Abstract
Background
Disruption of DNA methylation (DNAm) is one of the key signatures of cancer, however, detailed mechanisms that alter the DNA methylome in cancer remain to be elucidated.
Methods
Here we present a novel integrative analysis framework, called MeLncTRN (Methylation mediated LncRNA Transcriptional Regulatory Network), that integrates genome-wide transcriptome, DNA methylome and copy number variation profiles, to systematically identify the epigenetically-driven lncRNA-gene regulation circuits across 18 cancer types.
Finding
We show that a significant fraction of the aberrant DNAm and gene expression landscape in cancer is associated with long noncoding RNAs (lncRNAs). We reveal distinct types of regulation between lncRNA modulators and target genes that are operative in either only specific cancers or across cancers. Functional studies identified a common theme of cancer hallmarks that lncRNA modulators may participate in. The coupled lncRNA gene interactions via DNAm also serve as markers for classifications of cancer subtypes with different prognoses.
Interpretation
Our study reveals a vital layer of DNAm and associated expression regulation for many cancer-related genes and we also provide a valuable database resource for interrogating epigenetically mediated lncRNA-gene interactions in cancer.
Funding
National Natural Science Foundation of China [91959106, 31871255].
Keywords: DNA methylation, LncRNA, Transcriptional regulation, Omics-data integration, Regulatory network
Research in context.
Evidence before this study
LncRNAs have long been recognized an integral component of chromatin that involved in the process of epigenetic modification. Recent evidence from in vitro studies indicates that lncRNAs could regulate DNA methylation by interacting with many epigenetic regulators, thus regulating gene expression in the cell at different conditions. However, there is still lack of a comprehensive investigation of this regulatory phenomenon at system level. In addition, the general mechanisms underlying this phenotype and its implications in tumorigenesis have not been systematically explored.
Added value of this study
By integrative analysis of genome wide expression, DNA methylation and copy number variation profiles from 5,970 samples across 18 major human cancers from The Cancer Genome Atlas (TCGA), we for the first time showed that lncRNA-associated DNA methylation regulation is prevalent and conserved across multiple cancer types. We proposed and explored four potential regulatory relationships that can explain the DNA methylation of protein coding genes and associated lncRNAs. Furthermore, we also investigated the biological and clinical significance of lncRNA associated DNA methylation regulation in terms of essential biological processes they participate and cancer hallmarks they present.
Implications of all the available evidence
Our present study is the first comprehensive analysis of lncRNA associated DNA methylation change in pan-cancer wide. Our work sheds new light on the complex interaction between lncRNAs and protein coding genes from the perspective of epigenetic regulation and its implications in tumorigenesis. It may serve as the first step towards the identification of driver lncRNA genes and may help in understanding the role of particular lncRNAs in cancer. After further functional validation, some of the lncRNAs may be utilized as cancer biomarker.
Alt-text: Unlabelled box
1. Introduction
Cancer is a complex disease which characterized by uncontrolled cell growth reflecting multiple hallmarks [1]. Beneath the aberrant cell proliferation is the complex interactions between a striking diversity of genetic and epigenetic factors, which give rise to the activation of critical oncogenes and inactivation of tumor suppressor genes in a cancer tissue-specific manner [2,3]. Among these, DNA methylation (DNAm) marks at the cytosine-phosphate-guanine (CpG) dinucleotide sites is extensively documented that regulate gene expression, genome stability and cell fate [4,5]. By regulating the chromatin accessibility and blocking recruitment of transcription factors (TFs) to cis-regulatory elements, methylation status within promoter region could determine regulatory activity of the target genes [6,7]. Alteration of the methylation status are well known to influence transcript abundance of many cancer-related genes, thus may define different types of ‘driver’ events, such as cell growth, proliferation, differentiation, and apoptosis processes [8], [9], [10]. Although DNAm related transcriptional dysregulation is closely associated with cancer, the underlying molecular mechanisms on how the DNA methylome patterns are determined in the transcriptional regulation circuitry in cancer remains largely to be discovered.
Being a complex process, DNA methylation status at particular site not only determined by the activities of DNA methyltransferases, which have little sequence specificity [11], but also affected by the highly coordinated functions of chromatin-remodeling complexes and histone modification enzymes. For instance, the Polycomb Repressive Complex 2 (PRC2) protein EZH2 was shown to interact with DNA methyltransferases and is crucial for recruitment of DNA methyltransferases at EZH2-target promoters [12]. Mutation of IDH1 could establish a hypermethylator phenotype and reorganization of the methylome and transcriptome in glioma [13]. We have also demonstrated a universal expression deregulation of epigenetic enzymes that associated with genome-wide DNA methylation patterns in cancers, several key genes including the UHRF1, WHSC1 and CBX7 were identified to play key roles in this process [14]. In spite of these advances, the key questions on how a precise methylation regulation at particular loci is achieved and whether there are other layers of regulator involved in remain still unanswered. Currently, accumulating evidence has indicated that long noncoding RNAs (lncRNAs) could be a kind of important regulatory factor that defines the genome-wide DNA methylation level.
LncRNAs are important regulators of gene expression at different levels, including transcriptional and post-transcriptional control [15,16]. One of the major advances for functional study of lncRNAs over the past decade has been the participating in epigenetic control by interacting with genes involved in chromatin organization or histone modification [17,18]. Emerging evidence has also indicated the underlying crosstalk between lncRNA and DNA methylation. For instance, a lncRNA arising from the CEBPA gene locus (ecCEBPA) that could compete with DNMT1, which inhibit CEBPA gene methylation and facilitates CEBPA expression [19]. Besides DNMT1, lncRNAs may also interact with other DNA methyltransferases to modulate their activity and DNA methylation patterns. The well-known lncRNA HOTAIR for example, was shown to recruit DNMT3B and to increase HOXA5 promoter methylation [20]. In addition to those by physical interaction with DNMTs, many other lncRNAs may participate in DNAm regulation through other mechanisms indirectly. Another well-known lncRNA H19, could bind to S-adenosylhomocysteine hydrolase (SAHH) and inhibits its function of hydrolysing S-adenosylhomocysteine (SAH). As a feedback inhibitor, SAH blocks S-adenosylmethionine (SAM) dependent DNMT3B that methylate at numerous genomic loci [21], [22], [23]. By regulation of the EZH2 and EED, the core subunits of PRC2, lncRNA LINC00470 could enhance the expression of ELFN2 through the hypomethylation at core promoters in glioblastoma [24]. These findings indicated that lncRNAs are emerging as important regulators of DNA methylation and associated expression dysregulation in cancer. In this case, a systematic identification of methylation related lncRNA modulators in cancer are urgently needed.
In this study, we propose a computational framework of DNA Methylation mediated LncRNA Transcriptional Regulatory Network (MeLncTRN) to integrate this substantial atlas of pan-cancer mRNA, lncRNA, DNA methylome and copy number variation (CNV) data from The Cancer Genome Atlas (TCGA) [25]. we discovered common DNA methylation regulatory architecture across 18 cancer types. Analysis of the DNA methylation associated gene expression pattern in the context of lncRNA expression revealed the complex impact of lncRNA on the methylation regulatory activity. Expression dynamics of many cancer related genes can be largely attributed to these lncRNA modulated methylation regulatory circuits. This comprehensive investigation of the context-specific transcriptional regulatory circuits will be a valuable resource for dissecting the underlying mechanism of gene expression dysregulation in tumors and enhance our understanding the interplay between cancer genome, epigenome and transcriptome.
2. Methods
2.1. Datasets used in the study
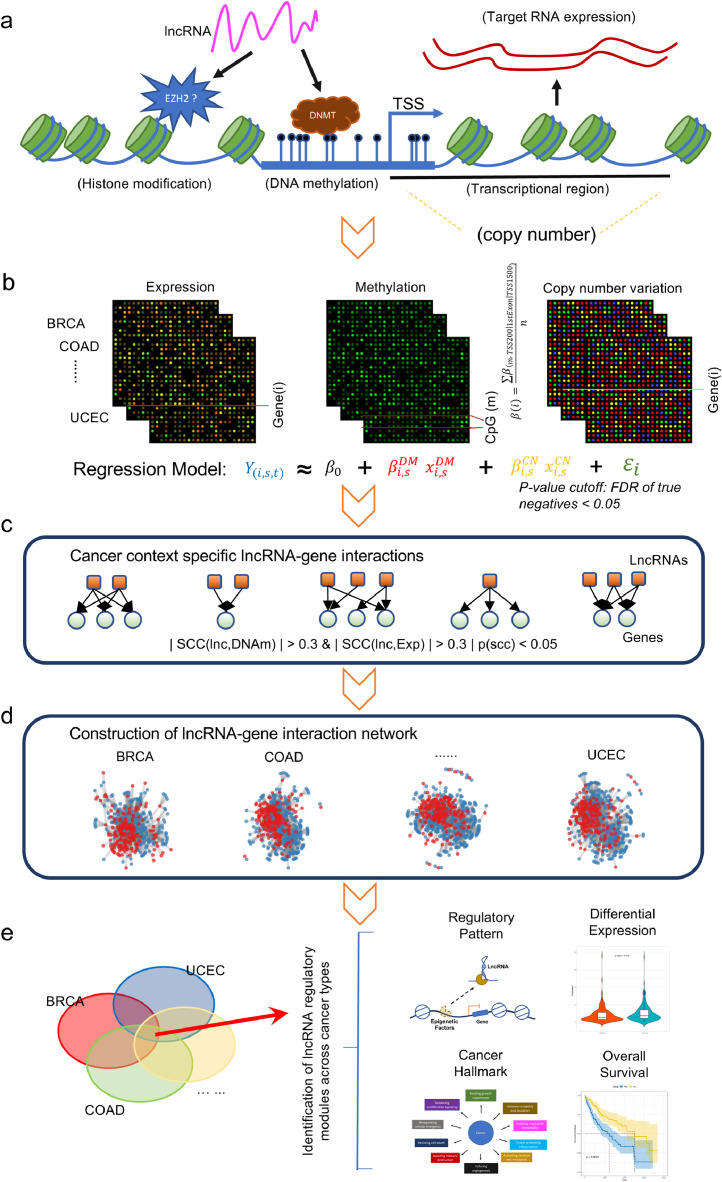
LncRNA and gene transcription profiling data: The genome-wide transcriptome data which includes both lncRNA and protein coding genes and quantified as FPKM (Fragments Per Kilobase per Million) were downloaded from TCGA. For propose of multiple omics data integration, we used the data for cancer types that had profiled sufficient numbers of samples for both RNA-Seq and DNAm data. This include bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney Clear Cell Carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC). Then we relied on the gene annotation of GENCODE (v22), which combines the HAVANA and Ensembl annotation pipelines to achieve an accurate classification of lncRNA and protein coding genes [26]. For lncRNAs, we collected those types of ‘antisense’, ‘sense_intronic’, ‘sense_overlapping’ and ‘lincRNA’ and ‘TEC’, which have sufficient members in each category. A total of 19,061 genes and 14,325 lncRNAs were identified in each cancer type. As the expression of lncRNAs are highly heterogeneous, we removed those lncRNAs whose PPKM value is equal to 0 in more than 10 percent of the samples to obtain the valid ones. For both genes and lncRNAs, the expression value was log2(FPKM + 1) transformed in order to regularize the data. Then we obtained the top components of data variation by using singular value decomposition (SVD) and correlated with normal/cancer status [27], in order to assess the inter-sample variability and quality of the data.
DNA methylation data: For those cancer types mentioned above, we downloaded the DNAm data generated with the Illumina Infinium HumanMethylation450 BeadChip array. The methylation level of each probe was measured as the beta-value, which ranges from 0 (unmethylated) to 1 (fully methylated). We removed the probes whose beta-values were missing in more than 30% of the samples, which result in 392,084 ~ 396,934 probes remain in these caner types with median number of 395,803, the remaining probes with missing values were imputed by using the k-nearest neighbors (KNN) method [28]. Finally, the BMIQ method was used to correct for the type II probe bias [29]. Similarly, we also performed the SVD quality control procedure for the DNA methylation data for each cancer type, as done for gene and lncRNA expression data. To obtain the DNAm value for each gene, we assign the methylation value of the probes within promoter region to that gene. Briefly, to assign the average beta value of probes mapping to within 200 bp of the transcription start site (TSS200) first, if no probe maps, then use the average beta value of probes mapping to the first exon of the gene (1stExon), if such probes are still absent, we use the average of probes mapping to upstream 1.5 kb from the TSS to the upstream 200 bp from the TSS (TSS1500). This procedure has been justified in previous publication [30]. In this way, we obtained methylation value for 16,131 genes in each cancer type.
Somatic copy number variation data: For the cancer types mentioned above, we obtained copy number segmentation data generated by Affymetrix SNP 6.0 platform from TCGA. The ‘nocnv.seg’ files for each sample were collected to capture the somatic CNV. Then we used the GISTIC2 with default parameters to obtain the focal copy number estimates at gene level [31].
2.2. Identification of DNA methylation regulated genes in cancer
Based on the methylation value at gene-level, we computed the moderated t-statistics using an empirical Bayes framework [32]. The same procedure was also applied to gene expression data. Methylation differences with false discovery rate (FDR) < 0.05 and with absolute difference of mean methylation levels between two groups of normal and cancer larger than 0.1 were considered statistically significant. Gene expression differences with FDR < 0.05 and with a log2 fold change between two groups more than 1 were considered statistically significant. To elucidate the individual DNAm-Exp relationship for each gene, we employed a multivariate linear regression model, which was fitted to the observed gene expression with DNAm and CNV as covariates. In this way, we guarantee that the relation between gene expression and DNAm is due to the same set of tumors, and rule out potential confounding gene expression change be explained by concomitant alterations at the CNV level. The resulting linear coefficients are indicative of the corresponding DNAm-Exp relationships in specific cancer type: the more negative they are the more likely the interactions are real. We filtered those genes with negative coefficients and FDR < 0.05 as confident genes with paired DNAm-Exp actions.
2.3. Define lncRNA modulators for methylation dysregulation
LncRNAs found to interact both with the DNA methylation and expression of protein coding genes were defined as candidate lncRNA modulators. To identify the lncRNA modulators with methylation regulatory function in different cancer types, we calculated the Spearman correlation coefficient (SCC) between the expression level of a lncRNA and the promoter methylation level, and also SCC between expression level of the lncRNA and the expression level of its candidate target genes. A functional lncRNA-gene interaction was defined if it meets all of the following criteria: SCC(lncRNA, DNAm) > 0.3 or SCC(lncRNA, DNAm) < -0.3, P < 0.05; and SCC(lncRNA, Exp) > 0.3 or SCC(lncRNA, Exp) < -0.3, P < 0.05, where SCC(lncRNA, DNAm) and SCC(lncRNA, Exp) represent the SCC of lncRNA-DNAm and lncRNA-Exp correlation, respectively. Using both coefficients for each lncRNA-gene pair, we then selected those with opposite signs of the coefficients, which indicates an anti-correlation between DNAm and gene expression. After assembling all identified lncRNA-gene pairs, we generated the DNA methylation mediated lncRNA transcriptional regulatory network for each cancer type.
For each lncRNA-gene interaction identified above, we classified the mode of action with respect to the effect of lncRNA regulators on target. LncRNA can activate or inhibit the activity of target genes, and lncRNAs can enhance or invert the activity of the target considering their differential expression status in cancer and normal samples. In total, there are four possible categories of actions were identified.
2.4. Topological measurements of the DNA methylation mediated lncRNA regulatory network
The DNA methylation mediated lncRNA regulatory networks were visualized by using Cytoscape [33]. Topological features were analysis by the package of ‘igraph’ in R language. For each node in the network, degree is defined as the number of edges incident to it. It is widely accepted that the hub genes with higher degrees in biological networks fundamentally determine the network's behavior and are more likely to be essential for network function [34]. We selected the top 10% of nodes with the highest degrees in the lncRNA regulatory network as the hub genes. To estimate the similarity of two networks between different cancers, we calculated the number of nodes and edges that are present in both networks (common lncRNA-gene interactions) and Jaccard index was calculated for similarity measure. In addition, a hypergeometric test was used to test if two networks significantly shared the common lncRNA-gene interactions.
2.5. Tissue specificity and conservation analysis of lncRNA modulators
To evaluate the tissue specificity of an lncRNA modulator, we employed the τ index to measure the tissue specificity of a given gene, which is represented as:
where n is the number of tissues examined, and E(i,max) is the highest expression signal of gene i across all tissues [35]. Here, we assembled a lncRNA transcriptome by averaging expression intensities across all normal samples in each particular human tissue from TCGA. Then we calculated the tissue specificity index of cancer specific lncRNA modulators, moderate lncRNA modulators and pan-cancer modulators and compared with Wilcox rank sum test. We also downloaded the transcriptome data for 31 human tissues from Genotype-Tissue Expression (GTEx) Program and performed the same analysis [36]. We also investigated the evolutionary conservation for these different lncRNA modulators. To do this, we downloaded the PhastCons scores for multiple alignments of 100 genomes to the human genome from UCSC Table Browser [37,38]. Then we calculated average PhastCons score for 200 nt at transcription start site as representative.
2.6. Collection of cancer related lncRNAs for functional validation
In order to explore the functional roles of these lncRNA modulators in tumorigenesis, we examined whether lncRNAs involved in the networks are intrinsic cancer driver genes or that are closely relevant with tumors. Thus, we collected the cancer related lncRNAs from LncRNADisease [39], Lnc2Cancer [40], MNDR (Mammalian ncRNA-Disease Repository) [41] and LncRNA Cancer Census [42] which are all manually curated databases for lncRNA dysregulation in human disease. Then, we used the hypergeometric test to evaluate whether these lncRNAs in the network are significantly enriched in our collected cancer related lncRNAs from public databases.
2.7. Collection of cancer hallmark genes for functional analysis
For functional annotation of the target genes of lncRNA modulators involved in network, we collected the cancer hallmark related Gene Ontology (GO) terms from a previous study [43]. Then, genes that annotated in these hallmark related GO terms were obtained from MsigDB database (v7.1), which hosts GO term sets for GSEA analysis [44]. The Gene Ontology enrichment analysis was performed by using TopGO, the enriched GO terms were obtained at the threshold of FDR < 0.05.
2.8. Identification of differentially expressed lncRNAs
It has been demonstrated that lncRNAs were expressed in a highly tissue specific manner and at considerable lower levels than protein coding genes [45,46]. To order to identify the differentially expressed lncRNAs reliably in each cancer type, we used two methods for this analysis: For the lncRNAs with expression level 0 in less than 30% of all samples were subjected to t-test. LncRNAs with fold change greater than two and FDR < 0.05 were identified as differentially expressed: up-regulated or down-regulated. If the lncRNA expression were 0 in more than 30% of all samples, for each lncRNA, we determined its expression status in binary mode: On (expression level > 0), Off (expression level = 0) in each sample. We next calculated the frequency of expression mode in both normal and cancer samples and also a fisher exact test was used to determine the differential expression status. LncRNAs expressed twice more frequently in cancer are determined as ‘switch-on’, whereas lncRNAs expressed twice more frequently in normal are determined as ‘switch-off’, and also the threshold of FDR adjusted fisher exact test p-value < 0.05 was used.
2.9. Identification of LncRNA regulatory modules
For the lncRNA regulatory network from each cancer, we identified biclique network modules which consist lncRNA modulators and their target genes regulated. Biclique module is a maximum network subgraph in which the vertices be partitioned into two disjoint sets, here it represents interaction between each vertex of one lncRNA to each vertex of target gene and no two vertices within the same set are adjacent. We used the Maximal Biclique Enumeration Algorithm (MBEA) implemented in R package Biclique to identify biclique modules [47].
2.10. Survival analysis of lncRNA regulatory networks
To identify survival associated biclique modules for each cancer type, we classified the samples into the discovery set and validation set randomly without demographic characteristic differences. We used the univariate Cox regression model to evaluate the association between the patient survival and expression level of each lncRNA/Gene within the network. Then we constructed a risk model to assess the relation between survival and combination of lncRNAs and target genes in network module. The risk score can be calculated as:
whereas where is the Cox regression coefficient of nodes (lncRNA/Gene) in the network module, Exp(i) is the normalized expression value of node i in the corresponding module. N is the number of nodes (lncRNAs/Genes) in the network module. Tumor samples were classified as two different groups based on the predicated risk score and the log-rank test was used to evaluate the survival difference between two groups.
2.11. Statistics
Unless stated otherwise, all statistical analyses were performed with R-3.5.1. For the heatmaps of P-values of association between each singular vector generated by SVD and biological and technical factors, if the biological and technical factors are numerical, the p-values were generated by liner regression, otherwise the factors are categorical, the p-values were generated by the Kruskal-Wallis test. The significance of differences between two groups was determined by Wilcoxon rank sum test. Hypergeometric test was used to determine the significance of overlap between two groups of lncRNAs or genes. For patient survival analyses, Kaplan-Meier plots were created, we used the Cox's proportional hazard model and a log-rank to determine the difference of survival between two group of the patients for specific cancer. Unless stated otherwise, a p-value < 0.05 was considered significant. If necessary, p-values were corrected for multiple tests with the Benjamini- Hochberg procedure.
2.12. Ethics
Ethical approval doesn't apply to this work due to this is a data mining of previously published data. The use of the data was approved by TCGA.
2.13. Role of the funding source
The funders had no role in the study design, data analysis, interpretation, preparation of the manuscript, and any aspect of the study.
3. Results
3.1. Overview of multi-omics data in human cancers
We initially obtained the normalized RNA-Seq, DNA methylation 450k and copy number variation data for 17 cancer types from TCGA. Expression profiles for lncRNAs and protein coding genes were generated by Gene ID annotation. Both expression profiles and DNA methylation profiles were subject to a quality control procedure to assess the relative data variation associated with biological and technical factors. The correlation p-value heatmaps indicated that for breast cancer, the inferred top component of data variation correlated strongly with hormone receptor status (estrogen receptor and progesterone receptor) as a confounding factor (Supplementary Fig. S1). Thus, we decided to divide the samples from breast cancer into two different cohorts, the ER+ (estrogen receptor positive) and ER- (estrogen receptor negative). In this case, a total of 18 cancer types across 5,970 samples were used for this study (Supplementary Table S1). We next sought to get an overview of the lncRNA profiles in each cancer type in order to guarantee the overall technical validity. By examine the expression level of lncRNAs in all cancer types, we obtained 14,325 lncRNAs that further classified as 7,150 ‘lincRNA’, 5,142 ‘antisense’, 882 ‘sense_intronic’, 186 ‘sense_overlapping’ and 957 ‘TEC’ (Supplementary Fig. S2a). The number of lncRNAs that have expression in each cancer ranges from 10,438 to 14,013 (Supplementary Fig. S2b). We also checked the average expression of both lncRNAs and protein coding genes across all samples, and found that lncRNAs were expressed at considerable lower levels than protein coding genes (Supplementary Fig. S2c), which is consistent with several observations [45].
3.2. DNA methylation mediated lncRNA regulatory landscape across 18 cancer types
By integration of matched gene Exp-DNAm-CNV profiles and genome-wide lncRNA-gene regulation across cancer types, we investigated the landscape of DNAm mediated lncRNA regulatory network for different cancer types (Fig. 1a). We first obtained differentially methylated genes and differentially expressed genes in each cancer type, then we identified methylation level-dependent transcriptional regulation circuits at gene level. Genes exhibit a hypermethylated promoter and underexpression and genes exhibit a hypomethylated promoter and overexpression in cancer were identified by using a multi-regression model (Fig. 1b). Furthermore, we obtained the lncRNA modulators whose expression correlated both with promoter methylation and expression of protein coding genes (Fig. 1c). In this way, we obtained the widespread lncRNA mediated methylation dysregulation in 18 cancer types (Fig. 1d, Supplementary Fig. S3). The LncRNA modulators were analyzed with distinct features, including tissue specificity and sequence conservation (Fig. 1e). As a result, we found a substantial of the genes (5-50%) whose promoter methylation change is attribute to candidate lncRNA regulation (Supplementary Fig. S4a), and also 16-48% of the lncRNAs that expressed are involved in this regulatory network within each cancer type (Supplementary Fig. S4b). In summary, these methylation mediated lncRNA-gene interactions constitute a large and uncharacterized lncRNA regulatory networks across cancers.
Fig. 1.
An integrative framework identifies widespread lncRNA mediated DNA methylation perturbations in pan-cancer. a). The regulatory mechanisms of DNA methylation by lncRNAs. b). The mRNA expression of gene i is modelled as a function of the DNA methylation and copy number variation. c). TF–gene regulations were identified based on correlation analysis. d). lncRNA mediated DNA methylation perturbations in each cancer type were discovered. e). The identified network modules were analyzed for different functional characteristics.
3.3. Properties of the methylation mediated lncRNA regulatory network
We first analyzed the topological features of the MeLncTRN across cancers (Fig. 2a). By examination of the degree distribution reveals a scale free properties of these lncRNA regulatory networks, which is similar to most other types of biological network. A power law distribution was presented, as most of the nodes (both for lncRNAs and genes) present few interaction partners, whereas a small subset has plenty of partners (Fig. 2b, Supplementary Fig. S5-S7). We also found that high degree lncRNAs are more likely to correlated with the promoter methylation and expression of their target genes (Fig. 2c,d, Supplementary Figs. S8–S9), which indicated that lncRNAs have more targets could have a stronger regulatory effect. This is also apples to protein coding genes (Supplementary Fig. S10-S11), as it could be more strongly regulated by other lncRNAs. In general, nodes in highly connected networks will have more neighbors and will thus have more strongly co-regulation effect.
Fig. 2.
A global view of the properties of the lncRNA-gene interaction network for 18 cancer types. a). Basic topological feature statistics of the network for 18 cancer types. The first and second columns represents the number of lncRNAs (LncNum) and genes (GeneNum) within each network, the third and fourth columns represents the average number of interacting nodes of lncRNAs (LncDegree) and genes (GeneDegree), respectively, the fifth to seventh columns are the number of the edges (EdgeNum), Median degree (MedianDegree) and the goodness of fit (R square) of degree distribution in each network. b). The degree distribution of the network from BLCA as example. c). The correlation between promoter of the targets and the total expression of lncRNAs is plotted as a function of the number of lncRNA regulators for BLCA. d). The correlation between expression of the genes and the total expression of lncRNAs is plotted as a function of the number of lncRNA regulators (red indicts high density whereas blue indicates low). e). The percentage of distance distribution between lncRNAs and genes across cancers. f). The proportion of lncRNA-gene pairs of different regulatory patterns in each cancer type. Different color lines indicate distinct regulatory patterns, including Enhance Activate, Enhance inhibit, Inverts Activate and Invert Inhibit.
LncRNAs have been found to regulate the protein coding gens either in cis or in trans, and lead to gene silencing or activation mode [48,49]. To further interpret the mechanisms of the lncRNA regulation, here we explored the distances between lncRNAs and target genes for the identified lncRNA-gene pairs across cancer types. We found that lncRNAs regulate their target genes on the different chromosomes accounted for about 94.3%. For those lncRNA-gene pairs located on the same chromosome, 81.6% are located beyond 10 Mb away, only 1.4% are less than 100 kb (Fig. 2e, Supplementary Fig. S12). Thus, methylation mediated lncRNA transcriptional regulation mainly act in trans. As lncRNAs could activate or inhibit the activity of target genes, it could also enhance or invert gene expression depending on specific context [50]. To parse this complexity, we assigned each lncRNA-gene pair to one of four patterns according to their differential expression status in cancer and normal tissues (Fig. 2f, Supplementary Fig. S13). Globally, the majority of lncRNAs inhibit the gene activity without the direction of expression changed. These identified lncRNA-gene regulatory landscape provides a novel perspective to investigate the role of lncRNAs across cancer types.
3.4. Conserved lncRNA modulators represent important functions across cancer
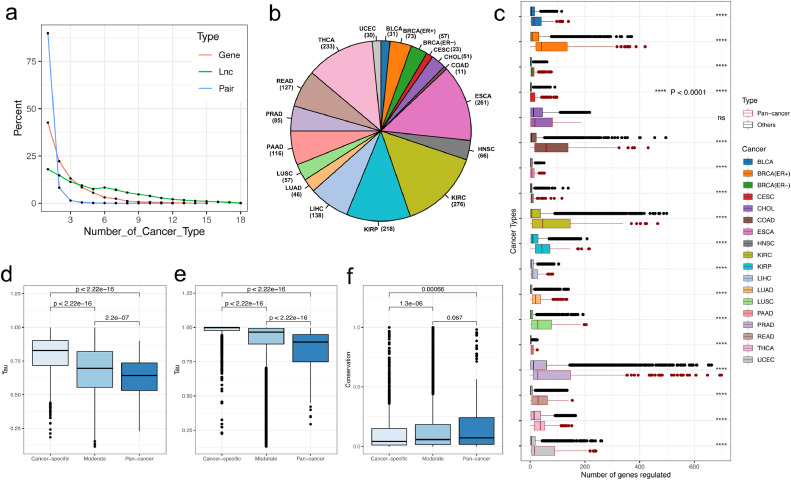
The investigation of the pan-cancer lncRNA regulatory networks presents several common features, whereas checking the network across cancers highlights a dynamic rewiring in the regulatory program between different cancers. We next analyzed the extent to which lncRNA mediated methylation dysregulation contributed to cancer specificity. We first calculated the number of cancer types in which the lncRNA regulation exits and found that most of the lncRNA related regulations occurs in cancer type-specific mode, and only a small proportion of the regulations occurs in pan-cancer wide mode (Fig. 3a). For lncRNA modulators, 17% occurs in only one type of cancer, whereas for lncRNA-gene interactions, 89.8% occurs in cancer specific manner. We found these tissue specific lncRNA modulators and associated targets mainly occurs in kidney cancers including the KIRC and KIRP, and also ESCA and THCA (Fig. 3b, Supplementary Fig. S14). This is probable due to the tissue specific expression of lncRNA and gene which suggests the significance of the pan-cancer interaction of the lncRNA modulators. At such a circumstance, we further classified the lncRNA modulators as three different types according to the number of the cancers it occurred: cancer specific (occurs in one cancer type), moderate (occurs in 2~14 cancer types) and pan-cancer (occurs in ≥15 cancer types). We first check the percentage of different lncRNA modulators across their categories and found ‘sense_overlapping’ lncRNA presents higher proportion of pan-cancer modulators (Supplementary Fig. S15). We also compared the degree of different lncRNA modulators, which is the number of transcriptional regulation they mediated. We found that pan-cancer modulators present significantly higher degree than other modulators (The Wilcoxon rank sum test P value < 0.05 in 17/18 cancer types) (Fig. 3c, Supplementary Fig. S16). This indicated that these pan-cancer lncRNA modulators play a key role in the regulation of gene expression during cancer development.
Fig. 3.
LncRNA modulator category and regulatory feature similarity across cancer types. a). Distribution of the number of cancer types that lncRNAs (green), genes (red) and lncRNA-gene pairs (blue) are involved in regulatory networks across 18 cancers. b). Pie chart of the numbers of lncRNAs that present in each cancer type. c). Boxplot of the degree distribution for different types of the lncRNA modulators across 18 cancer types. For each cancer type, red box is the degree distribution for pan-cancer modulators, whereas the gray one is the degree distribution for other two types, **** P < 0.0001 for Wilcox rank sum test. d). Tissue specify index differences for different types of lncRNA modulators from TCGA. e). Tissue specify index differences for different types of lncRNA modulators from GTEx. f). Promoter conservation differences for different types of lncRNA modulators.
As the tissue expression manner could give rise to the differences in the roles of lncRNA modulators played in cancer development, thus we calculated the tissue specificity index for lncRNA modulators based on their expression in normal tissues (details see Methods). We found that the pan-cancer modulator presents lower tissue specificity than other two types (Fig. 3d, P-value < 2.22e-16, Wilcoxon rank sum test). To further confirm this observation, we also obtained the transcriptome data from GTEx and performed the same analysis, finally a same conclusion was obtained (Fig. 3e). This indicated that pan-cancer modulators are more widely expressed across human tissues. Furthermore, we also found that pan-cancer lncRNA modulators present more sequence conservation in promoter regions than other two types of modulators (Fig. 3f). In summary, these important features of the lncRNA regulators highlights their important role in both normal and tumor tissues.
3.5. Similar tissue derived cancers exhibit comparable lncRNA mediated methylation dysregulation
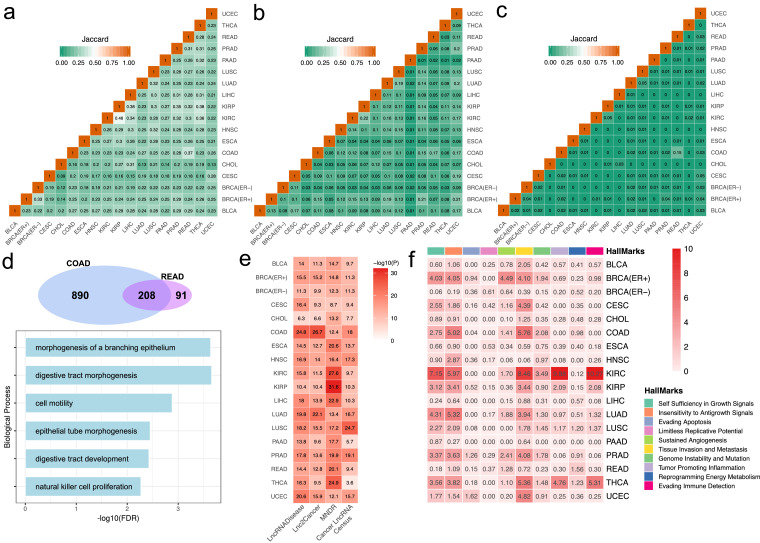
Plenty of evidences have indicated that similar tissue derived cancers may exhibit similar molecular profiles, including gene expression, microRNA or lncRNA expression [51]. It is still unknown whether similar tissue derived cancers may share similar methylation mediated lncRNA regulatory pattern. We thus calculated the paired Jaccard index between each cancer type based on the occurrence of lncRNA, gene and lncRNA-gene pair in the network to measure their similarities (Fig. 4a–c). We found that cancers derived from similar tissue are more likely to share common lncRNA modulators, and also the target genes and lncRNA-gene interactions in this network, such as COAD and READ, LUSC and LUAD, KIRC and KIRP, also the LIHC and CHOL. This observation indicated that cancers derived similar tissues may share related regulatory mechanisms. In addition, the similarities based on lncRNAs is generally higher than that based on genes (P-value < 2.22e-16, paired Wilcoxon rank sum test) and lncRNA-gene pairs (P-value < 2.22e-16, paired Wilcoxon rank sum test) (Supplementary Fig. S17). This indicated that some common lncRNA modulators may regulate different target genes thus may play a different role in cancer type specific context. We further investigated the functions of the lncRNA modulators derived from similar tissue types, by doing this, we performed the Gene Ontology enrichment analysis based on the common target genes of the lncRNA modulators between cancer types from the same tissue. The COAD and READ for example, a total of 208 genes were identified as shared targets (Fig. 4d, upper panel). Enrichment analysis indicated these genes are mainly involved in morphogenesis of the digestive tract and cell motility (Fig. 4d, lower panel). A similar functional enrichment result was observed for lung tissue derived cancers (Supplementary Fig. S18a). Whereas for kidney tissue derived cancers, functions are mainly about immune response pathways, for liver tissue derived cancers are mainly metabolic related functions (Supplementary Fig. S18b-c). These observations indicated that lncRNA mediated methylation dysregulation in cancers may fulfil diverse functions in tissue context manner, furthermore, it could help find patients the most appropriate treatment method, such as immune therapy.
Fig. 4.
Global comparison of lncRNA mediated networks in pan-cancer wide. a-c). The Jaccard coefficient matrices, which was determined from the shared number of lncRNA modulators, associated targets and lncRNA-gene pairs, respectively, measures the similarity of lncRNA mediated networks across 18 cancer types. Some similar tissue derived cancers present higher Jaccard coefficient within the matrix. d). Venn diagram indicated the number of shared lncRNA target genes (upper panel) and the biological processes enriched by these genes (lower panel) from COAD and READ. e). P-value heatmap generated from hypergeometric test evaluate the significance of lncRNA modulators enriched in the cancer related lncRNAs collected from public resources. f). P-value heatmap for lncRNA targets enriched in cancer hallmark across cancer types.
In order to further validate the functional roles of these lncRNAs and associated target genes in tumorigenesis. We first collected cancer related lncRNAs from public databases (see Methods). We performed a hypergeometric test to evaluate the enrichment significance of these lncRNA modulators to validated cancer lncRNAs. We found that lncRNA modulators from each cancer type are all enriched in the cancer related lncRNAs we collected (P-value < 0.05, hypergeometric test) (Fig. 4e). We then explored whether the lncRNA targets were enriched in cancer hallmark processes for each cancer [52]. This analysis revealed these lncRNA targets represent broad range of cancer hallmarks across cancer types (Fig. 4f). In particular, ‘Self Sufficiency in Growth Signals’, ‘Insensitivity to Antigrowth Signals’, ‘Sustained Angiogenesis’ and ‘Tissue Invasion and Metastasis’ are the most prevalent enriched hallmarks across different cancers, which suggests some common essential pathways these lncRNA related networks may participate in.
3.6. LncRNA modulators with aberrant expression exhibit biomedical significance
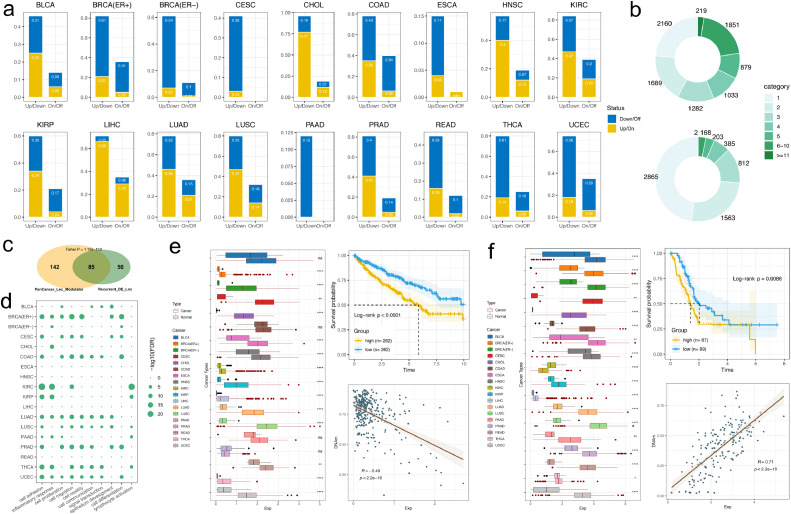
Expression aberration of lncRNAs are widespread observed during cancer development [45]. Here we analyzed the dysregulation pattern of lncRNA modulators based on the expression profiles in pan-caner wide with the aim to explore their biological significances. We found a substantial of the lncRNA modulators present a differential or switched expression pattern for each cancer type. In these 18 cancer types, a total of 2.8%–77.3% (average: 31.2%) of lncRNA modulators present significantly up-regulated or switch-on, and 8.9%~62.8% (average: 34.9%) of lncRNA modulators present down-regulated or switch-off, respectively (Fig. 5a). We also notice that the number of up/down regulated lncRNAs are far more surpass that of the switch on/off. We next investigated the distribution of the lncRNA modulators across cancer types and found that both the differentially expressed and switched lncRNAs are relatively tissue specific. A total of 2,160 (23.7%) present differential expression in only one cancer type, and 7,043 (77.3%) present differential expression in no more than five cancer types, only 219 (2.4%) present differential expression in more than 10 cancer types (Fig. 5b, upper panel). For the lncRNA present switched on/off, about one half (2,865, 47.8%) present only in one cancer type, and only 2 lncRNAs present in more than 10 cancer types (Fig. 5b, lower panel). In this case, we combined those differentially expressed lncRNAs and those have switched expression as a whole unit and investigated their roles in cancer development.
Fig. 5.
Landscape of aberrant expression of lncRNA modulators across cancer types. a). Proportion of lncRNA modulators present differential expression and switched expression for each cancer type. b). Distribution of the number of lncRNA modulators present differential expression in different numbers of cancer types. c). Overlap between pan-cancer lncRNA modulators and those present recurrent differential expressions. d). The GO terms enriched by recurrent differentially expressed pan-cancer lncRNA modulators. Rows are lncRNAs and each column are Gene Ontology terms. The circles size represent the FDR adjusted P-values from Fisher exact test. e). left: Differential expression pattern of ENSG00000227036 across cancer types; upper right: Kaplan–Meier plot of survival for lncRNA different expression level in KIRC; lower right: correlation between lncRNA expression and promoter methylation of target KRT15. f). left: Differential expression pattern of ENSG00000203499 across cancer types; upper right: Kaplan-Meier plot of survival for lncRNA different expression level in PAAD; lower right: correlation between lncRNA expression and promoter methylation of target CDKN1A.
We investigated those lncRNAs present differential or switched expression in multiple cancer types and a total of 141 lncRNAs showed recurrent differential expression in more than 12 cancer types. We found these recurrent differentially expressed lncRNAs were significantly enriched in pan-cancer modulators identified previously (Fig. 5d, p-value = 1.13e-110, hypergeometric test), and a total of 85 differentially expressed lncRNAs were also identified as pan-cancer regulators (Supplementary Table S2). Then we performed a Gene Ontology enrichment analysis to explore their functions of these key lncRNA modulators. This analysis revealed these lncRNA modulators are mainly participated in functions including cell adhesion, proliferation, differentiation, and also immune related functions, such as inflammatory response and lymphocyte activation (Fig. 5d). Further, our analysis discovered many lncRNA modulators that play a key role in particular cancer types. One of the example is the ENSG00000227036 (LINC00673/LINC00511). Our analysis indicated this lncRNA presents up-regulated expression in 14 cancer types (77.8%, 14/18) (Fig. 5e, left panel). Survival analysis indicated activated expression of this lncRNA is associated with poor prognosis of KIRC (P-value < 0.0001, log-rank test, Fig. 5e, right upper panel). This lncRNA was identified to interact with DNMT1 and also other histone modification proteins, such as EZH2 and histone deacetylase complexes of LSD1 to regulate cancer related pathways including cell proliferation and invasion and induced cell apoptosis [53], [54], [55]. We also found expression of this lncRNA is highly correlated with promoter methylation of serval key cancer genes, such as KRT15, a high negative correlation between expression of this lncRNA and the promoter methylation of KRT15 was observed in KIRC from our study (Fig. 5e, right lower panel, Supplementary Fig. 19). KRT15 is responsible for the structural integrity of epithelial cells, up-regulation and associated poor prognosis of this gene was observed in many cancer types [56,57]. Another example we identified is the ENSG00000203499 (FAM83H-AS1), which could interact with EZH2 to regulate expression of many cancer related genes, such as CDKN1A, thereby influencing the cell cycle and proliferation [58]. FAM83H-AS1 was presented here to have differential expression in all 18 cancer types (Fig. 5f, left panel). Up-regulation of FAM83H-AS1 is associated with poor prognosis of PAAD (P < 0.0086, log-rank test, Fig. 5f, right upper panel). One of the targets of FAM83H-AS1 identified is SLC39A6, which was also indicated to play a role in many cancer types [59,60]. Our analysis indicated FAM83H-AS1 expression is highly correlated with promoter methylation of SLC39A6 in PAAD (Fig. 5f, right lower panel, Supplementary Fig. S20). This interaction was also identified in multiple cancer types (Supplementary Fig. S21), which indicated this lncRNA related regulatory relationship may play a role in pan-cancer wide. In summary, our analysis indicated that these differentially expressed lncRNA modulators can have a role in cancer biology and motivated us to better understand of the functions of lncRNAs in tumorigenesis.
3.7. Diverse function of conserved network hubs across cancer types
Network hubs are highly connected nodes and are critical for maintaining network robustness, thus are expected to play a vital role in biological systems. We have shown that this methylation mediated lncRNA regulatory network is featured by variable degree distribution, which is the high degree for very few ‘hub’ genes and low degree for most other genes. We thus identified hub in this network to investigate if they can play important roles in cancer. We selected the top 10% of the lncRNAs and genes in the network as ‘hubs’, as a result, a total of 2,823 lncRNAs and 920 protein coding genes were selected (Fig. 6a,b). We found that many of the hub lncRNAs including TUG1, TARID, APTR, PVT1, PCAT6, TINCR and HULC, and protein coding genes as CEBPE, SMAD2 and HAND2 have been widely identified in tumorigeneses. Interestingly, we found most of these hub lncRNAs identified could interact with EZH2, which is the subunit of PRC2 and interact with DNMTs and associates with DNMT activity in vivo [12]. The TUG1 for example, was found to repress Kruppel-like factor 2 (KLF2) expression by interacting with PRC2 and recruiting it to KLF2 promoter region in HCC [61]. Another well-known lncRNA PVT1, was shown to bind EZH2 and inhibit the recruitment of EZH2 to the promoter region of MYC to promote its expression [62]. This lncRNA could also regulate miR-146a gene expression by inducing the CpG methylation level at its promoter during tumorigenesis of prostate cancer [63]. Other mechanism for the hub lncRNAs were also identified. The TARID for example, could interacts with both the GADD45A (growth arrest and DNA-damage-inducible, alpha), which a regulator of DNA demethylation, and TCF21 promoter to activates its expression by inducing its promoter demethylation [64]. For the hubs of protein coding gene, the HAND2 have been identified to present as the hub of the most highly ranked differential methylation hotspot in endometrial cancer [65].
Fig. 6.
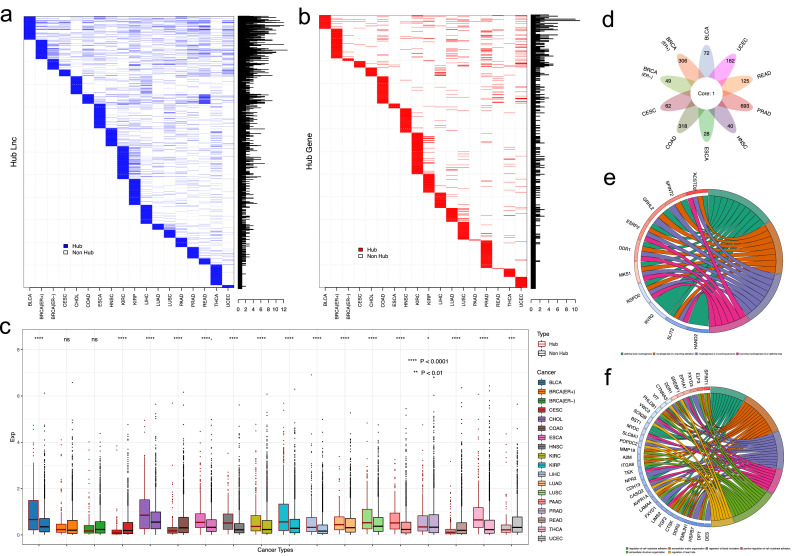
Hub analysis of methylation mediated lncRNA regulatory network across cancer types. a-b). The Distribution of hub lncRNAs and genes within network across different cancers. Histogram indicated the number of cancer types that the hubs was identified. c). Boxplot of the expression distribution for hub and non-hub lncRNA modulators across 18 cancer types. d). Venn diagram indicated the number of cancer specific and shared target genes of the Hub lncRNA modulator ADAMTS9-AS2 in 10 cancer types. e-f). Chord plot indicated the Gene Ontology terms that the ADAMTS9-AS2 targets enriched in BLCA and UCEC, respectively.
To further investigate the function of the hub lncRNAs across cancer types, we first examined the expression pattern of hub and non-hub lncRNAs. The result indicated that hub lncRNA have higher expression than these non-hubs in large proportion of cancer types (12/18, 66.7%) (Fig. 6c). This implies that hub lncRNAs may play more important roles in tumorigeneses than non-hub ones. Global comparison of the hub lncRNAs across cancer types indicated that many lncRNA hubs may present in multiple cancer types. Some lncRNA hubs could maintain this high degree in 12 cancer types. One of which, the ADAMTS9-AS2 (ADAMTS9 antisense RNA 2), have been identified to interact with DNMT1/DNMT3 to regulate the methylation at its target promoter [66]. Our analysis indicated that this lncRNA presents as hub in 10 cancer types. We found that ADAMTS9-AS2 may regulate different target genes in various cancer types but with only one gene of NKAPL (NFKB activating protein like) as common target (Fig. 6d). Functional annotation of the ADAMTS9-AS2 targets in respective cancer types indicated this lncRNA could have different functions in cancer specific manner. For instance, it mediated the cell morphogenesis, such as “epithelial tube morphogenesis” and “morphogenesis of a branching structure” in BLCA (Fig. 6e), whereas in UCEC, it mainly involves cell adhesion, including “regulation of cell−substrate adhesion”, “extracellular matrix organization”, “extracellular structure organization” and also function of “regulation of blood circulation” (Fig. 6f). In summary, this analysis indicated the hub lncRNAs may exert their functions in tissue dependent manner. Further detailed study may reveal undiscovered pathways for these lncRNAs regulated in different cancers.
3.8. Clinical relevance of lncRNA network modules as prognosis biomarkers
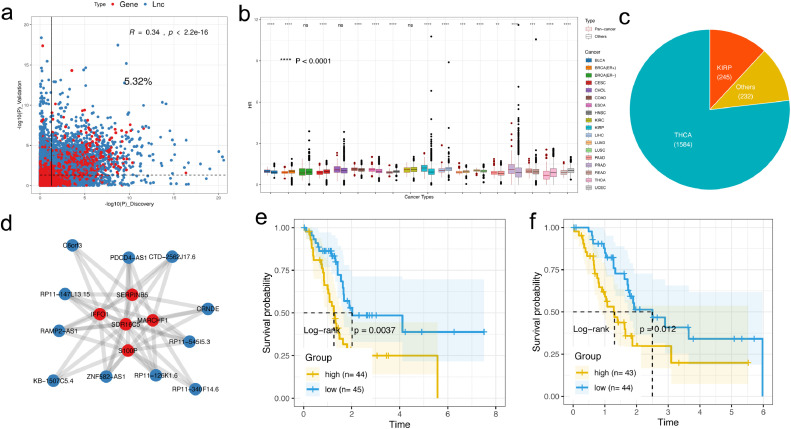
Studies have indicated lncRNAs present strong potential for predicting cancer prognosis as their expression is highly variable among different stages and tissues, thus could better represent disease features [67]. Thus, we integrated the survival information to evaluate the potential ability of the identified lncRNA modulators as candidate prognosis markers. For each cancer type, we randomly classified the tumor samples as discovery set and validation set with similar age and sex distributions in each set. Then we performed a cox regression analysis to identify the survival related lncRNA/genes in each network. For all the 18 cancer types, we found that a total of 5.32% of the lncRNA/genes are clinical-associated in both groups (Fig. 7a). Then we tested if there are differences for clinical relevance of different category of lncRNA regulators. We compared the Hazard Ratio distribution of the clinically related lncRNAs among different groups of pan-cancer regulator and others. We found that a substantial cancer types (9/18) present a higher Hazard Ratio of pan-cancer regulators than others (Fig. 7b), which indicated that methylation related lncRNA regulatory network is of great important for cancer prognosis.
Fig. 7.
Clinical analysis of lncRNA regulatory network across cancer types. a). The survival landscape of lncRNA and protein coding genes in network. X-axis represent the –log10(P_value) from discovery set whereas the Y-axis represents the –log10(P_value) in the validation set. lncRNA and genes are marked the blue and red respectively. b). Boxplot of the Hazard Ratio for different types of the lncRNA modulators across 18 cancer types. For each cancer type, red box is the degree distribution for pan-cancer modulators, whereas the gray one is the degree distribution for other two types, **** P < 0.0001 for Wilcox rank sum test. c). Pie chart of the numbers of survival related bipartite network modules that present in different cancer types. d). Survival related network module identified in PAAD. e-f). Kaplan–Meier plot of survival for PAAD samples with different risk scores. e). Discovery set; f). validation set; The survival difference among groups is calculated by log-rank test.
We and other studies have indicated that lncRNAs may collaborate to regulate individual genes to control their expression. Network module analysis provides us an important tool to investigate the cooperative interaction in pan-cancers. Thus we extensively identified the biclique modules across 18 cancer types. Then we calculated a risk score for each network module to evaluate their potential ability for prediction of the prognosis of cancer. As a result, 2,061 modules were identified that can be used to classify cancer samples into groups with significantly different overall survival rates in both discovery set and validation set (Fig. 7c), and the majority of the survival related modules were discovered in THCA (76.9%) and KIRP (11.9%). One such module for instance, which mainly involve the lncRNA CRNDE (Fig. 7d), was associated with patients’ survival both in discovery (Fig. 7e, log-rank test P=0.0037) and validation set (Fig. 7e, log-rank test P=0.012). CRNDE has been identified to interact with PRC2 component EZH2 and mediated their inhibition of tumor suppressor genes [68]. Elevated expression and the associated poor prognosis was found in many cancer types, including pancreatic cancer [69,70]. We also identified other prognostic related modules which was mediated by ZEB2-AS1 in multiple cancers including BLCA, LUSC and PAAD. (Supplementary Fig. 22a–c). ZEB2-AS1 was identified to regulate epithelial-mesenchymal transition in cancer development and their clinical roles in these cancer types have been validated [71], [72], [73]. However, we didn't observe that the expression of the individual lncRNA of ZEB2-AS1 alone have a discriminative effect for prognosis in these three cancer types. Such results indicated the critical roles of the combinative effect of lncRNAs in cancer methylome formation and provide potential clinical usage of these module related biomarkers.
3.9. A user-friendly database for exploring the methylation related lncRNA regulatory perturbation
For facilitating the users investigate the methylation mediated lncRNA regulatory pattern and associated transcriptional dysregulation of target genes in these cancer types, we constructed an online database MeLncTRN (http://compgenelab.info/MeLncTRN/) which allows users to query information about the lncRNA-gene interactions of interest. This platform provides a web interface for users to search and download all data sets. Users can query the database for exploring the interactions that involve the interested lncRNA or target gene in specific cancer type. The list of the matched entries will return after the interesting lncRNA or genes been submitted. In the entry detail page, the detailed information about the differential expression of lncRNA and genes, differential methylation in promoter of genes, and also the interaction information between lncRNA and genes were listed. All data in the platform can be downloaded from the ‘download’ page for further study. This database could serve as a useful tool for dissecting the interaction network between lncRNA and genes and identifying novel biomarkers for cancer.
4. Discussion
LncRNAs have long been recognized as an important kind of gene expression regulator that act in different mode. For the widely accepted “competitive endogenous RNA (ceRNA)” model, lncRNAs mainly act as “sponge” to combine with miRNAs and sequester its interaction with protein coding genes, which in turn to de-repressing the expression of targets that share the same group of miRNA binding sites (also referred to as miRNA response elements, MREs) [74,75]. However, this hypothesis is still in debate as discrepancies were found by experimental validation [76]. Besides, some other regulatory mechanisms have also been proposed, such as interact with transcription factors to regulate downstream gene expression [77]. LncRNAs have also been identified as the integral component of chromatin to play a role in biological processes with respect to epigenetic control [78,79]. DNA methylation at the promoter region of protein coding genes is one of the key components of epigenetic regulation. However, only limited number of examples that lncRNA regulate promoter methylation of target genes have been identified so far, this motivated us to perform a more in-depth exploration of the potential role of lncRNAs. Here we introduce a computational framework to perform an integrative pan-cancer-wide analysis of matched gene expression, DNAm and CNV data, in an attempt to identify lncRNAs which display regulatory effects of epigenomic and transcriptomic deregulation of protein coding genes in cancer. Our strategy identifies lncRNAs which exhibit universal patterns of correlation with genome-wide DNAm and expression levels of protein coding targets. It is likely that these lncRNAs constitute master regulators of the DNA methylome in cancer.
The prerequisite to identify the DNA methylation mediated lncRNA regulatory perturbation is to determine the genes whose expressional dysregulation driven by DNA methylation beforehand. Thus, the first two steps of our framework were focused on identifying the cancer context specific DNAm-Exp regulatory relations. Then we used the correlation changes to evaluate the lncRNA-gene regulation in both methylation and expression level. This may lead to overlook several genes that showed no correlation of DNAm-Exp. Our survey of the correlation analysis revealed significant numbers of genes that are not correlated between expression and methylation in promoters (Supplementary Fig. S23). It is speculated that methylation level at promoter of these gene would serve as a rate-limiting element when there is sufficient abundance of transcription factors. In this case, DNA methylation only determines the upper or lower limit that the gene expression level by controlling the maximum accessibility of TF [80]. On the other hand, distal CpG sites within enhancer regions may contribute to their activity change by promoting both active and passive DNA demethylation and by influencing chromatin architecture [81]. Whereas it is generally not easy to precisely locate this type of regulatory regions associated with particular genes. However, our analysis procedure can be easily extended to distal enhancers if large scale datasets in which precise identification of the regions are available.
Our analysis indicated that most lncRNAs may regulate multiple targets in a multimodal mode, acting as enhancers or invertors to activate or inhibit targets depending on the specific genes and cancer types, highlighting the complexity of DNA methylation mediated lncRNA regulatory pattern. Moreover, most of these regulatory interactions take place in trans and in cancer specific mode, while a substantial number of regulation by lncRNAs are common across cancer types. The pan-cancer lncRNA modulators presents many critical functional features, such as tissue-specific expression pattern and evolutionary conservation in promoter region. These results provide valuable resource for both computational and biological researchers, which will greatly widen our views about our understanding of the roles of lncRNAs in cancer.
Although the DNA methylation mediated lncRNA regulatory effects is prevalently identified in our study, the detailed mechanism on how these lncRNA modulators involved in this epigenetic regulation remains to be discovered. We notice that many lncRNA modulators in our study were documented to interact with EZH2, which is responsible for the methylation activity of PRC2, for instance, FAM83H-AS1, TUG1, PVT1 and LINC00511. EZH2 has been demonstrated that are universally overexpressed in cancer, and which are known to influence DNAm levels [82]. A wide spectrum of lncRNAs was found to act as scaffold to bind with EZH2 and recruit it to the promoter region of target genes and repress their expression [83]. Interestingly, many of these lncRNAs have also been identified to act as ceRNAs by interacting with miRNAs. For instance, the well-known cancer lncRNA PVT1 was demonstrated associated with miRNAs including miR-128, miR-214, miR-195, etc. in multiple cancer types, such as bladder cancer [84], colon cancer [85] and lung cancer [86], whereas the TUG1 targets including miR-145 [87], miR-26a [88], miR-29c [89]. This observation indicated that lncRNAs may play dual roles in a context specific manner by which to define a precise transcriptional regulation circuitry.
The lncRNA modulators that identified presents widespread transcriptional dysregulation and widely expressed in multiple tissues, which highlight their critical roles in cancer development. This was validated by functional annotation to the cancer hallmarks of their target genes. In addition, similar lncRNA mediated DNA methylation perturbations were observed for cancer types originated from same tissue. Functions of these cancer type specific lncRNA modulators manly involved in development of specific tissue, such as colon or lung. As one of the hallmarks of tumor cells is the lack of differentiation, it is not very surprise that many tissue specific genes were preferentially targeted in cancer. Furthermore, many lncRNAs were mainly involved in immune response pathways. The important role of epigenetics in determine immunity and immune therapy for cancers has been well demonstrated [90]. Our observation provides comprehensive understanding on how lncRNAs regulate immune response by epigenetic mechanisms and also novel insights into lncRNA based immune therapy.
In summary, our analysis indicated that lncRNAs represent an additional layer of genome wide DNA methylation modulator in cancer. Our results provide a valuable resource for dissecting the driving force of gene expression regulation related to tumorigenesis and cancer development. The lncRNA modulators predicted here deserve follow-up work on both experimental and computational study in order to elucidate their specific rules. Continued investigation will aid in the development of better therapies for human cancer and other diseases.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgments
Contributors
Zhen Yang conceived the study. Zhen Yang, Feng Xu and Haizhou Wang performed data analysis. Andrew E Teschendorff and Yungang He designed and supervised data analysis. Zhen Yang and Feng Xie constructed the database. Zhen Yang wrote the paper. Andrew E Teschendorff and Yungang He revised the paper. All authors read and approved the paper.
Acknowledgments
This work is supported by National Natural Science Foundation of China [91959106, 31871255]; We thank the support from the innovative research team of high-level local university in Shanghai.
Dr. Zhen Yang would like to dedicate this work to his father, who struggled with colon cancer and fade away in 2019.
Data sharing statement
The study was based on the data available at TCGA (https://www.cancer.gov/tcga). The code used for this analysis is publicly available in the GitHub repository (https://github.com/zyangx/MeLncTRN), All data supporting the findings of the current study are included in our data portal (http://compgenelab.info/MeLncTRN/).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103399.
Contributor Information
Zhen Yang, Email: zhenyang@fudan.edu.cn.
Yungang He, Email: heyungang@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 3.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 5.De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288(48):34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 9.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42(12):1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 10.Fialkova V, Vidomanova E, Balharek T, Marcinek J, Kudela E, Hanysova S. DNA methylation as mechanism of apoptotic resistance development in endometrial cancer patients. Gen Physiol Biophys. 2017;36(5):521–529. doi: 10.4149/gpb_2017032. [DOI] [PubMed] [Google Scholar]
- 11.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 13.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Jones A, Widschwendter M, Teschendorff AE. An integrative pan-cancer-wide analysis of epigenetic enzymes reveals universal patterns of epigenomic deregulation in cancer. Genome Biol. 2015;16:140. doi: 10.1186/s13059-015-0699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis MW, Li S, Franco HL. Transcriptional control by enhancers and enhancer RNAs. Transcription. 2019;10(4-5):171–186. doi: 10.1080/21541264.2019.1695492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55(2):171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang SL, Huang Y, Su R, Yu YY. Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 2019;19:114. doi: 10.1186/s12935-019-0808-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng J, Mueller M, Geng T, Shen Y, Liu Y, Hou P. H19 lncRNA alters methylation and expression of Hnf4alpha in the liver of metformin-exposed fetuses. Cell Death Dis. 2017;8(12):e3175. doi: 10.1038/cddis.2017.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Wang W, Li X, Liu Y, Niu Y, Zhang B. LncRNA H19 interacts with S-adenosylhomocysteine hydrolase to regulate LINE-1 Methylation in human lung-derived cells exposed to Benzo[a]pyrene. Chemosphere. 2018;207:84–90. doi: 10.1016/j.chemosphere.2018.05.048. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Fu H, Liu X, Lei Q, Zhang Y, She X. LINC00470 coordinates the epigenetic regulation of ELFN2 to distract GBM cell autophagy. Mol Ther. 2018;26(9):2267–2281. doi: 10.1016/j.ymthe.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teschendorff AE, Zhuang J, Widschwendter M. Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics. 2011;27(11):1496–1505. doi: 10.1093/bioinformatics/btr171. [DOI] [PubMed] [Google Scholar]
- 28.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 29.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao Y, Widschwendter M, Teschendorff AE. A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics. 2014;30(16):2360–2366. doi: 10.1093/bioinformatics/btu316. [DOI] [PubMed] [Google Scholar]
- 31.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 33.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 35.Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21(5):650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 36.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D8D8. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47(D1):D1034–D10D7. doi: 10.1093/nar/gky905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y, Wang P, Wang Y, Ma X, Zhi H, Zhou D. Lnc2Cancer v2.0: updated database of experimentally supported long non-coding RNAs in human cancers. Nucl Acids Res. 2019;47(D1):D1028–D1D33. doi: 10.1093/nar/gky1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui T, Zhang L, Huang Y, Yi Y, Tan P, Zhao Y. MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucl Acids Res. 2018;46(D1):D371–D3D4. doi: 10.1093/nar/gkx1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlevaro-Fita J, Lanzos A, Feuerbach L, Hong C, Mas-Ponte D, Pedersen JS. Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun Biol. 2020;3(1):56. doi: 10.1038/s42003-019-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plaisier CL, Pan M, Baliga NS. A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res. 2012;22(11):2302–2314. doi: 10.1101/gr.133991.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y, Phillips CA, Langston MA. Biclique: an R package for maximal biclique enumeration in bipartite graphs. BMC Res Notes. 2020;13(1):88. doi: 10.1186/s13104-020-04955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development. 2013;140(6):1184–1195. doi: 10.1242/dev.088849. [DOI] [PubMed] [Google Scholar]
- 51.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang E, Zaman N, McGee S, Milanese JS, Masoudi-Nejad A, O'Connor-McCourt M. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 53.Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8(56):95542–95553. doi: 10.18632/oncotarget.20980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25(4):1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5(11):e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao X, Wang J, Song HM, Deng B, Li JG. KRT15 overexpression predicts poor prognosis in colorectal cancer. Neoplasma. 2020;67(2):410–414. doi: 10.4149/neo_2019_190531N475. [DOI] [PubMed] [Google Scholar]
- 57.Shen YH, Xu CP, Shi ZM, Zhang YJ, Qiao YG, Zhao HP. Cytokeratin 15 is an effective indicator for progression and malignancy of esophageal squamous cell carcinomas. Asian Pac J Cancer Prev. 2016;17(9):4217–4222. [PubMed] [Google Scholar]
- 58.Bi YY, Shen G, Quan Y, Jiang W, Xu F. Long noncoding RNA FAM83H-AS1 exerts an oncogenic role in glioma through epigenetically silencing CDKN1A (p21) J Cell Physiol. 2018;233(11):8896–8907. doi: 10.1002/jcp.26813. [DOI] [PubMed] [Google Scholar]
- 59.Wu C, Li D, Jia W, Hu Z, Zhou Y, Yu D. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet. 2013;45(6):632–638. doi: 10.1038/ng.2638. [DOI] [PubMed] [Google Scholar]
- 60.Lue HW, Yang X, Wang R, Qian W, Xu RZ, Lyles R. LIV-1 promotes prostate cancer epithelial-to-mesenchymal transition and metastasis through HB-EGF shedding and EGFR-mediated ERK signaling. PLoS One. 2011;6(11):e27720. doi: 10.1371/journal.pone.0027720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang B, Yang B, Wang Q, Zheng X, Guo Y, Lu W. lncRNA PVT1 promotes hepatitis B viruspositive liver cancer progression by disturbing histone methylation on the cMyc promoter. Oncol Rep. 2020;43(2):718–726. doi: 10.3892/or.2019.7444. [DOI] [PubMed] [Google Scholar]
- 63.Liu HT, Fang L, Cheng YX, Sun Q. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Cancer Med. 2016;5(12):3512–3519. doi: 10.1002/cam4.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55(4):604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 65.Jones A, Teschendorff AE, Li Q, Hayward JD, Kannan A, Mould T. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med. 2013;10(11) doi: 10.1371/journal.pmed.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D, Wu K, Yang Y, Zhu D, Zhang C, Zhao S. Long noncoding RNA ADAMTS9-AS2 suppresses the progression of esophageal cancer by mediating CDH3 promoter methylation. Mol Carcinog. 2020;59(1):32–44. doi: 10.1002/mc.23126. [DOI] [PubMed] [Google Scholar]
- 67.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020;11(8):676. doi: 10.1038/s41419-020-02853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW, Li J. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci Rep. 2016;6:33535. doi: 10.1038/srep33535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017;50(6) doi: 10.1111/cpr.12389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Guo Y, Hu Y, Hu M, He J, Li B. Long non-coding RNA ZEB2-AS1 promotes proliferation and inhibits apoptosis in human lung cancer cells. Oncol Lett. 2018;15(4):5220–5226. doi: 10.3892/ol.2018.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X, Yan T, Wang Z, Wu X, Cao G, Zhang C. LncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and inhibits apoptosis by regulating miR-27b. Biomed Pharmacother. 2017;96:299–304. doi: 10.1016/j.biopha.2017.08.060. [DOI] [PubMed] [Google Scholar]
- 73.Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545–551. doi: 10.1016/j.ijbiomac.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 74.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, Stoffel M. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol Cell. 2016;64(3):565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Li L, Wang Z, Pan T, Sahni N, Jin X. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018;46(3):1113–1123. doi: 10.1093/nar/gkx1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodriguez-Campos A, Azorin F. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One. 2007;2(11):e1182. doi: 10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review) Oncol Rep. 2017;37(1):3–9. doi: 10.3892/or.2016.5236. [DOI] [PubMed] [Google Scholar]
- 80.Liu Y, Liu Y, Huang R, Song W, Wang J, Xiao Z. Dependency of the cancer-specific transcriptional regulation circuitry on the promoter DNA methylome. Cell Rep. 2019;26(12):3461–3474. doi: 10.1016/j.celrep.2019.02.084. e5. [DOI] [PubMed] [Google Scholar]
- 81.Angeloni A, Bogdanovic O. Enhancer DNA methylation: implications for gene regulation. Essays Biochem. 2019;63(6):707–715. doi: 10.1042/EBC20190030. [DOI] [PubMed] [Google Scholar]
- 82.Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su M, Xiao Y, Tang J, Wu J, Ma J, Tian B. Role of lncRNA and EZH2 interaction/regulatory network in lung cancer. J Cancer. 2018;9(22):4156–4165. doi: 10.7150/jca.27098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu C, Longfei L, Long W, Feng Z, Chen J, Chao L. LncRNA PVT1 regulates VEGFC through inhibiting miR-128 in bladder cancer cells. J Cell Physiol. 2019;234(2):1346–1353. doi: 10.1002/jcp.26929. [DOI] [PubMed] [Google Scholar]
- 85.Shang AQ, Wang WW, Yang YB, Gu CZ, Ji P, Chen C. Knockdown of long noncoding RNA PVT1 suppresses cell proliferation and invasion of colorectal cancer via upregulation of microRNA-214-3p. Am J Physiol Gastrointest Liver Physiol. 2019;317(2):G222–GG32. doi: 10.1152/ajpgi.00357.2018. [DOI] [PubMed] [Google Scholar]
- 86.Wu D, Li Y, Zhang H, Hu X. Knockdown of Lncrna PVT1 Enhances Radiosensitivity in Non-Small Cell Lung Cancer by Sponging Mir-195. Cell Physiol Biochem. 2017;42(6):2453–2466. doi: 10.1159/000480209. [DOI] [PubMed] [Google Scholar]
- 87.Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai) 2017;49(7):588–597. doi: 10.1093/abbs/gmx047. [DOI] [PubMed] [Google Scholar]
- 88.Yang B, Tang X, Wang Z, Sun D, Wei X, Ding Y. TUG1 promotes prostate cancer progression by acting as a ceRNA of miR-26a. Biosci Rep. 2018;38(5) doi: 10.1042/BSR20180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo P, Zhang G, Meng J, He Q, Li Z, Guan Y. Upregulation of long noncoding RNA TUG1 promotes bladder cancer cell proliferation, migration, and invasion by inhibiting miR-29c. Oncol Res. 2018;26(7):1083–1091. doi: 10.3727/096504018X15152085755247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527(7577):249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.