Graphical abstract
Keywords: Ribosomal frameshifting, Single-molecule, Optical tweezers, smFRET, MD simulation, Cryo-EM
Abstract
Programmed −1 ribosomal frameshifting (−1 PRF) is a translation mechanism that regulates the relative expression level of two proteins encoded on the same messenger RNA (mRNA). This regulation is commonly used by viruses such as coronaviruses and retroviruses but rarely by host human cells, and for this reason, it has long been considered as a therapeutic target for antiviral drug development. Understanding the molecular mechanism of −1 PRF is one step toward this goal. Minus-one PRF occurs with a certain efficiency when translating ribosomes encounter the specialized mRNA signal consisting of the frameshifting site and a downstream stimulatory structure, which impedes translocation of the ribosome. The impeded ribosome can still undergo profound conformational changes to proceed with translocation; however, some of these changes may be unique and essential to frameshifting. In addition, most stimulatory structures exhibit conformational dynamics and sufficient mechanical strength, which, when under the action of ribosomes, may in turn further promote −1 PRF efficiency. In this review, we discuss how the dynamic features of ribosomes and mRNA stimulatory structures may influence the occurrence of −1 PRF and propose a hypothetical frameshifting model that recapitulates the role of conformational dynamics.
1. Introduction
Protein synthesis in cells is catalyzed by ribosomes. The ribosome and bound transfer RNA (tRNA) read and translate trinucleotide codons from messenger RNA (mRNA) into the corresponding amino acid sequences [1]. The mRNA reading frame is followed strictly during translation, but errors can still occur when the ribosome shifts one nucleotide in a codon toward the 5′ or 3′ end of mRNA, resulting in −1 or +1 frameshifting, respectively. Spontaneous ribosomal frameshifting occurs at a frequency of <10−5 (see [2]). Proteins synthesized by this process may not be functional because the produced amino acid sequence after the shifted codon differs from the native sequence. However, the ability of ribosomal frameshifting can be used to specifically modulate the expression of two proteins from the same mRNA [2], [3], [4], [5], [6], [7]. This modulation is called programmed −1 (or +1) ribosomal frameshifting (−1 or +1 PRF).
Utilization of +1 PRF is found in viruses [6] and specific cellular genes from some species [8], [9]. In general, not many organisms employ +1 PRF in translation regulation, but the ciliate Euplotes is an exception [10], [11]. A genome-wide study has identified 3,700 putative +1 PRF genes (more than 10% of the transcriptome) in Euplotes octocarinatus [9]. A common feature shared by most of the +1 PRF signals is that the frameshift site is followed by a stop codon or a rare/hungry codon (decoded by low abundance tRNAs). Nevertheless, the stimulating mechanisms of +1 frameshifting are rather diverse [4].
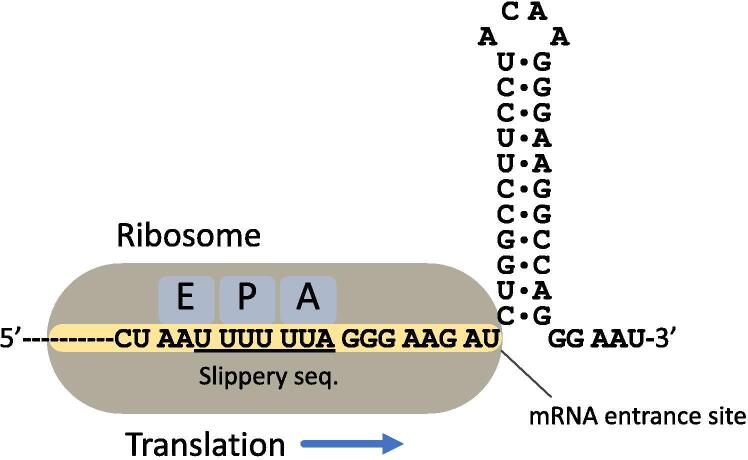
In contrast to +1 PRF, −1 PRF is widely spread in a variety of viruses [6]. Most −1 PRF occurs on a heptanucleotide slippery sequence with a pattern of X XXY YYZ (where XXX and YYY represent three identical nucleotides, and codons are separated by spaces) [6], [12]. This pattern allows for a diverse selection of codons in the slippery sequence and minimizes the difference between codons after the shift to the −1 frame (XXX YYY Z). In fact, the free energy difference of the codon-anticodon base-pairing between these two frames can quantitatively determine frameshifting efficiency [13]. Several factors are known to stimulate −1 frameshifting, including a hungry codon at the YYZ position [14], [15], [16], [17], mutant elongation factor G (EF-G) that retards translocation [18], and the cotranslational folding of nascent protein found in the Sindbis virus [19]. Importantly, the more general stimulator found in many −1 PRF signals is a structure (such as a hairpin or a pseudoknot) 5–9 nucleotides downstream of the slippery sequence [3], [6], [20] (see Fig. 1). The frameshifting signal (a slippery sequence with a stimulatory structure) from the human immunodeficiency virus type 1 (HIV-1; Fig. 1) can cause frameshifting in Escherichia coli (E. coli) cells [21] and cell-free lysates [22], and the signal from severe acute respiratory syndrome-associated coronavirus (SARS-CoV) can cause frameshifting in several eukaryotic translation systems with varying degrees of efficiency [23]. These studies suggest that the primary determinant of frameshifting is the cis-acting elements of mRNA and that efficiency is determined in part by architectural differences in ribosomes.
Fig. 1.
The −1 PRF signal from HIV-1. This signal consists of a slippery sequence (underlined), a stimulatory hairpin, and an 8-nucleotide spacer in between. Schematic presentation of the ribosome indicates relative positions between functional sites of the ribosome and mRNA during −1 PRF.
An increasing body of evidence has shown that in addition to structural stability, the conformational dynamics of an mRNA structure play a key role in the stimulatory effects on ribosomal frameshifting [24], [25], [26], [27], [28], [29], [30], [31], [32], [33]. In addition, ribosomes and tRNA also undergo profound conformational changes during translocation [34], [35], though how their structures are involved in −1 PRF remain unclear. Nevertheless, novel structures of ribosomes are visualized by cryogenic electron microscopy (cryo-EM) when translation is stalled by other regulatory signals [36], [37], and resolution-exchanged molecular dynamics (MD) simulations have also revealed that similar ribosomal conformations may play a role in −1 PRF [38].
Herein, we review recent advances in the study of −1 PRF and discuss its underlying molecular mechanisms from the perspective of the conformational dynamics of ribosomes and stimulatory structures of mRNA.
2. Choreography of ribosomal motions during translation elongation and regulation
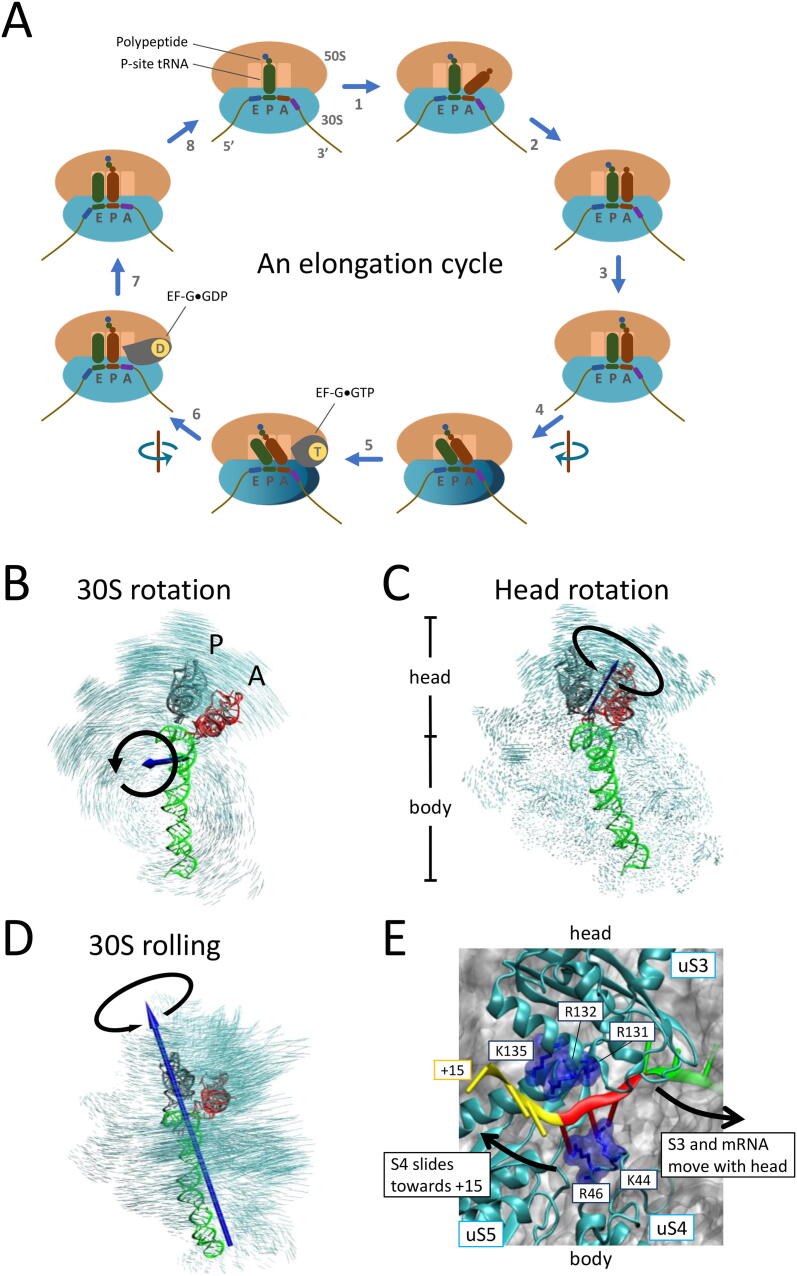
Minus-one PRF occurs during the elongation stage of translation, when the ribosome’s P (peptidyl) and A (aminoacyl) sites are localized at the slippery sequence and the mRNA entrance site encounters a downstream stimulatory structure (Fig. 1). This stage comprises cycles of aminoacyl-tRNA (aa-tRNA) selection, peptide bond formation, and translocation (Fig. 2A). Selection of the aa-tRNA for the A-site codon is conducted through binding of the aa-tRNA·EF-Tu·GTP ternary complex to the ribosome; EF-Tu is an elongation factor in bacteria, and GTP is guanosine triphosphate (step 1, Fig. 2A; please refer to this figure for other steps described in this paragraph). This process involves kinetic proofreading mechanisms with highly coordinated conformational rearrangements of the ribosome and tRNA [39], [40]. After hydrolysis of GTP into guanosine diphosphate (GDP) and dissociation of EF-Tu·GDP, the aa-tRNA is accommodated into the A site (step 2), followed by transfer of the polypeptide from the P- to the A-site tRNAs (step 3). Then, the small (30S in prokaryotes and 40S in eukaryotes) ribosomal subunit rotates counterclockwise with respect to the large (50S in prokaryotes and 60S in eukaryotes) subunit by 4–12° (step 4) (Fig. 2B) [41], [42]. The rotated state of the ribosome allows the 3′ acceptor ends of the tRNAs in the A and P sites to move into the P and E (exit) sites in the 50S subunit, respectively, whereas the anticodon stem-loops remain bound at the A and P sites in the 30S subunit, respectively. These tRNAs in the hybrid configurations are called A/P and P/E states [43]. To continue tRNA translocation, EF-G·GTP binds to the rotated/pre-translocation ribosome (step 5) [44], [45]. After hydrolysis of GTP, while the body domain of the 30S subunit rotates backward, the head domain of the 30S subunit swivels counterclockwise with respect to the body domain by 18–21° (step 6) (Fig. 2C) [46], [47], widening the steric gap between adjacent tRNA-binding sites [48], [49]. Finally, both tRNAs are positioned in the classical P/P and E/E states when the 30S subunit revert to the non-rotated/post-translocation states (step 7) [50], [51], [52], followed by release of the E-site tRNA (step 8) [46], [53].
Fig. 2.
Conformational changes of the bacterial ribosome during translation elongation. (A) Cycle of elongation. For clarity, only the aa-tRNA (red; instead of the ternary complex) is shown in step 1, and the swiveling of the 30S head is not shown in steps 5–7. See the text for details. (B) 30S subunit rotation during translocation. Calculated by superimposing the 50S subunit (not shown). (C) 30S head rotation (swiveling). Calculated by superimposing the 30S body. Rotational axes are calculated by pseudo-angular momentum, and the non-rotated and rotated states of the ribosome are modeled as described [38]. (D) 30S subunit rolling during pseudoknot-induced −1 PRF [38]. The 50S subunit (not shown) is superimposed. The rolling axis lies approximately along helix 44 (green ribbon). (B)–(D) are viewed from the solvent side. (E) mRNA entrance site of the 30S subunit (PDB ID: 6BY1) [67]. uS3 is on the 30S head, while uS4 and uS5 are on the 30S body. R131, R132, and K135 of uS3, as well as K44 and R46 of uS4, are highlighted in blue. The relative movement of the 30S head and body described in (C) are indicated with black arrows. The codons at positions +13 to +15, +10 to +12, and +7 to +9 are shown in yellow, red, and green ribbons, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition to regular translation, structural study of ribosomes has also focused on stalled states with −1 PRF stimulatory structures, such as the pseudoknots derived from the infectious bronchitis virus (IBV) [54], [55], human telomerase RNA [38], and SARS-CoV-2 [56], as well as the hairpin from HIV-1 [57]. An early study on cryo-EM revealed that the A/P-tRNA was distorted in IBV pseudoknot-stalled ribosomes [54], [55]; these results were recently recapitulated in our coarse-grained simulation study [38]. We reasoned that the tension built up during the structural unfolding process induced 30S subunit rolling (Fig. 2D), distorted tRNAs, and prompted tRNA slippage [38]. Although ribosomal rolling by −1 PRF stimulators has not been directly visualized in structural studies, it has been found in other translational regulations, including nascent peptide-induced stalling [36] and translational bypassing [37], in which the ribosome specifically skips 50 nucleotides of the T4 gene 60 transcript in a highly coordinated manner [58]. The rolled motion of the ribosomes in translational bypassing may appear only transiently before ribosomes transition to a hyper-rotated state [59]. A hyper-rotated state has also been inferred from measurements of single-molecule Förster resonance energy transfer (smFRET) [60], [61] in a hairpin-stimulated −1 PRF system [62], although similar conformation was not observed in a recent study using the same method [57]. In addition, both the frameshifting- and bypassing-programmed ribosomes undergo a long pause in rotated state [63], [64] and exhibit multiple rounds of EF-G binding and GTP hydrolysis [59], [63], [65], [66] (see below). These similarities suggest that the noncanonical ribosomal conformations may be generally involved in switching the ribosome into a nonregular yet well-controlled translation mode whereby recoding on the same mRNA can proceed. Further exploration of various types of ribosomal recoding is required to confirm this.
3. Coupling of ribosomal motions to mRNA structural unwinding
Because ribosomes translate mRNA in the single-stranded form, they must overcome obstacles formed on the template, such as RNA secondary structures [68], microRNA [69], and RNA-binding proteins [70]. Despite the widespread mRNA structures within transcriptomes [68], the intrinsic helicase activity of the ribosome is sufficiently active to translocate through secondary structures [71], [72], [73], [74] by biasing thermal fluctuations toward their open states and mechanically pulling apart closed junctions of the structure [71]. Nevertheless, as discussed in the following sections, several structural features are actively involved in stimulating −1 PRF.
Structural, biochemical, and computational studies have located potential ribosomal helicase in the positively charged residues that line up the mRNA entrance site at positions between +11 and +14 (the first nucleotide of mRNA at the P site is denoted by +1), including R131, R132, and K135 in ribosomal protein uS3, and K44 and R46 in uS4 [38], [67], [73], [75], [76] (Fig. 2E). A high-resolution X-ray structure [67] revealed that the residues in uS3 interact with and stabilize the single-stranded form of mRNA at +12 to +14 with a binding energy of approximately 9 kcal·mol−1 per codon [75]. Because uS3 is at the head and uS4 is at the body of the 30S subunit, uS3 and uS4 are pulled away from each other when the head rotates during translocation (Fig. 2E). While uS3 and mRNA move together and their interaction is relatively unchanged, uS4 slides in the opposite direction from +12 to +15 (from the red to the yellow regions in Fig. 2E) [67], [75]. This countermovement between the head and body of the 30S subunit effectively stretches the mRNA tunnel by one codon. Because the tunnel is too narrow to pass an mRNA duplex, forward head rotation requires the base pairs (bp) to be broken at least between +12 to +15 (around the yellow region in Fig. 2E) [67], [75]. Furthermore, simultaneous force and fluorescence measurements indicate that mRNA hairpin opening occurs after EF-G binding and during forward rotation of the 30S head domain [72].
Interestingly, the HIV-1 stimulatory hairpin was found to dock into the vacant A site and inhibit the aa-tRNA from binding to the ribosome [57]. Without A-site tRNA, the ribosome can switch to a slow −1 frameshifting pathway, which is independent of the downstream stimulator [17]. How the long and stable hairpin (with an 11-bp stem; see Fig. 1) is accommodated to the A site is not clear, but before translation can continue, it must be either unfolded or relocated to the mRNA entrance site, where ribosomal helicase activity can normally occur.
4. Role of mRNA structural stability in Stimulating −1 PRF
Although ribosomes can translocate through mRNA secondary structures, the translation rate is inversely correlated to structural stability [71], [74]. A stable structure can trigger the ribosome to switch to a slower kinetic pathway of translocation [72]. Thus, downstream mRNA structures can function as roadblocks impeding the translocation of ribosomes and may stimulate them to shift backward when localized at a slippery sequence.
Several early studies have used the stimulatory hairpin from HIV-1 as a model to suggest that the thermodynamic stability of the whole stimulator is a key determinant for frameshifting efficiency [77], [78]. However, a later study showed that by measuring a series of HIV-1 hairpin variants, only the local stability of the first 3–4 bp of the hairpin were positively correlated with frameshifting efficiency [79]. Given that the proximal moiety of mRNA structures closely interacts with the entrance site of the ribosome [75] and that the ribosome translates one codon and concomitantly unwinds 3 bp at a time [74], the observed correlation between the local thermostability of stimulatory hairpins and frameshifting efficiency is mechanistically reasonable.
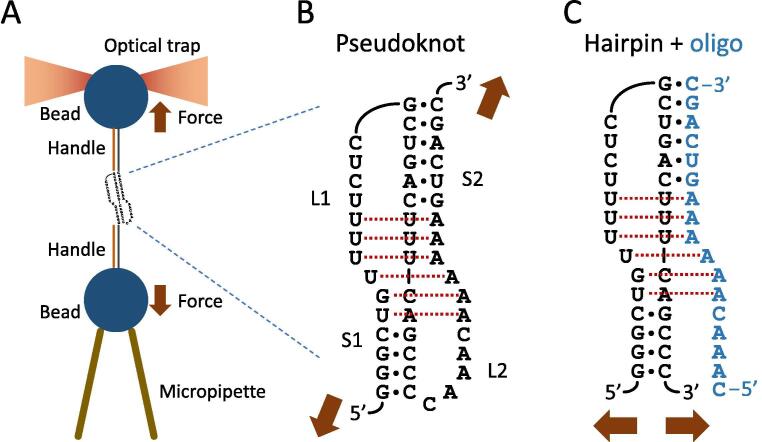
The unfolding of mRNA structures by ribosomes involves a mechanical process [71] in which the input work is generally greater than the free energy of the structure, especially for fast processes [80]. Thus, the mechanical stability of an RNA structure, rather than its thermostability, is a more relevant target for assessing the capacity to stimulate frameshifting. Optical tweezers [81], [82] are a convenient single-molecule tool for this task because they impose modulable tension on the two ends of a tethered RNA molecule to measure its unfolding force (Fig. 3A). Chen et al. [83] constructed a series of structural variants of the DU177 pseudoknot (Fig. 3B), an artificial yet highly efficient frameshifting stimulator derived from the human telomerase RNA [84], and altered the content of their internal base triples, which are crucial for maintaining structural stability. They found that the frameshifting efficiency (0%–50%) was positively correlated with the structural unfolding force [83], reaffirming the value of mechanical stability in stimulatory structures. Similar correlations have been observed in other stimulators, including pseudoknot variants derived from the IBV [85] and the simian retrovirus type 1 (SRV-1) [86], as well as a series of 32-mer RNA hairpins [87].
Fig. 3.
Mechanical unfolding of RNA structures by optical tweezers. (A) One type of optical tweezers. The RNA structure of interest is held between two micron-sized polystyrene beads by flanking the handles of RNA/DNA hybrids. One bead is placed in an optical trap for manipulation and force measurements. The figure is schematic and not to scale. (B) DU177 pseudoknot. The structure contains two stems (S1 and S2) and two loops (L1 and L2). Red dotted lines indicate five base triples and one noncanonical base pair. (C) Bimolecular design to mimic the DU177 pseudoknot. Pseudoknot is split into a hairpin (black) and an oligomer (blue) at the junction of S1 and L2. Expected direction of pulling force when the structure is placed on optical tweezers is indicated by arrows. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The unfolding force measured on optical tweezers is generally the force required to disrupt the whole RNA structure in one step and is dependent on the architecture and orientation of the structure during measurement. For those RNA variants derived from the same structure (such as DU177 and its mutants), their unfolding force is likely to reflect the mechanical barrier that translating ribosomes encounter and thus can more clearly demonstrate a correlation with frameshifting efficiency. However, the correlation does not necessarily hold when the comparison is among different stimulatory structures with varying sizes and architectures, especially for complex RNA pseudoknots [24]. A more relevant assessment of pseudoknot stimulators involves measuring their unzipping force for the 5′ helix (stem S1), which is the first structural moiety to interact with the translating ribosome and is less mechanically stable than the whole pseudoknot. Although a direct measurement of the secondary structural component within an intact tertiary RNA fold is technically challenging, a bimolecular design that preserves most of the original intramolecular interactions can provide a convenient assay platform on optical tweezers for this purpose (Fig. 3C) [88]. With this design, we demonstrated that stem S1 of the DU177 pseudoknot was involved in the interaction network of the whole structure and that the measured unfolding force of stem S1 could reflect the stability difference of its corresponding intact pseudoknot [88].
5. Mechanisms of −1 PRF involving conformational dynamics of mRNA
To identify a general feature that affects −1 PRF among various stimulatory structures, Woodside and coworkers used optical tweezers to measure nine RNA pseudoknots and found that the propensity to fold into alternative structures, known as “conformational plasticity,” was positively correlated with frameshifting efficiency [24]. Recently, the Shannon entropy was used to quantitatively define the conformational plasticity and showed a linear correlation with frameshifting efficiency in a certain force range [89]. These alternative structures may include partially-folded intermediates that are trapped during the folding process and are less stable than the native conformation. Similar results have also been observed in studies of other frameshifting stimulators, including the HIV-1 hairpin [29] and two complex pseudoknots from human CCR5 mRNA [26] and the West Nile virus (WNV) [32]. Remarkably, the WNV stimulatory structure, which causes an extremely high frameshifting efficiency (up to 70%) [27], shows extensive conformational plasticity [32]. The alternative folding propensity of stimulatory structures has also been identified in other viruses, such as the turnip crinkle virus [28], the potato leaf roll virus [30], and the recently identified SARS-CoV-2 [33], [90], through various methods. These results suggest that conformational plasticity is a common feature for most, if not all, frameshift-stimulating RNA structures.
How the propensity of folding into alternative structures is correlated to the frameshift-stimulating potency of an RNA is a pertinent concern. Several studies on smFRET have revealed that EF-G-catalyzed translocations slow down when the ribosome encounters the downstream stimulatory structures [31], [63], [65], [66], [72], [91]. During this prolonged translocation period, EF-G samples the ribosome repeatedly [63], [65], [66] and the ribosome undergoes multiple fluctuations between the classical and hybrid conformational states [91]. These ribosomal fluctuations may perturb and even partially unwind the tightly associated RNA structure. At this stage, it was proposed that the partially unwound structure can rapidly refold into another conformation having increased stability, which then imposes an elevated energy barrier to the ribosome in the late stage of translocation [31]. This is consistent with the finding that frameshifting occurs at a late stage of translocation [92]. In addition, profound transitions among intermediate structures under mechanical force (mimicking the action of ribosomes) were found in the highly efficient WNV frameshifting stimulator, and the most probable force under these intermediate interchanges fell within the range of 7–13 pN [32]. This force range is just below the maximum tension (13 pN) that an actively translating ribosome can generate [93], indicating that the ribosome is energetically capable of triggering structural rearrangements of RNA stimulators. Indeed, when the ribosome was translating along a hairpin-forming mRNA, the local RNA helix contacting the ribosome was observed to undergo cycles of unfolding-refolding transitions [94]. We recently found that a folding intermediate of the DU177 pseudoknot can be induced to refold when the first 2 nucleotides at the 5′ end of stem S1 were sequestered by a complementary DNA strand (mimicking the action of ribosomes), and then the refolded structure can retrieve the 2 nucleotides to restore the fully-folded conformation [95]. These observations support the idea that ribosomes can partially unwind the 5′ secondary structure of an RNA stimulator and prompt it to resample alternative (or the native) structures.
The alternative folding feature is required for an RNA structure to stimulate −1 PRF efficiently, but is not sufficient on its own. For example, the study by Chen et al. has demonstrated that alternative structures are the predominant folding products (greater than 50% of the population) for all 11 variants of the DU177 pseudoknot, including those with a frameshifting efficiency of approximately zero [83]. As discussed in previous sections, the ability to form a mechanically stable structure, especially in the 5′ secondary structure (e.g., the first 3–4 bp of a hairpin or stem S1 of a pseudoknot), is also required. The following examples offer additional evidence. By using a short peptide nucleic acid (PNA) [96] to invade stem S1 of the SRV-1 pseudoknot (mimicking ribosomal unwinding), Yang et al. showed that the PNA invasion was enhanced for the low-efficiency mutants, in which base triple formation involving stem S1 was disrupted [97]. Similarly, we found that stem S1 of the highly efficient DU177 pseudoknot could outcompete an invading complementary DNA strand, whereas a low-efficiency variant could not [95]. The study in which researchers used optical tweezers and steered MD simulations suggested that stem S1 stability of the pseudoknot from the beet western yellow virus affected frameshifting efficiency [98]. Finally, when an RNA strand was pulled into an α-hemolysin protein nanopore mimicking the mRNA entrance site of the ribosome, vectorial unfolding of pseudoknots from the 5′ to 3′ end were able to be measured, and the results showed that the unzipping of stem S1 was the rate-limiting step [99], [100].
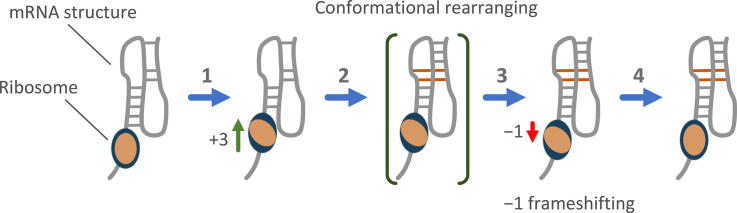
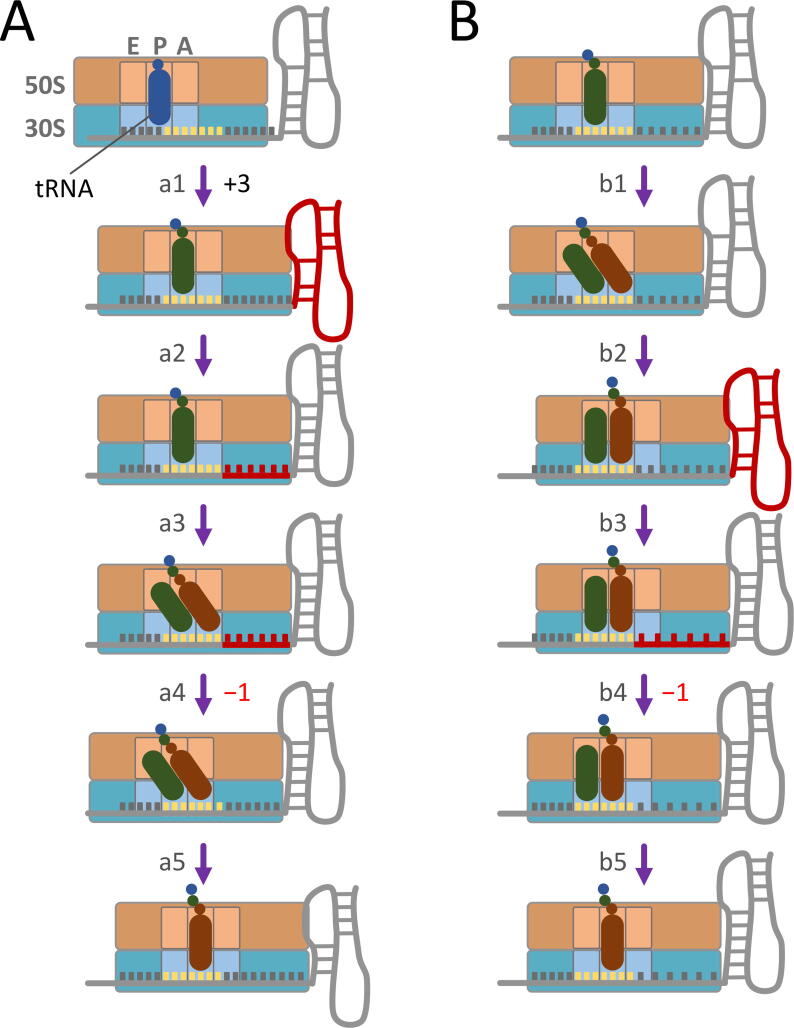
We propose two frameshifting models that can recapitulate the refolding dynamics of RNA intermediates discussed above (Fig. 4; also see [95]). In the first model (Fig. 4A), when the ribosome translocates to the first half of the slippery sequence, the downstream RNA intermediate structure is partially unwound by the ribosomal helicase (step a1) and then induced to refold and retrieve a few nucleotides from the ribosome to form a more stable conformation (step a2). This creates tension alone the mRNA inside the ribosome. After accommodation of the next aa-tRNA to the A site, the ribosome proceeds to the pre-translocation state (step a3). Minus-one frameshifting can occur at this step (step a4) or during a later stage progressing to the post-translocation state (step a5). In the second model (Fig. 4B), the RNA refolding occurs at a later step when the ribosome translocates from the first half of the slippery sequence (step b1) to the second half (step b2, corresponding to step 6 in Fig. 2A). The refolded stable structure creates tension on the mRNA (step b3), and the tension can cause backward slippage of the two tRNAs (step b4) before the head and body domains of the 30S subunit revert completely to the non-rotate state (step b5, corresponding to step 7 in Fig. 2A). In these two models, the mRNA tension built between the tRNA-binding sites and the mRNA entrance site is a key determinant of the following frameshifting. We hypothesize that the downstream RNA refolding accompanied by strand retrieving, as opposed to a rigid and well-folded structure, is a more effective way to create the mRNA tension under the action of the ribosome. Although some features proposed here are hypothetical, they consolidate the roles of ribosomal conformations and structural dynamics of mRNA in frameshifting and thus provides a convenient platform for future testing.
Fig. 4.
Hypothetical models for conformational changes of the ribosome and mRNA stimulatory structure during −1 PRF. Refolding of the downstream RNA intermediate structure occurs when the ribosome translocates to (A) the first half of the slippery sequence (step a1) or (B) the second half of the slippery sequence (step b2). The slippery sequence and the sequence under tension are shown in yellow and red, respectively. The partially-unwound downstream RNA structures before refolding are shown in red. See the text for details. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
6. Concluding remarks
In contrast with its prevalence and critical role in viruses, −1 PRF is rare in the human genome [101]. This fact suggests that −1 PRF is a potential target for drug- or host-factor-based antiviral therapeutic strategies [102], [103], [104], [105], [106], [107]. Annexin A2, a host RNA-binding protein, can bind the IBV pseudoknot and reduce its frameshifting efficiency [108]. Recently, Shiftless, the human interferon-stimulated protein, was identified as a broad-spectrum suppressor of various endogenous and viral −1 PRF signals [109], including that of SARS-CoV-2 [110]. Modulating the expression of these protein factors in infected cells may effectively suppress viral proliferation with minimal perturbation to the host cell.
Forging a deeper understanding of the molecular mechanism of −1 PRF will be a positive step torward realizing the goal of antiviral therapeutics. Although our research has elucidated this matter, additional insightful investigations based on state-of-the-art technology are still required. For example, cotemporal force and FRET detection at the single-molecule level [72], [111] may reveal how the mechanical strength of stimulatory structures perturb translocation of the ribosome. Time-resolved cryo-EM, which can reveal several states of the ribosome when aa-tRNA is delivered [112], may help identify various conformations of the stimulatory RNA structure when interacting with the ribosome. These future investigations can assist in identifying novel antiviral therapeutic strategies.
CRediT authorship contribution statement
Kai-Chun Chang: Conceptualization, Writing - original draft, Writing - review & editing, Visualization. Jin-Der Wen: Conceptualization, Writing - original draft, Writing - review & editing, Visualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Ministry of Science and Technology (MOST), grant no. 109-2311-B-002-009-MY3 and 109-2311-B-002-025. This manuscript was edited by Wallace Academic Editing.
References
- 1.Rodnina M.V. Translation in Prokaryotes. Cold Spring Harb Perspect Biol. 2018;10 doi: 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodnina M.V., Korniy N., Klimova M., Karki P., Peng B.Z. Translational recoding: canonical translation mechanisms reinterpreted. Nucl Acids Res. 2020;48:1056–1067. doi: 10.1093/nar/gkz783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierley I., Gilbert R.J.C., Pennell S. Pseudoknot-dependent programmed -1 ribosomal frameshifting: structures, mechanisms and models. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; New York: 2010. pp. 149–174. [Google Scholar]
- 4.Dinman J.D. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev-RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliskan N., Peske F., Rodnina M.V. Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem Sci. 2015;40:265–274. doi: 10.1016/j.tibs.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkins J.F., Loughran G., Bhatt P.R., Firth A.E., Baranov P.V. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucl Acids Res. 2016;44:7007–7078. doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dever T.E., Dinman J.D., Green R. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol. 2018;10 doi: 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Namy O., Rousset J.P., Napthine S., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang R., Xiong J., Wang W., Miao W., Liang A. High frequency of +1 programmed ribosomal frameshifting in Euplotes octocarinatus. Sci Rep. 2016;6:21139. doi: 10.1038/srep21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klobutcher L.A., Farabaugh P.J. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. doi: 10.1016/s0092-8674(02)01138-8. [DOI] [PubMed] [Google Scholar]
- 11.Lobanov A.V., Heaphy S.M., Turanov A.A., Gerashchenko M.V., Pucciarelli S. Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat Struct Mol Biol. 2017;24:61–68. doi: 10.1038/nsmb.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma V., Prere M.F., Canal I., Firth A.E., Atkins J.F. Analysis of tetra- and hepta-nucleotides motifs promoting -1 ribosomal frameshifting in Escherichia coli. Nucl Acids Res. 2014;42:7210–7225. doi: 10.1093/nar/gku386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bock L.V., Caliskan N., Korniy N., Peske F., Rodnina M.V. Thermodynamic control of -1 programmed ribosomal frameshifting. Nat Commun. 2019;10:4598. doi: 10.1038/s41467-019-12648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barak Z., Lindsley D., Gallant J. On the mechanism of leftward frameshifting at several hungry codons. J Mol Biol. 1996;256:676–684. doi: 10.1006/jmbi.1996.0117. [DOI] [PubMed] [Google Scholar]
- 15.Olubajo B., Taylor E.W. A -1 frameshift in the HIV-1 env gene is enhanced by arginine deficiency via a hungry codon mechanism. Mutat Res-Fund Mol M. 2005;579:125–132. doi: 10.1016/j.mrfmmm.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Temperley R., Richter R., Dennerlein S., Lightowlers R.N., Chrzanowska-Lightowlers Z.M. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- 17.Caliskan N., Wohlgemuth I., Korniy N., Pearson M., Peske F. Conditional Switch between Frameshifting Regimes upon Translation of dnaX mRNA. Mol Cell. 2017;66:558–567. doi: 10.1016/j.molcel.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 18.Peng BZ, Bock LV, Belardinelli R, Peske F, Grubmuller H, et al. Active role of elongation factor G in maintaining the mRNA reading frame during translation. Sci Adv 2019;5:eaax8030. [DOI] [PMC free article] [PubMed]
- 19.Harrington H.R., Zimmer M.H., Chamness L.M., Nash V., Penn W.D. Cotranslational folding stimulates programmed ribosomal frameshifting in the alphavirus structural polyprotein. J Biol Chem. 2020;295:6798–6808. doi: 10.1074/jbc.RA120.012706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J, O'Loughlin S, Atkins JF, Puglisi JD. The energy landscape of -1 ribosomal frameshifting. Sci Adv 2020;6:eaax6969. [DOI] [PMC free article] [PubMed]
- 21.Leger M., Sidani S., Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunelle M.N., Payant C., Lemay G., Brakier-Gingras L. Expression of the human immunodeficiency virus frameshift signal in a bacterial cell-free system: influence of an interaction between the ribosome and a stem-loop structure downstream from the slippery site. Nucl Acids Res. 1999;27:4783–4791. doi: 10.1093/nar/27.24.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plant E.P., Dinman J.D. Comparative study of the effects of heptameric slippery site composition on -1 frameshifting among different eukaryotic systems. RNA. 2006;12:666–673. doi: 10.1261/rna.2225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie D.B., Foster D.A., Woodside M.T. Programmed -1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc Natl Acad Sci USA. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie D.B., Soong J., Sikkema W.K., Woodside M.T. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J Am Chem Soc. 2014;136:2196–2199. doi: 10.1021/ja410344b. [DOI] [PubMed] [Google Scholar]
- 26.de Messieres M., Chang J.C., Belew A.T., Meskauskas A., Dinman J.D. Single-molecule measurements of the CCR5 mRNA unfolding pathways. Biophys J. 2014;106:244–252. doi: 10.1016/j.bpj.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moomau C., Musalgaonkar S., Khan Y.A., Jones J.E., Dinman J.D. Structural and Functional Characterization of Programmed Ribosomal Frameshift Signals in West Nile Virus Strains Reveals High Structural Plasticity Among cis-Acting RNA Elements. J Biol Chem. 2016;291:15788–15795. doi: 10.1074/jbc.M116.735613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhlmann M.M., Chattopadhyay M., Stupina V.A., Gao F., Simon A.E. An RNA Element That Facilitates Programmed Ribosomal Readthrough in Turnip Crinkle Virus Adopts Multiple Conformations. J Virol. 2016;90:8575–8591. doi: 10.1128/JVI.01129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie D.B., Cappellano T.R., Tittle C., Rezajooei N., Rouleau L. Conformational dynamics of the frameshift stimulatory structure in HIV-1. RNA. 2017;23:1376–1384. doi: 10.1261/rna.061655.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen K.K.Q., Gomez Y.K., Bakhom M., Radcliffe A., La P. Ensemble simulations: folding, unfolding and misfolding of a high-efficiency frameshifting RNA pseudoknot. Nucl Acids Res. 2017;45:4893–4904. doi: 10.1093/nar/gkx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu B., Zhang H., Sun R., Peng S., Cooperman B.S. Translocation kinetics and structural dynamics of ribosomes are modulated by the conformational plasticity of downstream pseudoknots. Nucl Acids Res. 2018;46:9736–9748. doi: 10.1093/nar/gky636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halma M.T.J., Ritchie D.B., Cappellano T.R., Neupane K., Woodside M.T. Complex dynamics under tension in a high-efficiency frameshift stimulatory structure. Proc Natl Acad Sci USA. 2019;116:19500–19505. doi: 10.1073/pnas.1905258116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omar S.I., Zhao M., Sekar R.V., Moghadam S.A., Tuszynski J.A. Modeling the structure of the frameshift-stimulatory pseudoknot in SARS-CoV-2 reveals multiple possible conformers. PLoS Comput Biol. 2021;17 doi: 10.1371/journal.pcbi.1008603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Tsai A., O'Leary S.E., Petrov A., Puglisi J.D. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol. 2012;22:804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noller H.F., Lancaster L., Zhou J., Mohan S. The ribosome moves: RNA mechanics and translocation. Nat Struct Mol Biol. 2017;24:1021–1027. doi: 10.1038/nsmb.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Pan X, Yan K, Sun S, Gao N, et al. Mechanisms of ribosome stalling by SecM at multiple elongation steps. eLife 2015;4:e09684. [DOI] [PMC free article] [PubMed]
- 37.Agirrezabala X., Samatova E., Klimova M., Zamora M., Gil-Carton D. Ribosome rearrangements at the onset of translational bypassing. Sci Adv. 2017;3 doi: 10.1126/sciadv.1700147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang K.C., Salawu E.O., Chang Y.Y., Wen J.D., Yang L.W. Resolution-exchanged structural modeling and simulations jointly unravel that subunit rolling underlies the mechanism of programmed ribosomal frameshifting. Bioinformatics. 2019;35:945–952. doi: 10.1093/bioinformatics/bty762. [DOI] [PubMed] [Google Scholar]
- 39.Rodnina M.V., Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 40.Geggier P., Dave R., Feldman M.B., Terry D.S., Altman R.B. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 42.Fischer N., Konevega A.L., Wintermeyer W., Rodnina M.V., Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 43.Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 44.Rodnina M.V., Savelsbergh A., Katunin V.I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 45.Wilden B., Savelsbergh A., Rodnina M.V., Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratje A.H., Loerke J., Mikolajka A., Brunner M., Hildebrand P.W. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohan S., Donohue J.P., Noller H.F. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci USA. 2014;111:13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J., Lancaster L., Donohue J.P., Noller H.F. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 50.Marshall R.A., Dorywalska M., Puglisi J.D. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J., Petrov A., Tsai A., O'Leary S.E., Puglisi J.D. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ermolenko D.N., Majumdar Z.K., Hickerson R.P., Spiegel P.C., Clegg R.M. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 53.Belardinelli R., Sharma H., Caliskan N., Cunha C.E., Peske F. Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol. 2016;23:342–348. doi: 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- 54.Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flanagan J.F.t., Namy O., Brierley I., Gilbert R.J.C. Direct observation of distinct A/P hybrid-state tRNAs in translocating ribosomes. Structure. 2010;18:257–264. doi: 10.1016/j.str.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatt P.R., Scaiola A., Loughran G., Leibundgut M., Kratzel A. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science. 2021 doi: 10.1126/science.abf3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bao C, Loerch S, Ling C, Korostelev AA, Grigorieff N, et al. mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding. eLife 2020;9:e55799. [DOI] [PMC free article] [PubMed]
- 58.Herr A.J., Atkins J.F., Gesteland R.F. Coupling of open reading frames by translational bypassing. Annu Rev Biochem. 2000;69:343–372. doi: 10.1146/annurev.biochem.69.1.343. [DOI] [PubMed] [Google Scholar]
- 59.Klimova M, Senyushkina T, Samatova E, Peng BZ, Pearson M, et al. EF-G-induced ribosome sliding along the noncoding mRNA. Sci Adv 2019;5:eaaw9049. [DOI] [PMC free article] [PubMed]
- 60.Joo C., Balci H., Ishitsuka Y., Buranachai C., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 61.Lerner E, Cordes T, Ingargiola A, Alhadid Y, Chung S, et al. Toward dynamic structural biology: Two decades of single-molecule Forster resonance energy transfer. Science 2018;359:eaan1133. [DOI] [PMC free article] [PubMed]
- 62.Qin P., Yu D., Zuo X., Cornish P.V. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 2014;15:185–190. doi: 10.1002/embr.201337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J., Petrov A., Johansson M., Tsai A., O'Leary S.E. Dynamic pathways of -1 translational frameshifting. Nature. 2014;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J., Coakley A., O'Connor M., Petrov A., O'Leary S.E. Coupling of mRNA Structure Rearrangement to Ribosome Movement during Bypassing of Non-coding Regions. Cell. 2015;163:1267–1280. doi: 10.1016/j.cell.2015.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C., Zhang H., Broitman S.L., Reiche M., Farrell I. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol. 2013;20:582–588. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim H.K., Tinoco I., Jr. EF-G catalyzed translocation dynamics in the presence of ribosomal frameshifting stimulatory signals. Nucl Acids Res. 2017;45:2865–2874. doi: 10.1093/nar/gkw1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amiri H., Noller H.F. Structural evidence for product stabilization by the ribosomal mRNA helicase. RNA. 2019;25:364–375. doi: 10.1261/rna.068965.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mustoe A.M., Busan S., Rice G.M., Hajdin C.E., Peterson B.K. Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell. 2018;173(181–95) doi: 10.1016/j.cell.2018.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belew A.T., Meskauskas A., Musalgaonkar S., Advani V.M., Sulima S.O. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Napthine S., Ling R., Finch L.K., Jones J.D., Bell S. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat Commun. 2017;8:15582. doi: 10.1038/ncomms15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qu X., Wen J.D., Lancaster L., Noller H.F., Bustamante C. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desai V.P., Frank F., Lee A., Righini M., Lancaster L. Co-temporal Force and Fluorescence Measurements Reveal a Ribosomal Gear Shift Mechanism of Translation Regulation by Structured mRNAs. Mol Cell. 2019;75:1007–1019. doi: 10.1016/j.molcel.2019.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 74.Wen J.D., Lancaster L., Hodges C., Zeri A.C., Yoshimura S.H. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amiri H., Noller H.F. A tandem active site model for the ribosomal helicase. FEBS Lett. 2019;593:1009–1019. doi: 10.1002/1873-3468.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurkcuoglu O., Doruker P., Sen T.Z., Kloczkowski A., Jernigan R.L. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys Biol. 2008;5 doi: 10.1088/1478-3975/5/4/046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bidou L., Stahl G., Grima B., Liu H., Cassan M. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 78.Telenti A., Martinez R., Munoz M., Bleiber G., Greub G. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem-loop structure. J Virol. 2002;76:7868–7873. doi: 10.1128/JVI.76.15.7868-7873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mouzakis K.D., Lang A.L., Vander Meulen K.A., Easterday P.D., Butcher S.E. HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome. Nucl Acids Res. 2013;41:1901–1913. doi: 10.1093/nar/gks1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collin D., Ritort F., Jarzynski C., Smith S.B., Tinoco I., Jr Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moffitt J.R., Chemla Y.R., Smith S.B., Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 82.Ritchie D.B., Woodside M.T. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr Opin Struct Biol. 2015;34:43–51. doi: 10.1016/j.sbi.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen G., Chang K.Y., Chou M.Y., Bustamante C., Tinoco I., Jr. Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc Natl Acad Sci USA. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Theimer C.A., Blois C.A., Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 85.Hansen T.M., Reihani S.N., Oddershede L.B., Sorensen M.A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhong Z., Yang L., Zhang H., Shi J., Vandana J.J. Mechanical unfolding kinetics of the SRV-1 gag-pro mRNA pseudoknot: possible implications for -1 ribosomal frameshifting stimulation. Sci Rep. 2016;6:39549. doi: 10.1038/srep39549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang L., Zhong Z., Tong C., Jia H., Liu Y. Single-Molecule Mechanical Folding and Unfolding of RNA Hairpins: Effects of Single A-U to A⋅C Pair Substitutions and Single Proton Binding and Implications for mRNA Structure-Induced -1 Ribosomal Frameshifting. J Am Chem Soc. 2018;140:8172–8184. doi: 10.1021/jacs.8b02970. [DOI] [PubMed] [Google Scholar]
- 88.Chen Y.T., Chang K.C., Hu H.T., Chen Y.L., Lin Y.H. Coordination among tertiary base pairs results in an efficient frameshift-stimulating RNA pseudoknot. Nucl Acids Res. 2017;45:6011–6022. doi: 10.1093/nar/gkx134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Halma M.T.J., Ritchie D.B., Woodside M.T. Conformational Shannon Entropy of mRNA Structures from Force Spectroscopy Measurements Predicts the Efficiency of -1 Programmed Ribosomal Frameshift Stimulation. Phys Rev Lett. 2021;126 doi: 10.1103/PhysRevLett.126.038102. [DOI] [PubMed] [Google Scholar]
- 90.Neupane K., Zhao M., Lyons A., Munshi S., Ileperuma S.M. Structural dynamics of the SARS-CoV-2 frameshift-stimulatory pseudoknot reveal topologically distinct conformers. bioRxiv. 2020 doi: 10.1101/2020.12.28.424630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim H.K., Liu F., Fei J., Bustamante C., Gonzalez R.L., Jr A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci USA. 2014;111:5538–5543. doi: 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caliskan N., Katunin V.I., Belardinelli R., Peske F., Rodnina M.V. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–1631. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu T, Kaplan A, Alexander L, Yan S, Wen JD, et al. Direct measurement of the mechanical work during translocation by the ribosome. eLife 2014;3:e03406. [DOI] [PMC free article] [PubMed]
- 94.Yan S., Wen J.D., Bustamante C., Tinoco I., Jr. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsu CF, Chang KC, Chen YL, Hsieh PS, Lee AI, et al. Formation of frameshift-stimulating RNA pseudoknots is facilitated by remodeling of their folding intermediates. Nucl Acids Res 2021;In press. https://doi.org/10.1093/nar/gkab512. [DOI] [PMC free article] [PubMed]
- 96.Puah R.Y., Jia H., Maraswami M., Kaixin Toh D.F., Ero R. Selective Binding to mRNA Duplex Regions by Chemically Modified Peptide Nucleic Acids Stimulates Ribosomal Frameshifting. Biochemistry. 2018;57:149–159. doi: 10.1021/acs.biochem.7b00744. [DOI] [PubMed] [Google Scholar]
- 97.Yang L., Toh D.K., Krishna M.S., Zhong Z., Liu Y. Tertiary Base Triple Formation in the SRV-1 Frameshifting Pseudoknot Stabilizes Secondary Structure Components. Biochemistry. 2020;59:4429–4438. doi: 10.1021/acs.biochem.0c00611. [DOI] [PubMed] [Google Scholar]
- 98.White K.H., Orzechowski M., Fourmy D., Visscher K. Mechanical unfolding of the beet western yellow virus -1 frameshift signal. J Am Chem Soc. 2011;133:9775–9782. doi: 10.1021/ja111281f. [DOI] [PubMed] [Google Scholar]
- 99.Zhang X., Xu X., Yang Z., Burcke A.J., Gates K.S. Mimicking Ribosomal Unfolding of RNA Pseudoknot in a Protein Channel. J Am Chem Soc. 2015;137:15742–15752. doi: 10.1021/jacs.5b07910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X., Zhang D., Zhao C., Tian K., Shi R. Nanopore electric snapshots of an RNA tertiary folding pathway. Nat Commun. 2017;8:1458. doi: 10.1038/s41467-017-01588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mikl M., Pilpel Y., Segal E. High-throughput interrogation of programmed ribosomal frameshifting in human cells. Nat Commun. 2020;11:3061. doi: 10.1038/s41467-020-16961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park S.J., Kim Y.G., Park H.J. Identification of RNA pseudoknot-binding ligand that inhibits the -1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J Am Chem Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 103.Hung M., Patel P., Davis S., Green S.R. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J Virol. 1998;72:4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brakier-Gingras L., Charbonneau J., Butcher S.E. Targeting frameshifting in the human immunodeficiency virus. Expert Opin Ther Targets. 2012;16:249–258. doi: 10.1517/14728222.2012.665879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dinman J.D., Ruiz-Echevarria M.J., Peltz S.W. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Y., Tao H., Shen S., Miao Z., Li L. A drug screening toolkit based on the -1 ribosomal frameshifting of SARS-CoV-2. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kelly J.A., Woodside M.T., Dinman J.D. Programmed -1 Ribosomal Frameshifting in coronaviruses: A therapeutic target. Virology. 2020;554:75–82. doi: 10.1016/j.virol.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kwak H., Park M.W., Jeong S. Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang X., Xuan Y., Han Y., Ding X., Ye K. Regulation of HIV-1 Gag-Pol Expression by Shiftless, an Inhibitor of Programmed -1 Ribosomal Frameshifting. Cell. 2019;176:625–635. doi: 10.1016/j.cell.2018.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat Microbiol. 2021;6:339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Comstock M.J., Ha T., Chemla Y.R. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Loveland A.B., Demo G., Korostelev A.A. Cryo-EM of elongating ribosome with EF-Tu·GTP elucidates tRNA proofreading. Nature. 2020;584:640–645. doi: 10.1038/s41586-020-2447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]