Graphical abstract
Keywords: Hi-C, Chromatin conformation, 3D nucleus, Inter-chromosomal interaction, Homologous chromosomes, Allele specific gene regulation
Abstract
The three-dimensional (3D) organization of chromatin in the nucleus of diploid eukaryotic organisms has fascinated biologists for many years. Despite major progress in chromatin conformation studies, current knowledge regarding the spatial organization of diploid (maternal and paternal) genomes is still limited. Recent advances in Hi-C technology and data processing approaches have enabled construction of diploid Hi-C contact maps. These maps greatly accelerated the pace of novel discoveries in haplotype-resolved 3D genome studies, revealing the role of allele biased chromatin conformation in transcriptional regulation. Here, we review emerging concepts and haplotype phasing strategies of Hi-C data in 3D diploid genome studies. We discuss new insights on homologous chromosomal organization and the interplay between allelic biased chromatin architecture and several nuclear functions, explaining how haplotype-resolved Hi-C technologies have been used to resolve important biological questions.
1. Introduction
Chromatin has a linear length spanning several meters but is compacted into the micrometer-sized nucleus of eukaryotic cells via an intricate and non-random folding process. High-throughput chromosome conformation capture (Hi-C) [1] is an essential technology providing genome-wide three dimensional (3D) chromatin organization by coupling proximity-based ligation with massively parallel sequencing [2]. The development of Hi-C and its derivatives [3], [4], [5] has revealed a complex genomic landscape of chromosomal structure across different cells and tissues of different species. Multilevel hierarchical chromatin structures are found in interphase cells of most higher eukaryotes, including chromosome territories (CTs), compartments of active (euchromatin, A-type) and inactive (heterochromatin, B-type) chromatin; chromosomal domains variably known as contact domains or topologically associating domains (TADs); chromatin loops; and promoter-enhancer interactions (PEIs) [1], [3], [6], [7], [8], [9], [10], [11], [12], [13]. The conformation of chromatin plays important roles in the regulation of gene expression, recombination, and DNA replication and repair [11], [14], [15], [16], [17], [18]. Hence, understanding 3D chromatin folding is essential to determining the mechanisms behind gene regulation in multicellular organisms across different cell types developing from the same genetic blueprint, as well as to reveal the genetic basis underlying distinct phenotypes and physical disorders.
Despite recent advances, at least two limitations affect the majority of 3D genome studies. The first is masking the differences in chromatin conformation that are inherent to individual cells. The other is assembling an averaged structure of the homologous chromosomes. The former has essentially been overcome by the development of single cell Hi-C technologies [19], [20], [21], whereas the latter remains a challenge. As a typical Hi-C experiment does not directly produce haplotype-resolved (phased) data, most Hi-C studies lack sufficient allele-specific information to distinguish the two parental haploids [22]. Apart from some genomic loci [23], [24], [25], little is known about the variability in higher order structures between homologous chromosomes in diploid genomes. In fact, substantial allelic differences in transcription, DNA methylation, and epigenetic chromatin states have already been found in diploid organisms [26], [27], [28], [29], [30], [31]. Hence, it has long been hypothesized that two homologous sets of chromosomes may differ in how they spatially fold and function.
Recently, significant research efforts have attempted to decipher 3D haplotype structures in diverse diploid organisms (Supplementary Table S1). In this review, we discuss recent methodological and biological advances in the search for 3D haploid-level genome architectures. We focus on the spatial organization of homologous chromosomes within the nucleus as well as their associations with different genomic functions. We also provide an outlook on the future trends and challenges for this field, as well as a glimpse into possible developments in 3D diploid genome research.
2. Reconstruction of the haplotype-resolved 3D genome
Conventional Hi-C data does not typically distinguish between different allelic chromosome copies. Hence, the main challenge in 3D haplotype-resolved genome studies is accurate allele assignment of each chromatin contact (pair of genomic loci that are joined by proximity ligation) [3], [32], [33], [34]. Observing a single Hi-C contact between locus i and j corresponds to one of four possible events, where either copy of locus i comes in contact with either copy of locus j [35]. When i and j come from the same chromosome, the contact represents a cis interaction. In contrast, when the two loci come from different chromosomes, it is considered a trans interaction. In particular, if i and j respectively belong to the homologous maternal and paternal chromosomes, the contact represents a trans-homolog (t-homo) interaction that reflects the spatial architecture of homologous chromosomes. Analogously, if i and j come from heterologs, the contact is classified as trans-heterolog (t-heter). Importantly, every 3D genome study at a true diploid scale must accurately account for this allelic uncertainty. In this section, we discuss the methodology currently used for reconstructing haplotype-resolved 3D genomes.
2.1. Phasing Hi-C data based on heterozygous variations
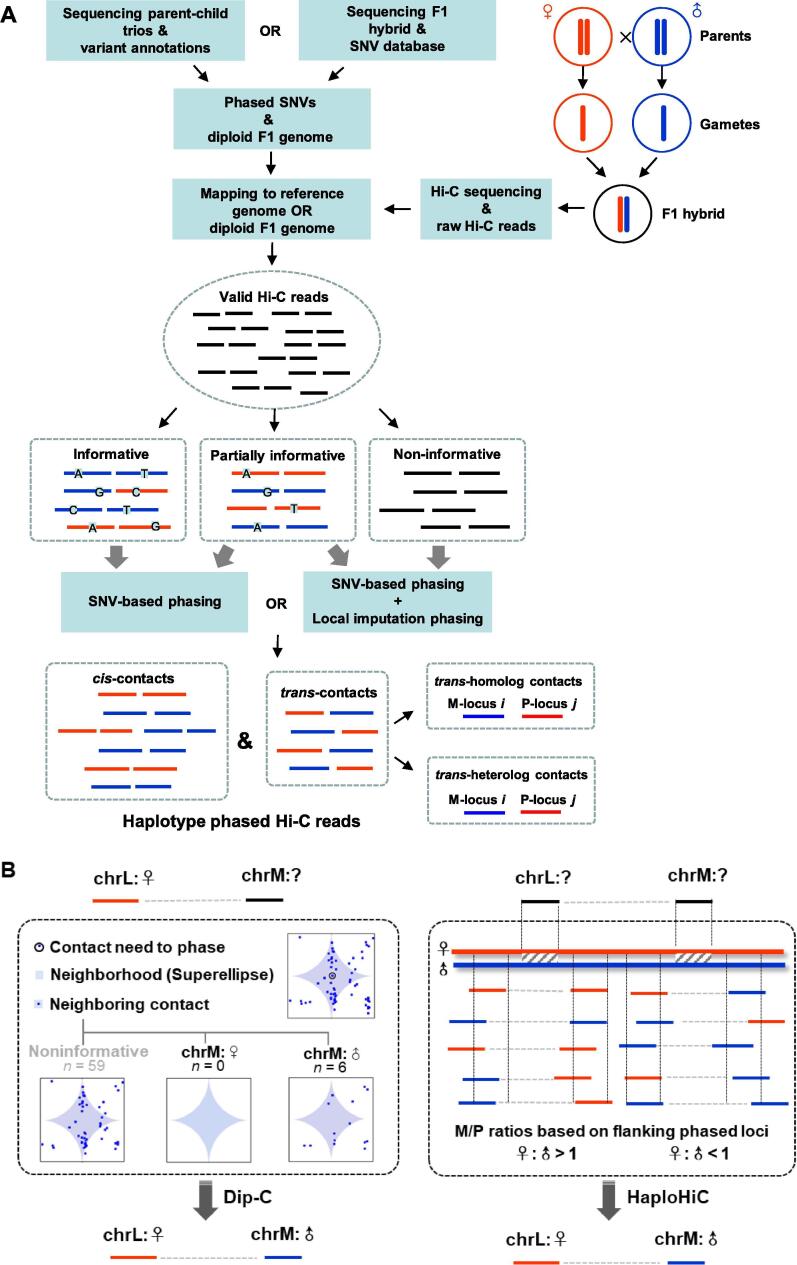
The most commonly used approach to assign alleles of each Hi-C contact is employing phased single nucleotide variations (SNVs) to define haplotypes from the Hi-C reads (Fig. 1). Parent-specific SNVs make it possible to resolve the haplotypes of Hi-C data by mapping reads containing allele-specific genetic variations (i.e., informative Hi-C reads) to their parental chromosome. Because many Hi-C reads overlap SNVs, this strategy efficiently assigns Hi-C contacts to specific chromosomal homologs in order to finally construct a ‘diploid’ Hi-C map comprising both maternal and paternal contact matrices. However, only in rare instances is it possible to obtain phased SNV data, as is the case of the International Hapmap Project [36] and the 1000 Genomes Project [37], [38], [39]. Such information is not available for the majority of studies. Genotyping parent–child trios can directly determine whole-genome haplotype information for the F1 hybrid based on heterozygous sites. However, in order to obtain a high density of heterozygous sites, and consequently high resolution of haplotype phasing, it is usually necessary to establish hybrids with divergent breeds or lineages. The hybrids should be sufficiently divergent to allow for straightforward sequence-level discrimination of homologs but maintain a nearly complete synteny in order to guarantee the mapping rates. Hi-C reads can be aligned to reference genome N – masking variable sites and then performing haplotype imputation based on phased variants. Alternatively, Hi-C reads can be mapped to the diploid F1 hybrid genome, which is then rebuilt with the reference genome and the phased variants (replacing the corresponding bases) to directly obtain allele assignments of the Hi-C reads.
Fig. 1.
Schematics of haplotype phasing strategies for Hi-C read pairs. (A) The strategy of haplotype phasing for paired-end Hi-C reads based on heterozygous variants. (B) Two local imputation phasing methods for Hi-C reads. Left: The haplotype imputation algorithm in Dip-C developed by Tan et al. [21]. An unknown haplotype of chromatin contact (circled blue dot) can be imputed based on the statistical properties of interchromosomal and long-range intrachromosomal contacts (blue dots) in the super-elliptical neighborhood (light blue shaded area) of the unphased contact (circled blue dot). The plot is modified from Tan et al. [21]. Right: A novel phasing technique for Hi-C data, named ‘HaploHiC’. A paired-end Hi-C read with no allele-specific SNV or short InDels (insertions and deletions) has its origin imputed using information from nearby reads. The ratios of paternally and maternally mapped reads in the flanking regions of the unknown-haplotype reads are used to determine the likelihood of a given parental origin of unknown-haplotype reads.
Paired-end Hi-C reads can be classified into three different groups based on SNVs: reads containing SNVs at a) one or b) both ends, and c) reads containing no SNVs, respectively termed partially informative, informative, and non-informative reads (Fig. 1A). Haplotype phasing for reads with allele-specific SNVs at both ends is simple and accurate: it is possible to unambiguously map these reads to their respective parental genome based on the SNVs present (Fig. 1A). All proposed methods can process this group of Hi-C reads, including the algorithm Haplotype-Assisted Read Parsing (HARP, https://github.com/dvera/harp) [40]. Notably, HARP performs allele-specific alignment of Hi-C reads to the two parental reference genomes, thus yielding a higher number of usable reads for phasing due to the separate alignment of each haploid. Rivera-Mulia et al. [40] took advantage of HARP methodology and the high SNV density between the genomes of mouse subspecies to explore the allele-specific control of genome organization and replication timing during development. This kind of approach has been widely used to obtain 3D diploid genomes [3], [41], [42].
With partially informative Hi-C reads, it is possible to determine parental origin by arbitrarily using the SNVs at one end of the read, or by combining information from local imputation phasing based on empirical probabilities or statistical modeling in order to increase accuracy (Fig. 1A). Some studies have adopted the former strategy, including Giorgetti et al. [34], Du et al. [41], Bonora et al. [43], and Greenwald et al. [44], while others have opted for the latter, such as the studies proposing Dip-C [21] and HaploHiC [45]. Dip-C, an algorithm originally designed for single-cell Hi-C data by Tan et al. [21], can impute unknown haplotypes from ‘neighboring’ contacts (measured as linear distances) by assuming that the two homologs would typically contact different partners (Fig. 1B). Briefly, considering the statistical properties of interchromosomal and long-range intrachromosomal contacts, Tan et al. [21] defined a contact neighborhood as a super-ellipse with an exponent of 0.5 and a radius of 10-Mb, where the haplotypes of nearby contacts were weighted to impute the origin of the unphased contacts. Then, the assignment of allele-ambiguous contacts was refined by the 3D structures. In this case, the imputation accuracy for each haplotype was estimated as ~ 96% by cross-validation.
Notably, a substantial proportion of total Hi-C reads are non-informative, ranging from 21.1% (patski) to 94.1% (GM12878) as reported in [46], and are typically discarded in most phasing methods. Even for hybrids, maternal and paternal haploids are often identical in most parts of the genome, and most phasing approaches can only infer the parental origin at a small subset of genomic loci [45]. This means that the segregated maternal and paternal contact matrices are extremely sparse. To tackle this problem, a few methods [21], [45], [46], [47] have attempted to reintroduce unphased reads to both parental contact maps. The Poisson-Gamma model developed by Deng et al. [47] can impute both partially informative and non-informative contacts using an iterative expectation–maximization (EM) algorithm. However, this model might not work robustly in fine-resolution analyses due to lack of diploid 3D structures to assist the assignment of allele-ambiguous contacts.
HaploHiC [45], a novel phasing algorithm, also allows for more Hi-C data to be imputed using local imputation combined with SNVs/InDels-based phasing, including Hi-C reads with only one or no informative read end, thus reducing the sparsity of the allele-specific Hi-C contact matrices (Fig. 1B). With HaploHiC, assigned reads containing informative SNVs/InDels can be employed to predict the origin of nearby unknown reads using a data-derived ratio. The haplotype-predicting accuracy of HaploHiC (97.66%) was comparable to Dip-C (~96%) for reads of unknown origin using simulation data. Notably, rigorous evaluations are required to further assess its efficiency and accuracy on more Hi-C data, given that HaploHiC is currently a preprint version.
More recently, Ye and Ma [46] proposed ASHIC methods, a hierarchical Bayesian framework to provide a probabilistic model of all possible allele origins, to impute allele-specific chromatin organizations and simultaneously infer allelic 3D structures in diploid genomes using both informative and non-informative Hi-C contacts. The ASHIC methods include the Poisson-multinomial (ASHIC-PM) model and the zero-inflated Poisson-multinomial (ASHIC-ZIPM) model. Both models outperformed the above-mentioned allele-certain methods [3], [42] as well as mate-rescue approaches in which allele-ambiguous read-ends are directly assigned to the same allele as with allele-certain mate-ends [34], [41], [43], especially under low coverage and low SNV density conditions using both simulated and real Hi-C data.
There at least two kinds of errors that may be introduced to allele assignment of Hi-C data based on heterozygous variants: homolog misassignment and systematic bias caused by variable genetic variant density in the genome. Erceg et al. [48] developed a computational approach, called Ohm (oversight of homolog misassignment), which can minimize homolog misassignment and thus robustly distinguish t-homo from cis contacts for haplotype-resolved Hi-C studies. This approach can be extended to any number of methods in order to improve the assignment of parental origin to Hi-C reads. As for the bias in diploid Hi-C contact maps caused by asymmetric genetic variant density along the genome, a novel strategy has been proposed by Luo et al. [49] to facilitate whole-genome identification of the 3D organization of diploid chromatin. This approach is included as an integrated tool called HiCHap.
2.2. Inferring the 3D structure of diploid genomes
Hi-C data allows for reconstruction of 3D genome structures with the aid of sophisticated computational algorithms. Even though Hi-C contact matrices yield valuable insights, modeling and visualizing 3D genome structures can unveil higher-order structural patterns and relationships that are not apparent from the raw data [11], [50], [51]. In particular, this might provide a 3D architecture that can be interpreted by humans, orienting genomic regions relative to various nuclear landmarks, and serving as a framework for integrating other data types [52]. Most inference methods have modeled a single structure without differentiating the two alleles, but many fewer algorithms have so far been developed to account for diploidy in Hi-C 3D modeling [6], [21], [46], [53], [54], [55], [56]. Hence, 3D structural inference of diploid genomes remains challenging.
Two separate research groups have developed modeling methods that can be exclusively applied to single cell Hi-C data to generate 3D physical models of diploid genomes [21], [54]. One approach [21] adopted an iterative modeling strategy, starting with coarse resolution modeling and gradually refining the structures to higher resolutions. For more widely available bulk Hi-C data, some genome modeling approaches [6], [50], [55] have been developed to generate a diploid 3D genome structure using a simulated annealing/molecular dynamics sampling method. Ay et al. [53] and Cauer et al. [35] proposed an extension of the PASTIS software to infer haploid 3D genome structures [57]. One published algorithm [35] can infer the 3D chromatin structure at a true diploid level from population Hi-C data. Specifically, it incorporates two constraints into structural inference – it enforces even spacing of the beads along the chromosome (the homolog separation constraint), and spatially separates the homologs (the bead connectivity constraint). Notably, the weight assigned to the homolog-separating constraint should be tuned for each organism based on prior knowledge of genome architecture, especially the relationship between the number of contacts and pairwise distances. Despite its good performance with real Hi-C data, this diploid PASTIS method can be further improved via joint estimation of the biophysical model alongside the 3D structure, or by incorporating a multiscale optimization strategy, in which a high-resolution structure is deduced in a stepwise fashion with gradually increasing resolution.
Coincidentally, Han et al. [22] have built an iteratively weighted adjusting model to fit each haploid chromosome into the 3D nuclear space from the allele-specific interaction matrices. The iterative process continuously minimizes the sum of errors between coordinate-based distance and the 'real' distance that was converted from the allele-specific interaction matrix. Briefly, Han et al. first initialized random 3D coordinates for each chromosome, and then iteratively adjusted the coordinates based on the distance errors between a specific chromosome and the remaining chromosomes. The model is thereby more likely to reach local optima and a steady state with finite iterations.
In addition, Ye and Ma [46] designed an iterative expectation–maximization (EM) algorithm to infer 3D structures of homologs. They developed an inter-homologous optimization strategy, that is, they estimated homologous chromosomes separately, then predicted the relative position between the two homologs to improve the final 3D structures. However, this method cannot be used to build an accurate genome-wide 3D model, due to lack of an additional estimation step to model the relative position of multiple haploid chromosomes.
3. Features of the spatial organization of homologous chromosomes
Dissection of 3D genome structures in diploid genomes has revealed homolog-specific features. In this section, we discuss the biophysical characteristics and links between these structural features.
3.1. Similar Hi-C contact patterns between homologous chromosomes
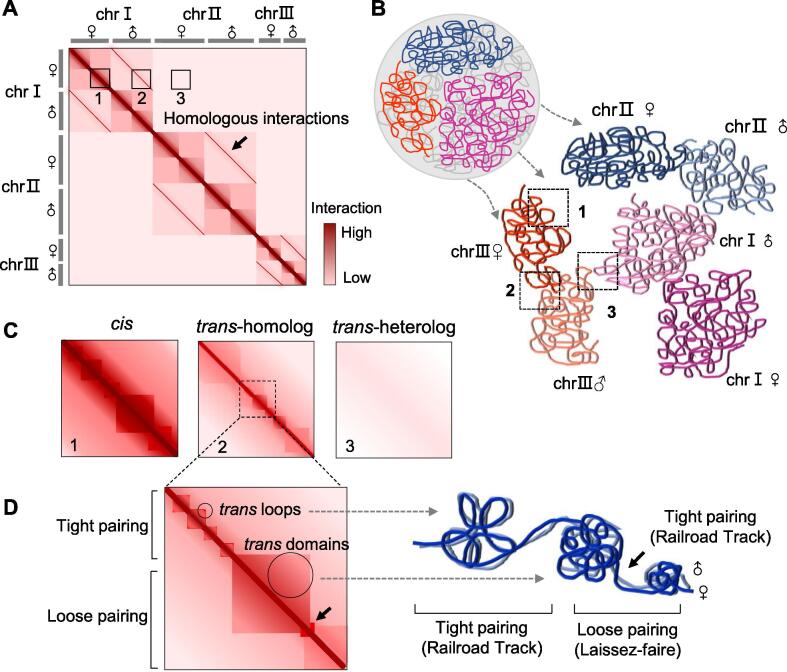
In interphase cells, homologous chromosomes have the most similar Hi-C contact patterns (Fig. 2A), a prominent characteristic of diploid genome architecture. Both inter- and intra-chromosomal contact profiles of a chromosome resemble those of its respective homolog during interphase in human and mouse cells, and this similarity is strongly correlated with allelic co-expression levels [3], [22]. Principal component analysis (PCA) on diploid Hi-C interaction matrices showed that homologous chromosomes are in close proximity on the PCA projection and that PCA distances between homologous pairs are significantly shorter compared to non-homologs [22].
Fig. 2.
Schematics of highly structured homolog pairing in diploid genomes. (A) Haplotype-resolved Hi-C map of a diploid F1 hybrid, which is assumed to have three pairs of chromosomes. The three boxes on the map show the chromatin interactions within the haploid chromosome (cis contacts, box 1), between homologous chromosomes (trans homolog contacts, box 2), and between heterologous chromosomes (trans heterolog contacts, box 3). The t-homo diagonals along the main cis diagonal of the map indicate the homolog pairing. (B) Spatial organization of haploid-level chromosomes in the 3D nucleus of diploid F1 hybrids obtained by 3D modeling of the haploid-resolved Hi-C map. Cis (box 1), t-homo (box 2), and t-heter (box 3) contacts are indicated on the map. (C) Zoomed in Hi-C maps of boxes 1, 2, and 3 shown in (A). (D) Local contact map and 3D organization of variable homolog pairing. Homolog pairing encompasses tightly and loosely paired regions, and displays highly structured trans-domains and trans-interaction peaks.
Global folding patterns of homologous chromosomes are highly and consistently similar during interphase (Fig. 2B) [32]. Studies on 3D nucleus reconstruction have shown that homologs are nearly equidistant from the center of the 3D nucleus, and are usually located next to each other, confirming the existence of homolog pairing based on Hi-C data. The distance of a chromosome to the nuclear center is highly correlated with that of its homologous counterpart, and the distance between any chromosome and a homologous pair is highly correlated, supporting similarity in the spatial organization of homologs [22]. This is in agreement with traditional chromatin conformation studies showing that loci with similar genomic composition and chromatin status tend to be located close to each other [1], [58], [59], which is expected given that homologous chromosomes are nearly identical in size, sequence composition, and associated proteins. However, there are exceptions. For example, maternal and paternal X chromosomes in mammalian cells are located far away from each other due to X chromosome inactivation (XCI) [34]. In a study of 3D diploid nuclei [22], larger chromosomes tended to locate in one pole, with smaller chromosomes in the opposite pole, which was consistent with observations in non-haplotype-resolved Hi-C studies.
3.2. Homolog pairing in diploid genomes
One of the most intriguing biological phenomena in nuclear architecture of diploid genomes involves the colocalization of homologous chromosomes, known as ‘homolog pairing’ (Fig. 2C, 2D). Homolog pairing was first found in Drosophila somatic cells nearly a century ago, occurring from embryogenesis to adulthood, and various degrees of somatic homolog pairing have since been observed during interphase in diploid yeasts and mammals. Such pairing has been implicated in DNA repair, V(D)J recombination, imprinting, XCI, and cell fate commitment across species [60], [61]. As attention towards this phenomenon grows, it is becoming increasingly clear that homolog pairing can facilitate regulatory interactions between different alleles of the same gene, a process known as transvection or intragenic complementation [61], [62]. In particular, allele-specific Hi-C technologies shed light on the 3D organization and formation of homolog pairing, and partially explain whether pairing is uniform across the genome or how homolog proximity correlates with alignment precision and genome function.
3.2.1. 3D organizational characteristics and their association with function
One of the main characteristics of homolog pairing is the remarkable variation in pairing intensity, which has been observed in diploid yeasts [63] and fruit flies [48], [64]. After achieving a high-resolution map of homolog pairing in the diploid Saccharomyces genome, Kim and colleagues found that the strength of pairing fluctuates across the genome and as a function of growth conditions [63]. Their results on condition-specific pairing also suggested a role for the nuclear pore complex in homolog pairing of Saccharomyces. Hence, the degree of pairing in Saccharomyces depends on both the genomic location and the external environment, and sometimes on an interaction between these factors.
Homolog pairing in Drosophila embryos is highly structured in different forms, including the well-aligned form ‘railroad track’, the less-aligned form ‘laissez-faire’, and highly disordered pairing [48]. By introducing pairing scores (PS) to quantify the variation of homolog colocalization, AlHaj Abed et al. [64] identified at least two distinct forms of homolog pairing in the somatic cells of Drosophila: tight and loose pairing (analogous to the ‘railroad track’ and ‘laissez-faire’ forms, respectively) (Fig. 2D), which have quite different 3D structural features. Tightly paired regions, usually spanning contiguous small TADs, are extensively interspersed with loosely paired segments typically consisting of single large domains. The two types of pairing also differ in the decay of cis and t-homo contact frequencies. Moreover, tighter pairing is more likely to be located at domain boundaries, suggesting that some insulator proteins and elements can contribute to homolog pairing and promote transvection, since the domain boundaries are enriched for insulators and architectural proteins [8], [65], [66].
Most recently, Hoencamp and colleagues [67] have determined that homologs tend to be separated or paired across species depending on whether they carry functional condensin II subunits. Species lacking all or partial condensin II subunits (e.g., fruit flies and mosquitoes), exhibit Rabl-like architectural features (centromere clustering, telomere clustering, and a telomere-to-centromere axis), and tend to be homolog pairing. In contrast, species with full sets of condensin II subunits (e.g., humans and mice), display typical chromosome territories and are more likely to show homolog separation or highly disordered pairing.
Besides the variation in pairing, significant genome-wide correlations were found between pairing, compartments, and gene expression in Drosophila [48], [64], [68]. Focusing on trans interactions, Erceg et al. [48] discovered that pairing correlates with the opening of the chromatin mediated by the transcription factor Zelda during zygotic genome activation. Taking this analysis further, AlHaj Abed and colleagues [64] uncovered complex associations between pairing and chromatin status, compartments, gene expression, and regulatory epigenetic states. First, the active compartment A or highly expressed regions were usually tightly paired, while compartment B showed a bimodal pairing distribution – inactive and repressed chromatin contained regions of both tight and loose pairing. Second, the pairing types were differentially associated with gene expression: highly expressed genes tended to be tightly paired, whereas genes expressed at low levels were associated with both high and low pairing. These observations demonstrate the complex relationship between homolog pairing and chromatin activity or, alternatively, the imperfect classification of pairing/chromatin status. Nevertheless, this suggests that pairing has functional implications.
Importantly, the close proximity of homologs may influence the way in which loops and domains are formed in Drosophila. As observed by AlHaj Abed et al. [64], the boundaries of loose pairings are tightly paired, supporting a model that integrates pairing, loop formation, and chromosome compaction via a mechanism in which the chromosomes are buckled out to form loops by extrusion, anti-pairing, or a combination of both [64], [69]. In the model, tightly paired regions form barriers to extrusion. A series of RNAi experiments showed that the disruption of pairing-promoting factors (Slmb and TopII) resulted in global changes in pairing and disruption of some interaction peaks, suggesting that the close proximity of homologs may influence the formation of loops and domains in Drosophila [64].
Potential somatic pairing in human and mouse, albeit rare, was captured on 3D models by Tan et al. [21]. Nonetheless, homolog pairing has not yet been systematically investigated in mammals. Joyce et al. [61] suggest in a review that pairing in mammals has so far remained unnoticed because of unknown mechanisms to prevent it. Thus, conducting studies on somatic paring of mammals can not only provide an overview of homolog positioning in these animals, but also clarify the inhibitory mechanisms underlying anti-pairing.
3.2.2. Mechanical models of pairing formation
Two main models explain the mechanisms driving colocalization of homologous chromosomes: the ‘zipper’ and ‘button’ models. In the ‘zipper’ model, the different regions of the genome have an equal pairing affinity based on sequence homology, and homolog pairing begins at the centromere and proceeds distally to the telomeres [62]. In contrast, the ‘button’ model proposes that regions of high pairing ability are interspersed across the chromosomes and come together through a random walk that launches pairing [70], [71]. Viets et al. [68] identified TADs (the ‘buttons’) interspersed across the fly genome that brought homologous chromosomes together in order to facilitate cell-type-specific inter-chromosomal gene regulation. These specialized TADs may take on unique chromatin conformations or uniquely bind different protein combinations to create nuclear microcompartments that enable homologous TAD association and pairing. In addition, using a button spanning the spineless gene as a paradigm, they revealed that pairing was necessary but not sufficient to enable transvection, indicating that these two processes were mechanistically independent. The ‘button’ model was also supported by the distribution and common features of tightly paired regions found by AlHaj Abed et al. [64].
However, there are many uncharacterized features of homolog pairing. For example, it is still unclear whether homologs become structurally similar because of pairing, or instead have comparable conformations prior to pairing [48]. Similarly, the existence of different pairing mechanisms remains unknown, in light of the contrary correlations between tight pairing and gene activity found by Li et al. [72] and AlHaj Abed et al. [64]. Present studies have not provided definitive evidence, meaning that these issues need to be addressed in the future.
3.3. Fundamental 3D architecture of homologous chromosomes
3.3.1. Multi-layered organizational structures
Allele-specific Hi-C research has not only validated the central cis diagonal (representing short-distance intra-chromosomal interactions) and hierarchical chromatin structures within chromosomes (Fig. 2C), but also revealed more detailed features of the multi-layered organization and the rich structure of t-homo interactions (Fig. 2D).
Haplotype-resolved chromosomes occupy distinct regions within the simulated 3D nucleus of most animals. These regions are known as chromosome territories (CTs) and their nuclear positioning mainly correlates with genomic properties and functions [21], [35]. Haploid-level Hi-C maps demonstrate the characteristic chromatin compartmentalization in diploid genomes [21]. As expected, broad similarity has been observed between the patterning of compartments and TADs in maternal and paternal genomes [22], [32], [45]. Moreover, a diploid Hi-C study revealed that A compartments have higher heterogeneity compared to B compartments in interphase cells of mice [22]. For example, the local boundary score (LBS) – a quantitative value with peaks representing TAD boundaries – within A compartments exhibit stronger fluctuations and higher variability than that in B compartments. The multiple TADs found in A compartments contrast with the often single TAD found in B compartments, demonstrating the complexity of A compartments in diploid genomes.
Furthermore, allele-specific Hi-C research on Drosophila has revealed the existence of t-homo diagonals in the diploid Hi-C maps, and showed that paired homologs can form t-homo compartments, TADs, and loops (Fig. 2D) [48], [64]. The hierarchical structures of t-homo interactions largely coincided with analogous features in cis contacts. In particular, the observed concordance between the t-homo, cis-maternal, and cis-paternal Hi-C maps in terms of the sizes and positions of domains and loops supports the idea that cis and trans interactions are structurally coordinated and may be formed by similar mechanisms [48], and demonstrates a high level of agreement between paired homologs [64].
Similar to what was found in haploid mouse embryonic stem cells (mESCs) [73], chromosomes in diploid mESCs preferred a more parallel conformation (i.e. Rabl configuration), whereas terminally differentiated cells displayed a more centromere-facing-out organization [21]. The differentiated cell type-specific chromatin organization has been confirmed by Han et al. [22], who showed that inter-chromosomal interactions increased along the chromosomes from the centromeres to the telomeres. These observations suggested that chromatin conformation may function in pluripotency and the characterization of cell identity.
3.3.2. Allelic-biased chromatin conformation
Rare genomic regions show changes in compartment status between alleles. Despite the high similarities in the patterns of compartments between homologs, ~0.6–2.3% of the genome exhibited different compartments between homologs in human embryonic stem cells (hESCs) and four hESC-derived lineages [32]. Furthermore, allele switching between active and inactive compartments was observed genome-wide in the cell cycle of human B-lymphocytes, and the switched alleles were not the same in the different cell cycle phases [74]. Moreover, the vast majority of AB switching occurred at the borders between compartments, instead of within entire compartments [45], [74]. This indicated that the boundaries may be more unstable compared to the inner parts of the compartments. The compartments identified in two alleles of a single cell can vary in an almost independent manner, with an average Spearman correlation close to zero [21]. Han et al. [22] found that the A/B compartment status of haploids in hybrid mice was highly correlated with its parent-of-origin due to cis-effects, and parental divergent compartments basically transited into the same status in hybrid mice due to a shared cellular environment, i.e., the trans-effect. Approximately 58% of the divergent A/B compartment bins between two parents retained the same status as the parent-of-origin in hybrid mice (cis-effect), while 95% of the remaining parentally divergent compartments converged into the same compartment status (trans-effect). These observations confirmed the essential function of micro-environments in epigenetic regulation of gene expression.
Even though TAD is considered to have a relatively conservative organizational structure among different species and across tissues, allele-specific TADs have been detected in diploid genomes. TADs with the largest allelic differences are smaller in size and contain higher gene density compared to randomly selected TADs [74]. Lindsly et al. [45] carefully identified sub-TADs and local structural changes, and determined that sub-domains were not always consistent between alleles or cell cycle phases, and were even more variable between cell types.
Several studies focusing on chromatin loop variation across haplotypes have provided conflicting conclusions. Rao et al. [3] studied allelic differences in looping with diploid human in situ Hi-C contact matrices and observed few disparities between the maternal and paternal 3D genome maps except for imprinted regions. In contrast, studies using dilution Hi-C and 4C-seq [32], ChIA-PET [15], and H3K27ac Hi-ChIP [75] technologies reported that allelic imbalance in chromatin looping occurred throughout the genome. Analyzing phased human multi-omics data including high resolution haplotype-resolved Hi-C maps, Greenwald et al. [44] quantitatively investigated chromatin looping across different haplotypes and cell types. Aided by the increased power of combined cell type data, the authors found subtle allelic differences in the majority of loops, and identified infrequent, large allelic imbalances in looping in the genome (only 114 haplotype-associated chromatin loops across all individuals [FDR < 0.05]), which were mainly driven by imprinting and/or copy number variants (CNVs). These contradictory results are likely a result of distinct experimental designs, detection power, and the types of effects examined in these studies.
4. From homologous chromatin conformation to genome function
Allelic differences have been detected in gene expression, protein binding, and 3D chromatin organization [76], [77], [78], [79], [80]. Given the basic biological principle that structure (form) determines function, it follows that 3D chromatin architecture contributes to allele-biased (or allele-specific) gene expression and regulation. At present, both direct and indirect evidence support an intimate relationship between allele-specific genome structures and nuclear functions.
4.1. Allele biased expression (ABE)
Diploid 3D genome studies showed that ABE can, to some extent, be attributed to allele-biased chromatin structures. By integrating haplotype-resolved 3D genome maps with epigenomic and transcriptomic data, Dixon et al. [32] observed that genomic regions containing allele biased or imprinted genes show a subtle but statistically significant increase in the variability of A/B compartment scores between alleles. However, A/B compartment switches between alleles constituted rare regions of the genome and were not enriched for either allele-biased or known imprinted genes. These results indicate that despite most allele-biased genes having no differential compartment status between alleles, subtle differences may exist in higher-order chromatin structure between homologs at ABE regions. In addition, they found that allele-biased enhancer activity possibly underlies ABE through either stable DNA looping between alleles or potential allele-specific PEIs.
Interaction domains also function in ABE. Focusing on the top 10% of bins with the largest degree of Hi-C change to fully characterize Hi-C allelic differences, Chen et al. [74] discovered a subset of ABE genes that were preferentially localized near TAD boundaries in lymphoblastoid cells. These genes were enriched with chromatin organization transcription factor binding sites and contained a higher number of SNVs/InDels in the CCCTC-binding factor (CTCF) binding sites, providing a view of how the two haploid genomes differ in form and function. Parental-specific domains participate in parentally biased gene expression in early murine embryogenesis, including parentally pre-formed domains associated with a lower average expression on the structured allele and a higher frequency of strongly biased genes (including some transiently imprinted loci) [81]. These findings offer important clues to understand how dynamic allele-specific chromatin organization relates to gene expression during embryogenesis.
More recently, a genome-wide proportional relationship between differential contact propensity and differential expression across haplotypes and cell types has been quantitatively investigated at the chromatin loop level [44]. Greenwald et al. [44] determined that haplotype-associated differences in chromatin looping were considerably smaller than cell-type-associated differences, and both were smaller than the range at which gene expression and H3K27ac fold changes occurred, indicating that subtle changes in chromatin contact propensity were associated with large functional effects. Additionally, haplotype-associated loops that were highly enriched for imprinted regions, were related to CNVs but not eQTLs, suggesting that regulatory genetic variation does not exert large effects on chromatin interaction but might affect gene expression through small modifications of contact frequency at chromatin loops. Hence, contact propensity is a mechanism likely involved in gene regulation in a similar way to enhancer activity or transcription factor binding strength. Notably, contact propensity may either affect, or be affected by, gene expression and/or regulation.
Interestingly, allelic imbalance in Hi-C contacts is more relevant to nascent gene transcripts than to mature mRNAs [74]. Hence, the form-function relationship can be more accurately captured when comparing Hi-C data with nascent RNA Bru-seq (bromouridine labeling and sequencing) data, instead of steady-state RNA-seq (RNA sequencing) data. However, the majority of the aforementioned studies employed mRNA-seq data to analyze the interplay between chromatin contact imbalance and gene expression. This may lead to inaccurate conclusions and requires further in-depth investigation using Bru-seq data.
4.2. Imprinting
Haplotype-resolved 3D genome assays enable an understanding of the genetic basis of imprinting, which is essential for mammal development. At imprinted loci, the two alleles can differ drastically in transcriptional activity [82]. Therefore, imprinted regions dictate allele-specific gene activity [83], [84], which can only be studied at the haploid chromosome level.
Rao et al. [3] observed many homolog-specific features within diploid Hi-C maps, including imprinting-specific loops. The 3D structural differences between alleles at imprinted loci were also captured by Tan et al. [21]. Through visualizing the maternally transcribed H19 gene and the paternally transcribed IGF2 gene in simulated 3D nuclei of single cells, Tan et al. found that, despite cell-to-cell heterogeneity, the maternal allele more frequently separated IGF2 from both H19 and the nearby HIDAD site, and disrupted the IGF2-HIDAD CTCF loop; in contrast, the paternal allele more frequently stayed fully intermingled, which was consistent with the observed allelic differences found in contact profiles of bulk Hi-C [3]. More recently, Collombet et al. [81] found that maternally pre-formed domains in early mice embryos encompassed most transiently maternally imprinted genes (19 out of 27 genes). They also observed a similar pattern of shifting, from maternal imprinted domains at the two-cell stage to TADs at the blastocyst stage, for some imprinted genes, including Enc1, Mbnl2, Tle3, and Xist.
4.3. X chromosome inactivation (XCI)
Similar to imprinting, XCI means that one of the X chromosomes in female mammals is expressed exclusively from a single allele in somatic cells, which represents a striking case of homolog differences [82]. In the 3D space, the active X chromosome tends to exhibit an extended chromatin morphology, whereas the inactive X chromosome is generally compacted [21]. This pair of homologs is characterized by distinct patterning of compartments defined based on chromatin contacts: the active X chromosome features clear compartmentalization of euchromatin and heterochromatin; by contrast, the silent X chromosome contains a more uniform compartment patterning with multiple ‘super-domains’ or ‘super-loops’ [3], [21], [34], [42]. In mice, the inactive X chromosome shows a global loss of the TAD structure, except in genes that are being expressed [34], [81], [85], which corroborates the association between chromatin architecture and nuclear activity. Furthermore, a study on DNA organization dynamics related to XCI during early development suggested that loss of the TAD structure on the silenced X chromosome follows gene silencing, rather than preceding it [81].
Additionally, allele-biased 3D genome organization, which is dynamically coordinated with transcriptional activity, can facilitate the programming of replication-timing (RT) during development. Rivera-Mulia et al. [40] showed that RT asynchrony in mESCs was strongly correlated with changes in Hi-C compartments between alleles but not with sequence variation, gene expression, or chromatin accessibility. Long-range PEIs in RT asynchronous domains were restricted to the early replicating allele. Furthermore, the RT asynchrony and allelic discordance observed in the genome organization of mESCs were gradually lost during cell fate commitment. Dynamic allelic imbalances in chromatin organization also function directly in early mammalian development, which has also been investigated in several 3D diploid genome studies [41], [81], [86], [87], [88]. These studies depicted notably asymmetric chromatin architectures between alleles as well as significant reorganization during early development, partially explaining how haploid chromosomal conformation flexibly facilitates genomic functions during different biological processes.
5. Summary and outlook
Many fundamental and long-standing biological questions are linked to 3D genome architecture [59]. Haplotype-resolved Hi-C studies provide new ways to answer these questions. In this review, we described some of the emerging strategies and algorithms that are used to phase genome-scale Hi-C data, and summarized the highly structured chromatin organization of the diploid genome, especially homolog pairing. We also discussed the functional implications of the recently reported allele-biased chromosomal conformation, which attempts to explain how differences in chromosome architecture between alleles affects differences in genome function.
One of the main challenges of haplotype-resolved Hi-C studies is to improve the efficiency of data generation and processing. In the coming years, the integration of comprehensive studies on diploid genome conformation, multi-omics data, and live-cell imaging will enable the generation of a panorama of the genome. This trend necessitates derivation of comprehensive models that allow for integrating haplotype imputation and 4D Nucleome (combining 3D genome with time series) with gene expression and epigenetic signals to investigate chromatin form-function dynamics under different contexts (e.g., cold stress or elevated glucose) or during important biological processes (e.g., cell reprogramming and early development). The dissection of the functional significance of haploid chromosomal conformation, especially higher-order structural elements, with genetic perturbations will likely be predominant in the near future.
CRediT authorship contribution statement
Jing Li: Visualization, Funding acquisition. Yu Lin: Methodology. Qianzi Tang: Methodology, Writing - original draft. Mingzhou Li: Conceptualization, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2020YFA0509500), the National Natural Science Foundation of China (U19A2036, 31872335, 31802044, 31772576), the China Postdoctoral Science Foundation (2020M683648XB), the Postdoctoral Research Project of Sichuan Province, and the Science and Technology Support Program of Sichuan (2021YFYZ0009 and 2021YFYZ0030).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belton J.M., McCord R.P., Gibcus J.H., Naumova N., Zhan Y., Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang Z., Li G., Wang Z., Djekidel M.N., Li Y., Qian M.P. BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat Commun. 2017;8:1622. doi: 10.1038/s41467-017-01754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin D., Hong P., Zhang S., Xu W., Jamal M., Yan K. Digestion-ligation-only Hi-C is an efficient and cost-effective method for chromosome conformation capture. Nat Genet. 2018;50:754–763. doi: 10.1038/s41588-018-0111-2. [DOI] [PubMed] [Google Scholar]
- 6.Kalhor R., Tjong H., Jayathilaka N., Alber F., Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2011;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou C., Li L., Qin Z.S., Corces V.G. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagano T., Lubling Y., Varnai C., Dudley C., Leung W., Baran Y. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547:61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain S, Ba Z, Zhang Y, Dai HQ, Alt FW. CTCF-binding elements mediate accessibility of RAG substrates during chromatin scanning. Cell 2018;174:102-16.e14. [DOI] [PMC free article] [PubMed]
- 13.Tan L., Xing D., Daley N., Xie X.S. Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat Struct Mol Biol. 2019;26:297–307. doi: 10.1038/s41594-019-0205-2. [DOI] [PubMed] [Google Scholar]
- 14.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 15.Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Northcott P.A., Buchhalter I., Morrissy A.S., Hovestadt V., Weischenfeldt J., Ehrenberger T. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitter M., Gasser C., Takacs Z., Langer C.C.H., Tang W., Jessberger G. Conformation of sister chromatids in the replicated human genome. Nature. 2020;586:139–144. doi: 10.1038/s41586-020-2744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagano T., Lubling Y., Yaffe E., Wingett S.W., Dean W., Tanay A. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10:1986–2003. doi: 10.1038/nprot.2015.127. [DOI] [PubMed] [Google Scholar]
- 20.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan L., Xing D., Chang C.H., Li H., Xie X.S. Three-dimensional genome structures of single diploid human cells. Science. 2018;361:924–928. doi: 10.1126/science.aat5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Z., Cui K., Placek K., Hong N., Lin C., Chen W. Diploid genome architecture revealed by multi-omic data of hybrid mice. Genome Res. 2020;30:1097–1106. doi: 10.1101/gr.257568.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Splinter E., de Wit E., Nora E.P., Klous P., van de Werken H.J., Zhu Y. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Wit E., Bouwman B.A., Zhu Y., Klous P., Splinter E., Verstegen M.J. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 25.Holwerda S.J., van de Werken H.J., Ribeiro de Almeida C., Bergen I.M., de Bruijn M.J., Verstegen M.J. Allelic exclusion of the immunoglobulin heavy chain locus is independent of its nuclear localization in mature B cells. Nucleic Acids Res. 2013;41:6905–6916. doi: 10.1093/nar/gkt491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie W., Barr C.L., Kim A., Yue F., Lee A.Y., Eubanks J. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McVicker G., van de Geijn B., Degner J.F., Cain C.E., Banovich N.E., Raj A. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasowski M., Kyriazopoulou-Panagiotopoulou S., Grubert F., Zaugg J.B., Kundaje A., Liu Y. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilpinen H., Waszak S.M., Gschwind A.R., Raghav S.K., Witwicki R.M., Orioli A. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov V., Xie D., Chen R., Pushkarev D., Ma Z., Blauwkamp T. Whole-genome haplotyping using long reads and statistical methods. Nat Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giorgetti L., Lajoie B.R., Carter A.C., Attia M., Zhan Y., Xu J. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cauer A.G., Yardimci G., Vert J.P., Varoquaux N., Noble W.S. In 19th International Workshop on Algorithms in Bioinformatics (WABI 2019). Schloss Dagstuhl-Leibniz-Zentrum fuer Informatik. 2019. Inferring diploid 3D chromatin structures from Hi-C Data. [Google Scholar]
- 36.International HapMap Consortium, Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.1000 Genomes Project Consortium, Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.1000 Genomes Project Consortium, Abecasis GR G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.1000 Genomes Project Consortium, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivera-Mulia J.C., Dimond A., Vera D., Trevilla-Garcia C., Sasaki T., Zimmerman J. Allele-specific control of replication timing and genome organization during development. Genome Res. 2018;28:800–811. doi: 10.1101/gr.232561.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Z., Zheng H., Huang B., Ma R., Wu J., Zhang X. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 42.Darrow E.M., Huntley M.H., Dudchenko O., Stamenova E.K., Durand N.C., Sun Z. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 2016;113:E4504–E4512. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonora G., Deng X., Fang H., Ramani V., Qiu R., Berletch J.B. Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome. Nat Commun. 2018;9:1445. doi: 10.1038/s41467-018-03694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenwald W.W., Li H., Benaglio P., Jakubosky D., Matsui H., Schmitt A. Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat Commun. 2019;10:1054. doi: 10.1038/s41467-019-08940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindsly S, Jia W, Chen H, Ronquist S, Chen C, Wen X, et al. Genome architecture and transcription data reveal allelic bias during the cell cycle. bioRxiv 2020.
- 46.Ye and Ma ASHIC: hierarchical Bayesian modeling of diploid chromatin contacts and structures. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkaa872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng X., Ma W., Ramani V., Hill A., Yang F., Ay F. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erceg J., AlHaj Abed J., Goloborodko A., Lajoie B.R., Fudenberg G., Abdennur N. The genome-wide multi-layered architecture of chromosome pairing in early Drosophila embryos. Nat Commun. 2019;10:4486. doi: 10.1038/s41467-019-12211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo H., Li X., Fu H., Peng C. HiCHap: a package to correct and analyze the diploid Hi-C data. BMC Genomics. 2020;21:746. doi: 10.1186/s12864-020-07165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjong H., Li W., Kalhor R., Dai C., Hao S., Gong K. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci U S A. 2016;113:e1663–e1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin D., Bonora G., Yardımcı G.G., Noble W.S. Computational methods for analyzing and modeling genome structure and organization. Wiley Interdiscip Rev Syst Biol Med. 2019;11 doi: 10.1002/wsbm.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunnik E.M., Cook K.B., Varoquaux N., Batugedara G., Prudhomme J., Cort A. Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nat Commun. 2018;9:1910. doi: 10.1038/s41467-018-04295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ay F., Vu T.H., Zeitz M.J., Varoquaux N., Carette J.E., Vert J.P. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics. 2015;16:121. doi: 10.1186/s12864-015-1236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carstens S., Nilges M., Habeck M. Inferential structure determination of chromosomes from single-cell Hi-C data. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua N., Tjong H., Shin H., Gong K., Zhou X.J., Alber F. Producing genome structure populations with the dynamic and automated PGS software. Nat Protoc. 2018;13:915–926. doi: 10.1038/nprot.2018.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paulsen J., Sekelja M., Oldenburg A.R., Barateau A., Briand N., Delbarre E. Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol. 2017;18:21. doi: 10.1186/s13059-016-1146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varoquaux N., Ay F., Noble W.S., Vert J.P. A statistical approach for inferring the 3D structure of the genome. Bioinformatics. 2014;30:i26–i33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibcus J.H., Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramani V., Shendure J., Duan Z. Understanding spatial genome organization: methods and insights. Genomics Proteomics Bioinformatics. 2016;14:7–20. doi: 10.1016/j.gpb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Apte M.S., Meller V.H. Homologue pairing in flies and mammals: gene regulation when two are involved. Genet Res Int. 2012;2012 doi: 10.1155/2012/430587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joyce E.F., Erceg J., Wu C.T. Pairing and anti-pairing: a balancing act in the diploid genome. Curr Opin Genet Dev. 2016;37:119–128. doi: 10.1016/j.gde.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis E.B. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat. 1954;88:225–239. [Google Scholar]
- 63.Kim S., Liachko I., Brickner D.G., Cook K., Noble W.S., Brickner J.H. The dynamic three-dimensional organization of the diploid yeast genome. Elife. 2017;6 doi: 10.7554/eLife.23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.AlHaj Abed J., Erceg J., Goloborodko A., Nguyen S.C., McCole R.B., Saylor W. Highly structured homolog pairing reflects functional organization of the Drosophila genome. Nat Commun. 2019;10:4485. doi: 10.1038/s41467-019-12208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blanton J., Gaszner M., Schedl P. Protein: protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowley M.J., Corces V.G. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoencamp C., Dudchenko O., Elbatsh M.O.A., Brahmachari S., Raaijmakers A.J., Schaik T. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science. 2021;372:984–989. doi: 10.1126/science.abe2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viets K., Sauria M., Chernoff C., Viales R.R., Echterling M., Anderson C. Characterization of button loci that promote homologous chromosome pairing and cell-type-specific interchromosomal gene regulation. Dev Cell. 2019;51:341–356.e7. doi: 10.1016/j.devcel.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joyce E.F., Williams B.R., Xie T., Wu C.T. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fung J.C., Marshall W.F., Dernburg A., Agard D.A., Sedat J.W. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gemkow M.J., Verveer P.J., Arndt-Jovin D.J. Homologous association of the Bithorax-Complex during embryogenesis: consequences for transvection in Drosophila melanogaster. Development. 1998;125:4541–4552. doi: 10.1242/dev.125.22.4541. [DOI] [PubMed] [Google Scholar]
- 72.Li Q., Tjong H., Li X., Gong K., Zhou X.J., Chiolo I. The three-dimensional genome organization of Drosophila melanogaster through data integration. Genome Biol. 2017;18:145. doi: 10.1186/s13059-017-1264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stevens T.J., Lando D., Basu S., Atkinson L.P., Cao Y., Lee S.F. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544:59–64. doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen H, Liu S, Seaman L, Najarian C, Wu W, Ljungman M, et al. Parental allele-specific genome architecture and transcription during the cell cycle. bioRxiv 2017;2017:201715.
- 75.Mumbach M.R., Satpathy A.T., Boyle E.A., Dai C., Gowen B.G., Cho S.W. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet. 2017;49:1602–1612. doi: 10.1038/ng.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 77.Rozowsky J., Abyzov A., Wang J., Alves P., Raha D., Harmanci A. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rivera C.M., Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng Q., Ramskold D., Reinius B., Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 80.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collombet S., Ranisavljevic N., Nagano T., Varnai C., Shisode T., Leung W. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature. 2020;580:142–146. doi: 10.1038/s41586-020-2125-z. [DOI] [PubMed] [Google Scholar]
- 82.Lee J.T., Bartolomei M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 83.Inoue A., Jiang L., Lu F., Suzuki T., Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547:419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Inoue A., Jiang L., Lu F., Zhang Y. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 2017;31:1927–1932. doi: 10.1101/gad.304113.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minajigi A., Froberg J., Wei C., Sunwoo H., Kesner B., Colognori D. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349 doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flyamer I.M., Gassler J., Imakaev M., Brandao H.B., Ulianov S.V., Abdennur N. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gassler J., Brandão H.B., Imakaev M., Flyamer I.M., Ladstätter S., Bickmore W.A. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. Embo j. 2017;36:3600–3618. doi: 10.15252/embj.201798083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017;170(367–81) doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.