Abstract
Limb and CNS expressed 1 like (LIX1L) is over-expressed in several types of tumors. However, the function of LIX1L in glucose metabolism and hepatocellular carcinoma (HCC) progression remains elusive. Here we report that LIX1L is over-expressed in human HCC tissues, which predicts unfavorable prognosis. LIX1L deficiency in vivo significantly attenuated liver cancer initiation in mice. Functional studies indicated that LIX1L overexpression elevated proliferation, migratory, invasive capacities of HCC cells in vitro, and promoted liver cancer growth and metastasis in vivo. LIX1L knockdown up-regulated fructose-1,6-bisphosphatase (FBP1) expression to reduce glucose consumption as well as lactate production. Mechanistically, LIX1L increased miR-21-3p expression, which targeted and suppressed FBP1, thereby promoting HCC growth and metastasis. MiR-21-3p inhibitor could abrogate LIX1L induced enhancement of cell migration, invasion, and glucose metabolism. Inhibition of miR-21-3p suppressed tumor growth in an orthotopic tumor model. Our results establish LIX1L as a critical driver of hepatocarcinogenesis and HCC progression, with implications for prognosis and treatment.
Key words: LIX1L, FBP1, miR-21-3p, Glucose metabolism, Hepatocellular carcinoma, Proliferation, Metastasis, Gluconeogenesis
Abbreviations: CCl4, carbon tetrachloride; DEN, diethylnitrosamine; ECAR, extracellular acidification rate; EMT, epithelial–mesenchymal transition; FBP1, fructose-1,6-bisphosphatase 1; HCC, hepatocellular carcinoma; LIX1L, Limb and CNS expressed 1 like; miRNA, microRNA; NASH, non-alcoholic steatohepatitis; Seq, sequencing; shRNA, short-hairpin RNA
Graphical abstract
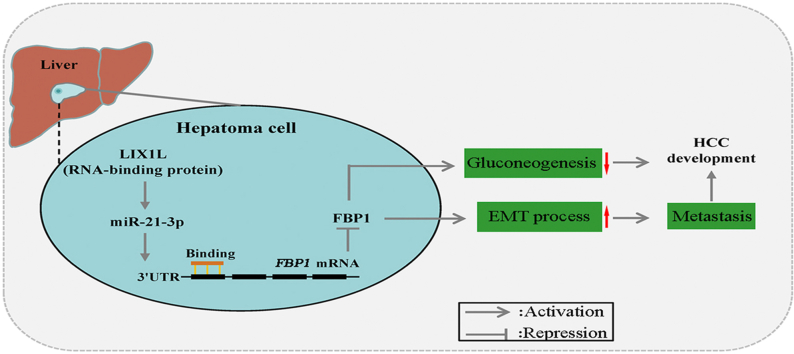
Limb and CNS expressed 1 like (LIX1L) promotes liver cancer progression via miR-21-3p-mediated inhibition of fructose-1,6-bisphosphatase. LIX1L and miR-21-3p may be important cellular targets for designing new therapies against liver cancer.
1. Introduction
Liver cancer is a worldwide health problem and has emerged as a leading cause of cancer-related death1. Although liver surgical resection, chemotherapy, radiotherapy, transplantation have become curative therapies that are used in the early stages of the disease, the long-term survival remains unsatisfactory due to high recurrence rates2. Thus, effective mechanism-based therapies and new prognostic markers are urgently needed.
An altered cell metabolism, particularly glucose metabolism, including aerobic glycolysis, is a characteristic feature of many cancers3. An increased glycolysis and lactate production, the so-called Warburg effect, is one of the characteristics of cancer cells, including hepatocellular carcinoma (HCC)4,5. Glycolysis and gluconeogenesis are reciprocal pathways that maintain glucose homeostasis6. In recent years, a glycolytic switch in cancer metabolism has been well described; however, little attention has been paid so far to gluconeogenesis, which regulates glucose metabolism in parallel with glycolysis, but in an opposite direction. Fructose-1,6-bisphosphatase (FBP1), a gluconeogenesis regulatory enzyme, mainly promotes gluconeogenesis and inhibits glycolysis7. FBP1 deficiency contributes to tumor progression by promoting glycolysis, leading to a poor prognosis in breast cancer8, renal cell carcinoma9, gastric cancer10, and liver cancer11. Epigenetic modifications, including promoter DNA methylation12,13 and histone deacetylation11, were reported to be the underlying mechanisms of FBP1 suppression. However, the mechanism of FBP1 dysregulation and the potential upstream molecules in HCC remain far from clear.
MicroRNAs (miRNAs) are approximately 18–24-nucleotide-long non-coding RNAs whose function is gene expression regulation14. Generally, miRNAs bind to the 3′-untranslated regions (3′-UTR) of mRNAs, leading to its degradation or translational inhibition15. Numerous studies suggest that miRNA dysregulation is involved in various liver diseases, including HCC16. For instance, miRNAs (miR-93, miR-221, miR-21, and miR-34a) are up-regulated in HCC tumors and are associated with dysregulated proliferation, metabolism, and immune system processes16, 17, 18. Among them, miR-21 is over-expressed in obesity and is further increased in steatohepatitis, fibrosis, and HCC. MiR-21 promotes tumor growth and chemotherapy resistance in cholangiocarcinoma19,20. The epithelial–mesenchymal transition (EMT), a process that facilitates cancer progression and fibrosis of multiple organs21, is regulated by miR-21. MiR-21 expression level was positively correlated with the hepatitis C virus (HCV) viral load and with fibrosis severity22. MiR-21-3p targets tuberous sclerosis 2 (TSC2) mRNA to regulate mTOR pathway in pheochromocytoma/paraganglioma, which is associated with a metastatic risk and tumor progression23. Under stress stimulation, in hepatocytes, miR-21 accelerates hepatic insulin resistance, glucose intolerance, and fatty degeneration24. Moreover, miR-21 was identified as one of the most increased microRNAs in the non-alcoholic steatohepatitis (NASH) patients25,26. The above evidence suggests that miR-21 could regulate glucose metabolism; however, the underlying mechanism via which miR-21 regulates glucose metabolism to affect tumorigenesis is still unknown.
Limb and CNS expressed 1 like (LIX1L) is over-expressed in various human cancers, as well as in The Cancer Genome Atlas (TCGA) tumor samples27,28. LIX1L is a putative RNA-binding protein (RBP) involved in post-transcriptional gene regulation. LIX1L binds to microRNAs and regulates their expression, thereby promoting cell proliferation27. LIX1L is associated with mesenchymal gene expression, cell migration, and invasion regulation28. Long noncoding RNA (LncRNA) TatD DNase domain containing 1 (TATDN1) promotes HCC progression by degrading miRNA-6089 to up-regulate LIX1L29. However, the potential role of LIX1L in regulating glucose metabolism and hepatocarcinogenesis as well as the underlying molecular mechanisms remain to be defined. Our interest in this fundamental biomedical question stems from publicly available database analyses which showed that LIX1L is aberrantly over-expressed in HCC tissues, suggesting a link between LIX1L and liver cancer.
In this study, we demonstrated that LIX1L promoted HCC development by regulating the glucose metabolism via down-regulation of FBP1. LIX1L is obviously up-regulated in HCC tissues, which is correlated with an unfavorable prognosis of HCC patients. Functional studies showed that LIX1L knockdown up-regulated FBP1, enhanced gluconeogenesis, and thereafter resulted in a reduced HCC development and metastasis. Mechanistically, LIX1L up-regulated miR-21-3p to inhibit FBP1 expression. In orthotopic HCC models, the inhibition of miR-21-3p increased gluconeogenesis and reduced tumor growth. Taken together, we identified novel correlations between LIX1L, miR-21-3p, and FBP1 signaling pathways, that contribute to HCC progression by reprogramming tumor glucose metabolism.
2. Materials and methods
2.1. Cell lines and plasmids
HepG2, Huh7, SNU449 and HCCLM3 of HCC cell lines (Shanghai Cell Bank of the Chinese Academy of Sciences) were cultured in the 10% fetal bovine serum basal medium. LIX1L stable knockdown or overexpression cell lines, including HCCLM3-shLIX1L, SNU449-shLIX1L, HepG2-LIX1L, Huh7-LIX1L and their control cells, were constructed by transfecting pGPU6/Neo-shLIX1L (GenePharma) or LIX1L-Myc, respectively. Stable HCCLM3-shLIX1L-Luc or control HCCLM3-Luc cell lines were constructed by transfecting luciferase reporter vector pGL6-Luc. Cell lines were routinely detected for mycoplasma. The LIX1L-Myc and FBP1-Flag plasmids were constructed in pCMV-Tag3B and pCMV-C-Flag, respectively.
2.2. Animal models
The Lix1l-knockout (KO) mice were generated with CRISPR-Cas9 system (Bioray Laboratories Inc., Shanghai, China), the sgRNA were designed to target LIX1L domain. Lix1l–/– with gRNA1: 5′-GAATGTGGTGGAGGCCCTTCAGG-3′; gRNA2: 5′-GTGACATAGCAGACGTAGGGAGG-3′. Only male mice were used to avoid interanimal variation caused by the difference in estrus stages of female mice. All randomly assigned experimental mice were matched sex and age.
For HCC mice model, diethylnitrosamine (DEN, dissolved in saline, 25 mg/kg) was intraperitoneally injected into 2-week-old wild-type (WT) and Lix1l-knockout C57BL/6J mice. Six weeks later, the mice were intraperitoneally injected with 2 mL/kg carbon tetrachloride (CCl4) twice per week for 12 weeks.
For xenograft mice model, 4 × 106 of HepG2 cells (LIX1L stable overexpression or control) and 3 × 106 of HCCLM3 cells (LIX1L stable knockdown or control) were suspended in 200 μL PBS and Matrigel (1:1; BD Biosciences) and then injected subcutaneously into the posterior flanks of 6-week-old athymic nude mice (Model Animal Research Center of Nanjing University). Meanwhile, tumor sizes were measured at indicated time and volumes were calculated by the Eq. (1):
| (1) |
Six weeks later, tumors were harvested followed by histologic examination.
To establish metastasis model in vivo, 1 × 106 cells (HCCLM3-shLIX1L-Luc or HCCLM3-Luc) were injected into the BALB/c nude mice (10 in each group) through the tail vein. Six weeks later, VivoGlo™ Luciferin (In Vivo Grade, Promega, USA) was injected intraperitoneally into mice at 3 mg dissolved in 200 μL saline per mouse. The noninvasive bioluminescence system was used to monitor lung metastases progression.
Orthotopic HCC mice model was established to examine the effect of miR-21-3p in tumor progression. HCCLM3-Luc cells (4 × 106) were inoculated into the left lobes of the livers of the nude mice. Tumorigenesis was imaged by bioluminescence after two weeks. Subsequently, the mice were randomly assigned to two experimental groups. Then miR-21-3p inhibitors or the control particle were delivered to liver with PEGylated cationic lipid nanoparticle (NP) via tail vein per three days for 3 weeks.
According to the ARRIVE guidelines, all experimental mice were approved by the Committee of the University of Animal Use and Care of China Pharmaceutical University, Nanjing, China.
2.3. Real-time RT-PCR
Total RNA was extracted by RNA-Quick Purification Kit (ES Science, Shanghai, China). MicroRNAs were reversely transcribed into cDNAs with Bulge-Loop™ miRNA qRT-PCR primer Reverse Transcription Kit. Real-time PCR was performed using Bulge-Loop™ miRNA qRT-PCR Starter Kit (RiboBio). MiR-21-3p primers, miR-21-3p mimics and inhibitors were from RiboBio. All primer sequences are presented in Supporting Information Table S1.
Human liver cancer cDNA arrays (Outdo Biotech, China) were used for quantitative RT-PCR verification. The information of the 30 patients has been reported previously30.
2.4. Cell proliferation, apoptosis assays
Cell growth and colony formation assays were conducted as previously described30. Lipofectamine RNAiMax (Invitrogen) was used to transfect LIX1L siRNA. Cell apoptosis was measured by Annexin V-FITC/PI staining method.
2.5. Transwell assays
Cell migration and invasion were evaluated via Transwell assay as previously described31.
2.6. Metabolic assays
The extracellular acidification rate (ECAR) was measured using a Seahorse Biosciences XF96 analyzer (North Billerica, MA, USA). The system includes Seahorse XFe assay medium (pH 7.4) which contains 2 mmol/L glutamine, 10 mmol/L glucose, 0.5 mmol/L oligomycin, and 100 mmol/L 2-deoxyglucose. Approximately, 2 × 104 cells were seeded into Seahorse XFe96 cell culture microplates. The data represent for the glycolytic capacity.
Glucose level was quantitated via Glucose Assay Kit (Jiancheng Bioengineering Institute, Nanjing, China). The lactate content was measured using lactic acid assay kit (KeyGen Biotechnology). All of the procedures were according to the manufacturer's instructions.
To measure glucose production, cells were seeded in six-well plate. The medium was replaced with 1 mL of glucose-free DMEM supplemented with 2 mmol/L sodium pyruvate and 20 mmol/L sodium lactate. After 3 h incubation, medium was collected and the glucose concentration was measured with Glucose Assay Kit.
2.7. Detection of ATP level
Intracellular ATP level was assayed by a bioluminescence method with the ATP Assay System Bioluminescence Detection Kit (Jiancheng Bioengineering Institute, Nanjing, China) according to the kit instructions.
2.8. Luciferase reporter assay
Wild-type (WT) or mutant 3′UTR of FBP1 luciferase reporter plasmids were cloned into pGL6-miR downstream of the luciferase coding region (Luc2) luciferase reporter vector (Beyotime). Luciferase activity was evaluated with Dual-Luciferase Reporter Assay kit (Promega).
2.9. Immunoblots, immunofluorescence and immunohistological staining
Immunoblots were analyzed using standard methods. Antibodies are presented in Supporting Information Table S2. Liver, lung and tumor specimens were embedded, deparaffinized, hydrated, stained and scanned by standard methods. Paired human liver cancer paraffin tissue sections (HLiv-HCC180Su-14) were obtained from Shanghai Outdo Biotech, and LIX1L expression was detected by immunofluorescence.
2.10. RNA-seq analysis
The strand-specific RNA-seq library was constructed using the NEBNext Ultra Directional RNA library kit for Illumina (NEB, Beverly, MA, USA). Meanwhile, the RiboMinus Eukaryote kit was used to remove ribosomal RNA. Quality control of the library was performed using Agilent 2200 TapeStation (Agilent), and sequenced using Illumina HiSeq 3000 from RiboBio.
2.11. Statistical analysis
ANOVA or Student's t test were applied to statistical analysis by GraphPad Prism version 5.0. Survival curves were analyzed by Kaplan–Meier log-rank (MentelCox) test. All statistics were presented as the mean ± standard error of mean (SEM). All experiments were performed at least 3 biological replicates. A P value < 0.05 is statistically significant.
3. Results
3.1. LIX1L is over-expressed and correlated with poor prognosis in HCC
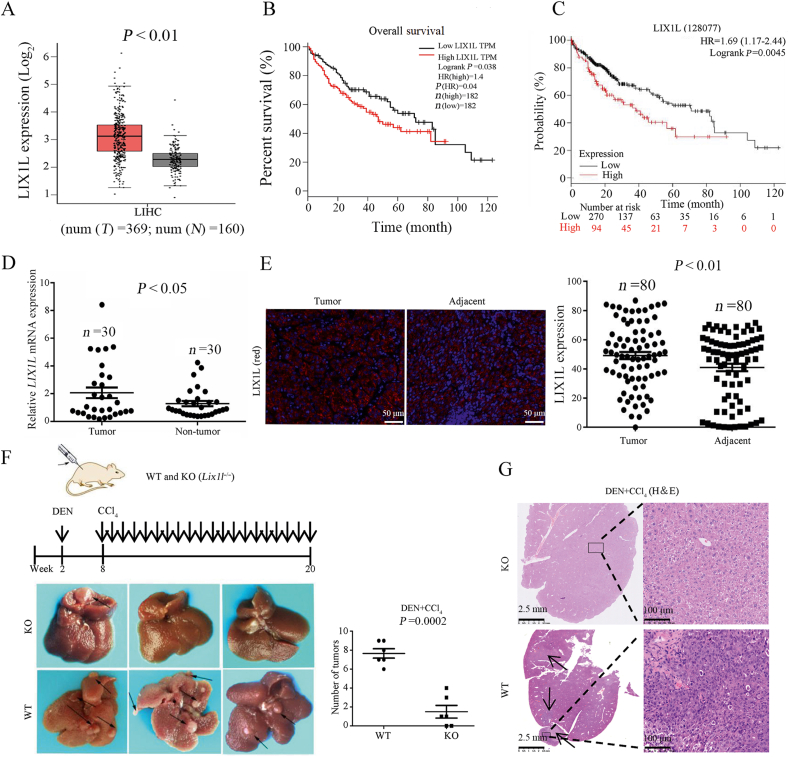
To reveal the relationship between LIX1L expression and HCC development, we analyzed the public TCGA transcriptome datasets and Gene Expression Omnibus (GEO) database to compare LIX1L expression between HCC tissues and normal tissues. We found that LIX1L expression was significantly up-regulated in HCC patients (Fig. 1A and Supporting Information Fig. S1A). Kaplan–Meier analysis of the TCGA dataset revealed that high LIX1L level was associated with poor overall survival (Fig. 1B). Meanwhile, according to the analysis of the public database KMploter (https://kmplot.com), we also found that the high expression of LIX1L did relate to a poor prognosis of liver cancer (Fig. 1C). To further validate our findings, we used human liver cancer tissue cDNA arrays for LIX1L mRNA expression detection. Comparing to adjacent normal tissues, the LIX1L mRNA expression level in the liver cancer tissues was much higher (Fig. 1D). Additionally, the HCC tissue microarray was evaluated using immunofluorescence, and the LIX1L protein expression level was also higher in HCC tumor tissues (Fig. 1E). The data set was divided into low and high LIX1L expression groups, and LIX1L expression was negatively correlated with the survival rate of patients, which was defined by Kaplan–Meier survival analysis (Fig. S1B). To further investigate the function of LIX1L in HCC development, LIX1L knockout (Lix1l–/–) and WT littermate mice were subjected to DEN and CCl4-induced mouse HCC model. In this model, LIX1L knockout mice generated fewer and smaller tumors than WT mice (Fig. 1F). LIX1L deficiency significantly suppressed tumor cell proliferation, as indicated by hematoxylin and eosin (H&E) staining (Fig. 1G). All of these results indicate the inducing role of LIX1L in HCC development.
Figure 1.
The expression of LIX1L positively correlates with unfavorable prognosis of human HCC. (A) The expression of LIX1L in HCC (n = 369) and non-tumour tissues (n = 160) from GEPIA database. Based on the selected TCGA tumors and TCGA normal + GTEx normal datasets performed difference analysis (one-way ANOVA). (B) Kaplan–Meier overall survival curve of HCC patients, using the TCGA datasets in the GEPIA database. High and low expression of LIX1L were stratified by the median. (C) Kaplan–Meier survival analysis of patients with HCC using KMplot (http://kmplot.com). (D) Relative expression of LIX1L from 30 HCC tumor and adjacent non-tumor tissues measured by qRT-PCR. (E) LIX1L expression of tumor and adjacent non-tumor tissues detected by immunofluorescence (left) with quantized analyses of paired clinical HCC samples (n = 80). Scale bar = 50 μm. (F) Left, the schematic diagram of DEN + CCl4 induced HCC model (top) and liver images (bottom). Right, tumor numbers (n = 6). (G) Representative hematoxylin and eosin (H&E) staining showed proliferation of tumor cells. Scale bar = 2.5 mm (left), 100 μm (right). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.2. LIX1L promotes hepatoma cell growth in vitro and in vivo
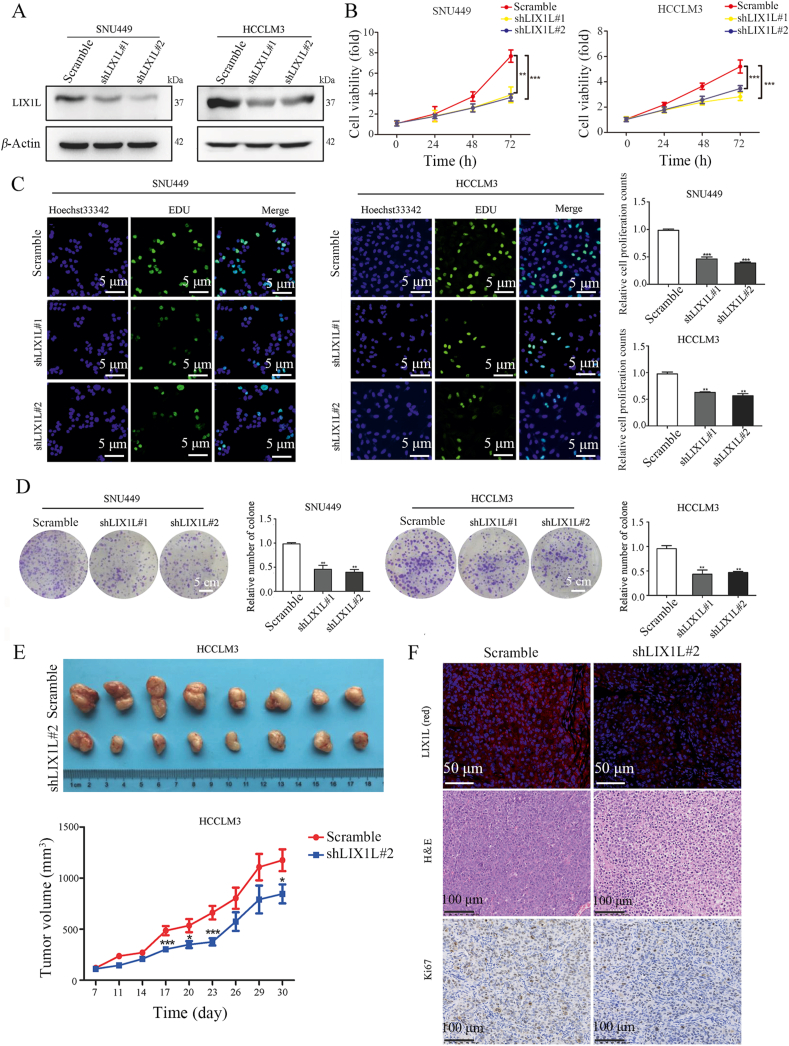
To further investigate the function of LIX1L in HCC, we first studied the effect of LIX1L on hepatoma cell growth. LIX1L was highly expressed in HCCLM3 and SNU449 cells, whilst a relatively low expression in HepG2 and Huh7 cells (Supporting Information Fig. S2A). According to the expression level of LIX1L in HCC cell lines, we stably knocked down LIX1L (shLIX1L#1 and shLIX1L#2) in HCCLM3 and SNU449 cells (Fig. 2A and Fig. S2B). LIX1L knockdown significantly hindered cell proliferation, which was analyzed using Cell Counting Kit-8 (CCK8) assay (Fig. 2B) and a 5-ethynyl-2′-deoxyuridine (EDU) incorporation assay (Fig. 2C), compared with cells transfected with scramble siRNA, which were used as controls. The colony formation assay further validated that the proliferation rate evidently decreased with LIX1L knockdown (Fig. 2D). However, knockdown of LIX1L showed no effect on apoptosis (Fig. S2C). Furthermore, LIX1L knockdown obviously inhibited HCCLM3 cell growth in a xenograft mice model (Fig. 2E). Additionally, compared to the control groups, knocking down LIX1L decreased the tumor cell proliferation, as observed using H&E staining and by analyzing Ki67 expression in the tumor tissues (Fig. 2F).
Figure 2.
Silencing of LIX1L suppresses hepatoma cell growth in vitro and vivo. (A) Western blot detection of LIX1L expression in stable knockdown cell lines. (B)–(D) Cell proliferation (CCK8, EDU assays) and colony formation assays were performed in LIX1L stable knockdown SNU449 and HCCLM3 cell lines. Scale bar = 5 μm (C), 5 cm (D). (E) Images (top) and volume (bottom) of xenograft tumor in nude mice (n = 8). (F) Expression of LIX1L was detected by immunofluorescence, scale bar = 50 μm. The proliferative status of xenograft tumor was evaluated by H&E and Ki67 staining in nude mice, scale bar = 100 μm. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
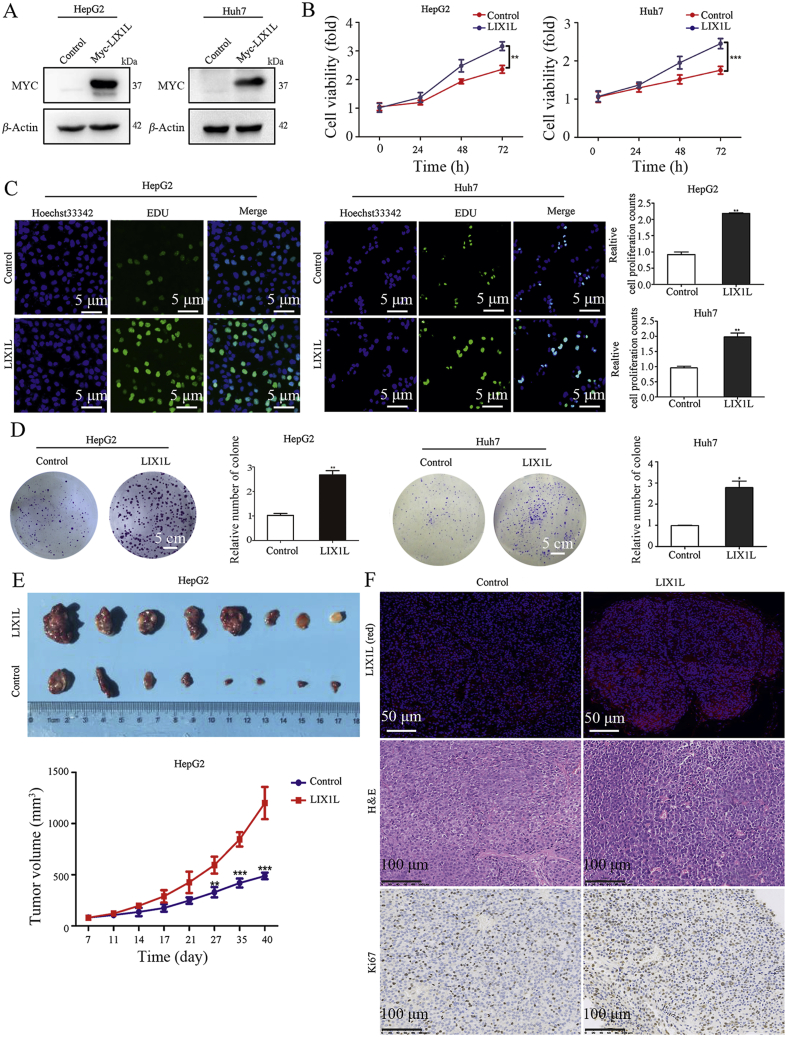
Conversely, we also established LIX1L stable overexpression HepG2 and Huh7 cell lines (Fig. 3A, and Fig. S2D). Cell proliferation (Fig. 3B–D) results show that LIX1L overexpression promoted cell growth in vitro. Similarly, LIX1L overexpression significantly promoted the HCC xenograft (Fig. 3E). The increased cell proliferation in LIX1L overexpression tumors was further confirmed by H&E and Ki67 staining (Fig. 3F). Accordingly, we propose that LIX1L promotes HCC growth in vitro and in vivo.
Figure 3.
LIX1L promotes HCC progression in vitro and vivo. (A) Western blot analysis of LIX1L expression in stable overexpression cell lines. (B)–(D) Cell proliferation and colony formation assays were performed in LIX1L stable overexpression HepG2 and Huh7 cell lines. Scale bar = 5 μm (C), 5 cm (D). (E) Images (top) and volume (bottom) of xenograft tumor in nude mice (n = 8). (F) Expression of LIX1L was detected by immunofluorescence, scale bar = 50 μm. The proliferative status of xenograft tumor was evaluated by H&E and Ki67 staining in nude mice, scale bar = 100 μm. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.3. LIX1L promotes migratory, invasive, and metastasis abilities of hepatoma cells
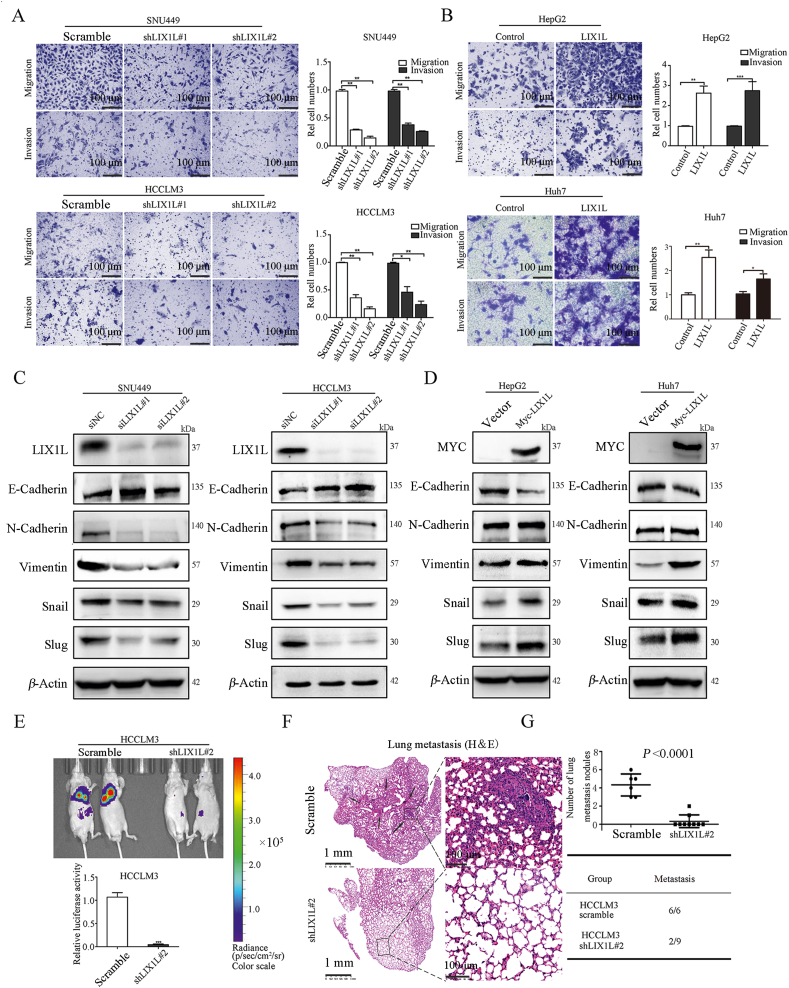
Next, we investigated the role of LIX1L on tumor metastasis. Transwell assay results show that LIX1L knockdown reduced cell migration and invasion in SNU449 and HCCLM3 cells (Fig. 4A). In contrast, LIX1L overexpression strongly increased the migration and invasion ability of HepG2 and Huh7 cells (Fig. 4B). In addition, LIX1L knockdown reduced the mesenchymal markers (N-cadherin, vimentin, snail and slug) and increased the epithelial marker (E-cadherin) in SNU449 and HCCLM3 cells (Fig. 4C). Conversely, the ectopic expression of LIX1L exhibited the opposite effects in HepG2 and Huh7 cells (Fig. 4D). Next, we tested the effect of LIX1L on metastasis in vivo. HCCLM3-shLIX1L#2-Luc stable cells and the control cells were injected into nude mice via their lateral tail veins. According to the metastatic foci detected using a noninvasive bioluminescence system (Fig. 4E) and histologic analysis (Fig. 4F), LIX1L knockdown significantly suppressed HCC metastasis (Fig. 4G). In addition, the incidence of lung metastases in the LIX1L knockdown group mice decreased (Fig. 4G). Hence, these results further demonstrate the important role of LIX1L in HCC metastasis.
Figure 4.
LIX1L promotes hepatoma cell migration, invasion and metastasis. (A) and (B) Transwell assay analyzed effects of LIX1L on cell migration and invasion. scale bar = 100 μm. (C) and (D) EMT-related proteins were assessed by Western blot. (E) Silencing of LIX1L inhibited lung metastases in vivo. Bioluminescence imaging of mice 6 weeks after the tail vein injected with HCCLM3-shLIX1L#2-Luc or HCCLM3-Luc cells (n = 10 per group). Quantitative evaluation of bioluminescence from color scale. Four mice in control group and one mouse in HCCLM3-shLIX1L#2-Luc group died naturally under SPF conditions. (F) H&E staining of lung tissues obtained from nude mice. Arrow indicates lung metastatic nodules. Scale bar = 1 mm (left), 100 μm (right). (G) The number of lung metastatic foci (upper) and incidence of lung metastases (bottom). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
3.4. LIX1L regulates HCC glucose metabolism and negatively affects gluconeogenesis
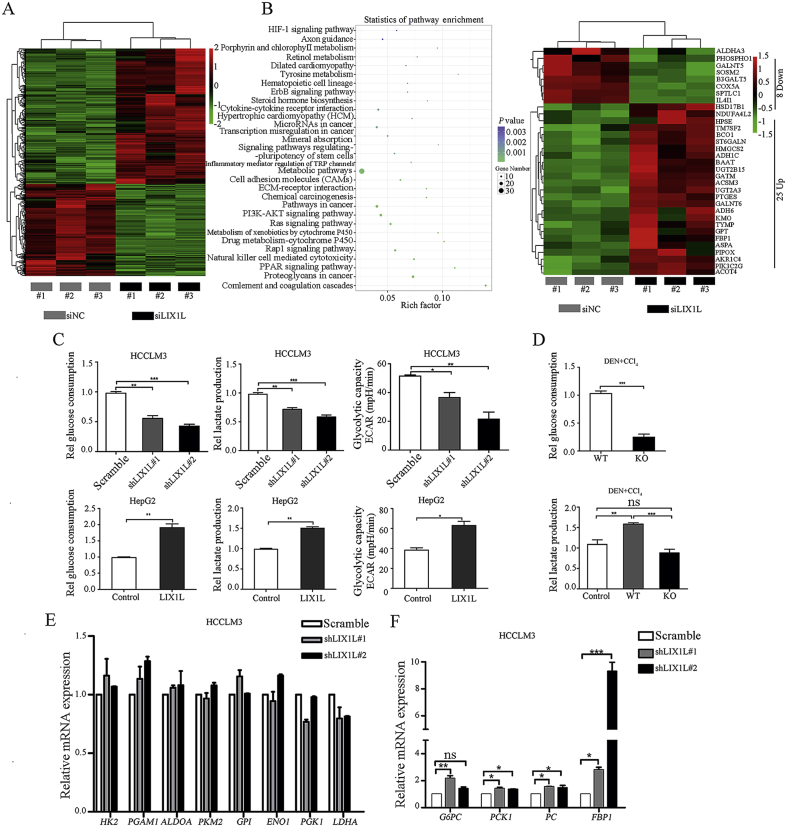
To explore the molecular mechanism underlying LIX1L-induced HCC progression, RNA-seq analysis was carried out to get insight into the transcriptional profiles of HCCLM3 cells transfected with LIX1L siRNAs or NC siRNAs. QRT-PCR analysis confirmed that LIX1L siRNA could successfully reduce more than 90% of LIX1L expression in HCCLM3 cells (Supporting Information Fig. S3A). As shown in Fig. 5A, the expression of 412 genes increased and that of 280 genes decreased. KEGG pathway analysis revealed that metabolic pathways were significantly altered upon LIX1L deficiency (Fig. 5B, left panel). Further metabolic gene expression analyses revealed that LIX1L knockdown significantly regulated glucose-metabolism-related gene expression, including FBP1 (Fig. 5B, right panel). Moreover, these regulatory connections were supported by a Gene Set Enrichment Analysis (GSEA) which identified that LIX1L knockdown positively correlated with gluconeogenesis (Fig. S3B). Indeed, LIX1L knockdown reduces the consumption of glucose and the production of lactate in HCCLM3 cells, and inhibits the extracellular acidification rate (ECAR). In addition, LIX1L knockdown increased glucose production and decreased ATP level (Fig. 5C and Fig. S3C). However, LIX1L overexpression in HepG2 cells led to an enhanced glucose consumption, lactate production, ECAR and ATP level, decreased glucose production, which implying that gluconeogenesis, an ATP-consuming process of glucose anabolism, could be decreased by LIX1L (Fig. 5C and Fig. S3D). In the DEN + CCl4 induced HCC mice model, LIX1L knockout mice showed a reduced glucose consumption and lactate production (Fig. 5D). Interestingly, we found that the knockdown or overexpression of LIX1L had no significant effect on glycolysis-related gene expression, including HK2, PKM2, PGK1, ALDOA, LDHA, PGAM1, GPI, and ENO1 (Fig. 5E and Fig. S3E). However, LIX1L knockdown enhanced the gluconeogenic gene expression, including that of G6PC, PCK1, PC, and FBP1, while LIX1L overexpression attenuated it (Fig. 5F and Fig. S3F). Gluconeogenesis is the main glucose metabolism pathway in liver cells, which can regulate whole-body glucose32. In liver cancer, gluconeogenesis plays a role in suppressing tumors33. Therefore, we assumed that LIX1L might promote HCC progression via switching gluconeogenesis toward glucose metabolism.
Figure 5.
LIX1L regulates HCC glucose metabolism. (A) RNA sequencing heat map of gene expression profile of HCCLM3 cells with LIX1L knockdown. (B) Left, gene ontology analysis of silencing LIX1L by KEGG Pathway Database. Right, a heat map showing the differentially expressed genes related to metabolic pathway. (C) Analysis of glucose consumption, lactate production and ECAR in HCCLM3 cells with LIX1L knockdown (top) or HepG2 cells with LIX1L overexpression (bottom). (D) The glucose and lactate level were tested in DEN + CCl4 induced HCC mice. The expression of glycolysis (E) and gluconeogenesis (F) related genes were measured by qRT-PCR. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant.
3.5. LIX1L promotes HCC metastasis via FBP1 down-regulation
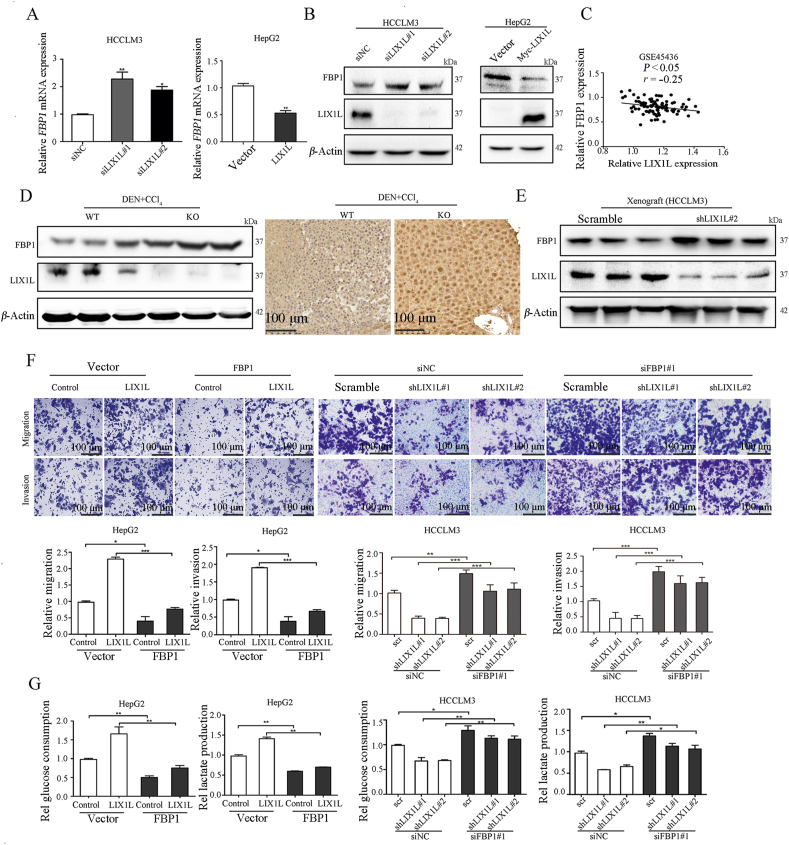
Among the four key gluconeogenesis enzyme genes that are affected by LIX1L, FBP1 was the most increased in HCCLM3-shLIX1L cells while decreased in HepG2-LIX1L cells (Fig. 5F and Fig. S3F). To confirm this result, we employed LIX1L siRNAs and plasmid to transiently knock down and overexpress LIX1L, respectively. Indeed, LIX1L knockdown increased FBP1 expression, while LIX1L overexpression decreased it (Fig. 6A and B). Notably, a negative correlation between the expression of LIX1L and FBP1 was observed in HCC samples (Fig. 6C). In addition, for the DEN + CCl4-treated HCC model, ablation of LIX1L resulted in a higher FBP1 expression level (Fig. 6D and Supporting Information Fig. S4A). Consistently, LIX1L knockdown increased FBP1 expression in BALB/c nude mice xenograft tumors, while LIX1L overexpression decreased it (Fig. 6E and Fig. S4A).
Figure 6.
LIX1L regulates glucose metabolism of HCC via FBP1. Gene (A) and protein (B) detection of FBP1 expression in HCCLM3 and HepG2 cells transfected with LIX1L siRNA or LIX1L plasmid. (C) The correlation between LIX1L and FBP1 in HCC patients from GEO database (GSE45436) was analyzed. Endogenous FBP1 expression in tissue samples from HCC mice (D) and xenograft models (E) were measured by Western blot analysis or immunohistochemical staining. Scale bar = 100 μm. The migration and invasion abilities (F), glucose and lactate levels (G) were measured in LIX1L stable overexpression HepG2 cells transfected with FBP1 plasmid or LIX1L stable knockdown HCCLM3 cells transfected with FBP1 siRNAs. Scale bar = 100 μm. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
FBP1 plays a pivotal role in tumor metastasis34. Consistent with the reported results, and in contrast with FBP1 overexpression, FBP1 knockdown promoted HCCLM3 cell migration, invasion, and EMT process (Figs. S4B–S4D). To elucidate the role of FBP1 in LIX1L-mediated HCC metastasis, we overexpressed FBP1 in HepG2-LIX1L cells and knocked down FBP1 in HCCLM3-shLIX1L cells. FBP1 could abrogate the cell migration and invasion induced by LIX1L overexpression in HepG2 cells (Fig. 6F). Moreover, the inhibitory effect of LIX1L knockdown on cell migration, invasion, and EMT processes could be reversed via FBP1 knockdown in HCCLM3 cells (Fig. 6F and Fig. S4E). These results indicate that FBP1 is involved in LIX1L loss-mediated inhibition of HCC metastasis. The metabolic analysis results showed that FBP1 could reverse the enhancement effect of LIX1L on glucose and lactate metabolisms (Fig. 6G). LIX1L knockdown no longer altered the glucose metabolism when FBP1 was concurrently knocked down (Fig. 6G). Together, these results demonstrate that LIX1L promotes HCC progression via FBP1 expression regulation.
3.6. LIX1L reduced FBP1 expression via miR-21-3p
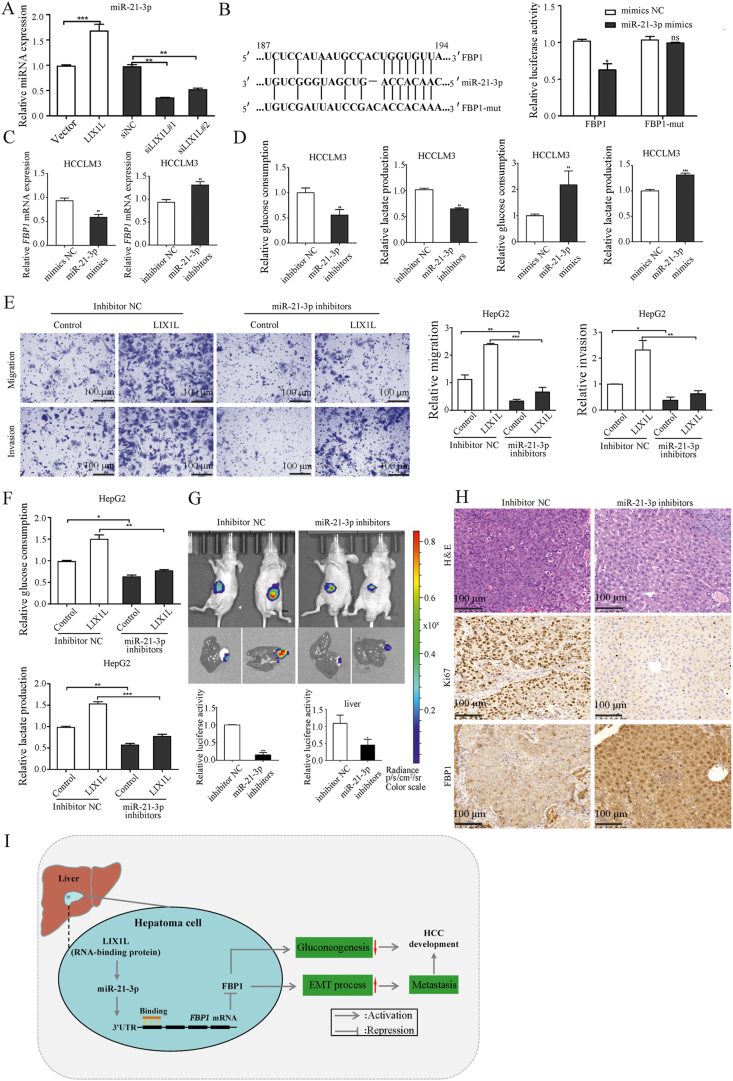
MicroRNAs participate in the regulation of tumor growth and metastasis by targeting and inhibiting gene expression35. LIX1L binds to microRNAs, regulates their expression, and promotes cell proliferation27. Thus, we speculated that LIX1L might regulate FBP1 expression via miRNAs. Integrating the miRanda and TargetScan predicted miRNAs that targeting FBP1, we found miR-21-3p expression was significantly reduced in LIX1L knockdown HCCLM3 cells, while it was increased in LIX1L overexpressed HCCLM3 cells (Fig. 7A). Indeed, the RNA-seq data also showed that knocking down LIX1L significantly reduced the expression of miR-21. Thus, we assumed that miR-21-3p might participate in LIX1L-mediated regulation of HCC growth, metastasis, and glucose metabolism. miRNA binds to the 3′-UTR of mRNA to inhibit gene expression; thus, we used 3′-UTR reporter gene analysis to verify whether miR-21-3p targets FBP1. MiR-21-3p mimic inhibits the luciferase activity of FBP1 3′-UTR in HCCLM3 cells. However, when the miR-21-3p binding site is mutated, the regulatory effect is weakened (Fig. 7B). In addition, miR-21-3p reduced FBP1 mRNA expression (Fig. 7C and Supporting Information Fig. S5A). These findings indicate that miR-21-3p targets the 3′UTR of FBP1 to inhibit its expression.
Figure 7.
FBP1 is a direct target of miR-21-3p. (A) MiR-21-3p was measured by qRT-PCR in LIX1L overexpression or knockdown HCCLM3 cells. (B) Left, schematic diagram of the binding site of miR-21-3p in FBP1 3′UTR. Right, the luciferase activity with transfecting the WT or Mut FBP1-3′UTR reporter under miR-21-3p mimics treatment. (C) Relative FBP1 mRNA expression upon treatment with miR-21-3p mimics or miR-21-3p inhibitors in HCCLM3 cells. (D) Analysis of glucose and lactate levels. (E) and (F) LIX1L stable overexpression HepG2 cells were transfected with inhibitor NC or miR-21-3p inhibitors. Abilities of migration and invasion were measured via Transwell assay. Scale bar = 100 μm (E). Levels of glucose and lactate were measured (F). (G) Bioluminescence images of luciferase expression obtained after receiving six times tail vein injections of miR-21-3p inhibitors or inhibitor NC co-encapsulated NP in the orthotopic liver tumor model. (H) The proliferative status of tumors was evaluated by H&E and Ki67 staining. The expression of FBP1 in the orthotopic liver tumor tissues was detected by immunohistochemistry. Scale bar = 100 μm. (I) Schematic hypothetical model for the LIX1L‒miR-21-3p-FBP1 axis in regulating proliferation, metastasis and glucose metabolism of HCC. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant.
Next, we investigated the function of miR-21-3p on proliferation, metastasis and metabolism in HCC. MiR-21-3p inhibitor reduced cell proliferation, while FBP1 knockdown could reverse this effect (Fig. S5B and S5C). Moreover, miR-21-3p inhibitor decreased cell migration and invasion (Fig. S5D), glucose metabolism (Fig. 7D), and inhibited the EMT process (Fig. S5E). In contrast, miR-21-3p mimic led to opposite effects. In addition, miR-21-3p inhibitor could abrogate LIX1L overexpression-induced cell migration, invasion, and glucose metabolism (Fig. 7E and F). MiR-21-3p mimic could reverse the effect of LIX1L knockdown on cell migration, invasion, EMT process, and glucose metabolism (Fig. S5F–S5H). Collectively, these results demonstrate that LIX1L up-regulated miR-21-3p expression, thereby inhibiting FBP1 expression to promote HCC metastasis and metabolism.
We next constructed an orthotopic model of HCC to study the significance of miR-21-3p in HCC development. The orthotopic model was successfully established as indicated in the bioluminescence images showing luciferase expression (Supporting information Fig. S6A). Nanoparticles enter the cell via the endocytic pathway, and the lysosomal escape is important for the gene silencing in the cytoplasm. We employed PEGylated cationic lipid nanoparticle (NP), which showed a greater stability and targeting ability to deliver miR-21-3p inhibitor36. We co-encapsulated DiD/miRNA inhibitors with NPs to investigate whether the nanoparticles could deliver miR-21-3p inhibitors to escape from endosomes/lysosomes, and then, we detected the location of the nanoparticles in the cells using Lyso Sensor staining after 3, 6, and 9 h of culture. After 9 h of incubation, we observed a great dissociation between the DiD and Lyso Sensor signals, which confirmed an efficient endosomal escape of DiD/miRNA co-encapsulated NP (Fig. S6B). The mice were then injected with miR-21-3p inhibitor or inhibitor NC co-encapsulated NPs through the tail vein, after inoculation for two weeks. After 3 weeks, compared with the mice injected with control inhibitors, the mice injected with miR-21-3p inhibitors emitted a significantly reduced bioluminescence and exhibited smaller tumors (Fig. 7G). IHC and qRT-PCR revealed that FBP1 expression was significantly elevated in miR-21-3p inhibitor-treated tumors, accompanied by a decreased Ki67 expression (Fig. 7H and Fig. S6C). Thus, these results demonstrate that miR-21-3p mediates LIX1L-induced HCC cell growth, invasion, and metastasis by suppressing FBP1 expression.
4. Discussion
Liver cancer is the third leading cause of cancer death worldwide37. Despite improvements in the treatments, the overall survival is still poor due to a high rate of metastasis38. Therefore, a deep understanding of the molecular mechanisms underlying HCC development and the discovery new targets to block cancer progression are urgently needed. An enhanced glycolysis is correlated with tumor progression and poorer clinical outcomes, and the involving molecules could be potential targets for drug development. Although it has been reported that LIX1L is associated with cancer cell proliferation, its role in HCC progression has not been delineated. In this study, we present novel evidence that LIX1L expression is dramatically increased in human HCC tissues, which predicts an unfavorable HCC prognosis. Mechanically, LIX1L up-regulates miR-21-3p, which targets and down-regulates FBP1 to promote tumorigenesis and metastasis by regulating glucose metabolism. The inhibition of miR-21-3p enhanced gluconeogenesis and suppressed liver cancer progression. Thus, integrated with the mechanistic studies and the clinical data, we believe that LIX1L has an important role in liver cancer progression.
A high glycolysis rate, which is one of most important characteristics of cancer, promotes cancer cell proliferation39. The expression of the three key enzymes (PEPCK, FBPase, and G6Pase) involved in gluconeogenesis are regulated by transcriptional factors and coactivators40. Gluconeogenesis is impaired in malignant hepatocytes. Therefore, the promotion of gluconeogenesis could be a promising strategy to retard HCC development. Here, we reveal that LIX1L deficiency reduced glucose consumption and lactate production, meanwhile suppressing the ECAR (Fig. 5). LIX1L showed no effect on most glycolytic enzymes, except for gluconeogenesis-regulatory enzyme, with FBP1 being the most affected enzyme gene. FBP1 restored altered the glucose metabolism and suppressed tumor progression, indicating that FBP1 may serve as a key molecule in the LIX1L-mediated regulation of glucose metabolic pathway in HCC.
FBP1 and FBP2 are the two FBP isoenzymes in mammalian cells. FBP1 is the key gluconeogenic enzyme, which is ubiquitously expressed, and FBP2 is restricted to the muscles41. FBP1 deficiency contributed to metabolic advantages and promoted the progression of cancer, including HCC34. FBP1 interacted and inhibited HIF1α activity to suppress renal carcinoma progression9. Loss of FBP1 is required for Snail-mediated EMT and cancer progression in breast cancer and HCC8,34. Our data present the evidence that there is an inverse relationship between LIX1L and FBP1 expression in HCC. LIX1L promotes cell migration, invasion, EMT process, and glucose metabolism dependent on the regulation of FBP1. Knockdown of FBP1 could block the inhibitory effect of LIX1L knockdown on cell migration, invasion, and EMT processes. FBP1 could also inhibit the enhancement effect of LIX1L on glucose and lactate metabolisms (Fig. 6). These results suggest that LIX1L inhibits gluconeogenesis by down-regulating FBP1, finally resulting in the promotion of HCC cell proliferation and metastasis.
Another critical question is how LIX1L down-regulated FBP1 expression. An increased DNA methylation in the promoter region of FBP1 suppressed its expression8,12. It is reported that Forkhead Box C1 (FOXC1) inhibited FBP1 transcription by binding to its promoter region42. Snail–G9a–Dnmts complex inhibited FBP1 expression via an increase in H3K9me2 modifications and DNA methylations on the FBP1 promoter in breast cancer8. Actually, LIX1L could bind to microRNAs and regulate their expression, which may regulate gene expression at the post-transcriptional level27. Considering the negative correlation between the expression of LIX1L and FBP1 in HCC samples, we supposed that LIX1L may regulate FBP1 expression via miRNAs. Indeed, LIX1L up-regulated oncogenic miR-21-3p expression, and miR-21-3p inhibitor could abrogate LIX1L-overexpression-induced cell migration, invasion, and glucose metabolism (Fig. 7). Wu et al.26 reported that miR-21 targets HMG-box transcription factor 1 (HBP1) and links nonalcoholic fatty liver disease (NAFLD) and HCC progression by regulating HBP1–P53–SREBP1C pathway. Our current study provides a potential direction for LIX1L in the progression of NASH and NASH-associated HCC. MiR-21 regulates the lipid and glucose metabolism in hepatocytes, which served as a promising target for fatty liver disease. However, the detailed mechanisms behind this phenotype were not yet investigated24. Here, we found that miR-21-3p targets and down-regulates FBP1, while miR-21-3p inhibition decreased cell migration, invasion, and glucose and lactate metabolisms (Fig. 7). Moreover, employing a lipid polymer hybrid nanoparticle delivery system to deliver miR-21-3p inhibitor to the livers of mice could inhibit orthotopic HCC growth. However, the mechanism on how LIX1L up-regulates miR-21-3p needs to be further studied. Since LIX1L is an RNA-binding protein, one attractive hypothesis is that LIX1L may be an intermediate player in miRNA processing.
5. Conclusions
In summary, the data provided establishes a glucose metabolism regulation novel mechanism (model, Fig. 7I), which involves the antagonism of LIX1L and FBP1 in regulating gluconeogenesis. First, LIX1L expression increased and favored a poor prognosis in HCC patients. Second, LIX1L up-regulated miR-21-3p expression to block FBP1 expression, thereby promoting HCC progression. Finally, inhibiting miR-21-3p increased FBP1 expression and suppressed liver cancer growth. The functional characterization of the LIX1L–miR-21-3p–FBP1 axis in HCC progression may provide insights for the treatment of liver cancer. Therefore, our studies identify LIX1L and miR-21-3p, which are important cellular targets for designing new therapies against liver cancer.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 82074068 and 81872889) and Natural Science Foundation of Jiangsu Province (BK20181332, China) to Hao Zhang. The Drug Innovation Major Project (2018ZX09711-001-007 and 2018ZX09735002-003, China), the “Double First-Class” University Project (CPU2018GF03, China) to Lingyi Kong.
Author contributions
Hao Zhang designed and conceived the experiments. Jie Zou and Xiaoyun Zhu developed methodology and conducted most of the experiments. Jie Zou, Xiaoyun Zhu, Dejuan Xiang, Yanqiu Zhang and Jie Li acquired the data. Hao Zhang and Jie Zou wrote and reviewed the manuscript. Zhigui Su provided technical or material support. Hao Zhang and Lingyi Kong conducted supervision.
Conflicts of interest
No potential conflict of interest are disclosed.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.02.005.
Contributor Information
Lingyi Kong, Email: cpu_lykong@126.com.
Hao Zhang, Email: hiaaron@sina.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Yarchoan M., Agarwal P., Villanueva A., Rao S., Dawson L.A., Llovet J.M. Recent developments and therapeutic strategies against hepatocellular carcinoma. Cancer Res. 2019;79:4326–4330. doi: 10.1158/0008-5472.CAN-19-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen L.Z., Ding K., Wang Z.R., Ding C.H., Lei S.J., Liu J.P. SHP-1 acts as a tumor suppressor in hepatocarcinogenesis and HCC progression. Cancer Res. 2018;78:4680–4691. doi: 10.1158/0008-5472.CAN-17-3896. [DOI] [PubMed] [Google Scholar]
- 3.Hsu P.P., Sabatini D.M. Cancer cell metabolism: warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 4.Levine A.J., Puzio-Kuter A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen M.C., Vatner D.F., Shulman G.I. Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong J., Wang X., Zheng X., Wang D., Fu B., Sun R. Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metab. 2018;28:243–255. doi: 10.1016/j.cmet.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Dong C., Yuan T., Wu Y., Wang Y., Fan T.W., Miriyala S. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Canc Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B., Qiu B., Lee D.S., Walton Z.E., Ochocki J.D., Mathew L.K. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Wang J., Xu H., Xing R., Pan Y., Li W. Decreased fructose-1,6-bisphosphatase-2 expression promotes glycolysis and growth in gastric cancer cells. Mol Cancer. 2013;12:110. doi: 10.1186/1476-4598-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J., Jin X., Yan Y., Shao Y., Pan Y., Roberts L.R. Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci Rep. 2017;7:43864. doi: 10.1038/srep43864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M., Zhang J., Li N., Qian Z., Zhu M., Li Q. Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Wang X., Zhang J., Lam E.K., Shin V.Y., Cheng A.S. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene. 2010;29:442–450. doi: 10.1038/onc.2009.332. [DOI] [PubMed] [Google Scholar]
- 14.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Iwakawa H.O., Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Otsuka M., Kishikawa T., Yoshikawa T., Yamagami M., Ohno M., Takata A. MicroRNAs and liver disease. J Hum Genet. 2017;62:75–80. doi: 10.1038/jhg.2016.53. [DOI] [PubMed] [Google Scholar]
- 17.Pineau P., Volinia S., McJunkin K., Marchio A., Battiston C., Terris B. MiR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thurnherr T., Mah W.C., Lei Z., Jin Y., Rozen S.G., Lee C.G. Differentially expressed miRNAs in hepatocellular carcinoma target genes in the genetic information processing and metabolism pathways. Sci Rep. 2016;6:20065. doi: 10.1038/srep20065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lampis A., Carotenuto P., Vlachogiannis G., Cascione L., Hedayat S., Burke R. MiR21 drives resistance to heat shock protein 90 inhibition in cholangiocarcinoma. Gastroenterology. 2018;154:1066–1079. doi: 10.1053/j.gastro.2017.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng F., Henson R., Lang M., Wehbe H., Maheshwari S., Mendell J.T. Involvement of human microRNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 21.Kumarswamy R., Volkmann I., Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquez R.T., Bandyopadhyay S., Wendlandt E.B., Keck K., Hoffer B.A., Icardi M.S. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 23.Calsina B., Castro-Vega L.J., Torres-Perez R., Inglada-Perez L., Curras-Freixes M., Roldan-Romero J.M. Integrative multi-omics analysis identifies a prognostic miRNA signature and a targetable miR-21-3p/TSC2/mTOR axis in metastatic pheochromocytoma/paraganglioma. Theranostics. 2019;9:4946–4958. doi: 10.7150/thno.35458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calo N., Ramadori P., Sobolewski C., Romero Y., Maeder C., Fournier M. Stress-activated miR-21/miR-21∗ in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut. 2016;65:1871–1881. doi: 10.1136/gutjnl-2015-310822. [DOI] [PubMed] [Google Scholar]
- 25.Loyer X., Paradis V., Henique C., Vion A.C., Colnot N., Guerin C.L. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARalpha expression. Gut. 2016;65:1882–1894. doi: 10.1136/gutjnl-2014-308883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H., Ng R., Chen X., Steer C.J., Song G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. 2016;65:1850–1860. doi: 10.1136/gutjnl-2014-308430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura S., Kahyo T., Tao H., Shibata K., Kurabe N., Yamada H. Novel roles for LIX1L in promoting cancer cell proliferation through ROS1-mediated LIX1L phosphorylation. Sci Rep. 2015;5:13474. doi: 10.1038/srep13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajapakse V.N., Luna A., Yamade M., Loman L., Varma S., Sunshine M. CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. iScience. 2018;10:247–264. doi: 10.1016/j.isci.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C., Xu Y., Lu T.F., Zhang J.J., Qian Y.B., Xu N. LncRNA TATDN1 induces the progression of hepatocellular carcinoma via targeting miRNA-6089. Eur Rev Med Pharmacol Sci. 2019;23:6459–6466. doi: 10.26355/eurrev_201908_18529. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Zhang Y., Zhu X., Chen C., Zhang C., Xia Y. DEAD box protein 5 inhibits liver tumorigenesis by stimulating autophagy via interaction with p62/SQSTM1. Hepatology. 2019;69:1046–1063. doi: 10.1002/hep.30300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Zhang Y., Chen C., Zhu X., Zhang C., Xia Y. A double-negative feedback loop between DEAD-box protein DDX21 and Snail regulates epithelial–mesenchymal transition and metastasis in breast cancer. Cancer Lett. 2018;437:67–78. doi: 10.1016/j.canlet.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Adeva-Andany M.M., Perez-Felpete N., Fernandez-Fernandez C., Donapetry-Garcia C., Pazos-Garcia C. Liver glucose metabolism in humans. Biosci Rep. 2016;36 doi: 10.1042/BSR20160385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma R., Zhang W., Tang K., Zhang H., Zhang Y., Li D. Switch of glycolysis to gluconeogenesis by dexamethasone for treatment of hepatocarcinoma. Nat Commun. 2013;4:2508. doi: 10.1038/ncomms3508. [DOI] [PubMed] [Google Scholar]
- 34.Liu G.M., Li Q., Zhang P.F., Shen S.L., Xie W.X., Chen B. Restoration of FBP1 suppressed Snail-induced epithelial to mesenchymal transition in hepatocellular carcinoma. Cell Death Dis. 2018;9:1132. doi: 10.1038/s41419-018-1165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartonicek N., Maag J.L., Dinger M.E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue J., Zhao Z., Zhang L., Xue L., Shen S., Wen Y. Neutrophil-mediated anticancer drug delivery for suppression of postoperative malignant glioma recurrence. Nat Nanotechnol. 2017;12:692–700. doi: 10.1038/nnano.2017.54. [DOI] [PubMed] [Google Scholar]
- 37.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 38.Heimbach J.K., Kulik L.M., Finn R.S., Sirlin C.B., Abecassis M.M., Roberts L.R. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z., Dong C. Gluconeogenesis in cancer: function and regulation of PEPCK, FBPase, and G6Pase. Trends Cancer. 2019;5:30–45. doi: 10.1016/j.trecan.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Huangyang P., Li F., Lee P., Nissim I., Weljie A.M., Mancuso A. Fructose-1,6-bisphosphatase 2 inhibits sarcoma progression by restraining mitochondrial biogenesis. Cell Metab. 2020;31:174–188. doi: 10.1016/j.cmet.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q., Wei P., Wu J., Zhang M., Li G., Li Y. The FOXC1/FBP1 signaling axis promotes colorectal cancer proliferation by enhancing the Warburg effect. Oncogene. 2019;38:483–496. doi: 10.1038/s41388-018-0469-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.