Abstract
Most COVID-19 vaccines are designed to elicit immune responses, ideally neutralizing antibodies (NAbs), against the SARS-CoV-2 spike protein. Several vaccines, including mRNA, adenoviral-vectored, protein subunit and whole-cell inactivated virus vaccines, have now reported efficacy in phase III trials and have received emergency approval in many countries. The two mRNA vaccines approved to date show efficacy even after only one dose, when non-NAbs and moderate T helper 1 cell responses are detectable, but almost no NAbs. After a single dose, the adenovirus vaccines elicit polyfunctional antibodies that are capable of mediating virus neutralization and of driving other antibody-dependent effector functions, as well as potent T cell responses. These data suggest that protection may require low levels of NAbs and might involve other immune effector mechanisms including non-NAbs, T cells and innate immune mechanisms. Identifying the mechanisms of protection as well as correlates of protection is crucially important to inform further vaccine development and guide the use of licensed COVID-19 vaccines worldwide.
Subject terms: Vaccines, Vaccines, SARS-CoV-2, Translational research, Adaptive immunity
This Progress article summarizes our current understanding of the immune mechanisms of protection induced by the available COVID-19 vaccines. The authors compare vaccine-induced antibody responses following one or two doses of different vaccines and consider the relative importance of neutralizing antibodies for vaccine-mediated protection against SARS-CoV-2.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), caused more than 3 million deaths worldwide in the 16 months since it was identified in December 2019 (refs1–3). It was evident early on that the pandemic could only be controlled with effective vaccines. This resulted in rapid vaccine development, with limited insight into what would constitute protective immunity. Currently licensed vaccines for COVID-19 are based on experience with SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV); but although multiple SARS-CoV and MERS-CoV vaccine candidates were developed, none had advanced beyond phase I clinical trials4. There are currently >270 candidate COVID-19 vaccines in development, including >90 in clinical trials5–7. These include nucleic acid vaccines (RNA and DNA)8–11, human and simian replication-deficient and replication-competent adenoviral-vectored vaccines12,13, whole-cell inactivated virus14,15, subunit protein vaccines16 and virus-like particles6. As of April 2021, 28 of these vaccines have entered phase III clinical trials, and 5 (Table 1) have reported efficacy in the peer-reviewed literature and/or through detailed publicly available reports submitted to regulatory authorities, resulting in emergency authorizations for their use in a large number of countries. These include the mRNA vaccines BNT162b2 (Pfizer/BioNTech) and mRNA-1273 (Moderna), and the three adenoviral-vectored vaccines ChAdOx1 nCoV-19 (University of Oxford/AstraZeneca), Gam-COVID-Vac (Gamaleya Research Institute) and Ad26.COV2.S (Janssen). One protein subunit vaccine (NVX-CoV2372; Novavax) and one whole-cell inactivated viral vaccine (BBV152; Bharat Biotech) have reported positive efficacy results via official company press releases (Table 1), and BBV152 has received emergency authorization in several countries. A further four vaccines have suggested positive efficacy via media reports — the adenoviral-vectored vaccine Ad5-nCoV (CanSino Biologics) and the whole-cell inactivated vaccines CoronaVac (Sinovac Biotech), BBIBP-CorV (Sinopharm) and WIBP-CorV (Sinopharm) (Table 1) — and these plus another three with no publicly available efficacy data — EpiVacCorona peptide vaccine (VECTOR Center of Virology, Russia), CoviVac inactivated vaccine (Chumakov Centre, Russia) and ZF2001 recombinant vaccine (Anhui Zhifei Longcom/Chinese Academy of Sciences) — have received emergency authorizations in some countries17. This represents a remarkable feat for biomedical science, but there are many outstanding issues. For example, most approved vaccines are believed to require two doses for optimal protection, as do the majority of the other vaccines that are still in clinical development, which translates into logistical challenges and a slower roll-out. In addition, logistical hurdles posed by the requirement for cold chains, and in particular the ultra-cold chains required for mRNA-based vaccines, impede the roll-out of the currently licensed vaccines in low and middle-income countries. Furthermore, the ongoing evolution of this virus generates mutations that can reduce vaccine-induced immunity18. Although there is no evidence to date of an ongoing ‘antigenic drift’, such as that observed with influenza virus, mutations affecting transmission and disease severity can occur19, and vaccine-induced immune selection pressure at a population level may accelerate the development of escape mutants as has been suggested for other pathogens20,21. Vaccines for COVID-19 must therefore continue to be optimized as a matter of urgency.
Table 1.
Human studies of COVID-19 vaccines with reported efficacy
| Vaccine (developer) (dosing regimen) | Formulation | Efficacy against symptomatic infection (phase III trials) | Effectiveness (post implementation) | Antibody responses in humans | T cell responses in humans |
|---|---|---|---|---|---|
| mRNA | |||||
| BNT162b2 mRNA (BioNTech/Pfizer) (30 μg mRNA, 2 doses, 21 days apart)81 | mRNA-lipid nanoparticle encoding full-length S protein, modified by two proline mutations to lock protein in the pre-fusion conformation110–112 | 95% after 2 doses; 52% after 1 dose81, although review of the data suggests efficacy of 93% 14 days after 1 dose113, 91% at 6 months post second dose114 |
Symptomatic infection: 94–96% (2 doses) and 46–80% (1 dose) Any infection: 86–92% (2 doses) and 46–72% (1 dose) Hospitalization: 87% (2 doses) and 71–85% (1 dose) Asymptomatic infection: 79% (1 dose) and 90% (2 doses)93–95,115–123 |
S1-binding antibody present after first dose, responses increased following the second dose124; significant NAb was only present after second dose124 | Increases in antigen-specific IFNγ+ CD4+ and CD8+ T cells after second dose125; predominance of IFNγ and IL-2 secretion, compared with IL-4, suggesting TH1 cell polarization |
| mRNA-1273 (Moderna) (100 μg mRNA, 2 doses, 28 days apart) | mRNA-lipid nanoparticle encoding full-length S protein, modified by two proline mutations to lock protein in the pre-fusion conformation126 | 95% after 2 doses; 92% after 1 dose82 | Symptomatic infection: 90% (2 doses) and 80% (1 dose)123 | S-binding antibody detected 14 days after first dose, levels increased slightly by 28 days, with marked increase after second dose126; minimal NAb present after first dose, peak at 14 days after second dose127 | Significant increases in CD4+ T cells secreting TH1 type cytokines (TNF > IL-2 > IFNγ) after second dose, small increases in TNF-secreting and IL-2-secreting cells after first dose126; minimal change in TH2 cell responses; low levels of CD8+ responses126 |
| Viral vector | |||||
| ChAdOx1 nCoV-19 (University of Oxford/Astra-Zeneca) (2.5–5 × 1010 viral particles, 2 doses, ≥28 days apart)87 | Recombinant, replication-deficient simian adenovirus vector expressing the full-length S protein with a tPA leader sequence128 | 62–67% after 2 doses87,76% after 1 dose88; 90% in participants who received a low dose followed by a high dose; interval between doses varied with a median of 36-69 days; 81% with ≥12-week interval, 55% with <6-week interval88 | Hospitalization: 80–94% after 1 dose95,115 | S-binding antibody present 14 days after first dose, levels increased by 28 days128; marked increase after second dose, peak at 14 days after second dose; predominantly IgG3 and IgG1 (ref.129); significant NAb detected after first dose, increased by 14 days after second dose; IgG avidity increased 28–56 days after single dose129; peak IgM and IgA responses at day 14 or 28 | Peak T cell responses 14 days after first dose, but slightly higher responses measured 28 days after second dose128; increase in TNF and IFNγ production by CD4+ T cells at day 14 |
| Gam-COVID-Vac (Gamaleya Research Institute) (1011 viral particles, 2 doses, 21 days apart)90 | Recombinant, replication-deficient human adenovirus 26 (dose 1) and human adenovirus 5 (dose 2) expressing full-length S protein130 | 91% after 2 doses; 74% after 1 dose (moderate to severe infection)90 | – | S-binding antibody detected in 85–89% and NAb in 61% of individuals 14 days after first dose130; S antibody levels (binding and neutralizing) boosted by second dose, with binding antibody in 98% and neutralizing antibody in 95% of individuals 14 days after second dose90 | CD4+ and CD8+ T cell responses observed by 14 days after first dose (based on proliferation assays and antigen-specific IFNγ secretion)130; all individuals had S-specific IFNγ responses 7 days after second dose based on in vitro stimulation of PBMCs |
| Ad26.COV2.S (Janssen) (5 × 1010 viral particles, 1 dose)89,131 | Recombinant, replication-deficient human adenovirus 26 expressing full-length S protein with two amino acid changes in S1/S2 junction that delete furin cleavage site and two proline substitutions in hinge region that lock protein in the pre-fusion conformation132 | 67% after 1 dose89 | – | S-binding and neutralizing antibody present by 28 days after vaccination in 99% of individuals and antibody levels sustained until at least 84 days post vaccination89,131 | CD4+ and CD8+ T cell responses present at 14 and 28 days post vaccination, based on presence of CD4+ and CD8+ T cells secreting IFNγ and/or IL-2 and not IL-4 or IL-3, suggesting TH1 cell polarization of the CD4+ T cell response89,131 |
| Ad5-nCoV (CanSino Biologics) (5 × 1010 viral particles, 1 dose)91 | Recombinant, replication-deficient human adenovirus 5 expressing full-length S protein with a tPA leader sequence12 | 66% after 1 dose, decreasing to 50% by 5–6 months post immunization133,134 | – | 14 days after vaccination, 44% of individuals had RBD-binding antibodies; 28 days after vaccination, 97% had anti-RBD binding antibodies and 47–50% had NAbs; individuals with pre-existing anti-Ad5 antibody titre >1:200 had reduced levels of both binding antibodies and NAbs12,91 | 28 days after vaccination, 78–88% of participants had T cell responses, based on IFNγ ELIspot, although peak T cell responses were observed at day 14 after vaccination12,91 |
| Protein subunit | |||||
| NVX-CoV2373 (Novavax) (5 µg protein, 2 doses, 21 days apart)135 | Recombinant nanoparticle of full-length S protein with mutations at the S1/S2 cleavage sites to confer protease resistance and two proline substitutions to stabilize protein in a pre-fusion conformation, with saponin-based adjuvant (Matrix-M1)136 | 90% by 7 days after second dose137 | – | S-binding antibody detected 21 days after first dose, with a marked increase after the second dose; some NAb present after the first dose, with a significant increase by 7 days after second dose136 | CD4+ T cell responses present by 7 days after second dose, based on IFNγ, IL-2 and TNF production in response to S protein stimulation, with a strong bias towards a TH1 cell phenotype; minimal TH2 cell responses (as measured by IL-5 and IL-13)136 |
| Whole-cell inactivated virus | |||||
| CoronaVac (Sinovac Biotech) (3 µg protein, 2 doses, 14–28 days apart)138,139 | SARS-CoV-2 grown in Vero cells, inactivated with β-propiolactone and adsorbed onto aluminium hydroxide138 | 50–84% after 2 doses140,141 | – | By day 28 day after second dose, RBD-specific binding antibody detected in 88–97% of participants with a 14-day dosing interval and 99–100% with a 28-day interval; NAb present in 94–100% of individuals 28 days after second dose138,139 | – |
| BBIBP-CorV (Sinopharm) (4 µg protein, 2 doses, 21 days apart)142 | SARS-CoV-2 grown in Vero cells, inactivated with β-propiolactone and adsorbed onto aluminium hydroxide142 | 86% after 2 doses143 | – | By day 14 after second dose, 46–87% of individuals had binding antibodies; this increased to 92–100% by day 28; all recipients had NAbs by 21 days after second dose142 | – |
| WIBP-CorV (Sinopharm) (5 µg protein, 2 doses, 21 days apart)144 | SARS-CoV-2 grown in Vero cells, inactivated with β-propiolactone and adsorbed onto aluminium hydroxide144 | 73% after 2 doses145 | – | By day 14 after second dose, 100% of participants had binding antibodies against whole inactivated SARS-CoV-2 and 98% had neutralizing antibodies144 | – |
| BBV152 (Bharat Biotech) (6 µg protein, 2 doses, 28 days apart)146 | SARS-CoV-2 grown in Vero cells, inactivated with β-propiolactone and adsorbed onto aluminium hydroxide and an imidazoquinoline molecule (TLR7/TLR8 agonist)146 | 78% after 2 doses147 | – | After first dose, 65% of participants had anti-S binding antibodies, increasing to 98% by day 14 after second dose; 48% had NAbs after first dose, increasing to 97% by day 14 after second dose; GMTs for binding and NAbs markedly increased by second dose100,146 | Strong bias towards a TH1 cell phenotype (IFNγ and TNF), with minimal TH2 cell responses (as measured by IL-5 and IL-13) after in vitro stimulation. Increase in CD4+CD45RO+ memory T cells by day 76 after second dose100,146 |
ELISpot, enzyme-linked immunosorbent spot; GMT, geometric mean titre; IFNγ, interferon-γ; IL-2, interleukin-2; NAb, neutralizing antibody; PBMC, peripheral blood mononuclear cell; RBD, receptor-binding domain; S, spike; TH1 cell, T helper 1 cell; TLR, Toll-like receptor; TNF, tumour necrosis factor; tPA, tissue plasminogen activator.
Here, we provide a brief overview of the immune response to SARS-CoV-2, followed by a discussion of the mechanisms of immune protection of the five vaccines for which detailed results from phase III trials are publicly available. We then discuss how insights into vaccine-induced immune protection and the identification of correlates of protection may be used to guide vaccine development and speed up the licensing of the next generation of vaccines.
Immune responses to SARS-CoV-2
Recovery following infection with SARS-CoV-2 in humans appears to involve both humoral and cell-mediated immunity22–24. In patients hospitalized with COVID-19, the early presence of broadly functional antibodies directed at the SARS-CoV-2 spike (S) protein (see also Box 1) correlated with survival25, and S-protein-targeted neutralizing antibodies (NAbs) are present in the majority of individuals following infection. The magnitude of these NAb responses appear to correlate with viral load, with higher responses reported in patients with more severe disease, and in older adults compared with younger adults26–29. In early studies of SARS-CoV-2 vaccine candidates in a rhesus macaque model, the amount of NAbs directed at the S protein, which mediates cellular binding, emerged as the strongest correlate of protection10,30. This led to a general acceptance that it was imperative for vaccines to elicit NAb responses. Non-NAbs can also have an important role in protection, however, via Fc-mediated effector functions including antibody-dependent phagocytosis, antibody-dependent cellular cytotoxicity and antibody-dependent natural killer cell activation10,25,31. On the other hand, antibodies promoting inflammatory responses may contribute to a cytokine storm resulting in severe disease32,33. The complexity of antibody-dependent effector functions and their relationships with the structure of the Fc component of IgG, including subclass and glycosylation, can be assessed by system serology approaches, allowing multivariate analyses of correlates of immunity and disease34.
Although mucosal immunity is likely key to the prevention of SARS-CoV-2 infection, relatively little is known regarding mucosal antibody responses in COVID-19. Historical studies of controlled human infection with endemic coronaviruses indicated that levels of nasal IgA correlate with protection against these infections35,36. SARS-CoV-2-specific IgA is detected in nasal washes and in saliva of patients who are in convalescence and could contribute to a reduced interpersonal spread through neutralization and Fc-dependent effector functions37.
SARS-CoV-2 is able to spread from cell to cell without exposure to the extracellular environment38, and it is therefore possible that antibodies that only target intact extracellular viral particles have a limited role in reducing viral spread within the host. As expected for a viral infection, T cells are also important mediators in the host response to SARS-CoV-2 infection, by killing infected cells, supporting B cell function and antibody responses, and, possibly, reducing the risk of vaccine-induced enhanced disease39,40 (see Box 2). Both reduced and increased CD8+ and CD4+ T cell responses have been observed following infection41. Milder disease and recovery have been associated with a more robust clonal expansion of CD8+ T cells in both the lungs and blood42,43, although whether this is the cause of milder disease or an effect of recovery is unclear. Virus-specific CD8+ and CD4+ T cells, including CD8+ memory T cells, are present in patients who have recovered from COVID-19 (refs44,45), but their importance in protection against future infection and/or severe disease remain uncertain44,46–48. Interferon-γ (IFNγ)-producing T helper 1 cells (TH1 cells) are produced during acute infection, and it has been suggested that this TH1 cell-biased phenotype is associated with less severe disease24,49 — an important consideration given that current COVID-19 vaccines have been designed to induce responses skewed towards the TH1 cell phenotype (Table 1). There are indications that individuals with higher levels of IFNγ-secreting T cells (measured by enzyme-linked immunosorbent spot) against the S protein, nuclear proteins and membrane proteins of SARS-CoV-2 may have better protection from disease50. Moreover, individuals with mild disease favour more efficient T follicular helper cell responses in the germinal centre, which supports an increase in plasmablast numbers and enhances antibody production51.
Studies showed that adoptive transfer of antigen-specific T cells protected immunodeficient mice from infection after challenge with the SARS-CoV-2-related coronaviruses SARS-CoV and MERS-CoV52. The passive transfer of NAbs was also found to be protective in non-human primate models, whereas removal of CD8+ T cells in the same models impaired protection, suggesting a role for both components53. Evidence from human and animal studies has suggested that in addition — or, possibly, instead of high titres of NAbs — a robust cytotoxic CD8+ T cell response and a TH1 cell-biased CD4+ T cell effector response would result in protective immunity against COVID-19 (ref.54).
Like other pathogenic respiratory RNA viruses (including other coronaviruses, respiratory syncytial virus and enteroviruses)55, SARS-CoV-2 can evade innate immune responses via multiple mechanisms55,56, indicating that innate immunity is likely crucial for host protection54,57. A predominant strategy appears to be the inhibition of the type I interferon response58–60 at multiple points, including impaired recognition of viral RNA61,62, decreased nuclear translocation of pro-inflammatory transcription factors (such as IRF3, IRF7 and STAT1)61,63 and suppression of STAT1 and STAT2 phosphorylation64,65. Furthermore, humans deficient in producing or responding to type I interferon have an increased risk of severe COVID-19 (refs66–68). Although it is likely that there are many innate immune components that are relevant to protection from COVID-19, type I and type III interferons appear centrally important69,70. The timing of induction of type I interferon (or type III interferon in mucosal tissue) is crucial as the presence of type I interferon early in infection appears to be protective, whereas its relevance for viral control at later time points may be reduced or may even contribute to immunopathology57,71,72.
Box 1 The SARS-CoV-2 spike protein as vaccine target.
Most candidate COVID-19 vaccines are designed to elicit immune responses, ideally mediated by neutralizing antibodies (NAbs), against the trimeric SARS-CoV-2 spike (S) protein. The S protein is a class I fusion protein that facilitates binding of the virus to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface, which triggers fusion between the virus and cell membrane110,148,149. Prior to contact with the host cell, the S protein is in a metastable pre-fusion conformation and the trimer undergoes substantial rearrangement at the time of virus–cell fusion150,151. Some COVID-19 vaccines include mutations that stabilize the S protein in this pre-fusion form, on the basis that this is the expected conformation prior to epithelial cell attachment, and thus immune responses directed against the pre-fusion S protein are more likely to be protective and reduce transmission. However, vaccines without this stabilization (for example, ChAdOx1 nCoV-19) have proven efficacy and so the importance of this for COVID-19 vaccines is uncertain. The S protein consists of an amino-terminal S1 subunit and a carboxy-terminal S2 subunit. Within the S1 subunit lies the receptor-binding domain (RBD), which binds the ACE2 receptor, and this domain can undergo conformational rearrangement that transiently hides or exposes the determinants of receptor binding. Eliciting an immune response that targets the RBD has been a major focus of vaccine development on the assumption that antibodies that bind this critical domain can prevent viral entry into host cells, thereby allowing for sterilizing immunity (that is, the complete prevention of infection)152–154, which would support herd immunity if vaccine coverage is sufficiently high155. Other S protein epitopes can also be valuable vaccine targets. A polyclonal antibody against multiple epitopes of the S protein beyond the RBD might, for example, inhibit viral attachment156–160, provide additional neutralizing activity161 and/or prevent post-attachment fusion162. A vaccine targeting multiple epitopes would also mitigate the possibility of immune escape by mutation156. There is currently no defined correlate of protection against COVID-19 or SARS-CoV-2 infection, and as such the immunological thresholds required for vaccine efficacy have not yet been defined163.
Box 2 Antibody-dependent enhancement and vaccine-induced enhanced disease.
One of the concerns that was raised early in the pandemic was that any vaccines used in humans could lead to enhanced disease in individuals who were infected following vaccination164. Postulated mechanisms for this were antibody-dependent enhancement (ADE), where SARS-CoV-2 is enabled to enter cells not expressing the angiotensin-converting enzyme 2 (ACE2) receptor via Fc-mediated attachment in the presence of binding, non-neutralizing antibody (non-NAb); or vaccine-associated enhanced respiratory disease (VAERD) via immune complex deposition or ‘aberrant’ T cell responses in the lungs of individuals who are vaccinated164. In animal models of infections with other coronaviruses and for previous whole-cell inactivated vaccines developed for respiratory syncytial virus and measles virus, VAERD has usually been associated with the development of T helper 2 cell (TH2 cell)-biased CD4+ T cell responses, with significantly increased levels of interleukin-4 (IL-4), IL-5 and/or IL-13 (refs12,126,165–174). By contrast, VAERD has not usually occurred if a TH1 cell-biased CD4+ T cell response is elicited, reflected by increased levels of interferon-γ (IFNγ), IL-2 and tumour necrosis factor (TNF)44–46,49. COVID-19 vaccines developed to date have therefore attempted to elicit either a TH1 cell-biased response or a balanced TH1 cell/TH2 cell response. Current evidence indicates that SARS-CoV-2 does not productively infect macrophages, making enhancement of infection unlikely, and ADE and VAERD have so far not been demonstrated for SARS-CoV-2 (refs175,176). However, the potential role of IgG with reduced Fc fucosylation and enhanced binding to FcγRIII in the development of severe COVID-19 suggests that the quality of antibodies induced by vaccination may also be important in minimizing the risk of ADE32,33. There is also a possibility that the risk of ADE may be higher in the presence of low to moderate amounts of antibody, which will occur as antibody levels wane post immunization177, although recurrent SARS-CoV-2 infections have not been consistently associated with severe disease.
Insights into vaccine-induced immunity
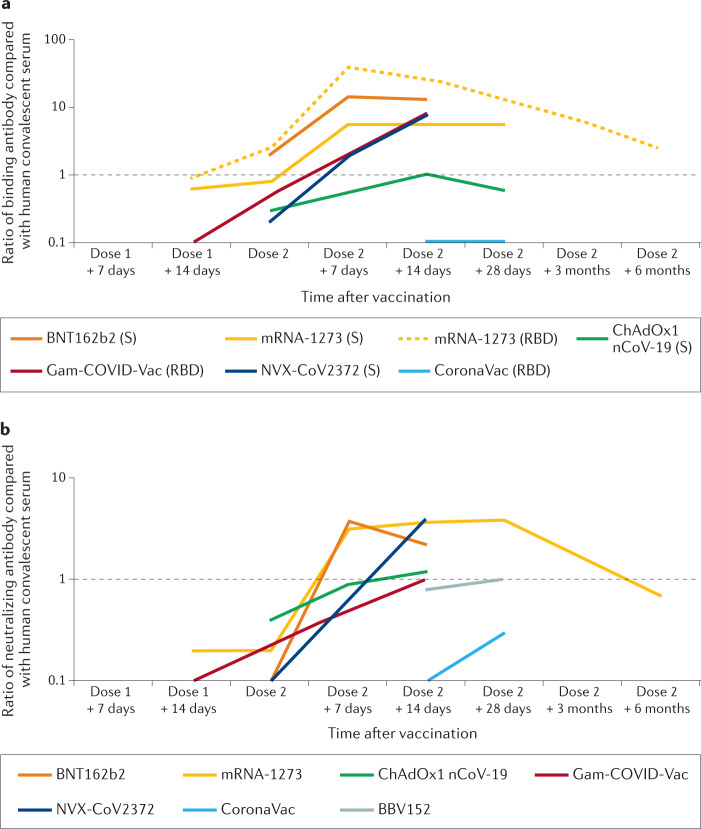
In order to understand how vaccine-induced immune responses relate to protection against disease for COVID-19, it is important to consider the available immunologic data within the context of vaccine efficacy from similar populations (Supplementary Figure 1) — this Progress article therefore focuses on the five vaccines for which both detailed efficacy and immunological data are available. For completeness, data from other vaccines with reported efficacy are included in the tables and figures. Few immunological data have so far been published from the phase III trials in which vaccine efficacy was determined, and the assessment of human immunologic responses to vaccination are therefore largely reliant on analyses from the earlier phase I/II clinical trials. Some of these studies used multiple different formulations and/or different antigens to the final formulations included in phase III trials, so the descriptions below are focused on the formulations that were used in subsequent clinical trials and for which efficacy has been established (Table 1). Although numerous studies have reported vaccine effectiveness and immunologic evaluations from initial mass vaccination campaigns, prioritization of older individuals and groups with high-risk medical conditions means that most of these data do not improve our ability to link the immunological data with clinical outcomes. Although antigen-specific antibodies (including NAbs) and T cell responses have been determined for all of the vaccines discussed here (Table 1), the specific assays have varied and, thus, are not directly comparable. However, most of the studies used previously established assays to analyse samples from patients who are in convalescence after SARS-CoV-2 infection. Although the source of the convalescent samples differed between studies (for example, asymptomatic versus mild versus severe disease), these data provide the basis on which comparisons between studies can be made (Fig. 1; see Supplementary Table 1).
Fig. 1. Comparison of antibody responses induced by different COVID-19 vaccines.
a | Vaccine immunogenicity based on antibody against the spike (S) protein of SARS-CoV-2 and/or against the receptor-binding domain (RBD) of the S protein relative to levels seen in convalescent serum. b | Vaccine immunogenicity based on neutralizing antibodies (NAbs) against SARS-CoV-2, again relative to levels seen in convalescent plasma. Relative antibody levels induced are indicated for seven COVID-19 vaccines where these data are available. To enable direct comparison, only vaccines tested in two-dose schedules are included. For antibody data, all comparisons are based on relative amount of antibody compared with human convalescent serum used in the same study (see data in Supplementary Table 1). Although the source of these samples differed between studies, these data enable reasonable direct comparisons between different vaccines, accounting for different assays used in different trials. In parts a,b, a ratio of one (black horizontal dashed line) indicates equivalence in amount of antibody between individuals who are vaccinated and average value for the relevant human convalescent serum — note logarithmic vertical axes. Where a range of data were reported for a specific parameter at a given time point (for example, between different age groups), the maximum reported value was used.
mRNA vaccines
Both BNT162b2 and mRNA-1273 have demonstrated very high efficacy in clinical trials, including >90% protection from symptomatic disease after only a single dose, when levels of NAbs are <5% of the post-second dose peak (Table 1; see Supplementary Figure 1). mRNA-1273 was shown to elicit TH1 cell responses after the first dose, with 0.05% of circulating CD4+ T cells secreting tumour necrosis factor (TNF) and/or interleukin-2 (IL-2) following in vitro stimulation with S protein peptides73 (Table 1; see Supplementary Table 1); and both vaccines, after just a single dose, induced levels of anti-S and/or anti-receptor-binding domain (anti-RBD) binding antibodies that were equivalent to or higher than those observed in patients who are in convalescence (Fig. 1; see Supplementary Table 1). By contrast, relatively low levels of CD8+ T cell responses are elicited after one or two doses (Table 1; see Supplementary Table 1). These data would suggest that protection after one dose of these vaccines either requires extremely low levels of NAbs, is the result of non-NAbs leading to other effector mechanisms and/or is mediated by a relatively low frequency of antigen-specific T cells. Alternatively, it is possible that there are non-adaptive (that is, innate) immune mechanisms that are responsible for this early protection after vaccination, for example via type I or type III interferons57,71,72, with the possibility of a ‘trained immunity’-type effect that has been described for the Bacillus Calmette–Guérin (BCG) vaccine in the COVID-19 context74. Both BNT162b2 and mRNA-1273, as well as the adenoviral-vectored vaccine ChAdOx1 nCoV-19, have been shown to induce type I interferon, thus potentially inducing pathogen-agnostic protection75–77. Unfortunately, given the urgency of the situation, the trials of the currently licensed COVID-19 vaccines did not include a vector control (such as scrambled mRNA or simian adenovirus without the S protein), and therefore the role of pathogen-agnostic immunity in humans cannot be assessed with the currently available data; however, antigen non-specific stimulation of type I interferon pathways has been demonstrated with other formulations of mRNA in animal models78,79. If such a mechanism was occurring, there would be a possibility of protection against pathogens other than SARS-CoV-2, and it is vital that any data collected on other infections in the trials are analysed to evaluate this possibility. It must also be considered that mechanisms of protection may differ after two doses versus one dose, where NAbs may mediate the predominant protective mechanism following subsequent doses of vaccine. Data from medium-term follow-up of individuals after infection suggest that T cell responses wane more rapidly than antibody responses80. Therefore, if there are different mechanisms of protection involved after one versus two doses, a complete understanding of this will enable decisions on intervals between doses to be made on a scientific basis — current guidelines already vary between countries, with intervals of 21–28 days used in the vaccine trials81,82, a recommendation of 6 weeks from the WHO (World Health Organization)83 and up to 12 or 16 weeks in the UK and Canada, respectively84,85. This will also be critical in determining the timing of any future booster doses that may be needed, to ensure that these can be given before protection has waned. The role of B cell memory is also critical — a study comparing individuals who are SARS-CoV-2 naive and individuals who have recovered from SARS-CoV-2 identified that either prior infection or vaccination with BNT162b2 was able to efficiently prime memory B cell responses, such that the second exposure (first vaccine dose after previous infection or second vaccine dose in individuals who are SARS-CoV-2 naïve) resulted in boosting of memory B cell responses86. This may enable sparing of vaccine doses by recommending only one dose in individuals who had been previously infected, although the critical interval between infection and efficient boosting requires further investigation. Finally, an additional potential advantage of mRNA vaccines compared with repeated homologous administration of viral-vectored vaccines is that anti-vector immunity will not be a potential issue that may result in attenuation of responses to booster doses.
Adenoviral-vectored vaccines
It is apparent from the data available that there are both similarities and differences between the mRNA vaccines and the adenoviral-vectored vaccines. For protection against symptomatic COVID-19 infection, the mRNA vaccines have an efficacy of ~95% in clinical trials after two doses81,82, whereas data from the viral-vectored vaccines are mixed. There was ~70% efficacy for ChAdOx-1 nCoV-19 (after one or two doses)87,88 and Ad26.COV2.S (after one dose)89 (Table 1). Additional benefit of a second dose is evident for Gam-COVID-Vac, when efficacy is ~90%87–90 (Table 1; see Supplementary Figure 1). This may be, in part, due to the fact that Gam-COVID-Vac uses different adenovirus vectors for each dose (adenovirus 26 for dose one and adenovirus 5 for dose two), thus circumventing the potential problem of anti-vector immunity that could inhibit anti-S responses, as has been identified for Ad5-nCoV (refs12,91) (Table 1) and non-COVID-19 adenovirus-based vaccines92. It should be noted that high effectiveness (>80%) against severe disease and hospitalization has been reported for both BNT162b2 (refs93,94) and ChAdOx1 nCoV-19 (ref.95). Both mRNA and adenoviral-vectored vaccines, after two doses, elicit levels of NAbs that are equivalent to or higher than those seen in patients who are in convalescence (Fig. 1), although the level of NAbs induced seems to be relatively higher with the mRNA vaccines. One dose of ChAdOx1 nCov-19 was shown to elicit polyfunctional antibodies, which are capable of mediating neutralization and multiple other antibody-dependent effector mechanisms — all of which may contribute to protection against disease. ChAdOx1 nCov-19-induced antibodies were shown to facilitate monocyte-mediated and neutrophil-mediated phagocytosis. Both functions were already induced following only one dose, although they were substantially increased by the second96. The first dose of ChAdOx1 nCov-19 also induces antibodies capable of antibody-dependent complement deposition; again, this functionality was increased following a second dose96. In addition, this vaccine induced potent T cell responses that peaked at 14 days after a single dose, based on production of TNF and IFNγ from CD4+ T cells upon antigen stimulation in vitro (Table 1; see Supplementary Figure 1). The similar efficacy after one and two doses of this vaccine, despite decreased T cell responses and increased antibody responses after the second dose, suggests that different protective mechanisms may therefore be prominent after one compared with two doses. Increased immunogenicity and efficacy was observed with increasing interval between doses for the ChAdOx1 nCoV-19 vaccine88, and this strategy may therefore result in better protection after two doses and could be considered for other vaccines. In the long term, a strategy involving homologous prime–boost with identical viral-vectored vaccines may be limited by anti-vector immunity12,91,92. Heterologous prime–boost strategies, such as that employed with Gam-COVID-Vac or based on using combinations of different vaccines, may be able to overcome this issue.
Future of COVID-19 vaccine development
Although the speed of vaccine development for COVID-19 already represents a remarkable landmark, the conceptual breakthroughs now appearing on the horizon — for example, data showing that protective mechanisms beyond NAb are likely to be important — will produce further monumental achievements. In order to identify correlates and mechanisms of protection without a massive financial outlay and substantial delay, we need to fully utilize the existing data via a data-driven approach to carefully assess which immunological pathways are associated with protection against COVID-19. Specifically, the trials leading to licensure of the current vaccines have already collected biological samples, analysis of which will usher in a revolution in our understanding of host responses. An initial analysis from trials of seven current vaccines has suggested that anti-S antibody is a reasonable correlate of protection — a robust correlation was reported between NAb titre and vaccine efficacy (rank correlation rs = 0.79) and between anti-S binding antibody titre and efficacy (rs = 0.93) after a complete vaccine series (one or two doses, depending on the vaccine)97. However, this analysis did not fully consider efficacy and immune responses after one dose for the two-dose vaccines, or T cell responses. In addition, these analyses were based on short-term efficacy over 2–3 months and correlation with longer-term outcomes will also be necessary. To accurately identify a correlate and/or mechanism of protection against COVID-19, trial samples will need to be analysed in an unbiased manner (that is, not just focused on NAbs or antibodies, or even just adaptive immunity)98. These correlates would then need to be validated in prospective cohort studies in different populations and controlled human infection models. In light of emerging viral variants with multiple mutations in the S protein — some of which are able to evade both natural and vaccine-induced immunity18 — it is paramount to target both humoral and T cell immunity, and potentially innate immune mechanisms. Reduced protection against any symptomatic COVID-19 disease caused by the B.1.351 variant of concern (now known as the Beta variant) has been reported from clinical trials18, but emerging data suggest that there remains high protection against the important end points of severe disease and hospitalization99. This indicates the importance of immune mechanisms other than NAbs, including T cell immunity. It may also be important to include SARS-CoV-2 antigens other than S, which are genetically much more stable, in the design of next-generation vaccines. For example, anti-nucleocapsid as well as anti-S binding antibodies are elicited by the whole-cell inactivated vaccine BBV152 (ref.100), and comparison of outcomes post vaccination stratified by both anti-S and anti-nucleocapsid responses will aid our understanding of the role of non-S responses in protection. This includes assessing areas of mammalian physiology far beyond classical immunology that have long been known to be central in host defence during viral infections, such as metabolism, via interrogation of proteomic and metabolomics changes that occur after vaccination and how they relate to vaccine efficacy101,102. Both the samples as well as the analytical pipelines to achieve this mammoth task are in place. Long-term follow-up of individuals who are vaccinated is needed to identify precisely how memory B and T cell responses correspond to the risk of infection and/or severe disease following vaccination. In addition, controlled human infection models may enable a more rapid evaluation of multiple vaccines and/or combinations of vaccines103. Moreover, such models will allow us to evaluate the role of reinfection in individuals who have previously been infected and/or are vaccinated — given the relative scarcity of natural reinfection, these studies will enable the interrogation of early immune responses and identify the relevant mucosal and systemic mechanisms that protect against reinfection. It will be important that discovery is not confined by what we expect, but allows the emerging data rather than dogma to guide formulating the hypothesis on how these vaccines protect.
Identifying correlates of protection will not only enable a pathway to licensure of additional vaccines based on immunogenicity, thus requiring smaller numbers of participants compared with efficacy trials, but would also allow one to rapidly investigate the effects of modified vaccination regimens (lower dose, single dose, dose spacing and heterologous vaccine) and predict protection in specific populations (such as pregnant women and patients who are immunocompromised) — all of which could result in more rapid global deployment of these precious resources. Therefore, there is significant urgency that these analyses will be undertaken by the custodians of the relevant data and samples. This has been highlighted globally, with the WHO identifying an urgent need to ‘accelerate research to establish correlates of protection from COVID-19 vaccines against infection and disease, including for variants of concern’104. It is important that such correlates enable both the licensure of vaccines using already approved platforms for vaccines targeting these variants and also the licensure of vaccines based on additional platforms that are still in development.
Concluding remarks
As effective vaccines for COVID-19 are deployed in some high-income countries, it will still be many months, possibly even years, before sufficient numbers of doses of these vaccines are available to supply the global population. In the meantime, vaccine trials must continue105,106. As effective vaccines are gradually rolled out, conducting large phase III efficacy trials will become increasingly difficult — for many reasons, including the ethical issues of a placebo-controlled trial in the context of an effective vaccine being available and also the likelihood of decreasing disease incidence in countries where vaccines are being used — which are usually the same countries that would be able to support large clinical trials105,106. It may therefore become even more important to establish an immunologic correlate of protection against COVID-19, which could be used as the basis for vaccine licensure in the future. The data from these early trials highlight the challenges associated with this — multiple immunologic parameters will likely need to be measured, and if there are multiple pathways of protection against disease, it is possible that correlates will be different at different time points after vaccination and/or with different vaccines. Comprehensive and careful analyses of all immunologic data, comparing initial post-vaccine responses in individuals who are infected after vaccination with those who are not, will ultimately provide an answer. Such analyses are expected in the first half of 2021, but the high vaccine efficacy means the number of individuals who are infected after vaccination is relatively low, limiting the statistical power of these analyses until sufficient cases occur. Uncertainties also remain around the importance of protecting not only against symptomatic disease but also against asymptomatic infection, and understanding this will be critically important when considering vaccines that may block transmission. Some early post-implementation analyses have suggested that the mRNA vaccines also reduce the incidence of asymptomatic infection and the nasopharyngeal viral load in individuals who become infected despite the vaccination93,107, but it is yet unclear how this translates to risk of transmission. Inhibition of disease transmission would allow for the protection of individuals who are not vaccinated, and additional analyses that specifically evaluate herd immunity in the context of vaccination campaigns are required to confirm this. In addition to the polyfunctionality of vaccine-induced antibodies and T cells, it is important that vaccines induce polyclonal responses against a range of epitopes on the S protein, in order to avoid viral escape mutations. Thus, high-quality post-implementation surveillance of vaccine effectiveness and a careful study of vaccine failures will be a vital part of ongoing monitoring once vaccines are introduced. This will require viral genetic and immunologic analysis of infecting strains in cases of vaccine failure, including sieve analyses whereby pathogen isolates are compared genetically (for example, using whole-genome or targeted sequencing) between subjects who are infected after vaccination and those who are infected and not vaccinated108. Finally, most individuals have pre-existing immune responses to other endemic human coronaviruses109. The impact of these baseline responses on vaccination needs to be fully assessed and understood, especially if COVID-19 vaccines become part of future routine immunization schedules worldwide. Understanding how responses to the initial series (one or two doses) of a COVID-19 vaccine further shape this immune response will be critical, including use of heterologous vaccines for second and/or third doses. Unprecedented insight has been extracted in only 16 months of this pandemic. But work must continue at the same pace in order to control the current COVID-19 pandemic and to be adequately prepared for the next pandemic.
Supplementary information
Acknowledgements
M.S. is supported via salary awards from the BC Children’s Hospital Foundation, the Canadian Child Health Clinician Scientist Program and the Michael Smith Foundation for Health Research. T.R.K. is supported through the Telethon Kids Institute and Perth Children’s Hospital Foundation. A.M. is Research Director of the Fund for Scientific Research (F.R.S.-FNRS), Belgium.
Author contributions
All authors researched data for the article and contributed substantially to discussion of the content. M.S. wrote the first draft of article. All authors reviewed and edited the manuscript before submission.
Competing interests
M.S. has been an investigator on projects funded by GlaxoSmithKline, Merck, Pfizer, Sanofi-Pasteur, Seqirus, Symvivo and VBI Vaccines. A.M. has been an investigator on projects funded by GlaxoSmithKline and Sanofi-Pasteur, and is a member of the Clinical Advisory Board of Curevac. T.R.K. is an investigator in the Human Vaccines Project. All funds have been paid to their respective institutes to support research, and the authors have not received any personal payments.
Footnotes
Peer review information
Nature Reviews Immunology thanks R. Kennedy, Z. Xing and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arnaud Marchant, Tobias R. Kollmann.
Supplementary information
The online version contains supplementary material available at 10.1038/s41577-021-00578-z.
References
- 1.Johns Hopkins University. Coronavirus Resource Centerhttps://coronavirus.jhu.edu/map.html (2020).
- 2.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callaway E, Cyranoski D, Mallapaty S, Stoye E, Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579:482–483. doi: 10.1038/d41586-020-00758-2. [DOI] [PubMed] [Google Scholar]
- 4.Diamond MS, Pierson TC. The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe. 2020;27:699–703. doi: 10.1016/j.chom.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thanh Le T, et al. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organisation. COVID-19 vaccine tracker and landscapehttps://www.who.int/who-documents-detail/draft-landscape-of-covid-19-candidate-vaccines (2020).
- 7.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulligan MJ, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 9.Corbett KS, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith TRF, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu FC, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Doremalen N, et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Q, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen WH, Strych U, Hotez PJ, Bottazzi ME. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop.Med. Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regulatory Focus. COVID-19 Vaccine Trackerhttps://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker (2021).
- 18.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants — clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvin MR, et al. Potentially adaptive SARS-CoV-2 mutations discovered with novel spatiotemporal and explainable AI models. Genome Biol. 2020;21:304. doi: 10.1186/s13059-020-02191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganusov VV, Antia R. Imperfect vaccines and the evolution of pathogens causing acute infections in vertebrates. Evolution. 2006;60:957–969. [PubMed] [Google Scholar]
- 21.Weigand MR, et al. Complete genome sequences of Bordetella pertussis isolates with novel pertactin-deficient deletions. Genome Announc. 2017;5:e00973–17. doi: 10.1128/genomeA.00973-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, et al. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Valle DM, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atyeo C, et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbiani DF, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee WT, et al. Neutralizing antibody responses in COVID-19 convalescent sera. J. Infect. Dis. 2020;223:47–55. doi: 10.1093/infdis/jiaa673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin AT, et al. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur. J. Epidemiol. 2020;35:1123–1138. doi: 10.1007/s10654-020-00698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrashekar A, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercado NB, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty S, et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen MD, et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2020;371:eabc8378. doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jennewein MF, et al. Fc glycan-mediated regulation of placental antibody transfer. Cell. 2019;178:202–215.e14. doi: 10.1016/j.cell.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callow KA. Effect of specific humoral immunity and some non-specific factors on resistance of volunteers to respiratory coronavirus infection. J. Hyg. 1985;95:173–189. doi: 10.1017/S0022172400062410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/S0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler SE, et al. Features and functions of systemic and mucosal humoral immunity among SARS-CoV-2 convalescent individuals. Preprint at. medRxiv. 2020 doi: 10.1101/2020.08.05.20168971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenrich M, et al. SARS-CoV-2 dissemination through peripheral nerves explains multiple organ injury. Front. Cell. Neurosci. 2020;14:229. doi: 10.3389/fncel.2020.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Excler JL, Ake J, Robb ML, Kim JH, Plotkin SA. Nonneutralizing functional antibodies: a new “old” paradigm for HIV vaccines. Clin. Vaccine Immunol. 2014;21:1023–1036. doi: 10.1128/CVI.00230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 43.Wen W, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekine T, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng Y, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neidleman J, et al. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.06.08.138826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni L, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977 e973. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiskopf D, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyllie D, et al. SARS-CoV-2 responsive T cell numbers are associated with protection from COVID-19: a prospective cohort study in keyworkers. Preprint at. medRxiv. 2020 doi: 10.1101/2020.11.02.20222778. [DOI] [Google Scholar]
- 51.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMahan K, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeyanathan M, et al. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JS, Shin EC. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee AK, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia H, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Konno Y, et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lei X, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed-Hassan H, et al. Innate immune responses to highly pathogenic coronaviruses and other significant respiratory viral infections. Front. Immunol. 2020;11:1979. doi: 10.3389/fimmu.2020.01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park A, Iwasaki A. Type I and type III interferons — induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Higgins PG, et al. Intranasal interferon as protection against experimental respiratory coronavirus infection in volunteers. Antimicrob. Agents Chemother. 1983;24:713–715. doi: 10.1128/AAC.24.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Q, et al. Interferon-α2b treatment for COVID-19. Front. Immunol. 2020;11:1061. doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson EJ, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Neill LAJ, Netea MG. BCG-induced trained immunity: can it offer protection against COVID-19? Nat. Rev. Immunol. 2020;20:335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hensley SE, et al. Type I interferon inhibits antibody responses induced by a chimpanzee adenovirus vector. Mol. Therapy. 2007;15:393–403. doi: 10.1038/sj.mt.6300024. [DOI] [PubMed] [Google Scholar]
- 77.Collignon C, et al. Innate immune responses to chimpanzee adenovirus vector 155 vaccination in mice and monkeys. Front. Immunol. 2020;11:579872. doi: 10.3389/fimmu.2020.579872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hartman ZC, Appledorn DM, Amalfitano A. Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Res. 2008;132:1–14. doi: 10.1016/j.virusres.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Devoldere J, Dewitte H, De Smedt SC, Remaut K. Evading innate immunity in nonviral mRNA delivery: don’t shoot the messenger. Drug Discov. Today. 2016;21:11–25. doi: 10.1016/j.drudis.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 80.Dan JM, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ledford H. How can countries stretch COVID vaccine supplies? Scientists are divided over dosing strategies. Nature. 2021;589:182. doi: 10.1038/d41586-021-00001-6. [DOI] [PubMed] [Google Scholar]
- 84.Public Health England. Immunisation Against Infectious Diseasehttps://www.gov.uk/government/collections/immunisation-against-infectious-disease-the-green-book (UK Government, 2021).
- 85.Quach C, Deeks S. COVID-19 vaccination: why extend the interval between doses? JAMMI. 2021 doi: 10.3138/jammi-2021-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goel RR, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Voysey M, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2020;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Voysey M, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Janssen Biotech. COVID-19 Vaccine Ad26.COV2.S. Sponsor Briefing Document (Vaccines and Related Biological Products Advisory Committee, 2021).
- 90.Logunov DY, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu FC, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dagan N, et al. BNT162b2 mRNA COVID-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haas, E. J. et al. Nationwide vaccination campaign with BNT162b2 in israel demonstrates high vaccine effectiveness and marked declines in incidence of SARS-CoV-2 infections and COVID-19 cases, hospitalizations, and deaths. Preprint at SSRNhttps://ssrn.com/abstract=3811387 (2021). [DOI] [PMC free article] [PubMed]
- 95.Hyams C, et al. Assessing the effectiveness of BNT162b2 and ChAdOx1nCoV-19 COVID-19 vaccination in prevention of hospitalisations in elderly and frail adults: a single centre test negative case–control study. Preprint at. SSRN. 2021 doi: 10.2139/ssrn.3796835. [DOI] [Google Scholar]
- 96.Barrett JR, et al. Phase 1/2 trial of SARS-CoV-2 vaccine ChAdOx1 nCoV-19 with a booster dose induces multifunctional antibody responses. Nat. Med. 2020;27:279–288. doi: 10.1038/s41591-020-01179-4. [DOI] [PubMed] [Google Scholar]
- 97.Kristen AE, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Preprint at. medRxiv. 2021 doi: 10.1101/2021.03.17.20200246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shannon CP, et al. Multi-omic data integration allows baseline immune signatures to predict hepatitis B vaccine response in a small cohort. Front. Immunol. 2020;11:578801. doi: 10.3389/fimmu.2020.578801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abu-Raddad LJ, Chemaitelly H, Butt AA, National Study Group for COVID-19 Vaccination. Effectiveness of the BNT162b2 COVID-19 vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ella R, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: a double-blind, randomised, phase 1 trial. Lancet Infect. Dis. 2021;21:637–646. doi: 10.1016/S1473-3099(20)30942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang A, et al. Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell. 2016;166:1512–1525.e12. doi: 10.1016/j.cell.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang A, Luan HH, Medzhitov R. An evolutionary perspective on immunometabolism. Science. 2019;363:eaar3932. doi: 10.1126/science.aar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kirby T. COVID-19 human challenge studies in the UK. Lancet Respir. Med. 2020;8:e96. doi: 10.1016/S2213-2600(20)30518-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.World Health Organisation. Statement on the seventh meeting of the International Health Regulations (2005) Emergency Committee regarding the coronavirus disease (COVID-19) pandemichttps://www.who.int/news/item/19-04-2021-statement-on-the-seventh-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (2021).
- 105.WHO Ad Hoc Expert Group on the Next Steps for COVID-19 Vaccine Evaluation et al. Placebo-controlled trials of COVID-19 vaccines — why we still need them. N. Engl. J. Med. 2020;384:e2. doi: 10.1056/NEJMp2033538. [DOI] [PubMed] [Google Scholar]
- 106.Wendler D, Ochoa J, Millum J, Grady C, Taylor HA. COVID-19 vaccine trial ethics once we have efficacious vaccines. Science. 2020;370:1277–1279. doi: 10.1126/science.abf5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Levine-Tiefenbrum M, et al. Decreased SARS-CoV-2 viral load following vaccination. Preprint at. medRxiv. 2021 doi: 10.1101/2021.02.06.21251283. [DOI] [Google Scholar]
- 108.Gilbert P, Self S, Rao M, Naficy A, Clemens J. Sieve analysis: methods for assessing from vaccine trial data how vaccine efficacy varies with genotypic and phenotypic pathogen variation. J. Clin. Epidemiol. 2001;54:68–85. doi: 10.1016/S0895-4356(00)00258-4. [DOI] [PubMed] [Google Scholar]
- 109.Huang AT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pardi N, et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karikó K, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Skowronski DM, De Serres G. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2021;384:1576–1578. doi: 10.1056/NEJMc2036242. [DOI] [PubMed] [Google Scholar]
- 114.Pfizer. Pfizer and BioNTech confirm high efficacy and no serious safety concerns through up to six months following second dose in updated topline analysis of landmark COVID-19 vaccine study https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-confirm-high-efficacy-and-no-serious (2021).
- 115.Vasileiou E, et al. Effectiveness of first dose of COVID-19 vaccines against hospital admissions in Scotland: national prospective cohort study of 5.4 million people. Preprint at. SSRN. 2021 doi: 10.2139/ssrn.3789264. [DOI] [Google Scholar]
- 116.Heymann AD, et al. BNT162b2 vaccine effectiveness in preventing asymptomatic infection with SARS-CoV-2 virus: a nationwide historical cohort study. Preprint at. SSRN. 2021 doi: 10.2139/ssrn.3796868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hall VJ, et al. Effectiveness of BNT162b2 mRNA vaccine against infection and COVID-19 vaccine coverage in healthcare workers in England, multicentre prospective cohort study (the SIREN Study). Preprint at. SSRN. 2021 doi: 10.2139/ssrn.3790399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chodick G, et al. The effectiveness of the first dose of BNT162b2 vaccine in reducing SARS-CoV-2 infection: real-world evidence. Preprint at. SSRN. 2021 doi: 10.2139/ssrn.3769977. [DOI] [Google Scholar]
- 119.Andrejko K, et al. Early evidence of COVID-19 vaccine effectiveness within the general population of California. Preprint at. medRxiv. 2021 doi: 10.1101/2021.04.08.21255135. [DOI] [Google Scholar]
- 120.Regev-Yochay G, et al. Decreased infectivity following BNT162b2 vaccination. Preprint at. SSRN. 2021 doi: 10.2139/ssrn.3815668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bernal JL, et al. Early effectiveness of COVID-19 vaccination with BNT162b2 mRNA vaccine and ChAdOx1 adenovirus vector vaccine on symptomatic disease, hospitalisations and mortality in older adults in England. Preprint at. medRxiv. 2021 doi: 10.1101/2021.03.01.21252652. [DOI] [Google Scholar]
- 122.Tande AJ, et al. Impact of the COVID-19 vaccine on asymptomatic infection among patients undergoing pre-procedural COVID-19 molecular screening. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thompson MG, et al. Interim estimates of vaccine effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines in preventing SARS-CoV-2 infection among health care personnel, first responders, and other essential and frontline workers — eight U.S. locations, December 2020–March 2021. MMWR. 2021;70:495–500. doi: 10.15585/mmwr.mm7013e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Walsh EE, et al. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pfizer. Pfizer-Biontech COVID-19 Vaccine (BNT162, PF-07302048) Vaccines And Related Biological Products Advisory Committee Briefing Document (Pfizer Inc, 2020).
- 126.Jackson LA, et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Widge AT, et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2020;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Folegatti PM, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ewer KJ, et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2020;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 130.Logunov DY, et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sadoff J, et al. Interim results of a phase 1–2a trial of Ad26.COV2.S COVID-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bos R, et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines. 2020;5:91. doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reuters. CanSinoBIO’s COVID-19 vaccine 65.7% effective in global trials, Pakistan official says. Reutershttps://www.reuters.com/article/us-health-coronavirus-vaccine-pakistan/cansinobios-covid-19-vaccine-65-7-effective-in-global-trials-pakistan-official-says-idUSKBN2A81N0 (2021).
- 134.Reuters. CanSinoBIO says COVID-19 shot may be less effective over time, booster shot promising. Reutershttps://www.reuters.com/article/us-health-coronavirus-vaccine-cansinobio-idUSKBN2BO4CG (2021).
- 135.National Institues of Health. A study to evaluate the efficacy, immune response, and safety of a COVID-19 vaccine in adults ≥ 18 years with a pediatric expansion in adolescents (12-17 years) at risk for SARS-CoV-2. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04611802 (2021).
- 136.Keech C, et al. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Novavax. Novavax confirms high levels of efficacy against original and variant COVID-19 strains in United Kingdom and South Africa trials. Novavaxhttps://ir.novavax.com/news-releases/news-release-details/novavax-confirms-high-levels-efficacy-against-original-and-0 (2021).
- 138.Zhang Y, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu Z, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:803–812. doi: 10.1016/S1473-3099(20)30987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.BBC. Sinovac: Brazil results show Chinese vaccine 50.4% effective. BBC Newshttps://www.bbc.com/news/world-latin-america-55642648 (2021).
- 141.Reuters. Turkish study revises down Sinovac COVID-19 vaccine efficacy to 83.5%. Reutershttps://www.reuters.com/article/us-health-coronavirus-turkey-sinovac/turkish-study-revises-down-sinovac-covid-19-vaccine-efficacy-to-83-5-idUSKBN2AV13A (2021).
- 142.Xia S, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21:39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reuters. UAE says Sinopharm vaccine has 86% efficacy against COVID-19. Reutershttps://www.reuters.com/business/healthcare-pharmaceuticals/uae-says-sinopharm-vaccine-has-86-efficacy-against-covid-19-2020-12-11/ (2020).
- 144.Xia S, et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA. 2020;324:951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Reuters. Sinopharm’s Wuhan unit reports 72.5% efficacy for COVID shot, seeks approval in China. Reutershttps://www.reuters.com/business/healthcare-pharmaceuticals/sinopharms-wuhan-unit-reports-725-efficacy-covid-shot-seeks-approval-china-2021-02-24/ (2021).
- 146.Ella R, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect. Dis. 2021;21:950–961. doi: 10.1016/S1473-3099(21)00070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bharat Biotech. Bharat Biotech and ICMR Announce Interim Results from Phase 3 trials of COVAXIN®; Demonstrates overall Interim Clinical Efficacy of 78% and 100% efficacy against Severe COVID-19 diseasehttps://www.bharatbiotech.com/images/press/covaxin-phase3-clinical-trials-interim-results.pdf (2021).
- 148.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rauch S, Jasny E, Schmidt KE, Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Duan J, et al. A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem. Biophys. Res. Commun. 2005;333:186–193. doi: 10.1016/j.bbrc.2005.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Coughlin M, et al. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology. 2007;361:93–102. doi: 10.1016/j.virol.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang L, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 2018;92:e02002-1. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang N, et al. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405.e6. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen Y, et al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg. Microbes Infect. 2017;6:e37. doi: 10.1038/emi.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rogers TF, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shi R, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 161.Wang W, et al. Characterization of anti-MERS-CoV antibodies against various recombinant structural antigens of MERS-CoV in an imported case in China. Emerg. Microbes Infect. 2016;5:e113. doi: 10.1038/emi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Widjaja I, et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg. Microbes Infect. 2019;8:516–530. doi: 10.1080/22221751.2019.1597644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vabret N, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Graham BS. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 165.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. JMD. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hou YJ, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Scobey T, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim HW, et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 169.Fulginiti VA, Eller JJ, Downie AW, Kempe CH. Altered reactivity to measles virus. Atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202:1075–1080. doi: 10.1001/jama.1967.03130250057008. [DOI] [PubMed] [Google Scholar]
- 170.Bolles M, et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Czub M, Weingartl H, Czub S, He R, Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lambert PH, et al. Consensus summary report for CEPI/BC March 12-13, 2020 meeting: assessment of risk of disease enhancement with COVID-19 vaccines. Vaccine. 2020;38:4783–4791. doi: 10.1016/j.vaccine.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sahin U, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 174.Sadoff J, et al. Safety and immunogenicity of the Ad26.COV2.S COVID-19 vaccine candidate: interim results of a phase 1/2a, double-blind, randomized, placebo-controlled trial. Preprint at. medRxiv. 2020 doi: 10.1101/2020.09.23.20199604. [DOI] [Google Scholar]
- 175.Halstead SB, Katzelnick L. COVID-19 vaccines: should we fear ADE? J. Infect. Dis. 2020;222:1946–1950. doi: 10.1093/infdis/jiaa518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Arvin AM, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 177.Sridhar S, et al. Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med. 2018;379:327–340. doi: 10.1056/NEJMoa1800820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.