Abstract
Resveratrol plays a crucial phytoalexin role in the grapevine and is beneficial to human health. However, the molecular mechanism of resveratrol accumulation in the enhancement of disease resistance is unclear. Here, we report that the transcription factor VqMYB154 from Vitis quinquangularis accession Danfeng-2 is strongly expressed under artificial inoculation with Uncinula necator and regulates resveratrol accumulation. Unlike its homolog, VqMYB154 has a pathogen-induced promoter and responds to stimulation by U. necator, Pseudomonas syringae, and other treatments. Yeast one-hybrid and GUS activity assays confirmed that VqMYB154 can activate the stilbene synthase genes VqSTS9, VqSTS32, and VqSTS42 by directly binding to their promoters. Overexpression of VqMYB154 in grape leaves resulted in activation of the stilbene pathway, upregulation of STS genes, and accumulation of stilbenoids. In addition, heterologous overexpression of VqMYB154 in Arabidopsis activated resistance-related genes and resulted in greater programmed cell death and accumulation of reactive oxygen species, which led to resistance against P. syringae. These results suggest that the transcription factor VqMYB154 from V. quinquangularis accession Danfeng-2 participates in the regulatory mechanism that improves the biosynthesis and accumulation of stilbenes and enhances resistance to disease in grapevine.
Subject terms: Plant signalling, Biotic, Microbe, Transcriptional regulatory elements
Introduction
Grapevine is one of the most prestigious economic fruit crops worldwide. According to data from the Food and Agriculture Organization, the total output of grapes ranked third among fruit crops in 2018. Of Vitis vinifera grown worldwide, few cultivars possess high resistance to phytopathogenic microorganisms, including Uncinula necator1. If left unchecked, pathogen-triggered diseases will seriously affect the growth and quality of grapevine, ultimately leading to a decline in yield. Long-term use of pesticides pollutes the environment, and hazardous residues in grape berries threaten human health2. Therefore, it is crucial to improve grapevine resistance to reduce the need for pesticide application. Some resistant grape species, such as V. labrusca, have an unacceptable foxy smell, which greatly limits their application in crossbreeding3. China is a vital area of origin for wild grapevine germplasm, with many disease-resistant Vitis spp4. These wild species do not have undesirable flavors, and can easily be crossed with V. vinifera cultivars5; thus, they provide critical resources for resistance breeding of grapevine.
As a well-known stilbene-producing plant, the grapevine has high levels of resveratrol. This bioactive compound was first extracted from Veratrum grandiflorum6 and was determined to be present in grape tissues7. Resveratrol has great research value. Furthermore, it is beneficial to human health due to its antioxidant activity8 and antiaging9, neuroprotective10, and cancer prevention properties11. In addition, resveratrol acts as an important stilbene phytoalexin, which has been widely reported to possess antimicrobial ability against pathogen invasion12–14. These positive effects of resveratrol regarding health benefits and phytopathology have led to worldwide research on its pharmacological properties. The biosynthesis of resveratrol involves numerous enzymatic reactions, with stilbene synthases (STSs) being the most closely related and essential enzymes15. In the grapevine, 48 STS genes have been identified by sequencing the Pinot Noir PN40024 genome16. Among them, 33 STSs can be divided into three subfamilies named A to C, and B subfamily genes exhibit the greatest response to pathogen infection17. Stable expression of STSs in multiple species using transgenics might increase the content of stilbenoids and improve plant disease resistance. Hence, these STSs from the grapevine are resistance-related genes responsible for the accumulation of stilbene phytoalexins18,19.
Because of the crucial roles of STS genes in resveratrol biosynthesis and plant disease resistance, research in recent years has focused on exploring their upstream regulatory mechanisms. In 2013, it was reported that two transcription factors (TFs) derived from Pinot Noir grapevine, MYB14 and MYB15, activate STS41 and STS29 to promote stilbenoid accumulation20, which suggests that specific TFs in plants closely participate in the regulation of STS genes. Genome sequencing provides extensive data to support in-depth research on the regulatory mechanisms of TFs in plants. In grapevine, 2004 TFs have been identified by sequencing of the Pinot Noir genome16,21. Extensive research has shown that these TF genes are inextricably linked to the growth and development of grapevine, including cell expansion22, seed morphogenesis23, and berry ripening24. Moreover, numerous regulators function in defense responses to exogenous stresses, such as drought25, cold26, high salinity27, and pathogens28. Members of the WRKY29, NAC28, MYB30, ERF31, and bZIP families32 have been reported to play critical roles in resistance against pathogens.
R2R3-MYB TFs participate extensively in phenylpropanoid metabolism33; they act in functional regulation by recognizing conserved MYB binding sites (MBSs), such as the AC-box (ACCA/TAA/CT/C)34, MYBCORE (CAGTTA and CTGTTG)35, and L5-box motif (GAGTTGGTGAGA)36. Genome sequencing has identified these regulators in various plant species, including 138 members in Arabidopsis37, 126 in rice38, and 134 in grape39. They were initially divided into 25 subgroups in Arabidopsis based on the motif at the C-terminus40, and a later study on grapevine increased the number of subgroups to 3439. The numerous R2R3-MYB members may contain resistance-related TFs that are still unknown. Some resistance-related MYB proteins have been identified in Arabidopsis. Overexpression of AtMYB30 leads to hypersensitive cell death in Arabidopsis, thereby enhancing resistance to Pseudomonas syringae41. Its homologous gene, MdMYB30, also has similar disease resistance properties42. Furthermore, AtMYB96 enhances resistance to P. syringae by regulating salicylic acid (SA) biosynthesis and pathogenesis-related (PR) genes43. Knockdown of AtMYB46 improves the resistance of Arabidopsis mutants to Botrytis cinerea44. In wheat, the MYB gene TaPIMP1 was shown to promote SA-related resistance genes PR1a and PR2, thereby enhancing resistance to Bipolaris sorokiniana45. In addition to VvMYB14 and VvMYB15 in grapevine, VvMYB13 was shown to respond to infection by downy mildew and plays a positive role in stilbenoid accumulation39. VqMYB35 positively regulates the expression of STS genes by interacting with VqERF11446. However, considering the complexity of the MYB superfamily, more potential factors still need to be determined to fully clarify the MYB-mediated phytoalexin metabolic network.
Based on our long-term observations in vineyards, the Chinese wild-growing grapevine accession Danfeng-2 has a higher content of resveratrol and disease resistance than V. vinifera4,47. Therefore, this resistant germplasm has been used for in-depth analysis of grapevine-pathogen interactions. Recently, we determined the expression levels of 106 R2R3-MYB members in Danfeng-2 under artificial inoculation with U. necator, 27 of which showed greater than 4-fold upregulated expression (Supplementary Fig. S1). We then performed transcriptome analysis using Danfeng-2 berries during different developmental periods (unpublished data). Coexpression analysis based on transcriptome data was performed to identify pathogen-induced MYB TFs that regulate STS genes. Here, a resistance-related MYB TF, VqMYB154, from the V. quinquangularis accession Danfeng-2, was screened and isolated. Our research indicated that VqMYB154 is a novel regulator that improves the accumulation of stilbene phytoalexins and enhances resistance to disease in transgenic Arabidopsis. These results are significant for elucidating the regulatory mechanisms involved in plant-pathogen interactions and provide valuable references for the long-term goal of disease-resistant grapevine breeding.
Results
VqMYB154 is a resistance-related TF that participates in plant defense responses
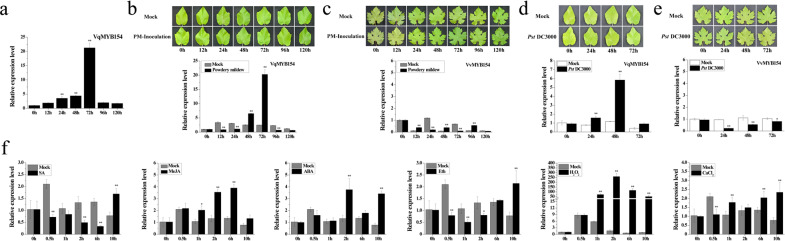
To screen out MYB genes involved in defense responses, we inoculated Danfeng-2 leaves with U. necator and performed qRT-PCR analysis. The results showed that the R2R3-type MYB gene VqMYB154 (this study) can be induced and significantly upregulated by U. necator (Fig. 1a, Supplementary Fig S1). To investigate whether MYB154 exhibits different expression patterns in disease-resistant and disease-susceptible grapes under pathogen inoculation, we inoculated the leaves of Danfeng-2 and Cabernet Sauvignon with U. necator and P. syringae (Pst DC3000). After artificial inoculation, the expression level of VqMYB154 from Danfeng-2 began to increase at 48 h and peaked at 72 h, increasing 8.2-fold, compared to the mock control at the same timepoint (Fig. 1b). In contrast to VqMYB154, MYB154 from Cabernet Sauvignon showed no clear expression pattern (Fig. 1c). On the other hand, after Pst DC3000 infection, VqMYB154 responded to induction at 24 h and then peaked at 48 h, increasing 5.0-fold (Fig. 1d). However, VvMYB154 exhibited a downward trend and reached its lowest abundance at 24 h (Fig. 1e). Therefore, we speculate that VqMYB154 is a resistance-related regulator in Danfeng-2.
Fig. 1. Specific expression of the transcription factor gene VqMYB154 under artificial inoculation conditions using U. necator and other exogenous treatments.
a VqMYB154 was screened from 106 MYB genes in Danfeng-2 leaves infected by U. necator artificial inoculation under field conditions. b–e Expression of MYB154 was determined by qRT-PCR. b Leaves of Danfeng-2 and (c) Cabernet Sauvignon were inoculated with U. necator and sampled at 0, 12, 24, 48, 72, 96, and 120 h. d Leaves of Danfeng-2 and (e) Cabernet Sauvignon were inoculated with Pst DC3000 and collected at 0, 24, 48, 72 h. Pst DC3000, Pst DC3000 infection; PM-Inoculation, U. necator infection; Mock, control treated with sterile water. f Leaves from Danfeng-2 were treated with 100 μM phytohormones, 1% (w/v) hydrogen peroxide, and 5 mM CaCl2. Leaves were collected at 0, 0.5, 1, 2, 6, and 10 h after spraying. Results are shown as the means (±SD) of three biological assays. Significance was determined with GraphPad Prism using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01)
To explore whether MYB154 responds to exogenous phytohormones, we treated the leaves of Danfeng-2 and Cabernet Sauvignon by spraying them with phytohormones. Compared to the control at the same time, the transcript level of VqMYB154 decreased at 0.5 h, 2 h, and 6 h but then increased 2.2-fold at 10 h after SA treatment. After MeJA treatment, the transcript level of VqMYB154 increased at 1 h and peaked at 6 h by 2.9-fold. The transcript level of VqMYB154 was upregulated at 2 h and peaked at 10 h by 4.4-fold after ABA treatment. Under Eth treatment, expression of VqMYB154 decreased from 0.5 h to 2 h but then increased 2.8-fold at 10 h (Fig. 1f). On the other hand, compared to the mock control, transcript levels of VvMYB154 were upregulated 2.4-fold at 10 h after SA treatment, 7.8-fold at 6 h after MeJA treatment, 3.3-fold at 2 h after ABA treatment, and 2.1-fold at 1 h after Eth treatment (Supplementary Fig. S3). These results demonstrate that the two MYB154 genes respond to exogenous phytohormone induction and exhibit different expression patterns in Danfeng-2 and Cabernet Sauvignon. VqMYB154 also responded to other exogenous signals, including H2O2 and CaCl2. After H2O2 treatment, expression of VqMYB154 was significantly upregulated, reached its highest level at 2 h by 162.2-fold, and then gradually decreased. Under the induction of CaCl2, the expression level of VqMYB154 was decreased at 0.5 h but increased at 1 h and 6 h and then reached its highest level at 10 h by 3.0-fold (Fig. 1f).
Expression profiles of VqMYB154 in ‘Danfeng-2’ grapevine
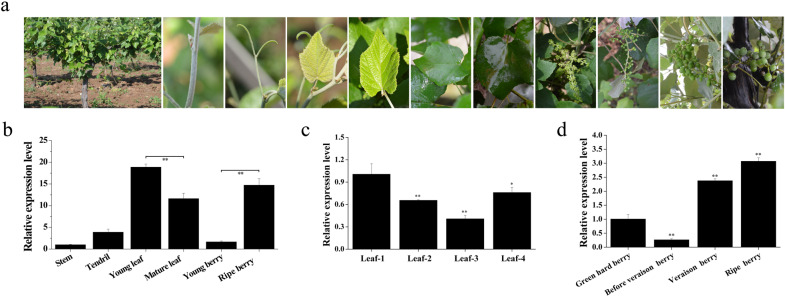
After exploring the response patterns of VqMYB154 under stress, we determined VqMYB154 transcripts in various organs of Danfeng-2 under natural conditions (Fig. 2a). In nutritive organs, including stems, tendrils, and leaves, the expression level of VqMYB154 in leaves was higher than that in other organs, and the expression level in young leaves was higher than that in mature leaves (Fig. 2b). The expression pattern of VqMYB154 showed a downward trend during leaf development (Fig. 2c). In reproductive organs containing berries at the four developmental stages, expression of VqMYB154 was higher in ripe berries than in other stages, showing an upward trend with the stage before veraison to the ripe stage (Fig. 2d). These observations indicate that young leaves and ripe berries in the Danfeng-2 grapevines are the primary sites where VqMYB154 is expressed. Furthermore, we analyzed the expression of VqSTS9, VqSTS32, and VqSTS42 in various organs of the Danfeng-2 grapevine. The results demonstrated that in nutritive organs, the three VqSTS genes showed the highest expression levels in young leaves; in reproductive organs, the three VqSTS genes were mainly expressed in ripe berries, which was similar to the expression pattern of VqMYB154. This may reflect coexpression relationships between VqMYB154 and VqSTS9, VqSTS32 and VqSTS42 under natural conditions (Supplementary Fig. S4).
Fig. 2. Differential expression of VqMYB154 in various organs of Danfeng-2 under natural conditions.
a From left to right are Danfeng-2 and its stems, tendrils, leaves during four different periods (from young to mature, leaf-1, leaf-2, leaf-3, and leaf-4), and berries of four developmental stages (green hard berry, before veraison berry, veraison berry, and ripe berry). b Expression analysis of VqMYB154 in various tissues. c Expression analysis of VqMYB154 in Danfeng-2 leaves in four different periods. d Expression analysis of VqMYB154 in Danfeng-2 berries at four developmental stages. Results are shown as the means (±SD) of three biological assays. Significance was determined with GraphPad Prism using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01)
Structure analysis and characteristics of VqMYB154
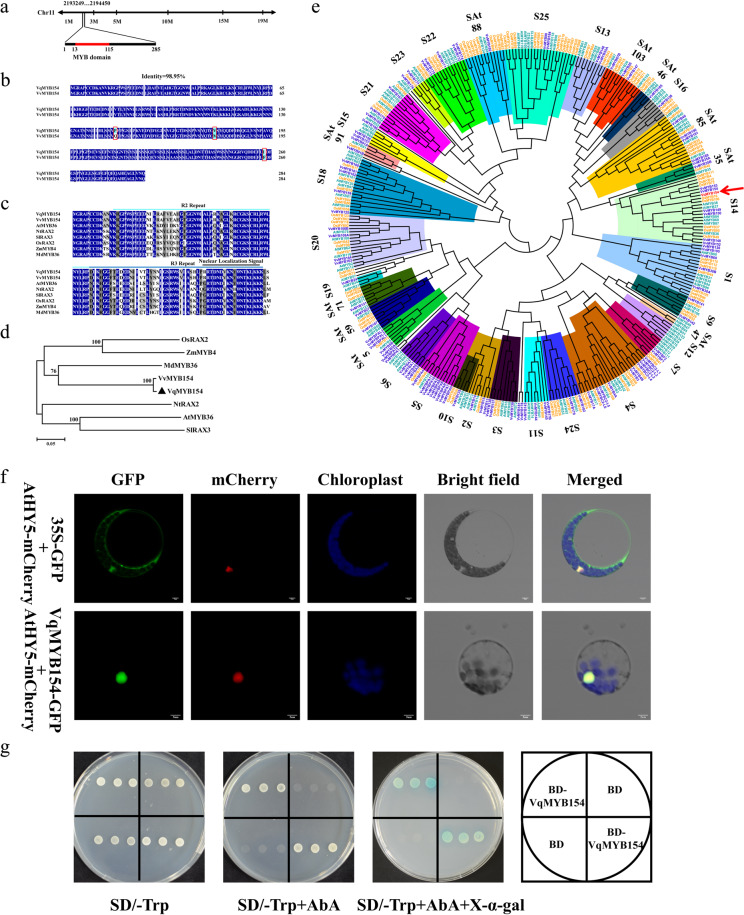
To understand the function of the resistance-related gene VqMYB154, we first isolated VqMYB154 and performed sequence analysis. The length of the VqMYB154 gDNA is 1202 bp, containing two introns at positions 137–360 bp and 491–610 bp. A BLAST search in Grape Genome Browser indicated that VqMYB154 is located on chromosome 11 (Fig. 3a). The coding sequence (CDS) of VqMYB154 was cloned from Danfeng-2. The CDS of VqMYB154 is 858 bp and encodes a 285 amino acid protein (Fig. 3a, b). VqMYB154 shares 98.95% amino acid identity with VvMYB154. Compared to VvMYB154, three mutations were found in the VqMYB154 protein, from glutamine to proline, methionine to valine, and phenylalanine to isoleucine (Fig. 3b). The N-terminus of VqMYB154 contains highly conserved R2 and R3 MYB domains (residues 13–115 aa) (Fig. 3a, c). An NLS motif is present at the N-terminus of VqMYB154, indicating its nuclear localization, and phylogenetic analysis of VqMYB154 with homologous proteins from multiple species demonstrated that it exhibits high homology with VvMYB154 and MdMYB36 (Fig. 3d). Cluster analysis of VqMYB154 with MYB subfamily proteins from the grapevine, Arabidopsis, and rice indicated that VqMYB154 is a member of subgroup 14 (S14) (Fig. 3e). VvMYB148 in this subfamily is coexpressed with STS genes48.
Fig. 3. Location and structural analysis of VqMYB154 isolated from V. quinquangularis accession Danfeng-2.
a Chromosomal location of VqMYB154. VqMYB154 is located on chromosome 11, from position 2193249 to 2194450. The MYB domain is located at the N-terminus of VqMYB154, from aa 13 to aa 115. b Amino acid sequence alignments between VqMYB154 and VvMYB154. Differences are marked with a red box. c Multiple sequence alignments among VqMYB154 and its homologs in other species. The R2 MYB domain is indicated with a light blue line, and the R3 domain is shown with a dark blue line. The sequences are from the following: VvMYB154 (XP_002280054.2), AtMYB36 (OAO95809.1), NtRAX2 (XP_016474887.1), SlRAX3 (XP_004238554.1), OsRAX2 (XP_015637875.1), ZmMYB4 (XP_008656763.1), and MdMYB36 (XP_008337621.2). d Cluster analysis of VqMYB154 with its homologous genes. VqMYB154 is marked with a black triangle. e Cluster analysis of VqMYB154 with MYB proteins in grape, Arabidopsis, and rice. VqMYB154 belongs to subgroup 14 (S14) and is highly homologous to VvMYB154, VvMYB153, OsMYB12, and OsMYB72. VqMYB154 is highlighted in red and shown with a red arrow. f The location of VqMYB154 is in the nucleus. GFP was ligated to VqMYB154 (VqMYB154-GFP), and mCherry was ligated to the Arabidopsis nuclear localization marker AtHY5 (AtHY5-mCherry). The mixed plasmids were transformed together into Arabidopsis protoplasts. 35S-GFP was adopted as the control. The GFP and mCherry signals were detected with a confocal microscope. Bars: 5 μm. g Transactivation assay of VqMYB154 in yeast. Yeast strains harboring BD-VqMYB154 and the BD control were grown on SD/-Trp, SD/-Trp with AbA, and SD/-Trp with AbA and X-α-Gal medium and cultivated at 28 °C for 3 days. Only the yeast strain of BD-VqMYB154 grew and turned blue on SD/-Trp with AbA and X-α-Gal medium
To determine the subcellular site where VqMYB154 functions, we performed a subcellular localization assay, in which the GFP signal of VqMYB154 overlapped with the mCherry signal of AtHY5, indicating that VqMYB154 is a nuclear protein (Fig. 3f). In addition, yeast cells harboring BD-VqMYB154 grew normally in all cultures and showed X-α-gal activation and AbA resistance (Fig. 3g). This indicates that VqMYB154 functions as a transcriptional activator.
The MYB154 promoter exhibits stronger pathogen-induced activity in the resistant grapevine Danfeng-2
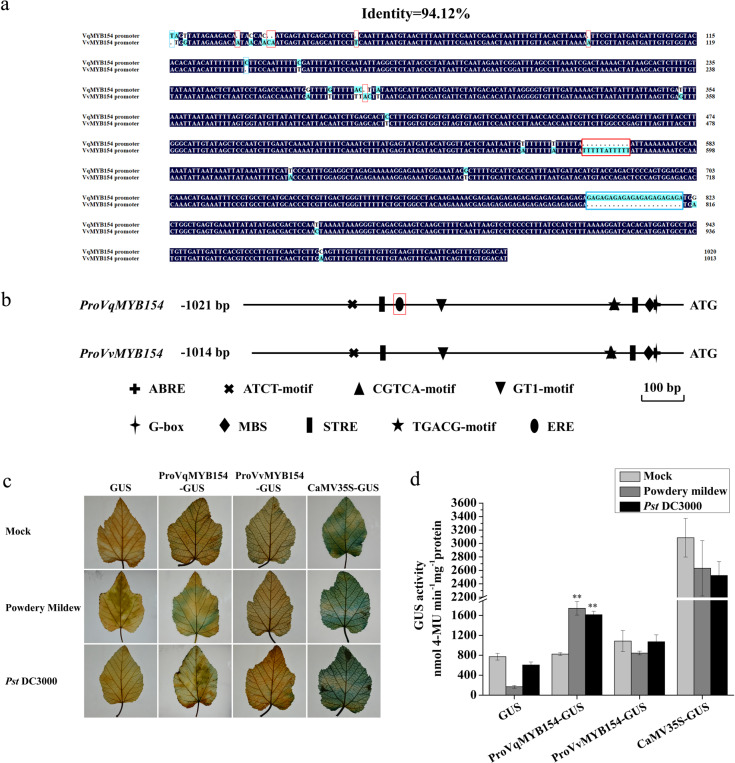
Previous results have suggested that MYB154 from disease-resistant and disease-susceptible grapevines shows different pathogen-induced response patterns. To further explore the basis for these differences in expression, we separately cloned and obtained the promoters of MYB154 in Danfeng-2 and Cabernet Sauvignon. Compared with the 1014 bp promoter of VvMYB154, we detected six deletion mutations and three insertion mutations in the 1021 bp promoter of VqMYB154 (Fig. 4a). We further discovered numerous motifs in the VqMYB154 promoter, including an MBS element, ABRE element (ABA-responsive), CGTCA motif, TGACG motif (MeJA-responsive), ERE element (ethylene-responsive), and STRE element (stress-responsive). This suggests a potential role for VqMYB154 in multiple stress responses (Fig. 4b).
Fig. 4. Expression activity analysis of MYB154 promoters between Danfeng-2 and Cabernet Sauvignon induced by U. necator.
a Sequence alignment between the VqMYB154 and VvMYB154 promoters. Deletion mutations in the VqMYB154 promoter are marked with a red box, and the insertion mutation is marked with a blue box. b Cis-element analysis between the VqMYB154 and VvMYB154 promoters. The difference in cis-element in the VqMYB154 promoter is marked with a red box. c Histochemical staining of GUS activity mediated by VqMYB154 and VvMYB154 promoters under U. necator and Pst DC3000 inoculation in grapevine leaves. Transiently transformed samples were cultivated for 48 h in a phytotron before inoculation with pathogens for 24 h. d Determination of GUS expression mediated by transient expression of VqMYB154 and VvMYB154 promoters under U. necator and Pst DC3000 inoculation in grapevine leaves. Results are shown as the means (±SD) of three biological assays. Significance was determined with GraphPad Prism using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01)
To investigate the effect of promoter differences on pathogen-induced response activity, we transiently transformed the two promoters into grape leaves and evaluated GUS activity in leaves inoculated with U. necator and P. syringae (Pst DC3000). Compared to the mock control, GUS activity driven by the VqMYB154 promoter was significantly enhanced after inoculation with U. necator and Pst DC3000, whereas GUS activity of the VvMYB154 promoter showed no significant change after pathogen infection (Fig. 4c, d). These results indicate that VqMYB154 possesses a pathogen-inducible promoter that responds to both fungal and bacterial pathogens, and the differential expression of MYB154 genes in pathogen-inoculated leaves is closely associated with the activity of their promoters.
VqMYB154 enhances the expression of STS genes and stilbenoid synthesis
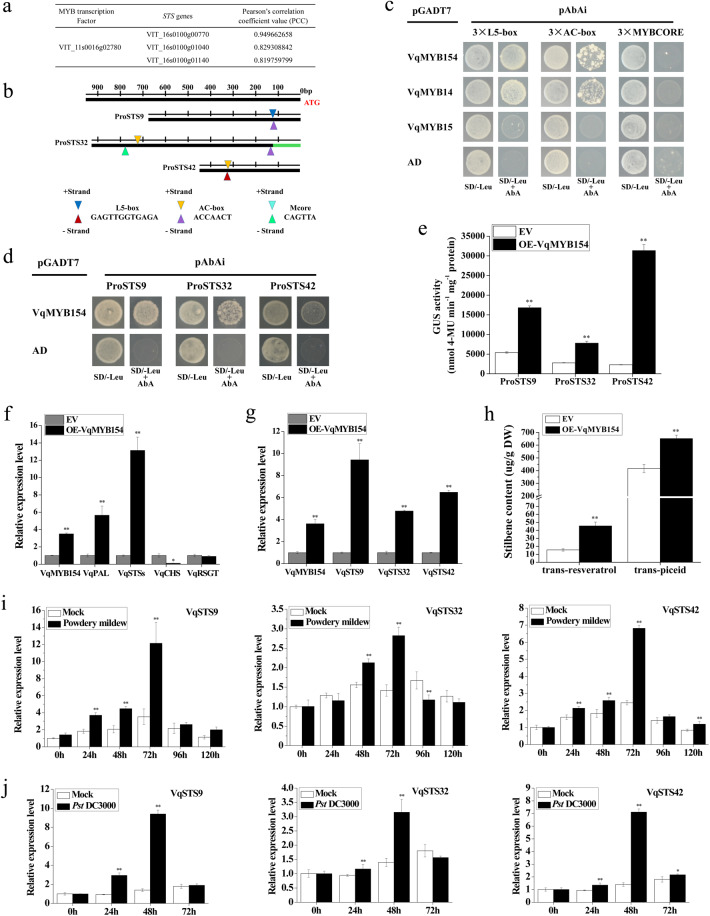
To identify the downstream target gene of VqMYB154, we conducted gene correlation analysis using Danfeng-2 transcriptome data. The Pearson correlation coefficient (PCC) was adopted as the key index for analyzing correlations49. In particular, we noticed that VqMYB154 (VIT_11s0016g02780) was coexpressed with STS9 (VIT_16s0100g00770), STS32 (VIT_16s0100g01040), and STS42 (VIT_16s0100g01140) according to high PCC indexes of 0.94, 0.82, and 0.81 (Fig. 5a), respectively. Thus, VqMYB154 may act as a regulator of the stilbene pathway. To test this hypothesis, we amplified and obtained these three promoters using Danfeng-2 genomic DNA. By analyzing the sequences, we found that the VqSTS9 promoter contains an MYB binding motif L5-box and AC-box and that the VqSTS32 promoter possesses two AC-boxes and an MYBCORE element. In addition, the VqSTS42 promoter contains an AC box and L5 box element (Fig. 5b). Therefore, we deduced that these elements are binding sites for VqMYB154. We then used a yeast one-hybrid (Y1H) assay to examine the ability of VqMYB154 to bind to these motifs. VqMYB14 and VqMYB15 from Danfeng-2, which have been proven to promote the expression of STS genes50, were used for comparison. The results indicated that VqMYB154 is able to bind to the L5-box and AC-box motifs but not the MYBCORE motif, which is consistent with the results for VqMYB14 but not VqMYB15 (Fig. 5c). A further Y1H assay demonstrated that VqMYB154 interacts with the promoters of VqSTS9, VqSTS32, and VqSTS42 (Fig. 5d). To determine whether the impact of these interactions is positive or negative, we next performed a GUS activity determination assay in tobacco. The results demonstrate that transient overexpression of VqMYB154 activated the promoters of these VqSTS genes in vivo (Fig. 5e).
Fig. 5. Expression of three VqSTS genes and the biosynthesis of stilbene phytoalexin regulated by VqMYB154.
a Coexpression of VqMYB154 and STS genes. b Graphical analysis of VqSTS9, VqSTS32, and VqSTS42. L5-box (GAGTTGGTGAGA), AC-box (ACCAACT), and MYBCORE element (CAGTTA) are MYB binding cis-elements. c A yeast one-hybrid experiment indicated that VqMYB154 has the ability to bind to L5 box and AC box elements. The CDS of VqMYB154 was integrated into pGADT7 to form AD-VqMYB154. AD-VqMYB14 and AD-VqMYB15 fusion vectors were used for the comparison of binding ability with VqMYB154. The pGADT7 vector was adopted for controls. The yeast strains harboring three tandem repeats of GAGTTGGTGAGA, ACCAACT, and CAGTTA were used as bait. Transformants were grown on medium with SD/ − Leu and AbA. d A yeast one-hybrid experiment indicating that VqMYB154 interacts with STS promoters. Y1H strains harboring the promoters of VqSTS9, VqSTS32, and VqSTS42 were used as bait. The pGADT7 vector was adopted for controls. e VqMYB154 activates VqSTS9, VqSTS32, and VqSTS42 promoters. The vectors PVqSTS9-GUS, PVqSTS32-GUS, and PVqSTS42-GUS were cotransformed with VqMYB154 into tobacco leaves. Samples were collected at 72 h after transformation. EV, the empty vector pCAMBIA1391, was used as the negative control. f Transient overexpression of VqMYB154 promotes the expression of genes related to the stilbene biosynthesis pathway. PAL, Phenylalanine ammonia lyase gene. VqSTSs represent the total STS genes in the grapevine, as amplified based on the homologous sequences of STS genes. RSGT, resveratrol glycosyl transferase gene. The chalcone synthase gene (CHS), which functions in the flavonoid pathway, was utilized as the control gene. Overexpression of VqMYB154 was performed in Agrobacterium-transformed leaves of Danfeng-2. EV, empty vector was adopted as the control. g Transient overexpression of VqMYB154 promotes expression of VqSTS9, VqSTS32, and VqSTS42. EV, the empty vector, was used as the control. h Overexpression of VqMYB154 promotes stilbenoid biosynthesis. Accumulation of trans-piceid and trans-resveratrol was determined by HPLC. i, j Expression of VqSTS9, VqSTS32, and VqSTS42 was determined by qRT-PCR. i Leaves of Danfeng-2 were inoculated with U. necator and sampled at 0, 24, 48, 72, 96, and 120 h. j Leaves of Danfeng-2 were inoculated with Pst DC3000 and collected at 0, 24, 48, 72 h. PM-Inoculation, U. necator infection; Pst DC3000, Pst DC3000 infection; Mock, control treated with sterile water. Results are shown as the means (±SD) of three biological assays. Significance was determined with GraphPad Prism using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01)
After determining the role of VqMYB154 in activating STS promoters, we investigated the effect of VqMYB154 on several node genes in the phenylalanine pathway and assessed the transcriptional levels of genes, including phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), total VqSTSs (total expression level of STS genes), and resveratrol glycosyl transferase (RSGT), by performing transient overexpression assays in Danfeng-2 leaves. The results showed that overexpression of VqMYB154 led to activation of PAL and total VqSTSs but that the CHS gene was inhibited. Expression of RSGT was unaffected by VqMYB154 (Fig. 5f). Moreover, as expected, VqSTS9, VqSTS32, and VqSTS42 were activated in VqMYB154-overexpressing leaves (Fig. 5g). The HPLC assay indicated that the contents of trans-resveratrol and trans-piceid increased 2.9-fold and 1.6-fold, respectively, after VqMYB154 overexpression (Fig. 5h). Together, VqMYB154 can promote phenylalanine metabolism and the downstream STS pathway while inhibiting the flavonoid pathway.
We further analyzed the expression patterns of VqSTS genes in Danfeng-2 leaves under pathogen inoculation and found that VqSTS9, VqSTS32, and VqSTS42 can respond to induction by U. necator. Compared to the mock control, transcriptional levels of VqSTS9 and VqSTS42 were significantly upregulated at 24 h and peaked at 72 h by 3.4-fold and 2.8-fold, respectively. The transcriptional level of VqSTS32 was increased at 48 h and peaked at 72 h by 2.0-fold (Fig. 5i). Under inoculation with Pst DC3000, expression of the three STS genes was enhanced at 24 h and peaked at 48 h by 6.7-fold, 2.2-fold and 5.1-fold, respectively. Furthermore, three VqSTS genes shared the same upregulated periods with VqMYB154 under the induction of U. necator (48 h and 72 h) and Pst DC3000 (24 h and 48 h), which indicates that VqMYB154 is coexpressed with VqSTS9, VqSTS32, and VqSTS42 under pathogen-induced conditions (Fig. 5j).
VqMYB154 is a positive regulator of resistance to Pseudomonas syringae in transgenic Arabidopsis
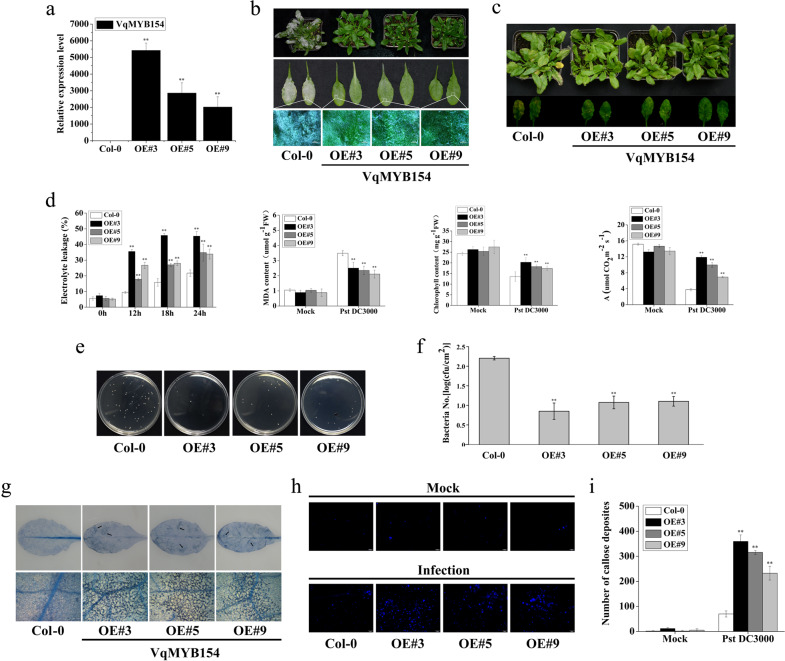
VqMYB154 and its promoter respond to induction by pathogens (Fig. 1, Fig. 4). To further explore its function in the defense response, we generated VqMYB154-overexpressing Arabidopsis lines. Wild-type (Col-0) plants and VqMYB154 transgenic lines (OE#3, OE#5, and OE#9) were used for disease assays (Fig. 6a). We first inoculated transgenic lines and Col-0 plants with Golovinomyces cichoracearum. At 168 h postinoculation, the leaves of three independent transgenic lines showed fewer fungal spores and hyphae than did Col-0 plants, indicating the transgenic lines to be more resistant to G. cichoracearum (Fig. 6b). In addition, VqMYB154-overexpressing transgenic lines showed stronger resistance to Pst DC3000. At 72 h postinoculation, the transgenic lines exhibited less severe chlorosis symptoms than did Col-0 plants (Fig. 6c). To further evaluate stress tolerance under Pst DC3000 inoculation, resistance-related physiological indexes were measured. Notably, electrolyte leakage from transgenic lines was significantly increased, higher than that of Col-0 plants, within 24 h postinoculation, which may indicate that a more intense hypersensitive reaction occurred in the transgenic lines. Furthermore, consistent with the phenotype of the inoculated plants, the transgenic lines had a lower malonaldehyde content and higher chlorophyll content and net photosynthetic rate than wild-type plants (Fig. 6d). We hypothesize that these phenotypic differences may be linked to the growth status of the pathogens in vivo. Therefore, we measured the abundance of bacteria in leaves at 72 h after Pst DC3000 inoculation and found them to be significantly lower than those of Col-0 plants (Fig. 6e, f). A trypan blue assay was performed to visualize areas of cell death in transgenic lines and Col-0 plants, showing more intense cell death at 72 h after Pst DC3000 inoculation (Fig. 6g). By staining leaf samples with aniline blue, we observed more intensive callose deposition in the transgenic lines at 72 h after Pst DC3000 inoculation than in wild-type plants. The amount of callose in the transgenic lines was also significantly increased (Fig. 6h, i). These results indicate that VqMYB154 can enhance disease resistance against Pst DC3000.
Fig. 6. Overexpression analysis of VqMYB154 in Arabidopsis under artificial inoculation with pathogens.
a VqMYB154 was overexpressed in OE#3, OE#5, and OE#9 Arabidopsis transgenic lines. b Phenotype of leaves from wild-type (Col-0) and transgenic lines (OE#3, OE#5, and OE#9) after inoculation with G. cichoracearum UCSC1 for 168 h. The figures from top to bottom are as follows: overall observation of Arabidopsis plants, close observation of leaf phenotype, and microscopic observation (Bars = 500 μm) of hyphae on the leaf surface. c Phenotype of leaves from wild-type (Col-0) and transgenic lines (OE#3, OE#5, and OE#9) after infection with Pst DC3000 (2 × 106 cfu ml−1 MgCl2 suspension) for 72 h. d Physiological indexes were measured in leaves of wild-type (Col-0) and transgenic lines (OE#3, OE#5, and OE#9) after infection with Pst DC3000 (8 × 106 cfu ml−1 MgCl2 suspension). The indexes from left to right are electrolyte leakage, malondialdehyde (MDA) content, total chlorophyll content, and assimilation rate. Electrolyte leakage was determined at 0 h, 12 h, 18 h, and 24 h, and other indexes were detected after inoculation for 72 h. The assimilation rate (A) represents the net photosynthetic rate. e, f The measurement of bacterial colonies in Arabidopsis leaf samples at 72 h after inoculation with Pst DC3000 (1 × 103 cfu ml−1 MgCl2 suspension). e Bacterial colonies from leaf samples were generated in Petri dishes, and (f) quantities were counted. g Trypan blue staining was performed to detect cell death after infection with Pst DC3000 (1 × 103 cfu ml−1 MgCl2 suspension) for 72 h. Bars: 200 μm. h Aniline blue staining was performed to detect callose deposition in the leaves 24 h after 1 × 103 cfu ml−1 Pst DC3000 inoculation. Bars: 100 μm. i Count of callose deposition in the microscopic field using ImageJ software. Results are shown as the means (±SD) of three biological assays. Significance was determined with GraphPad Prism using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01)
VqMYB154 stimulates the production of reactive oxygen species (ROS) in vivo and enhances resistance to P. syringae via the SA pathway
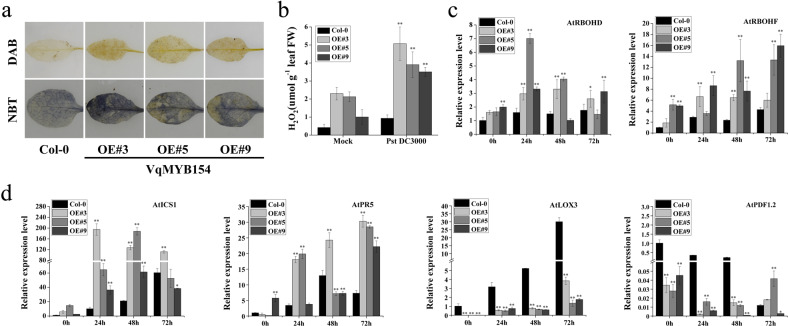
To investigate whether ROS participate in the defense response, we performed DAB and NBT staining to visualize the contents of H2O2 and O2−. We observed that more ROS accumulated in VqMYB154-overexpressing transgenic lines than in wild-type plants at 72 h after Pst DC3000 inoculation (Fig. 7a). We also quantified endogenous H2O2 content. As expected, the H2O2 content in vivo was higher in transgenic plants, which is consistent with the above results (Fig. 7b). The further qRT-PCR analysis demonstrated that transcriptional levels of the NADPH oxidase genes AtRBOHD and AtRBOHF, which are defense-related genes involved in ROS production51,52, were upregulated in transgenic plants and higher than those in Col-0 plants at 24 h and 48 h postinoculation (Fig. 7c).
Fig. 7. Overexpression of VqMYB154 in Arabidopsis stimulates ROS accumulation in vivo and enhances the expression of defense-related genes.
a DAB staining and NBT staining were utilized to detect ROS (H2O2 and O2−) production after infection with Pst DC3000 (1 × 103 cfu ml−1 MgCl2 suspension) for 72 h. From top to bottom: DAB staining and NBT staining. b Measurement of the H2O2 content in leaves after infection of Pst DC3000 (1 × 103 cfu ml−1 MgCl2 suspension) for 72 h. c qRT-PCR analysis at the transcriptional level of the NADPH oxidase genes AtRBOHD and AtRBOHF in Arabidopsis transgenic lines and wild-type at 0, 24, 48, and 72 h after 1 × 103 cfu ml−1 Pst DC3000 infection. d qRT-PCR analysis of expression levels of defense-related genes AtICS1, AtPR5, AtLOX3, and AtPDF1.2 in transgenic lines and wild-type at 0, 24, 48, and 72 h under 1 × 103 cfu ml−1 Pst DC3000 inoculation. Results are shown as the means (±SD) of three biological assays. Significance was determined with GraphPad Prism using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01)
Our results indicate that VqMYB154 can respond to the defense-related phytohormones SA and MeJA (Fig. 1f). To explore whether VqMYB154 can activate resistance signaling pathways that involve SA or JA, we analyzed SA-independent and JA-independent defense genes in transgenic lines and Col-0 plants under Pst DC3000 inoculation. AtICS1 participates in SA biosynthesis for plant defense responses53, and AtPR5 is an important SA-independent resistance-related gene54. After pathogen inoculation, the expression level of AtICS1 in transgenic lines increased at 24 h and 48 h compared to wild-type. Expression of AtPR5 in transgenic lines was more intense than that in Col-0 plants at 72 h postinoculation (Fig. 7d). However, transcript levels of AtLOX3 and AtPDF1.2, which are JA-independent defense genes55,56, were significantly lower than those of wild-type (Fig. 7d). These results demonstrate that overexpression of VqMYB154 stimulates ROS accumulation and enhances resistance to P. syringae via the SA signaling pathway.
Discussion
Grape diseases can cause yield loss and even depression in the grape industry. Disease-resistant grapevines possess unclarified grapevine-pathogen interaction mechanisms and elucidating transcription networks regulating plant resistance is vital for viticulture and vine breeding. In this study, we cloned and identified a novel resistance-related R2R3-MYB gene, VqMYB154, from the disease-resistant grapevine Danfeng-2. This research focused on elucidating its role in phytoalexin biosynthesis and defense responses. We found that pathogens are direct factors resulting in the activation of VqMYB154 and its promoter and further determined its involvement in the biosynthesis of stilbene phytoalexin by identifying its target STS genes. We then evaluated its resistance-related function using assays in VqMYB154-overexpressing Arabidopsis mutants. In summary, our results reveal that VqMYB154 positively contributes to plant defense responses.
VqMYB154 is a resistance-related transcription factor that participates in the plant defense response
The Chinese wild-growing V. quinquangularis accession Danfeng-2 shows excellent resistance to exogenous pathogens, including U. necator4. TFs involved in plant defense mechanisms can respond to pathogen induction57. In our study, VqMYB154, an MYB TF isolated from resistant Danfeng-2, significantly responded to U. necator and Pst DC3000 (Fig. 1a–c). However, its homolog in susceptible Cabernet Sauvignon is insensitive to pathogen invasion. Thus, VqMYB154 differs from VvMYB154, and there is a specific TF gene involved in the resistance mechanism of Danfeng-2. Furthermore, we found that various mutations, including insertions and deletions, exist in the VqMYB154 promoter (Fig. 4). Homologs may behave differently, even within the same species. For example, the ubiquitin ligase gene RH2 enhances resistance to powdery mildew in Chinese wild-growing grapevine Baihe-35-1 but not in susceptible cultivars because of mutations in the promoter sequence and differences in promoter elements58. OsMYBS1 binds to the promoter of C2H2-type TF Bsr-d1 in disease-resistant rice Digu to improve resistance to rice blast, whereas, in susceptible rice, such binding does not occur because of a single-nucleotide variation ‘G’ to ‘A’ in the promoter59. Therefore, we propose that the mutations between the two MYB154 genotypes are directly responsible for the different promoter activities. This is also the reason why the expression of VqMYB154 is more prominent in the disease response.
Previous studies have reported that P. syringae can trigger incompatible interactions and innate defense responses in grapevines60,61. It is speculated that Pst DC3000 is a nonadapted pathogen for grapevine. Furthermore, interactions of Pst DC3000 with grapevine induce expression of resistance-related TFs, including ERF112, ERF114, and ERF07231. Therefore, we combined U. necator and Pst DC3000 together to investigate defense-related gene expression in the grapevine. The results showed that VqMYB154 was induced and exhibited upregulated expression under artificial inoculation with Pst DC3000. This may reflect that VqMYB154 also plays a role in incompatible grapevine-pathogen interactions, which deserves further research.
VqMYB154 activates the STS pathway and enhances stilbenoid accumulation
Resveratrol is an attention-attracting metabolite because of its antimicrobial activity in plants and positive pharmacological effects on human health11,62. It is particularly abundant in grape berries and gradually accumulates from veraison to the ripe period63,64. In recent years, several MYB proteins have been identified as resveratrol regulators20,30. VviMYB14, VviMYB15, and VviMYB13, which regulate the STS pathway, belong to MYB subgroup 2. Other identified MYB genes that are coexpressed with STS genes include VviMYB139 (subgroup 3), VviMYB148 (subgroup 14), and VviMYB108A (subgroup 20)48, and VqMYB154 in this study belong to the same subfamily as VviMYB148 (Fig. 3e). According to the grapevine expression atlas from V. vinifera L. cv. Corvina, VviMYB14, and VviMYB15 are expressed during the developmental periods of berries, leaves, stems, and tendrils, while VviMYB154 is only expressed in young leaves and tendrils65. In contrast to its homolog in Corvina, VqMYB154 is expressed in multiple organs, and its mRNA accumulates significantly in veraison and ripe berries (Fig. 2). Expression levels of VqSTS9, VqSTS32, and VqSTS42 in ripe berries were higher than those in young berries (Supplementary Fig. S5). These results indicate that VqMYB154 is similar to VviMYB14 and VviMYB15 in the expression distribution in grapevine organs and provides evidence for the correlation between VqMYB154 and the STS pathway under natural conditions.
Specific binding elements act as a “bridge” between transcription factors and their target genes. In grapevines, MYB14 recognizes and binds to the L5-box element (GAGTTGGTGAGA) in the STS promoter to regulate stilbene accumulation36. In addition, the AC box (ACCA/TAA/CT/C) and MYBCORE (CAGTTA and CTGTTG) serve as cis-acting sites for MYB proteins34,35. In our study, the promoters of VqSTS9 and VqSTS42 were found to contain both an L5-box GAGTTGGTGAGA and AC-box ACCAACT. The promoter of VqSTS32 harbors the AC-box ACCAACT and MYBCORE CAGTTA (Fig. 5b). The existence of these specific sites indicates that STS genes may be regulated by MYB TFs. As a control TF for comparing binding properties, VqMYB14 can bind to the L5-box element in Danfeng-2, which is consistent with a previous study on grapevine36. Moreover, we confirmed that the AC box acts as a novel binding element for VqMYB14. Furthermore, VqMYB15 can weakly interact with the L5 box but cannot bind to the other two elements, indicating that it may bind to other cis-elements with better affinity in STS promoters. Interestingly, VqMYB154 shares the same binding preferences as VqMYB14. This suggests that VqMYB154 performs transcriptional regulatory functions in a manner similar to that of VqMYB14.
Because of their common evolutionary origin, STS and CHS compete for the same substrates, such as p-coumaroyl-CoA15. As an upstream enzyme, PAL participates in p-coumaroyl-CoA biosynthesis. Therefore, activating the PAL gene can create more substrates to be used for resveratrol biosynthesis20. Our results show that VqMYB154 not only enhances expression of the three STS genes but also upregulates VqPAL expression and downregulates that of VqCHS (Fig. 5f, g). More substrates weakened CHS competitiveness, and higher STS activity promotes the accumulation of stilbenes. As expected, we detected higher levels of trans-piceid and trans-resveratrol in VqMYB154-overexpressing grape leaves than in the control (Fig. 5h). Taken together, VqMYB154 is a transcriptional activator of resveratrol accumulation.
VqMYB154 contributes to resistance against P. syringae in transgenic Arabidopsis
Plants have developed a series of emergency mechanisms to resist pathogens. Programmed cell death (PCD) leads to acute necrosis in infected cells, interrupting pathogen spread66. To date, only a few MYB TFs have been shown to directly regulate pathogen-triggered PCD67,68. Our results showed that heterologous expression of VqMYB154 in Arabidopsis leads to more intense PCD upon Pst DC3000 infection (Fig. 6f). Moreover, we noted that regional cell necrosis is accompanied by more reactive oxygen species (ROS) generation (Fig. 7a–c). ROS, such as H2O2, can resist pathogen invasion by inducing resistance-related gene expression and further PCD69. In fact, it is commonly accepted that ROS and SA signals function synergistically in systemic acquired resistance70. Consistent with this viewpoint, we also detected upregulated expression of SA-dependent AtICS1 and AtPR5 following Pst DC3000 inoculation, indicating activation of the SA signaling pathway (Fig. 7d). Interestingly, VqMYB154 responded intensively to H2O2 induction in grapevines (Fig. 1f), and the endogenous H2O2 generated by pathogenic stimulation might further induce expression of VqMYB154, possibly constituting a positive feedback loop mediated by ROS. Furthermore, we observed that callose deposition was denser in transgenic lines upon Pst DC3000 inoculation (Fig. 6g). Overall, enhanced callose accumulation can more effectively protect cells against pathogen penetration71.
In summary, our research reveals that multiple resistance-related molecular mechanisms comprise the VqMYB154-mediated plant defense response. VqMYB154 is activated by pathogens and promotes stilbenoid biosynthesis by activating the promoters of STS genes. Moreover, VqMYB154 promotes the expression of PAL and inhibits that of CHS, further activating the resveratrol pathway. Thus, the accumulation of resveratrol contributes to plant disease resistance. In addition, VqMYB154 activates downstream resistance-related genes and ROS production, which leads to enhanced disease resistance. The ROS accumulated might further induce continuous expression of VqMYB154 and form a positive feedback pathway (Fig. 8). Overall, our research provides new insight into the mechanism of transcriptional regulation in phytoalexin metabolism and the plant defense response, offering valuable evidence for utilizing Danfeng-2 as an important resource for grapevine breeding.
Fig. 8. Proposed model of how VqMYB154 regulates stilbene phytoalexin biosynthesis and disease resistance. Pathogens induce expression of VqMYB154.
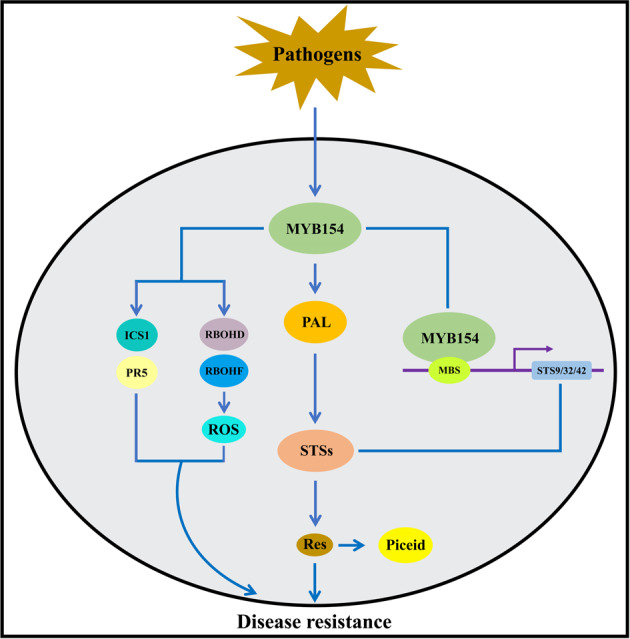
VqMYB154 can act in two pathways. First, VqMYB154 activates MBS elements in STS promoters to induce gene expression. Furthermore, VqMYB154 promotes the expression of the upstream PAL gene to provide more substrate for stilbene synthases, which biosynthesize more stilbenoids. The accumulation of stilbene phytoalexins enhances plant immunity. In addition, VqMYB154 stimulates ROS accumulation and upregulates the SA signaling-related genes AtICS1 and AtPR5. These defense-related genes and ROS accumulation also contribute to disease resistance
Materials and methods
Plant materials
Organs from Danfeng-2 and Cabernet Sauvignon were acquired from the Grape Germplasm Resources database at Northwest A&F University, Yangling, Shaanxi, China (34°20′N, 108°24′E). Grape berries were acquired during four developmental periods: green hard, before veraison, veraison, and ripe. The collection times were 25, 40, 50, and 80 days after flowering. The leaf collection periods were the same as those of berries. Based on the condition of the leaves, they were denoted Leaf-1, Leaf-2, Leaf-3, and Leaf-4. Tobacco (Nicotiana benthamiana) was cultivated in a climate chamber at 25 °C. Arabidopsis (Columbia ecotype) was cultivated in a plant incubator at 22 °C.
Gene isolation and bioinformatics analysis
cDNA from Danfeng-2 berries was used for gene amplification. With reference to the sequence of homologous genes in Pinot Noir, the VqMYB154 coding sequence (CDS) (VIT_11s0016g02780) was cloned using the primers VqMYB154-F/VqMYB154-R (Supplementary Table S1). Sequence alignment was analyzed using DNAMAN (Lynnon Biosoft, San Ramon, CA, USA). Cluster analysis was performed using MEGA 10.1.8 (Pennsylvania State University, University Park, USA) and FigTree (Andrew Rambaut, Institute of Evolutionary Biology, UK). Chromosomal localization was analyzed using Grape Genome Browser (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/). Conserved protein domains were determined on the website SMART (http://smart.embl-heidelberg.de/). The sequence of nuclear localization was analyzed using the SeqNLS website (http://mleg.cse.sc.edu/seqNLS/).
Artificial inoculation of U. necator and P. syringae under field conditions
U. necator was collected from the surface of susceptible grape leaves. Infection of Danfeng-2 and Cabernet Sauvignon leaves with U. necator was based on a previously described method4. Petioles of inoculated leaves were wrapped in moist, medical absorbent cotton and placed in flat trays with wet filter paper padded inside. The infected leaves were sampled at 0, 12, 24, 48, 72, 96, and 120 h, were wrapped in marked tin foil, immediately stored in liquid nitrogen, and placed in a cryogenic refrigerator for experiments. P. syringae (Pst DC3000) was cultivated in liquid medium72 with 25 mg/L rifampicin added in an orbital shaker (28 °C; 2 days). Healthy grape leaves were infiltrated with a suspension of Pst DC3000, and the inoculated leaves were collected at 0, 24, 48, and 72 h.
Treatments using phytohormone and abiotic stress
Leaves of Danfeng-2 and Cabernet Sauvignon were used for phytohormone treatment. SA, abscisic acid (ABA), methyl jasmonate (MeJA), and ethylene (Eth) were prepared with absolute ethanol and then diluted with double distilled water to 100 μM. Hydrogen peroxide (H2O2; 1% [w/v]) and 5 mM CaCl2 were used for the treatment of Danfeng-2 leaves. The leaves of the control group were treated with double-distilled water. Treated leaves were collected after 0, 0.5, 1, 2, 6, and 10 h.
Subcellular localization for VqMYB154 analysis
The VqMYB154 CDS (stop codon excluded) was ligated to the pCAMBIA2300 vector73, and the 35S-VqMYB154-GFP plasmid generated was used in this assay. The recombinant vector 35S-AtHY5-mCherry acted as a marker for the nucleus74. The pCAMBIA2300 vector was used as the control. The plasmids (35S-AtHY5-mCherry+35S-VqMYB154-GFP and 35S-AtHY5-mCherry+35S-GFP) were transfected into Arabidopsis leaf protoplasts and cultivated for 22 h based on a previous protocol75. GFP and mCherry signals were detected using a laser scanning confocal microscope (Olympus FV1000MPE, Tokyo, Japan). The color of the chloroplast signal in the figure is shown in blue to distinguish it from mCherry fluorescence.
Yeast transactivation assay of VqMYB154
The CDS of VqMYB154 was ligated to the pGBKT7 vector (Clontech, Mountain View, CA, USA). The BD-VqMYB154 plasmid generated was transferred into the Y2HGold strain (Clontech). The pGBKT7 vector served as the control. The transformed strains were cultured on SD/-Trp medium at 28–30 °C for 3 days, and transformants were grown on Petri dishes and cultivated at 28–30 °C for 3 days before observation. Three types of media were used: SD/-Trp, SD/-Trp with aureobasidin A (AbA), and SD/-Trp with AbA or X-α-Gal.
Yeast one-hybrid for screening promoter assays
Matchmaker™ Gold Yeast One-Hybrid System (Clontech, Palo Alto, USA) was adopted for experimental validation. The STS promoters of VqSTS9, VqSTS32, and VqSTS42 were integrated into the pAbAi vector to form pAbAi-ProVqSTS9, pAbAi-ProVqSTS32, and pAbAi-ProVqSTS42. Three tandem copy sequences of ACCAACT (AC-box), GAGTTGGTGAGA (L5-box), and CAGTTA (MYBCORE) were also integrated into the pAbAi vector. Then, linearized vectors were digested with a single endonuclease and transfected into the Y1HGold strain; the strains generated were used as bait. The CDS of VqMYB154 was integrated into pGADT7 to form AD-VqMYB154. The fusion vector was transfected into baits separately; the pGADT7 vector was also transfected into baits as a control. Transformants were grown on medium with SD/ − Leu with AbA.
Agrobacterium-mediated transient overexpression in grape leaves
The fusion vector 35S-VqMYB154-GFP and empty vector were ligated into Agrobacterium tumefaciens GV3101. The transformed strains were grown in a lysogeny broth (LB) liquid medium at 28 °C. After centrifugation, the pelleted bacteria were resuspended (OD600 = 0.6). Leaves of Danfeng-2 were immersed in a jar containing an Agrobacterium suspension. After vacuum infiltration for 30 min using a previously described method76, the samples were stored with the petioles wrapped in moist medical absorbent cotton in trays for 48 h before collection (Supplementary Fig. S2).
GUS activity analysis
The promoters of VqMYB154 and VvMYB154 were ligated into the pC0390-GUS vector, and the fusion vector was infused into GV3101 for transient expression in grape leaves77. After vacuum infiltration, the grape leaves were cultivated for 2 days and then infected with U. necator for one d before collection. GUS activity experiments were performed as previously described72. The CaMV35S-GUS vector was used as the positive control, and the negative control was the pC0390-GUS vector. For the stilbene regulation assay, the VqSTS promoters were integrated into the pC1391-GUS vector and then infused into the GV3101 strain. The vector 35S-VqMYB154-GFP was also infused into the GV3101 strain. Strains carrying various vector combinations were infiltrated into tobacco leaves based on Agrobacterium-mediated transient transformation78. After 72 h of cultivation, GUS activity was detected. The empty vector pC1391-GUS was used as a negative control. A TECAN Infinite M200 PRO Absorbance Microplate Reader (TECAN, Switzerland) was used in the above assays.
Stilbenoid extraction and analysis
The fully ground powder of grape leaves was dried at −105 °C for 24 h; the samples were weighed and then extracted in methanol (Tedia, Fairfield, USA) away from light at 4 °C overnight. The insoluble solid was discarded by low-temperature centrifugation at 4000 × g for 15 min. The clear methanol extracts were filtered through a 0.22 µm membrane film and stored in sample bottles. HPLC determination was conducted using an Agilent 1260 Infinity HPLC system (Agilent, USA). Stilbene was separated from the filtered samples (10 µL) using a binary gradient of solvent A (acetonitrile) and solvent B (ultrapure water). The wavelength for fluorimetric determination was 306 nm. The gradient conditions were based on a previous study79. The retention times were confirmed using standard samples of trans-resveratrol and trans-piceid (Sigma-Aldrich, USA). The stilbenoid concentration was determined based on the peak area.
Arabidopsis transformation and disease assays
The GV3101 strain carrying the 35S-VqMYB154-GFP construct was used for Arabidopsis transformation. T3 transgenic lines were adopted for disease assays. Four-week-old Arabidopsis leaves were infected with a suspension containing Pst DC3000 following a previously described method80. The samples were used for counting bacterial colonies after inoculation for 3 days. Leaves were acquired at 0, 24, 48, and 72 h postinfection and then used for quantitative RT-PCR. Callose deposition was observed using aniline blue. The transparent leaves decolorized with 95% ethanol were stained with aniline blue solution for 24 h and then observed under a fluorescence microscope (Olympus BX63, Tokyo, Japan) with UV irradiation. Cell death was detected using a trypan blue solution. The samples at 72 h postinfection were submerged in the solution and boiled for staining. The stained leaves were decolorized with chloral hydrate. DAB staining was performed to visualize levels of H2O2. Leaves to be observed were immersed in DAB, stained for 8 h, and then boiled in 95% ethanol for decolorization. NBT staining was used to visualize O2− levels in the leaves. Leaves at 72 h postinoculation were submerged in NBT solution for 2 h, soaked in 80% ethanol, and decolorized at 60 °C for 2 h.
To perform phenotypic analysis of Arabidopsis plants under artificial inoculation with Golovinomyces cichoracearum, the G. cichoracearum isolate UCSC1 was cultivated on pad4 Arabidopsis mutants. Four-week-old leaves were used for inoculation according to a previous method81. After inoculation for 7 days, the plants were used for phenotypic analysis.
Measurement of plant physiological indexes
Relative electrolyte leakage (REL) was analyzed following a previous method82. Approximately 0.1 g fresh leaves were immersed in 10 mL deionized water. After vacuum infiltration for 20 min, the leaves were allowed to stand for 3 h, and electrical conductivity (EC1) was recorded by a conductivity meter. Next, tubes containing leaf samples were boiled for 20 min and cooled down. The second electrical conductivity (EC2) was then recorded, and the REL ratio was determined (EC1/EC2 × 100%). The total chlorophyll content and malondialdehyde (MDA) content were analyzed as previously described83,84. The assimilation rate (A), which represents the net photosynthetic rate, was determined using a portable photosynthetic apparatus CIRAS-3 (PP Systems, USA). A commercial detection kit was used to determine H2O2 content (Suzhou Keming Bioengineering Institute, China).
Gene expression analysis by qRT-PCR
RNA from grapes and Arabidopsis was extracted according to the manufacturer’s instructions (Omega, Norcross, GA, USA). cDNA was acquired using the FastKing RT kit (Tiangen, Beijing, China). The sample mixture consisted of 7 μL sterile water, 0.8 μL each primer, 1 μL template, and 10 μL SYBR Taq (Takara, Japan). The 2−∆∆Ct method was used for calculations. Grapevine GAPDH (XM_002278316.4) and Arabidopsis Actin (AT3G18780) were used as standard controls. The assays were performed using Applied Biosystems QuantStudio 6 Flex System (Applied Biosystems, Foster City, USA). Data are presented as the mean (±SD) from three biological replicates. For assays with multiple timepoint controls, the fold change was calculated based on the rate between the treatment group and the mock control at the same timepoint. For screening assays of resistance-related MYB TFs, the fold change was acquired by comparison with the expression in inoculated samples at 0 h. The fold change of gene expression in grapevine organs was calculated by comparison with the expression level in stems. Significance analysis was conducted with GraphPad Prism 7.0 (GraphPad Software, La Jolla, USA) using one-way ANOVA with Fisher’s LSD test (*P < 0.05; **P < 0.01). The qRT-PCR primers used are listed in Supplementary Tables S1 and S2.
Supplementary information
Acknowledgements
This study was supported by the National Science Foundation of China (No. 31872055). This work was performed at the State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A&F University, Yangling 712100, Shaanxi, China. The authors thank Dr. Weirong Xu from Ningxia University for useful comments and Wiley Editing Services (https://wileyeditingservices.com/en/) for language editing, which has improved the manuscript.
Author contributions
Y.J.W. designed the study. C.Y.J. conducted the related experiments and data analysis and wrote the manuscript. D.W. and J.Z. participated in the experiments. Y.X., C.Z., J.Z., and X.W. revised the manuscript. Y.J.W. reviewed and revised the manuscript.
Conflict of interest
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00585-0.
References
- 1.Armijo G, et al. Grapevine pathogenic microorganisms: understanding infection strategies and host response scenarios. Front. Plant Sci. 2016;7:382. doi: 10.3389/fpls.2016.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deme P, Upadhyayula VV. Ultra performance liquid chromatography atmospheric pressure photoionization high resolution mass spectrometric method for determination of multiclass pesticide residues in grape and mango juices. Food Chem. 2015;173:1142–1149. doi: 10.1016/j.foodchem.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Alleweldt G, Possingham JV. Progress in grapevine breeding. Theor. Appl. Genet. 1988;75:669–673. doi: 10.1007/BF00265585. [DOI] [Google Scholar]
- 4.Wang Y, et al. Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis species. Vitis. 1995;34:159–164. [Google Scholar]
- 5.Luo S, He P. The inheritances of fruit skin and must colors in a series of interspecific and intraspecific crosses between V. vinifera and the wild grape species native to China. Sci. Hortic. 2004;99:29–40. doi: 10.1016/S0304-4238(03)00085-2. [DOI] [Google Scholar]
- 6.Takaoka M. Resveratrol, a new phenolic compound, from Veratrum grandiflorum. Nippon Kagaku Kaishi. 1939;60:1090–1100. doi: 10.1246/nikkashi1921.60.1090. [DOI] [Google Scholar]
- 7.Langcake P, Pryce RJ. The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol. Plant Pathol. 1976;9:77–86. doi: 10.1016/0048-4059(76)90077-1. [DOI] [Google Scholar]
- 8.Lee J, Rennaker C. Antioxidant capacity and stilbene contents of wines produced in the Snake River Valley of Idaho. Food Chem. 2007;105:195–203. doi: 10.1016/j.foodchem.2007.03.069. [DOI] [Google Scholar]
- 9.Park SJ, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Gong Q, Dong H, Shi J. Resveratrol, a neuroprotective supplement for Alzheimer’s disease. Curr. Pharm. Des. 2012;18:27–33. doi: 10.2174/138161212798919075. [DOI] [PubMed] [Google Scholar]
- 11.Jang M, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 12.Jeandet P, et al. Phytoalexins from the Vitaceae: biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J. Agric Food Chem. 2002;50:2731–2741. doi: 10.1021/jf011429s. [DOI] [PubMed] [Google Scholar]
- 13.Schnee S, Viret O, Gindro K. Role of stilbenes in the resistance of grapevine to powdery mildew. Physiol. Mol. Plant Pathol. 2008;72:128–133. doi: 10.1016/j.pmpp.2008.07.002. [DOI] [Google Scholar]
- 14.Hain R, et al. Disease resistance results from foreign phytoalexin expression in a novel plant. Nature. 1993;361:153–156. doi: 10.1038/361153a0. [DOI] [PubMed] [Google Scholar]
- 15.Chong J, Poutaraud A, Hugueney P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009;177:143–155. doi: 10.1016/j.plantsci.2009.05.012. [DOI] [Google Scholar]
- 16.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 17.Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M. Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 2012;12:130. doi: 10.1186/1471-2229-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu YJ, Agbayani R, Jackson MC, Tang CS, Moore PH. Expression of the grapevine stilbene synthase gene VST1 in papaya provides increased resistance against diseases caused by Phytophthora palmivora. Planta. 2004;220:241–250. doi: 10.1007/s00425-004-1343-1. [DOI] [PubMed] [Google Scholar]
- 19.Xu W, et al. VpSTS29/STS2 enhances fungal tolerance in grapevine through a positive feedback loop. Plant Cell Environ. 2019;42:2979–2998. doi: 10.1111/pce.13600. [DOI] [PubMed] [Google Scholar]
- 20.Höll J, et al. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell. 2013;25:4135–4149. doi: 10.1105/tpc.113.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velasco R, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim SD, et al. A Vitis vinifera basic helix-loop-helix transcription factor enhances plant cell size, vegetative biomass and reproductive yield. Plant Biotechnol. J. 2018;16:1595–1615. doi: 10.1111/pbi.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malabarba J, et al. The MADS-box gene Agamous-like 11 is essential for seed morphogenesis in grapevine. J. Exp. Bot. 2017;68:1493–1506. doi: 10.1093/jxb/erx025. [DOI] [PubMed] [Google Scholar]
- 24.Nicolas P, et al. The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes. Plant Physiol. 2014;164:365–383. doi: 10.1104/pp.113.231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou L, et al. Negative regulation by transcription factor VvWRKY13 in drought stress of Vitis vinifera L. Plant Physiol. Biochem. 2020;148:114–121. doi: 10.1016/j.plaphy.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Sun X, et al. The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J. Exp. Bot. 2018;69:1749–1764. doi: 10.1093/jxb/ery020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu D, et al. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019;280:132–142. doi: 10.1016/j.plantsci.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Le Henanff G, et al. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013;64:4877–4893. doi: 10.1093/jxb/ert277. [DOI] [PubMed] [Google Scholar]
- 29.Marchive C, et al. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J. Exp. Bot. 2007;58:1999–2010. doi: 10.1093/jxb/erm062. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, et al. The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2) BMC Plant Biol. 2019;19:478. doi: 10.1186/s12870-019-1993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Liu W, Wang Y. Heterologous expression of Chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to Botrytis cinerea. Plant Sci. 2020;293:110421. doi: 10.1016/j.plantsci.2020.110421. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Jiang C, Li R, Wang Y. VqbZIP1 isolated from Chinese wild Vitis quinquangularis is involved in the ABA signaling pathway and regulates stilbene synthesis. Plant Sci. 2019;287:110202. doi: 10.1016/j.plantsci.2019.110202. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant. 2015;8:689–708. doi: 10.1016/j.molp.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Shen H, et al. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. N. Phytol. 2012;193:121–136. doi: 10.1111/j.1469-8137.2011.03922.x. [DOI] [PubMed] [Google Scholar]
- 35.Ithal N, Reddy AR. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 2004;166:1505–1513. doi: 10.1016/j.plantsci.2004.02.002. [DOI] [Google Scholar]
- 36.Fang L, et al. Myb14, a direct activator of STS, is associated with resveratrol content variation in berry skin in two grape cultivars. Plant Cell Rep. 2014;33:1629–1640. doi: 10.1007/s00299-014-1642-3. [DOI] [PubMed] [Google Scholar]
- 37.Katiyar A, et al. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. 2012;13:544. doi: 10.1186/1471-2164-13-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smita S, Katiyar A, Chinnusamy V, Pandey DM, Bansal KC. Transcriptional regulatory network analysis of MYB transcription factor family genes in rice. Front. Plant Sci. 2015;6:1157. doi: 10.3389/fpls.2015.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong D, et al. A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 2016;23:451–466. doi: 10.1093/dnares/dsw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubos C, et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Marino, D. et al. Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun.4, 1–9 (2013). [DOI] [PubMed]
- 42.Zhang YL, et al. TheR2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 2019;19:362. doi: 10.1186/s12870-019-1918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo PJ, Park CM. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. N. Phytol. 2010;186:471–483. doi: 10.1111/j.1469-8137.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- 44.Ramirez V, et al. MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 2011;155:1920–1935. doi: 10.1104/pp.110.171843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, et al. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. N. Phytol. 2012;196:1155–1170. doi: 10.1111/j.1469-8137.2012.04353.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Wang Y. Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Rep. 2019;38:1347–1360. doi: 10.1007/s00299-019-02456-4. [DOI] [PubMed] [Google Scholar]
- 47.Shi J, et al. The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera) Plant Physiol. Biochem. 2014;74:24–32. doi: 10.1016/j.plaphy.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 48.Vannozzi A, et al. Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.) Plant Cell Physiol. 2018;59:1043–1059. doi: 10.1093/pcp/pcy045. [DOI] [PubMed] [Google Scholar]
- 49.Sedgwick P. Pearson’s correlation coefficient. N.Z. Med. J. 1996;109:38. [PubMed] [Google Scholar]
- 50.Wang D, Jiang C, Liu W, Wang Y. The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. J. Exp. Bot. 2020;71:3211–3226. doi: 10.1093/jxb/eraa097. [DOI] [PubMed] [Google Scholar]
- 51.Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015;56:1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- 52.Chaouch S, Queval G, Noctor G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012;69:613–627. doi: 10.1111/j.1365-313X.2011.04816.x. [DOI] [PubMed] [Google Scholar]
- 53.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 54.Blanco F, et al. Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 2009;70:79–102. doi: 10.1007/s11103-009-9458-1. [DOI] [PubMed] [Google Scholar]
- 55.Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porta H, Rocha-Sosa M. Plant lipoxygenases. physiological and molecular features. Plant Physiol. 2002;130:15–21. doi: 10.1104/pp.010787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu J, et al. ZmWRKY79 positively regulates maize phytoalexin biosynthetic gene expression and is involved in stress response. J. Exp. Bot. 2018;69:497–510. doi: 10.1093/jxb/erx436. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, et al. RING-H2-type E3 gene VpRH2 from Vitis pseudoreticulata improves resistance to powdery mildew by interacting with VpGRP2A. J. Exp. Bot. 2017;68:1669–1687. doi: 10.1093/jxb/erx033. [DOI] [PubMed] [Google Scholar]
- 59.Li W, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170:114–126.e15. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 60.Robert N, et al. Expression of grapevine chitinase genes in berries and leaves infected by fungal or bacterial pathogens. Plant Sci. 2002;162:389–400. doi: 10.1016/S0168-9452(01)00576-3. [DOI] [Google Scholar]
- 61.Bordiec S, et al. Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J. Exp. Bot. 2011;62:595–603. doi: 10.1093/jxb/erq291. [DOI] [PubMed] [Google Scholar]
- 62.Hart JH. Role of phytostilbenes in decay and disease resistance. Phytopathology. 1981;19:437–458. doi: 10.1146/annurev.py.19.090181.002253. [DOI] [Google Scholar]
- 63.Gatto P, et al. Ripening and genotype control stilbene accumulation in healthy grapes. J. Agric Food Chem. 2008;56:11773–11785. doi: 10.1021/jf8017707. [DOI] [PubMed] [Google Scholar]
- 64.Versari A, Parpinello GP, Tornielli GB, Ferrarini R, Giulivo C. Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J. Agric Food Chem. 2001;49:5531–5536. doi: 10.1021/jf010672o. [DOI] [PubMed] [Google Scholar]
- 65.Fasoli M, et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell. 2012;24:3489–3505. doi: 10.1105/tpc.112.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen B, et al. Identification, cloning and characterization of R2R3-MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive-like cell death. DNA Res. 2016;23:101–114. doi: 10.1093/dnares/dsv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vailleau F, et al. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl Acad. Sci. USA. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 70.Bolwell, G. P. & Daudi, A. Reactive Oxygen Species in Plant-Pathogen Interactions. (Springer Berlin Heidelberg, 2009).
- 71.Huckelhoven R. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 2007;45:101–127. doi: 10.1146/annurev.phyto.45.062806.094325. [DOI] [PubMed] [Google Scholar]
- 72.KING EO, WARD MK, RANEY DE. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab Clin. Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 73.Ma F, Wang L, Wang Y. Ectopic expression of VpSTS29, a stilbene synthase gene from Vitis pseudoreticulata, indicates STS presence in cytosolic oil bodies. Planta. 2018;248:89–103. doi: 10.1007/s00425-018-2883-0. [DOI] [PubMed] [Google Scholar]
- 74.Yao W, et al. VpPUB24, a novel gene from Chinese grapevine, Vitis pseudoreticulata, targets VpICE1 to enhance cold tolerance. J. Exp. Bot. 2017;68:2933–2949. doi: 10.1093/jxb/erx136. [DOI] [PubMed] [Google Scholar]
- 75.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 76.Xu W, Yu Y, Ding J, Hua Z, Wang Y. Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta. 2010;231:475–487. doi: 10.1007/s00425-009-1062-8. [DOI] [PubMed] [Google Scholar]
- 77.Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Report. 1987;5:387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- 78.Liu L, et al. An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 2010;61:893–903. doi: 10.1111/j.1365-313X.2009.04109.x. [DOI] [PubMed] [Google Scholar]
- 79.Zhou Q, Du Y, Cheng S, Li R, Wang Y. Resveratrol derivatives in four tissues of six wild Chinese grapevine species. N.Z. J. Crop Hortic. Sci. 2015;43:204–213. doi: 10.1080/01140671.2015.1010547. [DOI] [Google Scholar]
- 80.Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao S, Ellwood S, Findlay K, Oliver RP, Turner JG. Characterization of three loci controlling resistance of Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J. 1997;12:757–768. doi: 10.1046/j.1365-313X.1997.12040757.x. [DOI] [PubMed] [Google Scholar]
- 82.Cao WH, et al. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007;143:707–719. doi: 10.1104/pp.106.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H, et al. A newly isolated Na+/H+ antiporter gene, DmNHX1, confers salt tolerance when expressed transiently in Nicotiana benthamiana or stably in Arabidopsis thaliana. Plant Cell Tiss. Org. Cult. 2012;110:189–200. doi: 10.1007/s11240-012-0142-9. [DOI] [Google Scholar]
- 84.Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979;95:351–358. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.