Abstract
Plant glandular trichomes (GTs) are epidermal outgrowths with the capacity to biosynthesize and secrete specialized metabolites, that are of great scientific and practical significance. Our understanding of the developmental process of GTs is limited, and no single plant species serves as a unique model. Here, we review the genetic mechanisms of GT initiation and development and provide a summary of the biosynthetic pathways of GT-specialized metabolites in nonmodel plant species, especially horticultural crops. We discuss the morphology and classification of GT types. Moreover, we highlight technological advancements in methods employed for investigating GTs. Understanding the molecular basis of GT development and specialized metabolites not only offers useful avenues for research in plant breeding that will lead to the improved production of desirable metabolites, but also provides insights for plant epidermal development research.
Subject terms: Plant development, Plant molecular biology
Introduction
Trichomes, the specialized structures that cover most aerial plant tissues, are classified as glandular or nonglandular based on their morphology and secretion ability1. Glandular trichomes (GTs) are described as biofactories with the unique capacity to biosynthesize specialized metabolites, which are critical for the capacity of plants to adapt to their environment and to overcome biotic and abiotic stresses2,3. Moreover, several metabolites produced by medicinal plant GTs are exploited by pharmaceutical industries that benefit from their psychoactive, antiparasitic, antitumor, antimicrobial, antiviral, and antithrombotic properties4.
GT engineering is a plant breeding strategy that requires a detailed understanding of the genetic network controlling GT development3,5. Literature searches using the keyword “glandular trichome” generated approximately 4200 published articles on the Web of Science database between 1900 and 2020. Over 70% of these articles focused on the biochemistry, anatomy, and morphology of GTs, whereas 11% were related to their developmental biology (Fig. 1). Our understanding of the genetic mechanisms of GT development is still in its infancy. However, research progress has been sharply and steadily increasing over the past 15 years. This period coincides with the discovery of the first gene involved in the biosynthesis of metabolites from mint and the increased availability of genetic resources from plants, especially horticultural crops, such as Artemisia annua L. (sweet wormwood) and Cucumis sativus L. (cucumber)6,7.
Fig. 1. Numbers of glandular trichome-related publications from 1900 to 2020.
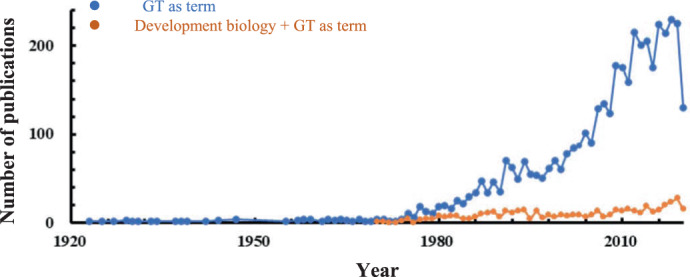
(Based on the Web of Science database using the search word “glandular trichome”)
Research on unicellular nonglandular trichomes in the model plant Arabidopsis thaliana has been abundant. As no GTs have been found in A. thaliana, research on GT development has been conducted in other plant species, including vegetable crops, floral crops, medical plants, aromatic crops and so on. However, no single plant species serves as a unique model for multicellular GTs. From careful analysis of recent literature, five plant species emerged as key working materials: tomato (Solanum lycopersicum), cucumber (C. sativus), sweet wormwood (A. annua), tobacco (Nicotiana tabacum), and cotton (Gossypium hirsutum). Recent studies on these plant species that have led to significant advances in our understanding of GT morphology and developmental biology are summarized in this review. Furthermore, we discuss current advances and innovations applied in the study of GT development and provide future perspectives on the application of current knowledge to enhance breeding efforts.
Morphology and classification of glandular trichomes
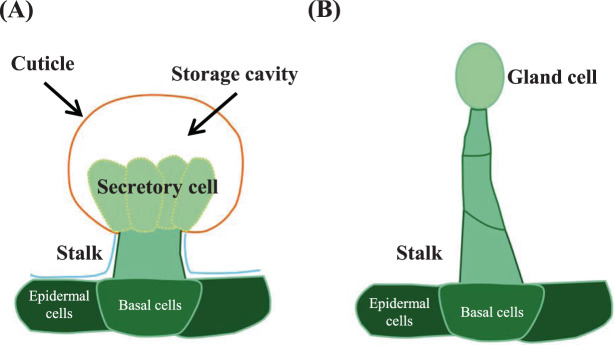
GTs are extremely diverse in terms of shape, cell number, and type of secreted metabolites and may be the result of different evolutionary events3. The morphology of GTs has been described in many plant species, and there are excellent reviews that highlight their diversity3. Briefly, GTs are typically multicellular, consisting of a head/gland that secretes specialized metabolites, a stalk that supports the head, and a differentiated base that connects the stalk to surrounding epidermal cells. There are two main types of GTs, namely, peltate and capitate, with differences in morphology and structure8. Peltate trichomes are short with unicellular or bicellular stalks and a large multicellular secretory head containing several secretory cells, whereas capitate trichomes typically consist of a multicellular stalk of variable cell number and length with a smaller unicellular head8 (Fig. 2). Here, we briefly describe the morphology and classification of GTs in relevant plant species.
Fig. 2. Types of glandular trichomes.
Schematic diagrams showing a peltate trichome (A) and capitate trichome (B)
Cultivated tomato and its wild relatives contain four types (I, IV, VI, and VII) of GTs9. Types I and IV are capitate trichomes, while types VI and VII are peltate trichomes. Type I trichomes are typically longer than type IV trichomes and are present in several tomato species, while type IV trichomes are commonly found in wild species such as S. pennellii and S. habrochaites10. Type VI GTs represent the most abundant trichome type on the leaves and stems of tomato plants and contain four-celled secretory heads with an intercellular storage cavity that can accumulate metabolites. The development and shape of the type VI glandular trichome head differ between cultivated tomato and the wild species S. habrochaites11.
A. annua contains two types of trichomes, known as glandular secreting trichomes (GSTs) and T-shaped nonglandular trichomes (TNGs)12. A. annua GST (AaGST) has received much attention due to its capacity to biosynthesize artemisinin, an effective medicine used in malaria treatment13–16. The AaGSTs in which artemisinin biosynthesis and accumulation occur in a 10-celled biseriate structure are composed of two apical cells, four subapical cells, two stalk cells and two basal cells16,17.
Cucumber fruit is covered by trichomes that combine with tubercles to form the warty fruit trait, which is an important quality trait in cucumber production18,19. Xue et al. examined fruit trichomes of 200 cucumber varieties and classified them into eight distinct types (I–VIII), of which types I and VI are GTs18. Type I trichomes are peltate with a short stalk composed of 3–4 cells and a four-celled or five-celled head18 and are the most widespread and studied GTs found on almost all cucumber species. Although most research on cucumber trichomes has focused on the fruit, type I and VI trichomes can also be found on leaves, stems, flowers, and tendrils18,19.
Tobacco contains two types of capitate trichomes: long trichomes with a multicellular stalk possessing unicellular or multicellular heads and short trichomes with a unicellular stalk and a multicellular head20. Long trichomes synthesize and secrete several exudates and metabolites21, including terpenoids and calcium oxalate crystals22, whereas short trichomes are hydathodes that secrete aqueous droplets under conditions of high atmospheric humidity21. Nicotine (dissolved in aqueous droplets) and heavy metals, including Cd and Zn, are extruded by short trichomes23. Short trichomes are also regarded as specialized biosynthetic structures that produce and secrete defense-related proteins known as T-phylloplanins to aerial leaf surfaces24.
Unlike other species, cotton species are characterized by the presence of darkly pigmented lysigenous glands, which are also called gossypol glands, black glands, or oil glands; these glands are located in the subepidermal layer of many plant tissues and originate from a cluster of cells in the ground meristem25–27. These glands contain high-density gossypol and related terpenoids, which defend the plant from biotic and abiotic stresses and are toxic to monogastric animals25–27. The whole gossypol gland is composed of modified nucleus-free secretory cells with a large vacuole and drastic cytoplasmic structural disorganization, that is, bounded by one layer of secretory cells and one to three layers of sheath cells27. Although glands on cotton show different structures from other species, in-depth knowledge of their molecular regulation is a good way to help obtain a clear view of plant GT development.
Cannabis (Cannabis sativa) and Japanese catnip (Schizonepeta tenuifolia Briquetare) also contain trichomes of biotechnological interest. The female cannabis flower contains three types of GTs: sessile, bulbous and stalked. Sessile trichomes are similar to peltate trichomes, while stalked trichomes are similar to capitate trichomes28,29. These GT types also differ in their fluorescent properties, the number of secretory cells and terpene metabolite profiles28. Japanese catnip, in traditional Asian medicine, contains three distinct GT types, namely, peltate, capitate, and digitiform, with peltate trichomes being the main site for the biosynthesis of essential oil30.
Genetic mechanisms of glandular trichome development
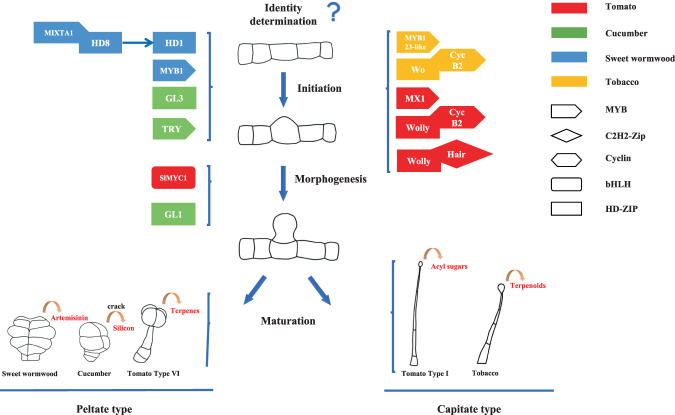
Considerable interest in bioactive compounds produced by GTs, combined with the application of multiomic technologies (genomics, transcriptomics, proteomics, and metabolomics), has greatly accelerated our understanding of gene regulatory networks that function in GT formation1. The development of GTs can be roughly divided into four stages: identity determination, initiation, morphogenesis, and maturation. Given their common organization scheme, it is suggested that some GTs share similar developmental events5. For example, the initiation of most GTs is regulated by MYB transcription factors. Furthermore, most capitate trichomes, such as tomato type I GTs and tobacco GTs, are typically regulated by the interaction of cyclins and homeodomain-leucine zipper (HD-ZIP) transcription factors (TFs). However, peltate trichomes, such as tomato type VI GTs, are also regulated by bHLH TFs (Fig. 3). Recent studies have characterized several genes involved in GT development in key plant species (Fig. 3 and Table 1), which are summarized in this review.
Fig. 3. Model of glandular trichome development.
Schematic diagrams showing GTs of tomato, cucumber, sweet wormwood and tobacco, as well as their main molecular development regulation pathway
Table 1.
Genes involved in the development of GTs
| Types | Species | TFs Name | Function | Metabolites production | Binding sites | Interaction Genes | Hormone involved | Reference |
|---|---|---|---|---|---|---|---|---|
| AP2/ERF | Sweet wormwood | TAR1 | GST morphogenesis | Artemisinin | 14 | |||
| bHLH | Cotton | GoPGF/CGF3 | Pigment gland formation | Gossypol | G-box | TPSs, WRKYs | JA | 25,61 |
| CGF1 | Pigment gland morphogenesis | 25 | ||||||
| Tomato | SlMYC1 | Type VI formation | Terpenoids | TPSs | 49 | |||
| MYB | Sweet wormwood | AaMYB1 | GST density | Artemisinin | GA | 31 | ||
| AaMIXTA1 | GST density | Artemisinin | cuticle biosynthesis genes | 32 | ||||
| Cotton | CGP1 | Pigment gland pigmentation | Gossypol | GoPGF | 26 | |||
| Tomato | SlMX1/SlMIXTA-like | Glandular trichome density (especially type I) | Terpenoids, carotenoids, and phenylpropanoids | MRE | 50–52 | |||
| Cucumber | CsTRY | Trichome dentsity | CsMYB6 | 43 | ||||
| Tobacco | NbMYB123-like | Glandular trichome density | 67 | |||||
| NAC | Cotton | CGF2 | Pigment gland density | Gossypol | 25 | |||
| HD-ZIP | Tomato | Wolly | Type I density | Terpenoids | SlCycB2 | 44,45 | ||
| Tobacco | Nbwo | Glandular trichome formation | L1-like | NbCycB2,Nbwo | 66 | |||
| Sweet wormwood | AaHD1 | GST density | Artemisinin | AaJAZ8 | JA | 34 | ||
| Sweet wormwood | AaHD8 | GST density | Artemisinin | L1-box/HZBS | AaHD1, AaMIXTA1, cuticle biosynthesis genes | 33 | ||
| Cucumber | CsGL3/ Tril | Trichome initiation | 40–42 | |||||
| Cucumber | CsGL1/Mict/Tbh | Trichome morphogenesis | CsMYB6, CsGA20ox1 | 36–39 | ||||
| ZFPs | Tomato | Hair | Type I formation | Wolly | 46 | |||
| Sweet wormwood | AaSAP1 | GST density | Artemisinin | JA, ABA, GA | 35 | |||
| Tobacco | NbGIS | Glandular trichome density | GA | 67 | ||||
| Aux ⁄ IAA | Tomato | SlIAA15 | Type I, VI density | auxin | 58 | |||
| ARF | Tomato | SlARF3 | Type I, VI density | auxin | 57 | |||
| WD-repeat protein | Cucumber | CsTTG1 | Trichome density | CsGL1/Mict/Tbh | 43 | |||
| Cyclin | Tomato | SlCycB2 | Glandular trichome density (especially Type I) | Terpenoids | 44,45 | |||
| Tobacco | NbCycB2 | Glandular trichome formation | Nbwo | 66 | ||||
| WAVE regulatory complex | Tomato | SRA1 | Glandular trichome morphogenesis | Flavonoids, sesquiterpenoids | 55 | |||
| CHI | Tomato | CHI1 | Glandular trichome density | Flavonoids | 54 | |||
| JA-responsive | Tomato | JAI-1 | Type I,VI formation | JA | 59 |
Mechanisms in sweet wormwood (A. annua)
In A. annua, TRICHOME AND ARTEMISININ REGULATOR 1 (TAR1), encoding an AP2 TF, plays a crucial role in regulating the development of GTs and the biosynthesis of artemisinin14. MYB TFs, including AaMYB1, are known to positively regulate the development of AaGSTs31. AaMIXTA1, encoding an R2R3MYB TF, interacts with AaHD8, an HD-ZIP IV TF, forming a regulatory complex that directly promotes AaHD1 expression and positively regulates the initiation of GTs32–34. Furthermore, AaSAP1 encodes stress-associated protein 1 (SAP1), which positively regulates the development and density of AsGSTs and the production of artemisinin35.
Mechanisms in cucumber (C. sativus)
Understanding the molecular genetic basis of fruit spine development is a key aspect of cucumber research18. In cucumber, several trichome-related mutants have been reported. Chen et al. characterized the tiny branched hair (tbh) mutant, which had no noticeable fruit spines but contained tiny branched trichomes with reduced cell numbers and aberrant cell shapes and organization36. Li et al. identified C. sativus Glabrous 1 (CsGL1), encoding an HD-Zip I TF, which plays a significant role in cucumber trichome formation. The csgl1 mutant had no noticeable spines but contained papillae on the leaf epidermis, as observed by scanning electron microscopy (SEM). Furthermore, CsGL1 indirectly regulates the expression of CsMYB6 and CsGA20ox137. Zhao et al. characterized a micro-trichome (mict) mutant with microsized stunted spines similar to those of csgl138,39. Notably, TBH, MICT, and CsGL1 are allelic and mapped to Csa3M74822037,39. Pan et al. characterized a completely glabrous mutant with a single recessive gene named C. sativus Glabrous 3 (CsGL3), encoding an HD-Zip IV TF40. Cui et al. identified the glabrous mutant NCG157 and postulated its candidate gene to be Csa6M514870, which is also the candidate gene for CsGL341. Wang et al. demonstrated that Tril (allelic to CsGL3) had a long segment insertion following the first exon and that the tril mutant displayed the same phenotype as csgl342. Moreover, CsGL3/Tril had an epistatic effect on TBH/CsGL1/Mict40,42. Trichome density was shown to be influenced by the expression of CsTTG1, which encodes a WD-repeat protein43. The genes listed above play a role in both cucumber GTs and nonglandular trichomes; however, little is known about genes directly involved in the development of cucumber GTs.
Mechanisms in tomato (S. lycopersicum)
In tomato, the formation of type I capitate GTs involves the Wolly (Wo) gene, encoding an HD-ZIP TF, together with SlCycB2. The repression of Wo has been shown to decrease the number of type I trichomes44,45. Additionally, the Hair (H) gene, encoding a C2H2 zinc finger protein, interacts with Wo to form an H-Wo dimer protein complex, which acts as an important regulator of type I trichomes46. This suggests that H-Wo-CycB2 may form a trimer protein complex that acts as an important regulator of type I trichomes. This differs from the formation of unicellular nonglandular trichomes in A. thaliana, which is regulated by an MYB-bHLH-WD trimer complex46,47. Recently, the long noncoding RNA (lncRNA) lncRNA000170 was reported to inhibit type I trichome formation. Overexpression of lncRNA000170 caused a decrease in type I trichomes by downregulating several trichome regulators, including Wo, H, and SlCycB248. The formation of type VI peltate trichomes involves SlMYC1, encoding a bHLH TF49. Overexpression of SlCycB2 also led to a decrease in type VI GTs44. Ectopic expression of SlMIXTA1, an R2R3MYB TF, increased the number of GTs, especially type I GTs, in tomato50–52. The identification and characterization of several tomato trichome-related mutants have aided the elucidation of the mechanisms of GT formation. The tomato odorless-2 (od-2) mutant had abnormal type I trichomes and a low density of type VI trichomes53. The anthocyanin-free (af) mutant, with mutation of SlCHI, an isoform of the flavonoid biosynthetic enzyme chalcone isomerase (CHI), exhibited a lower density of type VI trichomes54. Kang et al. used a map-based cloning approach to demonstrate that a previously reported hairless (hl) mutant, controlled by a highly conserved SRA1 (specifically Rac1-associated protein) subunit of the WAVE regulatory complex (WRC), showed a severely bent and shortened trichome phenotype55,56. Furthermore, several phytohormone-related genes are involved in the development of GTs, such as JAI-1, SlIAA5, and SlARF357–59, and treatment with exogenous JA was shown to increase the densities of type VI trichomes60.
Mechanisms in cotton (Gossypium spp.)
Understanding the molecular genetic basis of gossypol gland formation in cotton could provide additional methods for developing gossypol-free cotton seeds or decrease the density of gossypol glands61. Six independent loci, gl1–gl6, that regulate gossypol gland formation have been identified, with the combination of gl2gl2gl3gl3 producing a glandless phenotype62. A glandless mutant discovered in Egyptian cotton (G. barbadense) was formed due to the expression of a dominant allele at the Gl2 locus, which is epistatic to Gl3 and designated GL2e 63,64. Using the Gl2e mutant, Ma et al. characterized GoPGF (pigment gland formation) on chromosome A12 as Gl2 and its homeolog on chromosome D12 as Gl361. GoPGF encodes a basic helix-loop-helix domain-containing TF that positively regulates gland formation. The insertion of a single nucleotide into the coding sequence (CDS) of GoPGF resulted in premature translation termination, leading to the glandless phenotype, while silencing GoPGF led to a completely glandless phenotype61. The glandless gene Gl2e was fine mapped to a 15 kb region, and GoPGF was confirmed as the candidate gene65. RNA-seq analysis of embryos from near-isogenic gland (Gl2Gl2Gl3Gl3) vs. glandless (gl2gl2gl3gl3) cotton plants identified three cotton gland formation (CGF) genes that participate in gland formation25. The sequences of CGF1 and CGF2 in the glandular and glandless cotton plants were identical. However, the A subgenome of CGF3 (synonymous GoPGF) in the glandless cotton had a 5.1 kb transposon insertion, while the D subgenome homolog had two SNPs in the CDS, one SNP in the terminator, and several major differences in the upstream regulatory sequence (~4.2 kb). Silencing of CGF1 and CGF3 resulted in a dramatic reduction in gland numbers, while CGF2 had a mild effect on gland density25. In another recent study, Gao et al. performed comparative transcriptome analysis of several glandular and glandless cultivars and further characterized Cotton Gland Pigmentation 1 (CGP1), an MYB TF, involved in the regulation of gland pigmentation but not morphogenesis. CGP1 is located in the nucleus and interacts with GoPGF26.
Mechanisms in tobacco (N. tabacum)
Recent research in N. benthamiana has led to the discovery of a novel reciprocal regulation mechanism that is involved in GT formation66. Wu et al. cloned two tobacco genes, namely, NbCycB2 and NbWo (homologs of SlCycB2 and SlWo), and demonstrated that NbWo directly regulated the expression of NbCycB2 by binding to an L1-like box in its promoter region66. Wu et al. also suggested that NbCycB2 can inhibit trichome initiation by binding to the LZ domain of NbWo66. Furthermore, two C2H2 zinc finger TFs, namely, NbGIS and NbMYB123-like (homologs of AtMYB123), encoding an R2R3 MYB domain putative TF, also participate in the development of tobacco GTs67.
Biosynthesis pathways of glandular trichome specialized metabolites
GTs are sites for the biosynthesis and accumulation of a wide range of plant natural products, such as cannabinoids and terpenes in cannabis28, bitter acid in Humulus lupulus68, tanshinone in Salvia miltiorrhiza69, and artemisinin in A. annua70. The cracks of cucumber GTs have also been shown to induce the excretion of silicon71. Recent advancements in multiomic technologies and metabolic analysis have shed some light on key molecular pathways regulating the biosynthesis of GT-specialized metabolites.
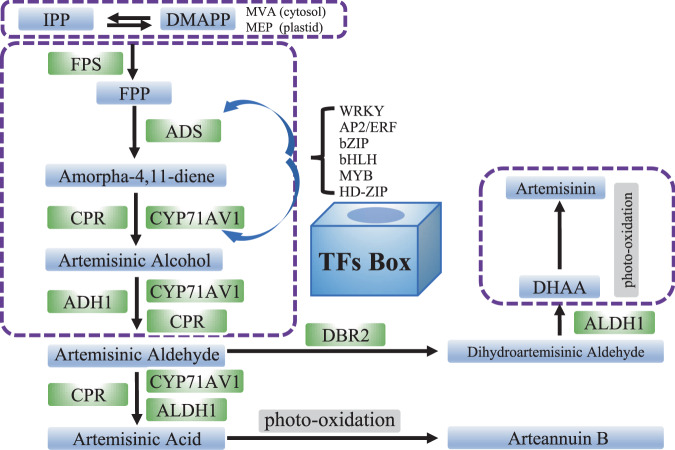
Due to its importance in the pharmaceutical industry, the mechanism of artemisinin biosynthesis is well described (Fig. 4)72–75. Artemisinin originates from isopentenyl diphosphate (IPP) or dimethylallyl diphosphate (DMAPP) via the methylerythritol phosphate (MEP) pathway in the plastid or via the mevalonate (MVA) pathway in the cytosol70. The first substrate of the artemisinin biosynthesis pathway is farnesyl diphosphate (FPP), which is synthesized from IPP and DMAPP by farnesyl diphosphate synthase (FPS)76,77. The cyclization of FPP to amorpha-4,11-diene by amorpha-4,11-diene synthase (ADS) is the initial step of artemisinin biosynthesis78,79. Cytochrome P450 monooxygenase (CYP71AV1), cytochrome P450 oxidoreductase (CPR) and alcohol dehydrogenase (ADH1) then convert ADS to artemisinic alcohol and eventually to artemisinic aldehyde80–82. Artemisinic aldehyde Δ11(13)-reductase (DBR) is then involved in the formation of dihydroartemisinic aldehyde (DHAAA)83, and aldehyde dehydrogenase 1 (ALDH1) converts DHAAA to dihydroartemisinic acid (DHAA)84. DHAA is then converted to artemisinin through photooxidation in the GT subcuticular space. Artemisinic acid is also converted to artemisinin B by photooxidation. Most of these key enzymes are specifically localized in AaGSTs17,85. The low yield of artemisinin (0.01–0.1% leaf DW) in A. annua severely restricts its supply86, and the overexpression of key genes in AaGSTs is an effective strategy for enhancing the artemisinin level. Several TF families are involved in the regulation of artemisinin biosynthesis, including the WRKY (AaWRKY1 and AaGSW1), AP2/ERF (AaORA, AaERF1, AaERF2, and TAR1), bZIP (AabZIP1 and AaHY5), bHLH (AaMYC2 and AabHLH1), MYB (AaMYB1 and AaMIXTA1), HD-ZIP (AaHD1 and AaHD8), and ZFP (AaSAP1) families. Many of these TFs are regulated by phytohormones such as GA, JA, MeJA, and ABA. These TFs regulate artemisinin biosynthesis by interacting with key enzymes, such as DBR2, ADS, and CYP71AV1, or with other TFs14,31–35,87–95.
Fig. 4. Regulation of artemisinin metabolism in sweet wormwood.
TFs involved in the biosynthesis of artemisinin by interaction with the key enzymes in A. annua
Understanding the genetic basis of gossypol biosynthesis could provide additional methods for the development of gossypol-free cotton seeds27. Several enzymes of the gossypol biosynthesis pathway have been characterized, including 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), farnesyl diphosphate synthase (FPS), (+)-δ-cadinene synthase (CDN), (+)-δ-cadinene-8-hydroxylase CYP706B1 and methyltransferase96. However, Ma et al. discovered that gossypol biosynthesis and pigment gland formation are uncoupled, as silencing CYP706B1 does not affect gland formation61.
Tomato type VI GTs are the main site of terpenoid, flavonoid and methyl ketone secretion, whereas type I and type IV GTs are involved in acyl sugar biosynthesis10,55,97. Overexpression of SlMX1/SlMIXTA-like increases the expression of genes involved in primary metabolic pathways, such as glycolysis, the pentose phosphate pathway and the shikimate pathway. SlMX1/SlMIXTA-like is also involved in the biosynthesis of terpenoids and carotenoids50,52. Genes that regulate glandular trichome formation, such as SlMYC1, Wolly, and SlCycB2, can also regulate terpenoid biosynthesis and the expression of terpene synthase (TPS) genes44,45,49. Mutations in Od-2, SlCH1, and SRA1 prevent the accumulation of both flavonoids and terpenoids in type VI glands53–55.
Specialized techniques applied in the study of glandular trichomes
Advanced biotechnologies employed in the study of GTs have assisted researchers in making breakthroughs in understanding the genetics, molecular basis, and functions of GTs and their metabolites. Early research on the separation and purification of GTs was generally ineffective and imprecise. Techniques such as freezing and powdering of plant samples followed by vortexing and mesh filtration were used to collect stalked GTs. In tobacco, direct extraction with tweezers was used to separate trichomes. In tomato, a pulled Pasteur pipette was used to collect type VI glands and their exudate on stems and leaves49,59. In peppermint and spearmint, a combination of chemical and physical methods was utilized, involving the application of an isolation buffer to protect the plant materials98. Gradually, this method was modified and then widely implemented in several other species99. In tomato, GTs were also isolated using glass beads in a chemical isolation buffer, followed by filtration and centrifugation to separate trichome types10. This method was adapted to harvest enriched cannabis trichome fractions28.
Single-cell data are useful for elucidating cell type-specific processes, cell differentiation and the evolution of cell states100. Currently, laser capture microdissection (LCM) is used to isolate and purify single trichomes or secretory cells from paraffin-embedded plant tissue sections. However, LCM is time consuming and inefficient at collecting data from a large number of trichomes. Single-cell RNA sequencing technology is considered an efficient method for identifying cell differentiation states and has been used to study root cell development, vascular cell lineage and stomatal lineage cells101–104. Laser microdissection and pressure catapulting (LMPC) is used to isolate single cells from trichomes, and further proteinase K treatment improves RNA yields for downstream analysis105. In tomato, fluorescence-activated cell sorting (FACS) based on the autofluorescence of trichomes was used to separate trichrome developmental stages100. To observe trichome density and morphology, SEMs and stereoscopes are typically used11,18. In A. annua, an Olympus fluorescence microscope was used to observe trichome density35, while light and fluorescence microscopy analyses were applied in tomato10. Furthermore, metabolites stored in GTs can be stained with chemical reagents to observe their secretion process106. Metabolites in GTs have been measured using analytical techniques such as GC-MS, HPLC, UPLC, LC-MS, and LC-ESI-MS/MS14,26,28,33,61. Currently, internal electrode capillary pressure probe electrospray ionization mass spectrometry (IEC-PPESI-MS) is used for single-cell metabolite profiling of stalk and glandular cells of intact trichomes in tomato, enabling high-spatial-resolution cell sampling, precise postsampling manipulation, and high detection sensitivity107.
Concluding remarks and future perspectives
Specialized metabolites biosynthesized by GTs are considered important reservoirs of high-value bioactive natural products with largely unexploited potential. Despite extensive studies on GT morphology and specialized metabolites, almost nothing is known about the genetics underlying their development. Furthermore, identity determination of GTs is a significant stage, but the developmental signals that initiate the transformation of an epidermal cell to an epidermal hair, and regulate the differentiation of an epidermal hair cell to glandular or nonglandular trichomes are still not clear in most species. Recent advancements in multiomic technologies, genetic resources and specialized techniques have increased our understanding of the genetic mechanisms controlling GT initiation and development in several key plant species. Single-cell metabolite profiling of GTs provides significant results but is difficult to widely apply in diverse plants. In addition, the marker genes related to each developmental stage of GTs are still relatively unknown. Increased knowledge of GT biology and further improvements in these technologies not only will improve our understanding of cell differentiation and the development of plant trichomes, but also could inspire breeding efforts to utilize plants as biofactories that produce desirable metabolites in their GTs.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (32020103014), Project of Beijing Agricultural Innovation Consortium (BAIC01), and The Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032).
Author contributions
Z.F. planned the manuscript outline, wrote the draft and created the figures and tables. E.B., H.R., and X.L. revised the manuscript and figures. Z.L. created the figures. Y.C., Y.D., S.L., and H.W. proofread the manuscript.
Conflict of interest
The authors declare no competing interests.
Contributor Information
Huazhong Ren, Email: renhuazhong@cau.edu.cn.
Xingwang Liu, Email: liuxw01@cau.edu.cn.
References
- 1.Singh, S. P., Upadhyay, S. K., Pandey, A. & Kumar, S. in Molecular Approaches in Plant Biology and Environmental Challenges (eds. Singh, S. P. et al.) 1–5 (Springer, 2019).
- 2.Fahn, A. in Advances in Botanical Research 3137-75 (Academic Press, 2000).
- 3.Huchelmann A, Boutry M, Hachez C. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol. 2017;175:6–22. doi: 10.1104/pp.17.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Jing SX, Luo SH, Li SH. Non-volatile natural products in plant glandular trichomes: chemistry, biological activities and biosynthesis. Nat. Prod. Rep. 2019;36:626–665. doi: 10.1039/C8NP00077H. [DOI] [PubMed] [Google Scholar]
- 5.Chalvin C, Drevensek S, Dron M, Bendahmane A, Boualem A. Genetic control of glandular trichome development. Trends Plant Sci. 2020;25:477–487. doi: 10.1016/j.tplants.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 6.Croteau RB, Davis EM, Ringer KL, Wildung MR. Menthol biosynthesis and molecular genetics. Naturwissenschaften. 2005;92:562–577. doi: 10.1007/s00114-005-0055-0. [DOI] [PubMed] [Google Scholar]
- 7.Schuurink R, Tissier A. Glandular trichomes: micro-organs with model status? N. Phytol. 2020;225:2251–2266. doi: 10.1111/nph.16283. [DOI] [PubMed] [Google Scholar]
- 8.Werker, E. in Advances in Botanical Research 311-35 (Academic Press, 2000).
- 9.Glas J, et al. Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 2012;13:17077–17103. doi: 10.3390/ijms131217077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergau N, Bennewitz S, Syrowatka F, Hause G, Tissier A. The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 2015;15:289. doi: 10.1186/s12870-015-0678-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennewitz S, Bergau N, Tissier A. QTL mapping of the shape of type VI glandular trichomes in tomato. Front. Plant Sci. 2018;9:1421. doi: 10.3389/fpls.2018.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira JFS, Janick J. Floral morphology of Artemisia annua with special reference to trichomes. Int. J. Plant Sci. 1995;156:807–815. doi: 10.1086/297304. [DOI] [Google Scholar]
- 13.Noorden RV. Demand for malaria drug soars. Nature. 2010;466:672–673. doi: 10.1038/466672a. [DOI] [PubMed] [Google Scholar]
- 14.Tan H, et al. Trichome and artemisinin regulator 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant. 2015;8:1369–1411. doi: 10.1016/j.molp.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Duke SO. Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. Int. J. Plant Sci. 1994;155:365–372. doi: 10.1086/297173. [DOI] [Google Scholar]
- 16.Duke SO, Paul RN. Development and fine structure of the glandular trichomes of Artemisia annua L. Int. J. Plant Sci. 1993;154:107–118. doi: 10.1086/297096. [DOI] [Google Scholar]
- 17.Olsson ME, et al. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry. 2009;70:1123–1128. doi: 10.1016/j.phytochem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Xue S, et al. Classification of fruit trichomes in cucumber and effects of plant hormones on type II fruit trichome development. Planta. 2019;249:407–416. doi: 10.1007/s00425-018-3004-9. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Bartholomew E, Cai Y, Ren H. Trichome-related mutants provide a new perspective on multicellular trichome initiation and development in cucumber (Cucumis sativus L) Front. Plant Sci. 2016;7:1187. doi: 10.3389/fpls.2016.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzelac, B., Stojičić, D. & Budimir, S. in Plant Cell and Tissue Differentiation and Secondary Metabolites: Fundamentals and Applications (eds. Ramawat, K. G. et al.) 1–37 (Springer, 2019).
- 21.Meyberg M, Krohn S, Brümmer B, Kristen U. Ultrastructure and secretion of glandular trichomes of tobacco leaves*. Flora. 1991;185:357–363. doi: 10.1016/S0367-2530(17)30495-4. [DOI] [Google Scholar]
- 22.Wagner GJ, Wang E, Shepherd RW. New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. 2004;93:3–11. doi: 10.1093/aob/mch011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi YE, et al. Detoxification of cadmium in tobacco plants: formation and active excretion of crystals containing cadmium and calcium through trichomes. Planta. 2001;213:45–50. doi: 10.1007/s004250000487. [DOI] [PubMed] [Google Scholar]
- 24.Shepherd RW. Phylloplanins of tobacco are defensive proteins deployed on aerial surface by short glandular trichomes. Plant Cell. 2005;17:1851–1861. doi: 10.1105/tpc.105.031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janga MR, et al. Genes regulating gland development in the cotton plant. Plant Biotechnol. J. 2019;17:1142–1153. doi: 10.1111/pbi.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao W, et al. The gland localized CGP1 controls gland pigmentation and gossypol accumulation in cotton. Plant Biotechnol. J. 2020;18:1573–1584. doi: 10.1111/pbi.13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu WZ, Zhou YF, Wang X, Jiao ZJ. Programmed cell death during pigment gland formation in Gossypium hirsutum leaves. Plant Biol. 2010;12:895–902. doi: 10.1111/j.1438-8677.2009.00291.x. [DOI] [PubMed] [Google Scholar]
- 28.Livingston SJ, et al. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 2019;101:37–56. doi: 10.1111/tpj.14516. [DOI] [PubMed] [Google Scholar]
- 29.Hammond CT, Mahlberg PG. Morphology of glandular hairs of Cannabis sativa from scanning electron microscopy. Am. J. Bot. 1973;60:524–528. doi: 10.1002/j.1537-2197.1973.tb05953.x. [DOI] [Google Scholar]
- 30.Liu C, et al. Morphology of glandular trichomes of Japanese catnip (Schizonepeta tenuifolia Briquet) and developmental dynamics of their secretory activity. Phytochemistry. 2018;150:23–30. doi: 10.1016/j.phytochem.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Matías-Hernández L, et al. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 2017;90:520–534. doi: 10.1111/tpj.13509. [DOI] [PubMed] [Google Scholar]
- 32.Shi P, et al. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. N. Phytol. 2017;217:261–276. doi: 10.1111/nph.14789. [DOI] [PubMed] [Google Scholar]
- 33.Yan T, et al. A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. N. Phytol. 2018;218:567–578. doi: 10.1111/nph.15005. [DOI] [PubMed] [Google Scholar]
- 34.Yan T, et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. N. Phytol. 2016;213:1145–1155. doi: 10.1111/nph.14205. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Stress associated protein 1 regulates the development of glandular trichomes in Artemisia annua. Plant Cell Tissue Organ Cult. 2019;139:249–259. doi: 10.1007/s11240-019-01677-5. [DOI] [Google Scholar]
- 36.Chen C, et al. Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.) J. Exp. Bot. 2014;65:4943–4958. doi: 10.1093/jxb/eru258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q, et al. The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 2015;66:2515–2526. doi: 10.1093/jxb/erv046. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J, et al. Transcriptome analysis in Cucumis sativus identifies genes involved in multicellular trichome development. Genomics. 2015;105:296–303. doi: 10.1016/j.ygeno.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, et al. Micro-trichome as a class I homeodomain-leucine zipper gene regulates multicellular trichome development in Cucumis sativus. J. Integr. Plant Biol. 2015;57:925–935. doi: 10.1111/jipb.12345. [DOI] [PubMed] [Google Scholar]
- 40.Pan Y, Bo K, Cheng Z, Weng Y. The loss-of-function GLABROUS 3 mutation in cucumber is due to LTR-retrotransposon insertion in a class IV HD-ZIP transcription factor gene CsGL3 that is epistatic over CsGL1. BMC Plant Biol. 2015;15:302. doi: 10.1186/s12870-015-0693-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui J, et al. A new glabrous gene (csgl3) identified in trichome development in cucumber (Cucumis sativus L.) PLoS ONE. 2016;11:e148422. doi: 10.1371/journal.pone.0148422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 2016;129:305–316. doi: 10.1007/s00122-015-2628-4. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, et al. The WD-Repeat protein CsTTG1 regulates fruit wart formation through interaction with the Homeodomain-Leucine Zipper I protein Mict. Plant Physiol. 2016;171:1156. doi: 10.1104/pp.16.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao, S. et al. The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci.262, 103–114 (2017). [DOI] [PubMed]
- 45.Yang C, et al. A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl Acad. Sci. 2011;108:11836–11841. doi: 10.1073/pnas.1100532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang J, et al. Hair, encoding a single C2H2 zinc‐finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018;96:90–102. doi: 10.1111/tpj.14018. [DOI] [PubMed] [Google Scholar]
- 47.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao X, et al. Transcriptomic and functional analyses uncover the regulatory role of lncRNA000170 in tomato multicellular trichome formation. Plant J. 2020;104:18–29. doi: 10.1111/tpj.14902. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, et al. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell. 2018;30:2988–3005. doi: 10.1105/tpc.18.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewas M, et al. RNA-seq reveals mechanisms of SlMX1 for enhanced carotenoids and terpenoids accumulation along with stress resistance in tomato. Sci. Bull. 2017;62:476–485. doi: 10.1016/j.scib.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 51.Ewas M, et al. Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci. Bull. 2016;61:1413–1418. doi: 10.1007/s11434-016-1108-9. [DOI] [Google Scholar]
- 52.Ying S, Su M, Wu Y, Zhou L, Zhang Y. Trichome regulator SlMIXTA-like directly manipulates primary metabolism in tomato fruit. Plant Biotechnol. J. 2019;18:354–363. doi: 10.1111/pbi.13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang J, et al. The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 2010;154:262–272. doi: 10.1104/pp.110.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang JH, et al. The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 2014;164:1161–1174. doi: 10.1104/pp.113.233395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang JH, et al. Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J. Exp. Bot. 2016;67:5313–5324. doi: 10.1093/jxb/erw292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rick CM, Butler L. Cytogenetics of the tomato. Adv. Genet. 1956;8:267–382. doi: 10.1016/S0065-2660(08)60504-0. [DOI] [Google Scholar]
- 57.Zhang X, et al. Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant Cell Physiol. 2015;56:2110–2124. doi: 10.1093/pcp/pcv136. [DOI] [PubMed] [Google Scholar]
- 58.Deng W, et al. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. N. Phytol. 2012;194:379–390. doi: 10.1111/j.1469-8137.2012.04053.x. [DOI] [PubMed] [Google Scholar]
- 59.Li L, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G, Klinkhamer PGL, Escobar-Bravo R, Leiss KA. Type VI glandular trichome density and their derived volatiles are differently induced by jasmonic acid in developing and fully developed tomato leaves: implications for thrips resistance. Plant Sci. 2018;276:87–98. doi: 10.1016/j.plantsci.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 61.Ma D, et al. Genetic basis for glandular trichome formation in cotton. Nat. Commun. 2016;7:10456. doi: 10.1038/ncomms10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMichael SC. Combined effects of glandless genes gl2 and gl3 on pigment glands in the cotton plant. Agron. J. 1960;51:148–152. [Google Scholar]
- 63.Kohel RJ, Lee JA. Genetic analysis of egyptian glandless cotton1. Crop Sci. 1984;24:1119–1121. doi: 10.2135/cropsci1984.0011183X002400060027x. [DOI] [Google Scholar]
- 64.Tang C, et al. Genetic analysis for Hai 1 strain of glandless cotton (G.barbadense L.) I. allelism test. Acta Agronom. Sin. 1994;20:8–12. [Google Scholar]
- 65.Cheng H, et al. Fine mapping and candidate gene analysis of the dominant glandless gene Gl e 2 in cotton (Gossypium spp.) Theor. Appl. Genet. 2016;129:1347–1355. doi: 10.1007/s00122-016-2707-1. [DOI] [PubMed] [Google Scholar]
- 66.Wu M, et al. NbCycB2 represses Nbwo activity via a negative feedback loop in tobacco trichome development. J. Exp. Bot. 2020;71:1815–1827. doi: 10.1093/jxb/erz542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, et al. NbGIS regulates glandular trichome initiation through GA signaling in tobacco. Plant Mol. Biol. 2018;98:153–167. doi: 10.1007/s11103-018-0772-3. [DOI] [PubMed] [Google Scholar]
- 68.Xu H, et al. Characterization of the formation of branched short-chain fatty acid: CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol. Plant. 2013;6:1301–1317. doi: 10.1093/mp/sst004. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X, et al. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 2015;5:11244. doi: 10.1038/srep11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 71.Samuels AL, Glass ADM, Ehret DL, Menzies JG. Mobility and deposition of silicon in cucumber plants. Plant Cell Environ. 2010;14:485–492. doi: 10.1111/j.1365-3040.1991.tb01518.x. [DOI] [Google Scholar]
- 72.Wang Z, et al. Anti-inflammatory properties and regulatory mechanism of a novel derivative of artemisinin in experimental autoimmune encephalomyelitis. J. Immunol. 2007;179:5958–5965. doi: 10.4049/jimmunol.179.9.5958. [DOI] [PubMed] [Google Scholar]
- 73.Thomas E, et al. The antiviral activities of artemisinin and artesunate. Clin. Infect. Dis. 2008;47:804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 74.Crespo-ortiz MP, Wei MQ, Shibata M. Antitumor activity of artemisinin and its derivatives: from a well-known antimalarial agent to a potential anticancer drug. J. Biomedicine Biotechnol. 2012;2012:247597. doi: 10.1155/2012/247597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Efferth T, Dunstan H, Sauerbrey A, Miyachi H, Chitambar RC. The anti-malarial artesunate is also active against cancer. Int. J. Oncol. 2001;18:767–773. doi: 10.3892/ijo.18.4.767. [DOI] [PubMed] [Google Scholar]
- 76.Schramek N, et al. Artemisinin biosynthesis in growing plants of Artemisia annua. A13CO2 study. Phytochemistry. 2010;71:179–187. doi: 10.1016/j.phytochem.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 77.Hassani D, et al. Parallel transcriptional regulation of artemisinin and flavonoid biosynthesis. Trends Plant Sci. 2020;25:466–476. doi: 10.1016/j.tplants.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen KT, Arsenault PR, Weathers PJ. Trichomes + roots + ROS = artemisinin: regulating artemisinin biosynthesis in Artemisia annua L. In vitro Cell Dev. Biol. Plant. 2011;47:329–338. doi: 10.1007/s11627-011-9343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mercke P, Bengtsson M, Bouwmeester HJ, Posthumus MA, Brodelius PE. Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch. Biochem. Biophys. 2000;381:173–180. doi: 10.1006/abbi.2000.1962. [DOI] [PubMed] [Google Scholar]
- 80.Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, et al. Cloning and characterization of trichome-specific promoter of cpr71av1 gene involved in artemisinin biosynthesis in Artemisia annua L. Mol. Biol. 2011;45:751–758. doi: 10.1134/S0026893311040145. [DOI] [PubMed] [Google Scholar]
- 82.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y, et al. The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J. Biol. Chem. 2008;283:21501–21508. doi: 10.1074/jbc.M803090200. [DOI] [PubMed] [Google Scholar]
- 84.Teoh KH, Polichuk DR, Reed DW, Covello PS. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany. 2009;87:635–642. doi: 10.1139/B09-032. [DOI] [Google Scholar]
- 85.Wang H, Han J, Kanagarajan S, Lundgren A, Brodelius PE. Trichome-specific expression of the amorpha-4,11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-GUS fusion. Plant Mol. Biol. 2013;81:119–138. doi: 10.1007/s11103-012-9986-y. [DOI] [PubMed] [Google Scholar]
- 86.Abdin MZ, Israr M, Rehman RU, Jain SK. Artemisinin, a novel antimalarial drug: biochemical and molecular approaches for enhanced production. Planta Med. 2003;69:289–299. doi: 10.1055/s-2003-38871. [DOI] [PubMed] [Google Scholar]
- 87.Chen M, et al. Glandular Trichome-Specific Wrky 1 promotes artemisinin biosynthesis in Artemisia annua. N. Phytol. 2017;214:304–316. doi: 10.1111/nph.14373. [DOI] [PubMed] [Google Scholar]
- 88.Han J, Wang H, Lundgren A, Brodelius PE. Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry. 2014;102:89–96. doi: 10.1016/j.phytochem.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 89.Hao X, et al. Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 2019;60:1747–1760. doi: 10.1093/pcp/pcz084. [DOI] [PubMed] [Google Scholar]
- 90.Ji Y, et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014;55:1592–1604. doi: 10.1093/pcp/pcu090. [DOI] [PubMed] [Google Scholar]
- 91.Lu X, et al. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. N. Phytol. 2013;198:1191–1202. doi: 10.1111/nph.12207. [DOI] [PubMed] [Google Scholar]
- 92.Ma D, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 93.Shen Q, Yan T, Fu X, Tang K. Transcriptional regulation of artemisinin biosynthesis in Artemisia annua L. Sci. Bull. 2016;61:18–25. doi: 10.1007/s11434-015-0983-9. [DOI] [Google Scholar]
- 94.Yu Z, et al. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant. 2012;5:353–365. doi: 10.1093/mp/ssr087. [DOI] [PubMed] [Google Scholar]
- 95.Zhang F, et al. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant. 2015;8:163–175. doi: 10.1016/j.molp.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 96.Tian X, et al. Gossypol: phytoalexin of cotton. Sci. China Life Sci. 2016;59:122–129. doi: 10.1007/s11427-016-5003-z. [DOI] [PubMed] [Google Scholar]
- 97.Fan P, Leong BJ, Last RL. Tip of the trichome: evolution of acylsugar metabolic diversity in Solanaceae. Curr. Opin. Plant Biol. 2019;49:8–16. doi: 10.1016/j.pbi.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gershenzon J, et al. Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal. Biochem. 1992;200:130–138. doi: 10.1016/0003-2697(92)90288-I. [DOI] [PubMed] [Google Scholar]
- 99.Sallets A, Beyaert M, Boutry M, Champagne A. Comparative proteomics of short and tall glandular trichomes of Nicotiana tabacum reveals differential metabolic activities. J. Proteome Res. 2014;13:3386–3396. doi: 10.1021/pr5002548. [DOI] [PubMed] [Google Scholar]
- 100.Bergau N, Navarette Santos A, Henning A, Balcke GU, Tissier A. Autofluorescence as a signal to sort developing glandular trichomes by flow cytometry. Front. Plant Sci. 2016;7:949. doi: 10.3389/fpls.2016.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rodriguez-Villalon A, Brady SM. Single cell RNA sequencing and its promise in reconstructing plant vascular cell lineages. Curr. Opin. Plant Biol. 2019;48:47–56. doi: 10.1016/j.pbi.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 102.Birnbaum KD. Power in numbers: single-cell RNA-seq strategies to dissect complex tissues. Annu. Rev. Genet. 2018;52:203–221. doi: 10.1146/annurev-genet-120417-031247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Z, et al. Global dynamic molecular profiles of stomatal lineage cell development by single-cell RNA sequencing. Mol. Plant. 2020;13:1178–1193. doi: 10.1016/j.molp.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 104.Zhang T, Xu Z, Shang G, Wang J. A single-cell RNA sequencing profiles the developmental landscape of arabidopsis root. Mol. Plant. 2019;12:648–660. doi: 10.1016/j.molp.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 105.Olsson L, Lundgren A, Brodelius PE. Trichome isolation with and without fixation using laser microdissection and pressure catapulting followed by RNA amplification: expression of genes of terpene metabolism in apical and sub-apical trichome cells of Artemisia annua L. Plant Sci. 2012;183:9–13. doi: 10.1016/j.plantsci.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 106.Sarria E, Palomares-Rius FJ, López-Sesé AI, Heredia A, Gómez-Guillamón ML. Role of leaf glandular trichomes of melon plants in deterrence of Aphis gossypii Glover. Plant Biol. 2010;12:503. doi: 10.1111/j.1438-8677.2009.00239.x. [DOI] [PubMed] [Google Scholar]
- 107.Nakashima T, et al. Single-cell metabolite profiling of stalk and glandular cells of intact trichomes with internal electrode capillary pressure probe electrospray ionization mass spectrometry. Anal. Chem. 2016;88:3049–3057. doi: 10.1021/acs.analchem.5b03366. [DOI] [PubMed] [Google Scholar]