Abstract
Polymicrobial biofilms are a hallmark of chronic wound infection. The forces governing assembly and maturation of these microbial ecosystems are largely unexplored but the consequences on host response and clinical outcome can be significant. In the context of wound healing, formation of a biofilm and a stable microbial community structure is associated with impaired tissue repair resulting in a non-healing chronic wound. These types of wounds can persist for years simmering below the threshold of classically defined clinical infection (which includes heat, pain, redness, and swelling) and cycling through phases of recurrent infection. In the most severe outcome, amputation of lower extremities may occur if spreading infection ensues. Here we take an ecological perspective to study priority effects and competitive exclusion on overall biofilm community structure in a three-membered community comprised of strains of Staphylococcus aureus, Citrobacter freundii, and Candida albicans derived from a chronic wound. We show that both priority effects and inter-bacterial competition for binding to C. albicans biofilms significantly shape community structure on both abiotic and biotic substrates, such as ex vivo human skin wounds. We further show attachment of C. freundii to C. albicans is mediated by mannose-binding lectins. Co-cultures of C. freundii and C. albicans trigger the yeast-to-hyphae transition, resulting in a significant increase in neutrophil death and inflammation compared to either species alone. Collectively, the results presented here facilitate our understanding of fungal-bacterial interactions and their effects on host-microbe interactions, pathogenesis, and ultimately, wound healing.
Subject terms: Fungi, Biofilms, Microbial ecology, Pathogenesis
Introduction
Diverse microbial communities colonize nearly every ecosystem across the human body. Within specific niches, microbe-microbe interactions can play a significant role in driving community assembly and subsequent structural and functional properties. However, the forces governing these processes within the context of tissue microenvironment and host responses are largely undefined. Although a diverse microbiome is often associated with human health [1], chronic wounds frequently harbor diverse microbial communities. An archetypal example is the diabetic foot ulcer (DFU). The development of DFUs can be attributed to numerous host-associated factors such as hyperglycemia, vascular disease, and neuropathy [2–5] leading to the colonization and assembly of a distinct and diverse wound microbiome within the tissue, often without clinical signs of infection [6–16]. The wound microbiome is hypothesized to exist as a polymicrobial biofilm and it has been shown that up to 60% of all chronic wounds contain a biofilm [17–19]. Up to 25% of all persons with diabetes will develop a DFU in their lifetime [20] equating to ~9 million people in the United States alone. Beyond the staggering healthcare costs of up to $19 billion per year, the 5-year mortality rate is between 43 and 55% and increases to 74% if an amputation occurs [21–25].
Longitudinal studies in DFU patients have demonstrated that microbial community stability, or less change over time, is associated with worse wound healing outcomes [6, 12, 14, 26]. The majority of these studies have focused on bacteria, yet fungi have been reported to be present alongside diverse bacterial communities in up to 75% of DFUs [8, 26, 27]. The presence of fungi within these communities has been shown to be associated with poorer wound outcomes and higher amounts of necrosis or dead tissue [26], suggesting that antifungal treatment may be beneficial. Thus, fungal-bacterial infection can complicate DFU treatment by requiring both antifungal and antibacterial antibiotics [27, 28]. Furthermore, both bacteria and fungi from wounds are reported to be underestimated via standard culture-based methods [7, 26], presenting an obstacle to effective diagnosis and targeted treatment. Cross-kingdom fungal-bacterial interactions are of interest as they may be critical in shaping microbial community structure and effects on physiology, pathogenesis, and host responses [29–32].
The most common fungal and bacterial species detected in DFU are Candida albicans and Staphylococcus aureus, found in 47 and 95% of DFUs respectively [6–8, 13, 15, 16, 26, 33]. Interactions between these species are synergistic and enabled via cell-cell adhesion and cross-feeding mechanisms [34–36]. Attachment of S. aureus to C. albicans in biofilms is well studied [37, 38] and serve as model for studying cross-kingdom interactions [39–42]. These data further suggest that fungi may act as keystone species that can stabilize microbial communities by providing physical scaffolding for bacterial attachment and growth [43–45]. Such networks can be highly complex and dependent on microbe-microbe interactions. For example, the Gram-negative bacterium Pseudomonas aeruginosa can have both synergistic and antagonistic effects on C. albicans, even resulting in fungal death [29, 46–50], signifying the complicated and dynamic interactions occurring within microbial communities. Furthermore, the physical orientation of fungal-bacterial biofilms suggests that their assembly and growth likely involves a temporal component [51–55].
Since DFU microbiomes are more complex, comprised of multiple species alongside C. albicans and S. aureus, and can persist for weeks or months, we hypothesize that during the community assembly and succession process, fungal and bacterial interactions, especially through priority effects (effect of early colonizers on later colonizers [56–58]) and competition, can change the physical and compositional structure of a biofilm community. To address this question, we have developed a simple community of microbes isolated from a single DFU sample with established C. albicans colonization [26]. From this sample, C. albicans was cultivated alongside S. aureus and the Gram-negative bacterium Citrobacter freundii. Here, we study community assembly in ex vivo human skin wounds and in vitro biofilm models. We show that ecological interactions, including priority effects and interbacterial competition, shape community structure and pathogenesis.
Results
Fungal-bacterial interactions alter biofilm structure and spatial organization within ex vivo human skin wounds
Unlike uniform in vitro models utilizing synthetic materials, a human skin ex vivo wound model allows us to investigate biofilm architecture of single, dual, or three-member communities across the spatially structured environment and heterogenous biotic substrate represented by human skin [59–64] using scanning electron microscopy (SEM). With this model, priority effects, or the impact of an early colonizer on a later colonizer within a community [65–67] were tested under three conditions. The first condition represents neutral or no priority, where both microbes are coinoculated simultaneously and incubated for 48 h. Then, priority effects were tested by staggering inoculation, where one partner was given priority and grown for 24 h before the second was inoculated and allowed to grow for an additional 24 h.
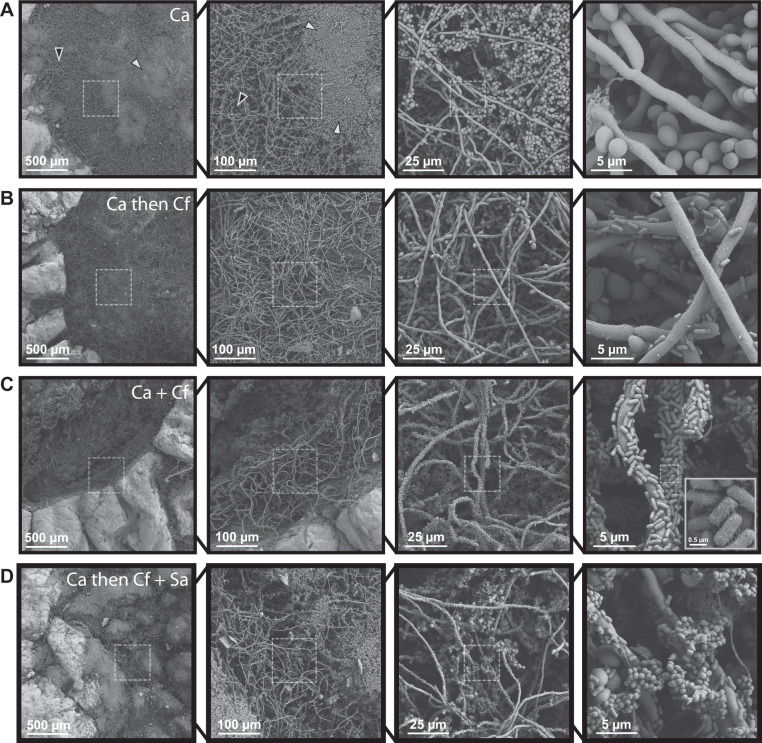
Human skin was obtained from donors undergoing elective surgery and used to create 6 mm excisional wounds within a 12 mm biopsy of full-thickness tissue. We directly observed physical interactions between C. albicans, C. freundii, and S. aureus with this human ex vivo wound model. C. albicans mono-infected wounds were covered with dense aggregates of yeast cells nested among open hyphal networks (Fig. 1A). C. freundii mono-infected wounds featured a dense layer of bacteria and small aggregates associated with collagen fibers and extracellular polymeric substances (Fig. S1A). In the S. aureus mono-infected wounds, sparse S. aureus aggregates adhering to both collagen and aggregated red blood cells were observed (Fig. S1C). We then imaged wounds co-infected with C. albicans and C. freundii under neutral priority (i.e., coinoculation). Under this condition, the wound bed was covered in extensive C. albicans hyphal networks with cells of C. freundii substantially attaching to and colonizing the fungal structures, clearly binding to C. albicans as opposed to forming clusters in the interstitial space (Fig. 1C). Furthermore, structural features such as putative pili were observed on the surface of individual rod-shaped bacterial cells (Fig. 1C inset). Collagen fibers coated in C. freundii were also clearly visible, indicating that both collagen and C. albicans are viable substrates for C. freundii attachment. In contrast to the C. albicans mono-infected wounds, few aggregates of yeast cells were observed (Fig. 1C). Under conditions giving priority to C. albicans prior to the addition of C. freundii, a similar phenotype was observed. However, fewer aggregates of yeast cells and pseudohyphae were present compared to C. albicans-only wounds (Fig. 1B). This suggests that C. freundii may trigger the C. albicans yeast-to-hyphae phenotypic transition. Conversely, when C. freundii had priority over C. albicans, no hyphae were observed, and aggregates of C. albicans yeasts were seen on dense beds of C. freundii (Fig. S1B). As expected based on the literature, when C. albicans has priority, we observed S. aureus aggregates bound to pre-formed C. albicans biofilms (Fig. S1D).
Fig. 1. Fungal-bacterial interactions and morphological heterogeneity within wound environments.
Scanning electron micrographs of ex vivo wounds at four different magnifications (×100, ×500, ×2000, ×10000). Fungal-bacterial biofilms were grown using both staggered and simultaneous inoculation models in a subset of combinations to illustrate effects of priority and interbacterial competition. Microbes were growth for up to 48 h before SEM processing in 6 mm excisional wounds on 12 mm punch biopsies of human skin suspended in a DMEM-agarose gel at 37 °C, 5% CO2. A C. albicans mono-infection. White arrowheads point to examples of yeast aggregates while black arrowheads point to hyphal networks. B C. albicans as early colonizer and C. freundii as late colonizer. C C. albicans and C. freundii simultaneously coinoculated. For imaging of putative pili on C. freundii, magnification was increased to ×20,000 as needed. D C. albicans as early colonizer and C. freundii + S. aureus as late colonizers. Dashed outlines represent region magnified.
Both S. aureus and C. freundii are reported to physically attach to C. albicans biofilms comprised of yeast and hyphae [26, 38, 40, 68]. We asked if bacteria could coadhere to the fungal scaffold, forming an integrated three-species biofilm. To test this, C. albicans biofilms were grown for 24 h, followed by the addition of both bacterial species. We observed few S. aureus aggregates adhered to C. albicans hyphae and found extensive C. freundii colonization and adhesion to both yeast and hyphal forms of C. albicans (Fig. 1D), suggesting that C. freundii may compete with S. aureus to adhere to C. albicans biofilms.
Fungal-bacterial interactions exhibit priority effects in an ex vivo human skin wound model
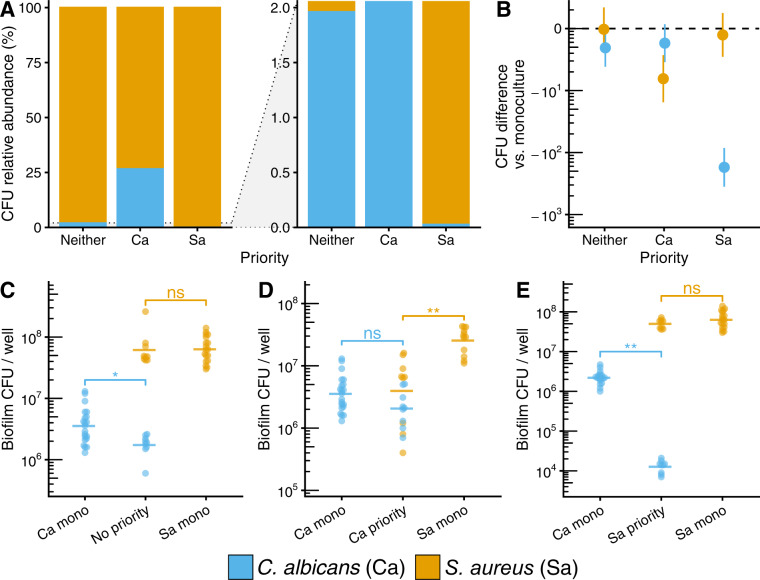
Viable cell counts were used to quantify absolute abundances and proportional abundance (i.e., community structure) of each microbe within the wound biofilms. We broadly found that priority effects led to increases in the relative abundance of the early colonizer and decreases in the late colonizer. For the C. albicans–C. freundii pairing, C. albicans proportional abundance increased tenfold from 0.056% within a neutral priority model to 0.59% when given priority to C. freundii, but decreased to 0.002% when C. freundii had priority (Fig. 2A).
Fig. 2. Fungal-bacterial interactions within ex vivo wound model.
Fungal-bacterial biofilms grown using both staggered and simultaneous inoculation models. Microbes were grown for up to 48 h in 6 mm excisional wounds on 12 mm punch biopsies of human skin suspended in a DMEM-agarose gel at 37 °C, 5% CO2. A Relative abundance (full scale and zoomed) of C. albicans, C. freundii, and S. aureus across priority effect models. Stacked bars calculated from means of CFU data shown in (C–E). B Summary of CFU differences between priority effects models and time-matched mono-cultures. Data points show median differences of microbes in co-infections to mono-infections with 95% confidence intervals calculated from CFU data shown in (C–E) for each microbe using the Mann–Whitney U test. Note that the non-parametric confidence intervals are asymmetric around the median and that C. freundii and S. aureus confidence intervals that do not include 0 are not significantly different due to multiple comparisons corrections. C C. albicans CFUs across inoculation conditions. D C. freundii CFUs across inoculation conditions. E S. aureus CFUs across inoculation conditions. Each data point represents one replicate bisect of a biopsy; horizontal bars show means of ≥6 replicates from ≥2 skin donors. * p < 0.05, ** p < 0.01.
To identify the changes in absolute microbial counts driving these compositional changes, we compared viable cell counts of mixed-cultures to time-matched mono-culture controls. Priority effects favoring a higher overall relative abundance of the early colonizer were the result of a decrease in the absolute abundance of the late colonizer while having neutral or no effect on the early colonizer. Compared to mono-cultures, C. albicans had 1.3 log10 lower cell counts when C. freundii had priority (p adj. <0.01) and 0.5 log10 lower cell counts when S. aureus had priority (p adj. < 0.05; Fig. 2B, C). We noted that C. albicans had significantly lower absolute abundances under neutral and priority conditions with C. freundii (1.3 and 0.7 log10 CFU decrease respectively, p adj. < 0.01, Fig. 2B, C), which could be due to hyphal induction (Fig. 1A–C). Across all conditions, C. freundii and S. aureus viable counts were not significantly different from mono-cultures (Fig. 2B, D, E).
For the C. albicans–S. aureus pairing, C. albicans proportional abundance was 99.98% when neutral, 99.15% when C. albicans had priority, and 99.88% when S. aureus had priority. Interestingly, S. aureus exhibited increased proportional abundance ex vivo when inoculated onto C. albicans biofilms; S. aureus proportional abundance was 0.017% when neutral, 0.12% when S. aureus had priority, and 0.85% when C. albicans had priority. Finally, we found that the tri-culture biofilms (0.36% C. albicans, 99.58% C. freundii, 0.067% S. aureus) resembled the composition of biofilms when C. albicans is given priority to C. freundii (0.59% C. albicans, 99.41% C. freundii), further suggesting that C. freundii competes with S. aureus. However, we observed high inter-donor variability in S. aureus colonization (Fig. 2E) that could not be explained by inhibition of growth in the tissue culture media (Fig. S2A). We tested S. aureus colonization of aged biopsies (5 days post-collection) and found colonization was higher than 1-day-old biopsies from the same donor utilized for our experiments (Fig. S2B). Not unexpectedly, this suggests host-factors in the local tissue environment may influence colonization in this model. Nonetheless, our results demonstrate that priority effects and inter-species competition are important factors influencing community assembly and biofilm architecture.
Priority effects alter biofilm species composition and growth interactions
To follow up on our ex vivo studies and better understand how priority effects impact community composition under controlled conditions, we used an in vitro biofilm model, first evaluating C. albicans-S. aureus interactions. We found S. aureus growth was consistent and reproducible in this model. Under the condition of neutrality, C. albicans made up 2.4% of the community, increasing to 26.6% when given priority, and decreasing 1000-fold to 0.026% as a late colonizer. When C. albicans is given priority, its absolute abundance is equivalent to a time-matched 48 h mono-culture control. However, as a late colonizer to S. aureus, C. albicans growth significantly decreased by 2.2 log10 relative to the time-matched 24 h mono-culture control (p adj. <0.0001; Fig. 3B, D, E). Similarly, S. aureus growth as an early colonizer is equivalent to the mono-culture but decreases significantly by 0.81 log10 CFU when C. albicans has priority (p adj. <0.05; Fig. 3B, D, E). When neither partner is given priority, C. albicans growth was 0.31 log10 CFU lower (p adj. <0.05), while S. aureus growth was unaffected (Fig. 3B, C). These results demonstrate that although C. albicans and S. aureus form robust mixed-species biofilm, priority effects can affect overall community composition and alter fungal-bacterial growth dynamics.
Fig. 3. C. albicans–S. aureus growth interactions are altered by priority effects.
A Relative abundance plots (full scale and zoomed) of in vitro C. albicans–S. aureus biofilms growth in RPMI-1640 media at 37 °C using both staggered and simultaneous inoculation models. Stacked bars represent means of CFU quantification shown in (C–E). B Summary of overall priority effects on mixed-species culture. Co-cultures were subtracted from time-matched mono-culture controls. Data points show mean differences with 95% confidence intervals calculated from CFU data shown in (C–E) for each microbe using a one-way ANOVA followed by Tukey’s HSD test. Differences are significant if confidence intervals do not include 0. C CFUs for C. albicans–S. aureus biofilms where C. albicans and S. aureus were inoculated simultaneously (no priority effect) and grown for 48 h, and time-matched mono-culture controls (48 h). D CFUs for C. albicans–S. aureus biofilms where C. albicans was inoculated 24 h before S. aureus (C. albicans exerts priority effect) and grown for 48 h, and time-matched mono-culture controls (C. albicans 48 h, S. aureus 24 h). E CFUs for C. albicans–S. aureus biofilms where S. aureus was inoculated 24 h before C. albicans (S. aureus exerts priority effect) and grown for 48 h, and time-matched mono-culture controls (S. aureus 48 h, C. albicans 24 h). For panels (C–E), each data point represents one replicate well; horizontal bars show means of ≥9 replicates; data are pooled from n ≥ 3 independent experiments. * p < 0.05, ** p < 0.0001, ns: not significantly different.
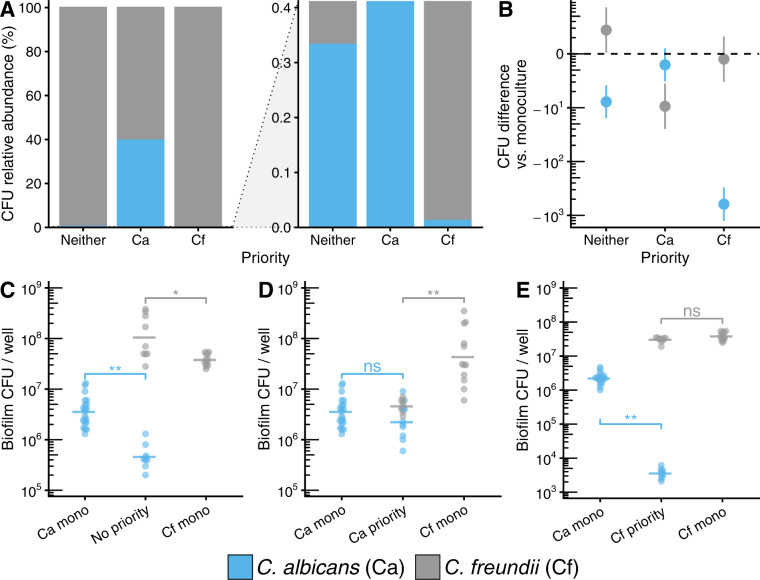
These experiments were repeated with C. albicans and C. freundii. Similarly, we found that priority effects increased relative abundance of the early colonizer (Fig. 4A), driven by a lower absolute abundance of the late colonizer (Fig. 4B). Within the C. albicans–C. freundii pairing, C. albicans made up 0.33% of the community under neutral priority with C. freundii, and increased to 39.6% with priority, with cell counts matching the mono-culture. As a late colonizer, C. albicans’s community proportion decreased to 0.012%, driven by a decrease of 2.8 log10 CFU compared to the mono-culture (p adj. <0.0001; Fig. 4B, D, E), supporting a competitive exclusion model (Fig. S1B). Under neutral priority conditions, C. freundii, made up 99.67% of the community and 99.99% as an early colonizer. However, as a late colonizer, C. freundii’s community proportion decreased to 60.4%, due to a 0.9 log10 CFU reduction as compared to the mono-culture (p adj. <0.0001; Fig. 4B, D, E). As observed with C. albicans–S. aureus interactions, priority effects can alter the composition of fungal-bacterial biofilms. Furthermore, we note that low proportional representation in a community (i.e., low relative abundance) does not necessarily correspond to a low absolute abundance. This is especially relevant for C. albicans, where a community relative abundance of less than 1% may still equate to an absolute abundance of more than 105 CFUs (Figs. 3, 4).
Fig. 4. C. albicans–C. freundii growth interactions are altered by priority effects.
A Relative abundance plots (full scale and zoomed) of in vitro C. albicans–C. freundii biofilms growth in RPMI-1640 media at 37 °C using both staggered and simultaneous inoculation models. Stacked bars calculated from means of CFU data shown in (C–E). B Summary of priority effects on growth in co-culture subtracted from time-matched mono-culture controls. Data points show mean differences with 95% confidence intervals calculated from CFU data shown in (C–E) for each microbe using a one-way ANOVA followed by Tukey’s HSD test. Differences are significant if confidence intervals do not include 0. C CFUs for C. albicans–C. freundii biofilms inoculated simultaneously (no priority effect) and grown for 48 h and time-matched mono-culture controls (48 h). D CFUs for C. albicans–C. freundii biofilms where C. albicans was inoculated 24 h before C. freundii (C. albicans exerts priority effect) and grown for 48 h, and time-matched mono-culture controls (C. albicans 48 h, C. freundii 24 h). E CFUs for C. albicans–C. freundii biofilms where C. freundii was inoculated 24 h before C. albicans (C. freundii exerts priority effect) and grown for 48 h, and time-matched mono-culture controls (C. freundii 48 h, C. albicans 24 h). For panels (C–E), each data point represents one replicate well; horizontal bars show means of ≥9 replicates; data are pooled from n ≥ 3 independent experiments. * p < 0.05, ** p < 0.0001, ns: not significantly different.
S. aureus and C. freundii compete for adhesion to C. albicans in mixed-species biofilms
To determine if bacterial competition for attachment sites to the fungal scaffold occurs as suggested by our ex vivo model, C. albicans biofilms were grown for 48 h to ensure biofilm maturity, followed by the addition of each bacterial species alone or together, with growth quantified after 24 h. When added alone, S. aureus grows to a cell density of 5.6 ± 0.7 (SD) log10 CFU/well, representing 28.3% of the community, while C. freundii grows to a density of 6.8 ± 0.2 log10 CFU/well representing 68.5% of the community. C. albicans, as the early colonizer was not affected in growth by late bacterial colonizers (Fig. 5C). When C. freundii and S. aureus are introduced simultaneously to C. albicans biofilms, S. aureus growth is reduced by 1.6 log10 CFU (p adj. <0.001), resulting in an altered community structure comprised of 0.71% S. aureus (Fig. 5A–C). Coinoculation with S. aureus does not affect the growth of C. freundii compared to C. freundii alone, offering further support of bacterial competition for fungal attachment sites. Compositionally, the tri-culture biofilms (31.9% C. albicans, 67.4% C. freundii, 0.71% S. aureus) were similar to C. albicans–C. freundii biofilms when C. albicans has priority (31.5% C. albicans, 68.5% C. freundii; Fig. 5A). Together, our results demonstrate that priority effects and inter-species competition are important factors influencing community assembly and biofilm formation.
Fig. 5. C. freundii and S. aureus compete when attaching to established C. albicans biofilms.
A Relative abundance plots (full scale and zoomed) of in vitro biofilms in RPMI-1640 media at 37 °C where bacteria (C. freundii, S. aureus, or C. freundii + S. aureus) are inoculated onto pre-established 48 h C. albicans biofilms and are allowed to grow for an additional 24 h. Stacked bars calculated from means of CFU data shown in (C). B Summary of interbacterial competition effects on growth on pre-established C. albicans biofilms subtracted from mono-colonizer controls. Data points show mean differences with 95% confidence intervals calculated from CFU data shown in (C) for each microbe using a one-way ANOVA followed by Tukey’s HSD test. Differences are significant if confidence intervals do not include 0. C CFUs for fungal-bacterial biofilms where bacteria (C. freundii, S. aureus, or C. freundii + S. aureus) are inoculated onto pre-established C. albicans 48 h biofilms. For panel (C), each data point represents one replicate well; horizontal bars show means of ≥9 replicates; data are pooled from n ≥ 3 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001. D Correlation plot between CFU counts from ex vivo and in vitro models. Data points are colored by organism for each condition and represent means with error bars showing standard deviation. Dashed line represents line where y = x. # indicates S. aureus counts in tri-culture with C. albicans and C. freundii.
We compared attachment and colonization efficiencies between in vitro conditions and ex vivo wounds and found that C. freundii and C. albicans colonize the wound tissue at cell densities similar to in vitro. S. aureus grew to lower absolute abundances ex vivo as compared to in vitro conditions across both mono- and co-cultures due to high inter-donor variability, except in the tri-culture condition (Figs. 5D, 2E).
C. freundii adheres to C. albicans via mannose-specific type I fimbriae, induces hyphae formation, and enhances neutrophil killing
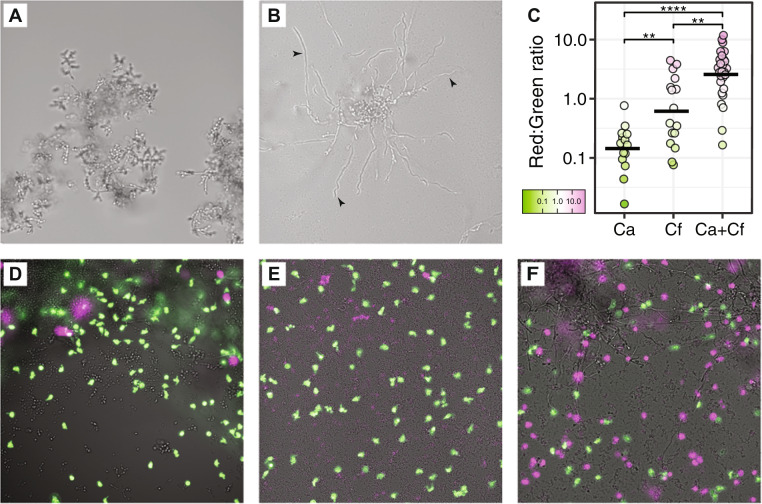
Our data suggest that C. freundii does not inhibit C. albicans growth or cause death, as is the case with other Gram-negative bacteria such as Pseudomonas aeruginosa and Acinetobacter baumanii [47, 69]. Based on our SEM observations, it appears C. freundii may alter C. albicans biofilm structure and morphology through induction of the yeast-to-hyphae transition. We used a chambered coverslip to permit observation of in vitro biofilms in situ with light microscopy. Similar to the observations in ex vivo wounds, C. albicans mono-culture biofilms exhibit a more globular phenotype primarily consisting of yeast cells and pseudohyphae (Fig. 6A). In contrast, C. albicans has a marked increase in hyphal growth when co-cultured with C. freundii (Fig. 6B). Bacterial species such as S. aureus are known to primarily bind C. albicans hyphae via protein-protein interactions, such as C. albicans Als3p that is primarily expressed in hyphae [38, 68]. Our data show C. freundii is able to bind to both yeast and hyphal cells, so we sought to determine the mechanism of C. freundii adherence to C. albicans cells. C. freundii and other members of the Enterobacteriaceae family are known to encode several pili, including type 1 fimbriae that are mannose-specific. Mannose residues exist as a core component of fungal cell wall mannans and mannoproteins, and therefore are present across all morphologies of C. albicans [70–72]. Yeast agglutination assays are used to detect and study sugar-specificities of lectin activity such as of type I fimbriae [73–76]. As a proxy for C. freundii adhesion to C. albicans within our model, we used C. albicans yeast agglutination by C. freundii cell suspensions to determine that C. albicans–C. freundii physical interactions are mannose-sensitive, and that agglutination of C. albicans by C. freundii can both be inhibited and reversed by mannose but not galactose (Fig. S3). These data support mannose-binding type I fimbriae as the likely mechanism of adhesion between C. freundii and C. albicans.
Fig. 6. C. albicans–C. freundii interactions increase hyphal induction and neutrophil death.
Mono- and co-culture biofilms of C. albicans and C. freundii were grown in RPMI-1640 media for 24 h at 37 °C in chambered coverslips set on a 30° angle to expose the biofilm edge for imaging. C. albicans has increased hyphal biofilms when co-cultured with C. freundii. A C. albicans mono-culture biofilm presents a globular phenotype. B C. albicans and C. freundii co-culture biofilm develops long C. albicans hyphae. Black arrowheads point to examples of hyphae. Micrographs are representative of at least three independent experiments. C Human neutrophils were stained with calcein AM (green) and loaded onto chambered coverslips and allowed to interact with the biofilms for 4 h. Propidium iodide (false-colored magenta) was then added to stain extracellular DNA and cells with permeabilized membranes. Panel shows C. albicans mono-culture biofilm (D) C. freundii mono-culture biofilm (E) C. albicans and C. freundii co-culture biofilm. Micrographs are representative of two independent experiments. F Image quantification of red:green fluorescence ratio. A Kruskal-Wallis test followed by pairwise Mann–Whitney U tests with the Benjamini-Hochberg correction were used to compare between groups. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
The hyphal morphology of C. albicans, along with the yeast-to-hyphae transition, are important virulence traits [77–79]. Based on the observation that C. freundii appears to promote the switch to C. albicans hyphal growth, we hypothesized that this fungal-bacterial interaction could also alter interactions with host cells. To determine if mixed-species biofilms of C. freundii and C. albicans alter the inflammatory response, we tested neutrophil responses to mono- or mixed-species biofilms.
We exposed calcein-AM-labeled human neutrophils to 24 h biofilms of C. albicans, C. freundii, and C. albicans–C. freundii co-cultures for 4 h within a chambered coverslip. We then stained for membrane-permeabilized neutrophils and extracellular DNA as a proxy for neutrophils that have released NETs. We then imaged the neutrophils across each culture condition and quantified the ratio of the area of red (dead) to green (viable) fluorescence. We found a significant increase in fluorescent staining associated with neutrophil death in the co-culture conditions, compared to both mono-culture of C. albicans (p adj. <0.0001) and C. freundii (p adj. <0.01; Fig. 6C-F). These results suggest that fungal-bacterial interactions may result in an increased neutrophil death response, leading to pro-inflammatory phenotypes.
Discussion
The structure of microbial communities in chronic wounds correlates with wound healing outcomes [6, 10, 13, 26, 80, 81]. Here, we utilized a three-member community comprised of a fungal pathogen and two bacterial species isolated from a DFU to probe community assembly and succession under different conditions. We demonstrate that cooperative and competitive species interactions are influenced by temporal factors to shape the overall community structure and pathogenicity. We show that priority effects can significantly alter the compositional structure of biofilm communities of both the well-characterized C. albicans–S. aureus pairing and the uncharacterized C. albicans–C. freundii pairing. We show these effects are consistent across both an in vitro biofilm model as well as an ex vivo human skin wound model. While fungi and bacteria both engage in niche competition for attachment to the underlying substrate such as host tissue or an abiotic surface, bacteria have the additional advantage of being able to colonize fungal cells directly. As a result, interbacterial competition occurs for attachment and colonization to fungal structures. Using SEM, we qualitatively characterized biofilm morphology and spatial organization across ex vivo human skin wounds. We found that C. freundii adhesion to C. albicans is mediated by mannose-specific binding and triggers C. albicans hyphal growth both ex vivo and in vitro. Finally, we showed that the interaction between C. freundii and C. albicans tunes neutrophil responses leading to increased cell death as compared to mono-cultures. This supports the hypothesis that mixed-species biofilms contribute to enhanced inflammation. Collectively, these results illustrate how competition during community assembly and succession processes can drastically affect community structure and subsequent host response. Our results also underscore the importance of including a temporal lens in studying microbial interactions within wound biofilms.
Physical interactions between fungi and bacteria have been shown to result in enhanced persistence, virulence, and antimicrobial resistance across different disease contexts and environments, including chronic wounds, dental caries, and the cystic fibrosis lung [26, 44, 45, 50, 55, 82–84]. However, microbial communities also change over time through the cyclical process of assembly and succession, especially after disruptions to the community. In the context of chronic wounds, this occurs through standard care procedures such as wound cleansing and mechanical disruption by debridement [81, 85–87]. We found that priority effects alter the microbial community composition between C. albicans and two different bacterial species (C. freundii and S. aureus; Figs. 1–5). Late colonizers experience negative effects with a larger fitness cost to late colonizing C. albicans. We hypothesize that this is due to the physical nature of fungal-bacterial interactions. Fungi can be an order of magnitude larger in size than a typical bacterial cell. Thus, fungi like C. albicans can play a structural role in providing bacteria with a substrate to attach to, dampening the priority effect. Conversely, bacterial priority effects exclude C. albicans, removing potential fungal attachment sites and thereby de-stabilizing the community. When C. albicans biofilms are allowed to establish, we find that bacteria compete for adherence to fungal hyphal structures with C. freundii outcompeting the professional skin pathogen S. aureus. This is an intriguing finding, suggesting community dynamics can limit the expansion of S. aureus by competitive exclusion. Over 95% of DFU are colonized by S. aureus [13], but not all progress toward a spreading infection such as osteomyelitis or systemic bacterial infection. Our findings may offer insights into interactions suppressing S. aureus abundance within the chronic wound environment, which we hypothesize may limit its pathogenicity and virulence. The potential for microbial interactions is high, given the diversity of the wound microbiome. Understanding these interactions is complex as community structure can also be shaped by external factors such as the mechanism of injury, where a blunt trauma may introduce different environmental contaminants than an ulcer forming as the result of anatomical deformities [88].
Although S. aureus showed variable growth in ex vivo skin compared in vitro culture conditions, this is expected between different patient samples and the general trends remained consistent (Figs. 1, 2, S1). Candida albicans acted as a substrate for S. aureus attachment, leading to an increase in its proportional abundance in the ex vivo model. This observation illuminates the challenges translating in vitro data into clinically relevant models. With S. aureus, differences in the models may highlight the utility of ex vivo models to study skin-derived innate immunity while isolated from circulating immune cells [60, 89]. We confirmed that media components such as complement proteins from fetal bovine serum in the tissue culture media do not inhibit growth of S. aureus (Fig. S2A). We hypothesize that both strain- and donor-specific factors may result in the observed variation in S. aureus colonization. Bacterial colonization and adhesion factors, as well as antimicrobial functions of host innate immunity, such as through antimicrobial peptides, have been shown to contribute to variations in S. aureus colonization and carriage [90–93]. We note, however, that S. aureus colonization within our ex vivo model was most consistent with in vitro results within the tri-culture condition, showing increased colonization and decreased variability compared to mono- and co-culture conditions (Fig. 5D, data point marked by #), indicating that polymicrobial interactions may add to the complexity of host-microbe colonization dynamics.
We observed that C. freundii attaches to yeast, pseudohyphal, and hyphal forms of C. albicans. In contrast, S. aureus, P. aeruginosa, and A. baumanii have all been reported to preferentially adhere to C. albicans hyphae [38, 47, 69]. This may account for the ability of C. freundii to effectively outcompete S. aureus for binding if C. albicans biofilms of clinical isolates are phenotypically heterogenous [94]. Furthermore, we found that C. freundii attachment to C. albicans cells induce hyphae formation, leading to more surface area as hyphae expand across the tissue surface to create a tangled three-dimensional network. We observed cellular appendages on C. freundii that appear to mediate the adhesion to C. albicans (Fig. 1C inset) and hypothesize that these appendages are type I pili, consistent with our finding that C. freundii induced agglutination of C. albicans yeasts can be both inhibited and reversed by mannose (Fig. S3). Mannose residues are a key component of the fungal cell wall regardless of morphology [95] and mannose-binding type I fimbriae are ubiquitous among the species in the Enterobacteriaceae family, such as C. freundii [75, 96]. Furthermore, persistence of bacteria in the Enterobacteriaceae family has been reported as a microbial marker and predictor of poor wound healing in DFUs localized to the heel of the foot [10].
We used viable cell enumeration on selective plates to quantify growth within biofilms. A limitation of this technique is that adhesive cell clusters and fungal hyphae, while functionally different compared to a single cell, will also plate as one countable colony. Although we found that C. albicans CFUs were reduced when in co-culture with C. freundii (Figs. 2, 4), the magnitude of change may not be absolute because of induced morphological differences in C. albicans due to C. freundii (Figs. 1, 6A, B). Fungal hyphae in general are difficult to quantify [97]. However, we want to note that our viable counts were consistent and reproducible. Furthermore, our use of SEM to investigate the spatial structure of biofilms supports our observations of reduced C. albicans cell counts due to hyphal induction, such as during C. albicans and C. freundii co-colonization. These observations raise the question of what might be missed if we study pairwise interactions by quantifying absolute abundances. Morphological changes in C. albicans may have a far greater impact on virulence than overall viable cell counts [77, 98–100]. We reiterate that proportional representation in a community (i.e., relative abundance) does not necessarily correspond to absolute abundance, and furthermore cannot capture physical and functional characteristics of the resulting community [101].
We found that C. albicans and C. freundii mixed biofilms increase neutrophil death. Neutrophils are among the first responders during the inflammatory phase of wound healing. Their primary role is to clean the wound of debris and contaminating microbes through phagocytosis. Neutrophils also undergo a cell death process to release neutrophil extracellular traps (NETs), which are web-like structures of DNA and antimicrobial proteins that trap and kill microbes to prevent them from spreading. Mono-cultures of C. albicans biofilms have been shown to inhibit NETosis [102] but the increased length of hyphae compared to yeast cells increases incomplete or “frustrated” phagocytosis and also alters reactive oxygen signaling in the neutrophil response [78, 103]. Attachment of C. freundii induces a morphological change in C. albicans and results in increased hyphae and neutrophil cell death. In the context of diabetes, neutrophils in both diabetic mice models and diabetic patients are primed to undergo NETosis [104, 105]. It is thought that the increased inflammation triggered by NETosis likely contributes to the delayed wound healing associated with this disease. Thus, we hypothesize that a subset of neutrophil-associated inflammation may be due to fungal-bacterial interactions.
The presence of pathogenic fungi in wounds is correlated to poorer wound healing outcomes and can complicate treatment [26–28]. A major component of the chronic wound microbiome is community stability, or the lack of, as being a key factor for predicting wound outcomes [6, 14]. Fungi such as C. albicans, although representing a small proportion of a community, provide a scaffold to colonizing bacterial species and contribute to overall community stability. We have shown that this process is affected by ecological factors such as order of arrival to a community and subsequent priority effects can drastically alter the physical and compositional structure of biofilm communities. In turn, virulence traits and host responses are altered. We anticipate as we uncover more ecological principles relevant to microbial growth and biofilm formation within wounds, a combination of bottom-up analyses building complexity within our models and top-down approaches such as metatranscriptomics will add to our understanding of the microbial impact on wound healing with positive implications for future basic and translation research.
Materials and methods
Strains and culture conditions
(1) Fungal-bacterial biofilms in 96-well plates
Isolates were grown overnight at 37 °C on yeast extract-peptone-dextrose (YPD; C. albicans) or tryptone soy (bacteria) agar plates. Inoculums were made by suspending colonies into sterile PBS followed by dilution in RPMI-1640 with 2% glucose and 0.165 M MOPS (pH 7.0) to a final cell density of 1 × 105 CFU/mL. Biofilms were grown statically at 37 °C in non-treated polystyrene 96-well plates (CC7672-7596; USA Scientific, Ocala, FL). For mono-cultures, 200 μL of inoculum was added to each well and incubated for 24 h or 48 h with fresh media replaced at 24 h. For staggered inoculation, 200 μL of inoculum containing the early colonizer was added to each well and grown statically for 24 h (or 48 h for competitive bacterial binding to C. albicans biofilms). The supernatant was gently removed and 200 μL of inoculum of the late colonizer was added to each well and grown statically for another 24 h. To harvest biofilms, media was removed and each well washed with 2 ×200 μL PBS to remove non-adherent cells. Biofilms were resuspended in 200 μL of PBS before serial dilution and spot plating 20 μL for CFU counts on selective/differential media: YPD agar with 50 μg/mL kanamycin (C. albicans), TSA with 50 μg/mL nystatin (C. freundii and S. aureus), and TSA with 7.5% NaCl and 50 μg/mL nystatin (S. aureus in tri-culture).
(2) Ex vivo human skin wound model
Human skin was obtained from patients undergoing elective reconstructive surgeries. The deidentified samples were exempt from the regulation of University of Wisconsin-Madison Human Subjects Committee Institutional Review Boards. The tissue was rinsed with PBS and partial-thickness wounds were made by puncturing the epidermis with a 6 mm biopsy punch and removing the entire epidermis and a portion of the dermis. A 12 mm biopsy punch was then used to make full-thickness biopsies with the wound. Biopsies were placed into 12-well plates containing 3 mL of a DMEM-agarose gel (0.15:0.85 ratio of 1% agarose in PBS and Dulbecco’s modified Eagle medium [DMEM] supplemented with 10% fetal bovine serum [FBS]). Biopsies were incubated at 37 °C with 5% CO2 and transferred onto new media every 48 h. Inoculums were prepared as described above and diluted to a cell density of 1 ×107 CFU/mL each. Wounds were inoculated within 24 to 48 h of tissue collection for a final cell density of 1 ×105 CFU/wound. For staggered colonization, late colonizers were inoculated as described above, 24 h after inoculation of the early colonizer. Following incubation, biopsies were processed for SEM (see below) or bisected and processed for viable cell enumeration. Bisects were vortexed in 1 mL PBS with 0.2 g of 1 mm sterile glass beads for 10 m at full-speed on a Vortex-Genie 2 (Scientific Industries, Bohemia, NY) before serial dilution and spot plating as described above. To test effect of biopsy age on S. aureus colonization, a subset of biopsies were kept sterile and uninfected for 5 days before infection. Donor skin tissue is pre-treated with antiseptics prior to collection during surgery and has been independently confirmed using qPCR and viable cell counts to have no detectable microbial load (data not shown). For each experiment, negative (uninfected) controls were maintained and transferred onto new media every 48 h to monitor for contamination.
(3) Growth curves
S. aureus inoculums were prepared as described above. Media tested include RPMI-1640 with 2% glucose and 0.165 M MOPS (pH 7.0) and DMEM. Aliquots of FBS were heat-inactivated for 30 m at 56 °C for a subset of media conditions and added to the media at 10% (v/v). Liquid cultures were grown statically at 37 °C in non-treated polystyrene 96-well plates (CC7672-7596; USA Scientific, Ocala, FL) with OD600 readings taken every 15 m over 24 h of growth on a plate reader (EPOCH2, Biotek, Winooski, VT).
Scanning electron microscopy
The following protocol was adapted from ref [106]. Briefly, ex vivo human skin wounds were rinsed with PBS and fixed overnight in 5 mL of 1.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) at 4 °C. Samples were rinsed, treated with 1% osmium tetroxide for 1 h, and then washed again. Samples were dehydrated through a series of ethanol washes (30–100%) followed by critical point drying (14 exchanges on low speed) and were subsequently mounted on aluminum stubs with a carbon adhesive tab and carbon paint. Silver paint was applied around the perimeter for improved conductivity. Samples were left to dry in a desiccator overnight. Following sputter coating with platinum to a thickness of 20 nm, samples were imaged in a scanning electron microscope (Zeiss LEO 1530-VP) at 3 kV.
Yeast-agglutination assay with sugar competition
Inoculums of C. albicans and C. freundii were made as described above. 1:10 dilutions of the suspensions were used to quantify OD600nm of 1.0~1.5, corresponding to an undiluted OD600nm of 10~15. To induce agglutination, 100 μL of each microbe and 100 μL of PBS was added to a 1.5 mL microcentrifuge tube and shaken at 175 rpm in a 37 °C incubator for 15 m. To test for inhibition of agglutination, 500 mM D-mannose or D-galactose in PBS was added. To test for reversal of agglutination, 50 μL of PBS, 500 mM D-mannose or D-galactose in PBS was added to 50 μL of the agglutinated C. albicans and C. freundii suspension in PBS and briefly vortexed to mix. Mono-culture controls were 100 μL of cell suspensions and 200 μL of PBS. Images were take using on a Nikon Eclipse E600 microscope equipped with a Leica DFC420 camera using LAS v4.12 software. Objective used was the Nikon Plan Fluor 40x using the Ph2 annulus on the sub-stage condenser.
Fluorescence imaging of neutrophil interactions in vitro
Human neutrophils were collected as previously described [102]. For fluorescent imaging, the following protocol was adapted from ref. 102. In total, 100 μL of fungal and bacterial cells in RPMI-1640 (1 × 105 cells/mL) were loaded into the wells of a tissue culture-treated μ-Slide (8 wells, ibidi, Fitchburg, WI) and grown on a 30° degree angle using a well plate stand for 24 h at 37 °C with 5% CO2. Neutrophils, stained with calcein AM (Thermo Fisher Scientific, Waltham, MA) at 0.5 µg/ml in DPBS for 10 m at room temperature in the dark, were added at a concentration of 1 × 105 cells/well and allowed to incubate flat for 4 h at 37 °C with 5% CO2. Propidium iodide (3 µM) incubated with samples for 15 m at 37 °C was used to visualize extracellular DNA and neutrophils with disrupted membranes. Images were obtained with a Nikon eclipse-TI2 inverted microscope and ORCA-Flash 4.0 LT sCMOS camera using NIS elements imaging software on bright field, FITC, and TexasRed channels using a 20x objective. Images were taken from random fields of view along the biofilm leading edge. Exposure times and linear contrast (LUTs) for each channel were fixed and consistent within each independent biological experiment. Image channels were exported separately and analyzed using FIJI [107]. Single channel images were converted to grayscale and the Auto Threshold function using the IJ-IsoData algorithm on a dark background was used to identify neutrophils. The pixel area occupied by neutrophils was calculated for each channel separately and the percent area of the red channel divided by the green channel was reported as the red:green ratio to normalize for varying amounts of neutrophils in each field of view. Brightfield, FITC, and TexasRed channels were overlaid for display within Fig. 6, with additional modifications including manual adjustment of linear contrast and conversion of the red channel to magenta for visualization and accessibility purposes.
Data and statistical analysis
Information regarding sample size and replication are described in the figure legends. All statistical analysis was performed using R [108]. Multiple comparisons and estimation of mean (parametric) or median (non-parametric) differences between inoculation conditions for each microbe were evaluated using a one-way between subjects ANOVA with Tukey’s Honest Significant Differences test or the Kruskal-Wallis test with pairwise Mann–Whitney U tests with the Benjamini-Hochberg correction for normally or non-normally distributed data, respectively. We used an α level of 0.05 for all statistical tests.
Supplementary information
Acknowledgements
We gratefully acknowledge use of facilities and instrumentation at the UW-Madison Wisconsin Centers for Nanoscale Technology (wcnt.wisc.edu) partially supported by the NSF through the University of Wisconsin Materials Research Science and Engineering Center (DMR-1720415). We also acknowledge Nate Holly for training and assistance with human tissue processing, members of the Kalan Lab for helpful feedback and discussion, and Cameron Currie for microscope access.This work was supported by grants from the National Institutes of Health (NIGMS R35 GM137828 [LRK], NIAID R01 AI145939 [JN]), UW-Madison Department of Medicine William A Craig Research Award [LRK], the Burroughs Wellcome Fund (1012299 [JN]), and the Doris Duke Charitable Foundation (2017074, [JN]).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00901-5.
References
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalan LR, Brennan MB. The role of the microbiome in nonhealing diabetic wounds. Ann NY Acad Sci. 2019;1435:79–92. doi: 10.1111/nyas.13926. [DOI] [PubMed] [Google Scholar]
- 3.de Souza Ferreira C, Araújo TH, Ângelo ML, Pennacchi PC, Okada SS, de Araújo Paula FB, et al. Neutrophil dysfunction induced by hyperglycemia: modulation of myeloperoxidase activity. Cell Biochem Funct. 2012;30:604–10. doi: 10.1002/cbf.2840. [DOI] [PubMed] [Google Scholar]
- 4.Volmer-Thole M, Lobmann R. Neuropathy and diabetic foot syndrome. Int J Mol Sci. 2016;17:917. doi: 10.3390/ijms17060917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prompers L, Schaper N, Apelqvist J, Edmonds M, Jude E, Mauricio D, et al. Prediction of outcome in individuals with diabetic foot ulcers: focus on the differences between individuals with and without peripheral arterial disease. The EURODIALE Study. Diabetologia. 2008;51:747–55. doi: 10.1007/s00125-008-0940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loesche M, Gardner SE, Kalan L, Horwinski J, Zheng Q, Hodkinson BP, et al. Temporal stability in chronic wound microbiota is associated with poor healing. J Invest Dermatol. 2017;137:237–44. doi: 10.1016/j.jid.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhoads DD, Wolcott RD, Sun Y, Dowd SE, Rhoads DD, Wolcott RD, et al. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci. 2012;13:2535–50. doi: 10.3390/ijms13032535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowd SE, Delton Hanson J, Rees E, Wolcott RD, Zischau AM, Sun Y, et al. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J Wound Care. 2011;20:40–47. doi: 10.12968/jowc.2011.20.1.40. [DOI] [PubMed] [Google Scholar]
- 9.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloan TJ, Turton JC, Tyson J, Musgrove A, Fleming VM, Lister MM, et al. Examining diabetic heel ulcers through an ecological lens: microbial community dynamics associated with healing and infection. J Med Microbiol. 2019;68:230–40. doi: 10.1099/jmm.0.000907. [DOI] [PubMed] [Google Scholar]
- 11.min KR, Galvis A, Nole KLB, Sinha R, Clarke J, Kirsner RS, et al. Association between baseline abundance of Peptoniphilus, a Gram-positive anaerobic coccus, and wound healing outcomes of DFUs. PLoS One. 2020;15:1–20. doi: 10.1371/journal.pone.0227006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter P, Greco E, Cross K, Perry J. Topical oxygen therapy shifts microbiome dynamics in chronic diabetic foot ulcers. Wounds. 2020;32:81–85. [PubMed] [Google Scholar]
- 13.Kalan LR, Meisel JS, Loesche MA, Horwinski J, Soaita I, Chen X, et al. Strain- and species-level variation in the microbiome of diabetic wounds Is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe. 2019;25:641–55. doi: 10.1016/j.chom.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tipton CD, Mathew ME, Wolcott RA, Wolcott RD, Kingston T, Phillips CD. Temporal dynamics of relative abundances and bacterial succession in chronic wound communities. Wound Repair Regen. 2017;25:673–9. doi: 10.1111/wrr.12555. [DOI] [PubMed] [Google Scholar]
- 15.Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016;24:163–74. doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- 16.Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62:923–30. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malone M. The microbiome of diabetic foot ulcers and the role of biofilms. In: The microbiology of skin, soft tissue, bone and joint infections. Cambridge, MA, USA: Elsevier Academic Press; 2017, pp 41–56.
- 18.Johani K, Malone M, Jensen S, Gosbell I, Dickson H, Hu H, et al. Microscopy visualisation confirms multi-species biofilms are ubiquitous in diabetic foot ulcers. Int Wound J. 2017;14:1160–9. doi: 10.1111/iwj.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Percival SL, Malone M, Mayer D, Salisbury A-M, Schultz G. Role of anaerobes in polymicrobial communities and biofilms complicating diabetic foot ulcers. Int Wound J. 2018;15:776–82. doi: 10.1111/iwj.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh N, Armstrong DG, Lipsky BA. Preventing foot ulcers in patients with diabetes. JAMA. 2005;293:217–28. doi: 10.1001/jama.293.2.217. [DOI] [PubMed] [Google Scholar]
- 21.Robbins JM, Strauss G, Aron D, Long J, Kuba J, Kaplan Y. Mortality rates and diabetic foot ulcers: is it time to communicate mortality risk to patients with diabetic foot ulceration? J Am Podiatr Med Assoc. 2008;98:489–93. doi: 10.7547/0980489. [DOI] [PubMed] [Google Scholar]
- 22.Rice JB, Desai U, Cummings AKG, Birnbaum HG, Skornicki M, Parsons NB. Burden of diabetic foot ulcers for Medicare and private insurers. Diabetes Care. 2014;37:651–8. doi: 10.2337/dc13-2176. [DOI] [PubMed] [Google Scholar]
- 23.Olsson M, Järbrink K, Divakar U, Bajpai R, Upton Z, Schmidtchen A, et al. The humanistic and economic burden of chronic wounds: a systematic review. Wound Repair Regen. 2019;27:114–25. doi: 10.1111/wrr.12683. [DOI] [PubMed] [Google Scholar]
- 24.Raghav A, Khan ZA, Labala RK, Ahmad J, Noor S, Mishra BK. Financial burden of diabetic foot ulcers to world: a progressive topic to discuss always. Ther Adv Endocrinol Metab. 2018;9:29–31. doi: 10.1177/2042018817744513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nussbaum SR, Carter MJ, Fife CE, DaVanzo J, Haught R, Nusgart M, et al. An economic evaluation of the impact, cost, and Medicare policy implications of chronic nonhealing wounds. Value Heal. 2018;21:27–32. doi: 10.1016/j.jval.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, et al. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. mBio. 2016;7:e01058–16. doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chellan G, Neethu K, Varma AK, Mangalanandan TS, Shashikala S, Dinesh KR, et al. Targeted treatment of invasive fungal infections accelerates healing of foot wounds in patients with Type 2 diabetes. Diabet Med. 2012;29:255–62. doi: 10.1111/j.1464-5491.2012.03574.x. [DOI] [PubMed] [Google Scholar]
- 28.Townsend EM, Sherry L, Kean R, Hansom D, Mackay WG, Williams C, et al. Implications of antimicrobial combinations in complex wound biofilms containing fungi. Antimicrob Agents Chemother. 2017;61:e00672–17. doi: 10.1128/AAC.00672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien S, Fothergill JL. The role of multispecies social interactions in shaping pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol Lett. 2017;364:1–10. doi: 10.1093/femsle/fnx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewin GR, Stacy A, Michie KL, Lamont RJ, Whiteley M. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. Proc Natl Acad Sci USA. 2019;116:19685–94. doi: 10.1073/pnas.1907619116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner KH, Stacy A, Whiteley M, Murray JL, Connell JL. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52:188–99. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chellan G, Shivaprakash S, Ramaiyar SK, Varma AK, Varma N, Sukumaran MT, et al. Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with Type 2 diabetes. J Clin Microbiol. 2010;48:2097–102. doi: 10.1128/JCM.02035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Förster TM, Mogavero S, Dräger A, Graf K, Polke M, Jacobsen ID, et al. Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell Microbiol. 2016;18:1709–15. doi: 10.1111/cmi.12657. [DOI] [PubMed] [Google Scholar]
- 36.Bertolini M, Dongari-Bagtzoglou A. The dysbiosis and inter-kingdom synergy model in oropharyngeal candidiasis, a new perspective in pathogenesis. J Fungi. 2019;5:87. doi: 10.3390/jof5040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, Filler SG, et al. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiol. 2015;161:168–81. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. Microbial interactions and differential protein expression in Staphylococcus aureus -Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol. 2010;59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio. 2016;7:1–12. doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, Burgess KE, et al. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todd OA, Noverr MC, Peters BM. Candida albicans impacts staphylococcus aureus alpha-toxin production via extracellular alkalinization. mSphere. 2019;4:1–12. doi: 10.1128/mSphere.00780-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Todd OA, Fidel PL, Harro JM, Hilliard JJ, Tkaczyk C, Sellman BR, et al. Candida albicans augments Staphylococcus aureus virulence by engaging the Staphylococcal agr quorum sensing system. mBio. 2019;10:1–16. doi: 10.1128/mBio.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, et al. Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 44.Tipton L, Müller CL, Kurtz ZD, Huang L, Kleerup E, Morris A, et al. Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome. 2018;6:12. doi: 10.1186/s40168-017-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio. 2016;7:e01250–16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergeron AC, Seman BG, Hammond JH, Archambault LS, Hogan DA, Wheeler RT. Candida albicans and Pseudomonas aeruginosa interact to enhance virulence of mucosal infection in transparent zebrafish. Infect Immun. 2017;85:1–18. doi: 10.1128/IAI.00475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science (80-) 2002;296:2229–32. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 48.Hogan DA, Vik Å, Kolter R. A pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–23. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 49.Méar J-B, Kipnis E, Faure E, Dessein R, Schurtz G, Faure K, et al. Candida albicans and Pseudomonas aeruginosa interactions: More than an opportunistic criminal association? Médecine Mal Infect. 2013;43:146–51. doi: 10.1016/j.medmal.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Bisht K, Baishya J, Wakeman CA. Pseudomonas aeruginosa polymicrobial interactions during lung infection. Curr Opin Microbiol. 2020;53:1–8. doi: 10.1016/j.mib.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wake N, Asahi Y, Noiri Y, Hayashi M, Motooka D, Nakamura S, et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. npj Biofilms Microbiomes. 2016;2:16018. doi: 10.1038/npjbiofilms.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douterelo I, Fish KE, Boxall JB. Succession of bacterial and fungal communities within biofilms of a chlorinated drinking water distribution system. Water Res. 2018;141:74–85. doi: 10.1016/j.watres.2018.04.058. [DOI] [PubMed] [Google Scholar]
- 53.De Tender C, Devriese LI, Haegeman A, Maes S, Vangeyte J, Cattrijsse A, et al. Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ Sci Technol. 2017;51:7350–60. doi: 10.1021/acs.est.7b00697. [DOI] [PubMed] [Google Scholar]
- 54.Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci USA. 2020;117:12375–86. doi: 10.1073/pnas.1919099117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, Koo H. Spatial design of polymicrobial oral biofilm in its native disease state. J Dent Res. 2020;99:597–603. doi: 10.1177/0022034520909313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Gremberghe I, Vanormelingen P, Van Der Gucht K, Souffreau C, Vyverman W, De Meester L. Priority effects in experimental populations of the cyanobacterium Microcystis. Environ Microbiol. 2009;11:2564–73. doi: 10.1111/j.1462-2920.2009.01981.x. [DOI] [PubMed] [Google Scholar]
- 57.Peay KG, Belisle M, Fukami T. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc R Soc B Biol Sci. 2012;279:749–58. doi: 10.1098/rspb.2011.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsen NMC, Røder HL, Russel J, Madsen JS, Sørensen SJ, Burmølle M. Priority of early colonizers but no effect on cohabitants in a synergistic biofilm community. Front Microbiol. 2019;10:1–8. doi: 10.3389/fmicb.2019.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts AEL, Kragh KN, Bjarnsholt T, Diggle SP. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 2015;427:3646–61. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Yoon DJ, Fregoso DR, Nguyen D, Chen V, Strbo N, Fuentes JJ, et al. A tractable, simplified ex vivo human skin model of wound infection. Wound Repair Regen. 2019;27:421–5. doi: 10.1111/wrr.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kucera J, Sojka M, Pavlik V, Szuszkiewicz K, Velebny V, Klein P. Multispecies biofilm in an artificial wound bed—A novel model for in vitro assessment of solid antimicrobial dressings. J Microbiol Methods. 2014;103:18–24. doi: 10.1016/j.mimet.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 62.Sun Y, Smith E, Wolcott R, Dowd SE. Propagation of anaerobic bacteria within an aerobic multi-species chronic wound biofilm model. J Wound Care. 2009;18:426–31. doi: 10.12968/jowc.2009.18.10.44604. [DOI] [PubMed] [Google Scholar]
- 63.Roche ED, Woodmansey EJ, Yang Q, Gibson DJ, Zhang H, Schultz GS, et al. Cadexomer iodine effectively reduces bacterial biofilm in porcine wounds ex vivo and in vivo. Int Wound J. 2019;16:674–83. doi: 10.1111/iwj.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One. 2011;6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukami T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23. [Google Scholar]
- 66.Fukami T, Mordecai EA, Ostling A. A framework for priority effects. J Veg Sci. 2016;27:655–7. [Google Scholar]
- 67.Drake JA. Community-assembly mechanics and the structure of an experimental species ensemble. Am Nat. 1991;137:1–26. [Google Scholar]
- 68.Peters BM, Ovchinnikova ES, Krom BP, Schlecht LM, Zhou H, Hoyer LL, et al. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiol. 2012;158:2975–86. doi: 10.1099/mic.0.062109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Mylonakis E. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105:14585–90. doi: 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Machová E, Fiačanová L, Čížová A, Korcová J. Mannoproteins from yeast and hyphal form of Candida albicans considerably differ in mannan and protein content. Carbohydr Res. 2015;408:12–17. doi: 10.1016/j.carres.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 71.Shibata N, Suzuki A, Kobayashi H, Okawa Y. Chemical structure of the cell-wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem J. 2007;404:365–72. doi: 10.1042/BJ20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burnham-Marusich AR, Hubbard B, Kvam AJ, Gates-Hollingsworth M, Green HR, Soukup E, et al. Conservation of mannan synthesis in fungi of the Zygomycota and Ascomycota reveals a broad diagnostic target. mSphere. 2018;3:1–14. doi: 10.1128/mSphere.00094-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abraham SN, Sun D, Dale JB, Beachey EH. Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature. 1988;336:682–4. doi: 10.1038/336682a0. [DOI] [PubMed] [Google Scholar]
- 74.Ofek I, Mirelman D, Sharon N. Adherence of escherichia coli to human mucosal cells mediated by mannose receptors. Nature. 1977;265:623–5. doi: 10.1038/265623a0. [DOI] [PubMed] [Google Scholar]
- 75.Mirelman D, Altmann G, Eshdat Y. Screening of bacterial isolates for mannose-specific lectin activity by agglutination of yeasts. J Clin Microbiol. 1980;11:328–31. doi: 10.1128/jcm.11.4.328-331.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mrázková J, Malinovská L, Wimmerová M. Microscopy examination of red blood and yeast cell agglutination induced by bacterial lectins. PLoS One. 2019;14:1–23. doi: 10.1371/journal.pone.0220318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–48. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 78.Lewis LE, Bain JM, Lowes C, Gillespie C, Rudkin FM, Gow NAR, et al. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 2012;8:e1002578. doi: 10.1371/journal.ppat.1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukaremera L, Lee KK, Mora-Montes HM, Gow NAR. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front Immunol. 2017;8:1–12. doi: 10.3389/fimmu.2017.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacDonald A, Brodell JD, Daiss JL, Schwarz EM, Oh I. Evidence of differential microbiomes in healing versus non‐healing diabetic foot ulcers prior to and following foot salvage therapy. J Orthop Res. 2019;37:1596–603. doi: 10.1002/jor.24279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verbanic S, Shen Y, Lee J, Deacon JM, Chen IA. Microbial predictors of healing and short-term effect of debridement on the microbiome of chronic wounds. npj Biofilms Microbiomes. 2020;6:21. doi: 10.1038/s41522-020-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He J, Kim D, Zhou X, Ahn S-J, Burne RA, Richards VP, et al. RNA-Seq reveals enhanced sugar metabolism in Streptococcus mutans co-cultured with Candida albicans within mixed-species biofilms. Front Microbiol. 2017;8:1036. doi: 10.3389/fmicb.2017.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017;13:e1006407. doi: 10.1371/journal.ppat.1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Dijck P, Jabra-Rizk MA. Fungal–Bacterial Interactions: In Health and Disease. In: Candida albicans: Cellular and Molecular Biology. Springer International Publishing: Cham, 2017, pp 115–43.
- 85.Johani K, Malone M, Jensen SO, Dickson HG, Gosbell IB, Hu H, et al. Evaluation of short exposure times of antimicrobial wound solutions against microbial biofilms: from in vitro to in vivo. J Antimicrob Chemother. 2018;73:494–502. doi: 10.1093/jac/dkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolcott RD, Rumbaugh KP, James G, Schultz G, Phillips P, Yang Q, et al. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J Wound Care. 2010;19:320–8. doi: 10.12968/jowc.2010.19.8.77709. [DOI] [PubMed] [Google Scholar]
- 87.Black CE, Costerton JW. Current concepts regarding the effect of wound microbial ecology and biofilms on wound healing. Surg Clin North Am. 2010;90:1147–60. doi: 10.1016/j.suc.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 88.Bartow-McKenney C, Hannigan GD, Horwinski J, Hesketh P, Horan AD, Mehta S, et al. The microbiota of traumatic, open fracture wounds is associated with mechanism of injury. Wound Repair Regen. 2018;26:127–35. doi: 10.1111/wrr.12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller LS, Cho JS. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol. 2011;11:505–18. doi: 10.1038/nri3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Midorikawa K, Ouhara K, Komatsuzawa H, Kawai T, Yamada S, Fujiwara T, et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, β-Defensins and CAP18, expressed by human keratinocytes. Infect Immun. 2003;71:3730–9. doi: 10.1128/IAI.71.7.3730-3739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mulcahy ME, McLoughlin RM. Host–bacterial crosstalk determines Staphylococcus aureus nasal colonization. Trends Microbiol. 2016;24:872–86. doi: 10.1016/j.tim.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 92.Sollid JUE, Furberg AS, Hanssen AM, Johannessen M. Staphylococcus aureus: determinants of human carriage. Infect Genet Evol. 2014;21:531–41. doi: 10.1016/j.meegid.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 93.Johannessen M, Sollid JE, Hanssen A-M. Host- and microbe determinants that may influence the success of S. aureus colonization. Front Cell Infect Microbiol. 2012;2:56. doi: 10.3389/fcimb.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kean R, Delaney C, Rajendran R, Sherry L, Metcalfe R, Thomas R, et al. Gaining insights from Candida biofilm heterogeneity: one size does not fit all. J Fungi. 2018;4:12. doi: 10.3390/jof4010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meyer-Wentrup F, Cambi A, Figdor CG, Adema GJ. Immunology of fungal infections. Netherlands: Springer; 2007. Detection of fungi by mannose-based recognition receptors; pp. 293–307. [Google Scholar]
- 96.Jones CH, Pinkner JS, Roth R, Heuser J, Nicholes AV, Abraham SN, et al. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–5. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clemons KV, Stevens DA. Conventional or molecular measurement of Aspergillus load. Med Mycol. 2009;47:S132–S137. doi: 10.1080/13693780802213340. [DOI] [PubMed] [Google Scholar]
- 98.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–24. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 99.Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2011;10:112–22. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gow NAR, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol. 2012;15:406–12. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 101.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:1–6. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johnson CJ, Cabezas-Olcoz J, Kernien JF, Wang SX, Beebe DJ, Huttenlocher A, et al. The extracellular matrix of Candida albicans biofilms impairs formation of neutrophil extracellular traps. PLoS Pathog. 2016;12:e1005884. doi: 10.1371/journal.ppat.1005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Warnatsch A, Tsourouktsoglou TD, Branzk N, Wang Q, Reincke S, Herbst S, et al. Reactive oxygen species localization programs inflammation to clear microbes of different size. Immunity. 2017;46:421–32. doi: 10.1016/j.immuni.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, et al. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med. 2015;21:815–9. doi: 10.1038/nm.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fadini GP, Menegazzo L, Rigato M, Scattolini V, Poncina N, Bruttocao A, et al. NETosis delays diabetic wound healing in mice and humans. Diabetes. 2016;65:1061–71. doi: 10.2337/db15-0863. [DOI] [PubMed] [Google Scholar]
- 106.Horton MV, Johnson CJ, Kernien JF, Patel TD, Lam BC, Cheong JZA, et al. Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. mSphere. 2020;5:1–8. doi: 10.1128/mSphere.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.R Core Team. R: A language and environment for statistical computing. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.