Abstract
Allergic asthma that is caused by inhalation of house dust mites (HDMs) is mainly mediated by Th2 cells. Recently, the roles of Sox (SRY-related high-mobility-group (HMG)-box) family members in various immune responses have been investigated. However, the roles of Sox12, a member of the SoxC group, in Th2 cell differentiation and allergic airway inflammation, remain unknown. We showed that Sox12 mRNA was significantly increased during Th2 cell differentiation. In vivo, HDM-induced eosinophil infiltration into the lung and Th2 cell differentiation were exacerbated in Sox12−/− mice compared with those in control Sox12+/− mice. In vitro, Sox12−/− CD4+ T cells that were cultured under Th2 conditions had increased production of Th2 cytokines and GATA3 protein compared with those of control Sox12+/− CD4+ T cells. Importantly, forced expression of Sox12 decreased the protein levels of GATA3 in CD4+ T cells under Th2 conditions without affecting mRNA expression. Furthermore, Sox12 induced degradation of GATA3 through the proteasome pathway in CD4+ T cells. Consistently, Sox12 enhanced ubiquitination of GATA3, which was mediated by the E3 ligase Fbw7. Finally, we found that Fbw7 knockdown partly abrogated Sox12-mediated GATA3 suppression in CD4+ T cells. Taken together, these results suggest that Sox12 suppresses Th2 cell differentiation by accelerating Fbw7-mediated GATA3 degradation, and attenuates HDM-induced allergic inflammation.
Keywords: Sox12, Asthma, GATA3, Th2, ubiquitination
Subject terms: T-helper 2 cells, Circadian rhythms
Introduction
Allergic asthma is characterized by chronic eosinophilic airway inflammation caused by T helper 2 (Th2) cells that secrete cytokines such as IL-4, IL-5, IL-9, and IL-13.1,2 This notion is supported by the fact that adoptive transfer of in vitro-generated antigen-specific Th2 cells is sufficient to reproduce most asthma-like features.3 The activation of Stat6 and subsequent induction of GATA3, which is a master transcription factor of Th2 cells, are required for the development of Th2 cells.4,5 GATA3 induces the remodeling of chromatin at Th2-associated gene loci, and enhances the production of Th2 cytokines.6
The function of the GATA3 protein is regulated by several kinds of modifications, including phosphorylation,7 methylation,8 acetylation,9 ubiquitination10–13, and deubiquitination.14 As a positive regulator of GATA3 function, phosphorylation of GATA3 induces its nuclear translocation,7 methylation of GATA3 regulates transactivation of the IL-5 gene,8 and acetylation of GATA3 affects T-cell survival and activation.9 In addition, the E3 deubiquitinase USP21 promotes the stabilization of GATA3, and is involved in the expression of GATA3 in Treg cells.14 However, the E3 ubiquitin ligase Mdm2 is associated with GATA3 ubiquitination in Th2 cells, and induces its proteasomal degradation.10,12 Another E3 ubiquitin ligase, Fbw7 (the protein encoded by Fbxw7), ubiquitinates and destabilizes GATA3, and conditional inactivation of Fbw7 in the T-cell lineage results in a reduction in thymic CD4 single-positive cells and splenic CD4+ and CD8+ T cells.11,13
Sox (SRY-related high-mobility-group (HMG)-box) family proteins are a group of transcription factors that have a conserved HMG DNA-binding domain, and are divided into eight groups (A–H) according to structural characteristics.15 The SoxC group, which is composed of Sox4, Sox11, and Sox12, has a C-terminal transactivation domain in addition to the HMG-box domain.16,17 The SoxD group, which is composed of Sox5, Sox6, and Sox13, has a HMG-box domain in the C-terminal region and a group-specific coiled-coil domain in the N-terminal region, but lacks a transactivation domain.18 Recently, the roles of Sox family members in CD4+ T cells have been investigated. We previously showed that Sox5 physically associates with c-Maf and induces Th17 cell differentiation via the induction of RORγt.19 We also showed that Sox12 is induced in regulatory T (Treg) cells in colitic mice, and that Sox12 is involved in the development of peripherally induced Treg (pTreg) cells.20 However, Kuwahara et al.21 showed that Sox4, another member of the SoxC group, suppresses Th2 cell differentiation by physically associating with GATA3 and preventing its binding to DNA. However, the roles of Sox12 in Th2 cell differentiation and in allergic asthma remain unknown.
In this study, we showed that Sox12 deficiency results in enhanced Th2 cell differentiation and eosinophilic inflammation in a HDM-induced asthma model. Sox12 physically associated with both GATA3 and the E3 ligase Fbw7, and induced ubiquitination and destabilization of GATA3 by the ubiquitin–proteasome pathway.
Results
HDM-induced Th2 cell differentiation and eosinophilic airway inflammation are exacerbated in Sox12-deficient mice
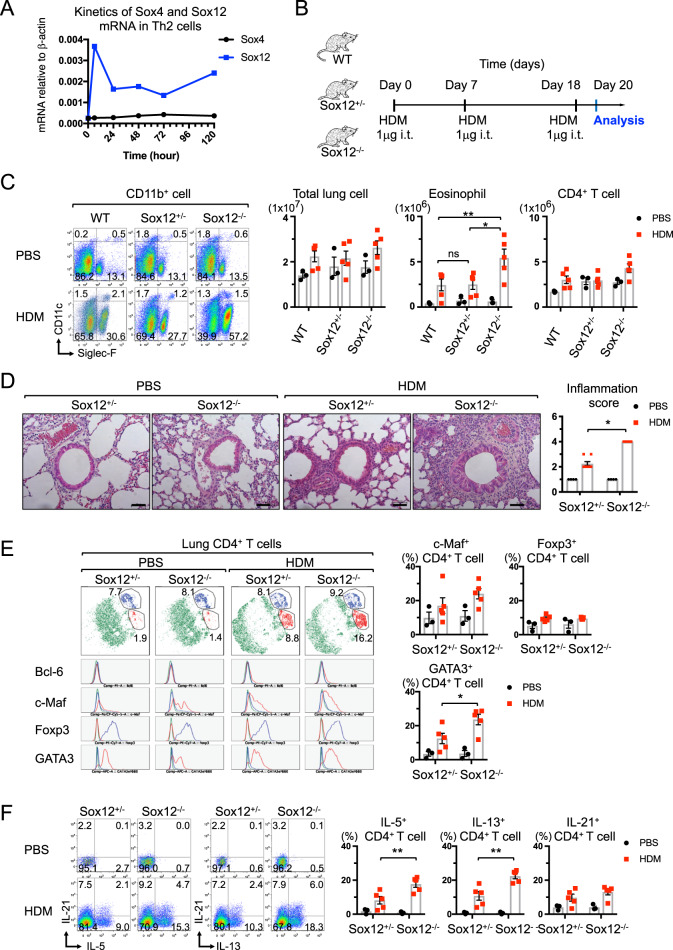
To address the roles of SoxC members in Th2 cell differentiation and allergic asthma, we first examined the kinetics of mRNA expression of SoxC members in Th2 cells. The mRNA level of Sox12 but not Sox4 was significantly increased during Th2 cell differentiation (Fig. 1a). However, Sox11 mRNA was not detected in CD4+ T cells (data not shown). We then examined Th2 cell-mediated immune responses in Sox12-deficient (Sox12−/−) mice by using a HDM-induced asthma model (Fig. 1b). Although the number of total lung cells and CD4+ T cells at 48 h after HDM challenge was not significantly different among Sox12−/−, Sox12-heterozygous (Sox12+/−), and wild-type (WT) mice, the number of eosinophils (Siglec-F+ CD11c− CD11b+ cells) was significantly higher in Sox12−/− mice than in WT or Sox12+/− mice (Fig. 1c). Histological analyses revealed that eosinophilic lung inflammation was significantly enhanced in Sox12−/− mice compared with that in Sox12+/− or WT mice (Fig. 1d and data not shown). Since eosinophilic inflammation was similar between Sox12+/− and WT mice, we hereafter used Sox12+/− littermates as controls.
Fig. 1.
HDM-induced Th2 cell differentiation and eosinophilic inflammation are exacerbated in Sox12−/− mice. a Kinetic analysis of mRNA expression of Sox4 and Sox12 in developing Th2 cells. b–f WT, Sox12+/−, and Sox12−/− mice were intratracheally sensitized and challenged with HDM (1 μg) or PBS (as control), as described in the “Materials and methods”. c Representative FACS profiles of CD11c vs. Siglec-F in CD45+ CD11b+ cells from the lung (left panels), and the means ± SEM of the numbers of indicated cells in the lung (right panels) are shown. d Representative photomicrographs of HE-stained lung sections and the means ± SEM of the inflammation scores are shown. Bars: 50 μm. e Representative t-SNE plots showing FACS analysis of CD4+ T cells from the lung (left panels). Treg cells and Th2 cells are depicted in blue and red, respectively. Histograms showing the indicated transcription factors of Treg cells (blue line) and Th2 cells (red line) in t-SNE plots. Means ± SEM of the percentages of cells expressing the indicated transcription factors (right panels). f Representative FACS profiles of the indicated cytokines in CD4+ T cells from the lung (left panels), and the means ± SEM of the percentages of cells expressing the indicated cytokines (right panels). Two-way ANOVA followed by Tukey’s test. *P < 0.05, **P < 0.01. ns not significant
To determine the characteristics of Sox12−/− CD4+ T cells in the lungs of HDM-challenged mice, we examined the expression of transcription factors (Bcl-6, GATA3, c-Maf, and Foxp3) in CD3ε+ CD4+ T cells that were isolated from the lungs (Fig. 1e). Two-dimensional visualization of cytometry data using t-distributed stochastic neighbor embedding (t-SNE) revealed that GATA3+ Bcl-6− c-Maf+ Th2 cells (depicted in red) were increased in Sox12−/− mice compared with those in Sox12+/− mice, whereas Foxp3+ Treg cells (depicted in blue) were similar between Sox12−/− and Sox12+/− mice (Fig. 1e). The frequency of GATA3-positive CD4+ T cells in the lung (Fig. 1e) and the mean fluorescence intensity (MFI) of GATA3 in the GATA3-positive CD4+ T cells (Fig. S1A) were significantly higher in Sox12−/− mice than in Sox12+/− mice. However, the frequency of c-Maf-positive CD4+ T cells (Fig. 1e) and the MFI of c-Maf in c-Maf-positive CD4+ T cells (Fig. S1A) were comparable between Sox12−/− and Sox12+/− mice. Consistent with the increased number of GATA3-positive CD4+ T cells in the lung, the number of IL-5- or IL-13-positive CD4+ T cells was increased in the lung in Sox12−/− mice compared with that of Sox12+/− mice (Fig. 1f). However, IL-21-positive CD4+ T cells in the lung were comparable between Sox12−/− and Sox12+/− mice (Fig. 1f), and IL-4-positive CD4+ T cells were undetectable in both Sox12−/− and Sox12+/− mice in this experimental setting (data not shown).
Contrary to the number of CD4+ T cells in the lung, the frequency of GATA3-positive CD4+ T cells in mediastinal lymph nodes (mLNs) was similar between Sox12−/− and Sox12+/− mice (Fig. S1B), which is consistent with the finding that the mRNA level of Sox12 in mLN CD4+ T cells was lower than that in lung CD4+ T cells (Fig. S1C). The frequency of CD11c+ DCs and the expression of CD80 on CD11c+ DCs in the lung (Fig. S1D) were also comparable between Sox12−/− and Sox12+/− mice. In addition, the number of group 2 innate lymphoid cells (ILC2s) in the lung and the MFI of GATA3 in lung ILC2s were comparable between Sox12−/− and Sox12+/− mice (Fig. S1E). Moreover, total IgE and HDM-specific IgG1 in sera were similarly upregulated in Sox12−/− and Sox12+/− mice (Fig. S1F), while HDM-specific IgE was undetectable in both Sox12−/− and Sox12+/− mice (data not shown). Taken together, these results suggest that Sox12 in lung CD4+ T cells is mainly involved in the suppression of eosinophilic inflammation and Th2 cell differentiation in this asthma model.
Sox12 suppresses the expression of GATA3 and production of cytokines in Th2 cells
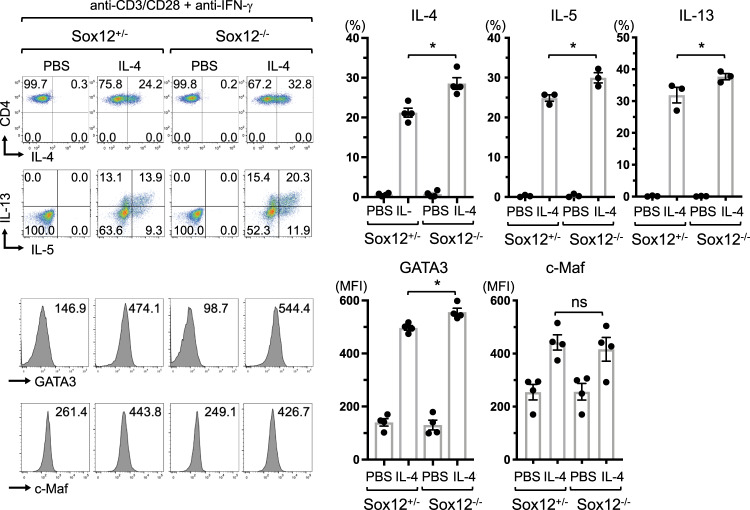
We next examined whether Sox12 in CD4+ T cells is involved in the regulation of Th2 cell differentiation. In this experiment, we utilized Foxp3-reporter mice (Foxp3YFP-cre) to minimize the influence of contaminating Treg cells. Foxp3YFP-naive CD4+ T cells were isolated from Foxp3YFP-cre Sox12–/– mice and littermate Foxp3YFP-cre Sox12+/− mice, and stimulated with anti-CD3/CD28 antibodies in the presence of anti-IFN-γ antibody with or without IL-4 (Fig. 2). The frequency of IL-4-, IL-5-, or IL-13-producing cells was significantly increased in Sox12−/− CD4+ T cells compared with that in Sox12+/− CD4+ T cells. Consistently, the MFI of GATA3 was significantly increased in Sox12−/− CD4+ T cells compared with that in Sox12+/− Th2 cells, whereas the MFI of c-Maf was similar between Sox12−/− and Sox12+/− Th2 cells (Fig. 2). These results indicate that Sox12 in CD4+ T cells inhibits the development of Th2 cells.
Fig. 2.
Sox12 suppresses the expression of GATA3 and production of cytokines in Th2 cells. Foxp3YFP-naive CD4+ T cells were isolated from Foxp3YFP-cre Sox12−/− mice and littermate Foxp3YFP-cre Sox12+/− mice, and stimulated with anti-CD3 + anti-CD28 in the presence of anti-IFN-γ mAb with/without IL-4 (10 ng/ml). On days 3 and 7, fresh medium supplemented with IL-2 (5 ng/ml) was added to the culture, and the cells were analyzed on day 11. Representative FACS profiles showing IL-4, IL-5, IL-13, GATA3, and c-Maf expression of CD4+ T cells, and the means ± SEM of the percentages of IL-4+ cells, IL-5+ cells, and IL-13+ cells, and the MFI of GATA3 and c-Maf is shown. One-way ANOVA followed by Dunnett’s test. *P < 0.05
Sox12 suppresses the expression of GATA3 independently of Foxp3
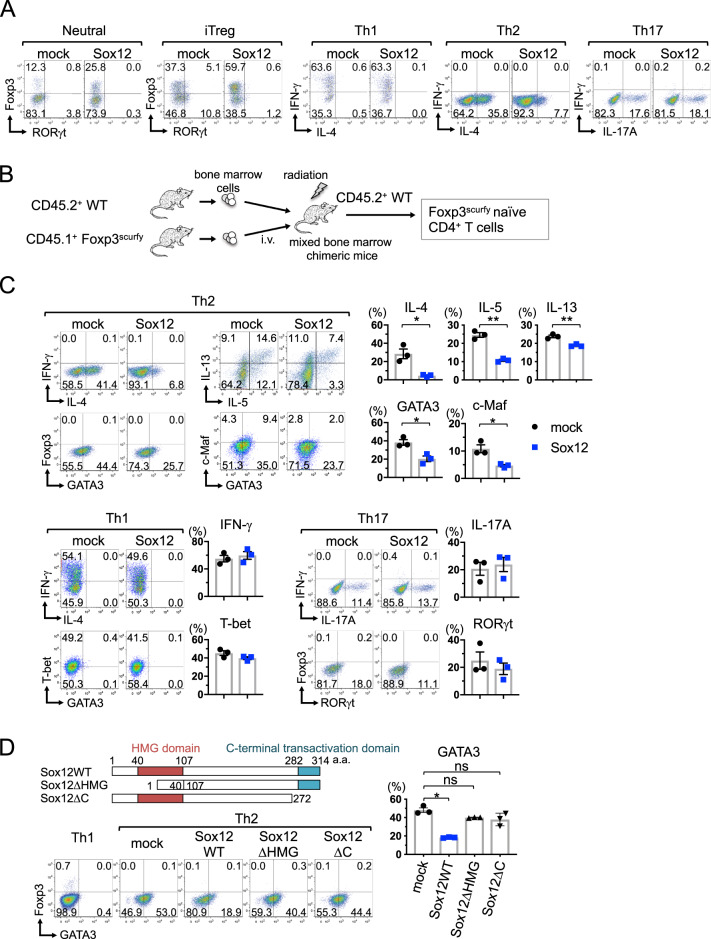
We next examined the effect of forced expression of Sox12 on the development of helper T cells. As shown in Fig. 3a, retrovirus-mediated expression of Sox12 inhibited the production of IL-4 under Th2 conditions. However, the forced expression of Sox12 did not affect the production of IL-17A under Th17 conditions, or the production of IFN-γ under Th1 conditions. Consistent with our previous report,20 the forced expression of Sox12 enhanced the development of Foxp3+ cells under neutral and iTreg conditions.
Fig. 3.
Sox12 suppresses the expression of GATA3 independently of Foxp3. a Naive CD4+ T cells were isolated from WT mice, stimulated under neutral, Th1, Th2, Th17, or iTreg conditions, and infected with retroviruses carrying either pMX-IRES-NGFR (mock) or pMX-Sox12-IRES-NGFR (Sox12). Representative FACS profiles of the indicated transcription factors and cytokines in NGFR+ cells are shown. b, c Naive CD45.1+ Foxp3scurfy CD4+ T cells were isolated from mixed bone marrow chimeric mice, stimulated under Th1, Th2, or Th17 conditions, and infected with retroviruses carrying either pMX-IRES-NGFR (mock) or pMX-Sox12-IRES-NGFR (Sox12). Representative FACS profiles of the indicated cytokines and transcription factors in infected NGFR+ cells, and the means ± SEM of the percentages of the indicated cells in each condition, are shown. Student’s t test. *P < 0.05. d Schematic representation of Sox12 mutants (upper panel). WT CD4+ T cells were stimulated under Th2 conditions and infected with retroviruses containing full-length Sox12 or Sox12 mutants. WT CD4+ T cells were stimulated under Th1 conditions and used as controls. Representative FACS profiles of GATA3 vs. Foxp3 in NGFR+ cells under Th2 conditions, and of live cells under Th1 conditions (left lower panels), and the means ± SEM of the percentages of GATA3+ cells in each condition (right panels), are shown. One-way ANOVA followed by Tukey’s test. *P < 0.05. ns not significant
Because Sox12-mediated Foxp3 induction affects the suppression of Th2 cells, we next examined the effect of Sox12 induction on the differentiation of naive CD4+ T cells that lack functional Foxp3. In this experiment, we prepared Foxp3-deficient naive CD4+ T cells from mixed bone marrow chimeric mice containing CD45.1+ Foxp3scurfy bone marrow cells and CD45.2+ WT bone marrow cells (Fig. 3b). Forced expression of Sox12 inhibited the production of IL-4, IL-5, and IL-13, and the expression of GATA3 and c-Maf even in Foxp3scurfy CD4+ T cells under Th2 conditions (Fig. 3c). However, forced expression of Sox12 did not inhibit the expression of IFN-γ or T-bet under Th1 conditions, or the expression of IL-17A or RORγt under Th17 conditions in Foxp3scurfy CD4+ T cells. Taken together, these results suggest that Sox12 inhibits Th2 cell differentiation by suppressing GATA3 expression independently of Foxp3 induction.
To determine which domain of Sox12 is involved in the suppression of GATA3, we prepared Sox12 mutants that lacked the HMG domain (Sox12ΔHMG) or the C-terminal transactivation domain (Sox12ΔC) (Fig. 3d). While wild-type Sox12 (Sox12WT) significantly inhibited GATA3 expression under Th2 conditions, neither Sox12ΔHMG nor Sox12ΔC inhibited GATA3 expression (Fig. 3d). These results suggest that both domains of Sox12 are involved in the suppression of GATA3 expression in Th2 cells, although we cannot exclude the possibility that conformational changes caused by the deletions may affect the function of the Sox12 protein.
Sox12 induces the degradation of GATA3 by the ubiquitin–proteasome pathway
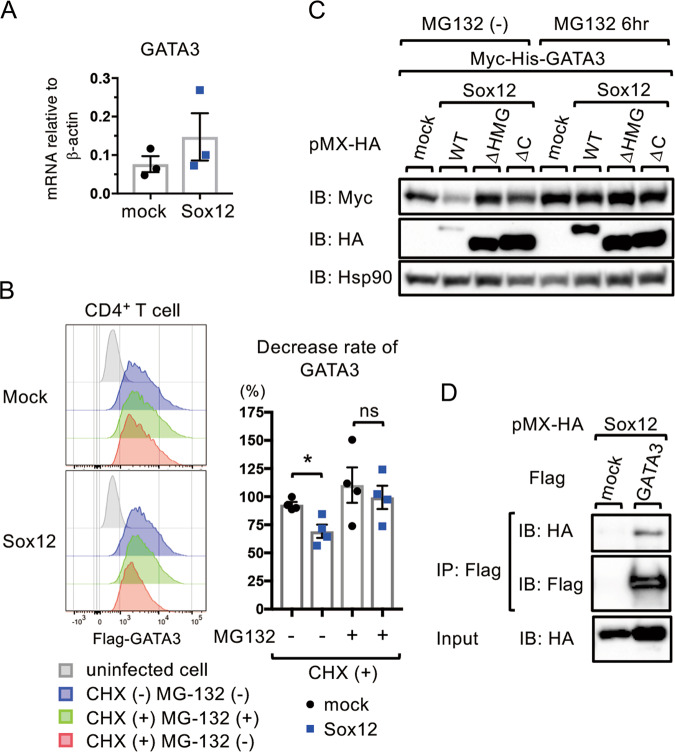
To address the mechanisms underlying Sox12-mediated GATA3 suppression under Th2 conditions, we first examined whether Sox12 suppressed the expression of GATA3 at the mRNA level. However, the mRNA expression of GATA3 was not inhibited by the forced expression of Sox12 in CD4+ T cells under Th2 conditions (Fig. 4a). We therefore analyzed whether Sox12 induced the degradation of GATA3 at the protein level in CD4+ T cells. In this experiment, Sox12−/− CD4+ T cells were infected with retroviruses carrying MSCV-Flag-GATA3-IRES-Thy1.1, along with pMX-IRES-NGFR (mock) or pMX-Sox12-IRES-NGFR (Sox12). Three days later, the cells were treated with the protein synthesis inhibitor cycloheximide (CHX) alone or in combination with the proteasome inhibitor MG-132 for 30 min, and the expression of Flag-GATA3 was evaluated by flow cytometry. As shown in Fig. 4b, the Sox12-mediated decrease in Flag-GATA3 was abrogated in the presence of MG-132, suggesting that Sox12 induces the degradation of GATA3 by the proteasome pathway in CD4+ T cells.
Fig. 4.
Sox12 induces degradation of GATA3 by the proteasome pathway. a Naive WT CD4+ T cells were stimulated under Th2 conditions and infected with retroviruses containing pMX-IRES-NGFR (mock) or pMX-Sox12-IRES-NGFR (Sox12). Two days after infection, NGFR+ cells were sorted, and the mRNA expression of GATA3 was evaluated. b Sox12−/− CD4+ T cells were infected with retroviruses containing MSCV-Flag-GATA3-IRES-Thy1.1, along with pMX-IRES-NGFR (mock) or pMX-Sox12-IRES-NGFR (Sox12). Three days after infection, the cells were treated with CHX alone or CHX+MG-132 for 30 min, and then analyzed by flow cytometry. Representative histograms of Flag-GATA3 in NGFR+ Thy1.1+ cells in untreated, CHX-treated, and CHX + MG-132-treated cells are shown in the left panels. The means ± SEM of the decreased rate of Flag-GATA3 in CHX-treated cells and CHX + MG-132-treated cells compared with those of untreated cells are shown in the right panels. Student’s t test. *P < 0.05, ns not significant. c 293T cells were transfected with Myc-His-GATA3, along with pMX-IRES-NGFR (mock) or pMX-HA-Sox12 mutants. Two days after transfection, the cells were treated with MG-132 or vehicle for 6 h, and subjected to immunoblotting (IB) with the indicated antibodies. d 293T cells were transfected with pMX-HA-Sox12, along with Flag-GATA3 or mock vector. Two days after transfection, the cell lysates were immunoprecipitated (IP) with the anti-Flag antibody and immunoblotted with anti-HA antibody
We next analyzed which domain of Sox12 is involved in the degradation of GATA3. In this experiment, 293T cells were transduced with GATA3, along with Sox12WT, Sox12ΔHMG, or Sox12ΔC, in the presence or absence of MG-132, and the protein levels of GATA3 were evaluated by western blotting. As shown in Fig. 4c, the transduction of Sox12WT, but not Sox12ΔHMG or Sox12ΔC, significantly decreased GATA3 expression in the absence of MG-132. In contrast, in the presence of MG-132, the transduction of Sox12WT did not significantly decrease GATA3 expression (Fig. 4c). These results suggest that both the HMG domain and the C-terminal domain of Sox12 are involved in proteasomal degradation of GATA3. Interestingly, the expression level of Sox12WT, but not Sox12ΔHMG or Sox12ΔC, was significantly reduced in the absence of MG-132 (Fig. 4c), suggesting that Sox12WT is also degraded by the proteasome pathway. We also examined the protein–protein interaction between GATA3 and Sox12 in 293T cells by coimmunoprecipitation assay and confirmed the interaction (Fig. 4d).
We next examined whether Sox12 enhances the ubiquitination of GATA3. 293T cells were transfected with pFlag-Ub and MSCV-Myc-His-GATA3, along with pMX-HA-Sox12 or pMX empty vector (mock), and ubiquitinated Myc-His-GATA3 proteins were purified by cobalt beads, followed by anti-Flag antibody, and visualized by immunoblotting with anti-Myc antibody. As shown in Fig. 5, ubiquitinated GATA3 was significantly increased in the presence of Sox12. Taken together, these results suggest that Sox12 physically associates with GATA3 and decreases GATA3 protein levels by the ubiquitin–proteasome pathway.
Fig. 5.
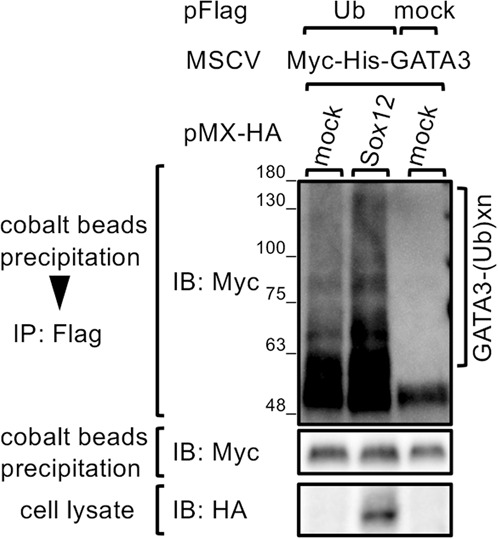
Sox12 is involved in the ubiquitination of GATA3. 293T cells were transfected with Myc-His-GATA3, along with either pFlag-Ub or pFlag-empty vector, and either pMX-HA-Sox12 or pMX empty vector. Two days after transfection, the cells were treated with MG-132 for 6 h. After the Myc-His-GATA3 proteins were purified by cobalt beads, ubiquitinated Myc-His-GATA3 proteins were immunoprecipitated with anti-Flag antibody and immunoblotted with HRP-conjugated anti-myc antibody
Sox12 enhances Fbw7-mediated ubiquitination of GATA3
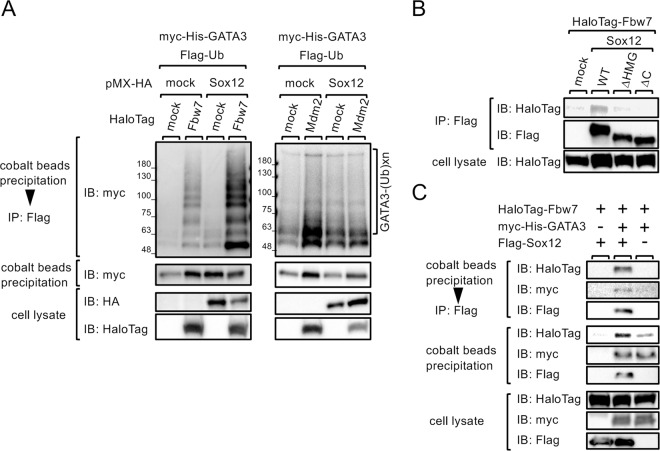
Because Sox12 does not have ubiquitin E3 ligase activity, it is likely that Sox12 indirectly induces the ubiquitination of GATA3 through the recruitment of some E3 ligases. Regarding the ubiquitination of GATA3, Mdm210 and Fbw711 have been reported to function as E3 ligases for GATA3.12,13 Therefore, we examined whether Sox12 enhances Fbw7- or Mdm2-mediated GATA3 ubiquitination. As shown in Fig. 6a (left panels), Fbw7 induced the ubiquitination of GATA3 (lane 2), and Sox12 enhanced Fbw7-mediated GATA3 ubiquitination (lane 4). However, Sox12 did not enhance Mdm2-mediated GATA3 ubiquitination (Fig. 6a, right panels). Consistently, coimmunoprecipitation assays revealed that Fbw7 was associated with Sox12WT, but not with the Sox12 mutants Sox12ΔHMG or Sox12ΔC in 293T cells (Fig. 6b). We also examined whether Sox12, GATA3, and Fbw7 form a ternary complex. As shown in Fig. 6c, sequential immunoprecipitation experiments demonstrated that GATA3 formed a ternary complex with Sox12 and Fbw7 in 293T cells. Taken together, these results suggest that Sox12 physically associates with Fbw7 and enhances the ubiquitination of GATA3.
Fig. 6.
Sox12 enhances Fbw7-mediated ubiquitination of GATA3. a 293T cells were transfected with MIT-Myc-His-GATA3 and pFlag-Ub, along with either pMX-HA-Sox12 or pMX empty vector, and pFN21-empty vector, pFN21A-HaloTag-Fbw7 (left panels), or pFN21A-HaloTag-Mdm2 (right panels). Two days after transfection, ubiquitinated Myc-His-GATA3 proteins were detected as described in Fig. 5. b 293T cells were transfected with pFN21A-HaloTag-Fbw7, along with pMX (mock) or pMX-Flag-Sox12 mutants. Two days after transfection, the cells were treated with MG-132 for 6 h. Cell lysates were then immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HaloTag antibody. c 293T cells were transfected with the indicated plasmids for 2 days, and then treated with MG-132 for 6 h. Cell lysates were precipitated with cobalt beads, and imidazole-eluted protein complexes were subsequently immunoprecipitated with anti-Flag antibody. Cell lysates (lower panels), imidazole-eluted protein complexes (middle panels), and anti-Flag immunoprecipitated protein complexes (upper panels) were immunoblotted with HRP-conjugated anti-HaloTag antibody, anti-myc antibody, or anti-Flag antibody
Fbw7 knockdown increases the production of IL-5 and IL-13 in Sox12-expressing CD4+ T cells
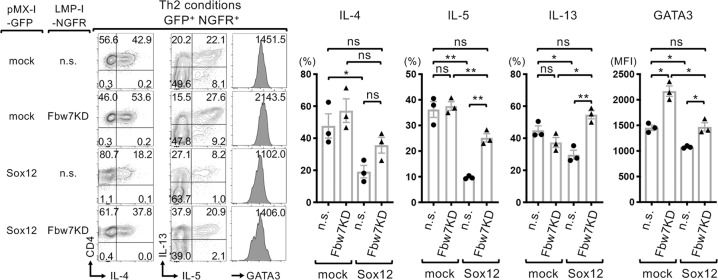
To assess the role of Fbw7 in Sox12-mediated suppression of Th2 cells, we examined the effect of Fbw7 knockdown on the expression of GATA3 and the production of Th2 cytokines in Sox12-expressing CD4+ T cells under Th2 conditions. Consistent with the data shown in Fig. 3, in the absence of Fbw7 knockdown [non-silencing (n.s.)], the forced expression of Sox12 significantly decreased the expression levels of IL-4, IL-5, IL-13, and GATA3 in CD4+ T cells (Fig. 7). Importantly, Fbw7 knockdown (Fbw7KD) increased the levels of IL-5, IL-13, and GATA3 in Sox12-expressing CD4+ T cells (Fig. 7). Fbw7 knockdown also increased the levels of GATA3 in CD4+ T cells even in the absence of Sox12 (Fig. 7). These results suggest that Fbw7 is involved in the suppression of Th2 cells via both Sox12-dependent and Sox12-independent mechanisms.
Fig. 7.
Sox12 enhances Fbw7-mediated ubiquitination of GATA3. Sox12−/− CD4+ T cells were cultured under Th2 conditions and coinfected with retroviruses containing either pMX-IRES-GFP (mock) or pMX-Sox12-IRES-GFP (Sox12), and either LMP-non-silencing-IRES-NGFR (n.s.) or LMP-Fbw7KD-IRES-NGFR (Fbw7KD), and were analyzed 6 days after infection. Representative FACS profiles of IL-4 vs. CD4, IL-5 vs. IL-13, histograms of GATA3 gated on GFP+ NGFR+ cells, the means ± SEM of percentages of the indicated cells in each condition (right panels), and the MFI of GATA3 are shown. One-way ANOVA followed by Tukey’s test. *P < 0.05. ns not significant
Discussion
In this study, we showed that among SoxC family members, Sox12 expression is increased during Th2 cell differentiation. We also showed that Sox12 deficiency results in the upregulation of Th2 cytokine production, as well as the expression of GATA3, leading to the exacerbation of HDM-induced allergic asthma. Regarding the underlying mechanism, we found that Sox12 induced the degradation of GATA3 through the proteasome-dependent pathway by associating with GATA3 and the E3 ligase Fbw7, and enhancing Fbw7-mediated ubiquitination of GATA3 (Fig. S2).
We found that eosinophilic inflammation was exacerbated in Sox12−/− mice compared with that in Sox12+/− or WT mice in the HDM-induced asthma model (Fig. 1c). We also found that the number of Th2 cells in the lung was increased in Sox12−/− mice compared with that in Sox12+/− mice in the asthma model (Fig. 1e, f), whereas the number of Th2 cells in draining LNs was comparable between these mice (Fig. S1B). Although we previously showed that Sox12 is expressed in Treg cells in colitic mice, and that Sox12 promotes the development of pTreg cells during experimental colitis,20 the proportion of Treg cells in the lung was much lower than that in the colon, and the number of Treg cells in Sox12−/− mice was similar to that in Sox12+/− mice in the asthma model (Fig. 1e, f). The number of IL-21-producing CD4+ T cells was also similar between Sox12−/− and Sox12+/− mice in the asthma model (Fig. 1e, f). These results suggest that Sox12 is primarily involved in the inhibition of the Th2 cell response in the lung in the HDM-induced asthma model.
We also showed that Sox12 in CD4+ T cells suppressed Th2 cytokine production and GATA3 expression under Th2 conditions. We found that in vitro differentiation of Th2 cells from highly purified naive CD4+ T cells was significantly enhanced by the absence of Sox12 (Fig. 2). We also found that forced expression of Sox12 markedly suppressed Th2 cell differentiation without affecting Th1 or Th17 cell differentiation (Fig. 3a). Although Sox12 promoted the development of Foxp3+ Treg cells (Fig. 3a), the forced expression of Sox12 suppressed Th2 cell differentiation even in the absence of Foxp3 (Fig. 3c), indicating that Sox12 suppresses Th2 cell differentiation independently of Foxp3 induction.
We found that Sox12 promoted proteasome-dependent degradation of GATA3. GATA3 expression is controlled by not only transcriptional activation of the GATA3 gene, but also modifications that affect the stability of the GATA3 protein. We found that Sox12 did not suppress the mRNA expression of GATA3, but reduced the protein levels of GATA3 in CD4+ T cells (Fig. 4). We also found that the reduction in GATA3 protein was inhibited by the proteasome inhibitor MG-132 (Fig. 4), suggesting that Sox12 promotes degradation of GATA3 via the proteasome pathway. However, Kuwahara et al.21 showed that Sox4, another member of the SoxC group, binds to GATA3 and prevents GATA3 binding to consensus sequences of target genes. Because the amino acid residues Arg61, Pro62, Phe66, and Met67, which are important for the suppression of GATA3 binding to the consensus sequences, are conserved between Sox4 and Sox12, Sox4 and Sox12 may share a mechanism of action regarding the inhibition of GATA3 function to some extent. Further studies are needed to determine whether Sox4 also promotes proteasome-dependent degradation of GATA3 in CD4+ T cells.
Our findings indicate that Sox12 enhances Fbw7-mediated ubiquitination and degradation of GATA3. Fbw7 ubiquitinates GATA3, and conditional inactivation of Fbw7 in the T-cell lineage results in a reduction in CD4 single-positive thymocytes, CD4+ splenocytes, and CD8+ splenocytes due to overexpression of GATA3 protein.11 We found that Sox12 formed a ternary complex with Fbw7 and GATA3, and enhanced Fbw7-mediated ubiquitination of GATA3 in 293T cells (Fig. 6). We also found that knockdown of Fbw7 in CD4+ T cells under Th2 conditions partly abolished Sox12-mediated downregulation of GATA3 expression and Th2 cytokine production (Fig. 7). Importantly, knockdown of Fbw7 increased the levels of GATA3 even in the absence of Sox12 (Fig. 7). These findings suggest that Fbw7 is involved in the suppression of Th2 cells via both Sox12-dependent and Sox12-independent mechanisms.
Unexpectedly, we found that Sox12, but not its mutants (Sox12ΔHMG or Sox12ΔC), was significantly degraded by the proteasome pathway (Fig. 4c). In addition, we found that Sox12, Fbw7, and GATA3 form a ternary complex when they were overexpressed (Fig. 6c). These findings suggest that the ternary complex could be subjected to Fbw7-mediated ubiquitination and subsequent degradation by the proteasome pathway (Fig. S2). Further studies evaluating endogenous Sox12 expression at the protein level and the ternary complex formed with endogenous GATA3 and Fbw7 in Th2 cells are required.
It is well established that GATA3 plays essential roles in the development of not only Th2 cells but also ILC2s. We found that the number of Th2 cells was increased in Sox12−/− mice compared with that in Sox12+/− mice (Fig. 1), but the number of ILC2s was comparable between Sox12−/− and Sox12+/− mice in the HDM-induced asthma model (Fig. E1). We also found that the levels of GATA3 were comparable between Sox12−/− ILC2s and Sox12+/− ILC2s (Fig. E1). These differences could be explained by the expression levels of Sox12. According to RNA-seq data in the immunological genome project (http://www.immgen.org), the expression of Sox12 mRNA in ILC2s was much lower than that in naive CD4+ T cells. In addition, we found that the expression of Sox12 was significantly induced upon TCR stimulation in CD4+ T cells.20 Therefore, although GATA3 is crucial for the development of both Th2 cells and ILC2s, Sox12-mediated GATA3 downregulation may occur only in Th2 cells.
In conclusion, we showed that Sox12 enhances Fbw7-mediated ubiquitination and degradation of GATA3 in differentiating Th2 cells, and that genetic loss of Sox12 results in enhanced Th2 cell differentiation and allergic inflammation in the HDM-induced asthma model. Although further studies are needed to clarify the pathophysiological roles of this pathway in more detail, our findings add new insights into the mechanism underlying the downregulation of Th2 cell-mediated immune responses in asthma.
Materials and methods
Mice
C57BL/6 mice, C57BL/6 background Sox12−/− mice, Foxp3YFP-cre mice, Foxp3YFP-cre Sox12−/− mice, and CD45.1+ background Scurfy mice were previously described.19,20,22 All mice were housed in microisolator cages under specific pathogen-free conditions. The Chiba University Animal Care and Use Committee approved the protocols for animal experiments.
RT-PCR analysis
Extraction of total cellular RNA, reverse transcription, and quantitative PCR analysis were performed as previously described.20 Quantitative PCR was performed with a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). The levels of target genes were normalized to the levels of β-actin. Sequences of the qPCR primers are shown in the supplementary information (Table S1).
HDM-induced allergic airway inflammation
The mice were sensitized and challenged with intratracheal administration of HDM extracts (Greer Laboratories) as previously described.23 In brief, the mice were sensitized intratracheally with HDM (1 μg in 25 μl of PBS) at days 0 and 7, and were challenged with HDM (1 μg in 25 μl of PBS) at day 18. Forty-eight hours after the HDM challenge, asthmatic responses were evaluated. The mice were perfused with 50 ml of ice-cold phosphate-buffered saline to remove blood cells. The total lung cells were prepared by using a gentleMACSTM Octo Dissociator (Miltenyi Biotec) with liberase TL (0.1 mg/ml, Roche, Basel, Schweiz) and DNase I (0.1 mg/ml, Roche) in 5 ml of RPMI 1640 medium. Single-cell suspensions of total lung cells were then obtained by passage through 50-μm cell strainers. Eosinophils were defined as CD45+ CD11b+ CD11c– Siglec-F+ cells, CD4+ T cells as CD45+ CD11b− CD4+ CD3+ cells, DCs as CD11c+ MHC ClassIIhigh cells, and ILC2s as Lin− CD45+ Thy1+ ST2+ cells. Lung cells (2 × 106 cells) were stimulated with PMA plus ionomycin in the presence of brefeldin A for 4 h, and the production of IL-5, IL-13, and IL-21 was evaluated by intracellular cytokine staining. Flow cytometric analyses were performed on a FACSCanto II (BD Biosciences) with FlowJo software (Tree Star, Ashland, OR). Histological analysis was performed as previously described.23,24
Reagents
Antibodies against CD3ε (145-2C11), CD28 (37.51), CD11b (M1/70), CD11c (HL3), CD44 (IM7), CD45 (30-F11), CD62L (MEL-14), Siglec-F (E50-2440), IL-4 (11B11), IL-4 (BDV4-1D11), IFN-γ (XMG1.2), RORγt (Q31-378), Bcl-6 (K112-91), and human NGFR (C40-1457) were purchased from BD Biosciences (San Diego, CA). Antibodies against CD16/32 (93), CD25 (7D4), CD25 (PC61), CD45.1 (A20), CD45.2 (104), DYKDDDDK (L5), IL-5 (TRFK5), T-bet (4B10), ST2 (DIH9), and IL-17A (TC11-18H10.1) were purchased from BioLegend (San Diego, CA, USA). Antibodies against Foxp3 (FJK-16s), c-Maf (sym0F1), GATA3 (TWAJ), and IL-13 (eBio13A) were purchased from eBioscience (San Diego, CA, USA). The IL-21R-Fc chimera was purchased from R&D Systems (Minneapolis, MN, USA). The antibodies used in western blot analyses were as follows: anti-Myc (9E10, Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-HA (ab9110, Abcam, Cambridge, UK), anti-Flag-M2 (Sigma), anti-Hsp90 (H114, Santa Cruz), and monoclonal anti-HaloTag (Promega). The anti-HaloTag monoclonal antibody was conjugated with HRP by using a HRP conjugation kit (Abcam). Cycloheximide and MG-132 were purchased from Sigma-Aldrich (St. Louis, MO).
Intracellular cytokine and transcription factor staining
Intracellular cytokine staining and transcription factor staining were performed as previously described.25–27
Cell isolation and cell culture
Naive CD4+ T cells were isolated from lymph nodes or the spleen by using a naive CD4+ T cell isolation kit (Miltenyi Biotec, Sunnyvale, CA) according to the manufacturer’s instructions. CD45.1+ CD62L+ naive CD4+ T cells or Foxp3YFP− CD25− CD62L+ CD44− CD4+ T cells were purified using a SH800 cell sorter (SONY, Kanagawa, Japan).
Naive CD4+ T cells were stimulated with plate-bound anti-CD3ε mAb (1 μg/ml) in the presence of anti-CD28 mAb (1 μg/ml) at 1 ×106 cells/ml in a 96-well plate under neutral conditions (anti-IL-4 mAb (10 μg/ml) and anti-IFN-γ mAb (10 μg/ml)), Th1 conditions (IL-12 (10 ng/ml), IL-2 (5 ng/ml), and anti-IL-4 mAb), Th2 conditions (IL-4 (10 ng/ml) and anti-IFN-γ mAb), Th17 conditions (IL-6 (10 ng/ml), TGF-β (1 ng/ml), anti-IL-4 mAb, and anti-IFN-γ mAb), or iTreg conditions (TGF-β (3 ng/ml), IL-2 (10 ng/ml), anti-IL-4 mAb, and anti-IFN-γ mAb). For Th1 and Th2 cell culture, cells were split in medium that was supplemented with IL-2 (5 ng/ml) on day 3.
Plasmids and retrovirus-mediated gene expression
MSCV-IRES-GFP (MIG), MSCV-IRES-Thy1.1 (MIT), and pMX-IRES-NGFR (pMX-IN) are retroviral vectors containing an internal ribosome entry site (IRES). pMX-Sox12-IRES-NGFR was a kind gift from Dr. M. Yamashita (Ehime University, Ehime, Japan). The HA tag was fused to Sox12 by PCR amplification, and HA-Sox12 was subcloned into pMX-IN to create pMX-HA-Sox12-IRES-NGFR. The Flag tag or Myc-His tag was fused to GATA3 by PCR amplification, and subcloned into MIT to create MSCV-Flag-GATA3-IRES-Thy1.1 or MSCV-Myc-His-GATA3-IRES-Thy1.1. Sox12 mutants lacking the HMG domain (Sox12ΔHMG) and C-terminal transactivation domain (Sox12ΔC) were generated using a KOD-Plus-mutagenesis kit according to the manufacturer’s instructions. Ubiquitin cDNA was generated by gene synthesis and subcloned into the pFlag-CMV-2 vector to make pFlag-Ub. pFN21A-HaloTag-FBXW7 (pFN21AB5050) and pFN21A-HaloTag-MDM2 (pFN21AB5561) were purchased from Promega. Retrovirus-mediated gene induction of CD4+ T cells was performed by a RetroNectin-bound viral infection method (Takara Bio, Otsu, Japan).25
Mixed bone marrow chimeras
Bone marrow cells were obtained from Foxp3scurfy mice (CD45.1+ background) and CD45.2+ background C57BL/6 mice (2 × 106 cells each), mixed, and injected intravenously into CD45.2+ background C57BL/6 mice after total body irradiation (9.5 Gy). Naive CD45.1+ Foxp3scurfy CD4+ T cells were purified from chimeric mice 10–12 weeks after bone marrow reconstitution.
Coimmunoprecipitation analysis
293T cells were transfected with the indicated vectors using Effectene transfection reagent (QIAGEN). Thirty-six hours after transfection, the cells were lysed in high-salt lysis buffer [240 mM NaCl, 50 mM Tris, 1% Nonidet P-40, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, and 3% protease inhibitor cocktail (Sigma, St. Louis, MO, USA)]. Whole-cell extracts were precleared with sepharose beads (Sigma) and then immunoprecipitated with anti-Flag-M2 magnetic beads (Sigma).
Ubiquitination assays
For the in vivo ubiquitination assay of GATA3, the cells were transfected with MSCV-Myc-His-tagged GATA3 (Myc-His-GATA3) and Flag-tagged Ub (Flag-Ub) and treated with/without MG-132 for 5 h. In total, 10% of the transduced cells were lysed in high- salt lysis buffer and used as an input sample. The residual 90% of transduced cells were lysed in 1 ml of buffer A [6 M guanidinium-HCl, 0.1 M sodium phosphate (pH 8.0), and 10 mM imidazole],28 sonicated for 10 s 3 times, and mixed with 50 μl of Dynabeads coated in cobalt-based immobilized metal affinity chromatography chemistry (cobalt beads) (Thermo Fisher Scientific, Waltham, MA) on a rotator for 30 min at room temperature. The cobalt beads were sequentially washed with 1 ml of buffer A, with 1 ml of buffer A diluted in binding/wash buffer at 1:4, and with 1 ml of binding/wash buffer [50 mM sodium phosphate (pH 8.0), 300 mM NaCl, and 0.02% Tween]. Purified myc-his-GATA3 proteins were eluted by boiling the cobalt beads in 2× sample buffer supplemented with 200 mM imidazole. The same amounts of Myc-His-GATA3 proteins were diluted in dilution buffer [50 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 5 mM EDTA] to adjust the SDS concentration to 0.1%. Two percent of the diluted solutions were left as a control sample, and 98% of the diluted solutions were mixed with 20 μl of anti-Flag M2 affinity beads (Sigma) on a rotator for 15 h at 4 °C to purify the ubiquitinated myc-his-GATA3 proteins. After the beads were washed three times with binding/wash buffer, the ubiquitinated Myc-His-GATA3 proteins were eluted by boiling in 2× sample buffer and analyzed by immunoblotting with HRP-conjugated anti-Myc antibody.
Short-hairpin RNA (shRNA) knockdown
A LMP retroviral vector that enables efficient expression of shRNAmir (shRNAs embedded within a natural microRNA backbone)29 was obtained from Thermo Fisher Scientific. To make the LMP-IRES-NGFR vector, the GFP cassette of the LMP vector was substituted with human NGFR. To make the Fbw7-knockdown retroviral vector (LMP-Fbw7KD-IRES-NGFR), Fbw7 shRNAmir was designed at the RNAi codex website (http://cancan.cshl.edu/cgi-bin/Codex/Codex.cgi) and subcloned into the LMP-IRES-NGFR vector according to the manufacturer’s instructions. The oligonucleotide DNA for LMP-Fbw7KD-IRES-NGFR was TGCTGTTGACAGTGAGCGCCCATGCAAAGTCTCAGATTATTAGTGAAGCCACAGATGTAATAATCTGAGACTTTGCATGGTTGCCTACTGCCTCGGA. The oligonucleotide DNA for LMP-non-silencing (n.s.)-IRES-NGFR was TGCTGTTGACAGTGAGCGATCTCGCTTGGGCGAGAGTAAGTAGTGAAGCCACAGATGTACTTACTCTCGCCCAAGCGAGATTGCCTACTGCCTCGGA.
Data analysis
The data are presented as the means ± SEM. The statistical analyses of the results were performed by unpaired Student’s t tests or one-way or two-way ANOVA, followed by Tukey’s test or Dunnett’s test. A value of P < 0.05 was considered significant.
Supplementary information
Acknowledgements
We thank Dr. S. Hori for the Foxp3scurfy mice, Dr. M. Yamashita for pMX-IN-Sox12, and Ms. K. Nemoto, J. Iwata, and M. Yoshino for technical help. This work was supported in part by Grants-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, the Japanese Government, the LGS (Leading Graduate School at Chiba University) Program, MEXT, and the Institute for Global Prominent Research, Chiba University, Japan. The sources of support for this work: this work was supported in part by Grants-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, the Japanese Government, the LGS (Leading Graduate School at Chiba University) Program, MEXT, and the Institute for Global Prominent Research, Chiba University, Japan. Dr. S. Hori provided Foxp3scurfy mice, and Dr. M. Yamashita provided pMX-Sox12-IRES-NGFR.
Author contributions
K. Suehiro, A.S., T.I., and S.T. conceived and designed the study; K. Suehiro, K. Suga, and A.S. acquired the data; K. Suzuki, K.H., H.F., A.I., and H.N. analyzed and interpreted the data; V.L. provided resources and interpreted the data; A.S. and H.N. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ken-Ichi Suehiro, Akira Suto
Contributor Information
Akira Suto, Email: suaki@faculty.chiba-u.jp.
Hiroshi Nakajima, Email: nakajimh@faculty.chiba-u.jp.
Supplementary information
The online version of this article (10.1038/s41423-020-0384-0) contains supplementary material.
References
- 1.Lambrecht BN, Hammad H. The immunology of asthma. Nat. Immunol. 2015;16:45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- 2.Martinez FD, Vercelli D. Asthma. Lancet. 2013;382:1360–1372. doi: 10.1016/S0140-6736(13)61536-6. [DOI] [PubMed] [Google Scholar]
- 3.Cohn L, Homer RJ, Marinov A, Rankin J, Bottomly K. Induction of airway mucus production By T helper 2 (Th2) cells: a critical role for interleukin 4 in cell recruitment but not mucus production. J. Exp. Med. 1997;186:1737–1747. doi: 10.1084/jem.186.10.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/S1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 5.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/S0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 6.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maneechotesuwan K, et al. Regulation of Th2 cytokine genes by p38 MAPK-mediated phosphorylation of GATA-3. J. Immunol. 2007;178:2491–2498. doi: 10.4049/jimmunol.178.4.2491. [DOI] [PubMed] [Google Scholar]
- 8.Hosokawa H, et al. Methylation of Gata3 protein at Arg-261 regulates transactivation of the Il5 gene in T helper 2 cells. J. Biol. Chem. 2015;290:13095–13103. doi: 10.1074/jbc.M114.621524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamagata T, et al. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J. 2000;19:4676–4687. doi: 10.1093/emboj/19.17.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita M, et al. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa K, et al. Fbw7 targets GATA3 through cyclin-dependent kinase 2-dependent proteolysis and contributes to regulation of T-cell development. Mol. Cell Biol. 2014;34:2732–2744. doi: 10.1128/MCB.01549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Layman AA, Oliver PM. Ubiquitin ligases and deubiquitinating enzymes in CD4+ T cell effector fate choice and function. J. Immunol. 2016;196:3975–3982. doi: 10.4049/jimmunol.1502660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao SF, Zhong B, Lin D. Regulation of T helper cell differentiation by E3 ubiquitin ligases and deubiquitinating enzymes. Int. Immunopharmacol. 2017;42:150–156. doi: 10.1016/j.intimp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, et al. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. J. Biol. Chem. 2013;288:9373–9382. doi: 10.1074/jbc.M112.374744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wegner M. All purpose Sox: the many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 2010;42:381–390. doi: 10.1016/j.biocel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Penzo-Méndez AI. Critical roles for SoxC transcription factors in development and cancer. Int. J. Biochem. Cell Biol. 2010;42:425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dy P, et al. The three SoxC proteins-Sox4, Sox11 and Sox12-exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefebvre V. The SoxD transcription factors-Sox5, Sox6, and Sox13-are key cell fate modulators. Int J. Biochem. Cell Biol. 2010;42:429–432. doi: 10.1016/j.biocel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka S, et al. Sox5 and c-Maf cooperatively induce Th17 cell differentiation via RORgammat induction as downstream targets of Stat3. J. Exp. Med. 2014;211:1857–1874. doi: 10.1084/jem.20130791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S, et al. Sox12 promotes T reg differentiation in the periphery during colitis. J. Exp. Med. 2018;215:2509–2519. doi: 10.1084/jem.20172082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwahara M, et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat. Immunol. 2012;13:778–786. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattaram P, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, et al. IL-22 induces Reg3γ and inhibits allergic inflammation in house dust mite-induced asthma models. J. Exp. Med. 2017;214:3037–3050. doi: 10.1084/jem.20162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaiss MM, et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity. 2015;43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suto A, et al. Development and characterization of IL-21-producing CD4+ T cells. J. Exp. Med. 2008;205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashiwakuma D, et al. B and T lymphocyte attenuator suppresses IL-21 production from follicular Th cells and subsequent humoral immune responses. J. Immunol. 2010;185:2730–2736. doi: 10.4049/jimmunol.0903839. [DOI] [PubMed] [Google Scholar]
- 27.Hiramatsu Y, et al. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J. Leukoc. Biol. 2010;87:703–712. doi: 10.1189/jlb.0909639. [DOI] [PubMed] [Google Scholar]
- 28.Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc. Natl Acad. Sci. USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.