Abstract
The molecular mechanism of muscle atrophy has been studied a lot, but there is no comprehensive analysis focusing on the denervated muscle atrophy. The gene network that controls the development of denervated muscle atrophy needs further elucidation. We examined differentially expressed genes (DEGs) from five denervated muscle atrophy microarray datasets and predicted microRNAs that target these DEGs. We also included the differentially expressed microRNAs datasets of denervated muscle atrophy in previous studies as background information to identify potential key microRNAs. Finally, we compared denervated muscle atrophy with disuse muscle atrophy caused by other reasons, and obtained the Den-genes which only differentially expressed in denervated muscle atrophy. In this meta-analysis, we obtained 429 up-regulated genes, 525 down-regulated genes and a batch of key microRNAs in denervated muscle atrophy. We found eight important microRNA-mRNA interactions (miR-1/Jun, miR-1/Vegfa, miR-497/Vegfa, miR-23a/Vegfa, miR-206/Vegfa, miR-497/Suclg1, miR-27a/Suclg1, miR-27a/Mapk14). The top five KEGG pathways enriched by Den-genes are Insulin signaling pathway, T cell receptor signaling pathway, MAPK signaling pathway, Toll-like receptor signaling pathway and B cell receptor signaling pathway. Our research has delineated the RNA regulatory network of denervated muscle atrophy, and uncovered the specific genes and terms in denervated muscle atrophy.
Subject terms: Cell biology, Genetics
Introduction
Denervated muscle atrophy refers to muscle atrophy caused by injury of nerves that innervate the muscle. Denervated muscle atrophy is the main cause of motor function loss after peripheral nerve injury. Due to the slow speed of nerve regeneration, it is difficult for regenerated axons to reach the target muscle in a short time. Skeletal muscle will atrophy and fibrosis, and eventually lead to poor recovery of muscle function or even failure to recover1–3. Over time, due to muscle fiber necrosis, connective tissue hyperplasia, muscle cell regeneration failure and a large loss of muscle cell, the muscles at nerve endings lose the ability to accept the regenerated motor axons4. In addition, depletion of skeletal muscle satellite cells, changes in related protein metabolism and enzyme activities, vascular bed remodeling and regulation of myogenic factors are all important reasons for denervated muscle atrophy5–9.
At present, the main methods of treating denervated muscle atrophy include electrical stimulation10,11, passive exercise12, and various drug treatments. Up-regulated breakdown of protein in skeletal muscle is a sign of atrophy, so all potential drugs target the proteolytic system to cure or prevent atrophy13. Nevertheless, the FDA has not recommended other drugs to treat muscle atrophy except for megestrol acetate (MA)14, because these drugs cannot effectively target whole proteolytic system, and the potential targets of denervated muscle atrophy have not been fully explored.
The balance between protein synthesis and degradation maintains muscle mass15,16. In atrophic muscles, muscle-specific E3 ubiquitin ligase and muscle-specific ring finger 1 (MuRF1) in the ubiquitin proteasome system (UPS) are activated, promoting protein degradation in an adenosine triphosphate-dependent manner17,18. The biological axis composed of histone deacetylases (HDACs) 4 and 5, transcription factors Dach2 and myogenin can control the expression of many denervation related genes, including atrogin and MuRF119–22. Myogenin directly mediates the transcriptional activation of the gene encoding E3 ubiquitin ligase, and the lockdown of HDAC4, HDAC5 or myogenin can alleviate denervated muscle atrophy23. In addition, the FoxO family is also an important regulator of atrogin and MuRF1. Overexpression of FoxOs can induce gene expression of atrogin and MuRF1, which will eventually lead to muscle atrophy24,25. Considering the complexity of these processes leading to denervated muscle atrophy, the identification of genes and pathways based on meta-analysis may help to understand the underlying molecular mechanisms.
Denervation, long-term bed rest, unloading of hind limbs, immobilization or microgravity can cause disuse muscle atrophy, of which the common features are decreased cross-sectional area of muscle fibers, decreased muscle strength, increased insulin resistance, and the transition of fiber types from slow to fast26. Decreased protein synthesis and increased protein degradation are the main reasons for the rapid loss of muscle protein due to denervation26. Current research focuses on the general signaling pathways and common key regulators that mediate disuse muscle atrophy26–28, but obviously, denervation is also accompanied by the loss of neurotrophic factors and a series of changes brought about by nerve injury. These denervation-specific pathological changes might be accompanied by specific transcriptome changes which remain to be discovered.
We systematically searched and integrated the gene expression data of denervated muscle atrophy to construct the microRNA-mRNA regulatory network. By comparing denervated muscle atrophy with disused muscle atrophy caused by other reasons, we have discovered the genes and terms that are specifically differentially expressed in denervated muscle atrophy.
Results
Data screening and inclusion criteria
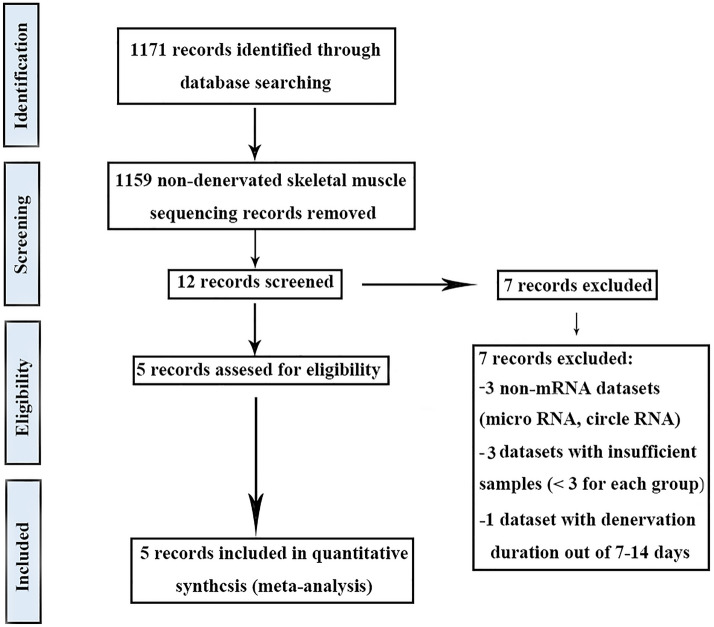
This study obtained five gene expression data of denervated muscle atrophy after screening (Fig. 1)29–31. The details of these data were aggregated into a table (Table 1) and they were all from mouse models in different muscles, including tibialis anterior muscle, gastrocnemius muscle, and gastrocnemius triceps muscle. We only included the data sets that denervated for 7–14 days, because the muscle with denervation greater than 7 days has entered the stage of pathological atrophy31, and the gene expression profiles denervated for 7/14 days have high similarity, which can ensure the homogeneity of the included data sets32.
Figure 1.
Flow chart of literature search in the meta-analysis.
Table 1.
Description of publicly available data sets used in the meta-analysis.
| Number | Gene expression platforms | Muscle | Species | Intervention duration | Ref. |
|---|---|---|---|---|---|
| 1 | Illumina mouse-6 v1.1 | Triceps surae muscle | Mus Musculus | 14 days | GSE4420529 |
| 2 | Affymetrix Mouse Exon 1.0 ST Array | Tibialis anterior muscles | Mus Musculus | 7 days | GSE39195 |
| 3 | Agilent-014868 | Tibialis anterior muscles | Mus Musculus | 14 days | GSE4982630 |
| 4 | Illumina HiSeq X | Gastrocnemius muscle | Mus Musculus | 7 days | 31 |
| 5 | Illumina HiSeq X | Gastrocnemius muscle | Mus Musculus | 14 days | 31 |
Ref reference.
DEGs in denervated muscle atrophy
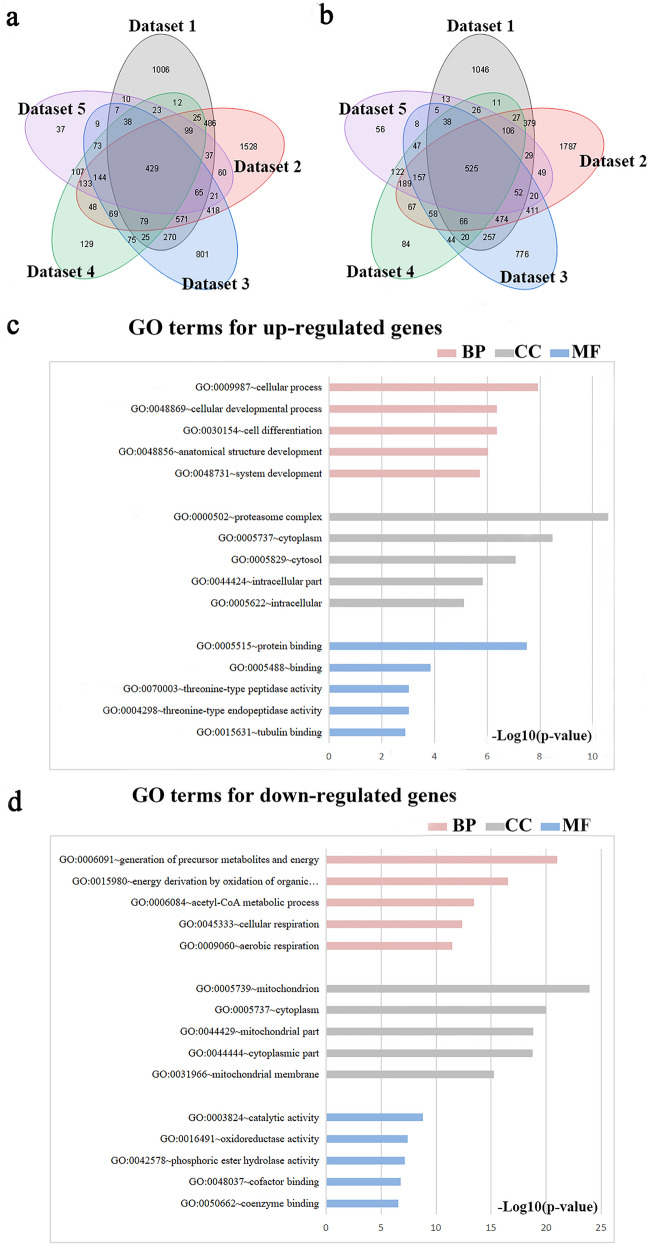
By crossing five data sets, we obtained 429 up-regulated mRNAs and 525 down-regulated mRNAs (Fig. 2a,b) (Table S1), GO analysis revealed the biological role of DEGs in denervated muscle atrophy (Table S2). The analysis showed the pathways enriched by up-regulated genes included cellular process, cellular developmental process, cell differentiation, anatomical structure development, system development, proteasome complex, cytoplasm, cytosol, intracellular part, intracellular, protein binding, binding, threonine-type peptidase activity, threonine-type endopeptidase activity and tubulin binding. The pathways enriched by down-regulated genes include generation of precursor metabolites and energy, energy derivation by oxidation of organic compounds, acetyl-CoA metabolic process, cellular respiration, aerobic respiration, mitochondrion cytoplasm, mitochondrial part, cytoplasmic part, mitochondrial membrane, catalytic activity, oxidoreductase activity, phosphoric ester hydrolase activity, cofactor binding and coenzyme binding (Fig. 2c,d). In order to avoid losing information, we performed GO analysis on the DEG of each data set, and then obtained GO terms that were commonly enriched in each data set (Table 2). We found that the up-regulated GO terms were mostly concentrated in cellular components, and the down-regulated GO terms were still dominated by energy metabolism.
Figure 2.
DEGs and GO terms in denervated muscle atrophy were identified. Venn diagrams for (a) up-regulated mRNAs and (b) down-regulated mRNAs were drawn based on 5 denervated muscle atrophy datasets. GO analysis of DEGs in denervated muscle atrophy were conducted for (c) up- and (d) down-regulated mRNAs respectively (Each figure shows the top10 terms with the lowest p value). BP biological process, CC cellular component, MF molecular function.
Table 2.
GO terms commonly altered in five data sets.
| Up-regulated GO terms | Down-regulated GO terms | |
|---|---|---|
| BP | GO:0071840 ~ cellular component organization or biogenesis | GO:0044710 ~ single-organism metabolic process |
| GO:0016043 ~ cellular component organization | GO:0055114 ~ oxidation–reduction process | |
| GO:0051128 ~ regulation of cellular component organization | GO:0044281 ~ small molecule metabolic process | |
| GO:0044267 ~ cellular protein metabolic process | GO:0015980 ~ energy derivation by oxidation of organic compounds | |
| GO:0048522 ~ positive regulation of cellular process | GO:0006091 ~ generation of precursor metabolites and energy | |
| GO:0044424 ~ intracellular part | GO:0005737 ~ cytoplasm | |
| GO:0005622 ~ intracellular | GO:0043227 ~ membrane-bounded organelle | |
| CC | GO:0043229 ~ intracellular organelle | GO:0044444 ~ cytoplasmic part |
| GO:0043226 ~ organelle | GO:0043226 ~ organelle | |
| GO:0043227 ~ membrane-bounded organelle | GO:0044424 ~ intracellular part | |
| GO:0005515 ~ protein binding | GO:0005515 ~ protein binding | |
| GO:0005488 ~ binding | GO:0003824 ~ catalytic activity | |
| MF | GO:0019899 ~ enzyme binding | GO:0019899 ~ enzyme binding |
| GO:0044822 ~ poly(A) RNA binding | GO:0005488 ~ binding | |
| GO:0044877 ~ macromolecular complex binding | GO:0048037 ~ cofactor binding |
BP biological process, CC cellular component, MF molecular function.
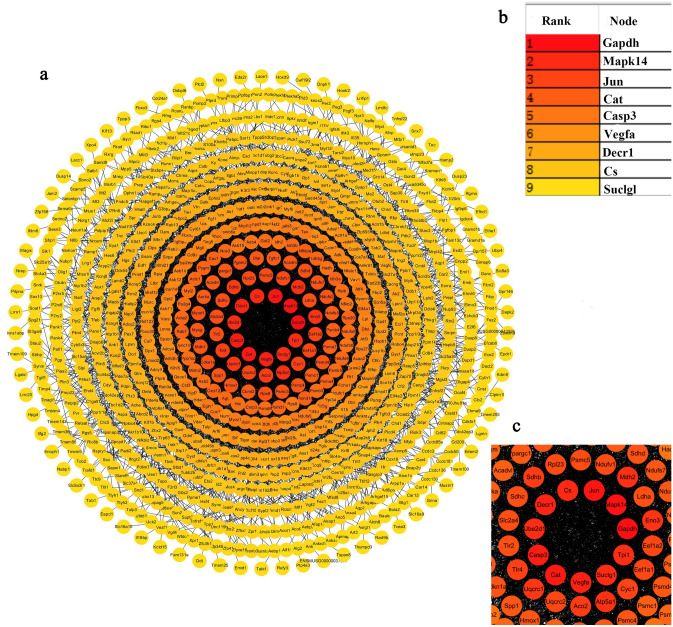
A protein–protein Interaction (PPI) network was mapped based on DEGs to bring in more function-related proteins, and the nodes were sorted according to the degree of interaction between the nodes (Fig. 3). The top10 proteins are Gapdh, Mapk14, Jun, Cat, Casp3, Vegfa, Decr1, CS and Suclg1. The function and location of these genes were annotated respectively, and most of these genes are related to energy metabolism (Table 3).
Figure 3.
PPI network in denervated muscle atrophy. (a) PPI network, the color was used to reflect the level of degree, the higher the degree, the darker the node color. (b) The top10 DEGs with the highest degree. (c) Partial enlarged view of PPI network. STRING v11.0 was used to generate protein interactions, and the resulting network was visualized using Cytoscape v3.7.2.
Table 3.
Description of top10 proteins in PPI.
| Official symbol | Molecular function | Biological process | Location |
|---|---|---|---|
| Gapdh | Oxidoreductase, transferase | Apoptosis, glycolysis, translation regulation | Plasma membrane and cytosol |
| Mapk14 | Kinase, serine/threonine-protein kinase, transferase | Apoptosis, stress response, transcription | Nuclear speckles and cytosol |
| Jun | Activator, DNA-binding | Transcription, transcription regulation | Nucleoplasm |
| Cat | Mitogen, oxidoreductase, peroxidase | Hydrogen peroxide | Vesicles |
| Casp3 | Hydrolase, protease, thiol protease | Apoptosis | Nucleoplasm and mitochondria |
| Vegfa | Developmental protein, growth factor, Heparin-binding, Mitogen | Angiogenesis, differentiation | Secreted to blood |
| Decr1 | Oxidoreductase | Fatty acid metabolism, lipid metabolism | Mitochondria and cytosol |
| Cs | Transferase | Tricarboxylic acid cycle | Mitochondria |
| Suclg1 | Ligase | Tricarboxylic acid cycle | Mitochondria and plasma membrane |
| Tpi1 | Isomerase, lyase | Gluconeogenesis, glycolysis | Nucleoplasm and vesicles |
Identification of potential microRNAs in denervated muscle atrophy
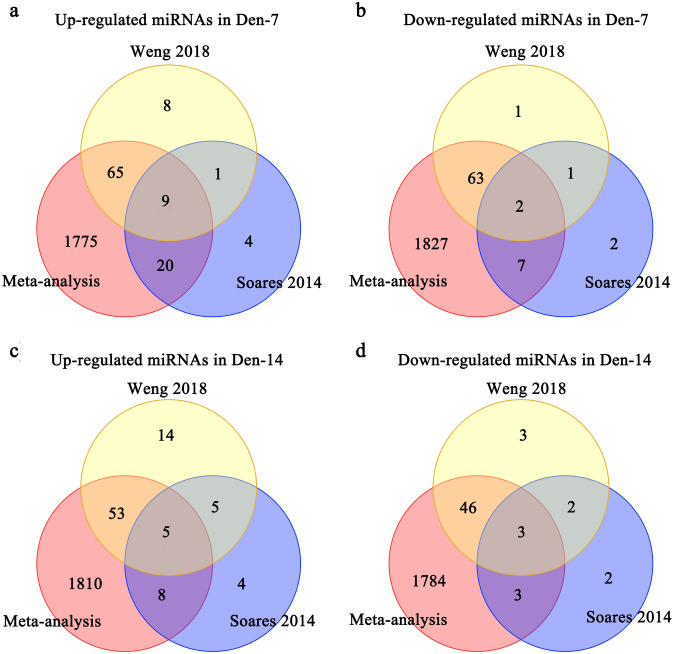
MiRNA-mRNA target prediction identified miRNAs targeting down-regulated mRNAs and those targeting up-regulated mRNAs for den-7 days and den-14 days respectively. By retrieving, we obtained microRNA data sets denervated 7/14 days31,32, we intersected the up-regulated miRNAs in these two data sets with the miRNAs targeting down-regulated mRNAs in this Meta-analysis to obtain 9 miRNAs for den-7 days. Similarly, we obtained 2 down-regulated miRNAs for den-7 days, 5 up-regulated miRNAs and 3 down-regulated miRNAs for den-14 days (Fig. 4, Table 4). We predicted the target genes of these key miRNAs and found eight microRNA-mRNA interactions of which the mRNAs are the top 10 denervation-related mRNAs identified in this research (miR-1/Jun, miR-1/Vegfa, miR-497/Vegfa, miR-23a/Vegfa, miR-206/Vegfa, miR-497/Suclg1, miR-27a/Suclg1, miR-27a/Mapk14). Notably, seven interactions showed an opposite direction of expression between microRNAs and mRNAs (Table 5).
Figure 4.
MicroRNAs identified in denervated muscle atrophy. MicroRNAs predicted as targeting mRNAs retrieved by our meta-analysis were further compared with microRNAs identified as differentially expressed in previous studies of denervated muscle atrophy for 7 days and 14 days respectively. (a) Venn for up-regulated miRNAs for Den-7 days. (b) Venn for down-regulated miRNAs for Den-7 days. (c) Venn for up-regulated miRNAs for Den-14 days. (d) Venn for down-regulated miRNAs for Den-14 days. Den-7 days denervation for 7 days, Den-14 days denervation for 14 days.
Table 4.
List of key miRNAs in denervated muscle atrophy.
| Den 7 | Den 14 |
|---|---|
| mmu-miR-23a↑ | mmu-miR-27b↑ |
| mmu-miR-497↑ | mmu-miR-27a↑ |
| mmu-miR-199b↑ | mmu-miR-24↑ |
| mmu-miR-199a↑ | mmu-miR-21↑ |
| mmu-miR-24↑ | mmu-miR-206↑ |
| mmu-miR-27b↑ | |
| mmu-miR-27a↑ | |
| mmu-miR-206↑ | |
| mmu-miR-21↑ | |
| mmu-miR-30c↓ | mmu-miR-30c↓ |
| mmu-miR-30b↓ | mmu-miR-497↓ |
| mmu-miR-1↓ |
↓, down regulated; ↑, up regulated.
Den 7 denervation for 7 days; Den 14 denervation for 14 days.
Table 5.
MicroRNA/mRNA interactions.
| MicroRNA | mRNA |
|---|---|
| mmu-miR-1↓ | Jun↑ |
| mmu-miR-1↓ | Vegfa↓ |
| mmu-miR-497↑ | Vegfa↓ |
| mmu-miR-23a↑ | Vegfa↓ |
| mmu-miR-206↑ | Vegfa↓ |
| mmu-miR-497 | Suclg1↓ |
| mmu-miR-27a↑ | Suclg1↓ |
| mmu-miR-27a↑ | Mapk14↓ |
↓, down regulated; ↑, up regulated.
Specific genes in denervated muscle atrophy
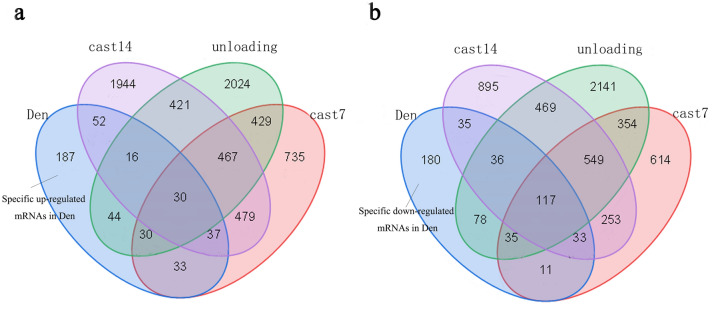
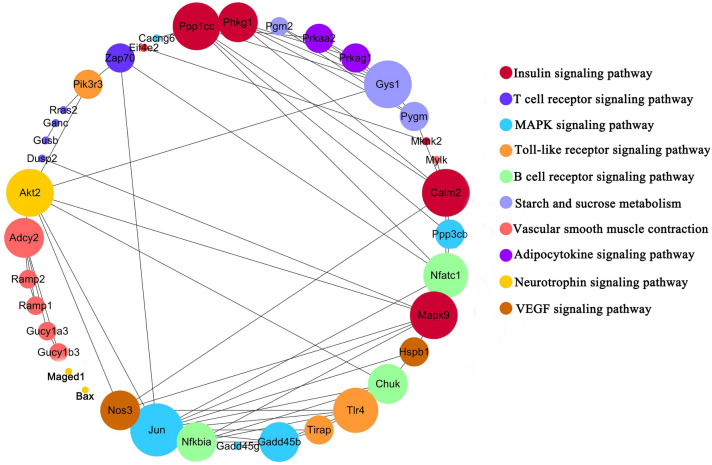
By retrieving, we got three data sets of disuse muscle atrophy caused by unloading or casting (Table 6), genes only differentially expressed in denervated muscle atrophy were regarded as denervation specific genes, called den-genes. We got 187 up-regulated den-genes and 180 down-regulated den-genes (Fig. 5), then performed KEGG enrichment analysis on these genes and got top10 most enriched pathways (Fig. 6). We searched and found the expression of neurotrophic factors (NGF, BDNF, NT-3, NT-4, CNTF, Neuregulin-1 and Neuritin) changed in nerves or muscles after nerve injury and there was evidence that Insulin signaling pathway, MAPK signaling pathway, Neurotrophin signaling pathway, T cell receptor signaling pathway, Toll-like receptor signaling pathway, and B cell receptor signaling pathway were regulated by the above neurotrophic factors (Table 7). Furthermore, most of these terms in skeletal muscle showed different expression after electrical stimulation and Insulin signaling pathway, MAPK signaling pathway, Toll-like receptor signaling pathway, Neurotrophin signaling pathway, and VEGF signaling pathway show regulatory effects on ion channels and connexins (Table 8).
Table 6.
Description of publicly available datasets of disuse muscle atrophy.
| Authors | Platforms | Muscle | Organism | Experiment | Intervention duration | Ref. |
|---|---|---|---|---|---|---|
| Zhang et al. 2018 | Illumina HiSeq 2500 | Soleus muscle | Mus Musculus | Unloading | 10 days | GSE102284 |
| Jelinsky et al. 2011 | Affymetrix Mouse Genome 430 | Gastrocnemius | Mus Musculus | Casting | 7 days | GSE25908 |
| Jelinsky et al. 2011 | Affymetrix Mouse Genome 430 | Gastrocnemius | Mus Musculus | Casting | 14 days | GSE25908 |
Ref reference.
Figure 5.
Specific mRNAs identified in denervated muscle atrophy. DEGs in denervated muscle atrophy retrieved by our meta-analysis (blue circle) were further compared with DEGs in disuse muscle atrophy caused by 7-day hindlimb casting (purple circle), 14-day hindlimb casting (red circle) and hindlimb unloading (green circle). (a) Venn for up-regulated genes. (b) Venn for down-regulated genes. Den denervation, cast 7 7-day hindlimb casting, cast 14 14-day hindlimb casting.
Figure 6.
KEGG analysis of den-genes. Lines highlight KEGG network, with Jun, Mapk9, Calm2, Gys1, Ppp1cc, and Akt2 presenting the highest degree of interactions. Colors highlight KEGG pathways of the circle network components. The larger the circle, the higher the interaction degree identified. Cytoscape v. 3.7.2 was used to generate the resulting network. Permission has been obtained from Kanehisa laboratories for using KEGG pathway database33.
Table 7.
The function of key microRNAs in skeletal muscle.
| MicroRNA | Function |
|---|---|
| mmu-miR-23a | Muscle atrophy (−)34 |
| mmu-miR-497 | Myoblast proliferation (−)35, myogenic differentiation (+)35 |
| mmu-miR-199a | Slow-to-fast muscle fiber type conversion (+)36 |
| mmu-miR-24 | Myoblast proliferation (−)37, myogenic differentiation (+)37 |
| mmu-miR-27b | Skeletal muscle satellite cells proliferation (−) and differentiation (+)38 |
| mmu-miR-27a | Skeletal muscle lipid use (+)39, muscle atrophy (+)40 |
| mmu-miR-206 | Myogenic differentiation (+)41, neuromuscular synapses regeneration (+)42 |
| mmu-miR-21 | Myogenic differentiation (+)43 |
| mmu-miR-497 | Skeletal muscle stem cells proliferation (−)44, myogenic differentiation (+)35 |
| mmu-miR-1 | Myogenesis (+)45 |
+ positive regulation, − negative regulation.
Table 8.
Altered neurotrophic factor expression in injured neurons or muscle.
| Neurotrophic factor | Source | Expression | Downstream pathway |
|---|---|---|---|
| NGF | Neurons46, satellite glial cell47, Schwann cell48, Skeletal muscle49 | Up | Insulin signaling pathway (IE), MAPK signaling pathway (DE)50, Neurotrophin signaling pathway (DE) |
| BDNF | Skeletal muscle51, Schwann cell52 | Up | MAPK signaling pathway (DE)53, Neurotrophin signaling pathway (DE) |
| NT-3, NT-4 | Neurons54, Skeletal muscle55 | Up | MAPK signaling pathway (DE)56, Neurotrophin signaling pathway (DE) |
| CNTF | Schwann cell57 | Down | T cell receptor signaling pathway (IE), Toll-like receptor signaling pathway (IE), B cell receptor signaling pathway (IE)58 |
| Neuregulin-1 | Schwann cell59 | Up | MAPK signaling pathway (DE)60 |
| Neuritin | Neurons61 | Up | Insulin signaling pathway (IE)62, MAPK signaling pathway (IE)62 |
NGF nerve growth factor, BDNF brain-derived neurotrophic factor, NT neurotrophin, DE direct evidence, IE indirect evidence.
Discussion
In this study, we collected and aggregated gene expression data sets of denervated muscle atrophy, and obtained 429 up-regulated mRNAs and 525 down-regulated mRNAs. We drew a PPI network, and sorted the nodes according to the interaction degree. The top10 proteins were Gapdh, Mapk14, Jun, Cat, Casp3, Vegfa, Decr1, Cs and Suclg1. Mapk14, Jun, and Casp3 have been confirmed to be important in denervated muscle atrophy. (1) Mapk14: the three main categories of mitogen-activated protein kinase (MAPK) family proteins are as follows: extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK) and p38 MAPK (Mapk14). The members of the p38 MAPK family (p38α, p38β, p38γ and p38δ MAPK) act as transducers of cellular stress and various non-stress-related stimuli. Therefore, the p38 MAPK pathway has multiple functions and is involved in various cellular processes, including aging, apoptosis, cell cycle arrest, inflammation, and tumorigenesis63. The p38 MAPK pathway can mediate the expression of MuRF1 and Atrogin164,65, and knockdown of p38αMAPK can inhibit muscle atrophy caused by denervation66. (2) Jun: the protein deacetylase HDAC4 is strongly induced in muscles affected by motor neuron diseases such as ALS19. Denervation-induced HDAC4 activates AP1 (Jun) transcription factor by stimulating MAPK signaling to promote denervated muscle atrophy67. (3) Casp3: in the ubiquitin–proteasome pathway and apoptosis process, the activation of Caspase-3 (Casp3) is a common phenomenon. Caspase-3 regulates denervation-induced signal transduction through the mitochondrial-related cell death/apoptosis pathway, resulting in the loss of muscle mass. The lack of caspase-3 has a protective effect on denervated muscle atrophy68. The other proteins Gapdh, Cat, Vegfa, Decr1, Cs, and Suclg1 are mostly important links in redox and energy metabolism pathways69–73, and they may be potential targets in denervated muscle atrophy for treatment.
We included the differentially expressed miRNA expression data of denervated muscle atrophy in previous studies as background information31,32, identified the potential key microRNAs and eight microRNA-mRNA interactions of which the mRNAs are the top 10 denervation-related mRNAs identified in this research (miR-1/Jun, miR-1/Vegfa, miR-497/Vegfa, miR-23a/Vegfa, miR-206/Vegfa, miR-497/Suclg1, miR-27a/Suclg1, miR-27a/Mapk14). Most of the Key miRNAs we obtained showed important regulatory effects on skeletal muscle (Table 7). After 7-day hind limbs unloading in mice, the serum levels of muscle-specific miRNAs such as miR-1, miR-23a, miR-206 increased significantly, and could induce severe muscle atrophy74. MiR-1 has been confirmed to play an important role in skeletal muscle development75,76. MiR-23a can target Atrogin-1 and MuRF1 and inhibit their translation, and the ectopic expression of miR-23a can protect muscles from atrophy in vitro and in vivo, indicating that miR-23a is a key regulator of muscle atrophy77. Overexpression of miR-27a in mice with chronic kidney disease attenuated muscle loss, improved grip strength and reduced the expression of FoxO1, MuRF1and Atrogin140.
Denervation, long-term bed rest, unloading of hind limbs, immobilization or microgravity can cause disuse muscle atrophy. Current research focuses on the general signaling pathways and common key regulators that mediate disuse muscle atrophy26–28, but obviously, denervation is also accompanied by the loss of neurotrophic factors and a series of changes brought about by nerve injury. These denervation-specific pathological changes might be accompanied by specific transcriptome changes which remain to be discovered.
The nervous system controls skeletal muscle through two mechanisms: (1) neuromotor control, which causes muscle contraction through excitation and contraction coupling. (2) Neurotrophic control, which regulates muscles by releasing soluble factors from the nerve endings of motor neurons on the NMJ (Neuromuscular junction)78. NGF, BDNF, NT-3, NT-4, CNTF, Neuregulin-1 and Neuritin are neurotrophic factors differentially expressed in nerves or skeletal muscle after nerve injury79, and they were found to regulate Insulin signaling pathway, MAPK Signaling pathway, Neurotrophin signaling pathway, T cell receptor signaling pathway, Toll-like receptor signaling pathway and B cell receptor signaling pathway which enriched by den-genes (Table 8). The specific changes in skeletal muscle after nerve transection include increased membrane permeability, decreased membrane potential and increased membrane excitability. Most of these are caused by changes in the expression of ion channels and the insertion of connexins 39, 43, and 45 into the muscle membrane, which mediate skeletal muscle atrophy78,80. Insulin signaling pathway, MAPK signaling pathway, Toll-like receptor signaling pathway, Neurotrophin signaling pathway and VEGF signaling pathway showed the potential to regulate ion channels and connexins. Insulin signaling pathway and Toll-like receptor signaling pathway can affect the formation of neuromuscular junctions (Table 9). Therefore, Insulin signaling pathway and Toll-like receptor signaling pathway, as metabolic or inflammation-related terms, have been seldom studied in the neuromuscular systerm, but this study found that they may have important research value in the denervated muscle atrophy.
Table 9.
The regulation of KEGG pathways enriched by den-genes on muscle components.
| Pathway | Altered after ECS81 | NMJ | Ion channel | Connexins |
|---|---|---|---|---|
| Insulin signaling pathway | + | + 82 | Kv4.283 | − |
| T cell receptor signaling pathway | + | − | − | − |
| MAPK signaling pathway | + | + 84 | K ATP85, Kv2.186, Nav1.587 | Connexin 4388 |
| Toll-like receptor signaling pathway | + | + 89 | Kv4.2/4.390, TRPV191, K ATP85 | Connexin 4392 |
| B cell receptor signaling pathway | + | − | − | − |
| Starch and sucrose metabolism | + | − | − | − |
| Vascular smooth muscle contraction | − | − | − | − |
| Adipocytokine signaling pathway | − | − | − | − |
| Neurotrophin signaling pathway | + | + 93 | CLC-494, Kv1.395 | − |
| VEGF signaling pathway | + | − | TRPV496, TRPM7 | − |
NMJ neuromuscular junction, ECS electrical Stimulation, CLC-4 chloride channel 4, TRPV4 transient receptor potential vanilloid 4, TRPM7 transient receptor potential melastatin-subfamily 7.
Current research focuses on the general signaling pathways and common key regulators that mediate disuse muscle atrophy, but obviously, denervation is also accompanied by the loss of neurotrophic factors and a series of changes brought about by nerve injury. These denervation-specific pathological changes must be accompanied by specific transcriptome changes which remain to be discovered. We systematically searched and integrated the gene expression data of denervated muscle atrophy to construct the microRNA-mRNA regulatory network. By comparing denervated muscle atrophy with disused muscle atrophy caused by other reasons, we have discovered the genes and terms that are specifically differentially expressed in denervated muscle atrophy.
Methods
Inclusion criteria for gene expression data
We performed a meta-analysis following the PRISMA Statement97, and found the gene expression data of denervated muscle atrophy by searching NCBI-GEO (http://www.ncbi.nlm.nih.gov/geo/) and PubMed (http://www.ncbi.nlm.nih.gov/pubmed). The keywords used were: “Denervated atrophy”, “Denervation AND muscle”, “nerve AND muscle”, and “Denervation AND muscle AND sequencing”. These meta-analysis searches comprised studies published between 2010 and 2020. Our inclusion criteria were (1) gene expression data in skeletal muscle with peripheral nerve injury, (2) the number of samples in each group should be greater than two, (3) the duration of denervation was 7 to 14 days, (4) all types of skeletal muscle were considered, (5) the inclusion of normal tissues for comparison, and (6) all gene expression analysis platforms were considered. Our exclusion criteria were (1) non-muscle samples, (2) non-mRNA datasets, and (3) review studies.
And we also searched NCBI-GEO (http://www.ncbi.nlm.nih.gov/geo/) and PubMed (http://www.ncbi.nlm.nih.gov/pubmed) to find the gene expression data of disuse muscle atrophy. The keywords used were: “Disuse atrophy”, “Disuse AND muscle”, “unloading AND muscle”, and “cast AND muscle”. These meta-analysis searches comprised studies published between 2010 and 2020. Our inclusion criteria were (1) gene expression data in skeletal muscle with disuse atrophy, (2) the number of samples in each group should be greater than two, (3) the duration of intervention was 7 to 14 days, (4) all types of skeletal muscle were considered, (5) the inclusion of normal tissues for comparison, and (6) all gene expression analysis platforms were considered. Our exclusion criteria were (1) non-muscle samples, (2) non-mRNA datasets, and (3) review studies.
In this study, each individual dataset was processed using edgeR packages to compare control samples and experiment samples to screen DEGs. The Benjamini & Hochberg false discovery rate method was used for p-value adjustment, and the default adj-p-value significance level cut-off is 0.05.
Meta-analysis of global gene expression data in denervated muscle atrophy
Meta-analysis was performed on the five datasets of denervated muscle atrophy using Vote counting generic ways of combining information98, and the DEGs that were common between all datasets were identified. The results were visualized using Venn diagrams by a graphing software (ORIGIN2019; OriginLab, Northampton, MA).
GO and KEGG enrichment analysis of DEGs
The DEGs were used to identify over-represented gene ontology categories and KEGG pathways by using DAVID 6.7 (https://david-d.ncifcrf.gov/). The p value has been corrected using FDR (false discovery rate). GO/KEGG enrichment with p < 0.05 was regarded as statistically significant and those with number of genes < 10 were removed. Bar chart of GO terms was drawn by Microsoft Excel 2016. KEGG network was visualized by Cytoscape software v. 3.7.2 54.
Protein–protein interaction (PPI) network construction for common DEGs
The DEGs were subjected to STRING v.11.0 database99 analysis to construct PPI networks. Minimum required interaction score was 0.4 (medium confidence). PPI networks were visualized by Cytoscape software v. 3.7.2100 and the degree which indicates the number of interactions of each node was calculated by cytoHubba101.
Identification of microRNAs as potential modulators of DEGs
The DEGs identified in our meta-analysis were used for microRNA prediction by DIANA-Tarbase102 to identify potential regulators in denervated muscle atrophy. Visualization of microRNA-gene interaction networks were generated using Cytoscape v3.7.2100 and the degree which indicates the number of interactions of each node was calculated by cytoHubba101.
Supplementary Information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 82072162), Beijing Municipal Natural Science Foundation (Grant 7192215), and National Natural Science Foundation of China (Grant 81971177).
Author contributions
X.Y. designed research, X.G. and B.J. analyzed data and performed research, X.G. wrote the paper, B.J. and Z.Q. recorded the data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xinyi Gu and Bo Jin.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92489-1.
References
- 1.Frostick SP. The physiological and metabolic consequences of muscle denervation. Int. Angiol. 1995;14:278–287. [PubMed] [Google Scholar]
- 2.Kraft GH. Fibrillation potential amplitude and muscle atrophy following peripheral nerve injury. Muscle Nerve. 1990;13:814–821. doi: 10.1002/mus.880130907. [DOI] [PubMed] [Google Scholar]
- 3.Muheremu A, Ao Q. Past, present, and future of nerve conduits in the treatment of peripheral nerve injury. Biomed. Res. Int. 2015;2015:237507. doi: 10.1155/2015/237507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veltri K, Kwiecien JM, Minet W, Fahnestock M, Bain JR. Contribution of the distal nerve sheath to nerve and muscle preservation following denervation and sensory protection. J. Reconstr. Microsurg. 2005;21:57–70. doi: 10.1055/s-2005-862783. [DOI] [PubMed] [Google Scholar]
- 5.Viguie CA, Lu DX, Huang SK, Rengen H, Carlson BM. Quantitative study of the effects of long-term denervation on the extensor digitorum longus muscle of the rat. Anat. Rec. 1997;248:346–354. doi: 10.1002/(SICI)1097-0185(199707)248:3<346::AID-AR7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. Vascular bed-specific regulation of the von Willebrand factor promoter in the heart and skeletal muscle. Blood. 2011;117:342–351. doi: 10.1182/blood-2010-06-287987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakao R, et al. Atypical expression of circadian clock genes in denervated mouse skeletal muscle. Chronobiol. Int. 2015;32:486–496. doi: 10.3109/07420528.2014.1003350. [DOI] [PubMed] [Google Scholar]
- 8.Voytik SL, Przyborski M, Badylak SF, Konieczny SF. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Dev. Dyn. 1993;198:214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues Ade C, Schmalbruch H. Satellite cells and myonuclei in long-term denervated rat muscles. Anat. Rec. 1995;243:430–437. doi: 10.1002/ar.1092430405. [DOI] [PubMed] [Google Scholar]
- 10.Modlin M, et al. Electrical stimulation of denervated muscles: First results of a clinical study. Artif. Organs. 2005;29:203–206. doi: 10.1111/j.1525-1594.2005.29035.x. [DOI] [PubMed] [Google Scholar]
- 11.Helgason T, et al. Monitoring muscle growth and tissue changes induced by electrical stimulation of denervated degenerated muscles with CT and stereolithographic 3D modeling. Artif. Organs. 2005;29:440–443. doi: 10.1111/j.1525-1594.2005.29073.x. [DOI] [PubMed] [Google Scholar]
- 12.Madsen K, Franch J, Clausen T. Effects of intensified endurance training on the concentration of Na, K-ATPase and Ca-ATPase in human skeletal muscle. Acta Physiol. Scand. 1994;150:251–258. doi: 10.1111/j.1748-1716.1994.tb09684.x. [DOI] [PubMed] [Google Scholar]
- 13.Dutt V, Gupta S, Dabur R, Injeti E, Mittal A. Skeletal muscle atrophy: Potential therapeutic agents and their mechanisms of action. Pharmacol. Res. 2015;99:86–100. doi: 10.1016/j.phrs.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Busquets S, et al. Megestrol acetate: Its impact on muscle protein metabolism supports its use in cancer cachexia. Clin. Nutr. 2010;29:733–737. doi: 10.1016/j.clnu.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman CA, Mayhew DL, Hornberger TA. Recent progress toward understanding the molecular mechanisms that regulate skeletal muscle mass. Cell Signal. 2011;23:1896–1906. doi: 10.1016/j.cellsig.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 18.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen TJ, et al. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Goldman D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16977–16982. doi: 10.1073/pnas.0601565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H, et al. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol. Biol. Cell. 2009;20:1120–1131. doi: 10.1091/mbc.E08-07-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macpherson PC, Wang X, Goldman D. Myogenin regulates denervation-dependent muscle atrophy in mouse soleus muscle. J. Cell Biochem. 2011;112:2149–2159. doi: 10.1002/jcb.23136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamei Y, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Chen X, Fan M. Signaling mechanisms involved in disuse muscle atrophy. Med. Hypotheses. 2007;69:310–321. doi: 10.1016/j.mehy.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 27.Fisher AG, et al. Transcriptomic and epigenetic regulation of disuse atrophy and the return to activity in skeletal muscle. FASEB J. 2017;31:5268–5282. doi: 10.1096/fj.201700089RR. [DOI] [PubMed] [Google Scholar]
- 28.Urso ML. Disuse atrophy of human skeletal muscle: Cell signaling and potential interventions. Med. Sci. Sports Exerc. 2009;41:1860–1868. doi: 10.1249/MSS.0b013e3181a6458a. [DOI] [PubMed] [Google Scholar]
- 29.Furlow JD, et al. Altered gene expression patterns in muscle ring finger 1 null mice during denervation- and dexamethasone-induced muscle atrophy. Physiol. Genomics. 2013;45:1168–1185. doi: 10.1152/physiolgenomics.00022.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sartori R, et al. BMP signaling controls muscle mass. Nat. Genet. 2013;45:1309–1318. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]
- 31.Weng J, Zhang P, Yin X, Jiang B. The whole transcriptome involved in denervated muscle atrophy following peripheral nerve injury. Front. Mol. Neurosci. 2018;11:69. doi: 10.3389/fnmol.2018.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soares RJ, et al. Involvement of microRNAs in the regulation of muscle wasting during catabolic conditions. J. Biol. Chem. 2014;289:21909–21925. doi: 10.1074/jbc.M114.561845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang A, et al. miRNA-23a/27a attenuates muscle atrophy and renal fibrosis through muscle-kidney crosstalk. J Cachexia Sarcopenia Muscle. 2018;9:755–770. doi: 10.1002/jcsm.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu H, et al. Regulatory axis of miR-195/497 and HMGA1-Id3 governs muscle cell proliferation and differentiation. Int. J. Biol. Sci. 2017;13:157–166. doi: 10.7150/ijbs.17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Beta-hydroxy-beta-methylbutyrate-induced upregulation of miR-199a-3p contributes to slow-to-fast muscle fiber type conversion in mice and C2C12 cells. J. Agric. Food Chem. 2020;68:530–540. doi: 10.1021/acs.jafc.9b05104. [DOI] [PubMed] [Google Scholar]
- 37.Hu X, et al. Bta-miR-24-3p controls the myogenic differentiation and proliferation of fetal, bovine, skeletal muscle-derived progenitor cells by targeting ACVR1B. Animals (Basel). 2019 doi: 10.3390/ani9110859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ling YH, et al. miR-27b regulates myogenic proliferation and differentiation by targeting Pax3 in goat. Sci. Rep. 2018;8:3909. doi: 10.1038/s41598-018-22262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chemello F, et al. Transcriptomic analysis of single isolated myofibers identifies miR-27a-3p and miR-142–3p as regulators of metabolism in skeletal muscle. Cell Rep. 2019;26:3784–3797. doi: 10.1016/j.celrep.2019.02.105. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, et al. MicroRNA-23a and MicroRNA-27a mimic exercise by ameliorating CKD-induced muscle atrophy. J. Am. Soc. Nephrol. 2017;28:2631–2640. doi: 10.1681/ASN.2016111213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Przanowska RK, et al. miR-206 family is important for mitochondrial and muscle function, but not essential for myogenesis in vitro. FASEB J. 2020;34:7687–7702. doi: 10.1096/fj.201902855RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgoulis D, et al. sPIF promotes myoblast differentiation and utrophin expression while inhibiting fibrosis in Duchenne muscular dystrophy via the H19/miR-675/let-7 and miR-21 pathways. Cell Death Dis. 2019;10:82. doi: 10.1038/s41419-019-1307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat. Commun. 2014;5:4597. doi: 10.1038/ncomms5597. [DOI] [PubMed] [Google Scholar]
- 45.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saika T, et al. Effects of nerve crush and transection on mRNA levels for nerve growth factor receptor in the rat facial motoneurons. Brain Res. Mol. Brain Res. 1991;9:157–160. doi: 10.1016/0169-328x(91)90142-k. [DOI] [PubMed] [Google Scholar]
- 47.Zhou XF, et al. Satellite-cell-derived nerve growth factor and neurotrophin-3 are involved in noradrenergic sprouting in the dorsal root ganglia following peripheral nerve injury in the rat. Eur. J. Neurosci. 1999;11:1711–1722. doi: 10.1046/j.1460-9568.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- 48.Abe S, et al. Induction of nerve growth factor mRNA in a rat dorsal root ganglion after application of a tourniquet. Acta Neuropathol. 2004;108:183–188. doi: 10.1007/s00401-004-0870-y. [DOI] [PubMed] [Google Scholar]
- 49.Amano T, Yamakuni T, Okabe N, Sakimura K, Takahashi Y. Production of nerve growth factor in rat skeletal muscle. Neurosci. Lett. 1991;132:5–7. doi: 10.1016/0304-3940(91)90418-s. [DOI] [PubMed] [Google Scholar]
- 50.Wu C, et al. A functional dynein-microtubule network is required for NGF signaling through the Rap1/MAPK pathway. Traffic. 2007;8:1503–1520. doi: 10.1111/j.1600-0854.2007.00636.x. [DOI] [PubMed] [Google Scholar]
- 51.Griesbeck O, Parsadanian AS, Sendtner M, Thoenen H. Expression of neurotrophins in skeletal muscle: Quantitative comparison and significance for motoneuron survival and maintenance of function. J. Neurosci. Res. 1995;42:21–33. doi: 10.1002/jnr.490420104. [DOI] [PubMed] [Google Scholar]
- 52.Meyer M, Matsuoka I, Wetmore C, Olson L, Thoenen H. Enhanced synthesis of brain-derived neurotrophic factor in the lesioned peripheral nerve: Different mechanisms are responsible for the regulation of BDNF and NGF mRNA. J. Cell Biol. 1992;119:45–54. doi: 10.1083/jcb.119.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen T, et al. Brain-derived neurotrophic factor increases synaptic protein levels via the MAPK/Erk signaling pathway and Nrf2/Trx axis following the transplantation of neural stem cells in a rat model of traumatic brain injury. Neurochem. Res. 2017;42:3073–3083. doi: 10.1007/s11064-017-2340-7. [DOI] [PubMed] [Google Scholar]
- 54.Omura T, et al. Different expressions of BDNF, NT3, and NT4 in muscle and nerve after various types of peripheral nerve injuries. J. Peripher Nerv. Syst. 2005;10:293–300. doi: 10.1111/j.1085-9489.2005.10307.x. [DOI] [PubMed] [Google Scholar]
- 55.Funakoshi H, et al. Differential expression of mRNAs for neurotrophins and their receptors after axotomy of the sciatic nerve. J. Cell Biol. 1993;123:455–465. doi: 10.1083/jcb.123.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye J, et al. Effect of NT-3 on repair of spinal cord injury through the MAPK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2165–2172. doi: 10.26355/eurrev_202003_20481. [DOI] [PubMed] [Google Scholar]
- 57.Smith GM, Rabinovsky ED, McManaman JL, Shine HD. Temporal and spatial expression of ciliary neurotrophic factor after peripheral nerve injury. Exp. Neurol. 1993;121:239–247. doi: 10.1006/exnr.1993.1091. [DOI] [PubMed] [Google Scholar]
- 58.Martin D, et al. Cachectic effect of ciliary neurotrophic factor on innervated skeletal muscle. Am. J. Physiol. 1996;271:R1422–R1428. doi: 10.1152/ajpregu.1996.271.5.R1422. [DOI] [PubMed] [Google Scholar]
- 59.Kopp DM, Trachtenberg JT, Thompson WJ. Glial growth factor rescues Schwann cells of mechanoreceptors from denervation-induced apoptosis. J. Neurosci. 1997;17:6697–6706. doi: 10.1523/JNEUROSCI.17-17-06697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim HG, et al. Neuregulin 1 up-regulates the expression of nicotinic acetylcholine receptors through the ErbB2/ErbB3-PI3K-MAPK signaling cascade in adult autonomic ganglion neurons. J. Neurochem. 2013;124:502–513. doi: 10.1111/jnc.12109. [DOI] [PubMed] [Google Scholar]
- 61.Azuchi Y, et al. Role of neuritin in retinal ganglion cell death in adult mice following optic nerve injury. Sci. Rep. 2018;8:10132. doi: 10.1038/s41598-018-28425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao JJ, Zhao QR, Lu JM, Mei YA. Functions and the related signaling pathways of the neurotrophic factor neuritin. Acta Pharmacol. Sin. 2018;39:1414–1420. doi: 10.1038/aps.2017.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Derbre F, et al. Inhibition of xanthine oxidase by allopurinol prevents skeletal muscle atrophy: Role of p38 MAPKinase and E3 ubiquitin ligases. PLoS ONE. 2012;7:e46668. doi: 10.1371/journal.pone.0046668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Childs TE, Spangenburg EE, Vyas DR, Booth FW. Temporal alterations in protein signaling cascades during recovery from muscle atrophy. Am. J. Physiol. Cell Physiol. 2003;285:C391–C398. doi: 10.1152/ajpcell.00478.2002. [DOI] [PubMed] [Google Scholar]
- 66.Yuasa K, et al. Targeted ablation of p38alpha MAPK suppresses denervation-induced muscle atrophy. Sci. Rep. 2018;8:9037. doi: 10.1038/s41598-018-26632-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi MC, et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol. Cell. 2012;47:122–132. doi: 10.1016/j.molcel.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plant PJ, Bain JR, Correa JE, Woo M, Batt J. Absence of caspase-3 protects against denervation-induced skeletal muscle atrophy. J. Appl. Physiol. 2009;107:224–234. doi: 10.1152/japplphysiol.90932.2008. [DOI] [PubMed] [Google Scholar]
- 69.Nakajima H, et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) aggregation causes mitochondrial dysfunction during oxidative stress-induced cell death. J. Biol. Chem. 2017;292:4727–4742. doi: 10.1074/jbc.M116.759084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nomura K, Ono K, Ono S. CAS-1, a C. elegans cyclase-associated protein, is required for sarcomeric actin assembly in striated muscle. J. Cell Sci. 2012;125:4077–4089. doi: 10.1242/jcs.104950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blomme A, et al. 2,4-dienoyl-CoA reductase regulates lipid homeostasis in treatment-resistant prostate cancer. Nat. Commun. 2020;11:2508. doi: 10.1038/s41467-020-16126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiegand G, Remington SJ. Citrate synthase: Structure, control, and mechanism. Annu. Rev. Biophys. Biophys. Chem. 1986;15:97–117. doi: 10.1146/annurev.bb.15.060186.000525. [DOI] [PubMed] [Google Scholar]
- 73.El-Hattab AW, Scaglia F. Mitochondrial DNA depletion syndromes: Review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics. 2013;10:186–198. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang F, et al. Serum miRNAs miR-23a, 206, and 499 as potential biomarkers for skeletal muscle atrophy. Biomed. Res. Int. 2017;2017:8361237. doi: 10.1155/2017/8361237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wust S, et al. Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab. 2018;27:1026–1039. doi: 10.1016/j.cmet.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 76.Kukreti H, et al. Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J. Biol. Chem. 2013;288:6663–6678. doi: 10.1074/jbc.M112.390369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sacheck JM, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 78.Cisterna BA, Cardozo C, Saez JC. Neuronal involvement in muscular atrophy. Front. Cell Neurosci. 2014;8:405. doi: 10.3389/fncel.2014.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kingham PJ, Terenghi G. Bioengineered nerve regeneration and muscle reinnervation. J. Anat. 2006;209:511–526. doi: 10.1111/j.1469-7580.2006.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cea LA, et al. De novo expression of connexin hemichannels in denervated fast skeletal muscles leads to atrophy. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16229–16234. doi: 10.1073/pnas.1312331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.La G, et al. Proteomics and transcriptomics analysis reveals clues into the mechanism of the beneficial effect of electrical stimulation on rat denervated gastrocnemius muscle. Cell Physiol. Biochem. 2019;52:769–786. doi: 10.33594/000000054. [DOI] [PubMed] [Google Scholar]
- 82.Hung WL, et al. Attenuation of insulin signalling contributes to FSN-1-mediated regulation of synapse development. EMBO J. 2013;32:1745–1760. doi: 10.1038/emboj.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yao JJ, et al. Neuritin activates insulin receptor pathway to up-regulate Kv42-mediated transient outward K+ current in rat cerebellar granule neurons. J. Biol. Chem. 2012;287:41534–41545. doi: 10.1074/jbc.M112.390260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Massaro CM, Pielage J, Davis GW. Molecular mechanisms that enhance synapse stability despite persistent disruption of the spectrin/ankyrin/microtubule cytoskeleton. J. Cell Biol. 2009;187:101–117. doi: 10.1083/jcb.200903166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeong Nam Y, Kim A, Sung Lee M, Suep Sohn D, Soo Lee C. KATP channel block inhibits the Toll-like receptor 2-mediated stimulation of NF-kappaB by suppressing the activation of Akt, mTOR, JNK and p38-MAPK. Eur. J. Pharmacol. 2017;815:190–201. doi: 10.1016/j.ejphar.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 86.Zhu J, et al. Involvement of the delayed rectifier outward potassium channel Kv2.1 in methamphetamine-induced neuronal apoptosis via the p38 mitogen-activated protein kinase signaling pathway. J. Appl. Toxicol. 2018;38:696–704. doi: 10.1002/jat.3576. [DOI] [PubMed] [Google Scholar]
- 87.House CD, et al. Voltage-gated Na+ channel activity increases colon cancer transcriptional activity and invasion via persistent MAPK signaling. Sci. Rep. 2015;5:11541. doi: 10.1038/srep11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Q, et al. The involvement of the ERK-MAPK pathway in TGF-beta1-mediated connexin43-gap junction formation in chondrocytes. Connect Tissue Res. 2019;60:477–486. doi: 10.1080/03008207.2019.1593394. [DOI] [PubMed] [Google Scholar]
- 89.Ballard SL, Miller DL, Ganetzky B. Retrograde neurotrophin signaling through Tollo regulates synaptic growth in Drosophila. J. Cell Biol. 2014;204:1157–1172. doi: 10.1083/jcb.201308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao X, et al. Toll-like receptor 3 controls QT interval on the electrocardiogram by targeting the degradation of Kv4.2/4.3 channels in the endoplasmic reticulum. FASEB J. 2019;33:6197–6208. doi: 10.1096/fj.201801464R. [DOI] [PubMed] [Google Scholar]
- 91.Min H, et al. Association of TRPV1 and TLR4 through the TIR domain potentiates TRPV1 activity by blocking activation-induced desensitization. Mol. Pain. 2018;14:1744806918812636. doi: 10.1177/1744806918812636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y, et al. Inhibition of connexin 43 hemichannels alleviates cerebral ischemia/reperfusion injury via the TLR4 signaling pathway. Front. Cell Neurosci. 2018;12:372. doi: 10.3389/fncel.2018.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steele AD, Yi CH. Neuromuscular denervation: Bax up against the wall in amyotrophic lateral sclerosis. J. Neurosci. 2006;26:12849–12851. doi: 10.1523/jneurosci.4086-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hur J, Jeong HJ, Park J, Jeon S. Chloride channel 4 is required for nerve growth factor-induced TrkA signaling and neurite outgrowth in PC12 cells and cortical neurons. Neuroscience. 2013;253:389–397. doi: 10.1016/j.neuroscience.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 95.Colley BS, Cavallin MA, Biju K, Marks DR, Fadool DA. Brain-derived neurotrophic factor modulation of Kv1.3 channel is disregulated by adaptor proteins Grb10 and nShc. BMC Neurosci. 2009;10:8. doi: 10.1186/1471-2202-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kanugula AK, et al. Novel noncanonical regulation of soluble VEGF/VEGFR2 signaling by mechanosensitive ion channel TRPV4. FASEB J. 2019;33:195–203. doi: 10.1096/fj.201800509R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moher D, Liberati A, Tetzlaff J, Altman DG, Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5:e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Mering C, et al. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 101.Chin CH, et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paraskevopoulou MD, Vlachos IS, Hatzigeorgiou AG. DIANA-TarBase and DIANA suite tools: Studying experimentally supported microRNA targets. Curr. Protoc. Bioinform. 2016;55:11–18. doi: 10.1002/cpbi.12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.