Abstract
Aims/hypothesis
The circadian clock influences both diabetes and immunity. Our goal in this study was to characterise more thoroughly the circadian patterns of immune cell populations and cytokines that are particularly relevant to the immune pathology of type 1 diabetes and thus fill in a current gap in our understanding of this disease.
Methods
Ten individuals with established type 1 diabetes (mean disease duration 11 years, age 18–40 years, six female) participated in a circadian sampling protocol, each providing six blood samples over a 24 h period.
Results
Daily ranges of population frequencies were sometimes large and possibly clinically significant. Several immune populations, such as dendritic cells, CD4 and CD8 T cells and their effector memory subpopulations, CD4 regulatory T cells, B cells and cytokine IL-6, exhibited statistically significant circadian rhythmicity. In a comparison with historical healthy control individuals, but using shipped samples, we observed that participants with type 1 diabetes had statistically significant phase shifts occurring in the time of peak occurrence of B cells (+4.8 h), CD4 and CD8 T cells (~ +5 h) and their naive and effector memory subsets (~ +3.3 to +4.5 h), and regulatory T cells (+4.1 h). An independent streptozotocin murine experiment confirmed the phase shifting of CD8 T cells and suggests that circadian dysrhythmia in type 1 diabetes might be an effect and not a cause of the disease.
Conclusions/interpretation
Future efforts investigating this newly described aspect of type 1 diabetes in human participants are warranted. Peripheral immune populations should be measured near the same time of day in order to reduce circadian-related variation.
Graphical abstract
Supplementary Information
The online version contains peer-reviewed but unedited supplementary material available at 10.1007/s00125-021-05468-6.
Keywords: Circadian rhythms, Clinical, Immune cells, Type 1 diabetes
Introduction
The circadian clock, a system of oscillating neuroendocrine hormonal signalling and cellular transcription factors distributed throughout the body, coordinates human physiology with the daily light/dark cycle, optimising organismal physiologic processes [1]. The clock is an important aspect of diabetes as well: it is essential for proper metabolic control and regulation of beta cell function [2] and the pancreatic islet transcriptome exhibits circadian variation [3]. Both insulin secretion and insulin action exhibit circadian patterns [4]. Qian et al [5] found that mice exposed to constant light had diminished glucose-stimulated insulin secretion due to a decrease in insulin secretory pulse mass. Marcheva and colleagues [6] found that conditional ablation of the pancreatic circadian clock caused diabetes in mice due to defective beta cell function during the late stage of stimulus–secretion coupling. Recently it was shown that, in human type 2 diabetes, alpha and beta cells have attenuated Clock expression disrupting insulin and glucagon releases [7].
Indeed, circadian disruption or dysrhythmia is associated in humans with diabetes and its complications as well [8]. Leproult et al [9] found that the circadian misalignment occurring in shift workers increases diabetes risk through mechanisms not related simply to sleep loss. These mechanisms likely involve the dysregulation of hormones and energetics as the circadian clock machinery controls and is itself controlled by both hormones and metabolic status [10–12]. Using murine diabetic models, circadian dysregulation has been associated with the appearance of complications such as peripheral neuropathies [11], accelerated progression of diabetic complications [13, 14] and reduced efficacy of immunotherapies [15].
The circadian clock is also an important and scientifically well-established aspect of immunity [16, 17]. Circadian patterns of immunity in humans have been previously described [18, 19] and peripheral circadian clocks have been found to regulate immune cell trafficking from peripheral blood to tissues, including the secondary lymph system [20]. Circadian-induced variation of some of the immune populations thought to be involved in type 1 diabetes has been previously described elsewhere [19, 21–27]. However, these studies were conducted in healthy people, were not specific to type 1 diabetes and were characterisations of broad populations, e.g. CD4 and CD8 T cells, and not of population subsets and cytokines that are components of type 1 diabetes autoimmunity (e.g. central memory, effector memory and regulatory T cell [Treg cell] subpopulations). We sought to address this gap in knowledge and more thoroughly characterise the circadian variation and rhythmicity in peripheral blood cell populations thought relevant to, or reflective of, the autoimmune process involved in type 1 diabetes and to compare these patterns with those previously observed in healthy control (HC) groups. We also sought to expand this circadian characterisation to include cytokines in order to capture a greater understanding than has heretofore been accomplished. Finally, we sought to fill another shortcoming in the literature by seeking to analyse and interpret our mechanistic findings in a clinically meaningful context.
Methods
Study participants
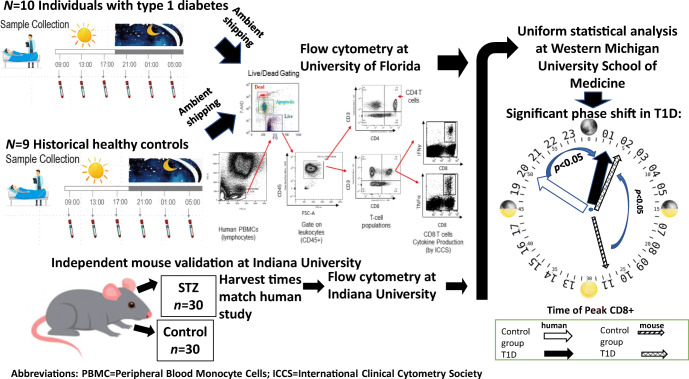
Participants were enrolled at the Indiana University (IU) School of Medicine. After informed consent, participants underwent a medical history check and physical examination. Participants were between 18 and 40 years old, had type 1 diabetes for at least 12 months without any other clinically significant diseases or illnesses, weighed at least 110 pounds (49.9 kg) and were willing to provide written informed consent, abstain from alcohol, maintain a regular sleep schedule for 1 week prior to the day of the trial and spend 24 h in the research facility in compliance with study requirements. Participants were excluded if they did shift work that required changes in sleep patterns, if they had travelled between time zones within 1 week prior to the study or if they were taking any immune modulatory medications. The IU Institutional Review Board approved all study procedures.
Study protocol
Participants were admitted to the Indiana Research Center between 07:00 and 08:00 hours on the day of the study. Participants were allowed an unrestricted diet during the study, including caffeinated beverages. Lights were turned off at 23:00. Blood samples were obtained via venipuncture at (±30 min) 09:00, 13:00, 17:00 and 21:00 hours on day 1, and 01:00 and 05:00 hours on the following day. Blood samples were aliquoted, with one sample processed immediately at each collection time at the IU Research Center (‘fresh’) and the other shipped (ambient) to University of Florida (UF) (‘shipped’) for processing. The shipped samples were analysed at UF in order to compare the participants with type 1 diabetes against HC samples that had been collected earlier and which had also been shipped (ambient) to UF (HC data reported by Beam et al [28]). Both the type 1 diabetes and HC samples were processed under the same protocol using the same cytometer and by the same UF personnel. All samples were handled in accordance with IU guidelines and regulations. Participants were discharged after the final sample collection. A schematic of the study design is shown in electronic supplementary material (ESM) Fig. 1.
Flow cytometry
All whole blood (200 μl) samples (HC, fresh type 1 diabetes and shipped type 1 diabetes) were stained with the same antibody cocktail designed to provide a broad profile of immune cell subsets [29].
All antibodies were purchased from Biolegend (San Diego, CA, USA) unless indicated otherwise: APC/H7-CD3 (SK7, BD Biosciences, San Jose, CA, USA), BB515-CD45RA (HI100, BD Biosciences), PE-Cy7-HLA-DR (LN243), BV650-CD4 (RPA-T4), BV711-CD8 (SK1), BV510-CD14 (M5E2), PerCPCCy5.5-CD16 (3G8), AlexaFluor647-CD25 (BC96), BV605-CD56 (HCD56), PE-CD127 (A019D5) and BV-421 CD197 (G043H7). Antibodies were mixed in appropriate concentrations in BD Horizon Brilliant stain buffer (BD Biosciences) and 50 μl was added for each sample and fluorescence minus one (FMO) control samples. Samples were incubated for 30 min at room temperature and protected from light. Then, 2 ml of Fix/Lyse 1X (eBioscience, San Diego, CA, USA) was added and incubated at room temperature for 5 min. Two successive washing/centrifugation (5 min, 450 g) steps were performed with BD FBS stain buffer (BD Biosciences). The pellet was resuspended in 300 μl of BD FBS stain buffer and stored at 4°C. Samples were acquired within 24 h on a BD Fortessa cytometer with the same configurations at the Flow Cytometry Resource Facility at IU and data were analysed by FlowJo (FlowJo, Ashland, OR, USA) using the same template as for the HC samples that were also processed at UF.
Cytokines
Plasma was isolated from EDTA tubes by centrifugation with the remaining sample after flow cytometry aliquots were dispensed. This was then immediately frozen until batch analyses using Luminex (Austin, TX, USA) technology and kits from Millipore (Burlington, MA, USA) as previously described [28].
Clock genes
Total RNA was isolated from each Tempus Blood RNA tube using the Tempus Spin RNA isolation kit (ThermoFisher, Waltham, MA, USA). RNA was reverse transcribed using the High-Capacity RNA-to-cDNA kit (ThermoFisher). Relative gene expression was quantified using a custom TaqMan array using validated gene expression assays (ESM Table 1).
Murine validation experiment
Male C57BL6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) at 8 weeks of age. Mice were housed in the institutional animal care facilities at the IU Medical School with a strict 12 h:12 h light/dark cycle. Diabetes was induced with an injection of streptozotocin (STZ) (50 mg/kg). Animals were confirmed to be diabetic after 1 month of housing and when the serum glucose level was above 13.9 mmol/l for at least two consecutive measurements. At 4 months of age, diabetic and control mice were randomly assigned to a time point of a 24 h cycle and terminated at ZT (‘zeitgeber’; hours since ‘lights on) times ZT1, ZT5, ZT9, ZT13, ZT17 and ZT21. Single-cell suspensions were prepared from whole blood at the termination of the experiment. Samples were fixed and data were acquired on an LSR II flow cytometer (Beckman Coulter Life Sciences, Indianapolis, IN, USA). Further details are provided in the ESM Methods.
Statistical methods
We followed study design protocols and analysis methodologies that are commonly found in human circadian studies, including sample size selection. In addition, we adjusted for individual participant-level differences in the dynamic range of immune factors via standardisation prior to analysis and adjusted for individual variation in the timing of individual participant-level circadian rhythms by incorporating the simultaneous measurement of cortisol in our statistical models. Statistical methods for systems immunology analysis of the data follow those used previously by the team [28]. Further details can be found in the ESM Methods.
Results
Demographics
Ten individuals with type 1 diabetes were recruited for the study. Six (60%) were female and four (40%) were male. Mean age was 27 years (range: 18–40 years). Average time post diagnosis was 11 years (minimum 1 year). A comparison with the baseline characteristics of the HC individuals is provided in ESM Table 2.
Several immune populations and serum cytokine levels exhibit appreciable daily variation and biologically significant circadian rhythmicity in type 1 diabetes patients with established disease
Daily variation (the daily difference between highest and lowest levels or ‘daily range’ when comparing the six timepoint measurements for an individual) in adaptive immune populations was often found to be appreciable (Table 1). B cells (expressed as percentage lymphocytes) varied by as much as 13.8 percentage points within one individual. For the average individual, B cells varied by 4%. In one individual, the CD4 effector memory subpopulation varied by 10.4%, while for another individual CD8 effector memory varied by as much as 31.9%. The average high–low differences for these populations were 5.8% and 10.7%, respectively. In contrast, CD4 Treg cells exhibited much less daily variation. The maximum range observed within a single participant was a daily difference of 2.4% with an average of 1.0%. Similarly, natural killer (NK) cells of the innate immune system were also found to be highly variable, with one participant having a daily difference of 9.4% (average difference=4.7%). On the other hand, dendritic cells (DCs) were found to have minor variation: the maximum daily difference was 0.7% with an average of 0.34%. The majority of the immune cells measured in circulation also exhibited a statistically significant circadian cosine pattern (Table 2).
Table 1.
Daily ranges of immune population frequencies and plasma cytokine levels in ten participants with type 1 diabetes
| Population | Minimum | Maximum | Mean | ICC |
|---|---|---|---|---|
| Leucocytes (%) | ||||
| Granulocyte | 8.4 | 43.3 | 19.36 | 43% |
| Monocytic leucocytes | 8.3 | 42.2 | 19.3 | 43% |
| Lymphocytes (%) | ||||
| B cell | 1.2 | 13.8 | 4.02 | 50% |
| DC | 0.2 | 0.7 | 0.34 | 53% |
| Monocyte | 2.9 | 11.2 | 7.93 | 69% |
| Classical monocyte | 3.9 | 12.8 | 7.82 | 49% |
| Non-classical monocyte | 3.9 | 12.8 | 7.82 | 49% |
| NK | 1.2 | 9.4 | 4.65 | 21% |
| CD56dim | 2.6 | 11.8 | 6.49 | 71% |
| CD56bright | 2.6 | 11.8 | 6.49 | 71% |
| NKT | 0.2 | 5.0 | 0.99 | 69% |
| CD4 T cell | 5.3 | 26.6 | 11.41 | 19% |
| Naive | 2.5 | 11.7 | 7.38 | 94% |
| Effector memory | 0.9 | 10.4 | 5.79 | 92% |
| Central memory | 1.5 | 9.7 | 5.56 | 97% |
| TEMRA | 0.1 | 5.3 | 0.93 | 59% |
| Treg | 0.4 | 2.4 | 0.95 | 52% |
| CD8 T cell | 1.7 | 15.3 | 5.64 | 72% |
| Naive | 3.3 | 27.1 | 12.07 | 85% |
| Effector memory | 3.6 | 31.9 | 10.69 | 71% |
| Central memory | 1.2 | 6.6 | 3.47 | 95% |
| TEMRA | 1.0 | 6.9 | 2.78 | 96% |
| CD4−CD8− | 0.5 | 4.0 | 1.39 | 81% |
| CD4CD8 | 0.1 | 0.7 | 0.34 | 61% |
| Unidentified | 7.3 | 66.9 | 20.42 | 0% |
| Plasma cytokines (pg/ml) | ||||
| IFNγ | 0.4 | 1.7 | 1.08 | 83% |
| IL-2 | 0.0 | 1.5 | 0.62 | 84% |
| IL-4 | 2.0 | 11.9 | 6.12 | 89% |
| IL-6 | 1.1 | 5.2 | 2.21 | 73% |
| IL-12 | 0.1 | 0.9 | 0.37 | 80% |
For each component, data presented are minimum, maximum and mean of the ranges found across all time points in each of the participants
ICC, intraclass correlation
Table 2.
Adaptive and innate immune population circadian peak level and time of peak in participants with type 1 diabetes
| Immune cell population | Circadian rhythmicity | Peak (SD units) | 95% CI | Time of peaka | 95% CI |
|---|---|---|---|---|---|
| Granulocytes | †‡ | 0.67 | 0.4015, 0.9699 | 12:54 | 10:48, 14:24 |
| Monocytic/lymphocytes | †‡ | 0.68 | 0.4245, 0.9868 | 00:24 | 00:04, 23:54 |
| B cells | †‡ | 0.76 | 0.5116, 1.0415 | 01:06 | 00:09, 23:48 |
| DCs | †‡ | 0.61 | 0.3484, 0.9135 | 00:42 | 00:08, 23:54 |
| Monocytes | †‡ | 0.51 | 0.2350, 0.8465 | 10:24 | 07:48, 12:56 |
| Classical monocytes | †‡ | 0.75 | 0.5147, 1.0212 | 03:54 | 02:24, 5:30 |
| Non-classical monocytes | †‡ | 0.75 | 0.5073, 1.0218 | 15:54 | 14:24, 17:30 |
| NK cells | 0.34 | 0.1028, 0.6932 | 13:30 | 08:42, 17:36 | |
| CD56dim NK cells | 0.37 | 0.1126, 0.7228 | 15:05 | 10:48, 18:57 | |
| CD56bright NK cells | 0.36 | 0.1082, 0.7175 | 03:06 | 00:24, 23:04 | |
| NKT cells | † | 0.45 | 0.1899, 0.7773 | 15:18 | 12:12, 16:12 |
| CD4 T cells | †‡ | 0.89 | 0.6418, 1.1502 | 00:18 | 00:02, 23:57 |
| CD4 naive T cells | †‡ | 0.74 | 0.4818, 1.0243 | 02:36 | 01:12, 4:12 |
| CD4 effector memory T cells | †‡ | 0.48 | 0.2039, 0.8316 | 11:48 | 08:58, 14:30 |
| CD4 TEMRA | 0.12 | 0.0453, 0.5306 | 11:36 | 22:24, 1:24 | |
| CD4 central memory T cells | 0.23 | 0.0618, 0.6129 | 16:24 | 02:54, 22:48 | |
| Treg cells | †‡ | 0.79 | 0.5150, 1.0817 | 23:30 | 00:03, 23:58 |
| CD8 T cells | †‡ | 0.86 | 0.6293, 1.1150 | 01:04 | 00:06, 23:36 |
| CD8 naive T cells | †‡ | 0.93 | 0.6974, 1.1822 | 01:06 | 00:12, 2:12 |
| CD8 effector memory T cells | †‡ | 0.95 | 0.7250, 1.1971 | 12:48 | 11:54, 13:36 |
| CD8 TEMRA | 0.30 | 0.0891, 0.6603 | 14:54 | 08:54, 19:03 | |
| CD8 central memory T cells | 0.31 | 0.0979, 0.6590 | 16:54 | 12:06, 21:24 | |
| CD4−CD8− T cells | 0.16 | 0.0495, 0.5478 | 15:18 | 02:06, 22:30 | |
| CD4CD8 T cells | †‡ | 0.50 | 0.2205, 0.8425 | 00:03 | 23:54, 00:05 |
| Unidentified cells | †‡ | 0.71 | 0.4408, 1.0231 | 13:30 | 12:06, 15:00 |
a24 h clock time
†Statistically significant, p < 0.05
‡Statistically significant at 10% FDR; see ESM Table 4
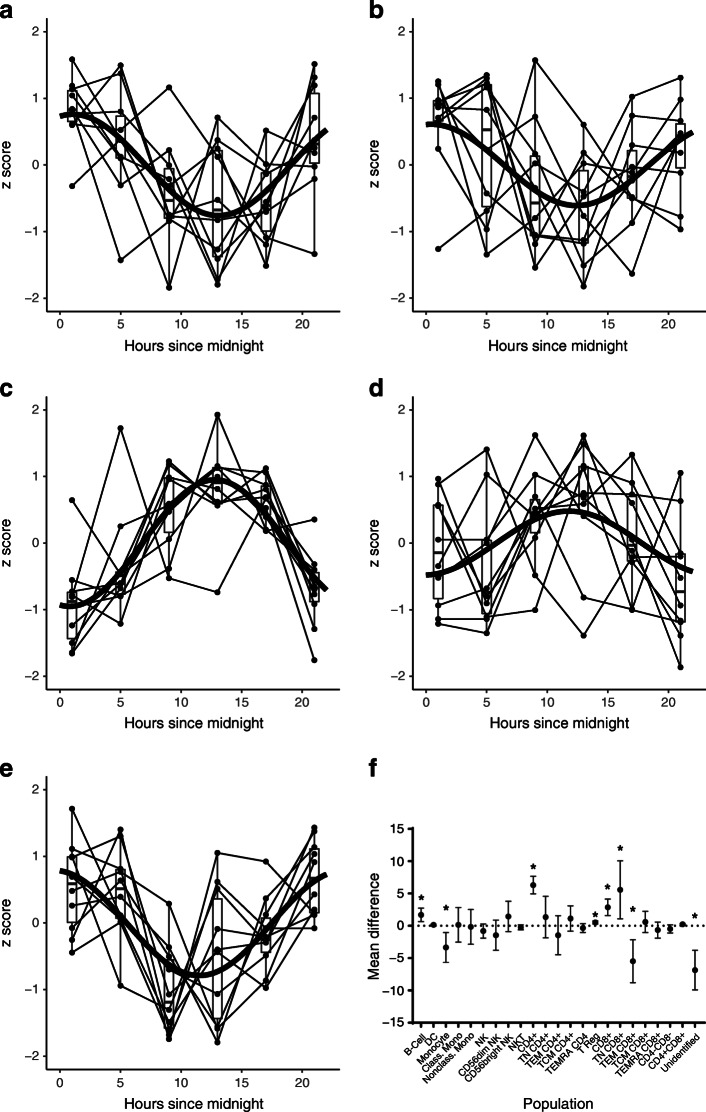
Inspection of circadian patterns (Table 2 and Fig. 1) confirms a well-known ‘chrono-phenotype’ demarcating two major subgroups of immune cell populations: one group that peaked during the ‘night’ (18:00 to 06:00 hours; Fig. 1a,b,e) and one group that peaked during the ‘day’ (06:00 to 18:00 hours; Fig. 1c,d). Night-peaking cell populations were lymphocytes, B cells, DCs, naive CD4 and CD8 T cells, Treg cells and classical monocytes. Day-peaking cell populations were granulocytes, non-classical monocytes, natural killer T (NKT) cells, effector memory CD4 and CD8 T cells, and other unidentified cells.
Fig. 1.
Circadian patterns in type 1 diabetes. (a–e) Daily variations observed in immune populations that have statistically significant circadian rhythmicity. A statistically fitted circadian curve is superimposed on data from each participant. Box-plots summarise data at each sampling time point, N=10. (a) B cells, (b) DCs, (c) effector memory CD4, (d) effector memory CD8 and (e) Treg . (f) Mean within-subject difference (95% CI) in cell populations sampled at 01:00 minus 09:00 hours in type 1 diabetes patients. Statistically significant differences (p < 0.05) are indicated by asterisks. Class. mono., classical monocytes; Nonclass. mono., non-classical monocytes; TN, naive T cells; TEM, effector memory T cells; TCM, central memory T cells
Among the plasma cytokines, IL-4 exhibited the largest daily difference (Table 1): one individual experienced a high–low difference of 11.99 pg/ml. The average daily difference of 6.12 pg/ml for this cytokine exceeded that of all other cytokines. Contrasted with IL-12, having an average daily difference of 0.37 pg/ml, the difference in variability with IL-4 is appreciable. Although only Il-6 was found to have statistically significant circadian rhythmicity, all cytokines were estimated to peak during the early evening (i.e. 17:00 to 21:00 hours) (Table 3).
Table 3.
Plasma cytokine circadian peak level and time of peak in participants with type 1 diabetes
| Plasma cytokine | Circadian rhythmicity | Peak (SD units) | 95% CI | Time of peaka | 95% CI |
|---|---|---|---|---|---|
| IFNγ | 0.20 | 0.01, 0.42 | 17:45 | 04:39, 23:38 | |
| IL-2 | 0.21 | 0.01, 0.43 | 17:14 | 04:45, 23:06 | |
| IL-4 | 0.15 | 0.01, 0.30 | 18:14 | 02:46, 23:50 | |
| IL-6 | † | 0.51 | 0.23, 0.85 | 20:22 | 17:30, 22:30 |
| IL-12 | 0.33 | 0.15, 0.78 | 18:07 | 09:20, 22:29 |
a24 h clock time
†Statistically significant, p < 0.05
In summary, daily ranges of immune variables in type 1 diabetes patients were large and several immune populations and cytokines thought to be relevant to type 1 diabetes exhibited statistically significant circadian rhythmicity.
Biologically and clinically significant circadian peak–trough differences
On the basis of our circadian pattern analysis we observed that, in general, peaks and troughs occur near 09:00 and 01:00 hours. We then analysed the within-subject differences (01:00 vs 09:00 hours) in each of the cell populations measured in this study (Fig. 1f). We first observed that in several populations these approximate peak and trough values differ significantly statistically from zero (p<0.05), suggesting that circadian variation is not inconsequential fluctuation, but is indicative of true biologically significant changes (i.e. true change in mean population frequency) in cell population frequencies during the day.
Second, we observed that some of the differences are of a magnitude that is considered clinically significant as well. For example, Herold and colleagues [30] found that a reduction as small as 1.7% in baseline CD8 effector memory populations discriminated participants treated with teplizumab (anti-CD3mAb) who subsequently ‘responded’ (preserved beta cell function) from those who did not, although they were also given the active treatment. As can be observed from Fig. 1f, CD8 effector memory and several other immune populations had peak–trough differences of this magnitude or larger. Thus, circadian variation can also be of a clinically significant magnitude.
Peripheral immune synchronisation
Correlations among the immune factors are presented in ESM Fig. 2. The data are ordered by acrophase (or time of peak) starting at 6:00 hours, which was approximately the time of daylight on the day of the study. Importantly, this arrangement of the data shows the succession in peak occurrence so that immune factors that peak after another factor appear sequentially and, along with the identification of biologically significant and false discovery rate (FDR)-significant correlations, documents the extent of phase synchronisation in the peripheral immune system. We define a ‘biologically significant’ correlation to occur whenever variation in one immune factor accounts for more than 10% of the variation seen in another factor, as indicated by the square of the correlation between them (known as ‘r2’ or ‘coefficient of determination’). An FDR-significant correlation is one that is statistically significant after controlling the FDR to be no greater than 10% among the many correlations generated by our data (see ESM Methods).
The 28 sequentially ordered immune factors give rise to 27 peak-adjacent correlations of which eight (30%) meet the criteria for biological and FDR significance, thereby establishing phase synchronisation between the following pairs: NK, CD8 T-effector memory cells (r=0.55); CD8 T-effector memory CD45RA-expressing cells (TEMRA), NK CD56dim (r=0.73); NKT, CD4−CD8− (r=0.69); CD4 central memory T cells, CD8 central memory T cells (r=0.54); IL-4, IFNγ (r=0.62); IL-4, IL-6 (r=0.57); B cells, CD8 (r=0.52); classical monocytes, NK56bright (r=0.54).
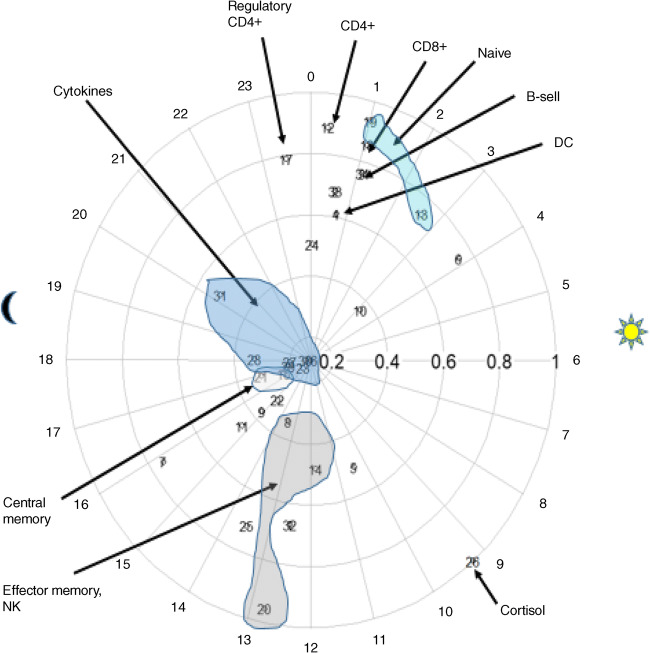
Further insight into synchronisation within the peripheral immune system in type 1 diabetes is given by Fig. 2, in which time of peak (‘acrophase’) and peak level (‘amplitude’) of the fitted circadian patterns are plotted on a 24 h clock face. From this figure it can be observed that phase synchronisation occurs within functional phenotypes.
Fig. 2.
Functional synchronisation. Chronograph of immune components measured in type 1 diabetes (shipped samples). Each numbered circle represents an immune component. Placement of the circle on the 24 h ‘clock’ indicates acrophase (time of peak). Amplitude, or height of peak (expressed in standard deviation units), is indicated by distance from the clock centre. Times of sunrise and sunset on the day of the study are indicated by sun and moon symbols. Some data points have been visually grouped by functional phenotype
Circadian rhythmicity of the Clock gene transcriptome
Clock genes are found in most cells and express proteins which act as transcription factors, producing circadian rhythmicity in cellular functioning via negative feedback looping. For example, clock gene PER (for Period) is a repressor of the expression of the transcription activator CLOCK/BMAL1 heterodimer [1]. Circadian variation in expression of the transcription factor PER, due to entrainment of the organism to external variation in light, is then thought to drive circadian oscillation in transcription activators CLOCK (circadian locomotor output cycles protein kaput) and BMAL1 [1] (brain and muscle ARNT [aryl hydrocarbon receptor nuclear translocator]-like protein 1). ESM Table 3 reports analysis of circadian rhythmicity in clock gene transcripts measured from peripheral whole blood sampled from the participants with type 1 diabetes. Seven (64%) of the 11 clock genes measured in this study displayed significant circadian rhythmicity.
Comparison with HC individuals
Data from a historical group of ten healthy individuals (six male participants, age: 22–38 years) who previously had participated in a circadian sampling study [28] were used for comparison. The HC samples had been shipped to the UF for processing and FACS analysis. These data were compared with aliquoted samples from participants with type 1 diabetes which had also been shipped to UF and processed in the same way as the HC samples (see Methods).
Table 4 reports the results of comparing the circadian features of the type 1 diabetes and historical HC cohorts (note that no cytokine comparisons were made because only one cytokine was found to have significant circadian rhythmicity, which was no longer significant after adjusting for false discovery). Differences in acrophase and their associated 95% CIs suggest that statistically significant (p < 0.05) phase shifts (CI does not include the value zero) occurred in the time of peak occurrence of B cells (+4.8 h from that observed in HC samples), CD4 and CD8 T cells (~+5 h) and their naive and effector memory subjects (~+3.3 to +4.5 h), and Treg cells (+4.1 h).
Table 4.
Circadian amplitude and acrophase differences between immune populations of HC samples and type 1 diabetes patients (shipped samples)
| Population | Amplitude | Acrophase | ||||
|---|---|---|---|---|---|---|
| T1D–HC | 95% CI | T1D–HC | 95% CI | |||
| Granulocytes | −0.26 | −0.73 | 0.2 | 3.06† | 0.34 | 5.36 |
| Monocytic/lymphocytes | −0.24 | −0.71 | 0.23 | 3.18† | 0.48 | 5.39 |
| B cells | 0.04 | −0.43 | 0.5 | 4.76† | 2.46 | 7.62 |
| DCs | 0.16 | −0.24 | 0.63 | 3.78 | −3.18 | 12.14 |
| Monocytes | −0.2 | −0.71 | 0.31 | −0.9 | −5.42 | 3.16 |
| Classical monocytes | 0.24 | −0.17 | 0.68 | 5.31† | 1.34 | 8.14 |
| Non-classical monocytes | 0.24 | −0.18 | 0.66 | 5.32† | 1.46 | 8.12 |
| NK cells | −0.07 | −0.54 | 0.38 | 3.96 | −4.44 | 9.22 |
| CD56dim NK cells | 0.12 | −0.4 | 0.56 | 4.14 | −9.69 | 10.08 |
| CD56bright NK cells | 0.12 | −0.39 | 0.57 | 4.15 | −9.82 | 10.14 |
| NKT cells | 0.11 | −0.39 | 0.65 | 4.21 | −1.32 | 7.35 |
| CD4 T cells | 0.25 | −0.17 | 0.69 | 5.38† | 2.75 | 7.76 |
| CD4 naive T cells | −0.14 | −0.51 | 0.18 | 4.54† | 2.44 | 6.56 |
| CD4 effector memory T cells | −0.33 | −0.77 | 0.05 | 3.32† | 0.42 | 6.03 |
| CD4 TEMRA | −0.06 | −0.53 | 0.39 | 1.08 | −4.35 | 8.59 |
| CD4 central memory T cells | −0.08 | −0.52 | 0.25 | −8.45 | −12.0 | 3.23 |
| Treg cells | 0.32 | −0.15 | 0.8 | 4.1† | 1.10 | 6.98 |
| CD8 T cells | 0.06 | −0.33 | 0.44 | 5.28† | 3.42 | 7.11 |
| CD8 naive T cells | 0.23 | −0.19 | 0.63 | 3.77† | 1.75 | 5.4 |
| CD8 effector memory T cells | 0.33 | −0.08 | 0.74 | 4.27† | 1.59 | 6.71 |
| CD8 TEMRA | 0.01 | −0.43 | 0.44 | 5.6 | −7.18 | 11.7 |
| CD8 central memory T cells | −0.18 | −0.67 | 0.25 | 3.31 | −8.63 | 9.96 |
| CD4−CD8− T cells | −0.3 | −0.77 | 0.17 | 6.76 | −7.67 | 11.69 |
| CD4CD8 T cells | 0.14 | −0.37 | 0.54 | 4.73† | 0.11 | 11.28 |
| Unidentified cells | 0.18 | −0.27 | 0.64 | 8.12† | 5.21 | 10.96 |
†Statistically significant difference, p < 0.05, according to 95% CI
T1D–HC, difference between samples from patients with type 1 diabetes and HC samples
Validation in the mouse STZ diabetes model
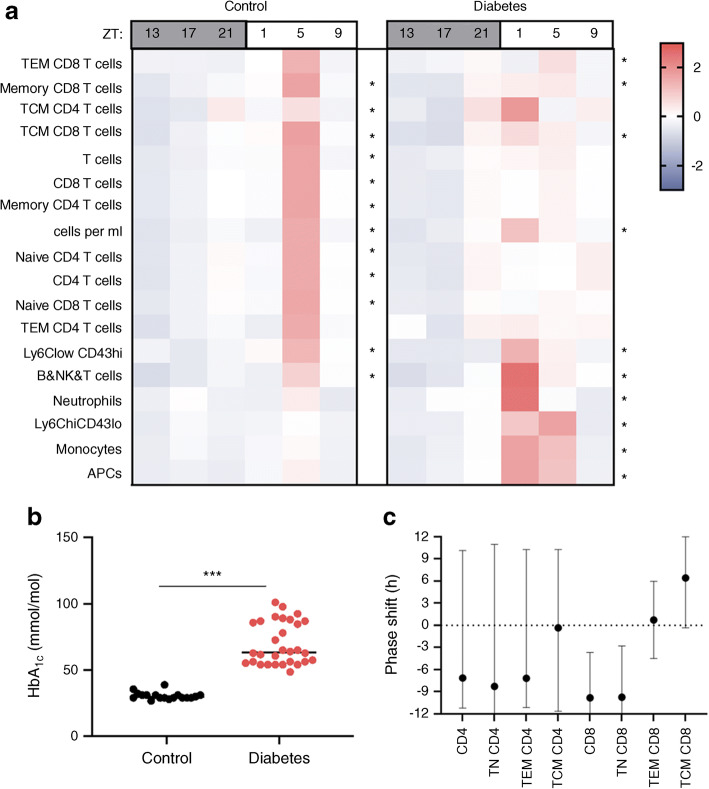
Results from an independent validation experiment using the STZ mouse model (see Methods) are presented in Fig. 3. Heatmaps of mean count levels across time (Fig. 3a) provide evidence of statistically significant circadian rhythmicity in both control and diabetic mice. As observed in our human study, statistically significant phase shifting was observed in the CD8 T cell population in diabetic mice (Fig. 3c). The extent of diabetes in the STZ cohort is depicted in Fig. 3b, which confirms that this experiment represents circadian aspects of autoimmune diabetes in the mouse model.
Fig. 3.
Dysrhythmia in the murine type 1 diabetes model. (a) Heatmap of z scores of cell population counts in blood from control (left) and diabetic (right) mice comparing circadian variation between the two. Asterisks indicate significant circadian rhythmicity, *p < 0.05. (b) HbA1c levels in STZ-induced diabetes. (c) Phase shift in STZ-induced diabetes compared with control mice. Data are mean phase difference (diabetic−control) and 95% interval on the difference using percentage data. TEM, effector memory T cells; TCM, central memory T cells; APC, antigen-presenting cells
Discussion
Circadian disruption has been found to be associated with diabetes and its complications [14, 31] as well as other autoimmune diseases such as arthritis [32] and lupus [33]. Our study now provides preliminary evidence of circadian dysrhythmia in type 1 diabetes. Importantly, significant dysrhythmia was observed in the CD8, CD4 and CD4 Treg cell populations for which, in all cases, the peak occurred significantly later in the day than was observed in HC samples.
In our parallel murine experiment, a significant phase shift was also observed in CD8 T cells. Since the STZ murine model reflects diabetes arising from induced pancreatic damage and subsequent altered immunity and glucose metabolism, we hypothesise that type 1 diabetes immune dysrhythmia is likely a result of, and not an early initiator of, this autoimmune disease.
Circadian ‘phase shifting’ has been implicated in other diseases [34] and so our discovery is supported by the literature. Yet, given the limitations of our study, further research is needed to challenge our finding of dysrhythmia before it is investigated as a possible actor in type 1 diabetes autoimmunity.
Although our study is not the first to characterise circadian rhythmicity in the human immune system, to the best of our knowledge, it is the first to provide such characterisation in immune populations and cytokine levels in a type 1 diabetes population. Moreover, our focused analysis of daily variations led to our novel discovery that circadian variation is not only biologically evident, but also is of a magnitude that has been determined to be clinically significant in some scientific investigations of therapies designed to intercede in type 1 diabetes disease progression. Importantly, we determined that mean daily peak–trough differences in CD8 effector memory populations are at a magnitude similar to the baseline difference between clinical ‘responders’ and ‘nonresponders’ to teplizumab as reported by Herold et al [30]. This finding raises a very important question related to baseline stratification efforts aimed at selecting individuals who are most likely to respond for either clinical trials or, eventually, for patient-tailored immunotherapies: if individuals with type 1 diabetes experience a daily change in CD8 effector memory populations as great as that determined to be associated with positive clinical response, how could single measurements taken at arbitrary points in time be maximally informative? Additionally, we hypothesise that the accumulated or continuous level of ‘exposure’ to immune populations across the day could improve our immune profiling at baseline and permit improved prediction of treatment response. Also, as we start to recognise and characterise endotypes of type 1 diabetes, differences in circadian patterns may identify an important new phenotype.
Druzd and colleagues [20] observed that adaptive immune responses to immunisation and pathogens depend on the time of day of exposure to the immunogen. Previous human studies also suggest that, due to circadian rhythmicity of immune populations, the timing of therapies can modulate the effectiveness of immune therapies [35]. For example, Long et al [36] show that administration of hepatitis A and flu vaccines, respectively, in the morning yielded the highest antibody titres compared with at other times. In a study of the immunosuppressive mycophenolate mofetil (MMF) in Wistar rats, Dridi et al [37] found that the level of renal toxicity was a function of the time of day of administration. This latter finding is of particular importance to type 1 diabetes since MMF has been previously tested in combination with daclizumab as a treatment for recent-onset type 1 diabetes [38]. Some therapies may be most effective when they are given in a way that maximises their effect on cell populations that are circulating; others may be better given when immune cells are resident in lymphatic tissues. Since our study has established the existence of circadian rhythmicity in immune factors considered relevant to the autoimmune process in type 1 diabetes, we therefore urge that further research be conducted of the efficacy of chrono-therapeutic approaches to immune modulation therapies.
In addition, since our data show that circadian variation leads to biologically and clinically significant differences in biomarker levels during the day, we observe that selection of the timing of biomarker measurement could modulate the range of possible immune responses. In addition, peripheral immune populations should be measured near the same time of day in order to reduce circadian-related variation and improve accuracy in the determination of responses to therapy.
Our finding of circadian rhythmicity in the clock gene transcriptome is in keeping with previous literature and, hence, provides another degree of external validation to our study. Previous murine models established the circadian rhythmicity of PER [39] and other clock genes [40], and our study has confirmed these findings in components of human whole blood.
Importantly, we also provide confirmatory evidence of significant circadian rhythmicity in human circulating cells of the clock genes CLOCK and ARNTL, whose mutations have been linked to the development of diabetes in mice [6]. Moreover, Lebailly and colleagues [41, 42] hypothesised that the clock gene ARNTL2 specifically is a candidate gene for type 1 diabetes as it controls expression of the IL21 gene, and the IL-21 cytokine is necessary for the development of diabetes in the NOD mouse [43]. We believe our work supports further investigation into this hypothesis.
Besides the use of historical control samples and the absence of technical replication to control variability, our study was limited in its precision to estimate the time of peak occurrence. Many of the CIs are wide, with some nearly covering a 12 h period. Since we based our sample size determination on previous studies, we conclude that more than ten participants are needed in order to ensure that adequate statistical precision is achieved in circadian studies of human immunity, even though ten participants proved sufficient to detect circadian rhythmicity in many populations and cytokines. Another limitation comes from our enrolment of individuals with established disease and who, therefore, have had long exposure to exogenous insulin and altered metabolism, each of which might alter circadian rhythmicity. Further research is needed to determine if there were confounding factors in our findings and interpretations. However, after adjusting for false discovery, we did not find sex or cortisol level to significantly alter circadian patterns. In addition, we note that the times of peak cortisol level were almost identical between the HC and type 1 diabetes cohorts. It is also very important to recognise that our findings are based on measurements taken from the peripheral immune system and that we have not directly measured the populations that are residing in lymph nodes or their rhythmic recruitment to pancreatic islets. Yet, recent research suggests that the day- and night-peaking immune compartments could be reciprocating manifestations of circadian-timed lymph trafficking [20], and so our observations might be translatable to the lymph system by ‘reflection’. Finally, we note that we used shipped samples in our comparison of HC and type 1 diabetes patterns, and differences in shelf-time or other factors related to shipping between the two cohorts might have confounded our results. A comparison using fresh samples might yield different results. For example, the mouse study used fresh samples and supported our human sample observation of phase shifting in CD8 populations. Yet, the mouse peak shifted in the opposite direction (earlier than control). Although this might be related to differences in the antibody panels themselves, we nonetheless recommend use of fresh samples from contemporary control groups to follow up on our initial finding in humans. However, future research into other methods of whole blood preservation could be conducted to find a substitute for fresh samples that also preserves the circadian architecture.
In conclusion, daily variation of immune populations relevant to type 1 diabetes autoimmunity is significant biologically and, in some cases, clinically. Often these variations are circadian in nature. Peripheral immune populations should be measured near the same time of day in order to reduce circadian-related variation. There is a possibility that type 1 diabetes is associated with circadian dysrhythmia which itself might be the result of metabolic (e.g. early morning dysglycaemia [44]) and immunologic perturbations related to the disease. Future efforts investigating this newly described aspect of type 1 diabetes are warranted.
Supplementary Information
(PDF 641 kb)
Acknowledgments
Data availability
Data are available by request to the authors.
Funding
We would like to acknowledge the support given to this project by JDRF (by grants INO-2017-458-A-N awarded to CAB and 1-FAC-2019-878-A-N awarded to EB) and the Indiana Clinical and Translational Sciences Institute funded, in part, by UL1TR001108.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Abbreviations
- DC
Dendritic cell
- FDR
False discovery rate
- HC
Healthy control
- IU
Indiana University
- MMF
Mycophenolate mofetil
- NK
Natural killer (cell)
- NKT
Natural killer T (cell)
- STZ
Streptozotocin
- Treg
Regulatory T (cell)
- TEMRA
T-effector memory CD45RA-expressing (cell)
- UF
University of Florida
- ZT
‘Zeitgeber’: hours since ‘lights on’
Contribution statement
Each author has (1) made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; (2) participated in drafting the article or revising it critically for important intellectual content; (3) given their final approval of the version to be published. CAB is the guarantor of this work.
Funding
We would like to acknowledge the support given to this project by JDRF (by grants INO-2017-458-A-N awarded to CAB and 1-FAC-2019-878-A-N awarded to EB) and the Indiana Clinical and Translational Sciences Institute funded, in part, by UL1TR001108.
Data availability
Data are available by request to the authors.
Footnotes
Craig A. Beam and Eleni Beli are co-first authors.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Craig A. Beam, Email: craig.beam@med.wmich.edu
Eleni Beli, Email: E.Beli@qub.ac.uk.
References
- 1.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science (80-) 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 2.Rakshit K, Qian J, Colwell CS, Matveyenko AV. The islet circadian clock: entrainment mechanisms, function and role in glucose homeostasis. Diabetes Obes Metab. 2015;17(Suppl 1):115–122. doi: 10.1111/dom.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakshit K, Qian J, Ernst J, Matveyenko AV. Circadian variation of the pancreatic islet transcriptome. Physiol Genomics. 2016;48(9):677–687. doi: 10.1152/physiolgenomics.00019.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saad A, Dalla Man C et al (2012) Diurnal pattern to insulin secretion and insulin action in healthy individuals. Diabetes 61(11):2691–2700. 10.2337/db11-1478 [DOI] [PMC free article] [PubMed]
- 5.Qian J, Block GD, Colwell CS, Matveyenko AV. Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes. 2013;62(10):3469–3478. doi: 10.2337/db12-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcheva B, Ramsey KM, Buhr ED et al (2010) Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466(7306):627–631. 10.1038/nature09253 [DOI] [PMC free article] [PubMed]
- 7.Petrenko V, Gandasi NR, Sage D, Tengholm A, Barg S, Dibner C. In pancreatic islets from type 2 diabetes patients, the dampened circadian oscillators lead to reduced insulin and glucagon exocytosis. Proc Natl Acad Sci U S A. 2020;117(5):2484–2495. doi: 10.1073/pnas.1916539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hergenhan S, Holtkamp S, Scheiermann C. Molecular interactions between components of the circadian clock and the immune system. J Mol Biol. 2020;432(12):3700–3713. doi: 10.1016/j.jmb.2019.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leproult R, Holmbäck U, Van Cauter E. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. 2014;63(6):1860–1869. doi: 10.2337/db13-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosby P, Hamnett R, Putker M et al (2019) Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 177(4):896–909.e20. 10.1016/j.cell.2019.02.017 [DOI] [PMC free article] [PubMed]
- 11.Amiya E, Watanabe M, Takata M et al (2014) Diurnal body temperature rise is reduced in diabetes with autonomic neuropathy. Clin Auton Res 24(2):95–97. 10.1007/s10286-013-0223-9 [DOI] [PubMed]
- 12.So AY-L, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci U S A. 2009;106(41):17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatwadekar AD, Beli E, Diao Y et al (2017) Conditional deletion of Bmal1 accentuates microvascular and macrovascular injury. Am J Pathol 187(6):1426–1435. 10.1016/j.ajpath.2017.02.014 [DOI] [PMC free article] [PubMed]
- 14.Busik JV, Tikhonenko M, Bhatwadekar A et al (2009) Diabetic retinopathy is associated with bone marrow neuropathy and a depressed peripheral clock. J Exp Med 206(13):2897–2906. 10.1084/jem.20090889 [DOI] [PMC free article] [PubMed]
- 15.Elliott WJ. Timing treatment to the rhythm of disease. Postgrad Med. 2001;110(2):119–129. doi: 10.3810/pgm.2001.08.999. [DOI] [PubMed] [Google Scholar]
- 16.Pick R, He W, Chen CS, Scheiermann C. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol. 2019;40(6):524–537. doi: 10.1016/j.it.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Scheiermann C, Kunisaki Y, Lucas D et al (2012) Article adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity 37:290–301. 10.1016/j.immuni.2012.05.021 [DOI] [PMC free article] [PubMed]
- 18.Abo T, Kawate T, Itoh K, Kumagai K. Studies on the bioperiodicity of the immune response. I. Circadian rhythms of human T, B, and K cell traffic in the peripheral blood. J Immunol. 1981;126(4):1360–1363. [PubMed] [Google Scholar]
- 19.Levi FA, Canon C, Touitou Y, Sulon J et al (1988) Circadian rhythms in circulating T lymphocyte subtypes and plasma testosterone, total and free cortisol in five healthy men. Clin Exp Immunol 71(2):329–335 [PMC free article] [PubMed]
- 20.Druzd D, Matveeva O, Ince L et al (2017) Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 46(1):120–132. 10.1016/j.immuni.2016.12.011 [DOI] [PMC free article] [PubMed]
- 21.Miyawaki T, Taga K, Nagaoki T, Seki H, Suzuki Y, Taniguchi N. Circadian changes of T lymphocyte subsets in human peripheral blood. Clin Exp Immunol. 1984;55(3):618–622. [PMC free article] [PubMed] [Google Scholar]
- 22.Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999;16(5):581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- 23.Mavroudis PD, Scheff JD, Calvano SE, Androulakis IP. Systems biology of circadian-immune interactions. J Innate Immun. 2013;5(2):153–162. doi: 10.1159/000342427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheiermann C, Kunisaki Y, Frenette PS (2013) Circadian control of the immune system. Nat Rev Immunol 13(3):190–198. 10.1038/nri3386 [DOI] [PMC free article] [PubMed]
- 25.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LAJ. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Man K, Loudon A, Chawla A. Immunity around the clock. Science (80-) 2016;354(6315):999–1003. doi: 10.1126/science.aah4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazzoccoli G, Sothern RB, De Cata A et al (2011) A timetable of 24-hour patterns for human lymphocyte subpopulations. J Biol Regul Homeost Agents 25(3):387–395 [PubMed]
- 28.Beam CA, Wasserfall C, Woodwyk A et al (2020) Synchronization of the normal human peripheral immune system: a comprehensive circadian systems immunology analysis. Sci Rep 10(1):672. 10.1038/s41598-019-56951-5 [DOI] [PMC free article] [PubMed]
- 29.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herold KC, Gitelman SE, Ehlers MR et al (2013) Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes 62(11):3766–3774. 10.2337/db13-0345 [DOI] [PMC free article] [PubMed]
- 31.Gubin DG, Nelaeva AA, Uzhakova AE, Hasanova YV, Cornelissen G, Weinert D. Disrupted circadian rhythms of body temperature, heart rate and fasting blood glucose in prediabetes and type 2 diabetes mellitus. Chronobiol Int. 2017;34(8):1136–1148. doi: 10.1080/07420528.2017.1347670. [DOI] [PubMed] [Google Scholar]
- 32.Petrovsky N, Socha L, Silva D, Grossman AB, Metz C, Bucala R. Macrophage migration inhibitory factor exhibits a pronounced circadian rhythm relevant to its role as a glucocorticoid counter-regulator. Immunol Cell Biol. 2003;81(2):137–143. doi: 10.1046/j.0818-9641.2002.01148.x. [DOI] [PubMed] [Google Scholar]
- 33.Cao Q, Zhao X, Bai J et al (2017) Circadian clock cryptochrome proteins regulate autoimmunity. Proc Natl Acad Sci 114(47):12548–12553. 10.1073/pnas.1619119114 [DOI] [PMC free article] [PubMed]
- 34.Copinschi G, Spiegel K, Leproult R, Van Cauter E. Pathophysiology of human circadian rhythms. Novartis Found Symp. 2000;227:143–157. doi: 10.1002/0470870796.ch9. [DOI] [PubMed] [Google Scholar]
- 35.Zee PC. A perfect time for circadian medicine. Neurol Clin. 2019;37(3):ix–x. doi: 10.1016/j.ncl.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Long JE, Drayson MT, Taylor AE, Toellner KM, Lord JM, Phillips AC. Morning vaccination enhances antibody response over afternoon vaccination: a cluster-randomised trial. Vaccine. 2016;34(24):2679–2685. doi: 10.1016/j.vaccine.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dridi I, Ben-Cherif W, Chahdoura H et al (2017) Dosing-time dependent oxidative effects of an immunosuppressive drug ‘Mycophenolate Mofetil’ on rat kidneys. Biomed Pharmacother 87:509–518. 10.1016/j.biopha.2016.12.087 [DOI] [PubMed]
- 38.Gottlieb PA, Quinlan S, Krause-Steinrauf H et al (2010) Failure to preserve beta-cell function with mycophenolate mofetil and daclizumab combined therapy in patients with new- onset type 1 diabetes. Diabetes Care 33(4):826–832. 10.2337/dc09-1349 [DOI] [PMC free article] [PubMed]
- 39.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343(6258):536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 40.Mazzoccoli G, Sothern RB, Greco A et al (2011) Time-related dynamics of variation in core clock gene expression levels in tissues relevant to the immune system. Int J Immunopathol Pharmacol 24(4):869–879. 10.1177/039463201102400406 [DOI] [PubMed]
- 41.Lebailly B, He C, Rogner UC. Linking the circadian rhythm gene Arntl2 to interleukin 21 expression in type 1 diabetes. Diabetes. 2014;63(6):2148–2157. doi: 10.2337/db13-1702. [DOI] [PubMed] [Google Scholar]
- 42.Lebailly B, Boitard C, Rogner UC. Circadian rhythm-related genes: implication in autoimmunity and type 1 diabetes. Diabetes Obes Metab. 2015;17:134–138. doi: 10.1111/dom.12525. [DOI] [PubMed] [Google Scholar]
- 43.McGuire HM, Walters S, Vogelzang A et al (2011) Interleukin-21 is critically required in autoimmune and allogeneic responses to islet tissue in murine models. Diabetes 60(3):867–875. 10.2337/db10-1157 [DOI] [PMC free article] [PubMed]
- 44.Bolli GB, Perriello G, Fanelli CG, De Feo P. Nocturnal blood glucose control in type I diabetes mellitus. Diabetes Care. 1993;16(3):71–89. doi: 10.2337/diacare.16.3.71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 641 kb)
Data Availability Statement
Data are available by request to the authors.
Data are available by request to the authors.