Abstract
Genome-wide selection (GWS) has been becoming an essential tool in the genetic breeding of long-life species, as it increases the gain per time unit. This study had a hypothesis that GWS is a tool that can decrease the breeding cycle in Jatropha. Our objective was to compare GWS with phenotypic selection in terms of accuracy and efficiency over three harvests. Models were developed throughout the harvests to evaluate their applicability in predicting genetic values in later harvests. For this purpose, 386 individuals of the breeding population obtained from crossings between 42 parents were evaluated. The population was evaluated in random block design, with six replicates over three harvests. The genetic effects of markers were predicted in the population using 811 SNP's markers with call rate = 95% and minor allele frequency (MAF) > 4%. GWS enables gains of 108 to 346% over the phenotypic selection, with a 50% reduction in the selection cycle. This technique has potential for the Jatropha breeding since it allows the accurate obtaining of GEBV and higher efficiency compared to the phenotypic selection by reducing the time necessary to complete the selection cycle. In order to apply GWS in the first harvests, a large number of individuals in the breeding population are needed. In the case of few individuals in the population, it is recommended to perform a larger number of harvests.
Subject terms: Agricultural genetics, Plant breeding, Plant genetics
Introduction
In recent decades, there has been exponential growth in demand for energy sources, which is linked to population expansion. It is estimated that the world population in 2050 will be 9.7 billion people, compared to approximately 7.3 billion people in 2015, i.e., the population will increase by about 32%1. This scenario has imposed challenges on society, as pertinent questions arise: How to increase the production of food to meet society's demand and still meet the environmental sustainability goals? In this context, science is the main ally, since it is possible to develop innovation and technologies to improve yields and restore natural balances throughout the food system simultaneously2.
Energy from fossil fuels is still crucial in the energy sector, but it is also known to be the primary source of greenhouse gas emissions and a finite source3. Renewable energy has grown fast in recent years, driven by policy support, advances in technology, and sharp reductions in production costs, and is at the heart of the transition to a less carbon-intensive and more sustainable energy system. In this context, using renewable energy is imperative in the world energy matrix. It is worth mentioning the use of biofuels, which has shown economic viability and some advantages compared to fossil fuels since they are non-toxic, biodegradable, and do not pollute the environment, have flash point and can be added to diesel due to similar properties4.
In Brazil, there are several potential sources of oilseeds for biodiesel production. Given the vast diversity of the national ecosystem, soybean (Glycine max L. Merril) presents highlight and supremacy as feedstock for biodiesel production, representing 69.8% of the Brazilian energy matrix5. Thus, there is a limitation in the number of raw materials composing the energy matrix. However, Brazil has the potential to expand the production of biofuels and other vegetable oil derivatives to meet both the domestic and global markets. One of the effective ways to increase the limited quantity of traditional raw materials and their high prices is to invest in the improvement of biodiesel production from inedible vegetable oil6, such as Jatropha (Jatropha curcas L.).
Jatropha has been the target of several studies as a potential source for biodiesel production7,8, since it has high oil production (30–48%)9,10, rusticity11,12 and simple spread10. Due to these characteristics, Jatropha qualifies as a potential candidate for the sustainable production of biofuels13. Despite the enormous potential, this species is in a domestication stage in Brazil, and because it is perennial, it needs several years to complete a breeding cycle. One of the objectives of the perennial plant breeders is to shorten the selection cycle. For this purpose, an alternative that has been used with considerable success is Genome-Wide Selection (GWS), which is one of the promising tools to increase selection efficiency, reduce costs in the launching of cultivars, reduce the breeding cycle through early selection, and increase the genetic gain between breeding generations14,15. For crops like Jatropha, it is estimated that GWS could shorten the breeding cycle14, which would have a high impact on the release of new cultivars for planting.
However, few studies have reported the use of GWS in Jatropha worldwide. A pilot evaluation of predictive model accuracy using only one harvest and demonstrated the potential of GWS in Jatropha breeding16. However, these authors recommended that the study should be validated over the years and by progeny evaluation. This research had a hypothesis that GWS is a tool that can decrease the breeding cycle in Jatropha. Thus, the objectives of this study were (1) to use the Jatropha training population evaluated in multiple harvests, and (2) to develop models for predicting breeding values between harvests. This paper presents unprecedented results in validating GWS in Jatropha for biofuel production.
Results
Initially, we estimated heritability in the restricted sense (h2a) for grain yield in the three harvests, to assess the extent to which phenotypic variation is genetically controlled and genomic selectable. Heritabilities ranged from 0.18 to 0.20 for the first and third harvests, respectively, and 0.25, when the average grain yield was considered (Table 1).
Table 1.
Narrow-sense heritability (h2a), phenotypic selection accuracy (ryy), predictive ability (ryg), and average grain yield (µ).
| Harvests | h2a | ryy | µ (g plant−1) | ryg |
|---|---|---|---|---|
| I | 0.18 | 0.17 | 173.76 | 0.22 |
| II | 0.19 | 0.20 | 760.85 | 0.24 |
| III | 0.20 | 0.37 | 1075.52 | 0.54 |
| Average grain yield | 0.25 | 0.37 | 667.09 | 0.57 |
Low selective accuracy was observed based on phenotypic information (ryy) on all harvests. Even using the mean of harvests, high magnitude accuracy values were not obtained17. The values varied from 0.17 to 0.37 for the first and third harvests, respectively.
On the other hand, the GWS (Table 2) accuracies were from low to high magnitude, ranging from 0.20 to 0.83 for the first and third harvests, respectively. As for the GWS analysis efficiency regarding phenotypic selection, when we apply genomic selection in the second year, gains of 66.74% become possible. Likewise, when genomic selection is performed in the third year, gains of 346% in relation to phenotypic selection are achieved.
Table 2.
Accuracy and efficiency of genomic selection compared to selection only with phenotypic data in Jatropha.
| Harvests | GWS accuracy | Efficiency | IRPS (%) |
|---|---|---|---|
| 1 | 0.20 | – | – |
| 2 | 0.31 | 1.667 | 66.74 |
| 3 | 0.83 | 4.464 | 346.45 |
| Average grain yield | 0.80 | 4.303 | 330.31 |
IRPS: Increase relative to phenotypic selection.
Regarding the estimate of the number of individuals required to obtain the desired selective accuracy (Table 3), it is observed the need to evaluate a larger number of individuals when larger estimates of selective accuracy are sought. In an antagonistic way, a smaller number of individuals will be necessary when the number of harvests to obtain accuracy is increased. Regarding the estimate of 0.8, which is considered as high magnitude17, it will be necessary to evaluate 1239 and 256 in the first and third harvests, respectively.
Table 3.
Number of individuals required to obtain the desired accuracy of GWS in the Jatropha population for grain yield.
| Desired accuracy | Number of individuals required | |||
|---|---|---|---|---|
| Harvest 1 | Harvest 2 | Harvest 3 | Average grain yield | |
| 0.40 | 207 | 175 | 43 | 51 |
| 0.50 | 310 | 263 | 64 | 77 |
| 0.60 | 465 | 394 | 96 | 115 |
| 0.70 | 723 | 613 | 149 | 179 |
| 0.80 | 1239 | 1051 | 256 | 307 |
| 0.90 | 2788 | 2364 | 576 | 692 |
In this case, it can be observed that to obtain high accuracy in GWS requires a large number of individuals. However, this fact is only justified if the trait under study has low heritability, however, if the trait has high heritability, the number of individuals can be reduced. This is evident when we look at Eq. (5), in which a direct relationship between the desired accuracy of GWS and heritability can be seen.
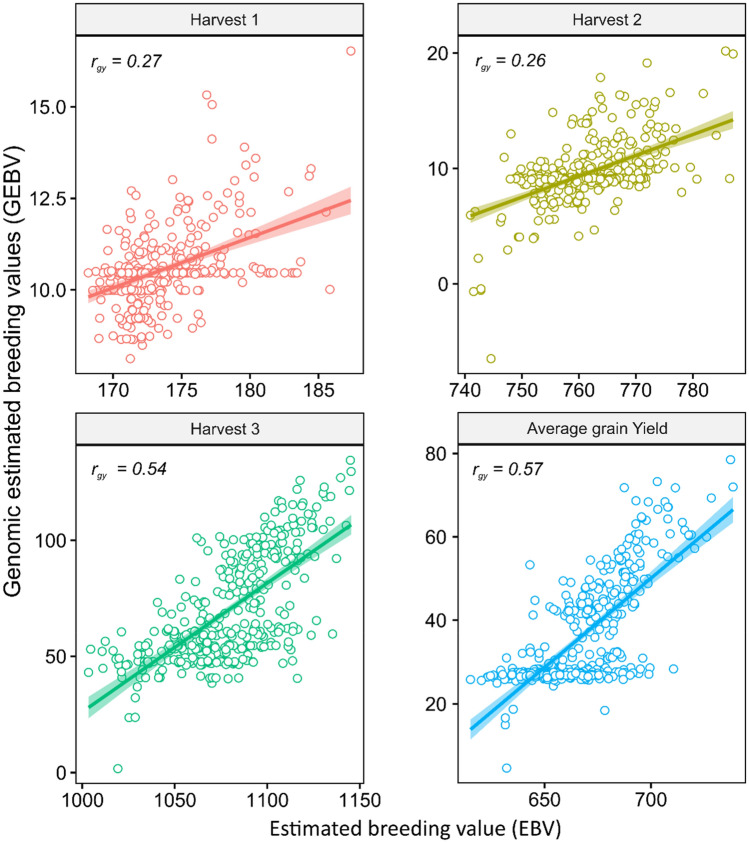
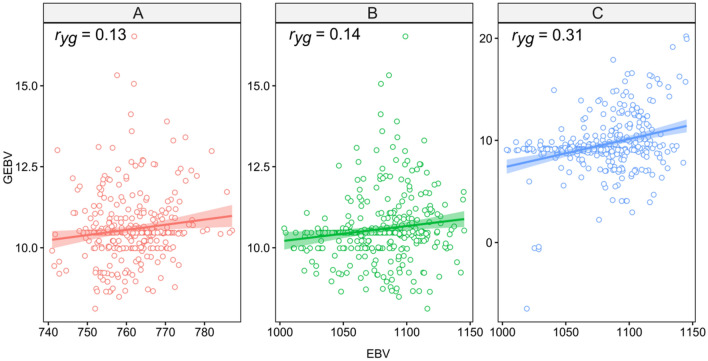
Predictive ability estimates of genomic selection (ryg) ranged from 0.27 to 0.57 for the first harvest and for average grain yield, respectively (Fig. 1). Jatropha demands at least 4 to 7 harvests for phenotypic selection with adequate accuracy. To verify if this time is also necessary for developing prediction models, we evaluated the accuracy of the models generated for fruit yield based on data collected in the first and second harvests, but validated in the same population, in the second and third harvests (Fig. 2).
Figure 1.
Scatterplots of genomic estimated breeding values (GEBVs) by RR-BLUP and unregressed phenotypes observed for grain yield. rgy: predictive ability of genomic selection. The package used of R to create this Figure was ggplot2 (v0.3.3, https://cran.r-project.org/web/packages/ggplot2/index.html).
Figure 2.
Accuracy of estimated prediction models in crop 1 validated in crop 2 (A), crop 1 validated in crop 3 (B), and crop 2 validated in crop 3 (C). The package used of R to create this Figure was ggplot2 (v0.3.3, https://cran.r-project.org/web/packages/ggplot2/index.html).
Regarding the models developed for grain yield based on data collected in the first harvest and validated in the same population at 2 and 3 years old (second and third harvest, respectively), it can be observed that in the validation of the second harvest data the accuracy reduced more than 50% (Fig. 2A) in relation to the accuracy of the second harvest (Table 1). The same result can be observed when models estimated in the first harvest were validated based on data from the third harvest (Fig. 2B). However, moderate precision was verified when models estimated in the second harvest were used and validated based on data from the third harvest (Fig. 2C).
Discussion
Based on narrow-sense heritability estimates (Table 1), the selection of superior Jatropha genotypes for grain yield based on phenotypic values will not provide selection gain for the next generation, since approximately 80% of the phenotypic variation is from a non-genetic origin. Grain yield, the main trait to be improved for biodiesel production, is very influenced by the environment. Other studies with Brazilian Jatropha genotypes have also found low heritability estimates over the harvests18–21. Thus, more accurate methodologies must be used to predict genetic effects. Therefore, GWS becomes more appropriate to select superior Jatropha genotypes than conventional methods based only on phenotypic data because this technique can efficiently capture small genetic differences between families.
However, even if the GWS allows adequate prediction of genetic effects, the heritability has importance in the model, because the lower the trait heritability the lower the phenotypic data accuracy and, therefore, lower the heritability of marker effects. Consequently, the lower will be the ability to reliably predict the phenotypes of individuals not sampled to compute the model. Several authors have demonstrated this theory through simulations22–24, in which increased trait heritability has resulted in increased GWS accuracy.
However, even with the impact of the low grain yield heritability throughout the harvests on selection, we can note that the predictive models performed well. This result corroborates those obtained by22, where accuracy increased only by 10–20% as heritability increased from 0.2 to 0.6, regardless of population size. Thus, unlike marker-assisted selection, GWS is efficient in selecting superior individuals even for low heritability traits as reported by25.
The observed values of accuracy based exclusively on phenotypic data showed low magnitude. This reveals that selection based only on phenotypic data has low accuracy in the first and second harvests, and moderate accuracy in the third harvests26. Similar findings were obtained by27, who found low accuracy for grain yield in Jatropha.
As for the GWS accuracy, low values were found in the first and second harvests, but the accuracy value was high in the third harvest. We highlight that these estimates were also obtained using genomic heritability as a proportion of phenotypic heritability, considering the efficiency of markers in capturing QTLs, i.e., considering the degree of imperfection of the linkage disequilibrium26.
The genomic selection efficiency depends on the correlation between the predicted and the actual genotypic value, i.e., the predictive capacity26. There was an increase in this correlation throughout the harvests, suggesting a higher additive genetic variation for this trait in this reproductive population. These findings show that rgy values were higher for the harvests with the highest h2a estimates. This indicates that phenotypes for low heritability traits contain higher environmental noise and hence will be less predictable by genomic models.
One of the principles of genomic selection is using a large number of markers able to cover the entire genome of the species to maximize the number of QTLs in linkage disequilibrium (LD) with at least one marker, allowing the maximization of the genetic variance explained by the QTLs28. However, the use of 811 SNPs allowed a satisfactory predictive ability. This result is consistent with those found by29 when assessing a Jatropha breeding population by GWS. These authors concluded that training models with 800 and 1000 markers are sufficient to capture the maximum genetic variation and hence the maximum predictive ability for grain yield in Jatropha families. In addition, not just the number of SNPs, but high LD with QTLs is the important factor in GWS success.
In genomic selection, the genotyping step is the costliest and often makes it impossible to apply genomic selection. However, given the results found here, as previously reported by20, reveal the possibility of using a small set of SNP markers in Jatropha breeding will enable significant gains in the short-term and at relatively low cost. The predictive ability of genomic selection (ryg) shows the potential of molecular information to consistently predict a phenotype30. The magnitudes obtained here were similar to those reported in forest species24 and coffee crop31.
Likewise32, by using canola as a model, reported that low-density marker sets comprising only a few hundred markers allow high accuracy of genomic prediction in breeding populations with strong linkage disequilibrium. The authors also mentioned that the breeder could obtain a significant advantage in selection by using reduced marker density, even when the prediction accuracy is lower than a high-density chip. This strategy provides substantial cost savings and thus enables phenotyping resources to be focused on pre-selected genotypes that, even with lower selection accuracy, will still allow significant gains for the breeding program.
Throughout the evaluation of the Jatropha harvests, many alleles will be acting with more or less expression on the expression of phenotypes. However, we do not know if the major effect alleles for grain yield in the first harvest will keep pronounced effect over the next harvests, i.e., over advanced ages. In this sense, considering that genomic selection requires phenotypic data for its calibration, a high correlation between the marker effects representing the alleles at different ages of the plant will allow the phenotype to be predicted earlier for prediction at future harvests. This would enable the early selection of superior genotypes by GWS.
Thus, grain yield was evaluated over three harvests, allowing the development of prediction models for each harvest. We tested how the models developed on the first harvest act in the prediction of phenotypes on the second and third harvests. However, the models developed for the first harvest showed limited accuracy in phenotype prediction in the second and third harvests. Given this result, we infer that there is a low genetic correlation between the first harvest and the second and third harvests. This low correlation is attributed to the lower yield stability of the Jatropha in the first harvest, since a significant part of the individuals in the population is still growing, making it impossible to express their productive potential.
When evaluating the agronomic performance of jatropha33, observed an increase in grain yield from 322 to 1972% from the first to the third harvest. On the other hand, the accuracy of the models developed based on the grain yield data of the second harvest was higher in the measurements of the third harvest, and there may be a higher genetic correlation between these harvests. Genotype by harvest interaction can affect the transferability of models between harvests when the first harvest is used. Therefore, the results obtained are essential to facilitate the ongoing use of genetic selection in Jatropha breeding programs.
Regarding the efficiency of wide-genome selection, considering the 50% reduction in the selection cycle, we observed an increased gain with GWS compared to the phenotypic selection of 66.74 and 346.45% for the second and third harvests, respectively. Our findings reveal that the use of GWS in Jatropha in an early manner is a reality to be explored. This strategy makes the applicability of GWS even more significant since late phenotyping would reduce the gain by time unit, i.e., the selection gain already achieved simply by conventional breeding.
By selecting superior individuals early through GWS analysis, the breeder will focus on potential genotypes and eliminating undesirable ones. For this reason, the costs of maintaining breeding populations in the field can be reduced. Furthermore, the early genomic selection makes it possible to carry out breeding populations with higher agronomic performance, which will maximize the genetic gains.
This possibility, as reported by15 and34, is due to the fact that genomic prediction and selection of superior genotypes can be performed at the seedling stage, and thus GWS becomes more time-efficient. Similar results were reported by several studies in pinus35, eucalyptus15, citrus34, and coffee31. An increase in breeding efficiency by using simulated data was also reported36.
Due to the significant population growth, the demand for energy sources, especially renewable energy, is intensified, since the use of energy from oil or coal is finite. Thus, encouraging the use of renewable energies, especially biofuels, has become one of the alternatives faced with the issue of global warming. Several studies have been carried out to consolidate new crops for biodiesel production.
In this sense, the use of Jatropha as a renewable energy source becomes a great alternative. However, to consolidate Jatropha as a new source of biodiesel, it is necessary to implement techniques able to assist the breeders in obtaining new cultivars, since the species is poorly improved. Genetic breeding programs have been started in several countries using conventional approaches to increase the grain and oil yield in Jatropha. However, by using conventional breeding, Jatropha yields are still low to make profitable and sustainable its growing10,37.
In this sense, clearing and using GWS allows access to genetic information, which is potentially useful to Jatropha breeding programs. Our study showed one of the first worldwide applications of GWS in a Jatropha breeding program. As Jatropha is a perennial crop, this tool becomes one of the most promising ways to promote the development of the crop for biodiesel production.
Conclusions
The wide-genome selection proved to be promising for Jatropha breeding since it allows the accurate obtention of GEBV and higher efficiency in relation to phenotypic selection, making it possible to reduce the time needed for selection cycle.
In order to apply genomic selection in the first harvests, a large number of individuals in the breeding population are needed. In the case of few individuals in the population, it is recommended to carry out a higher number of harvests.
Material and methods
Breeding and genotyping population
A total of 386 individuals from the Jatropha breeding population, which comes from the crossing between 42 parents, were evaluated. The population was evaluated in random block design with six replicates, three plants per plot, and a spacing of 4 × 2 m. The parents come from the Active Germplasm Bank (BAG) of Embrapa Agroenergia, which was composed of genotypes from several regions. The grain yield (g plant-1) was evaluated in the years 2015, 2016, and 2017, corresponding to 1, 2, and 3 years old, respectively.
The DNA extraction was performed from fully expanded young leaves, according to the protocol of the manufacturer's manual NucleoSpin Plant II (Macherey–Nagel), with modifications. The quantification and quality of the samples were performed using the NanoDrop Aspectrophotometer to evaluate the A260/A280 wavelength ratio, which represents the amount of nucleic acids by the amount of protein in the sample. Samples with A260/A280 between 1.80 and 2.10 were considered adequate, indicating a low amount of protein and RNA in the samples. The samples were genotyped using the Axiom myDesign Genotyping Arrays platform, using a chip developed by Embrapa Agroenergia based on Affymetrix's Axiom technology (Axiom ENERCHIP) selected for species with bioenergetic potential. SNPs were filtered based on multiple criteria that included: (1) consensus sequence size, (2) minimum and maximum reading depth, (3) SNP quality score, (4) Minor allele frequency (MAF), (5) presence of other SNPs in the adjacencies, (6) SNPs present in various populations (if they were sampled), (7) SNPs present in coding regions, coverage of genes of interest (in this case genes related to biotic and abiotic stresses, acidification and oil biosynthesis) and (8) coverage at the genomic level (assessed by the distribution of SNPs in the reference genome).
Genomic data corresponding to 12,598 SNPs were submitted to an initial quality control (QC), where the marker exclusion criteria were: Call Rate = 0.95 and MAF = 0.04. The Call Rate is used to eliminate markers with a large amount of lost values, whereas MAF is related to the polymorphism of marker loci in the population. The critical level for the MAF parameter was obtained through the equation, , where N refers to the number of genotypes in the population 38. After the QC, 811 SNPs were obtained that met the exclusion criteria.
Predicting the genomic model and cross-validation
Genomic selection analyses were performed using the RR-BLUP random regression method39. All statistical modeling was performed using software R. RR-BLUP was performed using the rrBLUP package (mixed.solve function). For estimating the marker effects by RR-BLUP methodology, the following mixed linear model was used:
| 1 |
wherein: y is the vector of phenotypic observations; β is the vector of fixed effects; is the vector of random marker effects; ε is the vector of random residuals; X and Z are the incidence matrices for β and a. The structure of means and variations in this model is described as defined by40:
The genomic mixed model equations for predicting a by the RR-BLUP method is equivalent to:
| 2 |
wherein is the total genetic variance of the trait.; is the residual variance; n: number of markers. The prediction of the individual's genomic breeding value (GBV) is given by:
| 3 |
The Z matrix was constructed from the number of alleles observed in each SNP marker (0, 1, or 2) and was standardized to have zero mean and variance 1, as described by41.
The k-fold cross-validation method was used. The set of observations was randomly divided into groups. In the analysis process, random samples of N1 = (9⁄10) × NT were used as the training population, while the remaining individuals in the population N2 = (1⁄10) × NT were used as the validation population, in which NT is the total number of individuals in the population. This process was repeated ten times (k = 10), using a different set of individuals as the validation population at a time. Thus, each fold did not overlap with the others, and at the end of the process (10 folds), all individuals had their phenotypes predicted by the genomic selection, as previously described by 42,43.
Accuracy and efficiency of genomic selection
The accuracy of genomic selection was estimated according to26, in which it considers genomic heritability as a proportion of phenotypic heritability, thus considering the efficiency of markers in capturing QTL, i.e., it considers the degree of imperfection of linkage disequilibrium:
| 4 |
wherein is the predictive ability of GWS, obtained through the correlation between predicted breeding values and observed phenotypic values; : is narrow-sense heritability, was obtained based on the following equation ; and is the genomic heritability, was calculated as the ratio of the additive variance to the phenotypic variance , .
The estimate of the number of individuals to be evaluated to achieve the desired accuracy was obtained by the expression:
| 5 |
where in it is GWS accuracy; nqtl is the number of QTLs controlling each trait given by
| 6 |
N is number of individuals in the population and h2 genomic heritability40.
The selective efficiency of GWS compared to selection based on phenotypes only was calculated using the expression:
| 7 |
wherein is selective accuracy of GWS; is the average time for the selection cycle based exclusively on the phenotypes; is the accuracy based on phenotypic selection, obtained through the equation: , where is the genotypic variation of the population and PEV is the variance of the prediction error; is the average time for the selection cycle based on GWS35.
Validating the models between harvests
The GWS models developed in each harvest were evaluated for accuracy in predicting breeding values, among other harvests. For this analysis, the accuracy was calculated by the correlation between GEBV derived from data collected in the first two harvests and EBV in the second and third harvests. As the same plant is compared between ages (harvests), there is a dependency between one plant on two different harvests. Therefore, tenfold cross-validation was performed as described previously. All analyses were performed in the R software 44.
Statements
The authors are allowed to do research with Jatropha curcas in Brazil.
The handling of plants were carried out in accordance with relevant guidelines and regulations.
Author contributions
A.S. performed the statistical analyzes and prepared figures 1-2; E.V. R. performed data collection; B.G.L. coordinated the research project; L.P.R.T., P.E.T. and L.L.B. contributed with a critical review of the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.FAO. The future of food and agriculture – Alternative pathways to 2050. Food and Agriculture Organization of the United Nations 224 (2018).
- 2.Schramski JR, Woodson CB, Brown JH. Energy use and the sustainability of intensifying food production. Nat. Sustain. 2020;3:257–259. doi: 10.1038/s41893-020-0503-z. [DOI] [Google Scholar]
- 3.Ong HC, et al. Production and comparative fuel properties of biodiesel from non-edible oils: Jatropha curcas, Sterculia foetida and Ceiba pentandra. Energy Convers. Manag. 2013;73:245–255. doi: 10.1016/j.enconman.2013.04.011. [DOI] [Google Scholar]
- 4.Takase M, et al. An expatiate review of neem, jatropha, rubber and karanja as multipurpose non-edible biodiesel resources and comparison of their fuel, engine and emission properties. Renew. Sustain. Energy Rev. 2015;43:495–520. doi: 10.1016/j.rser.2014.11.049. [DOI] [Google Scholar]
- 5.ANP. Anuário estatístico brasileiro do petróleo, gás natural e biocombustíveis 2019. Agência Nacional de Petróleo,Gás Natural e Biocombustível 264 http://www.anp.gov.br/arquivos/central-conteudos/anuario-estatistico/2019/2019-anuario-versao-impressao.pdf (2019).
- 6.Dharma S, et al. Optimization of biodiesel production process for mixed Jatropha curcas – Ceiba pentandra biodiesel using response surface methodology. Energy Convers. Manag. 2016;115:178–190. doi: 10.1016/j.enconman.2016.02.034. [DOI] [Google Scholar]
- 7.Fuentes A, García C, Hennecke A, Masera O. Life cycle assessment of Jatropha curcas biodiesel production: a case study in Mexico. Clean Technol. Environ. Policy. 2018;20:1721–1733. doi: 10.1007/s10098-018-1558-7. [DOI] [Google Scholar]
- 8.Baral, N. R. et al. Stochastic economic and environmental footprints of biodiesel production from Jatropha curcas Linnaeus in the different federal states of Nepal. Renew. Sustain. Energy Rev.120, 109619 (2020).
- 9.Laviola BG, et al. Desempenho agronômico e ganho genético pela seleção de pinhão-manso em três regiões do Brasil. Pesqui. Agropecu. Bras. 2014;49:356–363. doi: 10.1590/S0100-204X2014000500005. [DOI] [Google Scholar]
- 10.Mazumdar, P., Singh, P., Babu, S., Siva, R. & Harikrishna, J. A. An update on biological advancement of Jatropha curcas L.: new insight and challenges. Renew. Sustain. Energy Rev.91, 903–917 (2018).
- 11.Becker K, Makkar HPS. Jatropha curcas: a potential source for tomorrow’ s oil and biodiesel. Lipid Technol. 2008;20:104–107. doi: 10.1002/lite.200800023. [DOI] [Google Scholar]
- 12.Li Z, Lin B-L, Zhao X, Sagisaka M, Shibazaki R. System approach for evaluating the potential yield and plantation of Jatropha curcas L. on a Global Scale. Environ. Sci. Technol. 2010;44:2204–2209. doi: 10.1021/es903004f. [DOI] [PubMed] [Google Scholar]
- 13.Silitonga AS, Hassan MH, Ong HC, Kusumo F. Analysis of the performance, emission and combustion characteristics of a turbocharged diesel engine fuelled with Jatropha curcas biodiesel-diesel blends using kernel-based extreme learning machine. Environ. Sci. Pollut. Res. 2017;24:25383–25405. doi: 10.1007/s11356-017-0141-9. [DOI] [PubMed] [Google Scholar]
- 14.Alves AA, Laviola BG, Formighieri EF, Carels N. Perennial plants for biofuel production: bridging genomics and field research. Biotechnol. J. 2015;10:505–507. doi: 10.1002/biot.201400201. [DOI] [PubMed] [Google Scholar]
- 15.Resende MDV, et al. Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol. 2012;194:116–128. doi: 10.1111/j.1469-8137.2011.04038.x. [DOI] [PubMed] [Google Scholar]
- 16.Peixoto, L. de A. et al. Leveraging genomic prediction to scan germplasm collection for crop improvement. PLoS One12, e0179191 (2017). [DOI] [PMC free article] [PubMed]
- 17.Resende, M. D. V. & Duarte, J. B. Precisão E Controle De Qualidade Em Experimentos De Avaliação De Cultivares. Pesqui. Agropecuária Trop. (Agricultural Res. Trop.37, 182–194 (2007).
- 18.Alves RS, et al. Multiple-trait BLUP in longitudinal data analysis on Jatropha curcas breeding for bioenergy. Ind. Crops Prod. 2019;130:558–561. doi: 10.1016/j.indcrop.2018.12.019. [DOI] [Google Scholar]
- 19.Laviola BG, et al. Seleção de genitores em cruzamentos dialélicos em Jatropha curcas usando modelos mistos. Acta Sci. Agron. 2018;40:35008. doi: 10.4025/actasciagron.v40i1.35008. [DOI] [Google Scholar]
- 20.Peixoto, L. de A., Laviola, B. G., Alves, A. A., Rosado, T. B. & Bhering, L. L. Breeding Jatropha curcas by genomic selection: a pilot assessment of the accuracy of predictive models. PLoS One12, e0173368 (2017). [DOI] [PMC free article] [PubMed]
- 21.Junqueira, V. S. et al. Bayesian multi-trait analysis reveals a useful tool to increase oil concentration and to decrease toxicity in Jatropha curcas L. PLoS One11, e0157038 (2016). [DOI] [PMC free article] [PubMed]
- 22.Grattapaglia D, Resende MDV. Genomic selection in forest tree breeding. Tree Genet. Genomes. 2011;7:241–255. doi: 10.1007/s11295-010-0328-4. [DOI] [Google Scholar]
- 23.Heffner EL, Jannink JL, Iwata H, Souza E, Sorrells ME. Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci. 2011;51:2597–2606. doi: 10.2135/cropsci2011.05.0253. [DOI] [Google Scholar]
- 24.Resende MFR, Jr, et al. Accelerating the domestication of trees using genomic selection: accuracy of prediction models across ages and environments. New Phytol. 2012;193:617–624. doi: 10.1111/j.1469-8137.2011.03895.x. [DOI] [PubMed] [Google Scholar]
- 25.Calus MPL, Meuwissen THE, De Roos APW, Veerkamp RF. Accuracy of genomic selection using different methods to define haplotypes. Genetics. 2008;178:553–561. doi: 10.1534/genetics.107.080838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resende, M. D. V., Silva, F. F. E. & Azevedo, C. F. Estatística Matemática, Biométrica e Computacional: Modelos Mistos, Multivariados, Categóricos e Generalizados (REML/BLUP), Inferência Bayesiana, Regressão Aleatória, Seleção Genômica, QTL-GWAS, Estatística Espacial e Temporal, Competição, Sobrevivência. (Visconde do Rio Branco: Suprema, 2014).
- 27.Laviola, B. G., Rodrigues, E. V. V., Teodoro, P. E., Peixoto, L. de A. & Bhering, L. L. Biometric and biotechnology strategies in Jatropha genetic breeding for biodiesel production. Renew. Sustain. Energy Rev.76, 894–904 (2017).
- 28.Heffner EL, Sorrells ME, Jannink JL. Genomic selection for crop improvement. Crop Sci. 2009;49:1–12. doi: 10.2135/cropsci2008.08.0512. [DOI] [Google Scholar]
- 29.Peixoto, L. de A. et al. Breeding Jatropha curcas by genomic selection: A pilot assessment of the accuracy of predictive models. PLoS One12, e0173368 (2017). [DOI] [PMC free article] [PubMed]
- 30.Cavalcanti JJV, de Resende MDV, dos Santos FHC, Pinheiro CR. Predição simultânea dos efeitos de marcadores moleculares e seleção genômica ampla em cajueiro. Rev. Bras. Frutic. 2012;34:840–846. doi: 10.1590/S0100-29452012000300025. [DOI] [Google Scholar]
- 31.Sousa TV, et al. Early selection enabled by the implementation of genomic selection in coffea arabica breeding. Front. Plant Sci. 2019;9:1–12. doi: 10.3389/fpls.2018.01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner, C. R. et al. Effective Genomic selection in a narrow-genepool crop with low-density markers: Asian rapeseed as an example. Plant Genome11, 170084 (2018). [DOI] [PubMed]
- 33.Pereira JCDS, et al. Canopy growth and productivity of Jatropha genotypes. Semin. Agrar. 2017;38:135–141. doi: 10.5433/1679-0359.2017v38n1p135. [DOI] [Google Scholar]
- 34.Gois IB, et al. Genome wide selection in citrus breeding. Genet. Mol. Res. 2016;15:1–14. doi: 10.4238/gmr15048863. [DOI] [PubMed] [Google Scholar]
- 35.Resende Jr, M. F. R. et al. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics190, 1503–1510 (2012). [DOI] [PMC free article] [PubMed]
- 36.Valente MSF, Viana JMS, de Resende MDV, Silva FF, Lopes MTG. Genomic selection for plant breeding with different population structures. Pesqui. Agropecu. Bras. 2016;51:1857–1867. doi: 10.1590/s0100-204x2016001100008. [DOI] [Google Scholar]
- 37.Yue GH, Sun F, Liu P. Status of molecular breeding for improving Jatropha curcas and biodiesel. Renew. Sustain. Energy Rev. 2013;26:332–343. doi: 10.1016/j.rser.2013.05.055. [DOI] [Google Scholar]
- 38.Resende, M. D. V., Silva, F. F. & Azevedo, C. F. Atualidades da biometria no melhoramento de plantas perenes. in Desafios Biométricos No Melhoramento Genético (eds. Ludke, W., Andrade, A. & Volpato, L.) 166 (2017).
- 39.Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resende MDV, et al. Seleção genômica ampla (GWS) e maximização da eficiência do melhoramento genético. Pesqui. Florest. Bras. 2008;56:63. [Google Scholar]
- 41.Resende Jr, M. R. R. et al. Computação da Seleção Genômica Ampla (GWS). Série Documentos da EMBRAPA Florestas 78 (2010).
- 42.Legarra A, Robert-Granié C, Manfredi E, Elsen JM. Performance of genomic selection in mice. Genetics. 2008;180:611–618. doi: 10.1534/genetics.108.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verbyla KL, Calus MPL, Mulder HA, de Haas Y, Veerkamp RF. Predicting energy balance for dairy cows using high-density single nucleotide polymorphism information. J. Dairy Sci. 2010;93:2757–2764. doi: 10.3168/jds.2009-2928. [DOI] [PubMed] [Google Scholar]
- 44.R Core Team, D. R: a language and environment for statistical computing. https://www.r-project.org/ (2020).