Abstract
To investigate the influence of baseline enterotypes and dietary starch type on the concentration of short-chain fatty acids (SCFA), numbers of butyrate producing bacteria and the expression of genes related to intestinal barrier and inflammatory response in the colon of finishing pigs, a 60-d in vivo trial was conducted. A 2-wk pre-trial with 102 crossbred (Duroc × [Landrace × Yorkshire]) finishing barrows (90 d old) was conducted to screen enterotypes. Then, a total of 32 pigs (87.40 ± 2.76 kg) with high (HPBR, ≥ 14) and low (LPBR, ≤ 2) Prevotella-to-Bacteroides ratios (PBR) in equal measure were selected and randomly divided into 4 groups with 8 replicates per group and 1 pig per replicate. The trial was designed following a 2 (PBR) × 2 (amylose-to-amylopectin ratio, AMR) factorial arrangement. Pigs with different PBR were fed diets based on corn-soybean meal with high AMR (HAMR, 1.24) or low AMR (LAMR, 0.23), respectively. Results showed that neither PBR nor AMR influenced the growth performance of pigs. HPBR pigs fed HAMR diet had a higher number of colonic Clostridium cluster XIVa and higher gene expression of butyrate kinase compared to the LPBR pigs (P < 0.05). The HPBR pigs fed HAMR diets also had increased colonic concentrations of total SCFA and propionate compared to the LPBR pigs (P < 0.05). Comparing with other pigs, HPBR pigs fed HAMR diets showed a lower (P < 0.05) expression of histone deacetylases (HDAC) gene and higher (P < 0.05) expression of G protein-coupled receptor 43 gene (GPR 43) in the colonic mucosa. The interaction (P < 0.05) of HPBR and HAMR was also found to decrease the gene expression of interleukin (IL)-6, IL-12, IL-1β and tumor necrosis factor-α (TNF-α) in colonic mucosa. These findings show that HAMR diet increased the abundance and activity of butyrate-producing bacteria and the concentration and absorption of SCFA, which may be associated with the decreased gene expression of inflammatory cytokines in the colonic mucosa of pigs with Prevotella-rich enterotype. All these alterations are likely to have a positive effect on the intestinal health of finishing pigs.
Keywords: Enterotype, Amylose-to-amylopectin ratio, Gut health, Finishing pig
1. Introduction
It is well known that gut health is influenced by the microbial ecosystem in the gastrointestinal tract (GIT), which leads to an increasing demand by the animal industry to manage the gut microbiome. As the gut microbiome varies among individuals, the microbial community from different hosts can be stratified into several enterotypes based on the dominant genera (Arumugam et al., 2011). In the human GIT, the 2 main enterotypes are dominated by either Prevotella or Bacteroides, and in turn, the Prevotella-to-Bacteroides ratio (PBR) can be used to identify these 2 enterotypes (Hjorth et al., 2018, 2019; Christensen et al., 2019; Sandberg et al., 2019). In the GIT of swine, similar enterotypes have also been observed. Previous studies have shown the co-exclusion among Prevotella (Mach et al., 2015; Ramayo-Caldas et al., 2016; Lu et al., 2018; Le Sciellour et al., 2019), Ruminococcus (Mach et al., 2015; Ramayo-Caldas et al., 2016), Treponema (Ramayo-Caldas et al., 2016; Yang et al., 2018b), Clostridium (Lu et al., 2018; Le Sciellour et al., 2019) and Lactobacillus (Le Sciellour et al., 2019) in the GIT of pigs. The high abundance of dominant bacteria in each enterotype provides an opportunity to understand how the behavior of the dominant genera affects the whole community and how this influences the production of microbial metabolites such as short chain fatty acids (SCFA) which are important to gut health. In vitro studies have repeatedly demonstrated that gut microbiomes rich in Prevotella show higher complex polysaccharides utilizing capacity when compared to a Bacteroides-dominated population. Prevotella-dominated gut microbiomes have been shown to produce significantly higher concentrations of SCFA and propionate when compared to other enterotypes (Chen et al., 2017; Wu et al., 2017; Poeker et al., 2018), which may in part explain why Prevotella-driven enterotype is believed to benefit host health. Such a difference in metabolites, caused by the composition of intestinal microbes, may finally determine the individual responses to different diets. However, the health relevance of these enterotypes in vivo remains to be elucidated.
Starch is composed of α-glucan, polysaccharides, amylose, and amylopectin and is the predominant energy source for animals. Our previous studies have demonstrated that amylose-to-amylopectin ratio (AMR) in starch affects the digestive rate and degree of starch, as well as the site where the dietary starch is digested and absorbed. This in turn can have different impacts on the physiological function of the GIT system (He et al., 2010, 2011; Han et al., 2012; Luo et al., 2015; Yang et al., 2018a). Diets with a high AMR (HAMR) may offer potential benefits to the intestinal microbiota in pigs. Comparing with low AMR (LAMR) diets, the abundance of organic acid producers, such as Lactobacillus (Bird et al., 2007; Luo et al., 2015; Newman et al., 2018; Yu et al., 2019), Prevotella (Sun et al., 2016; Maier et al., 2017; Yu et al., 2019), Faecalibacterium (Wang et al., 2019; Yu et al., 2019), and Megasphaera (Newman et al., 2018; Yu et al., 2019) in the GIT of pigs, can be markedly increased by the consumption of diets with HAMR, resulting in higher concentrations of organic acids, such as SCFA and lactate (Yang et al., 2018a; Yu et al., 2019). Therefore, the modulation of microbiota composition and functions using HAMR diets could be regarded as an alternative strategy to promote the intestinal health of pigs. However, information on the relationship between the microbial community and the immune function of intestinal mucosa with the different treatments of AMR is very limited. Previous reports indicated that Prevotella, one of the predominant fiber-degrading bacteria in the intestine (Flint et al., 2012), is well known as a gut colonizer and is associated with an increase in long-term carbohydrate intake (Wu et al., 2011; Flint et al., 2012). But there is a paucity of information regarding the interaction between Prevotella-to-Bacteroides ratio (PBR) and AMR in pigs, especially on the response to dietary starch, amounts and profiles of bacteria metabolites, and the gut health of pigs.
Following on the results of our previous studies, an in vivo pig trial lasting for 60 d was conducted to investigate whether the baseline gut microbial composition, in particular the PBR, differentially responded to dietary AMR resulting in improved gut health. The influence of enterotypes and AMR on the abundance and activity of butyrate-producing bacteria, SCFA concentrations, the expression of genes associated with SCFA uptake, as well as the parameters indicative of intestinal barrier function and inflammatory response in the mucosa of the pigs were measured accordingly.
2. Materials and methods
This trial was performed in accordance with the Guide for the Care and Use of Laboratory Animals, and was approved by Sichuan Agricultural University Institutional Animal Care and Use Committee (DKYB20131704).
2.1. Animal management and experimental design
A two-week pre-trial with 102 crossbred (Duroc × [Landrace × Yorkshire]) finishing barrows (90 days old) was conducted to screen specific enterotypes based on the described method (Bergström et al., 2012; Sandberg et al., 2019). In brief, fecal samples were collected at baseline from the rectum of pigs in a commercial pig farm (New Hope Santai Agriculture and Animal Husbandry Co., Ltd.). Pigs were fed with commercial grower pig feed (corn-soybean meal based; the AMR is 0.47). Approximately 2 g of fecal sample from each pig was directly collected and transferred to a 2.5-mL CryoTube and stored at −80 °C until required. Genomic DNA of each sample was extracted from the frozen pellets using an E.Z.N.A Stool DNA Kit (Omega Bio-Tek, Doraville, GA) according to the manufacturer's instructions. The number of Prevotella and Bacteroides in each sample was quantified with a real-time PCR (qPCR) targeting the corresponding 16S rRNA gene, respectively. The sequences of primers used in the current study and the length of amplicons are listed in Table 1. The conditions for qPCR were previously reported (Bergström et al., 2012). During the pre-trial period (2 weeks), pigs in the pig farm were fed with the same feed. According to a previous study (Roager et al., 2014), PBR in swine gut is stable and can last for at least 6 months, thus it was reasonable to use the results of the pre-trial as the basis for grouping. Based upon the baseline PBR of each pig, a total of 16 barrows with high PBR (HPBR, PBR≥ 14) and another 16 barrows with low PBR (LPBR, PBR≤ 2) were finally selected for the formal study. The average body weight of these pigs was 87.37 ± 2.76 kg. All pigs were assigned into 4 groups with 8 replicates per group and 1 pig per replicate in a 2 (HPBR and LPBR enterotypes) × 2 (HAMR and LAMR diets) factorial arrangement.
Table 1.
The sequences of primers used for the real-time PCR analysis in the current study.
| Targeting gene or microorganism | Primer sequence (5′ to 3′) | References/Accession no. |
|---|---|---|
| Prevotella | F: CACCAAGGCGACGATCA | Bergström et al. (2012) |
| R: GGATAACGCCTGGACCT | ||
| Bacteroides | F: CGATGGATAGGGGTTCTGAGAGGA | Bergström et al. (2012) |
| R: GCTGGCACGGAGTTAGCCGA | ||
| β-Actin | F: TCTGGCACCACACCTTC | NM_001101 |
| R: TGATCTGGGTCATCTTC | ||
| GAPDH | F: TGAAGGTCGGAGTGAACGGAT | NM_001206359.1 |
| R: CACTTTGCCAGAGTTAAAAGCA | ||
| Clostridium cluster XIVa | F: AAATGACGGTACCTGACTAA | Mu et al. (2017) |
| R: CTTTGAGTTTCATTCTTGCGAA | ||
| Clostridium cluster IV | F: GCACAAGCAGTGGAGT | Mu et al. (2017) |
| R: CTTCCTCCGTTTTGTCAA | ||
| Faecalibacterium prausnitzii | F: CCCTTCAGTGCCGCAGT | Nielsen et al. (2014) |
| R: GTCGCAGGATGTCAAGAC | ||
| Butyryl-CoA:acetate CoA-transferase | F: GCIGAICATTTCACITGGAAYWSITGGCAYATG | Louis and Flint (2007) |
| R: CCTGCCTTTGCAATRTCIACRAANGC | ||
| Butyrate kinase | F: GTATAGATTACTIRYIATHAAYCCNGG | Louis et al. (2004) |
| R: CAAGCTCRTCIACIACIACNGGRTCNAC | ||
| SMCT1 | F: CGCAGATTCCTACTAACC | Haene et al. (2013a) |
| R: GATTGTCAGTTCCACCAT | ||
| MCT1 | F: CATCAACTACCGACTTCTG | Haene et al. (2013a) |
| R: TACTGGTCTCCTCCTCTT | ||
| Mucin 1 | F: GTGCCGCTGCCCACAACCTG | XM_001926883.4 |
| R: AGCCGGGTACCCCAGACCCA | ||
| Mucin 2 | F: GGTCATGCTGGAGCTGGACAGT | XM_003122394.1 |
| R: TGCCTCCTCGGGGTCGTCAC | ||
| OCLN | F: CTACTCGTCCAACGGGAAAG | NM_001163647.2 |
| R: ACGCCTCCAAGTTACCACTG | ||
| ZO-1 | F: CAGCCCCCGTACATGGAGA | XM_005659811 |
| R: GCGCAGACGGTGTTCATAGTT | ||
| GPR43 | F: GATCGTCTGTGCCCTCATGG | JX566881.1 |
| R: GAAAGCCACTCCCAGGTAGC | ||
| HDAC | F: TGACGAGTCCTATGAGGCCA | XM_013999116.2 |
| R: CAAACTCCACACACTTGGCG | ||
| IL-6 | F: GGGAAATGTCGAGGCTGTG | NM_214399 |
| R: AGGGGTGGTGGCTTTGTCT | ||
| IL-12 | F: CCTGACTGCCTCCCACTTTC | NM_214013 |
| R: AGGAGTGACTGGCTCAGAAC | ||
| IL-1β | F: ACGTGCAATGATGACTTTGTCTG | NM_214055.1 |
| R: AGAGCCTTCAGCATGTGTGG | ||
| TNF-α | F: CGTGAAGCTGAAAGACAACCAG | NM_214022.1 |
| R: GATGGTGTGAGTGAGGAAAACG |
GAPDH = glyceraldehyde-3-phosphate dehydrogenase; SMCT1 = sodium-coupled monocarboxylate transporter 1; MCT1 = monocarboxylate transporter 1; Mucin 1 = epithelial mucus 1; Mucin 2 = epithelial mucus 2; OCLN = epithelial tight junction protein occludin; ZO-1 = epithelial tight junction protein zonula occludens-1; GPR43 = G protein-coupled receptor 43; HDAC = histone deacetylases; IL-6 = interleukin-6; IL-12 = interleukin-12; IL-1β = interleukin-1β; TNF-α = tumor necrosis factor-α.
2.2. Diets and feeding management
Following the NRC (Nutrient requirements of swine, 2012) recommendation for the nutrient requirements of 75- to 100-kg finishing pigs, a corn-soybean meal-based diet was formulated. Experimental diets (Table 2) included the HAMR and LAMR diets. The HAMR diet (AMR = 1.24) was supplemented with 30% high amylose (the concentration of amylose was 68.50%) corn starch and the LAMR diet (AMR = 0.23) was supplemented with 30% high amylopectin (the concentration of amylopectin was 81.53%) corn starch. The high amylose starch was purchased from Shanghai Quanwang Biotechnology co., LTD (Shanghai, China) and the high amylopectin starch was purchased from Shandong Fuyang Biological Starch co., LTD (Qingdao, China). Diets were fed in mash form throughout the experiment. The 4 experimental groups in the current study were defined as HPBR + HAMR, HPBR + LAMR, LPBR + HAMR and LPBR + LAMR, respectively.
Table 2.
The ingredients and nutritional levels of diets (as-fed basis).
| Item | HAMR | LAMR |
|---|---|---|
| Ingredients, % | ||
| Corn1 (7.8% CP) | 42.13 | 42.13 |
| High amylose corn starch2 | 30.00 | – |
| High amylopectin corn starch3 | – | 30.00 |
| Soybean meal (44.2% crude protein) | 26.00 | 26.00 |
| Limestone | 0.55 | 0.55 |
| Dicalcium phosphate | 0.75 | 0.75 |
| NaCl | 0.25 | 0.25 |
| l-Lys·HCl | 0.01 | 0.01 |
| dl-Met | 0.01 | 0.01 |
| Chloride choline | 0.05 | 0.05 |
| Vitamin and mineral premix4 | 0.25 | 0.25 |
| Total | 100.00 | 100.00 |
| Nutrient levels, % | ||
| Digestible energy, Mcal/kg | 3.52 | 3.52 |
| Crude protein | 14.88 | 14.88 |
| Ca | 0.53 | 0.53 |
| Total P | 0.45 | 0.45 |
| Available P | 0.27 | 0.27 |
| Total Lys | 0.72 | 0.72 |
| Total Met + Cys | 0.40 | 0.40 |
| Total Thr | 0.49 | 0.49 |
| Amylose | 29.41 | 10.04 |
| Amylopectin | 23.73 | 43.21 |
| AMR of diet | 1.24 | 0.23 |
AMR = amylose-to-amylopectin ratio; HAMR = high AMR; LAMR = low AMR.
Corn contained 21.01% amylose and 44.51% amylopectin.
High amylose corn starch contained 68.50% amylose and 16.60% amylopectin.
High amylopectin corn starch contained 3.95% amylose and 81.53% amylopectin.
Provided the following per kilogram of complete diet: 100 mg of Fe (as ferrous sulfate); 15 mg of Cu (as copper sulfate); 120 mg of Zn (as zinc sulfate); 40 mg of Mn (as manganese sulfate); 0.3 mg of Se (as Na2SeO3); 0.25 mg of I (as KI); 13,500 IU of vitamin A; 2,250 IU of vitamin D3; 24 IU of vitamin E; 6.2 mg of riboflavin; 25 mg of nicotinic acid; 15 mg of pantothenic acid; 1.2 mg of vitamin B12; and 0.15 mg of biotin.
Each pig was housed in an individual metabolism cage (0.8 m × 1.5 m) with woven wire flooring in a temperature-controlled room (22 ± 1 °C) and the relative humidity was consistently maintained at 65% to 75%. The pigs were fed 3 times every day (08:00, 13:00 and 18:00) during the whole experimental period. All animals were allowed ad libitum access to feed and water, and the leftovers were collected in time to avoided wasting. The pigs were weighed at the beginning and the end of the trial to calculate the average daily gain (ADG). Feed consumption was recorded every 3 days and the average daily feed intake (ADFI) of each pig was calculated. The feed conversion ratio (FCR) was defined as the ratio of ADFI to ADG. All pigs were healthy and did not receive any antibiotic treatment in the current study. After 60 d, pigs were moved out from the cage and slaughtered for sampling.
2.3. Slaughter surveys and sample collection
Pigs were slaughtered according to a standard commercial procedure (Zhang et al., 2019). The abdomen of each pig was opened immediately after electrically stunned, the colon was dissected from the mesentery and immediately placed on ice. Approximate 5 g of the digesta in the middle colon were collected into a sterilized tube and frozen in liquid nitrogen. The colonic mucosa from the middle colon of each pig was firstly flushed with ice-cold saline and the mucosal layer was sequentially obtained through carefully scraping with a glass microscope slide. All the collected samples were stored at −80 °C for the following analysis.
2.4. Determination of bacteria by real-time PCR method
The genomic DNA from each colonic digesta sample was extracted using an E.Z.N.A Stool DNA Kit (Omega Bio-Tek, Doraville, GA) according to the manufacturer's instructions. The sequences of specific primers and probes for Prevotella, Bacteroides, the main butyrate-producing bacterial groups including Clostridium cluster IV, Clostridium cluster XIVa and Faecalibacterium prausnitzii are shown in Table 1 (Nielsen et al., 2014; Mu et al., 2017). The quantification of Prevotella and Bacteroides, as well as the calculation of PBR was conducted using the same method as that used in the pre-trial. The method for real-time PCR analysis of other bacterial groups has been described before (Mu et al., 2017). In brief, the copy numbers for each bacterial group were calculated according to the standard curve. Plasmids containings the insert of the responding specific gene for the standard curve of each bacterial group were set up first, then the standard curve was generated using a 10-fold serial dilution of the plasmid DNA. Quantification was performed in triplicates and copy numbers were transformed (log10) to allow for statistical analysis.
2.5. SCFA measurement by gas chromatography
The concentrations of the main SCFA including acetate, propionate, and butyrate were measured with a previously described method (Tang et al., 2019). A 2-mL tube was put on the electronic scales and zeroed. Then, 1 g of the thawed digesta sample was transferred into the tube using a sterilized weighing scoop. Then, it was brought to 1.5 mL by adding ultrapure water. The mixture was then voxtex mixed and incubated at 4 °C for 30 min. The prepared solution was centrifuged (12,000 × g, 4 °C) for 10 min. The supernatant (1 mL) was removed into another 1.5-mL centrifuge tube, and then mixed with 0.2 mL of metaphosphoric acid (25%) and 23.3 μL of crotonic acid (210 mmol/L) simultaneously, and centrifuged (12,000 × g, 4 °C) again for 10 min before being placed in an ice-bath for 30 min. A volume of 500 μL of the supernatant was mixed with isopyknic methanol and then homogenized for 10 min in another sterilzed tube. After that, the mixture was centrifuged at 12,000 × g for 10 min at 4 °C. Ten microliters of the supernatant was finally injected into a gas chromatographic system (VARIAN CP-3800, America) for the measurement of the SCFA. Peaks were identified by comparing their retention times with the standard references.
2.6. The quantity of genes in colonic mucosa by real-time PCR method
Total RNA from each mucosa sample was isolated by TRIzol reagent (Invitrogen) following the manufacturer's protocol. After measuring the quality, total RNA (1 μg) was reverse transcribed to cDNA in a 20 μL final volume using a PrimeScript RT reagent Kit (Takara, Dalian, China). The real-time PCR for each gene was carried out in triplicates using the SYBR Premix Ex Taq II (Takara, Dalian, China) on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). The relative expression of each gene was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) and normalized according to the expression of β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively. The sequences of primers and references for butyrate-producing genes including butyryl-CoA:acetate CoA-transferase and butyrate kinase (Louis et al., 2004; Louis and Flint, 2007), SCFA uptake transporters such as mucosa sodium-coupled monocarboxylate transporter 1 (SMCT1) and monocarboxylate transporter 1 (MCT1), SCFA signaling genes including mucosa histone deacetylases (HDAC) and G protein-coupled receptor 43 (GPR43), gut physical barrier involved genes such as epithelial mucus (Mucin 1 and Mucin 2), epithelial tight junction protein occludin (OCLN), and epithelial tight junction protein zonula occludens-1 (ZO-1), as well as the inflammatory cytokines including interleukin-6 (IL-6), IL-12, IL-1β and tumor necrosis factor-α (TNF-α) are listed in Table 1.
2.7. Statistical analysis
The effect of enterotypes (PBR) and starch types (AMR) (a 2 × 2 factorial design) on different variables was analyzed using the general linear model procedures of the SAS (Version 9.4; SAS Institute, Gary, NC). The factors of the models included the main effects of PBR (HPBR and LPBR) and dietary AMR (HAMR and LAMR) as well as their interaction, which was analyzed using the model: Yijk = μ + αi + βj + (αβ)ij + εijk, where Yijk is the dependent variable, μ is the overall mean, αi is the effect of enterotype, βj is the effect of starch type, (αβ)ij is the interaction between the enterotype and starch type, and εijk represents the unexplained random error. When P-value for interaction was less than 0.05, the multiple comparisons of the 4 treatments were analyzed using SAS adjusted by Tukey Kramer. Overall, P < 0.05 was defined to indicate a significant statistical difference, and 0.05 ≤ P < 0.10 was considered to indicate a statistical tendency.
3. Results
3.1. Enterotypes inferred by Prevotella/Bacteroides ratio
The growth performance (including ADG, ADFI and FCR) of pigs in the 4 treatments showed no significant difference (Table 3). To verify whether there were Prevotella or Bacteroides dominated enterotypes in the gut of the pigs, a real-time PCR analysis targeting these 2 genera was first conducted in 102 finishing pigs. A total of 69 subjects could be grouped by plotting the relative abundance of Prevotella against the relative abundance of Bacteroides, resulting in 2 clearly separated “clouds” in the 2-dimensional space (Fig. 1A). A kernel density plot, which can be considered a refinement of a frequency plot, showed a pronounced bimodal distribution with part of the subjects when plotting the PBR (Fig. 1B, P < 0.05). The 69 pigs in the 2 clearly separated “clouds” were the candidates of the formal trial. Of these candidates, 16 subjects with HPBR (≥14) and another 16 subjects with LPBR (≤2) (Fig. 1C, P < 0.05) were finally selected for the following animal trial. At the end of the 60-day trial, the PBR in HPBR + LAMR, HPBR + HAMR, LPBR + LAMR and LPBR + HAMR pigs was 5.07 ± 0.34, 6.29 ± 0.34, 0.48 ± 0.34 and 0.87 ± 0.34 (average PBR ± SEM), respectively.
Table 3.
Effects of PBR and AMR on growth performance of finishing pigs.
| Item | HPBR |
LPBR |
SEM |
P-value |
||||
|---|---|---|---|---|---|---|---|---|
| LAMR | HAMR | LAMR | HAMR | PBR | AMR | PBR × AMR | ||
| Initial weight, kg | 87.4 | 87.5 | 87.2 | 87.4 | 2.76 | 0.957 | 0.963 | 0.991 |
| Final weight, kg | 123.5 | 125.0 | 123.0 | 122.9 | 3.32 | 0.686 | 0.837 | 0.813 |
| ADG, g/d | 977 | 1014 | 966 | 959 | 49 | 0.514 | 0.763 | 0.654 |
| ADFI, g/d | 2869 | 3028 | 2928 | 2917 | 158 | 0.883 | 0.631 | 0.584 |
| FCR | 2.93 | 3.00 | 3.04 | 3.07 | 0.11 | 0.435 | 0.663 | 0.839 |
PBR = Prevotella-to-Bacteroides ratio; AMR = amylose-to-amylopectin ratio; HPBR = high PBR; LPBR = low PBR; LAMR = low AMR; HAMR = high AMR; ADG = average daily gain; ADFI = average daily feed intake; FCR = the ratio of ADFI to ADG.
Fig. 1.
Inferred Prevotella/Bacteroides groups. (A) The log-normalized abundances of Bacteroides spp. versus the log-normalized abundance of Prevotella spp. for all of the 102 pigs in the pre-selection period. Subjects fall into 2 groups, indicated with 2 circles. (B) Kernel density plots of log-normalized relative abundance of Prevotella-to-Bacteroides ratio (PBR). (C) Two discrete enterotype groups of pigs were selected based on their PBR (HPBR ≥ 14 or LPBR ≤ 2). HPBR, high PBR group; LPBR, low PBR group. ∗∗, P < 0.01, means an extremely significant difference.
3.2. The abundance of butyrate-producing bacteria and their functional butyrate related genes
According to the results of real-time PCR analysis, the copy numbers of Clostridium cluster IV and F. prausnitzii showed no significant difference among the 4 groups (Fig. 2A and B). The copy numbers of Clostridium cluster XIVa in HPBR pigs were significantly increased (P < 0.05) compared with the LPBR pigs (Fig. 2C). Meanwhile, the copy numbers of Clostridium cluster XIVa in HAMR pigs were significantly higher (P < 0.01) than that of LAMR pigs. Along with the higher copy numbers of Clostridium cluster XIVa, the expression of butyryl-CoA:acetate CoA-transferase and butyrate kinase (Fig. 2D and E) genes were also significantly higher (P < 0.01) in the HAMR pigs compared to the LAMR pigs. Furthermore, the pigs in the HPBR + HAMR group showed a significantly higher (P < 0.05) expression of butyrate kinase gene compared to the pigs in all other treatments.
Fig. 2.
The composition and activity of butyrate-producing bacteria. The 16S rRNA gene copies numbers of (A) Clostridium cluster IV, (B) Clostridium cluster XIVa, (C) Faecalibacterium prausnitzii. All of the copies were transformed (log10) before statistical analysis. The relative copy number of (D) butyryl-CoA:acetate CoA-transferase and (E) butyrate kinase. The cDNA copy number was calculated as the ratio of other treatment to HPBR + LAMR treatment. HPBR = high Prevotella-to-Bacteroides ratio, LPBR = low Prevotella-to-Bacteroides ratio, HAMR = high amylose-to-amylopectin ratio, LAMR = low amylose-to-amylopectin ratio. Data are the means ± SEM, n = 8. ∗, P < 0.05, means a significant difference, ∗∗, P < 0.01, means an extremely significant difference.
3.3. The concentration of SCFA in the colonic digesta of pigs in different groups
As shown in Fig. 3, the concentration of total SCFA, acetate, propionate and butyrate were influenced by both PBR and AMR. A significant interaction between the PBR and AMR on the concentration of total SCFA was observed (P < 0.05), and the highest total SCFA concentration was found in the HPBR + HAMR pigs. An interaction between PBR and AMR that tended to be significant was found in the concentration of acetate (P = 0.09) and propionate (P = 0.07), respectively. In addition, the concentrations of total SCFA and propionate in the HPBR pigs were significantly higher (P < 0.05) than that in the LPBR pigs. On the other hand, comparing the results in HAMR and LAMR pigs only, a main effect of AMR was detected on the concentration of propionate and butyrate, revealing a significantly higher (P < 0.05) concentration of propionate and butyrate in the HAMR pigs.
Fig. 3.
The concentrations of short chain fatty acids. (A) Total SCFA, (B) acetate, (C) propionate, (D) butyrate. SCFA = short chain fatty acids; HPBR = high Prevotella-to-Bacteroides ratio, LPBR = low Prevotella-to-Bacteroides ratio, HAMR = high amylose-to-amylopectin ratio, LAMR = low amylose-to-amylopectin ratio. Data are the means ± SEM, n = 8. ∗, P < 0.05, means a significant difference. A significant interaction between the PBR and AMR on the concentration of total SCFA was observed (P = 0.02). An interaction between PBR and AMR that tended to be significant was found in the concentration of acetate (P = 0.09) and propionate (P = 0.07), respectively.
3.4. The expression of genes involved in transport and utilization of SCFA in the colonic mucosa of pigs
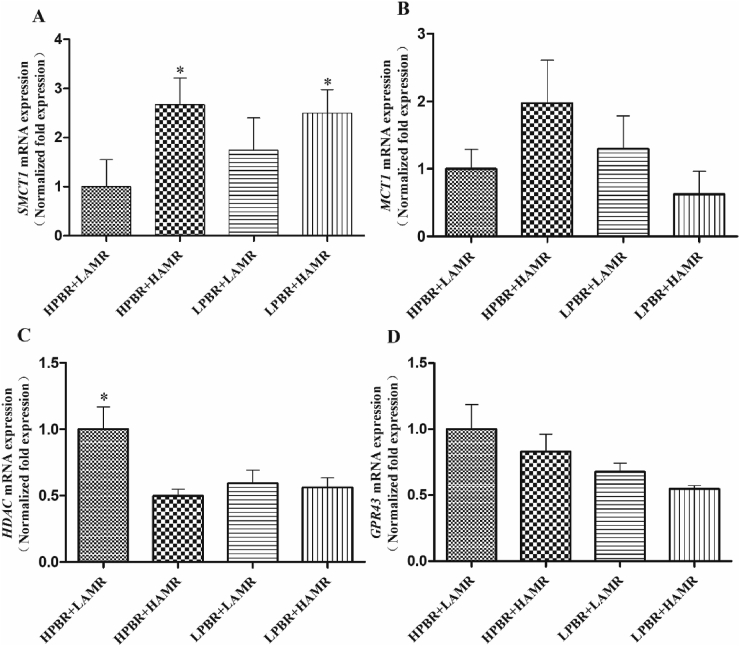
The real-time PCR analysis showed that the expression of SMCT1 in the HAMR pigs was significantly increased (P < 0.05) compared to the LAMR pigs (Fig. 4A). But the expression of another main SCFA transporter encoding gene, the MCT1, was shown to be neither influenced by PBR nor AMR (Fig. 4B). There was a tendency of interaction between PBR and AMR on the expression of HDAC (P = 0.05), while the expression of HDAC in the HPBR + HAMR pigs was decreased (P < 0.05) compared to the other treatment groups (Fig. 4C). Compared with the LAMR pigs, the expression of HDAC was found significantly increased (P < 0.05) in the HAMR pigs. Furthermore, PBR displayed a main effect (P < 0.05) on the expression of GPR43, which is one of the main SCFA receptors (Fig. 4D).
Fig. 4.
The gene expression of SCFA transporters and receptors. The relative copy number of SCFA transporters (A) SMCT1 and (B) MCT1. The relative copy number of SCFA receptors (C) HDAC and (D) GPR43. The cDNA copy number was calculated as the ratio of target gene to internal reference gene (β-actin and GADPH). SCFA = short chain fatty acids, SMCT1 = sodium-coupled monocarboxylate transporter 1 gene, MCT1 = monocarboxylate transporter 1, HDAC = mucosa histone deacetylases, GPR43 = G protein-coupled receptor 43, HPBR = high Prevotella-to-Bacteroides ratio, LPBR = low Prevotella-to-Bacteroides ratio, HAMR = high amylose-to-amylopectin ratio, LAMR = low amylose-to-amylopectin ratio. Data are the means ± SEM, n = 8. ∗, P < 0.05, means a significant difference. For the expression of HDAC (Fig. 4C), the interaction between PBR and AMR tended to be significant different (P = 0.05).
3.5. The expression of genes encoding tight junction proteins in the colonic mucosa of pigs
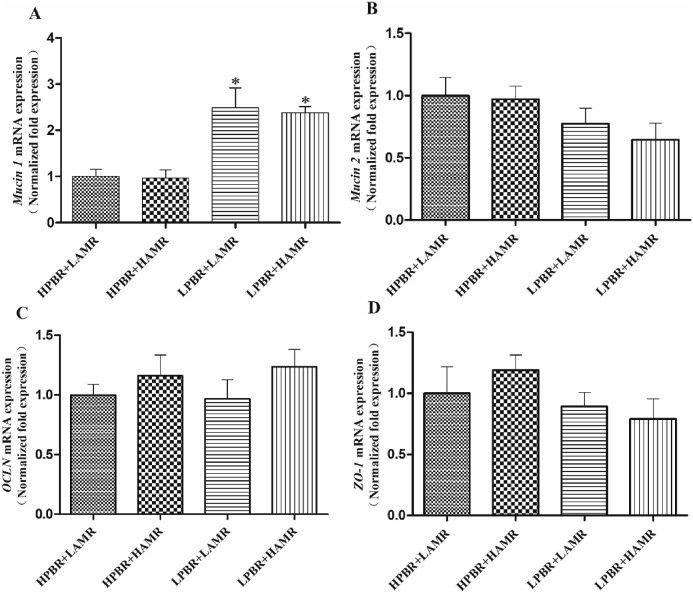
As shown in Fig. 5, the expression of Mucin1, but not Mucin2, in the HPBR pigs was significantly decreased (P < 0.05) compared to the LPBR pigs (Fig. 5A and B). PBR and AMR showed an interaction for the expression of Mucin2 (P < 0.05), but neither PBR nor AMR showed a main effect or interaction on the expression of ZO-1 or OCLN (Fig. 5C and D).
Fig. 5.
The gene expression of intestinal physical barriers. The relative copy number of epithelial mucus protein (A) Mucin 1 and (B) Mucin 2. The relative copy number of tight junction protein (C) OCLN and (D) ZO-1. The cDNA copy number was calculated as the ratio of target gene to internal reference gene (β-actin and GADPH). Mucin 1 = epithelial mucus 1, Mucin 2 = epithelial mucus 2, OCLN = epithelial tight junction protein occluding, ZO-1 = epithelial tight junction protein zonula occludens-1, HPBR = high Prevotella-to-Bacteroides ratio, LPBR = low Prevotella-to-Bacteroides ratio, HAMR = high amylose-to-amylopectin ratio, LAMR = low amylose-to-amylopectin ratio. Data are the means ± SEM, n = 8. ∗, P < 0.05, means a significant difference. PBR and AMR showed an interaction for the expression of Mucin 2 (P = 0.01).
3.6. The expression of genes encoding inflammatory cytokines in the colonic mucosa of pigs
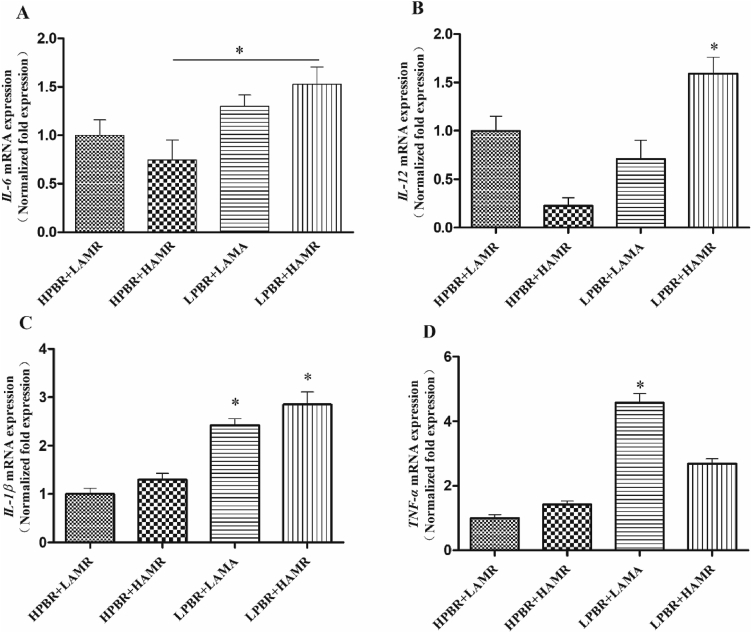
Results showed that the gene expression of IL-6 in the colonic mucosa of the pigs was significantly influenced by PBR (P < 0.01), and it was significantly decreased in the HPBR pigs compared to the LPBR pigs (Fig. 6A). There was an interaction between PBR and AMR on the gene expression of IL-12 (P < 0.01) which was found to be the lowest in the pigs from the HPBR + HAMR group (Fig. 6B). Compared with the LPBR pigs, the gene expression of IL-12 was significantly decreased (P < 0.05) in the HPBR pigs. Meanwhile, the expression of IL-12 was significantly increased (P < 0.01) in the HAMR pigs compared to the LAMR pigs. A significantly higher (P < 0.05) gene expression of IL-12 was found in the LPBR + HAMR pigs compared to the other pigs. The gene expression of IL-1β in the LPBR pigs was significantly increased (P < 0.01) compared to the HPBR pigs (Fig. 6C), while it was decreased in the LAMR pigs compared to the HAMR pigs. An interaction in the gene expression of TNF-α was found between PBR and AMR (P < 0.01). The gene expression of TNF-α in the HPBR pigs was significantly decreased (P < 0.01) compared to the LPBR pigs (Fig. 6D). Meanwhile, a lower gene expression of TNF-α was accompanied with an increasing AMR (P < 0.01).
Fig. 6.
The gene expression of mucosa inflammation. The relative copy number of mucosa inflammation relate genes (A) IL-6, (B) IL-12, (C) IL-1β and (D) TNF-α. The cDNA copy number was calculated as the ratio of target gene to internal reference gene (β-actin and GADPH). IL-6 = interleukin-6, IL-12 = interleukin-12, IL-1β = interleukin-1β, TNF-α = tumour necrosis factor-α, HPBR = high Prevotella-to-Bacteroides ratio, LPBR = low Prevotella-to-Bacteroides ratio, HAMR = high amylose-to-amylopectin ratio, LAMR = low amylose-to-amylopectin ratio. Data are the means ± SEM, n = 8. ∗, P < 0.05, means a significant difference. There was an interaction between PBR and AMR on the gene expression of IL-12 (P < 0.01) and TNF-α (P < 0.01).
4. Discussion
Different compositions of the gut microbiota have the potential to affect the gut health through the different individual responses to diets (Zeevi et al., 2015; Korem et al., 2017). Previous in vitro research has shown how individuals with different PBR led to distinct profiles of SCFA from the same carbohydrate substrates (Kovatcheva-Datchary et al., 2015; Chen et al., 2017; Wu et al., 2017; Poeker et al., 2018). On the other hand, more and more in vitro or in vivo research has confirmed that dietary HAMR could benefit host health. Accumulating evidence has indicated that diets containing starch with a HAMR can increase the mass of digesta in the distal intestine, the concentration of SCFA, and the SCFA producing populations in the gut (Bird et al., 2007, 2009; Regmi et al., 2011; Yu et al., 2019). Both Prevotella and Bacteroides have the capability to utilize complex carbohydrates in the diet (Chen et al., 2017), and Prevotella in the gut of most monogastric animals prefer to ferment dietary starch (Flint et al., 2012; Yu et al., 2019). However, the health relevance of enterotypes in pigs remains unknown. In the current study, we found that enterotypes dominated by Prevotella spp. or Bacteroides spp. could also be identified in the GIT of pigs, which is similar to those findings in the human gut. Here we also first report that the interaction between the indigenous PBR in swine gut and dietary AMR may affect the production of SCFA, intestinal barrier and immune function of the host. With the data of PBR after the 60-day trial, we could find that the PBR in HPBR pigs was still remarkably higher than that in LPBR pigs. On the other hand, AMR of diet may not be the main factor influencing PBR in the colon of the pigs.
Of all the SCFA in the hindgut of human and pigs, butyrate is recognized to play an important role in promoting intestinal barrier function and reducing inflammation (Koh et al., 2016), which prompted the investigation of composition and activity of butyrate-producing bacteria in the colon of pigs in the current study. It is reported that the main butyrate-producing microorganisms include F. prausnitzii, Clostridium cluster IV and XIVa (Mu et al., 2017). In the current study, the colon of pigs with HPBR (≥14) harbored a remarkably higher abundance of Clostridium cluster XIVa which could have also been increased by the HAMR. High amylose-to-amylopectin ratio has been proved to benefit the proliferation of butyrate-producing bacteria (Haenen et al., 2013a; Sun et al., 2016), but knowledge of PBR on the abundance of these beneficial microbes is very limited. It has been reported that some species belonging to Clostridium cluster XIVa are primary degraders of complex carbohydrates such as dietary fibers (Flint et al., 2008). Our results thus indicate that the enterotype dominated by Prevotella and higher dietary AMR may probably enrich the butyrate-producing bacteria such as Clostridium cluster XIVa. The 2 key enzymes, butyryl-CoA:acetate CoA-transferse and butyrate kinase, are related to the last steps of butyrate production (Louis and Flint, 2009) in most of the butyrate-producing bacteria. So both the expression of the genes encoding these 2 enzymes and the abundance of butyrate-producing bacteria provide practical indicators for the activity of butyrate production of the microflora (Mu et al., 2017). In the current study, along with the higher copy numbers of Clostridium cluster XIVa, the genes expression level of butyryl-CoA:acetate CoA-transferse and butyrate kinase in the colonic digesta of pigs with HPBR and HAMR was also found to be notably increased compared to other pigs, indicating an enhanced activity of butyrate production in these animals. However, we did not find a significant increase of butyrate in the colon of these pigs, which may be due to the rapid absorption of butyrate by the colonocytes. Butyrate is reported to be almost entirely used by colonocytes as their preferred energy substrate in a very short time (Haenen et al., 2013b), leading to difficulties in its measurement in collected digesta samples. But as an important regulator of the host immune system, butyrate has been shown to be increased by the high level of dietary amylose in previous studies (Topping and Clifton, 2001; Yang et al., 2018a; Yu et al., 2019), which are verified by the current study.
The PBR and dietary AMR also showed an interaction on the concentration of total SCFA. It is well known that SCFA are generally recognized as the marker of a healthy microbiome (Flint et al., 2012). Previous in vitro studies have demonstrated that the metabolic activity of intestinal microbiota differs among different enterotypes. Prevotella-driven enterotypes have repeatedly been observed to have a higher capacity for degrading the dietary fibers compared to other enterotypes, resulting in a higher production of total SCFA and propionate (Chen et al., 2017; Wu et al., 2017; Poeker et al., 2018; Christensen et al., 2019) and butyrate (Chen et al., 2017). These results are consistent with our finding that the concentration of total SCFA was significantly increased in the colon of pigs with HPBR. On the other hand, some reports demonstrate that HAMR can markedly increase the relative abundance of Prevotella, but not Bacteroides (Sun et al., 2016; Maier et al., 2017; Yu et al., 2019), suggesting a potential interaction between PBR and AMR, which was further confirmed by the current study. In addition, earlier reports have shown that higher fecal propionate are associated with an increased abundance of Prevotella species (Salonen and de Vos, 2014; De Filippis et al., 2016). The Prevotellaceae-dominated microbiota is characterized by high propionate production and can be explained by propionate–producing capacity of Prevotella spp. (Kolmeder et al., 2016; Louis and Flint, 2017). Similarly, we also found an increased concentration of propionate in the colon of pigs with HPBR compared with the other pigs.
The SCFA produced by the microorganisms in the colon are almost totally absorbed by colonocytes either through diffusion or by the help of monocarboxylate transporters like SMCT1 and MCT1 (Halestrap and Meredith, 2004; Ganapathy et al., 2013). In the current study, the gene expression of SMCT1 was found to be higher in the pigs fed diets with HAMR, which is consistent with the higher concentration of total SCFA in these pigs. The ability of SCFA to modulate the physiological process of intestinal epithelial cells in monogastric animals is thought to depend on 2 major mechanisms. The first one involves epigenetic modulation. SCFA, especially butyrate, acetate, and propionate, have been established as intrinsic inhibitors of HDAC via the inhibition of the HDAC-induced deacetylation of lysine residues within histones (MacDonald and Howe, 2009). According to our results, the gene expression of HDAC in the pigs with HPBR and fed diets with HAMR was decreased compared to the other pigs, indicating a possible reduced immune activation to promote the gut health. The second mechanism regarding SCFA effects is signaling through specific receptors such as the G-protein-coupled receptors (GPR) to affect the mucosal immune system (Kasubuchi et al., 2015; Rios-Covian et al., 2016). GPR43 is one of the predominant receptors of SCFA and is implicated in the maintenance of intestinal homeostasis (Sun et al., 2018), such as the motility of GIT (Shen et al., 2017). In the current study, the pigs with HPBR had a higher gene expression of GPR43 in the colonic mucosa than those pigs with lower PBR, implying that HPBR may be more important for the modulation of GPR43-invovled intestinal homeostasis.
The mucus layer (primarily composed of Mucin 1 and Mucin 2) and tight junction proteins (ZO-1 and OCLN) are crucial in maintaining the integrity of intestinal epithelial cells (Fan et al., 2019). Our findings indicate that PBR or dietary AMR may have almost no impact on the physical barrier in the colon of finishing pigs.
Inflammatory cytokines are a class of endogenous peptides mediating a variety of immune responses. Previous work has reported that SCFA receptors are regulators of inflammation (Sivaprakasam et al., 2016). In the current study, the genes expression of IL-1β, IL-6, IL-12 and TNF-α in colonic mucosa were analyzed, as they are important in the occurrence and development of intestinal inflammation. We found that compared with pigs that harbored lower Prevotella, the gene expression of certain pro-inflammatory cytokines in the colonic mucosa of pigs with HPBR was increased, which could be enhanced by the ingestion of HAMR diet. Such findings further indicate that the enterotype driven by Prevotella, rather than that driven by Bacteroides, may be more beneficial for the gut health of pigs. The interaction between PBR and dietary AMR should be considered in the feed formulation. Pigs with Prevotella dominated enterotype and simultaneously accepted HAMR diet may have stronger resistance to intestinal disease.
5. Conclusion
In conclusion, results in the current study provide evidence that pigs with Prevotella-rich enterotype fed high amylose-to-amylopectin ratio diet (HPBR + HAMR) increased the number and activity of butyrate-producing bacteria and the concentration of total SCFA and propionate, which may further result in the decreased expression of mucosal inflammation associated genes.
Author contributions
Daiwen Chen, Honglin Yan, Yuheng Luo and Bing Yu: conceptualization; Wen Ren, Jie Yu, Ping Zheng, Zhiqing Huang and Junqiu Luo: investigation; Xiangbin Mao, Jun He and Honglin Yan: resources; Yuheng Luo, Jun He and Honglin Yan: formal analysis; Wen Ren, Honglin Yan and Yuheng Luo: writing-original draft; Yuheng Luo, Daiwen Chen and Maria C. Walsh: writing-review & editing; Daiwen Chen, Bing Yu and Yuheng Luo: supervision and funding acquisition.
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that might inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
The work presented in this manuscript was supported by The National Natural Science Foundation of China (NSFC, grant number 31730091, 31872369 and 31672436).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Contributor Information
Daiwen Chen, Email: dwchen@sicau.edu.cn.
Yuheng Luo, Email: luoluo212@126.com.
References
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R. Enterotypes of the human gut microbiome. Nature. 2011;473:174. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A., Licht T.R., Wilcks A., Andersen J.B., Schmidt L.R., Grønlund H.A. Introducing gut low-density array (GULDA)–a validated approach for qPCR-based intestinal microbial community analysis. FEMS Microbiol Lett. 2012;337:38–47. doi: 10.1111/1574-6968.12004. [DOI] [PubMed] [Google Scholar]
- Bird A.R., Vuaran M., Brown I., Topping D.L. Two high-amylose maize starches with different amounts of resistant starch vary in their effects on fermentation, tissue and digesta mass accretion, and bacterial populations in the large bowel of pigs. Br J Nutr. 2007;97:134–144. doi: 10.1017/S0007114507250433. [DOI] [PubMed] [Google Scholar]
- Bird A.R., Vuaran M., Crittenden R., Hayakawa T., Playne M.J., Brown I.L. Comparative effects of a high-amylose starch and a fructooligosaccharide on fecal bifidobacteria numbers and short-chain fatty acids in pigs fed Bifidobacterium animalis. Dig Dis Sci. 2009;54:947–954. doi: 10.1007/s10620-008-0451-3. [DOI] [PubMed] [Google Scholar]
- Chen T.T., Long W.M., Zhang C.H., Liu S., Zhao L.P., Hamaker B.R. Fiber-utilizing capacity varies in Prevotella-versus Bacteroides-dominated gut microbiota. Sci Rep. 2017;7:2594. doi: 10.1038/s41598-017-02995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen L., Vuholm S., Roager H.M., Nielsen D.S., Krych L., Kristensen M. Prevotella abundance predicts weight loss success in healthy, overweight adults consuming a whole-grain diet ad libitum: a post hoc analysis of a 6-wk randomized controlled trial. J Nutr. 2019;149:2174–2181. doi: 10.1093/jn/nxz198. [DOI] [PubMed] [Google Scholar]
- De Filippis F., Pellegrini N., Vannini L., Jeffery I.B., La Storia A., Laghi L. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- Fan H., Chen Z., Lin R., Liu Y., Wu X., Puthiyakunnon S. Bacteroides fragilis strain ZY-312 defense against cronobacter sakazakii-Induced necrotizing enterocolitis in vitro and in a neonatal rat model. mSystems. 2019;4 doi: 10.1128/mSystems.00305-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H.J., Bayer E.A., Rincon M.T., Lamed R., White B.A. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Scott K.P., Louis P., Duncan S.H. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- Ganapathy V., Thangaraju M., Prasad P.D., Martin P.M., Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. 2013;13:869–874. doi: 10.1016/j.coph.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Haenen D., Souza da Silva C., Zhang J., Koopmans S.J., Bosch G., Vervoort J. Resistant starch induces catabolic but suppresses immune and cell division pathways and changes the microbiome in the proximal colon of male pigs. J Nutr. 2013;143:1889–1898. doi: 10.3945/jn.113.182154. [DOI] [PubMed] [Google Scholar]
- Haenen D., Zhang J., Souza da Silva C., Bosch G., van der Meer I.M., van Arkel J. A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. J Nutr. 2013;143:274–283. doi: 10.3945/jn.112.169672. [DOI] [PubMed] [Google Scholar]
- Halestrap A.P., Meredith D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflügers Archiv. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- Han G.Q., Xiang Z.T., Yu B., Chen D.W., Qi H.W., Mao X.B. Effects of different starch sources on Bacillus spp. in intestinal tract and expression of intestinal development related genes of weanling piglets. Mol Biol Rep. 2012;39:1869–1876. doi: 10.1007/s11033-011-0932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Chen D., Zhang K., Yu B. A high-amylopectin diet caused hepatic steatosis associated with more lipogenic enzymes and increased serum insulin concentration. Br J Nutr. 2011;106:1470–1475. doi: 10.1017/S0007114511001966. [DOI] [PubMed] [Google Scholar]
- He J., Chen D.W., Yu B. Metabolic and transcriptomic responses of weaned pigs induced by different dietary amylose and amylopectin ratio. PloS One. 2010;5 doi: 10.1371/journal.pone.0015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth M.F., Blaedel T., Bendtsen L.Q., Lorenzen J.K., Holm J.B., Kiilerich P. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int J Obes (Lond) 2019;43:149–157. doi: 10.1038/s41366-018-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth M.F., Roager H.M., Larsen T.M., Poulsen S.K., Licht T.R., Bahl M.I. Pre-treatment microbial Prevotella-to-Bacteroides ratio, determines body fat loss success during a 6-month randomized controlled diet intervention. Int J Obes (Lond) 2018;42:580–583. doi: 10.1038/ijo.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasubuchi M., Hasegawa S., Hiramatsu T., Ichimura A., Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015;7:2839–2849. doi: 10.3390/nu7042839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A., De Vadder F., Kovatcheva-Datchary P., Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Kolmeder C.A., Salojarvi J., Ritari J., de Been M., Raes J., Falony G. Faecal metaproteomic analysis reveals a personalized and stable functional microbiome and limited effects of a probiotic intervention in adults. PloS One. 2016;11 doi: 10.1371/journal.pone.0153294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korem T., Zeevi D., Zmora N., Weissbrod O., Bar N., Lotan-Pompan M. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metabol. 2017;25:1243–12453 e5. doi: 10.1016/j.cmet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P., Nilsson A., Akrami R., Lee Y.S., De Vadder F., Arora T. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metabol. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Le Sciellour M., Renaudeau D., Zemb O. Longitudinal analysis of the microbiota composition and enterotypes of pigs from post-weaning to finishing. Microorganisms. 2019;7 doi: 10.3390/microorganisms7120622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Louis P., Duncan S.H., McCrae S.I., Millar J., Jackson M.S., Flint H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Flint H.J. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Flint H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- Lu D., Tiezzi F., Schillebeeckx C., McNulty N.P., Schwab C., Shull C. Host contributes to longitudinal diversity of fecal microbiota in swine selected for lean growth. Microbiome. 2018;6:4. doi: 10.1186/s40168-017-0384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.H., Yang C., Wright A.-D.G., He J., Chen D.W. Responses in ileal and cecal bacteria to low and high amylose/amylopectin ratio diets in growing pigs. Appl Microbiol Biotechnol. 2015;99:10627–10638. doi: 10.1007/s00253-015-6917-2. [DOI] [PubMed] [Google Scholar]
- MacDonald V.E., Howe L.J. Histone acetylation: where to go and how to get there. Epigenetics. 2009;4:139–143. doi: 10.4161/epi.4.3.8484. [DOI] [PubMed] [Google Scholar]
- Mach N., Berri M., Estelle J., Levenez F., Lemonnier G., Denis C. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ Microbiol Rep. 2015;7:554–569. doi: 10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- Maier T.V., Lucio M., Lee L.H., VerBerkmoes N.C., Brislawn C.J., Bernhardt J. Impact of dietary resistant starch on the human gut microbiome, metaproteome, and metabolome. mBio. 2017;8 doi: 10.1128/mBio.01343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu C.L., Zhang L.L., He X.Y., Smidt H.K., Zhu W.Y. Dietary fibres modulate the composition and activity of butyrate-producing bacteria in the large intestine of suckling piglets. Anton Leeuw Int J G. 2017;110:687–696. doi: 10.1007/s10482-017-0836-4. [DOI] [PubMed] [Google Scholar]
- Newman M.A., Petri R.M., Grull D., Zebeli Q., Metzler-Zebeli B.U. Transglycosylated starch modulates the gut microbiome and expression of genes related to lipid synthesis in liver and adipose tissue of pigs. Front Microbiol. 2018;9:224. doi: 10.3389/fmicb.2018.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen T.S., Laerke H.N., Theil P.K., Sorensen J.F., Saarinen M., Forssten S. Diets high in resistant starch and arabinoxylan modulate digestion processes and SCFA pool size in the large intestine and faecal microbial composition in pigs. Br J Nutr. 2014;112:1837–1849. doi: 10.1017/S000711451400302X. [DOI] [PubMed] [Google Scholar]
- Nutrient requirements of swine . Eleventh revised edition Washington, DC. The National Academies Press; 2012. [Google Scholar]
- Poeker S.A., Geirnaert A., Berchtold L., Greppi A., Krych L., Steinert R.E. Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model (PolyFermS) Sci Rep. 2018;8:4318. doi: 10.1038/s41598-018-22438-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y., Mach N., Lepage P., Levenez F., Denis C., Lemonnier G. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016;10:2973–2977. doi: 10.1038/ismej.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regmi P.R., Metzler-Zebeli B.U., Ganzle M.G., van Kempen T.A., Zijlstra R.T. Starch with high amylose content and low in vitro digestibility increases intestinal nutrient flow and microbial fermentation and selectively promotes bifidobacteria in pigs. J Nutr. 2011;141:1273–1280. doi: 10.3945/jn.111.140509. [DOI] [PubMed] [Google Scholar]
- Roager H.M., Licht T.R., Poulsen S.K., Larsen T.M., Bahl M.I. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new Nordic diet. Appl Environ Microbiol. 2014;80:1142–1149. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., de Los Reyes-Gavilan C.G., Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., de Vos W.M. Impact of diet on human intestinal microbiota and health. Annu Rev Food Sci T. 2014;5:239–262. doi: 10.1146/annurev-food-030212-182554. [DOI] [PubMed] [Google Scholar]
- Sandberg J., Kovatcheva-Datchary P., Bjorck I., Backhed F., Nilsson A. Abundance of gut Prevotella at baseline and metabolic response to barley prebiotics. Eur J Nutr. 2019;58:2365–2376. doi: 10.1007/s00394-018-1788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Lu Z., Xu Z., Chen Z., Shen Z. Associations among dietary non-fiber carbohydrate, ruminal microbiota and epithelium G-protein-coupled receptor, and histone deacetylase regulations in goats. Microbiome. 2017;5:123. doi: 10.1186/s40168-017-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprakasam S., Prasad P.D., Singh N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol Therapeut. 2016;164:144–151. doi: 10.1016/j.pharmthera.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Wu W., Chen L., Yang W., Huang X., Ma C. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9:3555. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Su Y., Zhu W. Microbiome-metabolome responses in the cecum and colon of pig to a high resistant starch diet. Front Microbiol. 2016;7:779. doi: 10.3389/fmicb.2016.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Qian Y., Yu B., Zhang T., Gao J., He J. Effects of Bacillus subtilis DSM32315 supplementation and dietary crude protein level on performance, gut barrier function and microbiota profile in weaned piglets. J Anim Sci. 2019;97:2125–2138. doi: 10.1093/jas/skz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping D.L., Clifton P.M. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Wang Y., Leong L.E.X., Keating R.L., Kanno T., Abell G.C.J., Mobegi F.M. Opportunistic bacteria confer the ability to ferment prebiotic starch in the adult cystic fibrosis gut. Gut Microb. 2019;10:367–381. doi: 10.1080/19490976.2018.1534512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Liu W., Chen H., Yin Y., Yu H., Wang X. Fermentation properties of isomaltooligosaccharides are affected by human fecal enterotypes. Anaerobe. 2017;48:206–214. doi: 10.1016/j.anaerobe.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Yang C., He J., Yu B., Yu J., Mao X.B., Chen D.W. The effect of dietary amylose/amylopectin ratio on serum and hepatic lipid content and its molecular mechanisms in growing-finishing pigs. J Anim Physiol An N. 2018;102:1657–1665. doi: 10.1111/jpn.12884. [DOI] [PubMed] [Google Scholar]
- Yang H., Yang M., Fang S., Huang X., He M., Ke S. Evaluating the profound effect of gut microbiome on host appetite in pigs. BMC Microbiol. 2018;18:215. doi: 10.1186/s12866-018-1364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Li Z.M., Chen W.D., Rong T., Wang G., Ma X.Y. Microbiome-metabolomics analysis investigating the impacts of dietary starch types on the composition and metabolism of colonic microbiota in finishing pigs. Front Microbiol. 2019;10:1143. doi: 10.3389/fmicb.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu B., Yu J., Zheng P., Huang Z.Q., Luo Y.H. Butyrate promotes slow-twitch myofiber formation and mitochondrial biogenesis in finishing pigs via inducing specific microRNAs and PGC-1alpha expression. J Anim Sci. 2019;97:3180–3192. doi: 10.1093/jas/skz187. [DOI] [PMC free article] [PubMed] [Google Scholar]