Abstract
The recent appearance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected millions of people around the world and caused a global pandemic of coronavirus disease 2019 (COVID-19). It has been suggested that uncontrolled, exaggerated inflammation contributes to the adverse outcomes of COVID-19. In this review, we summarize our current understanding of the innate immune response elicited by SARS-CoV-2 infection and the hyperinflammation that contributes to disease severity and death. We also discuss the immunological determinants behind COVID-19 severity and propose a rationale for the underlying mechanisms.
Keywords: COVID-19, cytokine storm, immunoparalysis, inflammatory cytokines, innate immune response
INTRODUCTION
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, is an ongoing global pandemic. As of March 2021, more than 120 million patients have been confirmed, and the number of deaths from COVID-19 has risen to more than 2.7 million. Symptoms of COVID-19 can range from none to life-threatening diseases and are clinically heterogeneous among different patient groups (Del Valle et al., 2020; Merad and Martin, 2020; Vabret et al., 2020). Since its first appearance in Wuhan in December 2019, numerous efforts and studies have shown that host immune responses and immune-related symptoms are distinct between asymptomatic and severe COVID-19 patients, and dysregulation of the immune response may explicate the severity of COVID-19 (Brodin, 2021). Notably, emerging evidence suggests that severe COVID-19 presents with high levels of pro-inflammatory cytokines and is strongly associated with a hyperinflammatory response commonly referred to as a “cytokine storm” (Fajgenbaum and June, 2020). Inflammation is a beneficial part of the immune defenses against infection; nevertheless, excessive pro-inflammatory cytokines frequently exacerbate collateral tissue damages, increasing mortality and prolonging the disease (Consiglio et al., 2020; Fajgenbaum and June, 2020). Therefore, current treatments focus on managing the inflammation caused by SARS-CoV-2 infection, while vaccines and anti-viral agents are developed. Moreover, treatments targeting inflammatory mediators may have clinical benefits, as evidenced by the success of a recently trialed drug, dexamethasone (RECOVERY Collaborative Group, 2021). From this viewpoint, we summarize the changes in innate immune cells seen in moderate and severe COVID-19 and highlight the roles of pro-inflammatory cytokines and type I interferons (IFNs) in the clinical outcomes. We also discuss the potential mechanisms underlying the immunological determinants of COVID-19 severity in terms of cross-regulation and immunoparalysis.
INNATE IMMUNE SENSING AND INFLAMMATORY RESPONSE IN COVID-19
The innate immune cells serve as the first line of host defense and are essential for a rigorous immunity to viruses. These cells, which include macrophages, monocytes, neutrophils, and dendritic cells (DCs), respond to pathogens and produce cytokines that activate the adaptive immune response (Cho et al., 2020; McNab et al., 2015). RNA viruses such as SARS-CoV-2, SARS-CoV-1, and Middle East respiratory syndrome (MERS) is identified by various pattern-recognition receptors, including Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs), and induce the type I IFN responses and IFN-stimulated gene (ISG) expression. Most host cells express cytoplasmic RLRs, whereas TLRs (TLR3, 7, and 8) are generally expressed in the endosomes of innate immune cells. In addition, certain 2'-5'-oligoadenylate synthetase (OAS) and interferon induced protein with tetratricopeptide repeats (IFIT) family proteins can directly sense viral RNA and are associated with antiviral defenses (Park and Iwasaki, 2020; Schoggins and Rice, 2011). Innate immune sensing of virus triggers subsequent downstream signaling cascades through the adaptor molecules MyD88 (for TLR7, TLR8) and TRIF (for TLR3). Mice deficient in MyD88 or TRIF are highly susceptible to SARS-CoV and exhibit delayed viral clearance, indicating these adaptor proteins are required to protect the host from coronavirus infections (Sheahan et al., 2008; Totura et al., 2015). Activation of NF-κB and IRF3/7 signaling mediated by MyD88 and TRIF initiates the production of pro-inflammatory cytokines, such as tumor necrosis factor (TNF), interleukin (IL)-6, IL-1β and type I/III IFNs. Type I/III IFNs, including IFNα, IFNβ, IFNω and IFNλ, rapidly induce and orchestrate antiviral programs via the JAK and STAT signaling pathway and ISG expression (Ivashkiv and Donlin, 2014; McNab et al., 2015). Notably, the NF-κB–mediated antiviral response involves the activation, recruitment, and coordination of various immune cells orchestrated primarily by pro-inflammatory cytokines and chemokines (Sokol and Luster, 2015). Thus, in addition to IFN responses, pro-inflammatory responses are indispensable for a successful antiviral defense, and the balance between IFN and pro-inflammatory responses in target cells potentiates the adaptive immune response and may signify the severity of COVID-19 pathogenesis.
CYTOKINE STORMS AND INNATE IMMUNE PHENOTYPES IN SEVERE COVID-19
Increasing evidence indicates that a dysregulated inflammatory response to SARS-CoV-2 is responsible for severe pathology, virus-induced severe complications, and mortality. In patients with severe COVID-19, aggressive activation of the innate immune system results in a cytokine storm and contributes to lung injury, acute respiratory distress syndrome, and multi-organ failure (Consiglio et al., 2020; Fajgenbaum and June, 2020). While the molecular and mechanistic definitions of a cytokine storm are unclear, it is typically associated with a hyperinflammatory response that causes collateral tissue damages and is clinically characterized by hemodynamic instability, hyperferritinemia, and multiorgan failure (Fajgenbaum and June, 2020; Merad and Martin, 2020; RECOVERY Collaborative Group, 2021). Although the levels of pro-inflammatory cytokines differed to some extent in the studies reported to date, in general, circulating cytokines in patients with severe COVID-19 include the major pro-inflammatory cytokines such as IL-6, TNF, IL-1β, IL-18, IP-10, IFNγ, CCL2, CCL5, CXCL8, and CXCL10 (Abers et al., 2021; Blanco-Melo et al., 2020; Lee et al., 2020; Lucas et al., 2020; Schulte-Schrepping et al., 2020; Zhou et al., 2020a; 2020b) (Fig. 1, Table 1). For example, high serum IL-6 and TNF levels were found in most studies, and even after adjusting the disease severity for risk factors such as inflammation markers, age, sex, and hypoxia, elevated IL-6 and TNF have been shown to be independent and important predictors of disease severity and patient survival (Del Valle et al., 2020). In addition, inflammasome-mediated cytokines, such as IL-1β and IL-18, were also higher in patients with severe COVID-19 than in those with moderate disease (Rodrigues et al., 2021). Besides pro-inflammatory cytokines, both immune complement C5a-C5aR1 and long pentraxin 3 (PTX3) have been associated with adverse outcomes of SARS-CoV-2 infection (Brunetta et al., 2021; Carvelli et al., 2020). Plasma concentrations of soluble C5a and PTX3 were increased in proportion to the severity of COVID-19 and contribute to systemic inflammation and thrombosis. Corresponding to the high levels of circulating inflammatory mediators, gene expression levels of pro-inflammatory cytokines in blood immune cells were greatly enhanced in patients with severe COVID-19 (Arunachalam et al., 2020; Bernardes et al., 2020; Giamarellos-Bourboulis et al., 2020; Hadjadj et al., 2020; Lee et al., 2020; Lucas et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020; Wilk et al., 2020; Zhu et al., 2020). These transcriptional differences associated with various symptoms suggest that peripheral immune activation contributes to the hyperinflammatory phenotype of patients with severe COVID-19. A heightened inflammatory phenotype, which is proportional to lung injury and respiratory failure in critical cases, is also observed in both the lungs and bronchoalveolar lavage fluid (BALF) (Bost et al., 2020; Chua et al., 2020; Liao et al., 2020). Single-cell RNA-seq (scRNA-seq) data showed that SARS-CoV-2 primarily infects lung epithelial cells and macrophages, but only some subtypes of macrophages and monocytes in the BALF exhibited higher levels of pro-inflammatory cytokine expression (Bost et al., 2020; Chua et al., 2020; Liao et al., 2020). Therefore, innate immune cells appear to be responsible for the inflammatory environment in the lungs.
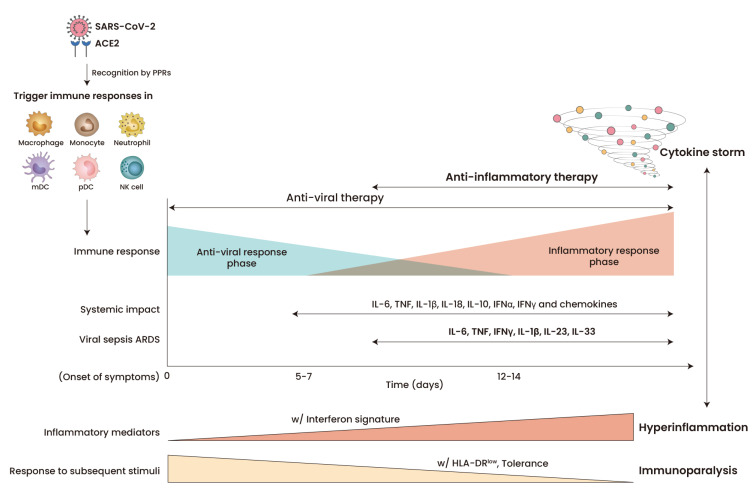
Fig. 1. Inflammatory mediators and innate immune responses during COVID-19 infection.
Patients with severe COVID-19 exhibit high levels of pro-inflammatory cytokines, causing a cytokine storm and collateral tissue damages in the late phase. Innate immune cells in severe COVID-19 have the prominent feature of higher gene expression of inflammatory mediators and impaired responses to subsequent stimuli. PPRs, pattern recognition receptors; ARSD, acute respiratory distress syndrome.
Table 1.
Inflammatory markers in COVID-19 patients
Innate immune cells, such as monocytes, macrophages, and neutrophils, are major producers of the pro-inflammatory cytokines associated with COVID-19 and are often implicated in the pathogenesis of excessive inflammation or cytokine storms in other human diseases (Merad and Martin, 2020). A number of scRNA-seq and FACS-based studies analyzing peripheral blood mononuclear cells (PBMCs) demonstrated that patients with COVID-19 exhibited increased numbers of inflammatory or classical monocytes, low-density neutrophils, eosinophils, and megakaryocytes (Ren et al., 2021), as well as a sharp decrease in the number and frequency of non-classical monocytes, myeloid DCs (mDCs), plasmacytoid DCs (pDCs), and lymphocytes such as CD4+ T cells, CD8+ T cells, and natural killer (NK) cells. For example, in the PBMCs of COVID-19 patients, non-classical monocytes (CD14low CD16hi) are dramatically depleted, while classical monocytes, which are marked by their low expression of human leukocyte antigen DR isotype (HLA-DR), are significantly increased, especially in severe cases (Lee et al., 2020; Mann et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020; Wilk et al., 2020). HLA-DRlow monocytes showed low induction of COX-2, the prostaglandin-producing enzyme, and enhanced expression of Ki-67, a cell cycle marker (Mann et al., 2020). Another independent study demonstrated that some monocyte subsets display high levels of MAFB, PLBD1, and CD163 (Schulte-Schrepping et al., 2020), revealing the phenotypic heterogeneity of monocytes within the microenvironment during SARS-CoV-2 infection. In many inflammatory diseases, inflammatory monocytes are actively recruited to the tissues via the high expression of multiple chemokine receptors, where they aid in maintaining tissue macrophage populations, whereas non-classical monocytes are recruited in fewer numbers (Kapellos et al., 2019). Thus, inflammatory monocytes are selectively recruited to the lungs by the increased chemokines, which may explain the increase in monocyte-derived macrophage counts in the lungs and BALF in severely ill patients that result in excessive inflammation, lung injury, and respiratory failure. Moreover, tissue-resident, alveolar macrophages have been shown to be greatly reduced in patients with severe COVID-19 (Bost et al., 2020; Grant et al., 2021; Liao et al., 2020), indicating the regulation of tissue homeostasis by monocytes and macrophages is profoundly disrupted during viral infection. The BALF and PBMCs of patients with severe COVID-19 comprise lower proportions of conventional DCs (cDCs) and pDCs, and the ratios of cDCs to pDCs are increased in critical cases (Bost et al., 2020; Laing et al., 2020; Liao et al., 2020; Zhou et al., 2020a). These DCs also exhibit functional impairments that can lead to dysregulated adaptive immune responses against SARS-CoV-2. The number of neutrophils also increases in patients with severe COVID-19; recent studies showed that immature CD10lowCD101-CXCR+/- neutrophils numbers were increased in the blood and lungs, and emergency myelopoiesis releases immature and dysfunctional neutrophils in these patients (Bernardes et al., 2020; Schulte-Schrepping et al., 2020). Neutrophil extracellular traps are also found in the lungs of COVID-19 patients and may contribute to thrombosis, tissue damage, and inflammation, indicating the potential role of neutrophils during SARS-CoV-2 infection (Middleton et al., 2020; Radermecker et al., 2020). Together, dysregulated innate immune responses and heightened inflammatory mediators contribute to the clinical observations of COVID-19 severity, cytokine storms, and critical tissue damage. However, it is not yet clear how ongoing SARS-CoV-2 replication modulates the innate immune responses and causes unresolving inflammation and severity.
IMBALANCE AND CROSS-REGULATION BETWEEN PRO-INFLAMMATORY CYTOKINES AND IFNs
Upon virus infection, the transient but high production of IFNs induces the expression of ISGs and mediates the protective immune response by inhibiting viral replication (McNab et al., 2015). While the induction of high levels of inflammatory cytokines is a commonly agreed COVID-19 trait according to many studies to date, the role of IFNs remains controversial. For example, recent studies showed that SARS-CoV-2, like SARS-CoV and MERS-CoV, has the ability to suppress and delay the production of type I IFN by infected cells (King and Sprent, 2021; Park and Iwasaki, 2020; Shin et al., 2020). Viral modulation of IFN productio can be mediated via multiple layers of regulation, such as inhibition of innate sensing, IFN signaling, and ISG effector function. Accordingly, the scRNA-seq data for various cell types show that SARS-CoV-2 triggers low IFNs and limited (or absent) ISG responses, which are accompanied by high viral loads, leading to severe COVID-19 phenotypes (Blanco-Melo et al., 2020; Combes et al., 2021; Hadjadj et al., 2020). The importance of impaired IFN responses has been further emphasized in recent studies, in which inborn errors of type I IFN cell-intrinsic immunity and type I IFN neutralizing autoantibodies with a lower viral load were strongly associated with patients with life-threatening COVID-19 (Bastard et al., 2020; Zhang et al., 2020).
By contrast, other research groups have reported that patients with severe COVID-19 exhibited higher levels of type I or III IFNs and a strong response to IFNs in the PBMCs and BALF, and postmortem, compared to moderate patients (Abers et al., 2021; Arunachalam et al., 2020; Bernardes et al., 2020; Chua et al., 2020; Galani et al., 2021; Lee et al., 2020; Lucas et al., 2020; Wilk et al., 2020; Zhu et al., 2020). Furthermore, the death rate was correlated with heightened levels of type I and III IFNs in patients’ BALF. A longitudinal profiling study has shown that the presence of high IFN levels for 12 days after the first symptom onset correlated with longer hospitalizations and higher mortality (Lucas et al., 2020). Although more work is needed, studies to date have discovered that ISG expression and IFN signatures are common, while type I IFN mRNA and protein levels are relatively low in the serum, PBMC, BALF, and lungs of severe COVID-19 patients. Thus, these observations suggest that patients are exposed to IFNs over time, but their IFN responses are dynamic and temporal. Additionally, the sustained production of IFN in patients with severe COVID-19 are more dependent on tissue-resident cells, such as lung epithelial cells, because IFN gene expression in immune cells is relatively low, and the proportion of pDCs, major IFN producers, is reduced. Acknowledging the complexity of the IFN response, heightened levels of IFNs and ISGs generally correlate with worse cases in the late and critical phases of COVID-19, similar to the finding of previous studies on SARS and MERS patients (Cameron et al., 2007; Park and Iwasaki, 2020). Importantly, untimely type I IFN signaling is known to be deleterious through harmful pro-inflammatory effects on immunopathology (Ivashkiv and Donlin, 2014). This was supported by a recent retrospective cohort study of patients with COVID-19, in which IFNα treatment in the late phase led to enhanced inflammation, longer hospital stays, and a higher death rate (Wang et al., 2020). The balance between pro-inflammatory and IFN responses is a critical determinant of severe COVID-19 immunopathology. The expression of IFN changes dynamically over time, and pro-inflammatory cytokines are maintained at high levels in the late phase during SARS-CoV-2 infection. Although the relative roles of NF-κB–mediated inflammatory and STAT/IRF-dependent IFN responses are still unclear in severe COVID-19, cross-regulation between pro-inflammatory cytokines and type I/II IFNs is likely to shape the pathogenic roles of cytokines and IFNs.
In infection and inflammation, priming by type I and II IFNs positively or negatively regulates the inflammatory response and vice versa (Ivashkiv, 2018; Ivashkiv and Donlin, 2014; McNab et al., 2015; Park and Iwasaki, 2020). The roles of IFNs via priming effects in enhancing the inflammatory response are well established (Fig. 2). Although IFN’s antiviral activities are mediated by the canonical JAK-STAT signaling pathway, other signaling mechanisms, including NF-κB, AP-1, PI3K-mTOR, and MAPK, are further activated by IFNs, leading to various potential inflammatory responses. This diversity of transcription factors activated by IFNs mediates the broad effects of IFN-induced signaling and increases the expression of genes other than ISG that encode pro-inflammatory cytokines and chemokines. For example, recent studies showed there was a positive feedback loop between IFNγ-expressing T cells and macrophages in patients with severe COVID-19, leading to adverse outcomes (Grant et al., 2021; Karki et al., 2021; Silvin et al., 2020; Zhu et al., 2020). Accordingly, STAT1/IRF3-dominant signaling pathways, which are key regulators of inflammatory and type II IFN responses, were more highly activated in COVID-19 compared with influenza A virus patients (Zhu et al., 2020). These IFNγ-primed macrophages have several additional salient features not seen in conventional ISG expression; they can become hyper-responsive to subsequent inflammatory stimuli such as TNF, IL-1β and ligands for TLRs (Ivashkiv, 2018). Thus, IFNγ-primed macrophages may contribute to the production of massive amounts of inflammatory cytokines and NF-κB target genes in patients with severe COVID-19. In addition, IFNγ-primed macrophages have been shown to be refractory to anti-inflammatory factors, such as glucocorticoids or IL-10, in a gene-specific manner, leading to unresolving inflammation and poor tissue healing. Similar to IFNγ, type I IFNs also can potentiate the inflammatory response in infection and inflammation. For example, most NF-κB–dependent genes of inflammatory cytokines are not activated by type I IFNs alone (Ivashkiv and Donlin, 2014; McNab et al., 2015), whereas cooperative actions between type I IFNs and TNF have been shown to enhance the responsiveness against subsequent stimuli and increase gene expression of pro-inflammatory cytokines in human macrophages (Park et al., 2017). Other studies demonstrated that gene expression of IL-6 and iNOS is activated by coordinate binding of STAT/IRFs and NF-κB to their promoters (Platanitis and Decker, 2018). As features of many infectious and inflammatory diseases, the IFN priming effects that increase inflammatory cytokine expression are mostly mediated by transcriptional and epigenetic reprogramming (discussed below).
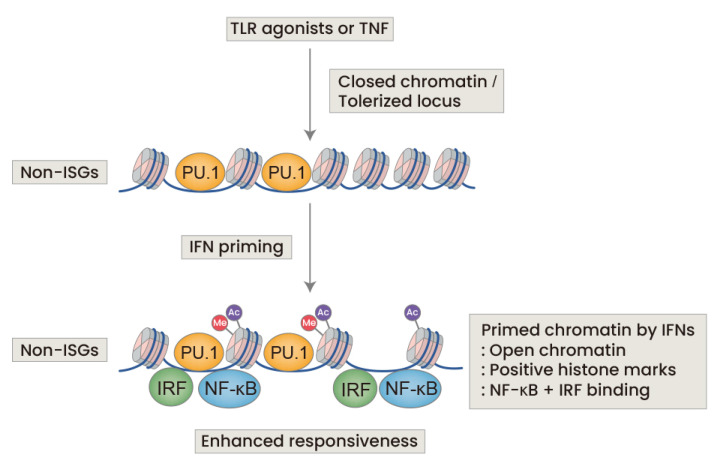
Fig. 2. IFN priming activates macrophages and reverses the state of tolerance.
TLR agonists and TNF induce a state of tolerance in which chromatin is closed, that is associated with immunoparalysis. IFN priming of basal or tolerized macrophages strongly increases responsiveness and pro-inflammatory responses by priming and opening the chromatin at the non-ISGs locus.
There may also be mechanisms by which the high levels of inflammatory cytokines in patients with severe COVID-19 conversely regulate the IFN response. For instance, TNF or macrophage colony-stimulating factor (M-CSF) induces the IRF1-mediated production of picomolar concentrations of IFNβ that results in the potent induction of ISGs (Venkatesh et al., 2013; Yarilina et al., 2008). It has been shown that IL-1β and prostaglandin E2 can inhibit the expression of type I IFN and ISGs during bacterial infection. These inflammatory mediators change the quantity and quality of IFN signaling and disrupt the timing and duration of the anti-viral response relative to virus loads (McNab et al., 2015), as shown in mouse models of MERS-CoV infections (Channappanavar et al., 2019). In summary, cross-regulation between IFNs and pro-inflammatory cytokines can contribute to excessive and unrestrained inflammation, which may result in severe clinical outcomes in COVID-19 patients. However, the related mechanisms involved in COVID-19 have not yet been studied and require further elucidation.
IMMUNOPARALYSIS AND INNATE IMMUNE MEMORY
It is interesting that, despite high levels of inflammatory mediators, innate immune cells in patients with severe COVID-19 are refractory to subsequent stimuli of cytokines or TLR agonists (Fig. 1). CD14+ monocytes isolated from the PBMCs of patients with severe COVID-19 exhibited a “tolerized state”, in which previous exposure to TLR ligands or cytokines led to a transient period of hypo-responsiveness upon subsequent stimuli (Arunachalam et al., 2020; Combes et al., 2021; Giamarellos-Bourboulis et al., 2020; Schulte-Schrepping et al., 2020; Zhou et al., 2020a). This “tolerization” involving innate immune memory (Netea et al., 2020) is an intrinsic biological process that prevents a surge in hyperinflammation and adjusts the magnitude and duration of the inflammatory response. It is worth noting that macrophage tolerance is not an anti-inflammatory or non-inflammatory state, but rather, an inflammatory state in which there is no further response to subsequent stimuli. Arunachalam et al. (2020) recently identified that CD14+ monocytes and myeloid DCs isolated from COVID-19 patients produced significantly less TNF and IL-6 in response to a bacterial cocktail of lipopolysaccharide (LPS), Pam3CSK4, and Flagellin or a viral cocktail of polyIC, R848. Similar to that seen in other macrophage tolerance models, phosphorylated p65 was decreased in the same cells, suggesting canonical inflammatory NF-κB signaling was suppressed. The response of pDCs to the viral cocktail was also considerably inhibited. Of note, Schulte-Schrepping et al. (2020) demonstrated that monocytes isolated from patients with severe COVID-19 exhibited stronger tolerization in response to LPS, suggesting that the inflammatory responses are related to the pathogenic hypo-responsiveness of monocytes. Other independent studies also showed that DCs, PBMCs, and CD14+ monocytes isolated from patients were less responsive to the pro-inflammatory cytokine cocktail (IL-1β, IL-6, TNF, and PGE2) as well as to SARS-CoV-2, respectively (Combes et al., 2021; Giamarellos-Bourboulis et al., 2020; Zhou et al., 2020a). The occurrence of innate immune cell tolerization was further supported by the finding that plasma from severe COVID-19 patients had increased levels of bacterial DNA (Arunachalam et al., 2020). These results collectively suggest that the severe phenotypes of patients, involving cytokine storms, are strongly associated with tolerization induced by infections and certain pro-inflammatory cytokines such as TNF. Interestingly, both type I and II IFNs have been shown to prevent and reverse macrophage tolerance. Ex vivo priming of IFNγg or IFNα2 abolished LPS- or TNF-induced tolerance in human macrophages and monocytes, respectively (Chen and Ivashkiv, 2010; Park et al., 2017). As shown in a report in which circulating endotoxin induced strong tolerance in sepsis (van der Poll et al., 2017) and abolished tolerance in systemic lupus erythematosus (SLE) (Shi et al., 2014), the magnitude and duration of IFNs in various human diseases are likely to be involved in determining macrophage/monocyte tolerance. Mechanistically, the induction or abrogation of tolerance is mediated by transcriptional, epigenetic, and metabolic reprogramming (as reviewed elsewhere: Ivashkiv, 2018; Netea et al., 2020), leading to the dysregulation of the inflammatory responses (Fig. 2). Accordingly, robust epigenetic and metabolic changes, which may be associated with macrophage tolerance and innate immune memory, have recently been reported in COVID-19 patients. Genome-wide CRISPR screens using SARS-CoV-2 showed that genes encoding the SWI/SNF chromatin-remodeling complex are pro-viral factors (Wei et al., 2021), and the distinct chromatin accessibility changes during virus infection were associated with COVID-19 severity (Giroux et al., 2020). Alterations of metabolic pathways, such as kynurenine, which is linked to COVID-19 severity (Nie et al., 2021; Shen et al., 2020) and sepsis (van der Poll et al., 2017), were also demonstrated. However, our current understanding of the immunoparalysis of patients with severe COVID-19 is very limited, and more work is needed to establish the immune determinants during infection and identify therapeutic targets.
CONCLUDING REMARKS
Despite the many unknowns, the limited success of the recent dexamethasone trial implies that excessive inflammation can be ameliorated in patients with severe COVID-19 to reduce their clinically critical immunopathology. Further clinical trials targeting hyperinflammation with antagonists against JAK, IL-1β, IL-6, TNF, and granulocyte-macrophage colony-stimulating factor (GM-CSF) in COVID-19 patients are ongoing, but it is hoped the results will present potential opportunities. The importance of age, sex, and comorbidity as determinants of severe COVID-19 is known, but our current understanding of the associations between COVID-19 and the immune response is very limited. It is also unclear how the inflammation observed in COVID-19 that causes long-term persistence and high mortality differs from the inflammation in other respiratory infections. Thus, broadening our understanding of the delicate balance between pro-inflammatory cytokines and IFNs will enable the development of effective treatments specifically for the hyperinflammation of severe COVID-19 without perturbing a wide range of anti-viral host defenses. As noted above, timing, duration, and qualitative regulation are important parameters in various cytokine activities and should be considered before targeting excessive inflammation in COVID-19. Importantly, studying the causal factors linking inflammatory mediators and clinical heterogeneity is a prerequisite for the development of truly predictive biomarkers and therapeutics for COVID-19. As more evidence emerges for several factors that manage inflammatory responses, drugs that target epigenetic markers on chromatin or metabolism pathways are also potential tools for reducing hyperinflammation and mortality in patients with COVID-19.
Footnotes
CONFLICT OF INTEREST
The author has no potential conflicts of interest to disclose.
REFERENCES
- Abers M.S., Delmonte O.M., Ricotta E.E., Fintzi J., Fink D.L., de Jesus A.A.A., Zarember K.A., Alehashemi S., Oikonomou V., Desai J.V., et al. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6:e144455. doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes J.P., Mishra N., Tran F., Bahmer T., Best L., Blase J.I., Bordoni D., Franzenburg J., Geisen U., Josephs-Spaulding J., et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53:1296–1314.e9. doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- Brunetta E., Folci M., Bottazzi B., De Santis M., Gritti G., Protti A., Mapelli S.N., Bonovas S., Piovani D., Leone R., et al. Macrophage expression and prognostic significance of the long pentraxin PTX3 in COVID-19. Nat. Immunol. 2021;22:19–24. doi: 10.1038/s41590-020-00832-x. [DOI] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvelli J., Demaria O., Vely F., Batista L., Chouaki Benmansour N., Fares J., Carpentier S., Thibult M.L., Morel A., Remark R., et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Jr., Perlman S., Jr. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ivashkiv L.B. IFN-gamma abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19438–19443. doi: 10.1073/pnas.1007816107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D.H., Kim J.K., Jo E.K. Mitophagy and innate immunity in infection. Mol. Cells. 2020;43:10–22. doi: 10.14348/molcells.2020.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thurmann L., Kurth F., Volker M.T., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Combes A.J., Courau T., Kuhn N.F., Hu K.H., Ray A., Chen W.S., Chew N.W., Cleary S.J., Kushnoor D., Reeder G.C., et al. Global absence and targeting of protective immune states in severe COVID-19. Nature. 2021;591:124–130. doi: 10.1038/s41586-021-03234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R., et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020;183:968–981.e7. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajgenbaum D.C., June C.H. Cytokine storm. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., Koukaki E., Fragkou P.C., Panou V., Rapti V., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux N.S., Ding S., McClain M.T., Burke T.W., Petzold E., Chung H.A., Palomino G.R., Wang E., Xi R., Bose S., et al. Chromatin remodeling in peripheral blood cells reflects COVID-19 symptom severity. BioRxiv. 2020 doi: 10.1101/2020.12.04.412155. https://doi.org/10.1101/2020.12.04.412155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., Abbott D.A., Donnelly H.K., Donayre A., Goldberg I.A., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B. IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018;18:545–558. doi: 10.1038/s41577-018-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapellos T.S., Bonaguro L., Gemund I., Reusch N., Saglam A., Hinkley E.R., Schultze J.L. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front. Immunol. 2019;10:2035. doi: 10.3389/fimmu.2019.02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C., Sprent J. Dual nature of type I interferons in SARS-CoV-2-induced inflammation. Trends Immunol. 2021;42:312–322. doi: 10.1016/j.it.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Munoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E.R., Menon M., Knight S.B., Konkel J.E., Jagger C., Shaw T.N., Krishnan S., Rattray M., Ustianowski A., Bakerly N.D., et al. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci. Immunol. 2020;5:eabd6197. doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E.A., He X.Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Giamarellos-Bourboulis E.J., Dominguez-Andres J., Curtis N., van Crevel R., van de Veerdonk F.L., Bonten M. Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell. 2020;181:969–977. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Qian L., Sun R., Huang B., Dong X., Xiao Q., Zhang Q., Lu T., Yue L., Chen S., et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184:775–791.e14. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A., Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Kang K., Giannopoulou E., Qiao Y., Kang K., Kim G., Park-Min K.H., Ivashkiv L.B. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 2017;18:1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanitis E., Decker T. Regulatory networks involving STATs, IRFs, and NFkappaB in inflammation. Front. Immunol. 2018;9:2542. doi: 10.3389/fimmu.2018.02542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermecker C., Detrembleur N., Guiot J., Cavalier E., Henket M., d'Emal C., Vanwinge C., Cataldo D., Oury C., Delvenne P., et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020;217:e20201012. doi: 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., et al. RECOVERY Collaborative Group, author. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues T.S., de Sa K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Goncalves A.V., Perucello D.B., Andrade W.A., Castro R., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Bassler K., Schlickeiser S., Zhang B., Kramer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., Heise M.T. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4:e1000240. doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Zhang Z., Yu A.M., Wang W., Wei Z., Akhter E., Maurer K., Costa Reis P., Song L., Petri M., et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One. 2014;9:e93846. doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., Almire C., Henon C., Kosmider O., Droin N., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol C.L., Luster A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015;7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183:1479–1495.e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Whitmore A., Agnihothram S., Schafer A., Katze M.G., Heise M.T., Baric R.S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. Bio. 2015;6:e00638–15. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Poll T., van de Veerdonk F.L., Scicluna B.P., Netea M.G. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 2017;17:407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- Venkatesh D., Ernandez T., Rosetti F., Batal I., Cullere X., Luscinskas F.W., Zhang Y., Stavrakis G., Garcia-Cardena G., Horwitz B.H., et al. Endothelial TNF receptor 2 induces IRF1 transcription factor-dependent interferon-beta autocrine signaling to promote monocyte recruitment. Immunity. 2013;38:1025–1037. doi: 10.1016/j.immuni.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., et al. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell Host Microbe. 2020;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarilina A., Park-Min K.H., Antoniv T., Hu X., Ivashkiv L.B. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K., Wong Y.C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.Y., Lau T.T., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020a;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020b;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Yang P., Zhao Y., Zhuang Z., Wang Z., Song R., Zhang J., Liu C., Gao Q., Xu Q., et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53:685–696.e3. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]