Abstract
Computer vision and artificial intelligence applications in medicine are becoming increasingly important day by day, especially in the field of image technology. In this paper we cover different artificial intelligence advances that tackle some of the most important worldwide medical problems such as cardiology, cancer, dermatology, neurodegenerative disorders, respiratory problems, and gastroenterology. We show how both areas have resulted in a large variety of methods that range from enhancement, detection, segmentation and characterizations of anatomical structures and lesions to complete systems that automatically identify and classify several diseases in order to aid clinical diagnosis and treatment. Different imaging modalities such as computer tomography, magnetic resonance, radiography, ultrasound, dermoscopy and microscopy offer multiple opportunities to build automatic systems that help medical diagnosis, taking advantage of their own physical nature. However, these imaging modalities also impose important limitations to the design of automatic image analysis systems for diagnosis aid due to their inherent characteristics such as signal to noise ratio, contrast and resolutions in time, space and wavelength. Finally, we discuss future trends and challenges that computer vision and artificial intelligence must face in the coming years in order to build systems that are able to solve more complex problems that assist medical diagnosis.
Keywords: Artificial intelligence (AI), computer vision (CV), medical image analysis, cardiology, oncology, microscopy, neurodegenerative disorders, respiratory diseases, gastroenterology
Introduction
Undoubtedly, it is intangible to think about all the technological advances that have been developed in the field of medicine in recent decades. Not only have they allowed us to understand more precisely the anatomy and physiology of the different organs that structure the human body, but they have also let us advance in the identification and, therefore, the treatment of several diseases from very early stages in different areas of medicine. This has been largely accomplished by the development of computer vision (CV) and artificial intelligence (AI). Briefly, these tools provide us the ability to acquire, process, analyze, and understand an infinite number of static and dynamic images in real time, which will represent a better characterization of each disease, and a better patient selection for early interventions.
Since many diagnostic methods available to date present the disadvantage of being invasive, expensive and/or very complex for their standardization in most parts of the world, assisted diagnosis through CV and AI represent a feasible solution that allows to identify a broad number of different diseases from initial stages, define better the treatment and follow-up, and decrease the health care costs associated with each patient.
The union of high-performance computing with machine learning (ML) offers the capacity to deal with big medical image data for accurate and efficient diagnosis. Moreover, AI and CV may reduce the significant intra- and inter-observer variability, which undermines the significance of the clinical findings. AI allows to automatically make quantitative assessments of complex medical image with increased diagnosis accuracy.
The objective of this review is to assess in an understandable and well-structured way recent advances in automatic medical image analysis in some of the diseases with higher incidence and prevalence rates worldwide.
Cardiovascular diseases (CVD), oncological disorders and pulmonary diseases such as chronic obstructive pulmonary disease (COPD) and coronavirus disease 2019 (COVID-19) have positioned themselves as the main causes of mortality in individuals 50 years of age and older (1-5). Moreover, CVD are also the main cause of premature death (4). Gastrointestinal and liver diseases account for some of the highest burden and cost in public health care worldwide (4,6). Moreover, the lack of timely attention at early stages of these diseases lead to high morbidity and mortality rates (3). Neurodegenerative diseases such as Alzheimer’s and Parkinson’s have increased their incidence in modern times. As life expectancy increases, so does the occurrence of these diseases (7). Although AI has contributed to develop a large number of methods that assist the diagnosis of these diseases (8,9), we limit this review to CV and ML methods applied on medical imaging, namely ultrasound, computer tomography, magnetic resonance, microscopy and dermoscopy. We show how these methods are used to detect, classify, characterize and enhance relevant information in order to aid clinical diagnosis and treatment.
In the next section, the relation of medical imaging with CV and AI is discussed from a general point of view. Subsequent sections show recent advances and applications of these two areas of computer science that have improved diagnosis performance of the above-mentioned diseases.
CV and AI in medical imaging
According to Patel et al. (10) some of the earliest works on AI applied to medicine date from the 1970s when AI was an already known discipline and the term was created at the very famous 1956 Dartmouth College conference (11). While many researchers created model-based image processing based on AI, they did not call them that, even when their algorithms were from this area (10). Since then, new applications started to emerge that led to the first conference of the new organization Artificial Intelligence in Medicine Europe (AIME), at Maastricht, The Netherlands on June 1991, that established the term AI in Medicine. At first, most methods were based on expert systems. More recently, Kononenko (12) punctuates that the goal of AI in medicine is to try to make computers more intelligent and one of the basic requirements of intelligence is the ability to learn. ML appeals to that concept and some of the first successful algorithms in medicine were naive Bayesian approaches (12).
CV deals with a large range of problems such as image segmentation, object recognition, detection, reconstruction, etc. It aims at modeling and understanding the visual world by extracting useful information from digital images, often inspired by complex tasks of human vision. Although it exists since the 1960s, it remains an unsolved and challenging task to the extent that only recently computers have been able to provide useful solutions in different application fields. It is a multidisciplinary subject closely related to AI. AI is a broad area of computer science that aims at building automatic methods to solve problems that typically require human intelligence. ML, in turn, is a part of AI that builds systems that are able to automatically learn from data and observations. Most successful methods of CV have been developed with ML techniques. Among the most widely used methods of ML are Support Vector Machines (SVM), Random Forests, Regression (linear and logistic), K-Means, k-nearest neighbors (k-NN), Linear Discriminant Analysis, Naive Bayes (NB), etc.
Deep learning (DL) is a subfield of ML that has become an attractive and popular tool in CV because of its amazing results in complex problems of visual information analysis and interpretation. The term deep refers to multiple-layer neural networks models. In recent years, there has been a rising interest in applying DL models to medical problems (12-14). For example, deep neural networks have shown amazing results in skin lesion classification tasks. Some outstanding examples can be found in annual challenges (15).
Examples of DL models are convolutional neural networks (CNNs), recurrent neural networks, long short-term memory, generative adversarial networks, etc. Recent advances in this field have shown impressive accuracies and measured results. A schematic relation between CV and AI in medical imaging is presented in Figure 1.
Figure 1.
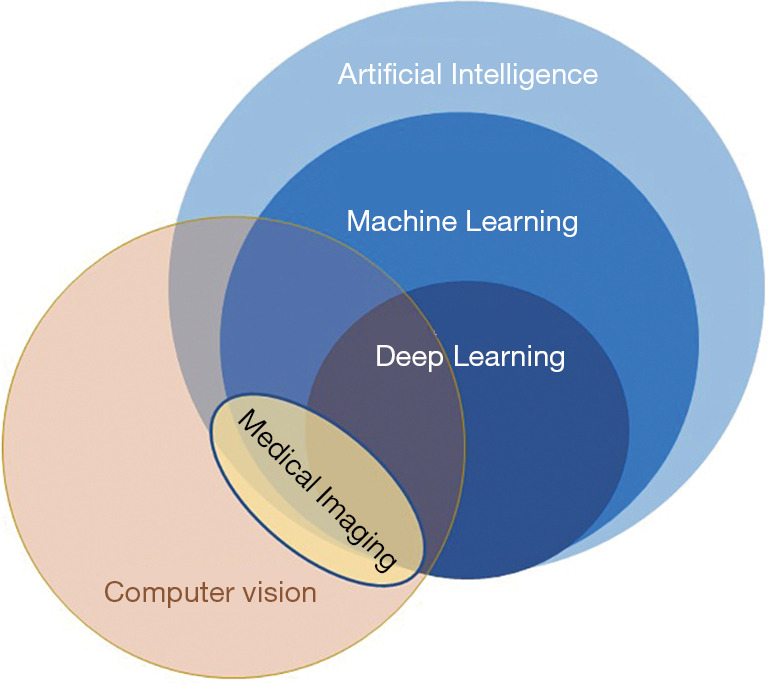
Relation between computer vision and artificial intelligence.
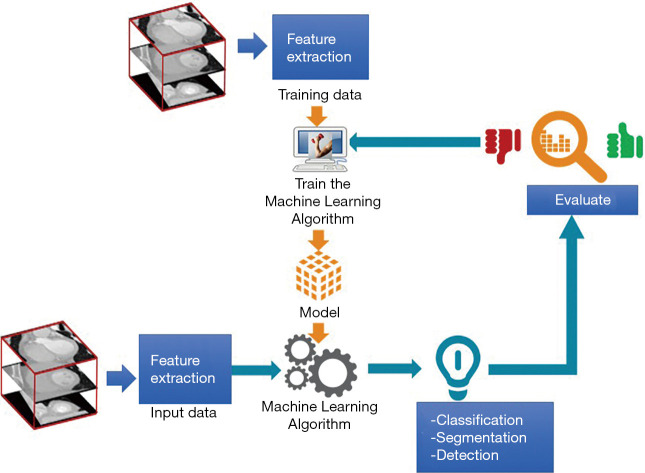
The interaction between CV and AI in medical imaging is shown in Figure 2. Relevant feature extraction in medical image databases is a first critical step to train a new ML model. The training process aims at obtaining a model that has learned a specific task such as segmentation, classification, detection, recognition, etc. on the specific training data. Next, the model is tested with new input data that undergoes the same feature extraction process. The results of the task achieved with this test data are evaluated with performance metrics. If results do not meet the user requirements, the process is repeated until a new combination of feature extraction and ML methods satisfies the required performance level.
Figure 2.
The flow of computer vision and machine learning interaction in medical imaging
CVD
Heart analysis is relevant for understanding cardiac diseases which are a major concern in the health area. They constitute a multifactorial and multidisciplinary clinical disease, leading to a great detriment to the quality of patients’ life. The World Heart Federation reports CVD as the main cause of morbidity and mortality in almost two thirds of the world population. Some of the most important diseases are related to the heart and blood vessels. They include coronary heart diseases, cerebrovascular disease, peripheral arterial disease, rheumatic heart disease, congenital heart disease, deep vein thrombosis and pulmonary embolism. Behavioral risk factors such as unhealthy diet, physical inactivity, alcohol consumption, and obesity represent major risk factors for heart failure (16).
Different medical imaging modalities are used for the assessment of heart failure such as computed tomography (CT), magnetic resonance imaging (MRI) and echocardiography (EG). Clinical analysis of these images comprises qualitative as well as quantitative examinations.
Applications of AI aiming at assessing heart function have ranged from localizing and segmenting anatomical structures to recognizing structural as well as dynamic patterns, and more recently to automatically identifying and classifying several diseases. Whenever heart failure occurs, the heart shows a reduced function. This may cause the left ventricle (LV) to lose its ability to contract or relax normally. In response, LV compensates for this stress by modifying its behavior, which creates hypertrophy and progresses to congestive heart failure (17). These patterns can be recognized and evaluated by AI.
Anatomical and dynamical parameter estimations are necessary to assess heart failure. These tasks related with imaging technology are so relevant that in the last decades their research and development allowed not only to search for better resolution and contrast in their studies, but also the possibility to merge anatomical and functional information. Moreover, new imaging protocols have been developed to comply with analysis and segmentation tasks. Chen et al. presented a complete study of different DL techniques in various image modalities to segment one, two or all cardiac structures (18), as well as coronary arteries, all of them supporting clinical measurements such as volume and ejection fraction.
Some of the more difficult problems of automatic segmentation include poor image quality, low contrast and poor structure border detection. Moreover, cardiac anatomy and gray value distribution of the images varies from person to person, especially when a cardiopathy exists. If we add the intra vendor and scanners differences, the challenge for the development of more sophisticated tools is considerable.
Many different methods for ventricle segmentation have been reported in the literature, where classical active contours are some of the more popular (19). CNNs often excel because of its improved segmentation performance despite their dependence on large amounts of training data (18). This is a serious issue in the case of the medical area, because of data availability restrictions in clinical environments and because of the lack of medical experts willing to annotate large amounts of data. This is a tedious task, causing fatigue as well as intra and inter variability between experts (20). In some areas such as CT and MRI challenges have emerged with annotated databases (18,21,22).
The use of DL has become common not only for cardiac segmentation but also to support classic algorithms, for example when searching for an initial localization of the cardiac structures to segment. Recently, this initialization has been achieved with a CNN that finds a coarse shape while a level set refines the contour, as shown on Figure 3. This hybrid method has the advantage of using only a small database to train a neural network, e.g., a UNet, since only a rough initial segmentation is needed; detail refining is a task left to the active contour method
Figure 3.
Modules of a segmentation step using UNet deep learning as initialization.
Different neural network models emerged for segmentation tasks, such as the UNet model or even Full Convolutional Networks (FCN) as the main algorithm (23,24). These methods have proved to obtain good results. Table 1 presents some of the relevant works in the cardiovascular area with a mention to the imaging modality.
Table 1. Cardiovascular applications. Selection of relevant works of different methods in cardiovascular area with different image modalities.
| Selected works | Description | Type of images | Year |
|---|---|---|---|
| Chen et al. (18) | Study of different deep learning techniques for analysis and segmentation tasks for different heart structures and coronary arteries with Convolutional neural networks | MR, CT, US | 2020 |
| Petitjean et al. (19) | Short axis view segmentation with active contours method | MR | 2011 |
| Carbajal-Degante et al. (23) | CNN and active contours for segmentation | CT | 2020 |
| Avendaño et al. (24) | UNet CNN segmentation of cardiac structures | MR | 2019 |
| Zhang et al. (25) | Creation of a CNN based tool that identifies cardiopathies in clinical practice | Echocardiogram | 2018 |
MR, magnetic resonance; CT, computer tomography; US, ultrasound.
Motion analysis as support for heart failure analysis
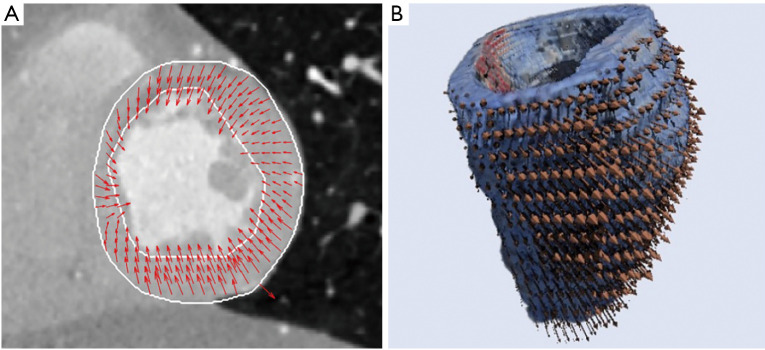
Most 2D segmentation methods analyze complete volumes on a slice by slice basis losing inter-slice interactions. They usually fail in some challenging slices located on the base or on the apex where contours are not well defined. To address these problems additional information, such as motion, has been introduced in many of these methods. Optical flow estimation comprises techniques that can be used to provide information of cardiac motion through the cardiac cycle. Dynamic changes of the cardiac walls can be characterized by motion vectors showing the heart deformation. Heart’s motion estimation can assist the study of different malformations and anomalies, and this information can be enhanced again with different image features. Over the years, the study of cardiac movement has been carried out mainly in two dimensional images, however, it is important to consider that the deformations due to the movement of the heart occur in a three-dimensional space. New 3D results can demonstrate the heart’s movement complexity as shown in Figure 4 (26).
Figure 4.
Motion estimation. (A) 2D motion estimation results for heart left ventricle on systole, short axis view. (B) Heart volume with 3D motion estimation results on diastole.
Future trends in the cardiac area aim at building fully automatic tools able to understand the images and relate them to specific pathologies. This is the case of Zhang et al. (25), who developed a complete tool that identifies different cardiopathies. Their integrated system analyses echocardiogram studies and is composed of modules that start from identifying metadata from Digital Imaging and Communications in Medicine format, followed by a pipeline of different DL algorithms with four tasks, (I) image classification, (II) image segmentation, (III) measurements of cardiac structure and function, and (IV) disease detection. Decisions are taken during the pipeline, ending in a differentiation of patients’ characteristics that indicate a disease detection, such as hypertrophic cardiomyopathy or pulmonary hypertension.
Oncological diseases
Breast ultrasound (BUS)
Breast cancer has become the most frequent cancer and the number one cause of cancer-related deaths among women around the world, with rates increasing in nearly every region globally. Early detection and screening are critical to improve the prognosis for the patient. Screening consists in identifying cancers before any symptoms appear and, although, mammography is recommended by the WHO for breast cancer screening, ultrasound has become a useful tool for assisting and complementing this procedure, especially in limited resource settings with weak health systems where mammography is not cost-effective (27). While mammography has a high sensitivity in the detection of microcalcifications, ultrasound is able to detect tumors in dense breast tissue, usually found in women under age 30, with higher sensitivity than mammography (28). Young women have low incidence rates of breast cancer; however, its incidence increases at a faster rate than in older women and their tumors tend to be larger, with a higher grade of malignancy and poorer prognostic characteristics, making ultrasound necessary as a complementary technique in breast screening. Also, ultrasound is the best imaging modality to identify if the lesion is solid, such as a benign fibroadenoma or cancer, or a fluid-filled benign cyst (29). Even with this advantage, the visualization of lesions in BUS images is a difficult task due to some intrinsic characteristics of the images like speckle, acoustic shadows and blurry edges (30). These drawbacks make ultrasound imaging and diagnosis highly operator-dependent and with a high inter-observer variation rate, increasing the risk of oversight errors. Therefore, computer-aided diagnosis (CAD) systems are desirable to help radiologists in breast cancer detection and classification. CAD systems using BUS images generally focus on solving three problems: lesion detection, lesion segmentation, and lesion classification.
Lesion detection and segmentation
Accurate automatic detection and segmentation methods of breast tumors can help the experts to achieve faster diagnoses, and it’s a key stage of fully automatic systems for breast cancer diagnosis using ultrasound images (31). Because of the mentioned inherent artifacts in BUS images the automatic detection and segmentation of lesions in this imaging modality is not an easy task (30).
Ultrasound gray-level intensities provide helpful information about the density of different tissues found in the images, however the poor quality of ultrasound images due to speckle noise makes the differentiation difficult. Because of this, most of the proposed methods for tumor detection and segmentation in BUS images use a pre-processing step to obtain more homogenous regions and enhance the contrast of the image, while preserving important diagnostic features (32,33). The internal echo pattern of structures has been used to depict the local variation of pixel intensities to detect abnormal regions within the BUS images, to distinguish lesion regions from normal tissue, acoustic shadows and glands. Texture information seems highly suitable for characterizing internal echo patterns in different structures. However, texture analysis in ultrasound images is not an easy task and many metrics have been used to describe the echo patterns in breast tumors (34-36).
Several studies report automatic methods (30-40), where modeling the knowledge of BUS and oncology as prior constraints is needed. Based on the literature, ML-based methods stand out among the most popular. Supervised methods like AdaBoost, Artificial Neural Networks and Random Forest are the most common methods used for lesion detection and segmentation in BUS (37). Image features used in these methods should be appropriately selected according to the application, and texture information seems highly suitable for ultrasound images, but making an appropriate Feature Selection is an important step specially when dealing with high dimensionality spaces (30). Feature selection methods like Principal Component Analysis and Genetic algorithms have been used to improve the results in the detection and segmentation of lesions, with true positive fraction (TPF) values of 84.48%; however, these approaches still rely on manually designated features, which depend on good understanding of the images and the lesions (38). DL, which is a new field of ML, can directly learn abstract levels of features directly from images, solving the need to propose an initial set of features to describe the problem. CNNs such as LeNet, UNet and FCN-AlexNet, have been successfully implemented in the tasks of lesion detection and segmentation with TPF values of 89.0%, 91.0% and 98.0% (39) respectively. A summary of representative works can be seen in Table 2.
Table 2. Lesion detection and segmentation. Selection of relevant works for lesion detection and segmentation in oncological diseases.
| Selected works | Description | Techniques | Year |
|---|---|---|---|
| Chen et al. (30) | Semi-automatic 3D lesion segmentation method | Discrete active contours with automatic initialization | 2003 |
| Moon et al. (31) | Automatic 3D lesion detection | 3D mean shift and fuzzy c-means clustering | 2014 |
| Madabhushi et al. (32) | Automatic 2D lesion segmentation | Directional gradient deformable shape-based model and probabilistic classification of image pixels | 2003 |
| Liu et al. (35) | Automatic 2D lesion detection | Kernel SVM and classification checkpoints | 2010 |
| Torres et al. (38) | Automatic 2D lesion detection | Genetic algorithms for feature and parameter selection and random forest classification | 2019 |
| Yap et al. (39) | Automatic 2D lesion segmentation | Patched and fully convolutional CNNs | 2008 |
| Huang et al. (41) | Automatic 3D lesion segmentation method | Neural networks applied to 2D texture patches | 2008 |
SVM, Support Vector Machines; CNN, convolutional neural network.
Classification of breast lesions
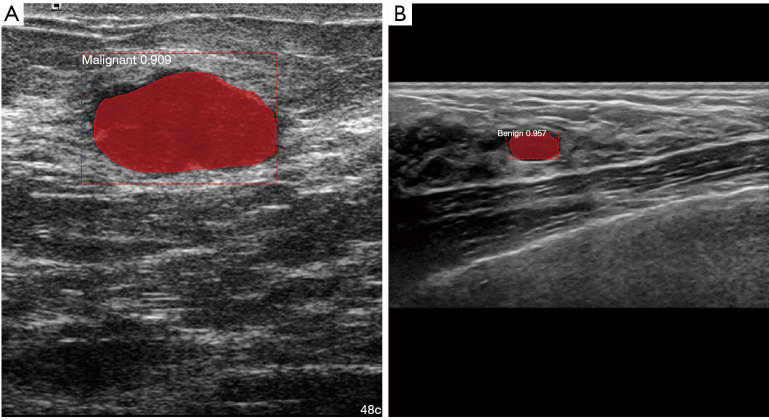
The malignancy of a tumor is estimated by the expert ultrasonographer mainly from its shape, echogenicity and the internal echo pattern. Morphological features such as roundness, aspect ratio, convexity and compactness have been used to characterize the shape of tumors. These descriptors along with texture analysis have shown good results in the classification of breast lesions using ML approaches (40). Most of the proposed methods used classical ML approaches including Linear Regression, Fuzzy Logic Systems and SVM, where the main difference between them are the chosen descriptors used to characterize lesions with accuracy values up to 95.45% (37). CNNs have been used for the classification of lesions by producing a set of transformation functions and image features directly from the data to characterize the malignancy of the lesion. Different CNN models, including GoogleNet, ResNet50, Inception V3, Xception and Mask-RCNN have been proposed for this task, having accuracy values up to 98.0% for the classification of breast lesions into benign and malignant (42,43). A summary of representative works can be seen in Table 3. Figure 5 shows a segmentation and classification of two different breast lesions.
Table 3. Breast lesion analysis. Selection of relevant works for detection and classification of breast lesions.
| Selected works | Description | Year |
|---|---|---|
| Chen et al. (36) | Texture classification of 3D ultrasound breast using run difference matrix with neural networks | 2005 |
| Cao et al. (42) | Evaluation of the performance of several state-of-the-art DL model for the detection and classification for breast lesions | 2019 |
| Chiao et al. (43) | Automatic detection, segmentation and classification of breast lesions using Mask RCNN | 2019 |
DL, deep learning.
Figure 5.
Detection, segmentation, and classification of breast lesions in US using Mask R-CNN. (A) Malignant lesion. (B) Benign lesion.
Fundamental issues and future directions
Although CV and AI methods have been successfully applied for breast lesion detection, segmentation, and classification, there are some fundamental issues that need to be solved before fully CAD systems can be developed taking advantage of these techniques. In fully automatic methods a key step is the lesion detection, which delivers candidate regions of interest (ROIs). Although ML approaches have shown good results in this task, it remains an open and challenging problem because the appearance of benign and malignant lesions are quite different and the quality of the images will also depend on the ultrasonic device, contributing to the difficulty of modeling the lesions. Another important aspect that should be considered is the presence of posterior acoustic shadowing, which appears as a dark region behind the lesion, often obfuscating lesion margins and making it difficult to determine the shape of the lesion. Since this is a challenging issue, this artifact has been overlooked in many BUS CAD systems, however there is an urgent to overcome this problem in order to improve these systems (44). Moreover, the increasing use of 3D ultrasound imaging calls for new lesion detection, segmentation and classification techniques that can be used in a 3D surgical context (41,45). For the development of these techniques, the creation of new datasets that consider ethical considerations is necessary (46).
Deep learning in dermatology
The skin is the largest organ in the human body (47) while skin cancer is the most common form of cancer in the United States (U.S.) (48). According to the American Academy of Dermatology, it is estimated that 1 in 5 Americans will develop skin cancer in their lifetime. As with any other type of cancer, the sooner it is spotted, the better the prognosis becomes. When skin cancer is caught early, it is highly treatable. This type of cancer is mainly diagnosed through a visual inspection. Among the different types of malignant neoplasia, melanoma is a rare form of skin cancer that develops from skin cells called melanocytes which are located in the layer of basal cells at the deepest part of the epidermis, they are also found in the iris, inner ear, nervous system, heart and hair follicles among other tissues. Melanoma represents fewer than 5% of all skin cancers in the U.S. However, melanoma is also the deadliest form of skin cancer, it is responsible for about 75% of all skin cancer related deaths (49). In Mexico, melanoma is responsible for 80% of deaths from skin cancer (50). It is increasing in the world, more than any other malignancy. According to the latest report of the National Institute of Cancerology (Instituto Nacional de Cancerología, INCan), the number of cases in Mexico increased 500% in recent years (51).
Dermatologists use different visual inspection methods to determine if a skin lesion under analysis may be melanoma (malignant). Some of these methods are the ABCDE rule, 7 points checklist, and the Menzies method (52) just to name a few. All these methods are based on the visual features that make a malignant melanoma lesion distinguishable. Dermatology is a medical specialty which can greatly benefit from the powerful feature extraction capabilities of deep neural networks.
State of the art
There is a variety of recent publications that utilize the tools and models of DL to address the problem of skin lesion classification, the use of these techniques has achieved a much better performance in terms of metrics such as accuracy, specificity and sensitivity in comparison with the results obtained with classical ML techniques. Some examples of them can be found in (53-59).
One of the most relevant articles about the applications and the potential that DL can offer in the field of dermatology was presented by Esteva et al. (54). The performance of this network was tested against 21 certified dermatologists in two critical cases: carcinomas vs. seborrheic keratosis and melanomas vs. nevus. Research concludes that the convolutional neuronal network achieves a superior performance than that reached by experts in dermatology. The use of the system in mobile devices to facilitate an early diagnosis is proposed.
Table 4 presents a review of some of the relevant works mentioned above. The last two columns respond to two important aspects: if the performance of the proposed method was compared with that of health professionals and if the dataset used to train the neural network models is publicly available, by “public” we mean whether the images belong to a well-known dataset or whether the authors collected their data from different public and private sources and ultimately made the final dataset public. This last aspect is important as it is one of the factors that allows the results to be reproduced and compared with other techniques.
Table 4. Skin lesion classification. Selection of relevant works of deep learning methods applied to skin lesion detection and classifications.
| Selected works | Description and main contributions | Compared against health-care professionals | Is the dataset public? | Year |
|---|---|---|---|---|
| Esteva et al. (54) | CNN trained with 129,450 images belonging to 757 skin lesion classes. Performance tested against 21 dermatologists. CNN outperforms dermatologists at skin cancer classification | Yes | No | 2017 |
| Fujisawa et al. (55) | CNN trained with 4,867 images belonging to 14 skin lesion classes. Performance tested against 13 dermatologists and nine dermatology trainees. CNN outperforms dermatologists at skin tumors classification | Yes | No | 2019 |
| DeVries et al. (56) | CNN trained on a public dataset for a skin lesion classification challenge. CNN uses two different scales of input images to obtain satisfactory results | No | Yes | 2017 |
| Mahbod et al. (57) | Automated computational method for skin lesion classification based on three CNNs that act as visual feature extractors. Features are combined with SVM whose outputs are merged to generate a final result. Training and validation steps are performed with a public dataset. Satisfactory performance on 150 validation images of the public dataset | No | Yes | 2017 |
| Harangi et al. (58) | Classification method for 3 different skin lesions based on an ensemble of 4 CNNs trained on public datasets. Method fuses outputs of classification layers of the four CNNs. Final classification is obtained by a weighted outputof each of the CNNs. Results are satisfactory | No | Yes | 2018 |
| Menegola et al. (59) | Ensemble of CNNs combining outputs with a SVM final decision. Several public datasets were combined into one to train the CNNs | No | Yes | 2017 |
| Results are satisfactory for two binary classification problems, melanoma vs. all and keratosis vs. all | ||||
| Liu et al. (60) | Deep learning system (DLS) identifies 26 common skin conditions in adults referred for teledermatology consultation (60). DLS provides full differential diagnosis across those 26 conditions | Yes | No | 2020 |
| The DLS consists of several CNN modules that process a variable number of input images. A secondary module processes patient metadata (demographic information and medical history) | ||||
| Dataset comes from a teledermatology service in the US | ||||
| System diagnostic accuracy was compared against clinicians. Its performance is comparable to dermatologists and more accurate than general practitioners |
SVM, Support Vector Machines; CNN, convolutional neural network.
CNNs for dermatology
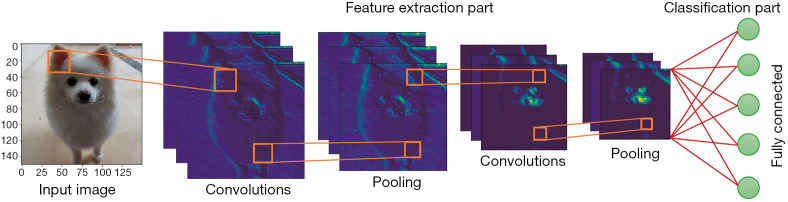
In Figure 6, we show the typical architecture of a CNN. CNNs are composed of several convolution and pooling layers that act as automatic feature extractors for images, the fully connected network receives the relevant features as input and acts as a classifier. CNNs can be trained to perform a widely different variety of tasks, they provide a way to train superhuman image classifiers (61).
Figure 6.
Simplified architecture of a Convolutional Neural Network.
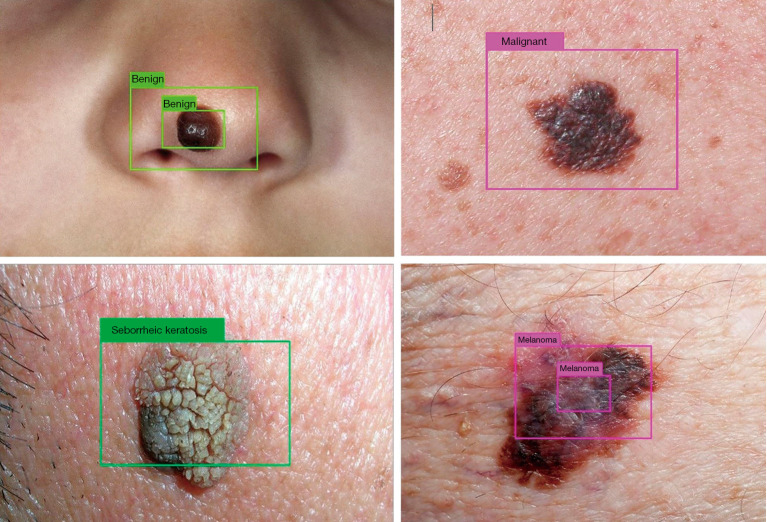
State of the art deep CNNs (YOLOv3) (62) have been trained in conjunction with image processing techniques (segmentation algorithms) to deploy a skin lesion detection system that learns to detect (localize and classify) three types of skin lesions: melanoma, nevus, and seborrheic keratosis. The more general case, detecting benign from malignant skin lesions has also been addressed. Some of the visual results are shown in Figure 7. The DL-based detection system can predict a confidence value (probability) for the nature of the skin lesion under analysis at three different scales.
Figure 7.
Processed test images by the trained deep neural network. Nevi correctly detected as benign and melanoma correctly detected as malignant (upper row). Seborrheic keratosis (benign lesion) and melanoma correctly detected (lower row).
Results of Figure 7 were verified by a dermatologist (to assure that the skin lesion in the image corresponded to the label attached to it). They belong to a test partition which was never seen by the deep neural network. The test partition was conformed from publicly available images from different healthcare websites.
For training the neural network, images from the International Skin Imaging Collaboration archive (ISIC archive) were used. The ISIC archive is a high quality dermoscopic image database, collected from international leading clinical centers and acquired from a variety of devices. At the time of preparing this work, the ISIC archive dataset consisted of 19,330 benign lesions and 2,286 malignant lesions. It is important to remark that DL models learn the most when they are trained with the largest possible number of images.
Future trends in AI for dermatology
One of the most recent works on the skin lesion classification problem was presented by Google Health in collaboration with other institutions like the University of California-San Francisco, the Massachusetts Institute of Technology and the Medical University of Graz. They developed a Deep Learning System (DLS) capable of delivering a differential diagnosis of the most common skin conditions seen in primary care. Unlike previous approaches, this system can exploit the information present in the images as well as in the metadata, achieving an accuracy across 26 skin conditions, on par with U.S. board-certified dermatologists (60).
DL-based systems can be of great use for dermatologists, they can be put to work as a desktop automatic diagnostic app or can be embedded into a smartphone attached to a mobile dermatoscope. We seek to deploy, in the near future, a real-time DL solution in a smartphone app that could be used by general practitioners and the general public. As stated by Liu et al. (60), a solution of this kind could be useful in regions where there is a lack of medical specialists in local clinics. This technology has the potential to have an impact on public health as it can increase the capabilities of general practitioners and help in the decision-making process.
Microscopy image analysis for cancer
Microscopy image acquisition modalities use the light spectrum of electromagnetic radiation and its interaction with internal tissues to evaluate molecular changes. In contrast to other acquisition techniques, microscopy takes advantage of various colors of light to visualize and measure several tissue features with improved contrast and resolution. These features also have potential applications in diagnosis, grading, identification of tissue substructures, prognostication and mutation prediction (63). One of the advantages of microscopy is high spatial resolution. The typical resolution is around hundreds of nanometers, useful for monitoring subcellular features. Superresolution imaging methods can further improve the resolution by one or two orders of magnitude (64). Microscopy takes advantage of a variety of contrast mechanisms, where the most popular stain is hematoxylin and eosin (H&E).
The pathologists have been beneficiated directly by these applications because they could propose tailored therapies based on individual profiles and spend more time focusing on more complicated cases that are not as quickly diagnosable. Even though the number of digital images generated for diagnostic and therapeutic purposes is increasing rapidly (65), extensive experimental comparisons and state-of-the-art surveys are difficult tasks because most data are not public (66) and because there are substantial differences in acquisition protocols, tissue preparation and stain reactivity (67).
In the field of CAD based on microscopy image assessment, an objective quantitative approach is required to increase reproducibility and predictive accuracy. Microscopy provides relevant information about the tissue since the structure is preserved in the preparation process, and it is considered the diagnosis gold standard for many diseases, including many types of cancer (68).
ML for cancer classification
In this section, we focus on two types of cancer: hematologic malignancies (HMs) and colorectal. The main idea is the classification of microscopy image databases that include different types of malignancies (hematologic) and tissues (colorectal). The whole classification process involves CV and ML algorithms, and the process consists of two significant steps: description and classification. The description of these databases is centered on textural information from the images to compute feature vectors (CV). Then, feature vectors are generated from textural information to be processed by the classifier; in other words, a machine-learning algorithm.
HMs are a collection of diseases that includes leukemia and lymphomas (69). Three types of malignancies will be addressed in this study: Chronic lymphocytic leukemia (CLL), follicular lymphoma (FL), and mantle cell lymphoma (MCL). The IICBU2008-lymphoma dataset (70) includes the three mentioned malignancies, and many automatic methods have been proposed to classify this dataset.
According to GLOBOCAN 2018 data, colorectal cancer (CRC) is the third most deathly and the fourth common type of cancer worldwide, with more than 1.8 million new cases registered in 2018 (71). The Epistroma database (72)is a collection of epithelial and stroma tissue samples taken from patients with histologically verified CRC at the Helsinki University Central Hospital, Helsinki, Finland, from 1989 to 1998.
A classification method based on textural features applied to both colorectal and HMs is based on discrete orthogonal moments (DOMs). The orthogonal moments are computed as the projection of the images on an orthogonal polynomial basis. One way of interpreting the projection is as the correlation measure between the image and the polynomial basis (73). The best known discrete polynomial bases (74), with a vast number of applications in a variety of research areas, are Tchebichef, Krawtchouk, Hahn, and Dual Hahn.
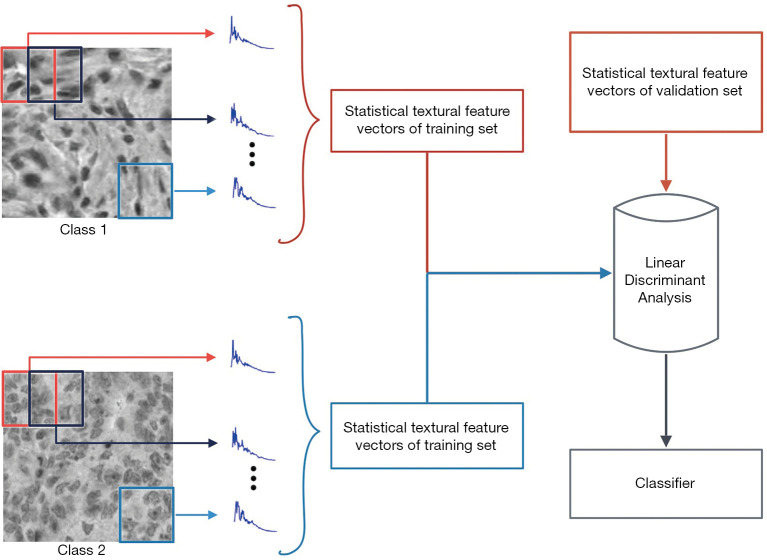
González et al. (75), based on previous papers (76,77), introduced and validated Shmaliy orthogonal moments as a statistical textural descriptor, and its performance is compared to the DOMs. This statistical textural descriptor is based on overlapping square windows and assure numerical stability. They also proposed a classification method with the most popular ML algorithms: SVM, k-NN rule, NB rule, and Random Forest. The general scheme of classification is depicted in Figure 8. This proposal and its detailed description have been published previously for CRC (78) and HMs (73,75) with state-of-art results at the time of publication. Recently, with the spread of neural networks, some papers (79,80) have worked on these databases using CNNs, e.g., Kather et al. (81) compared classical textural descriptors with the most known CNNs architectures applied to different microscopy image datasets. Table 5 summarizes a list of relevant classification methods with different algorithms applied to microscopy image datasets, ordered chronologically.
Figure 8.
General scheme of classification based on discrete orthogonal moments. After computing local DOMs using sliding windows, the statistical textural feature vectors are computed and classified.
Table 5. Microscopy image classification. List of representative classification methods on Lymphoma and Epistroma databases.
| Selected works | Description | Database | Year |
|---|---|---|---|
| Meng et al. (82) | Novel classification model called collateral representative subspace projection modeling (C-RSPM) | Lymphoma | 2010 |
| Orlov et al. (83) | Image description with Chebyshev and Fourier features. They used WND-CHARM, naïve Bayes, and RBF network classifiers. Color deconvolution in stained images (RGB to HE) | Lymphoma | 2010 |
| Xu et al. (84) | Introduction to Deep Convolutional Neural Networks based feature learning to segment or classify epithelium and stroma regions from microarrays. They compared their proposal with three handcraft feature extraction-based approaches | Epistroma | 2016 |
| Nava et al. (73) | Description based on Discrete Orthogonal Tchebichef Moments and classification with SVM and k-NN | Epistroma | 2016 |
| González et al. (75) | Description with discrete orthogonal moments and classification with classical ML algorithms. Introduction of Shmaliy polynomials as a texture descriptor | Lymphoma | 2018 |
| Bianconi et al. (85) | Novel image descriptor called improved opponent color local binary patterns (IOCLBP). It was compared with classical and neural-network-based textural features | Epistroma and Lymphoma | 2017 |
| Song et al. (86) | Description based in Fisher vector encoding with multiple handcrafted or learned local features, and a separation guided dimension reduction method | Lymphoma | 2017 |
| Kather et al. (81) | Performance comparison between classical textural descriptors with the most known CNNs architectures applied to different microscopy image datasets | Epistroma | 2020 |
SVM, Support Vector Machines; CNN, convolutional neural network; ML, machine learning.
DOMs, in conjunction with ML algorithms, have proven to be particularly useful methods for description and classification of microscopy images. From a future perspective, polynomial bases can be used as modulation filters in addition to CNNs, as Luan et al. (87) show in their paper.
As the area of microscopy imaging continues to develop, both in acquisition techniques and the application of new contrast mechanisms, the processing of these images will also grow. For example, through the cited cases, color processing (i.e., different color spaces and color deconvolution) in microscopy images is recurrent to obtain better classification results among tissues of interest. That improvement depends directly on the acquisition and the contrast mechanism. On the other hand, the more public databases there are, the better profitability from the latest DL classifiers, because more information, means better performance classifiers.
Neurodegenerative diseases and brain structure segmentation
In recent years, automatic detection of different brain structures has become a challenging task for scientists since the nature of image acquisition often yields issues as noise, blur or intensity inhomogeneities which depend on the specific medical image modality, affecting the correct brain localization and visualization process. The most popular image modalities used for brain analysis are positron emission tomography (PET), single photon emission computed tomography (SPECT) and MRI, the latter becoming a standard tool due to the good quality contrast of brain tissues in a non-invasive way and has proven very efficient for diagnosis, evaluation and monitoring.
Since the process of manual delineation of brain structures is exhausting and time consuming for experts, the need of automated segmentation is urgent, not only to improve accuracy but also to reduce inter and intra observer variability. For instance, the identification of subcortical brain structures, hippocampus, hypothalamus, amygdala, among others, is a challenging task due to their size and shape. Some of these structures possess a special interest for its association with neurodegenerative disorders (88,89).
This section reviews some representative semi-automatic methods based on CV and AI reported in the literature that tackle brain segmentation problems in medicine and its application nowadays.
Whole brain segmentation
Neurodegenerative disorders are usually related to brain alterations such as structure, shape and volume changes. Whole brain segmentation helps providing qualitative and quantitative comparison with other smaller brain structures. FreeSurfer is a software developed by Fischl et al. (90) which provides a set of algorithms to segment the whole brain from T1-weighted images. A particular approach based on Markov random fields is Local Cooperative Unified Segmentation (LOCUS) which was originally designed by Scherrer et al. (91) and includes subcortical segmentation in phantoms and real 3T brain scans. Other approaches extract relevant information such as high energy regions and oriented structures to ease the identification of white matter, gray matter and cerebrospinal fluid as stated in (92).
The role of hippocampus in dementia
Different types of neurodegenerative diseases are originated in different areas of the brain. For instance, hippocampus plays a key role in the prediction of diseases such as Alzheimer, schizophrenia and bipolar disorder, among others. Jorge Cardoso et al. (93) presented a technique for Alzheimer early detection named Similarity and Truth Estimation for Propagated Segmentation (STEPS). It segments the hippocampus with practical applications to Alzheimer’s Disease Neuroimaging Initiative (ADNI). ADNI stores a large data collection that attempts to keep record of the progression of Alzheimer disease, including MRI and PET images, blood biomarkers, genetic and cognitive tests. In order to reduce the number of atlases (labeled images) needed to guide the segmentation of the whole hippocampus, Multiple Automatically Generated Templates (MAGeT) is a method proposed by Pipitone et al. (94) to generate templates constructed via label propagation, this method achieves good performance compared to classic multi-atlas methods with a reduced training data.
Adapting segmentation methods to analyze shape and intensity information leads to a robust scheme to deal with common image degradations. An automatic segmentation of the caudate nuclei was constructed by using a probabilistic framework to estimate the relation between shape and pixel intensities. This leads to an important improvement for segmenting subcortical brain structures as proposed by Patenaude et al. (95) and originally applied on evaluating Alzheimer’s disease by the software FIRST. In order to improve the segmentation results in the accurate extraction of hippocampus from brain MR images, Gao et al. (96) exploit the capability of atlas-based methods which serves as the initialization for a further implementation in a semi-automatic model. Table 6 summarizes some of the more relevant segmentation methods in this area.
Table 6. Brain structure segmentation. A summary of representative methods for brain segmentation.
| Selected works | Description | Type of images | Structure(s) |
|---|---|---|---|
| Fischl et al. (90) | FreeSurfer software | T1-weighted MRI | Whole brain |
| Scherrer et al. (91) | LOCUS (Local Cooperative Unified Segmentation) based on Markov Random Fields | T3 MRI | Subcortical |
| Carbajal-Degante et al. (92) | Texture-based approach with Hermite coefficients and level sets | MRI | Brain tissue |
| Jorge Cardoso et al. (93) | STEPS (Similarity and Truth Estimation for Propagated Segmentation) | PET, MRI | Hippocampus |
| Pipitone et al. (94) | MAGeT (Multiple Automatically Generated Templates) | Templates | Hippocampus |
| Patenaude et al. (95) | Alzheimer’s evaluation by the software called FIRST | T1-weighted MRI | Subcortical |
| Gao et al. (96) | Semiautomatic atlas-based | MRI | Hippocampus |
| Li et al. (97) | Substantia nigra concentration in Parkinson’s patients | Ultrasound, TCS | Brainstem |
| Olveres et al. (98) | Deformable models based on local analysis | MRI | Midbrain |
| Milletari et al. (99) | Fully automatic localization and segmentation with a CNN | MRI, ultrasound | Deep brain regions |
| Kushibar et al. (100) | Spatial and deep convolutional features | MRI | Subcortical |
MRI, magnetic resonance imaging; PET, positron emission tomography; TCS, transcranial sonography; CNN, convolutional neural network.
Subcortical structures and brainstem linked to movement disorders
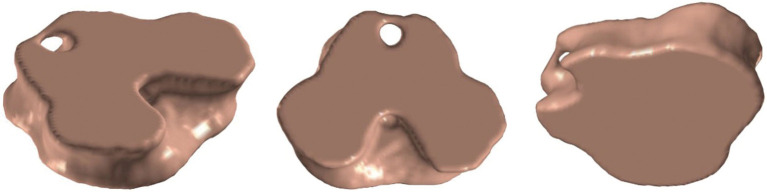
Electric stimulation of the functional brain regions has been suggested in the literature as a surgical treatment for some movement-related disorders (101). Surgical planning of these procedures implies segmenting automatically the appropriate region with high accuracy. For instance, brainstem is relevant to describe disorders as Parkinson or progressive supranuclear palsy. Brainstem includes structures like midbrain, pons, medulla oblongata and substantia nigra. Several diseases are directly related to atrophy, reduced nigra volume and structure deformations. Particularly, Li et al. (97) explored the literature to provide a conclusive relation of substantia nigra concentration in Parkinson’s patients. Complementary works have developed methods to increase accuracy in midbrain segmentation by including texture descriptors and performing local analysis as proposed by Olveres et al. (98) whose results are displayed in Figure 9.
Figure 9.
Segmentation results of midbrain in MRI by Olveres et al. (99).
Among the subcortical structures, the basal ganglia are located in the center of the brain and consist of several different groups of neurons. Neuronal death means that these groups of neurons can no longer work properly, causing stumbling and shaking. In order to improve classic segmentation techniques with subcortical applications, DL-based methods have emerged as successful alternatives. As shown by Milletari et al. (99), DL results may vary depending on the architecture used. Authors showed a segmentation comparative in MRI and transcranial ultrasound volumes of the basal ganglia and midbrain. The incorporation of probabilistic atlases as spatial features allows neural networks to segment the most difficult areas of subcortical brain structures, Kushibar et al. (100).
Respiratory diseases
COPD
There are several important respiratory diseases, such as COPD, asthma, occupational lung diseases and pulmonary hypertension. According to WHO (102), COPD is a progressive lung disease that causes breathlessness (initially with exertion) and predisposes to exacerbations and serious illness. It is characterized by a persistent reduction of airflow. Globally, it is estimated that it accounts for 5% of all deaths until 2019. More than 90% of COPD deaths occur in low and middle-income countries and the primary cause of COPD is exposure to tobacco smoke (either active smoking or secondhand smoke). Traditionally diagnosis of COPD is done by spirometry; however, this method might underdiagnose considerably (103).
Recently, several papers like the Bibault et al. and Mekov et al. (104,105), propose digital image modalities and AI using low dose CT images to detect signs of COPD with DL techniques. Even when the disease is not curable, results offer an opportune diagnosis which allows treatment to improve life quality in COPD patients. In order to pay attention to the lungs area, where lesions occur, several segmentation algorithms have been developed and, nowadays, this task can be executed with traditional as well as DL methods (106,107). COPD detection is treated in Rajpurkar et al. (108) and Shi et al. (109) reviews several methods of segmentation for severe COVID-19. These tasks are accomplished using X radiographies and chest CT images.
There is an increasing number of studies that detect pneumonia on chest radiographies like the one of Zech et al. (110). CT images have proven to have enough spatial resolution to obtain specific metrics of pneumonia lesions needed to estimate a damage level according to Castillo-Saldana et al. (111). They provide objective density-based and texture-based evaluation of the lungs. New methods of measuring airways have recently been developed that complete a prognosis and guide clinical decisions.
On the other hand, studies demonstrate that COPD may also increase the risk of contracting severe COVID-19 (112), another important respiratory disease to analyze. Table 7 presents some of the relevant works mentioned in this section.
Table 7. Respiratory diseases. Selection of relevant works of different methods in respiratory area.
| Selected works | Description | Type of images | Year |
|---|---|---|---|
| Bibault and Xing. (104) | Use artificial intelligence to detect signs of COPD with deep learning techniques | CT and X-ray | 2020 |
| Mekov et al. (105) | Use artificial intelligence and machine learning to detect signs of COPD | CT | 2020 |
| Afzali et al. (106) | Use of contour-based lung shape analysis for tuberculosis detection | Chest Xray | 2020 |
| Shi et al. (109) | Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19 | CT and X-ray | 2021 |
| Zech et al. (110) | Detect pneumonia on chest radiographies | Chest Xray | 2018 |
| Castillo-Saldana et al. (111) | Obtain specific metrics of pneumonia lesions to give a damage degree for chronic obstructive pulmonary disease and fibrotic interstitial lung disease | CT | 2020 |
| Li et al. (113) | Use of artificial Intelligence to distinguish between COVID-19 from Community Acquired Pneumonia on chest CT | Chest CT image | 2020 |
CT, computer tomography; COPD, chronic obstructive pulmonary disease.
COVID-19
The impact of the COVID-19 worldwide crisis has triggered efforts in research communities and interdisciplinary groups to collaborate and search for solutions to mitigate this disease effects. CT imaging and X-ray studies have been helpful in the prognosis for patients since they allow to detect characteristic lesions such as ground-glass opacity, consolidation, pleural effusion, bilateral involvement, peripheral and diffuse distribution (113). However, with the pandemic generated by COVID-19, the lack of radiological expertise and the need for precise interpretation chest medical images has become evident. AI solutions aim at helping the clinical area to face this worldwide crisis.
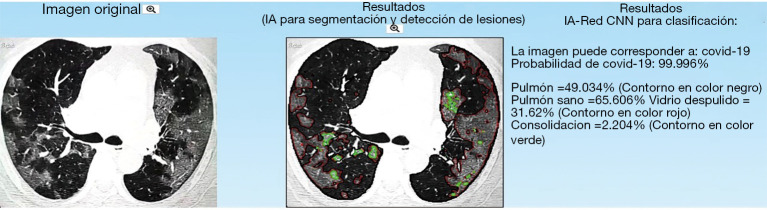
Image-analysis research groups have focused on building computer aided diagnosis systems to detect COVID-19 from the automatic analysis of lung chest radiography and CT images. These AI systems provide a probability of existence of COVID-19 as well as detect, segment and quantify lesions in the lungs (109,113). Figure 10 shows the results delivered by one of such systems that are already available online.
Figure 10.
AI-based online tool that predicts COVID-19 and segments (green and red contours) lung lesions associated to the disease (https://www.imagensalud.unam.mx).
Gastrointestinal and liver diseases
Another important area in which AI and CV have had impact is gastroenterology and hepatology. Le Berre et al. (114) made a ten-year compilation of contributions ranging from ML to DL technologies. In the endoscopic area AI has been used in lesions analysis, cancer detection and inflammatory lesions analysis. They present 53 studies, most of them related to endoscopy, that detect or improve diagnosis of intestinal lesions, polyp’s cancer, as well as premalignant and malignant analysis of the upper gastrointestinal tract, and/or the entire digestive tract. With the assistance of AI algorithms, performances are above 80%. Kudo et al. (115) identified neoplasms through endoscopic images with high accuracy and low interobserver variation. They identified colon lesions with 96.9% sensitivity, 100% specificity and 98% accuracy. Diagnostic accuracy of their EndoBRAIN system was calculated on a multi-center study using 69,142 endocytoscopic images.
AI has also been used in the study of inflammatory bowel diseases, ulcers. celiac disease, lymphangiectasia and hookworm (114), as well as in identifying the risk of possible bleeding of inflammatory bowel with an accuracy above 90%.
As seen above, most contributions of AI in gastroenterology have been applied on endoscopy images. In the case of liver diseases, AI has been found useful in other image modalities as described below. Table 8 presents some of the relevant works in the gastroenterology area.
Table 8. Gastrointestinal and liver diseases. Selection of relevant works of different methods in gastrointestinal area.
| Selected works | Description | Type of images | Year |
|---|---|---|---|
| Le Berre et al. (114) | Review of a 10-year compilation of contributions that include machine learning and deep learning technologies for diagnosis of intestinal lesions, polyp’s cancer, as well as premalignant and malignant analysis of the upper gastrointestinal tract | Endoscopy | 2020 |
| Kudo et al. (115) | Identification of neoplasms through endoscopic images with high accuracy and low interobserver variation | Endoscopy | 2020 |
| Vivanti et al. (116) | Automatic detection of tumors to evaluate liver CT scan studies | CT | 2017 |
| Liu et al. (117) | CNN method to detect liver cirrhosis with classification accuracy of 86.9% | US | 2017 |
| Biswas et al. (118) | Deep learning technique for detection of hypoechoic fatty liver diseases and stratification of normal and abnormal (fatty) ultrasound liver images | US | 2018 |
| Yasaka et al. (119) | CNN lesions classification method with an average accuracy of 84% for several liver lesions, malignant tumors, other mass-like lesions, and cysts | CT | 2018 |
| Ibragimov et al. (120) | CNN anatomical segmentation method for portal vein segmentation, with results of 83% for Dice coefficient | CT | 2017 |
US, ultrasound; CT, computed tomography; CNN, convolutional neural network.
Liver diseases
Nonalcoholic fatty liver disease (NAFLD) comprises a broad clinical spectrum which ranges from simple hepatic steatosis to more advanced stages such as non-alcoholic steatohepatitis (NASH), liver cirrhosis, and in certain cases it can lead to hepatocellular carcinoma (HCC) (121,122). Currently, it is considered the most prevalent chronic liver disease worldwide.
Predictions show a discouraging picture which brings greater costs associated with health, and a worse prognosis for patients. In this context, it is very important to identify patients who are at a higher risk of developing chronic liver disease in order to prevent cirrhosis progression or development of HCC (123,124).
AI has been used to assess patients with this disease, 13 studies used data from electronic medical records and/or biologic features to build the algorithms and three studies used data from elastography. These models identified their target factor with approximately 80% accuracy (114).
Several authors have applied AI to assist liver disease diagnosis, such as focal liver lesion detection, segmentation and evaluation, as well as diffuse liver staging. Most techniques use CT, MRI or Ultrasound images and the reported results are very promising.
In the case of tumor detection methods based on CNN with CT images have shown to increase the precision rate to 86% compared with the 72% of human performance (116). In the case of ultrasound Liu et al. (117) developed a CNN method that can detect liver cirrhosis with a classification accuracy of 86.9%. In the case of fat liver disease and stratification Biswas et al. (118) applied a DL technique for detection of hypoechoic fatty liver diseases and stratification of normal and abnormal (fatty) ultrasound liver images. They created a ML application for tissue characterization and risk assessment, composed of three different algorithms, namely SVM, extreme learning machines and a DL CNN model. The ultrasound images of 63 patients were annotated as normal and abnormal depending upon the biopsy laboratory tests. The report shows an astonishing accuracy of 100% with the DL CNN system.
Recent developments have been directed to specific problems such as detection of fatty liver disease, tumor detection, metastasis prediction with high performance rates, often above 90% of accuracy (125).
An advanced method (119) based on a CNN with CT images shows an average accuracy of 84% in the classification of several liver lesions, i.e., HCC, other malignant tumors, other mass-like lesions, and cysts.
Ibragimov et al. (120) showed a Dice coefficient of 83% with a CNN working on CT images in the case of portal vein segmentation.
The future of AI in liver medical care lies in the implementation of fully automated clinical tasks to help complex diagnosis. This implies to overcome present day limitations of ML technique such as multiple tasking. Massive efforts to gather annotated image and clinical data are also needed.
Discussion and conclusions
In this paper we presented several examples where CV and ML have been successfully used in medical applications in different organs, pathologies, and image modalities. These technologies have the potential to impact on public health as they can increase the capabilities of general practitioners and help in the standardization of the decision-making process, especially in regions where there is a lack of medical specialists in local clinics. Although these advantages have push towards constant update of the performance results across different applications, there exist some challenges that are commonly encountered, giving room for improvement.
Though multiple databases have emerged in the last decade there is still need for more information in the medical field, the need of databases with expert-annotated ground truth that supports the creation and performance of these methods is still an important issue. Benchmark datasets, accessible to the public, can be valuable to compare existing approaches, discover useful strategies and help researchers to build better approaches. The lack of up-to-date benchmark databases is due to several factors, among them, restrictions in clinical environments and lack of medical experts willing to annotate large amounts of data. The latter being a task that is challenging since manual segmentation is a tedious task that is prone to errors due to inter and intra observer variability. Although an incredible effort has been done, including many years of work and large quantities of spending resources, the amount of sufficient and balanced data to evaluate the performance of different techniques applied to the medical field is scarce, compared with the large amount of publicly available datasets in other areas such as ImageNet, COCO dataset, Google’s Open Images. There is a clear need of new medical workflows that enables to overcome the above-mentioned problems, but it is equally important to foster research on new AI techniques that are less dependent on big data quantities neither so computationally demanding.
Although transfer learning and data augmentation are commonly used to address the issue of small datasets, research on how to improve cross-domain and cross-modal learning and augmentation in the medical field is a most needed task. A promising paradigm in ML is meta-learning. This includes a series of methods that aim at using previously learned knowledge to solve related tasks. In medical imaging this opens new possibilities to overcome the problem of limited data sets. For instance, a model that learned to classify anatomical structures in a given modality, e.g., CT, can use that knowledge to classify the same structures in other modalities like MRI or ultrasound. Likewise, a model that learned to segment a given structure, e.g., a cardiac cavity, can use that knowledge to learn to segment other cavities without the need to train a new model from scratch.
Another topic that has recently called for attention in automatic methods for medical imaging is the need to account not only with high accuracy and precision but also with a measure of how certain the response given by an automatic method is. This is the case, for instance, of the Bayesian DL networks that include a probabilistic model that allows to quantify the uncertainty of a decision. An uncertainty measure provides the radiologist with an additional confidence parameter to emit a diagnosis assisted by an automatic method.
Finally, hybrid methods that combine conventional ML techniques with current state-of-the-art DL methods are also promising alternatives that have shown significant improvements over single methods. In addition, current computing capabilities have allowed these techniques to become increasingly popular, opening new challenges to the scientific community for developing and implementing fully automated real-time clinical tasks to help complex diagnosis and procedures.
Acknowledgments
Funding: This work has been sponsored by UNAM PAPIIT grants TA101121 and IV100420, and SECTEI grant 202/2019.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-1151). The authors have no conflicts of interest to declare.
References
- 1.GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117-71. 10.1016/S0140-6736(14)61682-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard JR, Officer A, de Carvalho IA, Sadana R, Pot AM, Michel JP, Lloyd-Sherlock P, Epping-Jordan JE, Peeters GMEEG, Mahanani WR, Thiyagarajan JA, Chatterji S. The World report on ageing and health: a policy framework for healthy ageing. Lancet 2016;387:2145-54. 10.1016/S0140-6736(15)00516-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marengoni A, Angleman S, Melis R, Mangialasche F, Karp A, Garmen A, Meinow B, Fratiglioni L. Aging with multimorbidity: a systematic review of the literature. Ageing Res Rev 2011;10:430-9. 10.1016/j.arr.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 4.Dagenais GR, Leong DP, Rangarajan S, Lanas F, Lopez-Jaramillo P, Gupta R, Diaz R, Avezum A, Oliveira GBF, Wielgosz A, Parambath SR, Mony P, Alhabib KF, Temizhan A, Ismail N, Chifamba J, Yeates K, Khatib R, Rahman O, Zatonska K, Kazmi K, Wei L, Zhu J, Rosengren A, Vijayakumar K, Kaur M, Mohan V, Yusufali A, Kelishadi R, Teo KK, Joseph P, Yusuf S. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. Lancet 2020;395:785-94. 10.1016/S0140-6736(19)32007-0 [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. WHO report on cancer: setting priorities, investing wisely and providing care for all. Geneva: World Health Organization, 2020. License: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 6.Wolff JL, Starfield B, Anderson G. Prevalence, expenditures, and complications of multiple chronic conditions in the elderly. Arch Intern Med 2002;162:2269-76. 10.1001/archinte.162.20.2269 [DOI] [PubMed] [Google Scholar]
- 7.Maresova P, Javanmardi E, Barakovic S, Barakovic Husic J, Tomsone S, Krejcar O, Kuca K. Consequences of chronic diseases and other limitations associated with old age - a scoping review. BMC Public Health 2019;19:1431. 10.1186/s12889-019-7762-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kontos D, Summers RM, Giger M. Special Section Guest Editorial: Radiomics and Deep Learning. J Med Imaging (Bellingham) 2017;4:041301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. Radiographics 2017;37:505-15. 10.1148/rg.2017160130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel VL, Shortliffe EH, Stefanelli M, Szolovits P, Berthold MR, Bellazzi R, Abu-Hanna A. The coming of age of artificial intelligence in medicine. Artif Intell Med 2009;46:5-17. 10.1016/j.artmed.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moor J. The Dartmouth College Artificial Intelligence Conference: The Next Fifty Years. AI Magazine 2006;27:87. [Google Scholar]
- 12.Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001;23:89-109. 10.1016/S0933-3657(01)00077-X [DOI] [PubMed] [Google Scholar]
- 13.Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. NeurIPS Proceedings (2018). Curran Associates, Inc., 2012:1097-105 [Google Scholar]
- 14.Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The ISIC 2020 Challenge Dataset. 2020 Jun. [accessed 29 Jun, 2020]. Available online: https://challenge2020.isic-archive.com/
- 16.World Health Organization. Cardiovascular diseases (CVD) 2019. Available online: https://www.who.int/health-topics/cardiovascular-diseases/. [Online; accessed: May 11th 2020].
- 17.Ventura-Clapier R. Encyclopedia of Exercise Medicine in Health and Disease. Springer Berlin Heidelberg, 2012. [Google Scholar]
- 18.Chen C, Qin C, Qiu H, Tarroni G, Duan J, Bai W, Rueckert D. Deep Learning for Cardiac Image Segmentation: A Review. Front Cardiovasc Med 2020;7:25. 10.3389/fcvm.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petitjean C, Dacher JN. A review of segmentation methods in short axis cardiac MR images. Med Image Anal 2011;15:169-84. 10.1016/j.media.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 20.Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, Bellomi M. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2018;2:36. 10.1186/s41747-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunnybrook Cardiac Data 2009. Cardiac MR Left Ventricle Segmentation Challenge. Available online: http://www.cardiacatlas.org/studies/sunnybrook-cardiac-data/. [Online; accessed: January 26th 2020].
- 22.Database CAMUS. Available online: https://www.creatis.insa-lyon.fr/Challenge/camus. 2019. [Online; accessed: December 1st 2019].
- 23.Carbajal-Degante E, Avendaño S, Ledesma L, Olveres J, Escalante-Ramírez B. Active contours for multi-region segmentation with a convolutional neural network initialization. SPIE Photonics Europe Conference, 2020:36-44. [Google Scholar]
- 24.Avendaño S, Olveres J, Escalante-Ramírez B. Segmentación de Imágenes Médicas mediante UNet. In: Reunión Internacional de Inteligencia Artificial y sus Aplicaciones RIIAA 2.0, Aug 2019 [Google Scholar]
- 25.Zhang J, Gajjala S, Agrawal P, Tison GH, Hallock LA, Beussink-Nelson L, Lassen MH, Fan E, Aras MA, Jordan C, Fleischmann KE, Melisko M, Qasim A, Shah SJ, Bajcsy R, Deo RC. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation 2018;138:1623-35. 10.1161/CIRCULATIONAHA.118.034338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mira C, Moya-Albor E, Escalante-Ramírez B, Olveres J, Brieva J, Venegas E. 3D Hermite Transform Optical Flow Estimation in Left Ventricle CT Sequences. Sensors (Basel) 2020;20:595. 10.3390/s20030595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO. WHO position paper on mammography screening. 2014. Available online: https://www.who.int/cancer/publications/mammography_screening/en/ [PubMed]
- 28.Devolli-Disha E, Manxhuka-Kërliu S, Ymeri H, Kutllovci A. Comparative accuracy of mammography and ultrasound in women with breast symptoms according to age and breast density. Bosn J Basic Med Sci 2009;9:131-6. 10.17305/bjbms.2009.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stavros AT, Thickman D, Rapp CL, Dennis MA, Parker SH, Sisney GA. Solid breast nodules: use of sonography to distinguish between benign and malignant lesions. Radiology 1995;196:123-34. 10.1148/radiology.196.1.7784555 [DOI] [PubMed] [Google Scholar]
- 30.Chen DR, Chang RF, Wu WJ, Moon WK, Wu WL. 3-D breast ultrasound segmentation using active contour model. Ultrasound Med Biol 2003;29:1017-26. 10.1016/S0301-5629(03)00059-0 [DOI] [PubMed] [Google Scholar]
- 31.Moon WK, Lo CM, Chen RT, Shen YW, Chang JM, Huang CS, Chen JH, Hsu WW, Chang RF. Tumor detection in automated breast ultrasound images using quantitative tissue clustering. Med Phys 2014;41:042901. 10.1118/1.4869264 [DOI] [PubMed] [Google Scholar]
- 32.Madabhushi A, Metaxas DN. Combining low-, high-level and empirical domain knowledge for automated segmentation of ultrasonic breast lesions. IEEE Trans Med Imaging 2003;22:155-69. 10.1109/TMI.2002.808364 [DOI] [PubMed] [Google Scholar]
- 33.Abd-Elmoniem KZ, Youssef AB, Kadah YM. Real-time speckle reduction and coherence enhancement in ultrasound imaging via nonlinear anisotropic diffusion. IEEE Trans Biomed Eng 2002;49:997-1014. 10.1109/TBME.2002.1028423 [DOI] [PubMed] [Google Scholar]
- 34.Bader W, Böhmer S, van Leeuwen P, Hackmann J, Westhof G, Hatzmann W. Does texture analysis improve breast ultrasound precision? Ultrasound Obstet Gynecol 2000;15:311-6. 10.1046/j.1469-0705.2000.00046.x [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Cheng HD, Huang J, Tian J, Tang X, Liu J. Fully automatic and segmentation-robust classification of breast tumors based on local texture analysis of ultrasound images. Pattern Recognit 2010;43:280-98. 10.1016/j.patcog.2009.06.002 [DOI] [Google Scholar]
- 36.Chen WM, Chang RF, Kuo SJ, Chang CS, Moon WK, Chen ST, Chen DR. 3-D ultrasound texture classification using run difference matrix. Ultrasound Med Biol 2005;31:763-70. 10.1016/j.ultrasmedbio.2005.01.014 [DOI] [PubMed] [Google Scholar]
- 37.Xian M, Zhang Y, Cheng HD, Xu F, Zhang B, Ding J. Automatic breast ultrasound image segmentation: A survey. Pattern Recognit 2018;79:340-55. 10.1016/j.patcog.2018.02.012 [DOI] [Google Scholar]
- 38.Torres F, Escalante-Ramirez B, Olveres J, Yen PL. Lesion Detection in Breast Ultrasound Images Using a Machine Learning Approach and Genetic Optimization. Lecture Notes in Computer Science, 2019:289-301. 10.1007/978-3-030-31332-6_26 [DOI] [Google Scholar]
- 39.Yap MH, Edirisinghe EA, Bez HE. A novel algorithm for initial lesion detection in ultrasound breast images. J Appl Clin Med Phys 2008;9:2741. 10.1120/jacmp.v9i4.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mainiero MB, Goldkamp A, Lazarus E, Livingston L, Koelliker SL, Schepps B, Mayo-Smith WW. Characterization of breast masses with sonography: can biopsy of some solid masses be deferred? J Ultrasound Med 2005;24:161-7. 10.7863/jum.2005.24.2.161 [DOI] [PubMed] [Google Scholar]
- 41.Huang SF, Chen YC, Woo KM. Neural network analysis applied to tumor segmentation on 3D breast ultrasound images. 2008 Proc IEEE Int Symp Biomed Imaging, 2008:1303-6. [Google Scholar]
- 42.Cao Z, Duan L, Yang G, Yue T, Chen Q. An experimental study on breast lesion detection and classification from ultrasound images using deep learning architectures. BMC Med Imaging 2019;19:51. 10.1186/s12880-019-0349-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiao JY, Chen KY, Liao KY, Hsieh PH, Zhang G, Huang TC. Detection and classification the breast tumors using mask R-CNN on sonograms. Medicine (Baltimore) 2019;98:e15200. 10.1097/MD.0000000000015200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Q, Luo Y, Zhang Q. Breast ultrasound image segmentation: a survey. Int J Comput Assist Radiol Surg 2017;12:493-507. 10.1007/s11548-016-1513-1 [DOI] [PubMed] [Google Scholar]
- 45.Fenster A, Parraga G, Bax J. Three-dimensional ultrasound scanning. Interface Focus 2011;1:503-19. 10.1098/rsfs.2011.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data Brief 2019;28:104863. 10.1016/j.dib.2019.104863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanitakis J. Anatomy, histology and immunohistochemistry of normal human skin. Eur J Dermatol 2002;12:390-401. [PubMed] [Google Scholar]
- 48.American Cancer Society. Cancer Facts & Figures 2019. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2019.html [Online; accessed: December 1st 2019].
- 49.Schadendorf D, Hauschild A. Melanoma in 2013: Melanoma—the run of success continues. Nat Rev Clin Oncol 2014;11:75. 10.1038/nrclinonc.2013.246 [DOI] [PubMed] [Google Scholar]
- 50.Camacho LCP, Gerson CR, Góngora JMÁ, Villalobos PA, Blanco VYC, López RO. Actualidades para el tratamiento del melanoma metastásico, estado del arte. Anales Médicos de la Asociación Médica del Centro Médico ABC 2017;62:196-207. [Google Scholar]
- 51.Orendain-Koch N, Ramos-Álvarez MP, Ruiz-Leal AB, Sánchez-Dueñas LE, Crocker-Sandoval AB, Sánchez-Tenorio T, Izquierdo-Álvarez J. Melanoma en la práctica privada en México: un diagnóstico oportuno. Dermatol Rev Mex 2015;59:89-97. [Google Scholar]
- 52.Johr RH. Dermoscopy: alternative melanocytic algorithms-the ABCD rule of dermatoscopy, menzies scoring method, and 7-point checklist. Clin Dermatol 2002;20:240-7. 10.1016/S0738-081X(02)00236-5 [DOI] [PubMed] [Google Scholar]
- 53.Codella NCF, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, Halpern A. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, 2018:168-72. [Google Scholar]
- 54.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. Erratum in: Nature. 2017 Jun 28;546(7660):686. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujisawa Y, Otomo Y, Ogata Y, Nakamura Y, Fujita R, Ishitsuka Y, Watanabe R, Okiyama N, Ohara K, Fujimoto M. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol 2019;180:373-81. 10.1111/bjd.16924 [DOI] [PubMed] [Google Scholar]
- 56.DeVries T, Ramachandram D. Skin lesion classification using deep multi-scale convolutional neural networks. arXiv preprint arXiv:170301402. 2017.
- 57.Mahbod A, Ecker R, Ellinger I. Skin lesion classification using hybrid deep neural networks. arXiv preprint arXiv:170208434. 2017.
- 58.Harangi B. Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 2018;86:25-32. 10.1016/j.jbi.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 59.Menegola A, Tavares J, Fornaciali M, Li LT, Avila S, Valle E. RECOD titans at ISIC challenge 2017. arXiv preprint arXiv:170304819. 2017.
- 60.Liu Y, Jain A, Eng C, Way DH, Lee K, Bui P, Kanada K, de Oliveira Marinho G, Gallegos J, Gabriele S, Gupta V, Singh N, Natarajan V, Hofmann-Wellenhof R, Corrado GS, Peng LH, Webster DR, Ai D, Huang SJ, Liu Y, Dunn RC, Coz D. A deep learning system for differential diagnosis of skin diseases. Nat Med 2020;26:900-8. 10.1038/s41591-020-0842-3 [DOI] [PubMed] [Google Scholar]
- 61.Osinga D. Deep Learning Cookbook: Practical Recipes to Get Started Quickly. 1st column. O’Reilly Media; 2018. [Google Scholar]
- 62.Redmon J, Farhadi A. Yolov3: An incremental improvement. arXiv preprintarXiv:180402767. 2018.
- 63.Kumar D, Hani AFM, editors. Optical imaging for biomedical and clinical applications. 1st editon. Boca Raton: CRC Press/Taylor & Francis, 2018 [Google Scholar]
- 64.Heintzmann R, Gustafsson MGL. Subdiffraction resolution in continuous samples. Nat Photonics 2009;3:362-4. 10.1038/nphoton.2009.102 [DOI] [Google Scholar]
- 65.Kothari S, Phan JH, Wang MD. Eliminating tissue-fold artifacts in histopathological whole-slide images for improved image-based prediction of cancer grade. J Pathol Inform 2013;4:22. 10.4103/2153-3539.117448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khan AM, Rajpoot N, Treanor D, Magee D. A Nonlinear Mapping Approach to Stain Normalization in Digital Histopathology Images Using Image-Specific Color Deconvolution. IEEE Trans Biomed Eng 2014;61:1729-38. 10.1109/TBME.2014.2303294 [DOI] [PubMed] [Google Scholar]
- 67.Rubin R. Rubin's pathology: clinicopathologic foundations of medicine. Strayer DS, Rubin E, McDonald JM, Michalopoulos GK, editors. 5th edition. Lippincott Williams & Wilkins, 2008 [Google Scholar]
- 68.Vardiman JW. The World Health Organization (WHO) classification of tumors of the hematopoietic and lymphoid tissues: An overview with emphasis on the myeloid neoplasms. Chem Biol Interact 2010;184:16-20. 10.1016/j.cbi.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 69.Shamir L, Orlov N, Mark Eckley D, Macura TJ, Goldberg IG. IICBU 2008: a proposed benchmark suite for biological image analysis. Med Biol Eng Comput 2008;46:943-7. 10.1007/s11517-008-0380-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sertel O, Kong J, Lozanski G, Arwa Sa, Catalyurek U, Saltz J, Gurcan M, editors. Texture classification using nonlinear color quantization: Application to histopathological image analysis. Proc IEEE Int Conf Acoust Speech Signal Process, 2008:507-600 [Google Scholar]
- 71.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol 2019;14:89-103. 10.5114/pg.2018.81072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linder N, Konsti J, Turkki R, Rahtu E, Lundin M, Nordling S, Haglund C, Ahonen T, Pietikäinen M, Lundin J. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn Pathol 2012;7:22. 10.1186/1746-1596-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nava R, González G, Kybic J, Escalante-Ramírez B. Characterization of hematologic malignancies based on discrete orthogonal moments. 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA). 12-15 Dec. 2016; Oulu, Finland. IEEE, 2016:1-6. [Google Scholar]
- 74.Flusser J, Suk T, Zitova B. 2D and 3D Image Analysis by Moments. John Wiley & Sons, 2016. [Google Scholar]
- 75.González G, Nava R, Escalante-Ramírez B. A Comparative Study on Discrete Shmaliy Moments and Their Texture-Based Applications. Math Probl Eng 2018;2018:1673283. 10.1155/2018/1673283 [DOI] [Google Scholar]
- 76.Marcos JV, Cristóbal G. Texture classification using discrete Tchebichef moments. J Opt Soc Am A Opt Image Sci Vis 2013;30:1580-91. 10.1364/JOSAA.30.001580 [DOI] [PubMed] [Google Scholar]
- 77.Mukundan R, Ong SH, Lee PA. Image analysis by Tchebichef moments. IEEE Trans Image Process 2001;10:1357-64. 10.1109/83.941859 [DOI] [PubMed] [Google Scholar]
- 78.Nava R, González G, Kybic J, Escalante-Ramírez B. Classification of Tumor Epithelium and Stroma in Colorectal Cancer Based on Discrete Tchebichef Moments. In: Laura CO, Shekhar R, Wesarg S, Gonzalez Ballester MÁ, Drechsler K, Sato Y, Erdt M, Linguraru MG. editors. Clinical Image-Based Procedures. Translational Research in Medical Imaging. CLIP 2015. Lecture Notes in Computer Science, vol 9401. Springer, Cham, 2016:78-87. [Google Scholar]
- 79.Huang Y, Zheng H, Liu C, Ding X, Rohde GK. Epithelium-Stroma Classification via Convolutional Neural Networks and Unsupervised Domain Adaptation in Histopathological Images. IEEE J Biomed Health Inform 2017;21:1625-32. 10.1109/JBHI.2017.2691738 [DOI] [PubMed] [Google Scholar]
- 80.Yu X, Zheng H, Liu C, Huang Y, Ding X. Classify epithelium-stroma in histopathological images based on deep transferable network. J Microsc 2018;271:164-73. 10.1111/jmi.12705 [DOI] [PubMed] [Google Scholar]
- 81.Kather JN, Bello-Cerezo R, Di Maria F, van Pelt GW, Mesker WE, Halama N, Bianconi F. Classification of Tissue Regions in Histopathological Images: Comparison Between Pre-Trained Convolutional Neural Networks and Local Binary Patterns Variants. Nanni L, Brahnam S, Brattin R, Ghidoni S, Jain LC, editors. Deep Learners and Deep Learner Descriptors for Medical Applications 2020. Intelligent Systems Reference Library. Springer, 2020:95-115 [Google Scholar]
- 82.Meng T, Lin L, Shyu ML, Chen SC, editors. Histology Image Classification Using Supervised Classification and Multimodal Fusion. 2010 IEEE International Symposium on Multimedia; 13-15 Dec. 2010; Taichung, Taiwan. IEEE, 2010:145-52. [Google Scholar]
- 83.Orlov NV, Chen WW, Eckley DM, Macura TJ, Shamir L, Jaffe ES, Goldberg IG. Automatic Classification of Lymphoma Images With Transform-Based Global Features. IEEE Trans Inf Technol Biomed 2010;14:1003-13. 10.1109/TITB.2010.2050695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 2016;191:214-23. 10.1016/j.neucom.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bianconi F, Bello-Cerezo R, Napoletano P. Improved opponent color local binary patterns: an effective local image descriptor for color texture classification. J Electron Imaging 2017;27:011002. 10.1117/1.JEI.27.1.011002 [DOI] [Google Scholar]
- 86.Song Y, Li Q, Huang H, Feng D, Chen M, Cai W. Low Dimensional Representation of Fisher Vectors for Microscopy Image Classification. IEEE Trans Med Imaging 2017;36:1636-49. 10.1109/TMI.2017.2687466 [DOI] [PubMed] [Google Scholar]
- 87.Luan S, Chen C, Zhang B, Han J, Liu J. Gabor Convolutional Networks. IEEE Trans Image Process 2018;27:4357-66. 10.1109/TIP.2018.2835143 [DOI] [PubMed] [Google Scholar]
- 88.Dolz J, Massoptier L, Vermandel M. Segmentation algorithms of subcortical brain structures on MRI for radiotherapy and radiosurgery: A survey. IRBM 2015;36:200-12. 10.1016/j.irbm.2015.06.001 [DOI] [Google Scholar]
- 89.González-Villà S, Oliver A, Valverde S, Wang L, Zwiggelaar R, Lladó X. A review on brain structures segmentation in magnetic resonance imaging. Artif Intell Med 2016;73:45-69. 10.1016/j.artmed.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 90.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002;33:341-55. 10.1016/S0896-6273(02)00569-X [DOI] [PubMed] [Google Scholar]
- 91.Scherrer B, Dojat M, Forbes F, Garbay C. LOCUS: LOcal Cooperative Unified Segmentation of MRI Brain Scans. Med Image Comput Comput Assist Interv 2007;10:219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carbajal-Degante E, Olveres J, Escalante-Ramírez B. A multiphase active contour model based on the Hermite transform for texture segmentation. Proceedings Volume 10679, Optics, Photonics, and Digital Technologies for Imaging Applications V; 106791H. 2018:364-72. [Google Scholar]
- 93.Jorge Cardoso M, Leung K, Modat M, Keihaninejad S, Cash D, Barnes J, Fox NC, Ourselin S, Alzheimer’s Disease Neuroimaging Initiative . STEPS: Similarity and Truth Estimation for Propagated Segmentations and its application to hippocampal segmentation and brain parcelation. Med Image Anal 2013;17:671-84. 10.1016/j.media.2013.02.006 [DOI] [PubMed] [Google Scholar]
- 94.Pipitone J, Park MT, Winterburn J, Lett TA, Lerch JP, Pruessner JC, Lepage M, Voineskos AN, Chakravarty MM, Alzheimer's Disease Neuroimaging Initiative . Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 2014;101:494-512. 10.1016/j.neuroimage.2014.04.054 [DOI] [PubMed] [Google Scholar]
- 95.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 2011;56:907-22. 10.1016/j.neuroimage.2011.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao Y, Corn B, Schifter D, Tannenbaum A. Multiscale 3D shape representation and segmentation with applications to hippocampal/caudate extraction from brain MRI. Med Image Anal 2012;16:374-85. 10.1016/j.media.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li DH, He YC, Liu J, Chen SD. Diagnostic Accuracy of Transcranial Sonography of the Substantia Nigra in Parkinson's disease: A Systematic Review and Meta-analysis. Sci Rep 2016;6:20863 10.1038/srep20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Olveres J, Carbajal-Degante E, Escalante-Ramírez B, Vallejo E, García-Moreno CM. Deformable Models for Segmentation Based on Local Analysis. Math Probl Eng 2017;2017:1646720. 10.1155/2017/1646720 [DOI] [Google Scholar]
- 99.Milletari F, Ahmadi SA, Kroll C, Plate A, Rozanski V, Maiostre J, et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput Vis Image Underst 2017;164:92-102. 10.1016/j.cviu.2017.04.002 [DOI] [Google Scholar]
- 100.Kushibar K, Valverde S, González-Villà S, Bernal J, Cabezas M, Oliver A, Lladó X. Automated sub-cortical brain structure segmentation combining spatial and deep convolutional features. Med Image Anal 2018;48:177-86. 10.1016/j.media.2018.06.006 [DOI] [PubMed] [Google Scholar]
- 101.Perlmutter JS, Mink JW. Deep brain stimulation. Annu Rev Neurosci 2006;29:229-57. 10.1146/annurev.neuro.29.051605.112824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.WHO. Chronic obstructive pulmonary disease (COPD). World Health Organization: WHO. [Online; accessed: October 4th 2020]. Available online: https://www.who.int/news-room/fact-sheets/detail/chronic-obstructive-pulmonary-disease-(copd)
- 103.Diab N, Gershon AS, Sin DD, Tan WC, Bourbeau J, Boulet LP, Aaron SD. Underdiagnosis and Overdiagnosis of Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med 2018;198:1130-9. 10.1164/rccm.201804-0621CI [DOI] [PubMed] [Google Scholar]
- 104.Bibault JE, Xing L. Screening for chronic obstructive pulmonary disease with artificial intelligence. Lancet Digit Health 2020;2:e216-7. 10.1016/S2589-7500(20)30076-5 [DOI] [PubMed] [Google Scholar]
- 105.Mekov E, Miravitlles M, Petkov R. Artificial intelligence and machine learning in respiratory medicine. Expert Rev Respir Med 2020;14:559-64. 10.1080/17476348.2020.1743181 [DOI] [PubMed] [Google Scholar]
- 106.Afzali A, Mofrad FB, Pouladian M. Contour-based lung shape analysis in order to tuberculosis detection: modeling and feature description. Med Biol Eng Comput 2020;58:1965-86. 10.1007/s11517-020-02192-y [DOI] [PubMed] [Google Scholar]
- 107.Selvan R, Dam EB, Detlefsen NS, Rischel S, Sheng K, Nielsen M, Pai A. Lung Segmentation from Chest X-rays using Variational Data Imputation. Lung Segmentation from Chest X-rays using Variational Data Imputation. Presented at the first Workshop on the Art of Learning with Missing Values (Artemiss) hosted by the 37th International Conference on Machine Learning (ICML). Jul 2020. [Google Scholar]
- 108.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren MP, Ng AY. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv:1711.05225 Nov 2017.
- 109.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Rev Biomed Eng 2021;14:4-15. 10.1109/RBME.2020.2987975 [DOI] [PubMed] [Google Scholar]
- 110.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med 2018;15:e1002683. 10.1371/journal.pmed.1002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Castillo-Saldana D, Hague CJ, Coxson HO, Ryerson CJ. Using Quantitative Computed Tomographic Imaging to Understand Chronic Obstructive Pulmonary Disease and Fibrotic Interstitial Lung Disease: State of the Art and Future Directions. J Thorac Imaging 2020;35:246-54. 10.1097/RTI.0000000000000440 [DOI] [PubMed] [Google Scholar]
- 112.Lippi G, Henry BM. Chronic obstructive pulmonary disease is associated with severe coronavirus disease 2019 (COVID-19). Respir Med 2020;167:105941. 10.1016/j.rmed.2020.105941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J. Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia on Chest CT. Radiology 2020;296:E65-71. 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology 2020;158:76-94.e2. 10.1053/j.gastro.2019.08.058 [DOI] [PubMed] [Google Scholar]
- 115.Kudo SE, Misawa M, Mori Y, Hotta K, Ohtsuka K, Ikematsu H, Saito Y, Takeda K, Nakamura H, Ichimasa K, Ishigaki T, Toyoshima N, Kudo T, Hayashi T, Wakamura K, Baba T, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Artificial Intelligence-assisted System Improves Endoscopic Identification of Colorectal Neoplasms. Clin Gastroenterol Hepatol 2020;18:1874-1881.e2. 10.1016/j.cgh.2019.09.009 [DOI] [PubMed] [Google Scholar]
- 116.Vivanti R, Szeskin A, Lev-Cohain N, Sosna J, Joskowicz L. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J Comput Assist Radiol Surg 2017;12:1945-57. 10.1007/s11548-017-1660-z [DOI] [PubMed] [Google Scholar]
- 117.Liu X, Song JL, Wang SH, Zhao JW, Chen YQ. Learning to Diagnose Cirrhosis with Liver Capsule Guided Ultrasound Image Classification. Sensors (Basel) 2017;17:149. 10.3390/s17010149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Biswas M, Kuppili V, Edla DR, Suri HS, Saba L, Marinhoe RT, Sanches JM, Suri JS. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed 2018;155:165-77. 10.1016/j.cmpb.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 119.Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 2018;286:887-96. 10.1148/radiol.2017170706 [DOI] [PubMed] [Google Scholar]
- 120.Ibragimov B, Toesca D, Chang D, Koong A, Xing L. Combining deep learning with anatomical analysis for segmentation of the portal vein for liver SBRT planning. Phys Med Biol 2017;62:8943-58. 10.1088/1361-6560/aa9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mendez-Sanchez N, Cruz-Ramon VC, Ramirez-Perez OL, Hwang JP, Barranco-Fragoso B, Cordova-Gallardo J. New Aspects of Lipotoxicity in Nonalcoholic Steatohepatitis. Int J Mol Sci 2018;19:2034. 10.3390/ijms19072034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Méndez-Sánchez N, Chavez-Tapia NC, Almeda-Valdes P, Uribe M. The management of incidental fatty liver found on imaging. What do we need to do? Am J Gastroenterol 2018;113:1274-6. 10.1038/s41395-018-0047-2 [DOI] [PubMed] [Google Scholar]
- 123.Valencia-Rodríguez A, Vera-Barajas A, Barranco-Fragoso B, Kúsulas-Delint D, Qi X, Méndez-Sánchez N. New insights into the association between non-alcoholic fatty liver disease and atherosclerosis. Ann Transl Med 2019;7:S300. 10.21037/atm.2019.11.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.López-Velázquez JA, Silva-Vidal KV, Ponciano-Rodríguez G, Chávez-Tapia NC, Arrese M, Uribe M, et al. The prevalence of nonalcoholic fatty liver disease in the Americas. Ann Hepatol 2014;13:166-78. 10.1016/S1665-2681(19)30879-8 [DOI] [PubMed] [Google Scholar]
- 125.Zhou LQ, Wang JY, Yu SY, Wu GG, Wei Q, Deng YB, Wu XL, Cui XW, Dietrich CF. Artificial intelligence in medical imaging of the liver. World J Gastroenterol 2019;25:672-82. 10.3748/wjg.v25.i6.672 [DOI] [PMC free article] [PubMed] [Google Scholar]