Abstract
Activated phosphoinositide 3-kinase δ syndrome (APDS) is an autosomal-dominant combined immunodeficiency disorder resulting from pathogenic gain-of-function (GOF) mutations in the PIK3CD gene. Patients with APDS display abnormal T cell homeostasis. However, the mechanisms by which PIK3CD GOF contributes to this feature remain unknown. Here, with a cohort of children with PIK3CD GOF mutations from multiple regions of China and a corresponding CRISPR/Cas9 gene-edited mouse model, we reported that hyperactive PI3Kδ disrupted TNaive cell homeostasis in the periphery by intrinsically promoting the growth, proliferation, and activation of TNaive cells. Our results showed that PIK3CD GOF resulted in loss of the quiescence-associated gene expression profile in naive T cells and promoted naive T cells to overgrow, hyperproliferate and acquire an activated functional status. Naive PIK3CD GOF T cells exhibited an enhanced glycolytic capacity and reduced mitochondrial respiration in the resting or activated state. Blocking glycolysis abrogated the abnormal splenic T cell pool and reversed the overactivated phenotype induced by PIK3CD GOF in vivo and in vitro. These results suggest that enhanced aerobic glycolysis is required for PIK3CD GOF-induced overactivation of naive T cells and provide a potential therapeutic approach for targeting glycolysis to treat patients with APDS as well as other immune disorders.
Keywords: Primary immunodeficiency disorders, Activated phosphoinositide3-kinase δ syndrome, PIK3CD, Naive T cells, Aerobic glycolysis
Subject terms: Primary immunodeficiency disorders, Immunogenetics, Mechanisms of disease
Introduction
As the lifetime reserve pool of effector/memory T cells, naive T (TNaive) cells circulate in the blood and peripheral lymphoid tissues in relatively stable numbers with a quiescent phenotype1–3. The quiescent state is essential for maintaining the homeostasis of TNaive cells and is also critical for ensuring vigorous immune competence in response to a wide variety of foreign antigens throughout life1–3. However, how quiescence is properly regulated in TNaive cells is not fully understood.
Primary immunodeficiency diseases always shed light on genes and signaling pathways critical for human immunity. Activated phosphoinositide 3-kinase δ syndrome (APDS; OMIM: 602839), also known as PI3Kδ-activating mutations causing senescent T cells, lymphadenopathy, and immunodeficiency (PASLI) disease, is an autosomal-dominant combined immunodeficiency disorder resulting from pathogenic gain-of-function (GOF) mutations in the PIK3CD gene, which encodes the phosphoinositide 3-kinase δ (PI3Kδ, also known as p110δ) protein4–8. This disease is clinically characterized by recurrent respiratory tract infections, persistent infection with herpesviruses such as Epstein-Barr virus (EBV) and cytomegalovirus (CMV), chronic nonneoplastic lymphoproliferation, hepatosplenomegaly, autoimmunity and an increased risk for lymphoma4–8. Despite recent studies showing that patients with APDS exhibit a decreased number of TNaive cells and an increased abundance of effector/memory T cells4–10, the mechanisms by which PIK3CD GOF contributes to this abnormal immune homeostasis of TNaive cells in patients remain unknown.
PI3Kδ, a catalytic subunit of class IA PI3K members, is predominantly expressed by lymphocytes11. In T cells, after the engagement of the TCR by the corresponding antigen as well as costimulatory molecule and cytokine/chemokine receptor signaling, activated PI3Kδ catalyzes the phosphorylation of phosphatidylinositol-(4,5)-bisphosphate to generate phosphatidylinositol (3,4,5)-trisphosphate (PIP3). As an intracellular lipid second messenger signaling molecule, PIP3 recruits and assembles a plethora of pleckstrin homology (PH) domain-containing signaling proteins, such as protein kinase B (PKB/AKT) and phosphoinositide-dependent kinase 1 (PDK1), which then phosphorylate downstream effector molecules, including mTOR, FoxO1 and GSK3β, that synergistically regulate T cell function and immunity12,13.
Gene-targeted deletion of PI3Kδ or introduction of a catalytically inactive form of PI3Kδ in mice has no discernable effects on T cell development in the thymus14–17. Genetic inactivation of PI3Kδ in mice results in impaired TCR signaling, reduced T cell proliferation, attenuated primary and secondary immune responses, and a compromised ability of naive CD4+ T to differentiate into Th1, Th2, and T follicular helper cell subsets16,18–23. Inactivation of p110δ blocks antigen-dependent T cell recruitment and trafficking to antigenic tissues24,25. Importantly, deletion of PI3Kδ represses alloimmune reactivity and thus prolongs graft survival26. Moreover, inhibition of PI3Kδ reduces cytokine production in naive and effector/memory T cells isolated from healthy donors and donors with allergy or inflammatory arthritis27,28. Interestingly, several studies reported that patients with PI3Kδ deficiency or biallelic loss-of-function mutations in the PIK3CD gene exhibited a normal or decreased number of T cell pools and normal T cell proliferative rates in response to different stimuli29–32, indicating that PIK3CD is dispensable for T cell proliferation. Collectively, PI3Kδ plays extremely important roles in numerous biological processes of T cells, but its role in T cell activation and proliferation has been controversial.
In response to cognate antigen stimulation and costimulatory molecule signaling, TNaive cells initiate a transcriptional program that contributes to the transition from quiescence to rapid cell growth, extensive clonal expansion, and differentiation into specific effector T cell subsets to supervise the immune response. Importantly, these processes are concomitant with the reprogramming of cellular metabolism from catabolism to anabolism to meet the increased bioenergetic and biosynthetic demands for effector function33–35. TNaive cells utilize relatively slow glucolipid oxidative phosphorylation for their survival, while activated T cells shift to aerobic glycolysis to provide energy and metabolic intermediates for cell growth, proliferation and effector responses33–35. Although studies found that T cells in APDS patients exhibited increased glucose uptake5, the precise metabolic phenotype and whether the impaired T cell homeostasis in patients with APDS is associated with glucose metabolism are not clear.
Here, we reveal that PI3Kδ is a key mediator of quiescence and homeostasis in TNaive cells. Peripheral TNaive cells from PIK3CD GOF mice displayed an activated phenotype with increased cell growth, proliferation, activation and cytokine production. PIK3CD GOF resulted in the loss of the quiescence-associated gene expression profile in TNaive cells and promoted TNaive cells to acquire an activated functional status by collectively coupling the cell cycle, cytokine and cytokine receptor signaling, nutrient metabolism and signal transduction. Moreover, TNaive cells from PIK3CD GOF mice presented an enhanced glucose demand and glycolytic capacity and decreased mitochondrial respiration in the resting or TCR ligation-induced activated state, which was concomitant with elevated expression of glycolytic genes and key transcription factors. Interestingly, blocking glycolysis obviously inhibited the activation of TNaive cells ex vivo. Most importantly, inhibition of glycolysis almost fully reversed the abnormal T cell pool in the spleen and inhibited the overactivated phenotype induced by PIK3CD GOF in vivo and in vitro. Together, these data from patients with APDS and genetic mouse models indicate that an enhanced capacity for aerobic glycolysis is required for PIK3CD GOF-induced TNaive cell overactivation and that targeting aerobic glycolysis can restore TNaive cell homeostasis ex vivo and in vivo.
Results
T cells from patients with GOF mutations in PIK3CD exhibited an activated phenotype
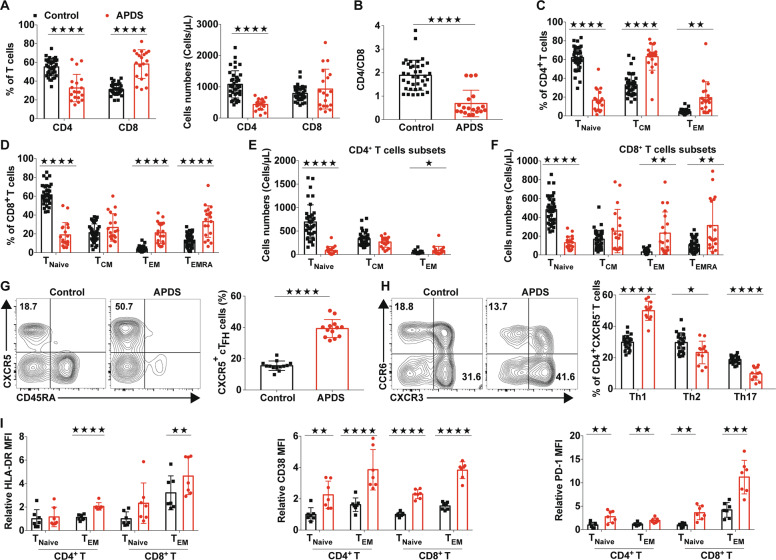
Consistent with previous reports4,5, we also found that T cells from patients with APDS had increased basal levels of phosphorylated AKT (Ser473) and ribosomal S6 protein (Supplementary Fig. 1A–B), a major downstream mediator of mTOR signaling, and these levels were constantly higher than those in T cells from healthy controls after stimulation with anti-CD3 and anti-CD28 antibodies, suggesting hyperactive PI3K-mTOR signaling. To determine the effects of PIK3CD GOF mutations on the homeostasis of T cells, we systemically evaluated the T cell compartment in the peripheral blood of patients with APDS. Despite the similar percentages of total T cells between the APDS patients and age-matched healthy control subjects, all patients exhibited obvious peripheral lymphopenia and reduced T cell numbers (Supplementary Fig. 1C-E). Consistent with recent reports in different cohorts of patients5,8–10, we found that the APDS patients from our Chinese cohort displayed decreased frequencies of CD4+ T cells and expanded CD8+ T cells, thereby yielding a reversed CD4/CD8 ratio (Fig. 1a, b). In addition, in line with previous reports5–10, in both the CD4+ and CD8+ T lymphocyte lineages from the patients, the numbers of TNaive cells were severely decreased with a corresponding increase in the numbers of effector memory T (TEM) cells or CD45RA+ effector memory T (TEMRA) cells (Fig. 1c–f and Supplementary Fig. 1F). Circulating follicular helper T (cTFH) cells and Th1 cells were increased in abundance, while the proportion of the Th17 subset was markedly reduced in the APDS patients compared to the healthy controls (Fig. 1g, h). Consistent with previous findings5–10,36, the expression of HLA-DR, CD38, and PD-1, which were increased in activated T cells, was higher in TNaive cell or/and TEM cell subsets from the APDS patients than in those from the healthy controls (Fig. 1i). However, the expression of CD62L and CCR7 in T cells was decreased compared with that in age-matched healthy controls (Fig. 1i and Supplementary Fig. 1G-I). Therefore, the T cells from the recruited patients with PIK3CD GOF displayed an activated phenotype.
Fig. 1.
The T cell compartment in patients with APDS showed altered homeostasis. a The frequencies (left) and absolute numbers (right) of T cells in blood from patients with APDS (n = 19) and healthy controls (n = 38). (b) The ratio of CD4+ to CD8+ T cells in blood from patients with APDS (n = 19) and healthy controls (n = 38). c, d Proportions of naive T cell (TNaive; CD45RA+CCR7+), central memory T cell (TCM; CD45RA−CCR7+), effector memory T cell (TEM; CD45RA−CCR7−), and CD45RA+ effector memory T cell (TEMRA; CD45RA+CCR7−) subsets in CD4+ and CD8+ T cell populations from patients with APDS (n = 19) and healthy controls (n = 38). e, f The absolute numbers of CD4+ and CD8+ T cell subsets from patients with APDS (n = 38) and healthy controls (n = 19). g Representative flow plots for the expression of CD45RA and CXCR5 within the CD3+ CD4+ T cells of the PBMCs and proportions of CD45RA−CXCR5+ cells (cTFH cells) within the CD3+CD4+ T cell populations from healthy controls and patients with APDS (n = 12). h Representative flow plots for the expression of CXCR3 and CCR6 (left) and the proportions of Th1 (CXCR3+CCR6−), Th2 (CXCR3−CCR6−) and Th17 (CXCR3−CCR6+) cells within the CD3+CD4+CXCR5−CD45RA− population (right) from healthy controls and untreated patients with APDS (n = 12 per group). i Expression of HLA-DR, CD38, PD-1 and CCR7 in CD4+ and CD8+ T cells from healthy controls (n = 7) and patients with APDS (n = 7). Numbers adjacent to outlined areas or in the indicated quadrants represent the percentage of cells in the area. Each symbol represents an individual throughout. Data are shown as the mean ± SD.★P < 0.05, ★★P < 0.01, ★★★P < 0.001, and ★★★★P < 0.0001 determined by Student’s unpaired t-test
T cells from PIK3CD GOF mice showed hyperactive PI3K/AKT and mTOR signaling
To further explore how activating mutations in PIK3CD impact the differentiation and functions of T cells, we generated a mouse model via the use of a CRISPR/Cas9 gene-targeting strategy with C57BL/6J mouse embryos to introduce a heterozygous E1024K mutation (ENSMUST 00000105690.8) in the PIK3CD gene, which is the common spot of substitution in PI3Kδ (E1021K) in patients with APDS7,8. Heterozygous mutant mice (hereafter referred to as PIK3CD GOF mice) were fertile and born at normal Mendelian ratios and did not show obvious anatomical or behavioral defects. Several studies have suggested that altering the level of one subunit of class IA PI3Ks can differentially affect the expression of the other heterodimeric subunits37–39. Therefore, we initially examined the protein expression of class IA PI3K components and found that before and following TCR stimulation with anti-CD3 and anti-CD28 antibodies, the expression of p110δ and p85α in T cells from PIK3CD GOF mice was comparable to that in wild-type (WT) cells, as was the level of the other class IA PI3K members (p110α, p100β, and p110γ) (Supplementary Fig. 2A, B). Moreover, the interaction between mutant p110δ and p85α was not impaired by delivering a retroviral vector expressing WT or mutant PIK3CD E1024K into HEK293T cells (Supplementary Fig. 2C), which was consistent with a previous study of patients40. Consistent with previous reports36,41, T cells from PIK3CD GOF mice had more basal and TCR-induced phosphorylation of AKT (Ser473 and Thr308) and the AKT target FoxO1 than those from WT mice (Supplementary Fig. 2D, E). In addition, compared with the control mice, the PIK3CD GOF mice showed dramatically increased phosphorylation of mTOR and the mTOR effector protein ribosomal protein S6 in T cells after TCR stimulation (Supplementary Fig. 2D, E)36,41. These results suggest that the E1024K mutation in mouse PIK3CD also leads to overactivation of PI3K/AKT and the mTOR signaling pathway36,41 and that the mouse model can partly recapitulate the pathogenesis of APDS9,10,36,41.
TNaive cells in PIK3CD GOF mice showed loss of quiescence and overactivation
In line with a previous report36, PIK3CD GOF mice had enlarged spleens with well-preserved anatomical structures, increased splenic cellularity, massive lymphocyte infiltration of the bronchioles and blood vessels of the lung, and scattered lymphocyte hyperplasia in the liver compared to WT mice (Supplementary Fig. 3A–C). To determine the effects of chronically hyperactivated PI3Kδ on the pathophysiological progress of APDS, we first examined T cell development in this mouse model. WT and PIK3CD GOF mice had comparable numbers of total thymocytes, except for decreases in the proportions of single-positive CD4+ and CD8+ T cells in the GOF mice (Supplementary Fig. 4A). In the spleen, despite the similar frequencies of CD8+ T cells, the percentage and absolute numbers of CD4+ T cells were elevated in the mutant mice (Supplementary Fig. 4B). Consistent with other reports9,10,36,41, among the T cell compartments in the spleen, the PIK3CD GOF mice had a significantly lower percentage of CD62LhiCD44lo TNaive cells than the control mice and a correspondingly expanded proportion of CD44hi cells with an activated or memory phenotype (Supplementary Fig. 4C). This alteration was further aggravated in aged mice (24 weeks, Supplementary Fig. 4D). In addition, both the CD4+ and CD8+ T cell populations in PIK3CD GOF mice displayed much higher expression of the activation markers CD25, CD44, and CD69 in both young (8–12 weeks) and aged mice than those in WT littermates (Supplementary Fig. 4E). These results suggested that, as seen in APDS patients, hyperactivation of PI3Kδ undermined the homeostasis of peripheral T cells in mice and promoted the accumulation of activated T cells.
Furthermore, to determine whether the changes in TNaive cell populations in PIK3CD GOF mice were a cell-autonomous defect, we generated mixed bone marrow (BM) chimeras by reconstituting irradiated wild-type mice (CD45.1) with a 50:50 mixture of wild-type or mutant mouse (CD45.2) bone marrow cells with wild-type (CD45.1) bone marrow cells. In line with a previous study by Julia Bier et al.9, 8 weeks after reconstitution, we also found that the splenic T cell populations of PIK3CD GOF donor cells in the reconstituted chimeras had fewer naive phenotype and greater activated/memory cells than those derived from wild-type donor bone marrow and retained higher expression of the activation markers CD44 and CD69 (Supplementary Fig. 5). However, wild-type CD4+ T cells (CD45.1) reconstituted in the presence of PIK3CD GOF bone marrow cells (CD45.2) exhibited a reduced percentage of naive T cells and elevated proportions of effector memory T cells compared to those (CD45.1) formed in the presence of wild-type hematopoietic cells (CD45.2), especially CD4+ T cells (Supplementary Fig. 5). These similar trends suggested that the altered peripheral T cell populations in PIK3CD GOF mice might not be fully cell-intrinsic but rather occur partially from extrinsic signals delivered by other cells, such as dendritic cells.
The diminished numbers of TNaive cells in PIK3CD GOF mice prompted us to determine the effects of hyperactivated PIK3CD on TNaive cell survival. Naive T cells were negatively isolated from splenocytes, with >95% purity in the isolated populations (Supplementary Fig. 6). The percentage of live TNaive cells was comparable between mutant mice and WT controls (Supplementary Fig. 7B). Despite the decreased level of CD127 (IL-7Ra) in the TNaive cells isolated from PIK3CD GOF mice compared with controls, there were no discerning differences in the survival rate of TNaive cells in the absence or presence of IL-7 (Supplementary Fig. 7A, B).
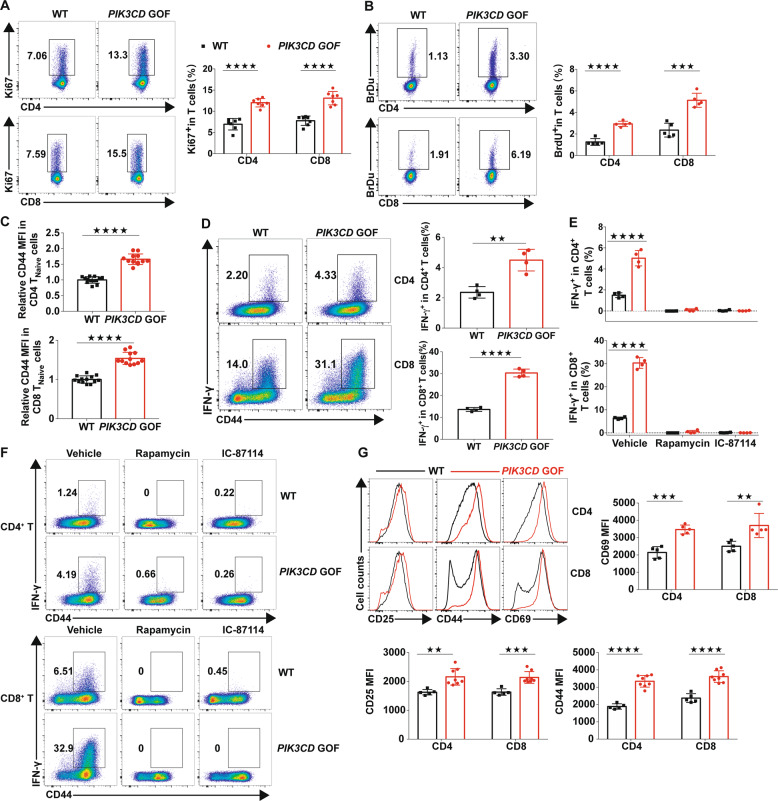
The homeostasis of TNaive cells mainly relies on the maintenance of a quiescent phenotype or resting state1–3. Based on this, we measured the expression of Ki67, a marker associated with the active phases of the cell cycle, in freshly isolated T cells. PIK3CD GOF mice had a greater frequency of Ki67+ cells than WT mice (Fig. 2a). In addition, we also examined the cycling of T cells by detecting the incorporation of bromodeoxyuridine (BrdU) in vivo. Similarly, an elevated proportion of splenic T cells from mutant mice incorporated BrdU (Fig. 2b), suggesting that PIK3CD GOF promoted an exit from the quiescent cellular state in T cells. Peripheral naive T cells newly emigrated from the thymus express low levels of the glycoprotein CD44, while mature, expanding and activated T cells accumulate more CD4442. Interestingly, we found that the expression of CD44 was significantly elevated in TNaive cells in PIK3CD GOF mice (Supplementary Fig. 8 and Fig. 2c), implying that TNaive cells in PIK3CD GOF mice have the potential to be easily activated.
Fig. 2.
TNaive cells from PIK3CD GOF mice showed loss of quiescence and overactivation. a Flow cytometry analysis of the proportion of Ki67+ cells in splenic T cells from WT or PIK3CD GOF mice (n = 7 per group). b Flow cytometry analysis of the proportion of BrdU+ T cells in splenocytes from WT or PIK3CD GOF mice (n = 5 per group). c Expression of CD44 in naive T cells from WT or PIK3CD GOF mice (n = 12–13 per group). d Proportion of IFN-γ-producing T cells from WT or PIK3CD GOF mice. Isolated total T cells were stimulated by anti-CD3 and anti-CD28 antibodies for 24 h, and IFN-γ+ T cells were analyzed by flow cytometry (n = 4 per group). e, f Naive T cells from WT and PIK3CD GOF mice were stimulated with anti-CD3 and anti-CD28 antibodies for 24 h in the presence or absence of rapamycin, IC-87114 or DMSO as a control (n = 4 per group), and the proportion of IFN-γ-producing cells in CD4+ T cells (top) and CD8+ T cells (bottom) was determined by flow cytometry. g Expression of CD25, CD44 and CD69 in TCR-stimulated TNaive cells from WT or PIK3CD GOF mice (n = 5–8 per group). Numbers adjacent to outlined areas represent the percentage of cells in the area. Each symbol represents an individual throughout. Data are shown as the mean ± SD.★P < 0.05, ★★P < 0.01, and ★★★★P < 0.0001 determined by Student’s unpaired t-test
Given the heightened activation state of TNaive cells in PIK3CD GOF mice, we next analyzed the frequency of cytokine-producing T cells. Total T cells from different groups were isolated and stimulated with anti-CD3 and anti-CD28 antibodies. After stimulation, PIK3CD GOF T cells displayed more IFN-γ production than control T cells (Fig. 2d). Moreover, to exclude the impact of high numbers of activated T cells on the expression of cytokines, we evaluated cytokine production in TCR-induced TNaive cells from mutant and control mice. The proportion of IFN-γ-producing cells detected upon activation in the cell population from mutant mice was greater than that from littermate controls (Fig. 2e, f). After TCR stimulation of TNaive cells, the expression of activation markers, including CD25, CD44, and CD69, was also increased in mutant T cells compared to WT cells (Fig. 2g). These results together indicated that PIK3CD GOF promoted TNaive cell acquisition of an activated functional status and predisposed these cells to overactivation in response to TCR stimuli.
Next, to assess the role of hyperactivated PI3Kδ activity in helper T cell differentiation, the capacity of CD4+ TNaive cells to differentiate into Th1, Th2, or Th17 effector cells was analyzed in vitro. Despite the similarities in polarization into Th2 cells, compared with the corresponding cells from littermate control mice, CD4+ TNaive cells from PIK3CD GOF mice had enhanced differentiation towards Th1 lineages accompanied by increased expression of the transcription factor T-bet (Supplementary Fig. 9). However, PIK3CD GOF T cells were less effective in Th17 polarization (Supplementary Fig. 9) concomitant with decreased RoRγt.
PIK3CD GOF resulted in a loss of quiescence-associated gene profiles in TNaive cells
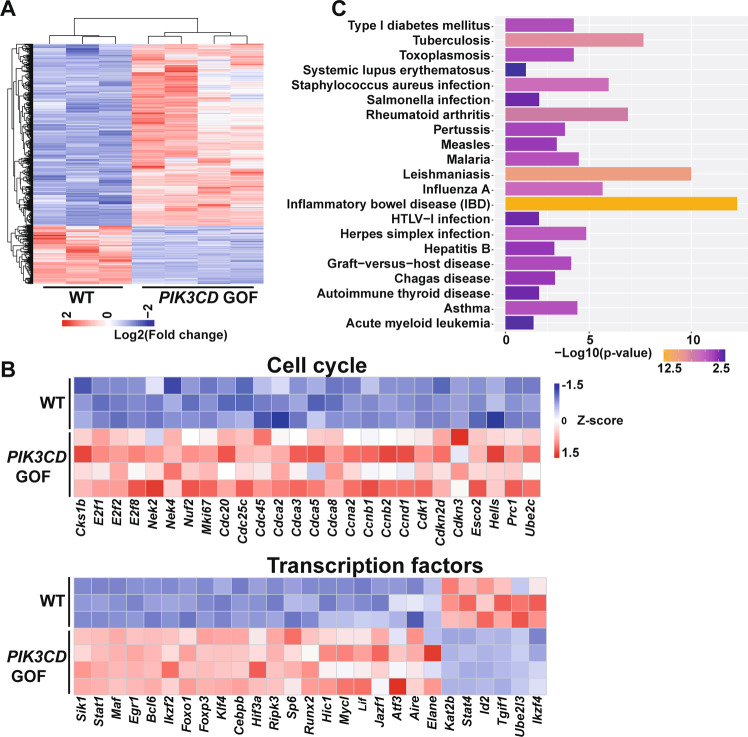
To further explore the molecular mechanisms by which PIK3CD GOF resulted in cells exiting quiescence and promoted the hyperactivation of peripheral T cells, we performed mRNA sequencing of sorted CD4+ TNaive cells from the spleens of PIK3CD GOF mice and WT littermate mice. Compared to the expression profiles in the WT cells, those in the mutant TNaive cells showed 2307 genes with differential expression (a greater than 0.5 log2-fold change compared to the expression in WT cells), of which 1749 genes exhibited increased expression and 558 exhibited decreased expression (Fig. 3a). Gene ontology analysis with the DAVID bioinformatic tool (version 6.8) showed that PIK3CD GOF TNaive cells upregulated the expression of genes encoding molecules involved in the cell cycle and mitosis, including Bub1, E2f1, E2f2, Nek2, Cdca2, cyclin A2, cyclin B2, Cdk1, Hells, Cdc20, Cdc45, and Nuf2, all of which might synergistically promote TNaive cell entry into the cycling phase (Fig. 3b). In addition, the mRNA expression levels of genes encoding T cell activation effectors (Cd44, Cd69, Cd86, and Icos), cytokines and cytokine receptors (Ifng, Il1b, Il18, and Il21), chemokines and chemokine receptors (Cxcr4, Cxcr5, Ccr3, Ccr4, and Bmp7), transcription factors (Tbx21, Foxp3, Bcl6, Myc, and Sik1), and metabolic regulators (Slc3a1, Slc7a5, Ldhd, Scd1, Ggdt1, and Hif3a) were differentially regulated in PIK3CD GOF mice compared to littermate control mice (Fig. 3b and Supplementary Fig. 10). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the altered genes in TNaive cells from PIK3CD GOF mice displayed significant enrichment of several pathways associated with infection, inflammation and autoimmune disease (Fig. 3c), which indicated that impaired TNaive cell homeostasis was associated with the disordered physiological processes of APDS. To further identify key signaling networks affected by PIK3CD GOF, we next conducted ingenuity pathway analysis (IPA) to analyze gene expression in TNaive cells from mutant and WT mice. IPA also revealed that signaling pathways regulating cellular growth, proliferation, activation, and metabolism and cell cycle pathways were upregulated in the TNaive cells from PIK3CD GOF T cells (Supplementary Table 1). In addition, the enriched networks with the highest scores among the differentially expressed genes were strongly associated with Th1 activation, T helper cell differentiation, T cell exhaustion, interferon signaling and the inflammatory process (Supplementary Table 1). Therefore, PIK3CD GOF results in a loss of quiescence-associated gene expression patterns in TNaive cells by collectively coupling the cell cycle, nutrient metabolism, cell trafficking, and signal transduction.
Fig. 3.
The gene expression patterns in naive PIK3CD GOF T cells were altered. a Heatmap analysis of genes differentially expressed in CD4+ TNaive cells from WT and PIK3CD GOF mice (differentially expressed genes were identified with a false discovery rate ≤0.05 and fold change ≥2, n = 3–4 per group). b Genes differentially expressed were analyzed by gene ontology and grouped as cell cycle and transcription factors. c KEGG analysis of the most enriched pathways for differentially expressed genes in isolated CD4+ TNaive cells between WT and PIK3CD GOF mice
PIK3CD GOF led to increased growth and glucose uptake of T cells
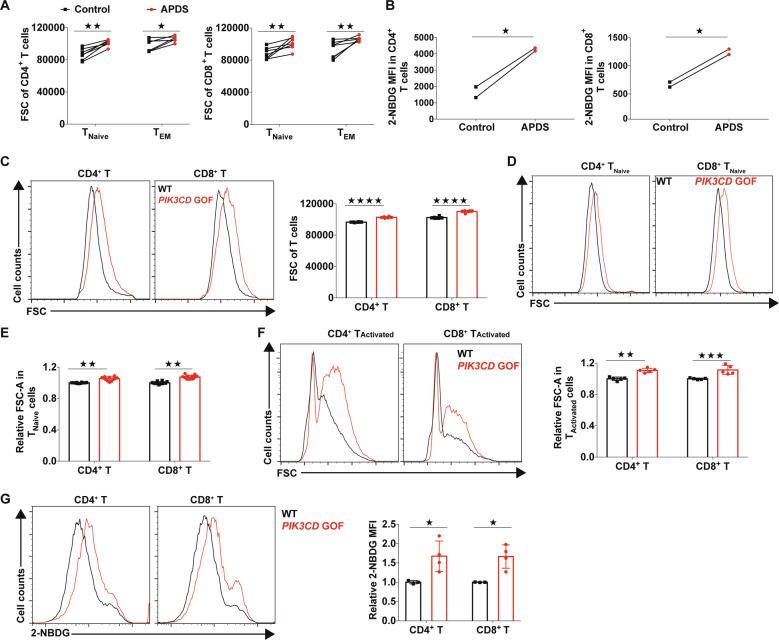
Given that several genes involved in glycolysis were regulated by PIK3CD GOF in the functional genomic analysis, we first examined the glucose metabolism of peripheral blood T cells from untreated patients with APDS. The size of freshly isolated T cells from newly diagnosed patients with APDS was much larger than that of T cells from healthy controls (Fig. 4a), suggesting that the T cells of APDS underwent more growth. Furthermore, we found that both CD4+ and CD8+ T cells from patients exhibited increased glucose uptake compared with the corresponding control cell populations, suggesting that the glucose demand of T cells in patients with APDS was enhanced (Fig. 4b), which was consistent with a previous report5. Consistently, the sizes of total T cells and even TNaive cell populations from PIK3CD GOF mice were larger than those from control mice (Fig. 4c–e). In addition, TCR-stimulated TNaive cells from PIK3CD GOF mice had larger cell sizes and more glucose uptake than those from wild-type cells (Fig. 4f, g). Taken together, these results indicated that PIK3CD GOF promoted cell growth and glucose utilization, which might be associated with the acquisition of an activated phenotype in PIK3CD GOF TNaive cells.
Fig. 4.
PIK3CD GOF led to increased growth and glucose uptake of T cells. a Size of isolated T cells from newly diagnosed patients with APDS and healthy controls. Cell size was evaluated by the forward-scatter (FSC) area. The patients shown here have not been treated with glucocorticoids or immunosuppressive agents (n = 7). b The glucose uptake of peripheral blood T cells was determined by staining with the glucose analog 2-NBDG. c–f Cell size of T cells from WT or PIK3CD GOF mice (n = 5–12 per group). Size of freshly isolated T cells (c), naive T cells (d, e), and T cells activated by anti-CD3/28 for 24 h (f). g Staining of 2-NBDG for assessing glucose uptake in naive T cells stimulated by anti-CD3 and anti-CD28 antibodies for 24 h (n = 3–4 per group). Each symbol represents an individual throughout. Data are shown as the mean ± SD. ★P < 0.05, ★★★P < 0.001 and ★★★★P < 0.001 determined by Student’s unpaired t-test
TNaive cells from PIK3CD GOF mice showed an elevated capacity for aerobic glycolysis
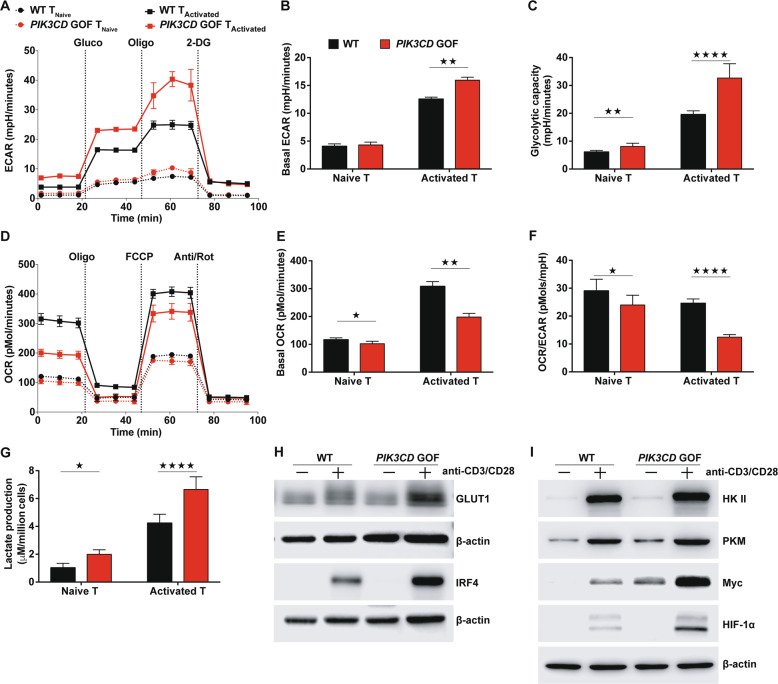
The diminished numbers of TNaive cells from untreated patients with APDS (<105 cell/ml) hampered further analysis of the metabolic phenotype (Fig. 1e, f). Therefore, to further interrogate the dynamics of metabolic programs caused by PIK3CD GOF, pan TNaive cells, including CD4+ and CD8+ T cell populations, from PIK3CD GOF and WT mice were isolated and either cultured in the presence of IL-7 or stimulated with anti-CD3 and anti-CD28 antibodies to maintain a resting phenotype or acquire an activated phenotype ex vivo, respectively. The extracellular acidification rate (ECAR), which mainly reflects glycolysis, and the mitochondrial oxygen consumption rate (OCR) were measured using an extracellular metabolic flux analyzer. We found that despite similar levels of basal glycolysis, glycolytic capacity was slightly higher in mutant TNaive cells than in WT control cells (Fig. 5a–c). Upon activation by TCR ligation, both the basal ECAR and capacity for glycolysis induced by injection of oligomycin were enhanced in T cells from WT mice and were further increased in those from PIK3CD GOF mice (Fig. 5a–c). In contrast, the baseline OCRs of both naive and activated T cells were decreased in PIK3CD GOF mice (Fig. 5d, e). In line with this finding, the ratio of the OCR to the ECAR was obviously reduced in naive and activated cells from mutant mice compared with those from control mice (Fig. 5f), which indicated that PIK3CD GOF TNaive cells rely largely on aerobic glycolysis rather than mitochondrial respiration. In addition, after stimulating TNaive cells with anti-CD3/CD28 antibodies for 72 h, despite the similar levels of basal glycolysis, the baseline OCRs and ratio of the OCR to the ECAR were obviously reduced in activated cells from PIK3CD GOF mice compared with those from control mice (Supplementary Fig. 11A–C). Consistent with this conclusion, compared with control cells, PIK3CD GOF T cells showed increased lactate production following activation (Fig. 5g). These results indicated that TNaive cells in PIK3CD GOF mice displayed an increased glycolytic capacity ex vivo and in response to activation mediated by TCR ligation in vitro.
Fig. 5.
Peripheral T cells from PIK3CD GOF mice showed an elevated glycolysis capacity. a–c The extracellular acidification rate (ECAR) was determined following consecutive injections of D-glucose (Gluco), the mitochondrial inhibitor oligomycin (Oligo) and the glycolytic inhibitor 2-DG. The time course of the ECAR (a), the basal ECAR (b) and calculations of glycolytic capacity (c) are shown. d–f The oxygen consumption rate (OCR) was determined following consecutive injections of the mitochondrial inhibitor oligomycin (Oligo), trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP), and antimycin A and rotenone (Act/Rot). The time course of the OCR (d), the basal OCR (e) and the ratio of the OCR to the ECAR (f) are shown. g Lactate production in medium from naive T cells or activated T cells obtained from WT or PIK3CD GOF mice. h, i Immunoblot analysis of the levels of Glut1, IRF4, HKII, PKM, Myc and HIF-1α in naive T cells from WT or PIK3CD GOF mice left unstimulated or stimulated with anti-CD3 and anti-CD28 antibodies for 14 h. Data are shown as the mean ± SD for 3–4 independent experiments.★P < 0.05, ★★P < 0.01, and ★★★★P < 0.0001 determined by Student’s unpaired t-test
Furthermore, the mRNA expression of the glycolytic genes Slc2a1, HkII, Pkm, and Ldha was much higher in PIK3CD GOF TNaive cells than in control cells and greater in activated T cells (Supplementary Fig. 11D). Likewise, PIK3CD GOF promoted the protein expression of GLUT1, HK II and PKM in both naive and activated T cells (Fig. 5h, i). In addition, the protein levels of HIF-1α, Myc, and IRF4, which are key transcription factors that orchestrate the expression of glycolytic enzymes, were also higher in activated T cells isolated from PIK3CD GOF mice than in those from control mice (Fig. 5h, i). These results suggested that PIK3CD played an important role in regulating the glycolytic demands of T cells and that global changes in glycolytic gene expression were associated with PIK3CD GOF-induced activation of naive T cells.
Inhibition of glycolysis repressed the overactivation and proliferation of TNaive cells induced by PIK3CD GOF
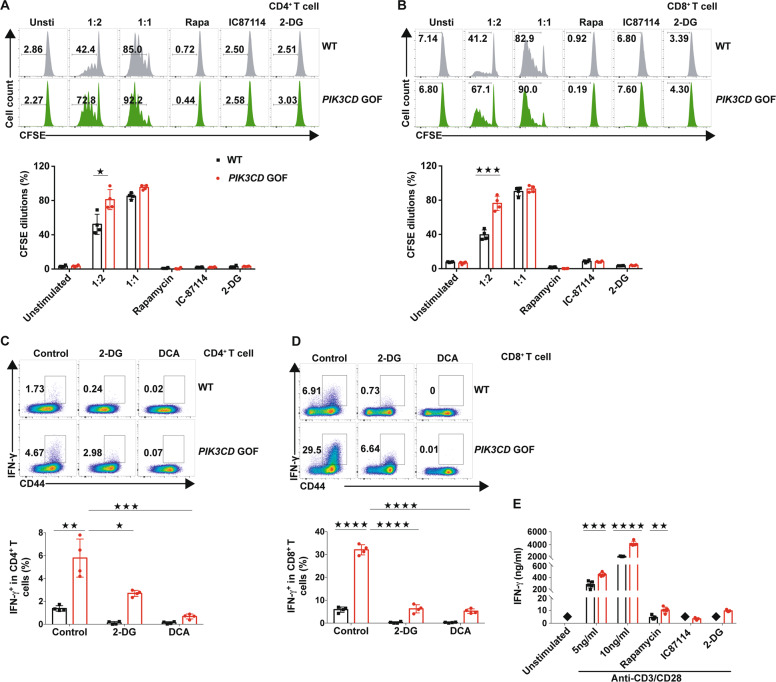
To analyze whether the elevated glycolytic capacity was involved in the hyperproliferation induced by PIK3CD GOF, isolated TNaive cells were stimulated with different doses of beads coated with anti-CD3 and anti-CD28 antibodies. When the ratio of bead-to-cell was 1:1, there was no significant difference in T cell proliferation between mutant and control mice (Fig. 6a, b). When the ratio of beads to cells was decreased to 1:2, the TNaive cell proliferation of PIK3CD GOF mice was much higher than that of the control (Fig. 6a, b), which indicated that PIK3CD GOF might lower the threshold for activation of TNaive cells. Similar to the selective PI3Kδ inhibitor IC87114 and the mTOR inhibitor rapamycin, inhibition of glycolysis by the hexokinase inhibitor 2-DG reduced the proliferation of TNaive cells activated by TCR ligation in wild-type cells, indicating that mTOR, PI3Kδ and enhanced glycolysis play important roles in T cell proliferation. In addition, these inhibitors prevented the proliferation of T cells from PIK3CD GOF mice, further suggesting that mTOR, PI3Kδ and glycolysis programs were also involved in the PIK3CD GOF-induced T cell proliferation (Fig. 6a, b). Interestingly, inhibition of glycolysis by the 2-DG or pyruvate dehydrogenase kinase inhibitor dichloroacetate (DCA) obviously repressed the overactivation of TNaive cells and reduced the expression of IFN-γ in PIK3CD GOF T cells (Fig. 6c, d). Furthermore, 2-DG also inhibited the production of IFN-γ in activated PIK3CD GOF T cells (Fig. 6e). These results suggested that PIK3CD GOF contributed to overactivation and hyperproliferation of TNaive cells by driving aerobic glycolysis (Fig. 6c–e).
Fig. 6.
Inhibiting glycolysis abolished T cell proliferation and activation caused by PIK3CD GOF. a, b CFSE-labeled TNaive cells from WT and PIK3CD GOF mice were stimulated with anti-CD3 and anti-CD28 antibodies for 72 h in the presence or absence of rapamycin, IC87114, 2-DG or vehicle as a control, and CFSE dilutions in (a) CD4+ T cells and (b) CD4+ T cells were determined by flow cytometry (n = 4 per group). c, d Naive T cells from WT and PIK3CD GOF mice were stimulated with anti-CD3 and anti-CD28 antibodies for 24 h in the presence or absence of 2-DG, DCA or vehicle as a control, and the proportion of IFN-γ-producing cells in c CD4+ T cells and d CD4+ T cells was determined by flow cytometry (n = 4 per group). e Naive T cells from WT and PIK3CD GOF mice were stimulated with anti-CD3 and anti-CD28 antibodies for 24 h in the presence of rapamycin, IC87114, 2-DG or vehicle as a control. After that, the concentration of IFN-γ in the medium was determined by ELISA (n = 5–6 per group). Solid black squares denote the concentration of IFN-γ in the medium below the detection limit. Numbers adjacent to outlined areas represent the percentage in the area. Data are shown as the mean ± SD for 3–6 independent experiments. ★P < 0.05, ★★P < 0.01, and ★★★★P < 0.0001 determined by two-way ANOVA
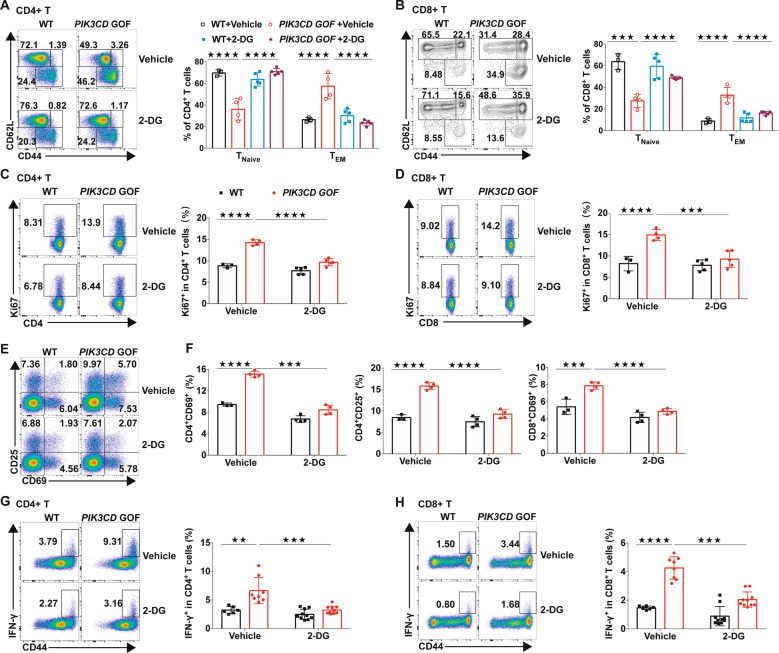
To further confirm this in vivo, we treated WT and PIK3CD GOF mice with normal or 2-DG-containing drinking water for 6 weeks. 2-DG treatment significantly decreased enlargement of the spleen in PIK3CD GOF mice, concomitant with a reduction in total splenocytes, but had little effect on body weight or blood glucose levels (Supplementary Fig. 12A–D). 2-DG treatment had no obvious effect on the viability of T cells from WT or mutated mice (data not shown). In addition, the frequency of naive T cells was increased, while the proportion and numbers of effector memory T cells were decreased in PIK3CD GOF mice after 2-DG treatment (Fig. 7a, b and Supplementary Fig. 12E). Importantly, 2-DG treatment also decreased the expression of activation markers (CD69, CD25, and CD44) and proliferation by PIK3CD GOF T cells to levels similar to those of age-matched WT mice (Fig. 7c–f). Furthermore, the production of IFN-γ in T cells derived from treated mutant mice was also decreased and was comparable to that of T cells from WT mice (Fig. 7g, h). Therefore, enhanced aerobic glycolysis was required for PIK3CD GOF-induced T cell overactivation, and targeting aerobic glycolysis could rescue the abnormal homeostasis induced by PIK3CD GOF ex vivo and in vivo.
Fig. 7.
Inhibiting glycolysis with 2-DG reversed abnormal T cell homeostasis caused by PIK3CD GOF in vivo. a, b Proportions of TNaive and TEM cells within splenic CD4+ T cells (a) and CD8+ T cells (b). c, d Proportions of Ki67+ cells within splenic CD4+ T cells (c) and CD8+ T cells (d). e, f Proportions of CD69+ and CD25+ cells within splenic CD4+ T cells and CD8+ T cells. g, h Splenocytes from WT or PIK3CD GOF mice treated with vehicle or 2-DG were stimulated with PMA and ionomycin for 5 h, and the proportions of IFN-γ-producing T cells were analyzed by flow cytometry. Proportion of IFN-γ+ cells within CD4+ (g) and CD8+ T cells (i). Each symbol represents an individual mouse. Data are shown as the mean ±SD. ★★P < 0.01, ★★★P < 0.001 and, ★★★★P < 0.0001 determined by two-way ANOVA
Discussion
Emerging evidence shows that PIK3CD GOF results in abnormal B cell development and differentiation in the BM and periphery, impaired T cell-independent immune responses and a defective capacity for the production of robust class-switched antigen-specific antibodies in response to T cell-dependent immunogen challenge9,36,41,43,44. Despite extensive research on APDS, the roles of PIK3CD GOF in T cells, especially for altered homeostasis of naive T cells, are not understood. With a cohort of children with PIK3CD GOF mutations from multiple regions from China and a corresponding CRISPR/Cas9 gene-edited mouse model, we reported that hyperactive PI3Kδ disrupted TNaive cell homeostasis in the periphery by promoting the growth, proliferation, and activation of TNaive cells. Therefore, our present study, coupled with lines of evidence from other reports5,9,10,16,18,36, strongly indicates that fine control of PI3Kδ activity is essential for maintaining the resting state of TNaive cell and producing optimal immune responses.
With a cohort of children with PIK3CD GOF mutations, we found that T cells from APDS patients displayed an activated phenotype and that CD4+ T cells in patients were biased to differentiate into cTfh subsets, which is consistent with other studies9,36. In addition, we found in the present cohort that the frequency of Th1 cell subsets was obviously increased, whereas the proportion of Th17 cell subsets was decreased in APDS patients. However, when we were preparing the manuscript, Julia Bier et al. reported that there were no significant differences in the proportions of non-Tfh memory CD4+ T cells with a Th1 or Th17 phenotype between healthy controls and APDS patients9. This discrepancy might be due to the heterogeneity in the study population. For example, the APDS patients recruited were much younger in our cohort (8 ± 4.8 years) than their subjects (20 ± 14.3 years). Using a mouse model, we also found that PIK3CD GOF drove differentiation of CD4+ TNaive cells into Th1 cells, which was consistent with recent studies9,45. In line with a previous study, we also found that there was a reduced trend in Th17 differentiation from CD4+ TNaive cells stimulated by anti-CD3/28 antibodies9. However, Silvia Preite and colleagues showed that CD4+ TNaive cells from PIK3CD GOF mice stimulated in the presence of antigen-presenting cells had increased production of IL-1745. This inconsistency might be due to the experimental context or more variable effects on Th17 differentiation45.
FOXO1, the major target of PI3K/AKT, promotes TNaive cell survival by regulating the expression of IL-7Rα46–48. We indeed found that hyperactive PI3Kδ decreased the expression of CD127 in TNaive cells. However, our results revealed that PIK3CD GOF had no discernible effect on the survival of naive T cells in the presence or absence of exogenous IL-7 in vitro. We speculated that FoxO1 expression in the nucleus was not completely ablated by PIK3CD GOF or that reduced CD127 expression can maintain T cell survival in the context of hyperactive AKT.
Multiple results in the present study suggested that hyperactive PI3Kδ abrogated TNaive cell quiescence. First, similar to patients with APDS, PIK3CD GOF mice exhibited aberrant T cell differentiation characterized by gradually decreased TNaive cell numbers and elevated effector T cell numbers, concomitant with exaggerated expression of activation markers. Second, TNaive cells from PIK3CD GOF mice displayed an activated phenotype, including a large cell size, enhanced homeostatic proliferation in vitro and in vivo, and increased expression of activation markers, which might be caused by increased cell size and a high capacity for glycolysis. Third, global gene expression profiling suggested that the expression of genes encoding positive regulators of T cell activation and entry into the cell cycle were obviously increased in TNaive cells from PIK3CD GOF mice. Fourth, the elevated expression levels of activation markers and cytokines induced by TCR stimulation in PIK3CD GOF mice were greatly decreased after inhibiting PI3Kδ activity. Thus, hyperactive PI3Kδ resulted in the loss of quiescence in TNaive cells by inducing the expression of activators of cellular growth, activation and proliferation.
The inability to control persistent viremia, including EBV and CMV viremia, is a common clinical manifestation in patients with APDS10,49–51. Paradoxically, the majority of APDS patients have relatively high proportions of total and EBV-specific CD8+ T cells, which are biased towards the TEM or TEMRA phenotype5,8,10,49–52. Although the involved mechanism is not clear, most researchers conclude that an exhausted and/or senescent state acquired by CD8+ T cells is a plausible contributor to impaired cytotoxicity and defective EBV control in APDS patients10,49–52. Although chronically recurrent EBV infections appear to be a powerful inducer of T cell activation, exhaustion or senescence in APDS, CD8+ T cells from patients without viremia still exhibit an exhausted and/or senescent phenotype with impaired function10,51. Therefore, we speculated that the exhaustion and/or senescence of CD8+ T cells in APDS patients was predominately caused by constitutive activation of PI3Kδ rather than chronic virus infection. Indeed, constitutively active PI3K/AKT induces senescence in various types of cells53,54. In addition, maintaining TNaive cell quiescence relies on the engagement of a tonic TCR by self-peptide MHC complexes1,2. We found that suboptimal TCR stimulation obviously promoted activation and proliferation of PIK3CD GOF TNaive cells, which had more basal AKT activity. Thus, it is also likely that overactivated PI3Kδ lowers the threshold for TCR signal strength to below the level required for maintaining quiescence, potentially allowing tonic self-peptides or foreign antigens to constantly activate TNaive cell and thereby contributing to the overactivation, exhaustion, senescence, or even death of T cells. Indeed, a recent study showed that PIK3CD GOF lead to CD8+ T cell susceptibility to reactivation-induced cell death4,10,49,51. Consistently, in this study, we further showed that after prolonged TCR activation, the viability of TNaive cells from PIK3CD GOF mice was obviously reduced (Supplementary Fig. 7C). These altered T cell states caused by PIK3CD GOF make the body susceptible to viral infection, which in turn chronically aggravates cell exhaustion or senescence with a positive feedback loop, ultimately leading to the pathophysiological progression of APDS. These findings are also supported by a recent study showing that tonic stimulation by the commensal microbiome induces hyperactivated and autoreactive phenotypes in APDS mouse models36. However, the detailed molecular basis of T cell exhaustion or senescence in APDS needs further elucidation.
mTOR, an evolutionarily conserved serine/threonine protein kinase, is activated downstream of PI3Kδ and has crucial roles in regulating T cell growth, proliferation, differentiation, survival, and metabolism by sensing and integrating multitudinous environmental signals55,56. PIK3CD GOF resulted in hyperactivation of the mTOR signaling pathway. Our results showed that the inhibition of activated mTOR rescued the abnormal T cell pool in PIK3CD GOF mice and largely restrained enhanced T cell proliferation, activation and cytokine production in vitro and in vivo (Fig. 6 and Supplementary Fig. 13). These results suggested that PI3Kδ modulated T cell homeostasis and function in an mTOR-dependent manner. Consistent with this finding, patients with APDS treated with the mTOR inhibitor rapamycin (sirolimus) experience decreases in hepatosplenomegaly and nonneoplastic lymphoproliferation. However, rapamycin has been found to be less effective in treating nonlymphoproliferative manifestations of APDS, particularly gastrointestinal disease and autoimmune cytopenia57–59. It is possible that there may be other mTOR-independent signaling pathways involved in the pathogenesis of APDS. Nevertheless, selective PI3Kδ inhibitors might be an effective approach to treat APDS5,7,60. However, PI3Kδ blockade in mice with PI3Kδ inhibitors enhances genomic instability61, and lymphocytes require balanced PI3Kδ activity to maintain homeostasis8. Thus, long-term follow-up in patients treated with these inhibitors is necessary, particularly given that long-term treatment is extremely likely.
As the major energy source, glucose is critical for T cell proliferation, activation, and effector function. Here, our results showed that T cells or even TNaive cells from PIK3CD GOF mice or patients with APDS had an increased cell size and elevated glucose uptake. We also found that PIK3CD GOF promoted glycolytic metabolism characterized by an enhanced glycolytic capacity, decreased mitochondrial oxidative respiration, and increased lactate production concomitant with elevated expression of rate-limiting enzymes and transcription factors for glycolysis in T cells. Thus, the overactive TNaive cells induced by PIK3CD GOF were strongly associated with an increased glycolysis capacity.
2-DG, a commonly used inhibitor of glycolysis, is a glucose analog that acts as a competitive inhibitor of hexokinase II, which catalyzes the initial phosphorylation of glucose into glucose-6-phosphate. 2-DG prevents the development of systemic autoimmunity in several lupus-prone mouse models by eliminating autoreactive TFH cells and normalizing increased CD4+ T cell metabolism62,63. Combined inhibition of glycolysis and glutaminolysis alleviates allograft rejection by inhibiting T cell proliferation and activation64. 2-DG also attenuates joint inflammation and the activation of adaptive and innate immune cells as well as the production of pathogenic autoantibodies in a mouse model of rheumatoid arthritis65. In addition, suppressing glycolysis with DCA protects animals against inflammation in both inflammatory bowel disease and experimental autoimmune encephalomyelitis66. In the present study, we found that systemic inhibition of glycolysis with 2-DG had no obvious effect on the viability of T cells (data not shown) and normalized the homeostasis of peripheral T cell pools. Following 2-DG treatment, T cell proliferation, overactivation and effector molecule expression in PIK3CD GOF mice also declined to levels comparable to those in WT mice. Intriguingly, 2-DG had no discernable effects on WT mice, suggesting that this treatment has increased effectiveness against T cells with active glycolysis. However, our results do not rule out the possibility that global treatment with 2-DG has additional effects on other immune cells, such as dendritic cells, B cells and macrophages. When combined with those of other studies, our findings further provide proof of concept that aerobic glycolysis is a viable therapeutic target in immune disorders. What needs to be considered is that systematic inhibition of glycolysis with 2-DG may weaken the immune response against infection. Therefore, how to target glycolysis for the treatment of APDS and other related diseases needs to be further explored from a clinical perspective.
In conclusion, in the present study, our findings emphasize the critical roles of PI3Kδ in the regulation of naive T cell quiescence and activation of peripheral naive T cells. Our data suggest that GOF mutations in PIK3CD disrupt TNaive cell homeostasis and promote the activation of TNaive cells by promoting the aerobic glycolysis program. These findings extend our knowledge on the function of PI3Kδ in T cell immunity, expand our understanding of the pathophysiological progression mechanism of APDS and provide a potential therapeutic approach to target glycolysis for the treatment of patients with activated PI3Kδ or other associated immune diseases.
Materials and methods
Human samples
From December 2013 to September 2018, a total of 22 patients (8 males and 14 females) with APDS from 18 unrelated Chinese families were enrolled in the present study. All of the patients carried heterozygous germline mutations in PIK3CD (c.3061 G>A, E1021K) that were identified by next-generation sequencing or whole-exome sequencing (MyGenostics, Beijing, China) and further confirmed by Sanger sequencing. Age- and sex-matched healthy controls (n = 44) were recruited from subjects who underwent routine medical check-ups. Human peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Hypaque (GE Healthcare, USA) gradient centrifugation. The absolute number of total lymphocytes was determined by an automatic hematology analyzer (Sysmex XE-2100, Sysmex Co., Ltd, Japan). Written informed consent was obtained from all the subjects included in this study and their legal guardians in accordance with the Declaration of Helsinki and with approval from the Ethics Committee of the Children’s Hospital of Chongqing Medical University.
Flow cytometry analysis of human cells
Whole-blood staining
Conventional T lymphocyte subsets in whole blood were analyzed as previously described67. The absolute subset number in the samples was obtained by multiplying subset percentages by the absolute lymphocyte count.
PBMC staining
Human PBMCs were incubated with fluorochrome-conjugated antibodies for 30 min at 4 °C in PBS containing 2% (wt/vol) fetal bovine serum (FBS). The antibodies used are listed in the Supplementary Information. Cell size was evaluated by assessing the forward-scatter area.
Mice
To generate PIK3CD E1024K mice, heterozygous PIK3CD c.3070G>A mutations (transcript ID: ENSMUST00000105690.8), which was also equivalent to the E1020K mutant in other studies 10,36,41,43,44, were produced in C57BL/6J mouse embryos with CRISPR/Cas9 technology by Shanghai Biomodel Organism Science & Technology Development Co., Ltd. (Shanghai, China). Two single guide RNAs (sgRNAs) were designed to ensure a protospacer adjacent motif sequence as close as possible to the targeted nucleotide change and to ensure that the Cas9 endonuclease would generate a double-strand break close to the nucleotide of interest, yielding the following sgRNAs: guide#1, 5′-GAGCTTCGTTGAACTTCACCCGG-3′, and guide#2, 5′-ACGAAGCTCTCCGAGAAAGCTGG-3′. A single-stranded oligo donor DNA template with the following sequence was chemically synthesized: 5′-CTCTGGCACTGGGGAAGACGGAGGAAGAGGCGCTAAAGCACTTTCGGGTGAAGTTCAACAAAGCTCTCCGAGAAAGCTGGAAAACCAAAGTCAACTGGCTGGCGCACAATGTGTCCAAGG-3′. In this study, all heterozygous PIK3CD E1024K mice (hereafter referred to as PIK3CD GOF mice) and littermates used were obtained by mating PIK3CD WT/mu mice with WT mice or PIK3CD WT/mu mice. Genomic DNA was genotyped by PCR and confirmed by Sanger sequencing (Sangon Biotech, China). The CD45.1 congenic mouse line was purchased from Shanghai Biomodel Organism Science & Technology Development Co., Ltd. All animal experiments were performed with sex- and age-matched littermate controls (8–12 weeks old, unless other indicated). Experimental analyses were nonblinded. All mice were kept in a specific pathogen-free environment. All procedures were approved by the Animal Care and Use Committee of Chongqing Medical University.
Histopathology
The liver, spleen, and lungs from sacrificed mice were fixed in 4% paraformaldehyde and embedded in paraffin. Five-micron sections were stained with hematoxylin & eosin and then evaluated with a microscope (Leica).
Flow cytometry analysis of mouse cells
For analysis of surface markers, in vitro-cultured cells or cells isolated from the indicated lymphoid organs were stained for 30 min at 4 °C in PBS containing 2% (wt/vol) FCS with fluorochrome-conjugated antibodies. For intracellular cytokine staining, stained cells were fixed and permeabilized with a Cytofix/Cytoperm kit (BD Biosciences), and intranuclear staining was conducted with a Foxp3/Fixation/Permeabilization kit according to the manufacturer’s instructions (eBiosciences). The antibodies used are listed in the Supplementary Information. For staining of phosphorylated proteins, cells were fixed in 2% PFA, permeabilized in 100% ice-cold MeOH and then stained with anti-p-AKT Ser473 (BD Biosciences) and anti-p-S6 Ser235/236 (Cell Signaling Technology) antibodies. All data were collected on a FACSCalibur or FACSCanto flow cytometer (BD Biosciences) and analyzed with FlowJo software (TreeStar, USA). Mean fluorescence intensity (MFI) quantifications were calculated by normalizing the mean fluorescence intensity values obtained for each mouse, with the mean of the values obtained from control mice set as 100% for each experiment. The specific gating strategies for each population are indicated in each figure legend.
In vivo BrdU labeling
Eight-week-old mice were injected intraperitoneally with BrdU (2 mg/mouse) in a DPBS solution. After 24 h, splenocyte-incorporated BrdU was evaluated with the BrdU Flow Kit (BD Biosciences) according to the manufacturer’s instructions.
Bone marrow (BM) chimeras
BM cells from PIK3CD GOF (CD45.2) or WT (CD45.2) and congenic C57BL/6J WT (CD45.1) donor mice were harvested and processed into single-cell suspensions. Recipient C57BL/6J WT (CD45.1) mice were lethally irradiated with 650 rad. BM chimeras were generated by reconstituting irradiated recipient WT (CD45.1) mice with a 50:50 mixture of WT (CD45.2) or mutant mouse (CD45.2) donor BM cells with WT (CD45.1) donor BM cells via intravenous injection (5 × 106 cells per mouse). Eight weeks after reconstitution, splenocytes from recipient mice were analyzed by flow cytometry.
Treatment of mice with 2-DG or rapamycin
Six-week-old PIK3CD GOF mice and sex-matched WT mice were randomized into treatment or vehicle groups. Animals were administered vehicle (0.2% carboxy-methylcellulose and 0.25% Tween-80 in sterile ultra-pure water) or rapamycin (4 mg/kg, Selleck) daily by intraperitoneal injection for 15 days. For glycolysis inhibitor treatment, mice were administered 2-DG (5 mg/ml, Selleck) dissolved in drinking water that was replaced every other day for 45 days. Age-matched control mice were given plain drinking water during the experiment. Body weight and blood glucose levels were monitored weekly and biweekly, respectively. At the end of treatment, spleens were collected, washed in ice-cold PBS to remove residual blood and accessory fascia, weighed and subjected to further analysis. All experiments were conducted according to protocols approved by the University of Chongqing Medical University.
Retrovirus construction and infection
Full-length cDNA sequences encoding mouse PIK3CD-His fusion proteins were amplified and cloned into pL-MCS vectors using NheI and AscI restriction sites. The cDNA sequence encoding the mouse PIK3CD E1024K mutation (muPIK3CD-His), which corresponds to the most common GOF substitution in PIK3δ (E1021K) in patients with APDS, was introduced by PCR-based site-directed mutagenesis. All constructs were confirmed by sequencing (Invitrogen). Retroviruses were prepared by transfection of HEK293T cells (ATCC) with retroviral vectors along with the psPAX2 and pMD2G plasmids by Lipofectamine 3000 (Invitrogen). HEK293T cells were maintained in DMEM (Gibco, USA) supplemented with 10% FBS, 100 U/ml penicillin G and 100 μg/ml streptomycin (Gibco) and infected with the prepared retroviruses. After 48 h, the cells were collected and lysed for protein extraction.
Cell isolation and activation
Total T cells were negatively enriched from splenocytes using an immunomagnetic selection kit (Stemcell Technologies) and stimulated with anti-CD3 (5 µg/ml, eBioscience) and anti-CD28 (5 µg/ml, eBioscience) antibodies, followed by cross-linking with goat anti-hamster IgG (10 µg/ml) antibodies for the indicated times in a 37 °C water bath. At different times, the phosphorylation of S6 and AKT (Ser473) was determined by flow cytometry as previously described. Mouse pan-naive T cells were negatively isolated from pooled single-cell suspensions of splenocytes (Stemcell Technologies) by removing the unwanted cells with antibodies directed against non-naive T cells (expressing CD11b, CD19, CD24, CD25, CD44, CD45R/B220, CD49b, and TER119) according to the manufacturer’s instructions, with >95% purity (Supplementary Fig. 4F-G) in the isolated populations, and cultured in RPMI 1640 medium (Gibco) supplemented with 10% (vol/vol) FBS and 1% penicillin-streptomycin at 37 °C in 5% CO2. Freshly isolated naive T cells were activated with plate-coated anti-CD3 (5 µg/ml) and anti-CD28 (5 µg/ml) antibodies for the indicated time. In addition, cell tracer (CFSE, Invitrogen, C34554)-labeled naive T cells were activated with Dynabeads® Mouse T-Activator CD3/CD28 (Gibco, 11453D) for 72 h, and proliferation was determined by CFSE dilutions. For specific chemical inhibitor treatment, cells were incubated with IC87114, rapamycin (Selleck), 2-DG (Selleck), DCA (Sigma) or vehicle before being stimulated. Freshly isolated naive T cells were maintained in culture medium or medium containing 5 ng/ml recombinant IL-7 (PeproTech) or stimulated by anti-CD3/CD28 antibodies, and cell viability was determined by Fixable Viability Dye eFluor™ 780 staining (eBioscience, USA) at different time points.
In vitro differentiation of mouse CD4+ TNaive cells
Naive CD4+ T cells were negatively isolated ex vivo (Stemcell) and cultured in flat-bottom 48-well plates previously coated with 5 μg/ml anti-CD3 in RPMI-1640 medium supplemented with 10% FBS (Excell Bio, China), 2 mM L-glutamine, 10 mM HEPES buffer solution, 1 mM sodium pyruvate, 100 μM nonessential amino acid solution, 50 μM β-mercaptoethanol, 100 U/ml penicillin and 100 µg/ml streptomycin at a density of 1 × 106 cells/well. The following cytokines were added to generate each subset: Th1, 5 µg/ml anti-CD28 antibody (eBioscience), 10 ng/ml IL-2 (R&D), 20 ng/ml IL-12 (PeproTech), and 10 µg/ml anti-IL-4 antibody (eBioscience); Th2, 5 µg/ml anti-CD28 antibody, 10 ng/ml IL-4 (PeproTech), 10 ng/ml IL-2, and 10 µg/ml anti-IFN-γ antibody (eBioscience); and Th17, 5 µg/ml anti-CD28 antibody, 20 ng/ml IL-6 (PeproTech), 10 ng/ml IL-23 (BioLegend), 10 ng/ml IL-1β (R&D), 2 ng/ml human TGF-β (R&D), 10 µg/ml anti-IL-4 antibody, and 10 µg/ml anti-IFN-γ antibody. After 5 days, the cells were restimulated with 50 ng/ml PMA and 500 ng/ml ionomycin for 5 h, and GolgiStop (BD Biosciences) and GolgiPlug (BD Biosciences) were added for the final 4 h. The cells were harvested, washed, fixed and permeabilized with a Foxp3/Fixation/Permeabilization kit (eBioscience), stained with the indicated antibody, and analyzed by flow cytometry.
Quantitative real-time PCR
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions, and first-strand cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative real-time PCR was conducted with a TB Green® Premix Ex Taq™ kit (Tli RNaseH Plus) (Takara Bio, China) and a CFX96 real-time system PCR system (Bio-Rad, USA) according to the manufacturers’ instructions. Gene expression levels were analyzed with the comparative Ct method and normalized to β-actin mRNA levels.
Primers for quantitative real-time PCR
The primers used are as follows: Slc2a1 forward, 5′-TCTCCAAACTGGGCAAGTCCT-3′; Slc2a1 reverse, 5′-ATGGAGTCTAAGCCAAACACCT-3′; Hk2 forward, 5′-ATCTACGCCATTCCCGAGGACA-3′; Hk2 reverse, 5′-ATCTCTGCCTTCCACGCCACT-3′; Pkm2 forward, 5′-GGAGCCACTCTGAAGATCACC-3′; Pkm2 reverse, 5′-CATTCTCCACCTCCGTCACCA-3′; Ldha forward, 5′-ATATCTTGACCTACGTGGCTT-3′; Ldha reverse, 5′-CTCTCCCATCAGGTAACGGAA-3′; Pfkl forward, 5′-CTACTGTGACCGGCATTGCT-3′; Pfkl reverse, 5′-TGCAGTCAAACACGCCCCTG-3′; and β-actin forward, 5′-ACATCCGTAAAGACCTCTATGCC-3′; β-actin reverse, 5′-ACCGATCCACACAGAGTACTTGC-3′.
Immunoblotting and immunoprecipitation
Cells were lysed in RIPA lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris, 1 mM EDTA and 1 mM EGTA) containing complete protease inhibitor cocktails (Roche). Whole-cell extracts were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore). The immunoblots were blocked in Tris-buffered saline/Tween 20 (TBST) and 5% skim milk for 1 h at room temperature and then incubated with the primary antibody at 4 °C overnight. Following three consecutive 10-minute washes with TBST, the blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (1:5000, Cell Signaling Technology, Boston, MA, USA) for 1 h at room temperature. The blots were scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA), and protein band intensities were quantified by densitometry using Bio-Rad analysis software (Bio-Rad, Hercules, CA, USA).
For immunoprecipitation, cells were lysed in 1% NP-40 buffer (25 mM Tris-HCl, 150 mM NaCl, pH 7.4, 1% NP-40, 1 mM EDTA, and 5% glycerol) supplemented with protease inhibitor cocktails (Roche). The lysates were immunoprecipitated with the indicated antibodies overnight at 4 °C before adding Dynabeads Protein G (Invitrogen) for 2 h. After extensive washing, the immunoprecipitates were denatured by boiling in sample buffer, resolved by SDS-PAGE, and then analyzed by immunoblotting with the antibodies indicated in the figures. Primary antibodies are listed in the Supplementary information.
Enzyme-linked immunosorbent assay (ELISA)
The concentration of IFN-γ in the culture medium of TNaive cells from different groups was determined by ELISA according to the manufacturer’s instructions (4A Biotech, Beijing, China; Cat. No. CME0003).
Metabolic assays
Sorted naive mouse T cells were either maintained in the resting stage without or with 5 ng/ml IL-7 or activated with 5 μg/ml plate-bound anti-CD3 antibody and 5 μg/ml anti-CD28 antibody. After 24 or 72 h, the live cells of resting or activated T cells were sorted (BD FACSAria II, USA) and then attached to culture microplates precoated with Cell-Tak (Corning) and maintained in an unbuffered assay medium (Seahorse Biosciences) in a non-CO2 incubator at 37 °C for 1 h before the assay was performed. The plating Seahorse medium comprised nonbuffered RPMI medium (Sigma) supplemented with 25 mM glucose, 2 mM L-glutamine, 1 mM pyruvate sodium and 1 mM HEPES (Seahorse Biosciences). After 1 h, the plate was loaded into the instrument to determine the ECAR and OCR using the XF24 Extracellular Flux Analyzer (Seahorse Bioscience) according to the manufacturer’s recommended protocols. Baseline ECAR and OCR values were averaged between replicates for the first 3 successive time intervals. Extracellular lactate production was measured with a colorimetric assay kit according to the manufacturer’s instructions (Sigma). Cellular glucose uptake was evaluated by flow cytometry with the fluorescently labeled deoxyglucose analog 2-NBDG (100 μg/ml, Cayman).
Bioinformatics—Library construction and sequencing
Total RNA from isolated naive CD4+ T cells was extracted using the RNeasy Plus Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions and checked for RNA integrity by an Agilent Bioanalyzer 2100 (Agilent Technologies, US). Qualified total RNA was further purified by the RNeasy Micro Kit (QIAGEN) and RNase-Free DNase Set (QIAGEN). Paired-end libraries were constructed using the TruSeq® RNA Sample Preparation Kit (Illumina, USA) according to the manufacturer’s instructions. The products were purified, enriched, and quantified by a Qubit® 2.0 Fluorometer (Life Technologies, USA) and validated by an Agilent 2100 bioanalyzer to confirm the insert size and calculate the molar concentration. Clusters were generated using cBot with the library diluted to 10 pM and then sequenced on the Illumina HiSeq 2500 (Illumina, USA). Library construction and sequencing were performed at Shanghai Biotechnology Corporation (Shanghai, China).
RNA-seq analysis
Sequenced raw reads were preprocessed by removing rRNA reads, sequencing adapters, short-fragment reads and other low-quality reads with Seqtk (https://github.com/lh3/seqtk). The obtained clean reads were mapped to the mouse mm10 reference genome with two mismatches by Hisat2 (version 2.0.4). After genome mapping, Stringtie (version: 1.3.0) was run to generate FPKM (fragments per kilobase of exon model per million mapped reads) values for known gene models. Differentially expressed genes (DEGs) were identified using edgeR with the following filter criteria: false discovery rate (FDR) ≤0.05 and fold change ≥2. For functional analysis, the selected DEGs were mapped to each term of the GO and KEGG databases, the number of genes of each item was calculated, and then a hypergeometric test was applied to screen the GO or KEGG pathway terms with significant enrichment. In addition, the data were analyzed using Ingenuity Pathway Analysis® (IPA, www.ingenuity.com) to further identify biological processes enriched for the DEGs.
Statistics
All data are presented as the mean ± standard deviation (SD) and were analyzed using Student’s unpaired t-test or two-way ANOVA with a post-hoc test for multiple comparisons with SPSS 24.0 (SPSS graduate pack; SPSS, USA). Differences were considered to be statistically significant at P < 0.05.
Supplementary information
Acknowledgements
This work was supported by grants from the National Science Foundation of China (81974255) and the Public Welfare Scientific Research Project of China (201402012) to X.Z.
Author contributions
Y.J., Q.Y., and Y.W. designed and performed experiments and analyzed the data; Y.J. wrote the initial paper; Q.Y., Y.W., X.C., and W.T. recruited and managed patients; W.L., X.C., T.X., Z.T., M.F., L.Z., N.T., and L.Z. performed experiments; W.S. contributed to scientific discussion and data interpretation and reviewed and revised the paper. X.Z. designed the research, supervised the study, and reviewed and revised the paper.
Data availability
The RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with the accession number GSE 134322.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version of this article (10.1038/s41423-020-0379-x) contains supplementary material.
References
- 1.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Takada K, Jameson SC. Naive T cell homeostasis: from awareness of space to a sense of place. Nat. Rev. Immunol. 2009;9:823–832. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 3.Silva SL, Sousa AE. Establishment and maintenance of the human naïve CD4+ T-cell compartment. Front Pediatr. 2016;4:1–10. doi: 10.3389/fped.2016.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angulo I, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342:866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucas CL, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat. Immunol. 2014;15:88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elgizouli M, et al. Activating PI3Kδ mutations in a cohort of 669 patients with primary immunodeficiency. Clin. Exp. Immunol. 2016;183:221–229. doi: 10.1111/cei.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulter TI, et al. Clinical spectrum and features of activated phosphoinositide 3-kinase δ syndrome: a large patient cohort study. J. Allergy Clin. Immunol. 2017;139:597–606.e4. doi: 10.1016/j.jaci.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes-Santos CJ, Uzel G, Rosenzweig SD. PI3K pathway defects leading to immunodeficiency and immune dysregulation. J. Allergy Clin. Immunol. 2019;143:1676–1687. doi: 10.1016/j.jaci.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Bier J, et al. Activating mutations in PIK3CD disrupt the differentiation and function of human and murine CD4+ T cells. J. Allergy Clin. Immunol. 2019;144:236–253. doi: 10.1016/j.jaci.2019.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards ESJ, et al. Activating PIK3CD mutations impair human cytotoxic lymphocyte differentiation and function and EBV immunity. J. Allergy Clin. Immunol. 2019;143:276–291.e6. doi: 10.1016/j.jaci.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 11.Vanhaesebroeck B, et al. p110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl Acad. Sci. USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev. Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucas CL, Chandra A, Nejentsev S, Condliffe AM, Okkenhaug K. PI3Kδ and primary immunodeficiencies. Nat. Rev. Immunol. 2016;16:702–714. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb LMC, Vigorito E, Wymann MP, Hirsch E, Turner M. Cutting edge: T Cell development requires the combined activities of the p110γ and and p110δ catalytic catalytic isoforms of phosphatidylinositol 3-kinase. J. Immunol. 2005;175:2783–2787. doi: 10.4049/jimmunol.175.5.2783. [DOI] [PubMed] [Google Scholar]
- 15.Clayton E, et al. A crucial role for the p110delta subunit of phosphatidylinositol 3-kinase in B cell development and activation. J. Exp. Med. 2002;196:753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okkenhaug K. Impaired B and T Cell antigen receptor signaling in p110delta PI3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 17.Jou S-T, et al. Essential, nonredundant role for the phosphoinositide 3-kinase p110delta in signaling by the B-cell receptor complex. Mol. Cell Biol. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okkenhaug K, et al. The p110delta isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J. Immunol. 2006;177:5122–5128. doi: 10.4049/jimmunol.177.8.5122. [DOI] [PubMed] [Google Scholar]
- 19.Rolf J, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J. Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 20.Pearce VQ, Bouabe H, MacQueen AR, Carbonaro V, Okkenhaug K. PI3Kδ regulates the magnitude of CD8+T cell responses after challenge with listeria monocytogenes. J. Immunol. 2015;195:3206–3217. doi: 10.4049/jimmunol.1501227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, Uzonna JE. The p110delta Isoform of phosphatidylinositol 3-kinase controls the quality of secondary anti-leishmania immunity by regulating expansion and effector function of memory T cell subsets. J. Immunol. 2010;184:3098–3105. doi: 10.4049/jimmunol.0903177. [DOI] [PubMed] [Google Scholar]
- 22.Gracias DT, et al. Phosphoinositide 3-Kinases p110δ isoform regulates CD8+ T cell responses during acute viral and intracellular bacterial infections. J. Immunol. 2016;196:1186–1198. doi: 10.4049/jimmunol.1501890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, et al. The p110 isoform of phosphatidylinositol 3-kinase controls susceptibility to leishmania major by regulating expansion and tissue homing of regulatory T cells. J. Immunol. 2009;183:1921–1933. doi: 10.4049/jimmunol.0901099. [DOI] [PubMed] [Google Scholar]
- 24.Jarmin SJ, et al. T cell receptor-induced phosphoinositide-3-kinase p110δ activity is required for T cell localization to antigenic tissue in mice. J. Clin. Invest. 2008;118:1154–1164. doi: 10.1172/JCI33267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinclair LV, et al. Phosphoinositide 3-kinase (PI3K) and nutrient sensing mTOR (mammalian target of rapamycin) pathways control T lymphocyte trafficking. Nat. Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uehara M, et al. Regulation of T cell alloimmunity by PI3Kγ and PI3Kδ. Nat. Commun. 2017;8:951. doi: 10.1038/s41467-017-00982-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soond DR, et al. PI3K p110delta regulates T-cell cytokine production during primary and secondary immune responses in mice and humans. Blood. 2010;115:2203–2213. doi: 10.1182/blood-2009-07-232330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nashed BF, et al. Role of the phosphoinositide 3-kinase p110δ in generation of type 2 cytokine responses and allergic airway inflammation. Eur. J. Immunol. 2007;37:416–424. doi: 10.1002/eji.200636401. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K, Husami A, Marsh R, Jordan MB. Identification of a phosphoinositide 3-kinase (PI-3K) P110delta(PIK3CD) deficient individual. J. Clin. Immunol. 2013;33:673–674. [Google Scholar]
- 30.Sharfe N, et al. Dual loss of p110δ PI3-kinase and SKAP (KNSTRN) expression leads to combined immunodeficiency and multisystem syndromic features. J. Allergy Clin. Immunol. 2018;142:618–629. doi: 10.1016/j.jaci.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 31.Sogkas G, et al. Primary immunodeficiency disorder caused by phosphoinositide 3–kinase δ deficiency. J. Allergy Clin. Immunol. 2018;142:1650–1653.e2. doi: 10.1016/j.jaci.2018.06.039. [DOI] [PubMed] [Google Scholar]
- 32.Cohen SB, et al. Human primary immunodeficiency caused by expression of a kinase-dead p110δ mutant. J. Allergy Clin. Immunol. 2019;143:797–799.e2. doi: 10.1016/j.jaci.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J. Exp. Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geltink RIK, Kyle RL, Pearce EL. Unraveling the complex interplay between T cell metabolism and function. Annu Rev. Immunol. 2018;36:461–488. doi: 10.1146/annurev-immunol-042617-053019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preite S, et al. Hyperactivated PI3Kδ promotes self and commensal reactivity at the expense of optimal humoral immunity. Nat. Immunol. 2018;19:986–1000. doi: 10.1038/s41590-018-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110α subunit of phosphoinositide 3-kinase. J. Biol. Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 38.Fruman DA, et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 39.Lu-Kuo JM, Fruman DA, Joyal DM, Cantley LC, Katz HR. Impaired Kit- but not FcεRI-initiated mast cell activation in the absence of phosphoinositide 3-kinase p85α gene products. J. Biol. Chem. 2000;275:6022–6029. doi: 10.1074/jbc.275.8.6022. [DOI] [PubMed] [Google Scholar]
- 40.Dornan GL, et al. Conformational disruption of PI3Kδ regulation by immunodeficiency mutations in PIK3CD and PIK3R1. Proc. Natl Acad. Sci. USA. 2017;114:1982–1987. doi: 10.1073/pnas.1617244114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark AK, et al. PI3Kδ hyper-activation promotes development of B cells that exacerbate Streptococcus pneumoniae infection in an antibody-independent manner. Nat. Commun. 2018;9:3174. doi: 10.1038/s41467-018-05674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley SR, et al. Rasgrp1 mutation increases naive T-cell CD44 expression and drives mTOR-dependent accumulation of Helios+ T cells and autoantibodies. Elife. 2013;2:e01020. doi: 10.7554/eLife.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wray-Dutra MN, et al. Activated PIK3CD drives innate B cell expansion yet limits B cell–intrinsic immune responses. J. Exp. Med. 2018;215:2485–2496. doi: 10.1084/jem.20180617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avery DT, et al. Germline-activating mutations in PIK3CD compromise B cell development and function. J. Exp. Med. 2018;215:2073–2095. doi: 10.1084/jem.20180010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preite S, Gomez-Rodriguez J, Cannons JL, Schwartzberg PL. T and B-cell signaling in activated PI3K delta syndrome: From immunodeficiency to autoimmunity. Immunol. Rev. 2019;291:154–173. doi: 10.1111/imr.12790. [DOI] [PubMed] [Google Scholar]
- 46.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the forkhead-Box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L- selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton RH, et al. Maintenance of CD4 T cell fitness through regulation of Foxo1. Nat. Immunol. 2018;19:838–848. doi: 10.1038/s41590-018-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cannons JL, Preite S, Kapnick SM, Uzel G, Schwartzberg PL. Genetic defects in phosphoinositide 3-kinase δ Influence CD8+ T Cell survival, differentiation, and function. Front Immunol. 2018;9:1758. doi: 10.3389/fimmu.2018.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpier J-M, Lucas CL. Epstein–Barr Virus susceptibility in activated PI3Kδ syndrome (APDS) immunodeficiency. Front Immunol. 2018;8:2005. doi: 10.3389/fimmu.2017.02005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wentink MWJ, et al. Exhaustion of the CD8+ T cell compartment in patients with mutations in phosphoinositide 3-kinase delta. Front Immunol. 2018;9:446. doi: 10.3389/fimmu.2018.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nunes-Santos CJ, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. J. Allergy Clin. Immunol. 2014;143:1676–1687. [Google Scholar]
- 53.Imai Y, et al. Crosstalk between the Rb Pathway and AKT signaling forms a quiescence-senescence switch. Cell Rep. 2014;7:194–207. doi: 10.1016/j.celrep.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Astle MV, et al. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: implications for targeting mTOR during malignancy. Oncogene. 2012;31:1949–1962. doi: 10.1038/onc.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng H, et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity. 2016;45:540–554. doi: 10.1016/j.immuni.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng H, Chi H. mTOR signaling in the differentiation and function of regulatory and effector T cells. Curr. Opin. Immunol. 2017;46:103–111. doi: 10.1016/j.coi.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maccari ME, et al. Disease evolution and response to rapamycin in activated phosphoinositide 3-kinase δ syndrome: The european society for immunodeficiencies-activated phosphoinositide 3-kinase δ syndrome registry. Front Immunol. 2018;9:543. doi: 10.3389/fimmu.2018.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coulter TI, Cant AJ. The treatment of activated PI3Kδ syndrome. Front Immunol. 2018;9:2043. doi: 10.3389/fimmu.2018.02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michalovich D, Nejentsev S. Activated PI3 kinase delta syndrome: from genetics to therapy. Front Immunol. 2018;9:369. doi: 10.3389/fimmu.2018.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao VK, et al. Effective “activated PI3Kδ syndrome”–targeted therapy with the PI3Kδ inhibitor leniolisib. Blood. 2017;130:2307–2316. doi: 10.1182/blood-2017-08-801191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Compagno M, et al. Phosphatidylinositol 3-kinase δ blockade increases genomic instability in B cells. Nature. 2017;542:489–493. doi: 10.1038/nature21406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin Y, et al. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015;7:274ra18. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi SC, et al. Inhibition of glucose metabolism selectively targets autoreactive follicular helper T cells. Nat. Commun. 2018;9:4369. doi: 10.1038/s41467-018-06686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee C-F, et al. Preventing allograft rejection by targeting immune metabolism. Cell Rep. 2015;13:760–770. doi: 10.1016/j.celrep.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abboud G, et al. Inhibition of glycolysis reduces disease severity in an autoimmune model of rheumatoid arthritis. Front Immunol. 2018;9:1973. doi: 10.3389/fimmu.2018.01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerriets VA, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Y, et al. Reference values for peripheral blood lymphocyte subsets of healthy children in China. J. Allergy Clin. Immunol. 2018;142:970–973.e8. doi: 10.1016/j.jaci.2018.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with the accession number GSE 134322.