Abstract
This study was conducted to investigate host–microbiota interactions and explore the effects of maternal gut microbiota transplantation on the growth and intestinal functions of newborns in a germ-free (GF) pig model. Twelve hysterectomy-derived GF Bama piglets were reared in 6 sterile isolators. Among them, 6 were considered as the GF group, and the other 6 were orally inoculated with healthy sow fecal suspension as fecal microbiota transplanted (FMT) group. Another 6 piglets from natural birth were regarded as the conventional (CV) group. The GF and FMT groups were hand-fed with Co60-γ-irradiated sterile milk powder, while the CV group was reared by lactating Bama sows. All groups were fed for 21 days. Then, all piglets and then were switched to sterile feed for another 21 days. Results showed that the growth performance, nutrient digestibility, and concentrations of short-chain fatty acids in the GF group decreased (P < 0.05). Meanwhile, the serum urea nitrogen concentration and digesta pH values in the GF group increased compared with those in the FMT and CV groups (P < 0.05). Compared with the CV group, the GF group demonstrated upregulation in the mRNA expression levels of intestinal barrier function-related genes in the small intestine (P < 0.05). In addition, the mRNA abundances of intestinal development and absorption-related genes in the small intestine and colon were higher in the GF group than in the CV and FMT groups (P < 0.05). The FMT group exhibited greater growth performance, lipase activity, and nutrient digestibility (P < 0.05), higher mRNA expression levels of intestinal development and barrier-related genes in the small intestine (P < 0.05), and lower mRNA abundances of pro-inflammatory factor in the colon and jejunum (P < 0.05) than the CV group. In conclusion, the absence of gut microbes impaired the growth and nutrient digestibility, and healthy sow gut microbiota transplantation increased the growth and nutrient digestibility and improved the intestinal development and barrier function of newborn piglets, indicating the importance of intestinal microbes for intestinal development and functions.
Keywords: Germ-free, Host microbiota interaction, Maternal gut microbiota transplantation, Growth performance, Intestinal function, Pig model
1. Introduction
The gut harbors several trillion microbes that play a crucial role in the host's health (Kamada,et al., 2013; Yano et al., 2015; Haiser et al., 2013; Yatsunenko et al., 2012). Destruction of gut bacterial communities have been linked to various health conditions (Hakansson,and Molin, 2011; Turnbaugh et al., 2009). Thus, defining the effects of gut microbiota on the host's health is beneficial for understanding the host microbiota interactions. The majority of studies using human subjects could not be conducted because of ethical considerations, and animal models could help identify gut microbes and the underlying mechanisms. Germ-free (GF) animals are free from living microorganisms, including bacteria, viruses, fungi, protozoa, and parasites throughout their life and reared in sterile environments (Delzenne,and Cani, 2011; Meyer et al., 1964). They provide an excellent experimental model for detecting host–microbiota interactions. Compared with GF and conventional (CV) rodents, the gut microbiota and hosts not only simply coexist but also have mutualistic relationships (Chow et al., 2010; Leser and Mølbak, 2009). However, the rodent models are limited by many vital physiological and metabolic differences from humans (Heinritz,et al., 2013). Moreover, mice and rats do not dependably have clinical manifestations seen in human diseases. Thus, more clinically relevant models of the human gastrointestinal tract are needed. The domestic pig (Sus scrofa) is a model of human health, it has similar anatomy, physiology, and genetics to that of humans (Meurens,et al., 2012; Odle et al., 2014). Also, pigs eat an omnivorous diet, and their developmental phase is similar to that of humans, especially during infancy (Garthoff,et al., 2002). Furthermore, 96% functional pathways observed in the human catalog were found in the porcine catalog, supporting the potential use of pig for biomedical research (Liang et al., 2016). Consequently, the absence of microbes in pigs makes them a suitable experimental model to explore the host–microbiota interactions in human health. The gut microbiota of newborns is characterized by low diversity and high instability, and previous studies revealed that the diversity of maternal gut microbiota is higher than that of infants (Parker et al., 2018; Yan Shao et al., 2019). Maternal microbiota is also the major microbial source of infant-acquired strains (Ferretti et al., 2018). It is notable that increased microbial diversity has been shown to benefit the metabolic health (Le Chatelier et al., 2013). Thus, in the present study, replacing background microbes with maternal gut microbiota transplantation was hypothesized to promote growth development and health in newborns.
Taken together, CV piglets from natural birth, GF piglets from hysterectomy, and fecal microbiota-transplanted (FMT) piglets, which were GF piglets orally infused with healthy sow fecal suspension, were established to explore the microbiota–host interactions and the effects of maternal gut microbiota on newborns in a GF pig model.
2. Materials and methods
Experimental protocols were approved by the Ethics Committee of Sichuan Agricultural University (Chengdu, China) under permit number DKY-B20131704.
2.1. Preparation of fecal microbiota suspension
In accordance with the standard for donor identification and screening described previously (Hamilton et al., 2012), healthy multiparous Bama sows (n = 6) without antibiotic and probiotic treatments for 3 months were used as fecal donors. Actinobacillus pleuropneumoniae, Brucellosis, Helminths, Eimeria, Coccidia, Serpulinhyodysen, porcine parvovirus, porcine epidemic diarrhea virus, and transmissible gastroenteritis virus were not detected in the feces of the donor sows (Xishan Biotechnology Inc. Suzhou China). Fresh feces were collected after 12 h of fasting, and the fecal suspension was prepared following the method of Zeng et al. (2013).
2.2. Experimental animals, design, and diets
The experiment was carried out at the Experimental Swine Engineering Center of the Chongqing Academy of Animal Sciences (CMA No. 162221340234; Chongqing, China). Twelve GF piglets were delivered via hysterectomy from 2 multiparous Bama sows (a native breed of China). At 112 days of gestation (full-term, 114 days), the pregnant Bama sows were anesthetized with 4% isoflurane. Uterus was excised from the anesthetized sow and transferred into a sterile isolator (DOSSY Experimental Animals Co., Ltd, Chengdu, China) through a tank including 120 L of 0.1% peracetic acid for decontamination. Then, 12 neonatal piglets were taken from the uterus in the isolator and transferred to 6 rearing isolators (2 piglets per isolator, Class Biologically Clean Ltd., Madison, Wisconsin, USA) depending on the litter of origin and sex. The isolator contained a checkboard, and the piglets were fed separately. The rearing isolators were sterilized by spraying with 1% peracetic acid in advance and maintained in sterile environments as described previously (Meyer et al., 1964). The sterile environments, piglet's skin, oral mucosa, and rectal swabs were checked via anaerobic (thioglycollate medium) and aerobic (brain-heart infusion broth) culture of samples at least every week as described by Chinese National Standard (GB/T 14926-41-2001). Colonic digesta was collected at the end of the experiment for further confirmation of sterile status.
Of 12 GF piglets, 6 piglets were treated as the GF group, 6 piglets were designated as FMT group. They were orally infused with the fecal suspension (1 mL/day) from a healthy donor sow on day 7 after birth , and it continued for 3 days. The GF and FMT groups were hand-fed with Co60-γ-irradiated sterile milk powder (Appendix Table 1) diluted with sterile water (1:4) for 21 days. Another 6 piglets (gilts and boars in half) generated by natural birth from a multiparous Bama sow were regarded as the CV group, reared by a lactating Bama sow for 21 days, and then transferred to single feeding cage respectively. A corn-soybean feed, formulated according to NRC (2012) requirements and Chinese feeding standards for local piglets (2004) (Appendix Table 2), was sterilized via Co60-γ-radiation and introduced to the GF, FMT, and CV groups for another 21 days. In the two 21-day periods, all piglets were allowed ad libitum access to water (including sterile water). Sterile milk, feed, and water were transferred into the rearing isolator by a transfer port in replacement containers, which were preliminarily decontaminated with 0.5% peracetic acid before sterilization with 1% peracetic acid in the transfer port in order to prevent microbial contamination.
2.3. Sample collection
The corn-soybean feed was sampled once and stored at −20 °C for chemical analysis. Feces were collected from each piglet on day 39 to 42, added with 10% hydrochloric acid to fix excreta nitrogen after collection, and dried in a forced-air oven (65 °C) for 72 h. Samples of feed and feces were ground through a 1-mm screen for further analysis. In the morning of day 42, blood samples were obtained from the anterior vena cava before the piglets were euthanized via isoflurane anesthesia, centrifuged at 3,000 × g for 15 min, and then stored at −80 °C for further analysis. The abdomen was opened in the laminar airflow clean benches. Samples from middle sections (4 cm) of the duodenum, jejunum, and ileum were collected and stored in 4% fresh paraformaldehyde solution for histomorphologic measurements. Then, the jejunum, ileum, and colon tissues were opened longitudinally, and then washed with cold saline solution.. The jejunal mucosa was collected by scraping the intestinal wall with a glass microscope slide. All tissue and mucosa samples were snap-frozen in liquid nitrogen, and then stored at −80 °C for further analysis, followed by collecting approximately 4 g of digesta from the ileum, caecum, and colon, keeping it in sterile tubes, and immediately freezing it at −80 °C until analysis of short chain fatty acids (SCFA) concentrations. The pH values of the cecal and colonic digesta were determined with a pH meter. Following euthanasia, the heart, lung, liver, spleen, and kidney were removed, rinsed with cold saline, and blotted dry with an absorbent paper before weighing.
2.4. Growth performance
The piglets were weighed individually on days 21 and 42, and the corn-soybean feed consumption per piglet was measured daily to determine the average daily feed consumption, average daily weight gain, and feed efficiency (weight gain/feed consumption).
2.5. Relative organ weight
The relative organ weight is the ratio of organ weight to the pre-slaughter weight of each piglet.
2.6. Blood parameters measurement
Blood routine parameters were measured using a blood cell analyzer (Sysmex XT-1800, Japan), and blood biochemical indices were measured using an automatic biochemistry analyzer (Hitachi 7020, Japan).
2.7. Determination of nutrient digestibility
The apparent total tract digestibility (ATTD) was determined using acid-insoluble ash (AIA) as the internal marker. The AIA content in the fecal and feed samples was measured in accordance with the Chinese National Standard (GB/T 23742). Chemical analysis of the feed and fecal sample was conducted as follows. Dry matter (method 930.15), crude ash (method 942.05), ether extract (method 945.16), crude protein (CP, method 990.03) were assessed according to the procedures of AOAC (1995). The gross energy of the feed and fecal samples was determined using adiabatic oxygen bomb calorimetry (Parr Instrument Co., Moline IL). The digestibility was calculated using the following formula:
| ATTD (%) = 100 - A1/A2 × F2/F1 × 100, |
where A1 represents the AIA content of the feed, A2 represents the feed AIA content, A2 denotes the fecal AIA content, F1 is the feed nutrient content, and F2 represents the fecal nutrient content.
2.8. Intestinal morphology analysis
Morphologic measurements of the villus height and crypt depth were conducted in accordance with Touchette et al. (2002). In brief, 4-cm of each sample from middle sections of the duodenum, jejunum, and ileum was washed with cold sterile saline, fixed with 4% paraformaldehyde solution, and dehydrated and embedded in paraffin wax before transverse sections were cut. The preserved samples were stained with hematoxylin and eosin. Twelve well-orientated sections of villi and their adjoint crypts in each sample were measured using Image-Pro Plus software (version 6.0, Media Cybernetics, USA) at 40× magnification.
2.9. Analysis of enzyme activity
The frozen sample of jejunal mucosa (approximately 1.0 g) was homogenized in ice-cold saline solution (1:9, wt/vol), centrifuged at 3,000 × g for 15 min at 4 °C, and stored at −80 °C for further analysis. The activities of trypsin, lipase, lactase, sucrase, maltase, sodium/potassium ATPase (Na+, K+-ATPase), calcium/magnesium ATPase (Ca2+, Mg2+-ATPase) and γ-glutamyl-transferase (γ-GT), creatine kinase in jejunal mucosa were determined using commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) in accordance with the manufacturer's instructions. The total protein content of the jejunal mucosa was detected using Bradford brilliant blue method. Each parameter was simultaneously measured in triplicate on the same plate. The differences among parallels must be small (coefficient of variation was less than 10%) to guarantee the reproducibility of repeated measurements.
2.10. Detection of mRNA expression
Total RNA was isolated from the frozen jejunum, ileum, and colon by using Trizol reagent (TaKaRa) in accordance with the manufacturer's instructions. The concentration and purity of the RNA were determined using a NanoDrop ND-2000 spectrophotometer (NanoDrop, Germany). The OD260:OD280 ratios ranging from 1.8 to 2.0 in all samples were regarded as suitable for further analysis. The RNA integrity was detected via agarose gel electrophoresis, and the 28S:18S ribosomal RNA band ratio was determined as ≥ 1.8. The RNA was reverse transcribed into cDNA by using the PrimeScript RT reagent kit (TaKaRa) according to the manufacturer's guidelines. Primers for the selected genes (Appendix Table 3) were designed using Primer 6 software (PREMIER Biosoft International, Palo Alto, CA, USA) and synthesized commercially by Sangon Biotech Ltd. (Shanghai, China). Quantitative real-time PCR was performed on an ABI Prism 7000 detection system in a two-step protocol with SYBR Green (Applied Biosystems, Foster City, CA, USA). Each reaction was performed at a volume containing 1 μL cDNA, 5 μL SYBR Premix Ex Taq TM (2 × ), 0.2 μL ROX reference dye (50 × ), 0.4 μL of each forward and reverse primer, and 3 μL PCR-grade water. The PCR conditions were as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 25 s, and a 72 °C extension step for 5 min. A melting curve was generated following each quantitative real-time PCR assay to verify the specificity of the reactions. The housekeeping gene β-actin was chosen as the reference gene to normalize the mRNA expression of the target genes. The gene expression data of the replicate samples were calculated using the 2–ΔΔCT method (Pfaffl, 2001). The relative expression of the target genes in the CV group was set at 1.0. Each sample was measured in triplicate.
2.11. SCFA measurement and determination of pH values
The sample pretreatment of serum and cecal and colonic digesta was according to the previous method (Luo et al., 2017). The SCFA (acetate, propionate, and butyrate) concentrations in the serum and digesta of ileum, cecum, and colon were determined using the gas chromatography system (CP-3800 GC, Varian, Inc., Walnut Creek, CA, USA), and following the instructions described by Franklin et al. (2002). Approximately 5 g digesta of the ileum, cecum, and colon were collected into a sterile centrifugal tube, and then the pH value was measured using the PHS-3C pH meter (Shanghai, China).
2.12. Statistical analysis
All data were analyzed in SAS 9.2 (SAS Institute, Inc., Cary, NC, USA) and Figures were generated using GraphPad Prism (La Jolla, CA, USA). Data were analyzed using Tukey's tests and presented as means ± SEM, with the individual piglet as the statistical unit (n = 6). All differences were considered significant at P < 0.05, and a tendency was declared with 0.05 < P < 0.10.
3. Results
3.1. Growth performance
Table 1 shows that the weight gain in the FMT group was numerically higher than that in the other 2 groups. The feed efficiency in the FMT group increased compared with that in the CV and GF groups (P < 0.05). In addition, no difference was observed in terms of feed efficiency between the GF and CV groups (P > 0.05).
Table 1.
Effects of gut microbiota on the growth performance in a pig model.1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| Body weight (d 21), kg | 2.51 ± 0.07 | 2.62 ± 0.13 | 2.49 ± 0.20 | 0.77 |
| Body weight (d 42), kg | 4.92 ± 0.30 | 5.11 ± 0.22 | 5.25 ± 0.27 | 0.71 |
| Feed consumption, g/d | 186.37 ± 20.45 | 177.78 ± 4.38 | 171.23 ± 6.35 | 0.72 |
| Weight gain, g/d | 114.68 ± 12.25 | 118.85 ± 4.62 | 131.11 ± 4.34 | 0.39 |
| Feed efficiency | 0.62 ± 0.01b | 0.67 ± 0.02b | 0.77 ± 0.03a | <0.01 |
a, b Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
3.2. Relative organ weight
As shown in Table 2, the relative weights of the heart, lung, and liver in the GF group were lower than those in the CV group (P < 0.05). Likewise, the relative weights of the lung and liver in the FMT group decreased compared with those in the CV group (P < 0.05), whereas no difference was found compared with those in the GF group (P > 0.05).
Table 2.
Effects of gut microbiota on the relative organ weight in a pig model (%).1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| Heart | 0.55 ± 0.02a | 0.46 ± 0.02b | 0.47 ± 0.02 ab | 0.03 |
| Lung | 1.32 ± 0.12a | 0.96 ± 0.03b | 0.98 ± 0.06b | 0.02 |
| Liver | 3.54 ± 0.08a | 2.42 ± 0.09b | 2.53 ± 0.09b | <0.01 |
| Spleen | 0.26 ± 0.03 | 0.22 ± 0.02 | 0.19 ± 0.02 | 0.11 |
| Kidney | 0.73 ± 0.04 | 0.61 ± 0.02 | 0.65 ± 0.04 | 0.16 |
a,b Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
3.3. Blood parameters
As presented in Table 3, Table 4, the white blood cell (WBC), neutrophils (NEUT), lymphocyte (LY), eosinophils (EOS), and basophilic granulocyte (BAS) counts in the CV group increased compared with those in the GF and FMT groups (P < 0.05), and no differences were observed between the GF and CV groups (P > 0.05). Likewise, the alanine transaminase (ALT), total protein, and globulin concentrations were higher in the CV group than in the GF group (P < 0.05). In addition, the total protein and globulin contents in the FMT group increased compared with those in the GF group (P < 0.05). Strikingly, the serum urea nitrogen content in the GF group was enhanced compared with that in the CV and FMT groups (P < 0.05).
Table 3.
Effects of gut microbiota on the blood routine indices in a pig model.1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| WBC, 109/L | 19.79 ± 2.19a | 7.63 ± 0.32b | 8.95 ± 0.34b | <0.01 |
| NEUT, 109/L | 7.59 ± 1.33a | 2.05 ± 0.36b | 4.16 ± 0.37b | <0.01 |
| LY, 109/L | 10.47 ± 0.86a | 4.22 ± 0.83b | 3.96 ± 0.23b | <0.01 |
| MON, 109/L | 1.15 ± 0.23 | 1.11 ± 0.72 | 0.59 ± 0.04 | 0.64 |
| EOS, 109/L | 0.38 ± 0.09a | 0.10 ± 0.02b | 0.15 ± 0.02b | <0.01 |
| BAS, 109/L | 0.20 ± 0.03a | 0.09 ± 0.01b | 0.09 ± 0.02b | <0.01 |
| RBC, 1012/L | 6.30 ± 0.14 | 6.68 ± 0.24 | 6.71 ± 0.21 | 0.25 |
| HBC, g/L | 120.67 ± 3.58 | 121.25 ± 4.63 | 119.83 ± 3.50 | 0.96 |
| PLT, 109/L | 560.42 ± 31.76 | 588.58 ± 44.52 | 560.75 ± 35.83 | 0.87 |
WBC = white blood cell; NEUT = neutrophils; LY = lymphocyte; MON = monocyte; EOS = eosinophils; BAS = basophilic granulocyte; RBC = red blood cell; HBC = hemoglobin concentration; PLT = platelet.
a, b Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
Table 4.
Effects of gut microbiota on the serum biochemical indices in a pig model.1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| ALT, U/L | 57.50 ± 7.86a | 38.17 ± 1.04b | 32.67 ± 1.69b | <0.01 |
| AST, U/L | 35.33 ± 5.83a | 22.33 ± 2.13 ab | 19.08 ± 1.70b | 0.03 |
| AST/ALT | 0.66 ± 0.13 | 0.59 ± 0.05 | 0.58 ± 0.04 | 0.74 |
| Total protein, g/L | 58.77 ± 2.02a | 44.28 ± 0.98c | 50.86 ± 0.95b | <0.01 |
| Albumin, g/L | 40.40 ± 1.86a | 34.74 ± 0.63b | 36.81 ± 1.01 ab | 0.01 |
| Globulin, g/L | 18.37 ± 1.40a | 9.54 ± 0.49c | 13.83 ± 0.90b | <0.01 |
| Serum urea nitrogen, mmol/L | 1.70 ± 0.13b | 2.96 ± 0.29a | 1.77 ± 0.30b | 0.02 |
| LDH, U/L | 651.42 ± 142.89 | 733.67 ± 37.30 | 635.83 ± 101.28 | 0.21 |
ALT = alanine transaminase; AST = aspartate transaminase; LDH = lactate dehydrogenase.
a,b,c Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
3.4. ATTD
As shown in Table 5, the digestibility of dry matter, gross energy, and CP were higher in the CV group than in the GF group (P < 0.05). Meanwhile, the digestibility of all nutrients in the FMT group increased compared with that in the GF group (P < 0.05). The digestibility of crude ash and ether extract in the FMT group was enhanced compared with that in the CV group (P < 0.05).
Table 5.
Effects of gut microbiota on the nutrient digestibility in a pig model (%).1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| Dry matter | 90.29 ± 0.29a | 76.52 ± 0.95b | 92.07 ± 0.26a | <0.01 |
| Crude ash | 69.47 ± 1.50b | 65.78 ± 1.31b | 80.61 ± 1.92a | <0.01 |
| Ether extract | 80.65 ± 2.18b | 75.91 ± 1.57b | 88.47 ± 0.59a | <0.01 |
| Gross energy | 90.39 ± 0.33a | 79.04 ± 1.05b | 92.46 ± 0.23a | <0.01 |
| Crude protein | 89.02 ± 0.45a | 84.28 ± 1.08b | 91.57 ± 0.79a | <0.01 |
a, b Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
3.5. Intestinal morphology
As presented in Table 6, the villus height of the ileum in the FMT and GF groups was lower than that in the CV group (P < 0.05). The crypt depth in the small intestine of the CV group was also deeper than that of the FMT and GF groups (P < 0.05). Importantly, the villi height-to-crypt depth ratio in the small intestine of the FMT group was greater than that of the CV and GF groups numerically.
Table 6.
Effects of gut microbiota on the intestinal morphology in a pig model.1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| Duodenum | ||||
| Villus height, μm | 540.9 ± 35.73 | 450.90 ± 47.92 | 486.5 ± 21.91 | 0.33 |
| Crypt depth, μm | 199.90 ± 4.48a | 156.8 ± 3.45b | 149.2 ± 10.28b | <0.01 |
| Villus height:crypt depth ratio | 2.71 ± 0.18 | 2.87 ± 0.28 | 3.30 ± 0.11 | 0.16 |
| Jejunum | ||||
| Villus height, μm | 458.2 ± 55.64 | 339.6 ± 38.71 | 449.2 ± 12.29 | 0.11 |
| Crypt depth, μm | 160.3 ± 9.44a | 102.1 ± 3.86b | 116.8 ± 6.17b | <0.01 |
| Villus height:crypt depth ratio | 2.94 ± 0.46 | 3.34 ± 0.39 | 3.87 ± 0.15 | 0.24 |
| Ileum | ||||
| Villus height, μm | 328.6 ± 11.27a | 257.9 ± 22.17b | 257.1 ± 25.19b | 0.01 |
| Crypt depth, μm | 147.4 ± 5.70a | 88.67 ± 6.18b | 101.2 ± 6.38b | <0.01 |
| Villus height: crypt depth | 2.23 ± 0.07 | 2.93 ± 0.26 | 2.56 ± 0.25 | 0.18 |
a,b Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
3.6. The enzyme activity in jejunum
As shown in Table 7, the activities of lipase, trypsin, maltase, sucrase, and lactase in the FMT group were the highest among the treatments. Moreover, the lipase activity of the FMT group increased compared with that of the CV group (P < 0.05).
Table 7.
Effects of gut microbiota on the jejunal enzyme activity in a pig model.1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| Lipase, U/g prot | 69.94 ± 15.53b | 122.92 ± 5.70 ab | 165.53 ± 9.85a | <0.01 |
| Trypsin, U/mg prot | 57.86 ± 6.06 | 86.70 ± 10.54 | 88.18 ± 14.25 | 0.15 |
| Maltase, U/mg prot | 596.95 ± 66.63 | 765.08 ± 192.25 | 775.80 ± 126.24 | 0.69 |
| Sucrase, U/mg prot | 145.29 ± 19.08 | 150.79 ± 26.15 | 155.33 ± 15.45 | 0.96 |
| Lactase, U/mg prot | 164.60 ± 15.93 | 150.80 ± 30.58 | 213.77 ± 29.35 | 0.37 |
| γ-GT, U/g prot | 75.76 ± 13.64 | 104.32 ± 5.40 | 95.42 ± 6.17 | 0.12 |
| Creatine kinase, U/mg prot | 1.85 ± 0.09 | 2.16 ± 0.33 | 1.91 ± 0.17 | 0.65 |
| Na+, K+-ATPase, mol Pi/mg prot per h | 8.21 ± 0.45 | 7.10 ± 0.35 | 6.81 ± 0.39 | 0.11 |
| Ca2+, Mg2+-ATPase, mol Pi/mg prot per h | 8.27 ± 0.50 | 6.45 ± 0.87 | 6.82 ± 0.40 | 0.16 |
γ-GT = γ-glutamyl-transferase; Na+, K+-ATPase = sodium/potassium ATPase; Ca2+, Mg2+-ATPase = calcium/magnesium ATPase.
a,b Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
3.7. Relative mRNA expression levels of intestinal function-related genes
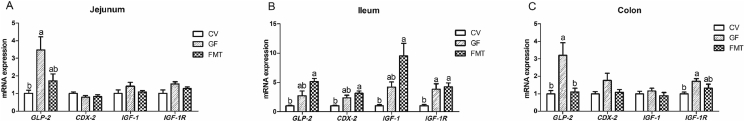
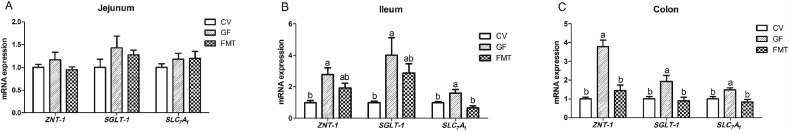
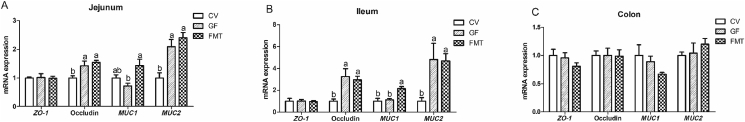
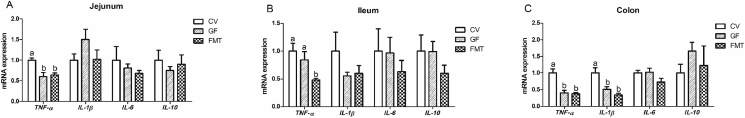
The GF group showed higher mRNA expression levels of glucagon-like peptide-2 (GLP-2) in the jejunum and colon and insulin-like growth factor-1 receptor (IGF-1R) in the colon than the CV group (P < 0.05, Fig. 1). In addition, the mRNA abundance of GLP-2 in the colon of the GF group was greater than that in the colon of the FMT group (P < 0.05). It is notable that the mRNA expression levels of GLP-2, caudal-related homeodomain transcription 2 (CDX-2), insulin-like growth factor-1 (IGF-1), and IGF-1R in the ileum of the FMT group increased compared with those in the ileum of the CV group (P < 0.05). Fig. 2 illustrates that compared with the CV and FMT group, the GF group had increased mRNA expression levels of zinc transporters 1 (ZNT-1), sodium/glucose cotransporter 1 (SGLT-1), and solute carrier family 7 (SLC7A1) in the colon and SLC7A1 in the ileum (P < 0.05). Moreover, the mRNA expression levels of ZNT-1 and SGLT-1 in the ileum of the GF group increased compared with those in the ileum of the CV group (P < 0.05). As shown in Fig. 3, the mRNA expression levels of occludin and mucin 2 (MUC2) in the jejunum and ileum of the GF group were higher than those of the CV group (P < 0.05). The mRNA expression of mucin 1 (MUC1) in the ileum and jejunum of the FMT group was higher than that in the ileum and jejunum of the GF group (P < 0.05). Moreover, the mRNA expression levels of occludin and MUC2 in the jejunum and ileum of the FMT group increased compared with those in the jejunum and ileum of the CV group (P < 0.05). In addition, the FMT group exhibited higher mRNA expression of MUC1 than the CV group (P < 0.05). As presented in Fig. 4, the mRNA expression levels of tumor necrosis factor α (TNF-α) and interleukin (IL)-1β in the colon and TNF-α in the jejunum of the CV group were higher than those of the GF and FMT groups (P < 0.05). The FMT group demonstrated a decrease in the mRNA level of TNF-α in the ileum compared with the CV and GF groups (P < 0.05).
Fig. 1.
Quantitative real-time PCR analysis reveals the differences in relative mRNA expression levels of intestinal development related genes in a pig model. The effects of gut microbiota on the relative mRNA expression levels of intestinal development related genes in jejunum (A), ileum (B) and colon (C). Values are means ± SEM; n = 6/group. Means without a common letter differ, P < 0.05. CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota; GLP-2 = glucagon-like peptide-2; CDX-2 = caudal-related homeodomain transcription 2; IGF-1 = insulin-like growth factor-1; IGF-1R = insulin-like growth factor-1 receptor.
Fig. 2.
Quantitative real-time PCR analysis reveals the differences in relative mRNA expression levels of intestinal transport and absorption related genes in a pig model. The effects of gut microbiota on the relative mRNA expression levels of intestinal transport and absorption related genes in jejunum (A), ileum (B) and colon (C). Values are means ± SEM; n = 6/group. Means without a common letter differ, P < 0.05. CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota; ZNT-1 = zinc transporters 1; SGLT-1 = sodium/glucose cotransporter 1; SLC7A1 = solute carrier family 7.
Fig. 3.
Quantitative real-time PCR analysis reveals the differences in relative mRNA expression levels of intestinal barrier function related genes in a pig model. The effects of gut microbiota on the relative mRNA expression levels of intestinal barrier function related genes in jejunum (A), ileum (B) and colon (C). Values are means ± SEM; n = 6/group. Means without a common letter differ, P < 0.05. CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota; ZO-1 = zonula occludens 1; MUC1 = mucin 1; MUC2 = mucin 2.
Fig. 4.
Quantitative real-time PCR analysis reveals the differences in relative mRNA expression levels of intestinal inflammatory cytokines genes in a pig model. The effects of gut microbiota on the relative mRNA expression levels of intestinal inflammatory cytokines genes in jejunum (A), ileum (B) and colon (C). Values are means ± SEM; n = 6/group. Means without a common letter differ, P < 0.05. CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota; TNF-α = tumor necrosis factor α; IL-1β = interleukin-1β; IL-6 = interleukin-6; IL-10 = interleukin-10.
3.8. SCFA concentrations and pH values in the intestinal tract
As presented in Table 8, compared with the FMT and CV groups, the GF group showed a decrease in the concentrations of acetate, propionate, and butyrate in the cecum and colon (P < 0.05). Meanwhile, the concentration of acetate in the ileum of the GF group was lower than that in the ileum of the CV and FMT groups (P < 0.05). It is notable that the FMT group demonstrated a numerically greater concentration of butyrate in the serum, cecum, and colon than the CV group. Moreover, the concentrations of acetate and butyrate in the serum of the FMT group were the greatest among the treatments, and they increased compared with those in the GF group (P < 0.05). Furthermore, the concentration of serum acetate was higher in the FMT group than in the CV group (P < 0.05). Correspondingly, the pH value in the cecum of the GF group increased compared with that in the cecum of the CV group (P < 0.05). The pH value of cecum digesta in the FMT group consistently decreased compared with that in the GF group (P < 0.05).
Table 8.
Effects of gut microbiota on the short-chain fatty acid concentrations and digesta pH values in a pig model.1
| Item | Groups2 |
P-value | ||
|---|---|---|---|---|
| CV | GF | FMT | ||
| Serum, μmol/L | ||||
| Acetate | 250.08 ± 17.47b | 195.31 ± 2.89b | 433.92 ± 62.03a | <0.01 |
| Propionate | 46.61 ± 2.15 | 53.34 ± 3.86 | 51.75 ± 6.84 | 0.51 |
| Butyrate | 4.66 ± 1.56 ab | 1.36 ± 0.15b | 7.04 ± 1.83a | <0.01 |
| Ileum, μmol/g | ||||
| Acetate | 8.48 ± 1.38a | 0.64 ± 0.10b | 5.50 ± 1.83a | <0.01 |
| Propionate | 0.27 ± 0.05 | 0.11 ± 0.05 | 0.28 ± 0.12 | 0.22 |
| Butyrate | 0.24 ± 0.08 | 0.05 ± 0.03 | 0.24 ± 0.16 | 0.30 |
| Cecum, μmol/g | ||||
| Acetate | 56.43 ± 5.36a | 1.43 ± 0.43b | 51.64 ± 6.19a | <0.01 |
| Propionate | 18.72 ± 3.13a | 0.11 ± 0.02b | 19.02 ± 3.18a | <0.01 |
| Butyrate | 6.25 ± 0.78a | 0.01 ± 0.00b | 8.00 ± 0.96a | <0.01 |
| Cecum pH | 4.73 ± 0.42c | 6.86 ± 0.28a | 5.77 ± 0.19b | <0.01 |
| Colon, μmol/g | ||||
| Acetate | 42.98 ± 6.53a | 1.64 ± 0.33b | 34.04 ± 3.28a | <0.01 |
| Propionate | 13.02 ± 1.13a | 0.15 ± 0.02b | 10.24 ± 1.58a | <0.01 |
| Butyrate | 3.86 ± 0.39a | 0.01 ± 0.00b | 4.34 ± 0.53a | <0.01 |
| Colon pH | 6.63 ± 0.09 | 6.64 ± 0.25 | 6.15 ± 0.16 | 0.09 |
a,b,c Labeled means with different superscripts within a row are significantly different at P < 0.05.
Values are means ± SEM, n = 6/group.
CV, conventional piglets; GF, germ-free piglets; FMT, germ-free piglets were transplanted with healthy sow fecal microbiota.
4. Discussion
The gut microbiota provides essential capacities to ferment non-digestible carbohydrates into SCFA and produces energy for the host (Turnbaugh,et al., 2006). In the present study, when there are no microbes in the gut, the SCFA concentrations in the ileum, cecum, and colon of the GF group were significantly reduced compared with those in the CV and FMT groups. Notably, SCFA performed various physiological functions in the gut, including promoting colonic mobility and colonic blood flow and decreasing gastrointestinal pH, which affected the absorption of electrolytes and nutrients (Tazoe,et al., 2008). The pH value in the cecal and colon digesta of the FMT group consistently decreased compared with that in the cecal and colon digesta of the GF group. The gut microbes provide numerous biological activities to the host (Turnbaugh,et al., 2006), and they have been recognized as a strong determinant factor of host digestion and carbohydrate metabolism (Backhed, 2005; Greenblum et al., 2012). The present study found that the nutrient digestibility in the GF group markedly decreased compared with that in the FMT and CV groups, and the feed efficiency in the GF group was apparently lower than that in the FMT group. A decrease in serum urea nitrogen is an indication of increased efficiency in the use of dietary nitrogen (Owusuasiedu et al., 2003), and an increase in the total protein and albumin in serum indicates increased protein synthesis. In the current study, the concentrations of total protein and albumin in the GF group apparently decreased compared with those in the CV group, while the concentration of serum urea nitrogen markedly increased. Moreover, the concentrations of total protein in the GF group significantly decreased compared with those in the FMT group, which was in agreement with the results of a previous study (Fouhse,et al., 2016). These findings may indicate that the absence of gut microbes damaged protein synthesis. GF animals have major shortcomings in their systemic and intestinal immune functions (Lee and Mazmanian, 2010). Indeed, in the present study, the concentration of globulin in the GF group significantly decreased compared with that in the CV and FMT groups, and the mRNA expression of TNF-α in the ileum was higher than that in the FMT group. These results suggested that gut microbiota is vital for host growth and health. The absence of gut microbes reduced the production of SCFA, decreased the nutrient digestibility and feed efficiency, and it may damage protein synthesis and the immune function.
In this study, the crypt depth in the small intestine of GF group was apparently reduced compared to that of the CV group, which was consistent with the results of previous studies (Shirkey,et al., 2006; Van Kessel and Willing, 2007). The increased of villus height and reduced crypt depth in the intestine indicated an enhancement of the surface area capable of greater absorption of available nutrients (Caspary, 1992). In the present study, the mRNA expressions levels of ZNT-1, SGLT-1, and SLC7A1 in the GF group markedly increased compared with those in the FMT and CV groups. SGLT-1 is an apical intestinal transporter responsible for the majority of luminal glucose transport across the intestinal epithelium, and it is the rate-limiting step for the absorption of dietary glucose (Zhang et al., 2016). ZNT-1 is extensively located at the intestinal mucosa, and it is considered an important transporter of zinc (Boudry,et al., 2010). Zinc is the component of multiple enzymes and proteins. SLC7A1 plays an important role in the transport and maintenance of homeostasis of the basic amino acids in the small intestine (Hyde et al., 2003). Similarly, a previous research reported that the activities of aminopeptidase nitrogen and lactase phlorizin hydrolase in the GF group were higher than those in the CV counterparts (Willing and Van Kessel, 2009). These findings showed that the mRNA expression levels of nutrient transporters in GF piglets may be upregulated to compensate for poor nutrient digestibility. GLP-2 plays a critical role in increasing the protein synthesis and villus height of the small intestine, improving nutrient digestion and absorption, and promoting intestinal development (Burrin,et al., 2000; Pedersen et al., 2008). IGF-1 and CDX-2 act as growth factors that could mediate intestinal proliferation, differentiation, and growth (Steeb,et al., 1994; Sanroman et al., 2015). The results of the present study showed the mRNA expression levels of GLP-2 and IGF-1R in the GF group were higher than those in the CV group. occludin, MUC1, and MUC2 are important for intestinal integrity and barrier function (Tornavaca,et al., 2015). In the present study, the mRNA expression levels of occludin and MUC2 in the GF group were upregulated compared with those in the CV group. This finding demonstrated that the intestinal development and barrier function of piglets may be improved when the gut microbiota is absent. Taken together, an in-depth understanding of the effects of microbiota on the intestinal functions needs further research, including in in vitro and in vivo experiments.
In this study, no difference in feed efficiency was found between the CV and GF groups, which was in agreement with the evidence observed in other GF animals (Dubos and Schaeler, 1960). Conversely, a previous study demonstrated that the growth rate of GF rats was lower than that of their CV counterparts (Gordon, 1959). The reason may be that the sanitary status differed from one laboratory to another. In unsanitary conditions, the pathogenic bacterial burden causes the nutrients used for growth to shift for fighting external stress and leads to growth depression (Dubos and Schaeler, 1960; Buchanan and Johnson, 2007). In the present study, the WBC count was significantly higher in the CV group than in the GF group. The mRNA expression levels of pro-inflammatory factors (TNF-α and IL-1β) in the CV group increased compared with those in the FMT and GF groups. Interestingly, the GF pigs colonized with one or more known strains of bacterial species and reared under sterile conditions also had higher abundances of IL-1β and IL-6 than their GF counterparts (Shirkey,et al., 2006). Thus, further well-controlled studies are urgently needed. In the current study, the relative weights of the heart, lung, and liver were markedly lower in the GF group than in the CV group, which was consistent with previous findings (Sun et al., 2016). However, Shurson et al. (1990) reported that GF pigs had larger spleen, lung, and heart than their CV counterparts. Bama pigs are a native breed of China, whose size is smaller and growth rate slower compared with other breeds. The inconsistent results on the relative weight of organs were possibly due to the differences in pig breed. The activities of AST and ALT in serum are commonly used as biomarkers for liver function (Goorden,et al., 2013). In the present study, the activities of AST and ALT in the GF group were lower than those in the CV group, whereas another work in GF piglets detected inconsistent results (Sun et al., 2016). In addition, the villus height in the ileum of the GF group decreased compared with that of the CV group, which was similar to the result of studies in GF mice (O'Hara and Shanahan, 2006; Smith et al., 2007). On the contrary, previous reports indicated that GF pigs had longer villus height than their CV counterparts (Shirkey,et al., 2006; Van Kessel and Willing, 2007). A previous observation found that GF pigs had longer ileal and duodenal villi but shorter jejunal villus than their CV counterparts (Shurson,et al., 1990). Therefore, further research on GF pigs is clearly warranted.
The pig is an excellent model because it is closely related to humans in terms of anatomy, genetics, and physiology. Studies with the pig as a model have drawn great interest, and they were extensively used in biomedical sciences (Chardon et al., 2006; Heinritz et al., 2013). By using 16s RNA sequencing (data unpublished), the microbiota diversity indices of Chao1 and Shannon in healthy Bama sows was found to be apparently increased compared with that in weaning Bama piglets. Moreover, the abundance of Spirochaetes was higher in donor sows than in weaning piglets. Spirochaetes was considered to degrade macromolecular polymers and improve the digestibility of non-starch polysaccharide (Lilburn et al., 1999; Turnbaugh et al., 2009). In addition, the relative abundance of Clostridiales (butyrate producers) in weaning piglets was reduced compared with that in healthy sows, while the relative abundances of Alistipes and Tyzzerella increased. Previous reports have shown that Alistipes and Tyzzerella were associated with health risks (Saulnier,et al., 2011; Kelly et al., 2016). Furthermore, the abundance of bacteria associated with human disease in healthy sows significantly decreased compared with that in weaning piglets. The gut microbiota community of newborns is characterized by low diversity and high instability (Parker et al., 2018), and fecal samples from babies and mothers revealed that the gut microbiota diversity increased with age (Yan Shao et al., 2019). Increased microbial diversity has been shown to improve metabolic health (Le Chatelier et al., 2013). Therefore, healthy donor sow fecal microbiota was transplanted into newborn GF piglets to investigate the contributions of maternal gut microbiota to the growth and intestinal functions of newborns. In the present study, the weight gain in the FMT group was higher than that in the CV group, and the feed efficiency and nutrient digestibility significantly increased. These findings demonstrated that recolonizing with maternal gut microbiota may promote growth in newborns. A higher villus height-to-crypt depth ratio means a larger absorptive area of total luminal villus and a more favorable mucosa structure, which could lead to adequate digestive enzyme development. In the current study, the villus height-to-crypt depth ratio in the small intestine was higher in the FMT group than in the CV group. Indeed, the activities of most digestive enzymes were greater in the FMT group than in the CV group. SCFA, particularly butyrate, are the major promoter of colonic health, they provide energy for colonocyte proliferation and differentiation (Suzuki et al., 2008). The results of the current study found that the concentration of butyrate in the serum, cecum, and colon of the FMT group was higher than that in the CV group. A previous study revealed that exogenously infusion of SCFA decreased the pro-inflammatory cytokine levels in CV pigs (Diao,et al., 2019). The mRNA expressions of pro-inflammatory factors (TNF-α and IL-1β) in the intestine of the FMT group were markedly lower than those in the intestine of the CV group. In addition, the mRNA abundances of intestinal development-related genes in the ileum and barrier function-related genes in the jejunum and ileum of the FMT group were apparently upregulated compared with those in the CV group. These observations demonstrated that sow fecal microbiota transplantation may improve the intestinal functions of newborn piglets. Overall, maternal gut microbiota intervention posed potential to enhance the growth and intestinal functions of newborns.
5. Conclusion
In summary, the absence of gut microbes in piglets impaired the feed efficiency and damaged the nutrient digestibility and SCFA production but showed some compensation in intestinal functions. The transplantation of healthy sow gut microbiota increased the growth and nutrient digestibility and improved the intestinal development and barrier function of newborn piglets. Therefore, gut microbiota and microbial maturity play important roles in the growth and intestinal functions of the host. Moreover, recolonizing with maternal microbiota may benefit the growth and health of newborns. This work also explored the microbiota–host interactions and contributed to the research of human maternal gut microbiota transplantation in a GF pig model.
Author contributions
Daiwen Chen, Bing Yu, Liangpeng Ge, and Zuohua Liu: obtained funding and contributed to experimental design; Hua Zhou and Jing Sun designed and performed the trial, and wrote the manuscript; Jun He, Xiangbing Mao, and Ping Zheng performed indicators analyses; Jie Yu, Junqiu Luo, and Yuheng Luo assisted with all of the data analyses and helped in drafting the manuscript; Hong Chen and Hui Yan revised the manuscript. All authors have read and approved the final manuscript.
Conflict interest
We declare that we have no financial and personal relationships with other people or organizations that might inappropriately influence our work, a there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This study was supported by National Natural Science Foundation of China (31730091) and the National Key Research and Development Program of China (2017YFD0500503).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2020.11.012.
Contributor Information
Liangpeng Ge, Email: geliangpeng1982@163.com.
Daiwen Chen, Email: dwchen@sicau.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- AOAC. Official . 16th ed. Association of Official Analytical Chemists; Washington, DC: 1995. Methods of analysis. [Google Scholar]
- Backhed F. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Boudry G., David E.S., Douard V., Monteiro I.M., Le Huerou-Luron I., Ferraris R.P. Role of intestinal transporters in neonatal nutrition: carbohydrates, proteins, lipids, minerals, and vitamins. J Pediatr Gastroenterol Nutr. 2010;51(4):380–401. doi: 10.1097/MPG.0b013e3181eb5ad6. [DOI] [PubMed] [Google Scholar]
- Buchanan J.B., Johnson R.W. Regulation of food intake by inflammatory cytokines in the brain. Neuroendocrinology. 2007;86(3):183–190. doi: 10.1159/000108280. [DOI] [PubMed] [Google Scholar]
- Burrin D.G., Stoll B., Jiang R., Petersen Y., Elnif J., Buddington R.K. GLP-2 stimulates intestinal growth in premature TPN-fed pigs by suppressing proteolysis and apoptosis. Am J Physiol Gastrointest Liver Physiol. 2000;279(6):G1249–G1256. doi: 10.1152/ajpgi.2000.279.6.G1249. [DOI] [PubMed] [Google Scholar]
- Caspary W.F. Physiology and pathophysiology of intestinal absorption. Am J Clin Nutr. 1992;55(1):299S–308S. doi: 10.1093/ajcn/55.1.299s. [DOI] [PubMed] [Google Scholar]
- Chardon P., Renard C., Vaiman M. The major histocompatibility complex in swine. Immunol Rev. 2006;167(1):179–192. doi: 10.1111/j.1600-065x.1999.tb01391.x. [DOI] [PubMed] [Google Scholar]
- Chow J., Lee S.M., Shen Y., Khosravi A., Mazmanian S.K. Vol. 107. 2010. 2010 Host-bacterial symbiosis in health and disease; pp. 243–274. (8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delzenne N.M., Cani P.D. Interaction between obesity and the gut microbiota: relevance in nutrition. Annu Rev Nutr. 2011;31(15):115–731. doi: 10.1146/annurev-nutr-072610-145146. [DOI] [PubMed] [Google Scholar]
- Diao H., Jiao A.R., Yu B., Mao X.B., Chen D.W. Gastric infusion of short-chain fatty acids can improve intestinal barrier function in weaned piglets. Genes Nutr. 2019;14:4. doi: 10.1186/s12263-019-0626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos R.J., Schaeler R.W. The effect of the intestinal flora on the growth rate of mice, and on their susceptibility to experimental infections. J Exp Med. 1960;111(3):407–411. doi: 10.1084/jem.111.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24(1):133–145. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhse J., Zijlstra R.T., Willing B. The role of gut microbiota in the health and disease of pigs. Anim Front. 2016;6(3):30–36. [Google Scholar]
- Franklin M.A., Mathew A.G., Vickers J.R., Clift R.A. Characterization of microbial populations and volatile fatty acid concentrations in the jejunum, ileum, and cecum of pigs weaned at 17 vs 24 days of age. J Anim Sci. 2002;80(11):2904–2910. doi: 10.2527/2002.80112904x. [DOI] [PubMed] [Google Scholar]
- Garthoff L.H., Henderson G.R., Sager A.O., Sobotka T.J., Khan M.A. The Autosow raised miniature swine as a model for assessing the effects of dietary soy trypsin inhibitor. Food Chem Toxicol. 2002;40(4):487–500. doi: 10.1016/s0278-6915(01)00120-x. [DOI] [PubMed] [Google Scholar]
- Goorden S.M., Buffart T.E., Bakker A., Buijs M.M. Liver disorders in adults: ALT and AST. Ned Tijdschr Geneeskd. 2013;157(43):A6443. [PubMed] [Google Scholar]
- Gordon H.A. Morphological and physiological characterization of germfree life. Ann N Y Acad Sci. 1959;78(1):208–220. doi: 10.1111/j.1749-6632.1959.tb53104.x. [DOI] [PubMed] [Google Scholar]
- Greenblum S., Turnbaugh P.J., Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109(2):594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser H.J., Gootenberg D.B., Chatman K., Sirasani G., Balskus E.P., Turnbaugh P.J. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341(6143):295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson A., Molin G. Gut microbiota and inflammation. Nutrients. 2011;3(6):637–682. doi: 10.3390/nu3060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M.J., Weingarden A.R., Sadowsky M.J., Khoruts A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent clostridium difficile infection. Am J Gastroenterol. 2012;107(5):761–767. doi: 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
- Heinritz S.N., Mosenthin R., Weiss E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr Res Rev. 2013;26(2):191–209. doi: 10.1017/S0954422413000152. [DOI] [PubMed] [Google Scholar]
- Hyde R., Taylor P., Hundal H. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem J. 2003;373(Pt 1):1–18. doi: 10.1042/bj20030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N., Chen G.Y., Inohara N., Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.N., Bazzano L.A., Ajami N.J., He H., Zhao J., Petrosino J.F. Gut microbiome associates with lifetime cardiovascular disease risk profile among bogalusa heart study participants. Circ Res. 2016;119(8):956–964. doi: 10.1161/CIRCRESAHA.116.309219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J.J., Prifti E., Hildebrand F., Falony G. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Mazmanian S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser T.D., Mølbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol. 2009;11(9):2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- Liang X., Estellé J., Kiilerich P., Ramayo-Caldas Y., Xia Z., Feng Q. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. 2016;1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- Lilburn T.G., Schmidt T.M., Breznak J.A. Phylogenetic diversity of termite gut spirochaetes. Environ Microbiol. 1999;1(4):331–345. doi: 10.1046/j.1462-2920.1999.00043.x. [DOI] [PubMed] [Google Scholar]
- Luo Y., Zhang L., Li H., Smidt H., Wright A.D.G., Zhang K. Different types of dietary fibers trigger specific alterations in composition and predicted functions of colonic bacterial communities in BALB/c mice. Front Microbiol. 2017;8:966. doi: 10.3389/fmicb.2017.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurens F.O., Summerfield A., Nauwynck H., Saif L., Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20(1):50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Bohl E., Kohler E. Procurement and maintenance of germ-free swine for microbiological investigations. Appl Microbiol. 1964;12(4):295–300. doi: 10.1128/am.12.4.295-300.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odle J., Lin X., Jacobi S.K., Kim S.W., Stahl C.H. The suckling piglet as an agrimedical model for the study of pediatric nutrition and metabolism. Annu Rev Anim Biosci. 2014;2(1):419–444. doi: 10.1146/annurev-animal-022513-114158. [DOI] [PubMed] [Google Scholar]
- Owusuasiedu A., Nyachoti C.M., Marquardt R.R. Response of early-weaned pigs to an enterotoxigenic (K88) challenge when fed diets containing spray-dried porcine plasma or pea protein isolate plus egg yolk antibody, zinc oxide, fumaric acid, or antibiotic. J Anim Sci. 2003;81(7):1790–1798. doi: 10.2527/2003.8171790x. [DOI] [PubMed] [Google Scholar]
- Parker A., Lawson M., Vaux L., Pin C. Host-microbe interaction in the gastrointestinal tract. Environ Microbiol. 2018;20(7):2337–2353. doi: 10.1111/1462-2920.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.B., Hjollund K.R., Johnsen A.H., Rskov C., Rosenkilde M.M., Hartmann B. Porcine glucagon-like peptide-2: structure, signaling, metabolism and effects. Regul Pept. 2008;146(1–3):310–320. doi: 10.1016/j.regpep.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):900–905. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanroman A.K., Tovaglieri A., Breault D.T., Shivdasani R.A. Distinct processes and transcriptional targets underlie CDX2 requirements in intestinal stem cells and differentiated villus cells. Stem Cell Rep. 2015;5(5):673–681. doi: 10.1016/j.stemcr.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier D.M., Riehle K., Mistretta T.A., Diaz M.A., Mandal D., Raza S. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirkey T.W., Siggers R.H., Goldade B.G., Marshall J.K., Drew M.D., Laarveld B. Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. J Anim Sci. 2006;231(8):1333–1345. doi: 10.1177/153537020623100807. [DOI] [PubMed] [Google Scholar]
- Shurson G.C., Ku P.K., Waxler G.L., Yokoyama M.T., Miller E.R. Physiological relationships between microbiological status and dietary copper levels in the pig. J Anim Sci. 1990;68(4):1061–1071. doi: 10.2527/1990.6841061x. [DOI] [PubMed] [Google Scholar]
- Smith K., Mccoy K.D., Macpherson A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Steeb C.B., Trahair J.F., Tomas F.M., Read L.C. Prolonged administration of IGF peptides enhances growth of gastrointestinal tissues in normal rats. Am J Physiol. 1994;266(Pt 1):G1090–G1098. doi: 10.1152/ajpgi.1994.266.6.G1090. [DOI] [PubMed] [Google Scholar]
- Sun J., Du L., Ding Y.C., Cao H.R., Wu M., Lin B.Z. Measurement of body weight, blood parameters and main organ coefficients of germ-free piglets. Acta Lab Anim Sci Sin. 2016;24(4):388–394. [Google Scholar]
- Suzuki T., Yoshida S., Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr. 2008;100(2):297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- Tazoe H., Otomo Y., Kaji I., Tanaka R., Kuwahara A. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol. 2008;59(Suppl 2):251–262. [PubMed] [Google Scholar]
- Tornavaca O., Chia M., Dufton N., Osuna L., Daniel A. ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J Cell Biol. 2015;208(6):821–838. doi: 10.1083/jcb.201404140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchette K.J., Carroll J.A., Allee G.L., Matteri R.L., Dyer C.J., Beausang L.A. Effect of spray-dried plasma and lipopolysaccharide exposure on weaned pigs: I. Effects on the immune axis of weaned pigs. J Anim Sci. 2002;80(2):494–501. doi: 10.2527/2002.802494x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Van Kessel A.G., Willing B.P. Enterocyte proliferation and apoptosis in the caudal small intestine is influenced by the composition of colonizing commensal bacteria in the neonatal gnotobiotic pig. J Anim Sci. 2007;85(12):3256–3266. doi: 10.2527/jas.2007-0320. [DOI] [PubMed] [Google Scholar]
- Willing B.P., Kessel A.G. Intestinal microbiota differentially affect brush border enzyme activity and gene expression in the neonatal gnotobiotic pig. J Anim Physiol Anim Nutr. 2009;93(5):586–595. doi: 10.1111/j.1439-0396.2008.00841.x. [DOI] [PubMed] [Google Scholar]
- Yan Shao S.C.F., Tsaliki E., Vervier K., Strang A., Simpson N., Kumar N. Stunted microbiota and opportunistic pathogen colonisation in caesarean section birth. Nature. 2019;574(7776):117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J., Yu K., Donaldson G., Shastri G., Ann P., Ma L. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguezbello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Li G., Yuan J., Li W., Tang H., Wei H. Effects of age and strain on the microbiota colonization in an infant human flora-associated mouse model. Curr Microbiol. 2013;67(3):313–321. doi: 10.1007/s00284-013-0360-3. [DOI] [PubMed] [Google Scholar]
- Zhang S., Yang Q., Ren M., Qiao S., He P., Li D. Effects of isoleucine on glucose uptake through the enhancement of muscular membrane concentrations of GLUT1 and GLUT4 and intestinal membrane concentrations of Na+/glucose co-transporter 1 (SGLT-1) and GLUT2. Br J Nutr. 2016;116(4):593–602. doi: 10.1017/S0007114516002439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.