Abstract
Simultaneous enrichment and fractionation of diverse proteins/peptides possessing different post-translational modifications (PTMs) from the same biological samples is highly desirable to reduce sample consumption, avoid complicated sample processing, and enable studies of potential crosstalks between different PTMs. In this work, we report a new approach to enable the simultaneous enrichment and separation of glycopeptides, phosphopeptides and mannose-6-phosphate (M6P) glycopeptides by using dual-functional Ti(IV)-IMAC material. Moreover, we also made the separation of neutral and sialyl glycopeptides, mono- and multi-phosphopeptides possible by performing different elution according to the differences in their electrostatic or hydrophilic properties. These separations are effective and efficient to eliminate the signal suppression from neutral glycopeptides for sialyl glycopeptide detection, allowing separation of mono-phosphopeptides from multi-phosphopeptides, as well as detection of M6P glycopeptides that are free from the above-mentioned modifications. This new strategy significantly improves the coverage and identification numbers of glycopeptides, phosphopeptides and M6P glycopeptides by 1.9, 2.3, and 4.3-fold compared with the conventional method, respectively. This is the first report on simultaneous enrichment and separation of neutral and sialyl glycopeptides, mono- and multi-phosphopeptides and M6P glycopeptides via dual-functional Ti(IV)- IMAC, revealing novel insights into potential crosstalk among these important PTMs.
Keywords: dual-functional Ti(IV)-IMAC material, neutral and sialyl glycopeptide separation, mono- and multi-phosphopeptide separation, mannose-6-phosphate (M6P) glycopeptide enrichment, posttranslational modifications
Introduction
Protein post-translational modifications (PTMs) are covalent and generally enzymatic modifications on specific protein sites, which increase the functional diversity of the proteins and affect almost all aspects of biological processes of a cell.1 It is reported that numerous proteins can bear multiple PTMs, which suggests the possibility of regulatory crosstalk between unique modification events.2 Liquid chromatography tandem mass spectrometry (LC-MS/MS) based proteomics strategy has become the crucial tool for the analysis of protein PTMs. To comprehensively profile protein PTMs, an efficient enrichment method is a prerequisite due to the typical low abundance and sub-stoichiometry of most protein PTMs in complex biological samples.3 Numerous enrichment methods have been developed, however, each method may only focus on one type of PTM.4 For example, immobilized metal affinity chromatography (IMAC) has been developed for phosphopeptide enrichment and hydrophilic interaction chromatography (HILIC) has been utilized primarily for enrichment of glycopeptides.5,6 Crosstalk analysis of different PTMs must perform different enrichment methods on different samples, which is not only tedious and time-consuming but also requires more precious samples that may not be available.3 Carr and coworkers developed a PTM crosstalk analysis strategy called serial enrichments of different post-translational modifications (SEPTM) that enabled enrichment of multiple PTMs from the same biological sample.7 Similar strategy have also been applied to enrich glycopeptides and phosphopeptides from plasma-derived extracellular vesicles.8 However, these methods still suffer from the requirement of different enrichment materials. Several research groups have developed new IMAC materials to simultaneous enrich and separate glycopeptides and phoshopetides.9-12 However, without proper fractionation, negatively charged sialylated glycopeptides will be supressed by more abundant neutral glycopetides in the positive ion mode LC-MS/MS analysis, and multi-phosphopeptides and M6P glycopeptides will be supressed by mono-phosphopeptides as well.
Previously, we demonstrated that M6P glycopeptide identifications increased significantly when using the dual-functional Ti-IMAC materials for enrichment while simultaneously eliminating the interference of phosphopeptides. 13 This strategy is based on the special properties of the Ti(IV)-IMAC materials we developed, which not only possesses substantial phosphate chelated Ti(IV) ions, enabling enrichment of phosphopeptides through electrostatic interaction, but also contain numerous hydroxyl, amine and phosphate groups on the material surface that are highly hydrophilic, enabling enrichment of glycopeptides through hydrophilic interactions.14,15 As M6P glycopeptides possess features of both phosphopeptides and glycopeptides, they could be captured by the dual-functional Ti(IV)-IMAC material through the synergistic electrostatic and hydrophilic interactions. Therefore, by performing HILIC-mode stepwise elution, M6P glycopeptides can be separated from phosphopeptides. However, in this strategy, glycopeptides were discarded together with the non-phosphopeptides.
In order to overcome the shortcomings of the above methods, in the current study, we improved and optimized the previous strategy and continually explored the hydrophilic characteristics of the Ti(IV)-IMAC material to enable simultaneous enrichment and separation of neutral and sialyl glycopeptides, mono- and multi-phosphopeptides and M6P glycopeptides (Figure 1). This strategy improves the quality of each individual PTM analysis significantly and can be effectively implemented and integrated to perform crosstalk analysis of these three important PTMs in a single experiment from the same biological sample.
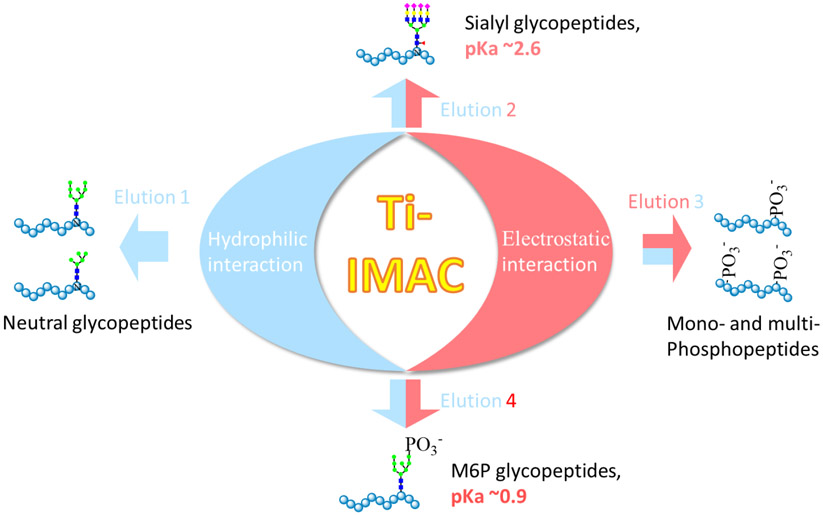
Figure 1.
Scheme of IMAC and HILIC dual-mode affinity enrichment approach for the simultaneous enrichment of glycopeptides, phosphopeptides and M6P glycopeptides. Elution is performed in four steps to separate different peptides according to their chemical properties: Elution 1 uses weak acidic buffer with decreased ACN gradient and mainly elutes neutral glycopeptides; Elution 2 uses strong acidic condition that allows protonating sialylated glycopeptides to be collected and makes them isolated from the IMAC beads that bind to the unprotonated phosphopeptides and M6P glycopeptides; Elution 3 uses basic buffer with high and medium ACN content, which can elute mono- and multi- phosphopeptides, respectively; Elution 4 uses basic buffer with low ACN content and M6P glycopeptides are mainly eluted in this fraction.
Experimental section
Sample preparation and protein digestion.
Details are provided in Supporting Information.
HILIC-mode Ti(IV)-IMAC enrichment and elution.
Typical enrichment and elution method for Ti(IV)-IMAC and HILIC are provided in Supporting Information. For HILIC-mode Ti(IV)-IMAC enrichment the standard (10 μg each sample) and mouse lung protein digests (500 μg each sample) were loaded onto Ti(IV)-IMAC materials with 80% ACN/3%TFA buffer and then Ti(IV)-IMAC materials captured samples were washed three times with the loading buffer, which was similar with the conventional HILIC enrichment. The flowthrough and washing buffer of the standard proteins were collected for MALDI MS analysis. After enrichment, the standard protein samples were eluted with following two elution procedures: 1)10% NH4OH (v/v) 10 min; 2) 0.1% FA (v/v) 10 min and 10% NH4OH (v/v) 10min, respectively. Samples were immediately dried down in SpeedVac. Samples were re-dissolved in 0.1% formic acid (FA) and load 5% of each sample for MALDI analysis (details are provided in Supporting Information).
For mouse lung protein sample, the elution was also performed with two methods: 1) direct eluted with 10% NH4OH (v/v); 2) sequentially eluted with the following four elution procedures: Elution 1 uses weak acidic buffer (0.1 TFA%) with decreased ACN gradient (1st fraction 60%, 2nd fraction 40%, and 3rd fraction 20% ACN (v/v)); Elution 2 uses strong acidic condition (6 TFA%) with decreased ACN gradient (40%, 20%, and 0% ACN (v/v) ), the three elution were combined for analysis (4th fraction); elution 3 uses basic buffer (10% NH4OH) with high and medium ACN content (5th fraction 60% and 6th fraction 40% ACN (v/v)); elution 4 uses basic buffer (10% NH4OH) with low ACN content (20%, 10%, and 0% ACN (v/v)), the three elution were combined for analysis (7th fraction). The elution was immediately dried down in SpeedVac. Samples were stored at −20 °C and re-dissolved in 1% formic acid (FA) and load 15% of each sample for LC-MS/MS analysis (details are provided in Supporting Information). It is worth mentioning that the number of the elution steps was determined based on the sample complexity. For standard samples containing only a few neutral glycopeptides and several phosphopeptides, 2-step elution was sufficient. For complex biological samples like mouse lung protein digests, 4 or more steps of elutions were needed to enable better separation and fractionation of multiple PTMs peptide.
Data processing.
Byonic software (Protein Metrics, San Carlos, CA) was used to analyze the acquired MS and MS/MS spectra of intact N-linked glycopeptides and M6P glycopeptides. Raw files were searched against Mus musculus protein database of reviewed (Swiss-Prot) sequences downloaded from Uniprot. Precursor ion mass tolerance of 10 ppm and fragment ion mass tolerance of 0.01 Da were selected. The phosphorylation of serine (S), threonine (T) and tyrosine (Y) and the oxidation of methionine (M) were set as variable modifications. Meanwhile, the carbamidomethylation of cysteine (C) was set as fixed modification. Common N-linked glycopeptides searching used a mammalian N-glycome database that contains 309 glycans in addition to M6P glycans. Peptide identifications were filtered at two-dimensional false discovery rate (2D FDR) <1%, PEP 2D <0.05, ∣Log Prob∣>1, and Byonic Score >150. Manual inspection of MS/MS spectra of M6P glycopeptides was performed to examine if Byonic identification results contained phosphorylated hexose diagnostic ions. With the same raw files and database, phosphopeptides were searched against MaxQuant software (Version, 1.5.8.3). Precursor ion mass tolerance of 4.5 ppm and fragment ion mass tolerance of 0.05 Da were selected. Phosphopeptides with the false discovery rate <0.01 and minimum score of 40 were accepted as confident identifications.
Results and Discussion
Conventional IMAC and HILIC approaches for glycopeptide, phosphopeptide and M6P glycopeptide enrichment.
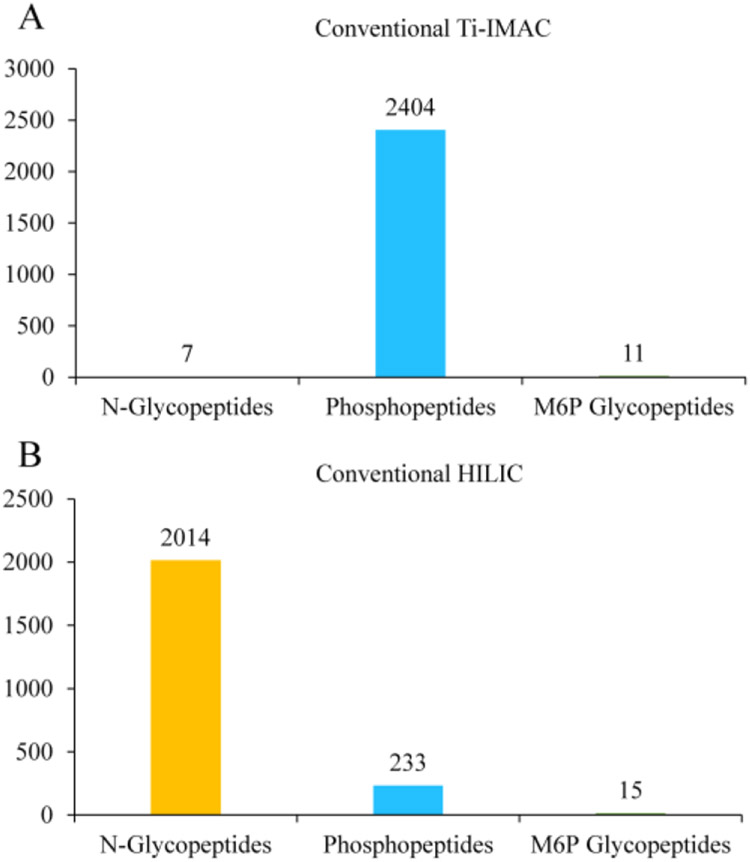
In general, IMAC is suitable for phosphopeptide and M6P glycopeptide enrichment as they both contain phosphate group and HILIC is applicable to enrich common glycopeptides and M6P glycopeptides as both types of peptides possess hydrophilic glycans. To investigate glycopeptide, phosphopeptide and M6P glycopeptide profiling status by these two conventional methods, we first performed typical Ti(IV)-IMAC and HILIC (homemade spin-tip) enrichment procedures using mouse lung tissue samples.16,17 As shown in Figure 2A, the conventional IMAC method enabled profiling of 2404 phosphopeptides, however, only resulted in the identification of 7 common N-glycopeptides and 11 M6P glycopeptides from 25 μg mouse lung protein digests. This result suggested that majority of glycopeptides cannot be effectively enriched by Ti(IV)-IMAC materials using its typical enrichment procedures. Furthermore, the M6P glycopeptide signals can be suppressed by the coeluted phosphopeptides in LC-MS/MS analysis. As comparison, conventional HILIC method enabled profiling of 2014 common N-glycopeptides and 15 M6P glycopeptides, however, only identified 233 phosphopeptides from the same amount of sample (Figure 2B). This result demonstrated that only a small fraction of phosphopeptides were enriched by HILIC materials due to their low hydrophilicity and the M6P glycopeptide signals were also suppressed by the coeluted common N-glycopeptides in LC-MS/MS analysis. Therefore, development of a new enrichment strategy to simultaneously enrich and separate glycopeptides, phosphopeptides, and M6P glycopeptides from the same biological samples would overcome ion suppression issue during MS analysis, which is highly beneficial for the global mapping of these PTMs.
Figure 2.
Common N-glycopeptide, phosphopeptide and M6P glycopeptide enrichment with conventional method. (A) conventional Ti-IMAC method. (B) conventional HILIC approach.
Exploring hydrophilicity of the Ti(IV)-IMAC material
The Ti(IV)-IMAC material we developed not only possesses substantial phosphate chelated Ti(IV) ions, which can be used to capture phosphopeptides through electrostatic interactions, but also contains numerous hydroxyl, amine and phosphate groups on the material surface. We previously demonstrated the high hydrophilicity of Ti(IV)-IMAC material and used this characteristic to separate the Ti(IV)-IMAC enriched phosphopeptides and M6P glycopeptides.13 However, this strategy was not able to enrich common glycopeptides due to non-HILIC mode loading procedures. To overcome this problem, we continually optimize the loading procedure and change it to HILIC mode. We aim to simultaneously capture glycopeptides during the phosphopeptide and M6P glycopeptide enrichment.
To test this possibility, we first used standard phosphoprotein β-casein and standard glycoprotein RNase B digests to perform the enrichment. Standard M6P glycoprotein was not found commercially available, so it was not included in the initial test experiment. The sample loading buffer was changed from 40% ACN/3%TFA to 80% ACN/3%TFA, which was similar with the conventional HILIC enrichment protocols. The increased ACN content can help to generate an aqueous layer across the substantial hydroxyl, amine groups and phosphate chelated Ti (IV) ions on the material surface, therefore, the glycopeptides can be captured on materials through hydrophilic interaction. Initially, the aqueous buffer of 10% ammonia was used as a single elution buffer because it can interrupt both electrostatic and hydrophilic interactions and should be able to elute captured phoshopeptides and glycopeptides simultaneously.
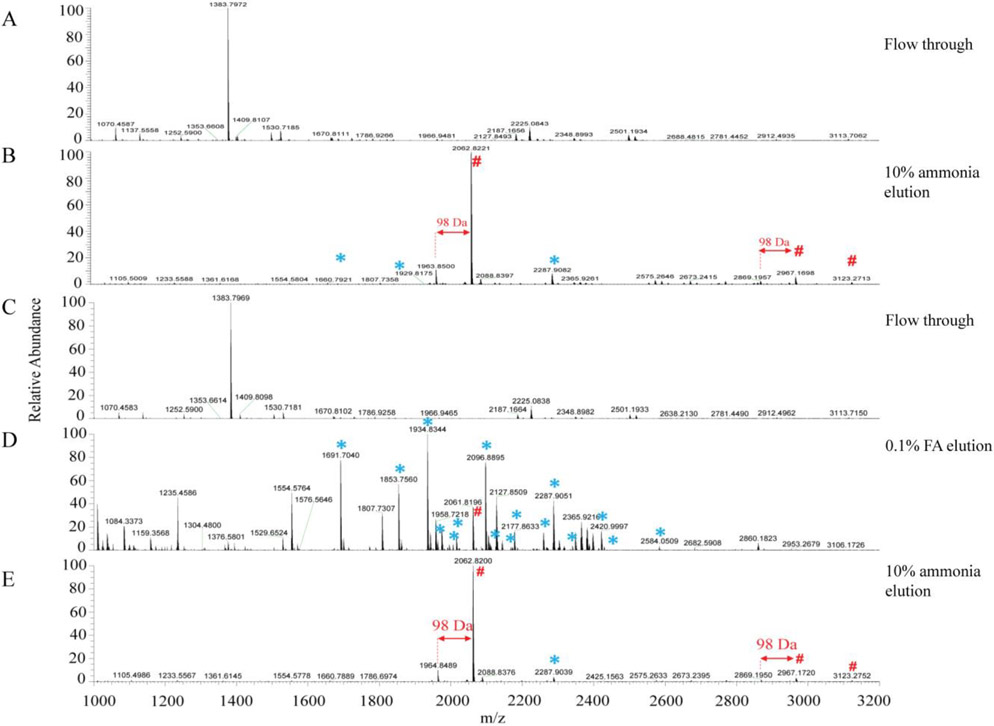
The flow-through and eluted samples were analyzed by MALDI-TOF MS and the results were shown in Figures 3A and 3B. Figure 3A showed that flow-through did not reveal the trace of phosphopeptides and glycopeptides, which demonstrated that majority of these modified peptides were captured on the materials. And in Figure 3B, the two intense peaks labeled with red pound signs can be annotated as the two phosphopeptides of β-casein. There are also some peaks that can be annotated to the glycopeptides of RNase B, however, their intensities are very low. We infer that the glycopeptides of RNase B are either not effectively enriched by Ti (IV)-IMAC materials or their signals are greatly supressed by the coeluted phosphopeptides. To evaluate these possibilities, we performed another elution experiment, that is the glycopeptides and phosphopeptides were eluted separately. As 0.1% FA can interrupt the hydrophilic interactions, we used it to elute the glycopeptides of RNase B, and then 10% ammonia was used to elute phosphopeptides. Figure 3C was similar with Figure 3A and no trace of phosphopeptides and glycopeptides was found. As shown in Figure 3D, significantly enhanced and increased glycopeptide peaks were observed with the 0.1% FA elution. And in Figure 3E, we detected the two phosphopeptides of β-casein without signals from glycopeptides (Annotated MALDI-TOF MS peaks were also shown in Table S1). These results demonstrated that Ti (IV)-IMAC materials can be used to simultaneously enrich glycopeptides and phosphopeptides from the same sample. And separating the glycopeptides from the phosphopeptides after enrichment can increase the MS signals for glycopeptides and improving their detection and coverage.
Figure 3.
MALDI-TOF MS analysis of Ti-IMAC enriched β-casein and RNase B digest mixture. (A, B) Ti-IMAC enrichment in HILIC-mode and elution without fractionation. (C-E) Ti-IMAC enrichment in HILIC-mode and elution with 0.1% FA (D) and 10% ammonia (E) fractionation. (# represents phosphopeptides and * represents glycopeptides)
Applying dual-functional Ti(IV)-IMAC material to enrichment of PTM peptides from complex biological samples
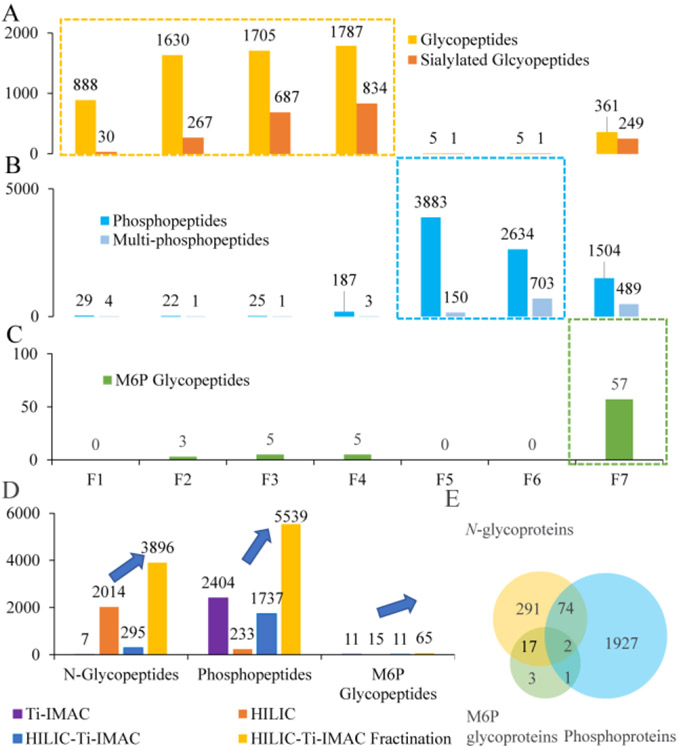
After demonstrating the potential of dual-functional Ti(IV)-IMAC material for the enrichment and separation of both glycopeptides and phosphopeptides. We then applied this novel strategy to the enrichment of N-glycopeptides, phosphopeptides and M6P glycopeptides from complex biological samples consisting of mouse lung protein digests. Initially, the samples were loaded onto the Ti(IV)-IMAC materials with conventional HILIC mode. In this way, neutral glycopeptides were captured on the materials through hydrophilic interaction, phosphopeptides were captured with electrostatic interaction and sialyl and M6P glycopeptides were captured by the synergistic electrostatic and hydrophilic interaction. We then performed stepwise elution of these captured peptides. Generally, as shown in Figure 4A, in Elution 1 (fractions 1-3), the numbers of identified glycopeptides were 888, 1630, and 1705, respectively, and majority of them were neutral glycopeptides. In Elution 2 (4th fraction), 1787 glycopeptides were identified, while the glycopeptides with sialylated glycan were continually increased to 834. Higher number of sialylated glycopeptides was profiled in this fraction. This observation is likely due to the fact that the pKa value of sialic acid is 2.1, therefore sialic acids may not be fully protonated in low TFA elution buffer and they can still be captured by Ti(IV)-IMAC materials through electrostatic interaction. However, if we increased the TFA content from 0.1% to 6%, more sialic acids can be protonated and continually be eluted. It is worth mentioning that this strong acid condition did not elute much phosphopeptides and M6P glycopeptides, as the pKa value of phosphate group is 0.7-1.0, which is much lower than sialic acid and is hard to be protonated.
Figure 4.
Dual-mode affinity enrichment approach enables the enrichment and separation of common N-glycopeptides, phosphopeptides, and M6P glycopeptides. (A) Common N-glycopeptide identification results. (B) Phosphopeptide identification results. (C) M6P glycopeptide identification results. (D) Comparison of identified N-glycopeptides, phosphopeptides and M6P glycopeptides across different enrichment methods. (E) Overlap analysis of N-glycoproteins, phosphoproteins and M6P glycoproteins profiled in the HILIC mode Ti-IMAC with fractionation method. (F1-7 represent fractions 1-7)
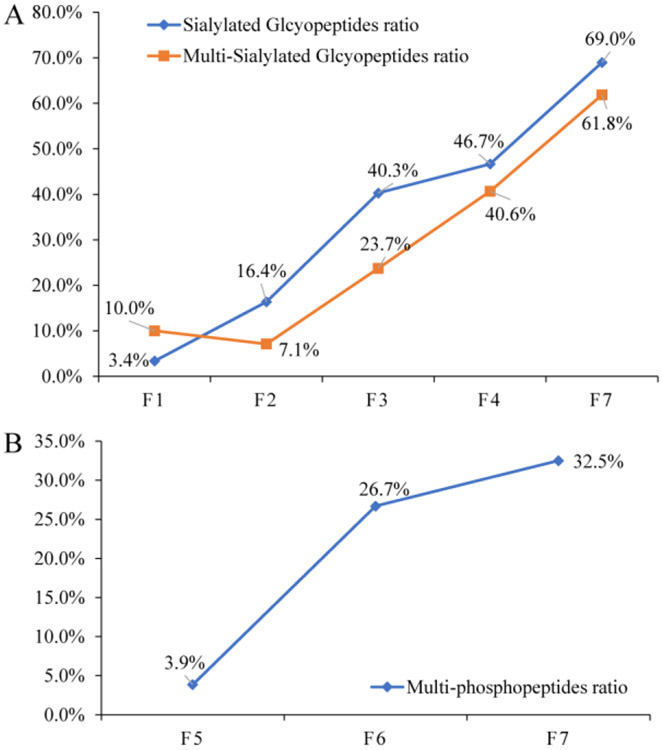
In fractions 1-4, in which majority of glycopeptides were identified, we found that the ratios of sialylated glycopeptides in all identified glycopeptides were 3.4%, 16.4%, 40.3%, and 46.7%, respectively, and the ratios were gradually increased in these four fractions. Moreover, the ratios of multi-sialylated glycopeptides in all identified sialylated glycopeptides in these four fractions were 10.0%, 7.1%, 23.7%, and 40.7%, respectively, and the ratios also have a general trend of increasing (Figure 5A, Figure S1). This is because the sialic acid is more hydrophilic than other monosaccharides and it is also negatively charged, eluting sialylated glycopeptides from dual-functional Ti(IV)-IMAC material is more difficult than other neutral glycopeptides due to stronger retention. The increasing trend of sialylated and multi-sialylated glycopeptide ratio corresponded well to the decreasing ACN content, increasing TFA content. These results demonstrated that the dual-functional Ti(IV)-IMAC materials can not only effectively enrich glycopeptides with excellent efficiency, it can also separate sialylated glycopeptides from neutral glycopeptides. As sialylation has been widely implicated in various biological processes. Therefore, comprehensive sialylation analyses will help to elucidate their roles in the related biological process and facilitate the discovery of novel biomarkers for diseases and drug targets.18 We assume that by performing negative ion mode MS,19 or explore sialic acid derivatization strategy20-22 and combine these approaches with our high efficiency enrichment and separation strategy, the coverage of sialylated glycopeptide analyses will continually be improved.
Figure 5.
Fractionation efficiency of different elution conditions. (A) Sialylated and multi-sialylated glycopeptides ratio. (B) Multi-phosphopeptides ratio. (F1-7 represent fractions 1-7)
As a comparison, we also performed fractionation for conventional HILIC enriched glycopeptides. In total, four fractions were collected, and the elution buffers all contained 0.1% FA and gradient decrease of ACN content at 60%, 40%, 20%, and 0%, respectively. The identified glycopeptides were 1684, 1404, 711, and 188, respectively, and the sialylated glycopeptides were 691, 572, 183, and 22 (Figure S2A). The data showed that conventional HILIC did not fractionate the glycopeptides like the dual-functional Ti-IMAC done. The greatest number of sialylated glycopeptides were identified in the first fraction and it decreased with the reduction of ACN content and the total number of glycopeptides in other three fractions, the sialylated glycopeptides ratio have the same trends (Figure S2B), which showed opposite trends with the dual-functional Ti-IMAC fractionation. This demonstrated that sialylated glycopeptides were not separated well from neutral glycopeptides in conventional HILIC fractionation. Moreover, the identified M6P glycopeptides in the four fractions were 15, 9, 2, and 0, respectively, which was also lower than the dual-functional Ti-IMAC approach profiled (Figure S2C).
After eluting most glycopeptides, materials were resuspended in 80% ACN to recover the aqueous layer and continually eluted with gradient ACN containing ammonia. In Elution 3 (5th and 6th fraction), only the majority of phosphopeptides were eluted, the results in Figure 4B demonstrated that in the 5th fraction, 3883 phosphopeptides were profiled, of which only 150 were multi-phosphopeptides, while in the 6th fraction, 2634 phosphopeptides were identified and the identified multi-phosphopeptides were increased to 703. In Elution 4 (7th fraction), the ACN content was continually decreased to the aqueous elution buffer, in this fraction, the total number of identified phosphopeptides was decreased to 1504, with 489 of them being multi-phosphopeptides. In the 5-7th fractions, the multi-phosphopeptides ratios were 3.9%, 26.7%, and 32.5%, respectively. It was contrary to the decreasing trend of phosphopeptides, the ratio of multi-phosphopeptides increased gradually (Figure 5B). This counter-intuitive result is likely due to the strong hydrophilicity of the phosphate group on phosphopeptides,23 and in general multi-phosphopeptides are more hydrophilic than mono-phosphopeptides, which will enhance their retention on materials under HILIC conditions.24 These results demonstrated dual-functional Ti(IV)-IMAC materials can not only effectively enrich phosphopeptides with high efficiency, but can also separate multi-phosphopeptides from mono-phosphopeptides according to their differences in hydrophilicity. Comprehensive isolation and characterization of both mono- and multi-phosphopeptides is important for understanding the graded regulation of phosphorylation signal that can more precisely modulate the switch of signal transduction pathways.25-27 Previously, separation of multi-phosphopeptides from mono-phosphopeptides was achieved by applying two enrichment materials with different mono- and multi-phosphopeptide selectivity.28-30 However, these methods cost additional enrichment material and often lead to greater sample loss during the sample transfer.31 Therefore, our simultaneous enrichment and separation strategy through dual-functional Ti(IV)-IMAC materials have unique advantages to enable more comprehensive profiling of mono- and multi-phosphopeptides. It is worth noting that, in the 7th fraction, we also identified 361 glycopeptides with 69.0% being sialylated glycopeptides and 61.9% being multi-sialylated glycopeptides (Figure 4A, Figure 5A), which were remnants of strong acid elution and were continually eluted by aqueous and basic elution conditions.
In Elution 1-3, only a handful of M6P glycopeptides were identified, if any. However, in Elution 4, we profiled 57 M6P glycopeptides (Figure 4C). These results demonstrated that M6P glycopeptides have the strongest retention on Ti(IV)-IMAC materials due to the unique properties of this class of glycopeptides possessing both hydrophilic glycans and negatively charged phosphate groups. Only the highest elution strength with aqueous buffer plus electrostatic interaction interruption reagent could effectively elute these peptides. Therefore, the separation of M6P glycopeptides from common N-glycopeptides and phosphopeptides based on their differences in hydrophilicity and electrostatic interactions using the dual-functional Ti(IV)-IMAC materials can be achieved with systematic optimization.
Through this sequential elution and separation, we identified 3896 N-glycopeptides, 5539 phosphopeptides, and 65 M6P glycopeptides in mouse lung and the identified glycopeptides, phosphopeptides and M6P glycopeptides by the new strategy were 1.9, 2.3, and 4.3 times higher than those achieved using the conventional method, respectively (Figure 4D). And we also performed the HILIC mode Ti-IMAC without fractionation, the result was similar with the standard protein result. The identified phosphopeptides and M6P glycopeptides were similar with the conventional Ti-IMAC method. However, the enriched N-glycopeptides only increased to 295, which was much lower than the conventional HILIC method and demonstrated that they were also suppressed by the co-eluted phosphopeptides in LC-MS/MS analysis. By analyzing the overlap of identified glycoproteins, phosphoproteins, and M6P glycoproteins, we observed that 2 proteins bear all three PTMs, 3 proteins have both M6P glycosylation and phosphorylation, 19 proteins have both common N-glycosylation and M6P glycosylation, and 76 proteins possess both glycosylation and phosphorylation sites, which shed light on the crosstalk analysis of these multiply modified peptides (Figure 4E).
Characterization of common N-glycoproteome and M6P glycoproteome datasets
Of the 3961 N-glycopeptides (including common N-glycopeptides and M6P glycopeptides), we mainly focus on the analysis of unique glycoforms and remove the redundant results. This analysis resulted in the identification of 3279 unique glycoforms, corresponding to 724 N-glycosites and 388 N-glycoproteins. We compared our current results with a published mouse lung glycoproteome.32 The comparison revealed that the two datasets profiled similar number of glycoproteins (388 vs. 394) and glycosites (724 vs. 686), and among the 724 glycosites, 335 were commonly identified in both datasets while 389 were newly identified in our dataset. Our dataset also contained more identifications of glycoforms, whereas 2433 glycoforms were uniquely identified in our dataset (Figure S3).
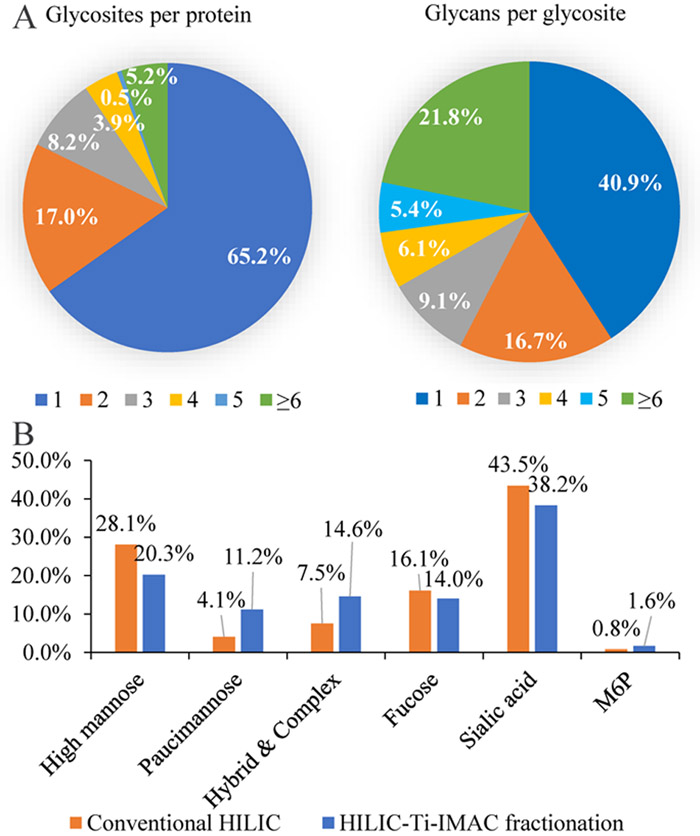
We then analyzed the heterogeneity of the profiled glycoproteome. In brief, 65.2% (253) of glycoproteins possess only one glycosite and each glycoprotein had an average of 1.9 N-linked glycosites (Figure 6A, left). However, the number of glycans per glycosite showed different trend, 59.1% (428) of profiled glycosites were modified with more than one glycan and 21.8% of them have more than six glycans, and each glycosite had an average of 4.5 different glycans (Figure 6A, right). These data demonstrated the significant heterogeneity of N-linked glycosylation.
Figure 6.
Glycoproteome heterogeneity analysis. (A) Distribution of the number of glycosites per glycoprotein (left) and the number of unique glycans per glycosites (right). (B) Glycan categorization of identified unique glycoforms.
In total, we identified 210 unique glycans and categorized the identified glycans into six groups based on the state of their biosynthetic process. The six categories represented a gradual maturation of glycans from mature high mannose, to paucimannose, mannose-6-phosphate, complex/hybrid, and finally fucosylated and sialylated. We found that 20.3% and 11.2% of the identified N-glycoforms possess high mannose and paucimannose glycans, respectively. And 1.6% of them corresponded to M6P glycosylation. 14.6% of the N-glycoforms possessed complex/hybrid glycans. Given our glycan categorization rules, any glycan with a NeuAc and/or NeuGc moiety was categorized as sialylated, therefore some glycans in the sialylated group were also fucosylated. However, the fucosylated glycan type group contained any glycans containing a fucose moiety that was not sialylated. Fucosylated and sialylated glycans corresponded to 14.0% and 38.2% of the identified N-glycoforms (Figure 6B). In total, 66.8% N-glycoforms were complex/hybrid glycans or decorated with fucose or sialic acid, which was in accordance with the results obtained with conventional HILIC method revealing 67.1% of mouse lung glycoproteome contained complex/hybrid glycans. These data demonstrated that, the majority of the glycans in mouse lung have been processed to their mature state.
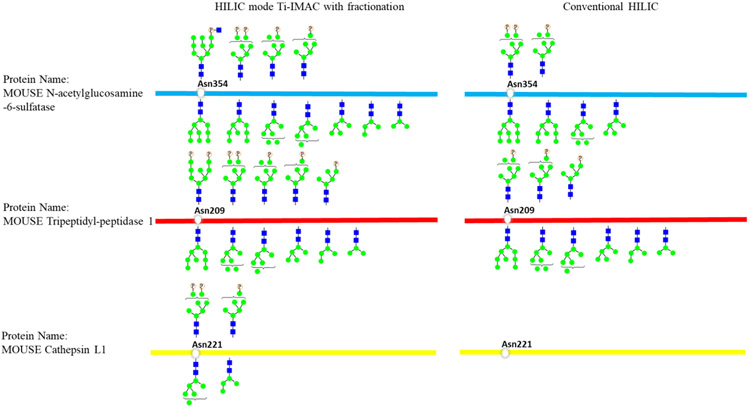
We identified 54 unique M6P glycoforms corresponding to 23 N-glycoproteins. It is interesting that 18 M6P glycoforms have their common N-glycoform counterparts. We selected some of them and showed in the left panel of Figure 7, N-acetylglucosamine-6-sulfatase (Gns) possesses the M6P glycans Man6GlcNAc2Phospho1, Man7GlcNAc2Phospho1, Man7GlcNAc2Phospho2, and Man9GlcNAc3Phospho1 at Asn354, and it also possesses Man6GlcNAc2, Man7GlcNAc2, and Man9GlcNAc2 at the same site. The situation for Tripeptidyl-peptidase 1 (Tpp1) and Cathepsin L1 (Cts1) was similar. These findings are consistent with literature reports that phosphate on M6P glycans can be removed by acid phosphatase after M6P glycoproteins are distributed to their appropriate cellular localization.33 With this in mind, to comprehensively profile the glycosylation of M6P glycoproteins, especially to quantify their changes during specific biological process, the non-M6P counterparts must be considered. Therefore, the simultaneous enrichment strategy is essential for this situation. As shown in the right panel of Figure 7, conventional HILIC method revealed fewer M6P glycoforms and their counterparts than the HILIC mode Ti-IMAC enrichment method, therefore our simultaneous enrichment and separation strategy can achieve more comprehensive profiling and quantification of M6P glycosylation in complex biological sample.
Figure 7.
Mouse lung M6P glycosylation and its phosphate free counterpart revealed by the HILIC mode with fractionation and conventional HILIC mode enrichment.
Characterization of phosphoproteome dataset and its crosstalk with glycoproteome
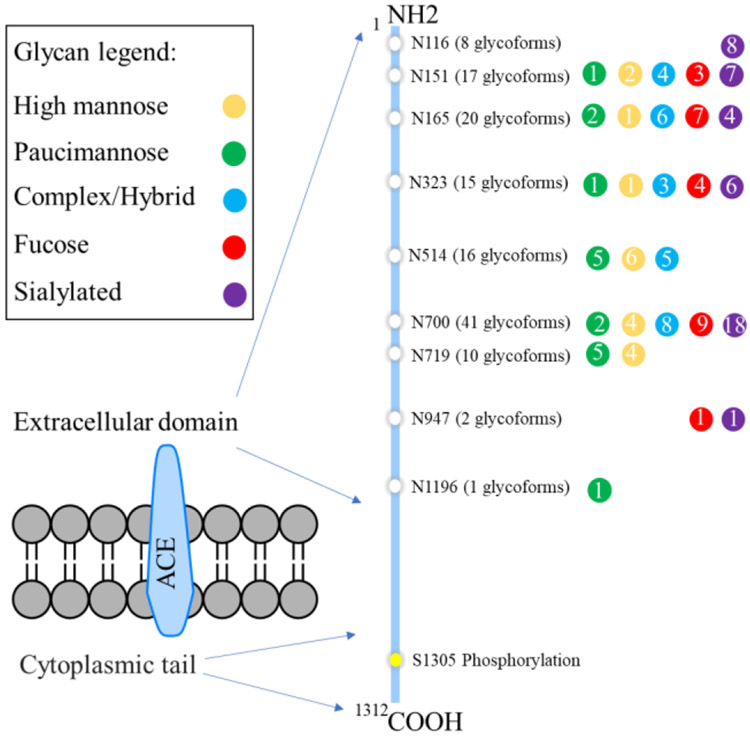
Another great advantage of our method is that we can also perform comprehensive profiling of the phosphoproteome in the same sample. In total, we identified 5539 phosphopeptides corresponding to 2004 phosphoproteins. To examine the potential cross-regulation between glycosylation and phosphorylation, we analyzed the proteins containing these two PTMs. A total of 76 proteins possesses both glycosylation and phosphorylation sites (Figure 4B). A representative example is angiotensin-converting enzyme (ACE), a zinc-dependent peptidase responsible for converting angiotensin I into the vasoconstrictor angiotensin II, which plays a critical role in the blood pressure regulation.34 Moreover, as a relatively nonspecific peptidase that can cleave a wide range of substrates, it also affects many other physiological processes including hematopoiesis, reproduction, renal development, renal function, and immune response.35 It has been demonstrated that N-linked glycosylation is crucial for maintaining the stability and enzymatic activity of ACE, since the expressed ACE without glycosylation is catalytically inactive and subjects to rapid degradation.36 Phosphorylation of ACE cytoplasmic tail at site Ser1305 regulates its retention in the endothelial cell plasma membrane and affect it secretion to extracellular space to perform its function.37 As shown in Figure 8, with our HILIC mode Ti-IMAC enrichment method, we successfully profiled 129 unique glycoforms of ACE across its nine glycosites and identified a phosphopeptide with phosphorylation at site Ser1305. As a comparison, in a published mouse lung glycoproteome, 133 unique glycoforms were mapped on the same nine glycosites in ACE, which was similar with our data (Table S2), however the phosphorylation site was not reported for ACE in that study.32 Our simultaneous enrichment strategy will facilitate more in-depth investigation of the cross-regulation of glycosylation and phosphorylation on ACE function in future studies. This new strategy will also shed light on the large-scale crosstalk analysis of protein glycosylation and phosphorylation based on the simultaneously enriched glycoproteome and phosphoproteome data collected from the same biological sample.
Figure 8.
Glycan diversity and phosphorylation on angiotensin-converting enzyme (ACE).
Conclusion
We explore and demonstrate the potential of Ti-IMAC for the simultaneous enrichment of common N-glycopeptides, phosphopeptides, and M6P glycopeptides from the same complex biological sample. Moreover, as majority of glycopeptides were captured on materials with only hydrophilic interaction, they were eluted first with aqueous buffer containing formic acid (FA) or trifluoracetic acid (TFA). Phosphopeptides were mainly captured on the materials with electrostatic interaction. Therefore, they can be eluted with high acetonitrile (ACN) content buffer containing ammonia. Finally, M6P glycopeptides captured on materials with synergistic electrostatic and hydrophilic interaction can be eluted with low ACN content buffer containing ammonia. The three classes of PTM peptides were successfully separated with appropriate fractionation, therefore, the interference among these three classes of peptides can be substantially eliminated and the coverage of N-glycopeptides, phosphopeptides and M6P glycopeptides were significantly increased to 1.9, 2.3, and 4.3-fold respectively compared with conventional enrichment method. The enrichment and fractionation of three different PTMs from the same biological sample can also facilitate the crosstalk analysis of these three PTMs.
Supplementary Material
Acknowledgements
This work was supported, in part, by the National Institutes of Health Grants U01CA231081, RF1AG052324, and R01 DK071801 (to L.L.), and funds from the National Key R&D Program of China (2016YFA0501402, to M.Y.), the National Natural Science Foundation of China (21535008 and 21525524, to M.Y.). The MS instruments were purchased through the support of NIH Shared Instrument Grants (NIH-NCRR S10RR029531 and S10OD025084 to L.L.) and the University of Wisconsin-Madison, Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy. The authors wish to pay tribute to the late Prof. Hanfa Zou in memory of his great contribution that led the development of Ti(IV)-IMAC materials.
Footnotes
The authors declare no conflict of interest.
References
- (1).Beltrao P; Albanèse V; Kenner Lillian R.; Swaney Danielle L.; Burlingame A; Villén J; Lim Wendell A.; Fraser James S.; Frydman J; Krogan Nevan J. Systematic Functional Prioritization of Protein Posttranslational Modifications. Cell 2012, 150, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Swaney DL; Beltrao P; Starita L; Guo A; Rush J; Fields S; Krogan NJ; Villen J Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Huang J; Wang F; Ye M; Zou H Enrichment and separation techniques for large-scale proteomics analysis of the protein post-translational modifications. J. Chromatogr. A 2014, 1372, 1–17. [DOI] [PubMed] [Google Scholar]
- (4).Doll S; Burlingame AL Mass Spectrometry-Based Detection and Assignment of Protein Posttranslational Modifications. ACS Chem. Biol 2015, 10, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chen Z; Huang J; Li L Recent advances in mass spectrometry (MS)-based glycoproteomics in complex biological samples. Trends Analyt. Chem 2019, 118, 880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Huang J; Qin H; Dong J; Song C; Bian Y; Dong M; Cheng K; Wang F; Sun D; Wang L; Ye M; Zou H In Situ Sample Processing Approach (iSPA) for Comprehensive Quantitative Phosphoproteome Analysis. J. Proteome Res 2014, 13, 3896–3904. [DOI] [PubMed] [Google Scholar]
- (7).Mertins P; Qiao JW; Patel J; Udeshi ND; Clauser KR; Mani DR; Burgess MW; Gillette MA; Jaffe JD; Carr SA Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 2013, 10, 634–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Andaluz Aguilar H; Iliuk AB; Chen IH; Tao WA Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat. Protoc 2020, 15, 161–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hong Y; Zhao H; Pu C; Zhan Q; Sheng Q; Lan M Hydrophilic Phytic Acid-Coated Magnetic Graphene for Titanium(IV) Immobilization as a Novel Hydrophilic Interaction Liquid Chromatography–Immobilized Metal Affinity Chromatography Platform for Glyco- and Phosphopeptide Enrichment with Controllable Selectivity. Anal. Chem 2018, 90, 11008–11015. [DOI] [PubMed] [Google Scholar]
- (10).Tang R; Yu Y; Dong J; Yao Y; Ma S; Ou J; Ye M Facile preparation of bifunctional adsorbents for efficiently enriching N-glycopeptides and phosphopeptides. Anal. Chim. Acta 2021, 1144, 111–120. [DOI] [PubMed] [Google Scholar]
- (11).Zou X; Jie J; Yang B Single-Step Enrichment of N-Glycopeptides and Phosphopeptides with Novel Multifunctional Ti4+-Immobilized Dendritic Polyglycerol Coated Chitosan Nanomaterials. Anal. Chem 2017, 89, 7520–7526. [DOI] [PubMed] [Google Scholar]
- (12).Wang Z; Wang J; Sun N; Deng C A promising nanoprobe based on hydrophilic interaction liquid chromatography and immobilized metal affinity chromatography for capture of glycopeptides and phosphopeptides. Anal. Chim. Acta 2019, 1067, 1–10. [DOI] [PubMed] [Google Scholar]
- (13).Huang J; Dong J; Shi X; Chen Z; Cui Y; Liu X; Ye M; Li L Dual-Functional Titanium(IV) Immobilized Metal Affinity Chromatography Approach for Enabling Large-Scale Profiling of Protein Mannose-6-Phosphate Glycosylation and Revealing Its Predominant Substrates. Anal. Chem 2019, 91, 11589–11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhou H; Ye M; Dong J; Han G; Jiang X; Wu R; Zou H Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J. Proteome Res 2008, 7, 3957–3967. [DOI] [PubMed] [Google Scholar]
- (15).Yu Z; Han G; Sun S; Jiang X; Chen R; Wang F; Wu R; Ye M; Zou H Preparation of monodisperse immobilized Ti(4+) affinity chromatography microspheres for specific enrichment of phosphopeptides. Anal. Chim. Acta 2009, 636, 34–41. [DOI] [PubMed] [Google Scholar]
- (16).Glover MS; Yu Q; Chen ZW; Shi XD; Kent KC; Li LJ Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. Int. J. Mass spectrom 2018, 427, 35–42. [Google Scholar]
- (17).Zhou H; Ye M; Dong J; Corradini E; Cristobal A; Heck AJR; Zou H; Mohammed S Robust phosphoproteome enrichment using monodisperse microsphere–based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc 2013, 8, 461–480. [DOI] [PubMed] [Google Scholar]
- (18).Li F; Ding J Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein & Cell 2019, 10, 550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Nwosu CC; Strum JS; An HJ; Lebrilla CB Enhanced Detection and Identification of Glycopeptides in Negative Ion Mode Mass Spectrometry. Anal. Chem 2010, 82, 9654–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).de Haan N; Yang S; Cipollo J; Wuhrer M Glycomics studies using sialic acid derivatization and mass spectrometry. Nature Reviews Chemistry 2020, 4, 229–242. [DOI] [PubMed] [Google Scholar]
- (21).Feng Y; Li M; Lin Y; Chen B; Li L Multiplex Quantitative Glycomics Enabled by Periodate Oxidation and Triplex Mass Defect Isobaric Multiplex Reagents for Carbonyl-Containing Compound Tags. Anal. Chem 2019, 91, 11932–11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nishikaze T Sialic acid derivatization for glycan analysis by mass spectrometry. Proceedings of the Japan Academy, Series B 2019, 95, 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Alpert AJ Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. A 1990, 499, 177–196. [DOI] [PubMed] [Google Scholar]
- (24).McNulty DE; Annan RS Hydrophilic Interaction Chromatography Reduces the Complexity of the Phosphoproteome and Improves Global Phosphopeptide Isolation and Detection. Molecular & Cellular Proteomics 2008, 7, 971–980. [DOI] [PubMed] [Google Scholar]
- (25).Park K-S; Mohapatra DP; Misonou H; Trimmer JS Graded Regulation of the Kv2.1 Potassium Channel by Variable Phosphorylation. Science 2006, 313, 976–979. [DOI] [PubMed] [Google Scholar]
- (26).Lee CW; Ferreon JC; Ferreon ACM; Arai M; Wright PE Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 19290–19295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Samarasimhareddy M; Mayer G; Hurevich M; Friedler A Multiphosphorylated peptides: importance, synthetic strategies, and applications for studying biological mechanisms. Org. Biomol. Chem 2020, 18, 3405–3422. [DOI] [PubMed] [Google Scholar]
- (28).Zhong H; Xiao X; Zheng S; Zhang W; Ding M; Jiang H; Huang L; Kang J Mass spectrometric analysis of mono- and multi-phosphopeptides by selective binding with NiZnFe2O4 magnetic nanoparticles. Nature Communications 2013, 4, 1656. [DOI] [PubMed] [Google Scholar]
- (29).Tsai C-F; Hsu C-C; Hung J-N; Wang Y-T; Choong W-K; Zeng M-Y; Lin P-Y; Hong R-W; Sung T-Y; Chen Y-J Sequential Phosphoproteomic Enrichment through Complementary Metal-Directed Immobilized Metal Ion Affinity Chromatography. Anal. Chem 2014, 86, 685–693. [DOI] [PubMed] [Google Scholar]
- (30).Choi S; Kim J; Cho K; Park G; Yoon JH; Park S; Yoo JS; Ryu SH; Kim YH; Kim J Sequential Fe3O4/TiO2 enrichment for phosphopeptide analysis by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom 2010, 24, 1467–1474. [DOI] [PubMed] [Google Scholar]
- (31).Long X.-y.; Zhang Z.-j.; Li J.-y.; Sheng D; Lian H.-z. A combination strategy using two novel cerium-based nanocomposite affinity probes for the selective enrichment of mono- and multi-phosphopeptides in mass spectrometric analysis. Chem. Commun 2017, 53, 4620–4623. [DOI] [PubMed] [Google Scholar]
- (32).Liu MQ; Zeng WF; Fang P; Cao WQ; Liu C; Yan GQ; Zhang Y; Peng C; Wu JQ; Zhang XJ; Tu HJ; Chi H; Sun RX; Cao Y; Dong MQ; Jiang BY; Huang JM; Shen HL; Wong CCL; He SM; Yang PY pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun 2017, 8, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Makrypidi G; Damme M; Müller-Loennies S; Trusch M; Schmidt B; Schlüter H; Heeren J; Lübke T; Saftig P; Braulke T Mannose 6 Dephosphorylation of Lysosomal Proteins Mediated by Acid Phosphatases Acp2 and Acp5. Mol. Cell. Biol 2012, 32, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bernstein KE; Khan Z; Giani JF; Cao D-Y; Bernstein EA; Shen XZ Angiotensin-converting enzyme in innate and adaptive immunity. Nature Reviews Nephrology 2018, 14, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bernstein KE; Ong FS; Blackwell W-LB; Shah KH; Giani JF; Gonzalez-Villalobos RA; Shen XZ; Fuchs S A Modern Understanding of the Traditional and Nontraditional Biological Functions of Angiotensin-Converting Enzyme. Pharmacol. Rev 2013, 65, 1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sadhukhan R; Sen I Different Glycosylation Requirements for the Synthesis of Enzymatically Active Angiotensin-converting Enzyme in Mammalian Cells and Yeast. J. Biol. Chem 1996, 271, 6429–6434. [DOI] [PubMed] [Google Scholar]
- (37).Kohlstedt K; Shoghi F; Müller-Esterl W; Busse R; Fleming I CK2 Phosphorylates the Angiotensin-Converting Enzyme and Regulates Its Retention in the Endothelial Cell Plasma Membrane. Circul. Res 2002, 91, 749–756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.