the basic understanding of the replicative cycle of these viruses as well as their host interactions (Prompetchara et al., 2020). The development of clinically effective interventions for the latest pandemic depends on the molecular characteristics, propagation, gene ontology, pathophysiology, and host receptors interaction of the SARS-CoV-2 (Sanders et al., 2020). This review provides an update on the immunotherapeutic interventions implemented to combat COVID-19 along with the molecular understanding of SARS-CoV-2, including its replication, immune evasion, basic pathophysiology, and interactions with the host.
HOST INTERACTION WITH SARS-CoV-2
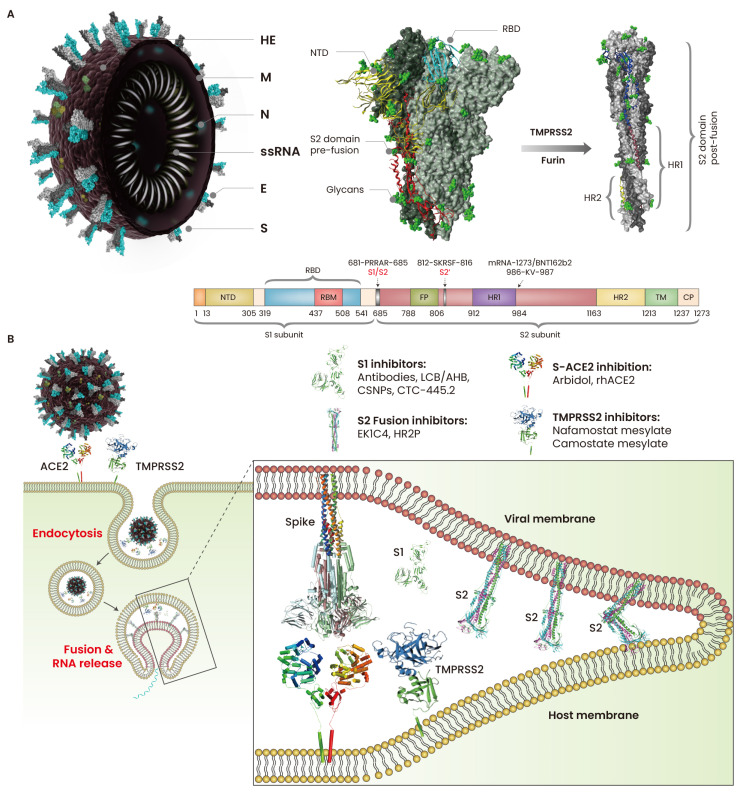
SARS-CoV-2 is a positive single-stranded RNA virus with a ~30 kb genome. SARS-CoV-2 binds to host cells by specifically recognizing angiotensin-converting enzyme 2 (ACE2) receptors via spike (S) proteins (Letko et al., 2020). ACE2 binding has been observed in a range of mammals, including dogs, pangolins, rabbits, civets, cats, rhesus macaques, pigs, and ferrets (Conceicao et al., 2020), suggesting a broad-range host opportunity for SARS-CoV-2. S is a class 1 fusion glycoprotein that is functionally divided into two subdomains, S1 and S2 (Ke et al., 2020). Typically, the S protein exists in a trimeric pre-fusion state where all three receptor-binding domains (RBDs) in the S1 subunit remain in the ACE2-inaccessible “down” conformation. The S2 domain is primed for membrane fusion by the proteolytic cleavage of the polybasic (681-PRRAR-685) site at the S1/S2 junction by furin (Shang et al., 2020) and S2’ cleavage site 812-SKRSF-816 by transmembrane protease serine 2 (TMPRSS2) (Hoffmann et al., 2020). The S2 subdomain contains two heptad repeats, HR1 and HR2, a transmembrane (TM) region, and a fusion peptide (FP) that facilitates the virus–host membrane fusion (Fig. 1A). The PRRAR site in the S1/S2 junction provides a better advantage to SARS-CoV-2 than SARS-CoV, where furin-dependent cleavage enhances the infectivity of the former (Shang et al., 2020). After S1 dissociation by S2’ and S1/S2 cleavage, the S protein acquires a post-fusion state, releases the structural constraints on the FP and other motifs of S2, and then undergoes a cascade of conformational changes. The FP at the top of the elongated, coiled-coil S2 anchor the host and viral membrane, and the conformational changes in the HR2 motifs bring the two membranes closer, facilitating fusion pore formation and viral entry (Fig. 1B). More recently, the virus entry cofactor neurophilin-1 (NRP1) has been reported to facilitate SARS-CoV-2 entry. Proteolytic cleavage of S at the S1/S2 site generates the C-terminal-end sequence TQTNSPRRAROH at the S2 domain, which binds to NRP1 and enhances cell entry (Cantuti-Castelvetri et al., 2020).
Fig. 1. Cell attachment and fusion of SARS-CoV-2.
(A) SARS-CoV-2 particles are spherical (60 to 140 nm in diameter) with spike proteins (S) protruding from the surface of virion. The S is densely glycosylated by O- and N-linked glycans (green color) that potentially mask the immunogenic nature of the S protein during infection. The S1 domain of S anchors the virus to the host cells, and the S2 domain of S establishes membrane fusion. TMPRSS2 and furin cleave the S1 domain of S, thereby priming the S2 domain for membrane fusion. (B) The virus is engulfed through endocytosis or the direct membrane fusion at the cell surface. After S1 cleavage, S2 domain undergoes conformational re-arrangement and exposes fusion peptides (FPs) for anchoring into the host cell membrane; the HR1 and HR2 motifs fold and bring the viral and host membrane close for fusion. HE, hemagglutinin-esterase; M, membrane protein; N, nucleocapsid; E, envelope; NTD, N-terminal domain; RBD, receptor-binding domain; HR, heptad repeat; RBM, receptor-binding motif; TM, transmembrane; CP, cytoplasmic domain; TMPRSS2, trans-membrane protease, serine 2.
The indispensability of S–ACE2 binding and membrane fusion makes these processes crucial targets for vaccines and drugs. The transition of S proteins from pre-fusion to post-fusion is facilitated by the PRRAR motif and the hinge region (985-DKVE-988) between the N-terminus of HR1 and the central helix of the S2 domain. The S-2P antigen was created by mutating the furin-recognizing S1/S2 site RRAR into GSAS and 986-KV-987 into two proline residues (PP) (Jackson et al., 2020; Kuo et al., 2020). Therefore, many researchers are interested in implementing the S-2P antigen in the development of mRNA-based vaccines such as BNT162b2 (Pfizer, USA/BioNTech, Germany) and mRNA-1273 (Moderna, USA) (Jackson et al., 2020; Kuo et al., 2020). In addition, many other SARS-CoV-2 vaccines, including inactivated, live attenuated, recombinant S, recombinant RBD, mRNA, DNA, and virus vector-based vaccines, are under different phases of clinical trials (Krammer, 2020). However, host adaptability of SARS-CoV-2 and fast mutation of the S protein are alarming. A SARS-CoV-2 mutant, D614G, was initially identified in March 2020; within three months, this mutant was found in nearly all COVID-19 patients worldwide (Korber et al., 2020). D614 has been associated with the increased infectivity in human airway tissues and the enhanced viral replication in human lung epithelial cells. The His69 and Val70 deletion at the N-terminal of the S protein in B.1.1.7 strain causes a conformational change that enhances the viral infectivity twofold, compared to the D514G strain (Brookman et al., 2021). Deletion of amino acids 242–244 in the B.1.351 strain has been associated with reduced binding ability with S-neutralizing antibodies (McCarthy et al., 2021). Monitoring the newly arising mutants is therefore necessary to help contain the spread of the mutant viruses, track their evolution, and inform public health practices such as vaccine development and diagnostics.
Cell entry blockers
Over the last few months, we observed that SARS-CoV-2 is adapting to the host environment by rapidly mutating its S protein. The complementarity-determining regions of antibodies specifically recognize a conformational epitope on the antigen, where a single mutation in the hotspot residues could abrogate antibody-mediated S neutralization (Shah et al., 2020). This S-dependent host adaptability of the virus can put the whole development of neutralizing antibodies at risk. REGN-COV2 is a cocktail of two monoclonal antibodies (mAbs) of imdevimab and casirivimab, which have been designed to prevent the escape-mutants of SARS-CoV-2. REGN-COV2 has been evaluated for its safety, efficacy, and tolerability in phase I and II trials in adults (NCT04426695) and is currently undergoing phase III Randomized Evaluation of COVID-19 Therapy (RECOVERY) trial. The cocktail has been found to be effective in ambulatory adult and pediatric COVID-19 patients (NCT04425629) (Weinreich et al., 2021). Although the individual antibodies in the REGN-COV2 cocktail bind to distinct and non-overlapping epitopes, a recent study has shown that a single amino acid mutation in the RBD could escape the antibody cocktail (Starr et al., 2021). These mutants are already presented in the circulating SARS-CoV-2 strains and are also found in the patients with persistent infection who have been treated with REGN-COV2 (Starr et al., 2021). Another S-neutralizing mAb, CT-P59 (Celltrion, Korea), has been shown to be safe for healthy subjects (NCT04525079) and is currently under clinical evaluation for the COVID-19 patients with mild to moderate symptoms (NCT04602000) (Kim et al., 2021). In addition, dozens of antibody-based therapeutics are under clinical investigation, including convalescent plasma, human mAbs VIR-7831 and LY-CoV555, and human polyclonal neutralizing antibody SAB-185 (Jiang et al., 2020; Weinreich et al., 2021; Zhou et al., 2021).
In addition, peptide biologics that block the S–ACE2 binding or S2–cell fusion may have great therapeutic potential, because all the SARS-CoV-2 variants utilize the same receptor and the fact that vaccines are only effective as a prophylactic agent for uninfected personnel. Recently, we identified short and structurally stable peptides those can abolish the S1–hACE2 interaction in vitro (data not published). Other groups implemented similar strategies and reported mini-proteins that effectively blocked S–ACE2 binding in pre-clinical investigations (Cao et al., 2020; Linsky et al., 2020).
Fusion blockers
The small peptide EK1 (OC43-HR2P) derived from HR2 motif has a potential to inhibit the fusion of CoVs by targeting the HR1 motif in the S2 subunit (Xia et al., 2019). EK1C4, derived from EK1, has shown to inhibit the fusion of the SARS-CoV-2 S-expressing pseudovirus as well as other lethal CoVs, including MERS-CoV and SARS-CoV (Xia et al., 2020b). On the contrary, S-neutralizing antibodies have been shown to be less effective against the fusion of SARS-CoV-2 S proteins (Shah et al., 2020; Tai et al., 2020), suggesting that the S-derived fusion blockers and ACE2-derived decoys eventually blocked the S–ACE2 interaction. Thymosin α1 (Tα1), a naturally occurring peptide, is not a fusion blocker, but can trigger the lymphocyte maturation and enhance the immune response by activating T cells. Administration of Tα1 can reduce the mortality of COVID-19 patients (Liu et al., 2020b). These results strongly support that the peptide-based biologics have great therapeutic potential for COVID-19.
THE ROLE OF REPLICATION AND TRANSCRIPTION COMPLEX IN SARS-CoV-2
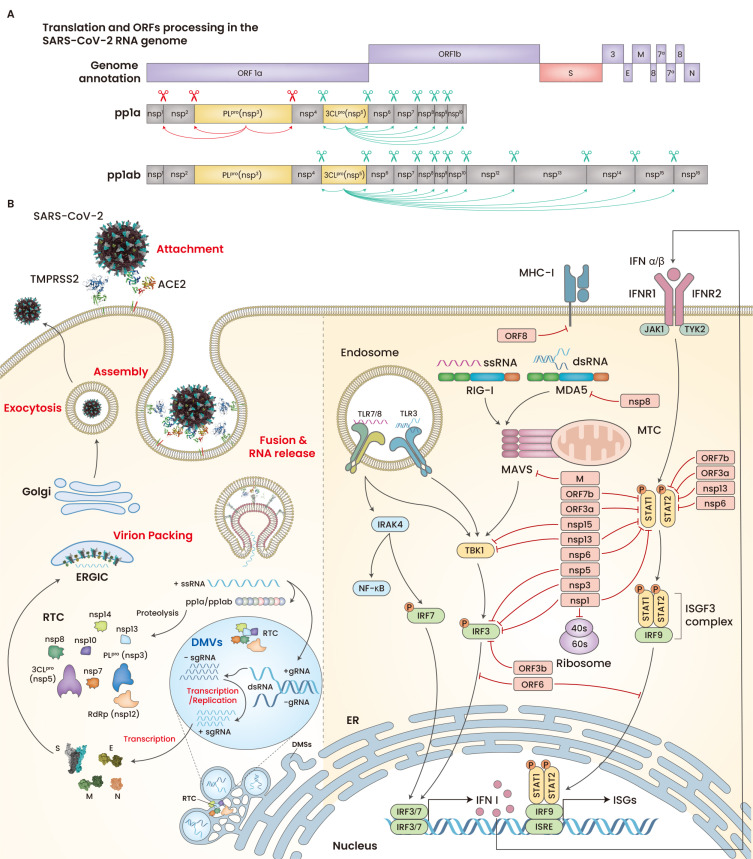
After membrane fusion and subsequent RNA release into the cytoplasm, SARS-CoV-2 translates non-structural proteins (nsps) that make the replication and transcription complex (RTC) and initiate the biogenesis of replication organelles (ROs), including double membrane vesicles (DMVs) and double membrane spherules (DMSs) arising from the endoplasmic reticulum (ER) in host cells (Yan et al., 2020). Although not fully elucidated yet, CoVs utilize nsp3, nsp4, and nsp6 to generate ROs, which conceal the double-stranded RNA intermediates from host cell immune sensors and provide building block molecules during viral RNA replication/transcription (Scutigliani and Kikkert, 2017; Snijder et al., 2020). During the initial course of replication, SARS-CoV-2 produces the polyproteins pp1a and pp1ab from ORF1a and ORF1b, respectively (Klein et al., 2020; Yan et al., 2020). These polyproteins produce 16 nsps by the enzymatic cleavage of papain-like protease (PLpro) and chymotrypsin-like protease (3CLpro), which are self-cleaved from nsp3 and nsp5, respectively (Fig. 2A). One-third of the 3’ end of the viral genome encodes five structural proteins and a similar number of accessory proteins, including 3a, 3b 6, 7a, 7b, 8, and 9b (Gordon et al., 2020; Yan et al., 2020). Replication and transcription occur within the ROs and the translated structural proteins are translocated to the ER-to-Golgi intermediate compartments (ERGICs), where they are packed with the RNA-producing progeny virions (Fig. 2B, left).
Fig. 2. Genome annotation, polyprotein processing, stepwise replication, and host immune evasion mechanism of SARS-CoV-2.
(A) The ORF1a and ORF1b are transcribed into pp1a and pp1ab; the latter is transcribed as the result of –1 ribosomal frameshift at the overlapping point between IRF1a and ORF1b. Nsp3 (PLpro) cleaves pp1a at three points (red arrows), and Nsp5 (3CLpro) releases nsp4–nsp16 by cleaving pp1a and pp1ab (green arrows). (B) Following membrane fusion, the viral RTC members generated by pp1a and pp1ab are involved in the biogenesis of ROs (left). The SARS-CoV-2 proteins antagonizes IFNs (red lines) (right). ORF, open reading frames; S, spike; E, envelope; M, membrane; N, nucleocapsid; ssRNA, single strand RNA; sgRNA, subgenomic RNA; gRNA, genomic RNA; dsRNA, double strand RNA.
Many SARS-CoV-2 proteins have been proven to counter the immune response by interferons (IFNs), such as nsp1, nsp3, nsp8, nsp12, nsp13, nsp14, ORF3, ORF6, ORF8, and M proteins (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Konno et al., 2020; Lei et al., 2020; Park, 2020; Xia et al., 2020a; Yang et al., 2020). For example, nsp6 and nsp13 modulate the effect of TBK1, and nsp8 binds MDA5, thereby suppressing the type I interferon (IFN-I) response (Xia et al., 2020a; Yang et al., 2020). SARS-CoV-2 ORF8 directly binds and downregulates the expression of MHC-I molecules, whereas PLpro and 3CLpro cleave IRF3, TAB1, and NLRP12 (Moustaqil et al., 2021; Park, 2020) (Fig. 2B, right). Moreover, SARS-CoV-2, compared to SARS-CoV, has been found to have the same or better capacity to counter the IFN response (Blanco-Melo et al., 2020; Hadjadj et al., 2020). SARS-CoV-2 nsp1, nsp6, and ORF3b can suppress IFN-I signaling more efficiently than SARS-CoV (Konno et al., 2020). This efficient anti-IFN ability of SARS-CoV-2 causes the delayed IFN-I responses in severe COVID-19 infection (Acharya et al., 2020).
RTC inhibitors
Three key enzymes, RNA-dependent RNA polymerase (RdRp), PLpro, and 3CLpro, make RTC a promising therapeutic target that can stop viral replication. RdRp is the core of the SARS-CoV-2 RTC and is involved in both replication and transcription, sharing ~95% similarity with that of SARS-CoV. Hence, anti-SARS-CoV RdRp drugs (remdesivir and favipiravir) have been evaluated for COVID-19 treatment. Remdesivir has been approved by the U.S. Food and Drug Administration (FDA) for the treatment of adult and pediatric COVID-19 patients, owing to its efficacy in animal models of COVID-19 (Williamson et al., 2020) and in vitro viral inhibitory potential (Wang et al., 2020b). Remdesivir was shown to have some benefits in hospitalized patients who require supplemental oxygen; however, no clinical benefits were observed in patients who were on high oxygen supply and noninvasive and mechanical ventilation (Beigel et al., 2020; Williamson et al., 2020). In a multicenter randomized, blind trial conducted in China, the use of remdesivir showed no clinical benefits in mortality and viral clearance in COVID-19 patients (Wang et al., 2020d), although other studies reported better clinical outcomes (Spinner et al., 2020). In addition, favipiravir has been reported to eliminate the viral load, showing significant patient recovery (NCT04600999 and NCT04359615). Other members of the RTC, 3CLpro, or Mpro are equally important in combating viral propagation. For example, Mpro enzymatically releases other members of the RTC and structural entities of the virus (Kim et al., 2020). Researchers are now developing effective anti-3CLpro drugs by leveraging the substrate-specific knowledge of enzymes (Ma et al., 2020; Sacco et al., 2020).
IMMUNE RESPONSES IN COVID-19
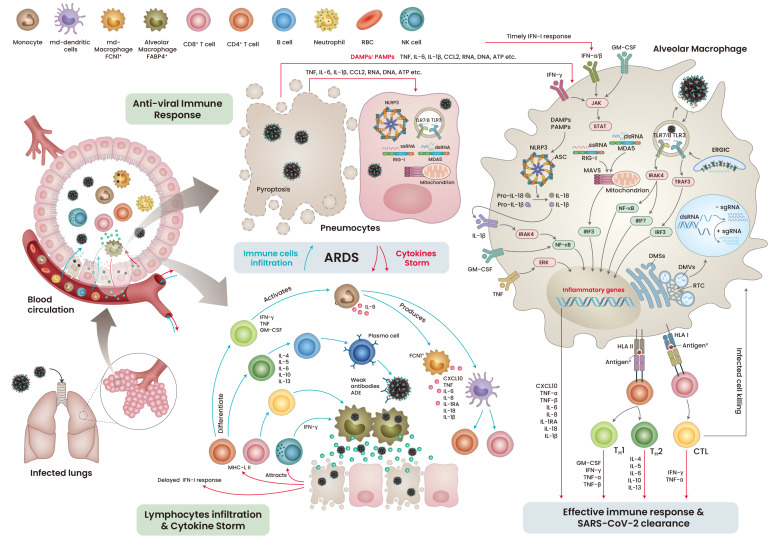
Unlike SARS-CoV, which infects the lower respiratory tract, SARS-CoV-2 primarily targets the ACE2-expressing respiratory epithelial cells in the upper respiratory tract and later infects type I and type II pneumocytes and alveolar macrophages (Zeng et al., 2020). The extensive replication of SARS-CoV-2 leads to pyroptosis of the infected cells, releasing pro-inflammatory cytokines, danger-associated molecular patterns (DAMPs), and pathogen-associated molecular patterns (PAMPs) (Yap et al., 2020). These factors are detected by neighboring cells through pattern recognition receptors (PRRs), which further release a wave of pro-inflammatory cytokines, recruiting monocytes and T-lymphocytes to the infection site (Merad and Martin, 2020). Antigen presenting cells (APCs) display viral antigenic components to the CD4+ and CD8+ T cells, which further boost the viral specific humoral and T cell responses (Chen and John Wherry, 2020; Cox and Brokstad, 2020). These cellular and humoral immune responses are sufficient to fight and eliminate the infection (Fig. 3, right). However, like other viruses, SARS-CoV-2 utilizes its immune-masking strategies to escape the host immune response, resulting in severe immune complications.
Fig. 3. The chronological pathophysiological events during mild and severe COVID-19 infection.
RBC, red blood cell; NK cells, natural killer cells; TNF, tumor necrosis factor; IL, interleukins; CCL, chemokine (C-C motif) ligand; IFN, interferon; GM-CSF, granulocyte-macrophage colony-stimulating factor; ssRNA, single strand RNA; dsRNA, double strand RNA; RIG-I, retinoic acid-inducible gene I; MDA5, melanoma differentiation-associated protein 5; NLRP3, NLR family pyrin domain containing 3; DAMPs, danger/damage associated molecular pattern; PAMP, pathogen associated molecular pattern; MAVS, mitochondrial antiviral signaling protein; TLR, toll-like receptor; ERGIC, endoplasmic-reticulum–Golgi intermediate compartment; DMV, double membrane vesicle; DMS, double membrane spherules; RTC, replication and transcription complex; ADE, antibody-dependent disease enchantment; MHC, major histocompatibility complex; CXCL, chemokine (C-X-C motif) ligand; HLA, human leukocyte antigen; ASC, apoptosis-associated speck-like protein containing a CARD; CTL, cytotoxic lymphocytes.
As seen in SARS-CoV infection, a considerable loss of ACE2 function by SARS-CoV-2 infection can dysregulate the renin-angiotensin system, leading to an enhanced vascular permeability and an imbalance in the electrolyte and immune cell homeostasis (Kuba et al., 2005). This could partly be the reason for the higher mortality in older COVID-19 patients, as they are more susceptible to fluctuations in blood homeostasis (Bonanad et al., 2020). In about 20% of the infected patients with acute respiratory distress syndrome (ARDS), the hype in the cytokine profile was associated with an increase in levels of tumor necrosis factor (TNF), interleukin (IL)-6 and IL-7, chemokines (CCL2 and CCL3), and CXC motif chemokine ligand 10 (CXCL10). The patients with severe COVID-19 requiring intensive care exhibited higher levels of cytokines such as TNF, IL-2, IL-7, IL-10, and IL-1β (Huang et al., 2020). During active infection, T lymphocytes, particularly CD4+ T cells, were found to accumulate at infection site, which may be due to the infected APCs (Song et al., 2020). The activated TH1 cells released IFN-γ, TNF, and granulocyte-macrophage colony-stimulating factor (GM-CSF), which increased the levels of CD14+ and CD16+ monocytes at the infection sites in the patients with severe COVID-19 (Zhou et al., 2020).
The severity of COVID-19 is the result of uncontrolled viral replication, which leads to cytolysis and the release of aggravated cytokines. Typically, viral infection is cleared by cellular and humoral immune responses (Tay et al., 2020); however, uncontrolled rapid replication of the virus may trigger aggravated immune responses and paradoxically delay the IFN-I immune response, leading to ARDS approximately 8 to 9 days after the onset of symptoms (Grasselli et al., 2020). The patient’s condition can be further worsened by weak neutralizing antibodies against SARS-CoV-2 antigens, which leads to antibody-dependent enhancement of disease. The severity of the disease is reinforced by the infiltration of lymphocytes and monocytes into the inflamed lungs, resulting in lung collapse and death (Fig. 3, left).
IMMUNOMODULATORS
Although tremendous efforts have been made to collect insights into the SARS-CoV-2 itself and the demographics and pathophysiological manifestation of COVID-19, effective drugs that could uproot the disease have not made their way to clinical evaluation yet. Thus, the drugs in current clinical consideration are for those indications that overlap with the symptoms of COVID-19. In particular, the interplay between the dysregulated host immune response and the severity of COVID-19 has led to the rapid worldwide clinical evaluation of immunomodulators. The following immune-modulating drugs are currently under therapeutic evaluation for COVID-19.
Chloroquine (CQ) and hydroxychloroquine (HCQ)
Though known as antimalarial drugs, HCQ and CQ are widely used in the treatment of autoimmune disorders such as systemic lupus erythematosus or rheumatoid arthritis. CQ and its less toxic derivative HCQ have been repurposed, showing their therapeutic efficacy against SARS-CoV-2 in vitro (Wang et al., 2020b). In particular, HCQ was thought to be more efficient than CQ (Liu et al., 2020a); however, when tested at a clinically approved dosage, HCQ was not efficient in vitro (Kang et al., 2020). Many clinical trials have confirmed the ineffectiveness of HCQ in the treatment of COVID-19 (Rosenberg et al., 2020; Skipper et al., 2020); therefore, the FDA revoked the use of both CQ and HCQ in June 2020.
Dexamethasone and other corticosteroids
Dexamethasone was used in a RECOVERY trial, which revealed a substantial one-third decrease in mortality in patients on ventilators and one-fifth in patients on oxygen supply (RECOVERY Collaborative Group, 2021; WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group, 2020). Thus, the FDA has added dexamethasone sodium phosphate to its list of drugs that have been temporarily compounded during the COVID-19 pandemic to prevent supply disruption. Additionally, the World Health Organization has recommended corticosteroids, including hydrocortisone and prednisone, for the treatment of severe COVID-19 patients. There are some differences in the clinical efficacy of dexamethasone in patients with severe disease and in those who do not require air support. Severe patients who received dexamethasone for more than 7 days after the onset of symptoms were benefited, whereas the patients without severe inflammation did not respond well to the treatment, resulting in an increased mortality rate. The systemic use of corticosteroids can impair the clearance of the virus in SARS-CoV- and MERS-CoV-infected patients (Arabi et al., 2018; Lee et al., 2004); therefore, the use of dexamethasone in mildly infected patients may decrease the antiviral response and worsen the severity and mortality of the disease.
IFN
The induction of IFN-I response is vital for stimulating antiviral IFN-stimulating genes (ISGs) in both autocrine and paracrine modes. However, the results regarding the role of IFN-I and IFN-I response in the development of severe COVID-19 complications are contradictory. Peripheral blood evaluation of COVID-19 patients of varying severity has shown that the IFN-I response is significantly impaired in patients with severe COVID-19 (Hadjadj et al., 2020). In contrast, the transcriptome data of peripheral blood mononuclear cells and single-cell RNA-sequencing data of bronchoalveolar lavage fluid from COVID-19 patients showed the increased expression of various types of ISGs, pro-inflammatory cytokines, and chemokines (Wilk et al., 2020). Another study has demonstrated that the hyper-IFN-I response is associated with the TNF/IL-1β levels in severe COVID-19 but not in mild COVID-19 (Lee et al., 2020). IFN-I abolishes TNF tolerability in monocytes, macrophages, and animal models to respond to toll-like receptors (TLR) signals (Israelow et al., 2020). In patients with COVID-19, contradictory IFN-I responses can be attributed to the severity of the disease, the number of immune cells combating viral invasion and overcoming the inflammatory state, and the type and time of sample collection. This notion was addressed by a recent retrospective cohort study of 446 COVID-19 patients in China, wherein the early administration of IFN-α2b with or without umifenovir reduced mortality and accelerated patient recovery (Wang et al., 2020c). Moreover, IFN-β-based trials have shown promising results using a combination with the antiviral drugs (NCT04276688 and NCT04291729) or HCQ (NCT04350281) or a combination of an antiviral drug and HCQ (NCT04291729) (Bosi et al., 2020; Hung et al., 2020; Wang et al., 2020a). Furthermore, IFN-III-based clinical trials are ongoing to study how to overcome IFN-induced inflammatory effects and elicit effective antiviral responses (NCT04354259, NCT04343976, NCT04344600, and NCT04388709).
IL-6 inhibitors
Since the pathophysiology of COVID-19 overlaps with most of the complications in rheumatic diseases (Fajgenbaum and June, 2020), targeting the inflammatory pathways related to rheumatic diseases has been reported with some promising results in COVID-19 therapeutics. IL-6 is released at a considerably higher level in severe COVID-19 patients than in mild infection (Narayan et al., 2021). Inhibition of the IL-6 receptor (IL-6R) has been effective in relieving chronic rhinosinusitis (CRS) (Tanaka et al., 2016). Thus, the FDA has approved two classes of the IL-6 pathway inhibitors: one that blocks the IL-6R (e.g., tocilizumab and sarilumab) and others that block IL-6 itself (e.g., siltuximab). Tocilizumab has been approved by the FDA for CRS and rheumatologic disorders (Le et al., 2018). The administration of tocilizumab along with an off-label anti-retroviral protease inhibitor was effective against COVID-19 (Sciascia et al., 2020). Siltuximab, a recombinant mAb that binds to both soluble and membrane-bound IL-6, has shown promising clinical outcomes in patients with ARDS (NCT04322188). The combined use of siltuximab with anakinra and tocilizumab (NCT04330638) or normal saline (NCT04616586) is under evaluation, although the results have not been published yet. In contrast, other studies have shown that tocilizumab and sarilumab are not effective against COVID-19 (NCT04320615, NCT04327388, and NCT04315298).
IL-1R blockers
IL-1β is secreted by mature caspase-1 through the TLR-activated NLRP3 pathway loop, which further aggravates cytokine induction through IL-1R in an autocrine and paracrine manner. Anakinra, a recombinant IL-1R blocker used for the treatment of rheumatoid arthritis and cryopyrin-associated periodic syndromes (Cavalli and Dinarello, 2018), is currently being investigated in more than 10 clinical trials. Few studies have demonstrated that the administration of anakinra is safe and associated with the clinical improvement of COVID-19 (Cavalli et al., 2020; Huet et al., 2020).
Bruton’s tyrosine kinase (BTK) inhibitors
BTK relays the signaling of TLRs, IL-1R, CD19, BCR, CXCR4, and Fcγ-R1 (Pal Singh et al., 2018; Sharma et al., 2009), thus prompting studies on the use of BTK inhibitors against excessive host inflammation in severe COVID-19. The use of BTK inhibitors was further encouraged by the overlapping cytokine profiles of B-cell malignancies and chronic graft versus host disease (Dubovsky et al., 2014). The use of acalabrutinib was associated with the normalization of IL-6 and C-reactive protein (Roschewski et al., 2020). Ibrutinib is also currently undergoing two clinical trials (NCT04375397 and NCT04439006), and acalabrutinib has completed phase I and II randomized clinical trials (NCT04564040 and NCT04380688).
CONCLUSIONS
With the accumulation of comprehensive clinical data, the spectrum of effective therapeutics for the treatment of COVID-19 is rapidly growing and evolving, including therapeutics targeting cell entry, fusion, RTC production, and immune responses. In this review, we provide a concise update on the current therapeutic interventions for COVID-19 with perspectives on their molecular mechanistic targets (Table 1).
Table 1.
Molecular targets and their therapeutic interventions for COVID-19
| Category | Target | Drug/biologic | Trials ID | Phase |
|---|---|---|---|---|
| Cell entry blockers | ACE | Arbidol | NCT04260594 | Phase 4 |
| rhACE2 APN01 | NCT04335136 | Phase 2 | ||
| rbACE2 | NCT04375046 | Phase 1 | ||
| Spike | REGN-COV2 (REGN10933 + REGN10987) Placebo |
NCT04425629 NCT04426695 NCT04452318 |
Phase 2 Phase 3 |
|
| XAV-19 | NCT04453384 | Phase 2 | ||
| CT-P59 Placebo |
NCT04602000 | Phase 2 | ||
| SAB-185 Normal Saline |
NCT04469179 NCT04468958 |
Phase 1 Phase 2 |
||
| VIR-7831 Placebo |
NCT04545060 | Phase 2 Phase 3 |
||
| VIR-7831 BRII-196/BRII-198 LY3819253 Remdesivir Placebo |
NCT04501978 | Phase 3 | ||
| LY3819253 LY3832479 Placebo |
NCT04427501 NCT04497987 NCT04634409 |
Phase 2 Phase 3 Phase 2 Phase 2 |
||
| Fusion blockers | TMPRSS2 | Nafamostat mesilate |
NCT04390594 NCT04418128 |
Phase 3 Phase 2 Phase 3 |
| Nafamostat mesilate Placebo |
NCT04352400 | Phase 2 Phase 3 |
||
| Nafamostat mesilate TD139 Standard care |
NCT04473053 | Phase 2 Phase 3 |
||
| RTC inhibitors | RdRp | Remdesivir Standard care |
NCT04292899 NCT04292730 |
Phase 3 |
| Remdesivir Placebo |
NCT04280705 | Phase 3 | ||
| Favipiravir Standard care |
NCT04542694 NCT04600999 |
Phase 3 | ||
| Favipiravir Hydroxychloroquine |
NCT04359615 | Phase 4 | ||
| IFN | IFNR | Interferon β-1b | NCT04465695 | Phase 2 |
| IFNR/3CLpro/immune system | Hydroxychloroquine Lopinavir/ritonavir Interferon β-1a Interferon β-1b |
NCT04343768 | Phase 2 | |
| IFNR/RdRp | Interferon β-1b Remdesivir |
NCT04647695 | Phase 2 | |
| IFNR/RdRp | Lopinavir/ritonavir Ribavirin Interferon β-1b |
NCT04276688 | Phase 2 Phase 3 |
|
| Interferon α-2b Rintatolimod |
NCT04379518 | Phase 1 Phase 2 |
||
| Interferon α-1b Thymosin α1 |
NCT04320238 | Phase 3 | ||
| Peginterferon λ-1A Placebo |
NCT04354259 NCT04343976 NCT04344600 NCT04388709 |
Phase 2 | ||
| Cytokines inhibitors | IL-6R | Tocilizumab Placebos |
NCT04356937 | Phase 3 |
| Tocilizumab | NCT04331795 | Phase 2 | ||
| Tocilizumab | NCT04363736 | Phase 2 | ||
| Tocilizumab Placebos |
NCT04320615 | Phase 3 | ||
| Tocilizumab Anakinra |
NCT04339712 | Phase 2 | ||
| Sarilumab | NCT04327388 | Phase 3 | ||
| Sarilumab Placebos |
NCT04315298 | Phase 2 Phase 3 |
||
| Sarilumab | NCT04661527 | Phase 2 | ||
| Sarilumab | NCT04359901 | Phase 2 | ||
| Siltuximab | NCT04329650 | Phase 2 | ||
| Siltuximab | NCT04330638 | Phase 3 | ||
| IL-1 | Anakinra Tocilizumab |
NCT04339712 | Phase 2 | |
| Anakinra | NCT04443881 | Phase 2 Phase 3 |
||
| Anakinra Placebos |
NCT04680949 | Phase 3 | ||
| Anakinra Tocilizumab Standard care |
NCT04412291 | Phase 2 | ||
| Anakinra | NCT04357366 | Phase 2 | ||
| Anakinra Standard care |
NCT04643678 | Phase 2 | ||
| Anakinra Normal saline |
NCT04362111 | Phase 3 | ||
| Anakinra Tocilizumab Siltuximab |
NCT04330638 | Phase 3 | ||
| BTK | Anakinra | NCT04148430 | Phase 2 | |
| Acalabrutinib |
NCT04564040 NCT04380688 |
Phase 1 Phase 2 |
||
| Ibrutinib |
NCT04375397 NCT04665115 |
Phase 2 | ||
| Corticosteroids | Immune suppressor | Dexamethasone Hydroxychloroquine |
NCT04347980 | Phase 3 |
| Dexamethasone (IV, nasal) | NCT04513184 | Phase 2 | ||
| Dexamethasone | NCT04509973 | Phase 3 | ||
| Dexamethasone Methylprednisolone |
NCT04603729 | Phase 3 | ||
| Dexamethasone Methylprednisolone |
NCT04499313 | Phase 3 | ||
| Dexamethasone | NCT04395105 | Phase 3 | ||
| Dexamethasone Remdesivir Baricitinib Placebo |
NCT04640168 | Phase 3 | ||
| Dexamethasone (early) Dexamethasone (late) |
NCT04530409 | Phase 4 | ||
| NA-831 NA-831 & atazanavir NA-831 & dexamethasone Atazanavir & dexamethasone |
NCT04452565 | Phase 2 Phase 3 |
||
| Corticosteroids | Dexamethasone Tocilizumab |
NCT04476979 | Phase 2 | |
| CQ and HCQ | TLRs/NLRs | HCQ | 40 trials are withdrawn or terminated, 52 are completed, and ~100 studies are active | |
| CQ | 3 trials are terminated and few are active |
The pathogenesis of COVID-19 is driven by two main processes; the manifestation of disease is primarily driven by the rapid replication of the virus during the early course of infection, whereas the severity of the disease is driven by the exacerbated antiviral inflammatory response, which leads to tissue and organ damage. With respect to these, the use of antiviral therapies and IFNs in the early course of infection can provide optimal benefits. Antibody-based therapeutics against the viral S or other entities that can neutralize the virus will likely have the greatest benefit in the early stage of infection before the host immune system responds to the virus. During the late stage, the use of anti-inflammatory drugs and corticosteroids has more benefits, whereas their use in the early course of infection can interfere with the antiviral immune system. In the absence of specific and effective drugs that can block SARS-CoV-2 replication, immunomodulatory drugs can be used at this stage to save lives. Therefore, it is important to understand the cytokine profile, stage of the disease, comorbid pathological conditions, and other important immune-related factors before administering immunomodulatory drugs. Undoubtedly, future efforts to deepen our understanding of the molecular mechanisms of SARS-CoV-2 will provide opportunities to develop more effective and promising therapeutics.
ACKNOWLEDGMENTS
This study was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT) (NRF-2019R1A5A2026045 and 2017M3C9A6047620), Republic of Korea.
Footnotes
AUTHOR CONTRIBUTIONS
M.S. and H.G.W. contributed to the overall study design and wrote the manuscript. H.G.W. directed the study.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., Jose J., Pinto R., Al-Omari A., Kharaba A., et al. Corticosteroid therapy for critically ill patients with Middle East Respiratory Syndrome. Am. J. Respir. Crit. Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanad C., Garcia-Blas S., Tarazona-Santabalbina F., Sanchis J., Bertomeu-Gonzalez V., Facila L., Ariza A., Nunez J., Cordero A. The effect of age on mortality in patients with COVID-19: a meta-analysis with 611,583 subjects. J. Am. Med. Dir. Assoc. 2020;21:915–918. doi: 10.1016/j.jamda.2020.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi E., Bosi C., Rovere Querini P., Mancini N., Calori G., Ruggeri A., Canzonieri C., Callegaro L., Clementi M., De Cobelli F., et al. Interferon beta-1a (IFNbeta-1a) in COVID-19 patients (INTERCOP): study protocol for a randomized controlled trial. Trials. 2020;21:939. doi: 10.1186/s13063-020-04864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman S., Cook J., Zucherman M., Broughton S., Harman K., Gupta A. Effect of the new SARS-CoV-2 variant B.1.1.7 on children and young people. Lancet Child Adolesc. Health. 2021;5:e9–e10. doi: 10.1016/S2352-4642(21)00030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.J., et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., Dinarello C.A. Anakinra therapy for non-cancer inflammatory diseases. Front. Pharmacol. 2018;9:1157. doi: 10.3389/fphar.2018.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao C., Thakur N., Human S., Kelly J.T., Logan L., Bialy D., Bhat S., Stevenson-Leggett P., Zagrajek A.K., Hollinghurst P., et al. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 2020;18:e3001016. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.J., Brokstad K.A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020;20:581–582. doi: 10.1038/s41577-020-00436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovsky J.A., Flynn R., Du J., Harrington B.K., Zhong Y., Kaffenberger B., Yang C., Towns W.H., Lehman A., Johnson A.J., et al. Ibrutinib treatment ameliorates murine chronic graft-versus-host disease. J. Clin. Invest. 2014;124:4867–4876. doi: 10.1172/JCI75328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum D.C., June C.H. Cytokine storm. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G., Tonetti T., Protti A., Langer T., Girardis M., Bellani G., Laffey J., Carrafiello G., Carsana L., Rizzuto C., et al. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir. Med. 2020;8:1201–1208. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J.M., Bezie Y., Laplanche S., et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. SSRN. 2020 doi: 10.1084/jem.20201241. https://doi.org/10.2139/ssrn.3628297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Zhang X., Yang Y., Hotez P.J., Du L. Neutralizing antibodies for the treatment of COVID-19. Nat. Biomed. Eng. 2020;4:1134–1139. doi: 10.1038/s41551-020-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.K., Seong M.W., Choi S.J., Kim T.S., Choe P.G., Song S.H., Kim N.J., Park W.B., Oh M.D. In vitro activity of lopinavir/ritonavir and hydroxychloroquine against severe acute respiratory syndrome coronavirus 2 at concentrations achievable by usual doses. Korean J. Intern. Med. 2020;35:782–787. doi: 10.3904/kjim.2020.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Ryu D.K., Lee J., Kim Y.I., Seo J.M., Kim Y.G., Jeong J.H., Kim M., Kim J.I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., et al. USFQ-COVID19 Consortium, author. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.Y., Lin M.Y., Coffman R.L., Campbell J.D., Traquina P., Lin Y.J., Liu L.T., Cheng J., Wu Y.C., Wu C.C., et al. Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci. Rep. 2020;10:20085. doi: 10.1038/s41598-020-77077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le R.Q., Li L., Yuan W., Shord S.S., Nie L., Habtemariam B.A., Przepiorka D., Farrell A.T., Pazdur R. FDA approval summary: tocilizumab for treatment of chimeric antigen receptor T cell-induced severe or life-threatening cytokine release syndrome. Oncologist. 2018;23:943–947. doi: 10.1634/theoncologist.2018-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Allen Chan K.C., Hui D.S., Ng E.K., Wu A., Chiu R.W., Wong V.W., Chan P.K., Wong K.T., Wong E., et al. Effects of early corticosteroid treatment on plasma SARS-associated Coronavirus RNA concentrations in adult patients. J. Clin. Virol. 2004;31:304–309. doi: 10.1016/j.jcv.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsky T.W., Vergara R., Codina N., Nelson J.W., Walker M.J., Su W., Barnes C.O., Hsiang T.Y., Esser-Nobis K., Yu K., et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science. 2020;370:1208–1214. doi: 10.1126/science.abe0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020a;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Pan Y., Hu Z., Wu M., Wang C., Feng Z., Mao C., Tan Y., Liu Y., Chen L., et al. Thymosin alpha 1 reduces the mortality of severe coronavirus disease 2019 by restoration of lymphocytopenia and reversion of exhausted T cells. Clin. Infect. Dis. 2020b;71:2150–2157. doi: 10.1093/cid/ciaa630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M., Ollivier E., Chiu H.P., Van Tol S., Rudolffi-Soto P., Stevens C., Bhumkar A., Hunter D.J.B., Freiberg A.N., Jacques D., et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microbes Infect. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A., Garg P., Arora U., Ray A., Wig N. Pathophysiology of COVID-19-associated acute respiratory distress syndrome. Lancet Respir. Med. 2021;9:e3. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal Singh S., Dammeijer F., Hendriks R.W. Role of Bruton's tyrosine kinase in B cells and malignancies. Mol. Cancer. 2018;17:57. doi: 10.1186/s12943-018-0779-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.D. Immune evasion via SARS-CoV-2 ORF8 protein? Nat. Rev. Immunol. 2020;20:408. doi: 10.1038/s41577-020-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. RECOVERY Collaborative Group, author. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschewski M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., Roshon M., Wrzesinski S.H., Desai J.V., Zarakas M.A., et al. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 2020;5:eabd0110. doi: 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E.S., Dufort E.M., Udo T., Wilberschied L.A., Kumar J., Tesoriero J., Weinberg P., Kirkwood J., Muse A., DeHovitz J., et al. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. JAMA. 2020;323:2493–2502. doi: 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci. Adv. 2020;6:eabe0751. doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Schalk A.F., Hawn M.C. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931;78:413–423. [Google Scholar]
- Sciascia S., Apra F., Baffa A., Baldovino S., Boaro D., Boero R., Bonora S., Calcagno A., Cecchi I., Cinnirella G., et al. Pilot prospective open, single-arm multicentre study on off-label use of tocilizumab in patients with severe COVID-19. Clin. Exp. Rheumatol. 2020;38:529–532. [PubMed] [Google Scholar]
- Scutigliani E.M., Kikkert M. Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth Factor Rev. 2017;37:17–27. doi: 10.1016/j.cytogfr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Ahmad B., Choi S., Woo H.G. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput. Struct. Biotechnol. J. 2020;18:3402–3414. doi: 10.1016/j.csbj.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Orlowski G., Song W. Btk regulates B cell receptor-mediated antigen processing and presentation by controlling actin cytoskeleton dynamics in B cells. J. Immunol. 2009;182:329–339. doi: 10.4049/jimmunol.182.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper C.P., Pastick K.A., Engen N.W., Bangdiwala A.S., Abassi M., Lofgren S.M., Williams D.A., Okafor E.C., Pullen M.F., Nicol M.R., et al. Hydroxychloroquine in nonhospitalized adults with early COVID-19: a randomized trial. Ann. Intern. Med. 2020;173:623–631. doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F., Koster A.J., Barcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Hu W., Yu H., Zhao L., Zhao Y., Zhao X., Xue H.H., Zhao Y. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. [2020 Dec 6 [Epub]];Cytometry A. 2020 doi: 10.1002/cyto.a.24285. https://doi.org/10.1002/cyto.a.24285. [DOI] [PubMed] [Google Scholar]
- Spinner C.D., Gottlieb R.L., Criner G.J., Arribas Lopez J.R., Cattelan A.M., Soriano Viladomiu A., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A., et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;71:850–854. doi: 10.1101/2020.11.30.405472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhang X., He Y., Jiang S., Du L. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020;179:104820. doi: 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Narazaki M., Kishimoto T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Li D., Liu T., Wang H., Luo F., Liu Y. Subcutaneous injection of IFN alpha-2b for COVID-19: an observational study. BMC Infect. Dis. 2020a;20:723. doi: 10.1186/s12879-020-05425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020b;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., et al. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell Host Microbe. 2020c;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020d;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne J.A.C., Murthy S., Diaz J.V., Slutsky A.S., Villar J., Angus D.C., Annane D., Azevedo L.C.P., Berwanger O., Cavalcanti A.B., et al. WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group, author. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA. 2020;324:1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Perez-Perez L., et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020a;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020b;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.K., Wang Q., Du L., Tan W., Wilson I.A., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhang X., Wang F., Wang P., Kuang E., Li X. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. BioRxiv. 2020 https://doi.org/10.1101/2020.08.12.247767. [Google Scholar]
- Yap J.K.Y., Moriyama M., Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID-19. J. Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Xu L., Xie X.Y., Yan H.L., Xie B.J., Xu W.Z., Liu X.A., Kang G.J., Jiang W.L., Yuan J.P. Pulmonary pathology of early-phase COVID-19 pneumonia in a patient with a benign lung lesion. Histopathology. 2020;77:823–831. doi: 10.1111/his.14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Wang X., Fu Y., Zhang X., Liu C. Letter to the editor: neutralizing antibodies for the treatment of COVID-19. Acta Pharm. Sin. B. 2021;11:304–307. doi: 10.1016/j.apsb.2020.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]